Abstract
Glucagon-like peptide-2 (GLP-2), secreted from enteroendocrine cells, attenuates gut motility, enhances barrier function, and augments nutrient absorption, actions mediated by a single GLP-2 receptor (GLP-2R). Despite extensive analyses, the precise distribution and cellular localization of GLP-2R expression remains controversial, confounded by the lack of suitable GLP-2R antisera. Here, we reassessed murine Glp2r expression using regular and real-time quantitative PCR (qPCR), in situ hybridization (ISH), and a Glp2rLacZ reporter mouse. Glp2r mRNA expression was detected from the stomach to the rectum and most abundant in the jejunum. Glp2r transcripts were also detected in cerebral cortex, mesenteric lymph nodes, gallbladder, urinary bladder, and mesenteric fat. Surprisingly, Glp2r mRNA was found in testis by qPCR at levels similar to jejunum. However, the testis Glp2r transcripts, detected by different primer pairs and qPCR, lacked 5′ mRNA coding sequences, and only a minute proportion of them corresponded to full-length Glp2r mRNA. Within the gut, Glp2r-driven LacZ expression was localized to enteric neurons and lamina propria stromal cells, findings confirmed by ISH analysis of the endogenous Glp2r mRNA. Unexpectedly, vascular Glp2rLacZ expression was localized to mesenteric veins and not arteries. Moreover, mesenteric fat Glp2rLacZ expression was detected within blood vessels and not adipocytes. Reporter LacZ expression was not detected in all tissues expressing an endogenous Glp2r transcript, such as gallbladder, urinary bladder, and mesenteric lymph nodes. Collectively, these findings extend our understanding of the cellular domains of Glp2r expression and highlight limitations inherent in application of commonly used technologies to infer analysis of gene expression.
The mammalian proglucagon gene is expressed in the pancreas, gut, and central nervous system, giving rise to multiple proglucagon-derived peptides (PGDPs) following tissue-specific post-translational processing of a common proglucagon precursor (1). The three best-studied PGDPs—glucagon and glucagon-like peptide-1 and -2 (GLP-1 and GLP-2)—exert overlapping yet distinct actions in the control of glucose homeostasis, appetite, gastrointestinal (GI) motility, energy absorption, and assimilation. These actions are mediated through three structurally distinct yet highly related receptors: the glucagon receptor (GCGR) and the GLP-1 and -2 receptors (GLP-1R and GLP-2R), members of the class B G protein–coupled receptor (GPCR) superfamily (2).
Glucagon, the most extensively studied and original member of the PGDP family, maintains glycemia in the fasting state, simultaneously acting as a counter-regulatory hormone in the context of hypoglycemia (3). Indeed, pharmaceutical preparations of glucagon are approved for the treatment of severe hypoglycemia in people with diabetes. Glucagon also exhibits a number of actions beyond glucose control mediated by the GCGR, including regulation of amino acid metabolism, lipolysis, heart rate, appetite, and GI motility (3, 4). The second-most studied PGDP, GLP-1, was originally described as an incretin hormone that stimulates glucose-dependent insulin secretion (5). Subsequent studies demonstrated a broad spectrum of GLP-1 actions encompassing control of food intake, gut motility, glucagon secretion, heart rate, natriuresis, gut growth, and immune function—actions transduced through a single canonical GLP-1R (1, 6, 7). Collectively, the profile of GLP-1 action supported development of multiple GLP-1R agonists for the treatment of type 2 diabetes and a single agent, liraglutide, for the therapy of obesity (7, 8).
GLP-2, cosecreted with GLP-1 from enteroendocrine L cells, exerts important actions regulating control of nutrient assimilation, including attenuation of GI motility, rapid enhancement of nutrient absorption, and the stimulation of intestinal crypt cell proliferation leading to expansion of the absorptive mucosal epithelium, predominantly in the small bowel (9, 10). As is the case for GLP-1, GLP-2 also exerts pleiotropic activities in distal tissues, including stimulation of mesenteric blood flood, control of appetite, inhibition of gallbladder emptying, and reduction of bone resorption (5, 9, 11). These actions are mediated by activation of a single GLP-2R, expressed within the gut, brain, and some peripheral tissues (12).
To understand how the PGDPs mediate their diverse actions in multiple cell types, considerable effort has been focused on cellular identification of receptor expression using antisera, RNA analyses, and reporter gene expression. The use of antisera for localization of GLP-1R immunoreactivity has been highly problematic, with incorrect assignment of receptor localization arising as a result of widespread use of poorly characterized antisera with inadequate sensitivity and impaired specificity (13–15). Similar problems have been encountered with multiple commercially available antisera against the GLP-2R, which following characterization in our laboratory, have been shown to be inappropriate for use in accurate detection of GLP-2R expression (9). Indeed, our laboratory has yet to detect a sufficiently validated GLP-2R antisera with high sensitivity and specificity (9).
To circumvent the challenges inherent in validation of antisera sensitivity and specificity for detection of PGDP receptors, complementary analyses have used regular and real-time quantitative PCR (qPCR), single-cell RNA sequencing, ligand binding, and in situ hybridization (ISH) to identify expression of mRNA transcripts encoding GLP-1R in different cell types and species (16–21). In contrast, less effort has been directed at the identification of GLP-2R expression in cell types beyond the GI tract (12, 22, 23). Surprisingly, a recent study using qPCR to characterize murine GLP-2R expression demonstrated a widespread receptor in extraintestinal tissues, including fat, lymph nodes, bladder, spleen, and liver, including hepatocytes (24). Accordingly, to reconcile the contrasting reports of GLP-2R expression in various mouse organs (12, 22, 24), we have now re-examined Glp2r expression using several complementary techniques, including qPCR, regular PCR to establish the presence of full-length mRNA transcripts, ISH, and expression of a LacZ reporter gene under the control of the endogenous Glp2r locus.
Materials and Methods
Animals
The mouse strain Glp2rtm1(KOMP)Vlcg, where KOMP is the Knockout Mouse Project on a C57BL/6J background, was obtained from the KOMP Repository (www.komp.org). The mouse carries a Glp2r null allele in which the coding region of the Glp2r gene has been replaced by the Escherichia coli LacZ gene. These mice were mated with B6.C-Tg(CMV-cre)1Cgn/J (stock 006054; The Jackson Laboratory, Bar Harbor, ME) to delete the LoxP site-flanked antibiotic selection transcriptional unit, creating the Glp2rtm1.1(KOMP)Vlcg/Ddr strain, henceforth abbreviated Glp2rLacZ, used in the current study.
Heterozygous Glp2r+/LacZ breeders were used to generate Glp2r wild-type (WT; Glp2r+/+), heterozygous Glp2r+/Z, and homozygous Glp2rZ/Z littermates at the Toronto Centre for Phenogenomics Animal Facility. Except for analysis of female reproductive system tissues, all experiments were performed in male mice aged 13 to 18 weeks, housed up to five per cage, with free access to food (2018 Teklad global; Envigo Corp., Mississauga, ON, Canada) and water. All animal experiments were conducted according to protocols approved by the Animal Care and Use Subcommittee at the Toronto Centre for Phenogenomics, Mt. Sinai Hospital, and were consistent with the Animal Research: Reporting In Vivo Experiments guidelines.
β-Galactosidase staining
The stomach, small intestine, and colon were flushed with PBS, cut open along their cephalocaudal axes, and pinned on silicone elastomer (SylgardTM 184; Dow Silicones Corp., Midland, MI)-coated plates. To clear the blood from the liver and lung before subjecting those organs to fixation and staining, mice were transcardially perfused with PBS, supplemented with sodium heparin (10 U/mL). Tissues were fixed in ice-cold 1% paraformaldehyde, 0.2% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA), and 0.02% Nonidet P-40 in PBS with 10 mM HEPES, pH 7.7, for 2 hours. The tissue whole-mount preparations were stained for β-galactosidase overnight at room temperature using 0.1% 5-Br-4-Cl-3-indolyl-β-d-galactopyranoside substrate (catalog no. 15520034; Thermo Fisher Scientific, Markham, ON, Canada; prepared from a 2.5% stock of 5-Br-4-Cl-3-indolyl-β-d-galactopyranoside in dimethyl-formamide) in 2 mM MgCl2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.02% Nonidet P-40, and 0.01% sodium deoxycholate in PBS with 10 mM HEPES, pH 7.7 (25). After digital image acquisition using a Leica MZ16 FA stereomicroscope, tissue whole mounts used for the analysis of β-galactosidase staining were processed for paraffin embedding before sectioning at 5 μm and counterstaining with Neutral Red.
RNA extraction, cDNA synthesis, and RT-PCR
Total RNA was extracted from the different mouse tissues by the guanidinium thiocyanate method using TRI Reagent (Molecular Research Center Inc., Cincinnati, OH). Mouse islets were isolated following pancreas digestion with collagenase type V, as previously described (26). The digest was fractionated using a Histopaque (Sigma-Aldrich Canada, Oakville, ON, Canada) discontinuous density gradient, and the islets were collected from the interface. Individual islets were handpicked and immediately lysed in TRI Reagent for subsequent RNA isolation. RT was performed with 500 ng total RNA, treated with DNase I (catalog no. EN0521; Thermo Fisher Scientific), using random hexamers (catalog no. 58875) and SuperScript III (catalog no. 18080-044) from Thermo Fisher Scientific. The resulting cDNA was used both for regular PCR and for real-time qPCR purposes. PCR amplification of the murine Glp2r was accomplished using Platinum Taq DNA polymerase (catalog no. 10966018; Thermo Fisher Scientific) and the primer pair 5′-GCCCAGTAGATGCAGAGAGG-3′ and 5′-AGTTGCCAAGCTGTGGTGAT-3′ at an annealing temperature of 60°C, giving rise to a 1660-bp product spanning the entire Glp2r open-reading frame (ORF). PCR amplification of the LacZ reporter from Glp2r+/Z mouse tissue cDNA was done using the primer pair 5′-GCGAATACCTGTTCCGTCAT-3′ and 5′-TACGCCAATGTCGTTATCCA-3′, generating a 467-bp product. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) PCR amplification was performed, as previously described (27). PCR products were analyzed by agarose gel electrophoresis. Southern blot analysis, using an internal, Glp2r-specific, 32P-labeled oligonucleotide probe, was used to verify further the identity of the 1660-bp PCR product as corresponding to a genuine, full-length Glp2r transcript. Real-time qPCR was performed on a QuantStudio 5 System (Thermo Fisher Scientific) with TaqMan Fast Advanced Master Mix (catalog no. 4444557; Thermo Fisher Scientific) and TaqMan Gene Expression Assays (Thermo Fisher Scientific) for Glp2r (Mm01329471_m1, Mm01329473_m1, Mm01329475_m1, and Mm00558835_m1), Glp1r (Mm00445292_m1), Gcgr (Mm00433546_m1), and Gipr (Mm01316344_m1). The following endogenous control RNA transcripts were tested to evaluate their suitability for the purpose of normalization of Glp2r expression across the multiple extraintestinal tissues analyzed: 18S rRNA (Hs99999901_s1), Ppia (Mm02342430_g1), Tbp (Mm01277042_m1), and Rpl32 (Mm02528467_g1). Relative quantification of transcript levels in selected tissue sample comparisons was performed by the comparative threshold cycle method using Rpl32 (mouse GI tract tissue), Tbp (mouse pancreatic islets), or Ppia (jejunum and urinary bladder) for normalization.
ISH
Following fixation in 10% neutral-buffered formalin (Sigma-Aldrich Canada) for 24 hours at room temperature and paraffin embedding, tissues were cut into 5-μm sections and mounted onto Superfrost Plus (Thermo Fisher Scientific) glass slides. ISH was performed using the RNAscope 2.5 HD Detection Reagent kit (Advanced Cell Diagnostics Inc., Newark, CA), according to the manufacturer’s instructions. Specific hybridization probes (Advanced Cell Diagnostics Inc.) included mouse Glp2r (Mm-Glp2r, catalog no. 447061) and Ppib positive control (Mm-Ppib; catalog no. 313911). Tissue sections were counterstained with Mayer Hematoxylin (Dako Canada, Mississauga, ON, Canada) and coverslipped before microscopy examination and digital image acquisition.
Statistical analysis
Results are presented either as bar diagrams or scatter plots with overlapping means ± SD. Summary statistics were calculated using GraphPad Prism, v.7.0 (GraphPad Software, San Diego, CA).
Results
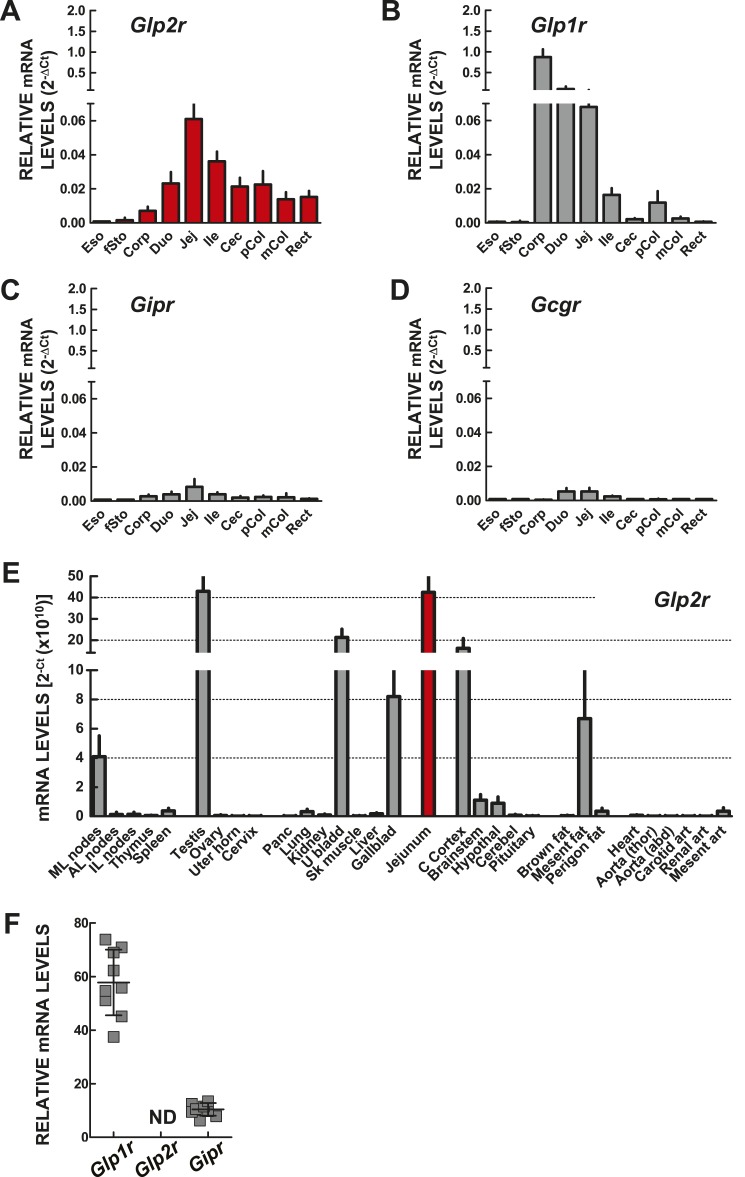
Following the cloning of the cDNAs encoding the GLP-2R (12), we initially used Northern blotting (as qPCR technology was not yet available) to delineate the distribution of Glp2r mRNA transcripts in the murine gut, simultaneously obtaining an estimation of relative Glp2r mRNA abundance in various tissues (22). Several reports have subsequently described GLP-2R RNA and protein expression within and outside the GI tract in different species, sometimes yielding conflicting results with regard to the tissue and cellular sites of Glp2r expression (9, 24). Accordingly, we reanalyzed quantitative Glp2r mRNA expression in the mouse GI tract as well as in 30 additional extraintestinal tissues using qPCR with TaqMan primers (Fig. 1A, 1E, and 1F). For comparative purposes, we simultaneously analyzed the expression of transcripts for the structurally related Class B GPCRs (2) Glp1r, Gcgr, and Gipr (Fig. 1B–1D), which like the Glp2r, are linked to the control of gut motility (1) and in the case of Glp1r, also to stimulation of intestinal growth (5).
Figure 1.
Expression of mRNA transcripts for the PGDP receptors (A) Glp2r, (B) Glp1r, (D) Gcgr, and (C) Gipr along the murine GI tract, assessed by quantitative RT-PCR analysis of total RNA. Transcript abundance is expressed relative to the levels of the Rpl32 mRNA, which was used as endogenous normalization control. Data are means ± SD (n = 6 mice). (E) Glp2r mRNA expression, assessed by quantitative RT-PCR in total RNA from the indicated mouse extra-GI tissues. Glp2r transcript abundance is reported as threshold cycle (Ct) values without endogenous control normalization. Levels of jejunum Glp2r mRNA transcripts without normalization (red filled bar) are shown for comparison. Data are means ± SD (n = 6 mice). (F) Glp1r, Glp2r, and Gipr expression in isolated pancreatic islets from WT mice. Transcript abundance is expressed relative to the levels of Tbp mRNA. Shown are individual data points with overlapping means ± SD (n = 9 independent islet preparations). 2−ΔCt, comparative threshold cycle method; AL, axillary lymph node; Aorta (abd), abdominal aorta; Aorta (thor), thoracic aorta; art, artery; C Cortex, cerebral cortex; Cec, cecum; Cerebel, cerebellum; Corp, stomach corpus; Duo, duodenum; Eso, esophagus; fSto, forestomach; Gallblad, gallbladder; Hypothal, hypothalamus; IL, inguinal lymph node; Ile, ileum; Jej, jejunum; mCol, medial colon; Mesent, mesenteric; ML, mesenteric lymph node; ND, not detectable; Panc, pancreas; pCol, proximal colon; Perigon, perigonadal; Rect, rectum; Sk muscle, skeletal muscle (quadriceps); U bladd, urinary bladder; Uter horn, uterine horn.
Glp2r transcripts were detected, albeit at extremely low levels, in esophagus and forestomach (Fig. 1A). Relative levels of Glp2r expression increased progressively along the GI tract and were highest in proximal jejunum, a segment of the gut highly sensitive to the intestinotrophic action of GLP-2 (11, 28). Glp2r expression was lower in more distal segments of the gut, with relative colonic Glp2r transcript abundance approximately one-third of the levels found in jejunum (Fig. 1A). Similar to findings for Glp2r, levels of Glp1r mRNA transcripts were nearly undetectable in esophagus and forestomach; however, Glp1r mRNA abundance increased markedly in the corpus of the stomach (Fig. 1B), consistent with previous findings (19, 29), and then gradually declined from the duodenum to colon (Fig. 1B). In contrast, Gipr and Gcgr expression was very low or undetectable along the GI tract, with peak values in the small intestine (Fig. 1C and 1D). However, even in the small intestine, Gipr and Gcgr transcript levels were nearly 10-fold lower than those of Glp2r (Fig. 1A).
We next assessed the relative expression of Glp2r mRNA transcripts across 30 different extraintestinal mouse tissues, including jejunum as reference for transcript abundance (Fig. 1E). To facilitate the direct comparison of the level of Glp2r mRNA between tissues, Glp2r transcript abundance is reported without endogenous control RNA normalization, as we recently described for analysis of human GLP1R expression (15). Notably, none of the commercially available TaqMan endogenous control gene-expression assays that we tested, including 18S rRNA, Ppia, Tbp, and Rpl32, were suitable for the purpose of normalization across the 30 different tissues analyzed. This was not a surprising finding considering the intrinsic variability from tissue to tissue in expression of mRNA transcripts commonly used for normalization purposes (30).
Overall, Glp2r expression ranged from very low to undetectable in most of the mouse tissues examined (Fig. 1E). Although the rat Glp2r cDNA was first cloned from a hypothalamic cDNA library (12), Glp2r expression was surprisingly low in mouse hypothalamus, and only the cerebral cortex, among the multiple regions of the mouse brain assessed, including the pituitary gland, exhibited levels of Glp2r mRNA transcripts comparable with jejunum. Consistent with our recent findings of a functional GLP-2-gallbladder axis (31), Glp2r mRNA transcripts were relatively abundant in the gallbladder (Fig. 1E), as well as in the urinary bladder. Unexpectedly, testis, but not the female reproductive organs, expressed Glp2r transcripts at robust levels, similar to those detected in jejunum (Fig. 1E).
The main lymphoid organs, with the exception of the mesenteric lymph nodes, were nearly devoid of Glp2r mRNA transcripts (Fig. 1E). Levels of Glp2r mRNA transcripts were barely detectable in interscapular brown adipose tissue and very low in the perigonadal white adipose tissue depots (Fig. 1E). Notably, Glp2r expression was relatively higher but more variable within mesenteric adipose tissue depots, whereas Glp2r mRNA transcripts were negligible in lung, kidney, liver, skeletal muscle, whole heart, and major arteries (Fig. 1E). Finally, Glp2r expression was undetectable in whole pancreas and with the corroboration of our previous observations (32), equally undetectable in isolated mouse islets, where both Glp1r and Gipr transcripts were clearly expressed (Fig. 1F).
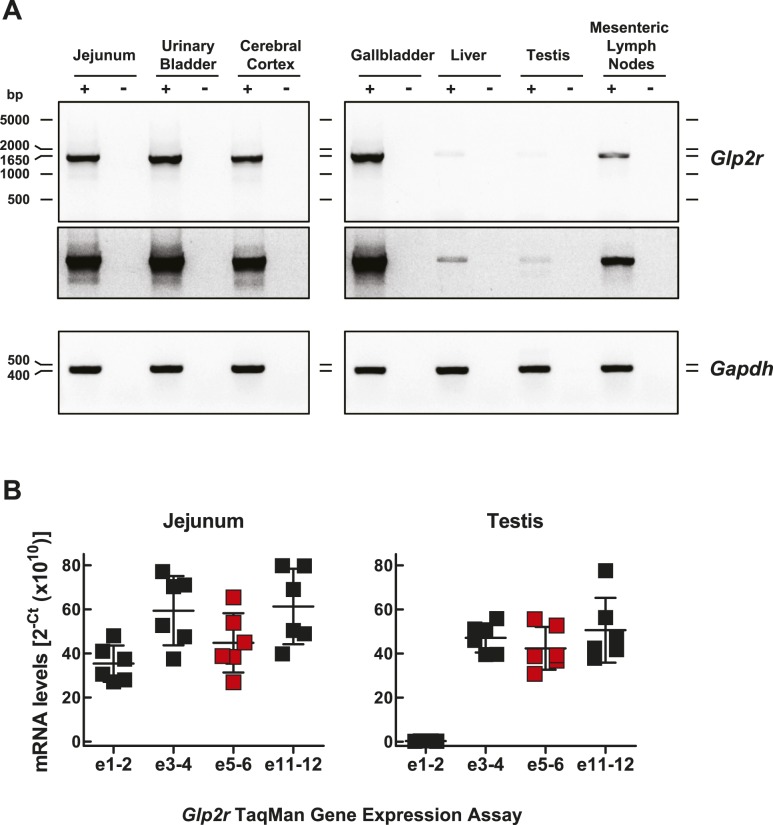
To verify that results obtained with qPCR reflected expression of a full-length Glp2r mRNA transcript, we carried out conventional RT-PCR using primer pairs spanning the entire ORF, followed by gel electrophoresis. A full-length, 1660-bp Glp2r mRNA transcript encompassing the entire mouse Glp2r ORF was detected in jejunum and also in the gallbladder, urinary bladder, and cerebral cortex (Fig. 2A). A less-abundant, full-length Glp2r PCR product was also clearly present in mesenteric lymph node RNA (Fig. 2A), as well as in liver (Fig. 2A), albeit at much lower levels, consistent with the very low expression of the hepatic Glp2r, as assessed by qPCR (Fig. 1E). Surprisingly, despite the robust expression of Glp2r in testis RNA identified by qPCR analysis (Fig. 1E), regular RT-PCR amplified a very low-abundance, full-length Glp2r product from the same testis RNA (Fig. 2A). To ascertain the reason for the discrepancy between qPCR and the conventional RT-PCR analyses, we compared Glp2r expression in jejunum and testis using various TaqMan gene-expression assays spanning multiple exons along the murine Glp2r transcript (Fig. 2B). Relative Glp2r abundance in jejunum was fairly consistent, independent of the TaqMan assay used. In contrast, we detected a marked divergence between results obtained using TaqMan assays when analyzing Glp2r mRNA transcripts in testis (Fig. 2B). TaqMan assays targeting exons 3-4, 5-6, and 11-12 revealed relative levels of testis Glp2r mRNA expression that were comparable with jejunum (Fig. 2B). Remarkably, Glp2r mRNA abundance was ∼200-fold lower in testis when measured with qPCR primer pairs targeting exons 1-2. Collectively, these findings indicate that the Glp2r TaqMan assay targeting exons 5-6 largely detects an incomplete Glp2r transcript in testis, missing either exon 1 or 2 or perhaps both. These observations exemplify the potential pitfalls of the reliance only on qPCR as proof of gene expression in any particular tissue without independent confirmation that the partial PCR product actually reflects the presence of a full-length mRNA transcript capable of undergoing translation into a functional protein (33).
Figure 2.
(A) Demonstration of a full-length Glp2r mRNA transcript in selected mouse tissues. Glp2r and Gapdh mRNA expression was assessed by RT-PCR in total RNA from mouse jejunum, urinary bladder, cerebral cortex, gallbladder, liver, testis, and mesenteric lymph nodes. PCR products (Glp2r 1660 bp, spanning the entire mouse Glp2r ORF, and Gapdh 450 bp) were analyzed by agarose gel electrophoresis using the SYBR Safe DNA Gel Stain. The specificity of each RT reaction (+) was monitored by control reactions using samples in which the RT was omitted from the RT reaction mixture (−). A longer exposure of the gel illustrated in the top blot is shown in the middle blot. Agarose gel images are shown in reverse contrast. (B) Glp2r mRNA expression in jejunum and testis assessed by qPCR using TaqMan gene-expression assays targeting the specified Glp2r transcript exon boundaries. The red filled squares correspond to the expression data using the Glp2r TaqMan assay targeting exons (e)5-6, used to generate the data shown in Fig. 1. Glp2r transcript abundance is reported without endogenous control normalization. Shown are individual data points with overlapping means ± SD (n = 6 mice).
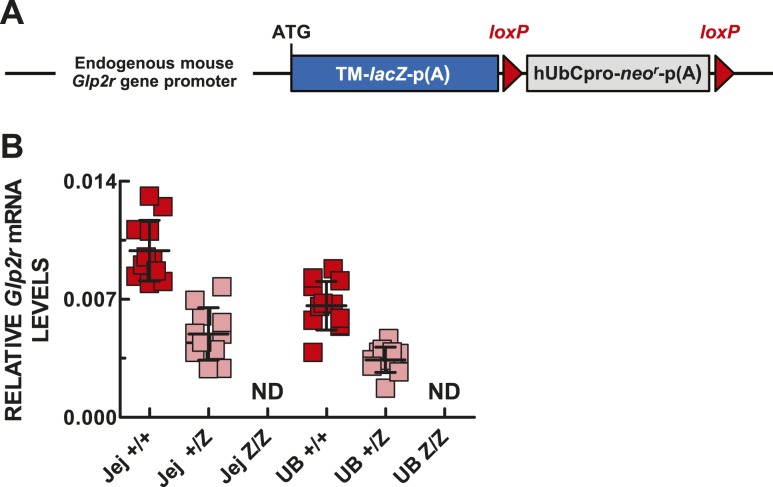
Critical to understanding the biology of the GLP-2 action is the knowledge of the spatial and cellular expression of the GLP-2R within tissues. Accordingly, we investigated the use of a Glp2r reporter mouse available from the KOMP Repository, in which the expression of the bacterial β-galactosidase reporter gene (LacZ) is driven by the endogenous regulatory elements of the Glp2r gene (Fig. 3A). As expected, whereas Glp2r expression was robust in jejunum and urinary bladder of Glp2r WT mice, Glp2r mRNA levels were reduced by one-half in the heterozygous and were undetectable in the same tissues of Glp2rZ/Z littermate mice (Fig. 3B).
Figure 3.
(A) Schematic of the KOMP Regeneron-targeted mutation Glp2rtm1(KOMP)Vlcg allele, based on the information posted at www.komp.org. (B) Glp2r expression in jejunum (Jej) and urinary bladder (UB) of WT Glp2r+/+ (+/+), heterozygous Glp2r+/Z (+/Z), and homozygous Glp2rZ/Z (Z/Z) mice. Glp2r transcript abundance is expressed relative to the levels of Ppia mRNA, which were not statistically different between tissues. Shown are individual data points with overlapping means ± SD (n = 11 +/+, 10 +/Z, and 5 Z/Z mice). hUbCpro, promoter of the human ubiquitin C gene; loxP, loxP sites; neor, coding sequence of the neomycin phosphotransferase; p(A), polyadenylation signal; TM-LacZ, transmembrane domain-LacZ coding fusion protein.
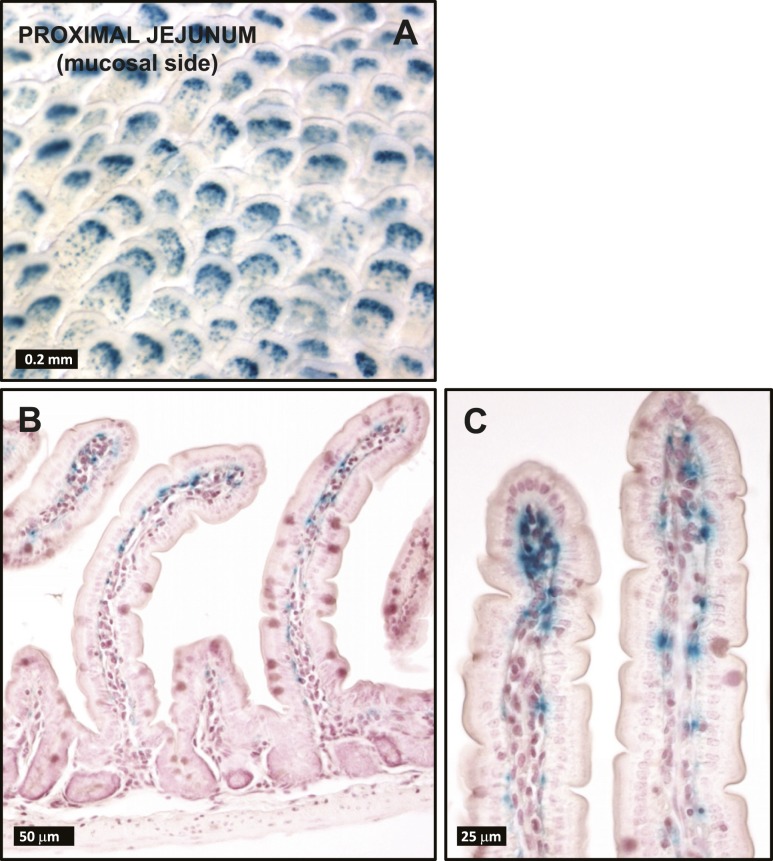
In small intestine whole-mount tissue preparations, no β-galactosidase-positive signal was discernible in the serosal side of the gut wall except for a few scattered fibers in the proximal duodenum running parallel to the cephalocaudal axis. In contrast, strong β-galactosidase staining was evident in the mucosal surface of the small intestine (Fig. 4A), distributed uniformly throughout its entire length. As shown both in the jejunal whole mount, as well in the corresponding histological sections (Fig. 4A–4C), the β-galactosidase positivity was more concentrated toward the tip of the villi and originated from stromal cells lying immediately underneath the epithelium. No β-galactosidase staining was detected in the stroma surrounding the crypts or in the small intestine muscularis nerve plexi.
Figure 4.
(A) Whole mounts and (B and C) histological sections from Glp2rZ/Z mouse jejunum following histochemical detection of β-galactosidase activity. Images are representative of independent whole-mount preparations and multiple tissue sections from three Glp2rZ/Z mice.
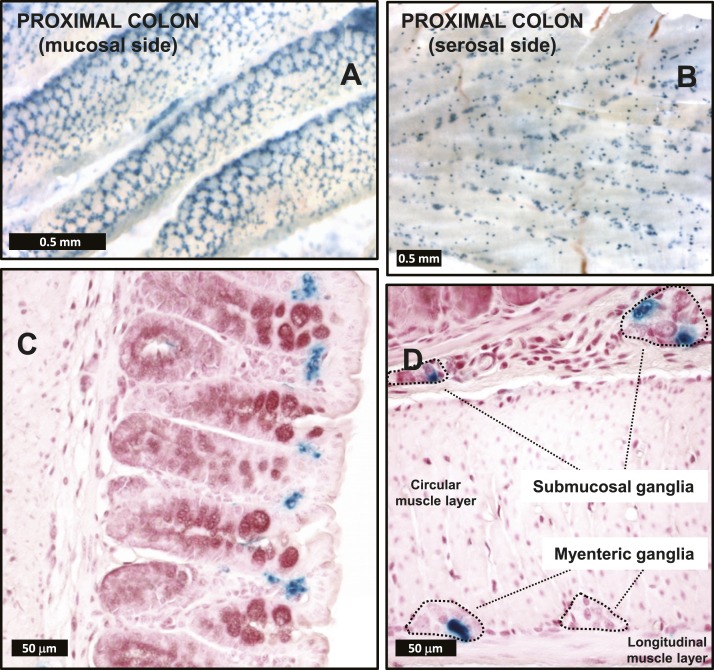
Whole-mount preparations of the proximal one-third of the colon exhibited strong β-galactosidase staining associated to both the mucosal and the serosal surfaces (Fig. 5A and 5B). On the mucosal surface, the staining resembled a honeycomb lattice pattern, created, as can be appreciated in the histological sections, by the combination of β-galactosidase-negative colonic crypt epithelial cells surrounded by β-galactosidase-positive stromal cells concentrated at the luminal end of the colonic crypts (Fig. 5A and 5C). A similar yet slightly more disorganized honeycomb lattice pattern of β-galactosidase staining was also observed in the mucosal surface of the rectum (not shown). The serosal surface of the proximal colon whole-mount preparations revealed a regular pattern of punctate β-galactosidase staining (Fig. 5B). Upon microscopy examination, it became obvious that the pattern was attributable to the presence of numerous enteric nervous-system ganglia, often containing β-galactosidase-positive cells (Fig. 5D).
Figure 5.
(A and B) Whole mounts and (C and D) histological sections from Glp2rZ/Z mouse colon following histochemical detection of β-galactosidase activity. Images are representative of independent whole-mount preparations and multiple tissue sections from three Glp2rZ/Z mice.
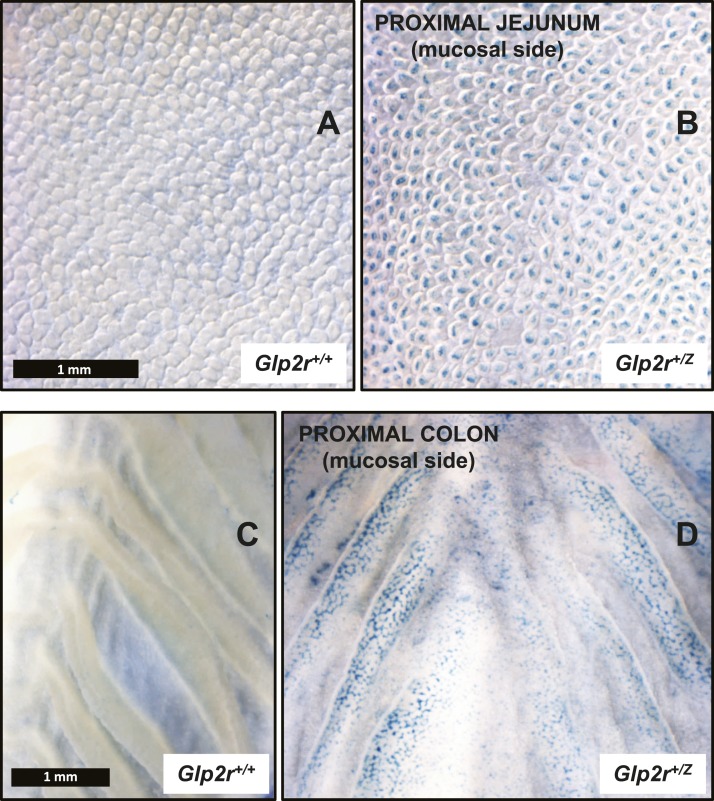
Although the Glp2r-driven LacZ cellular localization, illustrated in Figs. 4 and 5, corresponds to the β-galactosidase-staining pattern observed in the intestine of homozygous Glp2rZ/Z mice with undetectable gut Glp2r expression (Fig. 3B), identical observations were found in analyses of the heterozygous Glp2r+/Z intestine (Fig. 6B and 6D) but not in Glp2r+/+ mice (Fig. 6A and 6C). Hence, the LacZ expression-staining pattern detected in Glp2rZ/Z mice does not appear to be influenced by the presence or absence of functional GLP-2R signaling. Nevertheless, as one might predict, the intensity of the β-galactosidase staining in the Glp2r+/Z gut was weaker and particularly in the proximal colon, tended to be more patchy (Fig. 6B and 6D).
Figure 6.
(A and B) Whole mounts from the jejunum (both images shown at the same magnification) and (C and D) colon (both images shown at the same magnification) of (A and C) Glp2r+/+ and (B and D) Glp2r+/Z mice following histochemical detection of β-galactosidase activity. Images are representative of independent tissue whole mounts from two Glp2r+/+ and six Glp2r+/Z mice.
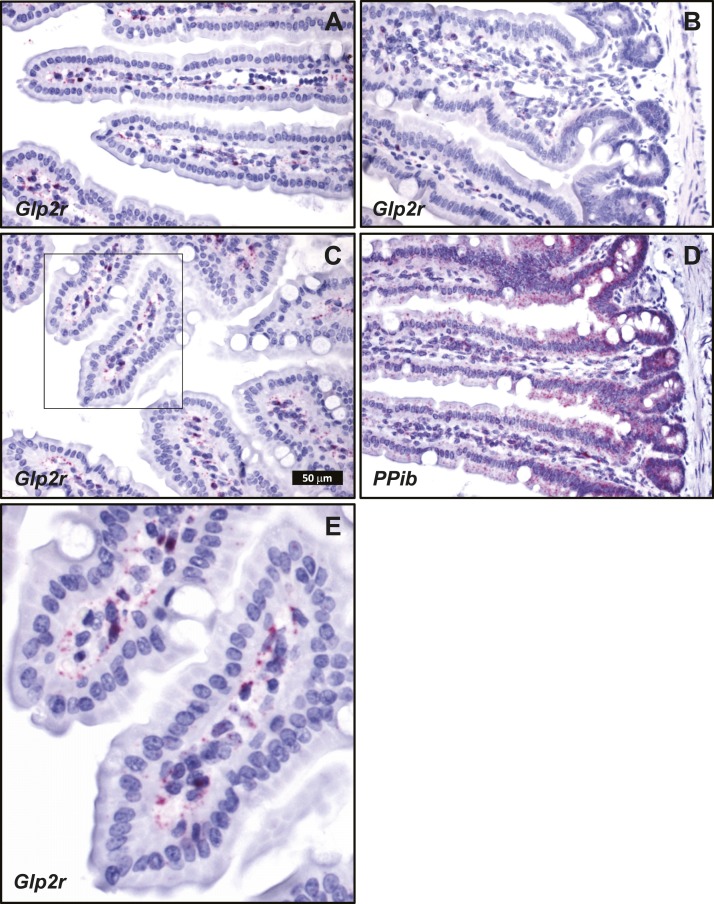
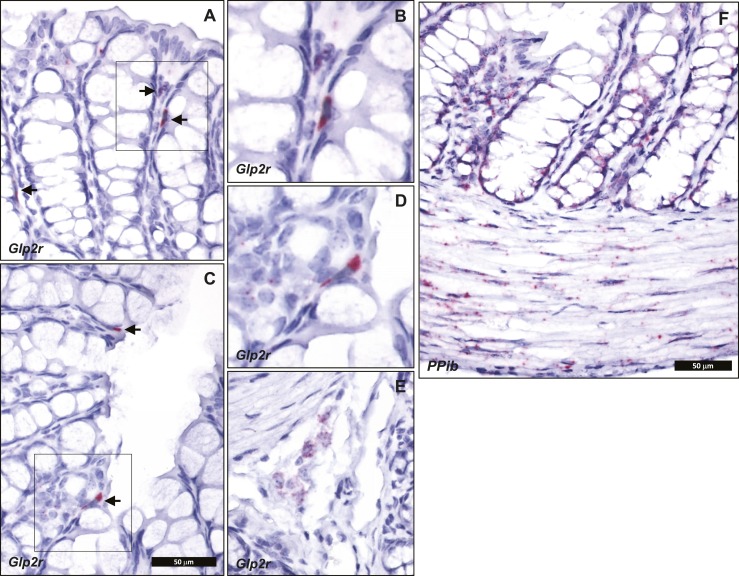
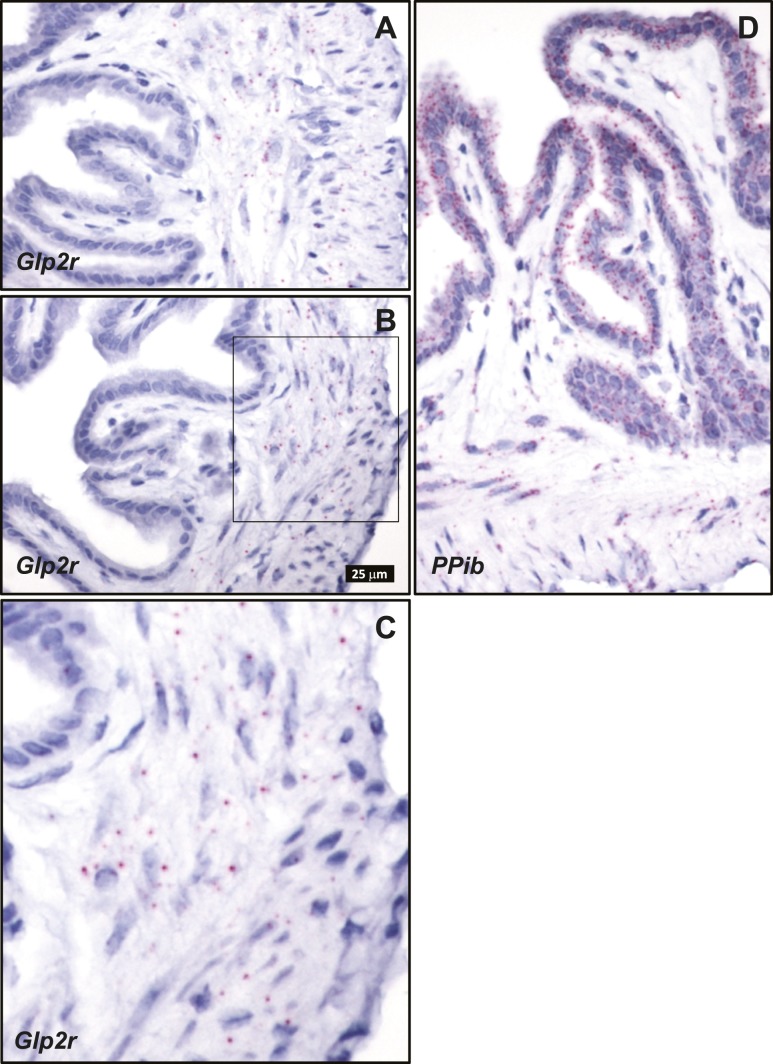
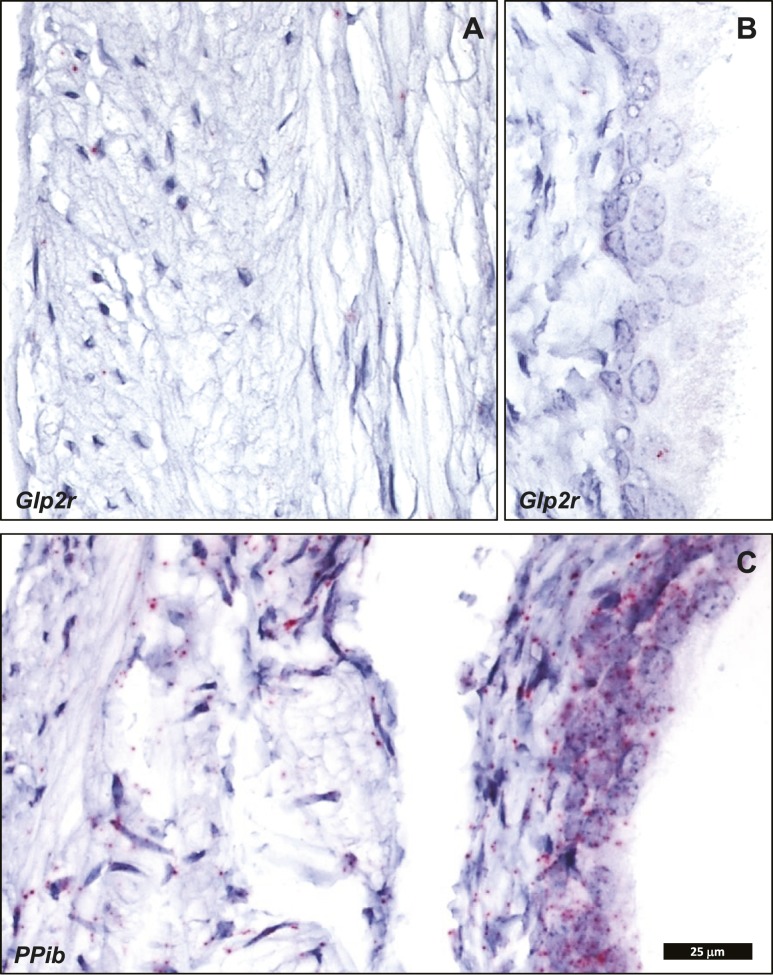
Cellular localization of endogenous intestinal Glp2r mRNA transcripts by ISH using RNAscope corroborated the key findings defined with the Glp2rLacZ reporter mouse. Consistent with a previous report, also using ISH with RNAscope (29), Glp2r mRNA was detected in the jejunum associated to scattered cells within the lamina propria, mainly concentrated in the upper half of the villi (Fig. 7). This distribution was similar to that found for β-galactosidase staining driven by the endogenous Glp2r locus in the reporter mouse (Fig. 4). Consistent with the observations in the Glp2rLacZ tissues, no Glp2r ISH signal was discernible in the villus epithelium, in the stroma surrounding the crypts, or in the muscularis nerve plexi of the small intestine (Fig. 7). Whereas the β-galactosidase-positive stromal cells were conspicuous in the colonic mucosa of the Glp2rLacZ mouse (Fig. 5A and 5C), the ISH signal reporting Glp2r expression in the colon was found associated only to rare, strongly positive cells located in the lamina propria adjacent to the epithelium (Fig. 8A–8D). Finally, consistent with the Glp2rLacZβ-galactosidase staining (Fig. 5B and 5D), the Glp2r mRNA ISH signal was also frequently found associated to ganglia from the colonic enteric nervous system (Fig. 8E). As a control for RNA quality, similar sections were hybridized with a probe for Ppib as a positive control (Fig. 8F).
Figure 7.
Localization of (A–C and E) Glp2r- and (D) Ppib-positive control mRNA in mouse jejunum using RNAscope 2.5 ISH. (A–D) All images correspond to the same magnification. (E) A higher magnification of C, inset. Images are representative of five independent ISH trials using tissue sections from four Glp2r WT mice.
Figure 8.
Localization of (A–E) Glp2r- and (F) Ppib-positive control mRNA in mouse colon using RNAscope 2.5 ISH. (E) Glp2r expression in a colon submucosal ganglion. (A and C) Arrows point to Glp2r mRNA-positive colon stromal cells. (A, C, and E) All images correspond to the same magnification. (B and D) A higher magnification of A and C insets, respectively. Images are representative of five independent ISH trials using tissue sections from four Glp2r WT mice.
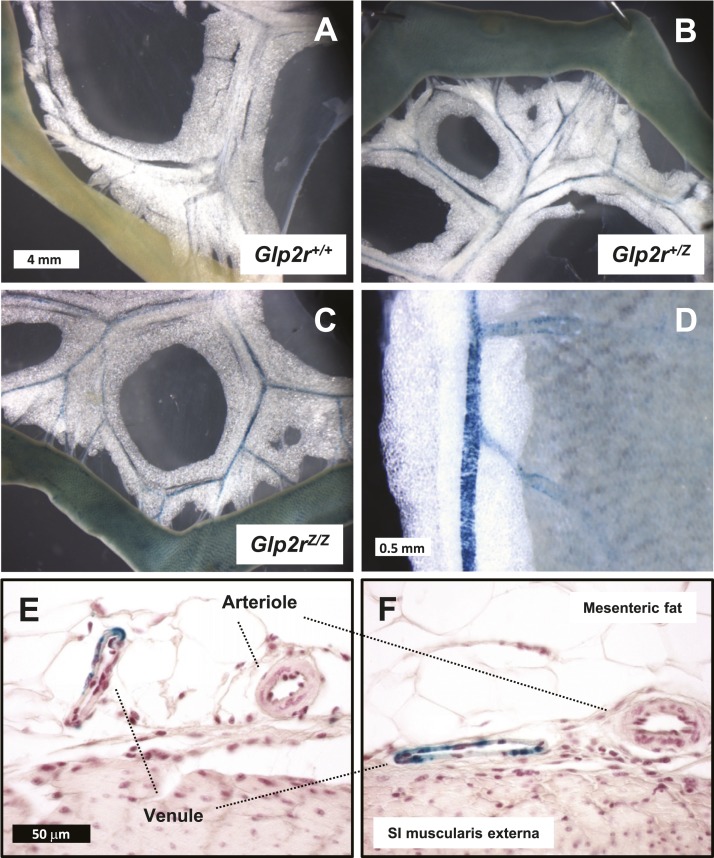
GLP-2 stimulates intestinal blood flow in animals and humans (34, 35); however, the cellular localization of GLP-2R expression within blood vessels has not been extensively studied. β-Galactosidase positivity was not detectable in WT blood vessels (Fig. 9A) but clearly detectable in blood vessels from the mesenteric vascular bed of both Glp2r+/Z and Glp2rZ/Z mice (Fig. 9B and 9C), yet the blue signal was relatively diminished within vascular structures soon after the vessels enter the gut wall (Fig. 9D). Unexpectedly, examination of the histological sections demonstrated that the staining was associated with the walls of veins but not arteries (Fig. 9E and 9F), consistent with the very low Glp2r transcript expression in isolated mesenteric arteries (Fig. 1E). Notably, perivascular adipocytes within mesenteric fat were devoid of β-galactosidase staining, in contrast with the clear β-galactosidase-positive signal from the adjacent mesenteric blood vessels (Fig. 9D). These findings are consistent with the possibility that the variable extent of Glp2r mRNA expression, observed within “mesenteric fat,” as shown by the qPCR data displayed in Fig. 1E, is likely a consequence of the heterogeneous composition of those tissues, with variable amounts of adipose tissue intermingled with blood vessels.
Figure 9.
(A–D) Whole mounts and (E and F) histological sections from (A) Glp2r+/+, (B) Glp2r+/Z, and (C–F) Glp2rZ/Z mouse mesenteric vascular bed following histochemical detection of β-galactosidase activity. (D) Higher magnification of a whole mount from a Glp2rZ/Z mouse mesenteric blood vessel embedded in mesenteric fat. (A–C) All images are shown at the same magnification. (E and F) Histological sections are at the same scale. Images are representative of independent whole-mount preparations from two Glp2r+/+, three Glp2r+/Z, and two Glp2rZ/Z mice and multiple histological tissue sections from two Glp2rZ/Z mice. SI, small intestine.
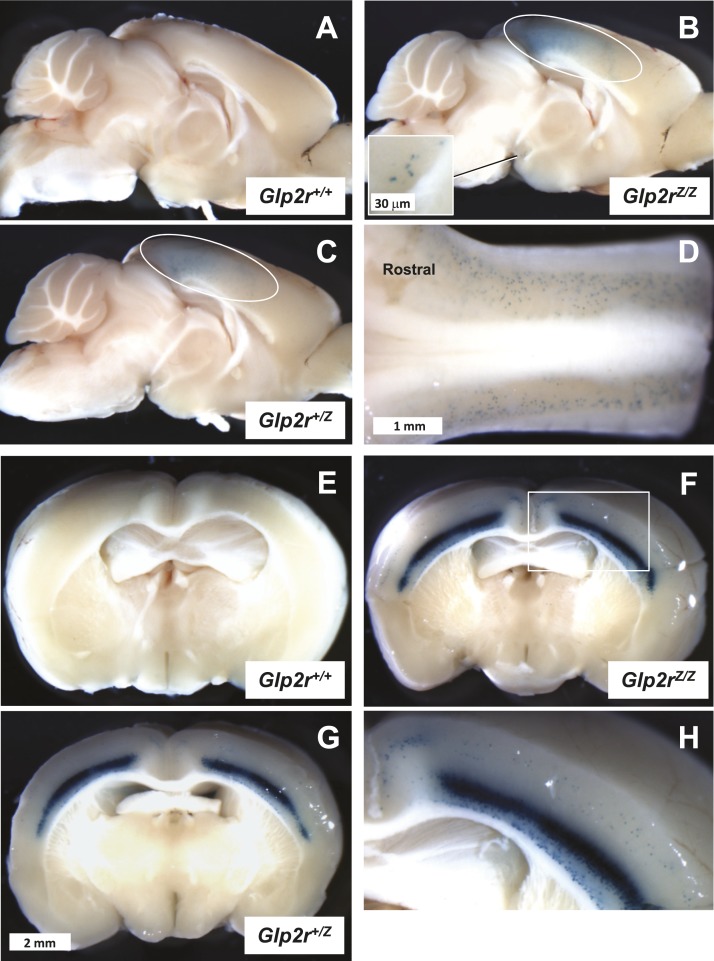
GLP-2R expression has been detected within the rat and mouse brain (12, 22), consistent with described roles for hypothalamic GLP-2R signaling in the control of food intake and glucose homeostasis (36–38). In sagittal sections of the brain (Fig. 10A–10C), β-galactosidase positivity was detected in the neocortex (Fig. 10B and 10C) and more clearly visible within brain coronal sections (Fig. 10E–10G). The β-galactosidase staining was largely restricted to the visual and somatosensory cortex, which displayed a high density of positive cell bodies, mainly concentrated in the inner cell layers (Fig. 10H). β-Galactosidase-positive cell bodies were also noticeable in the outer cell layers of the cortex (Fig. 10H), but they were sporadic in the hypothalamus and thalamus. The cerebellum was devoid of any positivity, whereas the brainstem exhibited numerous β-galactosidase-stained cell bodies scattered in the region of the nucleus of the solitary tract (Fig. 10D).
Figure 10.
Whole-mount (A–C) midsagittal and (E–H) coronal sections from (A and E) Glp2r+/+, (C and G) Glp2r+/Z, and (B, F, and H) Glp2rZ/Z mouse brain following histochemical detection of β-galactosidase activity. (A–C) All images are shown at the same magnification, as they are in (E–G). (D) Dorsal view of the Glp2rZ/Z mouse brainstem demonstrating the presence of β-galactosidase-positive cells in the nucleus of the solitary tract. The cerebellum has been removed to visualize the brainstem. (B and C) The area enclosed by the ovals outlines β-galactosidase positivity in the region of the visual and somatosensory cortex. (B) The inset illustrates a few scattered β-galactosidase–positive cell bodies noticeable in the hypothalamus. (H) A close view of the Glp2rZ/Z mouse brain coronal section of (F), inset, featuring strong β-galactosidase activity in the inner layers of the somatosensory cortex. Images are representative of independent whole-mount preparations from two Glp2r+/+, three Glp2r+/Z, and two Glp2rZ/Z mice.
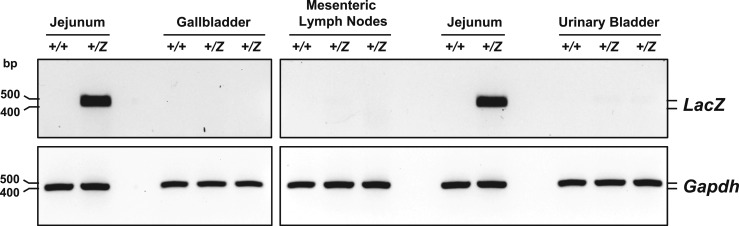
Despite the presence of endogenous full-length Glp2r transcripts in the gallbladder, urinary bladder, and mesenteric lymph nodes (Fig. 2A), β-galactosidase staining was absent in these tissues (data not shown). Consistent with these findings, whereas the LacZ reporter mRNA was strongly expressed in Glp2r+/Z mouse jejunum (assessed by RT-PCR analysis; Fig. 11), LacZ expression was virtually undetectable in the urinary bladder, gallbladder, and mesenteric lymph nodes of Glp2r+/Z mice (Fig. 11). Tissue-selective reporter gene silencing, likely attributable to increased promoter CpG island methylation, has been reported for a subset of LacZ reporter mutant mice from the KOMP Repository (39), thus providing a simple explanation for the failure to detect specific β-galactosidase staining in the corresponding tissues from Glp2r+/Z mice.
Figure 11.
LacZ mRNA expression was evaluated by RT-PCR in total RNA from Glp2r+/+ and Glp2r+/Z mouse jejunum, gallbladder, mesenteric lymph nodes, and urinary bladder. PCR products (LacZ 467 bp long, Gapdh 450 bp) were analyzed by agarose gel electrophoresis using SYBR Safe DNA Gel Stain. Agarose gel images are shown in reverse contrast.
ISH analysis localized Glp2r mRNA expression in the gallbladder to cells within the lamina propria and smooth muscle layers, whereas no Glp2r ISH signal was detected in the epithelium lining the gallbladder lumen (Fig. 12A–12C). mRNA transcripts for the control gene Ppib were detected in gallbladder sections (Fig. 12D). Surprisingly, the RNAscope ISH failed to identify Glp2r mRNA transcripts within cells of the urinary bladder (Fig. 13A and 13B), despite detection of a full-length Glp2r transcript in this organ (Fig. 2A). In contrast, the mRNA for the ISH-positive control Ppib was easily detectable, particularly in the urothelium within the same tissue samples (Fig. 13C).
Figure 12.
Localization of (A–C) Glp2r- and (D) Ppib-positive control mRNA in mouse gallbladder using RNAscope 2.5 ISH. (A, B, and D) All images are shown at the same magnification. (C) A higher magnification of the area selected within (B), inset. Images are representative of four independent ISH trials using tissue sections from four Glp2r WT mice.
Figure 13.
Localization of (A and B) Glp2r- and (C) Ppib-positive control mRNA in mouse urinary bladder using RNAscope 2.5 ISH. (A–C) All images are shown at the same magnification. (B) The image illustrating the urothelium is shown split from the image corresponding to the lamina propria and smooth muscle layers in (A), as they often detached from each other during tissue processing, creating an artifactual empty gap in the tissue section [partly noticeable in (C)]. Images are representative of two independent ISH trials using tissue sections from three Glp2r WT mice.
Discussion
The current studies re-examining Glp2r expression in mouse tissues were prompted, in part, by multiple published reports of GLP-2R protein expression using antisera, subsequently shown to be insufficiently validated for accurate detection of GLP-2R expression (9). Furthermore, other groups have described GLP-2R mRNA or protein expression in the heart (40), skeletal muscle (41), islets (42, 43), and hepatocytes (24), findings inconsistent with our current analyses and the emerging results of single-cell RNA sequencing (44). Similar challenges have emerged surrounding the accuracy of some studies describing localization of GLP-1R expression, where nonvalidated antisera have contributed to confusion regarding the identity of the precise cells and tissues expressing the endogenous canonical GLP-1R (13, 14). Accordingly, to circumvent the limitations of the commercially available GLP-2R antisera (9), we focused on assessment of endogenous Glp2r expression using multiple complementary techniques to provide independent validation and in the case of qPCR, relative quantitation.
As the GI tract is widely regarded as the primary target for GLP-2 action, we focused our studies on assessment of Glp2r mRNA transcripts along the GI tract. Relative Glp2r expression within the GI tract was originally found to be highest in the jejunum, and levels progressively declined in the distal small bowel and colon, as assessed by quantitative RNase protection assays (12). Indeed, a similar pattern of GLP-2R mRNA expression, with highest levels detected in jejunum, was reported in studies of the bovine GI tract (45). Our current findings, using qPCR to assess relative Glp2r expression along the mouse GI tract, are consistent with these previous observations, with higher levels of jejunal Glp2r expression and somewhat lower levels of Glp2r mRNA in the distal small bowel and colon. In contrast, other studies have reported relatively higher levels of mouse Glp2r expression in the rectum and colon and lower levels in the jejunum (24) using endogenous β-actin expression to normalize Glp2r expression across a range of tissues. Our experience, supported by others, is that the expression of genes commonly used to normalize relative gene expression across tissues may vary considerably in various organs and tissues (30), and this commonly used practice of the normalization of gene expression may account, in part, for reported differences in relative Glp2r expression.
The majority of studies examining the cellular localization of murine and rat GLP-2R expression in the intestine has identified enteric neurons as a key cell type that expresses the endogenous GLP-2R (29, 46, 47). Glp2r mRNA transcripts were also detected by ISH within the lamina propria of the mouse gut, consistent with the pattern of Glp2rLacZ expression detected here and confirmed by ISH. The anatomical localization of Glp2r expression to the lamina propria within the small and large intestine is consistent with functional observations linking GLP-2 action to the liberation of keratinocyte growth factor (48), insulin-like growth factor-1 (49), and ErbB ligands (50), growth factors known to be produced in and secreted from stromal cells residing within the lamina propria.
The analyses reported here also highlight common limitations of techniques widely used to analyze gene expression. First, in regard to data obtained using the Glp2rLacZ reporter mouse, we were unable to detect LacZ expression and corresponding β-galactosidase staining in several tissues that expressed robust levels of the endogenous full-length Glp2r mRNA transcript, notably gallbladder, mesenteric lymph nodes, and urinary bladder. The silencing of reporter transgene expression has been attributed to differential tissue-specific methylation of promoter sequences (39), highlighting the importance of using multiple independent techniques to assess the tissue and cellular localization of gene expression.
We also report a marked discrepancy between the abundance of Glp2r mRNA transcripts quantified in testis mRNA using qPCR vs regular PCR for detection of a full-length mRNA transcript. Indeed, a combination of qPCR assays using different primer pairs revealed markedly different expression of exons at the 5′-end vs downstream Glp2r coding sequences. As qPCR has become the most common technique used for detection and quantitation of gene expression, our observations re-emphasize the importance of establishing the presence of full-length mRNA transcripts using multiple primer pairs or regular PCR (33) before assuming that results from a single qPCR assay reflect expression of authentic full-length mRNA transcripts. Indeed, alternative RNA splicing is a common process in the testis, and aberrant alternative RNA splicing in the testis giving rise to noncoding RNAs has been described for vasopressin (51), pro-opiomelanocortin (52), and the follicle-stimulating hormone receptor (53).
In summary, these findings extend previous observations localizing GLP-2R expression to enteric neurons and lamina propria stromal cells within the small and large bowel and provide an extensive, quantitative survey of Glp2r expression in several dozen murine tissues. We highlight several findings, including differential Glp2r expression in adipose tissue depots and predominant vascular expression of the Glp2r within mesenteric veins and not arteries. We also describe and characterize a reporter Glp2rLacZ mouse that should prove useful for more detailed cellular analysis of Glp2r expression during development and in response to GI injury. Finally, we highlight potential pitfalls in assignment of gene expression, including discrepant endogenous Glp2r vs LacZ expression and differential expression of Glp2r mRNA transcripts lacking 5′ coding sequences, emphasizing the need for caution in the interpretation of data generated using a single pair of qPCR primers. Collectively, these findings should prove useful for future studies examining the cellular basis for the understanding of GLP-2 action in mouse tissues.
These studies have several limitations. First, we did not simultaneously report levels of GLP-2R protein expression, and it cannot be assumed that the Glp2r mRNA transcripts are uniformly translated to the same extent in each tissue. Second, we focused on studies of murine tissues, and a considerable gap remains in the quantitative assignment of GLP-2R expression to specific human cells and tissues. The limitations surrounding use of GLP-2R antisera extend to other members of the class B GPCR family. Indeed, we have described multiple problems with antibody sensitivity and specificity for the three major members of this class: the GLP-1R, GLP-2R, and gastric inhibitory polypeptide receptor (9, 14, 54). Moreover, it seems evident that these challenges with antibody validation are similarly relevant to a range of GPCRs within structurally related classes (55). Taken together, it remains prudent to assess RNA and protein expression using multiple quantitative, independently validated techniques to diminish the likelihood of experimental error.
Acknowledgments
Financial Support: These studies were supported, in part, by Canadian Institutes of Health Research Foundation Grant 154321 and an investigator-initiated grant to Mount Sinai Hospital from Shire Inc, a member of the Takeda group of companies.
Glossary
Abbreviations:
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GCGR
glucagon receptor
- GI
gastrointestinal
- GLP-1
glucagon-like peptide-1
- GLP-1R
glucagon-like peptide-1 receptor
- GLP-2
glucagon-like peptide-2
- GLP-2R
glucagon-like peptide-2 receptor
- GPCR
G protein–coupled receptor
- ISH
in situ hybridization
- KOMP
Knockout Mouse Project
- ORF
open-reading frame
- PGDP
proglucagon-derived peptide
- qPCR
quantitative PCR
- WT
wild-type
Additional Information
Disclosure Summary: D.J.D. has served as an advisor, consultant, or speaker within the past 12 months to Forkhead Biotherapeutics, Heliome, Inc., Intarcia Therapeutics, Kallyope, Merck Research Laboratories, Novo Nordisk Inc., Pfizer Inc., and Sanofi Inc. Neither D.J.D. nor his family members hold stock directly or indirectly in any of these companies. GLP-2 is the subject of a patent license agreement among Shire Inc.; the University of Toronto, Toronto General Hospital (University Health Network); and D.J.D. The remaining authors have nothing to disclose.
Data Availability: All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References and Notes
- 1. Sandoval DA, D’Alessio DA. Physiology of proglucagon peptides: role of glucagon and GLP-1 in health and disease. Physiol Rev. 2015;95(2):513–548. [DOI] [PubMed] [Google Scholar]
- 2. Mayo KE, Miller LJ, Bataille D, Dalle S, Göke B, Thorens B, Drucker DJ. International Union of Pharmacology. XXXV. The glucagon receptor family. Pharmacol Rev. 2003;55(1):167–194. [DOI] [PubMed] [Google Scholar]
- 3. Campbell JE, Drucker DJ. Islet α cells and glucagon--critical regulators of energy homeostasis. Nat Rev Endocrinol. 2015;11(6):329–338. [DOI] [PubMed] [Google Scholar]
- 4. Müller TD, Finan B, Clemmensen C, DiMarchi RD, Tschöp MH. The new biology and pharmacology of glucagon. Physiol Rev. 2017;97(2):721–766. [DOI] [PubMed] [Google Scholar]
- 5. Drucker DJ, Habener JF, Holst JJ. Discovery, characterization, and clinical development of the glucagon-like peptides. J Clin Invest. 2017;127(12):4217–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 2018;27(4):740–756. [DOI] [PubMed] [Google Scholar]
- 7. Andersen A, Lund A, Knop FK, Vilsbøll T. Glucagon-like peptide 1 in health and disease. Nat Rev Endocrinol. 2018;14(7):390–403. [DOI] [PubMed] [Google Scholar]
- 8. Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368(9548):1696–1705. [DOI] [PubMed] [Google Scholar]
- 9. Drucker DJ, Yusta B. Physiology and pharmacology of the enteroendocrine hormone glucagon-like peptide-2. Annu Rev Physiol. 2014;76(1):561–583. [DOI] [PubMed] [Google Scholar]
- 10. Brubaker PL. Glucagon-like peptide-2 and the regulation of intestinal growth and function. Compr Physiol. 2018;8(3):1185–1210. [DOI] [PubMed] [Google Scholar]
- 11. Drucker DJ, Erlich P, Asa SL, Brubaker PL. Induction of intestinal epithelial proliferation by glucagon-like peptide 2. Proc Natl Acad Sci USA. 1996;93(15):7911–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Munroe DG, Gupta AK, Kooshesh F, Vyas TB, Rizkalla G, Wang H, Demchyshyn L, Yang ZJ, Kamboj RK, Chen H, McCallum K, Sumner-Smith M, Drucker DJ, Crivici A. Prototypic G protein-coupled receptor for the intestinotrophic factor glucagon-like peptide 2. Proc Natl Acad Sci USA. 1999;96(4):1569–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pyke C, Knudsen LB. The glucagon-like peptide-1 receptor--or not? Endocrinology. 2013;154(1):4–8. [DOI] [PubMed] [Google Scholar]
- 14. Panjwani N, Mulvihill EE, Longuet C, Yusta B, Campbell JE, Brown TJ, Streutker C, Holland D, Cao X, Baggio LL, Drucker DJ. GLP-1 receptor activation indirectly reduces hepatic lipid accumulation but does not attenuate development of atherosclerosis in diabetic male ApoE(−/−) mice. Endocrinology. 2013;154(1):127–139. [DOI] [PubMed] [Google Scholar]
- 15. Baggio LL, Yusta B, Mulvihill EE, Cao X, Streutker CJ, Butany J, Cappola TP, Margulies KB, Drucker DJ. GLP-1 receptor expression within the human heart. Endocrinology. 2018;159(4):1570–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pyke C, Heller RS, Kirk RK, Ørskov C, Reedtz-Runge S, Kaastrup P, Hvelplund A, Bardram L, Calatayud D, Knudsen LB. GLP-1 receptor localization in monkey and human tissue: novel distribution revealed with extensively validated monoclonal antibody. Endocrinology. 2014;155(4):1280–1290. [DOI] [PubMed] [Google Scholar]
- 17. Körner M, Stöckli M, Waser B, Reubi JC. GLP-1 receptor expression in human tumors and human normal tissues: potential for in vivo targeting. J Nucl Med. 2007;48(5):736–743. [DOI] [PubMed] [Google Scholar]
- 18. Richards P, Parker HE, Adriaenssens AE, Hodgson JM, Cork SC, Trapp S, Gribble FM, Reimann F. Identification and characterisation of glucagon-like peptide-1 receptor expressing cells using a new transgenic mouse model. Diabetes. 2014;63(4):1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunphy JL, Taylor RG, Fuller PJ. Tissue distribution of rat glucagon receptor and GLP-1 receptor gene expression. Mol Cell Endocrinol. 1998;141(1–2):179–186. [DOI] [PubMed] [Google Scholar]
- 20. Merchenthaler I, Lane M, Shughrue P. Distribution of pre-pro-glucagon and glucagon-like peptide-1 receptor messenger RNAs in the rat central nervous system. J Comp Neurol. 1999;403(2):261–280. [DOI] [PubMed] [Google Scholar]
- 21. Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, Smith DM, Kasper M, Ämmälä C, Sandberg R. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24(4):593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yusta B, Huang L, Munroe D, Wolff G, Fantaske R, Sharma S, Demchyshyn L, Asa SL, Drucker DJ. Enteroendocrine localization of GLP-2 receptor expression in humans and rodents. Gastroenterology. 2000;119(3):744–755. [DOI] [PubMed] [Google Scholar]
- 23. Guan X, Karpen HE, Stephens J, Bukowski JT, Niu S, Zhang G, Stoll B, Finegold MJ, Holst JJ, Hadsell D, Nichols BL, Burrin DG. GLP-2 receptor localizes to enteric neurons and endocrine cells expressing vasoactive peptides and mediates increased blood flow [published correction appears in Gastroenterology 2006;130(3):1019–1021]. Gastroenterology. 2006;130(1):150–164. [DOI] [PubMed] [Google Scholar]
- 24. El-Jamal N, Erdual E, Neunlist M, Koriche D, Dubuquoy C, Maggiotto F, Chevalier J, Berrebi D, Dubuquoy L, Boulanger E, Cortot A, Desreumaux P. Glugacon-like peptide-2: broad receptor expression, limited therapeutic effect on intestinal inflammation and novel role in liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2014;307(3):G274–G285. [DOI] [PubMed] [Google Scholar]
- 25. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. [DOI] [PubMed] [Google Scholar]
- 26. Lamont BJ, Li Y, Kwan E, Brown TJ, Gaisano H, Drucker DJ. Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. J Clin Invest. 2012;122(1):388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yusta B, Baggio LL, Estall JL, Koehler JA, Holland DP, Li H, Pipeleers D, Ling Z, Drucker DJ. GLP-1 receptor activation improves β cell function and survival following induction of endoplasmic reticulum stress. Cell Metab. 2006;4(5):391–406. [DOI] [PubMed] [Google Scholar]
- 28. Scott RB, Kirk D, MacNaughton WK, Meddings JB. GLP-2 augments the adaptive response to massive intestinal resection in rat. Am J Physiol. 1998;275(5):G911–G921 [DOI] [PubMed] [Google Scholar]
- 29. Wismann P, Barkholt P, Secher T, Vrang N, Hansen HB, Jeppesen PB, Baggio LL, Koehler JA, Drucker DJ, Sandoval DA, Jelsing J. The endogenous preproglucagon system is not essential for gut growth homeostasis in mice. Mol Metab. 2017;6(7):681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee PD, Sladek R, Greenwood CM, Hudson TJ. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002;12(2):292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yusta B, Matthews D, Flock GB, Ussher JR, Lavoie B, Mawe GM, Drucker DJ. Glucagon-like peptide-2 promotes gallbladder refilling via a TGR5-independent, GLP-2R-dependent pathway. Mol Metab. 2017;6(6):503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bahrami J, Longuet C, Baggio LL, Li K, Drucker DJ. Glucagon-like peptide-2 receptor modulates islet adaptation to metabolic stress in the ob/ob mouse. Gastroenterology. 2010;139(3):857–868. [DOI] [PubMed] [Google Scholar]
- 33. Drucker DJ. The ascending GLP-1 road from clinical safety to reduction of cardiovascular complications. Diabetes. 2018;67(9):1710–1719. [DOI] [PubMed] [Google Scholar]
- 34. Stephens J, Stoll B, Cottrell J, Chang X, Helmrath M, Burrin DG. Glucagon-like peptide-2 acutely increases proximal small intestinal blood flow in TPN-fed neonatal piglets. Am J Physiol Regul Integr Comp Physiol. 2006;290(2):R283–R289. [DOI] [PubMed] [Google Scholar]
- 35. Bremholm L, Hornum M, Henriksen BM, Larsen S, Holst JJ. Glucagon-like peptide-2 increases mesenteric blood flow in humans. Scand J Gastroenterol. 2009;44(3):314–319. [DOI] [PubMed] [Google Scholar]
- 36. Tang-Christensen M, Larsen PJ, Thulesen J, Rømer J, Vrang N. The proglucagon-derived peptide, glucagon-like peptide-2, is a neurotransmitter involved in the regulation of food intake. Nat Med. 2000;6(7):802–807. [DOI] [PubMed] [Google Scholar]
- 37. Lovshin J, Estall J, Yusta B, Brown TJ, Drucker DJ. Glucagon-like peptide (GLP)-2 action in the murine central nervous system is enhanced by elimination of GLP-1 receptor signaling. J Biol Chem. 2001;276(24):21489–21499. [DOI] [PubMed] [Google Scholar]
- 38. Shi X, Zhou F, Li X, Chang B, Li D, Wang Y, Tong Q, Xu Y, Fukuda M, Zhao JJ, Li D, Burrin DG, Chan L, Guan X. Central GLP-2 enhances hepatic insulin sensitivity via activating PI3K signaling in POMC neurons. Cell Metab. 2013;18(1):86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirov JV, Adkisson M, Nava AJ, Cipollone A, Willis B, Engelhard EK, Lloyd KC, de Jong P, West DB. Reporter gene silencing in targeted mouse mutants is associated with promoter CpG island methylation. PLoS One. 2015;10(8):e0134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Angelone T, Filice E, Quintieri AM, Imbrogno S, Amodio N, Pasqua T, Pellegrino D, Mulè F, Cerra MC. Receptor identification and physiological characterisation of glucagon-like peptide-2 in the rat heart. Nutr Metab Cardiovasc Dis. 2012;22(6):486–494. [DOI] [PubMed] [Google Scholar]
- 41. Chai YL, Ma HM, Jiang J. Molecular characterization, tissue expression profile and SNP analysis of porcine GLP2R. Genet Mol Res. 2015;14(4):12931–12941. [DOI] [PubMed] [Google Scholar]
- 42. de Heer J, Pedersen J, Orskov C, Holst JJ. The alpha cell expresses glucagon-like peptide-2 receptors and glucagon-like peptide-2 stimulates glucagon secretion from the rat pancreas. Diabetologia. 2007;50(10):2135–2142. [DOI] [PubMed] [Google Scholar]
- 43. Khan D, Vasu S, Moffett RC, Irwin N, Flatt PR. Differential expression of glucagon-like peptide-2 (GLP-2) is involved in pancreatic islet cell adaptations to stress and beta-cell survival. Peptides. 2017;95:68–75. [DOI] [PubMed] [Google Scholar]
- 44. Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I, Itzkovitz S. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver [published correction appears in Nature 2017;543(7647):742]. Nature. 2017;542(7641):352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Connor EE, Baldwin RL VI, Capuco AV, Evock-Clover CM, Ellis SE, Sciabica KS. Characterization of glucagon-like peptide 2 pathway member expression in bovine gastrointestinal tract. J Dairy Sci. 2010;93(11):5167–5178. [DOI] [PubMed] [Google Scholar]
- 46. Bjerknes M, Cheng H. Modulation of specific intestinal epithelial progenitors by enteric neurons. Proc Natl Acad Sci USA. 2001;98(22):12497–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pedersen J, Pedersen NB, Brix SW, Grunddal KV, Rosenkilde MM, Hartmann B, Ørskov C, Poulsen SS, Holst JJ. The glucagon-like peptide 2 receptor is expressed in enteric neurons and not in the epithelium of the intestine. Peptides. 2015;67:20–28. [DOI] [PubMed] [Google Scholar]
- 48. Ørskov C, Hartmann B, Poulsen SS, Thulesen J, Hare KJ, Holst JJ. GLP-2 stimulates colonic growth via KGF, released by subepithelial myofibroblasts with GLP-2 receptors. Regul Pept. 2005;124(1-3):105–112. [DOI] [PubMed] [Google Scholar]
- 49. Dubé PE, Forse CL, Bahrami J, Brubaker PL. The essential role of insulin-like growth factor-1 in the intestinal tropic effects of glucagon-like peptide-2 in mice. Gastroenterology. 2006;131(2):589–605. [DOI] [PubMed] [Google Scholar]
- 50. Yusta B, Holland D, Koehler JA, Maziarz M, Estall JL, Higgins R, Drucker DJ. ErbB signaling is required for the proliferative actions of GLP-2 in the murine gut. Gastroenterology. 2009;137(3):986–996. [DOI] [PubMed] [Google Scholar]
- 51. Foo NC, Carter D, Murphy D, Ivell R. Vasopressin and oxytocin gene expression in rat testis. Endocrinology. 1991;128(4):2118–2128. [DOI] [PubMed] [Google Scholar]
- 52. Garrett JE, Collard MW, Douglass JO. Translational control of germ cell-expressed mRNA imposed by alternative splicing: opioid peptide gene expression in rat testis. Mol Cell Biol. 1989;9(10):4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Song GJ, Park YS, Lee YS, Lee CC, Kang IS. Alternatively spliced variants of the follicle-stimulating hormone receptor gene in the testis of infertile men. Fertil Steril. 2002;77(3):499–504. [DOI] [PubMed] [Google Scholar]
- 54. Ussher JR, Campbell JE, Mulvihill EE, Baggio LL, Bates HE, McLean BA, Gopal K, Capozzi M, Yusta B, Cao X, Ali S, Kim M, Kabir MG, Seino Y, Suzuki J, Drucker DJ. Inactivation of the glucose-dependent insulinotropic polypeptide receptor improves outcomes following experimental myocardial infarction. Cell Metab. 2018;27(2):450–460.e6. [DOI] [PubMed] [Google Scholar]
- 55. Michel MC, Wieland T, Tsujimoto G. How reliable are G-protein-coupled receptor antibodies? Naunyn Schmiedebergs Arch Pharmacol. 2009;379(4):385–388. [DOI] [PubMed] [Google Scholar]