Abstract
Pili on the surface of Sulfolobus islandicus are used for many functions, and serve as receptors for certain archaeal viruses. The cells grow optimally at pH 3 and 80° C, exposing these extracellular appendages to a very harsh environment. These pili, when removed from cells, resist digestion by trypsin or pepsin, and survive boiling in SDS or 5M guanidinium-HCl. We have used cryo-EM to determine the structure of these filaments at 4.1 Å resolution. An atomic model was built by combining the map with bioinformatics without prior knowledge of the pilin sequence, an approach that should prove useful for assemblies where all of the components are not known. The atomic structure of the pilus was unusual, with almost a third of the residues being either threonine or serine, and with many hydrophobic surface residues. While the map showed extra density consistent with glycosylation for only three residues, mass measurements suggested extensive glycosylation. We propose that this extensive glycosylation renders these filaments soluble and provides the remarkable structural stability. We also show that the overall fold of the archaeal pilin is remarkably similar to archaeal flagellin, establishing common evolutionary origins.
Introduction
Extremophiles are organisms that have evolved to thrive under extremely harsh environmental conditions, such as high temperature, acidic or alkaline pH, near-saturating salt concentrations, etc. The question of how macromolecular assemblies remain stable in such settings has mystified scientists ever since the discovery that life can exist in such harsh habitats. Particularly puzzling is the robustness of proteinaceous assemblies, such as virus particles, which have to maintain stability while outside of the host cell1, and various appendages, which commonly decorate the surface of the cells and mediate diverse interactions with the environment2. Both types of structures are assembled from a limited number of symmetrically arranged protein components. Recent structural studies on several viruses infecting hyperthermophilic archaea have provided valuable insights into how these viruses protect their genomes and maintain structural integrity3–7. However, the principles underlying the stability of cellular appendages remain poorly understood.
Type 4 pili (T4P) are among the most commonly found surface appendages in extremophilic archaea, with relatively small pilin proteins assembling into functionally diverse, stable structures of several μm in length2,8,9. T4P are conserved across both prokaryotic domains of life, suggesting their antiquity10. T4P have evolved to perform a host of functions, including motility, adhesion, aggregation, natural transformation, host cell signaling and more8,11. Although some functions of T4P are conserved in both bacteria and archaea, such as adhesion to various biotic and abiotic surfaces, there are also domain-specific T4P activities. The archaeal flagellum, which is evolutionarily unrelated to either the bacterial or eukaryotic flagellum, has evolved from T4P in the domain Archaea12.
All structurally characterized T4P display a characteristic two-domain organization, with the highly hydrophobic N-terminal α-helix, which is typically melted between residues ~14 to ~23 in the filament13–15 but a continuous helix in crystal structures of detergent-solubilized individual subunits16–18, and the C-terminal globular domain that can be quite variable or almost entirely absent19. The precursors of T4P, prepilins, have N-terminal class III signal peptides that target them for transport across the plasma membrane in a Sec-dependent manner20, which are then processed by a prepilin peptidase (PilD in bacteria, PibD in archaea) and subsequently incorporated into the pilus21,22. The only other components necessary for T4P assembly appear to be the PilB ATPase, which powers the extrusion of pilin subunits from the membrane, and PilC, a transmembrane protein that is believed to anchor the assembled T4P to the membrane8.
Here we studied the structure of a pilus from Sulfolobus islandicus LAL14/1, a polyextremophilic (hyperthermophilic acidophile) archaeon growing optimally at ca. 80 °C and pH 323. Previous comprehensive comparative genomics analysis of the distribution of loci encoding flagella and other T4P in archaea has shown that LAL14/1, unlike other S. islandicus strains, does not carry genes for a functional flagellum9. LAL14/1 cells contain only one discernable type of filamentous appendages, which are ~10 nm in diameter when negatively-stained or ~5 nm when observed using electron cryo-tomography, and their length is highly variable, reaching up to 12.5 μm24. Multiple pili are typically expressed on every cell (Supp. Fig. 1). These pili serve as the main receptor recognized by S. islandicus rod-shaped virus 224, a model virus in the family Rudiviridae1.
Results
The pili resist degradation and characterization
The pili were purified from Sulfolobus islandicus LAL14/1 cells. Initial attempts to characterize these pili by mass spectrometry (MS) in both Paris and Charlottesville failed, as no proteins that were likely pilins could be identified after in-solution digestion using trypsin. Incubating the pili with a high concentration of pepsin (at pH 2.2, near the peak activity for pepsin) for seven days resulted in a peptide mixture analyzed by mass spectrometry where over 90% were fragments of pepsin. No likely pilins were found in the remainder. Boiling the pili in 1% SDS failed to produce any bands seen by Coomassie Blue staining of gels, despite the high concentration of filaments, but trace amounts could be seen by silver staining. In fact, the pili were not depolymerized after boiling for 30 minutes in 5M guanidinium-HCl at pH 5.9 (Sup. Fig. 2).
Cryo-EM of the pili
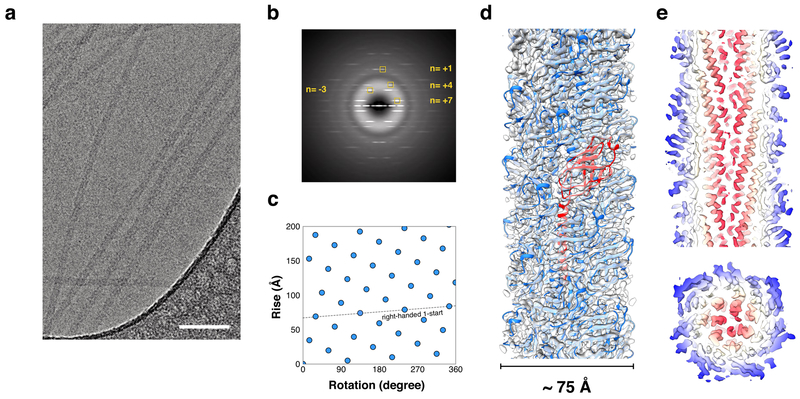
We used cryo-EM (Fig. 1a) to determine the structure of these filaments and were able to generate a 4.1 Å resolution reconstruction as judged by both a map:map Fourier Shell Correlation (FSC) and a subsequent map:model FSC (Sup. Fig. 3). The subunits in the filament are related to each other by a rotation of ~105° about the helical axis and a translation of 4.9 Å along the helical axis, generating a right-handed 1-start helix with a pitch of ~17 Å (Fig. 1b,c). The filaments are ~75 Å in diameter and have a solid core (Fig. 1d). Normally, this resolution would immediately allow for building a de novo atomic model25. However, the sequence of the component pilin was unknown, and we therefore attempted to use bioinformatics to determine it. The map revealed that the core consisted of α-helical segments each containing ~35 residues, and an atomic model without sidechains for the α-helical core of the Igniococcus hospitalis adhesion filament26 fit remarkably well into this density.
Figure 1. Cryo-EM of the LAL14/1 pilus.
a, Representative cryo-EM image of the pili, taken from 1,824 micrographs recorded. Scale bar, 50 nm.
b, Averaged power spectrum of the segments used in the final reconstruction. The layer-lines that were used for determining helical symmetry are labeled.
c, Helical net of the LAL14/1 pilus using the convention that the surface is unrolled and we are viewing from the outside. One turn of the ~ 17 Å pitch 1-start helix that passes through every subunit is shown.
d, Cryo-EM reconstruction of the LAL14/1 pilus at 4.1 Å resolution with fitted atomic model. One subunit is highlighted in red.
e,f, Thin slices parallel (top) and perpendicular (bottom) to the helical axis of the pilus are shown, colored by the helical radius. The tight packing of the all α-helical core can be seen.
It has previously been shown27 that this core for the I. hospitalis adhesion filament is composed of a homolog of the bacterial T4P N-terminal domain. Such homology is also found in the archaeal flagellar filament N-terminal domain28–32. In contrast to the partial melting of this α-helix in the assembled bacterial T4P filaments13,14,33, this region is all α-helical in the I. hospitalis adhesion filament26, the archaeal flagellar filaments28,29 and in the density for the LAL14/1 pilus. The helix-breaking residues Gly14 and Pro22 that surround the unstructured region in the bacterial T4P filaments are not found in the archaeal sequences27.
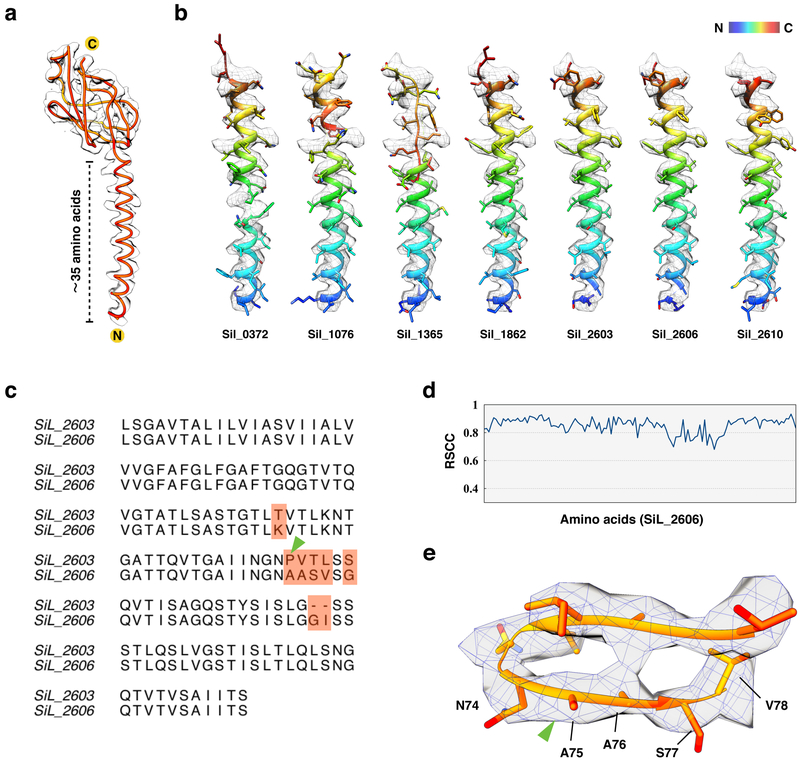
An unambiguous Cα trace was made through the density map, yielding ~125 residues (Fig. 2a). Given that the signal sequence might contain as many as 15 residues, and that there could be some uncertainty in the estimate of 125 residues, a search was made of the LAL14/1 proteome for all proteins containing between 120–145 residues with a single predicted transmembrane domain near the N-terminus and a β-strand rich C-terminal domain. Only seven such sequences were found. RosettaCM34 was then used to thread the first 50 amino acids of these seven candidates through the α-helical core densities (Fig. 2b). This model building approach screened the best 35 out of 50 residues and fit them into the map, thus eliminating the uncertainty of the length of the signal peptides. SiL_0372, SiL_1076 and SiL_1365 could not be fitted into the helical density; SiL_1862 and SiL_2610 could roughly fit into the helical density but had some side chain density mismatches; SiL_2603 and SiL_2606 have identical N-terminal sequences and fit into the map of the helical core very well (Fig. 2b). A full length model could not be built for either SiL_1862 nor SiL_2610. In addition, neither of them had signal peptides present assuming the helical fit was correct, which is very unlikely for a pilus that needs to be transported by the conserved secretion system. A full-length model could be built for both SiL_2603 and SiL_2606, and their sequences only differ by eight residues (Fig. 2c). It is challenging to distinguish them by cryo-EM at the available resolution as some of the amino acid changes involve leucine to valine or valine to alanine, and the two amino acid insertion/deletion occurs within a poorly defined loop in the density map. SiL_2606 matches the cryo-EM very well with overall 0.80 real-space correlation coefficient (Fig. 2d). If one assumes SiL_2603 to be the correct sequence, then there will be a proline in the middle of a well defined β-stand, which is extremely unlikely (Fig. 2e) as this would break the backbone hydrogen bonds holding the β-sheet together. Our structural model also indicates that the first 10 residues in both SiL_2603 and SiL_2606 correspond to the signal sequence, which is cleaved from the mature pilin protein. The cleavage thus occurs between Ala10 and Leu11 residues (KA↓LS) and the cleavage site matches perfectly the previously established21 consensus recognition motif of the signal peptidase PibD ([K/R][G/A][L/I/F][S/T/A]). A BLASTP search has shown that orthologs of SiL_2603 and SiL_2606 are conserved in other Sulfolobus species. Notably, orthologs in S. acidocaldarius have been characterized experimentally and shown to be involved in formation of the adhesive pili35. Consistent with our structural data, SiL_2606 is orthologous to the major pilin, Saci_2319 (40% identity over 140 aa alignment).
Figure 2. De novo atomic model building of LAL14/1 pilin.
a, The protein Cα trace in orange with N- and C-termini of the pilin subunit labeled.
b, RosettaCM models of the first 50 amino acids of seven bioinformatically-identified sequences which matched the Cα trace in (a), fit into the N-terminal helix density.
c, Sequence alignment of SiL_2603 and SiL_2606. The pink shading shows the regions where the two sequences differ. The green arrowhead indicates the proline present in Sil_2603.
d, The per-residue real space correlation coefficient (RSCC) plot of the atomic model from Sil_2606 against the 4.1 Å cryo-EM map.
e, The cryo-EM map of the region indicated in (c) by green arrowhead, with SiL_2606 atomic model fit into the map.
Understanding the basis for the remarkable stability
The first question was whether these filaments contained amyloid, since amyloid filaments have also been shown to resist extremely harsh conditions36. The β-sheets in the pili appear to be typical β-sheets, and not the steric zippers associated with amyloid fibers37. Consistent with this, the filaments failed to stain with either Congo Red or thioflavin T (Sup. Fig. 4). Long Range Order (LRO) parameters have been invoked to explain the unusual thermal and kinetic stability of some proteins38,39. We compared the LRO score for the LAL14/1 pili with a number of bacterial T4P and archaeal flagellar filaments (Sup. Fig. 5). It can be seen that while the predicted LRO for the LAL14/1 pilus is higher than most others in the comparison, it is comparable to that of an archaeal flagellar filament28 which does not display such anomalous stability.
We asked whether other measures of stability could explain the anomalous properties of the pili. Using the Rosetta interface analyzer40, the LAL14/1 pili interfaces were compared (Supplementary Table 1) to the structures of eight archaeal and bacterial flagellar filaments, and T4P (PDB ids: 5o4u, 5tfy, 5kyh, 5vxx, 5vxy, 5wjt, and 5wk6). While the interface size is average, or even a little below-average, when compared to these reference assemblies, the interfaces of LAL14/1 pili are markedly more hydrophobic (69% hydrophobic surface area, compared to a reference average and standard devation of 60±4%), and have a lower predicted interface energy per square Ångstrom (−0.037 kcal/mol Å−2), compared to a reference average and standard deviation of −0.031±0.002 kcal/mol Å−2. While some of these metrics, such as interface energy per Å2 or energy per residue, are lower for the LAL14/1 pilus than any of the references, the differences do not appear to be large enough to explain the anomalous properties.
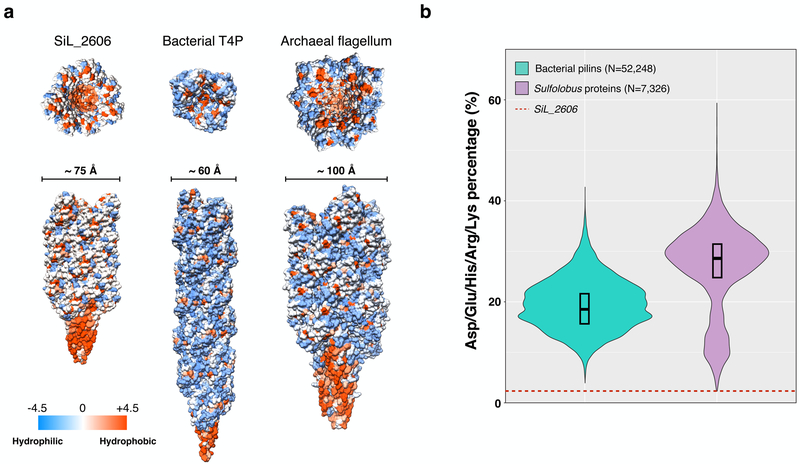
We next looked at the overall sequence of the pilin, which is quite unusual in some respects. There are very few hydrophilic residues on the surface of the LAL14/1 pilus (Fig. 3a), in contrast to the surfaces for a bacterial T4P and an archaeal flagellar filament. The percentage of charged residues (Asp+Glu+His+Arg+Lys) within the mature protein (after cleavage of the signal sequence) is only 1.5% (Fig. 3b). This is lower than all other bacterial and Sulfolobus proteins containing between 100–200 residues. This is also true when compared with archaeal flagellins and pilins (Sup. Fig. 6a), but the number of such annotated entries is limited. Considering that many of these other 100–200 residue proteins are integral-membrane proteins (while only the N-terminal α-helix in Sil_2606 is integrated into the membrane before filament formation) this is quite remarkable, and raises questions about how such a filament could be soluble.
Figure 3. LAL14/1 pilus contains an unusually high percentage of hydrophobic residues.
a, Hydrophobicity surface of 21 subunits of LAL14/1 pilus (left), bacterial (N. gonorrhoeae) type IV pilus (middle) and archaeal (M. hungatei) flagellar filament (right).
b, Volin plot showing the distributions of the percentage of charged residues (Asp+Glu+Lys+Arg+His) in bacterial pilins (100–200 amino acids long, N=52,248, colored in green) and Sulfolobus proteins (100–200 amino acids long, N=7,326, colored in purple). The width of the distribution corresponds to the frequency of occurrence. The box indicates the interquartile range, and the central line in the box indicates the median of the data. The red dashed line indicates the percentage in LAL14/1 pilin Sil_2606.
Glycosylation of the filament
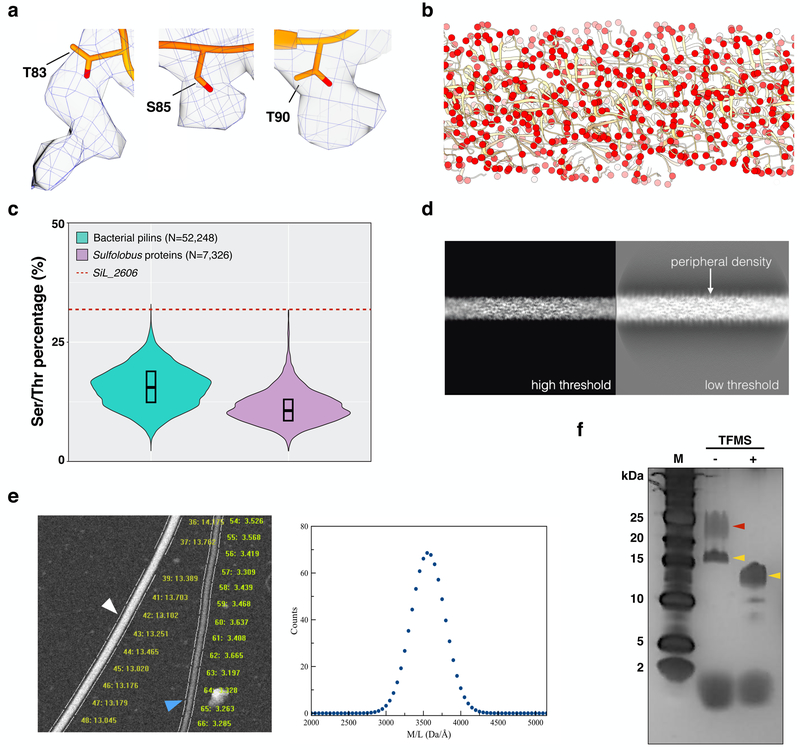
The cryo-EM map revealed additional mass associated with three surface residues: Thr83, Ser85 and Thr90 (Fig. 4a). Since these residues are potential targets for O-linked glycosylation, and it is known that many archaeal cell surface proteins are extensively glycosylated41, the most reasonable assumption is that this extra mass is due to glycosylation. Other post-translational modifications42 such as phosphorylation (+80 in mass), acetylation (+42 in mass) or methylation (+14 in mass) are much too small to explain the extra densities, while ubiquitination would generate a much greater density. The additional density for Thr83 (Fig. 4a) is best explained by two sugars, but at the available resolution we cannot distinguish among the many possible sugar groups. The potential targets for glycosylation on the pilus surface are extensive (Fig. 4b) given the huge number of threonine and serine residues on the surface. In fact, the frequency of threonine and serine in the SiL_2606 sequence is extremely anomalous when compared with similarly sized bacterial pilins or Sulfolobus proteins (Fig. 4c). This anomaly remains when comparing with archaeal flagellins and pilins (Sup. Fig. 6b). When the transmembrane N-terminal domain is removed from the Sil_2606 sequence, 37% of the residues are threonines and serines. Looking at a projection of the reconstruction at a low threshold reveals a fuzzy coat surrounding the filament, absent when a higher threshold is used (Fig. 4d). As a control, projections from bacterial T4P reconstructions13,14 were examined which did not exhibit this behavior (Sup Fig. 7). Such a fuzzy coat on the LAL14/1 pili would be consistent with extensive amounts of glycosylation.
Figure 4. O-linked sugar modifications of the LAL14/1 pilus.
a, Extra densities on residues threonine 83, serine 85 and threonine 90 indicate post-translational modifications.
b, Atomic model of the LAL14/1 pilus, with side chain oxygen atoms of serine and threonine shown as red spheres.
c, Violin plot showing the distributions of the sum of serine and threonine percentage in bacterial pilins (100–200 amino acids long, N=52,248, colored in green) and Sulfolobus proteins (100–200 amino acids long, N=7,326, colored in purple). The red dashed line indicates the corresponding percentage in LAL14/1 pilin Sil_2606. The box indicates the interquartile range, and the central line in the box indicates the median of the data.
d, Projections of the 3D reconstruction under high threshold (left) and low threshold (right). The low threshold projection reveals a fuzzy coat of peripheral density.
e, STEM analysis of LAL14/1 pili. Within one representative micrograph from 150 recorded (left), the white arrowhead indicates tobacco mosaic virus (control) and the blue arrowhead indicates a LAL14/1 pilus. The mass per length measurement for each segment are shown. The distribution of all mass/length values for the pili (right) yields an average of ~ 3.5 kDa/Å.
f, LAL14/1 pilus samples on silver stained SDS-PAGE before and after deglycosylation using trifluoromethanesulfonic acid (TFMS). This was performed twice, and the same bands were seen each time. Yellow arrowheads indicate the bands which contain SiL_2603 and SiL_2606 detected by MS/MS analysis. The red arrowhead indicates a higher mass band that was shown by MS/MS analysis to not contain either SiL_2603 or Sil_2606 (but contained a membrane-associated ESCRT-III-2 protein) and thus could not be due to a more heavily glycosylated species of Sil_2603 or Sil_2606. This figure (f) is reproduced in Supp Fig. 9 for purposes of comparison.
To quantify the additional mass that is most likely due to glycosylation, we employed Scanning Transmission Electron Microscopy (STEM)43 to measure the mass per unit length of the pili (Fig. 4e). The average value found was ~3.5 kDa/Å, while the predicted value of the protein alone (12.679 kDa/4.9 Å) is only 2.6 kDa/Å. This corresponds to a ~35% increase in the mass of each subunit, presumably due to glycosylation. Given the huge amount of glycosylation, why is additional density not seen in the three-dimensional reconstruction on many other sites for potential O-linked or N-linked glycosylation on the surface of the filaments? It is possible that many of the potential sites are only glycosylated on some subunits and not on others, so that this density is diminished after helical averaging. The three sites that are seen (Fig. 4a) must arise from both a very fixed orientation of these sugars as well as a glycosylation of these same three residues on almost every subunit.
We attempted to remove the glycosylation using a mix of deglycosylation enzymes, but without any effect as judged by MS of the enzyme-digested products. This may be due to the fact that most of the commercial deglycosylases work mainly on N-linked glycans. We next used trifluoromethanesulfonic acid (TFMS), described as a powerful method to remove both N-linked and O-linked glycans from proteins44. Conditions were found (Fig. 4f) where a significant shift to lower mass could be obtained for a silver-stained band that ran at ~17 kDa before acid treatment and at ~ 13 kDa after acid treatment. The shift seen is almost exactly what would be predicted from the STEM analysis for a removal of glycosylation. These two bands were excised from the gel and analyzed by MS, which found that both contained SiL_2603 and SiL_2606 (Sup. Fig. 8). A higher mass band that was apparent before the acid treatment but was absent after it cannot be due to a more heavily glycosylated SiL_2606 or SiL_2603 as neither protein was found within this band by MS. This higher mass band was shown by MS to contain protein SiL_1195 (CdvB2), a paralog of CdvB (ESCRT-III), which likely comes from membrane vesicles trapped between pili during the sample preparation, consistent with the previous proteomic analysis of Sulfolobus vesicles45. The glycosylation of the protein band running at ~17 kDa was directly demonstrated using a stain specific for glycoproteins (Supp. Fig. 9). A consequence of the deglycosylation reaction was an aggregation of the pili, not surprising as the absence of many hydrophilic residues from the surface of the filament (Fig. 3a) previously raised questions about how such a filament could be soluble.
Evolutionary relationships
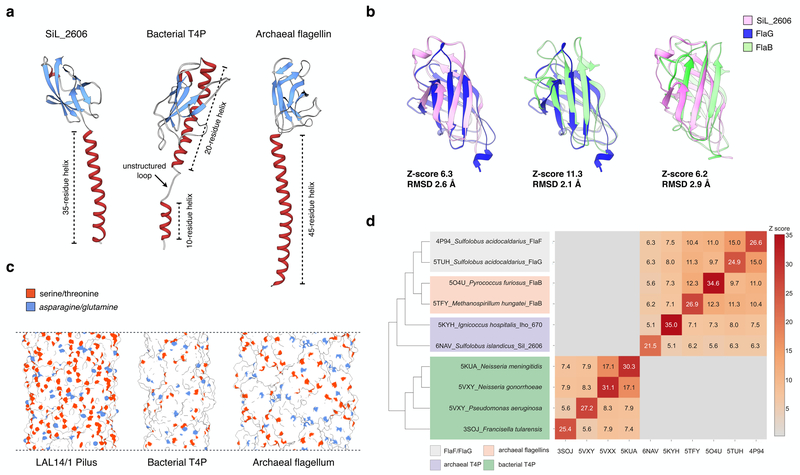
Sequence comparisons between SiL_2606 and the exsiting sequence databases of all proteins with known structures using BLASTP46 failed to find any hits, consistent with the sparse sampling of proteins from organisms living in the most extreme environments4. We therefore employed structure-based comparisons using the DALI server47 to find potential homologs of SiL_2606 (Fig. 5a). The most significant hit for SiL_2606 was with the FlaG protein from Sulfolobus acidocaldarius (PDB 5TUH), which yielded a Z-score of 6.3. The FlaG protein is part of the archaeal flagellum29, and FlaG shows clear homology with the archaeal flagellin, FlaB, with a Z-score of 11.3 (Fig. 5b). Thus, SiL_2606 shares a highly similar fold with FlaG, and FlaG shares an even more similar fold with FlaB. Accordingly, SiL_2606 and FlaB also share similarities (Z-score 6.2; Fig. 5b). However, due to differences in the angle between the β-sheets forming the β-sandwiches in all of these proteins, SiL_2606 is more similar to FlaG than it is to FlaB.
Figure 5. Comparison of LAL14/1 pilus with bacterial type IV pilus and archaeal flagellar filament.
a, ribbon display of a single subunit of Lal14 pilin, type IV pilin of N. gonorrhoeae (PDB 5VXX), and archaeal flagellin of M. hungatei (PDB 5TFY).
b, Structural alignment of the globular β-sandwich domains: LAL14/1 SiL_2606, FlaG (PDB 5TUH) and archaeal flagellin FlaB (PDB 5TFY). Z-scores are calculated by the DALI server.
c, Surface display of the filaments composed of pilins/flagellins shown in (a). The side chain oxygen atoms of serines and threonines are shown in orange, and the side chain nitrogen atoms of asparagines and glutamines are shown in blue.
d, All-against-all comparison of the bacterial and archaeal structures having N-termal T4P domains. The matrix (right) and cluster dendrogram (left) are based on the pairwise Z-score comparisons calculated using DALI. The color scale indicates the corresponding Z-scores. Based on their functions and origins, the proteins are partitioned into four groups, which are indicated with different colors.
Discussion
Archaea and the viruses that infect them have evolved to survive in the most extreme environments, including nearly boiling acid3,23. Quite strikingly, the pili isolated from Sulfolobus islandicus LAL14/1 were able to survive the conditions and treatments that are normally a first step to biochemical characterization of an assembly: boiling in 5M guanidinium-HCl, and digestion with trypsin or pepsin. The near-atomic resolution that we achieved by cryo-EM led to an atomic model for the filament, possible only by using a bioinformatics-based approach to search the LAL14/1 genome for all potential candidates and then using the cryo-EM map to exclude all but two proteins: SiL_2603 and SiL_2606. Both of these proteins were subsequently identified in the pilus preparation by MS. Since it would be difficult to imagine how a proline in SiL_2603 within a β-strand would maintain the hydrogen-bonding that holds the β-sheet together, we have assumed that the dominant pilin in the filament is actually SiL_2606. Subunits in a growing T4P must add at the base16 and the disrupted β-sheet in SiL_2603 might allow the protein to bind at the base of a growing pilus but prevent additional subunits from being added. It might also be the first protein assembled by the secretory system and be found at the pilus tip.
The atomic model that we have generated for the S. islandicus pilus revealed an unusual surface (Fig. 3a) that raised questions about how such a filament would be soluble. In fact, knocking out a particular glycosylation pathway in an archaeon has previously been shown to cause the pili to aggregate into large bundles48. A similar phenomenon has been observed in bacteria, where eliminating glycosylation of pili led to greatly enhanced aggregation of these filaments49. The density map also showed extra mass consistent with glycosylation for three residues, and a cloud of low density surrounding the filament suggestive of more extensive glycosylation. In addition, compared with baterial T4P and archaeal flagellum, there are significantly more serine and threonine residues present on the pilus surface (Fig. 5c), potential targets for O-linked sugars. STEM showed that the mass per unit length of the filament was ~35% greater than expected for unmodified protein subunits.
Taken together, our results suggest that extensive glycosylation of the pili both allows for their solubility and provides the basis for their extreme stability. Such stability is not seen for homologous structures, which we now know to be the archaeal flagellar filaments, showing that the extensive glycosylation appears to be an evolutionary modification that gives rise to the unusual physicochemical properties. A previous study50 showed that the glycosylation of a protein by a single fucose moiety led to a stabilization of the protein by ~1 kcal/mol as judged by thermal denaturation. Another study51 showed that the degree of thermal stabilization was directly proportional to the number of attached glycan chains. A review summarizes the extensive data on stabilization of proteins by glycosylation52, which yields resistance to proteolysis, thermal denaturation, and unfolding by guanidinium-HCl.
We have shown that the overall fold of this pilin is quite similar to that of archaeal flagellin, while previous work only suggested that this structural homology existed for the N-terminal domain32. The first indication that the archaeal flagellin fold might be more widespread came from a structure of an “adhesion filament” from Igniococcus hospitalis26. This organism is non-motile and the protein (Iho_670) forming the adhesion filament showed no homology at the sequence level to archaeal flagellin outside of the N-terminal T4P-like domain. It was suggested26 that the fold of Iho_670 would be the same as in true archaeal flagellin, and this has subsequently been confirmed28,29.
Pairwise structural comparisons of the bacterial and archaeal T4P proteins suggests a likely scenario of the T4P evolution. Although the N-terminal α-helical domains of bacterial and archaeal T4Ps are homologous, the globular β-strand-rich domains do not display appreciable structural similarity (Fig. 5d). Indeed, whereas in bacterial T4P the β-strands form a rather flat sheet with a juxtaposed α-helix extending from the N-terminal domain, in all archaeal T4P, including flagellins and pilins, the C-terminal domain adopts an immunoglobulin-like fold, a β-sandwich consisting of two β-sheets that surround a central hydrophobic core (Fig. 5a). It is thus likely that the globular domains in bacterial and archaeal T4P proteins have evolved independently. Many more structures of bacterial and archaeal T4Ps will be needed to substantiate this hypothesis. However, it is already clear that FlaB, FlaF and FlaG components of archaeal flagella are more closely related to each other than they are to archaeal pilins (Fig. 5d), suggesting that these functionally different flagellar components have evolved from a single ancestor, likely a pilin, by gene duplication and subsequent subfunctionalization.
Remarkably, the features underlying the stability of filamentous viruses of hyperthermophilic archaea appear to be very similar to those of LAL14/1 pili. Namely, the contacts between the capsid protein subunits in the helically symmetrical virions are largely mediated by hydrophobic interactions and the virion surfaces are extensively glycosylated3,5. This suggests that at least some viruses and pili of hyperthermophilic archaea have converged on similar adaptive solutions allowing them to maintain stability in extremely hot environments.
Methods
Strain cultivation and filament purification
A preculture of Sulfolobus islandicus strain LAL14/1 (PMID: 23594878) was grown in rich medium53 at 76 °C for 24h and then diluted into fresh medium. The cells were grown until mid-logarithmic phase (OD600 0.5), collected by centrifugation (6000×g, 15min, 15 °C) and resuspended in rich medium, 1/10 of the original volume. The pili were isolated as described previously (PMID: 24089554). Briefly, the filaments were mechanically sheared from the cells by vortexing at maximum speed for 15 min. The cells were removed by centrifugation (6000×g, 60min, 15 °C). The cleared supernatant containing pili was ultracentrifuged (144,000×g, 60min, 15 °C) and the resultant pellet was resuspended in 20 mM Tris-acetate buffer (pH 6).
Cryo-electron microscopy and image processing
The LAL14/1 pilus sample (4 μL) was applied to discharged lacey carbon grids and plunge frozen using a Vitrobot Mark IV (FEI, Inc.). Frozen grids were imaged in a Titan Krios at 300 keV and recorded with a Falcon III camera at 1.4 Å per pixel. Micrographs were collected using a defocus range of 1.5–2.5 μm, with a total exposure time of 2.4 s (amounting to ~ 56 electrons/Å2) distributed into 24 fractions. A total of 1,824 micrograph movies were analyzed. All the micrographs were first motion corrected (ignoring the first fraction) using MotionCorr v2.154 and then used for CTF estimation by the CTFFIND3 program55. Filament images were extracted using the e2helixboxer program within EMAN256 from the dose-weighted fractions 2–10 (amounting to ~ 20 electrons/Å2), after the images were corrected for the CTF through multiplication by the theoretical CTF. A total of 881,252 overlapping 384-px long segments (with a shift of 6 pixels between adjacent subunits, ~ 1.5 times the axial rise per subunit) were generated. The helical symmetry was determined as a 4.9 Å rise and 105.0 degrees rotation, after searching through a number of possible symmetries by trial and error. A ~ 4.5 Å reconstruction was generated using the IHRSR method implemented in Spider57, and this volume was subsequently filtered to 10 Å as the starting reference used in Relion58. The same micrographs and box coordinates used in the Spider reconstruction were imported into Relion. A comparable ~ 4.5 Å reconstruction was generated after class2D and refine3D steps, and it was then further improved to 4.1 Å after movie-refinement and particle-polishing steps. The final volume was estimated to have a resolution of 4.1 Å based on the model:map FSC and d9959 (Sup. Fig. 3, Sup. Table 1), and sharpened with a negative B-factor of −300.
De novo model building of LAL14/1 pilus
First, the density corresponding to a single LAL14/1 pilin (pilin-map) was segmented from the experimental filament density using Chimera60, and then the density corresponding to the N-terminal helix (long-helix map) was further segmented from the pilin-map. About 120 Cα atoms could be successfully traced from the pilin-map, including a ~35 amino acid N-terminal α-helix and a ~85 amino acids β-sandwich globular domain. Those patterns were used to search for possible pilin candidates in the S. islandicus LAL14/1 proteome23. The proteome (n=2,601) was downloaded from the NCBI GenBank and filtered to retain only proteins of 120–145 aa in length using the Galaxy platform maintained at Institut Pasteur (https://galaxy.pasteur.fr). The 263 extracted proteins were analyzed for the presence of transmembrane domain using TMHMM61 and only those containing a single N-terminal membrane-spanning domain were retained. For the 12 proteins satisfying the above characteristics, secondary structure was predicted using Jpred462 and the seven proteins (SiL_0372, SiL_1076, SiL_1365, SiL_1862, SiL_2603, SiL_2606 and SiL_2610) containing β-strand-rich C-terminal domains were retained for further analyses.The first 50 amino acids of those seven candidates were then built into the long-helix map using the RosettaCM protocol63. Candidates with good fit to the long-helix map were then built into the pilin-map with their full sequences using the RosettaCM protocol. SiL_2603 and SiL_2606 could be successfully built into the pilin-map, but SiL_2603 is extremely unlikely to be the correct candidate because it would have a proline in the middle of an ordered β-strand. Therefore SiL_2606 was selected to generate a filamentous model and refined against the full cryo-EM map, using real-space refinement in PHENIX64. MolProbity65 was used to evaluate the quality of the filament model. The refinement statistics are given in Table 1.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| LAL14/1 pilus model (EMDB-0397) (PDB 6NAV) |
|
|---|---|
| Data collection and processing | |
| Magnification | 59,000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 20 |
| Defocus range (μm) | −1.5 to −2.5 |
| Pixel size (Å) | 1.4 |
| Symmetry imposed | Helical (rise 4.94 Å, rotation 104.97°) |
| Initial particle images (no.) | 881,252 |
| Final particle images (no.) | 181,113 |
| Map resolution (Å) | |
| model:map FSC (0.38) | 4.1 |
| ”gold-standard” map:map FSC (0.143) | 4.0 |
| d99 | 4.1 |
| Refinement | |
| Initial model used (PDB code) | N.A. |
| Map sharpening B factor (Å2) | −300 |
| Model composition | |
| Non-hydrogen atoms | 891 |
| Protein residues | 131 |
| Ligands | N.A. |
| B factors (Å2) | |
| Protein | 61.4 |
| Ligand | N.A. |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.823 |
| Validation | |
| MolProbity score | 1.52 |
| Clashscore | 4.1 |
| Poor rotamers (%) | 0 |
| Ramachandran plot | |
| Favored (%) | 95.4 |
| Allowed (%) | 4.6 |
| Disallowed (%) | 0 |
Long-Range Order calculations
A Long-Range Order (LRO) parameter was defined for a protein from the knowledge of potential contacts between two residues that are close in space (< 8 Å) and far apart in sequence (> 12 in amino acid register)38. Potential contacts between residues were computed with the pairdist tool from GROMACS package66 using the equation LRO = Σnij/N (nij = 1 if |i-j| > 12; otherwise nij = 0) where i and j are two residues for which the Cα-Cα distance is ≤ 8 Å and N is the total number of residues in a protein. The dataset consisted of two archaeal pili from S. islandicus LAL14/1 and I. hospitalis, two archaeal flagellins from P. furiosus and M. hungatei, two bacterial T4P from N. gonorrhoeae and P. aeruginosa, and two bacterial flagellins from B. subtilis and P. aeruginosa. Since these are all polymers, any residue in a different chain that is within 8 Å from a given residue was automatically assumed to be a distant residue.
Scanning Transmission Electron Microscopy
STEM data were acquired at Brookhaven National Laboratory (BNL). The STEM instrument operates at 40 keV with a scanning probe of <0.3 nm diameter produced from a cold field-emission source. Specimen quality and mass calibration were checked by comparison of the images to the known structure of tobacco mosaic virus (TMV, theoretical M/L 13.1 kDa/Å). Samples containing LAL14/1 pili were deposited on thin carbon. TMV was added to the grid first as an internal control, followed by injection buffer, then specimen solution (in 20 mM TRIS/acetate buffer, pH 6.0) for 1 min, then 10 washes of 20 mM TRIS/acetate buffer. Excess solution was wicked from the edge with filter paper between each injection. After the last wash the grid was wicked to a thin layer, fast-frozen by plunging into liquid nitrogen slush and stored under liquid nitrogen. Grids were freeze-dried overnight in an ion-pumped chamber with an efficient cold trap and transferred under vacuum to the STEM cold stage (−160 °C). Imaging typically used a dose of 20 el/Å2 (causing <5% mass loss, corrected by comparison to TMV). Mass measurements were performed off-line with the customized software PCMass67.
LAL14/1 Deglycosylation Reaction
Initially, Protein Deglycosylation Mix II (New England BioLabs), which contains a mixture of different enzymes, was used unsuccessfully to remove the glycosylation from the LAL14/1 pili. Subsequently, deglycosylation of LAL14/1 pili was performed using the Glycoprofile IV™ chemical deglycosylation kit (Sigma Aldrich). Briefly, 50 μL of concentrated pili were lyophilized in a small glass tube. Then, 150 μL of Trifluoromethansulfonic acid (TFMS) was added to the tube and the mixture was incubated at 4 °C for 1 hour. Following this, 150 μL of 60 % pyridine solution, cooled to ~15 °C with a methanol-dry ice bath, was added drop-wise to the reaction tube, neutralizing the TFMS acid. Using a 2000 molecular mass cut off Slide-A-Lyzer® dialysis cassete (Thermo Scientific), the reaction solutions were removed from the deglycosylated pili sample with overnight dialysis at 4 °C into TRIS/HCl pH 7.8 buffer. After dialysis, small amounts of aggregates, presumably deglycosylated pili, were observed in the cassette. Centrifugation for 15 min at 4 °C and 20,000 RCF was used to pellet the aggregates. The pellet was re-suspended with 50 μL of tris buffer pH 7.8.
Analysis of Deglycosylation Reaction by SDS-PAGE
The pellet from the deglycosylation reaction and the glycosylated LAL14/1 stock were separately diluted 3-fold into Tricine Sample Buffer (Bio-RAD: 200 mM TRIS/HCl, pH 6.8, 40% glycerol, 0.04% Coomassie Blue G-250) containing 2% sodium dodecyl sulfate (SDS). The samples were then boiled at 100° C for 6–10 minutes. SDS-PAGE was then performed at a constant voltage of 120 V using 16.5 % precast polyacrylamide Mini-PROTEAN Tris-Tricine precast gels (Bio-RAD) and Tris/Tricine/SDS running buffer (BIO-RAD). For each sample, 15 μL of boiled sample in SDS were added to separate wells in the gel, and 5 μL of Precision Plus Protein™ Dual Xtra protein standard (Bio-RAD) was added to a third well. No protein bands were detectable using standard Coomassie staining methods. As a result, more sensitive silver staining was performed using a Pierce™ Silver Stain for Mass Spectrometry (Thermo Scientific) kit. The detected gel bands were then excised and mass spectrometry was performed to analyze the composition of each band.
Mass Spectrometry
The gel pieces from the band were transferred to a siliconized tube and washed in 200 μL 50% methanol. The gel pieces were dehydrated in acetonitrile, rehydrated in 30 μL of 10 mM dithiolthreitol (DTT) in 0.1 M ammonium bicarbonate and reduced at room temperature for 0.5 h. The DTT solution was removed and the sample alkylated in 30 μL 50 mM iodoacetamide (IA) in 0.1 M ammonium bicarbonate at room temperature for 0.5 h. The IA reagent was removed and the gel pieces dehydrated in 100 μL acetonitrile. The acetonitrile was removed and the gel pieces rehydrated in 100 μL 0.1 M ammonium bicarbonate. The pieces were dehydrated in 100 μL acetonitrile, the acetonitrile removed and the pieces completely dried by vacuum centrifugation. The gel pieces were rehydrated in 20 ng/μL chymotrypsin in 50 mM ammonium bicarbonate on ice for 30 min. Any excess enzyme solution was removed and 20 μL 50 mM ammonium bicarbonate added. The sample was digested overnight at 37 °C and the peptides formed extracted from the polyacrylamide in a 100 μL aliquot of 50% acetonitrile/5% formic acid. This extract was evaporated to 15 μL for MS analysis.
The LC-MS system consisted of a Thermo Electron Q Exactive HF-X mass spectrometer with an Easy Spray ion source (set to 40 °C) connected to a Thermo 75 μm × 15 cm (3μm C18) Easy Spray column. 3 μL of the extract was injected and the peptides eluted from the column by an acetonitrile/0.1 M formic acid gradient at a flow rate of 0.3 μL/min over 1.0 hours. The nanospray ion source was operated at 1.8 kV and 250 °C. The digest was analyzed using the rapid switching capability of the instrument acquiring a full scan mass spectrum (lock mass 445.12006, 120K resolution, 3e6 target, 60ms max IT) to determine peptide molecular weights followed by product ion spectra (10 HCD, 30K resolution, 1e5 target, 60ms max IT, 2.0 m/z, trigger 3e4, NCE 27) to determine amino acid sequences in sequential scans. Exclusions were made for +1 and unassigned ions as well as standard dynamic exclusion for 20s. This mode of analysis produces approximately 20000 MS/MS spectra of ions ranging in abundance over several orders of magnitude.
The data were analyzed by database searching using the Sequest search algorithm contained within Proteome Discoverer 2.2 against Uniprot S. Islandicus (11/20/18 with 16,206 entries). Parent mass was set to 10 ppm and fragment masses to 0.02 Da. The following parameters were used – chymotrypsin, fixed carbamidomethyl C, dynamic oxidation M, 1 missed cleavage. Raw data generated from the search were loaded into Scaffold 4.8.8 with initial filtering set to the following – xcorr+1>1.8, +2>2.0, +3>2.2, +4>3.0, peptide prophet >60%, protein prophet >90%. The search and data analysis settings produce a FDR<1%. Any peptides meeting these criteria are manually examined by Dr. Sherman to further eliminate false positives before data were reported. The Thermo RAW files as well as Scaffold analysis file have been deposited into PRIDE (PXD012799).
Pro-Q Emerald 488 Glycoprotein Staining
LAL14/1 pili in 20 mM Tris-acetate buffer pH 6 were diluted three-fold into Tricine Sample Buffer (Bio-RAD: 200 mM TRIS/HCl, pH 6.8, 40% glycerol, 0.04% Coomassie Blue G-250) containing 2% SDS. The sample was then boiled at 100° C for 8 minutes. SDS_PAGE was performed using the same conditions that were used to analyze the deglycosylation reactions. The CandyCane Glycoprotein Molecular Weight Standards (Invitrogen) were run on the gel with the LAL014/1 pili, in a separate lane. The gel was then stained with the Pro-Q Emerald 488 Glycoprotein Stain kit (Invitrogen). The Pro-Q Emerald stain was visualized using a FluorChem Q system (proteinsimple) with a light source of 475 nm and the emission filter set to 537 nm.
Supplementary Material
Acknowledgments
This work was supported by NIH GM122510 (to E.H.E.) and GM123089 (to F.D.), and l’Agence Nationale de la Recherche project ENVIRA, #ANR-17-CE15-0005-01 (to M.K.). M.A.B.K. was supported by NIH T32 GM080186. The cryo-EM imaging conducted at the Molecular Electron Microscopy Core facility at the University of Virginia was supported by the School of Medicine and built with NIH grant G20-RR31199. The Titan Krios and Falcon II direct electron detector were obtained with NIH S10-RR025067 and S10-OD018149, respectively. We thank Dr. Vincent Conticello for the suggestion of TFMS. We are also grateful to the Ultrastructural BioImaging (UTechS UBI) unit of Institut Pasteur for access to electron microscopes.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability Statement
The three-dimensional reconstruction has been deposited in the Electron Microscopy Data Bank with accession code EMD-0397. The atomic model has been deposited in the Protein Data Bank with accession code 6NAV. The MS data have been deposited in PRIDE with accession code PXD012799.
References
- 1.Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV & Krupovic M The enigmatic archaeal virosphere. Nature reviews. Microbiology 15, 724–739, (2017). [DOI] [PubMed] [Google Scholar]
- 2.Chaudhury P, Quax TEF & Albers SV Versatile cell surface structures of archaea. Mol Microbiol 107, 298–311, (2018). [DOI] [PubMed] [Google Scholar]
- 3.DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D & Egelman EH A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA. Science 348, 914–917, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Osinski T, Wang F, Krupovic M, Schouten S, Kasson P, Prangishvili D & Egelman EH Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile. Nature communications 9, 3360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ptchelkine D, Gillum A, Mochizuki T, Lucas-Staat S, Liu Y, Krupovic M, Phillips SEV, Prangishvili D & Huiskonen JT Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging. Nature communications 8, 1436, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasson P, DiMaio F, Yu X, Lucas-Staat S, Krupovic M, Schouten S, Prangishvili D & Egelman EH Model for a novel membrane envelope in a filamentous hyperthermophilic virus. eLife 6, doi: 10.7554/eLife.26268, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veesler D, Ng TS, Sendamarai AK, Eilers BJ, Lawrence CM, Lok SM, Young MJ, Johnson JE & Fu CY Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc Natl Acad Sci U S A 110, 5504–5509, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry JL & Pelicic V Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39, 134–154, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, Koonin EV & Albers SV Diversity and Evolution of Type IV pili Systems in Archaea. Front Microbiol 7, 667, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohlschroder M, Ghosh A, Tripepi M & Albers SV Archaeal type IV pilus-like structures--evolutionarily conserved prokaryotic surface organelles. Curr.Opin.Microbiol 14, 357–363, (2011). [DOI] [PubMed] [Google Scholar]
- 11.Pohlschroder M & Esquivel RN Archaeal type IV pili and their involvement in biofilm formation. Front Microbiol 6, 190, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albers SV & Jarrell KF The Archaellum: An Update on the Unique Archaeal Motility Structure. Trends in microbiology 26, 351–362, (2018). [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, Nassif X, Egelman EH & Craig L Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution. Structure 25, 1423–1435 e1424, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH & Craig L Structure of the Neisseria meningitidis Type IV pilus. Nature communications 7, 13015, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Castilla A, Thomassin JL, Bardiaux B, Zheng W, Nivaskumar M, Yu X, Nilges M, Egelman EH, Izadi-Pruneyre N & Francetic O Structure of the calcium-dependent type 2 secretion pseudopilus. Nat Microbiol 2, 1686–1695, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH & Tainer JA Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell 23, 651–662, (2006). [DOI] [PubMed] [Google Scholar]
- 17.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT & Tainer JA Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Molecular Cell 11, 1139–1150, (2003). [DOI] [PubMed] [Google Scholar]
- 18.Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L & Tainer JA Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J Biol Chem 286, 44254–44265, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reardon PN & Mueller KT Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J Biol Chem 288, 29260–29266, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ & Pohlschroder M Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. Journal of Bacteriology 189, 772–778, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albers SV, Szabo Z & Driessen AJ Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. Journal of Bacteriology 185, 3918–3925, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strom MS, Nunn DN & Lory S A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci U S A 90, 2404–2408, (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaubert C, Danioux C, Oberto J, Cortez D, Bize A, Krupovic M, She Q, Forterre P, Prangishvili D & Sezonov G Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open biology 3, 130010, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quemin ER, Lucas S, Daum B, Quax TE, Kuhlbrandt W, Forterre P, Albers SV, Prangishvili D & Krupovic M First insights into the entry process of hyperthermophilic archaeal viruses. J Virol 87, 13379–13385, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y & Baker D Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nature methods 12, 361–365, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun T, Vos MR, Kalisman N, Sherman NE, Rachel R, Wirth R, Schroder GF & Egelman EH Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc Natl Acad Sci U S A 113, 10352–10357, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, Goforth C, Meyer C, Rachel R, Wirth R, Schroder GF & Egelman EH Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-Terminal Type IV Pilin Helices. Journal of Molecular Biology 422, 274–281, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poweleit N, Ge P, Nguyen HH, Loo RR, Gunsalus RP & Zhou ZH CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pili. Nat Microbiol 2, 16222, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daum B, Vonck J, Bellack A, Chaudhury P, Reichelt R, Albers SV, Rachel R & Kühlbrandt W Structure and in situ organisation of the Pyrococcus furiosus archaellum machinery. eLife 6, doi: 10.7554/eLife.27470, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayley DP & Jarrell KF Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. J.Mol.Evol 46, 370–373, (1998). [PubMed] [Google Scholar]
- 31.Ng SY, Chaban B & Jarrell KF Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol 11, 167–191, (2006). [DOI] [PubMed] [Google Scholar]
- 32.Faguy DM, Jarrell KF, Kuzio J & Kalmokoff ML Molecular analysis of archael flagellins: similarity to the type IV pilin-transport superfamily widespread in bacteria. Can J Microbiol 40, 67–71, (1994). [DOI] [PubMed] [Google Scholar]
- 33.López-Castilla A, Thomassin J-L, Bardiaux B, Zheng W, Nivaskumar M, Yu X, Nilges M, Egelman EH, Izadi-Pruneyre N & Francetic O Structure of the calcium-dependent type 2 secretion pseudopilus. Nature Microbiology, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J & Baker D High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henche AL, Ghosh A, Yu X, Jeske T, Egelman E & Albers SV Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environmental microbiology 14, 3188–3202, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryndushkin D, Pripuzova N, Burnett B & Shewmaker F Non-targeted identification of prions and amyloid-forming proteins from yeast and mammalian cells. Journal of Biological Chemistry, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R & Eisenberg D Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gromiha MM & Selvaraj S Comparison between long-range interactions and contact order in determining the folding rate of two-state proteins: application of long-range order to folding rate prediction1. Journal of molecular biology 310, 27–32, (2001). [DOI] [PubMed] [Google Scholar]
- 39.Broom A, Ma SM, Xia K, Rafalia H, Trainor K, Colon W, Gosavi S & Meiering EM Designed protein reveals structural determinants of extreme kinetic stability. Proc Natl Acad Sci U S A 112, 14605–14610, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stranges PB & Kuhlman B A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein Science 22, 74–82, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohlschroder M, Pfeiffer F, Schulze S & Halim MFA Archaeal cell surface biogenesis. FEMS Microbiol Rev 42, 694–717, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann M & Jensen ON Proteomic analysis of post-translational modifications. Nat Biotechnol 21, 255–261, (2003). [DOI] [PubMed] [Google Scholar]
- 43.Wall JS & Hainfeld JF Mass Mapping with the Scanning Transmission Electron Microscope. Annu.Rev.Biophys.Biophys.Chem 15, 355–376, (1986). [DOI] [PubMed] [Google Scholar]
- 44.Sojar HT & Bahl OP Chemical deglycosylation of glycoproteins. Methods Enzymol 138, 341–350, (1987). [DOI] [PubMed] [Google Scholar]
- 45.Ellen AF, Albers SV, Huibers W, Pitcher A, Hobel CF, Schwarz H, Folea M, Schouten S, Boekema EJ, Poolman B & Driessen AJ Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13, 67–79, (2009). [DOI] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W & Lipman DJ Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402, (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holm L & Rosenstrom P Dali server: conservation mapping in 3D. Nucleic Acids Res 38, W545–549, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esquivel RN, Schulze S, Xu R, Hippler M & Pohlschroder M Identification of Haloferax volcanii Pilin N-Glycans with Diverse Roles in Pilus Biosynthesis, Adhesion, and Microcolony Formation. J Biol Chem 291, 10602–10614, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gault J, Ferber M, Machata S, Imhaus AF, Malosse C, Charles-Orszag A, Millien C, Bouvier G, Bardiaux B, Pehau-Arnaudet G, Klinge K, Podglajen I, Ploy MC, Seifert HS, Nilges M, Chamot-Rooke J & Dumenil G Neisseria meningitidis Type IV Pili Composed of Sequence Invariable Pilins Are Masked by Multisite Glycosylation. PLoS pathogens 11, e1005162, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mer G, Hietter H & Lefevre JF Stabilization of proteins by glycosylation examined by NMR analysis of a fucosylated proteinase inhibitor. Nat.Struct.Biol 3, 45–53, (1996). [DOI] [PubMed] [Google Scholar]
- 51.Shental-Bechor D & Levy Y Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci U S A 105, 8256–8261, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sola RJ & Griebenow K Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci 98, 1223–1245, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendörfer M & Kristjansson JK Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Systematic and Applied Microbiology 16, 609–628, (1993). [Google Scholar]
- 54.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA & Cheng Y Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods 10, 584–590, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mindell JA & Grigorieff N Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology 142, 334–347, (2003). [DOI] [PubMed] [Google Scholar]
- 56.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I & Ludtke SJ EMAN2: an extensible image processing suite for electron microscopy. Journal of Structural Biology 157, 38–46, (2007). [DOI] [PubMed] [Google Scholar]
- 57.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M & Leith A SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology 116, 190–199, (1996). [DOI] [PubMed] [Google Scholar]
- 58.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD & Urzhumtsev A New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC & Ferrin TE UCSF Chimera--a visualization system for exploratory research and analysis. J.Comput.Chem 25, 1605–1612, (2004). [DOI] [PubMed] [Google Scholar]
- 61.Krogh A, Larsson B, von Heijne G & Sonnhammer EL Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580, (2001). [DOI] [PubMed] [Google Scholar]
- 62.Drozdetskiy A, Cole C, Procter J & Barton GJ JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43, W389–394, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang RY, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D & DiMaio F De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature methods 12, 335–338, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC & Zwart PH PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography 66, 213–221, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS & Richardson DC MolProbity: More and better reference data for improved all-atom structure validation. Protein Science 27, 293–315, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B & Lindahl E GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wall JS & Simon MN Scanning transmission electron microscopy of DNA-protein complexes. Methods Mol Biol 148, 589–601, (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.