Abstract
Quantitative reverse transcription PCR (RT-qPCR) is one of the most efficient, reliable and widely used techniques to quantify gene expression. In this study, we evaluated the performance of six southern corn rootworm, Diabrotica undecimpunctata howardi (Barber), housekeeping genes (HKG), β-actin (Actin), β-tubulin (Tubulin), elongation factor 1 alpha (EF1α), glyceraldehyde-3 phosphate dehydrogenase (GAPDH), 40 S ribosomal protein S9 (RpS9) and ubiquitin-conjugating protein (Ubi), under different experimental conditions such as developmental stage, exposure of neonate and adults to dsRNA, exposure of adults to different temperatures, different 3rd instar larva tissues, and neonate starvation. The HKGs were analyzed with four algorithms, including geNorm, NormFinder, BestKeeper, and delta-CT. Although the six HKGs showed a relatively stable expression pattern among different treatments, some variability was observed. Among the six genes, EF1α exhibited the lowest Ct values for all treatments while Ubi exhibited the highest. Among life stages and across treatments, Ubi exhibited the least stable expression pattern. GAPDH, Actin, and EF1α were among the most stable HKGs in the majority of the treatments. This research provides HKG for accurate normalization of RT-qPCR data in the southern corn rootworm. Furthermore, this information can contribute to future genomic and functional genomic research in Diabrotica species.
Subject terms: Biotechnology, Plant sciences, RNAi
Introduction
The southern corn rootworm (SCR), Diabrotica undecimpunctata howardi (Barber), is a polyphagous/omnivorous plant pest found in the United States east of the Rocky Mountains, southern Canada, and northern Mexico1,2. SCR larvae feed on a wide variety of plants especially grasses, but its host range also includes cucurbits, peanuts, soybeans, cotton seedlings, dry beans, and maize1–4. SCR neonates can cause significant damage to corn seedling by feeding on the roots and drilling into the stem, subsequently killing the bud and reducing the maize stand1 to as much as 75%5. Furthermore, SCR has been used as a model insect to evaluate toxicity to different pesticides, including Bacillus thuringiensis (Bt) Berliner6–8, synthetic insecticides9, and double-stranded RNA (dsRNA)10–14.
RT-qPCR has become an important research tool for the quantification of gene expression in functional genomics and more recently for the evaluation of RNAi-mediated gene knockdown15. This efficient PCR-based assay is increasingly being utilized to compare gene expression, for which normalization through housekeeping genes (HKGs) is required. It is well known that the sensitivity, specificity and accuracy of RT-qPCR depend on several factors, including primer efficiency, the number of replications, but most importantly the choice of appropriate reference genes16,17. HKGs are expected to be stably expressed in all cells of an organism regardless of the physiological conditions18,19. Several studies suggest that some HKGs are differentially expressed depending on the experimental condition and no single reference gene is stably expressed and appropriate for all biological tissues or experimental conditions18–22. Therefore, identification of the characteristics of different HKGs under different environmental conditions, life stages and in different tissues is essential for accurate quantification of gene expression by RT-qPCR23.
In the last few years, several studies validating reference genes for genomic and molecular research in insects have been performed in a variety of biotic and abiotic experimental conditions especially developmental stage, tissue, and temperature range15. Most of the insects used in the studies are within the orders Hemiptera (i.e. aphids and whiteflies), followed by Lepidoptera (i.e. fall armyworm, Monarch butterfly, and the silk moth), Diptera (Drosophila spp., fruit flies, and mosquitoes) and Coleoptera (Colorado potato beetle, western corn rootworm, and lady beetles)15. Several plant pests have been used in these validation studies, including the sweet potato whitefly, Bemisia tabaci Gennadius18, the lepidopterans beet armyworm, Spodoptera exigua Hübner, silk moth, Bombyx mori L.24, tobacco cutworm, S. litura Fabricius25, oriental armyworm, Mythimna separate Walker26, the cotton and soybean aphids, Aphis gossypii Glover27 and A. glycines Matsumura28, respectively, the Asian longhorned beetle, Anoplophora glabripennis Motschulsky29, the leaf beetles Chrysomela populi L. and Phaedon cochleariae Fabricius30, the desert locust, Schistocerca gregaria Forsskål31, the cotton leafhopper, Amrasca biguttula biguttula Ishida32, and the pink spotted ladybeetle, Coleomegilla maculata De Geer33. The most used HKGs in these studies include β-actin (Actin), β-tubulin (Tubulin), glyceraldehyde-3 phosphate dehydrogenase (GAPDH), elongation factor 1 alpha (EF1α), and ribosomal proteins15,17.
The present study evaluated the efficiency of HKGs for SCR RT-qPCR-based studies. The selected genes are among the six most commonly reported genes in the literature15,17: Actin34,35, Tubulin34, EF1α34, GAPDH34,36, 40 S ribosomal protein S9 (RpS9)34 and ubiquitin-conjugating protein (Ubi)17. These HKGs were tested across different larval tissues (head, midgut, and carcass [integument + fat body]) and developmental stages (egg, first, second and third instar larvae, pupa, adult male and female). Additionally, SCR neonates and adults were exposed to different stressors, including ingested Snf7 dsRNA, starvation, and a range of temperatures (8 °C, 24 °C and 36 °C).
Results
Primer specificity and efficiency
Primer efficiency values ranged between 94.8% and 104.8% for all primers tested and correlation coefficients (R2) ranged around 0.99 (Table 1). RT-qPCR products generated with each set of primers were confirmed by the presence of a single peak in melting curve analyses (Supp. Fig. S1) and visualized in a 1% agarose gel (Supp. Fig. S2).
Table 1.
Primer sequences and accession number of the six candidate reference genes in SCR.
| Gene | Primer sequences (5’-3’) | NCBI Accession No. | Function | Amplicon (bp) | E (%) | R2 |
|---|---|---|---|---|---|---|
| β-actin (Actin) |
F: CCAGCTGCTTCCATACCCAA R: TGCCAGTTCCAGTTCCCTAG |
KX981996 |
Cell mortality, structure, and |
129 | 94.8 | 0.996 |
| 40 S ribosomal protein S9 (RpS9) |
F: GTCCATATGAGAAGGCCCGTT R: GGACCAATCGACGTAGGAGTG |
KX981997 | Component of the 40 S subunit of ribosome34 | 195 | 99.5 | 0.997 |
| β-tubulin (Tubulin) |
F: TCTTCATGCCCGGATTTGCT R: CACAAGCCGCCATCATGTTC |
KX981998 | Microtubule structures34 | 117 | 102.5 | 0.987 |
| Elongation factor 1α (EF1α) |
F: AAGCACTCCAGGAAGCAGTAC R: TGGGTTGTTCTTGGTGTCTCC |
KX981999 | Bring aminoacyl-transfer RNA to the ribosome34 | 110 | 99.8 | 0.999 |
|
glyceraldehydes-3-phosphate dehydrogenase (GAPDH) |
F: CCTCTGGAAAATTGTGGCGTG R: GCCCAAACGAACAGTCAAGTC |
KX982000 | Carbohydrate metabolism34,36 | 179 | 97.8 | 0.996 |
| Ubiquitin conjugating enzyme E2 (Ubi) |
F: AGATGCTGTAGTCGCTAGGC R: TGATGACAACGACACCCTGG |
KX982001 | Cell lysis by ubiquitination17 | 178 | 104.8 | 0.984 |
F: forward; R: reverse; E: percentage of amplification efficiency; R2: correlation efficiency.
Cycle threshold (Ct) values and expression studies of the six reference genes
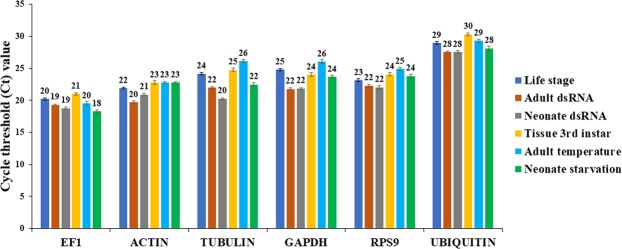
The Ct values generated in the RT-qPCR ranged between 18 and 30 throughout the treatments (Fig. 1). EF1α gene exhibited the lowest Ct values ranging from 18 to 20 for all treatments, and Ubi showed the highest Ct values in all treatments ranging from 28 to 30 (Fig. 1). Actin exhibited the second lowest Ct values (20–23), and Ct values for the genes GAPDH, Tubulin, and RpS9 ranged from 20 to 26 (Fig. 1). To perform the HKG validation study, we verified and compared the expression levels of each HKG among different SCR developmental stages (Supp. Fig. S3). In general, all HKGs were expressed in each stage at different levels, with the highest expression levels observed in adults especially in males (except RpS9) followed by eggs (GAPDH, Actin, and EF1α) and pupa (GAPDH, Ubi, and RpS9) (Supp. Fig. S3). RpS9 was the only HKG gene assayed that showed lower expression in males compared to females (Supp. Fig. S3).
Figure 1.
Cycle threshold (Ct) values generated with RT-qPCR for the HKGs in life stage, adult and neonate exposure to dsRNA, 3rd instar larval tissues, adult exposure to different temperatures, and neonate starvation. HKGs evaluated: Elongation factor 1α (EF1α), β-Actin (Actin), β-Tubulin (Tubulin), 40 S Ribosomal protein S9 (RpS9), Glyceraldehydes-3-phosphate dehydrogenase (GAPDH), and Ubiquitin conjugating protein (Ubi).
Stability of candidate reference genes
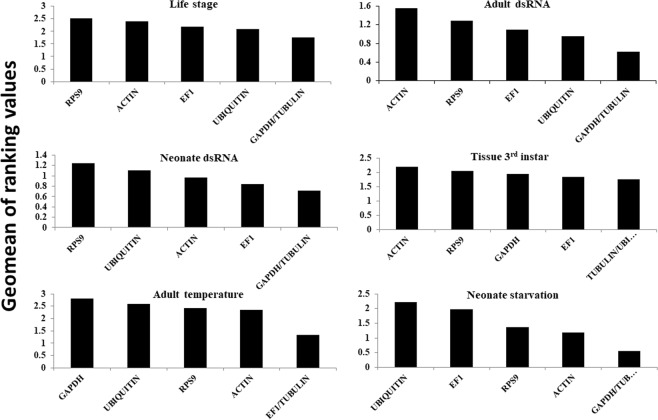
Four different algorithms were used (geNorm, NormFinder, BestKeeper, and delta-CT) for the validation of stability of the six candidate HKGs evaluated under six different experimental conditions (i.e. developmental stages, adult exposure to dsRNA, neonate exposure to dsRNA, 3rd instar tissue types, effect of temperature range to adults, and neonate starvation), using the web-based tool RefFinder which provides ranking for HKGs. The final ranking of the HKGs was calculated from the geometric mean of all the four different programs used in this study (Fig. 2). A lower geometric mean value represents higher stability of the HKGs.
Figure 2.
Stability of candidate reference genes according to geNorm in each bioassay: life stage, adult dsRNA exposure, neonate dsRNA exposure, 3rd instar larval tissues, adult exposed to different temperatures, and neonate starvation. A lower Geomean value suggests stable expression.
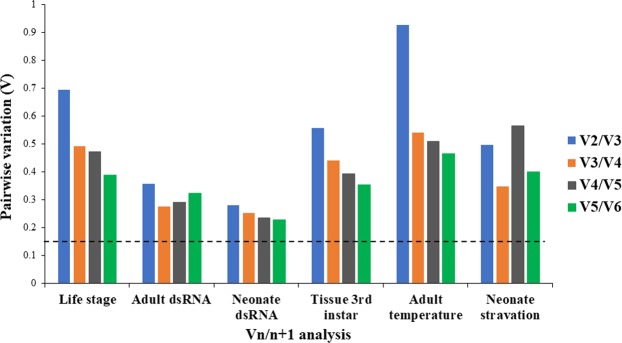
For the developmental stages, GAPDH and Actin were the most stable HKGs based on RefFinder ranking values, 1.32 and 1.68, respectively (Table 2). In the adults exposed to Snf7 dsRNA, EF1α and GAPDH were the most stable HKGs based on RefFinder ranking values, 1.19 and 1.57, respectively (Table 3). Whereas in the neonates exposed to Snf7 dsRNA, GAPDH and Tubulin were the most stable HKGs with RefFinder ranking values 1.57 and 2.21, respectively (Table 4). For the 3rd instar larval tissues, GAPDH and Actin were the most stable HKGs with RefFinder ranking values 2.00 and 2.06, respectively (Table 5). For exposure of adults to different temperature (8, 24, or 36 °C), RpS9 and Actin were the most stable HKGs with RefFinder ranking values 1.19 and 1.41, respectively (Table 6). For neonate starved for 48 h, Actin exhibited the lowest value followed by GAPDH with RefFinder ranking values 1.32 and 1.68, respectively (Table 7). One interesting observation obtained from this study was that the Ubi gene exhibited the highest stability values, but was the least stable HKG across all treatments in all algorithms (Tables 2–7) except for BestKeeper in adults exposed to dsRNA (Table 3). The lowest index rankings of Ubi across all the treatments were consistent with the highest delta-Ct values generated from RT-qPCR (Fig. 1). We also calculated the optimal HKG number based on geNorm algorithm analysis of reference genes necessary for accurate normalization for each bioassay. We found that the recommended number of HKGs for SCR is at least two for each treatment based on the pairwise values that were >0.15, performed in geNorm (Fig. 3).
Table 2.
Ranking of candidate HKGs based on stability values performed by RefFinder, Delta Ct, BestKeeper, NormFinder, and geNorm in SCR life stage (egg, neonate, 2nd instar, 3rd instar, pupa, adult male and female).
| HKG Gene | Stability (RefFinder) |
Rank | Stability (delta Ct) |
Rank | Stability (best keeper) |
Rank | Stability (normFinder) |
Rank | Stability (geNorm) |
Rank |
|---|---|---|---|---|---|---|---|---|---|---|
| GAPDH | 1.32 | 1 | 3.33 | 1 | 2.02 | 3 | 2.01 | 1 | 3.11 | 1 |
| β-Actin | 1.68 | 2 | 3.55 | 2 | 1.81 | 2 | 2.49 | 2 | 3.11 | 2 |
| EF1α | 2.45 | 3 | 3.60 | 4 | 1.78 | 1 | 2.57 | 3 | 3.16 | 3 |
| β-tubulin | 4.16 | 4 | 3.57 | 3 | 2.43 | 5 | 2.59 | 4 | 3.48 | 5 |
| RpS9 | 4.47 | 5 | 3.65 | 5 | 2.25 | 4 | 2.67 | 5 | 3.20 | 4 |
| Ubi | 6.00 | 6 | 3.78 | 6 | 2.64 | 6 | 2.96 | 6 | 3.58 | 6 |
Table 3.
Ranking of candidate HKGs based on stability values performed by RefFinder, Delta Ct, BestKeeper, NormFinder, and geNorm in SCR adults exposed to Snf7 dsRNA.
| HKG Gene | Stability (RefFinder) |
Rank | Stability (delta Ct) |
Rank | Stability (best keeper) |
Rank | Stability (normFinder) |
Rank | Stability (geNorm) |
Rank |
|---|---|---|---|---|---|---|---|---|---|---|
| EF1α | 1.19 | 1 | 1.67 | 1 | 1.17 | 1 | 0.43 | 2 | 0.87 | 1 |
| GAPDH | 1.57 | 2 | 1.68 | 2 | 1.46 | 3 | 0.34 | 1 | 0.87 | 2 |
| β-tubulin | 2.71 | 3 | 2.02 | 3 | 1.20 | 2 | 1.38 | 3 | 1.22 | 3 |
| β-Actin | 4.43 | 4 | 2.20 | 4 | 1.94 | 6 | 1.66 | 4 | 1.49 | 4 |
| RpS9 | 4.73 | 5 | 2.24 | 5 | 1.58 | 4 | 1.68 | 5 | 1.69 | 5 |
| Ubi | 5.73 | 6 | 3.06 | 6 | 1.73 | 5 | 2.80 | 6 | 2.14 | 6 |
Table 4.
Ranking of the candidate HKGs based on stability values performed by RefFinder, Delta Ct, BestKeeper, NormFinder, and geNorm in SCR neonates exposed to Snf7 dsRNA.
| HKG Gene | Stability (RefFinder) |
Rank | Stability (delta Ct) |
Rank | Stability (best keeper) |
Rank | Stability (normFinder) |
Rank | Stability (geNorm) |
Rank |
|---|---|---|---|---|---|---|---|---|---|---|
| GAPDH | 1.57 | 1 | 2.60 | 1 | 1.11 | 2 | 0.74 | 1 | 1.33 | 3 |
| β-tubulin | 2.21 | 2 | 2.95 | 3 | 1.04 | 1 | 1.24 | 2 | 2.05 | 4 |
| β-Actin | 2.21 | 3 | 2.91 | 2 | 1.49 | 4 | 1.95 | 3 | 1.26 | 1 |
| EF1α | 2.78 | 4 | 3.08 | 4 | 1.31 | 3 | 2.26 | 5 | 1.26 | 2 |
| RpS9 | 4.73 | 5 | 3.32 | 5 | 2.27 | 5 | 2.18 | 4 | 2.38 | 5 |
| Ubi | 6.00 | 6 | 5.36 | 6 | 2.38 | 6 | 5.08 | 6 | 3.37 | 6 |
Table 5.
Ranking of the candidate HKGs based on stability values performed by RefFinder, Delta Ct, BestKeeper, NormFinder, and geNorm in SCR 3rd instar larval tissues.
| HKG Genes | Stability (RefFinder) |
Rank | Stability (delta Ct) |
Rank | Stability (best keeper) |
Rank | Stability (normFinder) |
Rank | Stability (geNorm) |
Rank |
|---|---|---|---|---|---|---|---|---|---|---|
| GAPDH | 2.00 | 1 | 4.30 | 2 | 2.06 | 2 | 2.96 | 4 | 2.23 | 2 |
| β-Actin | 2.06 | 2 | 4.16 | 1 | 2.15 | 3 | 2.46 | 2 | 2.33 | 3 |
| EF1α | 2.11 | 3 | 4.58 | 4 | 1.42 | 1 | 3.48 | 5 | 2.23 | 1 |
| β-tubulin | 4.16 | 5 | 4.59 | 5 | 2.41 | 4 | 2.90 | 3 | 3.87 | 5 |
| RpS9 | 2.78 | 4 | 4.39 | 3 | 2.69 | 5 | 2.41 | 1 | 3.53 | 4 |
| Ubi | 6.00 | 6 | 6.57 | 6 | 3.76 | 6 | 6.01 | 6 | 4.77 | 6 |
Table 6.
Ranking of the candidate HKGs based on stability values performed by RefFinder, Delta Ct, BestKeeper, NormFinder, and geNorm in SCR adults kept at different temperatures for 8 h (8, 24, or 36 °C).
| HKG Genes | Stability (RefFinder) |
Rank | Stability (delta Ct) |
Rank | Stability (best keeper) |
Rank | Stability (normFinder) |
Rank | Stability (geNorm) |
Rank |
|---|---|---|---|---|---|---|---|---|---|---|
| RpS9 | 1.19 | 1 | 4.21 | 1 | 1.79 | 2 | 2.48 | 1 | 1.94 | 2 |
| β-Actin | 1.41 | 2 | 4.30 | 2 | 1.60 | 1 | 2.75 | 2 | 1.94 | 1 |
| EF1α | 3.22 | 3 | 4.54 | 3 | 2.62 | 4 | 2.94 | 3 | 2.87 | 3 |
| GAPDH | 3.72 | 4 | 4.72 | 4 | 2.31 | 3 | 3.14 | 4 | 3.41 | 4 |
| β-tubulin | 5.00 | 5 | 5.00 | 5 | 2.67 | 5 | 3.44 | 5 | 4.08 | 5 |
| Ubi | 6.00 | 6 | 6.45 | 6 | 3.79 | 6 | 5.80 | 6 | 4.87 | 6 |
Table 7.
Ranking of the candidate HKGs based on stability values performed by RefFinder, Delta Ct, BestKeeper, NormFinder, and geNorm in SCR neonates starved for 48 h.
| HKG Genes | Stability (RefFinder) |
Rank | Stability (delta Ct) |
Rank | Stability (best keeper) |
Rank | Stability (normFinder) |
Rank | Stability (geNorm) |
Rank |
|---|---|---|---|---|---|---|---|---|---|---|
| β-Actin | 1.32 | 1 | 2.53 | 1 | 1.00 | 1 | 0.78 | 1 | 1.60 | 3 |
| GAPDH | 1.68 | 2 | 2.90 | 2 | 1.35 | 2 | 1.62 | 2 | 1.56 | 1 |
| RpS9 | 2.28 | 3 | 2.96 | 3 | 1.80 | 3 | 1.76 | 3 | 1.56 | 2 |
| β-tubulin | 4.47 | 4 | 3.77 | 4 | 2.13 | 5 | 2.97 | 4 | 2.89 | 5 |
| EF1α | 4.47 | 5 | 3.86 | 5 | 1.84 | 4 | 3.22 | 5 | 2.27 | 4 |
| Ubi | 6.00 | 6 | 4.46 | 6 | 2.51 | 6 | 3.94 | 6 | 3.41 | 6 |
Figure 3.
Optimal number of reference genes required for accurate normalization of gene expression for each bioassay. Based on geNorm analysis, average pairwise variations are calculated between the normalization factors NFn and NFn + 1. Values < 0.15 indicate that additional genes are not required for the normalization of gene expression.
Discussion
Real Time-qPCR is one of the most valuable and reliable research tools used to quantify the expression of a target gene under different experimental conditions. Proper normalization of reference genes is necessary to get a robust and reliable estimate of gene expression under different experimental conditions and avoid unwanted variation. The HKGs evaluated in this study are reported to express constitutively to maintain various cellular functions. However, no HKG has been identified to be stably expressed in all tissues or cell types across different environmental conditions15. Therefore, the selection of appropriate HKGs under a particular treatment is important when using RT-qPCR to quantify gene expression. Many gene expression studies use a single endogenous control for different treatments and life stages, which can influence the statistical results and may lead to erroneous data interpretation37. Several genes have been selected as HKGs across different species, life stages, and tissues in various treatments38. Lack of stable expression of reference genes has been reported among different variables16,39 and suggests that no single gene (“universal reference gene”) can be selected for all variables18,35,40,41.
The results obtained in this study demonstrate that the stability of a HKG (among all six SCR HKGs studied) can be different under diverse experimental conditions including developmental stage, exposure of neonates and adults to dsRNA, exposure of adults to different temperatures, different 3rd instar larva tissues, and neonate starvation (Tables 2–7; Figs 1–3). The HKGs evaluated in this study have been used in a variety of organisms for evaluating the expression of target genes and can be found in several peer-reviewed publications reviewed in Lü et al.15. Among the six HKGs studied in this manuscript, we found that Actin and GAPDH were the most stable in different experimental conditions. Actin and GAPDH have been widely used as reference genes in different organisms ranging from insects to human tissues15,42–44. However, many studies have reported these genes to be unsuitable for RT-qPCR because of the variability of expression in different experimental settings and tissues43–45. In our study, GAPDH was found the most stable HKG in three of the total six experimental conditions (Tables 2,5,7) and exhibited the second highest Ct values (22–25) across four of the six treatments after Ubi (Fig. 1). Actin was the second most stable HKG in four treatments and the most stable in one of the six treatments (Tables 2, 5–7). The role of Actin is to control cellular mobility and growth and helps in regulating G-protein pool34,35, while the GAPDH enzyme is a key Glycolysis/neo-glucogenic enzyme34,36. The results of this study suggest that the role of Actin and GAPDH seems to remain stable under different environmental conditions and tissues in SCR. Therefore, Actin and GAPDH can be used as reliable HKGs in RNAi-based experimental approaches and gene expression bioassays in SCR.
On the contrary, both Ubi and RpS9 were the two least stable HKGs across all treatments and in five treatments, respectively (Tables 2–7) and are not recommended to be used for SCR gene normalization under the conditions mentioned above. In the 3rd instar larval tissue, adult exposed to different temperatures and in neonates exposed to starvation, Tubulin was identified as the most stable HKG with the lowest calculated Geomean values using geNorm algorithm (Fig. 2). Although Tubulin has been reported to be used for gene expression studies in multiple organisms, its low stability in various life stages and different treatment conditions used in this study make it unsuitable as a “universal” HKG in SCR. The last HKG evaluated in this study, EF1α, was identified as the most stable gene in SCR adults exposed to Snf7 dsRNA with lowest RefFinder ranking value (1.19) (Table 3).
Similar studies have been performed in other organisms, including insects, to evaluate the choice of reference gene selection for studying gene expression using RT-qPCR. Pan et al.46 found GAPDH as the HKG with the second highest Ct values across six different treatments performed in the lady beetle, Hippodamia convergens Guérin-Méneville, although was ranked as the most stable HKG in different treatments using different algorithms. Pan et al.47 also reported GAPDH as having the highest Ct values in the Monarch butterfly, Danaus plexippus L, but was ranked the most stable HKG in only one treatment (dsRNA). On the other hand, Yang et al.48 reported Actin as one of the least stable HKGs in the lady beetle Harmonia axyridis Pallas under biotic and abiotic conditions, while Dai et al.44 reported Actin as the most stable HKG under short-term thermal stress in the white fly B. tabaci. Rodrigues et al.49 validated the same HKGs used in this study (except Ubi) in western corn rootworm (WCR) and reported similar RT-qPCR Ct values, with EF1α exhibiting the lowest Ct values and RpS9 exhibiting the highest Ct values in all four experiments (larval tissues, developmental stages, dsRNA exposure, and Bt protein exposure). However, Rodrigues et al.49 reported EF1α as the most stable among the five HKGs in four algorithms for two of the four experiments (3rd instar larva tissue and life stage) and Actin as the least stable among five HKGs in one experiment (larval tissue) and the second least stable HKG in two (RNAi and Bt exposure) of the four experiments conducted. Even though SCR and WCR are similar and congeneric species, WCR is a pest that feeds almost exclusively on maize plants50, while SCR is polyphagous and is reported to feed on more than 200 different plant species from several families1,2. In addition, WCR is univoltine and eggs undergo diapause during the winter2, while SCR can have 3–4 generations per year in the field. These biological discrepancies can potentially play a role in the differences in HKG expressions reported in our study. Therefore, because of the differences in biology, feeding habits, oviposition, environmental factors and the differences in the methodology used between the two studies could lead to different results. Other studies performed with WCR have used similar HKGs to compare the expression of potential genes involved in Bt resistance (EF1α, Actin, and GAPDH)51, to compare candidate markers for behavioral resistance to crop rotation (EF1α)52, and to compare gene expression after gene knockdown by dsRNA (Actin)53. The examples provided above suggest that all of these HKGs can be selected for gene normalization, but only under specific condition(s) and should not be used as universal across all the different experimental conditions in any particular organisms.
Similar to the results of the studies listed above, our analyses also provide different results, for all four algorithms, under different experimental conditions, although the results were similar among some of the treatments. However, the most common observation from the analyses showed that Ubi was the least stable HKG across all evaluated treatments and in most algorithms (Tables 2–7). Our results suggest that the recommended number of HKG is at least two for each treatment based on the pairwise values (>0.15) obtained with geNorm (Fig. 3). A single HKG is not recommended for SCR, similar to what has been suggested in most of the studies reported so far with other insects45,54. Based on our results and the results obtained in other organisms, it is evident that the expression of these HKGs varies based on experimental conditions. Therefore, there is no universal reference gene/internal control that can serve as the perfect gene in all experimental conditions which firmly implicates the necessity for the custom case-specific selection and evaluation of endogenous reference genes using RT-qPCR in different organisms including the SCR15.
In conclusion, we tested six different reference candidate genes in six different experimental conditions using four different statistical algorithms and the additional web-based computational platform, RefFinder, that integrates the four algorithms to offer the overall ranking of the stability of the six HKGs. The current study not only demonstrates a standardized procedure for studying SCR gene expression but also the selection of appropriate internal control for efficient gene normalization under different experimental conditions. GAPDH and Actin were the most stable HKGs under different experimental conditions and can be recommended for RNAi studies in SCR. Actin has been used as a standard HKG in most of the RNAi studies with corn rootworms, and has proved to generate reliable results13,14,53,55–61. Ubi and RpS9 were considered as the least stable HKGs across the treatments which coincided with the higher Ct values generated in all treatments. Given that corn hybrids expressing Snf7 dsRNA will soon be available to growers for the management of WCR62, and since SCR Snf7 shares > 97% gene identity13 with WCR Snf7, it is likely that the hybrids will also affect SCR, and having a stable HKG will be necessary for studies in the upcoming years. This study can also be used as a steppingstone for providing detailed functional and genomic insights on SCR, an emerging model among belowground corn pests and several other economically important crops. Finally, two HKGs are recommended for normalization of SCR gene expression for each bioassay based on average pairwise variations (>0.15) calculated from geNorm analysis. Furthermore, the availability of HKGs and their stability will allow performing future studies focusing on genes essential for SCR biology, and choosing a reliable and appropriate HKGs will provide more accurate assessment of gene expressions for both the WCR and SCR.
Materials and Methods
Biological samples and experimental conditions
Newly emerged SCR adults were purchased from French Agricultural Research, Inc. (Lamberton, Minnesota) and kept in 30 cm3 BugDorm® cages (MegaView Science Co., Ltd., Taichung, Taiwan) at 24–26 °C, 50–70% relative humidity (RH), and 14:10 (light:dark) photoperiod, provided with artificial diet and water until used for bioassays. Eggs were also purchased from French Agricultural Research, Inc. and kept in a dark incubator at 28 °C and 90% RH until hatching.
Treatments
The treatments used to test the HKGs in this research were (1) developmental stage (egg, neonate, 2nd instar, 3rd instar, pre-pupa, pupa, and adult male and female), (2) adult exposure to Snf7 dsRNA on artificial diet, (3) neonate exposure to Snf7 dsRNA on artificial diet, (4) adult exposure to different temperatures (8 °C, 24 °C and 36 °C), (5) 3rd instar larvae tissue (dissection of head, midgut, and carcass), and (6) neonate starvation for 48 h.
In all bioassays, three tubes (1.5 ml or 2 ml microcentrifuge tubes) containing the insects were collected per each treatment from each of the three different cohorts, having a total of 9 biological replicates. For developmental stages, approximately 100 eggs and 30 neonates were collected in 1.5 ml microcentrifuge tubes; 10–2nd instar larvae, 5–3rd instar larvae, 2-pupae, and 2-adults were collected in 2 ml microcentrifuge tubes, flash-frozen in liquid nitrogen, and placed at −80 °C until used for RNA extraction. For larval body tissues, 3rd instar larvae were dissected to extract head, midgut, and carcass. Twenty heads, 5-midguts, and 5-carcasses were collected in 2.0 ml microcentrifuge tubes and flash-frozen in liquid nitrogen and subsequently stored at −80 °C until used for RNA extraction.
In dsRNA exposure bioassays, the gene used as target for dsRNA treatment was Snf713, which is a key operator gene of the endosomal sorting complex (ESCRT) required for unconventional secretion in Eukaryotes63 and related to membrane molecule transport and membrane stability64. For adults, 12 artificial diet pellets (modified from Branson & Jackson65 and described in Khajuria et al.57) cut with a cork borer (4.0 mm diameter × 2.0 mm height; area = 0.1256 cm2) were placed in each of three 5.0 cm × 2.5 cm × 2.5 wells (one well per treatment) in a 16-well tray (C-D International, Pitman, NJ). The diet pellets in each well were treated with 3 µl of dsRNA solution to yield a concentration of 20 ng/cm2 of Snf7 dsRNA or 20 ng/cm2 of Green Fluorescent Protein (GFP) dsRNA, or nuclease-free water only, and allowed to air dry. Ten SCR adults were placed in each of the three wells and allowed to feed for five days. Adults were transferred to clean trays with freshly treated diet on day 3. After five days, three 1.5 ml microcentrifuge tubes each containing 1–2 SCR adults were collected per well (treatment), flash-frozen in liquid nitrogen and stored at −80 °C until used for RNA extraction. For larvae, three 5.0 cm diameter petri dishes (PALL Corporation, Port Washington, NY, # 7242), each containing approximately 1 ml of SCR larval artificial diet (Frontier Agricultural Sciences®, Newark, DE) were treated with 450 µl of dsRNA solution that yielded 5.0 ng/cm2 of Snf7 dsRNA or 5.0 ng/cm2 of GFP dsRNA, or nuclease-free water only, and allowed to air dry. One hundred SCR neonates (<24 h old) were transferred to each of the three petri dishes and allowed to feed for three days. After three days, three 1.5 ml microcentrifuge tubes each containing 20 neonates were collected per each petri dish (treatment), flash-frozen in liquid nitrogen and stored at −80 °C until used for RNA extraction. The Snf7 dsRNA concentrations used for adults (20 ng/cm2) and neonates (5 ng/cm2) feeding assays were previously calculated and are within the concentration range that kills 50% (LC50) of the insects tested13.
To study the effect of starvation on gene expression of HKGs, SCR neonates (<24 h of hatching) were left starving in a 9.0 cm diameter × 1.4 cm height petri-dishes (Fisher Scientific, Pittsburgh, PA) lined with moist filter-paper for 48 h. Starved neonates were compared with newly hatched neonates. Three 1.5 ml microcentrifuge tubes containing 20–30 neonates each were collected in each treatment, flash-frozen in liquid nitrogen and stored at −80 °C until used for RNA extraction.
To elucidate the effect of heat and cold, SCR mixed adults (both males and females) were placed in 6.0 × 6.0 × 8.0 cm clear plastic boxes (Althor Products LLC, Windsor Locks, CT) and exposed to 8 °C, 24 °C, or 36 °C for 8 h before flash-frozen in liquid nitrogen and stored at −80 °C until used for RNA extraction.
Housekeeping genes
The HKGs used in this research were the same evaluated by Rodrigues et al.49 in WCR: Tubulin, EF1α, GAPDH, Actin, and RpS9 plus Ubi (Table 1). The primers for qPCR of the six reference genes were generated using Primer366 and the primer efficiencies (E) and correlation coefficients (R2) were also calculated (Table 1). For primer designing, SCR nucleotide sequences were obtained from NCBI using the accession numbers mentioned in Table 1. The length for RT-qPCR primers for all six genes were kept between 100–200 bp. Efficiency (E) of all these primers were calculated according to the equation: E = (10[−1/sl°pe] −1) × 100 and relative standard curves for the transcripts were generated with serial dilutions of cDNA (1/5, 1/25, 1/125 and 1/625). Efficiencies for all the primers were found to be in the ideal range (90%-105%) for proper function of primer pairs for all six reference genes (Table 1). We also checked for the possibility of primer dimer or secondary structure formation using Gene Runner v. 3.05 software (Hastings Software, Inc.)67.
RNA extraction and quantitative real-Time PCR (RT-qPCR)
Insects and insect tissues (i.e., eggs, larvae, and adults) were placed in 1.5 (eggs and neonates) or 2.0 ml tubes (other samples) containing one metal bead. The tubes were placed in metal blocks dipped in liquid nitrogen and appropriately balanced before being placed in SPEX SamplePrep 2010 Geno/Grinder® machine (Metuchen, NJ). The machine was then switched on for 30 seconds at 1400 strokes min−1 to grind insect tissue materials into a fine powder for RNA extraction. Different SCR tissue samples (50–100 mg) were used for total RNA extraction using Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA), following the manufacturer’s protocol and stored at −80 °C until use. The quantity of extracted RNA was estimated on a NanoDrop1000 (Thermo-Fisher Scientific, Wilmington, DE) and the quality was evaluated using 1% denaturing agarose gel electrophoresis. cDNAs were synthesized from 1 µg of total SCR RNA using the high capacity cDNA Reverse Transcriptase Kit (Applied Biosystems Inc., Foster City, CA). cDNA samples were diluted (1:50) before being used in RT-qPCR. Three independent biological replicates, each from three different insect cohorts were included in each RT-qPCR run. RT-qPCR was performed with SYBR Green Master Mix (iTaq™ Universal SYBR® Green Supermix) on a Real-Time PCR System (Applied Biosystems Inc., Foster City, CA). The master mix (30 µl) contained 15 µl SYBR green, 9 µl water, 3 µl diluted (1:10) HKG primers (F + R) and 3 µl diluted (1:50) cDNA and was replicated in three wells of a 96-well plates using 10 μl/well. RT-qPCR was run at 95 °C for 30 seconds (Holding stage), at 95 °C for 15 seconds, then 60 °C for 1 minute (Cycling stage); followed by 95 °C for 15 seconds, 60 °C for 1 minute, and 95 °C for 15 seconds (Melt curve). The comparative 2−ΔΔCt method68, which is calculated based on the difference between Ct values, was used to calculate the relative expression level of the six SCR HKGs in the samples as compared to control.
Data analyses
Four different statistical algorithmic models were currently used for HKGs evaluation17: geNorm16, NormFinder69, BestKeeper70 and delta-Ct71. GeNorm assesses the expression stability value (M) for each gene and identifies the best pair of reference genes. This program is based on the mean pairwise variation between genes across all samples and the gene with the lowest M value is considered the most stable16. NormFinder estimates the standard deviation for each gene and compares it with the expression of the other genes. The gene with the lowest variation between intra- and inter-group comparisons is then considered the most stable69. BestKeeper is a data processing method based on crossing points that compares all genes across all samples and generates a stability index for each HKG70. The comparative delta-Ct method compares Ct values and the relative expression of ‘gene pairs’ with each sample68,71.
The mean Ct values of each sample generated in RT-qPCR for each HKG in each experiment were used as input data and analyzed using the web-based tool RefFinder (http://leonxie.esy.es/RefFinder/)72. The website integrates all four software algorithms, GeNorm16, NormFinder69, BestKeeper70, and the comparative delta-Ct method71 which provides a comprehensive stability index that ranks each HKG72. The lower the rank value, the more stable the HKG is. Pairwise variation (V), determined by geNorm, was used to determine the optimal number of reference genes for accurate RT-qPCR normalization. Vn/Vn + 1 indicated the pairwise variation between two sequential normalization factors, and a cutoff threshold of Vn/Vn + 1 = 0.15 was used for valid normalization16.
Supplementary information
Acknowledgements
The authors would like to thank Dr. Thais B. Rodrigues for suggestions on the experimental design and to Molly Darlington for helping with sample collections. The work in Dr. Vélez Lab was supported by start-up research funds by the University of Nebraska-Lincoln. The work in Dr. Joe Louis lab was partially supported by the Nebraska Agricultural Experiment Station with funding from the Hatch Act (Accession # 1007272) through the USDA National Institute of Food and Agriculture and start-up research funds by University of Nebraska-Lincoln.
Author Contributions
A.E.P., S.B., B.D.S., J.L. and A.M.V. designed the study. A.E.P., S.B., D.H.P., H.W. and A.V.J. carried out the experiments. A.E.P. and X.J.Z. performed the statistical analysis. A.E.P. prepared the tables. A.E.P., S.B. and A.M.V. prepared the figures. A.E.P., S.B., A.V.J., B.D.S., J.L., X.J.Z. and A.M.V. interpreted the results and composed the manuscript. B.D.S, J.L. and A.M.V. provided resources. All authors edited and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Saumik Basu and Adriano E. Pereira contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47020-y.
References
- 1.Isely D. The southern corn rootworm. Arkansas Agricultural Experiment Station. 1929;232:31p. [Google Scholar]
- 2.Krysan, J. L. In Methods for the study of pest Diabrotica. (eds James, L.; Miller Krysan, Thomas, A.) 1–23 (Springer, New York, 1986).
- 3.Grayson J, Poos F. Southern corn rootwornl as a pest of peanuts. J. Econ. Entomol. 1947;40:251–256. doi: 10.1093/jee/40.2.251. [DOI] [PubMed] [Google Scholar]
- 4.Meinke LJ, Gould F, Van Duyn J. Soybean: a larval host for the southern corn rootworm (Coleoptera: Chrysomelidae) Fla. Entomol. 1985;68:496–498. doi: 10.2307/3495145. [DOI] [Google Scholar]
- 5.Arant FS. Biology and control of the southern corn rootworm. Agricultural Experiment Station of the Alabama Polytechnic Institute. 1929;230:48p. [Google Scholar]
- 6.Rupar M, et al. Two novel strains of Bacillus thuringiensis toxic to coleopterans. App. Environ. Microbiol. 1991;57:3337–3344. doi: 10.1128/aem.57.11.3337-3344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan WP, et al. Characterization of two genes encoding Bacillus thuringiensis insecticidal crystal proteins toxic to Coleoptera species. App. Environ. Microbiol. 1992;58:3921–3927. doi: 10.1128/aem.58.12.3921-3927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park Y, Abdullah MAF, Taylor MD, Rahman K, Adang MJ. Enhancement of Bacillus thuringiensis Cry3Aa and Cry3Bb toxicities to coleopteran larvae by a toxin-binding fragment of an insect cadherin. App. Environ. Microbiol. 2009;75:3086–3092. doi: 10.1128/AEM.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chio H, Chang C-S, Metcalf RL, Shaw J. Susceptibility of four species of Diabrotica to insecticides. J. Econ. Entomol. 1978;71:389–393. doi: 10.1093/jee/71.3.389. [DOI] [Google Scholar]
- 10.Baum JA, et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 11.Bolognesi R, et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte) PLoS One. 2012;7:e47534. doi: 10.1371/journal.pone.0047534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine SL, et al. Independent action between DvSnf7 RNA and Cry3Bb1 protein in southern corn rootworm, Diabrotica undecimpunctata howardi and Colorado potato beetle, Leptinotarsa decemlineata. PloS One. 2015;10:e0118622. doi: 10.1371/journal.pone.0118622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira AE, Carneiro NP, Siegfried BD. Comparative susceptibility of southern and western corn rootworm adults and larvae to vATPase-A and Snf7 dsRNAs. J. RNAi Gene Silencing. 2016;12:528–535. [Google Scholar]
- 14.Pereira AE, Vélez AM, Meinke LJ, Siegfried BD. Sublethal effects of vATPase-A and Snf7 dsRNAs on biology of southern corn rootworm, Diabrotica undecimpunctata howardi Barber. J. Econ. Entomol. 2017;110:2545–2553. doi: 10.1093/jee/tox263. [DOI] [PubMed] [Google Scholar]
- 15.Lü J, Yang C, Zhang Y, Pan H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 2018;9:1560. doi: 10.3389/fphys.2018.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shakeel M, Rodriguez A, Tahir UB, Jin F. Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 2018;40:227–236. doi: 10.1007/s10529-017-2465-4. [DOI] [PubMed] [Google Scholar]
- 18.Li R, et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) PloS One. 2013;8:e53006. doi: 10.1371/journal.pone.0053006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu X, et al. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae) PloS One. 2014;9:e84730. doi: 10.1371/journal.pone.0084730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X-Q, et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say) BMC Res. Notes. 2013;6:93. doi: 10.1186/1756-0500-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu W, et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae) Int. J. Biol. Sci. 2013;9:792–802. doi: 10.7150/ijbs.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan M, et al. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PloS One. 2014;9:e86503. doi: 10.1371/journal.pone.0086503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Docimo T, Consonni R, Coraggio I, Mattana M. Early phenylpropanoid biosynthetic steps in Cannabis sativa: link between genes and metabolites. I. J. Mol. Sci. 2013;14:13626–13644. doi: 10.3390/ijms140713626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teng X, Zhang Z, He G, Yang L, Li F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J. Insect Sci. 2012;12:1–17. doi: 10.1673/031.012.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu B, et al. Evaluation of reference genes for real-time quantitative PCR analysis in larvae of Spodoptera litura exposed to Azadirachtin stress conditions. Front. Physiol. 2018;9:372. doi: 10.3389/fphys.2018.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H-B, et al. Screening potential reference genes for quantitative real-time PCR analysis in the oriental armyworm, Mythimna separata. PloS One. 2018;13:e0195096. doi: 10.1371/journal.pone.0195096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, K.-S. et al. Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16 (2016). [DOI] [PMC free article] [PubMed]
- 28.Bansal R, Mamidala P, Mian MR, Mittapalli O, Michel AP. Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae) J. Econ. Entomol. 2012;105:1432–1438. doi: 10.1603/EC12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues TB, Dhandapani RK, Duan JJ, Palli SR. RNA interference in the Asian longhorned beetle: identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 2017;7:8913. doi: 10.1038/s41598-017-08813-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pentzold S, et al. Silencing cuticular pigmentation genes enables RNA FISH in intact chemosensory appendages. J. Exp. Biol. 2018;221:185710. doi: 10.1242/jeb.185710. [DOI] [PubMed] [Google Scholar]
- 31.Van Hiel MB, et al. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 2009;10:56. doi: 10.1186/1471-2199-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh S, et al. Selection of housekeeping genes and demonstration of RNAi in cotton leafhopper, Amrasca biguttula biguttula (Ishida) PloS One. 2018;13:e0191116. doi: 10.1371/journal.pone.0191116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C, et al. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae) Sci. Rep. 2015;5:18201. doi: 10.1038/srep18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stürzenbaum SR, Kille P. Control genes in quantitative molecular biological techniques: the variability of invariance. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2001;130:281–289. doi: 10.1016/S1096-4959(01)00440-7. [DOI] [PubMed] [Google Scholar]
- 35.Ruan W, Lai M. Actin, a reliable marker of internal control? Clin. Chim. Acta. 2007;385:1–5. doi: 10.1016/j.cca.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Kozera B, Rapacz M. Reference genes in real-time PCR. J. Appl. Genet. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson BS, Nam H, Hopkins RG, Morrison RF. Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PloS One. 2010;5:e15208. doi: 10.1371/journal.pone.0015208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bustin SA. Developments in real-time PCR research and molecular diagnostics. Expert Rev. Mol. Diagn. 2010;10:713–715. doi: 10.1586/erm.10.65. [DOI] [PubMed] [Google Scholar]
- 39.Klie M, Debener T. Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida) BMC Res. Notes. 2011;4:518. doi: 10.1186/1756-0500-4-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutierrez L, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheng D, Zhang Z, He X, Liang G. Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS One. 2013;8:e57718. doi: 10.1371/journal.pone.0057718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thellin O, et al. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 1999;75:291–295. doi: 10.1016/S0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 43.Radonić A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 44.Dai T-M, Lü Z-C, Liu W-X, Wan F-H. Selection and validation of reference genes for qRT-PCR analysis during biological invasions: The thermal adaptability of Bemisia tabaci MED. PloS One. 2017;12:e0173821. doi: 10.1371/journal.pone.0173821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lü, J. et al. Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9 (2018). [DOI] [PMC free article] [PubMed]
- 46.Pan H, Yang X, Siegfried BD, Zhou X. A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae) PloS One. 2015;10:e0125868. doi: 10.1371/journal.pone.0125868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan H, et al. Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bio-indicator. PloS One. 2015;10:e0129482. doi: 10.1371/journal.pone.0129482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Pan H, Yuan L, Zhou X. Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 2018;8:2689. doi: 10.1038/s41598-018-20612-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodrigues TB, et al. Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera) PloS One. 2014;9:e109825. doi: 10.1371/journal.pone.0109825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark TL, Hibbard BE. Comparison of nonmaize hosts to support western corn rootworm (Coleoptera: Chrysomelidae) larval biology. Env. Entomol. 2004;33:681–689. doi: 10.1603/0046-225X-33.3.681. [DOI] [Google Scholar]
- 51.Rault, L. C. et al. Investigation of Cry3Bb1 resistance and intoxication in western corn 516 rootworm by RNA sequencing. J. Appl. Entomol. 142, 921-936 (2018).
- 52.Knolhoff, L. M., Walden, K. K., Ratcliffe, S. T., Onstad, D. W. & Robertson, H. M. 518 Microarray analysis yields candidate markers for rotation resistance in the western corn 519 rootworm beetle, Diabrotica virgifera virgifera. Evol. Appl.3, 17-27 (2010). [DOI] [PMC free article] [PubMed]
- 53.Rangasamy M, Siegfried BD. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) adults. P. Manag. Sci. 2012;68:587–591. doi: 10.1002/ps.2301. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Z, et al. Evaluation of reference genes for normalization of RT-qPCR gene expression data for Trichoplusia ni cells during Antheraea pernyi (Lepidoptera: Saturniidae) multicapsid Nucleopolyhedrovirus (AnpeNPV) infection. J. Insect Sci. 2019;19:4. doi: 10.1093/jisesa/iey133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alves AP, Lorenzen MD, Beeman RW, Foster JE, Siegfried BD. RNA interference as a method for target-site screening in the western corn rootworm, Diabrotica virgifera virgifera. J. Insect Sci. 2010;10:162. doi: 10.1673/031.010.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fishilevich E, et al. Use of chromatin remodeling ATPases as RNAi targets for parental control of western corn rootworm (Diabrotica virgifera virgifera) and Neotropical brown stink bug (Euschistus heros) Insect Biochem. Mol. Biol. 2016;71:58–71. doi: 10.1016/j.ibmb.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Khajuria C, et al. Parental RNA interference of genes involved in embryonic development of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochem. Mol. Biol. 2015;63:54–62. doi: 10.1016/j.ibmb.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Tan SY, et al. RNAi induced knockdown of a cadherin-like protein (EF531715) does not affect toxicity of Cry34/35Ab1 or Cry3Aa to Diabrotica virgifera virgifera larvae (Coleoptera: Chrysomelidae) Insect Biochem. Mol. Biol. 2016;75:117–124. doi: 10.1016/j.ibmb.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 59.Vélez AM, Khajuria C, Wang H, Narva KE, Siegfried BD. Knockdown of RNA interference pathway genes in western corn rootworms (Diabrotica virgifera virgifera Le Conte) demonstrates a possible mechanism of resistance to lethal dsRNA. PloS One. 2016;11:e0157520. doi: 10.1371/journal.pone.0157520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vélez AM, et al. Parameters for successful parental RNAi as an insect pest management tool in western corn rootworm, Diabrotica virgifera virgifera. Genes. 2016;8:7. doi: 10.3390/genes8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vélez AM, et al. Developing an in vivo toxicity assay for RNAi risk assessment in honey bees, Apis mellifera L. Chemosphere. 2016;144:1083–1090. doi: 10.1016/j.chemosphere.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 62.EPA. Pesticide registration: EPA registers innovative tool to control corn rootworm, https://www.epa.gov/pesticide-registration/epa-registers-innovative-tool-control-corn-rootworm (2016).
- 63.Godinho RMDC, et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci. Rep. 2014;4:6198. doi: 10.1038/srep06198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat. Cell Biol. 2012;14:38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- 65.Branson T, Jackson J. An improved diet for adult Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae) J. Kans. Entomol. Soc. 1988;61:353–355. [Google Scholar]
- 66.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 67.Saiki RK, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.239.4839.487. [DOI] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 70.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 71.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.