Abstract
MicroRNAs from the miR-200 family are commonly associated with the inhibition of the metastatic potential of cancer cells, following inhibition of ZEB transcription factors expression and epithelial-to-mesenchymal transition. However, previous studies performed in pancreatic adenocarcinoma revealed a more complex picture challenging this canonical model. To gain better insights into the role of miR-200 family members in this disease, we analyzed the expression of miR-200a, miR-200b, miR-200c, miR-141, miR-429, and miR-205, and ZEB1, ZEB2, and CDH1 in pancreatic tumors and matching normal adjacent parenchyma and patient-derived xenografts. We found that miR-200a, miR-429, and miR-205 are frequently overexpressed in pancreatic tumors, whereas CDH1 is downregulated, and ZEB1 and ZEB2 levels remain unchanged. Furthermore, we measured a positive correlation between miR-200 family members and CDH1 expression, and a negative correlation between ZEB1 and miR-200c, miR-141, and miR-205 expression, respectively. Interestingly, we identified significant changes in expression of epithelial-to-mesenchymal transition regulators and miR-200 members in patient-derived xenografts. Lastly, functional studies revealed that miR-141 and miR-429 inhibit the tumorigenic potential of pancreatic cancer cells. Taken together, this comprehensive analysis strongly suggests that miRNAs from the miR-200 family, and in particular miR-429, may act as a tumor suppressor gene in pancreatic cancer.
Keywords: pancreatic cancer, patient-derived xenograft, miR-200 family, miR-429
Introduction
Pancreatic ductal adenocarcinoma (PDAC) cancer is the most common pancreatic neoplasm and is currently the third cause of cancer-related mortality in Western countries.1 PDAC is particularly aggressive because it easily invades adjacent tissues and metastasizes to distant organs.2 Moreover, PDAC is resistant to standard chemotherapies.3 The most frequent genetic abnormalities in PDAC generally follow a characteristic pattern where KRAS oncogene activation is an early event, with alterations in CDKN2A, TP53, SMAD4, and BRCA2 accumulating in later stages.2
Tumor progression and metastases formation in carcinomas are associated with a series of changes including epithelial-to-mesenchymal transition (EMT) to increase cell motility and invasiveness.4, 5 Downregulation of E-cadherin (CDH1) expression leading to loss of cell adhesion is considered a key feature of EMT.5 Transcription factors ZEB1 and ZEB2 directly inhibit E-cadherin expression and are often upregulated in cancer cells.6 Other transcription factors that regulate CDH1, such as Snail and Twist families, can be also upregulated.7
MicroRNAs (miRNAs) are a class of highly conserved, small non-coding RNAs that regulate gene expression by targeting mRNAs for cleavage or translational repression.8 The miR-200 family is composed of five members distributed in two clusters. The first cluster is located in chromosome 1 and consists of miR-200b, miR-200a, and miR-429. The second cluster is hosted by chromosome 12 and includes miR-200c and miR-141.9 In most tumors, miR-200 family members, as well as miR-205, which does not belong to this family, inhibit the expression of ZEB1 and ZEB2 transcription factors, notably following binding to their UTR.10 This is relevant for the maintenance of the epithelial integrity because it prevents EMT.11, 12, 13, 14
Although several studies have reported the downexpression of miR-200 family members in lung,15 colon,16 or breast tumors,11 upregulation of miR-200 is upregulated in non-small-cell lung cancer.17 Furthermore, enforced miR-200 expression promotes metastatic spread in breast cancer models,18, 19 but overexpression of miR-141 or miR-429 has been associated with a suppressor gene function in several tumor types.20, 21, 22, 23
However, the picture is seemingly more complex for PDAC. miR-200 family members are frequently overexpressed in precursor lesions24 and PDAC,18, 25 where they may act as oncogenes26 or as suppressor genes.27 A complex crosstalk between miR-200 and ZEBs has been suggested. Even though miR-200 family members negatively regulate ZEBs, ZEB1 expression may result in the downregulation of miR-141 and miR-200c levels in pancreatic, colorectal, and breast cancer cells.28 Recently, similar studies described ZEB1 as a key factor in precursor lesions formation, but also in tumor invasion and metastasis.29
The goal of this study was to get better insights into the complex crosstalk between miR-200 family members and EMT regulators, in PDAC primary tumors and in experimental metastases derived from patient-derived xenografts (PDXs). Moreover, the potential effect of miR-200 family members was explored in experimental models of PDAC. Our results depict a specific pattern of deregulation of miR-200 family members, as well as miR-205, in primary PDAC tumors. This pattern of expression was positively correlated with CDH1 expression and negatively correlated with ZEB transcription factors expression, respectively. Notably, miRNA and EMT regulators expression remained stable during PDX dissemination in mice, regardless of the site of metastasis, demonstrating the importance of this network in PDAC carcinogenesis. Lastly, functional studies performed in vivo demonstrated that miR-141 and miR-429 may act as tumor suppressor genes in this disease.
Results
Expression of miR-200 and Selected Genes Involved in the EMT in a Cohort of Pancreatic Tumors
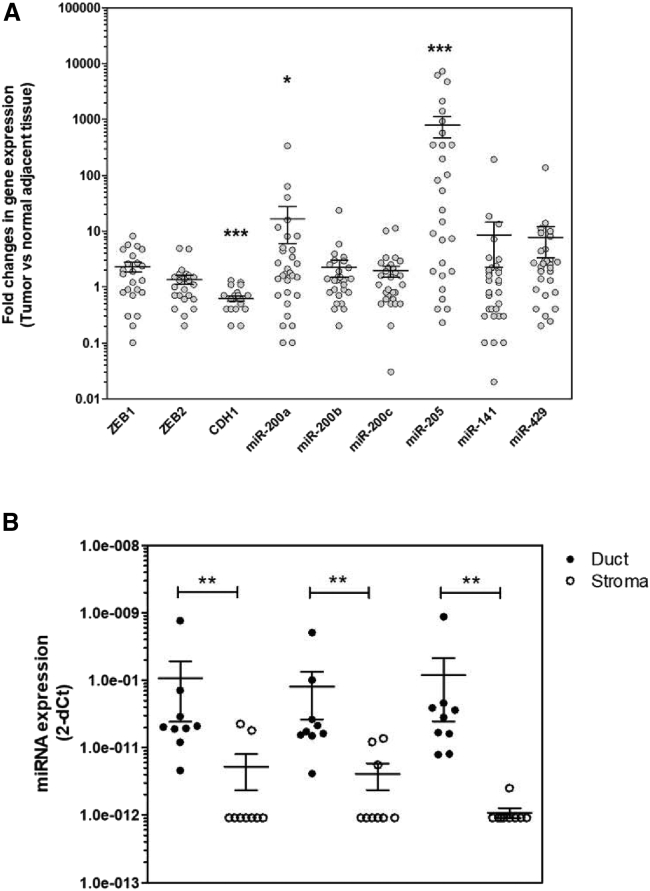
We first assessed by quantitative real-time PCR the expression levels of selected mRNAs and miRNAs involved in EMT in a cohort of 31 primary pancreatic tumors with matching adjacent macroscopically normal parenchyma that was selected 1–3 cm distal from the macroscopic tumor margin. We found that miR-200a (14/31 tumors [45%], p = 0.03), miR-429 (19/31 tumors [61%], p = 0.001), and miR-205 (20/31 tumors [65%], p = 0.0001) are overexpressed in PDAC primary tumors compared with adjacent normal tissues (Figure 1A; Table 1; Tables S1 and S2). We further analyzed the expression levels of miR-200a, miR-141, and miR-429 in nine paired microdissected ductal and stroma samples to better address the cell compartment of origin for these miRNAs because these miRNAs were constantly detected, at least in samples from ductal origin. The results presented in Figure 1B demonstrate that miRNAs are overexpressed in ductal cells as compared with the stromal compartment of pancreatic tumors (**p < 0.01). Specifically, miR-200a expression is detectable in 9 out of 10 ductal samples (90%), but in only 3 out of 9 (33%) in the corresponding stromal compartment, while miR-141 is detected in all ductal samples but in only 4 out of 9 (44%) of the paired stromal components. Lastly, miR-429 is expressed in 7 out of 9 (78%) ductal cells and in 3 out of 9 (33%) of corresponding stroma (Figure 2B; Table S3). Collectively, these results indicate that epithelial cancer cells are likely the main source for the candidate miRNAs expression in PDAC tumors.
Figure 1.
Expression Levels of miR-200 Family Members and ZEB1, ZEB2, and CDH1 in Primary Tumors of PDAC
(A) Target genes expression was measured in duplicates by quantitative real-time PCR in primary tumors and matching adjacent normal tissues. Results are represented as fold changes in gene expression. *p < 0.05; **p < 0.01; ***p < 0.001 (Wilcoxon test). (B) Expression of candidate microRNAs in microdissected ductal cancer cells and pancreatic stromal cells. Ductal and stromal compartments were microdissected in nine PDAC primary tumors as described in the Materials and Methods. miR-200, miR-141, and miR-429 miRNAs were quantified by quantitative real-time PCR in triplicates. Relative miRNA expression is expressed as 2−(CtmiRNA-CtRNU48). Forty cycles were attributed when miRNA was undetectable.
Table 1.
Expression Levels of Members of the miR-200 Family and ZEB1, ZEB2, or CDH1 in PDAC Tumors as Compared with Adjacent Normal Tissue
| miR-200a | miR-200b | miR-429 | miR-200c | miR-141 | miR-205 | ZEB1 | ZEB2 | CDH1 | |
|---|---|---|---|---|---|---|---|---|---|
| Overexpressiona | 14/31, 45% | 9/31, 29% | 19/31, 61% | 9/31, 29% | 8/31, 26% | 20/31, 65% | 13/31, 42% | 5/31, 16% | 2/31, 6.5% |
| Underexpressiona | 7/31, 23% | 4/31, 13% | 5/31, 16% | 12/31, 39% | 5/31, 16% | 6/31, 19% | 9/31, 29% | 9/31, 29% | 11/31, 35.5% |
| Unchangeda | 10/31, 32% | 18/31, 58% | 7/31, 23% | 10/31, 32% | 18/31, 58% | 5/31, 16% | 9/31, 29% | 17/31, 55% | 18/31, 58% |
| Ct values (range): normal tissue | 23–32 | 25–33 | 31–36 | 21–29 | 24–32 | 26–37 | 27–40 | 27–36 | 24–28 |
| Ct values (range): tumoral tissue | 22–30 | 25–30 | 30–34 | 21–26 | 23–29 | 26–37 | 27–40 | 27–35 | 24.3–28 |
Fold change (2−ΔΔCt) > 2: overexpression; fold change (2−ΔΔCt) < 0.5: underexpression.
The proportion of tumors overexpressing (FC > 2) and underexpressing (FC < 0.5) the target gene is shown as a percentage.
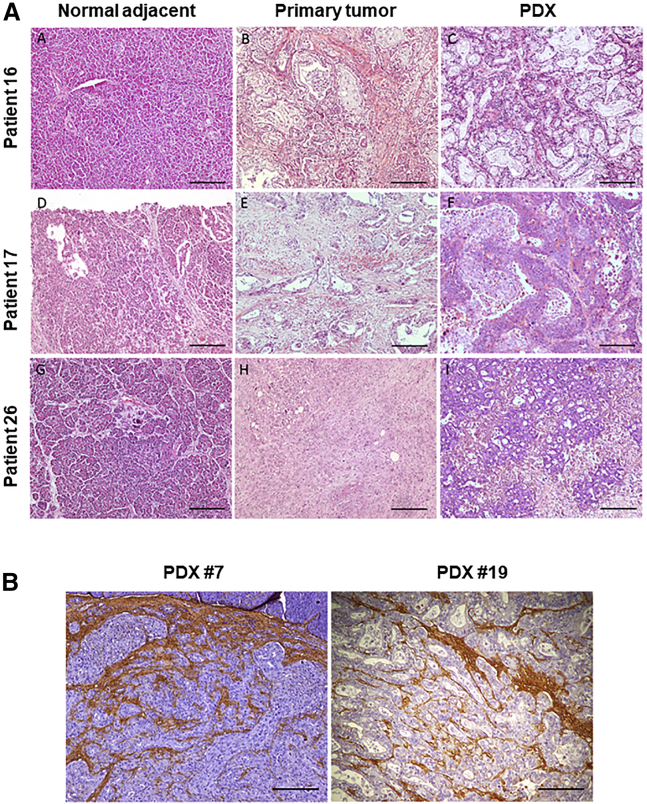
Figure 2.
Characterization of the Patient-Derived Xenografts (PDXs) Used in This Study
(A) Cases TP16, TP17, and TP26 are shown. H&E staining of normal tissue (subpanels A, D, and G), paired tumor tissue (subpanels B, E, and H), and corresponding PDXs (subpanels C, F and I). All images are original magnification ×100. (B) Hyaluronic acid staining of orthotopic PDXs is shown as a surrogate marker of desmoplastic reaction (left: PDX TP7; right: PDX TP19).
We next addressed the expression of selected genes governing EMT in the same cohort of samples. We found that CDH1 mRNA expression is significantly decreased in PDAC tissues as compared with normal adjacent parenchyma (Figure 1A). CDH1 levels are low in 11 of 31 primary tumors (range = 0.16–0.49, p = 0.0001; Table 1; Table S1), suggesting that EMT is frequent in this cohort of PDAC samples. Only two tumors (6.5%) demonstrate CDH1 overexpression as compared with normal parenchyma (Table 1; Tables S1 and S2). On the other hand, ZEB1 and ZEB2 mRNA levels remain unchanged between tumor and normal tissues (Figure 1), although variability is high among samples (Table 1; Table S1).
As stated before, the miR-200 family members repress EMT, the initiating step of metastasis, by maintaining the epithelial phenotype through direct targeting of ZEB1 and ZEB2, the well-characterized transcriptional repressors of CDH1. Consequently, we investigated whether the levels of miR-200 family members correlated with ZEB1, ZEB2, or CDH1 expression in this cohort of patients. We found a significant positive correlation between miRNAs from the miR-200 family and CDH1 expression levels (Table 2). On the other hand, no correlation exists between miR-205 and CDH1 expression levels. As for ZEB1, we identified a negative correlation with miR-200a, miR-200c, miR-141, and miR-205 expression in these samples. Finally, a negative correlation between ZEB2 and miR-200c, miR-429, and miR-205 was also demonstrated. These results suggest that miR-200 family members target ZEB1 and ZEB2, and restrain EMT in this cohort of PDAC samples. We next address whether DNA methylation could also account for EMT-related gene expression in these samples. CDH1 promoter hypermethylation was not detected in these samples, whereas half of tumors (4/11) demonstrated, albeit low, methylation levels of ZEB1 and ZEB2 promoters (Table 3). However, no correlation was found between ZEB1 or ZEB2 expression levels and DNA promoter methylation (data not shown). Thus, our results strongly suggest that ZEB1 and ZEB2 expression are mainly controlled by miRNA in these samples.
Table 2.
Gene Expression Correlation between miR-200 Family Members and ZEB1, ZEB2, and CDH1 mRNAs in Primary Pancreatic Tumors
| ZEB1 | ZEB2 | CDH1 | |
|---|---|---|---|
| miR-200a | −0.375 | −0.419 | 0.425 |
| p = 0.038a | p = 0.019a | p = 0.017a | |
| miR-200b | −0.263 | −0.333 | 0.642 |
| p = 0.151 | p = 0.067 | p = 0.0001a | |
| miR-200c | −0.460 | −0.298 | 0.580 |
| p = 0.009a | p = 0.102 | p = 0.0007a | |
| miR-141 | −0.41 | −0.375 | 0.610 |
| p = 0.021a | p = 0.038a | p = 0.0003a | |
| miR-429 | −0.2512 | −0.227 | 0.368 |
| p = 0.170 | p = 0.218 | p = 0.041a | |
| miR-205 | −0.374 | −0.2088 | 0.298 |
| p = 0.038a | p = −0.208 | p = 0.102 |
Gene expression is calculated using the 2−ΔCt formula as described before. Correlation coefficient values were assessed by Spearman analysis.
Significant p values, p < 0.05.
Table 3.
Expression and Promoter Methylation of ZEB1 and ZEB2 in PDAC
|
ZEB1 |
ZEB2 |
|||
|---|---|---|---|---|
| Tumor Sample | 2−ΔCT | Methylation | 2−ΔCT | Methylation |
| TP1 | 8.33 | M* | 8.6 | U |
| TP5 | ND | U | ND | M |
| TP7 | ND | U | ND | U |
| TP9 | 11.055 | U | 11.4 | U |
| TP10 | 12.06 | U | 10 | U |
| TP11 | 11.86 | U | 11.9 | U |
| TP16 | 7.60 | M* | 7.5 | U |
| TP18 | 11.96 | M* | 10.4 | U |
| TP19 | ND | M* | ND | M* |
| TP26 | 12.31 | U | 9.5 | M* |
| TP32 | 9.64 | U | 9.2 | M* |
Gene expression analyzed by quantitative real-time PCR is expressed as 2−ΔCt versus normal adjacent tissue. M, methylated; M*, low intensity of the methylation signal; ND, not determined; U, unmethylated.
Development of a Mouse Cohort of PDAC PDXs
To gain further insights into the expression and the role of miR-200 family members and ZEB1, ZEB2, and CDH1 in EMT in PDAC, we generated a collection of PDXs from a cohort of 32 patients who underwent surgery for PDAC. An expert pathologist selected the tumor fragments to be implanted from the invading front of the tumor. Eighteen of the 32 tumors were successfully perpetuated (56%), 11 of which were used in the present study. The histological analysis confirms the remarkable histological similarities between primary tumors and corresponding PDX (Figure 2A). Hematoxylin, eosin, and hyaluronic acid staining analysis performed by an expert pathologist reveals that tumor cellularity is seemingly higher in PDXs than in primary tumors, although some degree of heterogeneity is observed. Low (0%–6%) and moderate (10%–18%) stroma levels were measured in 2 out of 11 and 9 out of 11 PDXs, respectively (Figure 2B; Table S3), reflecting the well-characterized deprivation of the stromal component following serial passaging in mice.30 We next analyzed the genetic landscape of primary PDAC tumors and corresponding PDXs. We found that the proportion and the combination of genetic aberrations measured in PDAC primary tumors is perfectly mirrored by PDXs (Table 4) and is in agreement with previous reports from the literature.2 The latter demonstrates that no significant loss or accumulation of the analyzed driver genetic alterations occurred during passaging of PDXs. Taken together, we consider that the PDXs used in this study are a bona fide model of human pancreatic tumors. Because of the paucity of paired human primary tumor and metastasis available for research, we used this collection of orthotopic PDXs to generate a paired library of 25 experimental metastases. Similar to what was observed during PDX perpetuation, no additional genetic alterations were found in the metastases (Table 4).
Table 4.
Growth and Dissemination Patterns of Pancreatic PDXs and Genetic Characterization of Xenograft Samples (Orthotopic and Corresponding Metastases)
| PDX TP1 | PDX TP5 | PDX TP7 | PDX TP9 | PDX TP10 | PDX TP11 | PDX TP16 | PDX TP18 | PDX TP19 | PDX TP26 | PDX TP32 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Timea | 4.5 | 2.5–3 | 3 | 2.5 | 1–1.5 | 2.75 | 4 | 8.5 | 2.5–3 | 2 | 8 |
| Differentiationb | mod | mod | poor | poor | poor | poor | poor | poor | mod | poor | mod |
| Local invasion | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Metastasis | |||||||||||
| Peritoneal | × | × | × | × | × | × | × | × | × | × | |
| Lymph node | × | × | × | × | × | × | × | ||||
| Diaphragm | × | × | × | × | |||||||
| Lung | × | ||||||||||
| Liver | × | ||||||||||
| Genetic Alterations | |||||||||||
| KRAS | c.35G>A (p.G12D) | c.35G>A (p.G12D) | c.35G>C (p.G12R) | c.35G>A (p.G12D) | WT | c.35G>T (p.G12V) | c.35G>A (p.G12D) | WT | c.35G>A (p.G12D) | c.35G>A (p.G12D) | WT |
| BRAF | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| PI3KCA | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| EGFR | WT/del | amp | WT/amp | WT | del/amp | WT | WT | WT/amp | WT/amp | WT/del | WT |
| CDKN2A | c.330G>A (p.W110*) | WT | c.238C>T (p.R80*) | e1-3Del | WT | c.47-50del (p.L16P fs*9) | e1-3Del | e1-3Del | WT | e1-3Del | e1Del |
| TP53 | c.725G>C (p.C242S) | c.214dup (p.V73Rfs*76) | c.818G>A (p.R273H) | c.818G>T (p.R273L) | c.727A>G (p.M246V) | c.214dup (p.V73R fs*76) | c.949del (p.Q317Sfs*28) | WT | WT | WT | c.526T>C (p.C176R) |
| SMAD4 | WT | e1-11Del | WT | c.1052-1054del p.D351del | c.354G>A (p.A118 =) | WT | WT | WT | c.344G>A (p.C115Y) | WT | e8-11Del |
amp, amplification; del, deletion; mod, moderate; WT, wild type.
Time elapsed between passages (months).
Dissemination: no, absence; yes, presence.
Expression of miR-200 Family Members and ZEB1, ZEB2, and CDH1 in PDX and Experimental Metastases
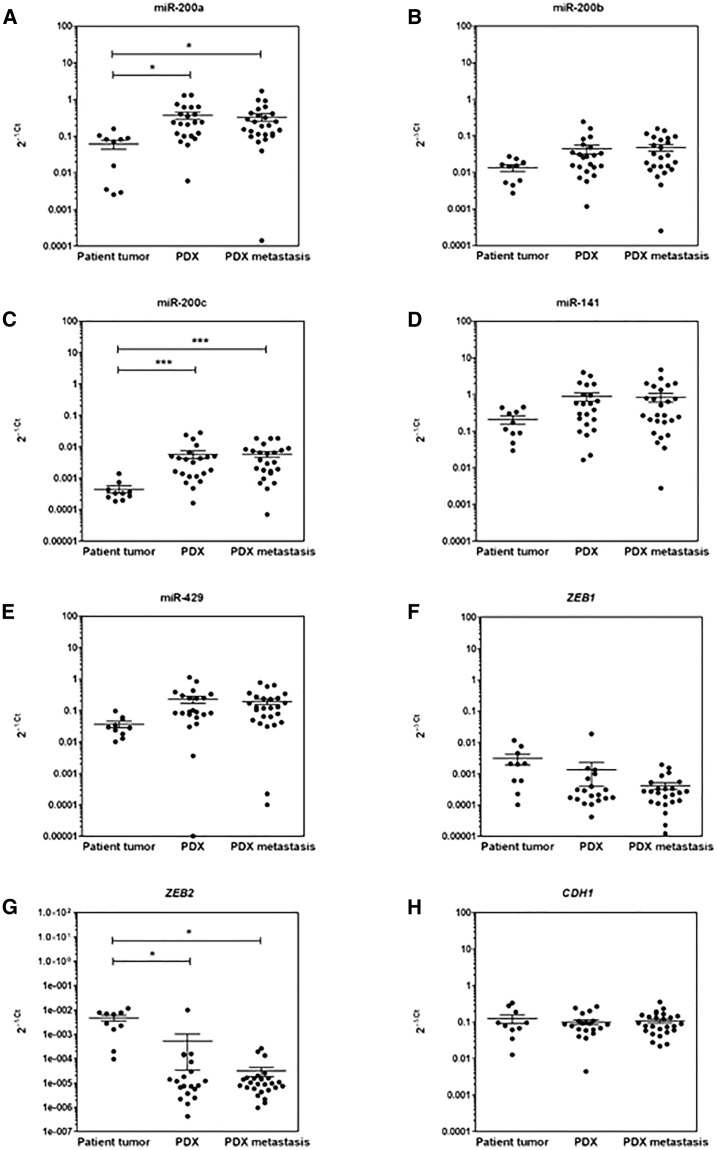
Next, we used this model system to better understand the pattern of expression and the impact of miR-200 family members in the regulation of the epithelial state of pancreatic tumors, and the expression of ZEB1/ZEB2 epithelial gene transcriptional repressors in vivo. We first investigated whether the expression of these candidate genes was altered following PDX propagation and distal dissemination. For this purpose, 22 independent samples from 11 original PDXs were analyzed with paired primary tumors. Also, 25 paired experimental metastases were studied. Two of the five miRNAs analyzed, miR-200a (2.93-fold increase) and miR-200c (14-fold increase), were overexpressed in PDXs when compared with primary tumors (p = 0.017 and p = 0.0001, respectively, Kruskal-Wallis and Tukey’s honestly significant tests; Figures 3A and 3C). On the other hand, ZEB2 expression levels were decreased in PDXs as compared with primary tumors (p = 0.019; Figure 3G), whereas ZEB1 and CDH1 RNA levels remained unchanged (Figures 3F and 3H). No correlation was found between the expression levels of candidate miRNAs, EMT effectors, and KRAS, EGFR, CDKN2A, TP53, and SMAD4 status (data not shown). Notably, the RNA levels of all target miRNAs and EMT regulators remained stable during PDX dissemination in mice, regardless of the site of metastasis (Figures 3A–3H).
Figure 3.
Expression Levels in PDX Model
(A–H) Expression levels (2−ΔCt) of miR-200 family members (A, miR-200; B, miR-2006b; C, miR-200c; D, miR-141; E, miR-429) and ZEB1 (F), ZEB2 (G), and CDH1 (H) in 11 primary tumors (PT) paired with PDXs and corresponding metastases (M1) (the p value calculated with Tukey multiple comparisons of means compares the mean of each group in pairs was shown).
Role of miR-200 Family Members in Experimental Models of PDAC
Our next goal was to address the functional significance of the expression of the miR-200 family members in experimental models of PDAC. Using quantitative real-time PCR, we first characterized the expression levels of the candidate miRNAs in a panel of human PDAC-derived cell lines that recapitulate the molecular heterogeneity of these tumors. We selected PANC-1 cells because these cells: (1) express low levels of miR-200 family members; and (2) show no evidence of promoter methylation of CDH1, ZEB1, and ZEB2 genes (data not shown).31 Thus, PANC-1 cells were selected for functional studies and were transduced with lentiviral vectors encoding for miR-200 family members, alone or in combination, as reported before.32 By quantitative real-time PCR, we found that transduced cells readily overexpressed miR-200 clusters (miR-200b-a-429 and miR-200c-141) or individual miRNAs (miR-141 and miR-429), and to a lesser extent miR-200c, in cells transduced with the miR-200c-141 cluster (Table S4). Robust and reproducible expression of the candidate miRNAs was also detected following transduction of other PDAC cell lines (data not shown).
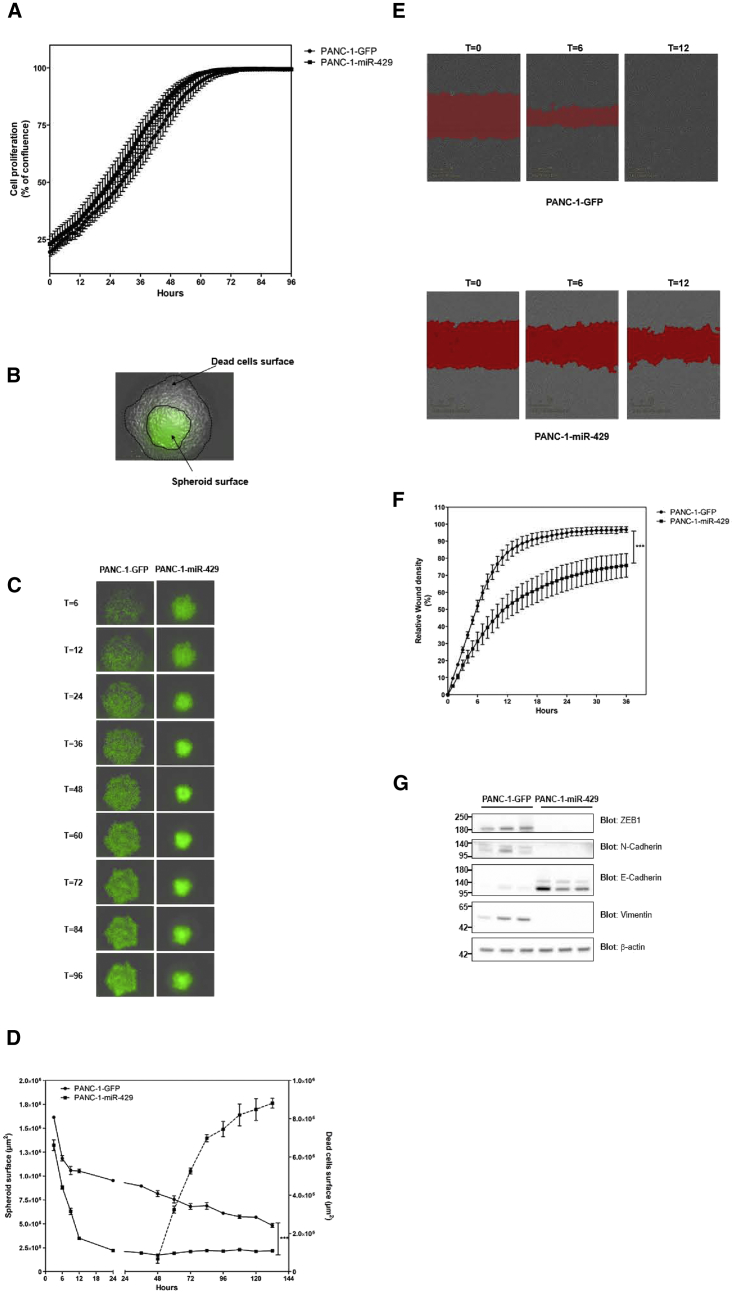
We next investigated the functional consequences of the overexpression of candidate miRNAs in experimental models of PDAC. PANC-1 cells transduced with a lentiviral vector encoding for GFP were used as a control (PANC-1-GFP). We found that PANC-1 cell proliferation remained largely unchanged following expression of candidate miRNAs, as compared with control-transduced cells (Figure 4A). On the other hand, overexpressing miR-429 strongly impaired PANC-1 cell 3D growth and migration. Indeed, spheroids from PANC-1 cells expressing miR-429 were significantly smaller as compared with control spheroids after 6 days in culture (2.23 ± 0.28-fold reduction; p < 0.001, two-way ANOVA test; Figure 4C). Interestingly, real-time imaging using IncuCyte Zoom revealed that miR-429 not only rapidly diminished the size of spheroids, but also strongly impacted cell viability because we evidenced an increase of dying cells after 60 h in culture (Figures 4B and 4D) that were easily distinguishable from GFP-positive, healthy cells. miR-429 expression also has a profound inhibitory effect on PDAC cell migration. Real-time imaging of gap-closure assay indicated that control PANC-1 cells migrate with high efficacy into the wound region (Figure 4E). Complete wound closure was achieved after 12 h (Figures 4E and 4G). On the other hand, expressing miR-429 strongly inhibited the gap-closure activity of PANC-1 cells (1.6 ± 0.4-fold reduction, p < 0.001, two-way ANOVA test; Figures 4F and 4G), and PANC-1-miR-429 cells failed to complete wound closure. These data indicate that miR-429 strongly inhibits the migration of PDAC cells. Molecular examination of PANC-1 cells indicated that overexpression of miR-429 decreases ZEB1, N-cadherin, and Vimentin expression, whereas E-cadherin expression was upregulated (Figure 4G). Thus, miR-429 induces a mesenchymal-to-epithelial (MTE) switch that is associated with inhibition of 3D tumor growth and migratory capacities of PDAC cells.
Figure 4.
miR-429 Expression Inhibits 3D Growth and Migration, and Inhibits EMT of PDAC Cells
PANC-1 cells were transduced by lentiviral vectors encoding for miR-429, as described in the Materials and Methods. As a control, PANC-1 cells were transduced with vectors encoding for GFP. (A) Cell confluence was measured using the IncuCyte Zoom. Results are mean ± SD of triplicates from two independent sets of transduction. (B) Description of the surface of spheroids and dead cells used for the analysis. (C) Representative images of spheroid formation using control of miR-429-expressing PANC-1 cells, at the time indicated, using the IncuCyte Zoom. (D) Quantification of spheroid surface and cell death in real-time using the IncuCyte Zoom. Results are mean ± SD of triplicates from two sets of transduction. ***p < 0.001. Cells were seeded in 96-well ImageLock plate, and homogeneous scratch wounds were performed using the WoundMaker mechanical device from Essen BioScience (Sartorius) 24 h later. (E) Representative wound closure in control (top) and miR-429-expressing (bottom) PANC-1 cells at the time indicated. Red area: mask used to calculate the relative wound density, as a measure (%) of the density of the wound region relative to the density of the cell region. (F) Real-time quantification of wound closure in both groups using the IncuCyte Zoom. Results are mean ± SD of quadruplicates from two sets of transduction. ***p < 0.001. (G) Western blotting for ZEB1, N-cadherin, Vimentin, and E-cadherin in PANC-1 cells expressing or not miR-429. β-Actin was used as a loading control. Molecular weights (in kDa) are indicated on the left side. Results are representative of two different sets of transduction.
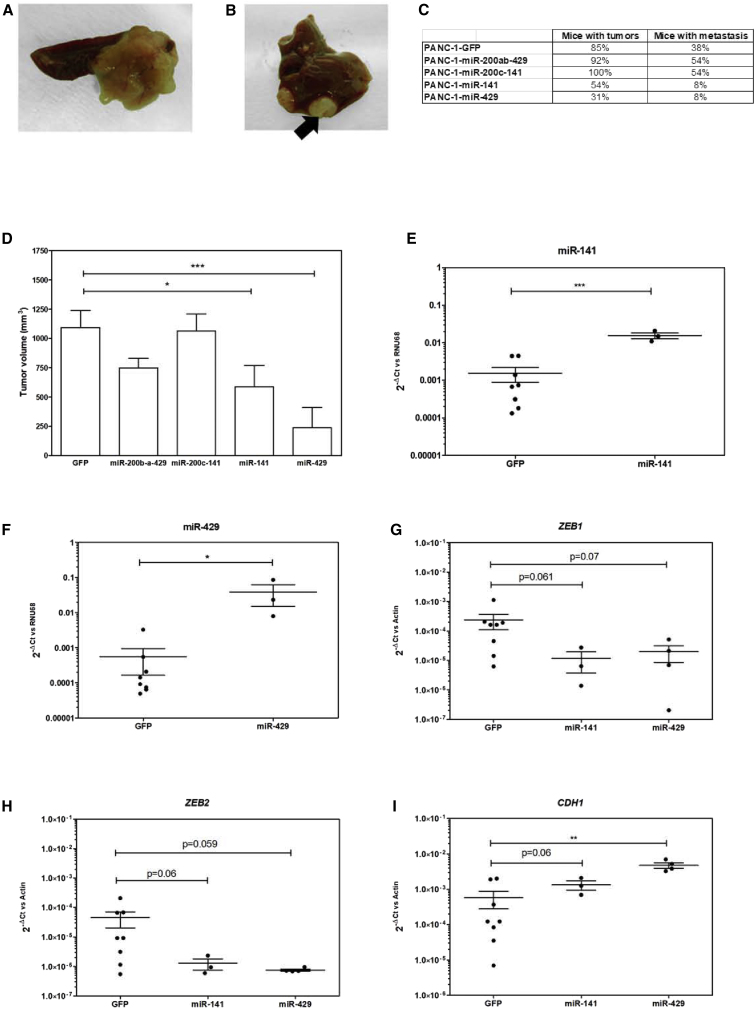
We next engrafted candidate miRNA-expressing cells that were in the pancreas of nude mice to address the tumorigenic potential of miRNA from the miR-200 family. No macroscopic histological differences were observed among the orthotopic tumors (Figures 5A–5F) or the metastases (Figures 5G–5L) generated from the control of miRNA-expressing cells, respectively. Figures 6A and 6B are representative of orthotopic and metastatic tumor growth, respectively. miR-200c-141 and miR-200b-a-429 miRNA clusters failed to inhibit pancreatic tumor growth, as compared with GFP-expressing PANC-1 cells; indeed 100% (13/13) and 92% (12/13) of mice engrafted with PANC-1/miR-200c-141 and PANC-1/miR-200b-a-429 developed orthotopic PDAC tumors, respectively, as compared with control cells (85% tumor formation, 11/13; Figure 6C). The proportion of mice engrafted with PANC-1 cells expressing either miRNA cluster displays a similar incidence of distal metastases (7/13, 53%) as compared with control tumors (5/13, 30%; Figure 6C). On the other hand, overexpression of miR-429, and to a lesser extent of miR-141, in PANC-1 cells strongly inhibits pancreatic carcinogenesis because 30% (4/13) and 54% (7/13) of mice receiving these cells developed tumors, respectively (Figure 6C). Furthermore, expressing miR-429 significantly inhibited PDAC tumor dissemination (1/13, 8%, p < 0.05; Figure 6C). Along the same line, the median tumor volume at sacrifice was also strongly decreased only in miR-429, and again to a lesser extent in miR-141-expressing tumors, as compared with control (Figure 6D). Tumors collected at the completion of the experiment demonstrated increased levels of the corresponding miRNAs as compared with control (Figures 6E and 6F). In addition, we found that CDH1 levels were significantly increased in the miR-429 transduced group (Figure 6I). We also identified a trend indicating that ZEB1 and ZEB2 expression are downregulated in miR-141- and miR-429-expressing tumors, which failed short to reach statistical significance because of the low tumor burden in these experimental groups (Figures 6G and 6H). Taken together, our results demonstrate that miR-429, and to a lesser degree miR-141, decreases PDAC tumor growth and metastatic spread, and modulates the level of expression of key regulators of EMT.
Figure 5.
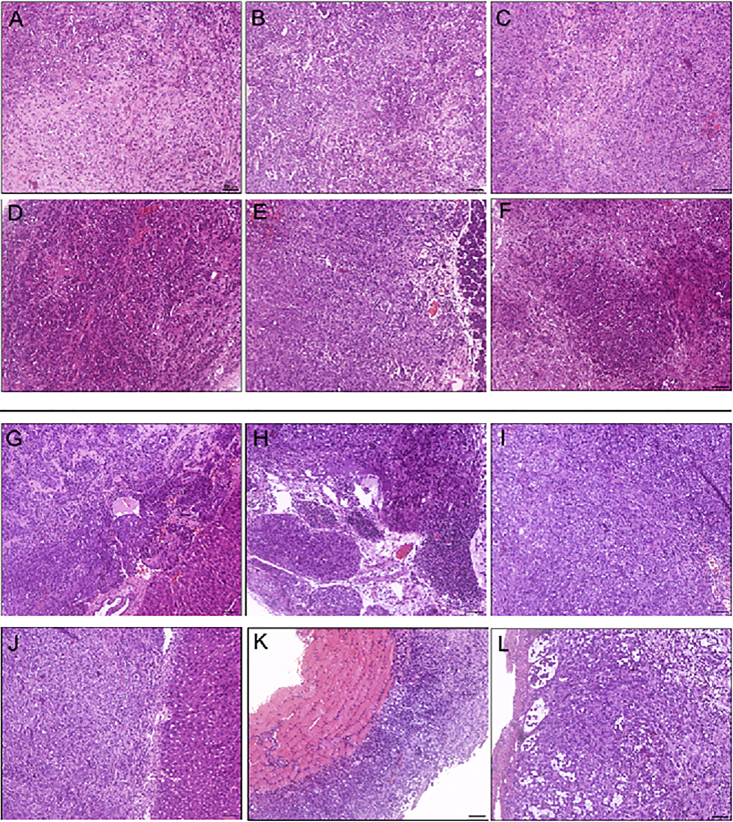
Experimental Tumors Generated from PDAC Cell Lines Expressing Candidate miRNAs from the miR-200 Family
Panc-1 cells were transduced with lentiviral vectors encoding for GFP (A, D, G, and J), miR-200a-b-429 (B and H), miR-200c-141 (C and I), miR-141 (E and K), or miR-429 (F and L) and implanted in the pancreas of athymic mice as described in the Materials and Methods. Mice were sacrificed, and tumors were sampled and stained using H&E. (A–F) Primary tumors. (G–L) Metastatic growth. Results are representative of 13 mice per group.
Figure 6.
In Vivo Activity of miR-200 Family Members on Experimental Tumor Growth and Dissemination
PANC-1 cells transduced with miR-200b-a-429, miR-200c-141, miR-141, miR-429, or control GFP cells were injected orthotopically in nude mice (n = 13 per group), as described in the Materials and Methods. (A and B) Illustrative pictures of the orthotopic grafts (A) and liver metastases (B) obtained from animals of the control group. (C) Number of mice with orthotopic grafts (left) and distal metastases (right) are indicated among the different study groups. (D) Bar graph of the orthotopic tumor volume (in mm3). (E–I) Expression levels of mIR-200 family (E, miR-141; F, miR-429) and target genes (G, ZEB1; H, ZEB2; and I, CDH1) (median of 2−ΔCT) in tumors generated from cell lines transduced with miR-141 or miR-429, or with GFP-encoding vector as a control.
Discussion
The miR-200 family members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) and miR-205 are commonly associated with EMT,11 a fundamental process in the early steps of the metastatic dissemination. The canonical hypothesis states that the miR-200 family inhibits the EMT process through the targeted silencing of ZEB proteins, resulting in increased CDH1 expression and the acquisition of a more differentiated epithelial phenotype.18, 33 Despite the importance of this process in PDAC carcinogenesis, the status of expression of miR-200 family members has been insufficiently investigated to date. In this work, we found that miR-200a, miR-429, and miR-205 are frequent in PDAC tissue samples when compared with match-paired normal parenchyma. Most tumors analyzed (84%) overexpress at least one of the candidate miRNAs, whereas a significant proportion (45%) express three or more. Importantly, the current work suggests that most of the miRNAs detected within PDAC tumors can be attributed to epithelial cancer cells, rather than the stromal compartment. In previous reports, miR-205 overexpression in PDAC has been observed in a limited set of cases,34 whereas miR-429 and miR-200a overexpression have been reported in metastatic PDAC cell line models.18, 25 Also, miR-200b overexpression has been described for cell lines and biopsies.35 In contrast, Dhayat et al.36 described the decreased expression of members of the miR-200 family in PDAC tumors. This result, however, must be interpreted with caution because the authors used a normal pancreas from benign, non-inflammatory pancreatic specimens as the control in their study, instead of paired adjacent normal parenchyma. Lastly, miRNAs that are organized in clusters, such as miR-11 and miR-998, usually share a co-dependent expression pattern.37 Interestingly, in our sample cohort, miR-200c and miR-141, and miR-200b, miR-200a, and miR-429, share the same expression pattern in only one-third of the tumors, regardless of RNA quality status, technical deviations, or genetic abnormalities. This later finding strongly suggests that miRNAs from the miR-200 family may differ in stability and/or post-transcriptional processing in PDAC tumors.
We demonstrate herein that downexpression of CDH1, a bona fide marker of EMT, is a frequent event in PDAC tumors as compared with matching normal parenchyma. In contrast, expression of ZEB1 and ZEB2 is low and unchanged in most paired samples analyzed during this study (58%), confirming previous findings from the literature.38 During this study, promoter methylation of CDH1, ZEB1, and ZEB2 genes was detected in only 40% of samples. This finding partly contradicts previous work reporting ZEB2 promoter methylation in 90% of PDAC samples.31 Such discrepancy can be easily explained by differences in the methods used, and advocate for alternative expression silencing modalities for these genes. Indeed, several studies demonstrated that CDH1 expression can be directly regulated by ZEB1 and ZEB2.5, 29 Furthermore, ZEB1 suppresses the transcription of miR-200 family members, in particular the expression of miR-141 and miR-200c.28 During this study, we confirmed these findings because we measured a negative correlation between the expression levels of ZEB1 and ZEB2 and the miR-200 family, and a positive regulation between miR-200 and the expression levels of CDH1 in primary PDAC tumors. Altogether, our results further suggest that miR-200 family members actively participate to maintain the epithelial state of a proportion of pancreatic tumors by suppressing ZEB1 and ZEB2, two well-described transcriptional repressors of CDH1.
Orthotopic PDXs provide a library of tumors that not only reflects the spectrum of primary tumors in the clinical setting, but also partially reproduces their dissemination propensity.39, 40 This preclinical model maintains key genetic aberrations present in primary tumors, although the expression levels of other key molecules may differ.41 During this study, we confirmed that PDXs perfectly mirror the genetic alterations found in primary tumors. On the other hand, we found that miR-200a and miR-200c levels were higher in orthotopic xenografts as compared with primary tumors, whereas ZEB2 presented significantly lower levels. This latter finding can be explained by the increase in tumor cellularity in PDX caused by the progressive loss of human stroma during passage in mouse, because we found that epithelial tumor cells mostly express the candidate miRNAs detected in primary tumors. Using this PDAC PDX cohort, we were able to compare gene expression patterns not only in primary tumors avatars but also during metastatic dissemination, a setting out of reach in clinical studies. We found that the levels of CDH1, ZEBs, and members of the miR-200 family remained unchanged in the 47 experimental metastases tested when compared with the corresponding orthotopically implanted PDXs. Although EMT is expected to play a role in tumor invasion and metastases formation,42 bulk analysis of metastatic material failed to highlight changes in expression of EMT regulators during this study. However, these results were obtained from late-stage metastasis; future works should address miRNAs and ZEB expression in early disseminated cancer cells. Considering the high heterogeneity of PDAC tissues,43 further studies based on single-cell analysis may help better understand the molecular processes underlying early and late distal dissemination events in these tumors.
We next addressed the functional consequences of overexpressing members from the miR-200 family into pancreatic cancer cell lines. We selected PANC-1 cells in which EMT gene regulators gene expression is not controlled by DNA methylation. We found that miRNA overexpression did not significantly impact cancer cell proliferation (data not shown). Molecular investigations revealed that miR-429 reverses EMT in PDAC-derived cell lines. Notably, our work is in agreement with recently published data showing that ZEB1 is a direct target of miR-429,10 and that ZEB targeting provokes a molecular cascade inducing mesenchymal-to-epithelial transition of PDAC cells. This results in smaller and denser spheroids with poor viability and in strong inhibition of cell migration. The functional importance of miR-429 was further validated in vivo, because this miRNA was the most effective to decrease tumor occurrence, tumor size, and metastatic spread. This latter effect is associated with an increase of CDH1 expression and a decrease of ZEB1 and ZEB2 expression, a pattern that was confirmed in vitro. Notably, miR-429 expression within the miR-200a-b cluster failed to inhibit tumor growth. To date, we do not have clear explanations for this discrepancy; in transduced cells, miR-429 expression levels are similar, whether expressed alone or as part of a cluster. Further studies are needed to determine whether miR-200a and miR-200b may counteract miR-429 antitumoral activity in PDAC experimental tumors. However, our results are in agreement with results from the literature describing miR-429, but also miR-141, as tumor suppressors in papillary thyroid cancer20 and PDAC27 and in gastric cancer,23 respectively. The interpretation of these observations is challenging because miR-429 and miR-141 are frequently overexpressed in primary tumors. Two potential explanations emerge: (1) a transient deregulation of miR-429 or miR-141 exclusively occurs during the early stages of tumorigenesis, and (2) the cellular context may significantly impact in the functional role of miRNAs including miR-429 and miR-141.11, 44 Further work is needed to determine whether delivering miR-429 in established tumors using either viral vectors45 or exosomes46 may impede experimental PDAC tumor growth. In conclusion, we demonstrated during this work that the miR-200 miRNA family is overexpressed in a comprehensive biological cohort of PDAC, and that members from this family, and especially miR-429, may have significant impact as nucleic acid-based therapies for this disease.
Materials and Methods
Tumor Biopsies
Tumor tissues and normal adjacent pancreatic parenchyma were collected from 48 patients undergoing surgical resection for PDAC between 2003 and 2011 at the Bellvitge Hospital, Barcelona, Spain. The study was approved by the Ethical Committee of University Hospital of Bellvitge CEIC 02/04, and written informed consent was obtained from all patients for the use of their tissues for research purposes.
Animals Studies
Six-week-old male nude Harlan mice weighing 18–22 g were used for tumor perpetuation. Animals were housed in cages with autoclaved bedding, food, and water in a sterile environment. Mice were maintained on a daily 12-h light, 12-h dark cycle. All experiments with mice were approved by the IDIBELL Animal Care and Use Committee (CEEA4591).
PDXs Cohort
Fresh surgical fragments of PDAC were minced into small fragments of 2 mm3 in size, and macroscopically viable tumor tissue fragments were implanted in the body tail of the pancreas (orthotopic implantation [OT]) of two animals per tumor under anesthesia with isoflurane. Meloxicam was administered subcutaneously just before and 24 and 48 h after surgery, to avoid the mice suffering. After implantation, tumor formation was monitored weekly by palpation. The first tumor passage was performed when an intra-abdominal mass measuring approximately 1 cm in diameter was palpated. The mice were anesthetized and euthanized by exsanguination with cardiac puncture and ulterior pneumothorax. Successive OT passages were performed in two animals until the fifth passage when the tumor was considered perpetuated.
Laser Capture Microdissection
Laser capture microdissection (LCM) of tumor cells and corresponding stroma was performed with the Applied Biosystems ArcturusXT LCM System, using Arcturus polyethylene naphthalate (PEN) Membrane Glass slides (Applied Biosystems, Life Technologies, UK) following staining with free RNase H&E. An average of 8,00,000–10,00,000 μm2 of tumor cells and stroma surface were separately captured and immediately frozen at −80°C prior to subsequent manipulation.
Molecular Characterization of Tumor Samples
Genetic aberrations in the KRAS, p16/CDKN2A, TP53, SMAD4, EGFR, BRAF, and PI3KCA genes were assessed using a variety of techniques including PCR and Sanger sequencing (TP53, PI3KCA, and CDKN2A), RT-PCR and Sanger sequencing (SMAD4 and EGFR), restriction fragment length polymorphism (RFLP)-PCR (KRAS),30 and single-nucleotide primer extension (SNuPE) (BRAF).31 Epidermal growth factor receptor (EGFR) copy number was determined by fluorescence in situ hybridization (FISH). The methylation status of the promoters of CDH1, ZEB1, and ZEB2 was studied by methylation-specific melting curve analysis (MS-MCA) as described previously.30 A different test to assess the methylation status had been described,17, 32, 33 but not with MS-MCA; for this reason, we designed specific primers.
Cell Lines
Pancreatic cancer-derived cell line PANC-1 was obtained from ATCC LGC Standards (Molsheim, France) and grown in DMEM containing +1 g/L glucose (Invitrogen, Carlsbad, CA, USA). The HEK293FT cell line used for production of the vectors was obtained from Invitrogen (Carlsbad, CA, USA) and maintained in DMEM containing 4.5 g/L glucose (Invitrogen, Carlsbad, CA, USA). All media were supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (complete medium) (all reagents were from Invitrogen). Cell lines were grown in a humidified incubator at 37°C in 5% CO2 and kept free from mycoplasma contamination using plasmocin (InvivoGen, Toulouse, France).
Lentiviral Vectors Production and Titration
The plasmids pLenti 4.1 Ex, pLenti 4.1 Ex miR-200c-141, and pLenti4.1 Ex miR-200b-a-429 co-expressing the indicated miRNA clusters and puromycin were a kind gift of P.A. Gregory and G.J. Goodall.12 Lentiviral backbones encoding for miR-141 or miR-429 precursors were purchased from System Biosciences (Palo Alto, CA). All vectors co-expressed GFP (plasmids pLenti 4.1 Ex EGFP and Lentiviral backbones copGFP). The lentiviral vectors were generated using the Lenti-SmartINT kit (InvivoGen, Toulouse, France) following the manufacturer’s instructions in a BSL-3 facility (Technology cluster; INSERM U1037, Toulouse, France). In brief, the lentiviral plasmid and the LENTI-Smart reagents were complexed and then added onto the 293FT cells following the manufacturer’s recommendations. Medium was changed 24 h later, and cell-free supernatant was collected 48 h after transfection. The supernatants were filtered using a 0.45-μm polyvinylidene fluoride (PVDF) filter, treated by DNase (Invitrogen), aliquoted, and stored at −80°C until use. Lentiviral vectors were titered by flow cytometry for GFP-positive cells (fluorescence-activated cell sorting [FACS]) and by ELISA for p24 (Innogenetics, Ghent, Belgium).
Cell Transduction with Lentiviral Vectors
One day before transduction, a total of 5 × 104 cells were cultured in 48-well plates in 500 μL of complete medium. Cells were transduced with 1,000 ng/mL p24 in the presence of protamine sulfate (4 μg/mL), in 250 μL of transduction medium (Quantum 263; PAA) overnight. A total of 5 μg/mL puromycin was added 48 h later (InvivoGen, Toulouse, France). Transduction efficiency was measured by FACS analysis.
RNA Isolation and Quantification
Total RNA from cell lines was isolated by the TRIzol method (TRIzol Reagent; Invitrogen, Carlsbad, CA, USA) or using the miRCURY RNA isolation kit (Exiqon, Vedbaek, Denmark) for primary tumors, according to the manufacturer’s instructions. RNA concentration was measured with ND-1000 NanoDrop spectrophotometer (Thermo Scientific). For laser microdissected samples, RNA was isolated with the Arcturus PicoPure RNA isolation Kit (Applied Biosystems, Life Technologies, UK). miRNAs and target genes expression were quantified using 1 μg or 500 ng of total RNA extracted from cell lines or primary tumors, respectively. The entire RNA eluate from laser capture-microdissected samples was used for reverse transcription. Simultaneous reverse transcription of mRNA and miRNA was performed using the miScript PCR System (QIAGEN, Cortaboeuf, France) following the manufacturer’s recommendations. qPCR was carried out using SYBRGreen PCR Master Mix (QIAGEN, Cortaboeuf, France). cDNAs were analyzed in triplicate using a StepOne-PlusReal-Time PCR System (Life Technologies, Carlsbad, CA, USA). Expression levels were calculated using the ΔCt (cycle threshold) formula. RNU48 and human actin were used as standard for miRNAs and target genes, respectively. The expression levels relative to control (paired normal tissue, control-transduced cell line), expressed as fold change (FC), were calculated using the formula 2−ΔΔCt.34 Infra-expression was defined as a FC value <0.5, whereas overexpression was achieved when FC > 2 (see Table S1 for qPCR primer sequences).
Western Blotting
Proteins were extracted from PANC-1 transduced cells, resolved on 6%–12% gradient SDS-polyacrylamide gels (50 μg/lane), and transferred to nitrocellulose membrane. After room-temperature blocking for 1 h, blots were incubated overnight at 4°C with antibodies against a panel of genes involved in EMT, following the manufacturer’s instructions (panel #9782; Cell Signaling Technology). β-Actin (clone C4; Santa Cruz) was used at a dilution of 1:2,000. Secondary HRP-conjugated antibodies (dilution 1:20,000; Bio-Rad) were added, and blots were incubated for 1 h at room temperature. Following several washes, immunoreactive proteins were visualized using Clarity ECL (Bio-Rad) and imaged with ChemiDoc XRS+ (Bio-Rad).
Real-Time Migration Analysis
A total of 35,000 PANC-1 cells overexpressing miR-429 were seeded in 100 μL of complete medium in a 96-well ImageLock plate (Essen BioScience, Sartorius). PANC-1-expressing GFPs were used as control. Twenty-hours later, homogeneous scratch wounds were performed using the WoundMaker mechanical device from Essen BioScience (Sartorius). Pictures of the scratch wound were taken every hour for 36 h using the IncuCyte Zoom (Essen BioScience, Sartorius) to assess cell morphology at every time point. A mask was applied to each picture to determine the relative wound density, as a measure (%) of the density of the wound region relative to the density of the cell region, according to the manufacturer’s instruction.
Real-Time Cell Proliferation and Spheroid Formation Analysis
A total of 10,000 or 25,000 PANC-1 cells overexpressing miR-429 were seeded in 100 μL of complete medium in 96-well plates or in Nunclon Sphera 96 U-well plates (Thermo Fisher Scientific) for cell proliferation and spheroid formation analysis, respectively. PANC-1-expressing GFPs were used as control. Pictures were taken at the time indicated using the IncuCyte Zoom (Essen BioScience, Sartorius). A mask was applied to each picture to determine the surface of live cells (GFP fluorescence) and cell debris.
In Vivo Tumorigenicity Assay
For orthotopic injection, a total of 5 × 106 transduced or control cells were injected in the body tail of the pancreas under anesthesia with isoflurane. Thirteen mice were used for each group. After injection, tumor formation was checked weekly by palpation. Mice were sacrificed after 3 months of implantation or if endpoint criteria were met, and tumor volume was calculated as previously described.45
Statistical Analysis
To compare the expression level of members of the miR-200 family and their target genes in paired primary tumor-adjacent nontumoral tissue, we used the Wilcoxon test (p < 0.05 was considered significant). The correlation between miRNAs and target genes expression was carried out by Spearman analysis (significant when p < 0.05). To compare the distributions (2−ΔCt) between primary tumor, adjacent normal tissue, orthotopic tumor, and metastasis for each miRNA and target genes, we used the Kruskal-Wallis test, considering differences when p < 0.05. Then, the Tukey multiple comparisons of means were applied for pairwise comparisons if significant differences were previously evidenced. For differences in the ability to generate tumors and metastases, contingency tables and chi-square test were utilized. Differences in tumor volume in the orthotopic in vivo experiment were analyzed using the Kruskal-Wallis and the Benjamini and Hochberg tests.
Author Contributions
Z.V.D-R., M.M.G., J.F., T.S., J.B., and P.C. did the experiments. All authors analyzed the results. Z.V.D-R., M.M.G., G.C., and P.C. designed the experiments and wrote the paper.
Acknowledgments
This work was supported by the Spanish Ministry of Economy and Competitiveness, co-funded by FEDER funds, A Way to Build Europe (grant SAF2015-68016-R to G.C.); the Carlos III National Health Institute (CIBERONC); the Government of Catalonia (Pla estratègic de recerca i innovació en salut 2014SGR338 2017SGR1282); and the Scientific Foundation of the Asociación Española Contra el Cáncer.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.015.
Contributor Information
Pierre Cordelier, Email: pierre.cordelier@inserm.fr.
Gabriel Capellá, Email: gcapella@iconcologia.net.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H., Neoptolemos J.P. Pancreatic cancer. Nat. Rev. Dis. Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos J.P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer D.H. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018;15:333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 4.Krantz S.B., Shields M.A., Dangi-Garimella S., Munshi H.G., Bentrem D.J. Contribution of epithelial-to-mesenchymal transition and cancer stem cells to pancreatic cancer progression. J. Surg. Res. 2012;173:105–112. doi: 10.1016/j.jss.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurahara H., Takao S., Maemura K., Mataki Y., Kuwahata T., Maeda K., Ding Q., Sakoda M., Iino S., Ishigami S. Epithelial-mesenchymal transition and mesenchymal-epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J. Surg. Oncol. 2012;105:655–661. doi: 10.1002/jso.23020. [DOI] [PubMed] [Google Scholar]
- 6.Arumugam T., Ramachandran V., Fournier K.F., Wang H., Marquis L., Abbruzzese J.L., Gallick G.E., Logsdon C.D., McConkey D.J., Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotz B., Arndt M., Dullat S., Bhargava S., Buhr H.-J., Hotz H.G. Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin. Cancer Res. 2007;13:4769–4776. doi: 10.1158/1078-0432.CCR-06-2926. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Mongroo P.S., Rustgi A.K. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010;10:219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G., Zheng H., Xu J., Guo Y., Zheng G., Ma C., Hao S., Liu X., Chen H., Wei S. miR-429 suppresses cell growth and induces apoptosis of human thyroid cancer cell by targeting ZEB1. Artif. Cells Nanomed. Biotechnol. 2019;47:548–554. doi: 10.1080/21691401.2018.1564320. [DOI] [PubMed] [Google Scholar]
- 11.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 12.Korpal M., Lee E.S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S.-M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Tilló E., Liu Y., de Barrios O., Siles L., Fanlo L., Cuatrecasas M., Darling D.S., Dean D.C., Castells A., Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell. Mol. Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons D.L., Lin W., Creighton C.J., Rizvi Z.H., Gregory P.A., Goodall G.J., Thilaganathan N., Du L., Zhang Y., Pertsemlidis A., Kurie J.M. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davalos V., Moutinho C., Villanueva A., Boque R., Silva P., Carneiro F., Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang Y., Xu S., Ma J., Wu J., Jin S., Cao S., Yu Y. MicroRNA-429 induces tumorigenesis of human non-small cell lung cancer cells and targets multiple tumor suppressor genes. Biochem. Biophys. Res. Commun. 2014;450:154–159. doi: 10.1016/j.bbrc.2014.05.084. [DOI] [PubMed] [Google Scholar]
- 18.Dykxhoorn D.M., Wu Y., Xie H., Yu F., Lal A., Petrocca F., Martinvalet D., Song E., Lim B., Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pecot C.V., Rupaimoole R., Yang D., Akbani R., Ivan C., Lu C., Wu S., Han H.D., Shah M.Y., Rodriguez-Aguayo C. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang M., Huang W., Wu X., Gao Y., Ou J., Zhang X., Li Y. MiR-141-3p Suppresses Tumor Growth and Metastasis in Papillary Thyroid Cancer via Targeting Yin Yang 1. Anat. Rec. (Hoboken) 2019;302:258–268. doi: 10.1002/ar.23940. [DOI] [PubMed] [Google Scholar]
- 21.Gao H., Liu C. miR-429 represses cell proliferation and induces apoptosis in HBV-related HCC. Biomed. Pharmacother. 2014;68:943–949. doi: 10.1016/j.biopha.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Wu S.-M., Ai H.-W., Zhang D.-Y., Han X.-Q., Pan Q., Luo F.-L., Zhang X.L. MiR-141 targets ZEB2 to suppress HCC progression. Tumour Biol. 2014;35:9993–9997. doi: 10.1007/s13277-014-2299-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M., Dong B.-B., Lu M., Zheng M.-J., Chen H., Ding J.-Z., Xu A.M., Xu Y.H. miR-429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. OncoTargets Ther. 2016;9:1123–1133. doi: 10.2147/OTT.S91879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.du Rieu M.C., Torrisani J., Selves J., Al Saati T., Souque A., Dufresne M., Tsongalis G.J., Suriawinata A.A., Carrère N., Buscail L., Cordelier P. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin. Chem. 2010;56:603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 25.Mees S.T., Mardin W.A., Wendel C., Baeumer N., Willscher E., Senninger N., Schleicher C., Colombo-Benkmann M., Haier J. EP300—a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int. J. Cancer. 2010;126:114–124. doi: 10.1002/ijc.24695. [DOI] [PubMed] [Google Scholar]
- 26.Kent O.A., Mullendore M., Wentzel E.A., López-Romero P., Tan A.C., Alvarez H., West K., Ochs M.F., Hidalgo M., Arking D.E. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol. Ther. 2009;8:2013–2024. doi: 10.4161/cbt.8.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y., Lu F., Xiong P., Pan M., Zhang Z., Lin X., Pan M., Huang H. WIPF1 antagonizes the tumor suppressive effect of miR-141/200c and is associated with poor survival in patients with PDAC. J. Exp. Clin. Cancer Res. 2018;37:167. doi: 10.1186/s13046-018-0848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebs A.M., Mitschke J., Lasierra Losada M., Schmalhofer O., Boerries M., Busch H., Boettcher M., Mougiakakos D., Reichardt W., Bronsert P. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 30.Mattie M., Christensen A., Chang M.S., Yeh W., Said S., Shostak Y., Capo L., Verlinsky A., An Z., Joseph I. Molecular characterization of patient-derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia. 2013;15:1138–1150. doi: 10.1593/neo.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li A., Omura N., Hong S.-M., Vincent A., Walter K., Griffith M., Borges M., Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70:5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravet E., Lulka H., Gross F., Casteilla L., Buscail L., Cordelier P. Using lentiviral vectors for efficient pancreatic cancer gene therapy. Cancer Gene Ther. 2010;17:315–324. doi: 10.1038/cgt.2009.79. [DOI] [PubMed] [Google Scholar]
- 33.Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., Goodall G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 34.Szafranska A.E., Davison T.S., John J., Cannon T., Sipos B., Maghnouj A., Labourier E., Hahn S.A. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Li M., Wang H., Fisher W.E., Lin P.H., Yao Q., Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J. Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhayat S.A., Traeger M.M., Rehkaemper J., Stroese A.J., Steinestel K., Wardelmann E., Kabar I., Senninger N. Clinical Impact of Epithelial-to-Mesenchymal Transition Regulating MicroRNAs in Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2018;10:e328. doi: 10.3390/cancers10090328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truscott M., Islam A.B.M.M.K., Frolov M.V. Novel regulation and functional interaction of polycistronic miRNAs. RNA. 2016;22:129–138. doi: 10.1261/rna.053264.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maier H.J., Wirth T., Beug H. Epithelial-mesenchymal transition in pancreatic carcinoma. Cancers (Basel) 2010;2:2058–2083. doi: 10.3390/cancers2042058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loukopoulos P., Kanetaka K., Takamura M., Shibata T., Sakamoto M., Hirohashi S. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas. 2004;29:193–203. doi: 10.1097/00006676-200410000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Reyes G., Villanueva A., García C., Sancho F.J., Piulats J., Lluís F., Capellá G. Orthotopic xenografts of human pancreatic carcinomas acquire genetic aberrations during dissemination in nude mice. Cancer Res. 1996;56:5713–5719. [PubMed] [Google Scholar]
- 41.Tentler J.J., Tan A.C., Weekes C.D., Jimeno A., Leong S., Pitts T.M., Arcaroli J.J., Messersmith W.A., Eckhardt S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brabletz S., Brabletz T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N., Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Gheldof A., Hulpiau P., van Roy F., De Craene B., Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell. Mol. Life Sci. 2012;69:2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sicard F., Gayral M., Lulka H., Buscail L., Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol. Ther. 2013;21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.