Abstract
Chromium (Cr) is a naturally occurring metallic element found in the Earth’s crust. While trivalent chromium ([Cr(III)] is considered non-carcinogenic, hexavalent chromium [Cr(VI)] has long been established as an IARC class I human carcinogen, known to induce cancers of the lung. Current literature suggests that Cr(VI) is capable of inducing carcinogenesis through both genetic and epigenetic mechanisms. Although much has been learned about the molecular etiology of Cr(VI)-induced lung carcinogenesis, more remains to be explored. In particular, the explicit epigenetic alterations induced by Cr(VI) in lung cancer including histone modifications and miRNAs, remain understudied. Through comprehensive review of available literature found between 1973–2019, this article provides a summary of updated understanding of the molecular mechanisms of Cr(VI)-carcinogenesis. In addition, this review identifies potential research gaps in the areas of histone modifications and miRNAs, which may prompt new niches for future research.
Keywords: Cr(VI), Carcinogenesis, Mechanism, Epigenetic
Introduction
Chromium (Cr) is a naturally occurring element found in soil, rocks and living organisms, and primarily exists in two stable valence states: trivalent chromium (Cr(III)) or hexavalent chromium (Cr(VI)) (Wilbur et al., 2012). Cr(III) is considered non-carcinogenic due to insufficient evidence in humans and animals. However, according to the International Agency for Research on Cancer (IARC) and U.S. Environmental Protection Agency (EPA), Cr(VI) compounds are classified as Group 1 and Group A human carcinogens, respectively(Stoss et al., 1983). Due to the potential adverse health effects resulting from exposure to Cr, the EPA established a maximum contaminant level of 0.1 mg/L total Cr in drinking water. However, Cr(VI) is often found in occupational settings as a result of industrial activities such as leather working, smelting, welding, and metal plating (Wilbur et al., 2012). Cr(VI) is also found in automobile exhaust and in tobacco products such as traditional and electronic cigarettes and hookahs (Williams et al., 2017). It has been estimated that 66% of current or former hazardous waste sites on the National Priorities List also contain Cr (Wilbur et al., 2012).
Cr(VI) is considered the biologically relevant form of Cr due to its ability to readily pass through the cell membrane via non-specific sulfate/phosphate anionic transporters. Cr(III) is capable of passing through cell membranes via diffusion or phagocytosis, albeit at much lower levels than Cr(VI) (Stout et al., 2009). Systemic toxicity attributable to Cr has been documented in the respiratory and pulmonary system, gastrointestinal system, dermis, and renal system (Wilbur et al., 2012). In addition, multiple mechanisms of carcinogenesis have been proposed involving oxidative stress, DNA damage and genomic instability, and epigenetic modulation. A comprehensive literature search was conducted through Pubmed using key words such as “Chromium”, “Cr(VI)”, “Carcinogenesis”, “Cancer”, “Epigenetic” and “Mechanisms”. In addition, the reference list of each article is also examined to prevent missing data. This review summarizes most current findings in the field of Cr(VI)-carcinogenesis, with a focus on epigenetic mechanisms. Furthermore, a comprehensive lists of genes, histone modifications, and miRNAs altered through Cr(VI) exposure is presented.
Chromate Exposure and Associated Health Risks
The primary non-occupational route of exposure to Cr is ingestion. However, for occupationally exposed individuals, exposure to Cr most often occurs via inhalation or dermal absorption (Wilbur et al., 2012). Adverse respiratory and pulmonary health effects due to Cr exposure include asthma (Bright et al., 1997), bronchitis and respiratory tract irritation (Khan et al., 2013), and nasal septum ulceration and perforation (Lindberg & Hedenstierna, 1983). Contact dermatitis is frequently documented following exposure to chromate and dichromate (Leijding et al., 2018), and more severe dermal reactions including skin burns, blisters, and skin ulcers have been reported (Wilbur et al., 2012). Gastrointestinal (GI) effects including chronic dyspepsia, gastric ulcers, and gastritis have all been documented following occupational exposure to Cr (Khan et al., 2013). Exposure to Cr(VI) can also cause acute tubular necrosis, which is localized to the proximal convoluted tubules and may result in rapid onset of renal failure (Wedeen et al., 1991).
As a prominent human carcinogen, Cr(VI) has long been reported to promote cancers of the lung and nasal and sinus cavities among occupationally-exposed workers. A study based on the Baltimore cohort which consisted of 2,357 participants, demonstrated a highly positive correlation between cumulative Cr(VI) exposure and lung cancer mortality rate (Gibb et al., 2000; Gibb et al., 2015). Another retrospective study based on 493 workers from a chromate production plant in Painesville further confirmed the correlation between lung cancer mortality and occupational Cr(VI) exposure (Luippold et al., 2003; Proctor et al., 2003). In addition, a meta-analysis of Cr(VI) exposure and GI cancers showed an increased risk of stomach cancer (relative risk = 1.27; 1.18 – 1.38) in Cr(VI)-exposed workers and an elevated stomach cancer mortality rate (rate ratio = 1.82; 1.11 – 2.91) in Cr contaminated regions (Beaumont et al., 2008; Welling et al., 2015; Zhang & Li, 1987). Despite the lack of conclusive evidence supporting a causal role of Cr(VI) in GI cancers in humans, associations between Cr(VI) ingestion and GI cancers have been documented in rodent models. In a two-year drinking water study conducted by the National Toxicology Program (NTP), oral exposure to sodium dichromate dihydrate in male and female rats increased the incidence of squamous cell neoplasms of the oral cavity. In mice there was an increase in incidence of neoplasms of the small intestine (duodenum, jejunum, or ileum) (NTP 2008). Together, these results suggest that Cr(VI) may act in a carcinogenic manner following ingestion, thereby raising concerns for non-occupationally exposed individuals. Molecular mechanisms involving oxidative stress and DNA damage are considered principal ways by which Cr(VI) exhibits its carcinogenic effects.
Molecular Mechanisms of Chromium Carcinogenesis
Oxidative Stress
Toxicity and carcinogenicity of Cr(VI) compounds is by virtue of their ability to readily enter the cells through isoelectric and isostructural anion transfer channels, which are used to transport HPO−4 and SO2−4 ions (Codd et al., 2001; Valko et al., 2005). Although Cr(VI) compounds do not bind directly to DNA, intermediates and byproducts of Cr(VI) metabolism can elicit a wide range of damages through DNA adducts and crosslinks (Kasprzak 1995). Notably, generation of reactive oxygen species (ROS) through detoxification is principally responsible for Cr(VI)-induced cellular damages such as DNA lesions, cytotoxicity, and tumor development (Chen et al., 2019; Shi et al., 1999; Wise et al., 2019; Zhitkovich 2005; Zhitkovich 2011). Cr species [(III), (IV), (V), and (VI)] are known to produce intracellular ROS. Specifically, ROS scavengers such as ascorbic acid and glutathione are able to detect and reduce Cr(VI) to Cr(III), thereby producing free radicals such as hydroxyl radicals and DNA-damaging intermediates such as Cr(V) and Cr(IV) (Arita & Costa, 2009; Chen et al., 2019; Jomova & Valko, 2011; Zhitkovich 2011).
During intracellular reduction of Cr(VI), hydroxyl radicals are generated through Fenton-like reactions in the presence of hydrogen peroxide (Sun et al., 2015). Endogenous superoxide anions and hydrogen peroxide produce hydroxyl radicals via Haber-Weiss-like reactions in the presence of Cr(VI) (Sun et al., 2015). Cr(VI) can also generate hydroxyl radicals through stimulated cells, which demonstrate upregulated activities of NADPH oxidase (Gao et al., 2002; Wang et al., 2000; Wang et al., 2004; Yao et al., 2008; Ye et al., 1995). Other reducing agents of Cr(VI) include flavoenzymes such as glutathione reductase and ferredoxin NADP+ oxidoreductase (Shi et al., 1999; Wise et al., 2019). In addition, Cr(VI) can disrupt the thioredoxin antioxidant system by irreversibly inhibiting thioredoxin reductase, which under normal conditions maintains thioredoxin in a reduced state (Myers & Myers, 2009). The thioredoxin antioxidant system promotes cell survival by defending against oxidative stress (Myers & Myers, 2009).
ROS including hydroxyl radicals, singlet oxygen, peroxides, and superoxides can serve as important secondary messengers and activators for various pathways including apoptosis, cell signaling, and homeostasis (Deyasagayam et al., 2004; Kwee 2014; Leonard et al., 2004; Valko et al., 2007; Wise et al., 2019). Specifically, Cr(VI) has been found to induce NF-kB, AP-1, and Nrf2 activation, each of which has important implications in cancer development (He et al., 2007; Klaunig et al., 2011; Valko et al., 2005; Ye et al., 1995). Hydroxyl radicals are able to react with guanine residues and generate radical adducts such as 8-hydroxy-deoxyguanosine (8-OH-dG), which is a prominent marker for oxidative damage in cancer (Valko et al., 2004). The accumulation of ROS can lead to oxidative stress and contribute to chronic inflammation, metabolic reprogramming, genetic instability, and development (Wang et al., 2016). Sprague-Dawley rats exposed to Cr(VI) for 5 days via intraperitoneal injection demonstrated enhanced antioxidant enzymes to combat oxidative stress in liver and kidneys as well as DNA damage in peripheral blood lymphocytes (Patlolla et al., 2009). In addition, adducts formed through Cr and ROS scavenger conjugation, including GSH-Cr-DNA, can generate bulky adducts and block proper DNA replication and repair (Chen et al., 2019; Quievryn et al., 2003; Zhitkovich 2005).
Cr(VI)-induced DNA Damage
It is widely accepted that Cr(VI) is capable of causing DNA damage following intracellular reduction in the form of apurinic/apyrimidinic sites (Casadevall & Kortenkamp, 1994) and by interacting with proteins (Wedrychowski et al., 1985), amino acids (Zhitkovich et al., 1995), or directly with DNA (DeLoughery et al., 2015), causing DNA single- (Christie et al., 1984) and double-strand breaks (DSBs) (DeLoughery et al., 2015). Upon intracellular reduction, Cr(VI) can form bulky Cr(III) binary adducts (i.e. Cr(III) – DNA) as well as ternary adducts (i.e. Cr(III)–ligand–DNA). Cr(VI) reduction in the presence of physiological concentrations of ascorbic acid demonstrated that ternary ascorbate-Cr(III)-DNA crosslinks were more mutagenic than their binary Cr(III)-DNA counterparts, with ternary complexes accounting for more than 90% of mutagenic damage (Quievryn et al., 2003). Cr(VI) is also capable of inducing 8-oxo-dG formation in proportion to Cr(VI) concentration in vitro, and 8-oxo-dG is often employed as a biomarker of exposure in chromate exposed individuals (Arakawa et al., 2012; Li et al., 2014). Using UvrABC and Fpg incision methods, Cr(VI) was found to induce bulky adducts and oxidative DNA damage at both dG’s as well as dA’s in the p53 gene (Arakawa et al., 2012).
More recently, DSBs caused by Cr-adducted DNA has been investigated in detail. Selective formation of Cr-induced DSBs occurred in euchromatin, despite the presence of Cr-DNA adducts in both euchromatin and heterochromatin (DeLoughery et al., 2015). It was further revealed that DSB repair signaling involving γH2AX, mono-, and di-ubiquitinated H2AX were dependent on ATR as opposed to the classical ATM mode of activation (DeLoughery et al., 2015). ATR is strongly activated by single-stranded tails as opposed to blunt-ended DSBs (Lukas et al., 2011). The authors further point out that mismatch repair operates by excision of one strand, producing a series of single-stranded DNA. This partly reveals that the nature of DNA damage following the formation of Cr-DNA adducts. Extended exposure (>48 h) to particulate Cr(VI), however, was shown to reduce Rad51 foci formation and protein expression, suggesting that the homologous recombination (HR) signaling pathway is repressed after longer exposures to Cr(VI) in particulate form (Qin et al., 2014). After 48 h, Rad51 was observed to aggregate in the cytoplasm, providing a possible explanation. It was later confirmed that prolonged exposure to particulate Cr(VI) does, in fact, inhibit HR repair (Browning et al., 2016). Rad51C is responsible for Rad51 nuclear import and stabilization of the Rad51 nucleofilament, and Rad51C foci formation was subsequently shown to be inhibited following prolonged exposure to particulate Cr(VI) (Browning et al., 2016).
Using formaldehyde-assisted isolation of regulatory elements and deep sequencing, Ovesen et al. reported, in vitro, that acute Cr(VI) versus chronic Cr(VI) treatment opened 3 times as many unique chromatin domains, and only eleven of these unique domains were shared between treatments (Ovesen et al, 2014). Chromatin domains surrounding both AP-1 and CTCF were found to become significantly more open after acute treatment whereas chromatin domains surrounding AP-1 and BAH2 were more open following chronic treatment. Furthermore, structural chromatin changes were not correlated with changes in global transcriptional response. Structural chromatin changes did, however, affect gene expression levels in target areas that vary with Cr(VI) concentration. Overall, the changes in chromatin structure in response to acute and chronic Cr(VI) suggests that the mechanisms governing Cr(VI)-induced transcriptional response are uniquely different depending on the dose, which may impact molecular events leading to carcinogenesis. Epigenetic Mechanisms of Chromium Carcinogenesis
Epigenetics refers to the reversible yet heritable changes in gene expression, independent of DNA sequence, caused by DNA hypo- or hypermethylation, histone tail post-translational modifications, and microRNAs (miRNAs). The 5 carbon of cytosine in DNA can be covalently methylated by DNA methyltransferases, or may be actively or passively demethylated. Histone post-translational modifications (PTMs) include, but are not limited to: acetylation, methylation, phosphorylation and citrullination, and can impact chromatin structure, thereby acting as gate-keepers for chromatin access (reviewed in Tessarz & Kouzarides, 2014). Histone PTMs also recruit or hinder epigenetic machinery or transcriptional regulators that direct gene expression. miRNAs, on the other hand, regulate gene expression by binding to complementary regions on target messenger RNAs (mRNAs) to dampen and fine-tune expression in the form of RNA degradation, and also reduce expression through induced decapping, induced deadenylation, altered cap protein binding, reduced ribosome occupancy, and sequestration of mRNA from translational machinery (reviewed in Mohr & Mott, 2015). Extensive studies have shown that Cr(VI) is capable altering gene expression and inducing cancer development through multiple epigenetic mechanisms. The following sections will examine these mechanisms in depth, and a comprehensive summary of genes altered by Cr(VI) exposure is listed in Table 1.
Table 1.
Summary of genes altered by Cr(VI)
| Gene Name | Direction of change | Chromium compound | Dose | Length of exposure | Tissue type | Reference |
|---|---|---|---|---|---|---|
| Angjogenin | Up | Na2Cr2O7 | 0.25 uM | 6 mo | BEAS-2B | Kim et al., 2016 |
| AP1 | Up | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| ATM | Down | K2Cr2O7 | L uM | 48 hr | BEAS-2B | Abreu et al., 2018 |
| ATR | Up | K2Cr2O7 | 1 uM | 48 hr | BEAS-2B | Abreu et al., 2018 |
| Bcl-xL | Up | Na2Cr2O7 | 100 nM | 3 mo | BEAS-2B | Dai et al., 2017 |
| Bcl2 | Up | Na2Cr2O7 | 100 nM | 3 mo | BEAS-2B | Dai et al., 2017 |
| Bcl2 | Up | Cr(VI) | 0.3 uM | 30 wk | BEAS-2B, keratinocytes | Ganapathy et al., 2017 |
| Bcl2 | Up | K2Cr2O7 | 0.5 uM | 4 wk | BEAS-2B | Huang et al., 2017 |
| Bcl2 | Up | Na2Cr2O7·2H2O | 5 uM | 24 wk | BEAS-2B | Medan et al., 2012 |
| Bcl2 | Up | Na2Cr2O7·2H2O | 10 uM | 24 hr | BEAS-2B | Son et al., 2017 |
| BTD | Down | K2CrO4 | 6.25–12.5 uM | 24 hr | 16HBE | Xia et al., 2011 |
| Capspase-9 | Up | K2Cr2O7 | 0.2 uM | 0–6 hr | A549 | Ge et al., 2019 |
| Caspase 3 | Up | K2Cr2O7 | 0.2 uM | 0–12 hr | A549 | Ge et al, 2019 |
| Caspase-8 | Up | K2Cr2O7 | 0.2 uM | 0–6 hr | A549 | Ge et al., 2019 |
| CDK4 | Down | K2Cr2O7; PbCrO4 |
5–15 uM; 1.25–5 uM |
2–24 hr | A549, human B lymphoblastoid | Lou et al., 2013 |
| CDK6 | Down | K2Cr2O7; PbCrO4 |
5–15 uM; 1.25–5 uM |
2–24 hr | A549, human B lymphoblastoid | Lou et al., 2013 |
| Cox-2 | Up | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| Cox-2 | Up | Na2CrO4 | 20 uM | 6, 12 hr | MEFs | Zuo et al., 2012 |
| Cox2 | Up | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Roy et al., 2016 |
| E-cadherin | Down | K2Cr2O7 | 1.5–15 uM | 10 hr-1 wk | BEAS-2B | Ding et al., 2013 |
| ERCC3 | Down | K2Cr2O7 | 0.6 uM | 24 hr | 16HBE | Hu et al., 2018 |
| ERK | Up | Na2Cr2O7 | 50–100 uM | 1–2 hr | A549 | Wang & Shi 2001 |
| EZH2 | Up | K2Cr2O7 | .25 uM | 20,40 wk | BEAS-2B, 16HBE | Wang et al., 2018 |
| FBP1 | Down | Na2Cr2O7 | 100 nM | 3 mo | BEAS-2B | Dai et al., 2017 |
| G9a | Up | K2CrO4 | 5–10 uM | 24 hr | A549 | Sun et al., 2009 |
| G9a | Up | K2Cr2O7 | .25 uM | 20,40 wk | BEAS-2B, 16HBE | Wang et al., 2018 |
| Gene 33 | Down | Na2CrO4 | 1–5 uM | 48–72 hr | BEAS-2B | Park et al., 2016 |
| Gli2 | Up | K2Cr2O7 | 0.5 uM | 4 wk | BEAS-2B | Huang et al., 2017 |
| GLP | Up | K2Cr2O7 | .25 uM | 20,40 wk | BEAS-2B, 16HBE | Wang et al., 2018 |
| GRO-a | Up | ZnCrO44Zn(OH)2 | 50 uL | 0–24 hr | BAL fluid | Beaver et al., 2008 |
| GRP78 | Up | K2Cr2O7 | 0.2 uM | 12 hr | A549 | Ge et al., 2019 |
| HCS | Up | K2CrO4 | 6.25–12.5 uM | 24 hr | 16HBE | Xia et al., 2011 |
| HDAC2 | Up | K2CrO4 | 2.5–5 uM | 24 hr | 16HBE | Xia et al., 2014 |
| HDAC3 | Up | K2CrO4 | 2.5–5 uM | 24 hr | 16HBE | Xia et al., 2014 |
| HIF-la | Up | K2Cr2O7 | 2.5–10 uM | 0–24 hr | DU145 | Gao et al., 2002 |
| HIF-la | Up | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| HIF-a | Up | Na2Cr2O7 | 0.25 uM | 6 mo | BEAS-2B | Kim et al., 2016 |
| HO-1 | Up | K2Cr2O7 | 5 uM | 24hr | Mycoplasma-free human dermal fbroblast | Joseph et al., 2008 |
| HO-1 | Up | Na2Cr2O7·2H2O | 1 uM acute, .125–5 uM chronic | 24 hr | BEAS-2B | Son et al., 2017 |
| HOGG1 | Down | K2Cr2O7 | 0.6 uM, 1.2 uM, 2.5 uM, 5.0 uM, 10.0 uM and 20.0 uM | 24 hr | 16HBE | Hu et al., 2018 |
| Hsp90a | Down | K2Cr2O7 | 1 uM | 48 hr | BEAS-2B | Abreu et al., 2018 |
| HSPA1A | Down | K2Cr2O7 | 1 uM | 48 hr | BEAS-2B | Abreu et al., 2018 |
| IL-6 | Up | ZnCrO44Zn(OH)2 | 50 uL | 0–24 hr | BAL fluid | Beaver et al., 2008 |
| IL-6 | Up | Na2Cr2O7 | 0.25 uM | 6 mo | BEAS-2B | Kim et al., 2016 |
| IL-6 | Up | K2Cr2O7 | 0–2 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| IL-8 | Up | K2Cr2O7 | 0–2 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| iNOS | Up | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| JNK | Up | K2Cr2O7 | 10–80 uM | l-12 hr | CL3 | Chuang et al., 2000 |
| MGMT | Down | K2Cr2O7 | 20 uM | 24 hr | 16HBE | Hu et al., 2018 |
| MMP-1 | Up | Na2Cr2O7 | 0.25 uM | 6 mo | BEAS-2B | Kim et al., 2016 |
| NQO1 | Up | Na2Cr2O7 | 100 nM | 3 mo | BEAS-2B | Dai et al., 2017 |
| Nrf2 | Up | K2Cr2O7 | 1–5 uM | 24 hr | BEAS-2B | Roy et al., 2016 |
| Nr£2 | Up | Na2Cr2O7·2H2O | 1 uM acute, .125–5 uM chronic | 24 hr | BEAS-2B | Son et al., 2017 |
| NUPR1 | Up | K2CrO4 | 5–10 uM | 24 hr | BEAS-2B | Chen et al., 2016 |
| OGG1 | Down | Na2Cr2O7 | 0–100 uM | 16 hr | A549 | Hodges & Chipman, 2002 |
| p-16 | Up | K2Cr2O7; PbCrO4 |
5–15 uM; 1.25–5 uM |
2–24 hr | A549, human B lymphoblastoid | Lou et al., 2013 |
| p-PERK | Up | K2Cr2O7 | 0.2 uM | 12 hr | A549 | Ge et al., 2019 |
| p38 | Up | K2Cr2O7 | 10–80 uM | l-12 hr | CL3 | Chuang et al., 2000 |
| p38 | Up | Na2Cr2O7 | 50–100 uM | 3 hr | A549 | Wang & Shi, 2001 |
| p53 | Up | Na2Cr2O7 | 20–100 uM | l-2 hr | A549 | Wang & Shi, 2001 |
| PDCD4 | Down | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2016 |
| RAD51 | Down | K2Cr2O7 | 0.6 uM, 1.2 uM, 2.5 uM, 5.0 uM, 10.0 uM and 20.0 uM | 24 hr | 16HBE | Hu et al., 2018 |
| Snail | Down | K2Cr2O7 | 1.5–15 uM | 10 hr-1 wk | BEAS-2B | Ding et al., 2013 |
| SOD1 | Up | Na2Cr2O7·2H2O | 1 uM acute, .125–5 uM chronic | 24 hr | BEAS-2B | Son et al., 2017 |
| SOD2 | Up | Na2Cr2O7 | 100 nM | 3 mo | BEAS-2B | Dai et al., 2017 |
| SUV39H1 | Up | K2Cr2O7 | .25 uM | 20,40 wk | BEAS-2B, 16HBE | Wang et al., 2018 |
| TNF-a | Up | K2Cr2O7 | 5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2014 |
| TNF-a | Up | K2Cr2O7 | 1–5 uM | 24 hr | BEAS-2B | Roy et al., 2016 |
| Twist | Down | K2Cr2O7 | 1.5–15 uM | 10 hr-1 wk | BEAS-2B | Ding et al., 2013 |
| VEGF | Up | K2Cr2O7 | 2.5 uM | 0–24 hr | DU145 | Gao et al., 2002 |
| VEGF | Up | Na2Cr2O7 | 0.25 uM | 6 mo | BEAS-2B | Kim et al., 2016 |
| Vimentin | Up | K2Cr2O7 | 1.5–15 uM | 10 hr-1 wk | BEAS-2B | Ding et al., 2013 |
| XRCC1 | Down | K2Cr2O7 | 2.5 uM, 10.0 uM and 20.0 uM | 24 hr | 16HBE | Hu et al., 2018 |
| yH2AX | Up | Na2CrO4 | 5 uM | 6 & 24 hr | BEAS-2B | Park et al., 2016 |
DNA Methylation
It was first reported that Cr(VI) could induce DNA methylation and silencing of the gpt transgene in G12 Chinese hamster lung cells (Klein et al., 2002). Since then, a number of studies have shown a wide range of epigenetic effects. Lou et al. found acute soluble Cr(VI) or particulate lead chromate induced global DNA hypomethylation, which maintained for 20 h (Lou et al., 2013). Aberrant DNA methylation, including global DNA hypomethylation as well as promoter specific DNA methylation, contributes to genomic instability and gene silencing, and has been identified in numerous cancer types and human diseases (reviewed in Pogribny & Beland, 2009). Hu et al. (2016) found that in human bronchial epithelial cells (16HBE), CpG sites on the p16 gene were significantly more methylated than controls following Cr(VI) exposure, and that p16 mRNA expression was negatively correlated with dose (Hu et al., 2016). The p16 tumor suppressor functions by inhibiting CDK4 and CDK6, which phosphorylate retinoblastoma protein and induces cell cycle arrest, and its inactivation via promoter methylation is common in lung cancers (Ohtani et al., 2004; Tam et al., 2013).
In lung tumors obtained from chromate workers in Japan, methylation of both DNA repair genes, hMLH1 (28%) and MGMT (20%), and tumor suppressor APC (86%), were detected using nested methylation-specific PCR (Ali et al., 2011). In non-chromate lung cancer tumors, methylation of hMLH1 was not detected, and methylation of APC was detected in only 44% of tumors. In a cross-sectional study examining CpG methylation of DNA repair genes, HOGG1, MGMT, XRCC1, ERCC3, and RAD53, methylation of HOGG1, MGMT, and RAD53 were significantly higher in chromate-exposed workers as well as in 16HBE cells treated with Cr(VI) (Hu et al., 2018). CpG sites within HOGG1, MGMT, and RAD51 were similarly modified in both lymphocytes from chromate-exposed workers and in Cr(VI)-treated 16HBE cells, revealing a distinct methylation pattern.
Recently, Cr was found to inhibit the levels of 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-foC), and 5-carboxylcytosine (5-caC) in mouse embryonic stem cells - all 10–11 translocation (TET) protein-induced derivatives of 5-methylcytosine (5-mC) (Xiong et al., 2017). While TET1–3 mRNA expression was found to be unchanged, TET protein activity decreased by 62.1% and 2-hydroxyglutarate (2-HG; endogenous TET inhibitor) was found dramatically increased. 2-HG can act as a competitive inhibitor of TET 5-methylcytocine hydroxylases due to structural similarities with α-KG, a necessary co-substrate for conversion of 5mC to 5-hmC (Xu et al., 2011). TET proteins and their 5-mC modifying activities are crucial for epigenetic reprogramming during both development and active DNA demethylation (reviewed in Kohli & Zhang, 2013).
Histone Posttranslational Modifications
The roles that histone modifications play in chromatin homeostasis are dynamic and dependent on factors such as the degree of modification (e.g. mono-, di-, and tri- methylation), location, and on histone cross-talk, which is context specific. Histone modification is tightly controlled by enzymatic activities of histone methyltransferases, demethylases, histone acetyltransferases (HATs), and deacetylases (HDACs), among others. For instance, HATs catalyze the transfer of acetyl groups from acetyl CoA to histone lysine residues, HDACs remove acetyl groups from histones. Acute high dose Cr(VI) exposure has been found to induce various histone modifications including increase in global histone 3 lysine 9 (H3K9) and histone 3 lysine 4 (H3K4) di- and trimethylation, and decrease global histone 3 lysine 27 (H3K27) trimethylation and histone 3 arginine 2 (H3R2) dimethylation, in vitro (Sun et al., 2009), see Table 2. Furthermore, H3K9 dimethylation was found enriched in the MLH1 promoter, and MLH1 promoter methylation was correlated with a decrease in MLH1 mRNA expression. H3K9 dimethylation and subsequent MLH1 silencing may be attributable to histone methyltransferase G9a, which specifically methylates H3K9, and was also found to increase at both the transcriptional and protein level. However, histone modifying enzymes including, but not limited to, SUV39H1, which methylates H3K9 (me2 and me3), and EZH2, which methylates H3K27, can also contribute to aberrant histone methylation following Cr(VI) exposure (Wang et al., 2018). Cr(VI) has also been shown to inhibit JHDM2A, a H3K9 demethylase, by reducing ascorbic acid availability (Sun et al., 2009). Specifically, the ascorbic acid reserve needed for JHDM2A demethylase activity is depleted upon intracellular reduction of Cr(III), which can subsequently lead to elevated H3K9me2 levels.
Table 2.
Summary of histone modifications altered by Cr(VI)
| Histone Mark | Direction of change | Chromium compound | Dose | Length of exposure | Tissue type | Reference |
|---|---|---|---|---|---|---|
| H4K16ac | Down | K2CrO4 | 10 uM | 24hr | BEAS-2B | Chen et al., 2016 |
| H3K27me3 | Down | K2CrO4 | 5–10 uM | 1 hr | A549 | Sun et al., 2009 |
| H3K4me2 | Up | K2CrO4 | 10 uM | 1 hr | A549 | Sun et al., 2009 |
| H3K4me3 | Up | K2CrO4 | 10 uM | 1 hr | A549 | Sun et al., 2009 |
| H3K9me2 | Up | K2CrO4 | 5–10 uM | 1 hr | A549 | Sun et al., 2009 |
| H3K9me3 | Up | K2CrO4 | 5–10 uM | 1 hr | A549 | Sun et al., 2009 |
| H3R2me2 | Down | K2CrO4 | 5–10 uM | 1 hr | A549 | Sun et al., 2009 |
| H3K27me3 | Up | K2Cr2O7 | .25 uM | 20,40 wk | BEAS-2B, 16HBE | Wang et al., 2018 |
| H3K9me2 | Up | K2Cr2O7 | .25 uM | 20,40 wk | BEAS-2B, 16HBE | Wang et al., 2018 |
| H3ac | Down | K2CrO4 | 2.5–5 uM | 24 hr | 16HBE | Xia et al., 2014 |
| H4ac | Down | K2CrO4 | 2.5–5 uM | 24 hr | 16HBE | Xia et al., 2014 |
| H3K4me3 | Up | K2CrO4 | 0.5–10 uM | 24 hr | A549 | Zhou et al., 2009 |
Cr(VI) not only interferes with histone methyltransferases and demethylases, but has also been shown to crosslink HDAC1-DNMT1 to the Cyp1A1 promotor (Schenekenburger et al., 2007). Total HDAC1 activity following Cr(VI) was shown to be unaffected by Cr(VI), yet local deacetylase activity was shown to be sufficient to impact histone acetylation levels in Cyp1A1 (Schenekenburger et al., 2007). In addition to HDAC1, both HDAC2 and HDAC3 protein expression levels in 16HBE cells were found to increase following 24 h exposure to Cr(VI) (Xia et al., 2014). Corresponding to this increase in HDAC2 and HDAC3, global levels of H3 and H4 acetylation were found to decrease (Xia et al., 2014). Xia et al. 2014 further showed that Cr(VI) significantly decreased biotinidase (BTD) at both the protein and mRNA levels, and this decrease was dependent on histone acetylation. Accordingly, histone biotinylation was shown to be inversely related to Cr(VI) at low (≤0.6 μM) doses (Xia et al., 2014). Lastly, Cr(VI) was found to significantly reduce histone 4 lysine 16 (H4K16) monoacetylation – a hallmark of cancers (Chen et al., 2016; Fraga et al., 2005).
Among many acetylated histone lysine residues, H4K16 acetylation is the most prevalent modification for controlling the formation of higher-order chromatin structure and modulating the functional interaction between non-histone proteins and chromatin fibers (Zhang et al., 2017). Located in the basic patch of the H4 N-terminal tail, acetylation of H4K16 can hinder the interaction between core histones such as H4 and H2A/H2B, and dynamically regulate chromatin related processes including chromatin condensation, DNA replication, transcription, repa ir, damage responses, as well as overall genome stability (Dorigo et al., 2003; Shogren-Knaak et al., 2006; Shogren-Knaak & Peterson, 2006). Due to considerable influence of this particular histone modification, it is not surprising that the loss of H4K16 acetylation has been reported as an important hallmark for many human cancers 35.
MOF, Males absent on the first, a member of the MYST (MOZ, Ybf2 (Sas3), Sas2, and Tip60) family of histone acetyltransferase (Chen et al., 2015; Li et al., 2010; Taipale et al., 2005), is an H4K16-specific HAT. Interestingly, although H4K16 can be acetylated by many different histone acetyltransferases, studies suggest that MOF may be the only HAT capable of acetylating H4K16 in intact cells. As shown in HeLa and HepG2 cell lines, knockdown of MOF led to reduction in H4K16 acetylation while other histone acetylation marks remained unchanged (Taipale et al., 2005; Smith et al., 2005). Loss of MOF and subsequent H4K16ac have been linked to greater genome instability and tumor development (Fraga et al., 2005).
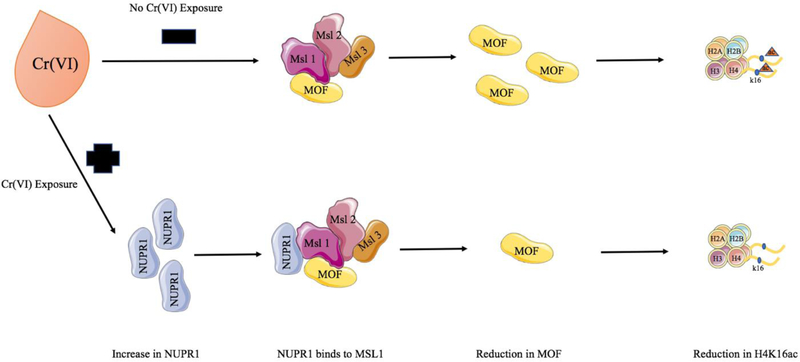
Stressor protein NUPR1 promotes tumorigenesis through mechanisms relating to cell cycle, apoptosis, and more recently uncovered, chromatin remodeling. Consistent with previous studies demonstrating the ability of Cr(VI) to reduce global H3 and H4 acetylation, Chen et al. showed that Cr(VI) induced NUPR1, see Figure 1 (Chen et al., 2016). NUPR1 induction resulted in the reduction of MOF and H4K16 acetylation, and subsequently led to cell transformation, as indicated by acquired ability for anchorage-independent growth in BEAS-2B cells (Chen et al., 2016). NUPR1-induced loss of H4K16ac and downregulation of MOF supports the assertion that the chromatin remodeling ability of NUPR1 is a mechanism driving Cr(VI)-induced lung carcinogenesis. However, NUPR1 and its relationship with other chromatin influencers, post-translational modifications governing protein expression and activity, and transcriptional regulation remain to be unveiled.
Figure 1.
Model illustrating Cr(VI)-induced reduction in H4K16ac through NUPR1 induction
The model depicts Cr(VI)-induced up-regulation of NUPR1, which can potentially bind to MSL complex (MSL1) and thereby hinder MOF transcription and subsequent histone H4K16 acetylation (Chen et al., 2015; Chen et al., 2016; Gironella et al., 2009).
MicroRNAs
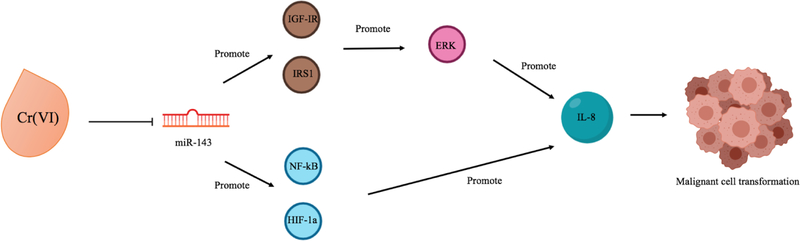
In comparison to DNA methylation and histone post-translational modifications, less is known about the influence of Cr(VI) on microRNAs (Table 3), yet they represent critical gene regulatory mechanisms and are often dysregulated in cancers. For instance, miR-143 was found to be downregulated in Cr(VI)-transformed cells, and was also found to be repressed in human lung cancer cells, see Figure 2 (He et al., 2013). Repression of miR-143 was capable of inducing cell transformation and angiogenesis via upregulation of insulin-like growth factor-1 receptor (IGF-IR) and insulin receptor substrate-1 (IRS1). IGF-IR/IRS1 was found to upregulate interleukin-8 (IL-8) as well as activate downstream ERK/HIF-1a/NF-kB signaling pathway to induce transformation and tumor angiogenesis. miRNA profile analysis of Drosophila melanogaster larva exposed to varying concentrations of Cr(VI) for 24–48 h showed 28 significantly dysregulated miRNAs targeting major biological processes (Chandra et al., 2015). Concurrent downregulation of miRNA gene targets, mus309 and mus312, acon, and pyd, which function in DNA repair, oxidation-reduction processes, and stress activated MAPK cascade, respectively, were also reported. A significant dose-dependent increase in miR-21 and dose-dependent decrease in mRNA and protein expression of its target gene, PDCD4, has also been observed (Pratheeshkumar et al., 2016). PDCD4 acts as a tumor suppressor, in part, via regulating E-cadherin. E-cadherin was subsequently found to be downregulated, and upregulation of active b-catenin and TCF4 was evident. Oncogenic c-MYC and uPAR, targets of b-catenin/TCF4-dependent transcription, were increased in a dose-dependent manner, and chromatin immunoprecipitation analysis showed their association with both uPAR and c-MYC promoters. uPAR expression has been shown to enhance tumor growth and metastasis, and has been associated with cancer stem-cell like property in small cell lung cancer (Gutova et al., 2007; Xing & Rabbani, 1996). In addition, the study revealed that Cr(VI) was able to induce phosphorylation and activation of the signal transducer and activator of transcription-3 (STAT3) as well as increased IL-6 secretion. Previous studies have shown that STAT3 can bind directly to the miR-21 promoter upon IL-6 activation (O’hara et al., 2007; Pratheeshkumar et al., 2016), thus Cr(VI) may promote miR-21 expression via the IL-6/STAT3 pathway.
Table 3.
Summary of miRNAs altered by Cr(VI)
| Gene Name | Direction of change | Chromium compound | Dose | Length of exposure | Tissue type | Reference |
|---|---|---|---|---|---|---|
| miR-1-3p | Up | K2Cr2O38 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-10-3p | Up | K2Cr2O29 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| mR-10-5p | Up | K2Cr2O28 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-1002-5p | Up | K2Cr2O16 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-12-5p | Up | K2Cr2O10 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-13b-3p | Up | K2Cr2O12 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-143 | Down | Na2Cr2O7·2H2O | 1 uM | 6 mo | BEAS-2B | He et al., 2013 |
| miR-184-3p | Up | K2Cr2O36 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-21 | Up | K2Cr2O7 | 2.5–5 uM | 24 hr | BEAS-2B | Pratheeshkumar et al., 2016 |
| miR-2493-5p | Up | K2Cr2O15 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-2494-5p | Up | K2Cr2O14 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-252-3p | Up | K2Cr2O40 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-276a-3p | Up | K2Cr2O25 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-277-3p | Up | K2Cr2O17 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-279-3p | Up | K2Cr2O11 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-281-2-5p | Up | K2Cr2O35 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-289-5p | Up | K2Cr2O19 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-2a-3p | Up | K2Cr2O30 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-2b-3p | Up | K2Cr2O31 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-306-5p | Up | K2Cr2O32 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-314-3p | Up | K2Cr2O8 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-31a-5p | Up | K2Cr2O27 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-34-5p | Up | K2Cr2O24 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-375-3p | Up | K2Cr2O26 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-389-3p | Up | K2Cr2O39 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-3940-5p | Down | Cr(VI) | 5 IQR | 3–10 yr | Human blood | Li et al., 2014 |
| miR-3940-5p | Down | Na2CrO4 | 5–10 uM | 0, 12 hr | 16HBE | Li et al., 2016 |
| miR-7-5p | Up | K2Cr2O33 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-79-3p | Up | K2Cr2O9 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-8-3p | Up | K2Cr2O23 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-954-5p | Down | K2Cr2O7 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-956-3p | Up | K2Cr2O18 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-959-3p | Up | K2Cr2O41 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-964-5p | Up | K2Cr2O42 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-970-3p | Up | K2Cr2O37 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al., 2015 |
| miR-986-5p | Up | K2Cr2O13 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-998-3p | Up | K2Cr2O34 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-9a-3p | Down | K2Cr2O20 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
| miR-9a-5p | Up | K2Cr2O21 | 5–20 ug/mL | 24, 48 hr | Drosophila mid-gut tissue | Chandra et al, 2015 |
| miR-9c-5p | Up | K2Cr2O22 | 5–20 ug/mL | 24, 48 hr | Drosophik mid-gut tissue | Chandra et al., 2015 |
Figure 2.
Model illustrating Cr(VI)-induced miR-143 reduction and subsequent cell signaling response
The model portrays Cr(VI)-induced reduction in miR-143, which has been shown to promote IL-8 and malignant cell transformation through two signaling pathways: IGF-IR/IRS1/ERK and HIF-1a/NF-kB (He et al., 2013).
Conclusion
Extensive research has solidified Cr(VI) as a potent human carcinogen, especially in the context of lung cancer. Although epigenetic alteration has been proposed as an important mechanism underlying Cr(VI) carcinogenicity, researchers are still trying to elucidate the specific changes as well as the epigenetic machineries responsible for mediating these alterations. This review provides an updated summary of current findings on mechanisms of Cr(VI) carcinogenesis, with a focus on epigenetic changes. In addition, this review also revealed that while much focus has been placed on the role of oxidative stress in Cr(VI) carcinogenesis, less focus has been given to the role of histone modifications and miRNAs. The current research gap in this area may provide a new niche for future research, which can generate new studies in the effort to provide a more comprehensive understanding of the mechanisms of Cr(VI) carcinogenesis.
Funding:
This research was funded by the following NIH grants: ES000260, ES022935, ES023174, ES026138
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abreu PL, Cunha-Oliveira T, Ferreira LMR, and Urbano AM (2018). Hexavalent chromium, a lung carcinogen, confers resistance to thermal stress and interferes with heat shock protein expression in human bronchial epithelial cells. Biometals 31, 477–487. [DOI] [PubMed] [Google Scholar]
- Ali AH, Kondo K, Namura T, Senba Y, Takizawa H, Nakagawa Y, Toba H, Kenzaki K, Sakiyama S, and Tangoku A (2011). Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol Carcinog 50, 89–99. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Weng MW, Chen WC, and Tang MS (2012). Chromium (VI) induces both bulky DNA adducts and oxidative DNA damage at adenines and guanines in the p53 gene of human lung cells. Carcinogenesis 33, 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A, and Costa M (2009). Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics 1, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont JJ, Sedman RM, Reynolds SD, Sherman CD, Li LH, Howd RA, Sandy MS, Zeise L, and Alexeeff GV (2008). Cancer mortality in a Chinese population exposed to hexavalent chromium in drinking water. Epidemiology 19, 12–23. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Stemmy EJ, Constant SL, Schwartz A, Little LG, Gigley JP, Chun G, Sugden KD, Ceryak SM, and Patierno SR (2009). Lung injury, inflammation and Akt signaling following inhalation of particulate hexavalent chromium. Toxicol Appl Pharmacol 235, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright P, Burge PS, O’Hickey SP, Gannon PF, Robertson AS, and Boran A (1997). Occupational asthma due to chrome and nickel electroplating. Thorax 52, 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning CL, Qin Q, Kelly DF, Prakash R, Vanoli F, Jasin M, and Wise JP Sr. (2016). Prolonged Particulate Hexavalent Chromium Exposure Suppresses Homologous Recombination Repair in Human Lung Cells. Toxicol Sci 153, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall M, and Kortenkamp A (1994). The generation of apurinic/apyrimidinic sites in isolated DNA during the reduction of chromate by glutathione. Carcinogenesis 15, 407–409. [DOI] [PubMed] [Google Scholar]
- Chandra S, Pandey A, and Chowdhuri DK (2015). MiRNA profiling provides insights on adverse effects of Cr(VI) in the midgut tissues of Drosophila melanogaster. Journal of hazardous materials 283, 558–567. [DOI] [PubMed] [Google Scholar]
- Chen D, Kluz T, Fang L, Zhang X, Sun H, Jin C, and Costa M (2016). Hexavalent Chromium (Cr(VI)) Down-Regulates Acetylation of Histone H4 at Lysine 16 through Induction of Stressor Protein Nupr1. PLoS One 11, e0157317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Costa M, and Sun H (2015). Structure and function of histone acetyltransferase MOF. Aims Biophys 2, 555–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, DesMarais T, and Costa M (2019). Metals and Mechanisms of Carcinogenesis. Annu Rev Pharmacol Toxicol 59, 537–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie NT, Cantoni O, Evans RM, Meyn RE, and Costa M (1984). Use of mammalian DNA repair-deficient mutants to assess the effects of toxic metal compounds on DNA. Biochemical pharmacology 33, 1661–1670. [DOI] [PubMed] [Google Scholar]
- Chuang SM, Liou GY, and Yang JL (2000). Activation of JNK, p38 and ERK mitogen-activated protein kinases by chromium(VI) is mediated through oxidative stress but does not affect cytotoxicity. Carcinogenesis 21, 1491–1500. [PubMed] [Google Scholar]
- Codd R, and Lay PA (2001). Chromium(V)-sialic (neuraminic) acid species are formed from mixtures of chromium(VI) and saliva. J Am Chem Soc 123, 11799–11800. [DOI] [PubMed] [Google Scholar]
- Dai J, Ji Y, Wang W, Kim D, Fai LY, Wang L, Luo J, and Zhang Z (2017). Loss of fructose-1,6-bisphosphatase induces glycolysis and promotes apoptosis resistance of cancer stem-like cells: an important role in hexavalent chromium-induced carcinogenesis. Toxicol Appl Pharmacol 331, 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoughery Z, Luczak MW, Ortega-Atienza S, and Zhitkovich A (2015). DNA double-strand breaks by Cr(VI) are targeted to euchromatin and cause ATR-dependent phosphorylation of histone H2AX and its ubiquitination. Toxicol Sci 143, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasagayam TP, Tilak JC, Boloor KK, Sane KS, Ghaskadbi SS, and Lele RD (2004). Free radicals and antioxidants in human health: current status and future prospects. J Assoc Physicians India 52, 794–804. [PubMed] [Google Scholar]
- Ding SZ, Yang YX, Li XL, Michelli-Rivera A, Han SY, Wang L, Pratheeshkumar P, Wang X, Lu J, Yin YQ, et al. (2013). Epithelial-mesenchymal transition during oncogenic transformation induced by hexavalent chromium involves reactive oxygen species-dependent mechanism in lung epithelial cells. Toxicol Appl Pharmacol 269, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, and Richmond TJ (2003). Chromatin fiber folding: Requirement for the histone H4N-terminal tail. Journal of Molecular Biology 327, 85–96. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. (2005). Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37, 391–400. [DOI] [PubMed] [Google Scholar]
- Ganapathy S, Li P, Lafontant J, Xiong R, Yu T, Zhang G, and Chen C (2017). Chromium IV exposure, via Src/Ras signaling, promotes cell transformation. Mol Carcinog 56, 1808–1815. [DOI] [PubMed] [Google Scholar]
- Gao N, Jiang BH, Leonard SS, Corum L, Zhang Z, Roberts JR, Antonini J, Zheng JZ, Flynn DC, Castranova V, and Shi X (2002). p38 Signaling-mediated hypoxia inducible factor 1alpha and vascular endothelial growth factor induction by Cr(VI) in DU145 human prostate carcinoma cells. J Biol Chem 277, 45041–45048. [DOI] [PubMed] [Google Scholar]
- Ge H, Li Z, Jiang L, Li Q, Geng C, Yao X, Shi X, Liu Y, and Cao J (2019). Cr (VI) induces crosstalk between apoptosis and autophagy through endoplasmic reticulum stress in A549cells. Chem Biol Interact 298, 35–42. [DOI] [PubMed] [Google Scholar]
- Gibb HJ, Lees PS, Pinsky PF, and Rooney BC (2000). Lung cancer among workers in chromium chemical production. Am J Ind Med 38, 115–126. [DOI] [PubMed] [Google Scholar]
- Gibb HJ, Lees PS, Wang J, and Grace O’Leary K (2015). Extended followup of a cohort of chromium production workers. Am J Ind Med 58, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutova M, Najbauer J, Gevorgyan A, Metz MZ, Weng Y, Shih C-C, and Aboody KS (2007). Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PloS one 2, e243–e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Qian X, Carpenter R, Xu Q, Wang L, Qi Y, Wang ZX, Liu LZ, and Jiang BH (2013). Repression of miR-143 mediates Cr (VI)-induced tumor angiogenesis via IGF IR/IRS1/ERK/IL-8 pathway. Toxicol Sci 134, 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XQ, Lin GX, Chen MG, and Ma Q (2007). Protection against Chromium (VI)-induced oxidative stress and apoptosis by Nrf2. recruiting Nrf2 into the nucleus and disrupting the nuclear Nrf2/Keap1 association by toxic metal. Faseb J 21, A1181–A1181. [DOI] [PubMed] [Google Scholar]
- Hodges NJ, and Chipman JK (2002). Down-regulation of the DNA-repair endonuclease 8-oxo-guanine DNA glycosylase 1 (hOGG1) by sodium dichromate in cultured human A549 lung carcinoma cells. Carcinogenesis 23, 55–60. [DOI] [PubMed] [Google Scholar]
- Hu G, Li P, Cui X, Li Y, Zhang J, Zhai X, Yu S, Tang S, Zhao Z, Wang J, and Jia G (2018). Cr(VI)-induced methylation and down-regulation of DNA repair genes and its association with markers of genetic damage in workers and 16HBE cells. Environ Pollut 238, 833–843. [DOI] [PubMed] [Google Scholar]
- Hu G, Li P, Li Y, Wang T, Gao X, Zhang W, and Jia G (2016). Methylation levels of P16 and TP53 that are involved in DNA strand breakage of 16HBE cells treated by hexavalent chromium. Toxicol Lett 249, 15–21. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu G, Zeng R, Wang J, Cai R, Ho JC, Zhang J, and Zheng Y (2017). Chromium contributes to human bronchial epithelial cell carcinogenesis by activating Gli2 and inhibiting autophagy. Toxicol Res (Camb) 6, 324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomova K, and Valko M (2011). Advances in metal-induced oxidative stress and human disease. Toxicology 283, 65–87. [DOI] [PubMed] [Google Scholar]
- Joseph P, He Q, and Umbright C (2008). Heme-oxygenase 1 gene expression is a marker for hexavalent chromium-induced stress and toxicity in human dermal fibroblasts. Toxicol Sci 103, 325–334. [DOI] [PubMed] [Google Scholar]
- Kasprzak KS (1995). Possible role of oxidative damage in metal-induced carcinogenesis. Cancer Invest 13, 411–430. [DOI] [PubMed] [Google Scholar]
- Khan DA, Mushtaq S, Khan FA, and Khan MQ (2013). Toxic effects of chromium on tannery workers at Sialkot (Pakistan). Toxicology and industrial health 29, 209–215. [DOI] [PubMed] [Google Scholar]
- Kim D, Dai J, Park YH, Fai LY, Wang L, Pratheeshkumar P, Son YO, Kondo K, Xu M, Luo J, et al. (2016). Activation of Epidermal Growth Factor Receptor/p38/Hypoxia-inducible Factor-1alpha Is Pivotal for Angiogenesis and Tumorigenesis of Malignantly Transformed Cells Induced by Hexavalent Chromium. J Biol Chem 291, 16271–16281. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Klaunig JE, Wang ZM, Pu XZ, and Zhou SY (2011). Oxidative stress and oxidative damage in chemical carcinogenesis. Toxicol Appl Pharm 254, 86–99. [DOI] [PubMed] [Google Scholar]
- Klein CB, Su L, Bowser D, and Leszczynska J (2002). Chromate-induced epimutations in mammalian cells. Environ Health Perspect 110 Suppl 5, 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, and Zhang Y (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee JK (2014). A Paradoxical Chemoresistance and Tumor Suppressive Role of Antioxidant in Solid Cancer Cells: A Strange Case of Dr. Jekyll and Mr. Hyde. Biomed Research International. [DOI] [PMC free article] [PubMed]
- Lejding T, Mowitz M, Isaksson M, Bruze M, Ponten A, Svedman C, Zimerson E, and Engfeldt M (2018). A retrospective investigation of hexavalent chromium allergy in southern Sweden. Contact dermatitis 78, 386–392. [DOI] [PubMed] [Google Scholar]
- Leonard SS, Harris GK, and Shi X (2004). Metal-induced oxidative stress and signal transduction. Free radical biology & medicine 37, 1921–1942. [DOI] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, and Dou Y (2010). MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol 30, 5335–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu G, Li P, Tang S, Zhang J, and Jia G (2016). miR-3940–5p enhances homologous recombination after DSB in Cr(VI) exposed 16HBE cell. Toxicology 344–346, 1–6. [DOI] [PubMed] [Google Scholar]
- Li Y, Li P, Yu S, Zhang J, Wang T, and Jia G (2014). miR-3940–5p associated with genetic damage in workers exposed to hexavalent chromium. Toxicology letters 229, 319–326. [DOI] [PubMed] [Google Scholar]
- Lindberg E, and Hedenstierna G (1983). Chrome plating: symptoms, findings in the upper airways, and effects on lung function. Archives of environmental health 38, 367–374. [DOI] [PubMed] [Google Scholar]
- Lou J, Wang Y, Yao C, Jin L, Wang X, Xiao Y, Wu N, Song P, Song Y, Tan Y, et al. (2013). Role of DNA methylation in cell cycle arrest induced by Cr (VI) in two cell lines. PLoS One 8, e71031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luippold RS, Mundt KA, Austin RP, Liebig E, Panko J, Crump C, Crump K, and Proctor D (2003). Lung cancer mortality among chromate production workers. Occup Environ Med 60, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, and Bartek J (2011). More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13, 1161–1169. [DOI] [PubMed] [Google Scholar]
- Medan D, Luanpitpong S, Azad N, Wang L, Jiang BH, Davis ME, Barnett JB, Guo L, and Rojanasakul Y (2012). Multifunctional role of Bcl-2 in malignant transformation and tumorigenesis of Cr(VI)-transformed lung cells. PLoS One 7, e37045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr AM, and Mott JL (2015). Overview of microRNA biology. Seminars in liver disease 35, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JM, and Myers CR (2009). The effects of hexavalent chromium on thioredoxin reductase and peroxiredoxins in human bronchial epithelial cells. Free radical biology & medicine 47, 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP (2008). Toxicology and carcinogenesis studies of sodium dichromate dihydrate (Cas No. 7789-12-0) in F344/N rats and B6C3F1 mice (drinking water studies). National Toxicology Program technical report series, 1–192. [PubMed] [Google Scholar]
- O’Hara KA, Vaghjiani RJ, Nemec AA, Klei LR, and Barchowsky A (2007). Cr(VI)-stimulated STAT3 tyrosine phosphorylation and nuclear translocation in human airway epithelial cells requires Lck. Biochem J 402, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Yamakoshi K, Takahashi A, and Hara E (2004). The p16INK4a-RB pathway: molecular link between cellular senescence and tumor suppression. The journal of medical investigation : JMI 51, 146–153. [DOI] [PubMed] [Google Scholar]
- Ovesen JL, Fan Y, Zhang X, Chen J, Medvedovic M, Xia Y, and Puga A (2014). Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) analysis uncovers broad changes in chromatin structure resulting from hexavalent chromium exposure. PLoS One 9, e97849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Li C, Zhao H, Darzynkiewicz Z, and Xu D (2016). Gene 33/Mig6 inhibits hexavalent chromium-induced DNA damage and cell transformation in human lung epithelial cells. Oncotarget 7, 8916–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlolla AK, Barnes C, Yedjou C, Velma VR, and Tchounwou PB (2009). Oxidative stress, DNA damage, and antioxidant enzyme activity induced by hexavalent chromium in Sprague-Dawley rats. Environ Toxicol 24, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, and Beland FA (2009). DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci 66, 2249–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratheeshkumar P, Son YO, Divya SP, Roy RV, Hitron JA, Wang L, Kim D, Dai J, Asha P, Zhang Z, et al. (2014). Luteolin inhibits Cr(VI)-induced malignant cell transformation of human lung epithelial cells by targeting ROS mediated multiple cell signaling pathways. Toxicol Appl Pharmacol 281, 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratheeshkumar P, Son YO, Divya SP, Turcios L, Roy RV, Hitron JA, Wang L, Kim D, Dai J, Asha P, et al. (2016). Hexavalent chromium induces malignant transformation of human lung bronchial epithelial cells via ROS-dependent activation of miR-21-PDCD4 signaling. Oncotarget 7, 51193–51210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DM, Panko JP, Liebig EW, Scott PK, Mundt KA, Buczynski MA, Barnhart RJ, Harris MA, Morgan RJ, and Paustenbach DJ (2003). Workplace airborne hexavalent chromium concentrations for the Painesville, Ohio, chromate production plant (1943–1971). Appl Occup Environ Hyg 18, 430–449. [DOI] [PubMed] [Google Scholar]
- Qin Q, Xie H, Wise SS, Browning CL, Thompson KN, Holmes AL, and Wise JP Sr. (2014). Homologous recombination repair signaling in chemical carcinogenesis: prolonged particulate hexavalent chromium exposure suppresses the Rad51 response in human lung cells. Toxicol Sci 142, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quievryn G, Peterson E, Messer J, and Zhitkovich A (2003). Genotoxicity and mutagenicity of chromium(VI)/ascorbate-generated DNA adducts in human and bacterial cells. Biochemistry 42, 1062–1070. [DOI] [PubMed] [Google Scholar]
- Roy RV, Pratheeshkumar P, Son YO, Wang L, Hitron JA, Divya SP, Zhang Z, and Shi X (2016). Different roles of ROS and Nrf2 in Cr(VI)-induced inflammatory responses in normal and Cr(VI)-transformed cells. Toxicol Appl Pharmacol 307, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M, Peng L, and Puga A (2007). HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Bba-Gene Struct Expr 1769, 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chiu A, Chen CT, Halliwell B, Castranova V, and Vallyathan V (1999). Reduction of chromium(VI) and its relationship to carcinogenesis. J Toxicol Environ Health B Crit Rev 2, 87–104. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, and Peterson CL (2006). Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, and Peterson CL (2006). Switching on chromatin - Mechanistic role of histone H4–K16 acetylation. Cell Cycle 5, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Smith ER, Cayrou C, Huang R, Lane WS, Cote J, and Lucchesi JC (2005). A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Molecular and Cellular Biology 25, 9175–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son YO, Pratheeshkumar P, Wang Y, Kim D, Zhang Z, and Shi X (2017). Protection from Cr(VI)-induced malignant cell transformation and tumorigenesis of Cr(VI)-transformed cells by luteolin through Nrf2 signaling. Toxicol Appl Pharmacol 331, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoss FBD; Blackburn K; Harris B; Neal M (1983). Health assessment document for chronium. In, (Research Triangle Park, N.C.: U.S. Environmental Protection Agency, Office of Research and Development; ), p. 328. [Google Scholar]
- Stout MD, Nyska A, Collins BJ, Witt KL, Kissling GE, Malarkey DE, and Hooth MJ (2009). Chronic toxicity and carcinogenicity studies of chromium picolinate monohydrate administered in feed to F344/N rats and B6C3F1 mice for 2 years. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association 47, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Brocato J, and Costa M (2015). Oral Chromium Exposure and Toxicity. Current environmental health reports 2, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhou X, Chen H, Li Q, and Costa M (2009). Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium. Toxicol Appl Pharmacol 237, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, and Akhtar A (2005). HMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Molecular and Cellular Biology 25, 6798–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam KW, Zhang W, Soh J, Stastny V, Chen M, Sun H, Thu K, Rios JJ, Yang C, Marconett CN, et al. (2013). CDKN2A/p16 inactivation mechanisms and their relationship to smoke exposure and molecular features in non-small-cell lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 8, 1378–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, and Kouzarides T (2014). Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 15, 703–708. [DOI] [PubMed] [Google Scholar]
- Valko M, Izakovic M, Mazur M, Rhodes CJ, and Telser J (2004). Role of oxygen radicals in DNA damage and cancer incidence. Mol Cell Biochem 266, 37–56. [DOI] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, and Telser J (2007). Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 39, 44–84. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, and Cronin MT (2005). Metals, toxicity and oxidative stress. Curr Med Chem 12, 1161–1208. [DOI] [PubMed] [Google Scholar]
- Wang L, Wise JT, Zhang Z, and Shi X (2016). Progress and prospects of reactive oxygen species in metal carcinogenesis. Curr Pharmacol Rep 2, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, and Shi X (2001). Mechanisms of Cr(VI)-induced p53 activation: the role of phosphorylation, mdm2 and ERK. Carcinogenesis 22, 757–762. [DOI] [PubMed] [Google Scholar]
- Wang SW, Leonard SS, Ye JP, Ding M, and Shi XL (2000). The role of hydroxyl radical as a messenger in Cr(VI)-induced p53 activation. Am J Physiol-Cell Ph 279, C868–C875. [DOI] [PubMed] [Google Scholar]
- Wang SW, Leonard SS, Ye JP, Gao N, Wang LY, and Shi XL (2004). Role of reactive oxygen species and Cr(VI) in Ras-mediated signal transduction. Molecular and Cellular Biochemistry 255, 119–127. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu J, Humphries B, Kondo K, Jiang Y, Shi X, and Yang C (2018). Upregulation of histone-lysine methyltransferases plays a causal role in hexavalent chromium-induced cancer stem cell-like property and cell transformation. Toxicol Appl Pharmacol 342, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen RP, and Qian LF (1991). Chromium-induced kidney disease. Environ Health Perspect 92, 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedrychowski A, Ward WS, Schmidt WN, and Hnilica LS (1985). Chromium-induced cross-linking of nuclear proteins and DNA. J Biol Chem 260, 7150–7155. [PubMed] [Google Scholar]
- Welling R, Beaumont JJ, Petersen SJ, Alexeeff GV, and Steinmaus C (2015). Chromium VI and stomach cancer: a meta-analysis of the current epidemiological evidence. Occup Environ Med 72, 151–159. [DOI] [PubMed] [Google Scholar]
- Wilbur S, Abadin H, Fay M, Yu D, Tencza B, Ingerman L, Klotzbach J, and James S (2012). Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological Profiles In Toxicological Profile for Chromium, (Atlanta (GA): Agency for Toxic Substances and Disease Registry (US)). [PubMed] [Google Scholar]
- Williams M, Bozhilov K, Ghai S, and Talbot P (2017). Elements including metals in the atomizer and aerosol of disposable electronic cigarettes and electronic hookahs. PLoS One 12, e0175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise J, Wang,., Xu J, Zhang Z, and Shi X (2019). Oxidative stress of Cr(III) and carcinogenesis. The nutritional biochemistry of chromium (III)., pp. 323–340. [Google Scholar]
- Xia B, Ren XH, Zhuang ZX, Yang LQ, Huang HY, Pang L, Wu DS, Luo J, Tan YL, Liu JJ, and Zou F (2014). Effect of hexavalent chromium on histone biotinylation in human bronchial epithelial cells. Toxicol Lett 228, 241–247. [DOI] [PubMed] [Google Scholar]
- Xia B, Yang LQ, Huang HY, Pang L, Hu GH, Liu QC, Yuan JH, Liu JJ, Xia YB, and Zhuang ZX (2011). Chromium(VI) causes down regulation of biotinidase in human bronchial epithelial cells by modifications of histone acetylation. Toxicol Lett 205, 140–145. [DOI] [PubMed] [Google Scholar]
- Xing RH, and Rabbani SA (1996). Overexpression of urokinase receptor in breast cancer cells results in increased tumor invasion, growth and metastasis. Int J Cancer 67, 423–429. [DOI] [PubMed] [Google Scholar]
- Xiong J, Liu X, Cheng QY, Xiao S, Xia LX, Yuan BF, and Feng YQ (2017). Heavy Metals Induce Decline of Derivatives of 5-Methycytosine in Both DNA and RNA of Stem Cells. ACS chemical biology 12, 1636–1643. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. (2011). Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Guo L, Jiang BH, Luo J, and Shi X (2008). Oxidative stress and chromium(VI) carcinogenesis. J Environ Pathol Toxicol Oncol 27, 77–88. [DOI] [PubMed] [Google Scholar]
- Ye J, Zhang X, Young HA, Mao Y, and Shi X (1995). Chromium(VI)-induced nuclear factor-kappa B activation in intact cells via free radical reactions. Carcinogenesis 16, 2401–2405. [DOI] [PubMed] [Google Scholar]
- Zhang JD, and Li XL (1987). [Chromium pollution of soil and water in Jinzhou]. Zhonghua Yu Fang Yi Xue Za Zhi 21, 262–264. [PubMed] [Google Scholar]
- Zhang R, Erler J, and Langowski J (2017). Histone Acetylation Regulates Chromatin Accessibility: Role of H4K16 in Inter-nucleosome Interaction. Biophys J 112, 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitkovich A (2005). Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium(VI). Chem Res Toxicol 18, 3–11. [DOI] [PubMed] [Google Scholar]
- Zhitkovich A (2011). Chromium in drinking water: sources, metabolism, and cancer risks. Chem Res Toxicol 24, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitkovich A, Voitkun V, and Costa M (1995). Glutathione and free amino acids form stable complexes with DNA following exposure of intact mammalian cells to chromate. Carcinogenesis 16, 907–913. [DOI] [PubMed] [Google Scholar]
- Zhou X, Li Q, Arita A, Sun H, and Costa M (2009). Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol 236, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Cai T, Li J, Zhang D, Yu Y, and Huang C (2012). Hexavalent chromium Cr(VI) up-regulates COX-2 expression through an NFkappaB/c-Jun/AP-1-dependent pathway. Environ Health Perspect 120, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]