Abstract
The Drosophila EAG (dEAG) potassium channel is the founding member of the superfamily of KNCH channels, which are involved in cardiac repolarization, neuronal excitability and cellular proliferation. In flies, dEAG is involved in regulation of neuron firing and assembles with CaMKII to form a complex implicated in memory formation. We have characterized the interaction between the kinase domain of CaMKII and a 53-residue fragment of the dEAG channel that includes a canonical CaMKII recognition sequence. Crystal structures together with biochemical/biophysical analysis show a substrate-kinase complex with an unusually tight and extensive interface that appears to be strengthened by phosphorylation of the channel fragment. Electrophysiological recordings show that catalytically active CaMKII is required to observe active dEAG channels. A previously identified phosphorylation site in the recognition sequence is not the substrate for this crucial kinase activity, but rather contributes importantly to the tight interaction of the kinase with the channel. The available data suggests that the dEAG channel is a docking platform for the kinase and that phosphorylation of the channel’s kinase recognition sequence modulates the strength of the interaction between the channel and the kinase.
Keywords: Channel-kinase interaction, NMDA receptor, kinase-substrate structure, isothermal calorimetry
Introduction
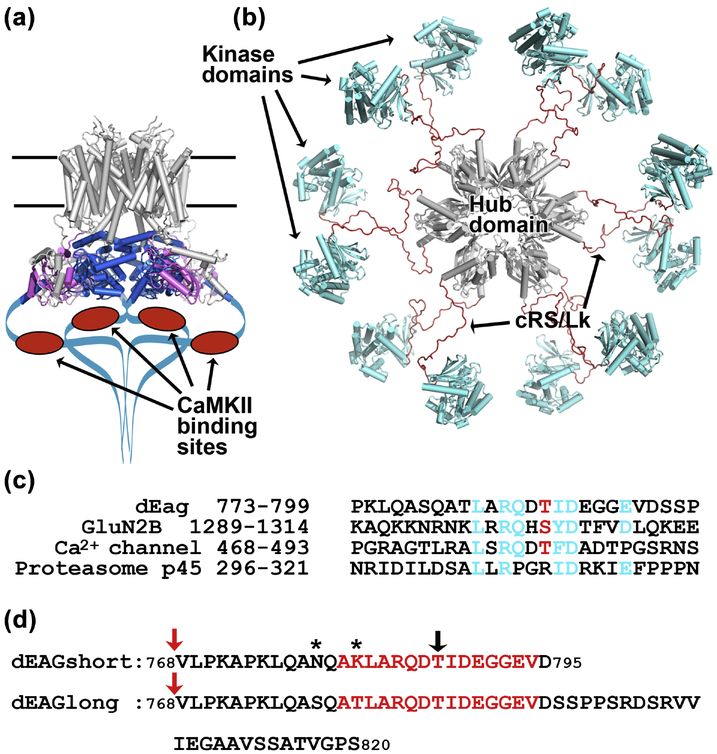
The KCNH voltage-gated potassium channels (EAG, ERG and ELK) are involved in important physiological processes like cardiac repolarization, neuronal excitability and cellular proliferation1; 2; 3; 4; 5; 6; 7. KCNH potassium channels have a typical tetrameric assembly, where each subunit has six transmembrane helices harboring a voltage sensor and a K+ selectivity filter8. These channels also include unique long N- and C-terminal cytoplasmic regions which are thought to function as interfaces with cellular signaling cascades including kinases (Figure 1a)9. The Drosophila EAG (dEAG) potassium channel is the founding member of the KCNH superfamily8; 10. Interestingly, it has been shown that this channel interacts with the Ca2+/calmodulin-dependent protein kinase II (CaMKII)11; 12.
Figure 1: Cartoon representations of EAG channel and CaMKII kinase.
a) Cryo-EM structure of the rat EAG channel (PDB: 5K7L). Membrane buried regions of channel are in grey at the top, PAS domain in magenta and CNBh domain in blue, C-terminal regions not observed in the structure are drawn as a blue line with CaMKII recognition sequence indicated as red oval. b) Pseudo-atomic EM model of the dodecameric CaMKII kinase (PDB: 5U6Y), probably corresponding to an activatable state18 with hub domain in grey at the center and kinase domains in blue tethered to the hub domain by brown cRS/Lk (C-terminal end of Regulatory Segment - residues 300–314, plus Variable Linker - residues 315–344); cRS/Lk region was not observed in the experimental determination of this structure. c) Amino acid alignment of the CaMKII recognition sequences in dEAG and in the NMDA receptor GluN2B subunit with similar sequences in the L-type voltage-dependent Ca2+ channel (GenBank: EAW60557.1) and 26S proteasome subunit p45 (GenBank: BAA07919.1). Sequences in the Ca2+ channel and p45 protein were found by PHI-BLAST search. In cyan are shown dEAG residues that interact with kinase domain (see main text and Figure 3) and homologous residues in the other sequences. Phosphorylatable residue is shown in red. d) Sequence of dEAG channels fragments used in this study. Residues highlighted in red correspond to sequence of channel fragment visible in crystal structures. Black arrow indicates T787, the residue that is phosphorylated by CaMKII. Stars indicate residues mutated in dEAGshort. Red arrows indicate position of MBP fusion.
CaMKII is a Ser/Thr protein kinase with a central role in the mechanism of long-term potentiation and synaptic plasticity13; 14. CaMKII is a dodecamer or tetradecamer15; 16; 17; 18 (Figure 1b); each subunit includes a protein kinase domain at the N terminus and a hub or multimerization domain at the C terminus19; 20; 21. These two domains are separated by a regulatory segment (RS) containing a Ca2+/CaM-binding motif and a variable region. Binding of Ca2+/CaM to the RS activates the kinase by releasing the inhibitory interaction between RS and the kinase domain22.
It has been proposed that phosphorylation of the dEAG potassium channel by CaMKII modulates channel activity11; 12 and reciprocally, that the functional properties of CaMKII are altered upon interaction with the channel23. CaMKII phosphorylates dEAG at residue T787, within a CaMKII recognition sequence present in the channel’s C-terminal cytoplasmic region. A similar sequence in the GluN2B subunit of the NMDA receptors has long been recognized as an interaction and phosphorylation site for CaMKII (Figure 1c)24. Other KCNH channels such as the human EAG and ERG (hERG) channels do not have this recognition sequence; in these channels the region corresponds to a section of the C terminus that in the recent cryo-EM structures was truncated as it is thought to be unstructured (Figure 1a)25; 26. In the fly, dEAG and CaMKII can be found at neuron synapses as part of a signaling complex. Importantly the two proteins can be co-purified and it has been proposed that the kinase recognition sequence is the major region of interaction23.
As in many other cases of regulation of potassium channel activity by kinases and phosphatases, the molecular details of the partnering between CaMKII and dEAG are not completely understood. In the present work we performed a biochemical, structural and functional characterization of determinants of the interaction between this potassium channel and the kinase. We found that the complex formed between a channel fragment, spanning the CaMKII recognition sequence in dEAG, and the kinase domain of CaMKII displays the expected features of a kinase-substrate complex but is unusually stable for a typical kinase-substrate pair, which have KDs around 200 μM27. The tight interaction involves residues previously defined for an ideal CaMKII substrate together with main-chain interactions and water-mediated contacts. Our functional data shows that interaction between the channel fragment and the kinase domain alters the activity of the kinase and that the presence of active CaMKII is crucial for a fully-functional channel. However, phosphorylation at the kinase recognition sequence in the channel does not alter the functional properties of the channel. Instead, the recognition sequence contributes to the stability of the channel/kinase complex with phosphorylation of the recognition site likely modulating complex stability. Overall, our analysis gives a detailed view of the interaction established between CaMKII and dEAG, providing molecular insights into the relationship established by CaMKII and other ion channels like the NMDA receptor and voltage-dependent calcium channels.
Results
Confirmation of CaMKII-dEAG interaction interface
It has been proposed that the protein regions mediating the interaction between CaMKII and the dEAG channel involve the kinase domain and a section of the channel spanning residues 761 to 803 which includes the phosphorylatable residue T787 (Figure 1a and 1d)23. To confirm this interaction and quantify its strength we used isothermal titration calorimetry (ITC) to determine the binding affinity of the wild type CaMKII kinase domain (residues 1–283) (Figure 1b), for two channel fragments (dEAGlong and dEAGshort, both including T787) fused to maltose-binding protein (MBP) (Figure 1d). The dEAGlong fragment is 53 amino acids long and spans residues 768 to 820. dEAGshort spans residues 768 to 795 and includes two mutations (S778N and T781K) leaving only T787 as a phosphorylatable residue and avoiding the possibility of nonspecific phosphorylation during mixture with the kinase. In the structure of the kinase and channel fragment described below the two mutated residues in dEAGshort (S778 and T781) are either not detected in the density or have the side-chain pointing away from the kinase indicating that these residues are not involved in the interaction with the kinase.
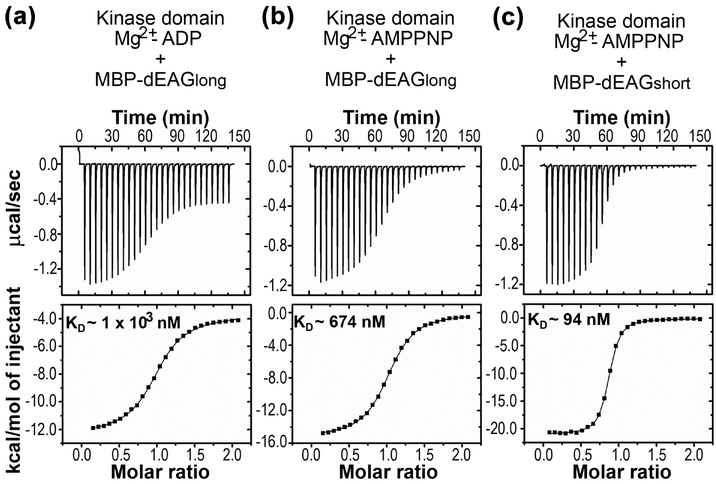
Titrations of the kinase domain with MBP-dEAGlong showed a constant of dissociation (KD) of ~1 μM in the presence of Mg2+-ADP (Figure 2a, Table 1). In the presence of Mg2+-AMPPNP, a nucleotide analog that is more stable than ATP, the KD is ~700 nM (Figure 2b, Table 1). In the presence of Mg2+-AMPPNP MBP-dEAGshort binds to the kinase domain with a KD ~90 nM (Figure 2c and Table 2). The difference in affinity between the two fragments results from a change in binding enthalpy (ΔH) for dEAGshort relative to dEAGlong, from −21 to −17 kcal/mol. The analysis showed that these channel fragments bind robustly to the kinase domain, supporting the proposal that this channel region is involved in the interaction with the kinase.
Figure 2: Interaction of dEAG fragments and CaMKII kinase domain.
Example of ITC experiments of kinase domain of CaMKII and dEAGlong channel fragment in the presence of a) Mg2+-ADP and b) Mg2+-AMPPNP. c) Example of ITC experiment of kinase domain of CaMKII and dEAGshort channel fragment in the presence of Mg2+-AMPPNP. Titration injection heats are shown in the upper panel and binding isotherm of integrated binding enthalpies in the lower panel. KD values for each protein combination are shown in lower panels.
Table 1:
ITC values for interaction between dEAGlong and CaMKII kinase domain.
| Nucleotide | In the cell | In the syringe | KD (nM) | N | ΔH (kcal/mol) | −TΔS (kcal/mol) | Cell/Syringe concentrations (μM) | n-value |
|---|---|---|---|---|---|---|---|---|
| Individually fitted titrations | ||||||||
| ADP | CaMKIIWT | EAGlongWT | 1.0 × 103 ± 0.1 × 103 | 0.94 ± 0.14 | −7.3 ± 1.7 | −0.8 ± 1.7 | 22.5/225; 22.5/225; 32/320 | 3 |
| AMPPNP | CaMKIIWT | EAGlongWT | 674.1 ± 82.4 | 0.96 ± 0.09 | −17.3 ± 2.4 | 8.9 ± 2.3 | 22.5/225; 22.5/225; 22.5/225; 33/330 | 4 |
| AMPPNP | CaMKIIWT | EAGlongR784A | 47.0 × 103 ± 27.1 × 103 | 0.85 ± 0.16 | −10.3 ± 4.3 | 4.4 ± 4.7 | 46/460; 96.5/965; 96.5/965 | 3 |
| AMPPNP | CaMKIIWT | EAGlongD789A | 7.3 × 103 ± 0.4 × 103 | 1.02 ± 0.03 | −12.7 ± 2.4 | 5.7 ± 2.3 | 36/360; 92/920; 95/950 | 3 |
| Globally fitted titrations | ||||||||
| AMPPNP | CaMKIIWT | EAGlongL782A | 24.6 × 103 ± 1.2 × 103 | 1.282 ± 0.000 0.645 ± 0.006 |
−3.75 ± 0.11 | −2.54 ± 0.11 | 15/280; 103.5/1035 | 2 |
| AMPPNP | CaMKIIWT | EAGlongQ785A | 12.6 × 103 ± 0.2 × 103 | 0.747 ± 0.002 0.664 ± 0.001 |
−18.47 ± 0.06 | 11.78 ± 0.06 | 45/450; 100/1000 | 2 |
| AMPPNP | CaMKIIWT | EAGlongI788A | 80.3 × 103 ± 0.2 × 103 |
0.667 ± 0.000 1.089 ± 0.001 |
−10.47 ± 0.02 | 4.88 ± 0.02 | 53/530; 140/1400 | 2 |
| AMPPNP | CaMKIIWT | EAGlongE790A | 2.43 × 103 ± 0.02 × 103 | 1.056 ± 0.001 | −15.73 ± 0.02 | 8.07 ± 0.02 | 20/200; 52/520 | 2 |
| AMPPNP | CaMKIIWT | EAGlongE793A | 1.68 × 103 ± 0.01 × 103 | 0.978 ± 0.001 | −16.72 ± 0.02 | 8.84 ± 0.02 | 45/450; 50/500 | 2 |
Mean and standard deviation of the mean of individually fitted titrations are presented. For globally fitted titrations the values presented are the fitted values and associated statistical error of fit. For the I788A mutant (underlined) the global fit did not converge to a unique solution; the solution shown had the lowest chi-square with solutions spanning KDs between 30 × 103 nM and 100 × 103 nM. Protein concentrations used in each titration are shown in the last column. Number of experimental replicas is shown as n-value.
Table 2:
ITC values for interactions between dEAGshort and CaMKII kinase domain.
| Nucleotide | In cell | In the syringe | KD (nM) | N | ΔH (kcal/mol) | −TΔS (kcal/mol) | Cell / Syringe concentrations (μM) | n-value |
|---|---|---|---|---|---|---|---|---|
| Individually fitted titrations | ||||||||
| AMPPNP | CaMKIIWT | EAGshort | 94.0 ± 7.3 | 0.86 ± 0.03 | −21.0 ± 0.9 | 11.4 ± 0.9 | 16/160; 17/170; 20/200 | 3 |
| AMPPNP | CaMKIIK43M,D136N | EAGshort | 173.2 ± 19.6 | 0.79 ± 0.04 | −18.0 ± 0.1 | 8.7 ± 0.1 | 13/130; 15/200; 23.5/370 | 3 |
| AMPPNP | CaMKIID136N | EAGshort | 196.7 ± 86.5 | 0.81 ± 0.12 | −14.5 ± 3.7 | 5.3 ± 3.8 | 20/200; 20/200; 20/200; 20/200 | 4 |
| ADP | CaMKIID136N | EAGshortP | 474.7 × 10−3 ± 111.0 × 10−3 | 1.07 ± 0.01 | −8.9 ± 0.3 | −3.8 ± 0.4 | 7/70; 7/70; 7/70 | 3 |
| ADP | CaMKIID136N | EAGshort | 216.3 ± 101.3 | 0.83 ± 0.04 | −16.3 ± 4.3 | 7.1 ± 4.5 | 20/200; 20/200; 20/200; 20/200 | 4 |
Mean and standard deviation of the mean are presented. Protein concentrations used in each titration are shown in the last column. Number of experimental replicas is shown as n-value.
Structure of CaMKII kinase domain in complex with the EAG channel fragment
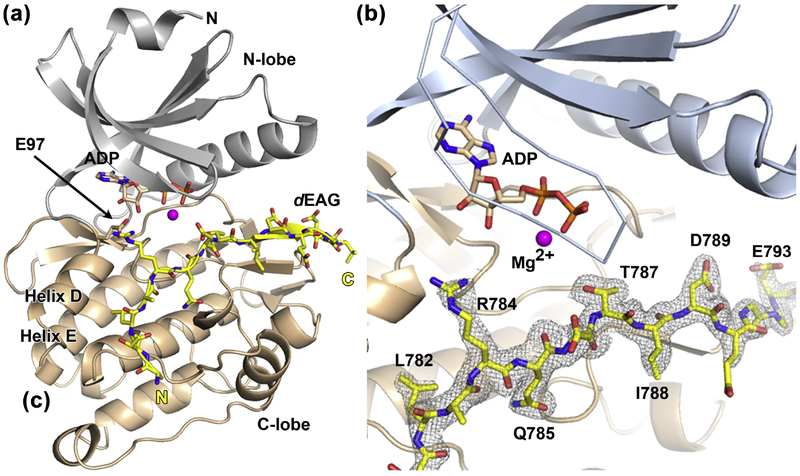
To understand the structural features of the interaction between the native kinase catalytic domain (residues 1–283) and the dEAGlong channel fragment we determined a 1.96 Å resolution structure of this complex (Figure 3a and Table S1). The structure shows residues 7–275 adopting the typical protein kinase fold that is formed by two lobes (N and C) with the active site positioned at the interface between the lobes, binding ADP. A section of the channel fragment (residues 779 to 794) is well defined in the electron-density map (Figure 3b). We were surprised to find that the channel fragment is un-phosphorylated at T787 since the crystals were grown from a mixture containing phosphorylated channel fragment and kinase. We observed a similar effect in another very similar structure, where the nucleotide (AMPPNP) bound in the kinase was hydrolyzed to AMPPN, lacking the γ-phosphate (Figure S1 and Table S1). We attribute dephosphorylation of the peptide and nucleotide to hydrolysis promoted in part by the low pH (pH 5.0) at which these crystals grew. In the structure, the channel fragment forms an extensive interaction surface with the kinase, for a total buried surface area of ~945 Å2, with the N-terminal half of the fragment interacting with the kinase C-lobe and the C-terminal half running along the periphery of the active site. Residue T787 is in the middle of the visible channel fragment, sitting at the edge of the active site pocket with its side-chain pointing into the pocket and towards the nucleotide, ADP in this case. The nucleotide is positioned deep in the pocket and is oriented so that the phosphate groups coordinate a single magnesium ion and are turned towards T787 (Figure 3b). The kinase has adopted the active-state conformation where the R- and C-spines are formed28; 29 and the α-helix D and loop DE optimize the positioning of E97 close to the ADP ribose moiety (Figure 3a)30; 31; 32. In addition, the structure shows the N-lobe collapsed on top of the nucleotide, closing the active site.
Figure 3: Structure of CaMKII kinase domain with dEAG bound.
a) Structure of the native kinase domain (residues 1–283) with the two lobes of CaMKII shown in gray (N-lobe) and wheat (C-lobe). Mg2+ is shown as a magenta sphere and ADP molecule in stick representation. dEAGlong channel fragment is displayed in yellow. Structure elements discussed in text are indicated. b) Close-up view of CaMKII active site. Color scheme as in previous figure, but showing the active site lid in ribbon and the 2Fo-Fc electron density map contoured around the dEAG fragment.
Overall, the crystallographic analysis reveals that the complex formed between the CaMKII kinase domain and the channel fragment displays the structural features expected for a kinase/substrate complex, with the substrate positioned at the active site and the kinase adopting an active conformation. Interestingly, the interaction surface observed is extensive and the affinity values measured above for the kinase domain and dEAGlong or dEAGshort are uncommonly tight, approximately 200 and 2000-fold tighter than expected for a kinase-substrate which typically displays a KD ~200 μM27.
Determinants of the interaction between the kinase and the channel fragment
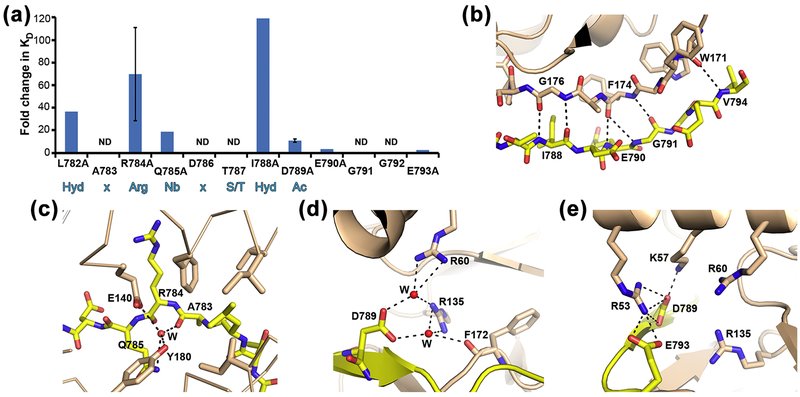
A first approach examination of the kinase/fragment structure identifies the side-chains of dEAG that are within interaction distance of the kinase domain: L782, R784, Q785, T787, I788, D789 and E793 (Figure 3b). We evaluated the side-chain contribution of the interacting dEAG residues to the stability of the complex by mutating these residues (except for T787) to alanine and determining the affinity of the fragment for the kinase domain in the presence of Mg2+-AMPPNP by ITC (Figure 4a, Table 1 and Figure S2). As a control we measured the impact of an alanine mutation in E790, that in the structure does not interact with the kinase, and which shows an unchanged affinity for the kinase (~2 μM). The large decrease in affinity and/or decrease in protein expression levels of some of the mutants (L782A, Q785A, I788A, E790A and E793A) allowed us to perform just two independent calorimetric assays for these mutants. In these cases we executed assays at two different concentrations and performed a global fit of the data using the Affinimeter software package. We show the global fit to both assays in Figure S2. The residues that have the largest impact on the stability of the association are L782, R784, and I788 (Figure 4a) with KD changes larger than 20-fold; Q785 and D789 also play a part in stabilizing the interaction, showing KD changes higher than 10-fold. Our data agrees with a qualitative study performed before, in which pull-downs showed that the double mutations L782K/A783K, R784K/Q785K, D789K/E790K and G792K/E793K reduce binding of dEAG channel fragment with full-length CaMKII23. The residues found to be determinants of the kinase/fragment interaction match with the residues previously determined to form the optimal sequence motif, or recognition sequence, for a CaMKII substrate: Hyd-x-Arg-Nb-x-Ser/Thr-Hyd-Ac; Hyd: hydrophobic, x: any residue, Nb: non-basic, Ac: acidic; (Figure 4a)33; 34.
Figure 4: Determinants of the interaction between channel fragment and kinase.
a) Bar graph representing changes in interaction stability (measured as KD) between kinase domain and mutants of dEAGlong channel fragment. Fold change of mutant KD relative to wild type is shown together with dEAG sequence. Standard deviation for mutants R784A and D789A is shown as error bars. Sequence positions marked ND (not determined) are included to allow comparison with optimal sequence motif for a CaMKII substrate shown in blue below graph: Hyd-x-Arg-Nb-x-Ser/Thr-Hyd-Ac; Hyd: hydrophobic, x: any residue, Nb: non-basic, Ac: acidic.. b) Main-chain hydrogen bond network established between dEAG and CaMKII is shown as dashed lines; c) and d) Water (W) mediated interactions between atoms in dEAG A783, Q785 and D789 with kinase domain are shown as dashed lines. e) Electrostatic cluster in CaMKII kinase/dEAG structure. Dashed lines indicate hydrogen bonding interactions but all positively charged residues are within 6 Å of negatively charged residues.
A more careful analysis of the structure reveals that besides the interactions mediated by side-chains (mainly present in the N-terminal section of the channel-fragment) there is also a network of interactions in the C-terminal end of the fragment that involve mainchain atoms. In particular, there is a network of main-chain hydrogen bonds formed between the stretch of dEAG residues I788 to V794 and the stretch W171-G176 in the kinase domain (Figure 4b). Additionally, the high-resolution structure clearly shows water molecules mediating interactions between the channel-fragment (both with main-chain and side-chain atoms) and the kinase (Figure 4c–4d). It is difficult to experimentally evaluate the impact of these interactions but it is likely that this extensive bond network will make a strong contribution to the binding of dEAG to the CaMKII.
In addition, we propose that some of the interactions established in an “electrostatic cluster” involving residues from the kinase domain and from the channel-fragment, also contribute to the stability of the complex (Figure 4e). This cluster is composed of four positively charged residues in the kinase (R53, K57 and R60 in the αC helix and R135 positioned between the catalytic loop and αE) and two negatively charged residues (D789 and E793) in the C terminus of the dEAG channel-fragment. Our ITC measurements support this claim as they show that mutation of D789 results in a 10-fold KD change but only a very small change for the E793A mutation (Figure 4a and Table 1). It is possible that besides contributing for the stabilization of the channel-fragment complex, the cluster also stabilizes the closure of the active site by promoting the downward movement of the αC helix towards the C-lobe.
Impact of the kinase/channel fragment interaction on ATP hydrolysis
A likely outcome of the tight interaction between the kinase and the channel fragment is a reduction in catalytic activity. We analyzed the effect of increasing concentrations of dEAGshort on ATP hydrolysis catalyzed by the CaMKII kinase domain during phosphorylation of syntide (Figure 5), a classical substrate of CaMKII with the amino acid sequence: PLARTLSVAGLPGKK (consensus residues underlined). The data show that the velocity of ATP hydrolysis is reduced by the presence of channel fragment. Fitting the data to an inhibition curve shows that in the presence of 250 μM syntide the channel fragment IC50, or the concentration at which the kinase activity has been reduced by 50%, is ~9 μM. This observation supports the idea that the strong interaction between CaMKII kinase domain and dEAG fragment, demonstrated by ITC, reduces the catalytic turnover of the enzyme. Interestingly, there is a close structural match between the dEAG channel fragment in complex with the kinase and a CaMKII inhibitor (CaMKII-Ntide) bound to the CaMKII kinase domain from C. elegans, despite conformational differences in the kinase domains35; 36 (Figure S3).
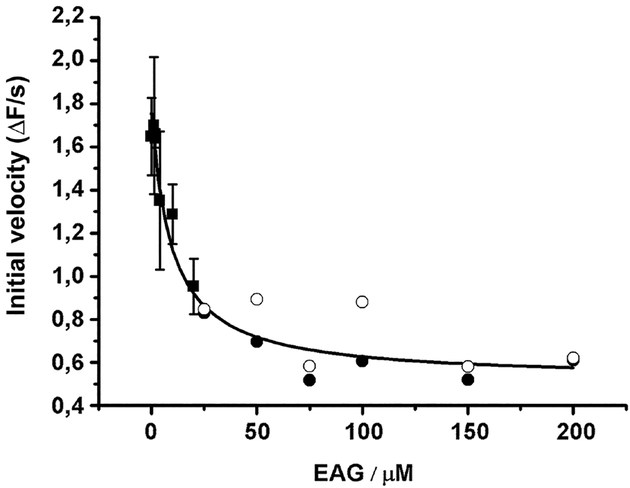
Figure 5: Impact of phosphorylation of dEAG fragment on interaction with kinase.
Plot of initial velocity of ATP hydrolysis catalyzed during phosphorylation of syntide-2 (at an initial concentration of 250 μM) in the presence of increasing dEAG concentrations. IC50 determined from curve fit is ~9 μM. In this coupled assay, ADP produced by the kinase is used in a parallel reaction with pyruvate kinase/lactate dehydrogenase to oxidize NADH, whose consumption results in a decrease of 340 nm absorbance. Points corresponding to more dilute concentrations of MBP-dEAGshort (≤20 μM) were measured in triplicate (mean ± SD shown), and those corresponding to higher concentrations of channel fragment were measured in duplicate (individual measurements are shown). Curve fitted to data is indicated in the material and methods section.
Role of CaMKII catalytic activity on the functional expression of dEAG channel
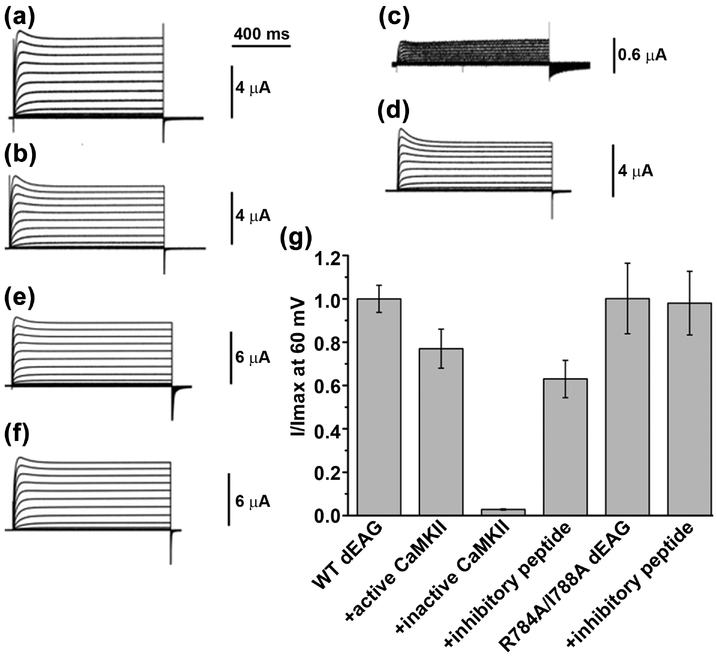
To examine the role of the CaMKII kinase activity in the functional properties of the dEAG channel we compared currents from the channel alone with currents from the channel co-expressed with full-length active and inactive forms of Drosophila CaMKII (Figure 6). The inactive form of CaMKII has two point mutations (K43M/D136N) in the active site that reduce kinase catalytic activity19; 35; 37; 38 and likely affect the mechanism of CaMKII activation by diminishing autophosphorylation at T286 and reducing Ca2+/calmodulin-independent activity22. In parallel we confirmed by ITC that these mutations in the active site do not affect the interaction between the kinase domain and dEAGshort, KD is ~170 nM versus ~90 nM for wild type (Table 2 and Figure S4). Co-expression of Drosophila CaMKII with wild type channel had a small effect of reducing steady-state current relative to oocytes without co-expressed kinase, to ~80% at 60 mV (Figure 6a, 6b and 6g). Strikingly, co-expression with inactive Drosophila kinase reduced channel current by more than 10-fold (Figure 6c and 6g).
Figure 6: Functional impact of CaMKII on dEAG channel currents.
Representative current traces of a) wild type dEAG channel, b) wild type channel + activated CaMKII, c) wild type channel + inactivated CaMKII, d) wild type channel + CaMKII inhibitory peptide, e) R784A-I788A dEAG channel and f) R784A-I788A dEAG co-expressed with CaMKII inhibitory peptide evoked by a series of depolarizing potentials ranging from −100 mV to +60 mV in 10 mV interval. e and f are exemplars from a separate experiment showing that functional properties are not obviously different from those of wild type. g) Comparison of steady state current recorded at the end of the 60-mV pulse. For WT (first bar), steady-state currents were averaged (WTmean) and normalized to 1. For WT+active CaMKII, averaged currents were divided by WTmean as were the averaged currents obtained with inactivate CaMKII and inhibitory peptide (bars 3 and 4). R784A/I788A currents were averaged (R784A/I788Amean) and normalized to 1 (5th bar); and R784A/I788A + inhibitory peptide currents were divided by R784A/I788Amean (6th bar). Data are mean ± SEM and n= 6–8.
We also applied an approach previously used by the Griffith lab12. We inhibited endogenous CaMKII activity with an inhibitory peptide derived from the autoinhibitory sequence of CaMKII that binds to the active site of CaMKII, competing substrates with an IC50 of 2–10 μM39; 40; 41. The inhibitory peptide was injected into oocytes at an estimated final concentration of 80 μM and currents were recorded 2–3 h later from injected and non-injected oocytes. The inhibitory peptide partially reduced steady-state current (at 60 mV) of the wild type channel to 60% (Figure 6d and 6g), mirroring the effect of inactive CaMKII.
Altogether, these results suggest that active kinase is crucial for the presence of active dEAG channels on the plasma membrane, either by affecting: 1) the functional properties of the channel; 2) the levels of channel protein in the membrane; and/or 3) by facilitating folding or processing of channel in the membrane.
Functional impact of the kinase domain interaction and of T787 phosphorylation
To specifically examine the functional role of the interaction between recognition sequence in the channel and the kinase domain we combined the mutations R784A and I788A in the full-length channel. Each one of these mutations reduces the affinity between the kinase domain and dEAGlong by more than 50-fold (Figure 4a and Table 1). If the interaction with kinase domain influences channel activity then we would expect to observe differences in mutant channel activity relative to wild type channel. Strikingly the mutations do not alter the functional properties of the channel, giving rise to currents that are very similar to wild type currents (Figure 6e and 6g). As a control we injected inhibitory peptide into oocytes expressing the R784A/I788A channel. Unlike for wild type channel (Figure 6d and 6g), the peptide had no effect on the currents of the double mutant channel. This is consistent with the interaction of the kinase with the recognition sequence having been altered by the double mutations so that the effect of the competing peptide is no longer detectable (Figure 6f and 6g).
Importantly, the lack of effect of the inhibitory peptide in this experiment also serves as control for all experiments where the inhibitory peptide was used, showing that the impact of the inhibitor in the activity of the dEAG channel does not result from a nonspecific effect caused from injecting a large amount of peptide into the oocyte.
The unexpected lack of functional impact of the double mutation could be simply explained by CaMKII playing no role on channel function or not interacting with channel. However, this contradicts our findings above that active kinase is crucial for channel activity and the findings of Griffith and colleagues showing that a complex is formed by CaMKII and dEAG23. An alternative explanation is that the interaction mediated by the kinase recognition sequence in the channel is not required either because the recognition sequence: 1) is not involved in the formation of the dEAG-CaMKII complex in the cell or 2) is part of a complement of interactions holding the channel-CaMKII complex together. In either case the data suggests that the CaMKII interaction with dEAG must involve other channel and kinase regions besides the one we analyzed above.
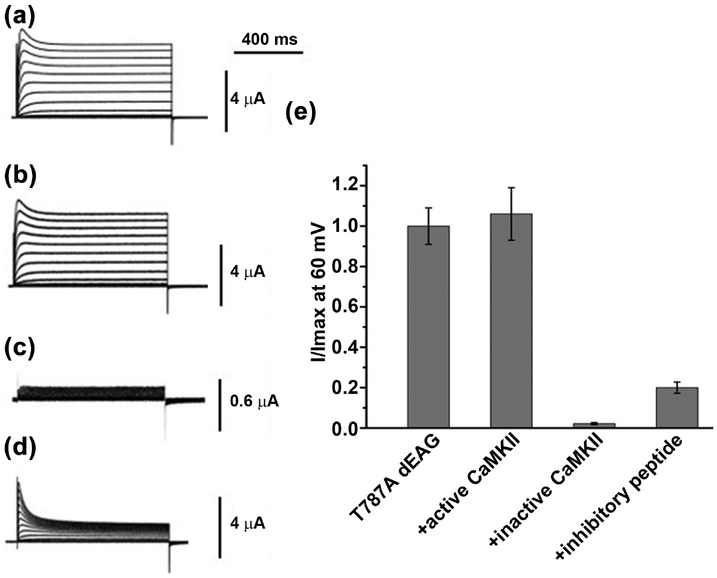
We also tested the effect of mutating the phosphorylatable residue (T787) from the CaMKII recognition sequence by repeating the electrophysiological experiments with the T787A mutant channel (Figure 7). Interestingly, the mutant channel resembles the wild type channel in several ways. First, there is little or no functional difference between the mutant and wild type channels (Figure 7a); this is clearly observed in the rates of activation or inactivation and in the current-voltage relationship (Figure S5). Second, co-expression of the mutant channel with active kinase did not affect channel activity and, like in the wild type channel, co-expression with inactive kinase resulted in loss of measurable currents (Figure 7b, 7c and 7e). Thus, since we have seen that CaMKII catalytic activity is required for dEAG channel activity, T787 is not its primary target.
Figure 7: Functional impact of phosphorylation at T787 in dEAG channel.
Representative current traces a) T787A dEAG channel, b) T787A channel + activated CaMKII, c) T787A channel + inactivated CaMKII and d) T787A channel + CaMKII inhibitory peptide evoked by a series of depolarizing potential ranging from −100 mV to +60 mV in 10 mV interval. e) Comparison of steady state current recorded at 60 mV among T787A channel alone and co-expressed with different regulators. Data are mean ± SEM and n=7–10
Strikingly, injection of the inhibitory peptide into the oocytes expressing the T787A channel mutant caused an accentuated reduction on the steady-state current levels compared with the wild type channel (cf. Figure 7d and 6d), down to 20% relative to non-injected oocytes (Figure 7e). This effect was accompanied by a clear increase in the extent of inactivation, with a small change in the time constant of inactivation (Figure S6). This mirrors the effect seen above for the double mutant channel, where alterations in recognition sequence altered the impact of the inhibitory peptide in the activity of the channel. This suggests that changes in the channel’s recognition sequence alter the functional relationship between the channel and CaMKII, supporting therefore a model where the kinase plays a role in the activity of the channel and the recognition sequence is involved in the formation of the CaMKII complex together with other channel and kinase regions. More work will be required to fully resolve the role of CaMKII catalytic activity in dEAG function and develop a comprehensive picture of all structural contacts between the two proteins.
Impact of T787 phosphorylation on complex stability
The tight interaction between the kinase domain and its recognition sequence in the dEAG potassium channel, together with the lack of a large functional impact of phosphorylation at T787 in the channel raises the possibility that the CaMKII recognition sequence in dEAG does not have a channel modulatory role but serves as a docking site for the kinase. In this perspective it is worthwhile considering the possibility that phosphorylation at T787 modulates the strength of the interaction between kinase and dEAG.
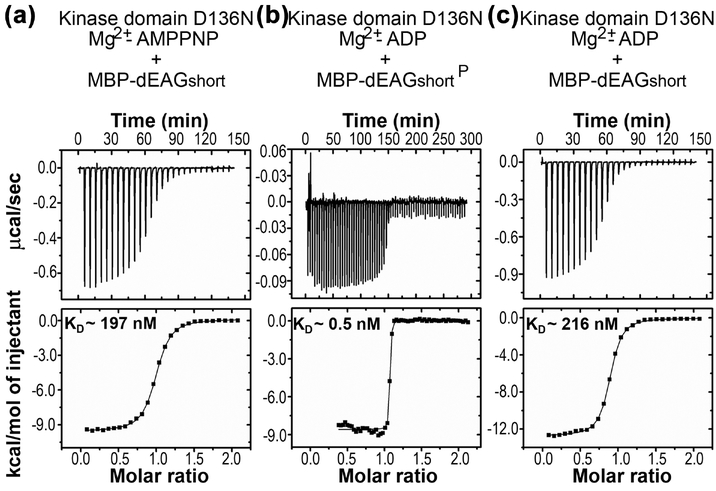
To test this idea we determined whether the stability of the complex changes during the phosphorylation reaction by comparing the affinity of the kinase for substrate in the presence of Mg2+-AMPPNP (ATP analog that inhibits the phosphorylation reaction) with that of the kinase for product in the presence of Mg2+-AMPCP (an ADP analog that inhibits the reverse catalytic reaction). The stability of the kinase-substrate complex was determined (KD of ~90 nM) by titrating unphosphorylated dEAGshort into wild type kinase domain, in the presence of Mg2+-AMPPNP (Table 2 and Figure 2c).
To measure the stability of the kinase-product complex we titrated phosphorylated dEAGshort (dEAGshortP) into wild type kinase domain (residues 1–283), in the presence of Mg2+-AMPCP42; 43; 44. The phosphorylated channel fragment at T787 was generated by incubating MBP-dEAGshort fusion protein with wild type kinase in the presence of Mg2+-ATP, followed by purification of the MBP fusion. Mass spectrometry analysis showed close to 100% phosphorylation of channel fragment. However, despite the use of the ADP analog to reduce the backward catalytic reaction, mass spectrometry analysis of the final mixture in the ITC experiment revealed that the channel fragment was dephosphorylated during the titration (Figure S7). Previous reports45; 46 have shown that CaMKII can promote dephosphorylation through the reverse reaction but no phosphorylated AMPCP was detected, suggesting that the loss of phosphoryl group from the channel fragment was due to kinase-mediated hydrolysis.
To avoid dephosphorylation of dEAGshortP by the kinase, we repeated the calorimetric titrations with partially inactive kinase domain (residues 1–283) which has the D136N active site mutation30; 47. We first measured the interaction of unphosphorylated dEAGshort with the kinase mutant in the presence of Mg2+-AMPPNP and obtained a KD of ~200 nM which is similar to the value determined for the wild type kinase domain (KD of ~90 nM), indicating that the mutation does not greatly alter the interaction with the channel fragment (Figure 8a, Table 2,). Strikingly, titration of dEAGshortP into the kinase mutant, in the presence of Mg2+-ADP, revealed a very tight association with the KD estimated to be ~0.5 nM, 260-fold tighter (Figure 8b, Table 2). Mass spectrometry analysis of the final mixture of an ITC experiment with the mutant kinase showed that the channel fragment remained fully phosphorylated (Figure S8). We also analyzed the interaction of mutant kinase domain with unphosphorylated fragment in the presence of Mg2+-ADP and obtained a KD ~220 nM (Figure 8c, Table 2), supporting the idea that it is the presence of a phosphoryl group in the channel fragment that causes a tightening of the interaction with the kinase.
Figure 8: ITC of phosphorylation effect on stability of kinase/channel fragment complex.
a) Titration of dEAGshort into the partially inactive form of CaMKII kinase domain mutant D136N in the presence of Mg2+-AMPPNP. b) Titration of dEAGshortP into CaMKII kinase domain mutant D136N in the presence of Mg2+-ADP. c) Titration of dEAGshort into CaMKII kinase domain mutant D136N in the presence of Mg2+-ADP. Titration injection heats are shown in the upper panel and binding isotherm of integrated binding enthalpies in the lower panel. KD values are shown for each mutant in lower panels.
Although generated with a mutant kinase, these data suggest that phosphorylation of T787 modulates the strength of the interaction between the kinase and the recognition sequence. Loss of the phosphoryl group reduces the strength of the interaction. Importantly, this is consistent with our electrophysiological findings above as the T787A channel, which cannot be phosphorylated, is especially susceptible to the competitive effects of the inhibitory peptide, as seen in the increased extent of inactivation for T787A relative to the wild type channel (cf. Figures 6d and 7d).
Structure of the kinase domain with phosphorylated channel fragment
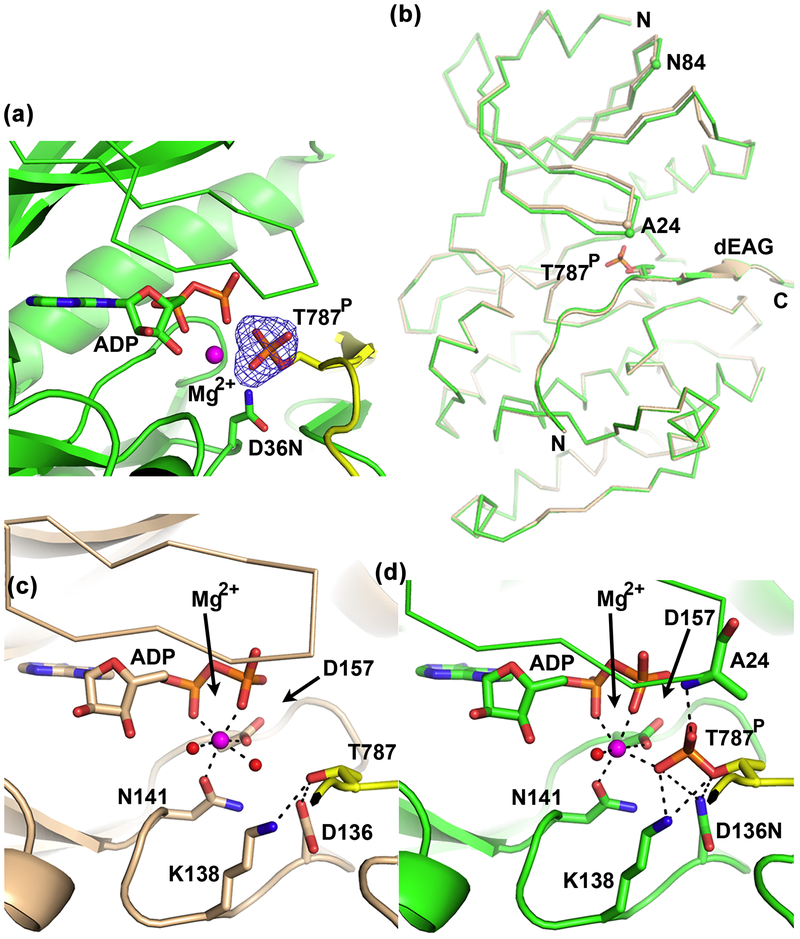
To understand the basis for the apparent increased stability of the kinase bound to phosphorylated channel fragment, we determined the structure of kinase domain D136N mutant (residues 1–283) bound to phosphorylated dEAGlong channel fragment (dEAGlongP) and Mg2+-ADP at better than 2 Å (Table S1). The electron-density map clearly shows the phosphate group in T787 of the channel fragment (Figure 9a).To assemble this complex with stably phosphorylated channel fragment we made use of the low levels of kinase activity displayed by the D136N mutant; the presence of the phosphoryl group in the structure suggests that the mutation has a larger impact on the reverse reaction relative to the forward reaction. Comparison of this structure with the structure described above of wild-type kinase bound with unphosphorylated dEAGlong and Mg2+-ADP shows that the kinase is still in the active conformation with the channel fragment bound in the same position. There is just a slight closing of the kinase N-lobe over the active site (Figure 9b), with Cα of A24 and N84 in the kinase N-lobe moving by 0.9 Å and 1.1 Å, respectively.
Figure 9: Structural changes in active site during phosphorylation of dEAG.
a) View of active site of CaMKII kinase domain D136N mutant (residues 1–283) bound to phosphorylated dEAGlong with Mg2+-ADP. Electron-density omit map for phosphoryl group in T787 shown as mesh; kinase shown in green, channel fragment in yellow, D136N shown as stick; b) Superposition of wild type CaMKII kinase bound to dEAGlong with kinase domain D136N bound to phosphorylated dEAGlong, both in the presence of Mg2+-ADP. Wild type (wheat) and mutant (green) domains shown as ribbon and channel fragments as cartoon with same color. Cα of A24 and N84 in both structures are shown as spheres. Detailed view of interactions between active-site residues and channel fragment in c) Wild type CaMKII kinase domain bound with dEAG and Mg2+-ADP and d) CaMKII kinase domain D136N mutant bound to dEAGlongP and Mg2+-ADP. Interactions described in main text are shown as dashed lines. Water molecules and Mg2+ are shown as red and magenta spheres, respectively.
The active site pockets of the two kinase domain structures, with unphosphorylated (representing the kinase-substrate complex) or with phosphorylated (representing the kinase-product complex) channel fragments, show interesting structural differences that result from the transfer of the phosphoryl group. In the structure of the substrate-complex (Figure 9c) the side-chain of dEAG-T787 is oriented towards the nucleotide by hydrogen bonding between the threonine hydroxyl group and the side-chains of two residues in the active site, D136 (O-O distance: 2.8 Å) and K138 (N-O distance: 3.1 Å). In this structure, the single Mg2+ is coordinated by N141, D157, the α- and β-phosphates of ADP and 2 waters. In the structure of product complex (Figure 9d) the interactions of T787 with N136 and K138, seen in the substrate complex, are maintained (2.9 Å and 3.3 Å bond-lengths, respectively). However, there is also a new set of interactions formed by the phosphoryl group in T787 with side-chains of N136 (O-N distance: 3.6 Å) and K138 (O-N distance: 2.8 Å) and main-chain of A24 (O-N distance: 2.6 Å). Additionally, the phosphoryl group coordinates the Mg2+ (O-Mg2+ distance: 2.4 Å), replacing one of the water molecules seen in the unphosphorylated dEAG structure.
Thus, the structure mimicking the kinase-product complex displays an extra set of interactions linking the channel fragment to the kinase that is established by the phosphoryl group. We suggest that these interactions underlie the increased stability of the kinase-product complex relative to the kinase-substrate complex observed in the ITC experiments, further supporting our proposal that phosphorylation of T787 in the CaMKII recognition sequence modulates the strength of the interaction between the kinase and the channel.
Discussion
In this study we have characterized the interaction between the kinase domain of CaMKII and a C-terminal fragment of the Drosophila EAG (dEAG) channel which spans the kinase recognition sequence. In particular we have found that this interaction displays the general features found in a kinase-substrate complex, with the channel fragment interacting with the active site of the kinase. The interaction is unusually tight for a kinase and its substrate, with a long interface between the two protein components, determined not only from specific contacts established by channel side-chains but also from contacts established by channel main-chain atoms and by water-mediated interactions. We also found that catalytically active CaMKII is required to observe active dEAG channels on the oocyte membrane but, interestingly, this does not involve phosphorylation of T787, a residue in the kinase recognition sequence. Instead, our results indicate that phosphorylation at T787, previously implicated in dEAG function, enhances the interaction between the channel fragment and kinase.
A question arising from our study is, what makes the CaMKII kinase domain-dEAG fragment an unusual kinase-substrate complex? Although there are many structures of protein kinases, structures of Ser/Thr protein kinases in complex with their cognate peptide substrates are not abundant probably because this interaction is typically of low affinity (KD ~ 200 μM;27). We analyzed the following kinase structures bound to high affinity substrates or high affinity inhibitory peptides: phosphorylase kinase (PHK) with a 7 residue optimal substrate (2PHK;); protein kinase B (PKB) in complex with a 10 residue substrate peptide derived from glycogen synthetase kinase 3β (GSK3β) (1O6K, 1O6L); protein kinase A (PKA) with a 20 residue protein kinase inhibitor (PKI) (1ATP); PKA with a PKI-derived substrate peptide (SP20) (1JBP, 1JLU). We also analyzed structures of CaMKII kinase bound in trans to the regulatory segment (RS) from another CaMKII subunit (3KK8, 2WEL) or in complex with the inhibitor CaMKII-Ntide (3KL8). In addition, we included in our analysis an example of a “normal affinity” peptide substrate, the structure of cyclin-dependent kinase 2 (CDK2)-cyclin A3 (1QMZ); this complex was obtained from co-crystallization of the kinase with large concentrations of a low affinity peptide substrate.
Comparison of these structures reveals that many of the side-chain interactions described for the CaMKII kinase domain bound the dEAG fragment (Figure 3) are specific to this kinase-substrate pair and not present in other kinase-substrate pairs. In contrast, the buried surface area for some of the high-affinity kinase-peptide complexes, around 1000 Å2, are comparable to that found in CaMKII-dEAG (~945 Å2). In a few high affinity structures, 2PHK and 1O6K, where the substrates are shorter (7 and 10 residues long, respectively), the buried surface area is smaller, ~600–700 Å2. However, for the low-affinity complex CDK2-cyclin A3-peptide substrate (7 residues long) the buried surface area is even smaller (~470 Å2). In addition, the high-affinity substrate structures display a main-chain hydrogen bond network equivalent to the one in CaMKII-dEAG and shown in Figure 4b; this network is absent from the low-affinity substrate structure. A general feature of the structures analyzed with high enough resolution is the presence of water molecules in the kinase-substrate interface and mediating interactions between the pair of molecules. This characteristic is independent of the kinase-substrate affinity. We can conclude that the combination of structural features described for the dEAG-CaMKII are also present in other high affinity kinase-substrate complexes, probably underlying the unusually tight interaction as they are absent from the “more common” low affinity complex structure.
What are the functional consequences of the tight interaction between the kinase domain and the channel? First, from the point of view of the kinase it has been demonstrated that the interaction with the channel fragment changes the properties of full-length CaMKII, giving rise to a sustained Ca2+/calmodulin-and autophosphorylation-independent activation of the kinase23. We have now shown that the interaction also reduces the rate of ATP hydrolysis in a single kinase domain. This observation can be explained by a slowing down of the catalytic turnover due to the tight binding of the channel fragment to the substrate binding-site in the kinase and the stabilization of the closed active site. The two outcomes of the tight interaction between the CaMKII and the dEAG channel fragment, sustained activation and reduction in catalytic activity, are compatible. Interaction between a channel kinase-recognition sequence with a kinase domain in the dodecameric CaMKII reduces catalytic activity of that particular domain while competing the kinase regulatory segment. This is likely to cause a rearrangement of the dodecamer, destabilize the inhibited state of other kinase domains in CaMKII and result in Ca2+-calmodulin and autophosphorylation independent activity.
Second, from the point of view of the channel we have shown that a catalytically active kinase is required for the display of channel activity on the membrane. The basis for this effect is not yet clear and it might result from impact of the kinase on the functional properties of the channel, on the levels of channel protein in the membrane or on the folding/processing of channel in the membrane or from a combination of these. Further studies are needed to dissect this effect. This finding contrasts with previous findings of the Griffith lab, where they found that T787A modestly increased the rate of channel inactivation and decreased channel current12. At present we do not have an explanation for the discrepancy.
What then is the role of the interaction between the kinase domain of CaMKII and its recognition sequence in the dEAG channel? Other proteins that harbor amino acid sequences similar to the CaMKII-binding region of dEAG provide clues about this question (Figure 1c). Such a sequence is present in the C-terminus of β1 and β2 subunits of human voltage-dependent calcium channel and has been shown to play a role in the formation of stable complexes between CaMKII and the calcium channel. This interaction, together with β2a phosphorylation, is required for the facilitation of L–type Ca2+ channels48; 49. CaMKII was described as a scaffold for proteasome recruitment to dendritic spines50 and was found to phosphorylate Ser120 (at the minimal R-x-x-S motif) of the Rpt6 regulatory subunit and concurrently enhance proteasome activity. A segment resembling the recognition sequence in dEAG but with a non-phosphorylatable residue (Arg instead of Thr/Ser, as in the CaMKII-Ntide inhibitor) can be found in the 26S proteasome p45/Rpt6 subunit. This protein segment is distal from the phosphorylation site and appears occluded in the structure of the fully assembled 26S proteasome particle. Nevertheless, it is tempting to envisage the identified segment playing a role in the interaction of CaMKII with the proteosome in some specific conformation.
Crucially, there is the well described interaction between CaMKII and the NMDA receptor through a C-terminal sequence that is very similar to the one studied here51. The interaction between the NMDA receptor subunit GluN2B and CaMKII is an important element of the long term potentiation mechanism at the synapse52; 53. There is a strong similarity in the properties of the CaMKII/dEAG channel complex and the CaMKII/NMDA receptor. Both channels contain CaMKII recognition sequences in their cytoplasmic C-terminal regions, forming very stable complexes with CaMKII which give rise to Ca2+/calmodulin- and autophosphorylation-independent activity of the kinase24; 54. The interaction between the two channels and CaMKII has a role in learning and memory formation11; 13; 14. In flies, the dEAG channel and CaMKII are localized to the synaptic compartment and both proteins are involved in a pathway that regulates neuronal plasticity and memory formation. In mammals, long-term potentiation (LTP) depends on the establishment and long-term stability of the interaction between CaMKII and the NMDA receptor at the post-synaptic density, where this interaction is a defining molecular event of synaptic plasticity that leads to enhanced synaptic strength.
These parallels strongly suggest that the interaction between the two ion channels and CaMKII follows similar rules and has related biochemical purposes. To explain the functional relationship between CaMKII and the NMDA receptor it has been proposed that the channel acts as a docking/recruiting platform for the kinase13; 14. Together with the demonstration that CaMKII and dEAG form a complex in Drosophila23 this leads us to propose that the dEAG channel is a docking or recruiting platform for CaMKII, where the contacts mediating the channel/kinase complex include the interaction between the kinase domain and the recognition sequence seen in our structures. Moreover, our data suggests that the role of phosphorylation at T787 is to modulate the strength of the interaction between the kinase and the channel. We found an apparent increased stability of the kinase/fragment complex upon phosphorylation of T787 and a competitor peptide has an enhanced functional effect on the mutant T787A channel relative to the wild type channel. Importantly, we reveal structural data that explain the stabilization effect of T787 phosphorylation by the formation of extra interactions between the phosphorylated channel fragment and the kinase. Overall, these results are consistent with a scenario where phosphorylation of the kinase recognition sequence in the protein does not alter the function of the channel but instead alters the action of the kinase by stabilizing its location.
Our functional results also suggest that the interaction between CaMKII and the full-length channel involves other contacts besides those characterized here. We speculate that the other interacting regions include stretches in the long disordered C-terminal region of the dEAG channel since these are the amino acid regions that differ the most between dEAG and either the rat EAG or hERG channels, both of which are not known to interact with CaMKII. Overall, to fully understand the functional and structural impact of CaMKII in the dEAG channel further work will be required.
The amino acid sequence similarity identified in the C-terminal stretches of dEAG and GluN2B subunit of the NMDA receptor involved in the interaction with CaMKII54 (Figure 1c) also strongly suggests that this region of GluN2B binds to the kinase domain of CaMKII in a similar manner to that observed in our structures, with the GluN2B region interacting as a substrate. In addition, the similarities also suggest that phosphorylation of the recognition sequence in GluN2B will lead to a tightening of the interaction with the kinase as observed for dEAG. In fact, phosphorylation of the GluN2B recognition sequence has been shown to alter its interaction with CaMKII but these results have been interpreted as indicating a destabilization of the complex upon phosphorylation55; 56. However, these experiments were performed differently from ours and do not directly explore the role of phosphorylation on the stability of the kinase/fragment complex formed at the end of the catalytic reaction, with ADP bound in the active site.
Faced with the multiple occurrences of CaMKII protein complexes where the kinase catalytic site appears to be an important determinant of the interaction it is worthwhile considering that CaMKII, like other protein kinases, may also have non-catalytic functions57. In the particular case of the dEAG channel, a simple consideration of the oligomeric structures of the channel and CaMKII together with the well-established dynamic nature of the structure of CaMKII raises two intriguing possibilities (Figure 1). First, the interaction between these two proteins may involve avidity and clustering effects. The EAG potassium channel is a homotetramer with four CaMKII recognition sites per channel, and CaMKII is a dodecamer (and sometimes a tetradecamer) and therefore the interaction of four kinase domains with the four channel subunits will result in increased stability of the complex through an avidity effect. Second, the dynamic architecture of CaMKII, where the 12 kinase domains project from the hub domain and display a “wing span” that can vary from 200 Å to 320 Å (Figure 1)18, naturally suggests the possibility that activated kinase can bind multiple channels simultaneously and promote clustering of EAG channels. Future studies will be required to assess the role of this interaction on the organization of the synaptic membrane.
Materials and Methods
Materials
Drosophila full-length CaMKII (R3 isoform;58) was a gift of Leslie Griffith (Brandeis University, Massachusetts, U.S.A.) while full-length Drosophila EAG (GenBank: M61157) was a gift of Dianne Papazian (UCLA, California, U.S.A.). Nucleotides (ATP (Sigma), AMPPNP (Jena Biosciences), ADP (Sigma) and AMPCP (Sigma)) were processed according to manufactures’ instructions. Stock solutions at 50 mM of ATP, AMPPNP and ADP were freshly prepared in either 1 M Tris pH 7.5 or 1 M HEPES pH 7.5. Stock solution of AMPCP at 50 mM in 1 M HEPES pH 7.5 was stored at −20 °C. λ-phosphatase (NEB) was processed according to manufacturer’s instructions, aliquoted and stored at −80 °C. Stock solutions of reagents used in the ATPase assay were prepared as follows: phosphoenolpyruvate (PEP; Sigma) at 50 mM in 50 mM HEPES pH 7.5; nicotinamide adenine dinucleotide, reduced (NADH; Sigma) at 6 mM in 50 mM HEPES pH 7.5; peptide syntide (GenScript) was prepared in water at 2.5 mM. All these solutions were kept aliquoted at −80 °C. The pyruvate kinase/lactate dehydrogenase mix (Sigma) was at 600–1000 units ml−1 and 900–1400 units ml−1 activity, respectively, and kept at −20 °C.
Cloning, expression and purification of the kinase domain of Drosophila CaMKII
CaMKII kinase domain (residues 1 to 283) was cloned into pETM-11 expression vector and overexpressed in the E. coli BL21(DE3) strain. Cells were grown at 37 °C in rich medium (20 g/L tryptone, 10 g/L yeast extract, 5 g/L glycerol, 50 mM K2HPO4, 10 mM MgCl2, 10 g/L glucose)59 with kanamycin; after induction with 350 μM IPTG growth proceeded overnight at 20 °C. Cells were lysed in buffer A (50 mM HEPES pH 7.5, 300 mM NaCl) with protease inhibitors. Cleared lysate was loaded into Co2+ Talon metal affinity resin (Clontech) in buffer A with 5 mM imidazole. Protein-bound resin was washed with high-salt buffer (50 mM HEPES pH 7.5, 500 mM NaCl) and buffer B (50 mM HEPES pH 7.5, 150 mM NaCl) with 12.5 mM imidazole and eluted with buffer B with 125 mM imidazole. Protein, with TEV protease, was dialysed at 4 °C overnight against buffer B with 1 mM DTT and then passed through a Ni2+ affinity gel to ensure removal of the 6xHis tag. Protein was further purified by size exclusion chromatography (Superdex 75 column) and ion-exchange chromatography (HiTrap Q HP, GE Healthcare). The Superdex column was in buffer C (25 mM HEPES pH 7.5, 150 mM KCl, 0.5 mM TCEP); protein loaded into the Hitrap Q column was in 50 mM HEPES pH 8.0, 50 mM KCl, 0.5 mM TCEP and eluted with a 50 mM – 1M KCl gradient. Protein batches used for crystallization experiments were treated with λ phosphatase (NEB) to remove auto-phosphorylation modifications of CaMKII before the final step of ion-exchange chromatography.
Preparation of the kinase D136N mutant had the following differences relative to the procedure described above. Cells were grown in LB medium with kanamycin and induced with 1 mM IPTG. Cell lysis was carried out in buffer D (50 mM Tris pH 7.5, 150 mM NaCl) supplemented with protease inhibitors and the cleared lysate was loaded into Ni2+ resin (Sigma) in buffer D containing 7.5 mM imidazole. Protein loaded beads were washed with a higher salt buffer (50 mM Tris pH 7.5, 300 mM NaCl), followed by buffer D supplemented with 15 mM imidazole. Protein was eluted with buffer D containing 200 mM imidazole and cleaved with TEV protease upon dialysis overnight at 4 °C against buffer D supplemented with 1 mM DTT. Protein was further purified by size exclusion chromatography with a Superdex 75 (GE Healthcare) column in 25 mM Tris pH 7.5, 150 mM KCl, 2.5 mM DTT after passage through a Ni2+ resin (Sigma) to ensure removal of the 6xHis tag.
Preparation of the kinase K43M/D136N mutant had the following differences relative to the procedure described above. Cells were grown in LB medium with kanamycin and induced with 1 mM IPTG. Cells were lysed in buffer B containing protease inhibitors and cleared lysate was loaded into Co2+ Talon metal affinity resin (Clontech) in buffer B supplemented with 7.5 mM imidazole. The protein-bound resin was washed with a higher salt buffer (buffer A) and then buffer B containing 15 mM imidazole. Protein was eluted with buffer B supplemented with 200 mM imidazole and cleaved with TEV protease at 4 °C overnight upon dialysis against 25 mM HEPES pH 7.5, 150 mM NaCl containing 1 mM DTT. Protein was further purified by size exclusion chromatography and ion-exchange chromatography as described above for the wild-type protein.
Cloning, expression, purification and modification of the CaMKII-binding fragments from Drosophila EAG
The coding sequence for the wild-type Drosophila EAG CaMKII-binding fragment (residues 768 to 820) was fused to Maltose-binding protein (MPB) with a linker sequence coding for the TEV protease cleavage site and cloned into the MCS1 of pRSF-Duet vector, forming the construct 6xHis-MBP-TEVsite-dEAGlong. The fusion protein was over-expressed in the BL21(DE3) strain. Cells were grown in LB medium at 37 °C and induced overnight at 25 °C with 0.5 mM IPTG. Cells were lysed in buffer E (50 mM HEPES pH 7.5, 250 mM KCl) containing protease inhibitors and the cleared lysate was loaded into Co2+ Talon metal affinity resin. The protein-bound resin was washed with buffer E followed by high-salt buffer (50 mM HEPES pH 7.5, 500 mM KCl) and eluted with buffer E with 25 mM imidazole. Protein was loaded into an MBPtrap (GE Healthcare) affinity column in buffer F (50 mM HEPES pH 7.5, 150 mM KCl, 0.5 mM TCEP), washed with the same buffer, and eluted with buffer F with 10 mM maltose. Protein was further purified using an anion-exchange column (HiTrap Q HP, GE Healthcare) in 50 mM HEPES pH 7.5, 50 mM KCl, 0.5 mM TCEP and eluted with a 50 mM – 1M KCl gradient.
The purification protocol for the MBP-channel fragment fusion protein used for assembly with D136N CaMKII was slightly different. Cells were lysed in buffer G (50 mM Tris pH 7.5, 250 mM KCl) containing protease inhibitors and the cleared lysate was loaded into Ni2+ beads (Sigma). The protein-bound resin was washed with buffer G followed by high-salt buffer (50 mM Tris pH 7.5, 500 mM KCl) and eluted with buffer G with 25 mM imidazole. Protein was subsequently loaded into an MBPtrap affinity column in buffer H (50 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT), washed with the same buffer and eluted with buffer H with 10 mM maltose and followed by dialysis overnight at 04 °C against buffer I (20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT).
Channel fragment single-site mutants used in the alanine-scanning experiments were generated by site directed mutagenesis of a 6xHis-TEVsite-MBP-linker-dEAGlong template, where the TEV site described in the MBP-fragment fusion above was altered to a non-cleavable sequence and a new TEV site was introduced immediately after the 6xHis tag. Expression and purification of these mutant dEAG fragments were carried out as for the wild type dEAG fragment, except for the introduction of an extra step to cleave off the His tag. For this purpose, before the MBPtrap step the protein was cleaved with TEV protease at 4 °C overnight during dialysis against 50 mM HEPES pH 7.5, 150 mM KCl,1 mM DTT. Just before loading into the MBPtrap column the protein solution was incubated shortly with Co2+ Talon metal affinity resin (Clontech) to remove the cleaved-off His tag.
The dEAGshort channel fragment (residues 768 to 795) was cloned into pRSF-Duet MCS1 forming the construct 6xHis-MBP-TEVsite-dEAGshort. Two residues in the sequence were mutated (S778N and T781K) by site directed mutagenesis. The resulting fusion protein MBP-dEAGshort was overexpressed and purified as described for the original dEAGlong version.
Phosphorylated dEAGshort was prepared by incubating overnight, at 4 °C, a 1:50 molar ratio mix of CaMKII kinase domain:MBP-dEAGshort in 50 mM HEPES pH ~7.5, ~200 mM KCl, 0.5 mM TCEP, 20 mM MgCl2, 3 mM ATP. The reaction product MBP-dEAGshortP was purified by loading the final reaction mix in Superdex 75 size exclusion column (GE Healthcare) in buffer F. The dEAGshortP fusion protein was further purified by affinity chromatography to ensure complete removal of kinase. For that purpose the protein was loaded into an MBPtrap (GE Healthcare) affinity column (pre-washed in buffer F) in buffer F with 0.5 mM AMPPNP. The column was washed with a small volume of the same buffer containing AMPPNP and afterwards washed longer with buffer F, until absorbance dropped to baseline. The protein was eluted with buffer F with 10 mM maltose.
Preparation of the kinase/dEAG fragment complexes for crystallization
The kinase domain/dEAG/Mg2+-AMPPNP and kinase domain/dEAGP/Mg2+-ADP complexes were assembled by mixing the purified proteins MBP-dEAGlong and CaMKII kinase domain at a molar ratio of 1.5. Kinase domain was pre-incubated in 50 mM HEPES pH ~8.0, ~200 mM KCl, 20 mM MgCl2, 1 mM ATP or AMPPNP and then incubated with MBP-dEAGlong on ice for 3 h or 2 h, respectively, before cleaving overnight at 4 °C with TEV protease for removal of MBP. In the case of the ATP mix this incubation generated dEAGP; to replace remaining ATP by ADP the final reaction mix was supplemented with 3 mM ADP before cleaving with TEV. In both mixes, concentration of TCEP was adjusted to 1 mM before adding the protease. Cleaved-off MBP was removed by passing protein mixes through an MBPtrap (GE Healthcare) column, pre-equilibrated in buffer F supplemented with either 1 mM ADP or AMPPNP, according to the complex, and washing column with Buffer F with 1mM ADP or AMPPNP until 280 nm absorbance reached a baseline. To ensure complete elimination of any contaminating MBP, purified complexes were supplemented with 5 mM imidazole and briefly incubated with Co2+ talon metal affinity resin (Clontech) beads, which was removed using Spin-X spin columns (Corning, Costar). Protein complexes were concentrated to about 10 mg ml−1 and dialyzed overnight at 4 °C against buffer C supplemented with 5 mM MgCl2, 0.5 mM ADP or AMPPNP. Nucleotide concentration was adjusted to 0.8 mM after dialysis.
The kinase domainD136N/dEAGlongP/Mg2+-ADP complex was assembled by mixing the purified proteins at a 1.5 molar ratio of MBP-dEAGlong and partially inactive kinase domain. The mutant kinase domain was pre-incubated in 25 mM Tris pH 7.5, 150 mM KCl, 2.5 mM DTT, 20 mM MgCl2, 1 mM ATP and then incubated with MBP-dEAGlong on ice for 31/2 h, resulting in phosphorylated channel fragment. The reaction was stopped by adding 3 mM ADP before cleaving with TEV protease overnight at 4 °C. The cleavage reaction mix was subjected to dialysis, to remove DTT, in buffer J (50 mM Tris pH 7.5, 300 mM KCl, 10 mM MgCl2) containing 0.45 mM ADP. To eliminate cleaved-off MBP, the assembled complex solution, in the dialysis buffer including 3 mM imidazole, was loaded into Co2+ Talon metal affinity resin (Clontech) pre-equilibrated in buffer J and washed with buffer J supplemented with 0.25 mM ADP. The kinaseD136N/dEAGlongP/Mg2+-ADP complex was further purified by size exclusion chromatography in a Superdex 75 column (GE Healthcare) equilibrated with buffer I containing 5 mM MgCl2, 60 μM ADP. Immediately after the chromatography step, the ADP concentration in the protein complex fractions was adjusted to 0.5 mM. The purified complex was concentrated to about 10 mg ml−1, adjusting the final buffer composition to 20 mM Tris pH 7.5, 150 mM KCl, 5 mM DTT, 5 mM MgCl2, 0.8 mM ADP.
Crystallization of kinase domain/dEAG fragment complexes
Crystals were grown at 293 K by the vapor diffusion method in sitting drops with 0.85 μl protein solution and 0.85 μl precipitant solution. For the kinase/dEAGlong/Mg2+-ADP structure the protein solution of kinase domain and dEAGlongP at 9.4 mg ml−1 in buffer C and containing 5 mM MgCl2, 0.8 mM ADP were mixed with 26 % PEG 4000, 100 mM sodium citrate pH 5.0, 200 mM ammonium acetate. Crystals were transferred to drops of 28 % PEG 4000, 100 mM sodium citrate pH 5.0, 200 mM ammonium acetate, 20 mM MgCl2, 1 mM ADP, 5% glycerol and flash-cooled in liquid nitrogen. For the kinaseD136N/dEAGlongP/Mg2+-ADP structure, protein solution at 10.4 mg ml−1 in 20 mM Tris pH 7.5, 150 mM KCl, 5 mM DTT, 5 mM MgCl2, 0.8 mM ADP was mixed with 19 % PEG 4000, 100 mM sodium citrate pH 5.0, 200 mM ammonium acetate. Crystals were transferred to drops of 25 % PEG 4000, 100 mM sodium citrate pH 5.0, 200 mM ammonium acetate, 5 mM MgCl2, 1 mM ADP, 5% glycerol and flash-cooled in liquid nitrogen. For the kinase/dEAGlong/Mg2+-AMPPN structure, protein solution was at 8.5 mg ml−1 in buffer C supplemented with 5 mM MgCl2, 0.8 mM AMPPNP and mixed with 26 % PEG 4000, 100 mM sodium citrate pH 5.0, 200 mM ammonium acetate. Crystals were transferred to drops of 28 % PEG 4000, 100 mM sodium citrate pH 5.0, 200 mM ammonium acetate, 5 mM MgCl2, 1 mM AMPPNP, 5 % glycerol and flash-cooled in liquid nitrogen.
Structure determination
X-ray diffraction data were collected at 100 K at ESRF synchrotron on beamline ID 14–4 with an ADSC QUANTUM 315r detector. Diffraction data were also collected at SOLEIL synchrotron on beamline PROXIMA 2-A with an EIGER X-9M detector. Data were processed with XDS and CCP460; 61; 62. Structure was determined by molecular replacement with PHASER63 using as search model a CaMKII kinase domain structure model generated using the SWISS-MODEL server64. Structure was refined with Refmac561, PHENIX65 and COOT66.
Isothermal calorimetry
ITC data were obtained in a MicroCal VP-ITC calorimeter (Malvern). Before each titration interaction partners were dialyzed together in two steps: first, against buffer C, for 2 h at 4 °C; second, overnight at 4 °C against buffer C with 1 mM MgCl2 + 250 μM AMPPNP or ADP or AMPCP. The final buffer composition of both samples was adjusted to 5 mM MgCl2, 1 mM nucleotide. Titrations were performed at 25 °C, with different MBP-dEAG proteins as titrant into the cell containing wild-type and mutant CaMKII kinase domain protein (concentrations are indicated in Tables 1 and 2). In general, titrations involved a first injection of 2 μl into the apparatus cell, followed by 27 injections of 10 μl. For titrations involving partners with very high affinity, 69 injections of 4 μl were applied after a first one of 2 μl. Data were initially processed with the ORIGIN software (OriginLab) using the final injections to define dilution heat baseline. This initially processed data, without baseline correction, was posteriorly subjected to individual or global fitting analysis with the AFFINIMETER software (S4Sd - Software for Science Developments) to obtain the final fitted parameters (Tables 1 and 2). In some cases, the N parameter could not be fitted globally and had to be fitted separately for each titration run (Tables 1 and 2).
Kinase assay
ATPase activity of CaMKII kinase domain in the presence of the peptide substrates (syntide - PLARTLSVAGLPGKK and dEAGshort channel fragment) was monitored using a continuous spectrophotometric assay protocol derived from previously described assays35; 67; 68. In this coupled assay, ADP produced by the kinase upon phosphorylation of substrates is used in a parallel reaction to oxidize NADH, whose consumption results in a decrease of 340 nm absorbance. Assays were performed at 25 °C in a Synergy-2 microplate reader (BioTek) by measuring the decrease in absorbance at 340 nm at equal time intervals. Reaction mix consisted of: 100 mM HEPES pH 7.5, 150 mM KCl, 10 mM MgCl2, 0.5 mM TCEP, 1 mM ATP, 1 mM PEP, 0.3 mM NADH, 30–50 units ml−1/45–70 units ml−1 pyruvate kinase/lactate dehydrogenase, 250 μM syntide, different concentrations of MBP-dEAGshort and 35 nM CaMKII kinase domain. Reactions were started by addition of the kinase to mixes immediately before measurements, with 100 μl of final mix per plate well. Both CaMKII and MBP-dEAGshort were dialyzed against the same buffer C overnight at 4 °C. Initial velocities were extracted from kinetic data using GraphPad software (Prism) and plotted against the concentration of dEAG fragment. Plot was fitted with formula y = Vmax ∗ 1/1 + ([EAG]/IC50) + Vmin (where y is the initial reaction velocity and [EAG] corresponds to the concentration of dEAG fragment) using the ORIGIN software (OriginLab).
Oocyte isolation and cRNA injection
To remove the follicle cell layer, Xenopus laevis oocytes were treated with 75 μg/ml Liberase TM research-grade enzyme (Sigma-Aldrich) for 40–60 min in Ca2+-free ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES, pH adjusted to 7.4 with NaOH). Stage V-VI oocytes were injected with cRNA encoding proteins of interest and incubated for 1–5 d at 16 °C in storage solution before recording. Storage solution contained 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 1 mM HEPES, and 10 mg/L gentamycin, with pH adjusted to 7.4 with NaOH. For experiments with dEAG channels co-expressed with CaMKII, cRNA was pre-mixed and injected into the oocytes. To apply CaMKII inhibitory peptide, diluted inhibitory peptide was injected into oocytes expressing dEAG channels 2–4 hours before recording. The final concentration of diluted inhibitory peptide in the oocytes is estimated to be 80 μM.
Two-electrode voltage-clamp protocols and data analysis
Oocytes expressing channels of interest were subjected to a standard two-microelectrode voltage clamp. The chamber was perfused with recording solution (96 mM NaCl, 2 mM KCl, 0.3 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES, with pH adjusted to 7.4 with NaOH) throughout the experiment. An OC-725C amplifier (Warner Instruments), Digidata 1440A data acquisition system, and pCLAMP 9.0 software (Molecular Devices) were used to produce command voltages and record current and voltage signals. The resistance of microelectrodes filled with 3 M KCl ranged from 0.2 to 0.6 MΩ. Experiments were performed at room temperature.
Amplitude measurements were taken at the end of the steady-state currents observed during the test pulses to the indicated voltages. Activation and inactivation time constants were determined by fitting traces with two exponentials. Normalization was carried out as indicated in Fig. 6 legend.
Mass spectrometry
Proteins and channel fragments were analyzed by Liquid Chromatography - High Resolution Mass Spectrometry. Experiments were performed on an UltiMate 3000 RSLC System (Dionex, Germering, Germany) connected to a 2-GHz maXis ultra-high resolution quadrupole-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with an electrospray ion source. The LC-MS setup was controlled by the Bruker HyStar™ software version 3.2. Proteins were on-line desalted on a Waters MassPREP™, 2.1 × 10 mm, phenyl 1000 Å reverse-phase cartridge and eluted at a flow rate of 500 μL/min using a 5 to 90% gradient of acetonitrile in 0.1% formic acid. The amount of injected protein was between 2 and 3 μg.
ElectroSpray Ionization - High Resolution Mass Spectrometry (ESI-HRMS) were acquired in positive ion MS mode over a 900–5000 m/z range with a nebulizer gas pressure of 2.0 bars. The drying gas flow was 9 liter/min, and the temperature was 200 °C. The ion source collision induced dissociation (isCID) parameter was adjusted at 70 eV to promote the observation of protonated forms of protein and limit the noncovalent adducts. The acquisition rate was 1 Hz corresponding to spectra summations of 5000. External calibration was performed with the ESI-L Low Concentration Tuning Mix (Agilent Technologies, Palo Alto, CA). Mass spectra were processed and charge-deconvoluted using DataAnalysis 4.0 software (Bruker Daltonics) and the MaxEnt algorithm.
Supplementary Material
Highlights:
The dEAG channel forms a complex with the CaMKII kinase in neuron synapses
Crystal structure shows the channel fragment bound to the active-site of kinase
Interaction is very tight for a kinase/protein-substrate pair (~200-fold tighter)
Observation of active dEAG channels requires catalytically active CaMKII
Phosphorylation at T787 in dEAG modulates the interaction with the kinase
Acknowledgments
We thank the i3S scientific platforms: Biochemical and Biophysical Technologies and Biointerfaces and Nanotechnology for technical support. We thank access to ESRF (ID14–4) and Soleil (PROXIMA1 and 2A) for collection of X-ray diffraction data. This work was supported by: Norte-01–0145-FEDER-000008 “Porto Neurosciences and Neurologic Disease Research Initiative at I3S”, a Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER); “Institute for Research and Innovation in Health Sciences” (POCI-01–0145-FEDER-007274) supported by FEDER and COMPETE 2020, Operational Program for Competitiveness and Internationalization (POCI), Portugal 2020, and Portuguese funds through FCT, Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Inovação; Marie Curie International Reintegration Grant (FP7-PEOPLE-IRG-2008: proposal 239251-StrucPer); SyMBioMS (FEDER #2699–33931), Region Centre Val de Loire, Université d’Orléans and CNRS for HRMS equipment, and by NIH RO1 grant NS081320.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Protein data bank accession numbers: 5FG8 - kinase domain with dEAG channel fragment and ADP-Mg2+; 5H9B - kinase domain with dEAG channel fragment and AMPPN-Mg2+; 5HU3 – D136N kinase domain mutant with phosphorylated dEAG channel fragment and ADP-Mg2+.
References
- 1.Crociani O, Guasti L, Balzi M, Becchetti A, Wanke E, Olivotto M, Wymore RS & Arcangeli A (2003). Cell cycle-dependent expression of HERG1 and HERG1B isoforms in tumor cells. J Biol Chem 278, 2947–55. [DOI] [PubMed] [Google Scholar]
- 2.Sanguinetti MC & Tristani-Firouzi M (2006). hERG potassium channels and cardiac arrhythmia. Nature 440, 463–9. [DOI] [PubMed] [Google Scholar]
- 3.Pardo LA & Stuhmer W (2014). The roles of K(+) channels in cancer. Nat Rev Cancer 14, 39–48. [DOI] [PubMed] [Google Scholar]
- 4.Ganetzky B, Robertson GA, Wilson GF, Trudeau MC & Titus SA (1999). The eag family of K+ channels in Drosophila and mammals. Ann N Y Acad Sci 868, 356–69. [DOI] [PubMed] [Google Scholar]
- 5.Vandenberg JI, Perry MD, Perrin MJ, Mann SA, Ke Y & Hill AP (2012). hERG K(+) channels: structure, function, and clinical significance. Physiol Rev 92, 1393–478. [DOI] [PubMed] [Google Scholar]
- 6.Cazares-Ordonez V & Pardo LA (2017). Kv10.1 potassium channel: from the brain to the tumors. Biochem Cell Biol 95, 531–536. [DOI] [PubMed] [Google Scholar]
- 7.Cui ED & Strowbridge BW (2018). Modulation of Ether-a-Go-Go Related Gene (ERG) Current Governs Intrinsic Persistent Activity in Rodent Neocortical Pyramidal Cells. J Neurosci 38, 423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warmke JW & Ganetzky B (1994). A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci U S A 91, 3438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morais-Cabral JH & Robertson GA (2015). The enigmatic cytoplasmic regions of KCNH channels. J Mol Biol 427, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warmke J, Drysdale R & Ganetzky B (1991). A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science 252, 1560–2. [DOI] [PubMed] [Google Scholar]
- 11.Griffith LC, Wang J, Zhong Y, Wu CF & Greenspan RJ (1994). Calcium/calmodulin-dependent protein kinase II and potassium channel subunit eag similarly affect plasticity in Drosophila. Proc Natl Acad Sci U S A 91, 10044–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Wilson GF & Griffith LC (2002). Calcium/calmodulin-dependent protein kinase II phosphorylates and regulates the Drosophila eag potassium channel. J Biol Chem 277, 24022–9. [DOI] [PubMed] [Google Scholar]
- 13.Herring BE & Nicoll RA (2016). Long-Term Potentiation: From CaMKII to AMPA Receptor Trafficking. Annu Rev Physiol 78, 351–65. [DOI] [PubMed] [Google Scholar]
- 14.Kim K, Saneyoshi T, Hosokawa T, Okamoto K & Hayashi Y (2016). Interplay of enzymatic and structural functions of CaMKII in long-term potentiation. J Neurochem 139, 959–972. [DOI] [PubMed] [Google Scholar]
- 15.Kolodziej SJ, Hudmon A, Waxham MN & Stoops JK (2000). Three-dimensional reconstructions of calcium/calmodulin-dependent (CaM) kinase IIalpha and truncated CaM kinase IIalpha reveal a unique organization for its structural core and functional domains. J Biol Chem 275, 14354–9. [DOI] [PubMed] [Google Scholar]
- 16.Morris EP & Torok K (2001). Oligomeric structure of alpha-calmodulin-dependent protein kinase II. J Mol Biol 308, 1–8. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya M, Stratton MM, Going CC, McSpadden ED, Huang Y, Susa AC, Elleman A, Cao YM, Pappireddi N, Burkhardt P, Gee CL, Barros T, Schulman H, Williams ER & Kuriyan J (2016). Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers JB, Zaegel V, Coultrap SJ, Miller AP, Bayer KU & Reichow SL (2017). The CaMKII holoenzyme structure in activation-competent conformations. Nat Commun 8, 15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao LH, Stratton MM, Lee IH, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H & Kuriyan J (2011). A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin-dependent kinase II holoenzyme. Cell 146, 732–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunkley PR (1991). Autophosphorylation of neuronal calcium/calmodulin-stimulated protein kinase II. Mol Neurobiol 5, 179–202. [DOI] [PubMed] [Google Scholar]
- 21.Hanson PI & Schulman H (1992). Neuronal Ca2+/calmodulin-dependent protein kinases. Annu Rev Biochem 61, 559–601. [DOI] [PubMed] [Google Scholar]
- 22.Stratton MM, Chao LH, Schulman H & Kuriyan J (2013). Structural studies on the regulation of Ca2+/calmodulin dependent protein kinase II. Curr Opin Struct Biol 23, 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun XX, Hodge JJ, Zhou Y, Nguyen M & Griffith LC (2004). The eag potassium channel binds and locally activates calcium/calmodulin-dependent protein kinase II. J Biol Chem 279, 10206–14. [DOI] [PubMed] [Google Scholar]
- 24.Bayer KU, De Koninck P, Leonard AS, Hell JW & Schulman H (2001). Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature 411, 801–5. [DOI] [PubMed] [Google Scholar]
- 25.Wang W & MacKinnon R (2017). Cryo-EM Structure of the Open Human Ether-a-go-go-Related K+ Channel hERG. Cell 169, 422–430 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whicher JR & MacKinnon R (2016). Structure of the voltage-gated K(+) channel Eag1 reveals an alternative voltage sensing mechanism. Science 353, 664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endicott JA, Noble ME & Johnson LN (2012). The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem 81, 587–613. [DOI] [PubMed] [Google Scholar]
- 28.Kornev AP, Haste NM, Taylor SS & Eyck LF (2006). Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A 103, 17783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornev AP, Taylor SS & Ten Eyck LF (2008). A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A 105, 14377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg OS, Deindl S, Sung RJ, Nairn AC & Kuriyan J (2005). Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell 123, 849–60. [DOI] [PubMed] [Google Scholar]
- 31.Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS & Sowadski JM (1993). Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry 32, 2154–61. [DOI] [PubMed] [Google Scholar]
- 32.Bossemeyer D, Engh RA, Kinzel V, Ponstingl H & Huber R (1993). Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5–24). EMBO J 12, 849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White RR, Kwon YG, Taing M, Lawrence DS & Edelman AM (1998). Definition of optimal substrate recognition motifs of Ca2+-calmodulin-dependent protein kinases IV and II reveals shared and distinctive features. J Biol Chem 273, 3166–72. [DOI] [PubMed] [Google Scholar]
- 34.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T & Cantley LC (1996). A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol 16, 6486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao LH, Pellicena P, Deindl S, Barclay LA, Schulman H & Kuriyan J (2010). Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation. Nat Struct Mol Biol 17, 264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang BH, Mukherji S & Soderling TR (1998). Characterization of a calmodulin kinase II inhibitor protein in brain. Proc Natl Acad Sci U S A 95, 10890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanson PI, Meyer T, Stryer L & Schulman H (1994). Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron 12, 943–56. [DOI] [PubMed] [Google Scholar]
- 38.Rich RC & Schulman H (1998). Substrate-directed function of calmodulin in autophosphorylation of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem 273, 28424–9. [DOI] [PubMed] [Google Scholar]
- 39.Fukunaga K, Miyamoto E & Soderling TR (1990). Regulation of Ca2+/calmodulin-dependent protein kinase II by brain gangliosides. J Neurochem 54, 103–9. [DOI] [PubMed] [Google Scholar]
- 40.Colbran RJ, Smith MK, Schworer CM, Fong YL & Soderling TR (1989). Regulatory domain of calcium/calmodulin-dependent protein kinase II. Mechanism of inhibition and regulation by phosphorylation. J Biol Chem 264, 4800–4. [PubMed] [Google Scholar]
- 41.Colbran RJ, Fong YL, Schworer CM & Soderling TR (1988). Regulatory interactions of the calmodulin-binding, inhibitory, and autophosphorylation domains of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem 263, 18145–51. [PubMed] [Google Scholar]
- 42.Melanson DL & Spencer MS (1981). Kinetics of ATP synthesis in pea cotyledon submitochondrial particles. Plant Physiol 68, 648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster SM, Reinhart GD & Lardy HA (1977). Studies on the kinetic mechanism of oxidative phosphorylation. J Biol Chem 252, 427–32. [PubMed] [Google Scholar]
- 44.O’Sullivan WJ, Reed GH, Marsden KH, Gough GR & Lee CS (1972). The binding of manganese-nucleoside diphosphates to creatine kinase as determined by proton relaxation rate measurements. J Biol Chem 247, 7839–43. [PubMed] [Google Scholar]
- 45.Kwiatkowski AP, Huang CY & King MM (1990). Kinetic mechanism of the type II calmodulin-dependent protein kinase: studies of the forward and reverse reactions and observation of apparent rapid-equilibrium ordered binding. Biochemistry 29, 153–9. [DOI] [PubMed] [Google Scholar]
- 46.Kim SA, Hudmon A, Volmer A & Waxham MN (2001). CaM-kinase II dephosphorylates Thr(286) by a reversal of the autophosphorylation reaction. Biochem Biophys Res Commun 282, 773–80. [DOI] [PubMed] [Google Scholar]
- 47.Skamnaki VT, Owen DJ, Noble ME, Lowe ED, Lowe G, Oikonomakos NG & Johnson LN (1999). Catalytic mechanism of phosphorylase kinase probed by mutational studies. Biochemistry 38, 14718–30. [DOI] [PubMed] [Google Scholar]
- 48.Grueter CE, Abiria SA, Wu Y, Anderson ME & Colbran RJ (2008). Differential regulated interactions of calcium/calmodulin-dependent protein kinase II with isoforms of voltage-gated calcium channel beta subunits. Biochemistry 47, 1760–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abiria SA & Colbran RJ (2010). CaMKII associates with CaV1.2 L-type calcium channels via selected beta subunits to enhance regulatory phosphorylation. J Neurochem 112, 150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bingol B, Wang CF, Arnott D, Cheng D, Peng J & Sheng M (2010). Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell 140, 567–78. [DOI] [PubMed] [Google Scholar]
- 51.Strack S & Colbran RJ (1998). Autophosphorylation-dependent targeting of calcium/calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem 273, 20689–92. [DOI] [PubMed] [Google Scholar]
- 52.Sanhueza M & Lisman J (2013). The CaMKII/NMDAR complex as a molecular memory. Mol Brain 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urakubo H, Sato M, Ishii S & Kuroda S (2014). In vitro reconstitution of a CaMKII memory switch by an NMDA receptor-derived peptide. Biophys J 106, 1414–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griffith LC (2004). Regulation of calcium/calmodulin-dependent protein kinase II activation by intramolecular and intermolecular interactions. J Neurosci 24, 8394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Leary H, Liu WH, Rorabaugh JM, Coultrap SJ & Bayer KU (2011). Nucleotides and phosphorylation bi-directionally modulate Ca2+/calmodulin-dependent protein kinase II (CaMKII) binding to the N-methyl-D-aspartate (NMDA) receptor subunit GluN2B. J Biol Chem 286, 31272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strack S, McNeill RB & Colbran RJ (2000). Mechanism and regulation of calcium/calmodulin-dependent protein kinase II targeting to the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem 275, 23798–806. [DOI] [PubMed] [Google Scholar]
- 57.Kung JE & Jura N (2016). Structural Basis for the Non-catalytic Functions of Protein Kinases. Structure 24, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffith LC & Greenspan RJ (1993). The diversity of calcium/calmodulin-dependent protein kinase II isoforms in Drosophila is generated by alternative splicing of a single gene. J Neurochem 61, 1534–7. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H & Wang JH (2012). Protein expression and purification of integrin I domains and IgSF ligands for crystallography. Methods Mol Biol 757, 101–10. [DOI] [PubMed] [Google Scholar]
- 60.Monaco S, Gordon E, Bowler MW, Delageniere S, Guijarro M, Spruce D, Svensson O, McSweeney SM, McCarthy AA, Leonard G & Nanao MH (2013). Automatic processing of macromolecular crystallography X-ray diffraction data at the ESRF. J Appl Crystallogr 46, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A & Wilson KS (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC & Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L & Schwede T (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42, W252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC & Zwart PH (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P, Lohkamp B, Scott WG & Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jama AM, Fenton J, Robertson SD & Torok K (2009). Time-dependent autoinactivation of phospho-Thr286-alphaCa2+/calmodulin-dependent protein kinase II. J Biol Chem 284, 28146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tzortzopoulos A, Best SL, Kalamida D & Torok K (2004). Ca2+/calmodulin-dependent activation and inactivation mechanisms of alphaCaMKII and phospho-Thr286-alphaCaMKII. Biochemistry 43, 6270–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.