The emergence of multidrug-resistant Klebsiella pneumoniae has rendered a large array of infections difficult to treat. In a high-throughput genetic screen of factors required for K. pneumoniae survival in the lung, amino acid biosynthesis genes were critical for infection in both immunosuppressed and wild-type (WT) mice.
KEYWORDS: Klebsiella pneumoniae, amino acid biosynthesis, aroA, glyphosate, multidrug resistant, neutropenic
ABSTRACT
The emergence of multidrug-resistant Klebsiella pneumoniae has rendered a large array of infections difficult to treat. In a high-throughput genetic screen of factors required for K. pneumoniae survival in the lung, amino acid biosynthesis genes were critical for infection in both immunosuppressed and wild-type (WT) mice. The limited pool of amino acids in the lung did not change during infection and was insufficient for K. pneumoniae to overcome attenuating mutations in aroA, hisA, leuA, leuB, serA, serB, trpE, and tyrA in WT and immunosuppressed mice. Deletion of aroA, which encodes 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase class I, resulted in the most severe attenuation. Treatment with the EPSP synthase-specific competitive inhibitor glyphosate decreased K. pneumoniae growth in the lungs. K. pneumoniae expressing two previously identified glyphosate-resistant mutations in EPSP synthase had significant colonization defects in lung infection. Selection and characterization of six spontaneously glyphosate-resistant mutants in K. pneumoniae yielded no mutations in aroA. Strikingly, glyphosate treatment of mice lowered the bacterial burden of two of three spontaneous glyphosate-resistant mutants and further lowered the burden of the less-attenuated EPSP synthase catalytic mutant. Of 39 clinical isolate strains, 9 were resistant to glyphosate at levels comparable to those of selected resistant strains, and none appeared to be more highly resistant. These findings demonstrate amino acid biosynthetic pathways essential for K. pneumoniae infection are promising novel therapeutic targets.
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative encapsulated bacterium that can reside in the gastrointestinal tract as a commensal microorganism (1); however, dissemination into extraintestinal sites causes a wide range of infections, including pneumonia, bacteremia, liver abscesses, and urinary tract infections (2–4). The rapid rise of multidrug resistance (MDR) in K. pneumoniae, including the expression of both extended-spectrum β-lactamases (ESBL) and carbapenemases (5) has transformed infection with K. pneumoniae from an easily treatable infection into an urgent public health threat (6, 7). Because of the shortage of effective drugs against MDR K. pneumoniae and a limited number of novel antibiotics in the clinical pipeline, new treatment strategies are needed (8–10).
Previous transposon library sequencing (TnSeq) screens have identified amino acid biosynthetic pathways as important for the growth and virulence of K. pneumoniae and other bacteria (11–14) in immunocompetent hosts. During K. pneumoniae lung infection in immunocompetent mice, neutrophils (polymorphonuclear cells [PMNs]) and inflammatory monocytes (iMOs) contribute to K. pneumoniae clearance in a strain-dependent manner (15, 16). Notably, immunosuppressed and/or immunocompromised hosts, a population susceptible to K. pneumoniae (17), may lack these cells and immune functions, changing the landscape of infection. As such, it is critical when identifying new strategies to treat K. pneumoniae infections that treatments work in both immunocompetent and immunosuppressed hosts.
In a genetic TnSeq screen (unpublished results), many genes involved in the biosynthesis of amino acids were identified as required for K. pneumoniae growth in both immunocompetent and immunosuppressed mice. Here, we validate these findings and test the hypothesis that inhibition of these pathways in K. pneumoniae may be a viable additional therapeutic strategy to use in combination with other treatments. As proof of principle, we used the inhibitor glyphosate to target aroA, a gene essential for tyrosine, phenylalanine, and tryptophan synthesis. Importantly, glyphosate treatment reduced bacterial colonization in lungs, even during infection with glyphosate-resistant mutants, and glyphosate resistance did not result in cross-resistance with other antibiotics.
RESULTS
Neutrophil-depleted mice have a higher bacterial burden despite similar limited bioavailability of amino acids in the lungs.
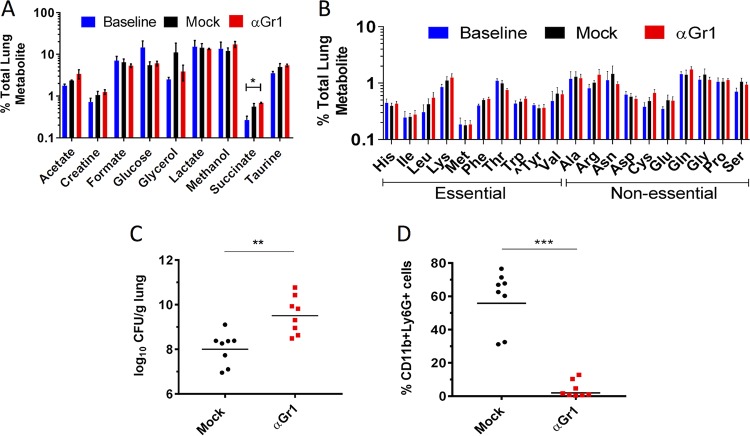
In a TnSeq screen of an ordered array of 13,056 transposon (Tn) insertion mutants in K. pneumoniae ATCC 43816, many K. pneumoniae genes with putative roles in amino acid biosynthetic pathways were identified as potentially attenuated during lung infection in both immunocompetent and neutropenic mice (see Table S1 in the supplemental material; also, unpublished results). This finding raised two possibilities: amino acids are limited in the mouse lung and/or amino acid biosynthetic genes are good targets for therapeutics against K. pneumoniae lung infection. Since some strains of K. pneumoniae are restricted by PMNs, iMOs, or both (15), we evaluated whether the relative abundances of individual amino acids change across three different immunological and infectious states. Bronchoalveolar lavage (BAL) fluid was collected and analyzed by nuclear magnetic resonance (1H-NMR) from mock-infected mice (i.e., mice given phosphate-buffered saline [PBS] intranasally), K. pneumoniae-infected immunocompetent (wild-type [WT]) mice, and K. pneumoniae-infected, α-Gr1-depleted (αGr1) mice lacking both PMNs and iMOs.
Only succinate levels varied between the uninfected, infected, and αGr1-depleted and infected mice (Fig. 1A). All amino acids were detected in the lung at <2% of total measured metabolites and, importantly, did not change in immunocompromised mice (Fig. 1B). Lactate was generated as by-product of using CO2 asphyxiation to sacrifice the mice (18), while glycerol and methanol were derived during collection and extraction of the samples (Fig. 1A). As expected (15), the bacterial load in the lungs of mice lacking PMNs was almost 100-fold larger than in mock-depleted mice (injected intraperitoneally with PBS), which retained a neutrophil population (Fig. 1C and D). These findings indicate that amino acids, relative to many other metabolites, were less available in the lung and that availability was regulated independently of infection level or innate immune status.
FIG 1.
Neutrophil-depleted mice have a higher bacterial burden despite similar limited bioavailability of amino acids in the lungs. (A to D) Mock-depleted (circles) and αGr1 (squares) mice were infected with 1 × 103 CFU of WT K. pneumoniae or PBS (baseline) and sacrificed at 40 hpi. (A and B) BAL fluid was analyzed by NMR, and the percentage of each metabolite normalized to the total measured metabolites in the lung is reported. “^Tyr” is conditionally essential in the absence of Phe in mammals. (C) CFU/g lung for each mouse. (D) PMN population (Ly6G+ CD11b+) assessed by flow cytometry. Each symbol represents an individual mouse, and the bar represents the geometric mean. Each condition was tested with two to four mice per cohort in at least two independent experiments. The data are combined from all experiments. Statistical significance was determined using two-way ANOVA on log-transformed values with Bonferroni’s posttest (A and B) and the Student t test on log-transformed (C) and (D) nontransformed values. **, P < 0.01; ***, P < 0.001.
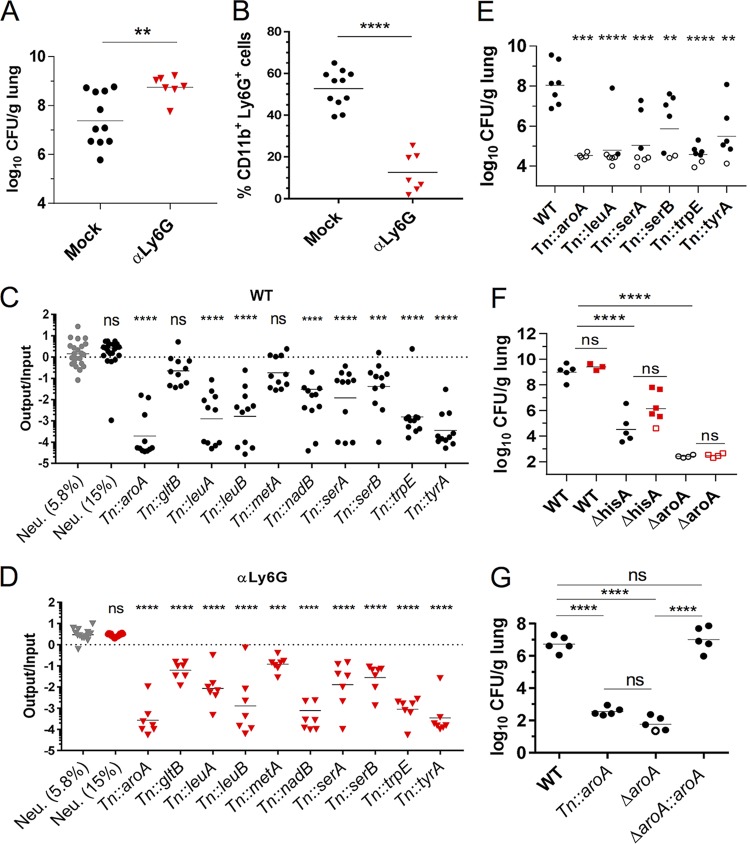
To evaluate the contribution of amino acid biosynthetic genes to the virulence of K. pneumoniae in the mouse lung in both WT and immunocompromised animals, a minilibrary of TnSeq mutants was assembled and used to infect cohorts of mice. This minilibrary contained four mutants with transposon insertions at neutral sites, where two neutral mutants each comprised 15%, and the other two comprised 5.8% of the pool to serve as controls for bottleneck effects. The remaining 58% of the pool consisted of 10 auxotrophic mutants with transposon (Tn) insertions in aroA, gltB, leuA, leuB, metA, nadB, serA, serB, trpE, and tyrA. These 10 Tn insertion auxotrophic mutants were recovered from our arrayed library and were confirmed for growth defects in M9 broth supplemented with glucose (M9+glucose) and growth was restored upon growth in low-salt Luria-Bertani medium (L), which contains amino acids (19), and upon growth in M9+glucose with supplementation with the appropriate cognate amino acids, indicating the transposon disrupted the predicted amino acid biosynthetic or metabolic pathway (see Fig. S1 in the supplemental material). Since K. pneumoniae 43816 is sensitive to neutrophils (15), we also infected α-Ly6G-depleted (αLy6G) mice depleted of PMNs with the minilibrary. Mice depleted with αLy6G lacked PMNs and had higher bacterial loads than mock-depleted mice (Fig. 2A and B), supporting previous findings that PMNs limit K. pneumoniae 43816 growth during lung infection (15, 20, 21). The aroA, leuA, leuB, nadB, serA, serB, trpE, and tyrA mutants were significantly attenuated in both mock-depleted mice (Fig. 2C) and αLy6G mice (Fig. 2D). Although the recoveries of the gltB and metA mutants were, on average, 5-fold lower than for the control K. pneumoniae strains (Fig. 2C, Neu., 5.8%) in mock-depleted mice, these mutants were only statistically significantly attenuated in αLy6G mice (Fig. 2D). Notably, fitness was not significantly restored for any of the mutants in αLy6G mice compared to mock-depleted mice (one-way analysis of variance [ANOVA] with Sidak’s posttest on log-transformed values). Combined these results indicate that K. pneumoniae relies on many amino acid biosynthetic synthesis genes during mouse lung infection regardless of host immune status. In single-strain infections, aroA, leuA, serA, serB, trpE, and tyrA mutants had significant fitness defects compared to WT K. pneumoniae (Fig. 2E), indicating that the presence of WT K. pneumoniae neither aids the growth of auxotrophic mutants by providing metabolites in trans nor impairs them by scavenging metabolites more efficiently.
FIG 2.
K. pneumoniae genes encoding enzymes required for amino acid biosynthesis are attenuated in the lungs of WT and neutropenic mice. (A to D) Mock-depleted and αLy6G mice were infected with 2 × 104 CFU of a minilibrary of K. pneumoniae Tn mutants and sacrificed at 33 hpi. (A) CFU/g lung for each mouse. (B) Percent PMN population (Ly6G+ CD11b+) per live cells. (C and D) Abundance of each Tn strain in the output versus the input pool in mock-depleted (C) and αLy6G (D) mice. (E) CFU in lungs of mice infected with 1 × 103 CFU of the indicated strains and sacrificed at 45 hpi. (F) CFU in lungs of Mock-depleted (circles) and αGr1 (squares) mice infected with 2 × 103 CFU of the indicated strains and sacrificed at 40 hpi. (G) CFU of mice infected with 2 × 104 CFU of the indicated strains and sacrificed at 33 hpi. Each symbol represents a mouse; open symbols indicate no CFU were recovered and limit of detection, and dashes represent geometric mean (A, C to G) or mean (B). Each strain was tested in two to four mice per cohort in at least two independent experiments. The data are combined from all experiments. Statistical significance was determined by Student t test (A and B) on log transformed (A) and nontransformed (B) values or by one-way ANOVA with Dunnett’s posttest (C and D) on log-transformed values by comparing each mutant against the neutral mutants present at 5.8% in the input pool. To assess whether Tn mutants had statistically significant fitness differences in αLy6G mice compared to WT, the fitness of each mutant in WT mice was compared to the fitness of the same mutant in αLy6G mice using one-way ANOVA with Dunnett’s posttest. No mutants had a significantly higher value in αLy6G mice than WT. (E to G) One-way ANOVA on log-transformed values using Dunnett’s (E) or Tukey’s (F and G) posttests. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The K. pneumoniae Tn::aroA mutant had the most severe defect in lungs (Fig. 2C to E). aroA encodes the metabolic lynchpin, EPSP synthase, which is required for the biosynthesis of three aromatic amino acids (22), and siderophores in other bacteria (23–25). In addition, multiple transposon-insertion mutants in the histidine biosynthesis operon, the fourth aromatic amino acid (26), were identified as attenuated in the TnSeq screen (see Table S1 in the supplemental material), but none were recoverable from our arrayed library due to ambiguity in matching and assigning locations for these strains in our arrayed library (data not shown). Thus, we generated in-frame deletions of aroA and hisA to test in lung infections. In-frame deletions of hisA (ΔhisA) and aroA (ΔaroA) were severely attenuated in the lung regardless of host immune status and inoculating dose (Fig. 2F; see Fig. S2A and B in the supplemental material), and the restoration of aroA at its native locus restored K. pneumoniae virulence (Fig. 2G). Combined, these results demonstrate that endogenous levels of amino acids were insufficient to support growth of amino acid auxotrophic mutants in both immunocompetent and immunosuppressed lungs.
aroA inhibition effectively attenuates K. pneumoniae growth in vitro and during lung infection.
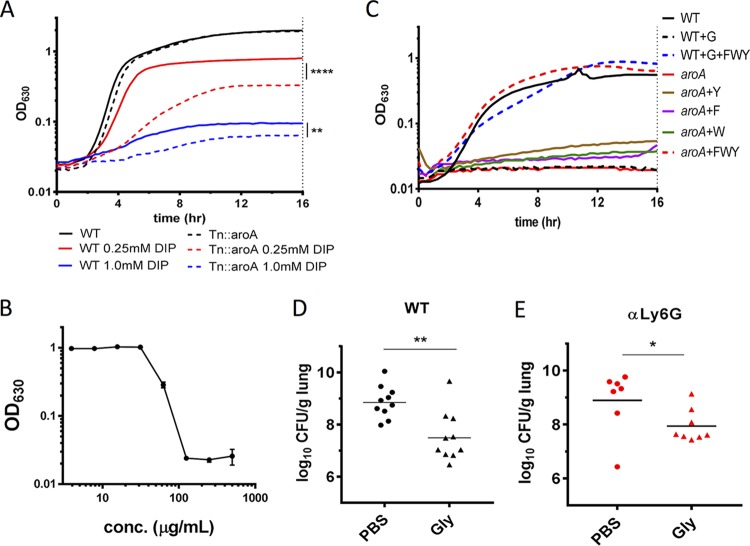
Metal scavenging is a critical requirement for K. pneumoniae pathogenesis (1), and chorismate, a product downstream of EPSP synthase in the shikimate pathway, is a precursor for some iron-acquiring siderophores (25). To test whether aroA is critical for iron acquisition in addition to its requirement for aromatic amino acid synthesis, WT and Tn::aroA strains were grown in L broth in the presence or absence of the iron-chelator 2,2′-bipyridyl (DIP) (Fig. 3A). While both strains were inhibited by 1 mM DIP, the Tn::aroA strain was more impaired for growth in iron-restricted conditions (0.25 mM DIP) than the WT in L broth, demonstrating a role for aroA in iron acquisition and growth under low-iron conditions.
FIG 3.
aroA inhibition attenuates K. pneumoniae growth in the lungs. (A) The indicated strains were grown in L with or without iron-chelator DIP (0.25 or 1 mM) for 16 h. (B) WT K. pneumoniae was grown in M9+glucose for 16 h in a dose curve of glyphosate. The OD630 after 16 h of growth is shown. (C) Strains were grown in M9+glucose with 10 mM phenylalanine (F), tryptophan (W), or tyrosine (Y) and/or 250 μg/ml glyphosate (G) as indicated. (D and E) Swiss-Webster (D) or αLy6G (E) mice were infected with 2 × 104 CFU of WT and mock treated with PBS (circles) or 0.5 mg of glyphosate (triangles) at 6 and 24 hpi and harvested at 30 hpi, and the CFU from the lungs were determined. Each symbol represents a mouse; the bar represents the geometric mean. Experiments were performed independently at least twice with three technical replicates (A to C) or at least three times (D and E) with two to four mice per cohort. The data are from representative experiments (A to C) or combined from all experiments (D and E). Statistical significance was determined by one-way ANOVA with Sidak’s posttest on EC50 values (A), Student t test on log-transformed values (D), or Mann-Whitney test (E). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
To test the idea that molecules targeting amino acid biosynthetic genes may be effective at controlling growth of K. pneumoniae during infection, we targeted EPSP I synthase in K. pneumoniae using glyphosate, an EPSP I synthase-specific competitive inhibitor (22, 27). Glyphosate inhibited K. pneumoniae growth in M9+glucose in a dose-dependent manner with an MIC of glyphosate (MICgly) of 125 μg/ml, defined as the lowest concentration of glyphosate inhibiting K. pneumoniae growth in M9+glucose at 16 h (Fig. 3B, Table 1). This inhibition was due to glyphosate targeting EPSP I synthase, since the addition of phenylalanine, tyrosine, and tryptophan in combination, but not individually, restored bacterial growth in glyphosate (Fig. 3C). Glyphosate was tested as an inhibitor of WT K. pneumoniae infection in the lungs by intranasally treating K. pneumoniae-infected mice with PBS (mock treated) or 0.5 mg of glyphosate/mouse at 6 and 24 h postinfection (hpi). At 30 hpi, glyphosate significantly inhibited K. pneumoniae growth in lungs, with the lungs of mock-treated mice containing almost 100-fold more bacteria than the glyphosate-treated mice (Fig. 3D). Notably, glyphosate inhibited K. pneumoniae growth in the lungs even in the absence of host PMNs (Fig. 3E and Fig. S2C), indicating the decrease in bacterial burden from AroA inhibition was not entirely dependent on PMNs. Together, these results show that AroA has multiple nonredundant roles during growth and virulence of K. pneumoniae infection and can be targeted by glyphosate to reduce bacterial burden regardless of host immune status.
TABLE 1.
MICs and EOPs of sGR, aroA-encoded, and clinical K. pneumoniae strains
| Straina | Genotype | Fold change in MICglyb | Mean EOP (%)c
± SEM on: |
||||
|---|---|---|---|---|---|---|---|
| 250 μg/ml glyphosate |
1,000 μg/ml glyphosate |
||||||
| 24 h | 48 h | 24 h | 48 h | 72 h | |||
| MKP220 | WT | 1 | 0 | 80 ± 13 | 0 | H | 103 ± 17 |
| RJS196 | sGR1 | 6 | 105 ± 18 | 105 ± 18 | TSTC | 71 ± 1 | 71 ± 1 |
| RJS197 | sGR2 | 10 | 119 ± 19 | 119 ± 19 | TSTC | 91 ± 15 | 91 ± 15 |
| RJS198 | sGR3 | 8 | 86 ± 20 | 86 ± 20 | TSTC | 65 ± 9 | 65 ± 9 |
| RJS199 | sGR4 | 8 | 94 ± 15 | 94 ± 15 | TSTC | 71 ± 7 | 71 ± 7 |
| RJS200 | sGR5 | 10 | 105 ± 14 | 105 ± 14 | TSTC | 77 ± 3 | 77 ± 3 |
| RJS201 | sGR6 | 4 | 80 ± 7 | 80 ± 7 | TSTC | 79 ± 8 | 79 ± 8 |
| ALM121 | aroAG96A | 4d | 0 | 89 ± 11 | 0 | 94 ± 12 | 94 ± 12 |
| ALM122 | aroAT97I | 2 | 79 ± 4 | 79 ± 4 | TSTC | 64 ± 4 | 64 ± 4 |
| ALM123 | aroAP101S | 2 | 86 ± 14 | 86 ± 14 | TSTC | 71 ± 5 | 71 ± 5 |
| ALM111 | aroATIPS | 8 | H | 114 ± 33 | H | 107 ± 32 | 107 ± 32 |
| R1 | ESBL | 2 | ND | ND | ND | ND | ND |
| R9 | ESBL | 4 | ND | ND | ND | ND | ND |
| R17 | KPC | 2.7 | ND | ND | ND | ND | ND |
| B1 | Non-ESBL/KPC | 4 | ND | ND | ND | ND | ND |
| B6 | KPC | 1.7 | ND | ND | ND | ND | ND |
| B17 | ESBL | 2.7 | ND | ND | ND | ND | ND |
| B18 | KPC | 5.3 | ND | ND | ND | ND | ND |
| B19 | Non-ESBL/KPC | 3.3 | ND | ND | ND | ND | ND |
| B20 | ESBL | 2 | ND | ND | ND | ND | ND |
R1, R9, etc., are respiratory isolates; B1, B6, etc., are blood isolates.
MICgly of WT = 125 μg/ml ± 12. Bacteria were grown overnight in minimal media and two-fold serial dilutions of glyphosate. Experiments were performed three independent times in technical triplicate, and values are reported as the average fold change for three experiments.
EOP, efficiency of plating measured in three independent experiments, as described in Materials and Methods. Technical replicates were averaged in each experiment, and the EOP was calculated as CFU+glyphosate/CFUM9+glucose. Values represent the average EOP and SEM, as calculated by ANOVA with Dunnett’s t test. TSTC, too small to count; H, hazy growth on concentrated spots where more than 1,000 colonies would be expected but no colonies detected on samples expected to generate 100 colonies. ND, not determined.
aroAG96A has a growth defect in minimal media; the MICgly was thus determined by evaluating growth in glyphosate at 24 h.
Selection of spontaneous glyphosate-resistant and generation of aroA catalytically active-site glyphosate-resistant mutants in K. pneumoniae.
As K. pneumoniae rapidly acquires resistance to antibiotics (28, 29), we evaluated the rate of spontaneous glyphosate resistance of K. pneumoniae (see Materials and Methods). WT K. pneumoniae was grown overnight in L and plated on M9+glucose containing 1,000 μg/ml glyphosate, 50 μg/ml rifampin (RIF), or 200 μg/ml streptomycin (STR). After 24 h, no glyphosate-resistant (glyr) colonies were seen above our limit of detection of 1 × 10−9 CFU on 1,000 μg/ml glyphosate plates (Fig. S3A). The rate of RIF resistance was 5 × 10−7 CFU, and the STR resistance was below our limit of detection (Fig. S3A). After 48 h, colonies were detected on glyphosate plates at an average frequency of 1 × 10−8 bacteria, while the frequency of RIF and STR resistance did not increase.
Six colonies, designated sGR1 to sGR6 for spontaneous glyphosate resistance (sGR), appeared as normal sized (sGR1 to sGR3) or small (sGR4 to sGR6) colonies on 1,000 μg/ml glyphosate plates after 48 h. These were streak purified on glyphosate plates before further characterization. All six sGR mutants had 4- to 10-fold higher MICgly values, defined by growth in M9+glucose with glyphosate at 16 h, than that of K. pneumoniae 43816 (Table 1). While all sGR mutants grew slightly slower than the WT in M9+glucose, none showed significant defects (Fig. S3B). The higher MICs indicate that these sGR mutants are resistant, versus tolerant, to glyphosate (30). Efficiency-of-plating (EOP) assays on M9+glucose plates with 250 or 1,000 μg/ml glyphosate revealed that all sGR mutants formed normal-sized colonies on the 250 μg/ml glyphosate and distinct, but too-small-to-count (TSTC) colonies on the 1,000 μg/ml glyphosate plates at 24 h. These all became normal-sized by 48 h. In contrast, WT K. pneumoniae did not form colonies until 48 and 72 h on 250 and 1,000 μg/ml plates, respectively (Table 1). The resistance of the sGR mutants to 14 clinically used antibiotics was measured using Vitek-2 analysis. This revealed that no additional antibiotic resistances occurred under glyphosate selection, indicating that the mechanism of glyphosate resistance did not confer cross-resistance to commonly used antibiotics (Table S2). Target modification is a common strategy used by bacteria to develop antimicrobial resistance (31, 32). Therefore, we sequenced the serC-aroA locus, but no base pair changes were detected, indicating resistance was unlinked.
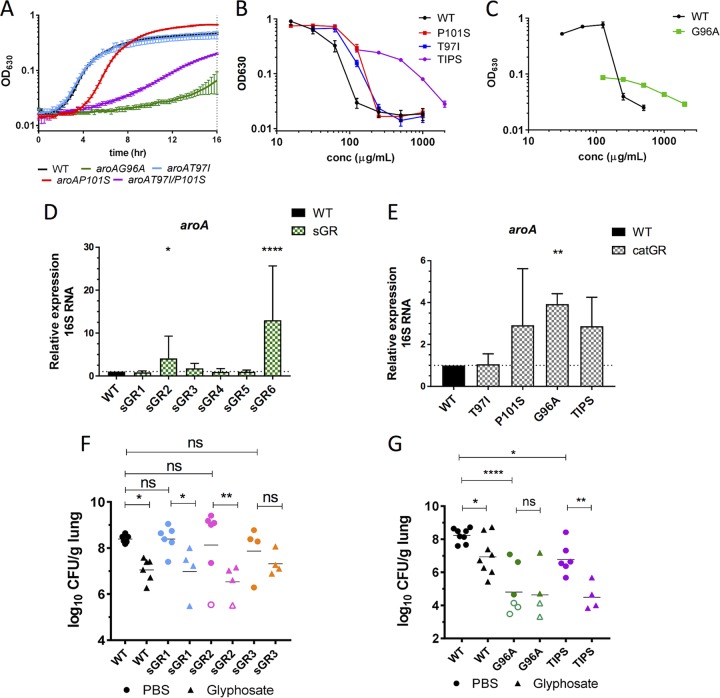
Although no sGR mutations were found in the aroA locus, previous work has shown that affecting the catalytic active site of AroA via a single amino acid substitution at aroAG96A (i.e., a G-to-A change at position 96) (27, 33) or a double mutation at aroAT97I/P101S (34, 35) gives rise to glyphosate resistance. Since the impact of these catalytic mutations on K. pneumoniae virulence has not been evaluated, strains containing mutations aroAG96A, aroAT97I, aroAP101S, and aroAT97I/P101S (TIPS) were generated (Table 1). aroAG96A and aroATIPS strains displayed significant growth defects in M9+glucose at 16 h (Fig. 4A and Table 1). Likewise, both had a delay in colony formation on M9+glucose plates compared to sGR mutants (Table 1). The MICgly values for these mutants were evaluated in M9+glucose, but because of the extremely impaired growth rate of aroAG96A, the MIC was quantified at 24 h rather than at 16 h (Table 1 and Fig. 4B and C). The aroAT97I and aroAP101S strains had MICglys of 250 μg/ml, 2-fold greater than for the WT, and the aroAG96A and aroATIPS strains had MICglys 4- and 8-fold greater than WT, respectively. Thus, mutations within the AroA catalytic site conferring high levels of glyphosate resistance come with unfavorable growth tradeoffs in amino acid poor environments, such as minimal media. This suggests that glyphosate-resistant active-site mutations, while advantageous under selective pressure of glyphosate, may be disadvantageous in vivo where aromatic amino acids are limited (Fig. 1B), and bacteria must synthesize them through the AroA pathway.
FIG 4.
Glyphosate-resistant mutants remain susceptible to glyphosate in infection. (A) The indicated strains were grown in M9+glucose for 16 h. (B and C) The indicated strains were grown in M9+glucose with various glyphosate, and the OD630 after 16 h (B) or 24 h (C) of growth is shown. (D and E) The aroA mRNA copy number of the indicated strains was quantified using qRT-PCR after growth to mid-log phase in M9+glucose. (F and G) CFU counts in lungs of mice infected with 2 × 104 CFU of the indicated strains, mock treated with PBS (circles), or treated with 0.5 mg of glyphosate (triangles) at 6 and 24 hpi and harvested at 30 hpi. Each symbol represents a mouse; open symbols indicate that no CFU were detected at limit of detection; bars indicate the geometric means. Experiments were done independently at least twice with groups of two to three mice. The data are from a representative experiment of three performed in triplicate (A to C) or are combined from all experiments (D to G). Significance was determined using one-way ANOVA on log-transformed values with Dunnett’s (D and E) or Fisher LSD (F and G) posttest. NS (not significant), P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Another reported mechanism of glyphosate resistance has been increased expression of aroA (31). We compared the aroA mRNA levels by quantitative reverse transcription-PCR (qRT-PCR) in each sGR mutant and the catalytic mutants at mid-log growth phase in M9+glucose. Two sGR mutants, sGR2 and sGR6, and the aroAG96A strain had significantly higher aroA mRNA levels compared to the WT (Fig. 4D and E), revealing a potential mechanism of glyphosate resistance. However, it is important to note that increased mRNA levels did not correlate with degree of glyphosate resistance (Table 1).
Glyphosate treatment lowers sGR and aroATIPS burden in vivo.
Three sGR mutants, sGR1 to sGR3, and two catalytic mutants (aroAG96A and aroATIPS) were evaluated for virulence in the mouse lung with and without glyphosate treatment (Fig. 4F and G). Both aroAG96A and aroATIPS mutants were significantly attenuated for growth in immunocompetent mouse lungs, consistent with their low growth rate in M9+glucose (Fig. 4C). Moreover, glyphosate significantly lowered the burden of the aroATIPS strain despite the MICgly being 8-fold higher than that of K. pneumoniae 43816. Although none of three sGR mutants were attenuated in untreated mice, intriguingly, treatment with glyphosate significantly lowered the bacterial burden of the sGR1 and sGR2 strains, which had 6- and 10-fold higher MICglys, respectively, than that of K. pneumoniae 43816 (Table 1). Similarly, the sGR3 bacterial burden was decreased on average by 10-fold, although this was not statistically significant. These data show glyphosate impedes growth of these glyphosate-resistant K. pneumoniae mutants and that the highly resistant catalytic mutations come with a significant virulence tradeoff in the lung, an amino acid-poor environment (Fig. 1A).
Sensitivity of clinical blood and respiratory K. pneumoniae isolates, including ESBL- and KPC-producing strains, to EPSP I inhibition.
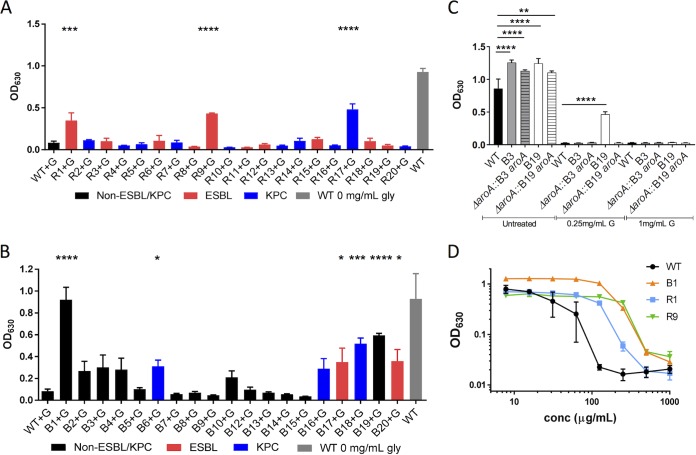
Thirty-nine clinical isolates from blood (B) or the respiratory tract (R) were characterized for their antibiotic resistance profile using Vitek-2 antibiotic susceptibility profiling (Table S3). Of these isolates, 12 B and 13 R strains were determined to be ESBL or KPC producers. To screen for glyphosate resistance, their growth in M9+glucose in the presence or absence of 250 μg/ml glyphosate was evaluated (Fig. S3D and E and Fig. 5A and B). All strains grew in M9+glucose, which demonstrated that none were auxotrophic, although the R14, R15, R19, B8, and B10 strains displayed significantly less growth than K. pneumoniae 43816 in M9+glucose (Fig. S3D and E). In M9+glucose with 250 μg/ml glyphosate, 30 strains were inhibited for growth, while R1, R9, R17, B1, B6, B17, B18, B19, and B20 grew significantly (Fig. 5A and B). Among these, there was no correlation as to whether they were ESBL or KPC producers (Fig. 5A and B).
FIG 5.
Sensitivity of clinical blood and respiratory K. pneumoniae isolates, including ESBL- and KPC-producing strains, to glyphosate. (A and B) Clinical K. pneumoniae strains were grown in M9+glucose containing 250 μg/ml glyphosate (black, non-EBSL/KPC; red, ESBL; or blue, KPC) or M9+glucose alone (gray, K. pneumoniae 43816; see also Fig. S3C and D in the supplemental material) for 20 h. (C) Mutations in aroA from B3 and B19 were introduced into the K. pneumoniae 43816 ΔaroA strain and grown in M9+glucose with the indicated concentrations of glyphosate for 16 h. (D) The indicated strains were grown in M9+glucose with glyphosate; the OD630 at 16 h is shown. Experiments were performed at least twice with three technical replicates. The data are combined from all experiments (A to C) or are from a representative experiment (D). Statistical significance was determined by using one-way ANOVA with the Fisher LSD test (A and B) or Sidak’s posttest (C). *, P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The serC-aroA locus and its upstream regulatory region were sequenced in the glyr clinical isolates. Only two isolates, B3 and B19, had differences in aroA sequence (Q419L and Q221K, respectively), but these differences were insufficient to cause glyphosate resistance when cloned into K. pneumoniae 43816 ΔaroA (Fig. 5C). To further evaluate glyphosate resistance of the clinical isolates, the MICglys of select strains that grew significantly in glyphosate (Fig. 5A and B) were determined. The MICgly of B1, the most resistant strain, was 4-fold higher than that of K. pneumoniae 43816 (Fig. 5D and Table 1). In addition, the MICglys of R1 and R9, two resistant respiratory strains, were 2- and 4-fold higher (Fig. 5D). Notably, all tested clinical isolate MICglys were within range of the sGR and aroATIPS mutant strains that remained sensitive to glyphosate treatment in the lungs (Fig. 4C to E).
DISCUSSION
Our findings strongly support the idea that targeting amino acid biosynthetic pathways is a promising strategy to contain MDR infections (36, 37). Importantly, regardless of immune status of the host, levels of amino acids in the lungs did not drastically change and amino acid biosynthetic K. pneumoniae mutants remained attenuated in the lung. This included all aromatic amino acid mutants, which were the most significantly attenuated, consistent with the fact that aromatic amino acids are the most energy-expensive amino acids to synthesize (38). In addition, some of these pathways share molecular precursors with iron-acquiring siderophores critical for K. pneumoniae virulence (1, 25). Importantly, BAL fluid in humans is similar in amino acid content to mice (39), suggesting that amino acids are too scarce in human lungs for K. pneumoniae to scavenge, and therefore targeting amino acid biosynthesis pathways could translate to the clinic. In addition, none of the clinical isolates were auxotrophic, indicating that amino acid biosynthetic pathways are intact and therefore potentially targetable in infectious strains. Using class I EPSP synthase as a proof of principle, we found that its inhibition by glyphosate limited the growth of K. pneumoniae in lungs and that glyr mutants arose infrequently and were either themselves attenuated and/or still sensitive to the dose of glyphosate during infection. In addition, glyr strains did not become cross-resistant to other commonly used antibiotics, and many clinical MDR strains remained sensitive to glyphosate, suggesting that the mechanisms of resistance in these six sGR strains are distinct from common antibiotic resistance mechanisms. Finally, studies by others show glyphosate can synergize with some antibiotics (40, 41), supporting the idea that essential amino acid enzymes could be targeted in concert with other therapies.
Although glyphosate is the active ingredient in the most common commercial and agricultural herbicide used globally since 1974, Roundup (42, 43), naturally occurring catalytic glyr EPSP synthase mutants in bacteria are reported infrequently, likely due to the adverse effects of mutations on the substrate Km (27, 34, 35) which impair bacterial growth under nutrient-limiting conditions. While bacterial EPSP has been studied for the design of glyr crops, our findings of significant growth and virulence defects in active-site EPSP mutants highlight the importance of an intact, WT aroA gene to retain virulence and are consistent with studies showing that aroA is important for virulence (14, 44, 45). Since aroA mutations were ruled out as the source of glyr in both the clinical and sGR strains, the resistance mechanisms of the sGR mutants could be one of many phenomena observed in other species. These include the presence of a metabolic enzyme that chemically modifies glyphosate, epigenetic changes to increase the copy number of EPSP synthase to overcome competitive inhibition, or by amplifying the aroA gene to increase transcription and/or translation (31, 46, 47). Upregulation of efflux pumps, an additional resistance mechanism, is an unlikely resistance determinant here since the MICs of clinical antibiotics were unchanged.
Our studies indicate that resistant mutants in catalytic sites of amino acid biosynthetic pathways may arise infrequently and/or be themselves attenuated, which are important considerations for antimicrobial therapeutics. While glyphosate is not recommended for human use, it is a well-studied, highly effective competitive inhibitor (34), making it a good molecule for a proof of principle to test our hypothesis that targeting amino acid enzymes would augment resolution of infection in difficult to treat MDR cases. In fact, the dose of glyphosate used in mice in this study is higher than the current U.S. Environmental Protection Agency (EPA) standard for human exposure of 2 mg/kg of body weight (48, 49), and there is currently much active research into the adverse effects of glyphosate and/or other additives in Roundup on a number of human health issues (42, 50–52).
In this study, we treated bacterial pneumonia by intranasal glyphosate delivery to mice, permitting high enough concentrations in the infected site to contain resistant mutants. Intranasal administration of therapies against lung infection models has been effective due to the high drug concentration achieved by direct administration to the lung (53, 54). However, this is not predictive of the ability to treat using systemic delivery, and therefore it remains uncertain whether glyphosate would be an effective inhibitor in a systemic infection such as K. pneumoniae bacteremia. Given our results, it is worthwhile to assess the efficacy of inhibitors of components other shikimate pathway enzymes (55, 56), as well as histidine (hisA) and leucine (leuA and leuB) biosynthesis (36, 57), in combination with each other and/or known antibiotics. Importantly, studies on other bacterial pathogens indicate that amino acid biosynthetic mutants are attenuated in the lung (44, 45, 58), gastrointestinal tract (59), mesenteric lymph nodes, spleen (14), and skin (13). Escherichia coli shows high upregulation of branched-chain amino acid and histidine biosynthetic genes during growth in urine (60, 61), and branched-chain amino acid biosynthetic inhibitors have shown promise as antifungal agents (36, 40). This study supports the design and development of safe molecules that target amino acid biosynthetic pathways to be tested in combination with other treatments as a promising strategy to improve the treatment of MDR K. pneumoniae infections.
MATERIALS AND METHODS
Generation of K. pneumoniae strains.
All strains and primers used in this study are listed in Tables S4 and S5, respectively, in the supplemental material. A WT K. pneumoniae strain (K. pneumoniae ATCC 43816; MKP203) was made spectinomycin resistant and verified as equally fit compared to the parental K. pneumoniae (see the supplemental material). K. pneumoniae transposon-insertion mutants were generated by introducing a kanamycin-resistant (Kanr) Himar1 transposon into K. pneumoniae by mating MKP220 with the DAP auxotrophic MFDpir-pSC189 (MKP216) (62), and insertion sites were confirmed by PCR as described in the supplemental material. All deletion mutants were generated using lambda red recombination as described in the supplemental material (63). Clinical isolates from blood (B) and respiratory (R) infections at Ohio State University were used. No human subjects were included in this study. The Tufts University Institutional Review Board (IRB) has approved the use of clinical isolates without patient information.
Mouse intranasal infections and flow cytometry analysis.
K. pneumoniae strains were cultured overnight at 37°C with aeration in 2 ml of low-salt (0.5 g/liter NaCl) Luria-Bertani medium (L). From the overnight cultures, bacteria were serially diluted in sterile PBS, and the infectious dose was quantified on L-agar plates. Female Swiss-Webster mice (7 to 12 weeks old; Taconic) were anesthetized with 3% isoflurane and intranasally infected with a 50-μl bolus of the indicated dose. Mock-infected mice intranasally received 50 μl of PBS (Corning). The K. pneumoniae minilibrary was prepared and assessed for fitness in lung infections as previously described (14) and in the supplemental material. Mice were depleted of PMNs by intraperitoneally injecting 100 μl of 0.5 mg/ml α-Ly6G antibody (1A8; Fisher Scientific) at 16 h preinfection or 100 μl of 0.5 mg/ml α-Gr1 antibody (RB6-8C5, Fisher Scientific) at 16 h preinfection and 24 h postinfection as previously described (14). Mock-depleted mice intraperitoneally received 100 μl of PBS. Mice treated with glyphosate were anesthetized with 3% isoflurane at 6 and 24 hpi, and 50 μl of PBS or 0.5 mg of glyphosate (Santa Cruz Biotechnology)/mouse in 50 μl of PBS was delivered intranasally. At the indicated times after infection, mice were sacrificed by CO2 asphyxiation. Lungs were harvested into sterile PBS, weighed, and homogenized by pushing tissue through a 70-μm-pore cell strainer. Homogenates were plated for CFU/g lung and collected for analysis by deep sequencing in the indicated experiments, and the PMN and iMO populations were quantified by flow cytometry using an LSRII. The lung homogenates were stained with α-Ly6G PE-Cy7 (eBioscience), α-Ly6C-APC (eBioscience), and α-CD11b-FITC (eBioscience) and evaluated with FlowJo (v10.0) by first gating on the live population and then quantifying the Ly6G+ CD11b+ (PMN) and Ly6C+ Ly6G– CD11b+ (iMO) cells. All mice were handled in accordance with protocols approved by the Institutional Animal Care and Use Committee of Tufts University.
Collection and analysis of BAL fluid by NMR.
Mice were sacrificed, and their tracheas were exposed and cut with fine-blade scissors. Plastic tubing was inserted into the tracheal slit, and 1 ml of sterile saline (0.9% NaCl in H2O) was injected into the lungs and collected. This was repeated twice for a total of 3 ml collected. A 2:1 volume of ice-cold methanol was added to the samples, which were incubated at –20°C for 20 min and centrifuged at 17,000 × g for 5 min at 4°C to pellet and remove proteins. Supernatants were collected and evaporated by spinning in a SpeedVac overnight. Dried metabolites were resuspended in 600 μl of deuterated water (D2O; Spectra). To serve as a standard, 1 μl of 100 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS; Cambridge Isotope Laboratories, Inc.) was added to each sample. 1H-NMR spectra of each sample were collected at 25°C on a Bruker Avance 600 spectrometer using 256 scans and a NOE1D pulse sequence. NMR spectral peaks were assigned, and the area of each peak was quantified (Chenomx NMR Suite software 8.0). The percentage of each metabolite out of the total measured metabolite levels in that sample was calculated. All amino acids, as well as several metabolites, including glucose, were measured.
Selection of spontaneous glyphosate-, rifampin-, and streptomycin-resistant mutants.
Overnight cultures of MKP220 grown in L were washed twice in M9+glucose and plated on M9+glucose or M9+glucose with 250 μg/ml of glyphosate, 1,000 μg/ml of glyphosate (Sigma or Santa Cruz), 50 μg/ml RIF, or 200 μg/ml STR. Glyphosate was added to M9+glucose agar at 50°C. The frequency of spontaneous resistance was determined by counting CFU after 24 and 48 h (CFU+drug/CFUM9glu). All glyphosate-resistant colonies were isolated after 48 h of incubation. Colonies were streak purified on M9+glucose plates containing 1,000 μg/ml glyphosate before characterization.
Antibiotic resistance profile of clinical and glyphosate-resistant isolates.
Spontaneous glyphosate-resistant mutants, catalytic aroA mutants, and clinical isolates were evaluated for susceptibility to a panel of 16 clinically used antibiotics using the Vitek-2 system (bioMérieux; Tufts Clinical Microbiology Lab). Vitek-2 assigned an MIC for each drug and flagged likely ESBL- and KPC-producing strains based on clinical laboratory standards.
Growth curves and calculation of MIC.
Strains were grown overnight in L, washed in PBS, and diluted to 1 × 107 cells/ml. Bacteria (100 μl) were placed in a 96-well plate in M9+glucose or L, with or without the indicated compound. Amino acids were added to a final concentration of 10 mM (Sigma) and 2,2′-bipyrydyl (DIP; Sigma) was added to the indicated concentration. MICgly values were determined by using 2-fold dilutions of glyphosate in M9+glucose. The 50% effective concentration (EC50) values were determined for DIP inhibition. Cultures were incubated at 37°C for 16 h with shaking, and the optical density at 630 nm (OD630) was measured every 15 min in a plate reader (BioTek Synergy HT). The resulting values were corrected for background and path length. MICgly was defined, using EUCAST standards (64, 65), as the lowest concentration of glyphosate in which a strain did not grow by 16 h with the exception of the aroAG96A mutant, which was measured at 24 h due to its slow growth in M9+glucose. Each experiment was performed in technical triplicate and averaged, and the average of at least three independent experiments was used to calculate the MICgly. The fold changes between the WT and mutant MICgly in each independent experiment were averaged to account for the 2-fold variation (ranging from 63 to 125 μg/ml) observed in the K. pneumoniae 43816 MICgly between experiments.
Efficiency of plating on glyphosate.
The EOP of K. pneumoniae strains was measured by culturing the indicated strains in L overnight with shaking at 37°C. Cultures were washed in PBS and diluted to 1 × 108 cells/ml in PBS, and serial dilutions were plated in triplicate on M9+glucose plates containing 0, 250, or 1,000 μg/ml glyphosate. Plates were incubated at 37°C overnight, and visible colonies counted after 24, 48, and 72 h. Technical replicates were averaged in each experiment, and the EOP was calculated as CFU+glyphosate/CFUM9+glucose.
RNA extraction and qRT-PCR analysis.
RNA from overnight cultures of each K. pneumoniae strain was isolated and analyzed as previously described (66). Briefly, cultures were back diluted 1:50 in M9+glucose and grown at 37°C for 3 h in L media. A volume of 1 ml was mixed with 200 μl of 1% Zwittergent–100 mM citric acid, followed by incubation for 10 min prior to pelleting and snap-freezing. The total RNA was extracted using TRIzol reagent (Ambion, Carlsbad, CA). Contaminating DNA was removed after a 1-h incubation in DNA-free Turbo (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol with up to 10 μg of RNA per sample. cDNA was synthesized with iScript cDNA kit (Bio-Rad, Hercules, CA) with 1 μg of RNA as the template. Samples were diluted 10-fold with RNA-free H2O. PCR was performed with 5 μl of cDNA, 500 nM concentrations of each primer designed with Primer3 (67), and 10 μl of SYBR green (Bio-Rad) with a CFX96 RealTime system (Bio-Rad). Relative aroA transcript levels (Table S5; RJS298 and RJS299) were normalized to 16S RNA transcript levels (Table S5; RJS259 and RJS260) and calculated using the 2–ΔΔCT method (68).
Statistical analysis.
Statistical significance was assessed using Prism 7.03 (GraphPad Software, La Jolla, CA) as indicated in each figure legend. Biological replicates are presented with the standard errors of the mean (SEM), and representative images are presented with the standard deviations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Urmila Powale and Efrat Hamami for help with strain construction, Dara Brena for help with rates of resistant mutants, Bernadette Chirokas and Cassandra Parker and their staff at the Tufts Medical Center Clinical Laboratory for Vitek-2 analysis, Albert Tai for TnSeq library analysis, and Bree Aldridge and members of the Mecsas lab for helpful discussions and critical review of the manuscript.
R.J.S. and M.K.P. were supported by National Institutes of Health/National Institute of Allergy and Infectious Disease (NIH NIAID) grants 2T32AI007077 (R.J.S. and M.K.P.) and T32AI007422 (M.K.P.). J.M. and A.L.M. were supported by NIH NIAID grants AI113166 and AI107055. This study used NMR instrumentation purchased with funding from NIH SIG grant S10OD020073. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
R.J.S., M.K.P., A.L.M., and J.M. designed research and wrote the paper. R.J.S., M.K.P., and A.L.M. performed research and analyzed data. J.M.B.-L. contributed clinical isolates. J.D.B. contributed analytic tools and NMR expertise.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02674-18.
REFERENCES
- 1.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis GS, Waits K, Nordstrom L, Weaver B, Aziz M, Gauld L, Grande H, Bigler R, Horwinski J, Porter S, Stegger M, Johnson JR, Liu CM, Price LB. 2015. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin Infect Dis 61:892–899. doi: 10.1093/cid/civ428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 8.Barczak AK, Hung DT. 2009. Productive steps toward an antimicrobial targeting virulence. Curr Opin Microbiol 12:490–496. doi: 10.1016/j.mib.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dersch P, Muehlen S. 2015. Anti-virulence strategies to target bacterial infections, p 147–183. In Stadler M, Dersch P (ed), How to overcome the antibiotic crisis, vol 398 Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 11.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5:e01163-14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HL. 2015. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775. doi: 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green ER, Clark S, Crimmins GT, Mack M, Kumamoto CA, Mecsas J. 2016. Fis is essential for Yersinia pseudotuberculosis virulence and protects against reactive oxygen species produced by phagocytic cells during infection. PLoS Pathog 12:e1005898. doi: 10.1371/journal.ppat.1005898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong H, Carter RA, Leiner IM, Tang YW, Chen L, Kreiswirth BN, Pamer EG. 2015. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun 83:3418–3427. doi: 10.1128/IAI.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, Keith JW, Samilo DW, Carter RA, Leiner IM, Pamer EG. 2016. Innate lymphocyte/Ly6Chi monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell 165:679–689. doi: 10.1016/j.cell.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meatherall BL, Gregson D, Ross T, Pitout JD, Laupland KB. 2009. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med 122:866–873. doi: 10.1016/j.amjmed.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 18.Traslavina RP, King EJ, Loar AS, Riedel ER, Garvey MS, Ricart-Arbona R, Wolf FR, Couto SS. 2010. Euthanasia by CO2 inhalation affects potassium levels in mice. J Am Assoc Lab Anim Sci 49:316–322. [PMC free article] [PubMed] [Google Scholar]
- 19.Sezonov G, Joseleau-Petit D, D’Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batra S, Cai S, Balamayooran G, Jeyaseelan S. 2012. Intrapulmonary administration of leukotriene B4 augments neutrophil accumulation and responses in the lung to Klebsiella infection in CXCL1 knockout mice. J Immunol 188:3458–3468. doi: 10.4049/jimmunol.1101985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye P, Garvey PB, Zhang P, Nelson S, Bagby G, Summer WR, Schwarzenberger P, Shellito JE, Kolls JK. 2001. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am J Respir Cell Mol Biol 25:335–340. doi: 10.1165/ajrcmb.25.3.4424. [DOI] [PubMed] [Google Scholar]
- 22.Rogers SG, Brand LA, Holder SB, Sharps ES, Brackin MJ. 1983. Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl Environ Microbiol 46:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao L-Y, Willis DK, Andrews-Polymenis H, McClelland M, Barak JD. 2012. Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl Environ Microbiol 78:4561–4570. doi: 10.1128/AEM.07867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha R, Saha N, Donofrio RS, Bestervelt LL. 2013. Microbial siderophores: a mini review. J Basic Microbiol 53:303–317. doi: 10.1002/jobm.201100552. [DOI] [PubMed] [Google Scholar]
- 25.Lamb AL. 2015. Breaking a pathogen’s iron will: inhibiting siderophore production as an antimicrobial strategy. Biochim Biophys Acta 1854:1054–1070. doi: 10.1016/j.bbapap.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao SM, Du QS, Meng JZ, Pang ZW, Huang RB. 2013. The multiple roles of histidine in protein interactions. Chem Cent J 7:44. doi: 10.1186/1752-153X-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sost D, Amrhein N. 1990. Substitution of Gly-96 to Ala in the 5-enolpyruvylshikimate-3-phosphate synthase of Klebsiella pneumoniae results in a greatly reduced affinity for the herbicide glyphosate. Arch Biochem Biophys 282:433–436. doi: 10.1016/0003-9861(90)90140-T. [DOI] [PubMed] [Google Scholar]
- 28.Bi W, Liu H, Dunstan RA, Li B, Torres VVL, Cao J, Chen L, Wilksch JJ, Strugnell RA, Lithgow T, Zhou T. 2017. Extensively drug-resistant Klebsiella pneumoniae causing nosocomial bloodstream infections in China: molecular investigation of antibiotic resistance determinants, informing therapy, and clinical outcomes. Front Microbiol 8:1230. doi: 10.3389/fmicb.2017.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, Xu YC, Zhang XJ, Zhang CX, Ji P, Xie Y, Kang M, Wang CQ, Wang AM, Xu YH, Shen JL, Sun ZY, Chen ZJ, Ni YX, Sun JY, Chu YZ, Tian SF, Hu ZD, Li J, Yu YS, Lin J, Shan B, Du Y, Han Y, Guo S, Wei LH, Wu L, Zhang H, Kong J, Hu YJ, Ai XM, Zhuo C, Su DH, Yang Q, Jia B, Huang W. 2016. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect 22(Suppl 1):S9–S14. doi: 10.1016/j.cmi.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 31.Sammons RD, Gaines TA. 2014. Glyphosate resistance: state of knowledge. Pest Manag Sci 70:1367–1377. doi: 10.1002/ps.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munita JM, Arias CA. 2016. Mechanisms of antibiotic resistance. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eschenburg S, Healy ML, Priestman MA, Lushington GH, Schonbrunn E. 2002. How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from Escherichia coli. Planta 216:129–135. doi: 10.1007/s00425-002-0908-0. [DOI] [PubMed] [Google Scholar]
- 34.Funke T, Yang Y, Han H, Healy-Fried M, Olesen S, Becker A, Schönbrunn E. 2009. Structural basis of glyphosate resistance resulting from the double mutation Thr97→Ile and Pro101→Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J Biol Chem 284:9854–9860. doi: 10.1074/jbc.M809771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutton KA, Breen J, Russo TA, Schultz LW, Umland TC. 2016. Crystal structure of 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase from the ESKAPE pathogen Acinetobacter baumannii. Acta Crystallogr F Struct Biol Commun 72:179–187. doi: 10.1107/S2053230X16001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia MD, Chua SMH, Low YS, Lee YT, Agnew-Francis K, Wang JG, Nouwens A, Lonhienne T, Williams CM, Fraser JA, Guddat LW. 2018. Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections. Proc Natl Acad Sci U S A 115:E9649–E9658. doi: 10.1073/pnas.1809422115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abrahams KA, Cox JAG, Futterer K, Rullas J, Ortega-Muro F, Loman NJ, Moynihan PJ, Perez-Herran E, Jimenez E, Esquivias J, Barros D, Ballell L, Alemparte C, Besra GS. 2017. Inhibiting mycobacterial tryptophan synthase by targeting the inter-subunit interface. Sci Rep 7:9430. doi: 10.1038/s41598-017-09642-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akashi H, Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A 99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruickshank-Quinn C, Powell R, Jacobson S, Kechris K, Bowler RP, Petrache I, Reisdorph N. 2017. Metabolomic similarities between bronchoalveolar lavage fluid and plasma in humans and mice. Sci Rep 7:5108. doi: 10.1038/s41598-017-05374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amorim Franco TM, Blanchard JS. 2017. Bacterial branched-chain amino acid biosynthesis: structures, mechanisms, and drugability. Biochemistry 56:5849–5865. doi: 10.1021/acs.biochem.7b00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurenbach B, Marjoshi D, Amabile-Cuevas CF, Ferguson GC, Godsoe W, Gibson P, Heinemann JA. 2015. Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. mBio 6:e00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesnage R, Bernay B, Seralini GE. 2013. Ethoxylated adjuvants of glyphosate-based herbicides are active principles of human cell toxicity. Toxicology 313:122–128. doi: 10.1016/j.tox.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Landrigan PJ, Belpoggi F. 2018. The need for independent research on the health effects of glyphosate-based herbicides. Environ Health 17:51. doi: 10.1186/s12940-018-0392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buzzola FR, Barbagelata MS, Caccuri RL, Sordelli DO. 2006. Attenuation and persistence of and ability to induce protective immunity to a Staphylococcus aureus aroA mutant in mice. Infect Immun 74:3498–3506. doi: 10.1128/IAI.01507-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Priebe GP, Brinig MM, Hatano K, Grout M, Coleman FT, Pier GB, Goldberg JB. 2002. Construction and characterization of a live, attenuated aroA deletion mutant of Pseudomonas aeruginosa as a candidate intranasal vaccine. Infect Immun 70:1507–1517. doi: 10.1128/iai.70.3.1507-1517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staub JM, Brand L, Tran M, Kong Y, Rogers SG. 2012. Bacterial glyphosate resistance conferred by overexpression of an Escherichia coli membrane efflux transporter. J Ind Microbiol Biotechnol 39:641–647. doi: 10.1007/s10295-011-1057-x. [DOI] [PubMed] [Google Scholar]
- 47.Healy-Fried ML, Funke T, Priestman MA, Han H, Schönbrunn E. 2007. Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. J Biol Chem 282:32949–32955. doi: 10.1074/jbc.M705624200. [DOI] [PubMed] [Google Scholar]
- 48.Mesnage R, Renney G, Seralini GE, Ward M, Antoniou MN. 2017. Multiomics reveal non-alcoholic fatty liver disease in rats following chronic exposure to an ultra-low dose of Roundup herbicide. Sci Rep 7:39328. doi: 10.1038/srep39328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barolo D. 1993. Reregistration eligibility decision (RED): glyphosate. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 50.Mesnage R, Antoniou MN. 2017. Ignoring adjuvant toxicity falsifies the safety profile of commercial pesticides. Front Public Health 5:361. doi: 10.3389/fpubh.2017.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarazona JV, Court-Marques D, Tiramani M, Reich H, Pfeil R, Istace F, Crivellente F. 2017. Glyphosate toxicity and carcinogenicity: a review of the scientific basis of the European Union assessment and its differences with IARC. Arch Toxicol 91:2723–2743. doi: 10.1007/s00204-017-1962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greim H, Saltmiras D, Mostert V, Strupp C. 2015. Evaluation of carcinogenic potential of the herbicide glyphosate, drawing on tumor incidence data from fourteen chronic/carcinogenicity rodent studies. Crit Rev Toxicol 45:185–208. doi: 10.3109/10408444.2014.1003423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aoki N, Tateda K, Kikuchi Y, Kimura S, Miyazaki C, Ishii Y, Tanabe Y, Gejyo F, Yamaguchi K. 2009. Efficacy of colistin combination therapy in a mouse model of pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. J Antimicrob Chemother 63:534–542. doi: 10.1093/jac/dkn530. [DOI] [PubMed] [Google Scholar]
- 54.Hua Y, Luo T, Yang Y, Dong D, Wang R, Wang Y, Xu M, Guo X, Hu F, He P. 2017. Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front Microbiol 8:2659. doi: 10.3389/fmicb.2017.02659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Díaz-Quiroz DC, Cardona-Félix CS, Viveros-Ceballos JL, Reyes-González MA, Bolívar F, Ordoñez M, Escalante A. 2018. Synthesis, biological activity and molecular modeling studies of shikimic acid derivatives as inhibitors of the shikimate dehydrogenase enzyme of Escherichia coli. J Enzyme Inhib Med Chem 33:397–404. doi: 10.1080/14756366.2017.1422125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies GM, Barrett-Bee KJ, Jude DA, Lehan M, Nichols WW, Pinder PE, Thain JL, Watkins WJ, Wilson RG. 1994. (6S)-6-fluoroshikimic acid, an antibacterial agent acting on the aromatic biosynthetic pathway. Antimicrob Agents Chemother 38:403–406. doi: 10.1128/aac.38.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdo MR, Joseph P, Mortier J, Turtaut F, Montero JL, Masereel B, Kohler S, Winum JY. 2011. Anti-virulence strategy against Brucella suis: synthesis, biological evaluation and molecular modeling of selective histidinol dehydrogenase inhibitors. Org Biomol Chem 9:3681–3690. doi: 10.1039/c1ob05149k. [DOI] [PubMed] [Google Scholar]
- 58.Arrach N, Cheng P, Zhao M, Santiviago CA, Hoffman RM, McClelland M. 2010. High-throughput screening for Salmonella avirulent mutants that retain targeting of solid tumors. Cancer Res 70:2165–2170. doi: 10.1158/0008-5472.CAN-09-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 60.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vejborg RM, de Evgrafov MR, Phan MD, Totsika M, Schembri MA, Hancock V. 2012. Identification of genes important for growth of asymptomatic bacteriuria Escherichia coli in urine. Infect Immun 80:3179–3188. doi: 10.1128/IAI.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crimmins GT, Mohammadi S, Green ER, Bergman MA, Isberg RR, Mecsas J. 2012. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS Pathog 8:e1002828. doi: 10.1371/journal.ppat.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang TW, Lam I, Chang HY, Tsai SF, Palsson BO, Charusanti P. 2014. Capsule deletion via a lambda-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res Notes 7:13. doi: 10.1186/1756-0500-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leclercq R, Canton R, Brown DF, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy CJ, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 19:141–160. doi: 10.1111/j.1469-0691.2011.03703.x. [DOI] [PubMed] [Google Scholar]
- 65.Brown DF, Wootton M, Howe RA. 2016. Antimicrobial susceptibility testing breakpoints and methods from BSAC to EUCAST. J Antimicrob Chemother 71:3–5. doi: 10.1093/jac/dkv287. [DOI] [PubMed] [Google Scholar]
- 66.Palacios M, Miner TA, Frederick DR, Sepulveda VE, Quinn JD, Walker KA, Miller VL. 2018. Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. mBio 9:e01443-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thornton B, Basu C. 2011. Real-time PCR (qPCR) primer design using free online software. Biochem Mol Biol Educ 39:145–154. doi: 10.1002/bmb.20461. [DOI] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.