Abstract
Background:
Asthma represents a significant public health burden; however, novel biological therapies targeting immunoglobulin E (IgE)-mediated pathways have widened clinical treatment options for the disease.
Objective:
In this study we sought to identify gene transcripts and gene networks involved in the determination of serum IgE levels in people with asthma that can help inform the development of novel therapeutic agents.
Methods:
We analyzed gene expression data from a cross-sectional study of 326 Costa Rican children with asthma, aged 6 to 12 years, from the Genetics of Asthma in Costa Rica Study and 610 young adults with asthma, aged 16 to 25 years, from the Childhood Asthma Management Program trial. We utilized differential gene expression analysis and performed weighted gene co-expression network analysis on 25,060 genes, to identify gene transcripts and network modules associated with total IgE, adjusting for age and gender. We used pathway enrichment analyses to identify key biological pathways underlying significant modules. We compared findings that replicated between both populations.
Results:
We identified 31 transcripts associated with total IgE that replicated between the two study cohorts. These results were notable for increased eosinophil-related transcripts (including IL5RA, CLC, SMPD3, CCL23, CEBPE). Pathway enrichment identified the regulation of T cell tolerance as important in the determination of total IgE levels, supporting a key role for IDO1.
Conclusions & Clinical Relevance:
These results provide robust evidence that biologically meaningful gene expression profiles (relating to eosinophilic and regulatory T cell pathways in particular) associated with total IgE levels can be identified in individuals diagnosed with asthma during childhood. These profiles and their constituent genes may represent novel therapeutic targets.
Keywords: Allergic asthma, computational biology, eosinophils, network medicine, gene expression
Introduction:
Asthma, allergies and related disorders have doubled in prevalence during recent decades and are among the most common chronic diseases in industrialized countries, with asthma affecting 26 million United States children and adults, and allergies affecting ~42.5% of the population (1–4). A large proportion of asthma cases are characterized by an increase in serum immunoglobulin E (IgE) levels (5), and a dose-response relationship between IgE levels and disease severity has been demonstrated (6). Total IgE is an important therapeutic target, and existing biologic therapies (including omalizumab and recently approved reslizumab and mepolizumab) target IL-4, IL-5, IL-13, and IgE-specific pathways (7). However, therapeutic trials of these biologic agents only demonstrate benefits among subjects with more severe asthma subtypes (7). These findings provide further evidence that asthma comprises a diverse range of immunologic and clinical phenotypes, which require an equally diverse variety of personalized therapies.
Given the high heritability of IgE (estimated to be up to 0.8 in cord blood) (8), a better understanding of the causal genetic factors underlying IgE levels among individuals with asthma may lead to novel IgE-focused treatments for a subset of cases. While many of the genetic and genomic studies of total IgE levels have been performed in genetically heterogeneous populations (9–12), we hypothesized that investigating a more homogeneous population, specifically a semi-population isolate (Costa Rican children with asthma), will enable the improved identification of novel biologic targets. A genetic study of this population previously identified an association between polymorphisms in IL13 and serum IgE, eosinophil count, and asthma exacerbations (13).
The aim of this study was to use a network genomics approach to identify novel differentially expressed genes and gene-networks associated with serum total IgE in two populations with asthma; the ‘Genetics of Asthma in Costa Rica Study’ (GACRS) and ‘The Childhood Asthma Management Program’ (CAMP) cohort. By comparing the semi-isolated GACRS population, which is characterized by a high asthma prevalence (24% in children), a more homogenous genetic background, and a highly allergic population, to the heterogeneous CAMP population, we aimed to improve the robustness and generalizability of our findings (14). These approaches using gene co-expression networks have been useful in identifying gene signatures associated with severe subtypes of asthma, but have not previously been employed in association with IgE levels in asthma(15, 16). Elucidating the biology underlying serum total IgE, particularly as it pertains to asthma severity, is vital to develop improved strategies for the prevention, management, and treatment of asthma and allergic conditions.
Methods:
GACRS population and study design
Participants in the GACRS are members of a Hispanic population semi-isolate from the Central Valley of Costa Rica, which has one of the highest rates of asthma worldwide (17). From February 2001 to August 2008, questionnaires were sent to the parents of 16,912 children (ages 6–14 years) enrolled in 140 Costa Rican schools; 9,180 (54.3%) questionnaires were returned. Children were eligible for the study if they had asthma (physician-diagnosed asthma and ≥2 respiratory symptoms or asthma attacks in the prior year) and a high probability of having ≥6 great-grandparents born in the Central Valley of Costa Rica. A total of 1,189 children with asthma and their parents (i.e. 383 parent-child trios) were enrolled. 383 children provided blood samples, on which whole-blood RNA gene expression profiling was performed, using the Illumina HumanHT-12v4 microarray. All participating children completed a protocol including a slightly modified and translated version of the questionnaire used in the Collaborative Study on the Genetics of Asthma (18). Serum total IgE level was determined using the UniCAP 250 system (Pharmacia & Upjohn, Kalamazoo, Mich), with samples measured in duplicate, conducted at the same time as gene expression profiling. Written parental consent and child’s assent were obtained. The study was approved by the Institutional Review Boards of the Hospital Nacional de Ninos (San Jose, Costa Rica) and Brigham and Women’s Hospital (Boston, MA, USA) (IRB Protocol Number 2000P001130).
CAMP population and study design
CAMP is a multi-center, randomized, double-masked, clinical trial designed to determine the long-term (~16.5 years of follow-up) effects of three inhaled treatments for mild to moderate asthma in children aged 5 to 12 at baseline: placebo, nedocromil, or budesonide.(19) Asthma was defined based on having both a clinical history (with presence of symptoms or use of an inhaled bronchodilator at least twice weekly or the use of daily medication for asthma) and airway responsiveness to methacholine (as measured by 20% decline in forced expiratory volume (FEV1) at doses ≤12.5 mg/mL of methacholine). A follow-up study to the primary trial extracted blood samples from 620 CAMP subjects at early adulthood (ages 16 to 25, a mean 11.6 years (range 10–13.5 years) after enrollment) for gene expression profiling using the same Illumina HumanHT chip and concurrently measured serum total IgE (two individuals for whom IgE was measured >18 months from gene expression profiling were removed from the analysis). Of these, 610 subjects had complete phenotype data for downstream analysis.
Gene expression profiling
In both populations, whole-blood gene expression was assayed with 47,009 probes on the Illumina HumanHT-12 v4 Expression BeadChip (Illumina, Inc., San Diego, CA, USA) that passed stringent and commonly used quality control metrics (20). Expression data were log2-transformed and quantile-normalized using the R package “lumi” (version 2.22) and Bioconductor (21). Principal components of gene expression were generated for both GACRS and for CAMP. Prior to downstream statistical analyses, a standard non-specific variance filter was applied to the expression data using the R package “genefilter”. Probes not annotated with a valid Entrez gene identifier or Human Genome Organization gene symbol and probes with inter-quartile ranges (IQR) of expression variance below the 50th percentile were excluded,(22) ensuring that the most informative probes were used for analysis. The data were then collapsed to assign a single probe to each gene based on the largest IQR of expression variance. This resulted in a total of 25,059 and 24,971 gene probes for the GACRS and CAMP analysis respectively. All 24,971 genes in the CAMP dataset were also represented in the larger GACRS dataset.
Statistical analyses
All analyses were first conducted in the GACRS, and then conducted independently in the CAMP population for validation of the significant findings using a parallel analysis design and identical methods. As in previous study designs (23, 24), we aimed to compare findings that replicated in both a relative genetic isolate (GACRS) as well as an outbred population (CAMP) (19).
Differential gene expression analysis
Generalized linear regression models (GLM)(25) were constructed to identify differentially expressed genes associated with log-transformed total serum IgE levels. We adjusted for age (due to the age differences in our two cohorts), sex (due to prior genetic associations with IgE (25)), and the first two principal components of gene expression to account for any batch effects (all concurrent with serum total IgE measurements). The Benjamini– Hochberg procedure was used to control the false discovery rate (FDR) with q-value set at 0.05 (26).
Gene network analysis
Gene expression modules have previously been shown to be enriched for biological functions. We hypothesized that by using Weighted Gene Co-expression Network Analysis (WGCNA) (27) we would be able to refine the results of the regression analysis to identify networks of highly significant genes that are expressed together in groups or ‘modules’ of co-expressed genes. These modules can then be related to phenotypic and baseline traits. WGCNA was used to generate a network describing correlation patterns between the genes. Using hierarchical clustering based on topological overlap and applying a soft thresholding power of 5, chosen to achieve a scale-free topology fitting index >0.9 (explaining over 90% of the variance), the interconnectedness between all possible gene pairs was quantified and they were assigned to gene co-expression modules. A dynamic clustering approach was utilized with a deep split of 0.8, and a minimum module size of 30 was specified, in order to further refine the degree of detail each module contained without influencing module structure, based on recommended defaults (27). We then removed all uncorrelated transcripts that were assigned to the grey module, a module typically reserved for transcripts that do not map to a discrete module. Each module was summarized by an eigenvalue based on its first principal component, and the association between the module eigenvalues and log10-transformed serum total IgE, after adjustment for age at gene expression profiling and gender, was computed using multivariable linear regression.
Pathway enrichment analysis
Pathway enrichment analysis using Gene Ontology (GO) terms was performed using an online tool from the GO Consortium (http://geneontology.org/page/go-enrichment-analysis), relying on the PANTHER Overrepresentation Test (release 20171205) (28) with an alpha<0.01, fold enrichment >5, and containing at least 2 genes transcripts from our study included in the GO category. We identified key biological pathways among (i) the differentially expressed genes identified using the multivariate linear regression models, (ii) the gene transcripts of WGCNA-generated modules associated with serum IgE.
Overrepresentation analysis
We conducted gene set overrepresentation analysis using replicated gene transcripts common to both the GACRS and CAMP population, in comparison with the Molecular Signatures Database version 6.1 (http://software.broadinstitute.org/gsea/msigdb/).(29) We specifically used hypergeometric tests with FDR to correct for multiple hypothesis testing and compared our dataset with the C7 database, which comprises 4872 immunologic gene sets that were curated based on published human and murine studies of the immune system from the Gene Expression Omnibus (30).
Results:
Study populations
The baseline characteristics of the GACRS and CAMP study populations are described in Table 1. The GACRS comprises a Hispanic/Latino population, while CAMP was predominantly white European (68%), and only 10% Hispanic/Latino. The proportion of males and females in the two cohorts were similar, with a higher percentage of males in both. The age of asthma onset was young in both cohorts: 2.4 years in the GACRS and 3.0 years in CAMP. Gene expression profiling and the measurement of serum IgE levels were performed on blood samples taken in childhood (mean: 9.1 years) in the GACRS, and in early adulthood in CAMP (mean: 20.4 years). Importantly, there was no significant difference in serum IgE levels between the two cohorts, despite the age difference.
Table 1:
Demographics of GACRS population and CAMP populations
| GACRS | CAMP | P-Value | |
|---|---|---|---|
| Number of participants | 326 | 610 | |
| Sex, n (%) | |||
| Female | 140 (43%) | 226 (37%) | 0.10 |
| Male | 186 (57%) | 384 (63%) | |
| Race/Ethnicity | |||
| Hispanic/Latino | 326 (100%) | 59 (10%) | na |
| American Indian or Alaskan Native | 0 (0%) | 4 (1%) | |
| Black or African American | 0 (0%) | 94 (15%) | |
| East/Southeast Asian | 0 (0%) | 4 (1%) | |
| European | 0 (0%) | 417 (68%) | |
| Multiple Races | 0 (0%) | 32 (5%) | |
| Age at gene expression profiling, mean (SD) | 9.1 (1.8) | 20.4 (2.2) | <0.001 |
| Age of asthma onset (years), mean (SD) | 2.4 (2.2) | 3.0 (2.4) | <0.001 |
| log10IgE (kU/L), mean (SD) | 2.5 (0.7) | 2.5 (0.6) | 0.74 |
| Atopic Dermatitis | 38 (25%) | 174 (29%) | 0.44 |
| Allergic Rhinitis | 300 (92%) | 310 (51%) | <0.001 |
| Inhaled Corticosteroid Use | 173 (53%) | 125 (23%) | <0.001 |
Differential gene expression analysis
A total of 81 gene transcripts were significantly associated with serum IgE levels in the GACRS cohort after adjustment for age at expression profiling, gender, and the first two principal components, and controlling for the FDR. 69 of these gene transcripts (85%) were positively associated with serum total IgE and 12 (15%) were negatively associated (Figure 1). Differential gene expression analysis showed no association of these genes with atopic dermatitis or allergic rhinitis. Of these 81 gene transcripts, 42 (54%) were replicated in CAMP with a concordant direction of effect; all 42 were overexpressed with increasing IgE levels (Figure 2, Supplementary Table 1). We then conducted a sensitivity analysis, adjusting for inhaled corticosteroid use in both populations, with 36 (86%) transcripts still significant (Supplementary Table 1). Of note, all of the transcripts identified in CAMP remained significantly associated with IgE after adjusting for original treatment group from the CAMP randomized trial. The 42 transcripts replicated in CAMP and GACRS were nominally enriched (p<0.01, fold enrichment>5, number of genes>1) for 32 biological processes (Supplementary Table 2). Although none of these 32 processes were robust to the most stringent correction for multiple testing, Bonferroni correction, a number had biologically plausible functional annotations, particularly those related to immune responses, such as ‘regulation of T cell tolerance induction’.
Figure 1; Number of genes significantly associated with serum total IgE in the GACRS and CAMP populations by differential gene expression.
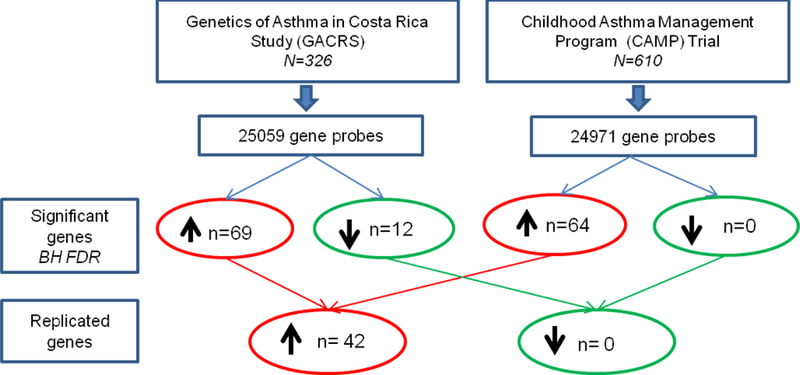
BH FDR = Benjamini-Hochberg adjusted false discovery rate.
Figure 2: Heatmap comparing differential gene expression of the 42 replicated genes by IgE (top versus bottom quartile) in the (A) GACRS and (B) CAMP populations.
All individuals were included in the differential gene expression analysis; however the top quartile IgE (green) and the bottom quartile IgE (yellow) were displayed for comparison purposes.
Gene network analysis
The gene expression network analysis was first performed in the GACRS. The samples were clustered, resulting in the exclusion of three samples identified as outliers. From the 25,059 transcripts, hierarchical clustering based on topological overlap then designated the 10,073 transcripts into 32 modules (43%) (Supplementary Figure 1A). Five modules, as summarized by their eigengenes, were associated with serum IgE level (alpha<0.05) (Supplementary Figure 1A). Only one module, containing 103 gene transcripts, was robust to correction for multiple testing (p=4.8×10−11). This module was also associated with the more stringent subtype of asthma (p=0.002), defined by physician diagnosis and the presence of airway hyperreactivity on methacholine challenge, but was not associated with allergic rhinitis or atopic dermatitis. The module was also not associated with inhaled corticosteroid use, but had a nominal association with oral steroid use in the last year (p=0.04) Pathway enrichment analysis of these 103 transcripts identified 52 GO-defined biological processes (Supplementary Table 3), including 12 of the 32 processes identified in the differential gene expression analysis and additional processes relating to immune function.
WGCNA was then applied to the CAMP dataset using the same network construction approach; one sample was excluded as an outlier. Within the remaining 611 samples the gene transcripts were clustered into 40 modules including 15,120 transcripts (61%). Fourteen modules were associated with serum IgE levels after adjustment for age and gender (p<0.05), and one module, was robust to correction for multiple testing (p=6.8×10−14) (Supplementary Figure 1B). This module was also associated with inhaled corticosteroid use (p=0.0003). This module contained 38 transcripts that were enriched for 25 biological processes (Supplementary Table 4).
To determine any genomic and biological similarities between the significant GACRS and CAMP modules, the overlap in terms of gene transcripts and enriched biological processes was computed. Of the 103 transcripts in the significant GACRS module and the 38 transcripts in the significant CAMP module, all 38 were common to both (Supplementary Table 5). Of the 52 and 25 enriched GO-defined biological processes from the GACRS and CAMP modules, respectively, 14 were common to both (Supplementary Table 5). Finally the similarities between the differential gene expression analysis and the WGCNA analyses were compared (Figure 3). Thirty-one transcripts (Table 2), and twelve enriched biological processes, including a number related to immune response, were common to both sets of analyses in both cohorts (Table 3), and further confirmed the similarities in results between the two independent populations. Based on the literature, eight of the thirty-one gene transcripts have been previously linked to IgE, while the remainder represent novel associations (Table 2).
Figure 3;
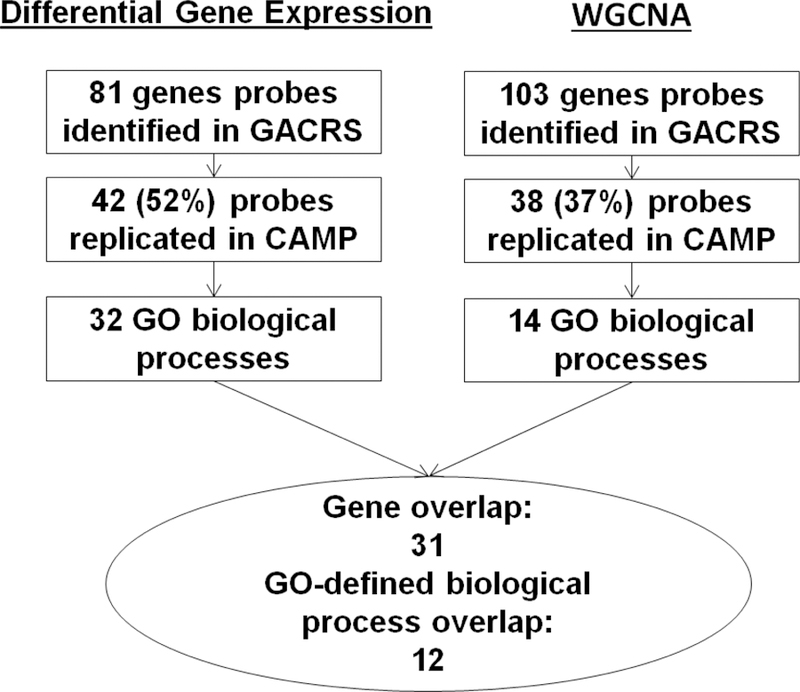
Comparison of the total IgE gene expression profiles generated using differential gene expression and network approaches
Table 2:
Replicated gene transcripts (n=31) associated with total IgE using two different statistical approaches in two distinct cohorts with asthma: GACRS and CAMP
| Gene Symbol | Gene Name | Chr | GACRS P-Value |
CAMP P-Value |
Sig. adjusted for ICS | Linked with eosinophils |
||
|---|---|---|---|---|---|---|---|---|
| Esnault (37) | Nakajima(38) | MSigDB(29) | ||||||
| PRSS33 | protease, serine, 33 | 16 | 2.8×10−9 | 2.4×10−10 | ■ | ● | ||
| OLIG2 | oligodendrocyte lineage transcription factor 2 | 21 | 1.7×10−8 | 2.3×10−12 | ■ | ● | ||
| TFF3 | trefoil factor 3 (intestinal) | 21 | 2.4×10−8 | 6.7×10−13 | ■ | ● | ||
| CLC* | Charcot-Leyden crystal protein | 19 | 3.8×10−8 | 4.0×10−9 | ■ | ● | ● | |
| CCL23 | chemokine (C-C motif) ligand 23; macrophage inflammatory protein 1-alpha (MIP-1-alpha) | 17 | 1.5×10−7 | 3.0×10−10 | ■ | ● | ● | |
| EMR4P | adhesion G protein-coupled receptor E4, pseudogene | 19 | 1.5×10−7 | 7.1×10−6 | ■ | ● | ||
| OLIG1 | oligodendrocyte transcription factor 1 | 21 | 1.5×10−7 | 7.8×10−7 | ■ | ● | ||
| GPR44* | prostaglandin D2 receptor 2, G protein-coupled receptor 44, (aka CRTH2) | 11 | 2.0×10−7 | 2.1×10−8 | ■ | ● | ● | |
| FLJ43093 | Unknown | 6 | 2.5×10−7 | 2.0×10−8 | ■ | |||
| CEBPE | CCAAT/enhancer binding protein (C/EBP), epsilon | 14 | 2.7×10−7 | 2.6×10−13 | ■ | ● | ||
| PYROXD2 | pyridine nucleotide-disulphide oxidoreductase domain 2 (PYROXD2) | 10 | 3.3×10−7 | 3.8×10−10 | ■ | ● | ||
| SORD2 | sorbitol dehydrogenase | 15 | 1.5×10−6 | 4.9×10−5 | ■ | |||
| SIGLEC8* | sialic acid binding Ig-like lectin 8 | 19 | 1.6×10−6 | 5.0×10−9 | ■ | ● | ||
| PIK3R6* | phosphoinositide-3-kinase, regulatory unit 6 | 17 | 2.9×10−6 | 2.0×10−8 | ■ | ● | ||
| LGALS12 | galectin 12 | 11 | 6.1×10−6 | 3.7×10−4 | ■ | ● | ||
| GFOD1 | glucose-fructose oxidoreductase domain containing 1 | 6 | 8.7×10−6 | 1.8×10−3 | ■ | |||
| CCR3* | chemokine (C-C motif) receptor 3 | 3 | 1.8×10−5 | 9.2×10−5 | ■ | ● | ● | |
| IL5RA* | interleukin 5 receptor, alpha | 3 | 3.5×10−5 | 0.020 | ■ | ● | ● | |
| SMPD3 | sphingomyelin phosphodiesterase 3, neutral membrane | 16 | 5.6×10−5 | 2.2×10−6 | ■ | ● | ||
| INPP1 | inositol polyphosphate-1-phosphatase | 2 | 1.5×10−4 | 6.4×10−5 | ■ | |||
| EMR1 | adhesion G protein-coupled receptor E1 (alias ADGRE1) | 19 | 2.5×10−4 | 5.5×10−3 | ■ | ● | ||
| ACOT11 | acyl-CoA thioesterase 11 | 1 | 2.6×10−4 | 2.1×10−4 | ■ | |||
| CAMK1 | calcium/calmodulin-dependent protein kinase I | 3 | 3.0×10−4 | 6.8×10−5 | ■ | ● | ● | |
| VSTM1 | V-set & transmembrane domain containing 1 | 19 | 4.6×10−4 | 1.9×10−6 | ■ | ● | ||
| RNF14 | ring finger protein 14 | 5 | 4.6×10−4 | 3.1×10−5 | ■ | |||
| SPNS3 | spinster homolog 3 (Drosophila) | 17 | 6.2×10−4 | 6.6×10−9 | ■ | ● | ||
| ZBTB42 | zinc finger and BTB domain containing 42 | 14 | 7.5×10−4 | 2.7×10−8 | ■ | |||
| IDO1* | indoleamine 2,3-dioxygenase 1 (aka INDO) | 8 | 1.1×10−3 | 2.4×10−10 | ■ | ● | ● | |
| LOC642639 | Unknown | 9 | 1.2×10−3 | 1.7×10−3 | ■ | |||
| LTC4S* | leukotriene C4 synthase | 5 | 2.5×10−3 | 6.9×10−6 | ■ | ● | ||
| CYP4F12 | cytochrome P450, family 4, subfamily F, polypeptide 12 | 19 | 4.2×10−3 | 2.0×10−5 | ■ | ● | ||
Known association with IgE.
known association with eosinophils.
GACRS=Genetics of Asthma in Costa Rica Study
CAMP=Childhood Asthma Management Program
MSigDB=Molecular Signature Database.
Significant in both cohorts after adjustment for recent inhaled corticosteroid (ICS) use.
Table 3;
Gene-ontology defined biological processes associated with total IgE using two different statistical approaches in two different cohorts: GACRS and CAMP cohorts.
| GO biological process | # Genes in GO Group | Differential Gene Expression | WGCNA | ||||
|---|---|---|---|---|---|---|---|
| GACRS & CAMP | GACRS | CAMP | |||||
| Fold Enrichment | Raw P value | Fold Enrichment | Raw P value | Fold Enrichment | Raw P value | ||
| regulation of leukocyte mediated immunity (GO:0002703): CLC, PIK3R6 | 167 | 9.7 | 0.0038 | 6.3 | 0.0014 | 10.5 | 0.0030 |
| regulation of leukocyte proliferation (GO:0070663) : IDO1, CLC | 219 | 7.4 | 0.0080 | 5.8 | 0.0007 | 8.0 | 0.0064 |
| regulation of lymphocyte mediated immunity (GO:0002706): CLC, PIK3R6 | 126 | 12.9 | 0.0017 | 6.7 | 0.0034 | 13.9 | 0.0014 |
| regulation of lymphocyte proliferation (GO:0050670) : IDO1, CLC | 210 | 7.7 | 0.0071 | 6.0 | 0.0006 | 8.4 | 0.0057 |
| regulation of mononuclear cell proliferation (GO:0032944) : IDO1, CLC | 211 | 7.7 | 0.0072 | 6.0 | 0.0006 | 8.3 | 0.0058 |
| regulation of T cell tolerance induction (GO:0002664): IDO1, CLC | 12 | 89.9 | 0.0003 | 35.1 | 0.0019 | 97.4 | 0.0003 |
| regulation of tolerance induction (GO:0002643): IDO1, CLC | 18 | 60.0 | 0.0006 | 23.4 | 0.0040 | 64.9 | 0.0005 |
| neuron fate commitment (GO:0048663): OLIG1, OLIG2 | 70 | 15.4 | 0.0079 | 12.0 | 0.0004 | 16.7 | 0.0067 |
| oligodendrocyte differentiation (GO:0048709) : OLIG1, OLIG2 | 65 | 16.6 | 0.0068 | 13.0 | 0.0003 | 27.0 | 0.0002 |
| positive regulation of glial cell differentiation (GO:0045687): OLIG2, CLC | 33 | 32.7 | 0.0019 | 19.1 | 0.0007 | 35.4 | 0.0016 |
| positive regulation of gliogenesis (GO:0014015) : OLIG2, CLC | 51 | 31.7 | 0.0001 | 16.5 | 0.0001 | 22.9 | 0.0037 |
| regulation of glial cell differentiation (GO:0045685) : OLIG2, CLC | 59 | 18.3 | 0.0057 | 14.3 | 0.0002 | 19.8 | 0.0049 |
Overrepresentation Analysis
To further clarify the immunologic relevance of the 31 validated transcripts, we compared our gene set to a database of immunologic signatures, yielding three gene sets that were enriched (Supplementary Table 6). These three gene sets encompassed gene transcripts upregulated in neutrophils (GPR44, CCR3, IL5RA, CLC, SIGLEC8; FDR q value<0.001), bone-marrow derived macrophages (EMR1, CCL23, OLIG2, OLIG1; q<0.011), and eosinophils (SMPD3, CCL23, IDO1, CEBPE; q<0.011).
Discussion:
Novel biological therapies targeting total IgE-mediated pathways have widened clinical treatment options for asthma. A better understanding of the biology underlying total IgE levels should help refine and improve these therapeutic approaches. We used differential gene expression approaches and network analysis, followed by a variety of pathway enrichment techniques, to identify novel gene transcripts co-regulated with determinants of serum IgE levels in individuals with asthma. The combination of statistical approaches allowed us to identify the most biologically relevant transcripts working in combination to influence total IgE. Crucially, we had access to two independent populations and achieved a high level of replication, with 52% of transcripts replicated using GLM, and 37% using WGCNA. Combining the results of these two methods yielded ultimately 31 biologically- relevant and informative transcripts that were associated with serum total IgE. It is also notable that the key gene co-expression network module associated with IgE, was also associated with the more stringent definition of asthma based on physician diagnosis and methacholine challenge, suggesting that these 31 replicated gene transcripts reflect not simply an IgE-related gene signature but also an allergic asthma signature. This network module was associated with the use of inhaled corticosteroid in the CAMP cohort, though not in the GACRS cohort. While it is difficult to determine if this is due to confounding with severity of asthma, we reported that all of transcripts reported in Table 2 remained significantly associated with IgE levels by differential gene expression even after adjusting for the use of inhaled corticosteroids.
Within the pathway enrichment analyses of the total IgE-associated transcripts, we identified the immune relevant GO category, “regulation of tolerance induction,” which was highlighted due to the upregulation of Charcot-Leyden Crystal galectin (CLC) and indoleamine-pyrrole 2,3-dioxygenase (IDO1) in association with increased total IgE levels. Both genes are expressed in eosinophils and their activity is stimulated following binding of IgE. CLC levels have been positively associated with eosinophilic airway inflammation and aspirin induced asthma (31, 32) and have also been associated with expansion of T regulatory cells(33). IDO1 encodes an enzyme involved in tryptophan catabolism and suppresses helper and effector T cells while activating regulatory T cells (34). These findings are notable given that regulatory T cell activity is thought to be deficient in allergic asthma (35), yet we found a positive association of increasing IgE with increased IDO1 and CLC. We hypothesize that this represents a regulatory mechanism induced to curb the allergic immune response in the setting of asthma. This hypothesis is supported by findings describing a spectrum of increased IDO1 activity that is higher among atopic individuals compared to healthy controls, and highest among asymptomatic atopic subjects compared to symptomatic ones, suggesting that IDO1 may be involved in inducing a state of clinical unresponsiveness in allergic individuals.(36) Further study is needed to clarify the roles of IDO1 and serum IgE in asthma.
Overrepresentation analysis suggested the presence of various immunologic signatures such as enrichment of neutrophil, macrophage, and eosinophil-associated genes. In particular, comparison with a gene set of eosinophils compared to Th1 cells showed that 4 of our 31 transcripts overlapped, including SMPD3, CCL23, IDO1, and CEBPE. These findings support the involvement of eosinophil-associated genes in total IgE regulation. At least 20 of these 31 validated transcripts were previously found to be upregulated in eosinophils (37, 38). Esnault et al. compared gene expression in sputum eosinophils from individuals with mild asthma before and after allergen challenge, and found 365 upregulated transcripts (37). Sixteen of our 31 transcripts overlapped with that set, providing further validation for the genes as being strongly associated with the asthmatic response to allergen (Table 2). Esnault et al. also conducted experiments comparing the changes in allergen-induced gene expression in broncheoalveolar lavage cells, before and after treatment with mepolizumab, in order to block IL-5 pathways. Of the gene transcripts that were upregulated after allergen exposure but down-regulated after mepolizumab treatment, 10 of our transcripts overlapped, suggesting involvement in IL-5 related pathways of inflammation in asthma (CYP4F12, EMR4P, IL5RA, LGALS12, LTC4S, PIK3R6, PRSS33, SIGLEC8, SPNS3, VSTM1). In another gene expression analysis of eosinophils compared with mast cells, neutrophils, and mononuclear cells, Nakajima et al. identified 30 eosinophil-specific genes (38); 9 of our 31 transcripts overlapped with that set, further supporting the association between eosinophilic inflammation and total IgE levels. While we found high levels of replication between our populations and with other microarray data, we also compared our findings with GWAS studies of IgE and were unable to find similarities (24, 39). However, given the limited overlap between IgE associated GWAS findings and those of asthma (39), it is not surprising that our replicated transcripts associated with IgE in asthmatics did not coincide with these prior association signals.
The most notable aspect of this analysis is the high degree of replication of the identified transcripts, using two distinct statistical methods in two different patient populations, as well as in comparison to gene sets described in the literature. Some of these genes, such as IL5RA and GPR44 (CRTH2, receptor for Prostaglandin D2), represent known therapeutic targets, with IL-5 receptor antagonists showing some efficacy in uncontrolled eosinophilic asthma (40). CCR3 (eotaxin-3), a chemokine receptor, has been associated with allergic asthma, though CCR3 targeted therapies appear to be less efficacious in humans than in mouse models (41, 42). In addition, the leukotriene pathway has been a therapeutic target for asthma for decades, though specific inhibitors for LTC4S (leukotriene C4 synthase) have only recently been developed and evaluated in mouse models (43, 44). The identification of these known targets using our analytical approach corroborates the possibility that the other transcripts in this set may be equally useful as therapeutic targets.
Other genes from our eosinophil-related gene sets have known associations with allergic asthma and may also have therapeutic or diagnostic potential, including CCL23, CLC, CEBPE, CAMK1, IDO1, SIGLEC8, SMPD3, and SPNS3 (45–52). Potential therapeutic targets include cell surface receptors, such as Siglec8, an inhibitory receptor on eosinophils with polymorphisms associated with asthma (46–48). CCL23 is an eosinophilic chemokine, that has been identified as a possible biomarker for anti-IL13 targeted therapies (49, 50). Several of these are enzymes expressed within eosinophils, such as CLC, IDO1, SMPD3, and SPNS3, and the latter two are both involved in the sphingolipid pathways, known to mediate airway hyperresponsiveness and mast cell activation (51, 52). Another gene CAMK1 has been implicated as an asthma control gene(20), but has not been identified previously with IgE or an allergy-related phenotype among individuals diagnosed with asthma. Of the calmodulin dependent kinases, CAMK2 and CAMK4 have been implicated in bronchoconstriction and smooth muscle proliferation (53, 54), suggesting that CAMK1 may also play a role in asthma pathogenesis.
We also report the expression of a number of novel genes, including EMR1, EMR4P, and PIK3R6, which were significant in our data and have been shown to be upregulated in eosinophilic asthma in other studies. These genes have no known mechanistic association with total IgE or asthma but have been implicated in allergic processes such as allergic rhinosinusitis and anaphylaxis (55, 56). EMR1 and EMR4P encode G-protein coupled receptors that may be particularly useful as therapeutic targets. While these genes do not have known roles in IgE or asthma, their replication in both of our cohorts as well as other published gene sets, implicates them as potentially novel biomarkers or therapeutic targets.
Our study design and analysis does have some limitations. The cross-sectional nature of the expression and phenotype data limit the ability to make true causal inferences. The use of whole blood gene expression profiles may not be as physiologically relevant as lung tissue- specific profiling; however, the use of less invasive sampling methods allows for greater sample sizes and more robust replication. While it could prove useful to split the whole blood expression data into specific cell populations, these techniques are in development and still face significant limitations, including concerns about accuracy and ability to detect expression profiles of cell subsets (57, 58). Recent studies support the utility of whole blood transcriptomics in the setting of asthma (59, 60). We cannot exclude the possibility that these transcripts play a role in atopic disease in general; however the WGCNA analysis showing an association with a stricter definition of asthma in the GACRS population, and a lack of association with either atopic dermatitis or allergic rhinitis, supports a more prominent role in allergic asthma. Also, GO based enrichment analyses are subject to annotation bias and can often yield high level information about biological processes that are ubiquitous to all cellular function; thus we attempted to improve the biological relevance of this analysis by placing more stringent criteria on fold enrichment and the number of genes required.
This analysis benefits from the GACRS population being a highly homogeneous relative genetic isolate with consistent and controllable environmental exposures, detailed clinical information, and minimal population stratification. We have abundant data demonstrating that the genetic findings in the GACRS cohort are generalizable to CAMP. There are over 15 studies of validated susceptibility loci replicated in GACRS and CAMP with similar effect size and direction of effect, whether identified by GWAS (61–67), by linkage and positional cloning (68–70), or by candidate gene testing (23, 65, 71–74). However, we acknowledge that there are ethnic, genetic and environmental differences between the GACRS population and the North American CAMP population, which may have limited our ability to find additional shared gene probes. Focusing on only replicated gene transcripts may have missed important gene probes relevant only in a single population and could have limited the detection of novel biological targets. In addition, the gene expression profiling was performed on childhood samples from GACRS and adulthood samples from CAMP. Given the striking overlap in our findings over both ethnicity and age, the generalizability of these results is a strength rather than a limitation.
In conclusion, we present robust and reproducible evidence for consistent gene expression profiles of serum total IgE in individuals with asthma. Indeed we found similar profiles with clear biological relevance regardless of the statistical approach used; multivariable regression or network-based approaches. Pathway enrichment identified an important role for regulation of T cell tolerance. In addition to gene transcripts with known or suspected relationships with IgE related pathways, we identified a number of novel transcripts. Many of the genes encoding these transcripts have been reported in other studies of asthma suggesting that they may represent novel therapeutic targets or biomarkers. Further functional validation of these transcripts and their roles in influencing total IgE and asthma will be essential to developing novel strategies for improved primary prevention, management and treatment of asthma.
Supplementary Material
Acknowledgments:
We thank the participants and families who have so generously participated in this study.
Funding: This manuscript is supported by an R01 grants from the National Heart, Lung, and Blood Institute of the National Institutes of Health to Brigham and Women’s Hospital, NHLBI 1R01HL123915–01 (JALS), the NIH grant R37 HL066289 (STW). D.C.C.-C. was supported by a grant from the National Heart, Lung, and Blood Institute of the U.S. National Institutes of Health (K01 HL127265). Y.V.V. was supported by a grant from the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (K23AI130408)
Abbreviations:
- IgE
Immunoglobulin E
- GACRS
Genetics of Asthma in Costa Rica Study’
- CAMP
The Childhood Asthma Management Program
- IQR
inter-quartile ranges
- GLM
Generalized linear regression models
- WGCNA
Weighted Gene Co-expression Network Analysis
- FDR
false discovery rate
- GO
Gene Ontology
Footnotes
Clinical Trial or Protocol number associated with this study: N/A
Conflict of Interest: None of the authors involved in this manuscript have any conflicts of interest relevant to this work
References
- 1.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy 2004;59(5):469–78. [DOI] [PubMed] [Google Scholar]
- 2.Devereux G The increase in allergic disease: environment and susceptibility. Proceedings of a symposium held at the Royal Society of Edinburgh, 4th June 2002. Clin Exp Allergy 2003;33(3):394–406. [DOI] [PubMed] [Google Scholar]
- 3.Global Initiative for Asthma Management and Prevention. U.S. Department of Health and Human Services, National Institutes of Health. NHLBI/WHO Workshop Report 1995;95–3659.
- 4.Akinbami Lara J.; Jeanne E Moorman CBHSZMKCAJaXL. Trends in Asthma Prevalence, Health Care Use, and Mortality in the United States, 2001–2010 Hyattsville, MD: National Center for Health Statistics; 2012. [Google Scholar]
- 5.Peng Z Vaccines targeting IgE in the treatment of asthma and allergy. Hum Vaccin 2009;5(5):302–9. [DOI] [PubMed] [Google Scholar]
- 6.Naqvi M, Choudhry S, Tsai HJ, Thyne S, Navarro D, Nazario S, et al. Association between IgE levels and asthma severity among African American, Mexican, and Puerto Rican patients with asthma. J Allergy Clin Immunol 2007;120(1):137–43. [DOI] [PubMed] [Google Scholar]
- 7.Bice JB, Leechawengwongs E, Montanaro A. Biologic targeted therapy in allergic asthma. Ann Allergy Asthma Immunol 2014;112(2):108–15. [DOI] [PubMed] [Google Scholar]
- 8.Husby S, Holm NV, Christensen K, Skov R, Morling N, Petersen PH. Cord blood immunoglobulin E in like-sexed monozygotic and dizygotic twins. Clin Genet 1996;50(5):332–8. [DOI] [PubMed] [Google Scholar]
- 9.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010;363(13):1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. The New England journal of medicine 362(1):36–44. [DOI] [PubMed] [Google Scholar]
- 11.Granada M, Wilk JB, Tuzova M, Strachan DP, Weidinger S, Albrecht E, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. The Journal of allergy and clinical immunology 2012;129(3):840–5 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasky-Su J, Himes BE, Raby BA, Klanderman BJ, Sylvia JS, Lange C, et al. HLA-DQ strikes again: genome-wide association study further confirms HLA-DQ in the diagnosis of asthma among adults. Clin Exp Allergy 2012;42(12):1724–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunninghake GM, Soto-Quiros ME, Avila L, Su J, Murphy A, Demeo DL, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol 2007;120(1):84–90. [DOI] [PubMed] [Google Scholar]
- 14.Asher MI, Montefort S, Bjorksten B, Lai CK, Strachan DP, Weiland SK, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 2006;368(9537):733–43. [DOI] [PubMed] [Google Scholar]
- 15.Bigler J, Boedigheimer M, Schofield JP, Skipp PJ, Corfield J, Rowe A, et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am J Respir Crit Care Med 2016. [DOI] [PubMed]
- 16.Modena BD, Bleecker ER, Busse WW, Erzurum SC, Gaston BM, Jarjour NN, et al. Gene Expression Correlated to Severe Asthma Characteristics Reveals Heterogeneous Mechanisms of Severe Disease LID - 10.1164/rccm.201607-1407OC [doi]. (1535–4970 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce N, Aït‐Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2007;62(9):758–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blumenthal MN, Banks-Schlegel S, Bleecker ER, Marsh DG, Ober C. Collaborative studies on the genetics of asthma–National Heart, Lung and Blood Institute. Clinical & Experimental Allergy 1995;25:29–32. [DOI] [PubMed] [Google Scholar]
- 19.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials 1999;20(1):91–120. [PubMed] [Google Scholar]
- 20.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF Jr., Liu AH, et al. Gene Expression Profiling in Blood Provides Reproducible Molecular Insights into Asthma Control. Am J Respir Crit Care Med 2016. [DOI] [PMC free article] [PubMed]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences of the United States of America 2010;107(21):9546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunninghake GM, Soto-Quiros ME, Avila L, Kim HP, Lasky-Su J, Rafaels N, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy 2010;65(12):1566–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010;363(13):1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raby BA, Soto-Quiros ME, Avila L, Lake SL, Murphy A, Liang C, et al. Sex-specific linkage to total serum immunoglobulin E in families of children with asthma in Costa Rica. Hum Mol Genet 2007;16(3):243–53. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 1995;57(1):289–300. [Google Scholar]
- 27.Horvath S. Extended Overview of Weighted Gene Co-Expression Network Analysis (WGCNA)
- 28.Starownik R, Michalak J, Bar K, Plaza P, Muc K, Rechberger T. An uncommon case of inflammatory infiltration of the urinary bladder in the long-term process of the purulent inflammation of the cervix and vaginal fornix, complicated with vesicovaginal fistula of unknown etiology. Central European journal of urology 2013;66(1):101–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, et al. Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity 2016;44(1):194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua JC, Douglass JA, Gillman A, O’Hehir RE, Meeusen EN. Galectin-10, a potential biomarker of eosinophilic airway inflammation. PloS one 2012;7(8):e42549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devouassoux G, Pachot A, Laforest L, Diasparra J, Freymond N, Van Ganse E, et al. Galectin-10 mRNA is overexpressed in peripheral blood of aspirin-induced asthma. Allergy 2008;63(1):125–31. [DOI] [PubMed] [Google Scholar]
- 33.Rabinovich GA, Toscano MA. Turning ‘sweet’ on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol 2009;9(5):338–52. [DOI] [PubMed] [Google Scholar]
- 34.Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, Munn DH, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol 2009;183(4):2475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd CM, Hawrylowicz CM. Regulatory T cells in asthma. Immunity 2009;31(3):438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Bubnoff D, Fimmers R, Bogdanow M, Matz H, Koch S, Bieber T. Asymptomatic atopy is associated with increased indoleamine 2,3-dioxygenase activity and interleukin-10 production during seasonal allergen exposure. Clin Exp Allergy 2004;34(7):1056–63. [DOI] [PubMed] [Google Scholar]
- 37.Esnault S, Kelly EA, Sorkness RL, Evans MD, Busse WW, Jarjour NN. Airway factor XIII associates with type 2 inflammation and airway obstruction in asthmatic patients. J Allergy Clin Immunol 2016;137(3):767–73 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima T, Matsumoto K, Suto H, Tanaka K, Ebisawa M, Tomita H, et al. Gene expression screening of human mast cells and eosinophils using high-density oligonucleotide probe arrays: abundant expression of major basic protein in mast cells. Blood 2001;98(4):1127–34. [DOI] [PubMed] [Google Scholar]
- 39.Potaczek DP, Kabesch M. Current concepts of IgE regulation and impact of genetic determinants. Clin Exp Allergy 2012;42(6):852–71. [DOI] [PubMed] [Google Scholar]
- 40.Molfino NA, Gossage D, Kolbeck R, Parker JM, Geba GP. Molecular and clinical rationale for therapeutic targeting of interleukin-5 and its receptor. Clin Exp Allergy 2012;42(5):712–37. [DOI] [PubMed] [Google Scholar]
- 41.Provost V, Larose MC, Langlois A, Rola-Pleszczynski M, Flamand N, Laviolette M. CCL26/eotaxin-3 is more effective to induce the migration of eosinophils of asthmatics than CCL11/eotaxin-1 and CCL24/eotaxin-2. J Leukoc Biol 2013;94(2):213–22. [DOI] [PubMed] [Google Scholar]
- 42.Ahmadi Z, Hassanshahi G, Khorramdelazad H, Zainodini N, Koochakzadeh L. An Overlook to the Characteristics and Roles Played by Eotaxin Network in the Pathophysiology of Food Allergies: Allergic Asthma and Atopic Dermatitis. Inflammation 2016;39(3):1253–67. [DOI] [PubMed] [Google Scholar]
- 43.Ago H, Okimoto N, Kanaoka Y, Morimoto G, Ukita Y, Saino H, et al. A leukotriene C4 synthase inhibitor with the backbone of 5-(5-methylene-4-oxo-4,5-dihydrothiazol-2-ylamino) isophthalic acid. J Biochem 2013;153(5):421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niegowski D, Kleinschmidt T, Ahmad S, Qureshi AA, Marback M, Rinaldo-Matthis A, et al. Structure and inhibition of mouse leukotriene C4 synthase. PloS one 2014;9(5):e96763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP epsilon and PU.1. Blood 2003;101(8):3265–73. [DOI] [PubMed] [Google Scholar]
- 46.Nutku E, Aizawa H, Hudson SA, Bochner BS. Ligation of Siglec-8: a selective mechanism for induction of human eosinophil apoptosis. Blood 2003;101(12):5014–20. [DOI] [PubMed] [Google Scholar]
- 47.Angata T Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology 2014;24(9):785–93. [DOI] [PubMed] [Google Scholar]
- 48.Gao PS, Shimizu K, Grant AV, Rafaels N, Zhou LF, Hudson SA, et al. Polymorphisms in the sialic acid-binding immunoglobulin-like lectin-8 (Siglec-8) gene are associated with susceptibility to asthma. Eur J Hum Genet 2010;18(6):713–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Youn BS, Zhang SM, Broxmeyer HE, Cooper S, Antol K, Fraser M Jr., et al. Characterization of CKbeta8 and CKbeta8–1: two alternatively spliced forms of human beta-chemokine, chemoattractants for neutrophils, monocytes, and lymphocytes, and potent agonists at CC chemokine receptor 1. Blood 1998;91(9):3118–26. [PubMed] [Google Scholar]
- 50.Syed F, Huang CC, Li K, Liu V, Shang T, Amegadzie BY, et al. Identification of interleukin-13 related biomarkers using peripheral blood mononuclear cells. Biomarkers 2007;12(4):414–23. [DOI] [PubMed] [Google Scholar]
- 51.Khavandgar Z, Murshed M. Sphingolipid metabolism and its role in the skeletal tissues. Cell Mol Life Sci 2015;72(5):959–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ono JG, Worgall TS, Worgall S. Airway reactivity and sphingolipids-implications for childhood asthma. Mol Cell Pediatr 2015;2(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Girodet PO, Ozier A, Bara I, Tunon de Lara JM, Marthan R, Berger P. Airway remodeling in asthma: new mechanisms and potential for pharmacological intervention. Pharmacol Ther 2011;130(3):325–37. [DOI] [PubMed] [Google Scholar]
- 54.Sanders PN, Koval OM, Jaffer OA, Prasad AM, Businga TR, Scott JA, et al. CaMKII is essential for the proasthmatic effects of oxidation. Sci Transl Med 2013;5(195):195ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Legrand F, Tomasevic N, Simakova O, Lee CC, Wang Z, Raffeld M, et al. The eosinophil surface receptor epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1): a novel therapeutic target for eosinophilic disorders. J Allergy Clin Immunol 2014;133(5):1439–47, 47 e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biethahn K, Orinska Z, Vigorito E, Goyeneche-Patino DA, Mirghomizadeh F, Foger N, et al. miRNA-155 controls mast cell activation by regulating the PI3Kgamma pathway and anaphylaxis in a mouse model. Allergy 2014;69(6):752–62. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Oh WK, Zhu J. Disease-specific classification using deconvoluted whole blood gene expression. Scientific reports 2016;6:32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol 2013;25(5):571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vijverberg SJ, Hilvering B, Raaijmakers JA, Lammers JW, Maitland-van der Zee AH, Koenderman L. Clinical utility of asthma biomarkers: from bench to bedside. Biologics 2013;7:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF Jr., Liu AH, et al. Gene Expression Profiling in Blood Provides Reproducible Molecular Insights into Asthma Control. Am J Respir Crit Care Med 2017;195(2):179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet 2009;85(3):377–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nature genetics 2011;43(9):887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forno E, Lasky-Su J, Himes B, Howrylak J, Ramsey C, Brehm J, et al. Genome-wide association study of the age of onset of childhood asthma. J Allergy Clin Immunol 2012;130(1):83–90 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lasky-Su J, Lange N, Brehm JM, Damask A, Soto-Quiros M, Avila L, et al. Genome-wide association analysis of circulating vitamin D levels in children with asthma. Hum Genet 2012;131(9):1495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA, et al. On the replication of genetic associations: timing can be everything! Am J Hum Genet 2008;82(4):849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Himes BE, Lasky-Su J, Wu AC, Wilk JB, Hunninghake GM, Klanderman B, et al. Asthma-susceptibility variants identified using probands in case-control and family-based analyses. BMC Med Genet 2010;11:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du R, Litonjua AA, Tantisira KG, Lasky-Su J, Sunyaev SR, Klanderman BJ, et al. Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect asthma exacerbations. J Allergy Clin Immunol 2012;129(2):368–73, 73 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hersh CP, Raby BA, Soto-Quiros ME, Murphy AJ, Avila L, Lasky-Su J, et al. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Respir Crit Care Med 2007;176(9):849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murphy A, Tantisira KG, Soto-Quiros ME, Avila L, Klanderman BJ, Lake S, et al. PRKCA: a positional candidate gene for body mass index and asthma. Am J Hum Genet 2009;85(1):87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hunninghake GM, Lasky-Su J, Soto-Quiros ME, Avila L, Liang C, Lake SL, et al. Sex-stratified linkage analysis identifies a female-specific locus for IgE to cockroach in Costa Ricans. Am J Respir Crit Care Med 2008;177(8):830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma S, Litonjua AA, Tantisira KG, Fuhlbrigge AL, Szefler SJ, Strunk RC, et al. Clinical predictors and outcomes of consistent bronchodilator response in the childhood asthma management program. J Allergy Clin Immunol 2008;122(5):921–8 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma S, Raby BA, Hunninghake GM, Soto-Quiros M, Avila L, Murphy AJ, et al. Variants in TGFB1, dust mite exposure, and disease severity in children with asthma. Am J Respir Crit Care Med 2009;179(5):356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bunyavanich S, Melen E, Wilk JB, Granada M, Soto-Quiros ME, Avila L, et al. Thymic stromal lymphopoietin (TSLP) is associated with allergic rhinitis in children with asthma. Clin Mol Allergy 2011;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma S, Murphy AJ, Soto-Quiros ME, Avila L, Klanderman BJ, Sylvia JS, et al. Association of VEGF polymorphisms with childhood asthma, lung function and airway responsiveness. Eur Respir J 2009;33(6):1287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


