Significance
The recent improvements in cryogenic electron microscopy (cryo-EM) caused a revolution in structural biology. However, 1) protein isolation and 2) sample preparation methods lag behind, and cryo-EM is performed at far from full efficiency. Here, we present a microfluidic method for the rapid isolation of a target protein from minimal amounts of cell lysate and for its direct preparation for high-resolution cryo-EM. Our technology opens more avenues for structural biology: High-throughput structure determination of proteins in a multitude of conditions, ultrafast isolation and structure determination of sensitive proteins, and the analysis of proteins that cannot be produced in sufficient amounts using conventional approaches.
Keywords: cryo-EM, protein purification, endogenous, sample preparation, microfluidics
Abstract
High-resolution structural information is essential to understand protein function. Protein-structure determination needs a considerable amount of protein, which can be challenging to produce, often involving harsh and lengthy procedures. In contrast, the several thousand to a few million protein particles required for structure determination by cryogenic electron microscopy (cryo-EM) can be provided by miniaturized systems. Here, we present a microfluidic method for the rapid isolation of a target protein and its direct preparation for cryo-EM. Less than 1 μL of cell lysate is required as starting material to solve the atomic structure of the untagged, endogenous human 20S proteasome. Our work paves the way for high-throughput structure determination of proteins from minimal amounts of cell lysate and opens more opportunities for the isolation of sensitive, endogenous protein complexes.
Knowledge of a protein’s architecture at high resolution is vital to understand its mechanics and chemistry. In recent years, cryogenic electron microscopy (cryo-EM) (1) has matured into a powerful method that can determine the architecture of biological macromolecules at the resolutions required to interpret the atomic fold of proteins (2, 3). In the single-particle cryo-EM approach (4, 5), an unsupported, thin layer of isolated protein complexes in amorphous (vitrified) ice is visualized at close to physiological conditions. Only several thousand to a few million imaged particles are needed to calculate a high-resolution, 3D structure. Nevertheless, protein production, purification, and sample preparation for cryo-EM are nowadays considered the bottleneck for structure determination (6–8). We have identified 2 dominating reasons for this: First, significant amounts of protein must be produced. Conventional sample preparation for cryo-EM requires several microliters of a purified protein solution at a concentration of ∼1 mg/mL per grid, from which extensive filter-paper blotting later removes the vast majority of protein particles (9–12). Second, both protein purification and cryo-EM sample preparation are lengthy and harsh procedures. Mostly, high-yield expression systems are used, and 1 or 2 chromatographic steps are needed to purify the protein particles. In addition, the classical cryo-EM sample preparation process that follows is a rough procedure (13), primarily because of the blotting step, and many proteins denature.
We recently developed a microfluidic cryo-EM grid preparation system termed cryoWriter, allowing the preparation of cryo-EM specimens from nanoliters of sample solution (14–16). Since the cryoWriter does not use paper blotting, it ensures that grid preparation is gentle and virtually lossless. Here, we report the combination of sample grid preparation using the cryoWriter with microfluidic protein purification (17), to determine the 3.5-Å cryo-EM structure of the untagged human 20S proteasome complex, which is the “catalytic core” of the ubiquitin–proteasome system involved in 80% of protein degradation (18) and an important drug target (19).
Results
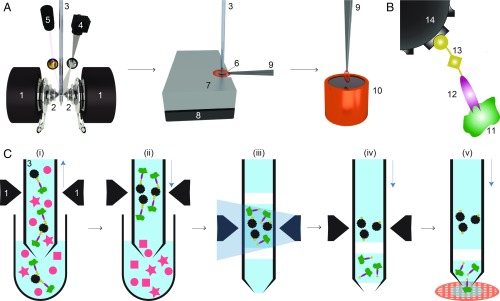
The microfluidic toolchain developed (Fig. 1A) consists of a module for affinity isolation of the untagged protein from small quantities of cell lysate, followed by modules to write the purified protein onto a cryo-EM grid and vitrify the sample (15–17). Briefly, antibody “fragment antigen binders” (Fabs) are used to recognize and extract untagged target proteins from cell lysate. These Fabs are biotinylated with a photo-cleavable cross-linker, which binds with high affinity to the streptavidin functionalization of superparamagnetic beads (Fig. 1B). First, the cell lysate is incubated with Fabs and superparamagnetic beads. A 1-L volume of this solution is then aspirated into the microcapillary of the cryoWriter system (Fig. 1C). The superparamagnetic particles are immobilized in a “magnetic trap” (SI Appendix, Fig. S1) isolating bound Fabs and their target proteins, while other cellular components are washed out. Ultraviolet (UV) light is used to break the photo-cleavable biotin cross-linker and the target proteins with the bound Fabs are eluted (17). This procedure results in a 25-nL eluate, which is directly used to prepare 1 or more cryo-EM grids, while the magnetic particles are retained in the microcapillary.
Fig. 1.
Schematic workflow for microfluidic protein isolation and cryo-EM grid preparation. (A) Hardware for protein isolation and cryo-EM grid preparation. The electromagnetic trap consists of 2 electromagnets (1) that produce a strong magnetic field gradient via their water-cooled iron tips (2). Sample processing in the capillary (3) is monitored by a camera (4), and a UV LED (5) allows photo-cleavage (B) of the sample, both via mirrors. After protein isolation, the capillary nozzle is moved above a cryo-EM grid covered with a holey carbon film (6). The cryo-EM grid is positioned on a stage (7) that is temperature controlled by a Peltier element (8) and held with a Peltier-cooled tweezer (9). The isolated protein is directly written onto the grid and plunge-frozen in liquid ethane (10). (B) Composite material for “protein fishing.” The target protein (11) is recognized by a Fab (12) that is covalently modified by a photo-cleavable cross-linker (13). The linker molecule ends with a biotin moiety, which strongly binds to the streptavidin-coated bead (14). (C) Protein isolation workflow. (i) Magnetic beads are incubated with biotinylated Fabs and cell lysate to capture the target structures (green). Less than 900 nL of sample is aspirated into the microcapillary for the protein isolation. (ii) The magnetic beads are immobilized in the magnetic trap (1). Nonbound lysate components (red) are flushed out. (iii) Illumination with UV light breaks the cross-linker. Before photoelution, 2 air bubbles are introduced and serve as boundaries to avoid dilution of the released proteins by diffusion (see SI Appendix, Figs. S1 and S3 for details). (iv) Separation of the capturing magnetic beads and the eluted proteins. (v) The isolated target proteins are directly deposited on a cryo-EM grid for vitrification. The blue vertical arrows indicate the pump direction.
We used Fabs instead of full-length antibodies to avoid the cross-linking and aggregation that occur when target proteins display more than 1 epitope. A linear cross-linker with 3 functional groups is used to connect the Fab to the superparamagnetic beads. One end of the linker covalently attaches to the Fab while the biotin at the other end binds to the streptavidin-coated bead. A nitrobenzene moiety in between allows photo-cleavage of the linker upon illumination with UV light at 365 nm (Fig. 1B). Preparation of Fab fragments from antibodies and biotinylation with the cross-linker took 7 h; the biotinylated Fabs could be stored in the dark for several weeks at 4 °C.
The cell lysate was incubated with the Fabs and beads outside of the microcapillary in a 5 -L well. The cytosol/Fab mixture was incubated for 5 h and then for an additional 1 h together with the superparamagnetic beads. The use of miniaturized sample wells rather than the microcapillary allowed several of these time-consuming incubations to be carried out in parallel.
After incubation, a volume of 900 nL was aspirated from the well into the microcapillary, which was precisely positioned between the iron tips of the particle trap system. In this setup, the trap is formed by electromagnets with water-cooled (4 °C) iron tips that concentrate the magnetic flux between the opposite poles and generate the field gradients required to trap the superparamagnetic particles (SI Appendix, Fig. S1). The cooling system prevents heating and denaturation of the sample. A camera and magnifying lens system allow the procedure to be monitored. The 1-m superparamagnetic beads used can be easily immobilized in the magnetic trap and only marginally scatter photons with 365-nm wavelength, allowing efficient photoelution in 15 min (17). Photoelution releases less nonspecifically bound protein than the competitive elution typically used in other methods, because it does not change the buffer composition (20). Just before elution, the beads were enclosed by two 6-nL air bubbles in a 25-nL buffer plug, to prevent diffusion and Taylor dispersion (21). After photoelution, the eluate was separated from the immobilized beads by the pump system and moved toward the apex of the microcapillary.
For cryo-EM specimen preparation, a sample carrier (grid) covered by a holey carbon film was placed on a temperature-controlled stage regulated to a temperature 7 °C above the environmental dew point. This temperature offset builds a microenvironment on the grid surface, allowing controlled evaporation of the sample liquid. In a first step, 20 nL of the eluate containing the purified protein was written onto the grid, covering an area of 0.75 mm2. Excess sample was reaspirated into the microcapillary, leaving a thin layer of the protein on the holey carbon film. This layer was left to settle for 50 ms with the grid still on the dew-point stage. The gentle evaporation that occurs during this time stabilizes (22) and thins the sample film. Finally, the written grid was rapidly removed from the stage and plunge-frozen, resulting in a vitrified ice layer (15, 16). In principle, several cryo-EM grids can be written with one 25-nL eluate. Interestingly, when this was done, the first cryo-EM grid contained more protein particles than later grids. For the analysis presented here, we used only 1 cryo-EM grid, and this was the first grid prepared from the eluate. The whole cryoWriter process starting from protein isolation and ending with cryo-EM grid preparation could be performed in less than 1 h.
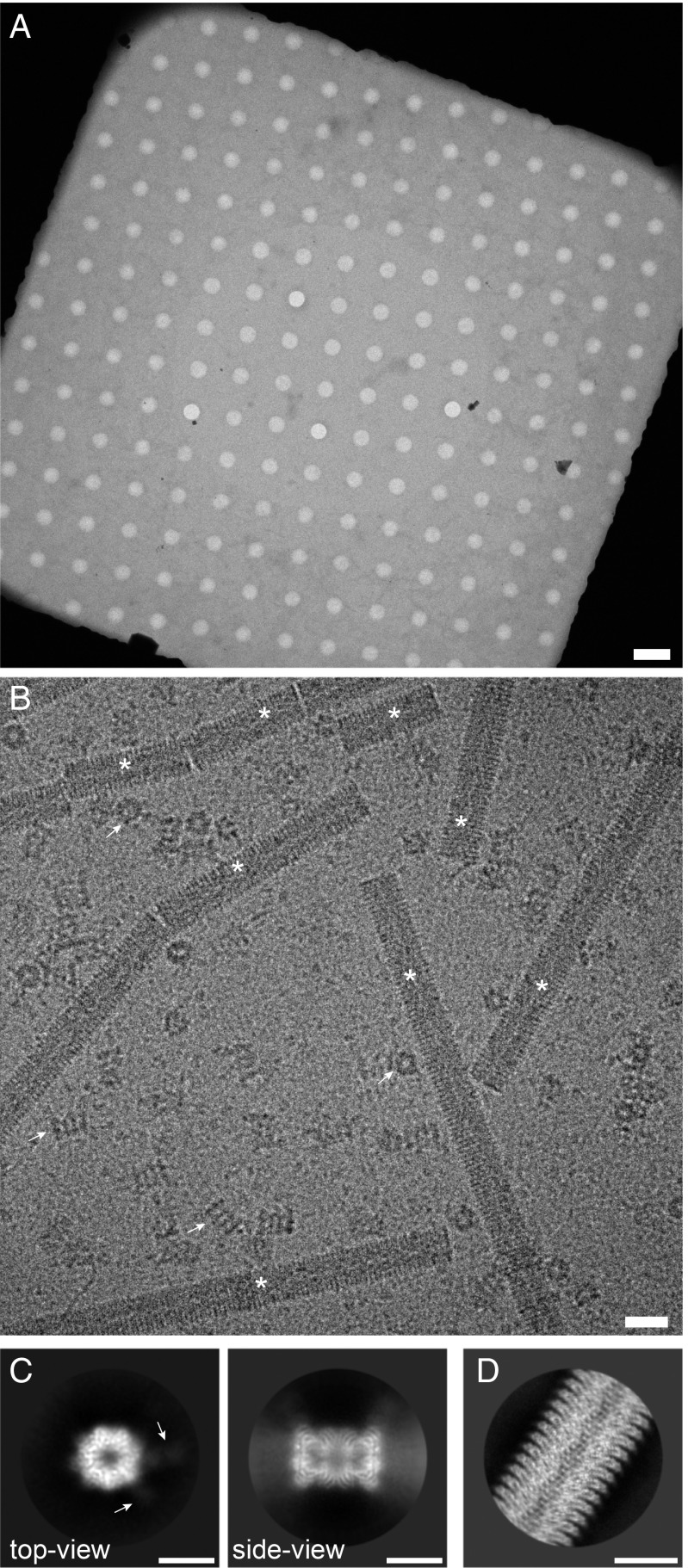
We used the cryoWriter toolchain to isolate endogenous and untagged human 20S proteasome from commercially obtained HeLa cell lysate using Fabs generated from an antibody against the subunit of the protein complex. As a positive control for the cryo-EM grid preparation, tobacco mosaic virus (TMV) particles were added to the elution buffer. The cryo-EM grid showed homogeneous ice layers (Fig. 2 A and B) with the protein embedded in thin, vitreous ice. TMV particles and randomly oriented 20S proteasomes are visible, as well as smaller protein particles that are most probably unbound Fabs.
Fig. 2.
Sample quality and data collection. (A) Overview image of a grid square and the holey carbon film, showing a thin film of vitreous ice. (Scale bar: 4 m.) (B) High-magnification image of the isolated 20S proteasome sample. Arrows indicate 20S proteasome as top and side views; asterisks denote TMV. Furthermore, small particles are visible in the background, most probably unbound Fabs. (Scale bar: 20 nm.) TMV was added to the elution buffer as a positive control for the cryo-EM grid quality (SI Appendix, Fig. S4). The contrast of the image was increased using a Gaussian blur and subsequent histogram adjustment. (C) Selected projection averages of the 20S proteasome. Arrows indicate 2 bound Fabs recognizing the subunit. (Scale bar: 10 nm.) (D) Typical projection average of TMV from the same cryo-EM grid as the 20S proteasome. (Scale bar: 10 nm.)
From only 1 grid, 523 dose-fractionated image stacks (movies) were recorded (23), yielding a total of 55,135 particles, which were processed with cryoSPARC v2 (Structura Biotechnology Inc.) (24) and RELION 3 (25) for structure determination. Fig. 2C shows typical projection class averages obtained from the 20S proteasome. A 7-fold pseudosymmetry is observed in the top view, with weakly visible Fab fragments attached to the 2 subunits. The side view exhibits the distinctive stack of rings. Fig. 2D shows a projection class average of TMV. The secondary structure was visible in all projection averages, demonstrating the excellent quality of the cryo-EM grid.
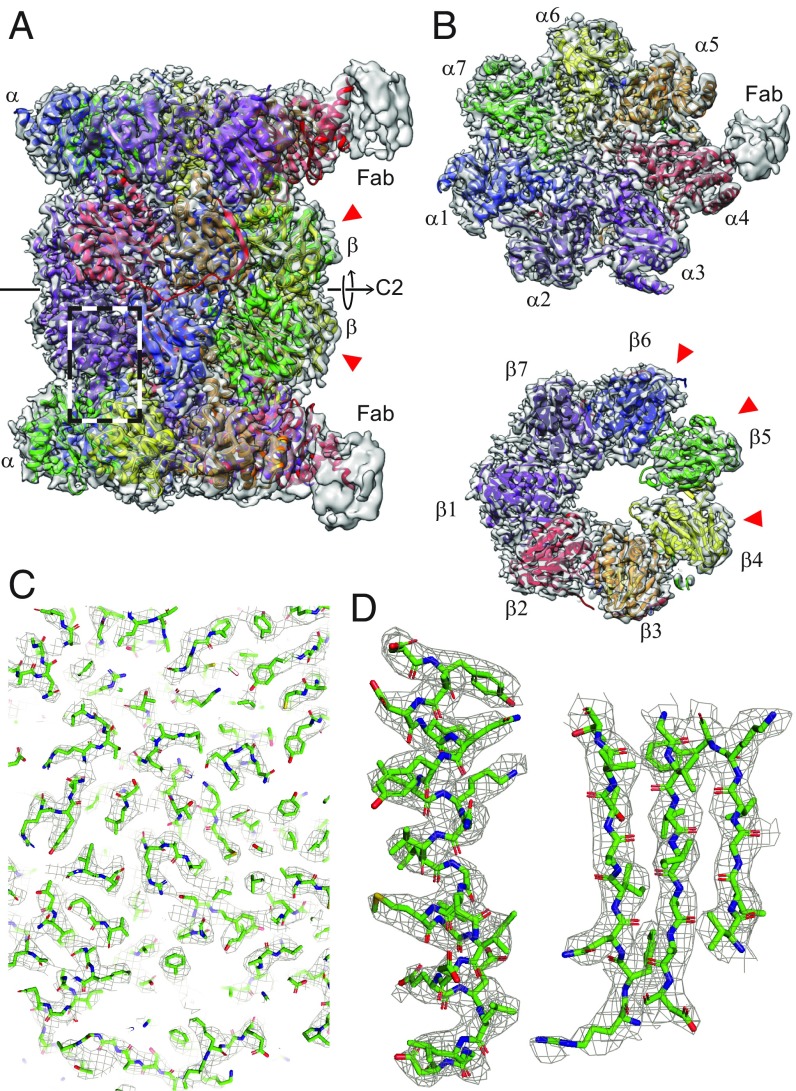
The 3D reconstruction of the human 20S proteasome (26) shown in Fig. 3 has a resolution of 3.5 Å (for details see SI Appendix, Fig. S4). The structure exhibits the typical dimeric, C2-symmetric arrangement of an and a ring pair, each ring with a pseudo-7-fold arrangement of the respective subunits. We refined the X-ray structure (27) into our density map (28). The 2 Fabs against the 4 subunits aided the assignment of the individual components. Fig. 3B shows the and rings with all 14 models fitting the experimental densities in good agreement (Fig. 3 C and D and SI Appendix, Table S5). The densities of subunits , , and are less well resolved (SI Appendix, Fig. S5), which can be attributed to their catalytic activity (29); we assume that the lower resolution reflects the greater flexibility they require to perform their biological function.
Fig. 3.
The 3D reconstruction of the human 20S proteasome. The red arrowheads indicate the catalytically active subunits. (A) Side view showing the 2 and 2 rings. All 14 subunits are fitted into the mass densities. Parts of the 2 bound Fabs are visible at lower resolution, due to the high flexibility of the attached Fabs. The C2 symmetry axis is indicated. (B) Top view of an and a ring. Both rings have a pseudo-7-fold symmetry. Different subunits are indicated by different colors. (C) A zoom-in into the side-view region indicated by the dashed box in A, documenting the quality of the data and model fitting. An enlarged view of 1 -helix and 3 strands of a -sheet is shown in D as an example.
The reconstruction of the added TMV particles (30) has a resolution of 1.9 Å (SI Appendix, Figs. S4 and S8; ref. 31), indicating that the microfluidic sample preparation did not limit the resolution of the human 20S proteasome. Unlike the archaeal T20S proteasome, which is often used as a cryo-EM test sample due to its 14-fold D7 symmetry, the human 20S proteasome is only 2-fold symmetric. The lower symmetry reduces the internal averaging by a factor of 7. Moreover, the similar subunits around the “pseudo-7-fold” axis can lead to misalignments of the particle projections. Together, these factors explain why existing cryo-EM maps are at the lower resolution of 3.5 Å (29, 32).
Conclusion
Several methods have been presented to improve sample preparation for cryo-EM, all having their own specific purpose. Examples include high-throughput grid preparation (33–35), time-resolved EM (36), and single-cell visual proteomics (14, 15). Furthermore, antibody-functionalized EM grids were proposed to “fish” target proteins (37, 38), and affinity grids designed to capture proteins with engineered his-tags (39). In both cases, the vitrified protein is supported by a continuous carbon film, which can limit the resolution obtained. Here, we combined microfluidics for protein isolation and purification with cryo-EM grid preparation, resulting in free-standing layers of vitrified protein samples that allow high-resolution cryo-EM.
Microfluidic methods could potentially overcome the current bottleneck in cryo-EM, resulting from the large amount of protein used for sample preparation and the long and harsh conditions used for protein purification. We demonstrated that microfluidics 1) can deliver cryo-EM grids of high quality allowing 2 Å resolution from 20 nL of a sample and 2) allows the isolation of endogenous proteins from less than 1 L cell lysate with subsequent structure determination at 3.5 Å. In addition, the method presented here significantly reduces both the amount of starting material and the time needed for the structural analysis of proteins, avoids harsh protein purification conditions, and eliminates the stringent and wasteful blotting steps otherwise used on cryo-EM grid preparation.
The high efficiency of cryo-EM combined with the microfluidic approach will allow the structure of proteins that cannot be produced in large quantities to be studied. Up to now, structural studies were possible only if the protein could be overexpressed or produced in a large numbers of cells. The small sample volumes used during the microfluidic purification process allow a high protein concentration to be maintained, which is helpful if protein complexes fall apart upon dilution. Furthermore, the small sample volume also allows for “buffer conditioning” (14), e.g., for the introduction of small ligands before sample vitrification. In addition, further optimization of the purification step and use of the cryoWriter will almost certainly make it possible to perform the whole preparation in less than 1 h, facilitating the investigation of sensitive protein targets. Finally, the toolchain presented here can also be combined with in vitro translation systems, which would enable high-throughput structure determination, or with a single-cell lysis device (14, 15, 40), which would bring the structural analysis of proteins originating from a single cell within reach.
Materials and Methods
Generation of Fabs.
Fabs were generated from an antibody against the -subunit of the human 20S proteasome (Enzo Life Sciences; BML-PW8120), using a commercial kit (Thermo Fisher Scientific Inc.; 44685) and following the provided protocol. After generation of Fabs, the buffer was changed to PBS (pH 7.4, 150 mM NaCl, 1.5 mM , 8.1 mM , 2.6 mM KCl) (Sigma; D8537), and the Fabs were concentrated to a concentration of 0.28 mg/mL using a centrifugal filter unit (Sigma; UFC501024).
Biotinylation of Fabs.
To biotinylate the Fabs, a 6-fold molar excess of photo-cleavable NHS-biotin cross-linker (Fisher Scientific; NC1042-383) was added to the Fabs (0.28 mg/mL, 5.6 mM) and incubated at room temperature for 1 h. Excess, unbound cross-linker was removed using a spin desalting column (Thermo Fisher Inc.; 87764) following the instructions from the kit protocol.
Binding of the 20S Proteasome to Magnetic Particles.
To fish the human 20S proteasome, HeLa cell lysate (human origin; Enzo Life Sciences; BML-SW8750) was incubated in the presence of 0.7 mM biotinylated Fabs at 4 °C for 5 h. Subsequently, superparamagnetic Dynabeads (Thermo Fisher Scientific Inc.; 65602) were added to achieve a final concentration of 5 pM and incubated additionally for 1 h at 4 °C. Glycerol and ATP which maintain proteasome stability were intentionally not added to the buffers to ensure that the regulatory complexes dissociated from the 20S proteasome core complex (41).
Extraction and Vitrification of 20S Proteasome.
After the incubation step, a 0.3-L volume of the cell lysate/Fabs/superparamagnetic beads mixture was aspirated into the microcapillary of the cryoWriter setup at a flow rate of 0.4 L/min and passed through the electromagnetic trap (SI Appendix, section A). The beads were retained by the magnetic field gradient to form a bead plug. Subsequently, the flow direction was inverted and the particle plug was washed with 4 L washing buffer (25 mM Hepes-KOH, pH 7.5, 5 mM ) at a flow rate of 3 L/min. The aspiration-wash cycle was repeated twice. In all, a total of 0.9 L cell lysate was loaded into the microcapillary. At the end, the capillary was washed further with 45 L of washing buffer at a flow rate of 3 L/min.
Before UV cleavage, the sample plug was enclosed between two 6-nL air bubbles that were introduced from the nozzle tip and were separated by a volume of 25 nL washing buffer containing 1 mg/mL of TMV. See also SI Appendix, section B for more details. TMV was kindly supplied by Ruben Diaz-Avalos, Howard Hughes Medical Institute, Chevy Chase, MD. The hydrophilicity of the inner surface of the microfluidic capillary retains a thin hydration layer, which allows magnetic beads with loaded samples to remain within the aqueous phase while the air bubbles are introduced. The sample plug was then exposed for 15 min to UV light emitted at a wavelength of 365 nm by a 190-mW UV LED (Thorlabs; M365L2).
After photo-cleavage, 20 nL of the 25-nL eluate containing the purified protein was primed on a 400-mesh copper grid covered with a holey carbon film (R1.2/1.3; Quantifoil) that was glow discharged for 45 s in air plasma immediately before use. The cryo-EM grids were prepared as described in ref. 15, applying protocol 1. However, we adjusted the parameters for the sample: The stage temperature was 7 °C above the dew-point temperature of the environment, i.e., 10 °C on most days, and the settling time was 50 ms.
Data Acquisition.
Cryo-EM image data were collected on an FEI Titan Krios (ThermoFisher Scientific) transmission electron microscope, operated at 300 kV and equipped with a Gatan Quantum-LS imaging energy filter (GIF, 20 eV zero loss energy window; Gatan Inc.). Micrographs were acquired using a K2 Summit direct electron detector (Gatan Inc.) operated in dose fractionation mode (superresolution, 30 frames, 0.2 s/frame, 6 s total exposure) and controlled by the SerialEM (42) software. The physical pixel size was 0.812 Å and the total electron dose was per recorded image stack (movie). Micrographs were drift corrected, dose weighted, and binned 2 times by Fourier cropping, using MotionCor2 (43) via the FOCUS interface (44). Additional data collection parameters are listed in SI Appendix, section C.
Image Processing.
An initial model for subsequent processing with RELION3 (25) was generated using cryoSPARC v2 (Structura Biotechnology Inc.) (24). Particles were picked in cryoSPARC v2 and then 2D classified. An ab initio model was generated using particles selected from the best 2D classes and subsequently refined to 4.1 Å resolution with C2 symmetry imposed.
For RELION3 processing, 55,135 particles were picked with Gautomatch (K. Zhang, http://www.mrc-lmb.cam.ac.uk/kzhang/) using templates projected from the map obtained using cryoSPARC v2 and low-pass filtered to 20 Å resolution. After 2D classification, 38,848 particles were 3D classified into 4 classes. The best class, containing 16,015 particles was selected for 3D refinement. A generous mask derived from the refined map low-pass filtered to 15 Å and with a soft edge of 6 voxels was used for postprocessing, yielding a resolution of 4.3 Å. Afterward, rounds of CTF refinement including defocus refinement per particle, astigmatism refinement per micrograph, and beam-tilt refinement per dataset were iterated with rounds of Bayesian polishing and 3D refinement. A final 3D refinement, using a soft mask that excluded the flexible Fabs and used solvent-flattened FSC curves to filter the 3D reference at every iteration, resulted in a map with a final global resolution of 3.5 Å after postprocessing. Image processing parameters are listed in SI Appendix, section C. For more details on resolution estimation please see SI Appendix, section D.
Helical processing of TMV was performed with RELION3 (25) from 481 micrographs of the same dataset that contained 20S proteasome, following standard helical refinement and reconstruction procedures (45). The final resolution of the TMV map was 1.9 Å (SI Appendix, section D).
Model Building.
The model for the native human 20S proteasome was built based on the X-ray structure from Schrader et al. (27) (PDB ID: 5LE5). Chimera (46) was used for the initial rigid-body fitting. The bound Fabs helped to identify the different subunits in the electron density map. For further processing, all heteroatoms were removed. Real space refinement with PHENIX (47) and manual adjustments in Coot (48) were done iteratively to obtain the final model, ending with a cycle of PHENIX real-space refinement. The graphics were generated using Chimera (46) and PyMOL (49).
Supplementary Material
Acknowledgments
We thank the workshop of the Biozentrum of the University Basel for technical support; A. Fecteau-LeFebvre, D. Caujolle-Bert, and K. Goldie for technical assistance; and A. Engel for discussions (all Center for Cellular Imaging and Nanoanalytics, University of Basel). RELION 3 processing was performed at sciCORE (http://scicore.unibas.ch/) scientific computing center at University of Basel. TMV was kindly provided by R. Diaz-Avalos (now at NanoImaging Services). We thank our former coworker Shirley Müller for critically reading the manuscript. We acknowledge support by the Swiss Nanoscience Institute (Project P1401 and Argovia Microfluidic Protein Isolation, Stabilization and Cryo-EM Preparation), the Swiss National Science Foundation (Projects 200021_162521 and 205320_166164), and the Swiss Commission for Technology and Innovation (Project 18272.1).
Footnotes
Conflict of interest statement: H.S. and T.B. declare the following competing financial interest: The cryoWriter concept is part of patent application PCT/EP2015/065398. A former student will found a spin-off for the commercialization of the cryoWriter (March 2019).
This article is a PNAS Direct Submission.
Data deposition: Image data are available at the Electron Microscopy Public Image Archive (EMPIAR) database under EMPIAR-10251. The reconstructed volumes are available at the The Electron Microscopy Data Bank (EMDB) under EMD-4738 (20S proteasome) and EMD-4628 (TMV). The atomic coordinates are available at the Protein Data Bank (PDB) under PDB-6R70 (20S proteasome) and PDB-6R7M (TMV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907214116/-/DCSupplemental.
References
- 1.Dubochet J., et al. , Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Kühlbrandt W., Biochemistry. The resolution revolution. Science 343, 1443–1444 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y., Single-particle cryo-EM-how did it get here and where will it go. Science 361, 876–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank J., Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy 1, 159–162 (1975). [DOI] [PubMed] [Google Scholar]
- 5.van Heel M., Frank J., Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6, 187–194 (1981). [DOI] [PubMed] [Google Scholar]
- 6.Glaeser R. M., How good can cryo-EM become? Nat. Methods 13, 28–32 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Thompson R. F., Walker M., Siebert C. A., Muench S. P., Ranson N. A., An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods 100, 3–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark H., Chari A., Sample preparation of biological macromolecular assemblies for the determination of high-resolution structures by cryo-electron microscopy. Microscopy 65, 23–34 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Taylor K. A., Glaeser R. M., Electron microscopy of frozen hydrated biological specimens. J. Ultrastruct. Res. 55, 448–456 (1976). [DOI] [PubMed] [Google Scholar]
- 10.Dubochet J., et al. , Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Kemmerling S., et al. , Connecting -fluidics to electron microscopy. J. Struct. Biol. 177, 128–134 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Arnold S. A., et al. , Miniaturizing EM sample preparation: Opportunities, challenges and “visual proteomics”. Proteomics 18, 1700176 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Glaeser R. M., Han B.-G., Opinion: Hazards faced by macromolecules when confined to thin aqueous films. Biophys. Rep. 3, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold S. A., et al. , Total sample conditioning and preparation of nanoliter volumes for electron microscopy. ACS Nano. 10, 4981–4988 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Arnold S. A., et al. , Blotting-free and lossless cryo-electron microscopy grid preparation from nanoliter-sized protein samples and single-cell extracts. J. Struct. Biol. 197, 220–226 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Schmidli C., et al. , Miniaturized sample preparation for transmission electron microscopy. J. Vis. Exp. 137, e57310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giss D., Kemmerling S., Dandey V., Stahlberg H., Braun T., Exploring the interactome: Microfluidic isolation of proteins and interacting partners for quantitative analysis by electron microscopy. Anal. Chem. 86, 4680–4687 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj R., Fang X., Attinger D., Pattern formation during the evaporation of a colloidal nanoliter drop: A numerical and experimental study. New J. Phys. 11, 075020 (2009). [Google Scholar]
- 19.Cromm P. M., Crews C. M., The proteasome in modern drug discovery: Second life of a highly valuable drug target. ACS Cent. Sci. 3, 830–838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giss D., Kemmerling S., Dandey V. P., Stahlberg H., Braun T., “Microfluidics to isolate untagged proteins from cell extracts for visual analysis by electron microscopy” in 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Zengerle R., Ed. (Chemical and Biological Microsystems Society, 2013), vol. 3, pp. 1785–1787. [Google Scholar]
- 21.Squires T. M., Quake S. R., Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 77, 977–1026 (2005). [Google Scholar]
- 22.Padmakar A. S., Kargupta K., Sharma A., Instability and dewetting of evaporating thin water films on partially and completely wettable substrates. J. Chem. Phys. 110, 1735–1744 (1999). [Google Scholar]
- 23.Schmidli C., et al. , CryoWriter: 3.5 Å structure of human 20S proteasome with bound Fabs from microfluidic protein isolation, and 1.9 Å TMV structure. Electron Microscopy Public Image Archive. https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10251. Deposited 21 February 2019.
- 24.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidli C., et al. , Endogeneous native human 20S proteasome with bound Fabs isolated from less than 1 ul cell lysate. The Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/emdb/EMD-4738. Deposited 28 March 2019.
- 27.Schrader J., et al. , The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 353, 594–598 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Schmidli C., et al. , Endogeneous native human 20S proteasome. Protein Data Bank. http://www.rcsb.org/structure/6R70. Deposited 28 March 2019.
- 29.da Fonseca P. C. A., Morris E. P., Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nat. Commun. 6, 7573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidli C., et al. , Cryo-EM structure of Tobacco Mossaic Virus from microfluidic grid preparation. The Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/emdb/EMD-4628. Deposited 22 February 2019.
- 31.Schmidli C., et al. , Tobacco Mosaic Virus (TMV). Protein Data Bank. http://www.rcsb.org/structure/6R7M. Deposited 29 March 2019.
- 32.Morris E. P., da Fonseca P. C. A., High-resolution cryo-EM proteasome structures in drug development. Acta Cryst. D73, 522–533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dandey V. P., et al. , Spotiton: New features and applications. J. Struct. Biol. 202, 161–169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razinkov I., et al. , A new method for vitrifying samples for cryoEM. J. Struct. Biol. 195, 190–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noble A. J., et al. , Reducing effects of particle adsorption to the air-water interface in cryo-EM. Nat. Methods 15, 793–795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng X., et al. , A fast and effective microfluidic spraying-plunging method for high-resolution single-particle cryo-EM. Structure 25, 663–670.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derrick K. S., Quantitative assay for plant viruses using serologically specific electron-microscopy. Virology 56, 652–653 (1973). [DOI] [PubMed] [Google Scholar]
- 38.Yu G., Li K., Jiang W., Antibody-based affinity cryo-EM grid. Methods 100, 16–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly D. F., Abeyrathne P. D., Dukovski D., Walz T., The affinity grid: A pre-fabricated EM grid for monolayer purification. J. Mol. Biol. 382, 423–433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemmerling S., et al. , Single-cell lysis for visual analysis by electron microscopy. J. Struct. Biol. 183, 467–473 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Leggett D. S., Glickman M. H., Finley D., Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Zheng S. Q., et al. , Motioncor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biyani N., et al. , Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017). [DOI] [PubMed] [Google Scholar]
- 45.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen E. F., et al. , UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Cryst. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrödinger, LLC , The PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, LLC, 2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.