Abstract
The Japanese archipelago is comprised of four main islands—Hokkaido, Honshu, Shikoku, and Kyushu—which contain high mountainous areas that likely allowed for lineage differentiation and population genetic structuring during the climatic changes of the late Pleistocene. Here, we assess the historical background of the evolutionary dynamics of herbivorous red-backed voles (Myodes) in Japan, examining the evolutionary trends of mitochondrial cytochrome b gene (Cytb) sequence variation. Four apparent signals from rapid expansion events were detected in three species, M. rufocanus and M. rutilus from Hokkaido and M. smithii from central Honshu. Taken together with results from previous studies on Japanese wood mice (Apodemus spp.), three of the expansion events were considered to be associated with predicted bottleneck events at the marine isotope stage (MIS) 4 period, in which glaciers are thought to have expanded extensively, especially at higher elevations. In the late Pleistocene, the possible candidates are transitions MIS 6/5, MIS 4/3, and MIS 2/1, which can be characterized by the cold periods of the penultimate glacial maximum, MIS 4, and the last glacial maximum, respectively. Our data further reveal the genetic footprints of repeated range expansion and contraction in the northern and southern lineages of the vole species currently found in central Honshu, namely M. andersoni and M. smithii, in response to climatic oscillation during the late Pleistocene. The time-dependent evolutionary rates of the mitochondrial Cytb presented here would provide a possible way for assessing population dynamics of cricetid rodents responding to the late Pleistocene environmental fluctuation.
Keywords: cytochrome b, evolutionary rate, mitochondrial DNA, Quaternary ice age impact, red-backed voles, the Japanese Islands
The Japanese archipelago is comprised of four main islands (from north to south: Hokkaido, Honshu, Shikoku, and Kyushu; Fig. 1), which include diverse habitats from a subarctic climate in the north to a temperate climate in the south. This range of conditions results in a pattern recorded in a number of species, including moles and voles, in which some forms are adapted to the northern habitats and other forms adapted to southern habitats (Tsuchiya et al. 2000; Harada et al. 2001; Iwasa and Suzuki 2002). In addition, the main islands harbor complex topographic features, with high mountain ranges such as the Japan Alps in central Honshu and the Kii Peninsula in southern Honshu (Fig. 1).
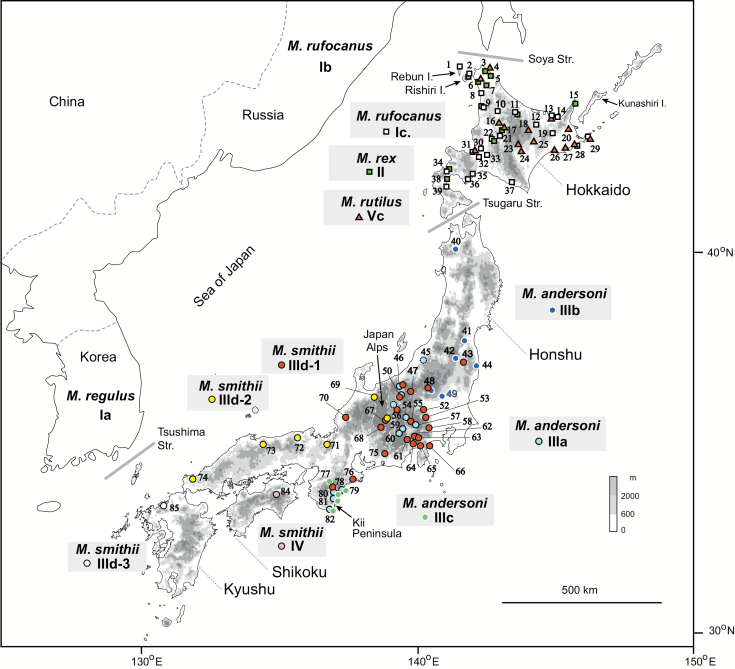
Fig. 1.
Collection locations for vole specimens used in this study. Myodes rex, M. rufocanus, and M. rutilus were collected from Hokkaido, and M. andersoni and M. smithii from Honshu, Shikoku, and Kyushu. Numbers correspond to the site numbers given in Supplementary Data SD1. Mitochondrial DNA clusters from phylogenetic and network analyses (Ic, II, IIId-1, IIId-2, etc.) are also indicated. Some specimens of cluster III (localities 45, 56, 77) for which exact locations are unknown are shown with dotted circles. An approximate location of the Japan Alps is shown with a dotted line.
The Japan Alps, which uplifted to their present heights during the Pleistocene (e.g., Research Group for Quaternary Tectonic Map 1968), are known to act as a physical barrier between northern and southern populations of several species (e.g., Tomozawa and Suzuki 2008; Oshida et al. 2009). The Kii Peninsula has a main body of mountainous blocks that sometimes harbors northern Honshu lineages of moles (Tsuchiya et al. 2000; Kirihara et al. 2013) and voles (Suzuki et al. 1999; Iwasa and Suzuki 2002; Iwasa et al. 2003). While the geographic distribution and genetic structuring in Japanese mammals are likely to have been affected by Pleistocene environmental fluctuations (Sato 2017), it remains unclear how and when the spatial genetic structures formed.
A primary factor predicted to drive the population dynamics of mammals in Japan is the influence of Pleistocene environmental changes accompanying glacial-interglacial cycles and sea level fluctuations (Tomozawa and Suzuki 2008; Hanazaki et al. 2017). In particular, the last (ca. 20,000 years ago) and penultimate (ca. 140,000 years ago) glacial maxima are reported to have made strong impacts on the present genetic structures of species with temperate origins (Oshida et al. 2009; Tomozawa et al. 2014; Suzuki et al. 2015). In addition, population expansion events have been deduced in a temperate population of small forest-dwelling rodents in Japan, in the intermediate period of 50,000–60,000 years ago (Hanazaki et al. 2017), at which time a warmer period (early marine oxygen isotope stage 3, MIS 3, 37,000–57,000 years ago—Lisiecki and Raymo 2005) had just started after a substantially colder period (MIS 4, 58,000–71,000 years ago; e.g., Sawagaki et al. 2004; Immonen et al. 2014). In central Honshu, glaciation is known to have expanded to its maximum extent, exceeding the extent of the last glacial maximum (LGM) 20,000 years ago (Sawagaki et al. 2004). During MIS 4, despite the extremely cold climate, the Tsushima Strait is thought to have been open, and a warm current entered the Sea of Japan, providing heat to cold and dry air masses that formed over Siberia, collecting water vapor from the sea surface, and creating heavy snowfall in the mountain ranges on the western coast of Japan during winter (Keigwin and Gorbarenko 1992; Igarashi and Oba 2006).Two congeneric species of wood mouse (Apodemus) inhabit the broad-leaf forests and show different patterns of responses to these environmental events. The large (A. specious) and small (A. argenteus) species on Honshu, Shikoku, and Kyushu showed responses to the penultimate glacial maximum and MIS 4, respectively. In contrast, these two species show similar rapid expansion associated with LGM in Hokkaido (Suzuki et al. 2015; Hanazaki et al. 2017). To better understand factors underlying these different response patterns, investigation of other species in the same geographic area may provide important insight (Jezkova et al. 2015).
Myodes, the vole genus found in a large area of northeastern Eurasia, including its easternmost peripheral insular domains of Sakhalin and the Japanese archipelago, is an ideal subject for assessing how the environmental factors described above affect genetic diversification and speciation processes in different species. In Japan, five Myodes species are known, including three from Hokkaido (M. rufocanus, M. rex, and M. rutilus), and two found on Honshu, Shikoku, and Kyushu (M. smithii and M. andersoni). On Hokkaido, M. rufocanus is the predominant species, while M. rex and M. rutilus have limited and fragmented distribution patterns (Iwasa and Nakata 2015). Myodes rufocanus and M. rex, although sympatric, show significant differences in microhabitat use; Myodes rex is more common in valley bottoms and on rather moderate slopes, with mixed undergrowth of Sasa bamboos and herbage (Nakata 1995). On Honshu, Shikoku, and Kyushu, M. andersoni and M. smithii are distributed in the north and the south, respectively, and their ranges overlap in central Honshu (Fig. 1). In contrast to the Hokkaido species, which occur from lowlands to mountainous areas, these herbivorous rodents are known to inhabit moist and rocky areas in mountainous terrain (e.g., Tabata and Iwasa 2013). Previous molecular phylogenetic and phylogeographic studies (Suzuki et al. 1999; Iwasa et al. 2000; Iwasa and Suzuki 2002, 2003; Luo et al. 2004; Abramson et al. 2012) have revealed an earlier divergence of M. rutilus and a close relationships among these five species of Myodes, referred to as “the rufocanus species group” (or independent genus Craseomys, e.g., Kohli et al. 2014), with estimated divergence times of two and one million years ago (Ma), respectively. The group includes M. rufocanus from the Asian continent and Hokkaido and M. regulus from the Korean Peninsula, in addition to the three species M. andersoni, M. smithii, and M. rex. Myodes smithii exhibits unique mitochondrial DNA (mtDNA) lineages from Shikoku that differ markedly from other lineages from parts of Honshu and Kyushu (Suzuki et al. 1999; Iwasa et al. 2000). In addition, the mtDNA composition of the Kii population of M. andersoni represents two distinct lineages (Iwasa and Suzuki 2002), perhaps reflecting past migration from central Honshu to the Kii Peninsula at different evolutionary times. Lastly, population expansion events have been noted in M. rex from previous mtDNA analysis (Kawai et al. 2013). Despite these previous studies, a comprehensive evolutionary history of Myodes in Japan has not yet been achieved, partly due to a lack of information about population dynamics and partly due to the lack of consensus from mtDNA data (e.g., Herman et al. 2014; Suzuki et al. 2015).
In studies using phylogenetic inference in rodent species, constant rates of mtDNA evolution based on calibration with fossil records, such as 0.024 substitutions/site/million years (My) for the cytochrome b gene (Cytb), have often been used as calibrations to estimate divergence times (Suzuki et al. 2003). However, growing evidence supports evolutionary rates that are not constant but time-dependent, especially in the early stage of divergence (Ho and Larson 2006; Howell et al. 2008; Galtier et al. 2009; Ho et al. 2011). Moreover, a consensus about these rates has not been reached to date. For example, in phylogeographic inference studies of vole species in the genera Myodes (Kohli et al. 2015) and Microtus (Herman et al. 2014) with Cytb sequences, different evolutionary rates of 0.0456 and 0.46 substitutions/site/My were used, respectively. Meanwhile, recent biogeographic studies of Japanese wood mice in the genus Apodemus led to additional molecular clock estimation based on multiple paleoclimatic calibration points, namely 1) lineage divergences between remote islands separated by deep sea in 100,000-year intervals, and 2) the bottleneck and expansion accompanying the three fluctuations of cooling and warming periods during the last 150,000 years (Suzuki et al. 2015; Hanazaki et al. 2017). The newly proposed values for time-dependent Cytb evolutionary rates for Apodemus are 0.028 (see “Materials and Methods”), 0.047, and 0.11 substitutions/site/My for estimation of divergence times > 130,000, 53,000, and 10,000 years ago, respectively. However, these values need to be tested across additional species to determine if the calibrations can be applied more broadly across multiple taxa in Japan (Hanazaki et al. 2017).
In this study, we examined the population dynamics of Myodes voles in Japan and address the factors driving these dynamics. With newly obtained Cytb sequences in addition to previously acquired sequences (Iwasa et al. 2000, 2002; Iwasa and Suzuki 2002, 2003), we assessed population expansion events in five Japanese vole species: M. rex, M. rufocanus, and M. rutilus from Hokkaido and M. andersoni and M. smithii from Honshu, Shikoku, and Kyushu. With these data, we also determined the temporal aspects of vole population dynamics in Japan, referring to previously described time-dependent molecular clocks from studies on Japanese wood mice (Suzuki et al. 2015; Hanazaki et al. 2017).
Materials and Methods
Sample collection
We used a total of 222 mtDNA Cytb sequences from six species of Myodes voles occurring in areas surrounding the Sea of Japan (Fig. 1; Supplementary Data SD1); 216 specimens of five species (M. andersoni, M. rex, M. rufocanus, M. rutilus, and M. smithii) from Japan and six specimens of four species (M. regulus, M. rufocanus, M. shanseius, and M. rutilus) from the continent (Supplementary Data SD1). Of these sequences, 133 (15, 14, 44, 25, and 35 specimens for M. andersoni, M. rex, M. rufocanus, M. rutilus, and M. smithii, respectively) were newly acquired as part of this study and another 89 sequences were obtained from GenBank (Supplementary Data SD1). Two species of Eothenomys (E. eva, GenBank KX354157 and E. chinensis, GenBank HM165437) were used as outgroup taxa.
Sequencing and sequence analysis
DNA was extracted using a DNeasy Blood & Tissue Kit (QIAGEN, Germantown, Maryland). Amplification of the Cytb gene (1,140 bp) was performed via polymerase chain reaction (PCR) using two primer sets. Primers were designed to target the 5′ half Cytb fragment (UF1: 5′-ACTTCAACTATGACCAATGACATGA-3′, UR1: 5′-TAGGCCTGTAGGGTTGTTGG-3′) and VOLE14 (Conroy and Cook 1999) was used with newly designed LF3 (5′-AGCCACCCTCACACGATTCT-3′) to target the 3′ half of Cytb. PCR was carried out with a preheating step of 10 min at 95°C and then 35 cycles of 30 s at 95°C, 30 s at 50°C, and 60 s at 72°C, followed by a final extension for 7 min at 72°C, using the Ampli Taq Gold 360 Master Mix kit (Invitrogen, Thermo Fisher Scientific Corp., Waltham, Massachusetts) and the primers above at final concentration of 0.25 pmol/µl. The PCR products were sequenced using the BigDye Terminator Cycle Sequencing kit v3.1 (Applied Biosystems, Thermo Fisher Scientific Corp., Waltham, Massachusetts) and an ABI3130 automated sequencer (Applied Biosystems). The sequences were edited with Proseq3.5 software (Filatov 2009). The Cytb sequences collected in this study were deposited in the DDBJ/EMBL/GenBank databases under accession numbers LC416877–LC416983 and LC458471–LC458497.
Phylogenetic analysis and estimation of divergence times
Phylogenetic trees of the mtDNA sequences were constructed using the maximum likelihood (ML) method with MEGA7 (Kumar et al. 2016) and the TN93 substitution model (Tamura and Nei 1993) with a gamma distribution (G) and the reliability of nodes was assessed with 1,000 bootstraps. The selection of the best fit nucleotide substitution model was selected based on the Akaike information criterion using jModelTest 2.1.9 (Darriba et al. 2012).
The time to the most recent common ancestor (TMRCA) of the Cytb sequences and 95% highest posterior density (HPD) were estimated using BEAST ver. 1.8.4 software (Drummond and Rambaut 2007) with representative sequences of each lineage. Analysis was carried out using the TN93 substitution model (Tamura and Nei 1993) and the strict-clock model, with an evolutionary rate of 0.03 substitutions/site/My, at which the curve of the time-dependent evolutionary rates is considered to reach to the steady state (Hanazaki et al. 2017). Bayesian searches were conducted using the Markov chain Monte Carlo (MCMC) method for 10 million generations. The first 1 million generations were discarded as a burn-in period. Tracer ver. 1.6 software (Rambaut et al. 2013) was used to assess the convergence of the MCMC chains. All parameters had effective sample sizes (ESS) > 200. The trees were summarized using TreeAnnotator v. 1.8.4 software (http://beast.community/treeannotator) with the settings “Maximum clade credibility tree” and “Mean heights” and displayed with FigTree v. 1.4.3 software (http://tree.bio.ed.ac.uk/software/figtree/).
Phylogeographic analysis
Median-joining haplotype networks were constructed using PopArt (Leigh and Bryant 2015) with the default settings. To calculate nucleotide and haplotype diversity of all data, ARLEQUIN ver. 3.5 software (Excoffier and Lischer 2010) was used.
To detect rapid expansion, mismatch distribution analysis and the neutrality tests of Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997) were performed using ARLEQUIN ver. 3.5. The mismatch distributions of the Cytb sequences were calculated based on the total number of mutations for each cluster of Myodes determined via phylogenetic and haplotype network analyses. The expected distribution was simulated under the sudden expansion model (Schneider and Excoffier 1999; Excoffier 2004) and evaluated using a parametric bootstrap approach (1,000 replicates). Populations that have experienced recent demographic expansion are expected to have smooth unimodal distributions in the mismatch distributions (Rogers and Harpending 1992). In this method, tests of deviation from a model of population expansion were undertaken using the sum of squares of the deviation (SSD) goodness-of-fit test in ARLEQUIN ver. 3.5. We used the raggedness index (r—Harpending 1994) as a test statistic for the estimated sudden expansion models. Significant SSD and r values (P ≤ 0.05) indicate a departure from the null model of population expansion. A confidence interval (CI) around the estimated tau (τ) parameter was obtained through 1,000 replicates of coalescent simulation.
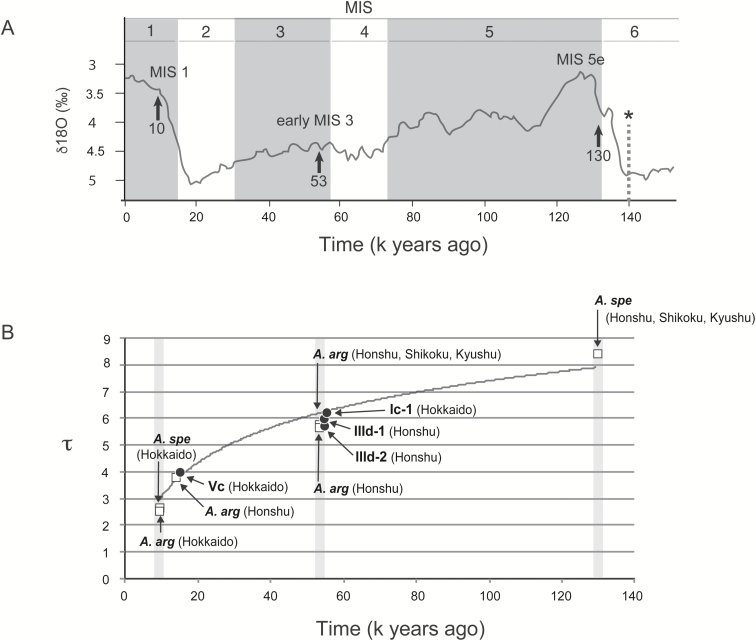
The time in generations (t) since expansion of each cluster was estimated using the formula t = τ/2u, where τ is a statistical value proportional to the time since expansion estimated using ARLEQUIN ver. 3.5 and u is the mutation rate per generation for the whole haplotype (Rogers and Harpending 1992). We estimated the time in millions of years (T) since expansion using the formula T = τ/2μk, where μ is the evolutionary rate (substitutions/site/My) and k is the sequence length of the haplotype. Molecular clock rates for intraspecific differentiation are known to decay when measured over increasing time scales (Ho et al. 2011). Therefore, we estimated time-dependent evolutionary rates of 0.11, 0.047, and 0.028 substitutions/site/My, for three levels of τ, ~2.3, ~5.6, and ~8.5, respectively (Suzuki et al. 2015; Hanazaki et al. 2017; this study). These correlate with times of ~ 10,000, ~ 53,000, and ~ 130,000 years ago, respectively. The τ value for the Japanese wood mouse (A. speciosus) estimated with our previous data set (Suzuki et al. 2015) was 9.2, which included haplotypes from the three main islands of Honshu, Shikoku, and Kyushu and the remote island of Tsushima.
Results
Phylogenetic analysis and estimation of divergence times
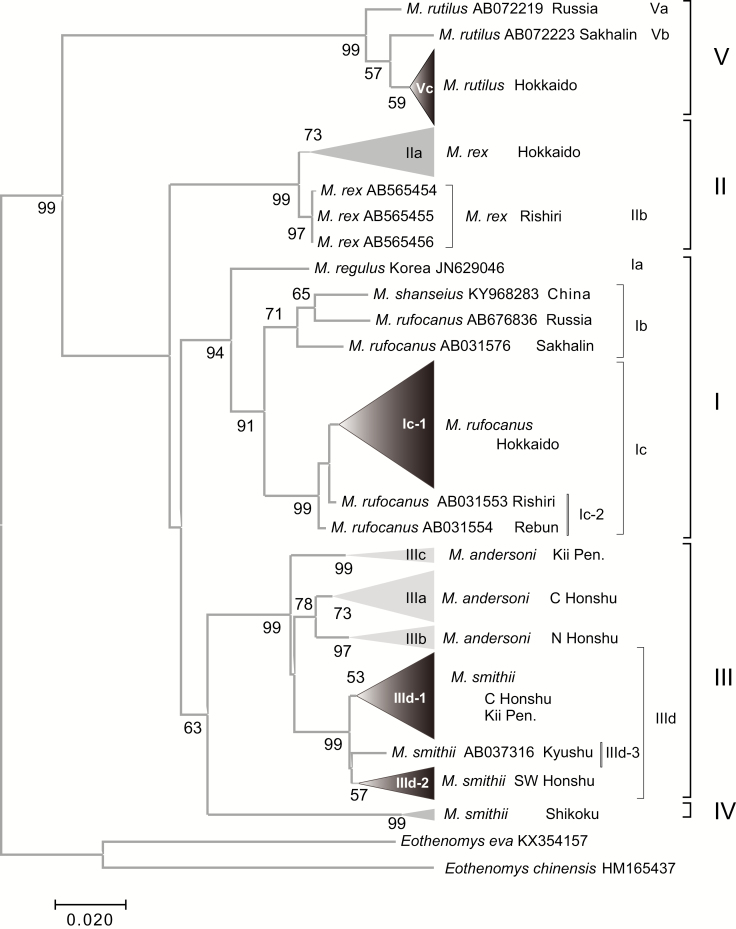
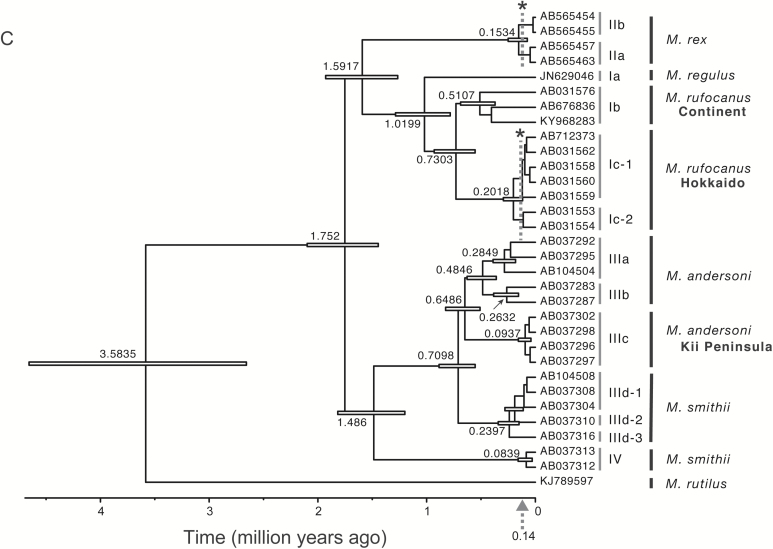
We constructed an ML tree to illustrate the relationships among the 216 sequences from five species of Japanese Myodes together with those of congeneric species from the continent, using two Eothenomys species as outgroup (Fig. 2). The phylogenetic analysis resulted in four distinct lineages (Lineages I, II, III, and IV) corresponding to the four M. rufocanus species groups (as suggested previously by Iwasa and Suzuki 2002), confirming the earlier divergence of M. rutilus (Lineage V, e.g., Luo et al. 2004). Lineage V contained a cluster represented by the haplotypes from the Hokkaido mainland (Vc), together with haplotypes from the Russian continent (Va) and Sakhalin (Vb), in accordance with previous work by Iwasa et al. (2002). Phylogenetic relationships among these lineages are undetermined, with only a node connecting lineages III and IV having weak bootstrap value (63%).
Fig. 2.
Maximum likelihood tree of cytochrome b sequences (1,140 bp) from Myodes rufocanus (lineage I), M. regulus (lineage I), M. rex (lineage II), M. andersoni (lineage III), M. smithii (lineage IV), and M. rutilus (lineage V). Clusters within lineages are also noted. Bootstrap support values >50% are indicated above the nodes.
Lineage I was comprised of three distinct clusters represented by haplotypes from M. regulus (Ia) from Korea, two species (M. rufocanus and M. shanseius) from the continent (Ib), and M. rufocanus from Hokkaido (Ic). Lineage II comprised two clusters of M. rex (IIa and IIb), showing geographic subdivision into the Hokkaido mainland and Rishiri Island, respectively. Lineage III contained four distinct clusters, with IIIa, IIIb, and IIIc represented by M. andersoni and IIId by M. smithii. Lineages IIIa and IIIb comprised haplotypes from central Honshu plus the Kii Peninsula and from northern Honshu, respectively. In addition to the IIIa haplotypes, voles from the Kii Peninsula included IIIc, in agreement with previous observations (Iwasa and Suzuki 2002). The database sequence of M. andersoni from Nagano prefecture, central Honshu (AB104505), was clustered into lineage IIId of M. smithii, as reported previously, indicating the possibility of interspecies genetic introgression (Iwasa and Suzuki 2003). Lastly, lineage IV was represented only by M. smithii from Shikoku.
Phylogeographic analysis
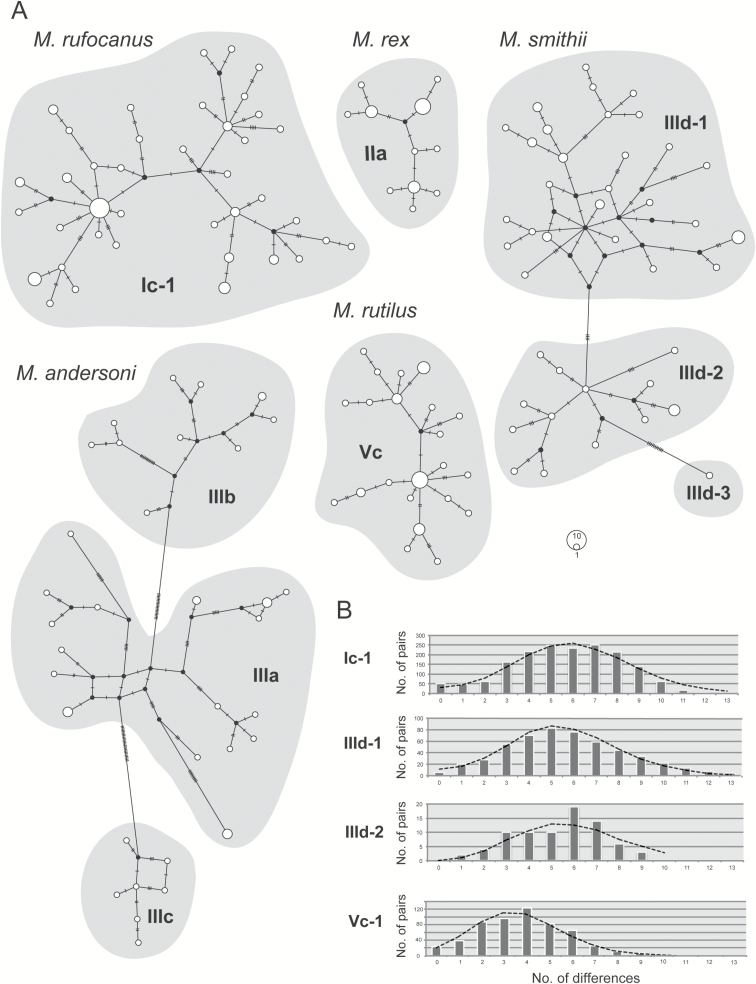
We constructed haplotype networks, using all available Cytb sequences to visualize levels of genetic diversity within each of the five species in Japan (Fig. 3A). Lineage IIId, representing M. smithii, was found to contain two clusters, IIId-1 and IIId-2 (Fig. 3).
Fig. 3.
(A) Median-joining haplotype network of Myodes mitochondrial DNA cytochrome b gene (Cytb) sequences. The number of mutations between haplotypes is indicated by dots and numbers. The size of the circles is related to haplotype frequency. The groups enclosed in a shaded area correspond to the haplotype groups listed in Table 1 and Fig. 2. (B) Mismatch distribution of the mitochondrial Cytb sequences. Only clades or subclades that showed significant evidence of expansion are shown. Bars indicate the observed frequencies of mutations between haplotypes and a line denotes the expected frequency under the sudden expansion model.
The cluster of M. rufocanus (Ic-1) from the Hokkaido mainland showed a star-like structure. Similarly, the haplotypes of M. rutilus from Hokkaido tended to show a star-like feature (Vc). Significantly negative values of Tajima’s D and Fu’s Fs tests were obtained for the four clusters with star-like network patterns (Fig. 3A), including the clusters of M. rufocanus from the Hokkaido mainland (Ic-1, n = 59), M. smithii (IIId-1, n = 33; IIId-2, n = 13) from Honshu, and M. rutilus from Hokkaido (Vc, n = 35; Table 1). Additionally, expansion signals of the four clusters were depicted in mismatch distribution analysis (Fig. 3B); nonsignificant SSD and r-values suggestive of population expansion were obtained. The τ-values ranged from 4.1 to 6.3 and the elapsed times since expansion (T) covering the last 140,000 years were estimated to be 16,000 and 54,000–59,000 years ago (using the recommended μ-values of 0.11 and 0.047 substitutions/site/My, respectively; Fig. 4A). These τ-values, together with those obtained in previous studies of wood mice (Suzuki et al. 2015; Hanazaki et al. 2017), were plotted against presumed initiation times of expansion events (Fig. 4B). Lineage IIa of M. rex (n = 23, τ = 3.63) and IIIa (n = 24, τ = 11.50), IIIb (n = 11, τ = 2.93), and IIIc (n = 7, τ = 4.58) of M. andersoni were not significant in the Tajima’s D test, while significant negative Fu’s Fs-values were observed in lineages IIa and IIIc.
Table 1.
Expansion time estimates and confidence intervals for Myodes voles from Japan with three potential evolutionary rates.
| Species | Haplotype group |
n | Tajima's D (P ) |
Fu's Fs (P ) |
τ
(CI) |
Elapsed times since expansion (My)a with three evolutionary rates (substations/site/My) | ||
|---|---|---|---|---|---|---|---|---|
| 0.028 | 0.047 | 0.11 | ||||||
| M. rufocanus | Ic-1 | 59 | −1.80 (0.013) |
−25.33 (0.000) |
6.28 (3.18–7.24) |
0.098 (0.050–0.114) |
0.058
(0.030–0.068) |
0.025 (0.012–0.028) |
| M. rex | IIa | 23 | −0.236 (0.452) |
−3.622 (0.028) | 3.63 | |||
| M. andersoni | IIIa | 24 | −0.41 (0.399) | −1.59 (0.265) |
11.50 | |||
| IIIb | 11 | −0.14 (0.489) | –1.99 (0.115) | 2.93 |
||||
| IIIc |
7 |
−0.11 (0.461) |
–3.58 (0.009) | 4.58 |
||||
| M. smithii | IIId-1 | 33 | −1.55 (0.045) |
–19.99 (0.000) | 5.90 (3.43–7.32) |
0.092 (0.054–0.115) |
0.055
(0.032–0.068) |
0.023 (0.014–0.029) |
| IIId-2 | 13 | −1.58 (0.048) |
−8.99 (0.000) |
5.82 (4.15–7.46) |
0.091 (0.065–0.117) |
0.054
(0.039–0.070) |
0.023 (0.017–0.030) |
|
| M. rutilus | Vc | 35 | −1.75 (0.018) | −14.19 (0.004) |
4.08 (2.06–5.02) | 0.063 (0.032–0.079) | 0.038 (0.039–0.047) | 0.016 (0.008–0.020) |
aConfidence intervals shown in parentheses. The most desirable values are shown in bold (see text). Expansion times were calculated in only clusters that showed rapid expansion signals in the Tajima’s D test. My, million years.
Fig. 4.
(A) The marine oxygen isotope curve for the last 140,000 years, adapted from Lisiecki and Raymo (2005). It highlights the three periods of rapid warming (arrows). In addition to the two prominent periods of marine isotope stages (MIS) 1 and 5e immediately after recovery from the impact of the glacial maxima, early MIS 3 is the third period of notably rapid warming (Suzuki et al. 2015; Hanazaki et al. 2017). (B) Five τ-values (2.6, 2.7, 5.7, 5.7, 8.5) with significant P-values from Tajima’s D test in our previous report (Hanazaki et al. 2017) are plotted. The points marked with “A. arg” and “A. spe” are τ-values from Apodemus argenteus and A. speciosus, respectively. The τ-value 8.5 is recalculated with the previous data set from the main islands of Honshu, Shikoku, and Kyushu (Suzuki et al. 2015), excluding data from the remote Tsushima Island. The four τ-values for the haplotype groups Ia-1, IIId-1, IIId-2, and Vc are plotted against the times of the expansion events, summarized in Table 1. (C) Bayesian phylogenetic tree with mitochondrial cytochrome b sequences (1,140 bp) of Myodes, showing means of the timing of divergence of nodes and horizontal bars of the interval of 95% HPDs estimated using BEAST dotted lines with asterisk indicate 140,000 years ago, when the lowest sea level of the penultimate glacial maximum occurred (Bintanja et al. 2005), supporting the view that divergence between the lineages representing the Hokkaido mainland and its surrounding islands of Rishiri and Rebun occurred before the predicted gene flow across the strait.
Estimation of divergence times between insular lineages
The genetic divergences (p-distance) between lineages I, II, III, and IV with representative sequences were determined to be 0.064–0.072 (0.073–0.082 in TN93+G distances). We assessed the divergence times between lineages using an evolutionary rate (μ) of 0.031 substitutions/site/My, with the nucleotide substitution model TN93+G, assuming 1.12 times higher than that of the p-distance evolutionary rate: 0.028). The resultant divergence times were estimated to be 1.49–1.75 million years ago (Ma; Fig. 4C). In M. rufocanus and M. rex, divergence times between the Hokkaido mainland (lineages Ic-1 and IIa) and its remote islands (lineages Ic-2 and IIb), Rebun and Rishiri, were estimated to be around or prior to the glacial maximum of 140,000 years ago (asterisks in Fig. 4C).
Discussion
In this study, we addressed the potential influence of Pleistocene climate changes on the population dynamics of vole species in Japan, covering subarctic (Hokkaido) and temperate (Honshu, Shikoku, and Kyushu) populations. From the present and previous (Suzuki et al. 2015; Hanazaki et al. 2017) studies, we are able to highlight three crucial periods to consider in assessing the impact of late Pleistocene climatic change.
While impacts of the paleoclimate have been suggested previously (e.g., Iwasa and Suzuki 2002; Kawai et al. 2013), here we provide evidence for signals of rapid expansion in four geographic lineages: Ic-1 (M. rufocanus, Hokkaido), IIId-1 (M. smithii, central Honshu), IIId-2 (M. smithii, southwestern Honshu), and Vc (M. rutilus, Hokkaido), revealing the presence of two ranges of the expansion indices, 4.1 and 5.8–6.3 (Table 1; Fig. 3). A similar trend was detected in our previous studies of Cytb (1,140 bp) variation in Japanese wood mice (Suzuki et al. 2015; Hanazaki et al. 2017), in which τ–values were classified as 2.7–3.9, ~ 5.7, and 8.5 (Fig. 4C). In Japanese large flying squirrels (Petaurista leucogenys), Oshida et al. (2009) identified two regional haplotype groups with expansion signals and expansion indexes of τ = 3.9 and 8.3. These lines of circumstantial evidence lead us to believe that three paleoclimatic stages are involved in the sudden expansion events of Japanese small mammals, including both granivorous and herbivorous rodents, while notable species-specific and region-specific factors are associated with the responses of these small rodents to paleoclimatic influences.
In our study, a population expansion event may be the consequence of the combination of a severe cold period and subsequent warmer period, which lead to a population bottleneck and population growth, respectively. In the late Pleistocene, such events are limited (Hanazaki et al. 2017) and the possible candidates are transitions MIS 6/5, MIS 4/3, and MIS 2/1, which can be characterized by the cold periods of the penultimate glacial maximum, MIS 4 (a cold stage of the last glacial period), and the LGM. According to the τ-values obtained in this study, the expansion events of Ic-1, IIId-1, and IIId-2 and Vc are likely to have occurred during the warming periods of MIS 4/3 and MIS 2/1, respectively (Fig. 4B). In the latter case, the τ-values differed substantially between voles (4.1) and wood mice (2.7; Suzuki et al. 2015), suggesting different onsets of population expansion at 16,000 (Table 1, immediately after LGM) and 10,000 years ago (Suzuki et al. 2015), respectively. The delayed onsets in the Apodemus species are likely to be correlated with the Younger Dryas cool reversal or the delayed recovery of Quercus forests that provide important food resources for wood mice (Igarashi 2016).
The aforementioned hypothetical scenario may explain the rapid population expansion events reported based on the population dynamics of M. rufocanus over its entire distributional range (Abramson et al. 2012). Abramson et al. (2012) detected the signal of rapid expansion in their Cytb sequence data (817 bp) of M. rufocanus from the mainland of Hokkaido and its peripheral islands, Kunashiri, Rebun, and Rishiri (Haplogroup A) with a calculated τ-value of 6.5, which corresponds to a τ-value of 9.0 for a data set of k = 1,140. This value is larger than our corresponding finding (τ = 8.5), probably due to their data containing sequences from both the mainland and peripheral islands, as the divergence of the islands likely occurred prior to the expansion event in the mainland. Abramson et al. (2012) further revealed two historical expansion events in M. rufocanus from the Eurasian continent. The first of these, in “the eastern clade (Haplogroup C2, τ = 4.4)” is estimated to have begun 57,000 years ago based on a transformed τ-value of 6.1 for k = 1,140 with an evolutionary rate of 0.047 substitutions/site/My. This event was followed by the expansion event of “the western clade (Haplogroup C1, τ = 1.7),” which is estimated to have begun 9,500 years ago with a molecular clock rate of 0.11 substitutions/site/My. The idea of an influence from the MIS 4/3 transition is less popular, compared to those related to glacial maxima, but has been suggested in both paleoclimatic (e.g., Sawagaki et al. 2004; Dalton et al. 2016; Stokes 2017) and phylogeographic (Kohli et al. 2015) studies. Kohli et al. (2015) considered the time period of 45,000 years for the expansion of M. rutilus from the Eurasian continent, accounting for paleoclimatic evidence in which the Verkhoyansk Mountain Range in northeastern Siberia was covered with ice during the glacial period of 80,000–55,000 years ago. While there are several instances of transitions between warmer and colder periods during the late Pleistocene (Hedges and Mann 1979; Igarashi and Oba 2006; Yamamoto et al. 2014), MIS 4 may be one of the most prominent in terms of the coldness that is essential to bottleneck events. Accordingly, the fairly strong influence of the paleoclimate on the two M. smithii haplotype groups distributing in central Honshu (IIId-1) and the coast of central-to-western Honshu (IIId-2) are predicted to have occurred during MIS 4. This is attributable to the topographical features of central Honshu, where multiple long mountain ranges are present including the southern and northern ranges of the Japan Alps.
In Hokkaido, the three species of vole examined have ancestral lineages hypothesized to have migrated in ancient times (Fig. 4B—see also Iwasa et al. 2000; Iwasa and Suzuki 2002; Luo et al. 2004), implying that they experienced the same environmental changes as sympatric species during at least half a million years. Nevertheless, the common and rare voles M. rufocanus and M. rutilus, respectively, have different time frames of rapid expansion (in the early MIS 3 and 1, respectively), as mentioned above. In addition, the other rare species, M. rex, which is a relic species displaced by the later M. rufocanus lineage (Nakata 1995), exhibited substantially less genetic diversity than did the predominant species M. rufocanus (Fig. 3). The consequences of the different responses to environmental changes can be explained in several ways. They may be more influential in rare species, increasing the strength of intraspecies competition and leading to bottlenecks in these species. Alternately, the ice age may have led to the formation of microhabitats on which vole species rely. Further studies are needed to specify the extent of contribution of factors affecting genetic diversity.
The Japanese archipelago, a “cradle swinging north and south.”
Understanding differential responses to paleoclimatic events, as shown in Myodes species in this study, is an important issue in phylogeography. Pleistocene climatic oscillations with 100,000-year intervals are known to have caused extinctions and repeated changes in the range of a given taxon with influences of latitude and topography, causing extensive extinction and recolonization of higher latitudes and elevations (Hewitt 2004). In mammals in Japan, past spatial dynamics associated with paleoclimatic oscillations have been suggested in a number of species of temperate origin, such as macaques (Macaca fuscata—Kawamoto et al. 2007), flying squirrels (Oshida et al. 2009), harvest mice (Micromys minutus—Yasuda et al. 2005), black bears (Ursus thibetanus—Ohnishi et al. 2009), and moles (Mogera wogura—Kirihara et al. 2013). In addition, in the Japanese archipelago, differential responses to paleoclimatic impacts between closely related species of arctic and temperate origin have been suggested in mammals based on fossil records, in particular elephants (the Palaeoloxodon-Sinomegaceroides complex) and bears (Hasegawa et al. 1972; Iwase et al. 2012). Notably, the two species of voles with a parapatric relationship, M. andersoni and M. smithii, may help clarify this phylogeographic issue.
Contrary to the response of the southern species M. smithii, the northern species M. andersoni showed no signal of rapid expansion events. However, repeated geographic extension and retreat of the northern species can be inferred from the presence of two mtDNA lineages in the currently geographically isolated population of the Kii Peninsula (Figs. 2 and 3 in this study—Iwasa and Suzuki 2002). The Kii Peninsula, exhibiting topographic complexity typical of the Japanese archipelago, may represent one of the southern limits of the distributional range of M. andersoni in colder geological times. The two Cytb lineages of M. andersoni found on the Kii Peninsula are considered to have branched off from the core range lineage of northern Honshu 0.55 and 0.20 Ma (Fig. 4A, B). Thus, the two vole species have conflicting distributions, with borders changing along with paleoclimatic environmental conditions; the northern and southern species extended their territories during colder and warmer periods, at fast and slow rates, respectively.
The current border between M. andersoni and M. smithii may reflect post-LGM events. If our hypothesis is correct, range expansion happened in early MIS 3. Accounting for these factors, it is possible that M. smithii experienced range expansion during both geological periods, but the influence of LGM had less impact on the genetic diversity of mtDNA, and thus is more difficult to recognize as a bottleneck event. Therefore, we emphasize that geographic expansion events are not always associated with mtDNA expansion signals, perhaps due to the weak influence of low temperature, which is not sufficient to cause bottleneck events.
Constructing evolutionary rate curves with multiple calibration points
There is considerable disagreement about the evolutionary rates of mtDNA. Evidence from fossils that can be used for calibration points is limited in number in general, especially for recent time periods. Hence, identifying calibration points in biogeographic events is a promising method for assessing evolutionary rates (e.g., Gratton et al. 2008; Ho et al. 2011). In this respect, small rodents in the Japanese archipelago provide fruitful information in several regards.
As noted above, extreme environmental changes in the late Pleistocene provide multiple efficient calibration points. Here, we categorized expansion indexes into three groups and hypothesized that the three expansion events could be attributed to the rapid warming periods beginning ~ 16,000 (MIS 1), ~ 53,000 (early MIS 3), and ~ 130,000 (MIS 5e) years ago resulting in evolutionary rate estimates of 0.11, 0.047, and 0.028 substitutions/site/My, respectively (Fig. 4B). These values for cricetid rodents are quite similar to those of murine rodents obtained previously (Hanazaki et al. 2017). The intermediate value is similar to the rate of 0.0456 substitutions/site/My derived from a demographic study of M. rutilus using Cytb sequences by Kohli et al. (2015), who noted the importance of the early MIS 3 period in demographic expansion of this species. We emphasize that the time-dependent evolutionary rates of the mitochondrial Cytb presented here are useful in assessing population dynamics during the late Pleistocene for not only cricetid rodents but also murine rodents and perhaps other rodents including squirrels (see Oshida et al. 2009).Some paleoclimatic events other than population expansion may contribute practical biogeographical calibration points for assessing the evolutionary rates of mtDNA. Hanazaki et al. (2017) assumed that land bridge structures appeared intermittently during glacial maxima at 100,000-year intervals, namely 140,000, 250,000, 350,000, and 440,000 years ago, and plotted possible evolutionary rates, showing a linear relationship among the plots. Regarding the divergence between the mainland and peripheral insular lineages, we suggest that the ancestral lineages of M. rufocanus (lineage Ic) and M. rex (lineage II) came from the Hokkaido mainland to Rishiri and Rebun when land bridges are assumed to have been present, around 140,000 years ago, which is harmonious with BEAST analysis using an evolutionary rate of 0.031 substitutions/site/My (TN93+G substitution model) inferred from the assumption of an expansion event 130,000 years ago (Fig. 4C).
The Japanese archipelago provides one additional option for determining biogeographic calibration points, namely the dispersal events of the ancestral lineages of Japanese mammals from the continent through land bridges across the deep straits (Hope et al. 2010; Kirihara et al. 2013). The Tsushima Strait, the entrance from the Asian continent to Kyushu, in particular, has attracted many researchers seeking calibration points for the time scale of about a million years (Luo et al. 2004; Hope et al. 2010; Kirihara et al. 2013). In this study, based on the possible evolutionary rate of 0.031 substitutions/site/My (TN93+G substitution model) and divergence between major lineages I, II, III, and IV, the ancestral lineage of all Japanese voles is suggested to have diverged 1.49–1.75 Ma (Fig. 4C). One possibility is that the ancestral lineage came to Japan 1.7 Ma, at which time the sea level of Tsushima Strait dropped (Kitamura and Kimoto 2006). Mammals living in the Japanese archipelago provide useful clues toward drawing the proper curves of mtDNA evolutionary rates, as they cover a variety of time periods in the Pleistocene with a variety of taxonomic groups. Finally, we stress that the intermittent connection between the continent and Japanese archipelago through a land-bridge and environmental changes at 100,000-year intervals during the early and middle Pleistocene, respectively, would provide valuable information as biogeographic calibration points in assessing the time-dependent evolutionary rates in a variety of taxonomic groups.
Supplementary Data
Supplementary data are available at Journal of Mammalogy online.
Supplementary Data SD1.—List of samples used in this study, collected from within and outside of Japan.
Acknowledgments
We thank M. Iijima, K. Hanazaki, T. Saitoh, J. J. Sato, Y. Suzuki, and M. Tomozawa for providing valuable comments on an earlier manuscript. We thank J. Light and three anonymous reviewers for their comments, which helped us to improve the manuscript. This study was conducted with the support of a JSPS KAKENHI Grant Number (JS18H05508).
Present address of GK: Laboratory of Forest Biology, Graduate School of Agriculture, Kyoto University, Sakyo-ku, Kyoto 606–8502, Japan
Literature Cited
- Abramson N. I., Petrova T. V., Dokuchaev N. E., Obolenskaya E. V., and Lissovsky A. A.. . 2012. Phylogeography of the gray red-backed vole Craseomys rufocanus (Rodentia: Cricetidae) across the distribution range inferred from nonrecombining molecular markers. Russian Journal of Theriology 11:137–156. [Google Scholar]
- Bintanja R., van de Wal R. S., and Oerlemans J.. . 2005. Modelled atmospheric temperatures and global sea levels over the past million years. Nature 437:125–128. [DOI] [PubMed] [Google Scholar]
- Conroy C. J., and Cook J. A.. . 1999. MtDNA evidence for repeated pulses of speciation within Arvicoline and murid rodents, Journal of Mammal 6:221–245. [Google Scholar]
- Dalton A. S., Finkelstein S. A., Barnett P. J., and Forman S. L.. . 2016. Constraining the Late Pleistocene history of the Laurentide Ice Sheet by dating the Missinaibi Formation, Hudson Bay Lowlands, Canada. Quaternary Science Reviews 146:288–299. [Google Scholar]
- Darriba D., G. L. Taboada R. Doallo, and Posada D.. 2012. Jmodeltest 2: more models, new heuristics and parallel computing. Nature Methods 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., and Rambaut A.. . 2007. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. 2004. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Molecular Ecology 13:853–864. [DOI] [PubMed] [Google Scholar]
- Excoffier L., and Lischer H. E.. . 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Molecular Ecology Resources 10:564–567. [DOI] [PubMed] [Google Scholar]
- Filatov D. A. 2009. Processing and population genetic analysis of multigenic datasets with proseq3 software. Bioinformatics (Oxford, England) 25:3189–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., B. Nabholz S. Glémin, and Hurst G. D.. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology 18:4541–4550. [DOI] [PubMed] [Google Scholar]
- Gratton P., M. K. Konopiński, and Sbordoni V.. 2008. Pleistocene evolutionary history of the clouded apollo (parnassius mnemosyne): genetic signatures of climate cycles and a ‘time-dependent’ mitochondrial substitution rate. Molecular Ecology 17:4248–4262. [DOI] [PubMed] [Google Scholar]
- Hanazaki K.,, et al. 2017. Estimation of evolutionary rates of mitochondrial DNA in two japanese wood mouse species based on calibrations with quaternary environmental changes. Zoological Science 34:201–210. [DOI] [PubMed] [Google Scholar]
- Harada M., Ando A., Tsuchiya K., and Koyasu K.. . 2001. Geographical variations in chromosomes of the greater Japanese shrew-mole, Urotrichus talpoides (Mammalia: Insectivora). Zoological Science 18:433–442. [Google Scholar]
- Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9:552–569. [DOI] [PubMed] [Google Scholar]
- Harpending H. C. 1994. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology 66:591–600. [PubMed] [Google Scholar]
- Hasegawa Y. 1972. Naumann’s elephant, Palaeoloxodon naumanni (Makiyama) from the late Pleistocene off Shakagahana, Shodoshima Is. In Inland Sea, Japan. Bulletin of the National Science Museum 15:513e591. [Google Scholar]
- Hedges J. I., and Mann D. C.. . 1979. The characterization of plant tissues by their lignin oxidation products. Geochimistry et Cosmoch Acta 43:1803–1807. [Google Scholar]
- Herman J. S., A. D. McDevitt A. Kawałko M. Jaarola J. M. Wójcik, and Searle J. B.. 2014. Land-bridge calibration of molecular clocks and the post-glacial colonization of scandinavia by the eurasian field vole Microtus agrestis. Plos ONE 9:e103949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. M. 2004. Genetic consequences of climatic oscillations in the quaternary. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359:183–195; discussion 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. Y.,, et al. 2011. Time-dependent rates of molecular evolution. Molecular Ecology 20:3087–3101. [DOI] [PubMed] [Google Scholar]
- Ho S. Y., and Larson G.. . 2006. Molecular clocks: when times are a-changin’. TRENDS in Genetics 22:79–83. [DOI] [PubMed] [Google Scholar]
- Hope A. G., et al. 2010. High-latitude diversification within Eurasian least shrews and Alaska tiny shrews (Soricidae). Journal of Mammalogy 91:1041–1057. [Google Scholar]
- Howell N., C. Howell, and Elson J. L.. 2008. Time dependency of molecular rate estimates for mtDNA: this is not the time for wishful thinking. Heredity 101:107–108. [DOI] [PubMed] [Google Scholar]
- Igarashi Y. 2016. Vegetation and climate during the LGM and the last deglaciation on Hokkaido and Sakhalin Islands in the northwest Pacific. Quaternary International 425:28–37. [Google Scholar]
- Igarashi Y., and Oba T.. . 2006. Fluctuations in the East Asian monsoon over the last 144 ka in the northwest Pacific based on a high-resolution pollen analysis of IMAGES core MD01-2421. Quaternary Science Reviews 25:1447–1459. [Google Scholar]
- Immonen N., Strand K., Huusko A., and Lunkka J. P.. . 2014. Imprint of late Pleistocene continental processes visible in ice-rafted grains from the central Arctic Ocean. Quaternary Science Reviews 92:133–139. [Google Scholar]
- Iwasa M. A., et al. 2000. Geographic patterns of cytochrome b and Sry gene lineages in the gray red-backed vole Clethrionomys rufocanus from Far East Asia including Sakhalin and Hokkaido. Zoological Science 17:477–484. [Google Scholar]
- Iwasa M. A., and Suzuki H.. . 2002. Evolutionary networks of maternal and paternal gene lineages in voles (Eothenomys) endemic to Japan. Journal of Mammalogy 83:852–865. [Google Scholar]
- Iwasa M. A., and Suzuki H.. . 2003. Intra- and interspecific genetic complexities of two Eothenomys species in Honshu, Japan. Zoological Science 20:1305–1313. [DOI] [PubMed] [Google Scholar]
- Iwasa M. A., and Nakata K.. . 2015. Conventionally and differentially stained karyotypes of the dark red-backed vole, Myodes rex. Mammal Study 40:181–185. [Google Scholar]
- Iwase A., Hashizume J., Izuho M., Takahashi K., and Sato H.. . 2012. Timing of megafaunal extinction in the late Late Pleistocene on the Japanese Archipelago. Quaternary International 255:114–124. [Google Scholar]
- Iwasa M. A., Kartavtseva I. V., Dobrotvorsky A. K., Panov V. V., and Suzuki H.. . 2002. Local differentiation of Clethrionomys rutilus in northeastern Asia inferred from mitochondrial gene sequences. Mammalian Biology-Zeitschrift für Säugetierkunde 67:157–166. [Google Scholar]
- Jezkova T., B. R. Riddle D. C. Card D. R. Schield M. E. Eckstut, and Castoe T. A.. 2015. Genetic consequences of postglacial range expansion in two codistributed rodents (genus Dipodomys) depend on ecology and genetic locus. Molecular Ecology 24:83–97. [DOI] [PubMed] [Google Scholar]
- Kawai K., F. Hailer A. P. de Guia H. Ichikawa, and Saitoh T.. 2013. Refugia in glacial ages led to the current discontinuous distribution patterns of the dark red-backed vole Myodes rex on Hokkaido, Japan. Zoological Science 30:642–650. [DOI] [PubMed] [Google Scholar]
- Kawamoto Y.,, et al. 2007. Postglacial population expansion of japanese macaques (Macaca fuscata) inferred from mitochondrial DNA phylogeography. Primates; Journal of Primatology 48:27–40. [DOI] [PubMed] [Google Scholar]
- Keigwin L. D., and Gorbarenko S. A.. . 1992. Sea level, surface salinity of the Japan Sea, and the Younger Dryas event in the northwestern Pacific Ocean. Quaternary Research 37:346–360. [Google Scholar]
- Kirihara T., A. Shinohara K. Tsuchiya M. Harada A. P. Kryukov, and Suzuki H.. 2013. Spatial and temporal aspects of occurrence of Mogera species in the Japanese Islands inferred from mitochondrial and nuclear gene sequences. Zoological Science 30:267–281. [DOI] [PubMed] [Google Scholar]
- Kitamura A., and K. Kimoto. 2006. History of the inflow of the warm Tsushima Current into the Sea of Japan between 3.5 and 0.8 Ma. Palaeogeography, Palaeoclimatology, Palaeoecology, 236:355–366. [Google Scholar]
- Kohli B. A.,, et al. 2014. Multilocus systematics and non-punctuated evolution of holarctic Myodini (Rodentia: Arvicolinae). Molecular Phylogenetics and Evolution 76:18–29. [DOI] [PubMed] [Google Scholar]
- Kohli B. A., Fedorov V. B., Waltari E., and Cook J. A.. . 2015. Phylogeography of a Holarctic rodent (Myodes rutilus): testing high latitude biogeographical hypotheses and the dynamics of range shifts. Journal of Biogeography 42:377–389. [Google Scholar]
- Kumar S., G. Stecher, and Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisiecki L. E., and Raymo M. E.. . 2005. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 20:PA1003 [Google Scholar]
- Luo J.,, et al. 2004. Molecular phylogeny and biogeography of oriental voles: genus Eothenomys (Muridae, Mammalia). Molecular Phylogenetics and Evolution 33:349–362. [DOI] [PubMed] [Google Scholar]
- Nakata K. 1995. Microhabitat selection in two sympatric species of voles, Clethrionomys rex and Clethrionomys rufocanus bedfordiae. Journal of the Mammalogical Society of Japan 20:135–142. [Google Scholar]
- Ohnishi N., R. Uno Y. Ishibashi H. B. Tamate, and Oi T.. 2009. The influence of climatic oscillations during the quaternary era on the genetic structure of asian black bears in Japan. Heredity 102:579–589. [DOI] [PubMed] [Google Scholar]
- Oshida T., Masuda R., and Ikeda K.. . 2009. Phylogeography of the Japanese giant flying squirrel Petaurista leucogenys (Rodentia: Sciuridae): implication of glacial refugia in an arboreal small mammal in the Japanese Islands. Biological Journal of the Linnean Society 98:47–60. [Google Scholar]
- Rambaut A., Suchard M., and Drummond A.. . 2013. Tracer v1. 5. 2009. Available from http://beast.bio.ed.ac.uk/ Accessed 12 February 2018
- Research Group for Quaternary Tectonic Map 1968. Quaternary tectonic map of Japan. The Quaternary Research (Daiyonki-Kenkyu) 7:182–187. (in Japanese with English abstract) [Google Scholar]
- Rogers A. R., and Harpending H.. . 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9:552–569. [DOI] [PubMed] [Google Scholar]
- Sato J. J. 2017. A review of the processes of mammalian faunal assembly in Japan-insights from molecular phylogenetics. Pp. 49–116 in Species diversity of animals in Japan (Motokawa M. and Kajihara H., eds.). Springer, Japan. [Google Scholar]
- Sawagaki T., Aoki T., Hasegawa H., Iwasaki S., Iwata S., and Hirakawa K.. . 2004. Late Quaternary glaciations in Japan. Developments in Quaternary Sciences 2:217–225. [Google Scholar]
- Schneider S., and Excoffier L.. . 1999. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes C. R. 2017. Deglaciation of the Laurentide Ice Sheet from the Last Glacial Maximum. Cuadernos de investigación geográfica 43:377–428. [Google Scholar]
- Suzuki H., et al. 1999. Molecular phylogeny of red-backed voles in Far East Asia based on variation in ribosomal and mitochondrial DNA. Journal of Mammalogy 80:512–521. [Google Scholar]
- Suzuki H., et al. 2003. Molecular phylogeny of wood mice (Apodemus, Muridae) in East Asia. Biological Journal of the Linnean Society 80:469–481. [Google Scholar]
- Suzuki Y., M. Tomozawa Y. Koizumi K. Tsuchiya, and Suzuki H.. 2015. Estimating the molecular evolutionary rates of mitochondrial genes referring to quaternary ice age events with inferred population expansions and dispersals in Japanese Apodemus. BMC Evolutionary Biology 15:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata M., and Iwasa M. A.. . 2013. Environmental factors for the occurrence of the Smith’s red-backed vole, Eothenomys smithii, in rocky terrains at the foot of Mt. Fuji in central Japan. Mammal Study 38:243–251. [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., and Nei M.. . 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512–526. [DOI] [PubMed] [Google Scholar]
- Tomozawa M., Nunome M., Suzuki H., and Ono H.. . 2014. Effect of founding events on coat colour polymorphism of Apodemus speciosus (Rodentia: Muridae) on the Izu Islands. Biological Journal of the Linnean Society 113:522–535. [Google Scholar]
- Tomozawa M., and Suzuki H.. . 2008. A trend of central versus peripheral structuring in mitochondrial and nuclear gene sequences of the Japanese wood mouse, Apodemus speciosus. Zoological Science 25:273–285. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K.,, et al. 2000. Molecular phylogeny of east Asian moles inferred from the sequence variation of the mitochondrial cytochrome b gene. Genes & Genetic Systems 75:17–24. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Ohira F., and Kumon F.. . 2014. Late Pleistocene variation in lignin and fatty acids from core TKN-2004 in a small mountain basin in central Japan. Geochemistry Journal 48:207–217. [Google Scholar]
- Yasuda S. P., Vogel P., Tsuchiya K., Han S. H., Lin L. K., and Suzuki H.. . 2005. Phylogeographic patterning of mtDNA in the widely distributed harvest mouse (Micromys minutus) suggests dramatic cycles of range contraction and expansion during the mid-to late Pleistocene. Canadian Journal of Zoology 83:1411–1420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.