Abstract
Purpose of the Review
To summarize current knowledge on interactions between genetic variants and lifestyle factors (G×L) associated with the development of coronary artery disease (CAD) and prioritize future research.
Recent Findings
Genetic risk and combined lifestyle factors and behaviors have a log-additive effect on the risk of developing CAD.
Summary
First, we describe genetic and lifestyle factors associated with CAD and then focus on G×L interactions. The majority of G×L interaction studies are small-scale candidate gene studies that lack replication and therefore provide spurious results. Only a few studies, of which most use genetic risk scores or genome-wide approaches to test interactions, are robust in number and analysis strategy. These studies provide evidence for the existence of G×L interactions in the development of CAD. Further G×L interactions studies are important as they contribute to our understanding of disease pathophysiology and possibly provide insights for improving interventions or personalized recommendations.
Keywords: Genetics, Lifestyle, Coronary artery disease, Genetic lifestyle interactions, Genome-wide association studies, Genetic risk scores
Introduction
Coronary artery disease (CAD) is a complex multifactorial disease leading to ischemic heart disease and myocardial infarction. Globally, CAD is an important cause of death and morbidity, with approximately 9 million deaths between 2007 and 2017 [1]. In line with the most complex and non-communicable diseases, the development of CAD is the result of an interplay between both lifestyle and genetic factors [2••].
Lifestyle factors can broadly be defined as behaviors, customs, and habits of persons or groups, generally considered in the context of consequences for health [3]. Lifestyle risk factors are often modifiable and so are their risks. Interventions to adhere to a healthy lifestyle as a means of prevention have the potential to greatly reduce incident CAD event rates [4, 5].
Over the past decade, large advances in technology have moved the boundary of our understanding of the genetics of CAD. Genome-wide association studies (GWAS’s) and next-generation sequencing have helped to identify a large number of genetic loci associated with CAD and helped to better appreciate the complexity of its genetic architecture [6, 7••]. Despite our progress in the understanding of the complex genetics and the many lifestyle factors involved in the development and progression of CAD, the interplay between genetic variants and lifestyle factors leading to CAD remains largely obscure. Large-scale, well-powered studies investigating combined genetic and lifestyle risks have only recently started to fill this knowledge gap with credible evidence [2••, 8••].
In this review, we first summarize important knowledge of genetic and lifestyle factors associated with CAD and then focus on the contribution of genetic and lifestyle (G×L) interactions in the development of CAD. We conclude with future research directions to progress the field of G×L in CAD.
Genetics of CAD
A heritable component to CAD has been well established, and recent studies estimate the heritability of CAD to range between 40 and 50 % [9]. Genetic analyses have been instrumental to progress our understanding of biological mechanisms involved in the development of CAD. Before the first well-powered GWAS in 2007, candidate gene studies were used to investigate common single nucleotide polymorphisms (SNPs) in genes coding for proteins with suspected biological importance in the pathophysiology of CAD. Although SNPs were frequently reported to be significantly associated with CAD, many candidate gene studies failed to achieve statistical significance after adjustment for multiple testing [10]. In addition, replication studies were often lacking. Since 2007, GWAS’s have become state-of-the-art to further our understanding of the genetics of complex diseases [6, 11]. GWAS’s investigate the association between millions of SNPs and a disease by comparing individuals with and without the disease [12]. In GWAS’s, common allele variants of SNPs that occur in at least 1–5% in the population are studied to determine their contribution to the disease [12]. To date, multiple GWAS’s have been performed on CAD in increasingly larger populations. Several SNPs have been identified to be strongly associated with CAD by both candidate gene and GWAS approaches, including APOB [13], PCSK9 [7••], and LPA [7••], which resulted in the development of drugs targeting these genes [14].
The most recent GWAS on CAD by van der Harst et al. included 122,733 cases and 424,528 controls and reported over 160 genome-wide significant (P < 5 × 10−8) loci associated with CAD. To understand the nature of these associations, possible shared genetic pathways with other traits or diseases were investigated and showed various associations with anthropometric measurements, lipids, inflammation markers, kidney function, diabetes mellitus, and blood pressure possibly providing an intermediate trait in the development of CAD [7••]. Insights into biology can also be obtained by studying gene expression patterns, for example with tools such as Data-driven Expression-Prioritized Integration for Complex Traits (DEPICT). DEPICT analyses on GWAS’s of CAD indicated important roles for platelets, blood vessel development, hemostasis, and a protein-protein interaction subnetwork [7••]. These findings provide reinforcement or novel evidence for the key roles of pathways and genes in the development of CAD and provide possible leads on how lifestyle factors might interact with them.
To further understand cumulative effects of biological pathways and outcomes associated with CAD, SNPs identified through GWAS’s have been summed to calculate genetic risk scores (GRS’s) as an estimation of an individual’s genomic risk of CAD. A weighted GRS counts the number of risk-increasing alleles (0, 1, or 2) per SNP for each individual and takes into account the effect size of each risk-increasing allele of each SNP as calculated in the GWAS. Since GRS’s are based on germline SNPs with alleles that are randomly allocated at conception, GRS’s are quantifiable from birth and potentially allow earlier risk stratification and primary prevention of events. GRS’s usually only include loci that reached genome-wide significance. For example, van der Harst et al. constructed a weighted GRS for CAD to investigate the risk of downstream cardiovascular diseases and observed associations with the development of atrial fibrillation and heart failure [7••].
Another strategy to estimate an individual’s genomic risk includes the creation of more extensive GRS’s using thousands or even millions of SNPs weakly or uncertainly associated with CAD. Another study constructed a CAD GRS with over 6 million SNPs and found individuals in the top 1% of the distribution were at almost 5-fold (odds ratio 4.83; 95% confidence interval, 4.25–5.46; P = 1 × 10−132) higher odds of CAD [15]. Inouye et al. used a GRS with 1.7 million SNPs linked to CAD and observed a 4-fold (hazard ratio 4.17; 95% confidence interval, 3.97–4.38) higher risk of CAD in individuals in the top quintile of the GRS compared with the lowest quintile [16]. A combination of the GRS with six conventional risk factors including diabetes, BMI, current smoking, hypertension, family history of heart disease, and high cholesterol led to a slight increase of 2.6% compared with the model with only the six conventional risk factors [16]. However, lipids and other biochemical variables were not available and a comparison with traditional risk scores such as the Framingham Risk Score could therefore not be performed [16].
So far, individual genetic variants identified by GWAS’s only explain ~ 15% of the estimated 40–50% heritability for CAD. The gap between currently explained CAD SNP–based heritability and other heritability estimates, the so-called missing heritability, may partly be found in rare variants which can be assessed using whole exome sequencing [17]. Using whole exome sequencing data, one recent study was able to increase SNP-based heritability estimates for height and BMI to pedigree heritability levels [18]. Rare variants in low linkage disequilibrium with neighboring variants, especially protein-coding variants, therefore likely contribute highly to heritability [18]. Another possibility is that underlying G×L interactions, in which the genetic component explains more variance depending on the lifestyle, remain to be elucidated. These G×L interactions could also possibly explain a proportion of the missing heritability.
Genetics of Lifestyle Factors Associated with CAD
The importance of lifestyle factors in the development and primary prevention of CAD is well established. Rappaport et al. studied 3229 Swedish twins and estimated 21.6% of CAD deaths were attributable to non-modifiable genetic factors, and the remaining 78.4% to lifestyle and environment exposures during an individuals’ lifetime [19]. The INTERHEART study investigated the importance of modifiable risk factors in 52 countries across the globe and found that raised apolipoprotein B/A1 ratio, current smoking status, no regular alcohol intake, hypertension, diabetes, abdominal obesity, psychosocial factors (depression, low locus of control, perceived stress, and major life events), lack of daily fruit and vegetable consumption, and no regular physical exercise accounted for most of the risk of acute myocardial infarction [20]. Although the identified lifestyle factors independently increase CAD risk, lifestyle risk factors tend to cluster in adults, with 20% of the individuals of the general population having at least three lifestyle risk factors [21]. The lifestyle factors highlighted by the INTERHEART study are discussed below.
Tobacco Smoking
Tobacco smoking is a major risk factor for CAD [22]. Since the average cigarette contains a complex and changing mix of poisonous compounds with various pathological effects [23], the exact mechanisms leading to CAD remain unknown [24]. Currently, it is known that smoking leads to atherosclerosis through endothelial dysfunction and damage, plaque vulnerability with increased risk of rupture, increased inflammatory and thrombotic state, and increased blood pressure [24]. Tabaco smoking is also under the influence of genetic factors, including several SNPs associated with smoking initiation, heaviness, and cessation [25, 26].
Alcohol and Coffee Consumption
Heavy alcohol consumption has been described to increase risk of CAD, whereas low to moderate intakes might reduce the risk [27, 28]. Similar to smoking, genetics also influence alcohol intake [26]. Liu et al. found 99 SNPs associated with the amount of drinks per week, but did not find a significant genetic correlation between drinks per week and phenotypic CAD [26].
A similar U-shaped risk pattern has been described for coffee consumption: excessive or no observational coffee and caffeine intake are associated with increased risks for CAD risk while moderate intakes appear to reduce risk [29, 30]. Genetic studies did not yield evidence for causal links between caffeine intake and CAD [29, 31], suggesting that reported beneficial observational findings may be confounded by the numerous non-caffeine constituents of coffee [30].
Physical Activity and Sedentary Behavior
Physical activity plays an important role in both the primary and secondary prevention of CAD [32]. Epidemiological studies show a dose-response relationship leading to a 20% reduction of cardiovascular events in individuals who practice leisure-time physical activity. Moreover, in secondary prevention, exercise training has been shown to improve endothelial function, halt the progression of coronary stenosis, and possibly induce collateral formation leading to improved myocardial perfusion [32]. However, despite these known protective effects, there is a dangerous trend towards less physical activity worldwide [33]. In addition, sedentary behavior has been established as an important driver of chronic diseases, independent of physical activity levels [34]. Although twin and family studies showed that both physical activity and sedentary behaviors are potentially heritable [35], GWAS’s were mostly performed on self-reported data of physical activity, yielding only a few associated loci [36]. A recent GWAS using accelerometer data found one locus associated with overall activity and 4 loci with sedentary behavior, explaining up to 21% of the heritability of physical activity and 12.9% of sedentary behavior [37]. Given the large economic and health burden [38, 39], more research into the genetic architecture of physical activity and sedentary behavior is needed.
Diet
The impact of diet on CAD has been studied mostly in observational studies, which have generally found Mediterranean and DASH (Dietary Approaches to Stop Hypertension) diets, which are both rich in fruits, vegetables, and nuts, to be associated with lower risk of MI and improved cardiometabolic factors with effects on blood pressure, blood lipids, inflammation, endothelial function, and thrombosis [40]. Robust genetic data to further understand the determinants of interindividual variation in response to diet is largely missing, although diet may influence epigenetic changes such as DNA methylation [40, 41].
Lifestyle behavior, especially smoking and alcohol use, can also be viewed as the willingness of an individual to take certain health risks. This risk tolerance or behavior may have a predisposing genetic architecture, as shown by Karlsson Linnér et al. who found hundreds of loci associated with risk tolerance and risky behaviors, including 124 SNPs associated with general risk tolerance [42]. The general risk tolerance SNPs were associated with genes that were highly expressed in brain regions such as the prefrontal cortex, basal ganglia, and midbrain [42].
Gene × Lifestyle Interactions
Genetic and lifestyle factors both contribute to each person’s risk of CAD and have a complex interplay. Lifestyle factors themselves are partly determined by genetic factors. In addition, increasingly more evidence indicates lifestyle can also modify the effects of genetic variants on CAD. This may be due to shared etiological pathways between the genetic variant and lifestyle risk factor, in turn leading to the disease. Another potential mechanism includes epigenetics, in which gene expression is altered by lifestyle or environmental factors through local chromatin environment changes affecting DNA accessibility despite the absence of DNA alterations [41, 43].
Some of the complex gene and lifestyle interplay can be better understood through G×L interaction studies. These interactions can be investigated using several approaches, as discussed below.
GRS Approach
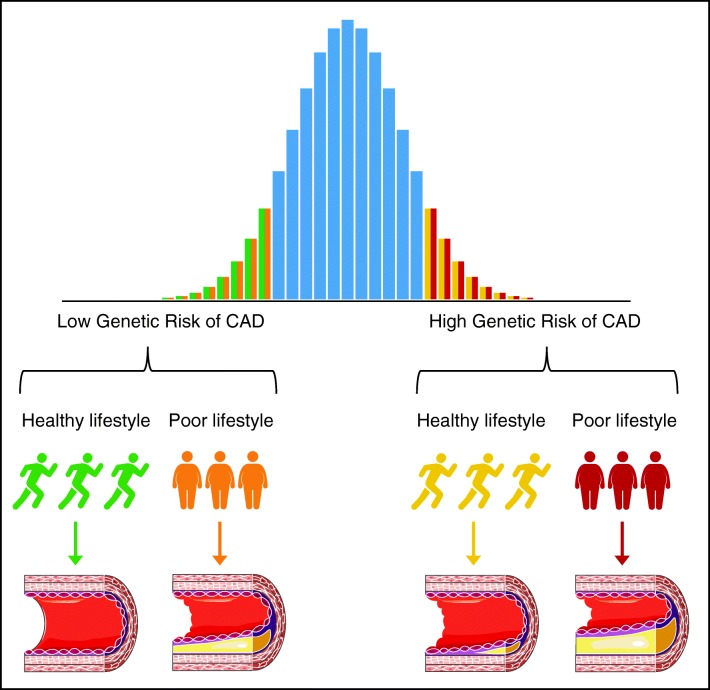
It is possible to construct a GRS based on the most highly associated variants of previous GWAS’s and analyze the interaction between this overall genetic risk and a lifestyle factor on CAD. However, the number of studies investigating the combined risks of both genetic and lifestyle factors is little. To date, two large studies investigated the combined risks of CAD associated with genetics and overall lifestyle [2••, 8]. The first study by Khera et al. included 55,685 individuals with 5,103 (9.2%) cases from four cohorts [8••], and the second study by Said et al. included 325,113 individuals with 9,771 (3.0%) cases from the UK Biobank [2••]. Both studies calculated a weighted GRS based on previous reports and categorized the population into quintiles of genetic risk. The lowest quintile was taken as a low genetic risk, the second to fourth quintile as intermediate risk, and the highest quintile as high genetic risk. Lifestyle was subsequently categorized as ideal, intermediate, or poor [44]. Individuals with poor lifestyle were at higher risk of CAD compared with individuals in the same genetic risk category but with an ideal lifestyle. The risk of CAD increased not only with less than ideal lifestyle, but also with increasing genetic risk. Importantly, both studies showed individuals with ideal lifestyle and high genetic risk were at nearly twice the risk of developing CAD compared with individuals with an ideal lifestyle but low genetic risk. These findings indicate individuals with high genetic risk have a higher starting risk of developing disease compared with individuals with similar lifestyle but lower genetic risk, and even higher risks of events if they have a poor lifestyle and high genetic risk (Fig. 1).
Fig. 1.
Combined genetic and lifestyle risks increase the risk of coronary artery disease. The risk of coronary artery disease (CAD) in individuals with low or high genetic risk is higher amongst individuals with poor lifestyle compared to a healthy lifestyle. Compared with individuals with low genetic risk, individuals with high genetic risk start off at higher risks of CAD, with the highest risks of CAD amongst individuals with poor lifestyle and high genetic risk
Although neither study observed statistically significant interactions between lifestyle categories and genetic risk, Said et al. estimated that with 80% power and an alpha of 0.005, interaction effects between genetic risk and intermediate or poor lifestyle would range from 1.21 to 1.50 [2••]. The risk of CAD in individuals with high genetic risk therefore possibly not only starts off at higher risk, but also increases more strongly with worse overall lifestyle (Fig. 1).
The importance of physical activity in genetic risk groups of CAD has been reported as well [45]. One study found that higher grip strength and cardiorespiratory fitness were associated with a lower risk of incident CAD events across tertiles of genetic risk of CAD [45].
Genome-Wide Interaction Studies
Instead of selecting lead variants of previous GWAS’s and testing these for interactions with lifestyle factors, SNPs across the whole genome can be scanned in a genome-wide interaction study. Because unveiling G×L interactions requires well-powered studies, the CHARGE Gene–Lifestyle Interactions Working Group was formed [46]. To date, this has led to three studies on the interaction between lipid levels with smoking [47•], physical activity [48•], and alcohol intake [49•]. These studies used a joint meta-analysis in which 2-degree-of-freedom test was adopted that jointly evaluates interaction and main effects to increase statistical power [50]. Bentley et al. studied the interaction between lipid levels with smoking and revealed 13 new loci associated with lipids, of which several appeared to be driven mainly by their interaction with smoking [47•]. The importance of interaction testing was further highlighted by another study in which 4 novel loci were discovered when testing for interactions between genetically determined lipid levels and physical activity, whereas not a single new locus was found in the test without interaction [48•]. The third study revealed 18 novel lipid loci, although none of which appeared to be driven by interactions with alcohol intake [49•]. Based on these studies, smoking and physical activity, but possibly not alcohol use, may be modifiable lifestyle risk factors of interest to alter blood lipid levels. Even if these interactions contribute little to the overall variance of blood lipid levels, insights in these interactions could contribute to our understanding of disease pathophysiology.
Twin Studies
One twin study assessed G×L interactions amongst 51,065 Swedish same-sex twins. This study found higher BMI was associated with lower genetic variance of CAD, suggesting a more important role for genetics in the development of CAD in individuals with low BMI [51]. Other lifestyle factors such as smoking and sedentary behaviors showed no significant G×L interactions, but rather seemed to increase CAD risk directly [51].
Candidate Gene Studies
The largest candidate gene study on G×L interactions is a meta-analysis which investigated the interaction between 45 CAD loci and smoking in 60,919 cases and 80,243 controls. In this study, a significant interaction was observed between rs7178051, located within ADAMTS7, and smoking [52]. Several studies including up to ~ 6,000 individuals reported interactions between ApoE and smoking as well [53–55]. However, larger meta-analyses did not find support for this conclusion [52, 56]. This stresses the need for large sample sizes in and replication of candidate gene studies investigating G×L interactions [57]. Many other candidate gene studies tested the interaction between CAD genes and lifestyle risk factors, including smoking [58–65], alcohol intake [62, 65, 66], diet [67–69], and physical activity [69]. However, these results should be interpreted with caution as sample sizes were small and results were not replicated in independent larger studies.
Future Perspectives
Only recently, studies with sufficient sample sizes have emerged and reported robust G×L interactions. Although the effect sizes are small and might add little to the explained variance of CAD heritability, they increase our knowledge on complex G×L interplays [15, 16]. This knowledge might be translated to strategies that pinpoint lifestyle risk factors with proven interactions and are therefore of increased interest to modify. Next, in the current era of huge biobanks, several cohorts will be well powered to perform genome-wide interaction analyses on CAD and modifiable risk factors of interest in the years to come.
In line with most GWAS’s, the GWAS’s on CAD included only or mostly white Europeans in order to reduce heterogeneity [70]. Logistic, systemic, or historical factors that make it easier to perform genetic studies in Europeans play a role as well [70]. As a consequence, little attention has been paid to genetic variants in other populations. Variants associated with G×L interactions may have different effects in non-Europeans, reducing its broader applicability in the clinic. As long as there is a lack of well-powered cohorts in other ethnicities, candidate gene studies offer an excellent approach to validate genes known to interact with lifestyle risk factors [71].
Conclusion
In this review, we summarized current knowledge on the genetics of CAD, lifestyle factors, and the genetics of lifestyle factors associated with CAD. We focused on the interplay of genetics and lifestyle, especially the effect modifications as determined by G×L interaction studies. The majority of G×L interaction studies are small-scale candidate gene studies, lacking replication and therefore providing spurious results. Only few studies are robust in number and analysis strategy. These studies provide evidence of the existence of G×L interactions. Current data suggest that genetics and G×L interactions contribute little to the overall risk prediction for CAD next to lifestyle and other phenotypic risk factors. However, well-powered G×L interactions studies are important as they contribute to our understanding of disease pathophysiology and may provide insights into improving interventions or personalized recommendations.
Compliance with Ethical Standards
Conflict of Interest
M. Abdullah Said, Yordi J. van de Vegte, Muhammad Mobeen Zafar, M. Yldau van der Ende, Ghazala Kaukab Raja, N. Verweij, and P. van der Harst declare that they have no conflict of interest.
Human/Animal Rights and Informed Consent
All reported studies/experiments with human or animal subjects performed by the authors have been previously published and complied with all applicable ethical standards.
Footnotes
This article is part of the Topical Collection on Lipid Abnormalities and Cardiovascular Prevention
M. Abdullah Said, Yordi J. van de Vegte, and Muhammad Mobeen Zafar shared first author
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
M. Abdullah Said, Email: m.a.said@umcg.nl.
Yordi J. van de Vegte, Email: y.j.van.de.vegte@umcg.nl
Muhammad Mobeen Zafar, Email: mobeenzafar@hotmail.com.
M. Yldau van der Ende, Email: m.y.van.der.ende@umcg.nl.
Ghazala Kaukab Raja, Email: ghazala@uaar.eedu.pk.
N. Verweij, Email: n.verweij@umcg.nl
Pim van der Harst, Phone: +31 (0)50 3612355, Email: p.van.der.harst@umcg.nl.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, Abdollahpour I, Abdulkader RS, Abebe HT, Abebe M, Abebe Z, Abejie AN, Abera SF, Abil OZ, Abraha HN, Abrham AR, Abu-Raddad LJ, Accrombessi MMK, Acharya D, Adamu AA, Adebayo OM, Adedoyin RA, Adekanmbi V, Adetokunboh OO, Adhena BM, Adib MG, Admasie A, Afshin A, Agarwal G, Agesa KM, Agrawal A, Agrawal S, Ahmadi A, Ahmadi M, Ahmed MB, Ahmed S, Aichour AN, Aichour I, Aichour MTE, Akbari ME, Akinyemi RO, Akseer N, al-Aly Z, al-Eyadhy A, al-Raddadi RM, Alahdab F, Alam K, Alam T, Alebel A, Alene KA, Alijanzadeh M, Alizadeh-Navaei R, Aljunid SM, Alkerwi A’, Alla F, Allebeck P, Alonso J, Altirkawi K, Alvis-Guzman N, Amare AT, Aminde LN, Amini E, Ammar W, Amoako YA, Anber NH, Andrei CL, Androudi S, Animut MD, Anjomshoa M, Ansari H, Ansha MG, Antonio CAT, Anwari P, Aremu O, Ärnlöv J, Arora A, Arora M, Artaman A, Aryal KK, Asayesh H, Asfaw ET, Ataro Z, Atique S, Atre SR, Ausloos M, Avokpaho EFGA, Awasthi A, Quintanilla BPA, Ayele Y, Ayer R, Azzopardi PS, Babazadeh A, Bacha U, Badali H, Badawi A, Bali AG, Ballesteros KE, Banach M, Banerjee K, Bannick MS, Banoub JAM, Barboza MA, Barker-Collo SL, Bärnighausen TW, Barquera S, Barrero LH, Bassat Q, Basu S, Baune BT, Baynes HW, Bazargan-Hejazi S, Bedi N, Beghi E, Behzadifar M, Behzadifar M, Béjot Y, Bekele BB, Belachew AB, Belay E, Belay YA, Bell ML, Bello AK, Bennett DA, Bensenor IM, Berman AE, Bernabe E, Bernstein RS, Bertolacci GJ, Beuran M, Beyranvand T, Bhalla A, Bhattarai S, Bhaumik S, Bhutta ZA, Biadgo B, Biehl MH, Bijani A, Bikbov B, Bilano V, Bililign N, Bin Sayeed MS, Bisanzio D, Biswas T, Blacker BF, Basara BB, Borschmann R, Bosetti C, Bozorgmehr K, Brady OJ, Brant LC, Brayne C, Brazinova A, Breitborde NJK, Brenner H, Briant PS, Britton G, Brugha T, Busse R, Butt ZA, Callender CSKH, Campos-Nonato IR, Campuzano Rincon JC, Cano J, Car M, Cárdenas R, Carreras G, Carrero JJ, Carter A, Carvalho F, Castañeda-Orjuela CA, Castillo Rivas J, Castle CD, Castro C, Castro F, Catalá-López F, Cerin E, Chaiah Y, Chang JC, Charlson FJ, Chaturvedi P, Chiang PPC, Chimed-Ochir O, Chisumpa VH, Chitheer A, Chowdhury R, Christensen H, Christopher DJ, Chung SC, Cicuttini FM, Ciobanu LG, Cirillo M, Cohen AJ, Cooper LT, Cortesi PA, Cortinovis M, Cousin E, Cowie BC, Criqui MH, Cromwell EA, Crowe CS, Crump JA, Cunningham M, Daba AK, Dadi AF, Dandona L, Dandona R, Dang AK, Dargan PI, Daryani A, Das SK, Gupta RD, Neves JD, Dasa TT, Dash AP, Davis AC, Davis Weaver N, Davitoiu DV, Davletov K, de la Hoz FP, de Neve JW, Degefa MG, Degenhardt L, Degfie TT, Deiparine S, Demoz GT, Demtsu BB, Denova-Gutiérrez E, Deribe K, Dervenis N, Des Jarlais DC, Dessie GA, Dey S, Dharmaratne SD, Dicker D, Dinberu MT, Ding EL, Dirac MA, Djalalinia S, Dokova K, Doku DT, Donnelly CA, Dorsey ER, Doshi PP, Douwes-Schultz D, Doyle KE, Driscoll TR, Dubey M, Dubljanin E, Duken EE, Duncan BB, Duraes AR, Ebrahimi H, Ebrahimpour S, Edessa D, Edvardsson D, Eggen AE, el Bcheraoui C, el Sayed Zaki M, el-Khatib Z, Elkout H, Ellingsen CL, Endres M, Endries AY, Er B, Erskine HE, Eshrati B, Eskandarieh S, Esmaeili R, Esteghamati A, Fakhar M, Fakhim H, Faramarzi M, Fareed M, Farhadi F, Farinha CSE, Faro A, Farvid MS, Farzadfar F, Farzaei MH, Feigin VL, Feigl AB, Fentahun N, Fereshtehnejad SM, Fernandes E, Fernandes JC, Ferrari AJ, Feyissa GT, Filip I, Finegold S, Fischer F, Fitzmaurice C, Foigt NA, Foreman KJ, Fornari C, Frank TD, Fukumoto T, Fuller JE, Fullman N, Fürst T, Furtado JM, Futran ND, Gallus S, Garcia-Basteiro AL, Garcia-Gordillo MA, Gardner WM, Gebre AK, Gebrehiwot TT, Gebremedhin AT, Gebremichael B, Gebremichael TG, Gelano TF, Geleijnse JM, Genova-Maleras R, Geramo YCD, Gething PW, Gezae KE, Ghadami MR, Ghadimi R, Ghasemi Falavarjani K, Ghasemi-Kasman M, Ghimire M, Gibney KB, Gill PS, Gill TK, Gillum RF, Ginawi IA, Giroud M, Giussani G, Goenka S, Goldberg EM, Goli S, Gómez-Dantés H, Gona PN, Gopalani SV, Gorman TM, Goto A, Goulart AC, Gnedovskaya EV, Grada A, Grosso G, Gugnani HC, Guimaraes ALS, Guo Y, Gupta PC, Gupta R, Gupta R, Gupta T, Gutiérrez RA, Gyawali B, Haagsma JA, Hafezi-Nejad N, Hagos TB, Hailegiyorgis TT, Hailu GB, Haj-Mirzaian A, Haj-Mirzaian A, Hamadeh RR, Hamidi S, Handal AJ, Hankey GJ, Harb HL, Harikrishnan S, Haro JM, Hasan M, Hassankhani H, Hassen HY, Havmoeller R, Hay RJ, Hay SI, He Y, Hedayatizadeh-Omran A, Hegazy MI, Heibati B, Heidari M, Hendrie D, Henok A, Henry NJ, Herteliu C, Heydarpour F, Heydarpour P, Heydarpour S, Hibstu DT, Hoek HW, Hole MK, Homaie Rad E, Hoogar P, Hosgood HD, Hosseini SM, Hosseinzadeh M, Hostiuc M, Hostiuc S, Hotez PJ, Hoy DG, Hsiao T, Hu G, Huang JJ, Husseini A, Hussen MM, Hutfless S, Idrisov B, Ilesanmi OS, Iqbal U, Irvani SSN, Irvine CMS, Islam N, Islam SMS, Islami F, Jacobsen KH, Jahangiry L, Jahanmehr N, Jain SK, Jakovljevic M, Jalu MT, James SL, Javanbakht M, Jayatilleke AU, Jeemon P, Jenkins KJ, Jha RP, Jha V, Johnson CO, Johnson SC, Jonas JB, Joshi A, Jozwiak JJ, Jungari SB, Jürisson M, Kabir Z, Kadel R, Kahsay A, Kalani R, Karami M, Karami Matin B, Karch A, Karema C, Karimi-Sari H, Kasaeian A, Kassa DH, Kassa GM, Kassa TD, Kassebaum NJ, Katikireddi SV, Kaul A, Kazemi Z, Karyani AK, Kazi DS, Kefale AT, Keiyoro PN, Kemp GR, Kengne AP, Keren A, Kesavachandran CN, Khader YS, Khafaei B, Khafaie MA, Khajavi A, Khalid N, Khalil IA, Khan EA, Khan MS, Khan MA, Khang YH, Khater MM, Khoja AT, Khosravi A, Khosravi MH, Khubchandani J, Kiadaliri AA, Kibret GD, Kidanemariam ZT, Kiirithio DN, Kim D, Kim YE, Kim YJ, Kimokoti RW, Kinfu Y, Kisa A, Kissimova-Skarbek K, Kivimäki M, Knudsen AKS, Kocarnik JM, Kochhar S, Kokubo Y, Kolola T, Kopec JA, Koul PA, Koyanagi A, Kravchenko MA, Krishan K, Kuate Defo B, Kucuk Bicer B, Kumar GA, Kumar M, Kumar P, Kutz MJ, Kuzin I, Kyu HH, Lad DP, Lad SD, Lafranconi A, Lal DK, Lalloo R, Lallukka T, Lam JO, Lami FH, Lansingh VC, Lansky S, Larson HJ, Latifi A, Lau KMM, Lazarus JV, Lebedev G, Lee PH, Leigh J, Leili M, Leshargie CT, Li S, Li Y, Liang J, Lim LL, Lim SS, Limenih MA, Linn S, Liu S, Liu Y, Lodha R, Lonsdale C, Lopez AD, Lorkowski S, Lotufo PA, Lozano R, Lunevicius R, Ma S, Macarayan ERK, Mackay MT, MacLachlan JH, Maddison ER, Madotto F, Magdy Abd el Razek H, Magdy Abd el Razek M, Maghavani DP, Majdan M, Majdzadeh R, Majeed A, Malekzadeh R, Malta DC, Manda AL, Mandarano-Filho LG, Manguerra H, Mansournia MA, Mapoma CC, Marami D, Maravilla JC, Marcenes W, Marczak L, Marks A, Marks GB, Martinez G, Martins-Melo FR, Martopullo I, März W, Marzan MB, Masci JR, Massenburg BB, Mathur MR, Mathur P, Matzopoulos R, Maulik PK, Mazidi M, McAlinden C, McGrath JJ, McKee M, McMahon BJ, Mehata S, Mehndiratta MM, Mehrotra R, Mehta KM, Mehta V, Mekonnen TC, Melese A, Melku M, Memiah PTN, Memish ZA, Mendoza W, Mengistu DT, Mengistu G, Mensah GA, Mereta ST, Meretoja A, Meretoja TJ, Mestrovic T, Mezgebe HB, Miazgowski B, Miazgowski T, Millear AI, Miller TR, Miller-Petrie MK, Mini GK, Mirabi P, Mirarefin M, Mirica A, Mirrakhimov EM, Misganaw AT, Mitiku H, Moazen B, Mohammad KA, Mohammadi M, Mohammadifard N, Mohammed MA, Mohammed S, Mohan V, Mokdad AH, Molokhia M, Monasta L, Moradi G, Moradi-Lakeh M, Moradinazar M, Moraga P, Morawska L, Moreno Velásquez I, Morgado-da-Costa J, Morrison SD, Moschos MM, Mouodi S, Mousavi SM, Muchie KF, Mueller UO, Mukhopadhyay S, Muller K, Mumford JE, Musa J, Musa KI, Mustafa G, Muthupandian S, Nachega JB, Nagel G, Naheed A, Nahvijou A, Naik G, Nair S, Najafi F, Naldi L, Nam HS, Nangia V, Nansseu JR, Nascimento BR, Natarajan G, Neamati N, Negoi I, Negoi RI, Neupane S, Newton CRJ, Ngalesoni FN, Ngunjiri JW, Nguyen AQ, Nguyen G, Nguyen HT, Nguyen HT, Nguyen LH, Nguyen M, Nguyen TH, Nichols E, Ningrum DNA, Nirayo YL, Nixon MR, Nolutshungu N, Nomura S, Norheim OF, Noroozi M, Norrving B, Noubiap JJ, Nouri HR, Nourollahpour Shiadeh M, Nowroozi MR, Nyasulu PS, Odell CM, Ofori-Asenso R, Ogbo FA, Oh IH, Oladimeji O, Olagunju AT, Olivares PR, Olsen HE, Olusanya BO, Olusanya JO, Ong KL, Ong SKS, Oren E, Orpana HM, Ortiz A, Ortiz JR, Otstavnov SS, Øverland S, Owolabi MO, Özdemir R, P A M, Pacella R, Pakhale S, Pakhare AP, Pakpour AH, Pana A, Panda-Jonas S, Pandian JD, Parisi A, Park EK, Parry CDH, Parsian H, Patel S, Pati S, Patton GC, Paturi VR, Paulson KR, Pereira A, Pereira DM, Perico N, Pesudovs K, Petzold M, Phillips MR, Piel FB, Pigott DM, Pillay JD, Pirsaheb M, Pishgar F, Polinder S, Postma MJ, Pourshams A, Poustchi H, Pujar A, Prakash S, Prasad N, Purcell CA, Qorbani M, Quintana H, Quistberg DA, Rade KW, Radfar A, Rafay A, Rafiei A, Rahim F, Rahimi K, Rahimi-Movaghar A, Rahman M, Rahman MHU, Rahman MA, Rai RK, Rajsic S, Ram U, Ranabhat CL, Ranjan P, Rao PC, Rawaf DL, Rawaf S, Razo-García C, Reddy KS, Reiner RC, Reitsma MB, Remuzzi G, Renzaho AMN, Resnikoff S, Rezaei S, Rezaeian S, Rezai MS, Riahi SM, Ribeiro ALP, Rios-Blancas MJ, Roba KT, Roberts NLS, Robinson SR, Roever L, Ronfani L, Roshandel G, Rostami A, Rothenbacher D, Roy A, Rubagotti E, Sachdev PS, Saddik B, Sadeghi E, Safari H, Safdarian M, Safi S, Safiri S, Sagar R, Sahebkar A, Sahraian MA, Salam N, Salama JS, Salamati P, Saldanha RDF, Saleem Z, Salimi Y, Salvi SS, Salz I, Sambala EZ, Samy AM, Sanabria J, Sanchez-Niño MD, Santomauro DF, Santos IS, Santos JV, Milicevic MMS, Sao Jose BP, Sarker AR, Sarmiento-Suárez R, Sarrafzadegan N, Sartorius B, Sarvi S, Sathian B, Satpathy M, Sawant AR, Sawhney M, Saxena S, Sayyah M, Schaeffner E, Schmidt MI, Schneider IJC, Schöttker B, Schutte AE, Schwebel DC, Schwendicke F, Scott JG, Sekerija M, Sepanlou SG, Serván-Mori E, Seyedmousavi S, Shabaninejad H, Shackelford KA, Shafieesabet A, Shahbazi M, Shaheen AA, Shaikh MA, Shams-Beyranvand M, Shamsi M, Shamsizadeh M, Sharafi K, Sharif M, Sharif-Alhoseini M, Sharma R, She J, Sheikh A, Shi P, Shiferaw MS, Shigematsu M, Shiri R, Shirkoohi R, Shiue I, Shokraneh F, Shrime MG, Si S, Siabani S, Siddiqi TJ, Sigfusdottir ID, Sigurvinsdottir R, Silberberg DH, Silva DAS, Silva JP, Silva NTD, Silveira DGA, Singh JA, Singh NP, Singh PK, Singh V, Sinha DN, Sliwa K, Smith M, Sobaih BH, Sobhani S, Sobngwi E, Soneji SS, Soofi M, Sorensen RJD, Soriano JB, Soyiri IN, Sposato LA, Sreeramareddy CT, Srinivasan V, Stanaway JD, Starodubov VI, Stathopoulou V, Stein DJ, Steiner C, Stewart LG, Stokes MA, Subart ML, Sudaryanto A, Sufiyan M’B, Sur PJ, Sutradhar I, Sykes BL, Sylaja PN, Sylte DO, Szoeke CEI, Tabarés-Seisdedos R, Tabuchi T, Tadakamadla SK, Takahashi K, Tandon N, Tassew SG, Taveira N, Tehrani-Banihashemi A, Tekalign TG, Tekle MG, Temsah MH, Temsah O, Terkawi AS, Teshale MY, Tessema B, Tessema GA, Thankappan KR, Thirunavukkarasu S, Thomas N, Thrift AG, Thurston GD, Tilahun B, To QG, Tobe-Gai R, Tonelli M, Topor-Madry R, Torre AE, Tortajada-Girbés M, Touvier M, Tovani-Palone MR, Tran BX, Tran KB, Tripathi S, Troeger CE, Truelsen TC, Truong NT, Tsadik AG, Tsoi D, Tudor Car L, Tuzcu EM, Tyrovolas S, Ukwaja KN, Ullah I, Undurraga EA, Updike RL, Usman MS, Uthman OA, Uzun SB, Vaduganathan M, Vaezi A, Vaidya G, Valdez PR, Varavikova E, Vasankari TJ, Venketasubramanian N, Villafaina S, Violante FS, Vladimirov SK, Vlassov V, Vollset SE, Vos T, Wagner GR, Wagnew FS, Waheed Y, Wallin MT, Walson JL, Wang Y, Wang YP, Wassie MM, Weiderpass E, Weintraub RG, Weldegebreal F, Weldegwergs KG, Werdecker A, Werkneh AA, West TE, Westerman R, Whiteford HA, Widecka J, Wilner LB, Wilson S, Winkler AS, Wiysonge CS, Wolfe CDA, Wu S, Wu YC, Wyper GMA, Xavier D, Xu G, Yadgir S, Yadollahpour A, Yahyazadeh Jabbari SH, Yakob B, Yan LL, Yano Y, Yaseri M, Yasin YJ, Yentür GK, Yeshaneh A, Yimer EM, Yip P, Yirsaw BD, Yisma E, Yonemoto N, Yonga G, Yoon SJ, Yotebieng M, Younis MZ, Yousefifard M, Yu C, Zadnik V, Zaidi Z, Zaman SB, Zamani M, Zare Z, Zeleke AJ, Zenebe ZM, Zhang AL, Zhang K, Zhou M, Zodpey S, Zuhlke LJ, Naghavi M, Murray CJL. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Said MA, Verweij N, van der Harst P. Associations of combined genetic and lifestyle risks with incident cardiovascular disease and diabetes in the UK biobank study. JAMA Cardiol. 2018;3(8):693–702. doi: 10.1001/jamacardio.2018.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porta M, Last JM. A dictionary of public health [Internet]. Vol. 1. Oxford University Press; 2018.
- 4.de Waure C, Lauret G-J, Ricciardi W, Ferket B, Teijink J, Spronk S, Myriam Hunink MG. Lifestyle interventions in patients with coronary heart disease. Am J Prev Med. 2013;45(2):207–216. doi: 10.1016/j.amepre.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Reid RD, McDonnell LA, Riley DL, Mark AE, Mosca L, Beaton L, et al. Effect of an intervention to improve the cardiovascular health of family members of patients with coronary artery disease: a randomized trial. CMAJ. 2014;186(1):23–30. doi: 10.1503/cmaj.130550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122(3):433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375(24):2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Won H-H, Natarajan P, Dobbyn A, Jordan DM, Roussos P, Lage K, Raychaudhuri S, Stahl E, Do R Disproportionate contributions of select genomic compartments and cell types to genetic risk for coronary artery disease. Snyder M, editor. PLoS Genet 2015;11(10):e1005622. [DOI] [PMC free article] [PubMed]
- 10.Ioannidis JPA, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 11.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschhorn JN. Genetic approaches to studying common diseases and complex traits. Pediatr Res. 2005;57(5 Part 2):74R–77R. doi: 10.1203/01.PDR.0000159574.98964.87. [DOI] [PubMed] [Google Scholar]
- 13.Spracklen CN, Chen P, Kim YJ, Wang X, Cai H, Li S, Long J, Wu Y, Wang YX, Takeuchi F, Wu JY, Jung KJ, Hu C, Akiyama K, Zhang Y, Moon S, Johnson TA, Li H, Dorajoo R, He M, Cannon ME, Roman TS, Salfati E, Lin KH, Guo X, Sheu WHH, Absher D, Adair LS, Assimes TL, Aung T, Cai Q, Chang LC, Chen CH, Chien LH, Chuang LM, Chuang SC, du S, Fan Q, Fann CSJ, Feranil AB, Friedlander Y, Gordon-Larsen P, Gu D, Gui L, Guo Z, Heng CK, Hixson J, Hou X, Hsiung CA, Hu Y, Hwang MY, Hwu CM, Isono M, Juang JMJ, Khor CC, Kim YK, Koh WP, Kubo M, Lee IT, Lee SJ, Lee WJ, Liang KW, Lim B, Lim SH, Liu J, Nabika T, Pan WH, Peng H, Quertermous T, Sabanayagam C, Sandow K, Shi J, Sun L, Tan PC, Tan SP, Taylor KD, Teo YY, Toh SA, Tsunoda T, van Dam RM, Wang A, Wang F, Wang J, Wei WB, Xiang YB, Yao J, Yuan JM, Zhang R, Zhao W, Chen YDI, Rich SS, Rotter JI, Wang TD, Wu T, Lin X, Han BG, Tanaka T, Cho YS, Katsuya T, Jia W, Jee SH, Chen YT, Kato N, Jonas JB, Cheng CY, Shu XO, He J, Zheng W, Wong TY, Huang W, Kim BJ, Tai ES, Mohlke KL, Sim X. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum Mol Genet. 2017;26(9):1770–1784. doi: 10.1093/hmg/ddx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner AW, Wong D, Dreisbach CN, Miller CL. GWAS reveal targets in vessel wall pathways to treat coronary artery disease. Front Cardiovasc Med. 2018;5:72. doi: 10.3389/fcvm.2018.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, Ye S, Webb TR, Rutter MK, Tzoulaki I, Patel RS, Loos RJF, Keavney B, Hemingway H, Thompson J, Watkins H, Deloukas P, di Angelantonio E, Butterworth AS, Danesh J, Samani NJ, UK Biobank CardioMetabolic Consortium CHD Working Group Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72(16):1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen B-S, Fredrich B, Hoeppner MP, Ellinghaus D, Franke A. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet. 2017;18(1):14. doi: 10.1186/s12863-017-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wainschtein P, Jain DP, Yengo L, Zheng Z, TOPMed Anthropometry Working Group T-O for PMC. Cupples LA, et al. Recovery of trait heritability from whole genome sequence data. bioRxiv. 2019;25:588020. [Google Scholar]
- 19.Rappaport SM. Genetic factors are not the major causes of chronic diseases. Scott RJ, editor. PLoS One 2016;11(4):e0154387. [DOI] [PMC free article] [PubMed]
- 20.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet (London, England) 2004;364(9438):937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 21.Schuit AJ, van Loon AJM, Tijhuis M, Ocké M. Clustering of lifestyle risk factors in a general adult population. Prev Med (Baltim) 2002;35(3):219–224. doi: 10.1006/pmed.2002.1064. [DOI] [PubMed] [Google Scholar]
- 22.Huxley Rachel R, Woodward Mark. Cigarette smoking as a risk factor for coronary heart disease in women compared with men: a systematic review and meta-analysis of prospective cohort studies. The Lancet. 2011;378(9799):1297–1305. doi: 10.1016/S0140-6736(11)60781-2. [DOI] [PubMed] [Google Scholar]
- 23.Talhout R, Schulz T, Florek E, Van Benthem J, Wester P, Opperhuizen A. Hazardous compounds in tobacco smoke. Int J Environ Res Public Health. 2011;8(2):613–628. doi: 10.3390/ijerph8020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messner B, Bernhard D. Smoking and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2014;34(3):509–515. doi: 10.1161/ATVBAHA.113.300156. [DOI] [PubMed] [Google Scholar]
- 25.Amos CI, Spitz MR, Cinciripini P. Chipping away at the genetics of smoking behavior. Nat Genet. 2010;42(5):366–368. doi: 10.1038/ng0510-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Liu DC, Wang QM, Long QQ, Zhao S, Zhang Z, Ma Y, Wang ZM, Chen LL, Wang LS. Alcohol consumption and risk of coronary artery disease: a dose-response meta-analysis of prospective studies. Nutrition. 2016;32(6):637–644. doi: 10.1016/j.nut.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Goel S, Sharma A, Garg A. Effect of alcohol consumption on cardiovascular health. Curr Cardiol Rep. 2018;20(4):19. doi: 10.1007/s11886-018-0962-2. [DOI] [PubMed] [Google Scholar]
- 29.Zhou A, Hyppönen E. Long-term coffee consumption, caffeine metabolism genetics, and risk of cardiovascular disease: a prospective analysis of up to 347,077 individuals and 8368 cases. Am J Clin Nutr. 2019;109(3):509–516. doi: 10.1093/ajcn/nqy297. [DOI] [PubMed] [Google Scholar]
- 30.Ding M, Bhupathiraju SN, Satija A, van Dam RM, Hu FB. Long-term coffee consumption and risk of cardiovascular disease. Circulation. 2014;129(6):643–659. doi: 10.1161/CIRCULATIONAHA.113.005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordestgaard AT, Nordestgaard BG. Coffee intake, cardiovascular disease and all-cause mortality: observational and Mendelian randomization analyses in 95 000-223 000 individuals. Int J Epidemiol. 2016;45(6):1938–1952. doi: 10.1093/ije/dyw325. [DOI] [PubMed] [Google Scholar]
- 32.Winzer EB, Woitek F, Linke A. Physical activity in the prevention and treatment of coronary artery disease. J Am Heart Assoc. 2018;7(4). [DOI] [PMC free article] [PubMed]
- 33.Althoff T, Sosič R, Hicks JL, King AC, Delp SL, Leskovec J. Large-scale physical activity data reveal worldwide activity inequality. Nature. 2017;547(7663):336–339. doi: 10.1038/nature23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekelund U, Steene-Johannessen J, Brown WJ, Fagerland MW, Owen N, Powell KE, Bauman A, Lee IM. Articles does physical activity attenuate, or even eliminate, the detrimental association of sitting time with mortality? A harmonised meta-analysis of data from more than 1 million men and women. Lancet. 2016;388:1302–1310. doi: 10.1016/S0140-6736(16)30370-1. [DOI] [PubMed] [Google Scholar]
- 35.De Vilhena E Santos DM, Katzmarzyk PT, Seabra AFT, Maia JAR Genetics of physical activity and physical inactivity in humans [Internet]. Vol. 42, Behavior Genetics. Springer US; 2012. p. 559–78. [DOI] [PubMed]
- 36.Zhang X, Speakman JR. Genetic factors associated with human physical activity: are your genes too tight to prevent you exercising? Endocrinology. 2019;160(4):840–852. doi: 10.1210/en.2018-00873. [DOI] [PubMed] [Google Scholar]
- 37.Doherty A, Smith-Byrne K, Ferreira T, Holmes MV, Holmes C, Pulit SL, Lindgren CM. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. doi: 10.1038/s41467-018-07743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding D, Lawson KD, Kolbe-Alexander TL, Finkelstein EA, Katzmarzyk PT, van Mechelen W, Pratt M. The economic burden of physical inactivity: a global analysis of major non-communicable diseases. Lancet. 2016;388(10051):1311–1324. doi: 10.1016/S0140-6736(16)30383-X. [DOI] [PubMed] [Google Scholar]
- 39.Heron L, O’Neill C, McAneney H, Kee F, Tully MA. Direct healthcare costs of sedentary behaviour in the UK. J Epidemiol Community Heal. 2019;73:625–629. doi: 10.1136/jech-2018-211758. [DOI] [PubMed] [Google Scholar]
- 40.Mozaffarian D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity. Circulation. 2016;133(2):187–225. doi: 10.1161/CIRCULATIONAHA.115.018585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Kutateladze TG. Diet and the epigenome. Nat Commun. 2018;9(1):3375. doi: 10.1038/s41467-018-05778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51(2):245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alegría-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–277. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd-Jones DM, Hong Y, Labarthe D, Mozaffarian D, Appel LJ, Van Horn L, et al. Defining and setting national goals for cardiovascular health promotion and disease reduction. Circulation. 2010;121(4):586–613. doi: 10.1161/CIRCULATIONAHA.109.192703. [DOI] [PubMed] [Google Scholar]
- 45.Tikkanen E, Gustafsson S, Ingelsson E. Associations of fitness, physical activity, strength, and genetic risk with cardiovascular disease. Circulation. 2018;137(24):2583–2591. doi: 10.1161/CIRCULATIONAHA.117.032432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao DC, Sung YJ, Winkler TW, Schwander K, Borecki I, Cupples LA, et al. Multiancestry study of gene–lifestyle interactions for cardiovascular traits in 610 475 individuals from 124 cohorts. Circ Cardiovasc Genet. 2017 Jun;10(3). [DOI] [PMC free article] [PubMed]
- 47.Bentley AR, Sung YJ, Brown MR, Winkler TW, Kraja AT, Ntalla I, et al. Multi-ancestry genome-wide gene–smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat Genet. 2019;51(4):636–648. doi: 10.1038/s41588-019-0378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilpeläinen TO. Bentley AR, Noordam R, Sung YJ, Schwander K, Winkler TW, et al. Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat Commun. 2019;10(1):376. doi: 10.1038/s41467-018-08008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.• de Vries PS, Brown MR, Bentley AR, Sung YJ, Winkler TW, Ntalla I, et al. Multi-ancestry genome-wide association study of lipid levels incorporating gene-alcohol interactions. Am J Epidemiol; 2019. This recent study performed an interaction GWAS between important lifestyle risk factors and blood lipid levels [DOI] [PMC free article] [PubMed]
- 50.Manning AK, LaValley M, Liu C-T, Rice K, An P, Liu Y, Miljkovic I, Rasmussen-Torvik L, Harris TB, Province MA, Borecki IB, Florez JC, Meigs JB, Cupples LA, Dupuis J. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet Epidemiol. 2011;35(1):11–18. doi: 10.1002/gepi.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song C, Chang Z, Magnusson PKE, Ingelsson E, Pedersen NL. Genetic factors may play a prominent role in the development of coronary heart disease dependent on important environmental factors. J Intern Med. 2014;275(6):631–639. doi: 10.1111/joim.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saleheen D, Zhao W, Young R, Nelson CP, Ho W, Ferguson JF, Rasheed A, Ou K, Nurnberg ST, Bauer RC, Goel A, Do R, Stewart AFR, Hartiala J, Zhang W, Thorleifsson G, Strawbridge RJ, Sinisalo J, Kanoni S, Sedaghat S, Marouli E, Kristiansson K, Hua Zhao J, Scott R, Gauguier D, Shah SH, Smith AV, van Zuydam N, Cox AJ, Willenborg C, Kessler T, Zeng L, Province MA, Ganna A, Lind L, Pedersen NL, White CC, Joensuu A, Edi Kleber M, Hall AS, März W, Salomaa V, O’Donnell C, Ingelsson E, Feitosa MF, Erdmann J, Bowden DW, Palmer CNA, Gudnason V, Faire UD, Zalloua P, Wareham N, Thompson JR, Kuulasmaa K, Dedoussis G, Perola M, Dehghan A, Chambers JC, Kooner J, Allayee H, Deloukas P, McPherson R, Stefansson K, Schunkert H, Kathiresan S, Farrall M, Marcel Frossard P, Rader DJ, Samani NJ, Reilly MP. Loss of cardioprotective effects at the ADAMTS7 locus as a result of gene-smoking interactions. Circulation. 2017;135(24):2336–2353. doi: 10.1161/CIRCULATIONAHA.116.022069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humphries SE, Talmud PJ, Hawe E, Bolla M, Day IN, Miller GJ. Apolipoprotein E4 and coronary heart disease in middle-aged men who smoke: a prospective study. Lancet. 2001;358(9276):115–119. doi: 10.1016/S0140-6736(01)05330-2. [DOI] [PubMed] [Google Scholar]
- 54.Grammer TB, Hoffmann MM, Scharnagl H, Kleber ME, Silbernagel G, Pilz S, Tomaschitz A, Lerchbaum E, Siekmeier R, März W. Smoking, apolipoprotein E genotypes, and mortality (the Ludwigshafen RIsk and Cardiovascular Health study) Eur Heart J. 2013;34(17):1298–1305. doi: 10.1093/eurheartj/eht001. [DOI] [PubMed] [Google Scholar]
- 55.Gustavsson J, Mehlig K, Leander K, Strandhagen E, Björck L, Thelle DS, Lissner L, Blennow K, Zetterberg H, Nyberg F. Interaction of apolipoprotein E genotype with smoking and physical inactivity on coronary heart disease risk in men and women. Atherosclerosis. 2012;220(2):486–492. doi: 10.1016/j.atherosclerosis.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Holmes MV, Frikke-Schmidt R, Melis D, Luben R, Asselbergs FW, Boer JMA, et al. A systematic review and meta-analysis of 130,000 individuals shows smoking does not modify the association of APOE genotype on risk of coronary heart disease. Atherosclerosis. 2014;237(1):5–12. doi: 10.1016/j.atherosclerosis.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt AF, Groenwold RHH, Knol MJ, Hoes AW, Nielen M, Roes KCB, de Boer A, Klungel OH. Exploring interaction effects in small samples increases rates of false-positive and false-negative findings: results from a systematic review and simulation study. J Clin Epidemiol. 2014;67(7):821–829. doi: 10.1016/j.jclinepi.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Hamrefors V, Hedblad B, Hindy G, Smith JG, Almgren P, Engström G, et al. Smoking modifies the associated increased risk of future cardiovascular disease by genetic variation on chromosome 9p21. PLoS One. 2014;9(1):1–10. doi: 10.1371/journal.pone.0085893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatzis G, Tousoulis D, Papageorgiou N, Bouras G, Oikonomou E, Miliou A, Siasos G, Toutouzas K, Papaioannou S, Tsiamis E, Antoniades C, Stefanadis C. Combined effects of smoking and interleukin-6 and C-reactive protein genetic variants on endothelial function, inflammation, thrombosis and incidence of coronary artery disease. Int J Cardiol. 2014;176(1):254–257. doi: 10.1016/j.ijcard.2014.06.058. [DOI] [PubMed] [Google Scholar]
- 60.Merhi M, Demirdjian S, Hariri E, Sabbah N, Youhanna S, Ghassibe-Sabbagh M, Naoum J, Haber M, Othman R, Kibbani S, Chammas E, Kanbar R, Bayeh H, Chami Y, Abchee A, Platt DE, Zalloua P, Khazen G. Impact of inflammation, gene variants, and cigarette smoking on coronary artery disease risk. Inflamm Res. 2015;64(6):415–422. doi: 10.1007/s00011-015-0821-1. [DOI] [PubMed] [Google Scholar]
- 61.Ren B, She Q. Study on the association between IL-1β, IL-8 and IL-10 gene polymorphisms and risk of coronary artery disease. Int J Clin Exp Med. 2015;8:7937–43. [PMC free article] [PubMed]
- 62.Bin WX, Di HY, Zhang S, Cui NH, Liu ZJ, Huang ZL, et al. Associations of polymorphisms in TXNIP and gene–environment interactions with the risk of coronary artery disease in a Chinese Han population. J Cell Mol Med. 2016;20(12):2362–2373. doi: 10.1111/jcmm.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song Y, Shan Z, Luo C, Kang C, Yang Y, He P, Li S, Chen L, Jiang X, Liu L. Glutathione S-transferase T1 (GSTT1) null polymorphism, smoking, and their interaction in coronary heart disease: a comprehensive meta-analysis. Hear Lung Circ. 2017;26(4):362–370. doi: 10.1016/j.hlc.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Iwanicka J, Iwanicki T, Niemiec P, Nowak T, Krauze J, Grzeszczak W, Górczyńska-Kosiorz S, Ochalska-Tyka A, Żak I. Relationship between rs854560 PON1 gene polymorphism and tobacco smoking with coronary artery disease. Dis Markers. 2017;2017:1–7. doi: 10.1155/2017/1540949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ying Y, Luo Y, Peng H. EBF1 gene polymorphism and its interaction with smoking and drinking on the risk of coronary artery disease for Chinese patients. Biosci Rep. 2018;38(3):BSR20180324. doi: 10.1042/BSR20180324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H, Ding S, Liu X, Wu Y, Wu X. Association of interleukin-6 genetic polymorphisms and environment factors interactions with coronary artery disease in a Chinese Han population. Clin Exp Hypertens. 2018;40(6):514–517. doi: 10.1080/10641963.2017.1403618. [DOI] [PubMed] [Google Scholar]
- 67.Liu F, Li Z, Lv X, Ma J. Dietary n-3 polyunsaturated fatty acid intakes modify the effect of genetic variation in fatty acid desaturase 1 on coronary artery disease. PLoS One. 2015;10(4):1–10. doi: 10.1371/journal.pone.0121255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng Y, Li Y, Huang T, Cheng HL, Campos H, Qi L. Sugar-sweetened beverage intake, chromosome 9p21 variants, and risk of myocardial infarction in Hispanics. Am J Clin Nutr. 2016;103(4):1179–1184. doi: 10.3945/ajcn.115.107177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang L, Cai X, Lian F, Zhang L, Kong Y, Cao C, et al. Interactions between ALDH2 rs671 polymorphism and lifestyle behaviors on coronary artery disease risk in a Chinese Han population with dyslipidemia: a guide to targeted heart health management. Environ Health Prev Med. 2018;23(1):1–8. doi: 10.1186/s12199-018-0719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McPherson R. A gene-centric approach to elucidating cardiovascular risk. Circ Cardiovasc Genet. 2009;2(1):3–6. doi: 10.1161/CIRCGENETICS.109.848986. [DOI] [PubMed] [Google Scholar]