Abstract
Intrinsically disordered proteins (IDPs) are ubiquitous in proteomes and serve in a range of cellular functions including signaling, regulation, transport and enzyme function. IDP misfunction and aggregation are also associated with several diseases including neurodegenerative diseases and cancer. During the past decade, single-molecule methods have become popular for detailed biophysical and structural studies of these complex proteins. This work has included recent applications to cellular liquid-liquid phase separation (LLPS), relevant for functional dynamics of membraneless organelles such as the nucleolus and stress granules. In this concise review, we cover the conceptual motivations for development and application of single-molecule fluorescence methods for such IDP studies. We follow with a few key examples of systems and biophysical problems that have been addressed, and conclude with thoughts for emerging and future directions.
Introduction
A long-standing paradigm in biology was that well-defined 3D structures are essential for proteins to perform their functions in the cell. A rapidly expanding body of work over the past couple of decades has challenged this idea 1,2, revealing that a substantial portion of the cellular proteome is comprised of proteins and protein regions that do not strongly encode well defined structures. Nevertheless, these intrinsically disordered proteins (IDPs) and regions (IDRs) are involved in numerous cell functions, with their flexibility and interaction promiscuity often thought to play critical roles in function. Note that in this review (as is often the case in the field), we will use the term IDP to refer to proteins that are predominantly unstructured throughout their sequence as well as proteins containing a mix of IDRs and structured regions. Virtually every class of cell function is represented by IDPs, including transcription and translation, signaling, transport, and enzyme function. Furthermore, many aspects of cell disfunction and disease are also associated with protein disorder, including neurodegenerative and other protein aggregation diseases, cancer, viral infection and heart disease.
In recent years, IDPs have also been shown to be important players in cellular liquid-liquid phase separation (LLPS) 3–6 Phase separation in biology has exploded onto the scene in the past 10 years due to the observation that LLPS underlies the formation and dynamics of many membrane-less organelles (MLOs) and their roles in essential cellular processes. These macromolecular assemblies generally contain a combination of proteins and nucleic acids, and their formation requires weak multivalent interactions between these component molecules. Of particular interest is that a substantial fraction of the phase-separating proteome contains intrinsically disordered proteins with low-complexity, often repetitive, amino-acid sequences. This is not coincidental, as the disordered nature of these proteins gives them the biochemical flexibility to interact, both specifically and non-specifically, with a wide range of phase-separating molecules. The physical and chemical properties of IDPs, which have been extensively studied over the past few decades, help give membrane-less organelles their diversity in composition, function, and physical properties. This has made the phase separation of IDPs an exciting area of study for biophysicists.
Given the broad importance of IDPs in biology, the physics and chemistry of these proteins are interesting and critical to understanding their function in biology and disease. However, the complexity and flexibility of these proteins makes them hard to study using traditional methods of structural biology and biophysics. Therefore, a major effort in the field has been towards adapting or developing alternative methodologies for studies of these systems.
Along these lines, single-molecule methods have revolutionized studies of complex systems over the past 3 decades and have been recently applied to IDPs. These methods can reveal detailed information about protein structure and dynamics that is usually hidden by averaging over an ensemble of molecules 7–9 Crick et al. reported in 2006 scaling behavior in the disordered poly-Q system studied using fluorescence correlation spectroscopy (FCS) 10, a method related to single-molecule fluorescence methods. A year later, Mukhopadhyay et al. reported single-molecule fluorescence studies of the yeast prion protein Sup35, revealing new structural and dynamic features of this system 11. Since then, other single-molecule studies have probed several such features in a number of IDP systems.
Single-molecule methods can be broadly categorized into fluorescence and manipulation techniques 9 Single-molecule fluorescence methods include measurements of energy transfer, intensity, fluctuations, and spatial variations which can provide equilibrium or dynamic information about conformation, binding, proximity, size, spatial arrangement, movement and other molecular parameters. For example, one of most commonly used methods is Single-molecule Förster resonance energy transfer (smFRET). This method can provide information about conformational distributions and dynamics as well as binding. smFRET has been widely used to study proteins and nucleic acids during the past two decades. The related method of fluorescence correlation spectroscopy (FCS) has used analyses of fluorescence fluctuations to provide measurements of dimensions and rapid molecular fluctuations in IDPs. These and other single-molecule fluorescence methods are discussed in more detail below in the context of specific problems and systems. Single-molecule manipulation methods such as optical and magnetic tweezers and atomic force microscopy (AFM) can also measure several molecular parameters, while simultaneously measuring the molecular responses to exerted forces 12–14 For folding measurements, all these methods typically attach molecules between two surfaces (beads, AFM tip, quartz, mica, etc.) and forces are generated on the molecule by changing the separation between the surfaces. Different methods are used to measure forces and separation, permitting a range of different types of experiments to be carried out 15.
In this short review, we focus on single-molecule fluorescence studies. The review is not meant to be comprehensive; rather it is meant to provide examples of key types of systems and insights that can be addressed using these methods. A series of examples of IDP studies are provided below, including a brief discussion about techniques used and emerging insights. Several of the systems discussed have been studied with respect to phase separation. However, application of single-molecule methods to LLPS is still in its infancy. Hence, the complementary examples provided of other IDP studies provide a good overview of the various kinds of experiments feasible using these methods. We conclude with our perspectives for emerging and future directions. Several other review articles16–19 and original papers are cited for broader and more detailed information.
A few examples – systems and biophysical problems
Poly-glutamine: Size scaling behavior, water solvent quality and phase separation
Several neurodegenerative diseases are linked with aggregation of expanded poly-glutamine (Q) repeat sequences. The 2006 work by Crick et al.10 probed polymer scaling behavior of several poly-Q polypeptides differing in amino acid repeat length using fluorescence correlation spectroscopy (FCS). FCS probes fluorescence fluctuations in a small ensemble (few) of molecules using confocal detection and was a precursor of single-molecule detection. Several molecular parameters can be extracted by analysis of these fluorescence fluctuations. One of the most basic parameters for diffusing molecules is translational diffusion, which results in signal fluctuations as molecules move in and out of the focal volume. Crick et al. used the fact that translational diffusion times are related to hydrodynamic radii of peptides to study how dimensions scaled as a function of Q repeat length. The dimensional scaling of long polymers in a solvent is influenced by a balance of polymer-polymer and polymer-solvent interactions. Accordingly, for long polymers in a solvent, the scaling constants assume three different values depending on whether the solvent is a good, neutral or poor solvent. This idea can be translated to the case of IDPs, which can (i) be extended (water is a good solvent) when chain-solvent interactions are favored over chain-chain interactions, (ii) behave like an ideal chain (water is a theta solvent) when chain-chain interactions balance chain-solvent interactions, or (iii) be collapsed (water is a poor solvent) when chain-chain interactions are favored. The measured scaling constant showed that water acts as a poor solvent and polyQ polypeptides assume an ensemble of collapsed structures. Based on the results, the authors discussed multiple possibilities for poly-Q aggregation, including that it proceeds through formation of disordered oligomeric species, with solvent quality playing a key role in determining which mechanism is dominant. This idea of disordered oligomers is related to droplet formation during LLPS that has subsequently become a topic of intense study20 as discussed in this review.
Sup 35-NM – Monomer structural and dynamic features, aggregation and phase separation
Amyloidogenic IDPs are involved in several diseases including neurodegenerative and heart diseases. One example where amyloid formation was postulated instead to have a functional cellular role is the yeast prion protein Sup35. This protein is a translational termination factor in yeast. Amyloid formation was previously postulated to act as a regulator of function that could be transmitted over generations 21, and recently phase separation has been linked to cellular fitness 22 (see below).
In 2007, Mukhopadhyay et al. reported the results of a series of single-molecule studies on the amyloid forming (NM) region of Sup35 11 (Figure 1). smFRET experiments were first used to probe the overall characteristics of the amyloid forming region of this protein. FRET is the non-radiative transfer of singlet excitation energy from a donor to an acceptor dye. The strong distance dependence of the FRET efficiency has been used to provide a measure of conformation in proteins and other biomolecules. In these experiments, donor and acceptor dyes were attached at positions that spanned this region, and confocal smFRET experiments were used to study the conformational features of freely diffusing single molecules. This experimental mode, which was first developed23 to test the distance dependence of FRET at single-molecule resolution, allowed studies of Sup35-NM without potential perturbations due to surface tethering, a mode discussed later that is often used in conjunction with imaging for single-molecule experiments. Here, sub-nM concentrations of dual labeled proteins were used in conjunction with the sub-fL detection volume to ensure that almost all signals originated from single molecules. The donor was excited using a laser, and donor and acceptor fluorescence photon counts were recorded as a function of time with sub-ms time-resolution. The single-molecule fluorescence bursts in the resulting time-trajectories were analyzed to produce smFRET histograms, which revealed populations of protein molecules with particular FRET efficiencies.
Figure 1.
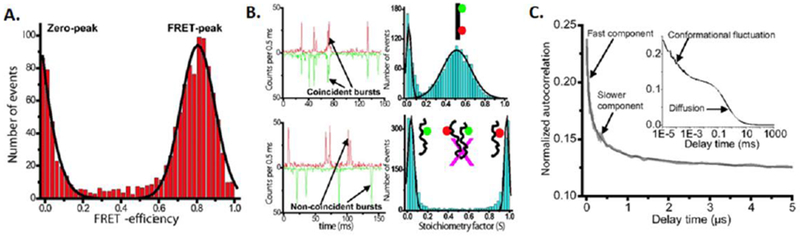
Single-molecule fluorescence data for Sup35-NM. A. smFRET histograms under non-denaturing conditions. B. 2-color single-molecule coincidence analysis to rule out oligomerization. C. FCS analysis reveal fast fluctuations. Adapted from Mukhopahyay et al. Proc. Natl. Acad. Sci. (2007) 104:2649. See text for additional details.
For Sup35-NM, the smFRET histograms revealed a relatively compact conformational distribution. The low concentrations used in these experiments (~100pM) should highly disfavor aggregation (Figure 1A). However, to directly study this issue, a complementary single-molecule technique, 2-color coincidence24, was used to test if the protein still formed small oligomers at these concentrations which might complicate the FRET analysis. Here, two protein samples were separately labeled with blue and red dyes. Once again, confocal detection was used, now with dual excitation. Since diffusion is random, these time traces would only show a significant population of 2-color bursts if the protein oligomerized. The results did not show such a distribution (Figure 1B), ruling out the existence of a significant fraction of oligomers, and confirming that the above FRET results were from monomers. An smFRET denaturation analysis was next carried out and showed a single smFRET histogram peak gradually moving to lower FRET efficiency (revealing continuous protein expansion) as a function of denaturant concentration. This was in contrast with results on folded proteins which showed multiple interconverting peaks corresponding to folded and unfolded states8,25, supporting the idea that Sup35-NM is intrinsically disordered and occupies a range of structures. The relatively narrow FRET peak (Figure 1A) also indicated that these structures were rapidly interconverting relative to the 0.5 ms data acquisition time. This idea was directly tested using FCS experiments and quenching of attached dyes by nearby tyrosines, showing rapid fluctuations in the ~100 ns range. Overall, this set of experiments showed that Sup35-NM is intrinsically disordered and relatively compact (compared to a typical denatured protein) yet is rapidly sampling a range of conformations.
The above work had implications for the aggregation mechanism of the protein, with the authors speculating that aggregation might proceed through the formation of disordered oligomeric species. Initial tests of this idea were made using a combination of single-molecule and ensemble studies 26 In fluorescence polarization experiments, samples are excited using polarized light and the depolarization of the emitted photons is measured. If the dyes rapidly tumble relative to the excited-state lifetime, the emitted light is depolarized, while increased residual polarization will be observed for slower tumbling dyes. Hence, this method can be used to probe the mobility imparted on the dye and attached IDP by its local environment. For Sup35-NM, the polarization data indicated that initial substantial changes in the environment occurred well before formation of amyloid structure was observed by conventional thioflavin-T fluorescence measurements. These early polarization changes coincided with changes in dye emission intensity due to changes in the environment of the dyes, presumably changing quenching by proximal tyrosine residues as discussed above. Together, these data were consistent with early formation of disordered oligomeric species, that eventually proceeded to form amyloids, one of the models discussed in the previous work by Crick et al.10
Recent work by Franzmann et al. has shed new light on the functional significance of phase separation of native Sup35 22 Using a variety of methods, the authors show that stress induced pH changes induce liquid-liquid phase separation in Sup35. A liquid-like dense phase is first formed that then changes to a protective gel-like state. This sequence of events is reminiscent of the putative aggregation mechanisms discussed above. The authors also showed that the N-terminal prion domain was a major determinant of phase separation, and that certain charged sequence regions in the M-domain can regulate these phase transitions. Moreover, the C-terminal GTPase domain by itself forms irreversible aggregates in cells under stress and reduced cell fitness. Single-molecule fluorescence methods could be used to probe various aspects of these interesting transitions, including inter- and intra-domain conformational properties in different phases of the protein system.
α-synuclein, a Parkinson’s disease protein: Monomer binding-folding complexity and aggregation
α-synuclein 27 is an IDP whose aggregation has been extensively implicated in Parkinson’s disease. It has also been suggested to have functions in synaptic vesicle fusion and vesicle transport. Furthermore, the protein folds upon binding to partners such as membranes and membrane mimics 27. This coupling of folding and binding is shared by many other IDPs and is often important for IDP function. In 2009, the Rhoades, Subramaniam and Deniz labs separately reported single-molecule FRET studies on this protein 28–30. These reports showed that smFRET could be used to study details of the coupled folding and binding of this protein. As an example, an ongoing debate in the field raised questions about the precise helical structure the protein assumed on binding to membrane-like surface - a hairpin-like shape or an extended helix. To test these possibilities directly, Ferreon et al. 30 labeled the protein in locations at the ends of the helical region to create a construct with which well-resolved high and low FRET efficiencies would be observed in the two possible aforementioned helical structures. A series of smFRET experiments encompassing titrations of several concentrations of the lipid-mimic SDS resulted in a 3D histogram that showed a striking series of complex transitions (Figure 2). Increasing SDS concentration below critical micelle concentration (CMC) of SDS showed that the system transitioned from an initial disordered state to a hairpin state and then to an extended state. Further titration to above CMC then resulted in a transition back to a hairpin state, and even higher SDS concentration finally pushed the system again to extended helical states. This work showed that binding to either small amphipathic molecules or membrane-like surfaces of different curvature could result in hairpin and extended helical forms. Follow-up smFRET work using novel microfluidic methods showed that folding to the extended helix occurred following initial binding to SDS and formation of collapsed species, analogous to formation of disordered encounter complexes 31. Nice examples of such dynamic intermediates have more recently been observed by smFRET in the folding pathways of other systems 32,33. Several other smFRET studies of this interesting protein have followed. For example, the Rhoades lab has used smFRET to study allosteric effects arising from the floppy C-terminal region of α-synuclein 34. They also combined smFRET with computational analysis to further characterize in detail the conformational ensembles of this protein 35,36.
Figure 2.
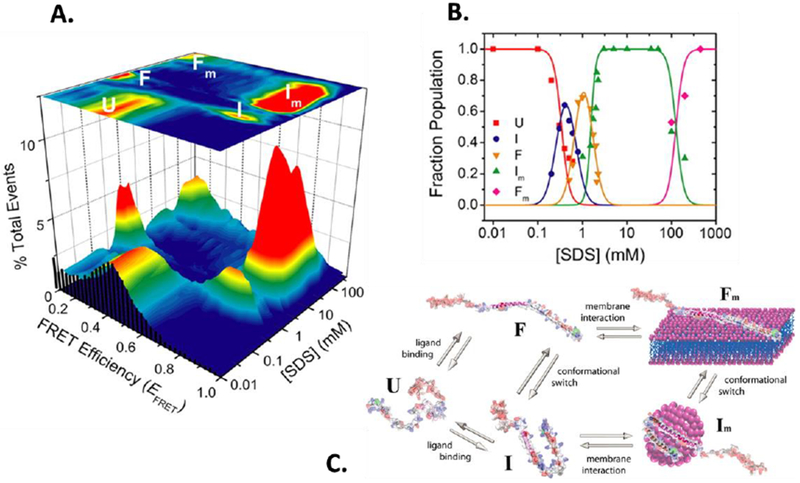
Single-molecule fluorescence data for α-synuclein. A. 3D smFRET histograms as a function of concentration of the lipid mimic showing a complex folding-binding landscape. B. Populations of different species derived form histograms in A, showing multiple transitions between different species. C. Cartoons of the different states revealed in A. Adapted from Ferreon et al. Proc. Natl. Acad. Sci. (2009) 14:5645. See text for additional details.
NMDA-sensitive glutamate receptor: effects of post-translational modifications
The effects of post-translational modifications such as phosphorylation on IDPs are an active area of study. These modifications can modulate the charge distribution and disorder-order balance, and corresponding binding and function in IDPs 37, but it is difficult to predict these effects. smFRET is a powerful tool to determine such effects in challenging IDP systems. One example is the study by Choi et. al. on the effect of phosphorylation on the C-terminal domain (CTD) of GluN2B, the major Tyr-phosphorylated protein in synapses and essential NMDA-sensitive glutamate receptor (NMDAR) regulator38. These studies used a different single-molecule detection geometry, total internal reflection (TIR). Because TIR can be used to illuminate a thin slice of surface and sample solution, it can eliminate a substantial amount of background signal from other portions of the sample.
GluN2B CTD’s function is regulated by Src kinase phosphorylation on the palmitoylation-motif-anchored C-terminal segment of CTD (CTD2). To elucidate the role of CTD2 phosphorylation, eight constructs of CTD2 were designed for donor-acceptor labeling throughout the peptide through site-directed mutagenesis. These peptides were then phosphorylated and encapsulated in biotin-containing liposomes before immobilization on quartz microscope slides. To study the effect of the physiological palmitoylated anchoring of CTD2, N-terminal biotinylated CTD2 constructs were similarly generated and directly immobilized onto the slides 39. Although a range of different regions of CTD2 were probed by the labeling design, all constructs produced broad distributions around mid-FRET-efficiency values, and a correlation between FRET efficiency and the fluorophore separation in the primary sequence was found in both phosphorylation states, consistent with a lack of structure. However, a small shift to lower median FRET efficiency after phosphorylation regardless of the labeling position, suggested an overall extension of the peptide. This led the authors to refute the induction of structure in CTD2 via phosphorylation, suggesting its allosteric effects on NMDA activity must arise from interactive or steric outcomes of this expansion.
CTD2 is natively rich in proline, a residue associated with intrinsically disordered proteins and polypeptide expansion 40. Although a completely Pro-depleted CTD2 mutant was found to lose its solubility, smFRET permitted an assessment of Pro’s role in the interactions between CTD2 and the second PDZ domain (PDZ2) of scaffold protein PSD-95, through which the receptor directs synaptic targeting 41. A FRET-acceptor-labeled Pro-depleted CTD2 construct was used with PDZ2 labeled with donor. The isolation of the Pro-depleted constructs as immobilized single molecules (at low density) prevented aggregation challenges of these aggregation-prone proteins. Using an analysis of the bound and unbound dwell-times, the authors observed that though Pro mutations do not directly affect the PDZ binding site of CTD2, they still reduce the binding affinity via increased dissociation rate constants. Additionally, using intramolecular smFRET, the authors observed that Pro-depletion changes CTD2 conformational dynamics. Overall, they concluded that the dynamics of the cytoplasmic IDR are an important component in allosteric regulation of receptor gating 41.
smFRET studies have also been used to study PTMs in other IDPs, like in alpha-synuclein 42. Another example is discussed below for the case of nucleophosmin.
Nucleophosmin: Structural changes and complex interplay of interactions - single molecules to phase separation
The nucleolus was one of the first MLOs discovered, with work during the past few years showing their liquid-like nature 43. It is an essential cellular body, as it is implicated in several important cellular processes including ribosome assembly and stress signaling. Nucleophosmin (NPM1) is one of the many functional IDPs within the nucleolus. It has been shown to interact with a variety of nucleolar biomolecules, including other signaling proteins, ribosomal RNA and DNA. NPM1 is an interesting protein to study in terms of phase separation, because it consists of multiple acidic, basic and low-complexity tracts as well as folded CTD and N-terminal domain (latter in the pentameric form), giving it variability in interaction modes and multivalency in terms of electrostatic interactions. The versatility of NPM1 is a function of its unique architecture.
Studies by Mitrea et al. showed that in vitro, higher salt concentrations were needed to bias the protein towards pentamers, and that phosphorylation could tune this equilibrium 44. In 2016, Banerjee et al. reported studies of the monomer-pentamer equilibrium of a truncated version of NPM1 that contained the C-terminal oligomerization domain and a part of the adjoining disordered region containing charge tracts 45. smFRET in conjunction with ensemble experiments provided more detailed information about this equilibrium. The overall reactions occurred in multiple discernible steps, and phosphorylation and partner binding differentially affected these steps. The findings pointed to different points of potential regulation in cells. smFRET studies of this protein were also used to probe aspects of its phase separation, as discussed below.
Work with NPM1 has revealed that it has the ability to phase-separate through several distinct mechanisms 46,47. It is capable of heterotypic interactions with both positively-charged repeat peptides and negatively-charged ribosomal RNAs. It is also capable of phase separation via homotypic interactions between its own component sequences. All of these properties bring up a notable question in the field of IDP phase separation: How does a variety of weak interactions coordinate the phase behavior of an MLO?
smFRET and ensemble turbidity (light scattering by absorbance to report on overall phase separation), SAXS (to understand overall structural features) and mutational analysis (to test molecular components affecting phase separation) were used to study the impact of these weak interactions on the phase separation of NPM1 on its own and in combination with partners 46,47. smFRET in particular showed changes in conformational features with ionic strength (supporting a model involving a complex interplay of intra- and inter-molecular interactions) and upon moving to phase-separation conditions. These studies revealed the promiscuity of interactions in NPM1, the importance of multivalency among its charged sequences for forming droplets, and how the electrostatic environment dictates the ability of NPM1 to phase separate. Electrostatic mediation of phase separation is important to consider when attempting to understand cellular mechanisms of phase separation. This is because many cellular conditions and stimuli can influence these interactions, whether it be pH, ionic strength, or other specific or non-specific interactions. Phase separation in this sense can also be altered by changes in PTMs and IDP interactions. The variety of molecular mechanisms involved in MLO assembly and dynamics are not yet fully understood, but a better understanding of the biophysical properties of the IDPs involved is essential to generate a complete picture of phase separation in the cell.
Molecular crowding and osmolytes
As discussed above, single-molecule fluorescence tools have been utilized to help understand fundamentals of the folding, dynamics and conformational heterogeneity of IDPs and proteins in general. Most experiments were performed in simple conditions in vitro, where cellular conditions were probed only in terms of very basic components, such as ionic strength, pH, and binding partners.
Several in vitro studies have aimed to probe the influence of key aspects of cellular conditions, while still preserving the tunable and well-defined conditions achievable in vitro. For example, as opposed to conditions in most in vitro studies, cells have crowded interiors that incorporate high concentrations of a variety of molecules as well as interaction surfaces such as membranes. Careful and extensive studies by Sorrano et. al. along with polymer-physics analysis indicated that crowding can cause different degrees of compaction for different IDPs, and this degree is related to the “non-crowded” expanded state of a given IDP 48. The authors discuss various examples of cellular IDPs where their results reveal factors that could play key functional roles. In another example, a study by Banerjee et al. showed that 2-dimensional crowding on the surface of model lipid membranes can result in the formation of “hidden” state that is not well-populated in the absence of crowding 49. Again, given that cellular membranes are very crowded, these results could have interesting implications for the folding, association and function of several membrane-associated IDPs and IDRs. In another related example, Ferreon et al. have shown interesting effects of high concentrations of osmolytes on IDPs. In one case, the osmolytes TMAO and urea could counteract 50 each other’s (compaction and expansion) effects on α-synuclein and showed a striking 1:2 counteraction ratio over a broad range of absolute osmolyte concentration 51. In a different case, TMAO was found to increase the phase separation propensity of the ALS-linked protein TDP-43, but decrease its aggregation propensity 52. Thus, these two types of phase transitions are related but can be differentially affected by solution conditions.
Cellular Studies
Direct in cell and in vivo applications of single-molecule fluorescence techniques require several criteria to be fulfilled such as (i) single-molecule emitters must be detectable over cellular autofluorescence; (ii) sample delivery to living cells should be optimized in terms of reproducibility and ideally in high precision in targeting the cellular compartments; (iii) fluorophores must be stable and bright, therefore allowing longer detection times before photobleaching; (iv) data acquisition must be prompt after sample delivery to overcome the depletion of material by degradation; (v) cells must maintain a viable state before and after the delivery of sample, and (vi) ideally, the experimental setup should allow multiparameter data analysis to perform controls and experiment concurrently for the limited number of molecules in question.
Several of these technical issues were first addressed by Sakon and Weninger in a 2010 study, where authors chose microinjection to deliver protein samples into live mammalian cells 53 With the aid of TIR fluorescence microscopy and single-particle tracking, this technical advance had been leveraged to study the conformational changes associated with SNAP-25, an IDP that folds upon forming a cellular membrane fusion (SNARE) complex. The authors found that SNAP-25, which is specific to neurons, forms promiscuous pairings in different cell lines where it is not endogenously expressed. Using the same method of sample delivery, Schuler and colleagues performed a comprehensive in vivo study that recapitulates well-characterized properties of several IDPs 54. The authors chose to circumvent the cellular autofluorescence problem by using a FRET pair with excitation wavelength above 520 nm. The disordered state of prothymosin-α was shown to be conserved in HeLa cells, by comparison of the radius of gyration in different compartments in the cell to in vitro dimensions. Similarly, heat- and cold-denaturation profile of a marginally stable protein, yeast frataxin was studied in cells and found to comply with in vitro stability profile. Finally, in combination with recurrence analysis of single-molecule bursts 55, interconversion rates between folded and unfolded forms of protein GB1 domain had been determined. Folding-unfolding relaxation time of protein GB1 domain marginally differed from in vitro rates, indicating a small effect of macromolecular crowding takes place within the cells.
While microinjection was proven to be an attractive method for protein delivery into mammalian cells, the large needle diameter and the existence of cell walls make the method unavailable for prokaryotic cells. Kapanidis and colleagues devised a strategy based on electroporation for delivery of folded proteins and DNA into E.coli 56,57. This method can be applicable for delivery into certain types of eukaryotic cells, as the authors have successfully shown that proteins can be delivered into S. cerevisiae.
Besides delivery of fluorophore-labeled proteins into cells, fluorophores can be readily expressed as fusion proteins in the form of fluorescent variants, labeling via modified enzymatic activity of native cellular proteins58. Due to the generally weaker photophysical properties of fluorophores exclusive to such schemes, the applications are limited. Incorporation of noncanonical amino acids (ncAA) via orthogonal tRNA/amino-acyl tRNA synthetase pairs to label proteins using click-chemistry provides high site-specificity. Moreover, many fluorophores that are frequently used for single-molecule fluorescence applications are available in “clickable” moieties. While this approach is widely used for in vitro investigations of protein conformation and dynamics with single-molecule fluorescence, low ncAA incorporation efficiency, labeling specificity and efficiency variance in different cellular locations specifically favoring cell surfaces, physical characteristics of ncAAs and fluorophores, together make his method challenging for in cell fluorophore labeling59. Lemke and colleagues leveraged this labeling strategy and investigated the conformational dynamics of folded influenza hemagglutinin trimers on viral envelope using smFRET imaging60. Particularly because of their subcellular location, IDPs pose a challenge to be labeled via click chemistry within cells to study their conformational populations and dynamics using single-molecule fluorescence.
Perspectives on emerging and future directions
We anticipate that the application of single-molecule fluorescence studies of IDPs and in particular phase-separation will continue to expand in the coming years. In terms of systems, we expect that both a larger array of systems and more in-depth studies of key systems will be carried out. Furthermore, concomitant advances in single-molecule and adjunct methods for such studies are expected to continue and expand.
Such studies in recent years have already begun to reveal new types of biophysical and biologically relevant insights. For example, higher order complexes with more complex IDPs have begun to reveal interesting information about allostery and cooperativity. For binding of viral oncoprotein E1A to two of its key cellular partners, Ferreon et al. used smFRET studies to show how this IDP can show dramatic shifts in the magnitude and even sign of the binding cooperativity depending on the available E1A regions 61. The results afforded insight into the complex layers of structural and functional regulation of such IDPs. In other examples, single-molecule fluorescence studies have directly probed the dynamics of disordered or fuzzy 62 complexes, where at least one partner is still unfolded and dynamic. Using a combination of smFRET and other methods, the Lemke lab has shown that nucleoporins, which are key to selective nuclear pore transport, bind nuclear transport receptors in a disordered state and with close to diffusion-limited rates 63. In a different study, Schuler lab showed that two IDPs, prothymosin-α and linker histone H1, bind each other with sub-nM affinity and yet remain disordered in the complex 64. Such charge interactions with dynamic and distributed interfaces are important in phase separation and will also make such binding partners amenable to phase separation as in the case of nucleoporins. In another example, Soranno et al. have studied the question of internal friction in IDPs 65,66 The authors used multiple methods including smFRET, photoinduced electron transfer mediated contact fluorescence quenching, correlation methods, as well as computation and comparison to theory to quantify friction in IDPs and unfolded proteins. The authors showed that internal friction can have important effects on the dynamics of IDPs and unfolded proteins, with more compact IDPs showing higher friction. This issue is particularly interesting from the point of view of phase separation since the high-density environments of many protein droplets will also likely give rise to friction effects on structural dynamics and potentially function.
Several directions for improving single-molecule methods can be envisioned. For example, further refinement of single-molecule methods such as FRET will be important for moving towards more quantitative measurements of IDP structure and dynamics in more complex droplet environments. Along these lines, multiparameter fluorescence detection as popularized by the Seidel lab integrate different types of fluorescence photon observables (including color, lifetime, and arrival times) that permit a more global analysis of molecular parameters 67. Of these, fluorescence lifetime measurements require more complex setups, but can provide important and complementary information especially when combined with other observables. These types of measurements will be very useful in the context of phase separation. Additionally, studies of molecules in droplets on surfaces would permit long time-trajectories of IDP properties to be studied. Myong and colleagues performed first phase separation studies in this geometry using TIR detection on immobilized RNA molecules within droplets 68,69. In these studies, smFRET of tethered RNA molecules was used to investigate the conformational changes and dynamics upon incorporation in protein-RNA droplets. Similar studies could also be performed on either immobilized or diffusing IDPs within such surface settled droplets. Since quartz/glass surfaces could influence the properties of proximal IDPs, careful testing of immobilization and comparisons to diffusing molecules would be important. Another direction is related to the fact that many single-molecule measurements need to be done at low (sub-nM) concentrations, which poses a problem for the high concentrations needed for studies of weak interactions and droplets. While studies of intramolecular IDP structure and dynamics in droplets can be performed by doping in a small fraction of dual-labeled protein in a large background of unlabeled protein, this is not the case for studies of intermolecular interactions. One interesting way to overcome this issue is to reduce the detection volume which then allows higher concentrations to be used. For instance, zero-mode waveguides (metallic film with an array of holes smaller than the wavelength of detection light) can reduce the focal volume by orders of magnitude down to the zL level 70,71. Also, extension of 2-color smFRET to 3 or more colors can provide powerful capabilities for more global analysis of IDPs and their complexes and condensates 72–78. For example, 3-color smFRET has been used to study the tetramerization domain of p53 77 as well as α-synuclein 73. Near-field scanning microscopy can also be used to study IDPs and their aggregates and oligomers with high resolution 79. Adjunct methods such as chemical biology labeling methods 72,80 and combination with microfluidics will also continue to contribute to improving experimental methods for understanding the molecular underpinnings of phase separation 31,81. Microfluidic methods that allow quick scanning of a range of interaction, temperature and other conditions 82 might be particularly useful for broadening detailed and controlled studies of IDP phase separation. Other methods could also be further developed or adapted for single-molecule detection of IDPs and droplets, including single-molecule manipulation and methods that combine single-molecule fluorescence with single-molecule manipulation83 or other structural methods such as cryo-EM and NMR as well as computational analysis.
We anticipate overall that such advances in single-molecule tools and applications to a larger number of IDP systems both in vitro and in vivo will continue to reveal and delineate additional biophysical principles related to IDP conformation, molecular-level interactions and phase separation. Novel additional understanding of allosteric 61,84, non-equilibrium and active-matter effects and their influence on function are certain to emerge 85. These emerging principles are also expected to continue to provide new insight into mechanisms of numerous cellular processes as well as associated diseases.
Highlights.
IDPs are dynamic proteins that are key for cell function and misfunction.
Single-molecule fluorescence can reveal novel information about IDP biophysics.
Several important IDP systems and biophysical problems are discussed.
IDP liquid-liquid phase separation underlies dynamic cellular compartments.
Acknowledgements.
We gratefully acknowledge Samrat Mukhopadhyay, Allan Ferreon, Edward Lemke, Yann Gambin, Priya Banerjee, Crystal Moran, Mahdi Moosa, Chris Lee, Anthony Milin, and Emily Bentley and several collaborators for their contributions to our work on IDPs and LLPS. We also gratefully acknowledge the US NIH (Grants R35 GM30375, RO1 GM066833, RO1 GM115634 and RO1 CA214054) and US National Science Foundation (Grant 1818385) for support of our lab’s related work. Irem Nasir is supported by a Swedish Research Council (Vetenskapsrådet) International Postdoctoral Fellowship (Grant number 2016-06672)
Biographies

Irem Nasir received her Ph.D. degree in Biochemistry from Lund University in Sweden, working on intrinsic and extrinsic factors that modulate amyloid formation in vitro as well as investigating protein unfolding/misfolding at nanoparticle surfaces. In 2016, she started her postdoctoral work in the Deniz lab at The Scripps Research Institute and currently a Swedish Research Institute International Postdoctoral Fellow. The main focus of her work is dedicated to deciphering the relationship between conformational heterogeneity and dynamics of intrinsically disordered proteins and their biological function by leveraging the single-molecule fluorescence toolbox.

Paulo Onuchic received his B.A. in Molecular and Cell Biology from the University of California, Berkeley in 2013. He began his Ph.D. work in 2015 at The Scripps Research Institute in San Diego. During his time as a graduate student in the Deniz Lab, Paulo has worked on questions surrounding the phase separation of disordered proteins, peptides, and RNA. Using fluorescence techniques, he has shown the sensitivity of in vitro phase-separated systems to rapidly changing biochemical environments, which has wide-ranging implications in the fields of biophysics and cell biology.

Sergio Labra received his B.S.E. in Chemical & Biomolecular Engineering and Master of Biotechnology from the University of Pennsylvania. In 2018, he began his Ph.D. studies at Scripps Research in La Jolla, where he worked on mathematical predictive models to characterize the effect of charge sequence clustering patterns in the in vitro liquid-liquid phase separation of charged peptides with RNA at the Deniz Lab. In 2019, he joined the lab of Jeffery Kelly and is currently developing novel in vitro human iPSC-derived models of neurodegeneration with Stuart Lipton, with a focus on testing the therapeutic potential of proteostasis network regulators.
Ashok Deniz received his PhD in Chemistry from the University of Chicago, then began postdoctoral work at the University of California, Berkeley. He is currently Associate Professor at Scripps Research in La Jolla. His lab studies mechanisms of protein intrinsic disorder, folding and assembly, while developing and using novel single-molecule tools for this purpose. Recent and emerging interests include dynamic complexity, active matter effects and cellular phase separation, important in biology and health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wright PE & Dyson HJ Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16, 18–29, doi: 10.1038/nrm3920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Lee R et al. Classification of intrinsically disordered regions and proteins. Chem Rev 114, 6589–6631, doi: 10.1021/cr400525m (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman AA, Weber CA & Julicher F Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30, 39–58, doi: 10.1146/annurev-cellbio-100913-013325 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Brangwynne CP, Tompa P & Pappu RV Polymer physics of intracellular phase transitions. Nat Phys 11, 899–904, doi: 10.1038/Nphys3532 (2015). [DOI] [Google Scholar]
- 5.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298, doi: 10.1038/nrm.2017.7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitrea DM & Kriwacki RW Phase separation in biology; functional organization of a higher order. Cell Commun Signal 14, 1, doi: 10.1186/s12964-015-0125-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joo C, Balci H, Ishitsuka Y, Buranachai C & Ha T Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem 77, 51–76, doi: 10.1146/annurev.biochem.77.070606.101543 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Schuler B & Eaton WA Protein folding studied by single-molecule FRET. Curr Opin Struct Biol 18, 16–26, doi: 10.1016/j.sbi.2007.12.003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deniz AA, Mukhopadhyay S & Lemke EA Single-molecule biophysics: at the interface of biology, physics and chemistry. J R Soc Interface 5, 15–45, doi: 10.1098/rsif.2007.1021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crick SL, Jayaraman M, Frieden C, Wetzel R & Pappu RV Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci U S A 103, 16764–16769, doi: 10.1073/pnas.0608175103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S & Deniz AA A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A 104, 2649–2654, doi: 10.1073/pnas.0611503104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuman KC & Block SM Optical trapping. Rev Sci Instrum 75, 2787–2809, doi: 10.1063/1.1785844 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenleaf WJ, Woodside MT & Block SM High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct 36, 171–190, doi: 10.1146/annurev.biophys.36.101106.101451 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins TT Angstrom-Precision Optical Traps and Applications. Annu Rev Biophys 43, 279–302, doi: 10.1146/annurev-biophys-042910-155223 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Neupane K et al. Direct observation of transition paths during the folding of proteins and nucleic acids. Science 352, 239–242, doi: 10.1126/science.aad0637 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Ferreon AC, Moran CR, Gambin Y & Deniz AA Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol 472, 179–204, doi: 10.1016/S0076-6879(10)72010-3 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Banerjee PR & Deniz AA Shedding light on protein folding landscapes by single-molecule fluorescence. Chem Soc Rev 43, 1172–1188, doi: 10.1039/c3cs60311c (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee T, Moran-Gutierrez CR & Deniz AA Probing protein disorder and complexity at single-molecule resolution. Semin Cell Dev Biol 37, 26–34, doi: 10.1016/j.semcdb.2014.09.027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brucale M, Schuler B & Samori B Single-molecule studies of intrinsically disordered proteins. Chem Rev 114, 3281–3317, doi: 10.1021/cr400297g (2014). [DOI] [PubMed] [Google Scholar]
- 20.Holehouse AS & Pappu RV Collapse Transitions of Proteins and the Interplay Among Backbone, Sidechain, and Solvent Interactions. Annu Rev Biophys, doi: 10.1146/annurev-biophys-070317-032838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halfmann R, Alberti S & Lindquist S Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol 20, 125–133, doi: 10.1016/j.tcb.2009.12.003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzmann TM et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654, doi: 10.1126/science.aao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Deniz AA et al. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc Natl Acad Sci U S A 96, 3670–3675 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Ying L, Green JJ, Balasubramanian S & Klenerman D Ultrasensitive coincidence fluorescence detection of single DNA molecules. Anal Chem 75, 1664–1670 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Deniz AA et al. Single-molecule protein folding: diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc Natl Acad Sci U S A 97, 5179–5184, doi: 10.1073/pnas.090104997 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnan R et al. Conserved features of intermediates in amyloid assembly determine their benign or toxic states. Proc Natl Acad Sci U S A 109, 11172–11177, doi: 10.1073/pnas.1209527109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snead D & Eliezer D Alpha-synuclein function and dysfunction on cellular membranes. Exp Neurobiol 23, 292–313, doi: 10.5607/en.2014.23.4.292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trexler AJ & Rhoades E Alpha-synuclein binds large unilamellar vesicles as an extended helix. Biochemistry 48, 2304–2306, doi: 10.1021/bi900114z (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veldhuis G, Segers-Nolten I, Ferlemann E & Subramaniam V Single-molecule FRET reveals structural heterogeneity of SDS-bound alpha-synuclein. Chembiochem 10, 436–439, doi: 10.1002/cbic.200800644 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Ferreon AC, Gambin Y, Lemke EA & Deniz AA Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A 106, 5645–5650, doi: 10.1073/pnas.0809232106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gambin Y et al. Visualizing a one-way protein encounter complex by ultrafast single-molecule mixing. Nat Methods 8, 239–241, doi: 10.1038/nmeth.1568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JY, Meng F, Yoo J & Chung HS Diffusion-limited association of disordered protein by non-native electrostatic interactions. Nat Commun 9, 4707, doi: 10.1038/s41467-018-06866-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturzenegger F et al. Transition path times of coupled folding and binding reveal the formation of an encounter complex. Nat Commun 9, 4708, doi: 10.1038/s41467-018-07043-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevcsik E, Trexler AJ, Dunn JM & Rhoades E Allostery in a disordered protein: oxidative modifications to alpha-synuclein act distally to regulate membrane binding. J Am Chem Soc 133, 7152–7158, doi: 10.1021/ja2009554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nath A et al. The conformational ensembles of alpha-synuclein and tau: combining single-molecule FRET and simulations. Biophys J 103, 1940–1949, doi: 10.1016/j.bpj.2012.09.032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrie JJ et al. Using a FRET Library with Multiple Probe Pairs To Drive Monte Carlo Simulations of alpha-Synuclein. Biophys J 114, 53–64, doi: 10.1016/j.bpj.2017.11.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bah A et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519, 106–109, doi: 10.1038/nature13999 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Choi UB, Xiao S, Wollmuth LP & Bowen ME Effect of Src kinase phosphorylation on disordered C-terminal domain of N-methyl-D-aspartic acid (NMDA) receptor subunit GluN2B protein. J Biol Chem 286, 29904–29912, doi: 10.1074/jbc.M111.258897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi Ucheor B., McCann James J., Weninger Keith R. & Bowen, Mark E. Beyond the Random Coil: Stochastic Conformational Switching in Intrinsically Disordered Proteins. Structure 19, 566–576, doi: 10.1016/j.str.2011.01.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsh JA & Forman-Kay JD Sequence determinants of compaction in intrinsically disordered proteins. Biophys J 98, 2383–2390, doi: 10.1016/j.bpj.2010.02.006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi UB et al. Modulating the intrinsic disorder in the cytoplasmic domain alters the biological activity of the N-methyl-D-aspartate-sensitive glutamate receptor. J Biol Chem 288, 22506–22515, doi: 10.1074/jbc.M113.477810 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trexler AJ & Rhoades E N-Terminal acetylation is critical for forming alpha-helical oligomer of alpha-synuclein. Protein Sci 21, 601–605, doi: 10.1002/pro.2056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brangwynne CP, Mitchison TJ & Hyman AA Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 108, 4334–4339, doi: 10.1073/pnas.1017150108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitrea DM et al. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc Natl Acad Sci U S A 111, 4466–4471, doi: 10.1073/pnas.1321007111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee PR, Mitrea DM, Kriwacki RW & Deniz AA Asymmetric Modulation of Protein Order-Disorder Transitions by Phosphorylation and Partner Binding. Angew Chem Int Ed Engl 55, 1675–1679, doi: 10.1002/anie.201507728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitrea DM et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat Commun 9, 842, doi: 10.1038/s41467-018-0255-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitrea DM et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5, 13571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soranno A et al. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc Natl Acad Sci U S A 111, 4874–4879, doi: 10.1073/pnas.1322611111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee PR, Moosa MM & Deniz AA Two-Dimensional Crowding Uncovers a Hidden Conformation of alpha-Synuclein. Angew Chem Int Ed Engl 55, 12789–12792, doi: 10.1002/anie.201606963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganguly P, Boserman P, van der Vegt NFA & Shea JE Trimethylamine N-oxide Counteracts Urea Denaturation by Inhibiting Protein-Urea Preferential Interaction. J Am Chem Soc 140, 483–492, doi: 10.1021/jacs.7b11695 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Ferreon AC, Moosa MM, Gambin Y & Deniz AA Counteracting chemical chaperone effects on the single-molecule alpha-synuclein structural landscape. Proc Natl Acad Sci U S A 109, 17826–17831, doi: 10.1073/pnas.1201802109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi KJ et al. A Chemical Chaperone Decouples TDP-43 Disordered Domain Phase Separation from Fibrillation. Biochemistry, doi: 10.1021/acs.biochem.1028b01051, doi: (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakon JJ & Weninger KR Detecting the conformation of individual proteins in live cells. Nat Methods 7, 203–205, doi: 10.1038/nmeth.1421 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konig I et al. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat Methods 12, 773–779, doi: 10.1038/nmeth.3475 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Hoffmann A et al. Quantifying heterogeneity and conformational dynamics from single molecule FRET of diffusing molecules: recurrence analysis of single particles (RASP). Phys Chem Chem Phys 13, 1857–1871, doi: 10.1039/c0cp01911a (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crawford R et al. Long-lived intracellular single-molecule fluorescence using electroporated molecules. Biophys J 105, 2439–2450, doi: 10.1016/j.bpj.2013.09.057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sustarsic M et al. Optimized delivery of fluorescently labeled proteins in live bacteria using electroporation. Histochem Cell Biol 142, 113–124, doi: 10.1007/s00418-014-1213-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12, 244–250, 243 p following 250, doi: 10.1038/nmeth.3256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikic I, Kang JH, Girona GE, Aramburu IV & Lemke EA Labeling proteins on live mammalian cells using click chemistry. Nat Protoc 10, 780–791, doi: 10.1038/nprot.2015.045 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Das DK et al. Direct Visualization of the Conformational Dynamics of Single Influenza Hemagglutinin Trimers. Cell 174, 926–937 e912, doi: 10.1016/j.cell.2018.05.050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreon AC, Ferreon JC, Wright PE & Deniz AA Modulation of allostery by protein intrinsic disorder. Nature 498, 390–394, doi: 10.1038/nature12294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuxreiter M Fold or not to fold upon binding - does it really matter? Curr Opin Struct Biol 54, 19–25, doi: 10.1016/j.sbi.2018.09.008 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Milles S et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 163, 734–745, doi: 10.1016/j.cell.2015.09.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borgia A et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66, doi: 10.1038/nature25762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soranno A et al. Integrated view of internal friction in unfolded proteins from single-molecule FRET, contact quenching, theory, and simulations. Proc Natl Acad Sci U S A 114, E1833–E1839, doi: 10.1073/pnas.1616672114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soranno A et al. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci U S A 109, 17800–17806, doi: 10.1073/pnas.1117368109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kilic S et al. Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1alpha. Nat Commun 9, 235, doi: 10.1038/s41467-017-02619-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langdon EM et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927, doi: 10.1126/science.aar7432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elbaum-Garfinkle S et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 112, 7189–7194, doi: 10.1073/pnas.1504822112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu P & Craighead HG Zero-mode waveguides for single-molecule analysis. Annu Rev Biophys 41, 269–293, doi: 10.1146/annurev-biophys-050511-102338 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Levene MJ et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686, doi: 10.1126/science.1079700 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Milles S, Koehler C, Gambin Y, Deniz AA & Lemke EA Intramolecular three-colour single pair FRET of intrinsically disordered proteins with increased dynamic range. Mol Biosyst 8, 2531–2534, doi: 10.1039/c2mb25135c (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee TC, Moran CR, Cistrone PA, Dawson PE & Deniz AA Site-Specific Three-Color Labeling of alpha-Synuclein via Conjugation to Uniquely Reactive Cysteines during Assembly by Native Chemical Ligation. Cell Chem Biol 25, 797–801 e794, doi: 10.1016/j.chembiol.2018.03.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gambin Y & Deniz AA Multicolor single-molecule FRET to explore protein folding and binding. Mol Biosyst 6, 1540–1547, doi: 10.1039/c003024d (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clamme JP & Deniz AA Three-color single-molecule fluorescence resonance energy transfer. Chemphyschem 6, 74–77, doi: 10.1002/cphc.200400261 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Hohng S, Joo C & Ha T Single-molecule three-color FRET. Biophys J 87, 1328–1337, doi: 10.1529/biophysj.104.043935 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chung HS et al. Oligomerization of the tetramerization domain of p53 probed by two- and three-color single-molecule FRET. Proc Natl Acad Sci U S A 114, E6812–E6821, doi: 10.1073/pnas.1700357114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoo J, Louis JM, Gopich IV & Chung HS Three-Color Single-Molecule FRET and Fluorescence Lifetime Analysis of Fast Protein Folding. J Phys Chem B, doi: 10.1021/acs.jpcb.8b07768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dalal V, Bhattacharya M, Narang D, Sharma PK & Mukhopadhyay S Nanoscale Fluorescence Imaging of Single Amyloid Fibrils. J Phys Chem Lett 3, 1783–1787, doi: 10.1021/jz300687f (2012). [DOI] [PubMed] [Google Scholar]
- 80.Lee TC et al. Dual Unnatural Amino Acid Incorporation and Click-Chemistry Labeling to Enable Single-Molecule FRET Studies of p97 Folding. Chembiochem 17, 981–984, doi: 10.1002/cbic.201500695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gambin Y, Simonnet C, VanDelinder V, Deniz A & Groisman A Ultrafast microfluidic mixer with three-dimensional flow focusing for studies of biochemical kinetics. Lab Chip 10, 598–609, doi: 10.1039/b914174j (2010). [DOI] [PubMed] [Google Scholar]
- 82.Vandelinder V, Ferreon AC, Gambin Y, Deniz AA & Groisman A High-resolution temperature-concentration diagram of alpha-synuclein conformation obtained from a single Forster resonance energy transfer image in a microfluidic device. Anal Chem 81, 6929–6935, doi: 10.1021/ac901008c (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hohng S et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science 318, 279–283, doi: 10.1126/science.1146113 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berlow RB, Dyson HJ & Wright PE Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature 543, 447–451, doi: 10.1038/nature21705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bentley EP, Frey BB & Deniz AA Physical Chemistry of Cellular Liquid-Phase Separation. Chemistry 25, 5600–5610, doi: 10.1002/chem.201805093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


