Abstract
Background:The fiddler crab Ucasindensis (Alcock, 1900) (Crustacea: Brachyura: Ocypodidae) is distributed in the northern coasts of the Arabian Sea (Pakistan, Iran, Iraq, and Kuwait). Its typical habitat is on high intertidal areas with higher salinity, which might restrict its distribution, especially within the Persian Gulf. The purpose of the present phylogeographicstudy is to understand whether the Strait of Hormuz acts as a barrier to the gene flow of this species.
Results:The genetic analyses of the mitochondrial 16S rRNA, cytochrome oxidase subunit I (COI), and control region (CR) of specimens from various localities showed that there was no genetic differentiation between the populations inside and outside of the Persian Gulf.
Conclusions:We conclude that the narrow Strait of Hormuz does not form a barrier for the larval dispersal in this species. Its restricted distribution in the northern Arabian Sea may instead be associated with its preference for higher salinity sediments present in the coasts of thisregion.
Keywords: Uca sindensis, Phylogeography, Arabian Sea, Persian Gulf, 16S rRNA, COI, Control region
BACKGROUND
Ucasindensis(Alcock, 1900) (Ocypodidae) is a species of fiddler crab with a relatively restricted distribution in the northern Arabian Sea. Described from Karachi, Pakistan (Alcock 1900), it has since been reported from Iran, Iraq, and Kuwait (Collins et al. 1984; Yamaguchi 1994; Apel and Türkay 1999; Naser et al. 2010; Mokhlesi et al. 2011; Naderloo and Türkay 2012). Its ecology and behavior have been studied in Kuwait (Collins et al. 1984), Iran (Mokhlesi et al. 2011), and Pakistan (Saher and Qureshi2012).
With regard to the taxonomy of U. sindensis, Alcock (1900) originally considered it as a variety of Ucainversa(Hoffmann, 1874), a decision followed by Crane (1975). She treated it as a subspecies of U. inversaand estab- lished a subgenus Amphiuca(= ParaleptucaBott, 1973) for the two taxa that also included Ucachlorophthalmus(H. Milne Edwards, 1837) and Ucacrassipes(White, 1847). However, Beinlich and von Hagen (2006) estab- lished a new subgenus, Cranuca, exclusively for U. inversaon the basis of its unusual pleonal structure and form of the dactylar tooth of the major chela. They also transferred the other taxa in Crane's (1975) Indo-West Pacific Celuca(= AustrucaBott, 1973) to the subgenus Paraleptuca, an action later followed by Ng et al. (2008). However, Beinlich and von Hagen's (2006) definition of Paraleptucaseems to be paraphyletic (Naderloo et al. 2010). Shih et al. (2013a,b) studied the phylogeny of the Indo-Pacific broad-fronted group and the results support the concept of the subgenera Austruca(= Crane's Celucafor Indo-West Pacific taxa), Parathelphuca, and Cranuca, withU.sindensisincludedinAustruca.Themorphological similarity of U. sindensisand U. inversa(cf. Alcock 1900; Crane 1975) thus appears to be superficial. In fact, Alcock (1900) had mentioned that U. sindensisresembles Ucaannulipes,andthishasbeensupportedgenetically(Shih et al.2013b).
Most phylogenetic and phylogeographic studies of marine crabs use the mitochondrial 16S rRNA and cyto- chrome oxidase subunit I (COI) genetic markers (e.g., Levinton et al. 1996; Sturmbauer et al. 1996; Shih and Suzuki 2008; Shih et al. 2009, 2010, 2012; Davie et al.
2010; Wong et al. 2010, 2011, 2012). In addition, the mitochondrial control region marker, with highly variable and fast divergence rates (Avise2000), has also recently been used for phylogeographic studies of crabs, e.g., Portunustrituberculatus(Miers, 1876) (Portunidae) (Cho et al. 2009; Guo et al. 2012), U. crassipes(Aoki andWada 2013), Ucaboninensis(Ocypodidae) (Shih et al. 2013a), Grapsusgrapsus(Linnaeus, 1758) (Grapsidae) (Ströheret al. 2011), Episesarma versicolor (Tweedie, 1940) (Sesarmidae) (Supmee et al. 2012), Ucidescordatus(Linnaeus, 1763) (Oliveira-Neto et al. 2007) (Ucididae), Cardisomaguanhumi(Latreille, in Latreille, Le Peletier, Serville&Guérin, 1828) (Oliveira-Neto et al. 2008), and Discoplax celeste Ng and Davie, 2012 (Gecarcinidae) (Turner et al.2013).
The Persian Gulf is a shallow semi-enclosed basin with average depth of only 35 m. It has a low-salinity current flowing towards the northwest from the Gulf of Omanand along the coast of Iran, forming a series of eddies around the Central Trough of the Gulf. It also has a high-salinity current flowing towards the southeast, mainly along the coast of Arabia and out through the Strait of Hormuz (Thoppil and Hogan 2010). The nar-row (approximate width 50 km) outlet of the Strait of Hormuz has a major influence on the circulation and salinity inside the Gulf, making it quite different from conditions outside (Johns et al. 2003). As such, it mayact as a barrier to the gene flow of marine organisms, resulting in different genetic structures inside and out- side the Gulf. Examples studied showing this pattern in- clude corals (Eghtesadi-Araghi 2011), barnacles (Tsang et al. 2012), sea cucumber (Claereboudt and Al-Rashdi 2011), and reef fishes (Burt et al. 2011). However, the Persian Gulf was completely dried in glacial periods due to its shallow depth, with marine organisms reestablish- ing themselves in the Gulf after that. This suggests that historic factors are less important in determining geo- graphical distribution of marine organisms (Tsang et al. 2012).
Compared with the Persian Gulf, another semi-enclosed basin,theRedSea,hasaverydifferentgeographyandgeo- logical history, with a prominent sill to constrain the out- flow and the formation of land bridge across the Arabian Sea during glaciation events (Johns et al. 2003; Sirocko 2003); resulting in a higher endemism (Tsang et al. 2012; DiBattista et al.2013).
To understand whether the larval dispersal of U. sin- densisis blocked by the narrow Strait of Hormuz, we examine the genetic variation of 16S, COI, and control region (CR) from different localities of its range. The possible biogeographic mechanisms for the limited dis- tribution of this species are discussed.
METHODS
The specimens of U. sindensis have been collected from Pakistan, Iran, and Iraq (Table 1; Figure 1), preserved in 70%to 95% ethanol and deposited in Zoological Collec- tions of the Department of Life Science,National Chung Hsing University, Taichung, Taiwan (NCHUZOOL) and Zoological Reference Collection, Raffles Museum ofBio- diversity Research, National University ofSingapore (ZRC). GenomicDNA was isolated from the muscle tissue of thelegs by using the GeneMark tissue and cell genomic DNA purification kit (Taichung, Taiwan). A region of ap- proximately 550 basepairs (bp) of the 5′-end of the 16S gene was selected for amplificationwith polymerase chain reaction (PCR) using the primers 1471and 1472 (Crandall and Fitzpatrick 1996). A portion ofthe COI genewas amplified with PCR using the primers LCO1490, HCO2198 (Folmer et al. 1994), and COH6(Schubart and Huber 2006). The PCR conditionsfor the above primers weredenaturation for 50 s at 94°C, annealing for 70 s at 45°Cto 47°C, and extension for 60 s at 72°C (40 cycles), followed by extension for 10 min at 72°C. The primers for aregion of approximately 960 bp of CR gene are the re- cently designed CR-F4 (Shih et al. 2013a) and ILEUCAR3 (Pie et al. 2008), with theannealing temperature of 45°C to50°C in PCR condition. Sequences were obtained by au- tomatedsequencing (Applied Biosystems 3730, Applied Biosystems Inc., Foster City, CA, USA) and were aligned manually, with the aid of ClustalW v 1.4 (Thompsonet al. 1994), after verification with thecomplementary strand. According to Shih et al.(2013b), U. sindensis belongs to themonophyletic Austruca, so other consubgeneric spe- ciesfrom Indian Ocean are used as outgroups, viz. Ucaalbimana (Kossmann, 1877), U. annulipes(H. Milne Edwards, 1837), Uca aff. annulipes, Ucabengali Crane, 1975,and Ucairanica, Pretzmann, 1971. Sequences of different haplotypes have been deposited inthe DNA DataBank of Japan (DDBJ), with other sequences which werepublished in earlier papers of author HTS (accession numbers see Table 1).
Table1 Haplotypes of 16S rRNA, COI, and CR genes.
| Species | Locality | Samplesize | Haplotypeof16S | DDBJ accession number | HaplotypeofCOI | DDBJ accession number | HaplotypeofCR | DDBJ accession number |
| U.sindensis | Pakistan:Sandspit, Karachi[1] | 1 | Usd-1 | AB813652 | Usd-C2 | LC015060 | Usd-CR1 | LC015065 |
| 1 | Usd-1 | AB813652 | Usd-C3 | LC015061 | Usd-CR2 | LC015066 | ||
| 1 | Usd-1 | AB813652 | Usd-C4 | LC015062 | Usd-CR3 | LC015067 | ||
| 1 | Usd-2 | LC015053 | Usd-C1 | AB813673 | Usd-CR4 | LC015068 | ||
| 2 | Usd-1 | AB813652 | Usd-C1 | AB813673 | Usd-CR5,6 | LC015069, LC015070 | ||
| 2 | Usd-1 | AB813652 | Usd-C2 | LC015060 | Usd-CR7,8 | LC015071, LC015072 | ||
| 1 | Usd-3 | LC015054 | Usd-C1 | AB813673 | Usd-CR9 | LC015073 | ||
| 1 | Usd-4 | LC015055 | Usd-C1 | AB813673 | - | |||
| Pakistan:Sonmiani, Balochistan[2] | 1 | Usd-1 | AB813652 | Usd-C1 | AB813673 | Usd-CR10 | LC015074 | |
| 1 | Usd-1 | AB813652 | Usd-C2 | LC015060 | Usd-CR11 | LC015075 | ||
| 1 | Usd-5 | LC015056 | Usd-C2 | LC015060 | Usd-CR12 | LC015076 | ||
| 1 | Usd-6 | LC015057 | Usd-C2 | LC015060 | Usd-CR13 | LC015077 | ||
| 1 | Usd-1 | AB813652 | Usd-C3 | LC015061 | - | |||
| Iran:Qeshm [3] | 1 | Usd-1 | AB813652 | Usd-C1 | AB813673 | Usd-CR14 | LC015078 | |
| 1 | Usd-1 | AB813652 | Usd-C2 | LC015060 | Usd-CR15 | LC015079 | ||
| 2 | Usd-1 | AB813652 | Usd-C1 | AB813673 | Usd-CR16,17 | LC015080, LC015081 | ||
| Iran:Bandar Abbas [4] | 1 | - | Usd-C2 | LC015060 | Usd-CR18 | LC015082 | ||
| 1 | Usd-7 | LC015058 | Usd-C1 | AB813673 | Usd-CR19 | LC015083 | ||
| 3 | Usd-1 | AB813652 | Usd-C2 | LC015060 | Usd-CR20,21, 23 | LC015084, LC015085, LC015087 | ||
| 1 | Usd-1 | AB813652 | Usd-C5 | LC015063 | Usd-CR22 | LC015086 | ||
| Iraq:Khur Al-Zubair [5] | 1 | Usd-8 | LC015059 | Usd-C2 | LC015060 | Usd-CR24 | LC015088 | |
| 1 | Usd-1 | AB813652 | Usd-C2 | LC015060 | Usd-CR24 | LC015088 | ||
| Outgroups | ||||||||
| U.bengali | Thailand:Ranong | AB813651 | LC015064 | - | ||||
| U.annulipes | India:Tamil Nadu, PortoNovo | AB471894 | AB471907 | - | ||||
| U.aff.annulipes | Madagascar | AB813648 | AB813669 | - | ||||
| U.iranica | Iran:Hormozgan, Qeshm | AB471897 | AB471911 | - | ||||
| U.albimana | Egypt:Sinai | AB471893 | AB471906 | - |
Haplotypesof 16S rRNA, COI, and control region (CR) genes for the specimens of Ucasindensis from the Arabian Sea and the outgroups. The numbers within localities correspond to those in Figure 1
Fig. 1.
Figure 1 Collection sites. Collection sites (red solid circles; nos. 1 to 4) for specimens of Uca sindensis used in this study. Red empty circle (no. 5) means the additional record of Kuwait (Collins et al. 1984).
For a combined analysis of 16S and COI markers, phylogenetic congruence among the two dataset parti- tions was tested under the maximum parsimony (MP) criterion using the incongruent length difference (ILD) test (Farris et al. 1994) implemented in the PAUP* pro- gram v 4.0b10 (Swofford 2003) as the partition homo- geneity test. The parameters included 1,000 reiterations of a heuristic search with 100 randomly added sequence replications and tree bisection-reconnection (TBR) branch- swapping, using Steepest Descent and the MULTREES option enabled, and total number of rearrangements for each search was limited to 500,000 to avoid excessive computation time. The topologies of the two datasets were congruent (p = 0.901) and as such, the sequences were combined.
Maximum likelihood (ML) analysis was conducted in RAxML v 7.2.6 (Stamatakis 2006) for the combined dataset. The model GTR + G (i.e., GTRGAMMA)was used for all subsets with 100 runs and found the best ML tree by comparing the likelihood scores. The robust- ness of the ML tree was evaluated by 1,000 bootstrap pseudoreplicates under the model GTRGAMMA. A consensus MP tree was constructed using PAUP* with 2,000 bootstrap replications of a simple heuristic search, TBR branch-swapping, and 100 random addition se- quence replications. Gaps in MP tree construction were treated as missing. All characters were equally weighted. To examine the relationships of the combined 16S and COI haplotypes in detail, a gene genealogy was con- structed using TCS v 1.20 (Clement et al. 2000), with the treatment of gaps as missing states.
To understand whether there is genetic differentiation between the populations inside and outside the Persian Gulf, we treated the specimens from Iran and Iraq (nos. 3 and 4; Figure 1) as one population, and another popula- tion is composed of specimens from Pakistan (nos. 1 and 2; Figure 1). The nucleotide diversity (π), number of hap- lotypes (nh), and haplotype diversity (h) for each popula- tion were estimated by using the program DnaSP v
5.10.01 (Librado and Rozas 2009). To determine whether patterns of sequence variation was consistent with predic- tion of the neutral model, Tajima's D (Tajima 1989) and Fu's FS (Fu 1997) tests were calculated by Arlequin v3.5.1.2 (Excoffier et al. 2005), with 1,000 permutationsfor the significance assessment of the neutrality test. To deter- mine whether a population expansion is valid, the mis- match distribution, with the Harpending's raggedness index (Hri, Harpending 1994), was computed by Arlequin using the demographic expansion model for pairwise dif- ferences with 1,000 bootstrap replicates. If the Hri value is significant (p < 0.05), the sudden population expansion model should be rejected (Schneider and Excoffier 1999). To estimate the degree of differentiation between popula- tions, pairwise FST between groups was calculated by Arlequin. Sometimes the value of FST is negative, which was discussed by several authors and we follow the usual treatment to round it to zero (Weir 1996; Roesti et al. 2012; Willing et al. 2012).
RESULTS
A 548-bp segment of the 16S was amplified and aligned; six positions are variable and none areparsimoniously informative. Among the total number of sequences, eight different haplotypes are distinguished (Table 1). The studied segment of 16S sequences is AT rich (68.7%) (T, 36.0%; A, 32.7%; G, 19.9%; and C, 11.4%). For the COI gene, a 658-bp segment was compared, resulting in five different haplotypes (Table 1). The studied segment of the COI sequence is also AT rich (58.4%) (T, 32.2%; A, 27.2%; G, 17.8%; and C, 23.8%). In this gene, five positions are variable and two parsimoniously informative. A 963-bp segment of CR was compared, and 24 differenthaplotypes were obtained (Table 1). The segment of CR is AT rich (75.8%) (T, 35.1%; A, 40.7%; G, 10.1%; C, 14.1%), with 75 positions variable and 34 parsimony informative.
The genealogic tree of the combined 16S and COI (Figure 2) shows that there is no subdivision of U. sindensis specimens from various localities. The haplo- type network of the combined dataset (Figure 3) shows the haplotype ‘Usd-1 + Usd-C1’ to be central, relative to all other haplotypes. As such, it is here regarded as the ancestral haplotype (cf. Clement et al. 2000). It is also clear that a lot of individuals, inside and outside the Gulf, share the same haplotypes. Al- though some haplotypes seem to exclusively appear in each locality, their difference from the common ones is only 1 or 2 bp.
Fig. 2.
Figure 2 A maximum likelihood (ML) tree for Uca sindensis and outgroups. The maximum likelihood (ML) tree is based on the combined 16S and COI markers. Probability values at the nodes represent support values for ML and maximum parsimony (MP). For haplotype names, see Table 1.
Fig. 3.
Figure 3 Genealogical network for the combined 16S and COI haplotypes observed from Uca sindensis. The specimens were taken from the Arabian Sea. The ancestral haplotype is indicated by a square. The unlabeled node indicates haplotype not found in the sampled populations. For haplotype names, see Table 1.
The molecular diversity indices for 16S, COI, and CR are shown in Table 2. For the 16S of all populations, the nucleotide diversity per site (π) = 0.076 and the haplo- type diversity (h) = 0.380; for COI, π = 0.13 and h = 0.63; and for CR, π = 1.35 and h = 0.996. The divergence of COI is about five times that of 16S and CR is about 3.5 times that of COI.
Table2 The measures of 16S, COI, and CR.
| n | nh | π (%) | h | |
| 16S | ||||
| Insidethe Gulf | 11 | 3 | 0.067 | 0.345 |
| Outsidethe Gulf | 13 | 4 | 0.084 | 0.423 |
| Total | 24 | 6 | 0.076 | 0.380 |
| COI | ||||
| Insidethe Gulf | 11 | 3 | 0.138 | 0.618 |
| Outsidethe Gulf | 13 | 4 | 0.129 | 0.679 |
| Total | 24 | 5 | 0.130 | 0.630 |
| CR | ||||
| Insidethe Gulf | 11 | 10 | 1.406 | 0.982 |
| Outsidethe Gulf | 13 | 13 | 1.290 | 1 |
| Total | 24 | 23 | 1.351 | 0.996 |
Themeasures of 16S, COI, and control region (CR) diversity for specimens of Ucasindensis from the Arabian Sea. Inside the Gulf includes Iraq and Iran; outside the Gulf includes only Pakistan. n, number of sampled individuals; nh, number of recovered haplotypes; π, nucleotide diversity; h, haplotype diversity
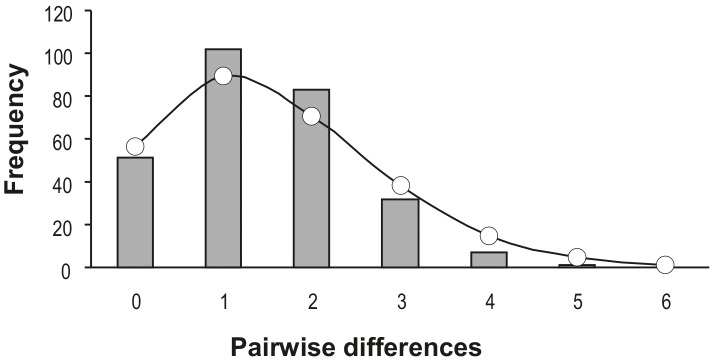
The pairwise FST value between populations is −0.0309 (and has rounded to zero) which implies that the gene flow between them is very high. As a result, the two populations are combined into a single population. Tajima's D is −1.7519 (p = 0.02) and Fu's FS value is −27.9037(p< 0.001). The significant negative values of the neutrality tests may imply that the population has ex- perienced a recent expansion (Tajima 1989; Fu 1997). The Hri value is 0.08172 (p= 0.29), which is not signifi- cant and also implies that the population might experi- ence sudden population expansion. In addition, the mismatch distribution is smooth and unimodal (Figure 4), with the simulated sum of squared deviation (SSD) being 0.00555 (p= 0.44), which also supports the recent popu- lation expansion.
Fig. 4.
Figure 4 Mismatch distribution for the populations of Uca sindensis from the Arabian Sea. The observed pairwise differences are shown in bars, and the expected values under the sudden expansion model are in solid line.
DISCUSSION
The results of the present study suggest that the Strait of Hormuz is not a barrier to gene flow between the populations of U. sindensis inside and outside the Gulf, at least on the basis of the low nucleotide diversities of 16S (0.08%), COI (0.13%), and CR (1.35%) (Table 2), genealogic tree (Figure 2), haplotype network (Figure 3), and the estimate of gene flow (FST).
Low genetic variability of COI has been reported in other Uca species, which is usually attributed to the high levels of larval dispersal, e.g., U. annulipes (= U. aff. annulipes, see Shih et al. 2013b) from East Africa (0.04%, Silva et al. 2010), U. crassipes from eastern Indian Ocean to Central Pacific Ocean (0.02%, Shih et al. 2012), and Ucamaracoani from Brazilian coast (0.291%, Wieman et al. 2014). However, some Uca species and other intertidal crabsare known to show relatively high genetic variability, e.g., Uca pugnax (Smith, 1870) from the Atlantic coast of USA (0.61%, Sanford et al. 2006), Uca splendida(Stimpson, 1858) from Vietnam and East Asia (0.82%, Shih et al. 2012), and Neohelice granulata (Dana, 1851) (Varunidae) from Brazil (0.706%, Ituarte et al. 2012), which may be explained by past historical barriers to theirlarval dispersal that may have affected their behav- ior, physiology,etc. (McEdward 1995; Brodie et al. 2007; Anger et al. 2008; López-Duarte et al. 2011; discussed below).
The genetic variability of U. sindensis, with a nucleo- tide diversity of 1.35% of CR, is 3.5 times that of the COI (Table 2). This CR divergence is relatively low compared with most crab species with wide distributions like Portunus trituberculatus (2.72%, Cho et al. 2009; 2.05%, Guo et al. 2012), Cardisoma guanhumi (3.0%, Oliveira-Neto et al. 2008; 3.1%, Pie et al. 2008), and Ucides cordatus (3.5%, Oliveira-Neto et al. 2007; 4.4%, Pie et al. 2008). Even a species with a restricted distribu- tion like U. boninensis (known only from the Ogasawara (= Bonin) Islands, Japan) has a high nucleotide diversity (3.18%, Shih et al. 2013a). The pattern is not always easy to decipher; some species with a low diversity like D. ce- leste (1.0%, Turner et al. 2013), has a small range and is endemic to Christmas Island in the Indian Ocean, while E. versicolor (0.7%, Supmee et al. 2012) (studied along the Andaman Sea Coast of Thailand) has awide Southeast Asian distribution. The highly variable CR marker may therefore provide another line of evidence to better understand the genetic structure and molecular diversity at the population and species level.
The neutrality tests and mismatch distribution (Figure 4) suggest that U. sindensis has undergone a recent expan- sion from a single source, with the most common haplotype ‘Usd-1 + Usd-C1’ likely to be the ancestral one (Figure 3) (Clement et al. 2000; Avise 2009). Rarer haplotypes, with only 1 or 2 bp difference from the most common haplotype (Figure 3), may have been de- rived relatively recently through separate mutations (Avise 2009).
Uca sindensis is restricted to the northern Arabian Sea (Pakistan, Alcock 1900) and the northern and north- western coasts of the Persian Gulf (Kuwait, Collins et al. 1984; Iran, Apel and Türkay 1999; Iraq, Naser et al. 2010). An analysis of the fiddler crab fauna in this region may help understand the factors affecting the distribu- tion of U. sindensis. In Pakistan, U. sindensis, Uca urvil- lei, U.annulipes, and U.iranica have been reported (Alcock 1900; Crane 1975; Saher and Qureshi 2012; Saheret al. 2014); Iran has U.sindensis, U.iranica, and
U. inversa (Pretzmann 1971; Apel and Türkay 1999; Shih et al. 2009; Naderloo and Türkay 2012), while Kuwait has U. sindensis and U. iranica (Collins et al. 1984; Apel and Türkay 1999). Interestingly, no Uca spe- cies has been reported from the southern coasts of the Persian Gulf, and Apel and Türkay (1999) speculated that this may be due to the very high salinity of the area. An exception is the sea in United Arab Emirates (UAE) which has relatively lower salinity, and here, five species are present: U. iranica, U. albimana, U. inversa, Uca hesperiae, and Uca tetragonon (Apel and Türkay 1999; see below). It is interesting that U.albimana, U.inversa,
U. hesperiae, and U. tetragonon also occur in the western part of the Indian Ocean (i.e., in the Red Sea and along East Africa) and are believed to be widespread because their larvae are dispersed by currents along the African coast (Apel and Türkay 1999). Only U. iranica has a comparatively restricted range.
The importance of salinity as a limiting factor for both larvae and adults of Uca species has previously been em- phasized (Crane 1975; Hedgecock 1986; Anger 2003; Spivak and Cuesta 2009; Hopkins and Thurman 2010; López-Duarte et al. 2011; Thurman et al. 2013). Salinity has also been proposed as a major factor controlling the crab distribution in the Persian Gulf, especially for Uca species by Apel and Türkay (1999). They hypothesized that areas in the Persian Gulf without Uca are because of the very high salinities present (>40‰), with the crabs preferring habitats with salinities <39‰. It is likely that the distribution of U. sindensis, from Pakistan to the northwestern Persian Gulf, is also influenced by salinity.
Although U. sindensis is sometimes sympatric with U. iranica, its preferred habitat is in higher intertidal areas or even in sheltered shores far from the sea (Crane 1975; Collins et al. 1984; Mokhlesi et al. 2011). These are gen- erally habitats with higher salinities than open waters. Within the Persian Gulf and the Gulf of Oman, U. sin- densis (and other Uca spp.) occur in the coast of Iran, Iraq, and Kuwait, areas that have salinities of between 36‰ to 37‰ at least in summer (Apel and Türkay 1999; Johns et al. 2003; Naderloo and Türkay 2012). In thehabi- tats in Pakistan (Sandspit and Korangi, Karachi), the salin- ity was always above 36.0‰ (Saher 2008). From the Gulf of Oman to Pakistan, the salinity is relatively lower (36.5‰ to 36‰) (Chodankar et al. 2005), but still high enough for U. sindensis to survive. Once we reach the western Indian subcontinent, the salinities become even lower (34‰ to 36‰) (Subrahmanyam et al. 2011), and interestingly, there are no records of U. sindensis here. From the known range at least, the preferred salinity for
U. sindensis appears to be between 36‰ to 39‰, and ac- cording to Thurman et al. (2013), it should be classified as a hypersaline species since this value exceed 35‰. Other hypersaline species are Uca burgersi Holthuis, 1967, Uca panacea Novak and Salmon, 1974, Uca rapax (Smith, 1870), and Uca subcylindrica (Stimpson, 1859) from America (Thurman 2002; Thurman et al. 2013), U. inversa (Hoffmann, 1874) from the Indian Ocean (Spaargaren 1977), and Uca formosensis Rathbun, 1921 from Taiwan (Shih et al. 1999, 2005; Shih 2008).
López-Duarte et al. (2011) found that the larvae of es- tuarine Uca species have a rhythmic vertical swimming behavior following hatching an action that promotes sea- ward transport (and dispersal) due to the physiological stresses to low salinities in estuaries. Coastal species with euhaline habitats, on the other hand, exhibit only weakly rhythmic or arrhythmic behavior. The habitats and the adjacent coastal areas of U. sindensis are highly saline (see above) which is presumably suitable for the larval development, there is no need to move too far, and as such, the larvae only have weak or no vertical migration behavior (López-Duarte et al. 2011). Surface currents therefore have a greater influence, with larvae trans- ported primarily by advection from adult habitats (Queiroga and Blanton 2005; López-Duarte et al. 2011). As such, populations may become localized due to the direction of surface currents and the hypersaline habits of the species; explaining the low genetic differentiation observed between localities. This habitat salinity hypoth- esis could explain the low genetic variability of U. sin- densis in this study as well as that of other coastal and oceanic species like U. crassipes (Shih et al. 2012). This is in contrast to the high genetic variability present in estuarine U. splendida (Shih et al. 2012), U. pugnax (Sanford et al. 2006; López-Duarte et al. 2011), and N. granulata (Ituarte et al. 2012). The current understand- ing is that the distribution of many marine organisms in the hypersaline Red Sea have been influenced by salinity (e.g., the barnacles, Tsang et al. 2012; reef fishes, DiBattista et al. 2013). This evidence suggests that salinity plays an important role in the distribution of intertidal and shallow subtidal organisms which face extreme environment stresses frequently.
Further studies on the larval development and behavior, as well as the salinity and temperature tolerance for larvae and adults, of U. sindensis will be important to better understand how these aspects affect their distribution.
CONCLUSIONS
Based on the low nucleotide diversities of mitochondrial 16S, COI, and control region (CR), genealogic tree, haplotype network and the estimate of gene flow, which suggest high levels of larval dispersal, we conclude there was no genetic differentiation between the populations of Uca sindensis inside and outside of the Persian Gulf; and the narrow Strait of Hormuz does not form a barrier for the larval dispersal in this species. Its restricted dis- tribution in the northern Arabian Sea may instead be as- sociated with its preference for sediments with higher salinity (36‰–39‰) present in the coasts of this region and U. sindensis is best classified as a hypersaline spe- cies. The neutrality tests and mismatch distribution also suggest that this species has undergone a recent expan- sion from a single source, with some rarer haplotypes the result of relatively recent but separate mutations.
Acknowledgments
Acknowledgements
This study was supported by grants from the National Science Council (NSC 98-2621-B-005-001-MY3, 101-2621-B-005-001-MY3), Executive Yuan, Taiwan, to HTS. Thanks are also due to the members of the HTS's laboratory for their help in molecular work. We acknowledge two anonymous referees who greatly improved this manuscript.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Authors’ contributions: HTS conceived this study, performed the molecular genetic analysis, and drafted the manuscript. NUS and EK collected and processed the samples and drafted the manuscript. PKLN participated in the discussion and drafted the manuscript. YCL carried out the molecular work and helped draft the manuscript. MYL conceived this study and carried out the molecular work. All authors read and approved the final manuscript.
References
- Alcock A. Materials for a carcinological fauna of India. J Asia Soc Bengal. 69:279–456. [Google Scholar]
- Anger K. Salinity as a key parameter in the larval biology of decapod crustaceans. Invertebr Reprod Develop. 43:29–45. [Google Scholar]
- Anger K, Spivak E, Luppi T, Bas C, Ismael D. Larval salinity tolerance of the South American salt marsh crab, Neohelice (Chasmagnathus) granulata: physiological constraints to estuarine retention, export and reimmigration. Helgol Mar Res. 62:93–102. [Google Scholar]
- Aoki M, Wada K. Genetic structure of the wide-ranging fiddler crab Uca crassipes in the west Pacific region. J Mar Biol Ass UK. 93:789–795. [Google Scholar]
- Apel M, Türkay M. Taxonomic composition, distribution and zoogeographic relationships of the grapsid and ocypodid crab fauna of intertidal soft bottoms in the Arabian Gulf. Estuar Coast Shelf Sci. 49:131–142. [Google Scholar]
- Avise J C. Phylogeography: the history and formation of species. Cambridge): Harvard University Press; 2000. [Google Scholar]
- Avise J C. Phylogeography: retrospect and prospect. J Biogeogr. 36:3–15. [Google Scholar]
- Beinlich B. Materials for a more stable subdivision of the genus Uca Leach. Zool Meded Leiden. 80:9–32. [Google Scholar]
- Brodie R J, Styles R, Borgianini S, Godley J, Butler K. Larval mortality during export to the sea in the fiddler crab Uca minax. Mar Biol. 152:1283–1291. [Google Scholar]
- Burt J A, Feary D A, Bauman A G, Usseglio P, Cavalcante G H, Sale P F. Biogeographic patterns of reef fish community structure in the northeastern Arabian Peninsula. ICES J Mar Sci. 68:1–9. [Google Scholar]
- Cho E M, Min G S, Kanwal S, Hyun Y S, Park S W, Chung K W. Phylogenetic analysis of mitochondrial DNA control region in the swimming crab, Portunus trituberculatus. Anim Cells Syst. 13:305–314. [Google Scholar]
- Chodankar A R, Banakar V K, Oba T. Past 100 ky surface salinity gradient response in the Eastern Arabian Sea to the summer monsoon variation recorded by δ18 O of G. sacculifer. Glob Planet Change. 47:135–142. [Google Scholar]
- Claereboudt M R, Km Al-Rashdi. Shallow-water sea cucumber inventory in the Sultanate of Oman. SPC Beche-de-mer. Inform Bull. 31:25–29. [Google Scholar]
- Clement M, Posada D, Crandall K A. TCS: a computer program to estimate gene genealogies. Mol Ecol. 9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Collins M J, Jones D A, Da Clayton. Redescription of Uca sindensis (Alcock, 1900) (Brachyura: Ocypodidae) with notes on the ecology of a population from Kuwait. J Crustacean Biol. 4:318–328. [Google Scholar]
- Crandall K A, Fitzpatrick Jfj. Crayfish molecular systematics: using a combination of procedures to estimate phylogeny. Syst Biol. 45:1–26. [Google Scholar]
- Crane J. Fiddler crabs of the world (Ocypodidae: genus Uca) Princeton): Princeton University Press; 1975. [Google Scholar]
- Davie Pjf, Shih H T, Chan Bkk. A new species of Mictyris (Decapoda, Brachyura, Mictyridae) from the Ryukyu Islands. Japan. Crustaceana Monogr. 11:83–105. [Google Scholar]
- Dibattista J D, Berumen M L, Gaither M R, Rocha L A, Eble J A, Choat J H, Craig M T, Skillings D J, Bowen B W. After continents divide: comparative phylogeography of reef fishes from the Red Sea and Indian Ocean. J Biogeogr. 40:1170–1181. [Google Scholar]
- Eghtesadi-Araghi P. Coral reefs in the Persian Gulf and Oman Sea: an integrated perspective on some important stressors. J Fish Aquat Sci. 6:48–56. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online. 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Farris J S, Källersjö M, Kluge A G, Bult C. Testing significance of incongruence. Cladistics. 10:315–319. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3:294–299. [PubMed] [Google Scholar]
- Fu Y X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo E M, Liu Y, Cui Z X, Li X L, Cheng Y X, Wu X G. Genetic variation and population structure of swimming crab (Portunus trituberculatus) inferred from mitochondrial control region. Mol Biol Rep. 39:1453–1463. doi: 10.1007/s11033-011-0882-3. [DOI] [PubMed] [Google Scholar]
- Harpending H C. Signature of ancient population growth in a low resolution mitochondrial DNA mismatch distribution. Hum Biol. 66:591–600. [PubMed] [Google Scholar]
- Hedgecock D. Is gene flow from pelagic larval dispersal important in the adaptation and evolution of marine invertebrates? Bull Mar Sci. 39:550–564. [Google Scholar]
- Hopkins M J, Thurman C L. The geographic structure of morphological variation in eight species of fiddler crabs (Ocypodidae: genus Uca) from the eastern United States and Mexico. Biol J Linn Soc. 100:248–270. [Google Scholar]
- Ituarte R B, 'anatro D, Luppi A, Ribeiro T A, Spivak P D, Iribarne E D, Lessa O O. Population structure of the SW Atlantic estuarine crab Neohelice granulata throughout its range: a genetic and morphometric study. Estuar Coast. 35:1249–1260. [Google Scholar]
- Johns W E, Yao F, Olson D B, Josey S A, Grist J P, Smeed D A. Observations of seasonal exchange through the Straits of Hormuz and the inferred heat and freshwater budgets of the Persian Gulf. J Geophys Res; 2003. 108 [Google Scholar]
- Levinton J S, Sturmbauer C, Christy J. Molecular data and biogeography: resolution of a controversy over evolutionary history of a pan-tropical group of invertebrates. J Exp Mar Biol Ecol. 203:117–131. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- López-Duarte P C, Christy J H, Tankersley R A. A behavioral mechanism for dispersal in fiddler crab larvae (genus Uca) varies with adult habitat, not phylogeny. Limnol Oceanogr. 56:1879–1892. [Google Scholar]
- Mcedward L ;, Mokhlesi A, Kamrani E, Backwell P, Sajjadi M. Ecology of marine invertebrate larvae. CRC Press. 91:245–249. [Google Scholar]
- Naderloo R, Türkay M. Decapod crustaceans of the littoral and shallow sublittoral Iranian coast of the Persian Gulf: faunistics, biodiversity and zoogeography. Zootaxa. 3374:1–67. [Google Scholar]
- Naderloo R, Türkay M, Chen H L. Taxonomic revision of the wide front fiddler crabs of the Uca lactea group (Crustacea: Decapoda: Brachyura: Ocypodidae) in the Indo-West Pacific. Zootaxa. 2500:1–38. [Google Scholar]
- Naser M D, Ali M H, Yasser A G. Basrah, Iraq): New record of the fiddler crab Uca (Paraleptuca) sindensis (Crustacea: Brachyura: Ocypodidae) from Khor Al Zubair; 2010. 3 [Google Scholar]
- Ng Pkl, Guinot D, Davie Pjf. Systema Brachyurorum: Part I. An annotated checklist of extant brachyuran crabs of the world. Raffles Bull Zool Suppl. 17:1–296. [Google Scholar]
- Oliveira-Neto J F, Pie M R, Boeger W A, Ostrensky A, Baggio R A. Population genetics and evolutionary demography of Ucides cordatus (Decapoda: Ocypodidae) Mar Ecol. 28:460–469. [Google Scholar]
- Oliveira-Neto J F, Pie M R, Chammas M A, Ostrensky A, Boeger W A. Phylogeography of the blue land crab, Cardisoma guanhumi (Decapoda: Gecarcinidae) along the Brazilian coast. J Mar Biol Ass UK. 88:1417–1423. [Google Scholar]
- Pie M R, Oliveira-Neto J F, Boeger W A, Ostrensky A, Baggio R A. The organization of the mitochondrial control region in 2 brachyuran crustaceans: Ucides cordatus (Ocypodidae) and Cardisoma guanhumi (Gecarcinidae) J Hered. 99:432–437. doi: 10.1093/jhered/esn024. [DOI] [PubMed] [Google Scholar]
- Pretzmann G. Ergebnisse einiger Sammelreisen nach Vorderasien 2. Teil: marine Brachyura. Ann Naturhist Mus Wien. 75:477–487. [Google Scholar]
- Queiroga H, Blanton J. Interactions between behaviour and physical forcing in the control of horizontal transport of decapod crustacean larvae. Adv Mar Biol. 47:107–214. doi: 10.1016/S0065-2881(04)47002-3. [DOI] [PubMed] [Google Scholar]
- Roesti M, Salzburger W, Berner D. Uninformative polymorphisms bias genome scans for signatures of selection. BMC Evol Biol; 2012. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saher N U. Population dynamics and biology of fiddler crabs in the mangrove areas of Karachi coast. Karachi, Pakistan): Ph.D. Dissertation; 2008. [Google Scholar]
- Saher N U, Qureshi N A. Spatial distribution of Uca sindensis (Crustacea, Ocypodidae) along the coast of Pakistan. Egypt Acad J Biol Sci Zool. 4:119–129. [Google Scholar]
- Saher N U, Sahir O, Shih H T, Kamal M, Qureshi N A. On a new record of the genus Uca (Decapoda, Brachyura, Ocypodidae) found along the coast of Pakistan. Crustaceana. 87:666–673. [Google Scholar]
- Sanford E, Holzman S B, Haney R A, Rand D M. Larval tolerance, gene flow, and the northern geographic range limit of fiddler crabs. Ecology. 87:2882–2894. doi: 10.1890/0012-9658(2006)87[2882:ltgfat]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates very among sites: application to human mitochondrial DNA. Genetics. 152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart C D, Huber Mgj. Genetic comparisons of German populations of the stone crayfish, Austropotamobius torrentium (Crustacea: Astacidae) Bull Fr Peche Piscic. 380:1019–1028. [Google Scholar]
- Shih H T. A study on the fiddler crab, Uca formosensis, and its habitat. Hsinchu City Government; 2008. [Google Scholar]
- Shih H T, Suzuki H. Taxonomy, phylogeny, and biogeography of the endemic mudflat crab Helice/Chasmagnathus complex (Crustacea: Brachyura: Varunidae) from East Asia. Zool Stud. 47:114–125. [Google Scholar]
- Shih H T, Mok H K, Chang H W, Lee S C. Morphology of Uca formosensis Rathbun. Crustacea: Decapoda: Ocypodidae), an endemic fiddler crab from Taiwan, with notes on its ecology. 38:164–177. [Google Scholar]
- Shih H T, Mok H K, Chang H W. Chimney building by male Uca formosensis Rathbun, 1921 (Crustacea, Decapoda, Ocypodidae) after pairing: a new hypothesis for chimney function. Zool Stud. 44:242–251. [Google Scholar]
- Shih H T, Kamrani E, Davie Pjf, Liu M Y. Genetic evidence for the recognition of two fiddler crabs. Uca iranica and U. albimana (Crustacea: Brachyura: Ocypodidae), from the northwestern Indian Ocean, with notes on the U. lactea species complex. 635:373–382. [Google Scholar]
- Shih H T, Naruse T, Ng Pkl. Uca jocelynae sp. nov., a new species of fiddler crab (Crustacea: Brachyura: Ocypodidae) from the Western Pacific. Zootaxa. 2337:47–62. [Google Scholar]
- Shih H T, Ng Pkl, Wong Kjh, Chan Bkk. Gelasimus splendidus Stimpson, 1858 (Crustacea: Brachyura: Ocypodidae), a valid species of fiddler crab from the northern South China Sea and Taiwan Strait. Zootaxa. 3490:30–47. [Google Scholar]
- Shih H T, Komai T, Liu M Y. A new species of fiddler crab from the Ogasawara (Bonin) Islands, Japan, separated from the widely-distributed sister species Uca (Paraleptuca) crassipes (White, 1847) (Crustacea: Decapoda: Brachyura: Ocypodidae) Zootaxa. 3746:175–193. doi: 10.11646/zootaxa.3746.1.8. [DOI] [PubMed] [Google Scholar]
- Shih H T, Ng Pkl, Liu M Y. Systematics of the Indo-West Pacific broadfronted fiddler crabs (Crustacea: Ocypodidae: genus Uca) Raffles Bull Zool. 61:641–649. [Google Scholar]
- Silva I C, Mesquita N, Paula J. Lack of population structure in the fiddler crab Uca annulipes along an East African latitudinal gradient: genetic and morphometric evidence. Mar Biol. 157:1113–1126. [Google Scholar]
- Sirocko F. Ups and downs in the Red Sea. Nature. 423:813–814. doi: 10.1038/423813a. [DOI] [PubMed] [Google Scholar]
- Spaargaren D H. On the water and salt economy of some decapod crustaceans from the Gulf of Aqaba (Red Sea) Neth J Sea Res. 11:99–106. [Google Scholar]
- Spivak E D, Cuesta J. The effect of salinity on larval development of Uca tangeri (Eydoux, 1835) (Brachyura: Ocypodidae) and new findings of the zoeal morphology. Sci Mar. 73:297–305. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Ströher P R, Firkowski C R, Freire A S, Pie M R. A molecular method for the detection of sally lightfoot crab larvae (Grapsus grapsus, Brachyura, Grapsidae) in plankton samples. Genet Mol Biol. 34:529–531. doi: 10.1590/S1415-47572011005000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmbauer C, Levinton J S, Christy J. Molecular phylogeny analysis of fiddler crabs: test of the hypothesis of increasing behavioral complexity in evolution. Proc Natl Acad Sci U S A. 93:10855–10857. doi: 10.1073/pnas.93.20.10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam B, Murty Vsn, Heffner D M. Sea surface salinity variability in the tropical Indian Ocean. Remote Sens Environ. 115:944–956. [Google Scholar]
- Supmee V, Ngernsiri L, Sriboonlert A, Wonnapinij P, Sangthong P. Population genetics of the violet vinegar crab (Episesarma versicolor) along the Andaman Sea coast of Thailand. Zool Stud. 51:1040–1050. [Google Scholar]
- Swofford D L. Phylogenetic Analysis Using Parsimony (*and other methods); 2003. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoppil P G, Hogan P J. A modeling study of circulation and eddies in the Persian Gulf. J Phys Oceanogr. 40:2122–2134. [Google Scholar]
- Thurman C L. Osmoregulation in six sympatric fiddler crabs (genus Uca) from the northwestern Gulf of Mexico. Mar Ecol. 23:269–284. [Google Scholar]
- Thurman C L, Faria S C, Mcnamara J C. The distribution of fiddler crabs (Uca) along the coast of Brazil: implications for biogeography of the western Atlantic Ocean. Mar Biodivers Rec; 2013. 6 [Google Scholar]
- Tsang L M, Achituv Y, Chu K H, Chan Bkk. Zoogeography of intertidal communities in the West Indian Ocean as determined by ocean circulation systems: patterns from the Tetraclita barnacles. PLoS One; 2012. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L M, Hallas J P, Smith M J, Morris S. Phylogeography of the Christmas Island blue crab, Discoplax celeste (Decapoda: Gecarcinidae) on Christmas Island, Indian Ocean. J Mar Biol Ass UK. 93:703–714. [Google Scholar]
- Weir B S. Genetic data analysis II: methods for discrete population genetic data. Sinauer, Sunderland): 1996. [DOI] [PubMed] [Google Scholar]
- Wieman A C, Berendzen P B, Hampton K R, Jang J, Hopkins M J, Jurgenson J, Mcnamara J C, Thurman C L. A panmictic fiddler crab from the coast of Brazil? Impact of divergent ocean currents and larval dispersal potential on genetic and morphological variation in Uca maracoani. Mar Biol. 161:173–185. [Google Scholar]
- Willing E M, Dreyer C, Van Oosterhout C. Estimates of genetic differentiation measured by F ST do not necessarily require large sample sizes when using many SNP markers. PLoS One; 2012. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Kjh, Chan Bkk, Shih H T. Taxonomy of the sand bubbler crabs Scopimera globosa De Haan, 1835, and S. tuberculata Stimpson, 1858 (Crustacea: Decapoda: Dotillidae) in East Asia, with description of a new species from the Ryukyus. 2345:43–59. [Google Scholar]
- Wong Kjh, Shih H T, Chan Bkk. Two new species of sand-bubbler crabs, Scopimera, from North China and the Philippines (Crustacea: Decapoda: Dotillidae) Zootaxa. 2962:21–35. [Google Scholar]
- Wong Kjh, Shih H T, Chan Bkk. Crustacea: Decapoda: Ocypodidae): redescription, distribution at its type locality, and the phylogeny of East Asian Ocypode species. Zootaxa. 3550:71–87. [Google Scholar]
- Yamaguchi T. Fiddler crabs of the genus Uca in the collections of three natural history museums in Europe. 1. The specimens held by the Nationaal Natuurhistorisch Museum, Leiden and the Natural History Museum. 11:151–189. [Google Scholar]