Abstract
Keto-carotenoids contribute to many important traits in animals, including vision and coloration. In a great number of animal species, keto-carotenoids are endogenously produced from carotenoids by carotenoid ketolases. Despite the ubiquity and functional importance of keto-carotenoids in animals, the underlying genetic architectures of their production have remained enigmatic. The body and eye colorations of spider mites (Arthropoda: Chelicerata) are determined by β-carotene and keto-carotenoid derivatives. Here, we focus on a carotenoid pigment mutant of the spider mite Tetranychus kanzawai that, as shown by chromatography, lost the ability to produce keto-carotenoids. We employed bulked segregant analysis and linked the causal locus to a single narrow genomic interval. The causal mutation was fine-mapped to a minimal candidate region that held only one complete gene, the cytochrome P450 monooxygenase CYP384A1, of the CYP3 clan. Using a number of genomic approaches, we revealed that an inactivating deletion in the fourth exon of CYP384A1 caused the aberrant pigmentation. Phylogenetic analysis indicated that CYP384A1 is orthologous across mite species of the ancient Trombidiformes order where carotenoids typify eye and body coloration, suggesting a deeply conserved function of CYP384A1 as a carotenoid ketolase. Previously, CYP2J19, a cytochrome P450 of the CYP2 clan, has been identified as a carotenoid ketolase in birds and turtles. Our study shows that selection for endogenous production of keto-carotenoids led to convergent evolution, whereby cytochrome P450s were independently co-opted in vertebrate and invertebrate animal lineages.
Keywords: carotenoid ketolase, convergent evolution, keto-carotenoids, lemon, CYP384A1
1. Introduction
Carotenoids are terpenoid pigments that are responsible for many of the bright yellow, orange and red colours observed in animals. These include the colours displayed in lizard throats, avian plumage, arthropod bodies and eggs [1,2]. In addition to coloration, carotenoids also contribute to multiple other traits in animals, such as the visual system [3–8]. For instance, in birds, carotenoids are deposited in retinal oil droplets where they function as long-pass cut-off filters to enhance colour discrimination [8]. Although carotenoid pigments are widespread in the natural world, their biosynthesis occurs primarily in plants, fungi, bacteria and archaea. While most animals lack the ability to biosynthesize carotenoids, many obtain, deposit and modify the carotenoids they encounter in their diets. Animal carotenoid ketolases add a double-bonded oxygen molecule at the C4 and/or C4′ position of the terminal rings of carotenoid structures, thereby converting more yellow-coloured carotenoids like β-carotene to more orange- and red-coloured keto-carotenoid derivatives [9]. The identification of the molecular mechanisms that underpin animal carotenoid metabolism is beginning to inform the evolutionary history and adaptive function of carotenoid-based traits [10]. For instance, recent work identified the cytochrome P450 CYP2J19 as the carotenoid ketolase responsible for keto-carotenoid production in both the integument and retinal oil droplets of birds where keto-carotenoids fulfil different biological roles [11,12].
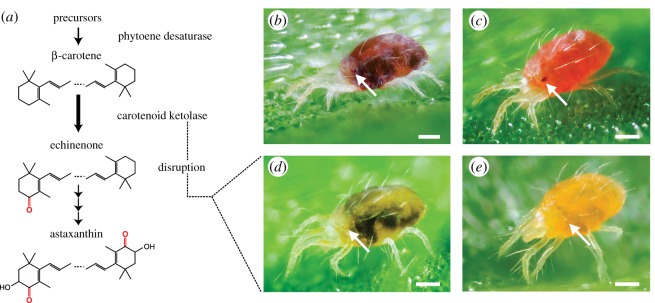
Studies on the pigmentation of spider mites (Chelicerata: Trombidiformes), from as early as 1914, revealed that the orange–red body and red eye colorations of mites within the Tetranychus genus depend solely on carotenoid pigments [13–18]. A largely conserved carotenoid metabolic pathway that produces red keto-carotenoids from β-carotene has been proposed for tetranychids (figure 1a) [13,15–18]. Here, a striking body colour change to deep orange–red is associated with diapause induction in adult females, and studies show that this change is the result of differential accumulation of endogenously produced keto-carotenoids (figure 1) [15,16]. Several spontaneous pigment mutants have been discovered in tetranychid populations and their carotenoid profiles have been characterized. Due to their indistinguishable carotenoid compositions, mutant phenotypes have been given identical descriptive names in different species. Two mutant phenotypes, albino and lemon, completely lack keto-carotenoid production [4,16,17,19]. Both albino and lemon mutants lack eye pigmentation, but whereas albino mites lack body pigmentation, the bodies of lemon mites display yellow coloration that markedly intensifies in diapause (figure 1).
Figure 1.
The conserved keto-carotenoid biosynthesis pathway in tetranychid mites and its disruption in lemon mutants. (a) The proposed pathway for carotenoid biosynthesis in spider mites [18] adapted to incorporate recent findings (endogenous synthesis of β-carotene by phytoene desaturase [4]). β-carotene is converted to echinenone, which leads to three major keto-carotenoids: 3-hydroxyechinenone and phoenicoxanthin (not shown), and astaxanthin [18]. Carotenoids are depicted in their de-esterified forms. (b) Wild-type T. kanzawai. (c) Wild-type diapausing T. kanzawai. (d) Lemon T. kanzawai. (e) Lemon diapausing T. kanzawai. Each panel depicts an adult female. Arrows highlight the anterior and posterior eye spots, which are red-coloured in wild-type individuals. The dark regions in feeding, non-diapausing mites are gut contents that are visible through the partially translucent cuticle. Feeding spots are absent (or nearly so) in diapausing mites that have ceased to actively feed. Scale bars represent 0.1 mm.
Genomes from several arthropod lineages, including spider mites and aphids (Hexapoda: Hemiptera), harbour horizontally transferred genes of fungal origin that code for carotenoid biosynthetic enzymes [4,20,21]. The discovery of these fungal genes embedded within arthropod genomes challenged the assumption that all animals lack the ability to biosynthesize carotenoids. Recently, inactivation of one of the horizontally acquired carotenoid biosynthetic genes, a phytoene desaturase, was shown to underlie albinism in Tetranychus urticae [4]. This finding, along with work on aphids [21], has revealed that the horizontally transferred biosynthetic genes remained active. Further, Bryon et al. [4] concluded that the carotenoid metabolic pathway of spider mites relies solely (or nearly so) on endogenously produced β-carotene.
Despite the recent study linking phytoene desaturase activity to endogenous β-carotene synthesis in spider mites, it has remained elusive how the orange and red keto-carotenoids that typify the body and eye colours of spider mites and related trombidiform mites are produced. In the early 1970s, Veerman suggested that lemon mutations disrupt carotenoid ketolase activity, inhibiting the formation of the keto-carotenoid echinenone and its downstream derivatives from β-carotene [16,17] (figure 1a). The lemon mutants studied by Veerman and co-authors no longer exist [3,16,17]; however, we recently recovered a new lemon pigment mutant in the spider mite Tetranychus kanzawai. Taking advantage of the chromosome-level genome assembly of the closely related species T. urticae [22], we performed bulked segregant analysis (BSA) genetic mapping to identify the locus underlying the lemon phenotype. As revealed by fine-mapping, the lemon phenotype resulted from an inactivating deletion in the fourth exon of a cytochrome P450, CYP384A1. CYP384A1 belongs to the CYP3 clan, and was conserved among sequenced genomes within the ancient Trombidiformes mite order. Our study shows that the rich orange and red colorations of many vertebrate and invertebrate animals have arisen by the convergent evolution of cytochrome P450s.
2. Material and methods
(a). Mite strains
The lemon mite strain, hereafter Jp-lemon, arose as a spontaneous mutant in a population of T. kanzawai collected from Japanese bindweed (Calystegia japonica Choisy) in Kyoto, Japan (hereafter Jp-WT). A second wild-type T. kanzawai population, hereafter Jp2-WT, was collected from Muskmelon (Cucumis melo) in Iwata, Japan. An inbred line from the Jp-lemon strain was generated by six sequential rounds of mother–son matings after Bryon et al. [4], and is hereafter referred to as Jp-inbred-lemon. Mites were reared on potted plants or detached leaves of Phaseolus vulgaris L. cv ‘Speedy’. Unless otherwise stated, mite cultures were maintained at 26°C, 60% RH and a 16 : 8 light : dark photoperiod.
(b). Mode of inheritance
The mode of inheritance of the Jp-lemon pigment phenotype was determined by performing reciprocal crosses with Jp-WT and Jp2-WT. In each cross, 20 virgin females were placed with 30 males on a detached bean leaf and allowed to mate. The pigment phenotype was scored in F1 females to identify a recessive or dominant mode of inheritance. During F1 development, 20 female teleochrysalids (nymphal females in their final quiescent stage) were selected and placed on a separate bean leaf. Upon eclosion, the unfertilized females were transferred to new detached bean leaves on a daily basis throughout their oviposition period. Due to the arrhenotokous mode of reproduction of T. kanzawai, F1 virgin females only produce haploid F2 males. The mutant and wild-type pigment phenotypes in the F1 and F2 generations were scored based on body and eye colour in adult mites. The hypothesis of a monogenic, recessive mode of inheritance was tested with χ2 goodness-of-fit tests in R (version 3.4.3) [23].
(c). Thin-layer chromatography and high-performance liquid chromatography analysis of carotenoid profiles
Extraction of bean and mite carotenoid pigments for thin-layer chromatography (TLC) analysis was based on previous work [16–18]. After collection, mite and bean leaf material were immediately transferred to 10 ml and 15 ml of ice-cold acetone, respectively. Mite and bean samples were homogenized using glass pestles. After sedimentation by gravity, the supernatants were transferred twice to a new glass vial. Five millilitres of hexane was added and the mixture was transferred to a glass separating funnel. Carotenoids were translocated to the hexane phase by washing the hexane–acetone mixture four times with 10 ml of water. Any residual water was carefully removed from the solution using the glass separating funnel. The solution was subsequently evaporated to complete dryness under vacuum at room temperature. Carotenoids were re-suspended in 100 µl of hexane and spotted on an HPTLC silica 60 plate (EMD Millipore). Mobile phases of 20% and 25% acetone in hexane were run to separate carotenoids with high and low retention values (Rf), respectively.
For high-performance liquid chromatography (HPLC) analysis, we followed previous work on carotenoid characterization [15]. Non-diapausing and diapausing adult females were obtained by placing 2-day-old eggs under long-day (25°C and 16 : 8 L : D photoperiod) and short-day (20°C and 9 : 15 L : D photoperiod) conditions, respectively. To identify and quantify astaxanthin and β-carotene, 30 females of each treatment were collected in three replicates and homogenized in 1 ml of acetone. The homogenate was filtrated using a glass syringe with a membrane possessing a pore size of 0.45 µm (Minisart RC4 17 822; Sartorius Stedim Biotech GmbH, Goettingen, Germany). The filtrate was dried under a nitrogen gas flow and dissolved in 300 µl of methanol. Five microlitres of the solution was used for HPLC analysis (electronic supplementary material). Carotenoids were quantified by monitoring the absorbance at 450 nm. External calibration curves were constructed with authentic standards (astaxanthin: AG Scientific, San Diego, CA; β-carotene: Wako Pure Chemical Industries, Osaka, Japan). Βeta-carotene levels were compared by a two-way ANOVA, followed by a Tukey test in R [23].
(d). Bulked segregant analysis experimental set-up, genomic sequencing and variant detection
A segregating mite population generated by crossing Jp-inbred-lemon to Jp2-WT was used to genetically map the lemon phenotype (electronic supplementary material). Approximately 10–12 generations after the initial cross, three replicates of 1100, 900 and 500 lemon females were collected, as were 1500 phenotypically wild-type females. For these four populations and the two parents, genomic DNA was prepared (electronic supplementary material). RNA was extracted from 110 adult females isolated from the segregating population using an RNeasy Minikit (Qiagen) per replicate. Two RNA replicates were collected for wild-type and lemon mites. Illumina libraries for the DNA and RNA samples were prepared and sequenced at the Huntsman Cancer Institute at the University of Utah to generate paired-end genomic DNA reads of 125 bp with library insert sizes of approximately 700 bp, and RNA-seq reads of insert sizes of approximately 335 bp. Genomic DNA reads were aligned to the three chromosomes of T. urticae [22] using the default settings of BWA (version 0.7.15-r1140) [24]. Alignments were sorted by position using SAMtools 1.3.1 [25]. Duplicate reads were marked using Picard Tools 2.6.0 (https://broadinstitute.github.io/picard/), and indel realignment was performed using GATK (version 3.6.0-g89b7209) [26] following GATK's best practices recommendations [27,28]. GATK's UnifiedGenotyper was used for joint variant calling across all samples to identify single-nucleotide differences and indels. RNA-seq reads were aligned to the T. urticae chromosomes using default settings of STAR (version 2.5.3a) [29], and the resulting alignments were indexed by SAMtools 1.3.1 [25].
(e). Bulked segregant analysis genetic mapping
Prior to BSA mapping, the 2 419 446 nucleotide variable positions predicted between the T. kanzawai DNA samples and the T. urticae reference sequence by GATK were filtered with quality control settings adapted from GATK Doc # 2806 (https://software.broadinstitute.org/gatk/documentation/article.php?id=2806) (electronic supplementary materials and methods). From the resulting 1 150 705 nucleotide positions, 196 214 single-nucleotide polymorphisms within T. kanzawai were identified as fixed for contrasting differences between the two parents and were retained. The locus responsible for the lemon phenotype was identified by comparing allele frequencies between the three lemon selected samples to the wild-type sample using previously published BSA genetic mapping methods with statistical testing for genotype–phenotype associations by permutation [22]; 75 kb windows with 5 kb offsets were used with the false discovery rate (FDR) set to 5%. Genome annotation of the BSA peak was based on Wybouw et al. [22].
(f). De novo assemblies
DNA sequence reads were imported into CLC Genomics Workbench 9.0.1 (https://www.qiagenbioinformatics.com/), trimmed using default settings, assembled into contigs using the default settings of the ‘De Novo Assembly’ tool, and the contigs were aligned to the T. urticae genome assembly using BLASR (version 1.3.1) [30]. CYP384A1 transcripts were assembled into contigs using Trinity 2.5.1 [31,32] with default settings and basic trimming by Trimmomatic [33]. Open reading frames in the Trinity-assembled contigs were predicted using TransDecoder v5.0.2. (https://github.com/TransDecoder/TransDecoder/wiki) [32].
(g). Fine-mapping the lemon locus
A second segregating population was generated by crossing Jp-inbred-lemon to Jp2-WT (electronic supplementary material). For genotyping, genomic fragments polymorphic between the two parents in the region of the BSA peak were PCR-amplified and sequenced using 429 phenotypically lemon and 50 wild-type single adult females from the segregating population (electronic supplementary material).
(h). CYP phylogenetic reconstruction
All CYP3 clan members were retrieved from nine arthropod and one tardigrade species with available annotated genome assemblies, and genomes of three mite species of the Acariformes superorder were additionally screened for close homologues to CYP384A1. Cytochrome P450s of the CYP2 clan were selected to root the phylogenetic tree (electronic supplementary material). After filtering (electronic supplementary material), the sequence set was aligned using the E-INS-i strategy of MAFFT, leaving ‘gappy’ regions, and with 1000 cycles of iterative refinement [34]. Identical amino acid sequences were removed from the final aligned dataset, resulting in a final set of 229 CYP sequences (electronic supplementary material, table S2). The LG + I + G + F model of protein evolution was used based on the corrected Akaike information criterion with PartitionFinderProtein (greedy search algorithm using RAxML and one datablock) [35,36]. Maximum-likelihood searches (n = 20, using randomized stepwise addition parsimony trees) and bootstrapping (n = 1000) were performed using RAxML (version 8.2.10) [36], with the random number seed set at 54 321.
3. Results
(a). The carotenoid profile and mode of inheritance of the lemon phenotype
A spontaneous pigment mutant, Jp-lemon, arose in the T. kanzawai Jp-WT population. Specifically, Jp-lemon mites lacked wild-type red pigmentation in the two eye spots, body and the distal segments of the front legs. Instead, Jp-lemon mites displayed yellow body coloration matching descriptions of the lemon phenotype, including marked intensification in diapausing females (figure 1d,e) [16,17,19,37]. Neither wild-type nor lemon females displaying diapausing coloration (figure 1c,e) laid eggs, the canonical reproductive criterion for diapause. Lemon females entered diapause at an incidence comparable to wild-type females on the same genetic background (electronic supplementary material, table S3).
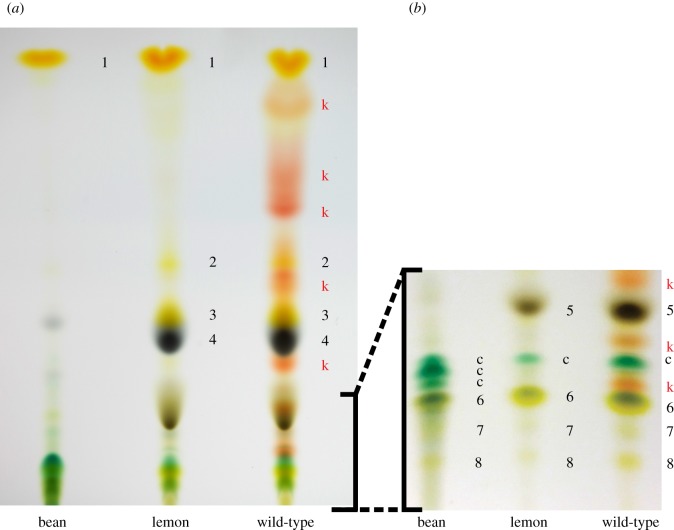
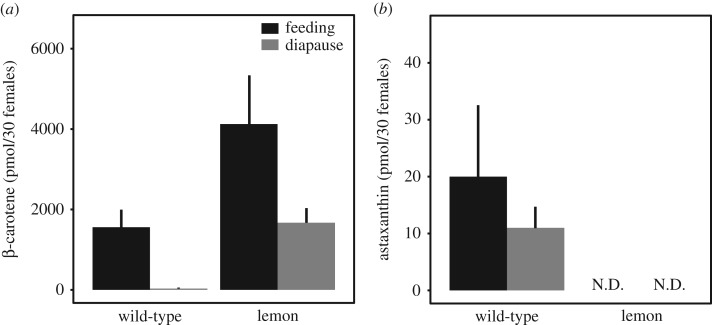
Biochemical characterization by TLC in previously identified but now extinct lemon mutants revealed the absence of endogenously produced keto-carotenoids [16,17]. Using the same approach, we compared the carotenoid pigment profiles of wild-type and lemon mites, and their diet (kidney bean), to assess the presence of mite-specific keto-carotenoids. Two mobile phases visibly separated the mite and bean carotenoid pigments on HPTLC silica plates (figure 2). As assessed in comparison to earlier TLC studies [16–18], the plant pigments α-carotene, chlorophyll a, chlorophyll b, chlorophyll derivatives, lutein, lutein 5,6-epoxide, violaxanthin and neoxanthin appeared to be present in both wild-type and lemon mite extracts, consistent with the expected ingestion of plant tissue. The profile of wild-type mites revealed the presence of endogenously produced red-coloured keto-carotenoids that are likely esterified in vivo [16–18] (figure 2, indicated by k). By contrast, TLC analysis showed that lemon mites entirely lacked these keto-carotenoids. To further examine the carotenoid content of the newly isolated lemon mutant, we quantified β-carotene and astaxanthin levels by absorption spectra in an HPLC system using exogenous standards (figure 3; electronic supplementary material, figure S1). Βeta-carotene was present in both lemon and wild-type mites, but was detected in significantly higher concentrations in the former (two-way ANOVA, F1,11 = 9.748, p = 0.0142; figure 3a). While astaxanthin accumulated in both feeding and diapausing wild-type mites, no free astaxanthin was detected in lemon mites using non-esterified astaxanthin as the exogenous standard (figure 3b).
Figure 2.
Lemon T. kanzawai lacks endogenously produced keto-carotenoids. Panels (a) and (b) show HPTLC plates for plant and mite extracts run with mobile phases of 20% and 25% acetone in hexane, respectively. Using previously determined Rf values and colour profiles [16–18], we tentatively identified the carotenoid pigments as: 1: α- and β-carotene; k: keto-carotenoid (esterified in vivo); 2: β-carotene-diepoxide; 3: unknown epoxide; 4: chlorophyll a; 5: chlorophyll b; c: chlorophyll derivatives; 6: lutein and lutein 5,6-epoxide; 7: violaxanthin; 8: neoxanthin.
Figure 3.
Lemon T. kanzawai accumulates higher levels of β-carotene. Panels (a) and (b) show the levels of β-carotene and astaxanthin, respectively, in wild-type and lemon T. kanzawai. Carotenoid levels were determined by HPLC for both feeding and diapausing adult female mites. N.D. stands for not detected. Error bars represent the standard errors, with a sample size of three.
To establish the mode of inheritance of lemon pigmentation, we performed reciprocal crosses between Jp-lemon and two wild-type populations, Jp-WT and Jp2-WT. All diploid female F1 progeny were phenotypically wild-type, and their haploid F2 sons were wild-type or lemon in phenotype in an approximately 1 : 1 ratio (table 1), establishing a monogenic, recessive genetic basis as reported previously for lemon mutants in tetranychids [19,38].
Table 1.
Lemon pigmentation has a recessive, monogenic mode of inheritance in T. kanzawai. The degrees of freedom for the χ2-tests were 1. For every cross, at least 70 F1 females were scored.
| cross (♀ × ♂) | % lemon in F1 ♀ (2n) | F2 ♂ (n) |
χ2 | p-value | |
|---|---|---|---|---|---|
| wild-type | lemon | ||||
| Jp-WT × Jp-lemon | 0 | 175 | 196 | 1.1887 | 0.2756 |
| Jp-lemon × Jp-WT | 0 | 365 | 372 | 0.066486 | 0.7965 |
| Jp2-WT × Jp-lemon | 0 | 198 | 210 | 0.35294 | 0.5525 |
| Jp-lemon × Jp2-WT | 0 | 187 | 174 | 0.46814 | 0.4938 |
(b). Localization of the lemon mutation
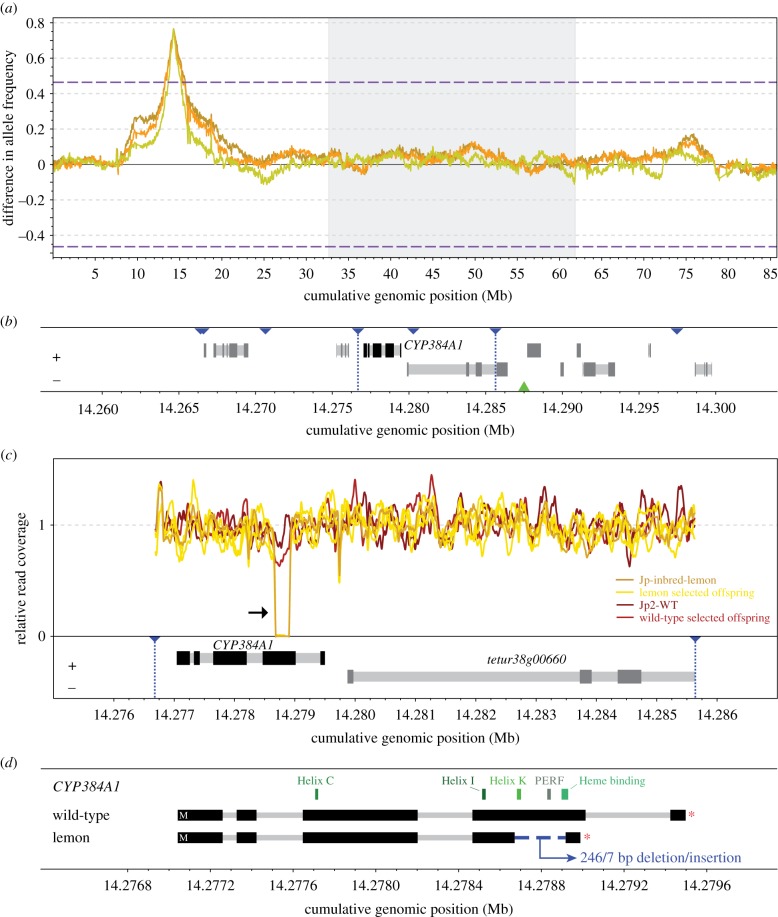
To identify the locus underlying lemon pigmentation, we crossed inbred, diploid lemon females to a single, haploid wild-type male of the non-related Jp2-WT population. After multiple generations of sib-mating, we performed high-throughput DNA sequencing of three replicates of lemon selected and one replicate of wild-type selected offspring from the resultant bulk population. Consistent with monogenic recessive inheritance, BSA genetic mapping revealed a single genomic region underlying the lemon phenotype (figure 4; electronic supplementary material, figure S2). Using the wild-type selected offspring pool as reference, the average allele frequency of the three lemon selected offspring pools peaked at position 14 287 500 bp on chromosome 1 (figure 4).
Figure 4.
Bulked segregant analysis locates the lemon locus and reveals a non-functional CYP384A1 as the genetic basis. (a) Differences in the frequencies of parental Jp-inbred-lemon alleles between each of the three lemon selected and one wild-type offspring pools are plotted in a sliding window analysis. The three T. urticae reference chromosomes are shown in alternating white and grey and are ordered by decreasing length. Dashed lines represent the 5% FDR for an association between parental Jp-inbred-lemon allele frequencies and the lemon phenotype. The maximal average allele frequency of the three replicates (i.e. the BSA peak) is located at cumulative genomic position 14 287 500. (b) CYP384A1 and a 3′ end fragment of its neighbouring gene reside in the minimal candidate region. Gene models and their genomic position are based on the T. urticae genome annotation, with exons and introns depicted as dark and light grey rectangles, respectively. Strands are represented as ‘+’ (forward) and ‘−’ (reverse). Blue triangles delineate the genomic position of the genetic markers used in the fine-mapping approach and the vertical dotted lines demarcate the 8.96 kb minimal candidate region. The green triangle highlights the location of the BSA peak. (c) Read coverage reveals a deletion within the CYP384A1 coding sequence in the three lemon selected offspring pools and parental Jp-inbred-lemon (black arrow). DNA sequence read coverage depth across the minimal candidate region is shown relative to the chromosome-wide average. (d) The deletion spans 246 bp within the fourth exon of the CYP384A1 coding sequence concomitant with 7 bp of inserted sequence. The five essential cytochrome P450 domains are plotted above the gene models.
The region surrounding this peak harboured a small number of genes, of which we identified the cytochrome P450 gene CYP384A1 (tetur38g00650 in the T. urticae annotation) as a likely candidate as cytochrome P450s have been implicated in keto-carotenoid production in other taxa [11,12,39–41]. To retain or exclude CYP384A1 as the gene responsible for keto-carotenoid synthesis, we performed fine-mapping with PCR-based markers using a population that segregated for the lemon mutation for approximately 10–12 generations. By genotyping with a set of markers flanking the peak BSA/CYP384A1 region, followed by iterative genotyping of informative recombinants, we established a minimal candidate region for the lemon phenotype of only 8.96 kb. This interval harboured the entire CYP384A1 gene, as well as a 3′ end fragment of a neighbouring gene (tetur38g00660 in T. urticae, which encodes a protein with an Immunoglobulin-like domain (IPR007110)) (figure 4b; electronic supplementary material, figure S3 and table S4).
(c). The lemon mutation and its impact on CYP384A1
Coverage of Illumina read alignments from the segregating populations, as well as that of the parents, to the T. urticae genome revealed a likely structural variant in T. kanzawai within the minimal candidate region that was specific to the lemon parent, and the three lemon selected populations (lack of read coverage at approximately 14.2788 Mb; black arrow in figure 4c). To investigate further, we generated de novo assemblies of the parents and segregating populations. In assemblies of the three lemon selected populations (contigs 3098, 441 and 996 from assembly lemonBSA1, lemonBSA2 and lemonBSA3, respectively), as well as parental Jp-inbred-lemon (contig 2665), a 246 bp deletion coupled with a 7 bp insertion (sequence CCTACCT) was present as compared to the T. urticae reference sequence and contig 1805 from the parental Jp2-WT strain. Cloning confirmed that the indel was located within the coding sequence of T. kanzawai CYP384A1, which was intact in wild-type T. kanzawai. The wild-type selected population used for BSA mapping is expected to segregate for the recessive lemon mutation, and two contigs were assembled from this population, one with the deletion and one without (contigs 487 and 21233, respectively). RNA-seq alignments, as well as localized de novo transcriptome assemblies for CYP384A1, revealed that CYP384A1 was expressed in both lemon and wild-type mites, and confirmed that the structural variant was unique to the former (electronic supplementary material, figure S4). With a PCR-based marker, we also found that the deletion segregated perfectly with the lemon phenotype in the individuals used for fine-mapping (electronic supplementary material, table S4). Apart from the 246/7 bp deletion/insertion, the genome and transcriptome assemblies revealed no other large structural variants, and no other fixed coding changes were found between the lemon and wild-type mites within the minimal 8.96 kb candidate region.
The 246/7 bp deletion/insertion was located internal to exon 4 of CYP384A1, and introduced a frameshift that results in a premature stop codon before the splice site at the 3′ end of exon 4. As a result of the deletion and frameshift, only the first 384 of the 497 amino acids present in the wild-type CYP384A1 of T. kanzawai are predicted to be encoded in lemon mites. The Helix K, PERF and heme binding motifs essential for CYP enzymatic activity were absent in the truncated CYP384A1 protein (figure 4d; electronic supplementary material, figure S4).
(d). Evolutionary history of CYP384A1
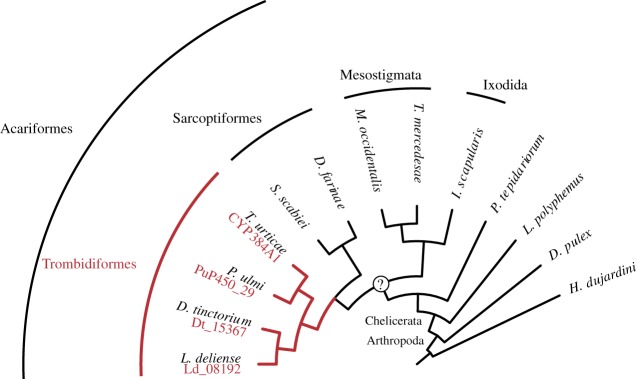
We investigated the evolutionary origin of CYP384A1 by a maximum-likelihood phylogenetic analysis using the CYP3 clans of 12 arthropods and one tardigrade, rooted by CYPs of the CYP2 clan (a total of 229 CYPs; figure 5; electronic supplementary material, figure S5). CYP384A1 exhibited a clear 1 : 1 : 1 : 1 orthology across all analysed mite species of the Trombidiformes order (T. urticae, Panonychus ulmi, Dinothrombium tinctorium and Leptotrombidium deliense). Three copies were initially identified in the D. tinctorium genome assembly, but were finally considered as putative allelic variants (lowest degree of sequence identity was 93.62% and the scaffolds that hold the copies did not code for additional proteins). CYP383A1, the closest homologue to CYP384A1 within T. urticae [20], also exhibited a clear 1 : 1 : 1 : 1 orthology across the four trombidiform mites. The CYP383A1 and CYP384A1 groups were clustered with strong bootstrap support and this well-supported clade only included CYP sequences from trombidiform mites (electronic supplementary material, figure S5).
Figure 5.
CYP384A1 is orthologous across mite species of the Trombidiformes order. The maximum-likelihood phylogenetic reconstruction uncovered a 1 : 1 : 1 : 1 orthology of CYP384A1 for the four trombidiform mite species with available genomic resources (Arthropoda: Chelicerata: Acari: Acariformes: Trombidiformes) (electronic supplementary material, figure S5). The gene IDs for the identified orthologues are given in red font below the species name. A monophyletic origin for mites (Chelicerata: Acari) remains under debate [42].
4. Discussion
As opposed to pigments like melanin, relatively little is known about the transport, modification and deposition of the diverse set of carotenoid pigments in animals [10]. Here, we identified a cytochrome P450 of the CYP3 clan, CYP384A1, as a likely carotenoid ketolase by showing that an inactivating deletion is strictly associated with the lemon pigment phenotype that lacks keto-carotenoids. Recent genomic research on zebra finches and canaries implicated CYP2J19, a cytochrome P450 of the CYP2 clan, as the main (or only) carotenoid ketolase enzyme in birds that produces keto-carotenoids in the integument and retinal oil droplets [11,12,43]. Although the exact enzymatic abilities of CYP2J19 and CYP384A1 remain unknown, it is very likely that these cytochrome P450s not only hydroxylate, but also oxidize carotenoid substrates at the C4 and/or C4′ positions of the terminal rings. Carotenoid profiles of the spider mite T. urticae and the bird Cardinalis cardinalis have been characterized in detail and no hydroxylated intermediates were identified [16,44]. In addition, in the fungus Xanthophyllomyces dendrorhous, a single cytochrome P450 is able to produce keto-carotenoids from a carotenoid precursor [39,40]. Our work now provides compelling evidence that birds and spider mites have addressed the biochemical challenge of producing keto-carotenoids by independent, convergent evolution across the CYP2 and CYP3 clans. Our findings add to several other examples of convergent evolution within this diverse multi-gene family, including the biosynthesis of cyanogenic glycosides in insects and plants [45], the production of growth-regulating gibberellins in plants and fungi [46], and syringyl lignin biosynthesis in lycophytes and flowering plants [47]. CYP383A1, the closest homologue to CYP384A1 in the T. urticae genome, is a potential candidate for an additional ketolase in spider mites, one that could potentially produce more oxygenated keto-carotenoids, such as astaxanthin, in which keto- and hydroxyl groups are present on both terminal cyclic rings of the β-carotene precursor backbone. It is also possible that CYP384A1, as well as CYP2J19, are multifunctional and able to produce different keto-carotenoids by multistep conversion, as observed in X. dendrorhous, where a single cytochrome P450 is strongly implicated in the conversion of β-carotene to astaxanthin [39,40].
Phylogenetic analysis suggests that CYP2J19 arose via gene duplication prior to the turtle-archosaur split and that a homologue has been maintained in turtles but lost in crocodiles [41]. It is also suggested that CYP2J19 originally played a role in colour vision by producing keto-carotenoids in retinal oil droplets and was independently co-opted in certain bird and turtle lineages for a role in integument coloration-based signalling [41,43]. Our phylogenetic reconstruction uncovered that CYP384A1, CYP383A1 and their orthologues are restricted to mites within the speciose Trombidiformes order, an ancient lineage that originated about 400 Ma [48,49]. Consistent with our hypothesis that the orthologous group containing CYP384A1 has a conserved carotenoid ketolase function, this order holds a high number of species that accumulate β-carotene and keto-carotenoid derivatives [50,51]. In contrast to the great majority of animals, including birds, previous work has strongly indicated that spider mites are able to biosynthesize β-carotene due to the lateral acquisition of fungal carotenoid biosynthetic genes and no longer depend on dietary salvage [4,52]. Interestingly, the horizontal transfer of carotenoid biosynthetic genes into mites occurred early in the evolution of the Trombidiformes order [52]. This raises the question of whether these cytochrome P450 enzymes were co-opted for keto-carotenoid production following the lateral acquisition. Although speculative, the horizontal transfer of carotenoid biosynthetic genes and the early evolution of CYP384A1 may have facilitated the appropriation of keto-carotenoids as the dominant pigments in many trombidiform mites.
Body coloration is known to have many adaptive functions in animals, including sexual, social and interspecific signalling [2,53]. Using water mites and fish predators as a biological system, Kerfoot [54] reasoned that the red-coloured, carotenoid-based body coloration serves an aposematic function in trombidiform mites, but more recent studies on the same system have reported contradictory results [55,56]. Carotenoid pigments have been implicated in protection against oxidative stress in various other animals [57], and currently the most popular hypothesis is that the keto-carotenoid accumulation in mite bodies is protective against light-associated oxidative damage in Trombidiformes. In support, Atarashi et al. [13] showed that albino Panonychus citri mites have a decreased non-enzymatic antioxidant capacity compared to wild-type mites. In addition, relative to non-diapausing T. urticae, diapausing T. urticae mites, which accumulate higher levels of keto-carotenoids [15,16], exhibit a higher resistance to UV light that induces reactive oxygen species [58]. Together, these studies suggest that carotenoids facilitate spider mite survival during long periods of increased UV exposure, although protection against other inducers of oxidative stress cannot be ruled out. Carotenoid metabolism is also critical for the animal visual system [10]. Visual chromophores, which function in phototransduction, rely on the enzymatic cleavage and modification of carotenoids into apo-carotenoids (vitamin A precursors) and disruption of these carotenoid metabolic pathways impairs vision [6,7]. In T. urticae, one of the horizontally acquired genes that allow for β-carotene biosynthesis is essential for diapause induction, reflecting the requirement of β-carotene as a precursory compound to perceive the inductive photoperiods [4]. We found that lemon T. kanzawai did not exhibit a decreased diapause incidence compared to its wild-type genetic background, supporting the hypothesis that light perception in spider mites depends on β-carotene but not its keto-carotenoid derivatives [3,4,37]. Biochemical and genetic work on previously recovered carotenoid pigment mutants strongly indicates that the spider mite eye spots owe their bright red coloration to an accumulation of esterified astaxanthin [16,17]. Astaxanthin might therefore act as a light filter, possibly protecting underlying photoreceptors from damage by intense light, a role also performed by the chemically unrelated ommochrome and pteridine pigments in the compound eyes of insects like Drosophila melanogaster [59,60]. Our findings shed light on the evolutionary history of keto-carotenoid production in trombidiform mites and open up new avenues to understand the potential adaptive value of keto-carotenoid-based traits in these invertebrates.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Xiaofeng Dong, Alistair Darby and Benjamin Makepeace for sharing the L. deliense and D. tinctorium proteomes ahead of peer-reviewed publication. We thank René Feyereisen for insightful discussions on cytochrome P450 function and phylogeny, and Jan van Arkel for technical assistance in photography. We are indebted to Tomoe Sekido and Tatsuya Sugawara for their help with the HPLC experiments. Sequencing data for this study were generated by the Genomics Core Facility of the Health Sciences Cores at the University of Utah.
Data accessibility
Read data and de novo assemblies are accessible at the NCBI SRA (PRJNA549514) and from the Dryad Digital Repository: https://doi.org/10.5061/dryad.qm364n4 [61], respectively. Coding sequences of T. kanzawai CYP384A1 are accessible at NCBI (MH879023-MH879024).
Authors' contributions
N.W., M.O., R.M.C. and T.V.L. designed experiments; N.W., A.B. and Y.M. performed experiments; N.W., A.H.K., R.G., O.K., Y.M., J.V., R.M.C. and T.V.L. analysed data; N.W. and A.H.K. produced figures; N.W. wrote the manuscript with input from all co-authors. All authors approved the final manuscript and agree to be held accountable for its content.
Competing interests
We declare we have no competing interests.
Funding
This project has received funding from the European Research Council (ERC) under the Horizon 2020 research and innovation program, grant agreement No 772026–POLYADAPT (to T.V.L.) and No 773902–SUPERPEST (to J.V. and T.V.L.) and by the USA National Science Foundation (award 1457346 to R.M.C.). N.W. was supported by a Research Foundation - Flanders (FWO) fellowship (12T9818N). R.G. was funded in part by the National Institutes of Health Genetics Training Grant T32GM007464. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.Goodwin TW. 1984. The biochemistry of the carotenoids, volume II: animals. London, UK: Chapman and Hall. [Google Scholar]
- 2.Hill GE, McGraw KJ. 2006. Bird coloration, vol. 2: function and evolution. Cambridge, MA: Harvard University Press. [Google Scholar]
- 3.Bosse TC, Veerman A. 1996. Involvement of vitamin A in the photoperiodic induction of diapause in the spider mite Tetranychus urticae is demonstrated by rearing an albino mutant on a semi-synthetic diet with and without p-carotene or vitamin A. Physiol. Entomol. 21, 188–192. ( 10.1111/j.1365-3032.1996.tb00854.x) [DOI] [Google Scholar]
- 4.Bryon A, et al. 2017. Disruption of a horizontally transferred phytoene desaturase abolishes carotenoid accumulation and diapause in Tetranychus urticae. Proc. Natl Acad. Sci. USA 114, E5871–E5880. ( 10.1073/pnas.1706865114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heath JJ, Cipollini DF, Stireman JO III. 2013. The role of carotenoids and their derivatives in mediating interactions between insects and their environment. Arthropod-Plant Interact. 7, 1–20. ( 10.1007/s11829-012-9239-7) [DOI] [Google Scholar]
- 6.von Lintig J. 2010. Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu. Rev. Nutr. 30, 35–56. ( 10.1146/annurev-nutr-080508-141027) [DOI] [PubMed] [Google Scholar]
- 7.von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K.. 2001. Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation in vivo. Proc. Natl Acad. Sci. USA 98, 1130–1135. ( 10.1073/pnas.98.3.1130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toomey MB, Collins AM, Frederiksen R, Cornwall MC, Timlin JA, Corbo JC. 2015. A complex carotenoid palette tunes avian colour vision. J. R. Soc. Interface 12, 20150563 ( 10.1098/rsif.2015.0563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britton G. 1995. Structure and properties of carotenoids in relation to function. FASEB J. 9, 1551–1558. ( 10.1096/fasebj.9.15.8529834) [DOI] [PubMed] [Google Scholar]
- 10.Toews DPL, Hofmeister NR, Taylor SA. 2017. The evolution and genetics of carotenoid processing in animals. Trends Genet. 33, 171–182. ( 10.1016/j.tig.2017.01.002) [DOI] [PubMed] [Google Scholar]
- 11.Lopes RJ, et al. 2016. Genetic basis for red coloration in birds. Curr. Biol. 26, 1427–1434. ( 10.1016/j.cub.2016.03.076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mundy NI, et al. 2016. Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr. Biol. 26, 1435–1440. ( 10.1016/j.cub.2016.04.047) [DOI] [PubMed] [Google Scholar]
- 13.Atarashi M, Manabe Y, Kishimoto H, Sugawara T, Osakabe M. 2017. Antioxidant protection by astaxanthin in the citrus red mite (Acari: Tetranychidae). Environ. Entomol. 46, 1143–1150. ( 10.1093/ee/nvx121) [DOI] [PubMed] [Google Scholar]
- 14.Ewing HE. 1914. The common red spider or spider mite. Or. Agric. Coll. Exp. Stn. 121, 33–39. [Google Scholar]
- 15.Kawaguchi S, Manabe Y, Sugawara T, Osakabe M. 2016. Imaginal feeding for progression of diapause phenotype in the two-spotted spider mite (Acari: Tetranychidae). Environ. Entomol. 45, 1568–1573. ( 10.1093/ee/nvw127) [DOI] [PubMed] [Google Scholar]
- 16.Veerman A. 1974. Carotenoid metabolism in Tetranychus urticae Koch (Acari: Tetranychidae). Comp. Biochem. Physiol. Part B Comp. Biochem. 47, 101–116. ( 10.1016/0305-0491(74)90095-9) [DOI] [PubMed] [Google Scholar]
- 17.Veerman A. 1972. Carotenoids of wild-type and mutant strains of Tetranychus pacificus McGregor (Acari: Tetranychidae). Comp. Biochem. Physiol. Part B Comp. Biochem. 42, 329–340. ( 10.1016/0305-0491(72)90277-5) [DOI] [Google Scholar]
- 18.Veerman A. 1970. The pigments of Tetranychus cinnabarinus boisd. (Acari: Tetranychidae). Comp. Biochem. Physiol. 36, 749–763. ( 10.1016/0010-406X(70)90530-X) [DOI] [Google Scholar]
- 19.Helle W, van Zon AQ.. 1967. Rates of spontaneous mutation in certain genes of an arrhenotokous mite, Tetranychus pacificus. Entomol. Exp. Appl. 10, 189–193. ( 10.1111/j.1570-7458.1967.tb00058.x) [DOI] [Google Scholar]
- 20.Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492. ( 10.1038/nature10640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran NA, Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328, 624–627. ( 10.1126/science.1187113) [DOI] [PubMed] [Google Scholar]
- 22.Wybouw N, et al. 2019. Long-term population studies uncover the genome structure and genetic basis of xenobiotic and host plant adaptation in the herbivore Tetranychus urticae. Genetics 211, 1409–1427. ( 10.1534/genetics.118.301803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org). [Google Scholar]
- 24.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. ( 10.1038/ng.806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Auwera GA, et al. 2013. From FastQ data to high-confidence variant calls: The Genome Analysis Toolkit best practices pipeline. In Current protocols in bioinformatics (eds Bateman A, Pearson WR, Stein LD, Stormo GD, Yates JR), pp. 11.10.1–11.10.33. Hoboken, NJ: John Wiley & Sons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. ( 10.1093/bioinformatics/bts635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinf. 13, 238 ( 10.1186/1471-2105-13-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26, 1899–1900. ( 10.1093/bioinformatics/btq224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. ( 10.1093/molbev/msw260) [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veerman A. 1980. Functional involvement of carotenoids in photoperiodic induction of diapause in the spider mite, Tetranychus urticae. Physiol. Entomol. 5, 291–300. ( 10.1111/j.1365-3032.1980.tb00237.x) [DOI] [Google Scholar]
- 38.Helle W, van Zon AQ.. 1970. Linkage studies in the pacific spider mite Tetranychus pacificus. II. Genes for white eye II, lemon and flamingo. Entomol. Exp. Appl. 13, 300–306. ( 10.1111/j.1570-7458.1970.tb00114.x) [DOI] [Google Scholar]
- 39.Álvarez V, Rodríguez-Sáiz M, de la Fuente JL, Gudiña EJ, Godio RP, Martín JF, Barredo JL.. 2006. The crtS gene of Xanthophyllomyces dendrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of β-carotene into astaxanthin and other xanthophylls. Fungal Genet. Biol. 43, 261–272. ( 10.1016/j.fgb.2005.12.004) [DOI] [PubMed] [Google Scholar]
- 40.Ojima K, Breitenbach J, Visser H, Setoguchi Y, Tabata K, Hoshino T, van den Berg J, Sandmann G. 2006. Cloning of the astaxanthin synthase gene from Xanthophyllomyces dendrorhous (Phaffia rhodozyma) and its assignment as a β-carotene 3-hydroxylase/4-ketolase. Mol. Genet. Genomics 275, 148–158. ( 10.1007/s00438-005-0072-x) [DOI] [PubMed] [Google Scholar]
- 41.Twyman H, Valenzuela N, Literman R, Andersson S, Mundy NI. 2016. Seeing red to being red: conserved genetic mechanism for red cone oil droplets and co-option for red coloration in birds and turtles. Proc. R. Soc. B 283, 20161208 ( 10.1098/rspb.2016.1208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunlop JA, Alberti G. 2007. The affinities of mites and ticks: a review. J. Zool. Syst. Evol. Res. 46, 1–18. ( 10.1111/j.1439-0469.2007.00429.x) [DOI] [Google Scholar]
- 43.Twyman H, Prager M, Mundy NI, Andersson S. 2018. Expression of a carotenoid-modifying gene and evolution of red coloration in weaverbirds (Ploceidae). Mol. Ecol. 27, 449–458. ( 10.1111/mec.14451) [DOI] [PubMed] [Google Scholar]
- 44.McGraw KJ, Hill GE, Stradi R, Parker RS. 2001. The influence of carotenoid acquisition and utilization on the maintenance of species-typical plumage pigmentation in male American goldfinches (Carduelis tristis) and Northern cardinals (Cardinalis cardinalis). Physiol. Biochem. Zool. 74, 843–852. ( 10.1086/323797) [DOI] [PubMed] [Google Scholar]
- 45.Jensen NB, Zagrobelny M, Hjernø K, Olsen CE, Houghton-Larsen J, Borch J, Møller BL, Bak S. 2011. Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nat. Commun. 2, 273 ( 10.1038/ncomms1271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. 2001. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20, 319–331. ( 10.1007/s003440010037) [DOI] [PubMed] [Google Scholar]
- 47.Weng J-K, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C. 2010. Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22, 1033–1045. ( 10.1105/tpc.109.073528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dabert M, Witalinski W, Kazmierski A, Olszanowski Z, Dabert J. 2010. Molecular phylogeny of acariform mites (Acari, Arachnida): strong conflict between phylogenetic signal and long-branch attraction artifacts. Mol. Phylogenet. Evol. 56, 222–241. ( 10.1016/j.ympev.2009.12.020) [DOI] [PubMed] [Google Scholar]
- 49.Xue X-F, Dong Y, Deng W, Hong X-Y, Shao R. 2017. The phylogenetic position of eriophyoid mites (superfamily Eriophyoidea) in Acariformes inferred from the sequences of mitochondrial genomes and nuclear small subunit (18S) rRNA gene. Mol. Phylogenet. Evol. 109, 271–282. ( 10.1016/j.ympev.2017.01.009) [DOI] [PubMed] [Google Scholar]
- 50.Czeczuga B, Czerpak R. 1968. Pigments occurring in Hydrachna geografica and Piona nodata (Hydracarina, Arachnoidea). Experientia 24, 218–219. ( 10.1007/BF02152777) [DOI] [PubMed] [Google Scholar]
- 51.Green J. 1964. Pigments of the hydracarine Eylais extendens (Acari: Hydrachnellae). Comp. Biochem. Physiol. 13, 469–472. ( 10.1016/0010-406X(64)90039-8) [DOI] [Google Scholar]
- 52.Dong X, et al. 2018. Genomes of trombidid mites reveal novel predicted allergens and laterally-transferred genes associated with secondary metabolism. GigaScience 7, giy127 ( 10.1093/gigascience/giy127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svensson Wong. 2011. Carotenoid-based signals in behavioural ecology: a review. Behaviour 148, 131–189. ( 10.1163/000579510X548673) [DOI] [Google Scholar]
- 54.Kerfoot WC. 1981. A question of taste: crypsis and warning coloration in freshwater zooplankton communities. Ecology 63, 538–554. ( 10.2307/1938969) [DOI] [Google Scholar]
- 55.Proctor HC, Garga N. 2004. Red, distasteful water mites: did fish make them that way? In Aquatic mites from genes to communities (ed. Proctor HC.), pp. 127–147. Dordrecht, The Netherlands: Springer. [DOI] [PubMed] [Google Scholar]
- 56.Walter DE, Proctor HC. 2013. Acari underwater, or, why did mites take the plunge? In Mites: ecology, evolution & behaviour, pp. 229–280. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 57.Esteban R, Moran JF, Becerril JM, García-Plazaola JI. 2015. Versatility of carotenoids: an integrated view on diversity, evolution, functional roles and environmental interactions. Environ. Exp. Bot. 119, 63–75. ( 10.1016/j.envexpbot.2015.04.009) [DOI] [Google Scholar]
- 58.Suzuki T, Watanabe M, Takeda M. 2009. UV tolerance in the two-spotted spider mite, Tetranychus urticae. J. Insect Physiol. 55, 649–654. ( 10.1016/j.jinsphys.2009.04.005) [DOI] [PubMed] [Google Scholar]
- 59.Cosens D, Briscoe D. 1972. A switch phenomenon in the compound eye of the white-eyed mutant of Drosophila melanogaster. J. Insect Physiol. 18, 627–632. ( 10.1016/0022-1910(72)90190-4) [DOI] [Google Scholar]
- 60.Summers KM, Howells AJ, Pyliotis NA. 1982. Biology of eye pigmentation in insects. Adv. Insect Physiol. 16, 119–166. [Google Scholar]
- 61.Wybouw N, Kurlovs AH, Greenhalgh R, Bryon A, Kosterlitz O, Manabe Y, Osakabe M, Vontas J, Clark RM, Leeuwen TV. 2019. Data from: Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals Dryad Digital Repository. (doi:10.5061/dryad.qm364n4) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wybouw N, Kurlovs AH, Greenhalgh R, Bryon A, Kosterlitz O, Manabe Y, Osakabe M, Vontas J, Clark RM, Leeuwen TV. 2019. Data from: Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals Dryad Digital Repository. (doi:10.5061/dryad.qm364n4) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Read data and de novo assemblies are accessible at the NCBI SRA (PRJNA549514) and from the Dryad Digital Repository: https://doi.org/10.5061/dryad.qm364n4 [61], respectively. Coding sequences of T. kanzawai CYP384A1 are accessible at NCBI (MH879023-MH879024).