Abstract
A p73 is a new member of p53 family of transcription factor, having two types. First is TAp73, transcriptionally active and expressed via upstream promoter as a tumor suppressor and vital apoptotic inductor, it also has a key role in cell cycle arrest/differentiation and Second is ΔNp73 that is transcriptionally inactive and expressed via downstream regulator as oncogenes. Both types are expressed in various isoforms, which originate from alternative splicing events at the C-terminus. Upon DNA damage, posttranslational modifications cause conformational changes in various amino acid residues via induction or inhibition of various proteins, which are present in the structural domains of p73. These modifications may cause up- or down-regulation of p73 expression levels, as well as alters the transcriptional activity and/or stability of the protein. In this review, we have made an effort to assemble all existing data regarding the role of p73, its modification and after effects in cancer.
Key words: p73, post translational modification, methylation, phosphorylation, acetylation, ubiqutination
Introduction
A monoallelically articulated tumor suppressor gene is found at 1p36 region in numerous tumor cell lines, someway similar to p53, named as p73 gene. It is rarely mutated in cancer, but its level increased in various cancers, including hepatocellular carcinomas, neuroblastomas, lung, prostate, colon, breast and ovarian cancer. 1,2 It allocates a noteworthy sequence resemblance with the p53 structure of DNA-binding, transactivation and oligomerization domain that is 63%, 29% and 38% respectively and intermingles with p53-binding proteins. Its wild type and mutant form bind with DNA more competently than p53.3 In this review, we have tried our best to summarize all information regarding p73 as a tumor suppressant as well as its oncogenic potential.
Structure and Isoforms of p73
p73 protein contains an amino-terminal Transactivation Domain (TA) which is having two Isoforms, TAp73 (tumor suppressant form, transcriptionally active because it includes exons 2 and 3, which encode the transactivation domain due to the presence of NH2-terminal) and ΔNp73 (oncogenic form, transcriptionally inactive due to lack of NH2-terminal)2 (Figure 1), A Proline Rich Domain (PR), a foundation and highest homology region with p53, DNA Binding Domain (DBD)4 and p73 protein contains an amino-terminal Transactivation Domain (TA) which is having two Isoforms. It is a globular domain, comprises of 4 α-helixes and finds in proteins those participate in the development and arbitrate in protein/protein and protein/RNA interactions.5
Figure 1.
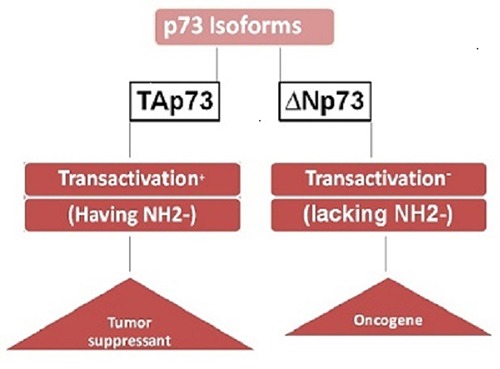
2 isoforms of p73.
The alternative splicing of p73 at the COOH - terminal generates six diverse TAp73 isoforms which includes α, a longer isoform with SAM domain, β, g, d, e and z (Figure 2) and NH2-terminal generates assorted ΔNp73 isoforms, Δ2p73, Δ3p73, Δ2, 3p73 etc. which are originating from alternative splicing events at the C-terminus.6
Figure 2.
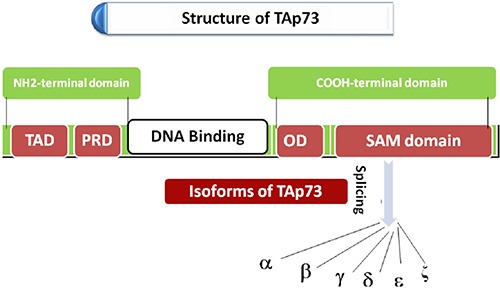
Splicing at COOH-terminal of TAp73.
Promoters of p73
Two promoters, P1 and P2 promote TAp73 upon DNA damage and trigger the transcription of p53-responsive genes, like p21 that control the cell-cycle arrest, NOXA, PUMA and BAX (apoptosis regulators), cause apoptosis and cell cycle arrest7 and ΔNp73 upon non apoptotic DNA damage correspondingly.
P1 is TAp73 promoter having 3 potential consensuses regions of E2F1 which is a fundamental apoptosis and cell cycle inductor and adjust the p73 expression in G1/S phase.8,9 It increases the level of p16 and p14ARF which inhibit the MDM2 and degrade p53 by forming MDM2-p53 complex and elevates its level in cancer cells.10 E2F1 influences p73 level and cause p73 up-regulation in a p53 dependent manner due to huge structural homology with p53 (Figure 3A). Another fact about MDM2 is that it causes p73 inactivation instead of degradation by E3 ubiquitin ligase.
Figure 3.
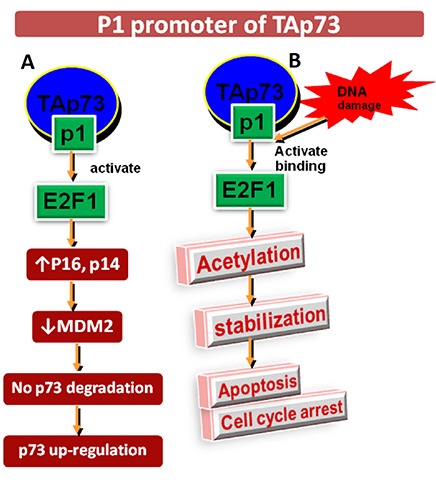
A) E2F1: p53 dependent p73 up-regulation by activating p16, p14ARF and inhibiting MDM2 complex like p53 manner. B) E2F1: p53 independent p73 up-regulation by initiating acetylation, stabilize E2F1/p73 complex and cause apoptosis and cell cycle arrest.
E2F1 also induces p53-independent cell death because it triggers the TAp73 expression at P1 promoter.11 E2F1 binding to TAp73 P1 promoter and triggers its acetylation after DNA damage. It stabilizes E2F1/p73 complex and induces apoptosis and cell cycle arrest12 (Figure 3B). P2 is ΔNp73 promoter with no E2F1 binding region, but having p53/p73 responsive element. The nonapoptotic DNA damage activates P2p73 in p53-dependent manner and cause abundance of ΔNp73 proteins that only causes cell cycle arrest but do not undergo apoptosis.13,14 It exhibits negative autoregulatory feedback by inhibiting p53 and TAp73 isoforms while both proteins induce ΔNp73 level.15,16 ΔNp73 deregulation promotes malignant transformation of fibroblasts and tumor growth.17
Regulation of p73
The comparative intensity of both forms TAp73 and ΔNp73 decide cell fortune either growth/development arrest or unrestrained propagation. After DNA damage in the steady state level of TAp73 isoforms is up-regulated but ΔNp73 degraded quickly.
E2F1 causes direct up-regulation of p73 via recognition and transactivation of TAp73 gene on P1 promoter site as discussed above while E1A and c-myc (oncogenes) promote p73-dependent apoptosis in tumor cells by binding with E1A and p300/CBP and RB (E2F1 regulators).18,19 So, E1A and c-myc put forth a synergistic effect on p73 activity which is sometimes not related to E2F1 activation.16
Post translational modification
These modifications establish p73 specificity/sensitivity to specific proteins and regulate its transcriptional activities like p53 by methylation, phosphorylation, acetylation, ubiquitination and sumoylation under normal circumstances and DNA damage.
Methylation: DNA methylation, which is a part of epigenetic alterations and histone modification, is a covalent chemical adaptation at CpG nucleotides in which CH3 group is shifted from donor S-adenosylmethionine (SAM) to C-5 cytosine via DNA cytosine-5-methyltransferases. It regulates the genomic expression and silencing of tumor suppressant genes which look after cells from malignant alteration called as TSG like p73, identified in 1997.20 Four isoenzymes of methyltransferase are DNMT1, DNMT2, DNMT3A and DNMT3B21 accountable for maintenance and de novo DNA methylation during embryogenesis and cause alterations in genomic methylation which pursue epigenetics and cancer in cells. For example, malignant tumor shows overall hypoleads to hypermethylation and silencing of TSG by directly blocking the specific binding sites of transcription factor by the methyl- CpG-binding proteins (MBDPs)22 (Figure 4).
Figure 4.
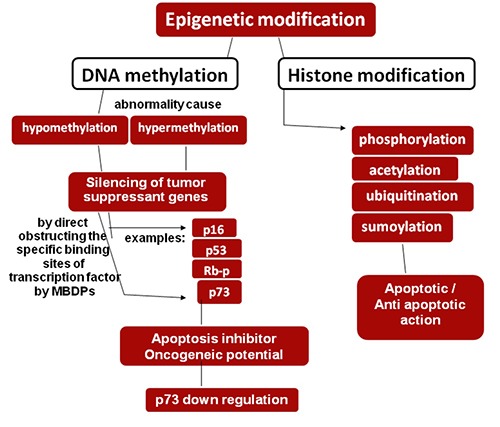
DNA methylation, a part of epigenetic modification leads to silencing of TSGs by direct obstructing the specific binding sites of transcription factor by MBDPs via aberrant methylation and leads to p73 down regulation resulting oncogenes and apoptosis inhibition.
These methylated TSGs play important role in regulation of cell cycle, DNA repairs, apoptosis, transcriptional regulation, angiogenesis and metastasis.23,24 TSGs become nonfunctional via mutation, deletion and in expression of promoter due to its hypermethylation which leads to epigenetic silencing and induces oncogenic potential.
Phosphorylation
Phosphorylation by the non-receptor tyrosine-kinase c-Abl was first revealed p73 post-translational modification. The c-Abl tyrosine kinase is dispersed in the nucleus and cytoplasm of proliferating cells. Nuclear c-Abl activity is pessimistic regulated by the retinoblastoma protein (RB) and optimistically regulated by DNA damaging agents like Cisplatin and ionizing radiation (gIR).25 CAbl tyrosine kinase interacts with the p73 proline-rich region in the SH3 domain via p73 PxxP motif at COOH-terminal OD and cause phosphorylation of p73 primarily at Tyr-99 then Tyr-121 and Tyr- 240. It leads to p73 stabilization, accumulation in the nucleus, halflife (T1/2), transcription, apoptosis and cell cycle arrest in growth phase.26
Other important kinases are mitogen-activated protein kinase (MAPK), Checkpoint 1 and 2 kinases, protein kinase C and cyclin dependent kinase. 1: Activated c-Abl tyrosine kinase also promotes MAPK after DNA damaging signals and causes p73 phosphorylation at thr-167 residues that are located adjacent to proline via p38 protein. p38 has ability to phosphorylate p73 threonine residue without c-Abl tyrosine kinase. So, it enhances the p73 transcriptional activity and apoptosis by phosphorylation pathways instead of cell cycle arrest.27 2: In response to DNA damage, Checkpoint kinase 1 (Chk1) causes phosphorylation of p73α at Ser-47 residues and causes TAp73α accumulation and apoptosis induction. Chk1 activates p73 expression, but Chk2 has a role in p73 up-regulation.
They contribute to E2F1-mediated p73 up-regulate transcription by phosphorylation and activation of E2F1 transcription factor28,29 (Figure 5). In fact, p73 does not require DNA damaging signals for phosphorylation. It becomes phosphorylated during cell cycle as well. So, it requires Protein kinase Cd and cyclin dependent kinase. During the cell cycle, PKCδ causes phosphorylation of p73 at Ser-388 residues at second TAD. It causes activation of second TAD and regulation of cell cycle progression but remains unable to induce apoptosis. But in response to DNA damage, PKCd is sliced by caspase-3 to active catalytic fragment (PKCδ CF) that causes phosphorylation of p73β at Ser-289 residues of DBD and leads to p73β accumulation and induce its transcriptional activity and apoptosis.30 During G2/M phase of cell cycle, cyclin/CDK complex causes p73 phosphorylation at Thr-86 residue via its cyclin recognition motifs. It inhibits the p73 transcriptional activity and activates the growth phase during cell cycle progression. It obstructs the p73 tumor suppressant activity as well. The cyclin dependent kinase inhibitor p16 causes cell cycle arrest and inhibits the phosphorylation at Thr-86 residues.31 This suggests that phosphorylation of p73 might not all time switch a suppressed type of p73 to the dynamic form. It may lead to p73 down regulation as well.
Figure 5.
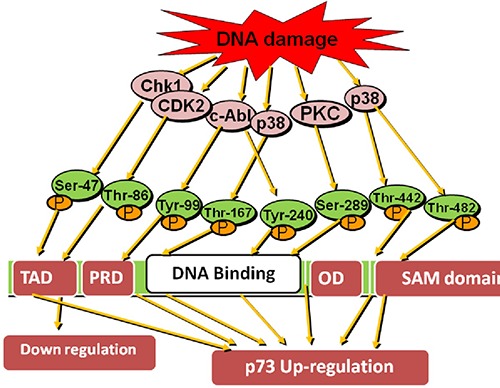
Phosphorylation of p73 via different kinases at Ser-47 and 289, Thr-86/167/442 and 482, Tyr-99 and 240 amino acid residues upon exposure with DNA damaging agents and up regulate p73 level else CDK2 which down regulate p73 via phosphorylating Thr-86 at TAD domain in p73 structure
Acetylation
p73 linked with p300 histone acetyltransferase at its NH2- transactivation domain. p300 provokes acetylation of p73α at Lys- 321, Lys-327 and Lys-331 with c-Abl kinase by DNA damaging agent doxorubicin. The prolil isomerase Pin1 (involve in G1/S and G2/M phase) promotes p73 acetylation with p300 that specifically identify phosphorylated serine/threonine residues caused by proline and bring conformational change in substrate structure.32 Pin1 stabilizes the p73 gene under normal and stress circumstances and increase p73 transcription.33 Upon stress circumstances, c-Abl tyrosine kinase and MAPKp38 pathway enhance affinity of p73a for Pin1.YAP (yes associated protein) also promotes p300-mediated acetylation of p73 and induce apoptosis. A non-acetylated p73 is transcriptionally inactive form (Figure 6).
Figure 6.
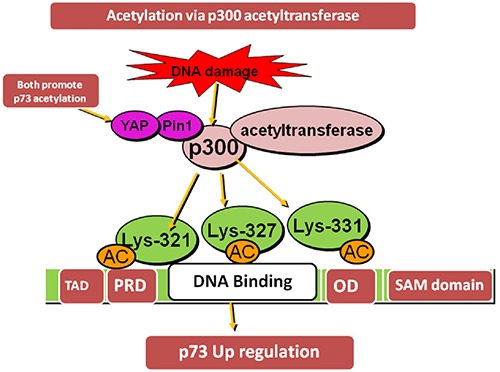
Acetylation of p73α via p300 acetyltransferase at NH2 terminal TAD at Lys-321/327 and 331 with c-Abl kinase in promotion with YAP and Pin 1 by DNA damaging agent doxorubicin resulting in p73 up-regulation
Ubiquitination.
Ubiquitination is responsible for proteasomal degradation of proteins. MDM2, the E3 ubiquitin (Ub) ligase is a key controller of p53 revenue. It unites with p53 NH2-domain and cause ubiquitination and proteasomal-dependent degradation. But p53 and p73 have the ability to promote transcription of gene encoding for MDM2. So, due to homology between p53 and p73, MDM2 correlates with p73 as well. E3 Ub ligase becomes down regulated and specifically binds to TAp73 α and β isoforms via PY motif and stabilize/ regulates their levels in the cell after exposure to DNA damaging agents like doxorubicin, cisplatin etc. instead of ubiquitination and degradation while ΔNp73 is degraded by proteasomal dependent pathway. So, TAp73 level increases and ΔNp73 level decreases.34 E3 Ub ligase regulates the ratio of TAp73/ΔNp73 that is important for the apoptosis.35 The TAp73 g and d isoforms lack PY motif domain and are not regulated by E3 Ub ligase. TAp73 acetylation provides shelter from ubiquitination and leads to p73 stabilization. Another factor, YAP1 (Yes-associated protein 1) fight with E3 Ub ligase for its binding to p73 and cause inactivation of E3 Ub ligase and ultimately up the p73 regulation36 (Figure 7).
Figure 7.
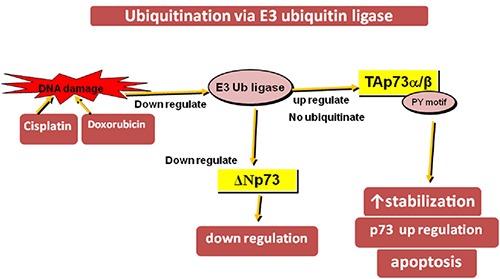
Ubiquitination of TAp73 α/β at PY motif domain via E3 Ub ligase upon doxorubicin or cisplatin exposure and leads to p73 up-regulation but ubiquitination of ΔNp73 via E3 Ub ligase down regulate it.
Sumoylation
Sumoylation by small ubiquitin-like modifier (SUMO) protein in p73 of COOH-terminal at Lys-627 takes place, particularly in p73a in vitro and triggers its degradation by proteasome but sumoylation in p53 stimulates its transcription.37 So, p73ß is more powerful than p73α in transcription because the sumoylation at Lys-627 of p73α influences its interactions with various proteins like c-Abl tyrosine kinase.38 Along with post-translational modifications the expression of p73 is keeping pace by physical interaction with numerous viral and cellular proteins.
p73 has an association with adenovirus E1A and the T-cell lymphotropic virus I-derived Tax which leads to its inactivation like p53.39 Various viral proteins such as adenovirus E1B, papilomavirus E6 and simian virus 40 T antigen has no interaction with p73 but all these cause inactivation of p53.
Importance of p73 expression in human cancer
The p73 genomic expression in all normal human tissues is at very low intensity. So, recognition is not so easy. All the findings show the adequate balance between two antagonistic classes of proteins TAp73 and ΔNp73 those are encoded by the same gene. Their balanced ratio determines the cell reaction upon growth stimuli, DNA damage or pathogenic attack. Due to depiction of a whole variety of p73 isoforms, it is well recognized by various agents and differentiates between TA and ΔN forms properly.
The 1p36.3 chromosomes for human gene Tp73 deficient in various types of cancers is enough for functional loss and persuades the cell to cancer like neuroblastoma, colorectal and breast cancer.12 A diversity of studies depicts the monoallelical expression in various tissues and cancers means when one allele is imprinted. This one allele deficient cell is sufficient to make a cell p73 void. It also demonstrates the loss of imprinting (LOI), biallelic expression or the allele switching of p73. The loss of function transmutations in p73 is very much rarer i.e. 0.6% when taking evidence from almost >1100 primary tumors.
But in spite of loss of expression the most common noticeable alteration is over expression of different isoforms of the wild type p73.39 A high occurrence of loss of heterozygosity for p73 has observed in neuroblastoma, lung, gastric and ovarian tumors.24 Mutations in TAp73 gene are seldom found in human malignant cells like P405R and P425L, a somatic and a germ line in primary neuroblastoma. P425L replacement decreases the p73α-dependent developmental suppression, but not p73β transcriptional activity, but exchange in P405R has no effect on p73α/β transcription.40
Chemo sensitivity with regards to p73
p73 level enhances by small spectrum chemotherapeutic anticancer drugs like anthracyclines, topoisomerase I and II inhibitors, microtubule inhibitors and alkylating agents such as cisplatin, adriamycin, texol, etoposide and doxorubicin treatment in different tumor derived cell lines.32 The reason of p73 up-regulation is its enhanced transcriptional activity and increased protein stability, which leads to induction of apoptosis via apoptosis commencement.
Chemo resistance increases by blocking TAp73 via inhibiting p73DD fragment or p73 RNA. Up-regulated ΔNp73 also has ability to stop the apoptosis in wild-type p53 cancer cells induced via various chemotherapeutic agents.14-20 But up-regulation of p53 mutant due to Arg-72 polymorphism40 blocks the p73 transcriptional activity and ultimately apoptosis.38 p73 has ability to induce apoptosis in the absence of p53 but p53 cannot do so when the fibroblast cells lacking p73. All these studies show the importance of p73 in regards to chemotherapeutic agents.
Concluding annotations
This review principally stresses the functions of p73, interacting proteins in human cancer and chemosenstivity with regards to p73 gene. p73 functions are mostly trans mutated in various cancers. One of p73 isoform, TAp73 potentially triggers upon DNA damage for cell cycle arrest and apoptosis and vice versa for other isoform ΔNp73. So, the balance between both types is vital to settle down the p73 apoptotic fate in cancerous cells. In spite of high structural and functional homology with p53, both have substantial dissimilarities as well. Post-translational modifications pin point their discrepancy behavior throughout growth and malignant makeover. Like E3 Ub ligase mdm2 causes degradation of p53 but stabilizes p73. So all the signals those mutate p53 are not able to infect p73. Eventually, improved perception of all these pathways may direct to the expansion of innovative p73 oriented therapies.
References
- 1.Melino G, Bernassola F, Ranalli M, et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem 2004;279:8076-83. [DOI] [PubMed] [Google Scholar]
- 2.Stiewe T, Putzer BM. Role of p73 in malignancy: tumor suppressor or oncogene. Cell Death Diff 2002;9:237-45. [DOI] [PubMed] [Google Scholar]
- 3.Kartasheva NN, Contente A, Lenz-Stoppler C, et al. p53 induces the expression of its antagonist p73 Delta N, establishing an autoregulatory feedback loop. Oncog 2002;21:4715-27. [DOI] [PubMed] [Google Scholar]
- 4.Dotsch V, Bernassola F, Coutandin D, et al. p63 and p73 the ancestors of p53. Cold Spring Harb Persp Biol 2010;2:a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishimoto O, Kawahara C, Enjo K, et al. Possible oncogenic potential of ΔNp73: a newly identified isoform of human p73. Canc Res 2002;62:636-41. [PubMed] [Google Scholar]
- 6.Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci 2003;12:625-8. [DOI] [PubMed] [Google Scholar]
- 7.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Canc 2002;2:605-15. [DOI] [PubMed] [Google Scholar]
- 8.Irwin MS, Kondo K, Marin MC, et al. Chemosensitivity linked to p73 function. Canc Cell 2003;3:403-10. [DOI] [PubMed] [Google Scholar]
- 9.Stevens C, La Thangue NB. E2F and cell cycle control: a double- edged sword. Arch Biochem Biophys 2003;412:157-69. [DOI] [PubMed] [Google Scholar]
- 10.Pediconi N, Ianari A, Costanzo A, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 2003;5:552-8. [DOI] [PubMed] [Google Scholar]
- 11.Polager S, Ginsberg D. E2F1 at the crossroads of life and death. Tre Cell Biol 2008;18:528-35. [DOI] [PubMed] [Google Scholar]
- 12.Stiewe T, Pützer BM. Role of the p53-homologue p73 in E2F1- induced apoptosis. Nat Genet 2000;26:464-69. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa T, Takahashi M, Ozaki T, et al. Autoinhibitory regulation of p73 by ΔNp73 to modulate cell survival and death through a p73-specific target element within the ΔNp73 promoter. Mol Cell Biol 2002;22:2575-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vossio S, Palescandolo E, Pediconi N, et al. ΔN-p73 is activated after DNA damage in a p53-dependent manner to regulate p53-induced cell cycle arrest. Oncog 2002;21:3796-803. [DOI] [PubMed] [Google Scholar]
- 15.Grob TJ, Novak U, Maisse C, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Diff 2001;8:1213-23. [DOI] [PubMed] [Google Scholar]
- 16.Zaika AI, Slade N, Erster SH. ΔNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med 2002;196:765-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allart S, Martin H, Detraves C, et al. Human cytomegalovirus induces drug resistance and alteration of programmed cell death by accumulation of ΔN-p73α. J Biol Chem 2002;277:29063-68. [DOI] [PubMed] [Google Scholar]
- 18.Putzer BM, Stiewe T, Parssanedjad K, et al. E1A is sufficient by itself to induce apoptosis independent of p53 and other adenoviral gene products. Cell Death Diff 2000;7:177-88. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe K, Ozaki T, Nakagawa T, et al. Physical interaction of p73 with c-Myc and MM1, a c-Myc-binding protein, and modulation of the p73 function. J Biol Chem 2002;277:15113-23. [DOI] [PubMed] [Google Scholar]
- 20.Bird A. DNA methylation patterns and epigenetic memory. Genet Devel 2002;16:6-21. [DOI] [PubMed] [Google Scholar]
- 21.Wang KY, James-Shen CK. DNA methyltransferase DNMT1 and mismatch repair. Oncog 2004;23:7898-902. [DOI] [PubMed] [Google Scholar]
- 22.Deng G, Chen A, Pong E, Kim YS. Methylation in hMLH1 promoter interferes with its binding to transcription factor CBF and inhibits gene expression. Oncog 2001;20:7120-7. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncog 2002;21:5427-40. [DOI] [PubMed] [Google Scholar]
- 24.Esteller M. Epigenetic in cancer. N Eng J Med 2008;358:1148-59. [DOI] [PubMed] [Google Scholar]
- 25.Tsai KK, Yuan ZM. c-Abl stabilizes p73 by a phosphorylationaugmented interaction. Canc Res 2003;63:3418-24. [PubMed] [Google Scholar]
- 26.Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncog 2000;19:5643-50. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez PR, Sanchez VJ, Servitja JM, Gutkind JS. Regulation of p73 by c-Abl through the p38 MAP kinase pathway. Oncog 2002;21:974-9. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez S, Prives C, Cordon C. p73α regulation by Chk1 in response to DNA damage. Mol Cell Biol 2003;23:8161-71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 2004;18:3041-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J, Datta R, Shioya H, et al. p73β is regulated by protein kinase CΔ catalytic fragment generated in the apoptotic response to DNA damage. J Biol Chem 2002;277:33758-65. [DOI] [PubMed] [Google Scholar]
- 31.Gaiddon C, Lokshin M, Gross I. Cyclin-dependent kinases phosphorylate p73 at threonine 86 in a cell cycle-dependent manner and negatively regulate p73. J Biol Chem 2003;278:27421-31. [DOI] [PubMed] [Google Scholar]
- 32.Costanzo A, Merlo P, Pediconi N, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell 2002;9:175-86. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani F, Piazza S, Gostissa M, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell 2004;14:625-36. [DOI] [PubMed] [Google Scholar]
- 34.Bernassola F, Salomoni P, Oberst A, et al. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med 2004;199:1545-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maisse C, Guerrieri P, Melino G. p73 and p63 protein stability the way to regulate function. Biochem Pharmacol 2003;66:1555-61. [DOI] [PubMed] [Google Scholar]
- 36.Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Diff 2007;14:743-51. [DOI] [PubMed] [Google Scholar]
- 37.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1, Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 2000;275:36316-23. [DOI] [PubMed] [Google Scholar]
- 38.Maisse C, Munarriz E, Barcaroli D, et al. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Diff 2004;11:685-7. [DOI] [PubMed] [Google Scholar]
- 39.Irwin M, Marin MC, Phillips AC, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nat 2000;407:645-8. [DOI] [PubMed] [Google Scholar]
- 40.Naka M, Ozaki T, Takada N, et al. Functional characterization of naturally occurring mutants (P405R and P425L) of p73alpha and p73beta found in neuroblastoma and lung cancer. Oncog 2001;20:3568-72. [DOI] [PubMed] [Google Scholar]


