Abstract
Objectives
Development of digital biomarkers to predict treatment response to a digital behavioural intervention.
Design
Machine learning using random forest classifiers on data generated through the use of a digital therapeutic which delivers behavioural therapy to treat cardiometabolic disease. Data from 13 explanatory variables (biometric and engagement in nature) generated in the first 28 days of a 12-week intervention were used to train models. Two levels of response to treatment were predicted: (1) systolic change ≥10 mm Hg (SC model), and (2) shift down to a blood pressure category of elevated or better (ER model). Models were validated using leave-one-out cross validation and evaluated using area under the curve receiver operating characteristics (AUROC) and specificity- sensitivity. Ability to predict treatment response with a subset of nine variables, including app use and baseline blood pressure, was also tested (models SC-APP and ER-APP).
Setting
Data generated through ad libitum use of a digital therapeutic in the USA.
Participants
Deidentified data from 135 adults with a starting blood pressure ≥130/80, who tracked blood pressure for at least 7 weeks using the digital therapeutic.
Results
The SC model had an AUROC of 0.82 and a sensitivity of 58% at a specificity of 90%. The ER model had an AUROC of 0.69 and a sensitivity of 32% at a specificity at 91%. Dropping explanatory variables related to blood pressure resulted in an AUROC of 0.72 with a sensitivity of 42% at a specificity of 90% for the SC-APP model and an AUROC of 0.53 for the ER-APP model.
Conclusions
Machine learning was used to transform data from a digital therapeutic into digital biomarkers that predicted treatment response in individual participants. Digital biomarkers have potential to improve treatment outcomes in a digital behavioural intervention.
Keywords: digital therapeutics, machine learning, hypertension, mobile health, behavioural therapy
Strengths and limitations of this study.
Proof-of-concept biomarkers demonstrated good power to predict treatment outcomes despite the small size of the training dataset.
Use of additional explanatory variables to develop the biomarkers may enhance the accuracy of predictions.
Generalisability of the biomarkers is unknown and may be limited by the demographics of the training dataset.
Introduction
Modifiable behaviours are responsible for 70% or more of all cardiometabolic diseases.1–3 Health systems are ill equipped to address the current epidemic of cardiometabolic diseases and, in particular, lack widely available behavioural therapies to address the common root causes of these conditions. Digital therapeutics, software designed to encourage changes in behaviours which treat disease, offer a means to deliver behavioural therapy to large populations and preliminary studies demonstrate their potential as a cost-effective treatment for cardiometabolic disease.4–7
Compared with pharmacotherapy, digital therapeutics offer potential advantages such as ease of access, ease of use, fewer side-effects and cost effectiveness.4 8–10 Digital therapeutics also generate readily accessible patient data without requiring an office or lab visit. The data generated are voluminous and vary in both type and quality. These can include remotely sensed measures of physiology (eg, blood pressure (BP), blood glucose, heart rate variability), behavioural data (eg, about eating, moving, thinking), medication adherence, as well as engagement parameters of unknown significance (eg, app use, geographic location, circadian patterns of use).
The best use of these data remains an open question. Feeding data directly into electronic medical records is of limited utility to providers or patients whereas transforming the data into markers of disease status, termed digital biomarkers11 could provide clinically actionable insights with or without conventional biometric data.12–14 Digital biomarkers afford a pragmatic approach to remotely monitor patients and intervene on a continuous rather than episodic basis. Greatly expanding opportunities to intervene means that patients have greater access to personalised care, which could improve treatment outcomes.15 16
Machine learning, a type of artificial intelligence (AI) used to make predictions with large and complex datasets, offers a novel approach for creating digital biomarkers. The exponential growth of smartphone use in the USA and advancing interoperability standards allow for digital biomarkers to be compiled across diverse populations and data sources. As a result, the opportunity to advance digital biomarker methodologies has never been greater.17 18
Machine learning is particularly valuable when there is ambiguity about what variables, or to what extent a set of variables predict an outcome of interest. Such ambiguity is inherent to behavioural interventions, like those used in the treatment of cardiometabolic disease. Human behaviour results from a complex interaction between the anthropogenic features of our living environment, genetic and epigenetic determinants of behaviour, neurobiology, social influences and to some degree chance events.19–22 While clinical experience and the scientific literature can identify many variables that influence behaviour, the interplay of these variables in a given individual and their environment is difficult for a clinician or patient to discern. This ambiguity limits enthusiasm for behavioural interventions because it makes it difficult for clinicians and patients to rely on behavioural therapy to achieve a predictable level of therapeutic response.
Digital biomarkers can reduce ambiguity by predicting current and forecasting future disease status during the course of treatment.23–25 Digital biomarkers that serve as markers of current disease status allow for tailoring or adjusting treatment between clinic visits (eg, when a patient is not doing as well as expected). Similarly, markers of future disease status enable pre-emptive action, such as adding or subtracting additional treatments, or taking preventive steps to avoid complications of the disease.14 25
In this paper, we present our ongoing work to develop digital biomarkers and aim to illustrate their utility in digitally delivered behavioural interventions to both the patient and prescribing clinician. We describe the analytic process to show that the development of digital biomarkers requires a hypothesis-driven approach, particularly when datasets are small. Finally, using actual examples of digital biomarkers intended to predict BP status, we discuss the practical and ethical considerations involved in both developing and applying digital biomarkers using machine learning.
METHODS
Digital therapeutic
The digital biomarkers discussed in this paper were generated using data from a digital therapeutic created by Better Therapeutics LLC (San Francisco, California, USA), a developer of prescription digital therapeutics for the treatment of cardiometabolic diseases.
The digital therapeutic integrates a mobile medical application (‘app’) that delivers behavioural therapy with the support of a remote multidisciplinary care team. The app delivers a personalised behavioural change intervention, including tools for goal setting, skill building, self-monitoring, biometric tracking and behavioural feedback designed to provide cognitive training and support the participant’s daily efforts to improve overall cardiometabolic disease status. The app facilitates the adoption of evidence-based behavioural strategies, such as planning and self-monitoring, to increase physical activity and consumption of vegetables, whole grains, fruits, nuts, seeds, beans and other legumes.26 27
Participants
Participants were over 18 years of age who self-identified with a diagnosis of hypertension, type 2 diabetes and/or hyperlipidaemia. Those reporting to have hypertension at enrolment were offered a free Omron 7 Series Upper Arm Blood Pressure Monitor (Omron Healthcare, Kyoto, Japan) to use during the intervention and to keep at its conclusion. This 12-week intervention was available to all participants at no cost.
This analysis of existing data from participants using the digital therapeutic was approved and overseen by Quorum Review Institutional Review Board28 and a waiver of informed consent was granted for this retrospective analysis.
Constraining the training dataset
The clinical intent of this biomarker exploration was to generate digital biomarkers that could improve treatment outcomes in future participants with hypertension for whom BP was not optimally controlled despite the use of pharmacological treatment. We first identified prior participants that met the following criteria: baseline BP was (1) at or above the cut-off for stage I hypertension (systolic ≥130 or diastolic ≥80)26 and 2) recorded no more than 2 weeks prior to or 2 weeks after the start of the intervention. The baseline value was calculated by taking an average of all values reported in a 6-day interval defined as starting with the date of the first BP value reported and all values reported in the following 5 days.
The development of a predictive model using a training dataset requires all participants to have known outcomes. This is referred to as the ‘ground truth’ in machine learning. In this case, the ground truth was change in BP from baseline. The follow-up BP values were calculated by taking an average over a 6-day interval ending with the last BP reported and all values in the previous 5 days. Of the participants identified, we only included those with a follow-up BP value in weeks 7–14 of the intervention.
The training set also needs to be a valid representation of future experience to make sure the data sources present are sufficiently representative of those that will be collected in future participants. This is an important consideration because of the evolving nature of digital therapeutics. As the therapeutic is modified over time, the data types collected in earlier versions of the app could be different from those collected in later versions. Therefore, for the purpose of developing biomarkers that predict BP status, we further constrained the training dataset to include participants who used versions of the app which enabled automatic BP tracking.
Choosing response variables and training window
The intent of machine learning is to train an algorithm that can predict a specific outcome, termed the ‘response variable’. The response variable must be both clinically relevant and sufficiently, but not universally, prevalent in the population of interest. For example, if the outcome is ‘any degree of improvement’, but any degree of improvement occurs in >95% of participants in the training dataset, then a predictive model may appear to work well, but could actually be invalid.29 Furthermore, ‘any improvement’ is arguably less clinically meaningful than ‘a specific degree of improvement’.
To address these concerns, we chose response variables that were well distributed in the training dataset, as detailed in the Results section, and were also clinically meaningful in the management of hypertension. Specifically, we chose to predict: (1) a systolic BP improvement of 10 mm Hg or higher near the end of a 12-week intervention period (defined as 7–14 weeks after start), because 10 mm Hg has been well demonstrated to be clinically meaningful30 31 and that degree of change is near the mean for participants in the training dataset (66/135 participants); and (2) a reduction in BP to the elevated range (ER) or below (systolic ≤130), because this level of BP control would signal a clinician to stop adding additional pharmaceuticals or consider reducing or deprescribing pharmaceuticals (48/135 participants).26
For a digital biomarker that predicts BP status to be clinically useful, it needs to compute with data collected in a short period of time, and in less time than typically occurs between clinic visits. This means the biomarker could be used to intervene between office visits and could play a role in addressing clinical inertia that limits primary care providers ability to optimise BP control in their patients.32 33 To demonstrate proof of concept, we chose a 28-day training interval, meaning that we trained machine learning models on the first 28 days of patient data to evaluate whether it could predict BP change in weeks 7–14. We hypothesised that data collected within this short training window could sufficiently represent changing behavioural patterns and treatment response, so as to predict future BP status.
Choosing modelling techniques and explanatory variables
There are many valid methodologies for machine learning. These methods can be categorised by the type of learning involved—supervised or unsupervised. While a discussion on the merits and nature of each type is beyond the scope of this paper, it is important to select modelling techniques appropriate for the size of the dataset and nature of response variables chosen. For the biomarkers presented herein, we used random forest models, which is a form of supervised classification learning known to reduce overfitting in small datasets.34 The models are supervised because the ground truth (ie, actual BP change) for each participant in the training dataset is labelled. The models are attempting to learn whether the classification was or was not achieved; for example, will a participant achieve a >10 mm Hg BP reduction, or not?
In addition to classification labels, random forest models must be trained on a set of explanatory variables. For a small training dataset, each model must have a limited number of variables so as to avoid excess noise and overfitting that can lead to reduced generalisability. These explanatory variables must be selected by hypothesis. For example, we hypothesised that baseline BP and achievement of behavioural goals would influence the degree of BP change observed and used these as explanatory variables. In a large dataset, feature engineering can be used to identify the most predictive explanatory variables.
For each biomarker model, denoted systolic change (SC of 10 mm Hg or more) or ER (ER achieved), we used 13 explanatory variables, which can be categorised as engagement or biometric variables. Engagement variables are counts of actions related to the use of the digital therapeutic, including count of all meals reported, plant-based meals reported, physical activity reported and length of exposure to the intervention. Biometric variables included baseline systolic, baseline diastolic, mean systolic and diastolic at training window end, initial systolic and diastolic change (end training mean—baseline), minutes of physical activity and baseline body mass index (BMI).
We trained each biomarker model on data generated during a 28-day training window, starting with participant day 0 (sign up) up to day 28 (a third of the way through the studied 12-week intervention period). We used hyperparameter optimisation35 to minimise overfitting and to achieve the maximal leave-one-out cross-validated area under the receiver operating characteristic curve for both models.
Model validation
Performance of each biomarker model was assessed using leave-one-out cross validation, which is a common and valid technique for use in samples of this size.36–38 This was done by training each model on N−1 samples of the data and then making a prediction on that one sample that was left out, producing an ‘out of sample’ prediction for all N samples. The N predictions were pooled to generate the classification variables of the receiver operator characteristic curve (ROC), the area under the curve of the ROC (AUROC) and a confusion matrix of true versus predicted values.39
For each biomarker model, the ROC curve illustrates predictive ability of the response variable (in this case SC of 10 mm Hg or move to a range of elevated BP) at different thresholds of discrimination. At each prediction discrimination threshold, the ROC displays the false positive rate (FPR) against the true positive rate (TPR). The FPR is the ratio of truly negative events categorised as positive (FP) to the total number of actual negative events (N). Specificity or true negative rate of a model is calculated as 1 − FPR and is an indication of how well a model does in correctly identifying those who do not achieve a successful outcome, as defined by the response variable.
Since the intended application of these biomarkers is analogous to a diagnostic test, which are traditionally evaluated based on their specificity, we evaluated model performance at a specificity of 90% (FPR=0.10). A low FPR minimises the number of participants who the model would predict to achieve a healthier state who actually will not. In turn, this minimises the number of participants who might be erroneously taken off BP medicine as a result of an erroneous prediction. It is less critical to avoid labelling participants who had achieved a healthier state as though they had not. This is why we choose a discrimination threshold with a low FPR, and then evaluate the TPR at that point.
As a further validation step, we examined the performance of each biomarker by excluding the four explanatory variables that capture BP change in the training window. While we hypothesised that these models would perform less well, they serve to test the concept that a digital biomarker that predicts BP status can be generated without using ongoing BP data. These validation models using only app engagement and other biometric variables are denoted SC-APP and ER-APP.
Making use of explainable AI
Digital biomarkers that are generated using machine learning do not need to be viewed as a black box. Instead, explainable AI techniques are available that can provide more granular details about the explanatory variables that influenced the prediction made. Explainable AI can afford both individual participant and population level insights.
We used the tree shapley additive explanation (SHAP) algorithm on the random forest models40 to generate more interpretable predictions at the participant level. The SHAP algorithm assigns each explanatory variable an importance value for each prediction. Using SHAP on a machine learning model is analogous to coefficient analysis in classical regression. Similar to coefficient analysis, it can be used to determine the relative importance of explanatory variables in addition to determining which explanatory variables drove a particular prediction. Predictions start at a base value that is the expectation of the response variable. For binary classification models, this is defined by the proportion of outcomes by class (eg, the proportion of participants who successfully reduced their BP). Then, SHAP values attribute to each explanatory variable the change in expected model prediction given the addition of that explanatory variable. This provides insight into how much each explanatory variable positively or negatively impacts the prediction made for each participant. The final prediction probability of whether the participant will achieve the response variable is the sum of the base value and all of the explanatory variable attributions.
Individual SHAP values can be used to provide specific behavioural feedback to participants, with the intent of motivating a change in behavioural pattern that may improve treatment outcomes. In particular, explanatory variables that are theoretically modifiable (such as minutes of exercise, or number of plant-based meals consumed) can be displayed to motivate changes, whereas fixed explanatory variables (such as baseline values) can be displayed to provide context. SHAP values for all participants can also be plotted to reveal the overall ranking of variables in the population studied. These variable rank lists can then inform hypotheses about how to further improve the design of the digital therapeutic to optimise clinical outcomes.
All machine learning model development was done using open-source packages in Python. The packages include but are not limited to Scikit-Learn, SHAP, Pandas and Numpy.
Patient and public involvement
We did not directly involve patients or public involvement (PPI) in the current study. However, the digital therapeutic that is the source of the database used in this study was developed with PPI input over the product development lifecycle.
RESULTS
Dataset
The training dataset contained 135 participants who met the inclusion criteria. The mean age was 54.9 years (95% CI 53.5 to 56.3), mean baseline BMI was 34.5 (95% CI 33.1 to 35.8) and 83% (112/135) were women. Based on the 2017 American College of Cardiology/American Heart Association definition,26 half of the participants (68/135) had stage 1 hypertension at baseline, with the other half (67/135) having stage II hypertension at baseline. Of those with stage 1 hypertension at baseline, 51.5% (35/68) had isolated diastolic hypertension (ie, diastolic BP 80–90 mm Hg). Of those with stage II hypertension at baseline, 14.9% (10/67) had isolated diastolic hypertension (ie, diastolic BP ≥89 mm Hg). On average, participants contributed three BP readings to the baseline value (95% CI 2.5 to 3.4) and 2.5 readings to the end-intervention value (95% CI 2.2 to 2.9). Baseline characteristics are listed in table 1.
Table 1.
Participant characteristics at baseline
| Participant characteristics | Total (n=135) |
Stage I BP (n=68) |
Stage II BP (n=67) |
| Age (years), mean (95% CI) | 54.9 (53.5 to 56.3) | 55.7 (53.7 to 57.7) | 54.2 (52.1 to 56.2) |
| Body mass index (kg/m2), mean (95% CI) | 34.5 (33.1 to 35.8) | 33.8 (31.9 to 35.6) | 35.2 (33.2 to 37.2) |
| Female, n (%) | 112 (83) | 54 (79.4) | 58 (86.6) |
| Systolic BP (mm Hg), mean (95% CI) | 138.9 (136.2 to 141.6) | 127.9 (126.1 to 129.7) | 150.0 (146.6 to 153.5) |
| Diastolic BP (mm Hg), mean (95% CI) | 87.8 (86.1 to 89.4) | 82.3 (81.1 to 83.6) | 93.3 (90.7 to 95.8) |
| Isolated diastolic hypertension, n (%) | 45 (33.3) | 35 (51.5) | 10 (14.9) |
| BP medications (count), mean (95% CI) | 1.3 (1.1 to 1.5) | 1.2 (1.0 to 1.4) | 1.4 (1.1 to 1.7) |
BP, blood pressure.
Over the intervention period examined, systolic BP changed by −12.7 mm Hg (95% CI −14.8 to 9.6), diastolic BP changed by −7.1 mm Hg (95% CI −9.0 to 5.2) and the mean duration of days between baseline and most recent value for BP was 79.3 days (95% CI 76.8 to 81.9). Of all participants, 35.6% (48/135) shifted to a BP range below stage I (<130/80). A shift to a normal range was seen in 16.4% (11/67) of those starting with stage II hypertension and 29.4% (20/68) of those starting with stage I hypertension.
Predictive models
The random forest classifier achieved optimal performance with 100 trees and a minimum of 3 samples per leaf node for the SC model. For the ER model, optimal performance was achieved with 400 trees and a minimum of 5 samples per leaf nodes.
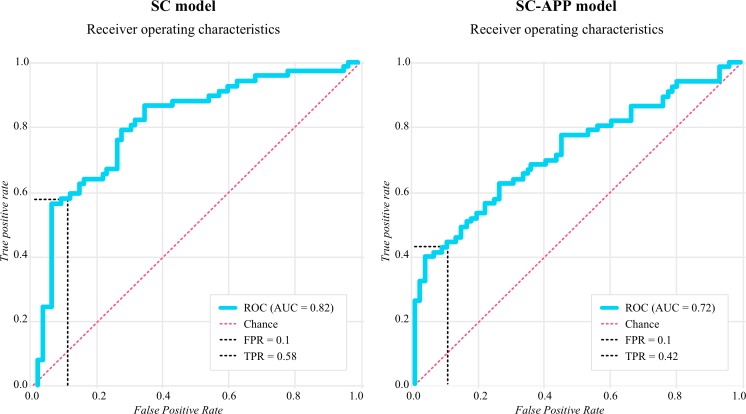
Biomarker models were assessed at the operating point on each ROC that was as close as possible to an FPR of 10%. The SC model (predicting an SC of 10 points) was assessed at an FPR of 10%, which means that 10% of participants who did not achieve a reduction in systolic BP of 10 mm Hg were labelled as though they had. Evaluating the model at 10% FPR, we were able to achieve a TPR of 58%. This means that 58% of participants who achieved a reduction in systolic BP of 10 mm Hg were labelled correctly. The AUROC was 0.82, model specificity (1 − FPR) was 90%, sensitivity (TPR) was 58% and accuracy ((TP+TN)/n) was 74%. In the SC-APP model, where variables related to changes in BP were removed, the AUROC was 0.72 and at an FPR of 10% (specificity of 90%), the TPR was 42%. The resultant receiver operator curves for these two models can be seen in figure 1.
Figure 1.
Receiver operator characteristics (ROC) curves for machine learning model predicting systolic change (SC) and a model predicting SC without use of ongoing blood pressure data (SC-APP). AUC, area under the curve; FPR, false positive rate; TPR, true positive rate.
The biomarker models exploring the ability to predict a shift down to a BP range of elevated or better (ER and ER-APP) also demonstrated predictive capacity, but less so than the SC models. For the ER model, the AUROC was 0.69 and at an FPR of 9% the TPR was 32%. When the BP change variables were removed, in the ER-APP model, the prediction ability was only slightly above chance (AUC=0.53, TPR 26% at FPR of 12%).
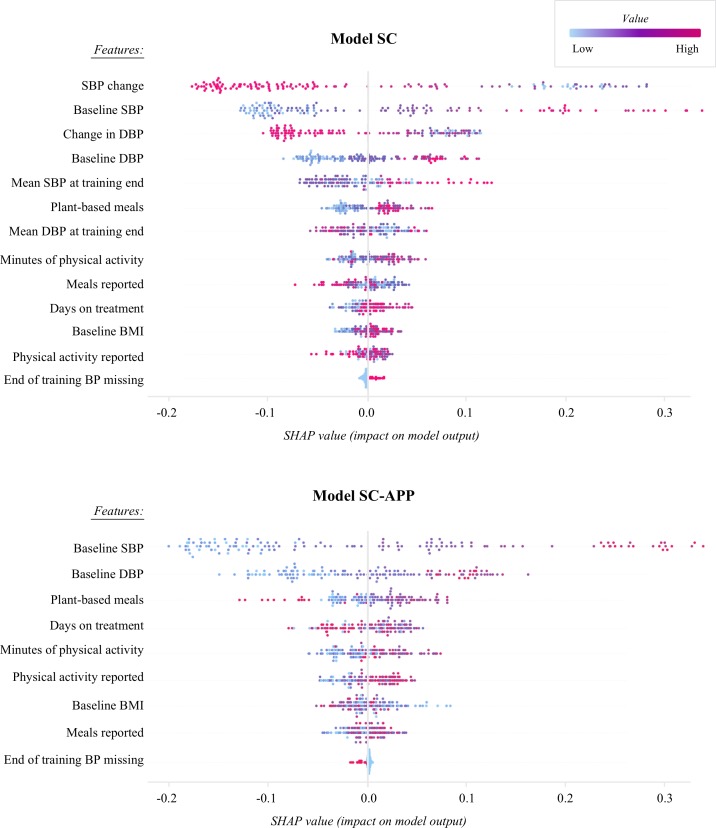
Plots of the tree SHAP algorithm results for the SC and SC-APP models are shown in figure 2. Explanatory variables on the y-axis are ordered from most to least predictive based on their average absolute contribution to the response variable. Each dot represents the SHAP value of that variable for one participant and the placement of the dots on the x-axis indicate if the contribution was subtractive or additive for a specific participant. The colour of the dot is indicative of the value for that variable, with highly positive values displayed as red and low or negative values showing up as light blue. The plot for the SC model reveals that explanatory variables related to BP were top contributors to the prediction. For example, the distribution of dots across the x-axis for the first variable listed shows that improvements in systolic BP early in the intervention, as seen by the blue and purple dots to the right of 0 on the x-axis, contributed positively to the prediction that a participant would succeed. Behavioural variables also had predictive power. For example, a high count of physical activity minutes and plant-based meals reported positively contributed to a prediction of success for most participants.
Figure 2.
Shapley values illustrate how explanatory variables contribute to success meeting the response variable (improvement in systolic blood pressure (SBP) ≥10 mm Hg). The feature list down the y-axis is in order of contribution to the model (most to least). Each dot represents the value for one participant. SBP change and diastolic blood pressure (DBP) change are the difference in measurements from baseline to the end of the 28-day training period. BMI, body mass index; BP, blood pressure; SC, systolic change; SHAP, shapley additive explanation.
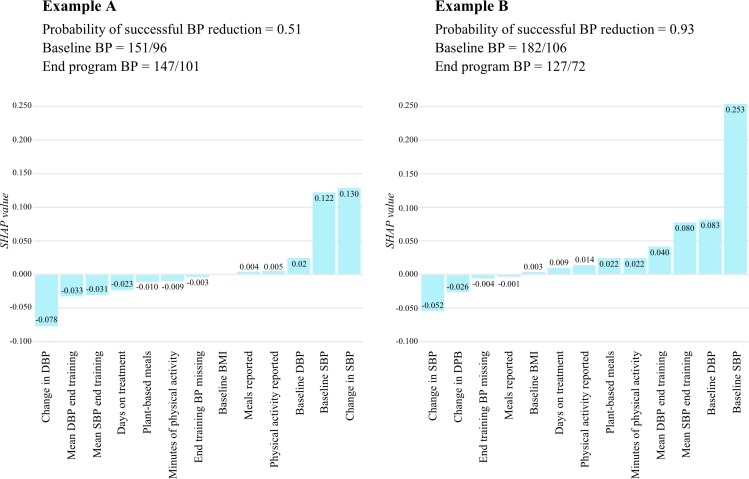
Shapley values can be aggregated and illustrated for every participant. A plot of the SHAP values helps to visualise which variables contributed most to a low or high prediction of success for an individual participant. In figure 3, we display the SHAP values for two participants, one with a lower than expected probability of success (example A), and one with a higher than expected probability of success (example B). In example A, the participant experienced a large improvement in their systolic BP in weeks 3 and 4 (−14 mm Hg), yet is given a low probability of sustaining this improvement at the end of the intervention period. This surprisingly low probability is explained by the SHAP values, which reveal low counts for several behavioural explanatory variables, such as the number of plant-based meals and minutes of physical activity reported. These data can be automatically translated into a simple explanation to the participant, that their probability for sustaining meaningful change could be higher if they made incremental improvements in their meal and activity pattern. Furthermore, the exact number of additional meals and activity minutes to accrue per week to sufficiently increase the probability of success could be calculated, to motivate the participant and to give meaning to the additional efforts prescribed.
Figure 3.
Shapley additive explanation (SHAP) values for explanatory variables for two participants. The SHAP value plotted on the y-axis indicates that amount the variable positively or negatively contributes to the prediction of success (the output value). The probability threshold (output value that assigns a prediction of success) is 0.66. BMI, body mass index; BP, blood pressure; DBP, diastolic blood pressure; SBP, systolic blood pressure.
In example B, the participant has evidenced no improvement in systolic BP at the end of week 4, yet they are predicted to meaningfully improve BP by the of the treatment period. This unexpected prediction is explained by the SHAP values, which show that the combined impact of their baseline BP and behavioural explanatory variables suggests a high likelihood of success. These data can be automatically translated to provide timely encouragement to the participant to maintain or advance their behavioural changes even though their BP has not yet responded.
Discussion
In this paper, we present a proof of concept that digital biomarkers can be developed using even a small training dataset from a digital therapeutic. We demonstrated that 28 days of data can be transformed, using machine learning, into a digital biomarker that predicts the degree of treatment response, in this case whether a meaningful drop in BP will occur at the end of the treatment period.
There are many ways to use such a biomarker in practice to tailor behavioural treatment and improve outcomes. For a patient, these biomarkers can be used as a continuous form of treatment feedback and behavioural reinforcement. The probability of a significant treatment response can be translated into a treatment score, much like a credit score. Since this score could be recalculated with every new engagement recorded in the digital therapeutic, it would serve to motivate app use and reinforce healing behaviours. In addition, the biomarker output can be made more meaningful using explainable AI techniques. For example, SHAP values can be translated into a prioritised list of behavioural actions to help a patient focus their attention on efforts that are most predictive of success.
For clinicians and health systems, digital biomarkers can function as a form of automated patient monitoring. The probability of a positive treatment response can be translated into a clinical alert by setting an acceptable specificity−sensitivity threshold for each biomarker paired with a duration of time above this threshold. Like any diagnostic test, the performance characteristics of the alert should be made known to those acting on it. Since such an alert would be intended to influence treatment decisions, for example, via a clinical decision support tool, specificity−sensitivity pairs need to be evaluated from a risk−benefit perspective. For instance, how do the risks associated with false positives and false negatives compare to the benefits of identifying true positives and true negatives? To accurately weigh these risks and benefits requires us to understand the context that the biomarker and therapeutics are used. Current clinical practice, for example, is plagued by high rates of clinical inertia (ie, a lack of timely and appropriate treatment decisions). Therefore, a higher FPR may be tolerated as a trade-off for an easier-to-access biomarker.
For a developer of digital therapeutics, digital biomarkers provide not just a way to personalise treatment and communicate clinical status to providers, but also a way to better understand what variables within the therapeutic are most predictive of clinical outcomes. These data can be used to guide the ongoing refinement of a digital therapeutic. When datasets are of sufficient size, the machine learning techniques used to generate digital biomarkers can also be applied to identify distinct digital phenotypes, that is, unique patterns of engagement with a behavioural intervention that represent meaningful subpopulations who share the same diagnosis. Identifying and targeting treatment to previously unknown subpopulations is thought of as meaningful step towards more personalised medicine.16 41
Limitations, practical and ethical considerations
The main limitation of the work presented here is the size of the training dataset used. It is likely that a larger dataset would improve the performance characteristics of biomarkers tested.15 It is noteworthy, given this limitation, that one of the biomarkers (SC) had an AUC greater than 0.8. This suggests the utility of beginning digital biomarker development early in the implementation of a digital therapeutic.
To lower the risk of prematurely taking patients off of medications, the digital biomarkers presented decreased the FPR (ie, a higher specificity), which resulted in a lower TPR (ie, a lower sensitivity). In this context, a lower sensitivity means that the digital biomarker will fail to identify some successful individuals and as a result they may not have their medications reduced as promptly as possible. This means that the current performance of the digital biomarker does not fully obviate the need for traditional biomarkers or in-office visits.
Other limitations include the omission of known predictors of treatment response (eg, time since diagnosis, medication adherence or change), the reliance on a small set of explanatory variables and the inclusion of self-reported variables that may be subject to human error. Addressing these limitations may enable more accurate biomarkers.
The ethical and practical implications of applying complex, ever-changing, predictive models generated by machine learning are only beginning to be appreciated. To pre-empt potential misuse of digital biomarkers, we must understand the true meaning of the data these biomarkers present. For example, a predictive model can identify variables that are predictive of a given outcome. This does not mean highly predictive variables caused the outcome, nor does it mean that poorly predictive variables are not causative. Instead, the predictive strength of each variable should be treated literally as ‘markers’. This does not preclude the automated use of explanatory variables to guide the personalisation of behavioural therapy. However, since the individual level of each variable could be influenced by unknown confounding factors, and since the degree of modifiability of each variable is not known, the impact of this form of behavioural therapy must be studied.
Finally, like any biomarker, a digital biomarker is only generalisable if the training dataset is truly representative of future patients. If the training dataset is biassed or overly skewed, it may produce a biomarker that underperforms at best, and is harmful at worst. To guard against this bias, revalidation of a digital biomarker should be performed if the treatment population or digital therapeutic change substantially. And when applying these novel biomarkers, we must appreciate that unknown sources of bias may exist, so that we avoid over-reliance on such biomarkers.
Future work
There are three areas of work that will extend this initial phase of digital biomarker development. As research expands into larger clinical trials, it will enable the revalidation (often called external validation) and to some degree reconstruction of these biomarkers using larger training datasets, creating even more robust biomarkers. A larger dataset enables inclusion of other potentially predictive variables, such as demographics, sociomarkers or omics data, and splitting of the dataset into a training and test set to minimise overfitting. External validation also gives the end-users of the biomarker greater confidence that the biomarker preforms well with varied individuals, settings or time of year.42–44
Second, it will be important to conduct an impact analysis to study whether the intended effects (eg, the improvement of treatment outcomes) are actualised when these biomarkers are put into practice and to observe whether there are any unintended consequences. Alongside empirical research, software usability testing must ensure that the practical application of biomarkers is interpreted by both patients and clinicians in the intended manner.
Third, similar machine learning methods can be applied to develop digital biomarkers that estimate present physiological status, for instance, current BP or fasting blood sugar. This will require a larger training dataset with frequent (ie, multiple times per week) measures of the ground truth (ie, resting BP, fasting blood sugar). A similar validation strategy can be used to determine the validity of the biomarkers with and without self-monitoring of the ground truth. Our aim is to develop cuff-less BP and stickless blood glucose biomarkers that would allow for more continuous patient care at a lower burden to patients and the health system. Our hypothesis is that these biomarkers will significantly reduce clinical inertia, enhance behavioural therapy delivery and further empower patients and providers, meaningfully increasing treatment outcomes at both the patient and population level.
CONCLUSIONS
Machine learning can be used to transform data from a digital therapeutic into actionable digital biomarkers. In this paper, we present a successful proof of concept for a biomarker that uses 28 days of patient-generated data to predict a clinically meaningful response to digitally delivered behavioural therapy. Many practical and ethical considerations arise in the development of digital biomarkers. Applying conventional clinical thinking to these novel computational processes provides the basis to identify and resolve these considerations. There is great potential to design digital biomarkers to enhance the delivery of medical care and improve treatment outcomes.
Supplementary Material
Acknowledgments
The authors would like to thank all of the participants whose use of the Better digital therapeutic contributed to the data that made this analysis possible. The authors would also like to thank the employees of Better Therapeutics, LLC (the study sponsor and intervention developer) for their many contributions and Val de Castro for graphics support.
Footnotes
Contributors: NLG, MAB, JC and SD prepared the data and conducted the analysis. All authors contributed to the conceptualisation of the project and the interpretations of results. NLG, MAB, KLE, KJA, JC and SD contributed to the writing of the paper.
Funding: This work was supported by Better Therapeutics, LLC.
Competing interests: NLG, KLE, KJA, DLK and MAB are employees and equity owners of Better Therapeutics, LLC; DME, JC and SD are independent paid scientific consultants of Better Therapeutics and JC was provided the raw data to perform all machine learning methods independently.
Patient consent for publication: Not required.
Ethics approval: Quorum Institutional Review Board, Seattle, Washington.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request.
References
- 1. Benjamin EJ, Muntner P, Alonso A, et al. Heart disease and stroke statistics—2019 update: a report from the American heart association. Circulation 2019;139:e56–66. 10.1161/CIR.0000000000000659 [DOI] [PubMed] [Google Scholar]
- 2. National Center for Chronic Disease Prevention and Health Promotion The power of prevention: chronic disease.the public health challenge of the 21st century. United States: Department of Health and Human Services, 2009. [Google Scholar]
- 3. Ford ES, Bergmann MM, Kröger J, et al. Healthy living is the best revenge. Arch Intern Med 2009;169:9. [DOI] [PubMed] [Google Scholar]
- 4. Turner RM, Ma Q, Lorig K, et al. Evaluation of a diabetes self-management program: claims analysis on comorbid illnesses, health care utilization, and cost. J Med Internet Res 2018;20:e207 10.2196/jmir.9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kvedar JC, Fogel AL, Elenko E, et al. Digital medicine's March on chronic disease. Nat Biotechnol 2016;34:239–46. 10.1038/nbt.3495 [DOI] [PubMed] [Google Scholar]
- 6. Charpentier G, Benhamou P-Y, Dardari D, et al. The Diabeo software enabling individualized insulin dose adjustments combined with telemedicine support improves HbA1c in poorly controlled type 1 diabetic patients. Diabetes Care 2011;34:533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J, Cai C, Padhye N, et al. A behavioral lifestyle intervention enhanced with Multiple-Behavior self-monitoring using mobile and connected tools for underserved individuals with type 2 diabetes and comorbid overweight or obesity: pilot comparative effectiveness trial. JMIR mHealth and uHealth 2018;6 10.2196/mhealth.4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Milani RV, Lavie CJ, Bober RM, et al. Improving hypertension control and patient engagement using digital tools. Am J Med 2017;130:14–20. 10.1016/j.amjmed.2016.07.029 [DOI] [PubMed] [Google Scholar]
- 9. Quinn CC, Clough SS, Minor JM, et al. WellDoc mobile diabetes management randomized controlled trial: change in clinical and behavioral outcomes and patient and physician satisfaction. Diabetes Technol Ther 2008;10:160–8. 10.1089/dia.2008.0283 [DOI] [PubMed] [Google Scholar]
- 10. Berman MA, Guthrie NL, Edwards KL, et al. Change in glycemic control with use of a digital therapeutic in adults with type 2 diabetes: cohort study. JMIR Diabetes 2018;3 10.2196/diabetes.9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coravos A, Khozin S, Mandl KD. Developing and adopting safe and effective digital biomarkers to improve patient outcomes. npj Digit. Med. 2019;2 10.1038/s41746-019-0090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meister S, Deiters W, Becker S. Digital health and digital biomarkers – enabling value chains on health data. Current Directions in Biomedical Engineering 2016;2 10.1515/cdbme-2016-0128 [DOI] [Google Scholar]
- 13. Wright J, Regele O, Kourtis L, et al. Evolution of the digital biomarker ecosystem. Digital Medicine 2017;3. [Google Scholar]
- 14. Fritz BA, Chen Y, Murray-Torres TM, et al. Using machine learning techniques to develop forecasting algorithms for postoperative complications: protocol for a retrospective study. BMJ Open 2018;8:e020124 10.1136/bmjopen-2017-020124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Westerman K, Reaver A, Roy C, et al. Longitudinal analysis of biomarker data from a personalized nutrition platform in healthy subjects. Sci Rep 2018;8 10.1038/s41598-018-33008-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minich DM, Bland JS. Personalized lifestyle medicine: relevance for nutrition and lifestyle recommendations. ScientificWorldJournal 2013;2013:1–14. 10.1155/2013/129841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darcy AM, Louie AK, Roberts LW. Machine learning and the profession of medicine. JAMA 2016;315:551–2. 10.1001/jama.2015.18421 [DOI] [PubMed] [Google Scholar]
- 18. Sun D, Liu J, Xiao L, et al. Recent development of risk-prediction models for incident hypertension: an updated systematic review. PLoS One 2017;12:e0187240 10.1371/journal.pone.0187240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thornton RLJ, Glover CM, Cené CW, et al. Evaluating strategies for reducing health disparities by addressing the social determinants of health. Health Aff 2016;35:1416–23. 10.1377/hlthaff.2015.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egger G, Dixon J. Beyond obesity and lifestyle: a review of 21st century chronic disease determinants. Biomed Res Int 2014;2014:1–12. 10.1155/2014/731685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gastil RD. The determinants of human behavior. Am Anthropol 1961;63:1281–91. 10.1525/aa.1961.63.6.02a00080 [DOI] [Google Scholar]
- 22. Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environ Mol Mutagen 2008;49:46–60. 10.1002/em.20357 [DOI] [PubMed] [Google Scholar]
- 23. Dagum P. Digital biomarkers of cognitive function. NPJ Digit Med 2018;1 10.1038/s41746-018-0018-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shin EK, Mahajan R, Akbilgic O, et al. Sociomarkers and biomarkers: predictive modeling in identifying pediatric asthma patients at risk of hospital revisits. NPJ Digit Med 2018;1 10.1038/s41746-018-0056-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Billings J, Blunt I, Steventon A, et al. Development of a predictive model to identify inpatients at risk of re-admission within 30 days of discharge (PARR-30). BMJ Open 2012;2:e001667 10.1136/bmjopen-2012-001667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whelton PK, Carey RM, Aronow WS, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults. J Am Coll Cardiol 2018;71:e127–248. 10.1016/j.jacc.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 27. Williams B, Mancia G, Spiering W, et al. ESC/ESH guidelines for the management of arterial hypertension. Eur Heart J 2018;2018:3021–104. [DOI] [PubMed] [Google Scholar]
- 28. Quorum Review IRB Quorum review IRB: independent ethics review board. Available: https://www.quorumreview.com/ [Accessed 6 Dec 2017].
- 29. Kuhn M, Johnson K. Applied predictive modeling. 5th ed Springer, 2016. [Google Scholar]
- 30. Ettehad D, Emdin CA, Kiran A, et al. Blood pressure lowering for prevention of cardiovascular disease and death: a systematic review and meta-analysis. Lancet 2016;387:957–67. 10.1016/S0140-6736(15)01225-8 [DOI] [PubMed] [Google Scholar]
- 31. Thomopoulos C, Parati G, Zanchetti A. Effects of blood pressure lowering on outcome incidence in hypertension. 1. overview, meta-analyses, and meta-regression analyses of randomized trials. J Hypertens 2014;32:2285–95. 10.1097/HJH.0000000000000378 [DOI] [PubMed] [Google Scholar]
- 32. Milman T, Joundi RA, Alotaibi NM, et al. Clinical inertia in the pharmacological management of hypertension. Medicine 2018;97:e11121 10.1097/MD.0000000000011121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogedegbe G. Barriers to optimal hypertension control. J of Clin Hypertens 2008;10:644–6. 10.1111/j.1751-7176.2008.08329.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Breiman L. Random forests. Mach Learn 2001;45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 35. scikit-learn developers 3.2. tuning the hyper-parameters of an estimator, 2007. Available: https://scikit-learn.org/stable/modules/grid_search.html [Accessed 31 May 2019].
- 36. Liu M-X, Chen X, Chen G, et al. A computational framework to infer human disease-associated long noncoding RNAs. PLoS One 2014;9:e84408 10.1371/journal.pone.0084408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 2007;8 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moore RG, Brown AK, Miller MC, et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol Oncol 2008;108:402–8. 10.1016/j.ygyno.2007.10.017 [DOI] [PubMed] [Google Scholar]
- 39. Airola A, Pahikkala T, Waegeman W, et al. An experimental comparison of cross-validation techniques for estimating the area under the ROC curve. Comput Stat Data Anal 2011;55:1828–44. 10.1016/j.csda.2010.11.018 [DOI] [Google Scholar]
- 40. Lundberg SM, Lee SI. Consistent feature attribution for tree ensembles, 2017. Available: https://arxiv.org/abs/1706.06060 [Accessed 19 Nov 2018].
- 41. Shaban-Nejad A, Michalowski M, Buckeridge DL. Health intelligence: how artificial intelligence transforms population and personalized health. NPJ Digit Med. 2018;1 10.1038/s41746-018-0058-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alba AC, Agoritsas T, Walsh M, et al. Discrimination and calibration of clinical prediction models: users' guides to the medical literature. JAMA 2017;318 10.1001/jama.2017.12126 [DOI] [PubMed] [Google Scholar]
- 43. Moons KGM, Kengne AP, Woodward M, et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart 2012;98:683–90. 10.1136/heartjnl-2011-301246 [DOI] [PubMed] [Google Scholar]
- 44. Moons KGM, Kengne AP, Grobbee DE, et al. Risk prediction models: II. external validation, model updating, and impact assessment. Heart 2012;98:691–8. 10.1136/heartjnl-2011-301247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.