Abstract
Guanine-rich DNA can fold into highly stable non-canonical four-stranded DNA structures called G-quadruplexes. These structures present obstacles for the DNA replication machinery, and it has been hypothesized that both eukaryotic DNA helicases and polymerases have evolved to resolve G4 DNA in vivo. Since the discovery of G-quadruplex DNA in the early 1960’s, a number of studies have emerged reporting G-quadruplex DNA unfolding by helicase enzymes and DNA synthesis past G4 by specialized translesion polymerase enzymes. Recently, the discovery of the primase-polymerase PrimPol and its role in G4 bypass has sparked even more excitement in the G-quadruplex and DNA replication fields. This review presents an overview of the molecular interactions of G-quadruplexes with DNA helicases and polymerases implicated in their resolution, with an emphasis on how the regulation and coordination of these enzymes is critical for genome homeostasis. Targeting G4-interacting DNA helicases and polymerases for therapeutic strategies is discussed.
Keywords: G-quadruplex, replication, helicase, polymerase, translesion synthesis, G4 DNA, PrimPol
1. DISCOVERY OF G-QUADRUPLEX DNA
A timeline marking some of the key events in the discovery of G-Quadruplex (G4) DNA, depicted in Fig. (1), shows the relatively slow awakening from the first observation in 1910 that guanine (G)-rich DNA sequences form a “gel” [1] to its description in 1962 at the sub-atomic level as a tetrad stack of coordinated guanines [2], bridging a time-period of over 50 years. Interestingly, during this period the DNA double helix structure of DNA was discovered [3], leaving doubt as to the relevance of a four-stranded DNA conformation. After another hiatus of 16 years, researchers in the late 1980’s and 1990’s began to recognize and document that G-rich DNA sequences found at telomeres, gene promoters, and chromosome recombination hotspots spontaneously fold into unique molecular G-quadruplex DNA structures in vitro [4, 5], and that G4 DNA can alter the activities of DNA metabolic enzymes such as telomerase [6, 7]. The dissension from the canonical B-form DNA double helix paralleled discoveries of other alternatively folded DNA structures including DNA triplexes, hairpins, and Z-DNA (for review, see [8–10]). What has perhaps uniquely driven the G-quadruplex field over the last decade and a half is the increasing number of experimental findings that G4 DNA exists in vivo and has important biological consequences for nuclear and mitochondrial genomic DNA metabolism and cellular homeostasis [11–20]. Moreover, the development of algorithms which have been used to analyze DNA sequence computationally and predict the formation of G4 has sparked large-scale genome analysis efforts [21, 22]. In more recent years, the characterization of G4 and its effects on cellular processes of replication and transcription, as well as its impact on genome stability, have provided exciting new directions for the G-quadruplex field [23–27]. Beyond the distilled timeline shown in Fig. (1), the reader is referred to several excellent review articles which chronicle the research efforts entailing G4 DNA discovery [28, 29]. While we do not address G4 RNA structures here, the reader is referred to several recent articles in this new and emerging field as well [30, 31].
Fig. (1).
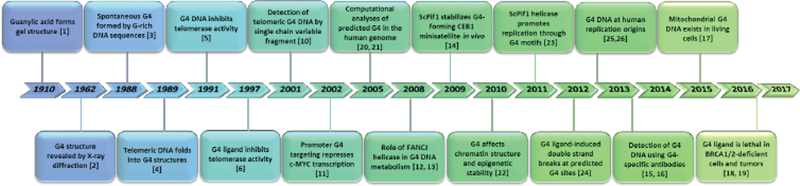
A timeline of G-quadruplex nucleic acid discovery.
In this review, we will focus the discussion on DNA helicases and polymerases implicated in G4 DNA metabolism, placing an emphasis on how their regulation and coordination are critical for genome homeostasis. Throughout the review, we bring attention to potential areas of translational research in which the unique G-quadruplex DNA structure and its interacting proteins may represent suitable targets for emerging therapeutic strategies.
2. COMPUTATIONAL TOOLS AND ALTERNATIVE STRATEGIES TO DETECT G4-PRONE SEQUENCES
Increased attention to G4-forming sequences and their roles in various biological contexts has led to the development of many computational tools. These instruments are meant to help in the identification and characterization of G4 sequences. Computational analyses use the DNA sequence itself to predict G4-forming regions and can effectively leverage the vast amounts of publicly available genome data.
Early algorithms used biophysical and biochemical experiments to construct a search pattern that could be applied to DNA sequences. Todd et al. [22] and Huppert and Balasubramanian (authors of QuadParser [21]) agreed on such a pattern based on previous literature as well as their own experiments: G3+ N1–7 G3+ N1–7 G3+ N1–7 G3+. The groups noted that guanine runs of 3 or more in the guanine tetrads led to significant increases in thermodynamic stability of the G4 structure. They also noted that loop sizes tended to be constrained between one and seven nucleotides [21, 22]. Both groups acknowledged that the pattern required further experimental validation, and noted some identified sequences were likely false positives that would not form G4 structures [22]. A significant conclusion from these two computational analyses was that predicted G-quadruplexes are relatively abundant in the human nuclear genome, with an estimate of ~375,000 in total. Sixty percent of these predicted G-quadruplexes were mapped to intergenic regions [22]. Of the remaining 40% found in genes, they were split evenly between the two DNA strands. Approximately 3.7% of the total predicted G4-forming sequences in the human nuclear genome were found in exons. The significant percentage of predicted G4 structures identified in gene promoter elements has garnered interest that G4 plays a major regulatory role in gene expression.
Using the idea of the consensus search sequence, other groups created tools such as QGRS Mapper [32], designed to provide a more customizable approach for further research. QGRS Mapper utilized a search pattern similar to that used in QuadParser. However, it allowed a moderate level of user customization, such as selecting a total sequence length of anywhere between 30 and 45 nucleotides, loop size specification, and loop sequence specification. QGRS Mapper assigned a G-score to sequences based on how likely they were to form G4 structures. It considered factors such as shorter loops being more common than longer loops, loops of equal size being most common, and the greater stability generated from a larger number of guanines in the tetrads. Interestingly, the authors even at this early stage in the field of computational approaches to study G-quadruplexes appreciated that such an unusual nucleic acid conformation has implications for not only G-rich DNA sequences such as promoters and telomeres, but also for RNA sequences. Indeed, as cited above in some recent reviews [30, 31], the RNA G-quadruplex field is blossoming.
QuadBase [33] investigated conservation of G4 sequences between species to highlight those that may have been evolutionarily relevant. The QuadBase project generated two databases, EuQuad and ProQuad, focusing on eukaryotic and prokaryotic genomes, respectively. EuQuad searched for orthologs of putative G4-forming motifs in the human genome to those in the mouse, rat, and chimpanzee genomes. ProQuad compared G4 sequences in a query genome to 145 other prokaryotic genomes for its ortholog search. This work was extended by QGRS Homology and QGRS Conserve [34, 35], which included the G-score function of QGRS Mapper to predict stability as well as an improved scoring system for conservation of G4 sequences between species.
A new algorithm, G4Hunter [36], diverged from the long-held search pattern constraint and instead gave sequences a G4 propensity score as a measure of thermodynamic stability. G4Hunter was based on literature that called for a focus on a scoring system for thermodynamic stability at physiological temperature, as sequences likely to form at physiological temperature are most likely to have a biologically relevant function [37]. Unlike a pattern search for a consensus sequence, G4Hunter searches windows of a user-specified size for guanine richness and guanine skewness (guanine-cytosine asymmetry between complementary strands). The tool assigns each of these windows a score, and selects windows that meet the user-specified propensity threshold. This tool showed a higher sensitivity and lower false discovery rate as compared to other programs such as QuadParser [36]. A striking conclusion from this study was that the number of DNA sequences in the human genome predicted to form G4 was significantly greater than earlier predictions, by a factor of 2- to 10-fold. A unique aspect of the work first describing G4Hunter was the extensive experimental validation of a large dataset of predicted G4-forming sequences using an array of biophysical techniques to determine if the corresponding oligonucleotides indeed formed G4 in vitro. Overall, the ambitious nature of the investigation and development of a G4 predictor algorithm that provides a propensity score make the Bedrat et al. [36] publication a highly significant contribution to the field. Consistent with the conclusion that the number of human genomic G4 structures may have been underestimated, a recently published high-resolution sequencing-based method revealed over 700,000 distinct G4 structures, over half of which had not been predicted by the earlier G4 algorithms [38]. The observation that many of these G-quadruplex structures were mapped to oncogenes, tumor suppressors, and somatic copy number alterations led the authors to propose that such G4’s may contribute to the genomic instability prevalent in cancers. Furthermore, small molecules that specifically bind the novel G4 DNA structures with high affinity may prove to be useful for anti-cancer therapeutic strategies in the future.
Experimental studies using G4 prediction algorithms which take into account variability in the length and nucleotide composition of G-tract and intervening loop sequences, and by inference G4 formation probability, stability and topography, are ongoing [39–41]. Interfacing the computational approach with quantitative and biophysical analysis of G4 formation will be useful in this regard [42]. Moreover, alternative strategies to map G4-forming sequences in the genome, such as the high-resolution sequencing technique [38], are rapidly moving the field forward. Genomic mapping of G4 structures has been advanced quite recently by the development of a G4-antibody-based chromatin immunoprecipitation and high-throughput sequencing procedure (ChIP-Seq) using fixed chromatin as the genomic DNA source [43]. This study suggested that G4 formation in human nuclei is strongly influenced by transcriptional state in which nucleosome-depleted chromatin that is highly transcribed has the greatest abundance of G4 structures. Other ChIP-Seq-based approaches that exploit the use of G4-specific binding proteins are also being used to inform the predictive G4 algorithm analysis (for review, see [28]). The development of a database (designated G4IPDB) which catalogs G4-forming and interacting proteins and associated parameters in a single platform may help to devise 3D G4 predictors (e.g., basket, propeller, etc.) for future efforts in drug discovery based on G4-interacting ligands [44].
3. DNA HELICASES THAT SMOOTH DNA REPLICATION THROUGH G4-FORMING SEQUENCES
Several recent reviews have described an emerging class of nucleoside triphosphate dependent DNA unwinding enzymes known as helicases that are capable of resolving various G-quadruplex DNA structures in vitro [45–50]. Presumably, if these G4-resolving heli-cases were to efficiently act upon their targets in vivo, their effects on cellular nucleic acid metabolism of G4-forming sequences would be profound. Indeed, cell-based assays have invoked unique and important roles of certain eukaryotic DNA helicases in replication and genome stability maintenance that are likely based at least in part on their ability to resolve G-quadruplexes (Table 1). There are a number of DNA helicases which behave very inefficiently to resolve various G4 topologies in vitro, suggesting that the biological functions of G4-resolving helicases are specialized and play unique roles in nucleic acid metabolism [51]. A provocative area of study is the investigation of coordinate action between G4-resolving helicases and proteins that are involved in the synthesis past or downstream of G4 obstacles. In the following sections, we will discuss some of the DNA polymerases which appear to have specialized roles in DNA synthesis of nuclear or mitochondrial DNA templates. We believe that concentrated efforts to decipher the coordinate action of such DNA polymerases with G4-resolving helicases may provide tremendous insight to how such G4 impediments to the replication machinery are overcome in vivo.
Table 1.
G4-resolving eukaryotic helicases and their proposed biological roles in G4 DNA metabolism.
| 5’ to 3’ DNA helicases | ||
|---|---|---|
| Helicase | Cellular Function | References |
| FANCJ | DNA replication | [13, 14, 58–60] |
| Epigenetic regulation | [61] | |
| RTEL1 | Telomeric G4 resolution | [62, 63] |
| PIF1 | Genomic G4 DNA stability | [15, 64] |
| DNA replication | [24, 65] | |
| DNA2 | Telomeric G4 resolution | [66] |
| XPD | G4-regulated gene expression | [67] |
| 3’ to 5’ DNA helicases | ||
| Helicase | Cellular Function | References |
| WRN | Telomeric G4 resolution | [68] |
| G4-regulated gene expression | [69–71] | |
| Epigenetic regulation | [61] | |
| BLM | Telomeric G4 resolution | [72] |
| G4-regulated gene expression | [69, 71, 73] | |
| Epigenetic regulation | [61] | |
| RECQL1* | G4-regulated gene expression | [74] |
| XPB* | G4-regulated gene expression | [67] |
DNA helicases shown to bind but not unwind G4 DNA
Molecular insights into the orchestration of G4-resolving helicases and DNA replication proteins may offer opportunities for therapeutic strategies, particularly to combat cancer. A number of G4-resolving helicases (e.g., human RecQ enzymes) are up-regulated in various cancers, suggesting their requirement for rapidly proliferating transformed cells to cope with abundant replicative lesions [52]. If such helicases can be targeted for inhibition by pharmacological means, then the cancerous cells may become hypersensitive to DNA damaging therapies. Such a chemical synthetic lethality approach has gained momentum in other areas of DNA repair and replication stress targeted therapies [53]. Small molecule inhibitors of human DNA helicases have been identified and characterized biochemically in vitro and in cell-based models [54–57], but their efficacy in pre-clinical organismal models has been largely under-studied. Further studies in this area are of paramount importance in order for translational approaches to develop. Given that some of the potential targets of G4-resolving helicases include telomeres and G-rich promoters of proto-oncogenes (Table 1), it seems reasonable that these cancer-relevant chromosomal regions would be good targets for anti-cancer strategies.
A good example of a potential G4-resolving helicase to target in a synthetic lethality approach is FANCJ. Previously, it was reported that cellular deficiency of FANCJ resulted in elevated sensitivity to a G4-stabilizing ligand known as Telomestatin (TMS) [13]. The premise of the study was that a greater number of G4 structures would persist in vivo when the G4-resolving helicase FANCJ was deficient; consequently, the cells would become hypersensitive to the DNA damage-inducing and anti-proliferative effects of a G4-specific DNA binding compound. Indeed, this was observed. Moreover, a subsequent study using a G4-specific antibody showed that FANCJ-deficient cells exposed to TMS displayed elevated immunofluorescent foci representing G4 DNA recognized by a G4 antibody [17]. Thus, it is reasonable to postulate that a FANCJ-specific helicase inhibitor may subject human cancer cells or tumors susceptible to a G4 ligand; however, this remains to be seen. A similar scenario may exist for Pif1, as it was reported that a yeast strain deficient in the G4-resolving helicase was hypersensitive to a G4 ligand [64]. However, to our knowledge a similar finding has not yet been reported for PIF1-deficient human cells.
The efficacy of a G4-specmc helicase inhibitor in an anti-cancer therapy regime is challenged by the probability that G4 structures themselves act as a driving force for mutagenesis that may be very well a determinant for carcinogenesis in a tissue-specific manner [75]. Therefore, finding a therapeutic threshold for a putative G4 helicase inhibitor or drug delivery mechanism that targets the tumor would be imperative to reducing toxic side effects for normal tissues.
4. NUCLEAR DNA REPLICATION OF G4-FORMING DNA SEQUENCES
4.1. DNA Polymerases are Required for Genome Replication
In eukaryotes, three DNA polymerases are responsible for carrying out faithful replication of the nuclear genome: polymerase alpha (pol α), polymerase delta (pol δ), and polymerase epsilon (pol ε). Pol α is known to synthesize a DNA/RNA hybrid primer, thereby initiating DNA synthesis on both the leading and lagging strands; Pol δ and pol ε then carry out synthesis by extending the primer [76].
Early experiments on the differential functions of pol δ and pol ε suggested that each is involved in synthesis of one strand only. Chromatin immunoprecipitation assays have shown that pol ε is bound at DNA origins during fork establishment and remains associated with the fork as it migrates from the origin [77–79]. These early findings suggested that pol ε is responsible for replicating only one strand at the replication fork. Furthermore, the observations that both polymerases can be purified as monomers [80], [81] and that pol δ and pol ε proofread opposite strands [82] have helped confirm the one-polymerase-one-strand hypothesis. It has now become widely accepted that pol δ is solely responsible for synthesis of the lagging strand; studies in yeast have shown that pol δ is known to pause during replication of the lagging strand in order to maintain a nick that can later be ligated [83]. Pol ε, mean-while, has been implicated in leading strand synthesis; a 2007 study in yeast utilizing a constructed pol ε derivative with an active site mutation documented a pattern indicating that the enzyme carries out replication of the leading strand [84]. The tailored functions of the replicative DNA polymerases raise the question of whether they are differentially affected by DNA lesions or unusual DNA conformations such as G-quadruplexes that they encounter in the respective template strands.
4.2. G4-forming Sequences Present Obstacles for Replicative Polymerases
Some of the earliest pioneer research to assess the effect of G4 DNA structure on DNA synthesis was from the Usdin lab. They developed an in vitro DNA synthesis arrest to show that G-rich sequences in the DNA template strand blocked DNA synthesis catalyzed by bacterial or bacteriophage DNA polymerase in a K+ ion-dependent manner, consistent with the requirement of a monovalent cation (e.g., K+, Na+) to reside in the central channel of the G4 tetrad to stabilize it [85]. It was also shown in this work that blocking the N7 positions of guanine residues which would be predicted to disallow the Hoogsteen hydrogen bonding of the G4 structure abolished polymerase arrest. In subsequent work from the Usdin lab, they determined using the DNA polymerase arrest assay the size and base composition of the G4 DNA stem and loop sub-elements [85]. Furthermore, their work demonstrated that increasing the length of the G tracts greatly increased arrest of DNA synthesis, suggesting that the number of G4 structures which form enhances the blocking effect. Importantly, their work with DNA templates harboring different putative G4-forming sequences suggested that a much greater number of G-quadruplexes were likely to exist under physiological conditions that what was previously thought.
A key aspect of replicative DNA polymerases is their high fidelity, since accurate replication of the genome is required to faithfully maintain genetic information over many generations and to avoid potentially deleterious mutations that can lead to human disease. While the replicative polymerases possess proofreading and exonuclease abilities to maintain fidelity, G-quadruplexes pose problems for the replication machinery that DNA polymerases do not tolerate well. A 2001 study by the Fry lab demonstrated that G4 structures directly block synthesis by all three replicative polymerases, pol α, pol δ and pol ε [86]. A more recent study by the Opresko lab investigated G4-induced stalling of pol δ at templates mimicking telomeric TTAGGG repeats and found that it was greater on the G-rich strand compared to the C-rich strand [87]. Similar experiments have shown that pol ε fails to replicate G4 sites as well; the Eoff lab has shown that pol ε exhibits both decreased efficiency and fidelity when trying to copy the first guanine it encounters in a G4 motif [88]. Even the primase/polymerase pol α is blocked by G-quadruplexes, as the Drosophila pol α has been shown to stall at G4 sites in vitro [89]. Thus, G4 sites present obstacles for all three classical DNA polymerases (Fig. 2A).
Fig. (2).
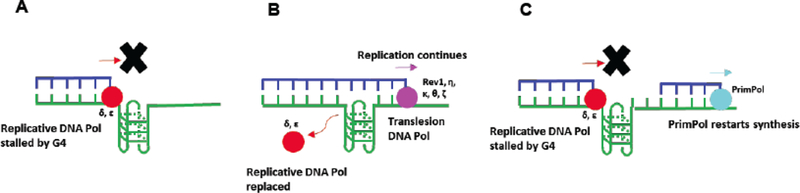
Processing of G4 by replicative and translesion DNA polymerases. A) A G4 site in template strand stalls the replicative polymerases pol δ and pol ε. B) Pol δ and pol ε are replaced by a translesion polymerase. The translesion polymerase adds a nucleotide opposite the G4 site and synthesis continues without resolving the G4. C) A G4 site in the template strand stalls pol δ and pol ε. Synthesis resumes when PrimPol synthesizes a short dNTP primer downstream of the G-quadruplex.
An important consideration regarding G4-prone sequences is whether they behave symmetrically on both strands. A 2011 yeast study by the Nicolas lab provided significant data to support the notion that the processing of G4 motifs by replicative polymerases in vivo depends largely upon which strand is more G-rich [65]. They inserted a G4-prone CEB1 tandem array near a replication origin and saw that it was frequently destabilized upon treatment with a G4 ligand or by knocking out the Pif1 helicase, but only when the more G-rich strand was the template of leading-strand replication. These results indicate that a G-quadruplex certainly constitutes a physical barrier to replication; however, the actual degree to which it impedes replication may depend on the polymerase that encounters it, the template strand in which the G4 resides, as well as on the ligand-induced stability of the G4 and the status of G4-resolving helicases.
While the immediate consequence of G4 formation in the nuclear genome is replicative polymerase stalling, a single G-quadruplex can interfere with replication over many generations. A recent study by the Tijsterman lab found that unresolved G-quadruplex structures in the DNA can retain their conformation through multiple mitotic divisions [90]. Furthermore, the study went on to show that failure to replicate across a G4 site causes gaps of single-stranded DNA that can give rise to double-strand breaks (DSBs) in future cell divisions. These results indicate that G4 sequences pose such significant problems for replicative polymerases that the persistence of a single G-quadruplex in the nuclear genome can have serious mutagenic consequences, including genetic heterogeneity and genomic rearrangements. The observation that just a single G4 can have such a profound effect on mutagenesis is striking, and it suggests that multiple tandem G4 structures are not required to affect replication fidelity in vivo. Moreover, even a poorly processive DNA helicase that can remove just one G-quadruplex may greatly influence DNA replication and potential errors that arise.
5. BYPASS OF G4 BY TRANSLESION DNA POLYMERASES
5.1. Translesion Synthesis Employs Specialized Polymerases to Synthesize past G4 Sites
Translesion synthesis (TLS) is a DNA damage tolerance process by which the DNA replication machinery bypasses a DNA lesion through the incorporation of a nucleotide opposite the lesion. Since many lesions (e.g., apurinic sites, UV-induced pyrimidine dimers, hydrolyzed bases, bulky adducts, G4 sites, etc.) are impermissible for replicative DNA polymerases 5 and s [86], a specialized class of translesion polymerases is employed to bypass the damage site. TLS polymerases can use damaged DNA as a template to bypass the lesion regardless of the lesion’s potential conformational constraints but do not correct the lesion (Fig. 2B); thus, they are typically regarded as reduced fidelity polymerases, employed strictly to mitigate the more deleterious effects of fork stalling and collapse. G-quadruplex-forming sequences fold into large and highly stable secondary structures in vivo [47] and much evidence has arisen that TLS plays an important role in the replication of these structures. A simple representation of the scenario in which a translesion polymerase replaces a replicative polymerase that is stalled by a G4 structure is shown in (Fig. 2B). In eukaryotes, TLS of G4 DNA is carried out by polymerases Rev1, eta (pol η), kappa (pol κ), theta (pol θ) and potentially zeta (pol ζ). The general function and localization of these enzymes is summarized in Table 2.
Table 2.
Genes, localization and function of eukaryotic G4 DNA translesion polymerases.
| Translesion Polymerases Known to Act on G4* | ||||||
|---|---|---|---|---|---|---|
| Rev1 | Pol η | Pol κ | pol θ | pol ζ* | ||
| Gene encoding catalytic subunit | S. cerevisiae | REV1 | RAD30 | REV3 | ||
| D. melanogaster | rev1 | DNApol-η | mus308 | mus205/dmREV3 | ||
| Mouse | Rev1 | Polη | DinB1 | Polq | Rev3 | |
| Human | REV1 | RAD30AF | DINB1 | POLQ | REV3L | |
| Localization | Nuclear and mitochondrial | Nuclear, potentially mitochondrial | Nuclear | Nuclear | Nuclear and mitochondrial | |
| Function | Scaffold protein, deoxycytidyl transferase | TLS of helix-distorting lesions | TLS, nucleotide excision repair | DSB repair |
TLS, mitochondrial genome maintenance | |
| References | [91–93] | [94–96] | [88, 97] | [98–100] | [101, 102] | |
Pol ζ is a potential candidate for G4 TLS
5.2. REV1 has Dual Roles in G4 Translesion Synthesis
Rev 1 has been implicated in both catalytic and non-catalytic roles in TLS of G4 in both the nucleus and the mitochondria [103]. It has been previously shown that Rev1 acts as a scaffolding protein to help coordinate the TLS process through protein-protein interactions with the pol δ DNA clamp PCNA and ubiquitin, as well as through direct interactions with pols ε and κ [92, 93, 104, 105]. Additionally, Rev1 is known to play a restricted catalytic role in TLS by acting as a deoxy-cytidyl transferase opposite G4 sites [106, 107]. A 2014 study by the Eoff lab showed that Rev1 binds G4 DNA with Kd values that are 4 to 15-fold lower than those of non-G4 DNA substrates and that the presteady-state rate constant of deoxycytidine monophosphate insertion opposite G4 by Rev1 is about half as fast as that opposite non-G4 DNA [91]. These results suggest that Rev1 promotes bypass of G4 by actively aiding in the unfolding of G4 structures, either by actively dislodging tetrad guanines and allowing extension by other polymerases or by binding to unfolded G4 DNA and preventing refolding. A recent review by Wickramasinghe et al. proposes an interesting speculative model for G4 replication by Rev1 in which the deoxycytidyl transferase activity of Rev1 provides a ‘bait’ to compete for the dG bases at the start of the quadruplex [98]. The model proposes that catalysis is then carried out through handoff to another polymerase, since Rev1 is known to interact directly with pol κ and pol ζ [92]. However, it is not known whether Rev1 behaves synergistically with pol κ or pol ζ to catalyze DNA synthesis at an elevated rate using G4-forming sequences as a DNA template, or if the fidelity is affected.
Thus, it is likely that Rev1 plays more than one role in G4 replication in vivo, as it has been shown to act both directly and indirectly on G4 structures, as well as help coordinate replication by other polymerases. It is also likely that Rev1 functions in conjunction with DNA helicases that help unwind the G4 structure [98], a suggestion requiring further biochemical studies. A study by Yeom et al. suggested that certain germline missense variations in Rev1 may either inhibit or enhance the ability of Rev1 to bypass G4 motifs, thereby affecting an individual’s susceptibility toward carcinogenesis [108], a finding which warrants further study of Rev 1 as a potential drug target.
5.3. G4 Replication by Translesion Polymerases η and κ
Two additional polymerases eta (η) and kappa (κ) have also been shown to replicate past G4 sites [94]. Pol η was originally identified as the protein defective in the disorder Xeroderma pigmentosum and characterized as being able to synthesize past UV-generated cyclobutane thymine dimers [95]. Kinetic analysis conducted by the Eoff lab has shown that pol n is capable of maintaining more than 25% activity on G4 substrates compared to non-G4 substrates, with a fidelity ~100-fold more accurate than the replicative polymerase pol ε when comparing the frequency of misinsertion opposite a G4 site [96]. Additionally, in another study, pol η was shown to copy damaged DNA with higher fidelity than the equivalent undamaged sequence; it can even ‘sense’ when the lesion has been successfully bypassed and triggers its own dissociation from the DNA [109]. Such findings reaffirm the notion that replicative polymerases fail to faithfully replicate past G4 sites and that the replication temporarily calls in translesion polymerases to do the job. These analyses also support a model in which the replication machinery carries out a simple kinetic switch between replicative and translesion polymerases to help govern fork progression during the replication of G4 [96].
Pol κ, in addition to acting as a translesion polymerase, has also been shown to play a role in the nucleotide excision repair pathway [97]. Past research on the crystal structure of pol κ in a ternary structure with DNA revealed that pol κ possesses a unique “N-clasp” structure at its terminus [110], a structure that is distinct from pol n and absent in other Y-family polymerases. The clasp region restricts the enzyme’s active site, limiting its affinity for certain lesions and increasing its fidelity. The N-clasp also serves to “lock in” a nucleotide after incorporation across from a lesion to promote efficient replication [110]. Another study was conducted by the Eoff lab to elucidate how the structure and biochemical properties of pol κ affect how it behaves on G4 substrates. Their group conducted several kinetic analyses of pol κ and determined that it exhibits both enhanced activity and decreased fidelity two nucleotides before a G4 motif [88], a finding which they interpreted to mean that pol κ may be involved in priming synthesis just upstream of the G4 motif.
While both are members of the same polymerase family, pol η and pol κ appear to differ slightly in their interactions with G4. A study from the Eoff lab presented data from polymerase extension assays that are consistent with the notion that G4 sites present a greater obstacle to DNA synthesis by pol κ as compared to pol η [88]. It has also been shown that an increase in DNA breakage associated with other alternative structures besides G4, such as repetitive GA-DNA, occurs after downregulation of pol κ but not pol η [94]. This suggests that while both act on G4, pol κ likely plays a more general role in maintaining genomic stability by also acting on less bulky lesions, while pol η may function on helix-distorting lesions.
5.4. Pol θ Joins Breaks Arising at G-quadruplexes
DNA polymerase theta (pol θ), which is encoded by the POLQ gene in eukaryotes, contains both a polymerase-like domain and a helicase-like domain [100]. Both domains have been shown to play important functional roles in DNA damage tolerance [111]; one study showed that the helicase domain binds its substrate downstream of the polymerase domain, thereby setting up the strand for subsequent processing by the polymerase [112]. However, it is not yet known if the helicase-like domain serves to actively aid in the unwinding of G4.
As a holoenzyme, Pol θ plays a unique role in the replication of G4 structures; it is responsible for repairing genomic DSBs that arise when the replication barrier persists at G4 sites [99]. This noncanonical DSB repair pathway, termed theta-mediated end joining (TMEJ), appears to be the preferred method to cope with DSBs occurring at structural replication fork barriers at G4 sequences. Evidence has arisen to suggest that a replication fork stalled at a G-quadruplex is subject to collapse, resulting in large-scale deletions when double stranded DNA breaks [113]. In a study by Koole et al., pol θ was shown to carry out end-joining of breaks by creating an overlap of one base pair and extending this to create a template on flanking DNA [99]. The study also showed that C. elegans lacking both the FANCJ homolog dog-1 (an important helicase for unwinding DNA structures) and pol θ displayed large deletions of several kilobases at G4 motifs while worms lacking only dog-1 displayed much smaller deletions (50–300 base pairs), suggesting that pol 0 is utilized as a backup for the failure to unwind G4 when dog-1 is nonfunctional. Thus, theta-mediated end joining of DSBs arising from G4 DNA structures seems to be a “last resort” translesion process that is potentially mutagenic but prevents catastrophic large-scale deletions.
5.5. Pol ζ Carries out Translesion Synthesis in the Mitochondria
Until very recently the mitochondrial DNA polymerase γ, which has both replicative and repair functions, was the only known mitochondrial polymerase [114]. A 2015 study by the Singh lab demonstrated that not only is there another human polymerase, pol ζ, that localizes to the mitochondria, but that this enzyme functions therein as a translesion polymerase to maintain and protect the mitochondrial genome from harmful lesions like UV photoproducts [101]. Pol ζ is composed of a catalytic subunit REV3 as well as a structural subunit REV7 that helps to stabilize the polymerase [115].
Work carried out by the Chang lab used fluorescence lifetime imaging microscopy in live cells using a G4 ligand to verify that G4 DNA exists in the mitochondrial genome [18]. There is little evidence in the literature implicating pol ζ as acting on G4 DNA substrates, since most experiments on the catalytic function of REV3 focus on UV-induced damage. However, the enzyme’s localization to the mitochondria and its known role as a translesion polymerase suggest it is a possible candidate for mitochondrial G4 TLS. Pol ζ has already been shown to work alongside Rev1 to replicate ‘difficult’ sequences, such as hairpin structures [116] and is required for the majority of damage-induced mutagenesis in budding yeast [117]. Thus, while pol ζ is a good candidate for G4 TLS, further research is required to confirm that it acts on G4.
6. SPECIALIZED ROLE OF PRIMPOL DNA POLYMERASE TO PRIME DNA SYNTHESIS DOWNSTREAM OF G4
With the 2013 discovery of a novel eukaryotic DNA polymerase and primase designated PrimPol [118–121], there was renewed excitement in the field of DNA metabolism. Previously, Pri1/Pri2 associated with DNA polymerase alpha was the only known nuclear primase, and it was shown to be responsible for replication initiation of DNA synthesis [119]. For mitochondria, the mitochondrial RNA polymerase (POLRMT) was known to prime at major initiation sites on either the light or heavy strands of the organelle’s circular genome [122]. Although another primase activity in mitochondria of human cells was identified, the actual protein was unknown [123]. Unlike polymerase alpha or POLRMT, PrimPol was found to be capable of initiating polynucleotide synthesis with deoxyribonucleotides and not be confined to the use of ribonucleotides as was the case for Pri1/Pri2 or POLRMT. PrimPol was determined to reside in both the nuclear and mitochondrial compartments of human cells [118, 119]. Interestingly, PrimPol depletion by RNA interference in human cells negatively affected mitochondrial DNA synthesis [119], providing the first insight to its biological importance. Although a PRIMPOL−/− mouse is viable, the redundancy with other mitochondrial DNA polymerases is imperfect as attested to by the observation of mitochondrial DNA depletion in PRIMPOL−/− mouse embryonic fibroblasts.
Several labs acquired scientific data implicating a specialized role of PrimPol to facilitate replication of unusual DNA sequences or bypass DNA damage. Research from the Doherty lab showed that PrimPol binds G4 DNA structure tightly and catalyzes DNA synthesis downstream of DNA structures [124]. This in vitro finding coupled with an observed G4-dependent transcriptional instability at a defined locus in a chicken DT40 primpol cell-based model led them to propose a model in which PrimPol promotes restart of DNA synthesis downstream of leading strand G4 DNA replication roadblocks (shown schematically in Fig. 2C), thereby ensuring efficient coupling of DNA synthesis with histone recycling [125]. Thus, PrimPol aids in the restart of replication forks perturbed by alternate DNA structure (e.g., G4) to preserve normal epigenetic gene expression patterns. PrimPol is also important to restart replication downstream of DNA synthesis-blocking lesions or when forks are stalled by chain-terminating nucleosides. The elevated sensitivity of PrimPol-deficient DT40 cells to UV irradiation is characterized by a pronounced checkpoint arrest [126], indicating that PrimPol plays an encompassing role at stalled forks. Indeed, PrimPol was shown by a variety of experimental approaches and model systems to enable bypass of UV photoproducts encountered by the eukaryotic chromosomal replication machinery. As mentioned above, in addition to nuclear replication, evidence also suggests a specialized role of PrimPol in mitochondrial DNA replication [118]. This is highly likely to involve the bypass of G-quadruplexes, as they are predicted to be highly abundant in the mitochondrial genome [36, 127, 128]. From data mining, the prevalence of mitochondrial deletion breakpoints in the vicinity of predicted G4-forming sequences suggests that PrimPol’s ability to bypass such alternate DNA structures is highly relevant to their potential involvement in mitochondrial genome instability associated with mitochondrial genetic disorders, cancer, and aging. Although a formal role of PrimPol in replicating past mitochondrial G4 obstacles has not yet been documented, it seems probable based on its involvement in nuclear G4 bypass and interaction with mitochondrial DNA metabolic proteins (for review, see [129]). A strong candidate to partner with PrimPol is the human mitochondrial transcription factor A (TFAM), a factor that was recently shown to bind G4 with a high affinity [130]. Interestingly, the nuclear single-stranded DNA binding protein Replication Protein A (RPA) significantly stimulates PrimPol activity [131], and helps to recruit it to nuclear replication forks through its physical interaction [132].
As pointed out in a recent review by Guilliam and Doherty [133], PrimPol may be a good candidate for chemotherapeutics because it is implicated in the tolerance of DNA damage, which is typically induced by many anti-cancer drugs. Indeed, the induction of genomic instability and disruption of normal epigenetic regulation by targeted inhibition of PrimPol DNA synthesis could be highly deleterious to the metabolism and growth of rapidly dividing cancer cells, which are prone to the formation of replicative DNA lesions on their own right. Computational analysis of PrimPol’s protein sequence indicates extensive homology throughout the Kingdoms of life, with several structural and catalytic domains including a primase active site and Zn2+ finger. Virtual docking and high-throughput biochemical screens of small molecule libraries may identify lead compounds that bind to these and other domains of the protein and serve as potent and specific PrimPol inhibitors that could be further developed and used for studies in cells and pre-clinical organismal models.
7. COORDINATE ACTION OF G4-RESOLVING HELICASES AND DNA POLYMERASES
The likely encounter of DNA polymerases with G-rich genomic DNA sequences prone to form G-quadruplexes prompts attention to G4-resolving helicases that would facilitate DNA synthesis. Early work from the Fry and Loeb labs established that the WRN helicase collaborates with DNA polymerase δ in vitro to copy G4-forming sequences derived from genomically unstable trinucleotide repeats [86]. The ability of WRN to bind tightly to G4 DNA structures may aid in efficiently resolving them in a manner to allow efficient DNA synthesis past the G4 obstacle [134]. In vivo data from a chicken cell-based system suggests that FANCJ collaborates with WRN, or the sequence-related BLM helicase, to enable smooth replication past G4. FANCJ is also believed to collaborate with REV1 translesion DNA polymerase in this capacity [61]. A role for FANCJ in replication fork progression through G4 DNA was substantiated by studies using Xenopus egg extracts and G-rich single-stranded DNA templates [58].
In addition to their more general role in replication of G4-forming sequences in the general genome, G4-resolving helicases may be particularly required for more specialized regions of the genome such as telomeres. BLM-deficient cells are significantly compromised in telomeric DNA synthesis, and G4 ligands exasperate the effect [72]. It remains to be seen if WRN and BLM have unique or overlapping functions to resolve telomeric G4 DNA structures. Aside from WRN and BLM, the Fe-S cluster helicase RTEL1 is proposed to resolve telomeric G4 DNA to maintain telomeric stability [63]. It is hard to reconcile how so many G4-resolving helicases could play a role in telomere metabolism. In general, the field now demands sophisticated cellular assays, perhaps involving single molecule analyses, to delineate the biological context in which these helicases operate with DNA replication and repair proteins to perform their functions. One idea in G4 DNA metabolism is that helicases may have specificity in targeting certain G4 topologies; however, more research in this area is required to establish if G4 substrate specificity is biologically relevant [51]. Interestingly, recent evidence suggests that non-canonical G4 DNA structures that vary in their connecting loops and thermal stability is related to their genome-destabilizing effects in vivo, raising the question if the G4 topology specificity of specialized DNA helicases, polymerases, and G4-binding proteins plays a key role in cellular DNA metabolism [135]. A fundamental distinction among families of DNA helicases is their directionality of translocation along single-stranded DNA. For example, the RecQ helicases (e.g., WRN, BLM) translocate in the 3’ to 5’ direction, whereas the Fe-S helicases (e.g., FANCJ, RTEL1) translocate in the 5’ to 3’ direction. As shown in (Fig. 3), it is conceptually easier to visualize a 3’ to 5’ helicase collaborating with DNA polymerase to replicate past a G4 obstacle. However, as noted in the G4 DNA discovery timeline (Fig. 1), evidence suggests that the 5’ to 3’ DNA helicase Pif1 plays a prominent role in resolving G4 DNA structures to allow smooth replication fork progression in yeast [15, 24]. The take-home message is that as far as scientific research has taken us in the study of G-quadruplexes, we may still be at the tip of the iceberg in our understanding of even the most basic questions, such as the mechanisms whereby DNA sequences prone to form G4 structures are efficiently copied in the cell.
Fig. (3).
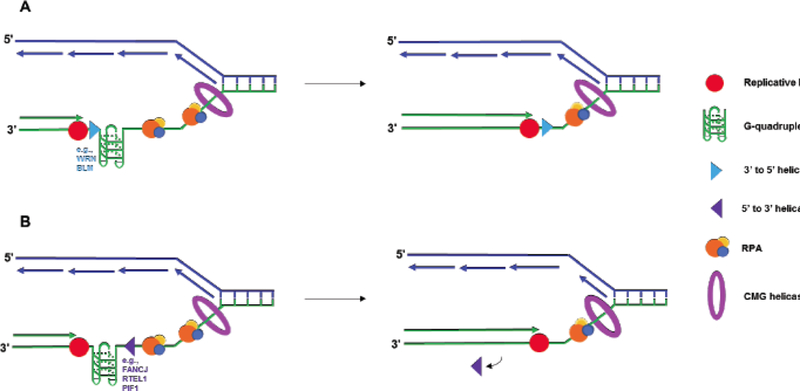
Coordinated action of DNA replication proteins during synthesis past G4 obstacle. A) A G-quadruplex DNA structure impedes leading strand synthesis during replication. As the CMG complex unwinds duplex DNA at the replication fork, RPA binds to stabilize the exposed single-stranded DNA. A 3′ to 5′ helicase, such as WRN or BLM, helps smooth over the G4 site, allowing the polymerase to synthesize the complementary strand. RPA heterotrimer is represented by spheres of orange (RPA70), blue (RPA32), and yellow (RPA14). B) Leading strand replication is again stalled by a G-quadruplex. The action of a 5’ to 3’ helicase, such as FANCJ, RTEL1, or PIF1, helps smooth the G-quadruplex in the direction opposite synthesis, resolving the replication block. The helicase dissociates from the DNA to allows the polymerase to proceed as synthesis continues.
A potentially informative approach will be to correlate G4-associated mutations in the nuclear or mitochondrial genomes with the propensity of respective DNA polymerases to misincorporate (or extend by mispairing) nucleotides across from G4-forming templates. In addition, characterizing the precise mechanisms (likely to involve G4-resolving helicases) in mammalian cells whereby DNA is replicated past G4 obstacles in the leading versus lagging strand, similar to what has been done in yeast [65], should be useful in understanding the mutagenicity and genomic instability imposed by G4. In this manner, G4 stability and architecture are likely to play key roles.
8. CONCLUDING REMARKS
In this review, we have provided the reader an emerging sense of the G-quadruplex field, emphasizing the unique and important roles of specialized DNA helicases and polymerases to resolve and copy sequences which are quite abundant based on algorithmic and genomic analyses, and have been shown to readily form G4 in vitro. The growing evidence that G4 DNA exists in vivo has attracted many researchers to experimentally address their impact on genomic stability, and the mechanisms which exist to deal with such structures during replication, recombination, and repair of the nuclear and mitochondrial genomes. The DNA G- quadruplex field has also sparked interest in potential medicinal approaches that exploit the unusual genomic architecture for targeting by G4 ligands and small molecule inhibitors of DNA helicases and polymerases; however, the field is still in its infancy and there is a significant amount of research required to assess its potential translational relevance.
Structure-based virtual screening to identify therapeutic G4-interacting compounds [136–138], as alluded to in Section 2, combined with interdisciplinary approaches involving biophysical and functional analysis in vitro, genome G4 mapping, and in vivo cell-based and animal models represent an exciting new area of investigation that offers clinical potential. Since the first molecular description of G4 DNA over 50 years ago, it remains an exciting journey in the coming years to unveil a deeper understanding of what was once a controversial DNA structure that is projected to have medicinal value.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health, Laboratory of Molecular Gerontology and Laboratory of Genetics and Genomics.
LIST OF ABBREVIATIONS
- CMG
Cdc45p, MCM, GINS complex
- DSB
Double-strand break
- G4
G-quadruplex
- Pol
Polymerase
- RPA
Replication Protein A
- TLS
Translesion synthesis
- TMS
Telomestatin
Footnotes
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- [1].Bang I, Untersuchungen über die Guanylsaure. Biochem., 1910. 26: p. 293–231. [Google Scholar]
- [2].Gellert M, Lipsett MN, and Davies DR, Helix formation by guanylic acid. Proc Natl Acad Sci U S A, 1962. 48: p. 2013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Watson JD and Crick FH, Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature, 1953. 171(4356): p. 737–8. [DOI] [PubMed] [Google Scholar]
- [4].Sen D and Gilbert W, Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature, 1988. 334(6180): p. 364–6. [DOI] [PubMed] [Google Scholar]
- [5].Sundquist WI and Klug A, Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 1989. 342(6251): p. 825–9. [DOI] [PubMed] [Google Scholar]
- [6].Zahler AM, et al. , Inhibition of telomerase by G-quartet DNA structures. Nature, 1991. 350(6320): p. 718–20. [DOI] [PubMed] [Google Scholar]
- [7].Sun D, et al. , Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem, 1997. 40(14): p. 2113–6. [DOI] [PubMed] [Google Scholar]
- [8].Zhao J, et al. , Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci, 2010. 67(1): p. 43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Choi J and Majima T, Conformational changes of non-B DNA. Chem Soc Rev, 2011. 40(12): p. 5893–909. [DOI] [PubMed] [Google Scholar]
- [10].Wang G and Vasquez KM, Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst), 2014. 19: p. 143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schaffitzel C, et al. , In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylony-chia lemnae macronuclei. Proc Natl Acad Sci U S A, 2001. 98(15): p. 8572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Siddiqui-Jain A, et al. , Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A, 2002. 99(18): p. 11593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu Y, Shin-ya K, and Brosh RM Jr., FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol, 2008. 28(12): p. 4116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].London TB, et al. , FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem, 2008. 283(52): p. 36132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ribeyre C, et al. , The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet, 2009. 5(5): p. e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Biffi G, et al. , Quantitative visualization of DNA G- quadruplex structures in human cells. Nat Chem, 2013. 5(3): p. 182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Henderson A, et al. , Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res, 2014. 42(2): p. 860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huang WC, et al. , Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res, 2015. 43(21): p. 10102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu H, et al. , CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA½ deficient tumours. Nat Commun, 2017. 8: p. 14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zimmer J, et al. , Targeting BRCA1 andBRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol Cell, 2016. 61(3): p. 449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huppert JL and Balasubramanian S, Prevalence of quad-ruplexes in the human genome. Nucleic Acids Res, 2005. 33(9): p. 2908–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Todd AK, Johnston M, and Neidle S, Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res, 2005. 33(9): p. 2901–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sarkies P, et al. , Epigenetic instability due to defective replication of structured DNA. Mol Cell, 2010. 40(5): p. 703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Paeschke K, Capra JA, and Zakian VA, DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell, 2011. 145(5): p. 678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rodriguez R, et al. , Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol, 2012. 8(3): p. 301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Besnard E, et al. , Unraveling cell type-specific and repro-grammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol, 2012. 19(8): p. 837–44. [DOI] [PubMed] [Google Scholar]
- [27].Hoshina S, et al. , Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem, 2013. 288(42): p. 30161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hansel-Hertsch R, Di Antonio M, and Balasubramanian S, DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol, 2017. 18(5): p. 279–284. [DOI] [PubMed] [Google Scholar]
- [29].Maizels N, G4-associated human diseases. EMBO Rep, 2015. 16(8): p. 910–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cammas A and Millevoi S, RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res, 2017. 45(4): p. 1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fay MM, Lyons SM, and Ivanov P, RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J Mol Biol, 2017. 429(14): p. 2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kikin O, D’Antonio L, and Bagga PS, QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res, 2006. 34(Web Server issue): p. W676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yadav VK, et al. , QuadBase: genome-wide database of G4 DNA--occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res, 2008. 36(Database issue): p. D381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Menendez C, Frees S, and Bagga PS, QGRS-H Predictor: a web server for predicting homologous quadruplex forming G-rich sequence motifs in nucleotide sequences. Nucleic Acids Res, 2012. 40(Web Server issue): p. W96–w103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Frees S, et al. , QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs. Hum Genomics, 2014. 8: p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bedrat A, Lacroix L, and Mergny JL, Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res, 2016. 44(4): p. 1746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stegle O, et al. , Predicting and understanding the stability of G-quadruplexes. Bioinformatics, 2009. 25(12): p. i374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chambers VS, et al. , High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Bio-technol, 2015. 33(8): p. 877–81. [DOI] [PubMed] [Google Scholar]
- [39].Tradigo GCF, ; Alcaro S; Greco S; Pollastri G; Veltri P; Prosperi M, G-qaudruplex Structure Prediction and integration in the GenData2020 data model, in Proceedings of the 7th ACM International Conference on Bioinformatics, Computation Biology, and Health Informatics. 2016, ACM: New York, NY, USA: p. 663–670. [Google Scholar]
- [40].Tradigo GM,L; Veltri P, Assessment of G-quadruplex prediction tools. Computer-Based Medical Systems, 2014: p. 243–246. [Google Scholar]
- [41].Wong HM, et al. , A toolbox for predicting g-quadruplex formation and stability. J Nucleic Acids, 2010. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim M, et al. , Quantitative analysis and prediction of G-quadruplex forming sequences in double-stranded DNA. Nucleic Acids Res, 2016. 44(10): p. 4807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hansel-Hertsch R, et al. , G-quadruplex structures mark human regulatory chromatin. Nat Genet, 2016. 48(10): p. 1267–72. [DOI] [PubMed] [Google Scholar]
- [44].Mishra SK, et al. , G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep, 2016. 6: p. 38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Geronimo CL and Zakian VA, Getting it done at the ends: Pifl family DNA helicases and telomeres. DNA Repair (Amst), 2016. 44: p. 151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Leon-Ortiz AM, Svendsen J, and Boulton SJ, Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair (Amst), 2014. 19: p. 152–62. [DOI] [PubMed] [Google Scholar]
- [47].Bochman ML, Paeschke K, and Zakian VA, DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet, 2012. 13(11): p. 770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wu Y and Brosh RM Jr., G-quadruplex nucleic acids and human disease. Febs j, 2010. 277(17): p. 3470–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bharti SK, et al. , Getting Ready for the Dance: FANCJ Irons Out DNA Wrinkles. Genes (Basel), 2016. 7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mendoza O, et al. , G-quadruplexes and helicases. Nucleic Acids Res, 2016. 44(5): p. 1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bharti SK, et al. , Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J Biol Chem, 2013. 288(39): p. 28217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Brosh RM Jr., DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer, 2013. 13(8): p. 542–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Puigvert JC, Sanjiv K, and Helleday T, Targeting DNA repair, DNA metabolism and replication stress as anti-cancer strategies. Febs j, 2016. 283(2): p. 232–45. [DOI] [PubMed] [Google Scholar]
- [54].Liu W, et al. , A Selective Small Molecule DNA2 Inhibitor for Sensitization of Human Cancer Cells to Chemotherapy. EBioMedicine, 2016. 6: p. 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Aggarwal M, et al. , Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Res, 2013. 73(17): p. 5497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Aggarwal M, et al. , Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A, 2011. 108(4): p. 1525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nguyen GH, et al. , A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem Biol, 2013. 20(1): p. 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Castillo Bosch P, et al. , FANCJ promotes DNA synthesis through G-quadruplex structures. Embo j, 2014. 33(21): p. 2521–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Schwab RA, et al. , FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J Cell Biol, 2013. 201(1): p. 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cheung I, et al. , Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet, 2002. 31(4): p. 405–9. [DOI] [PubMed] [Google Scholar]
- [61].Sarkies P, et al. , FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res, 2012. 40(4): p. 1485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vannier JB, et al. , RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science, 2013. 342(6155): p. 239–42. [DOI] [PubMed] [Google Scholar]
- [63].Vannier JB, et al. , RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell, 2012. 149(4): p. 795–806. [DOI] [PubMed] [Google Scholar]
- [64].Piazza A, et al. , Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res, 2010. 38(13): p. 4337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lopes J, et al. , G-quadruplex-induced instability during leading-strand replication. Embo j, 2011. 30(19): p. 4033–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lin W, et al. ,Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. Embo j, 2013. 32(10): p. 1425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gray LT, et al. , G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat Chem Biol, 2014. 10(4): p. 313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Crabbe L, et al. , Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science, 2004. 306(5703): p. 1951–3. [DOI] [PubMed] [Google Scholar]
- [69].Smestad JA and Maher LJ 3rd, Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med Genet, 2015. 16: p. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tang W, et al. , The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum Mol Genet, 2016. 25(10): p. 2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Johnson JE, et al. , Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res, 2010. 38(4): p. 1114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Drosopoulos WC, Kosiyatrakul ST, and Schildkraut CL, BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol, 2015. 210(2): p. 191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nguyen GH, et al. , Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc Natl Acad Sci U S A, 2014. 111(27): p. 9905–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li XL, et al. , Identification of RECQ1-regulated transcriptome uncovers a role of RECQ1 in regulation of cancer cell migration and invasion. Cell Cycle, 2014. 13(15): p. 2431–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].De S and Michor F, DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol, 2011. 18(8): p. 950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hubscher U, Maga G, and Spadari S, Eukaryotic DNA polymerases. Annu Rev Biochem, 2002. 71: p. 133–63. [DOI] [PubMed] [Google Scholar]
- [77].Masumoto H, Sugino A, and Araki H, Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol, 2000. 20(8): p. 2809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Feng W, et al. , Schizosacchromycespombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication. Mol Biol Cell, 2003. 14(8): p. 3427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hiraga S, et al. , DNA polymerases alpha, delta, and epsilon localize and function together at replication forks in Saccharomyces cerevisiae. Genes Cells, 2005. 10(4): p. 297–309. [DOI] [PubMed] [Google Scholar]
- [80].Johansson E, Majka J, and Burgers PM, Structure of DNA polymerase delta from Saccharomyces cerevisiae. J Biol Chem, 2001. 276(47): p. 43824–8. [DOI] [PubMed] [Google Scholar]
- [81].Chilkova O, Jonsson BH, and Johansson E, The quaternary structure of DNA polymerase epsilon from Saccharomyces cerevisiae. J Biol Chem, 2003. 278(16): p. 14082–6. [DOI] [PubMed] [Google Scholar]
- [82].Karthikeyan R, et al. , Evidence from mutational specificity studies that yeast DNA polymerases delta and epsilon replicate different DNA strands at an intracellular replication fork. J Mol Biol, 2000. 299(2): p. 405–19. [DOI] [PubMed] [Google Scholar]
- [83].Garg P, et al. , Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev, 2004. 18(22): p. 2764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pursell ZF, et al. , Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science, 2007. 317(5834): p. 127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Weitzmann MN, Woodford KJ, and Usdin K, The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J Biol Chem, 1996. 271(34): p. 20958–64. [DOI] [PubMed] [Google Scholar]
- [86].Kamath-Loeb AS, et al. , Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem, 2001. 276(19): p. 16439–46. [DOI] [PubMed] [Google Scholar]
- [87].Lormand JD, et al. , DNA polymerase delta stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res, 2013. 41(22): p. 10323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Eddy S, et al. , Human Translesion Polymerase kappa Exhibits Enhanced Activity and Reduced Fidelity Two Nucleotides from G-Quadruplex DNA. Biochemistry, 2016. 55(37): p. 5218–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kaguni LS and Clayton DA, Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci U S A, 1982. 79(4): p. 983–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lemmens B, van Schendel R, and Tijsterman M, Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat Commun, 2015. 6: p. 8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Eddy S, et al. , Human Rev1 polymerase disrupts G- quadruplex DNA. Nucleic Acids Res, 2014. 42(5): p. 3272–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Guo C, et al. , Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. Embo j, 2003. 22(24): p. 6621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Waters LS, et al. , Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev, 2009. 73(1): p. 134–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Betous R, et al. , Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol Carcinog, 2009. 48(4): p. 369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Song Q, et al. , Preparation of site-specific T=mCG cis-syn cyclobutane dimer-containing template and its error-free bypass by yeast and human polymerase eta. J Biol Chem, 2012. 287(11): p. 8021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Eddy S, et al. , Evidence for the kinetic partitioning of polymerase activity on G-quadruplex DNA. Biochemistry, 2015. 54(20): p. 3218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ogi T and Lehmann AR, The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide- excision repair. Nat Cell Biol, 2006. 8(6): p. 640–2. [DOI] [PubMed] [Google Scholar]
- [98].Wickramasinghe CM, et al. , Contributions of the specialised DNA polymerases to replication of structured DNA. DNA Repair (Amst), 2015. 29: p. 83–90. [DOI] [PubMed] [Google Scholar]
- [99].Koole W, et al. , A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun, 2014. 5: p. 3216. [DOI] [PubMed] [Google Scholar]
- [100].Seki M, Marini F, and Wood RD, POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res, 2003. 31(21): p. 6117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Singh B, et al. , Human REV3 DNA Polymerase Zeta Localizes to Mitochondria and Protects the Mitochondrial Genome. PLoS One, 2015. 10(10): p. e0140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhang H, Chatterjee A, and Singh KK, Saccharomyces cerevisiae polymerase zeta functions in mitochondria. Genetics, 2006. 172(4): p. 2683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Andersen PL, et al. , Sequential assembly of translesion DNA polymerases at UV-induced DNA damage sites. Mol Biol Cell, 2011. 22(13): p. 2373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Murakumo Y, et al. , Interactions in the error-prone pos-treplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem, 2001. 276(38): p. 35644–51. [DOI] [PubMed] [Google Scholar]
- [105].Ohashi E, et al. , Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells, 2004. 9(6): p. 523–31. [DOI] [PubMed] [Google Scholar]
- [106].Nair DT, et al. , Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science, 2005. 309(5744): p. 2219–22. [DOI] [PubMed] [Google Scholar]
- [107].Swan MK, et al. , Structure of the human Rev1-DNA- dNTP ternary complex. J Mol Biol, 2009. 390(4): p. 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yeom M, et al. , Effects of Twelve Germline Missense Variations on DNA Lesion and G-Quadruplex Bypass Activities of Human DNA Polymerase REV1. Chem Res Toxicol, 2016. 29(3): p. 367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].McCulloch SD, Kokoska RJ, and Kunkel TA, Efficiency, fidelity and enzymatic switching during translesion DNA synthesis. Cell Cycle, 2004. 3(5): p. 580–3. [PubMed] [Google Scholar]
- [110].Lone S, et al. , Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell, 2007. 25(4): p. 601–14. [DOI] [PubMed] [Google Scholar]
- [111].Beagan K, et al. , Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslinkrepair. PLoS Genet, 2017. 13(5): p. e1006813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Newman JA, et al. , Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway. Structure, 2015. 23(12): p. 2319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lambert S and Carr AM, Impediments to replication fork movement: stabilisation, reactivation and genome instability. Chromosoma, 2013. 122(1–2): p. 33–45. [DOI] [PubMed] [Google Scholar]
- [114].Krasich R and Copeland WC, DNA polymerases in the mitochondria: A critical review of the evidence. Front Biosci (Landmark Ed), 2017. 22: p. 692–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Nelson JR, Lawrence CW, and Hinkle DC, Thymine- thymine dimer bypass by yeast DNA polymerase zeta. Science, 1996. 272(5268): p. 1646–9. [DOI] [PubMed] [Google Scholar]
- [116].Northam MR, et al. , DNA polymerases zeta and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res, 2014. 42(1): p. 290–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lawrence CW and Maher VM, Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philos Trans R Soc Lond B Biol Sci, 2001. 356(1405): p. 41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bianchi J, et al. , PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell, 2013. 52(4): p. 566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Garcia-Gomez S, et al. , PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell, 2013. 52(4): p. 541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Mouron S, et al. , Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol, 2013. 20(12): p. 1383–9. [DOI] [PubMed] [Google Scholar]
- [121].Zhao F, et al. , Exome sequencing reveals CCDC111 mutation associated with high myopia. Hum Genet, 2013. 132(8): p. 913–21. [DOI] [PubMed] [Google Scholar]
- [122].Wanrooij S, et al. , Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci U S A, 2008. 105(32): p. 11122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Wong TW and Clayton DA, Isolation and characterization of a DNA primase from human mitochondria. J Biol Chem, 1985. 260(21): p. 11530–5. [PubMed] [Google Scholar]
- [124].Schiavone D, et al. , PrimPol Is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells. Mol Cell, 2016. 61(1): p. 161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Kobayashi K, et al. , Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides. Cell Cycle, 2016. 15(15): p. 1997–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Bailey LJ, et al. , PrimPol-deficient cells exhibit a pronounced G2 checkpoint response following UV damage. Cell Cycle, 2016. 15(7): p. 908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Dong DW, et al. , Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics, 2014. 15: p. 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Bharti SK, et al. , DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J Biol Chem, 2014. 289(43): p. 29975–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Bailey LJ and Doherty AJ, Mitochondrial DNA replication: a PrimPol perspective. Biochem Soc Trans, 2017. 45(2): p. 513–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lyonnais S, et al. , Corrigendum: The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Sci Rep, 2017. 7: p. 45948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Martinez-Jimenez MI, Lahera A, and Blanco L, Human PrimPol activity is enhanced by RPA. Sci Rep, 2017. 7(1): p. 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Guilliam TA, et al. , Molecular basis for PrimPol recruitment to replication forks by RPA. Nat Commun, 2017. 8: p. 15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Guilliam TA and Doherty AJ, PrimPol-Prime Time to Reprime. Genes (Basel), 2017. 8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kamath-Loeb A, Loeb LA, and Fry M, The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures. PLoS One, 2012. 7(1): p. e30189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Piazza A, et al. , Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Musumeci D, et al. , Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes. Biochim Biophys Acta, 2017. 1861(5 Pt B): p. 1341–1352. [DOI] [PubMed] [Google Scholar]
- [137].Rocca R, et al. , Identification of G-quadruplex DNA/RNA binders: Structure-based virtual screening and biophysical characterization. Biochim Biophys Acta, 2017. 1861(5 Pt B): p. 1329–1340. [DOI] [PubMed] [Google Scholar]
- [138].Salvati E, et al. , Lead Discovery of Dual G-Quadruplex Stabilizers and Poly(ADP-ribose) Polymerases (PARPs) Inhibitors: A New Avenue in Anticancer Treatment. J Med Chem, 2017. 60(9): p. 3626–3635. [DOI] [PubMed] [Google Scholar]


