Abstract
The E3 ubiquitin-protein ligase TRIM21, of the RING-containing tripartite motif (TRIM) protein family, is a major autoantigen in autoimmune diseases and a modulator of innate immune signaling. Together with ubiquitin-conjugating enzyme E2 E1 (UBE2E1), TRIM21 acts both as an E3 ligase and as a substrate in autoubiquitination. We here report a 2.82-Å crystal structure of the human TRIM21 RING domain in complex with the human E2-conjugating UBE2E1 enzyme, in which a ubiquitin-targeted TRIM21 substrate lysine was captured in the UBE2E1 active site. The structure revealed that the direction of lysine entry is similar to that described for human proliferating cell nuclear antigen (PCNA), a small ubiquitin-like modifier (SUMO)-targeted substrate, and thus differs from the canonical SUMO-targeted substrate entry. In agreement, we found that critical UBE2E1 residues involved in the capture of the TRIM21 substrate lysine are conserved in ubiquitin-conjugating E2s, whereas residues critical for SUMOylation are not conserved. We noted that coordination of the acceptor lysine leads to remodeling of amino acid side-chain interactions between the UBE2E1 active site and the E2–E3 direct interface, including the so-called “linchpin” residue conserved in RING E3s and required for ubiquitination. The findings of our work support the notion that substrate lysine activation of an E2–E3-connecting allosteric path may trigger catalytic activity and contribute to the understanding of specific lysine targeting by ubiquitin-conjugating E2s.
Keywords: ubiquitin-conjugating enzyme (E2 enzyme), substrate specificity, E3 ubiquitin ligase, ubiquitin, allosteric regulation
Introduction
Tripartite motif (TRIM)5 proteins constitute the largest subfamily of RING-type E3 ubiquitin ligases, with around 100 members in humans, and are associated with pathological conditions (1, 2). RING-type E3s catalyze the direct transfer of ubiquitin (Ub), or a ubiquitin-like (Ubl) entity such as SUMO or NEDD8, from a thioester-linked E2-conjugating enzyme to specific substrates in the ubiquitination pathway (3, 4). The multimodular TRIMs comprise an N-terminal RING domain, one or two B-box domains, a coiled-coil region, and a C-terminal substrate-binding domain (5) and predominantly support ubiquitination (6).
TRIM21 functionality appears to rely upon its ability to specifically catalyze the formation of multiple Ub chain types, with several distinct E2s, in both nuclear and cytosolic cell compartments and onto a variety of different substrates. TRIM21 (also denoted Ro52 or SSA) was first identified as a major autoantigen in systemic lupus erythematosus and Sjögren's syndrome (7), and RING-domain specific patient autoantibodies impair TRIM21-mediated autoubiquitination by blocking the E2–E3 interaction (8). TRIM protein autoubiquitination in general has been shown to inhibit viral DNA synthesis, direct interferon regulatory factor signaling (9), and steer cellular differentiation (10). We and others have shown that both the cytosolic UBE2D1 (UbcH5a) and the nuclear UBE2E1 (UbcH6) (11, 12) collaborate with TRIM21 in mediating polyubiquitination (13, 14). Nuclear translocation of TRIM21 has been observed as a result of inflammatory signaling (12, 15), and a splice variant, TRIM21β, lacking part of the coiled-coil domain also demonstrated a predominantly nuclear localization (12). TRIM21 negatively regulates innate immune signaling by promoting Lys48-linked substrate ubiquitination of nuclear interferon regulatory factors (16–19). TRIM21 also polyubiquitinates cytoplasmic targets such as the DDX41 DEAD-box protein (20) and mediates monoubiquitination of cytoplasmic substrates, including IKKβ (21) and GMP synthase (22). Finally, TRIM21 autoubiquitination by consecutive Ube2W and Ube2N/Ube2V2 activity produces Lys63-linked Ub chains, both free and anchored to the TRIM21 N terminus, with a suggested role in virus neutralization (23).
Direct interaction between an E2 and a corresponding E3 is essential for RING-mediated ubiquitination, where the “linchpin” arginine residue in the RING domain (24) and the conserved “SPA” motif in E2 loop 7 (25) have been shown to be critical for enzymatic activity (3, 4). Motifs flanking the E3 RING domain have been shown to stabilize the donor ubiquitin in a “closed state” most favorable for ubiquitin transfer (24, 26–31). Such motifs without interacting with the substrate are able to turn on and off ubiquitination activity entirely in response to other signaling factors such as phosphorylation and/or multimodular domain interactions.
Knowledge of substrate-targeting modes and E3-catalyzed substrate transfer mechanisms in Ub–substrate conjugation is scarce because no structures of captured substrates have been determined for a Ub-conjugating E2–E3 complex. However, structural studies of larger multidomain complexes, including SUMO- and NEDD8-targeting E2s UBC9 and UBCH12 trapped in action with their substrates, have revealed how key residues around the E2 active site support SUMO/NEDD8 conjugation at specific substrate residues (32–36). Catalytically inactive modules can assist in positioning specific acceptor lysines from substrates into the E2 active site, thus placing substrate specificity partly outside of the direct E2–substrate interaction (34, 37) and for human PCNA have been shown to support an alternate substrate entry path for SUMOylation (33). Whether Ub-conjugating E2s use the same specificity mechanisms is unclear, in particular because several key residues required for conjugation activity in SUMO- and NEDD8-ylating E2s are not conserved in Ub-conjugating E2s. Indeed, lack of detailed structures hampers the advancement of knowledge required to specifically target pathological conditions related to ubiquitination (1, 2).
In this work, we have investigated the UBE2E1–TRIM21 interaction, where TRIM21 acts both as an E3 catalyst and as a substrate in autoubiquitination. Our resulting TRIM21-bound UBE2E1 crystal structure together with that of free UBE2E1 present structural snapshots that reveal an acceptor Ub–lysine recognition mode that is similar to the lysine entry path for human PCNA (33). Finally, we show how the presence of an acceptor lysine at the E2 active site confers substrate-induced conformational changes that extend to the E2–E3 direct interface, and we propose a model for how this could activate linchpin-mediated ubiquitination.
Results
Structural and functional assembly of the UBE2E1C–TRIM21R complex
TRIM21 α (cytosolic) and β (nuclear) isoforms are identical within the TRIM21 RING (residues 1–91; TRIM21R) fragment used in the complex crystal structure. Corresponding E2s UBE2D1 (cytosolic) and UBE2E1 (nuclear) are closely homologous. We affirmed experimentally that the respective cellular localization of TRIM21 α and β isoforms are indeed compatible with both UBE2D1 (cytosolic) and UBE2E1 (nuclear) E2 enzymes in HeLa cells, transfected with GFP/JRed-tagged constructs or stained with specific antibodies (Fig. S1, A and B).
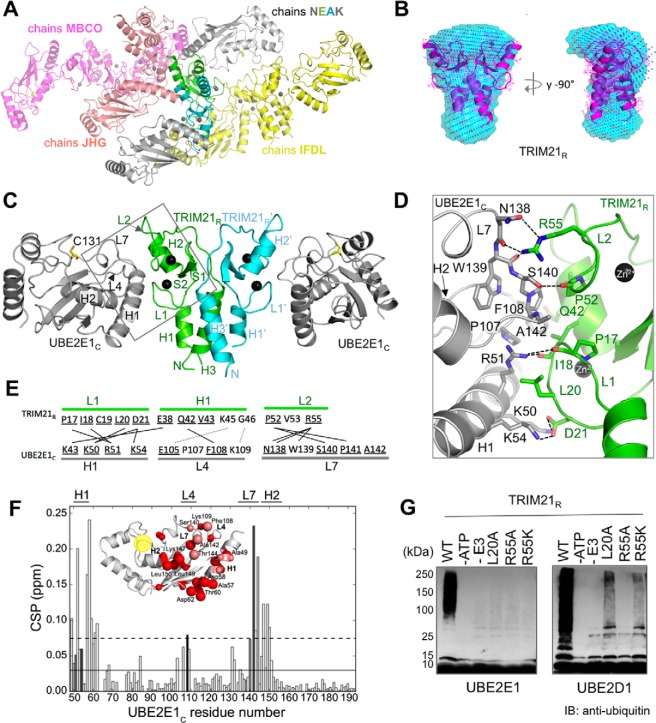
The structure of a complex comprising the TRIM21R domain with flanking helices and the catalytic core domain of UBE2E1 (residues 36–193; UBE2E1C) was determined by crystallography to a resolution of 2.82 Å (Fig. 1A and Table 1, PDB code 6FGA). The complex crystal structure includes four homodimeric TRIM21R and seven UBE2E1C entities in the unit cell, which together form four TRIM21R:UBE2E1C assemblies: two well-defined 2:2 complexes (chains IFDL and MBCO), one 2:2 complex with poor density in one of the E2 entities (chains NEAK), and one 2:1 complex (chains JHG) (Fig. 1A). TRIM21R is predominantly a dimer both in crystal and in solution as estimated by small-angle X-ray scattering (SAXS) and analytical gel filtration (Figs. 1B and S3 and Tables S2 and S3). SAXS measurements of TRIM21R reveal an overall similar shape in solution as in the corresponding crystal structure dimer (Fig. 1B). The quaternary arrangement of E2 and E3–RING dimer modules is highly similar to that in previously determined E2–RING–Ub assemblies (24, 26–31) (Figs. 1C and S2B and Table S1). Finally, to compare free and bound E2 states in this study, we obtained a crystal structure (1.4 Å) of similarly prepared free UBE2E1C (Table 1, PDB code 5LBN), which is similar to matching residues within full-length UBE2E1 (PDB code 3BZH, r.m.s.d. 0.47 Å (38)) (Fig. S2C).
Figure 1.
Structural assembly of the TRIM21R–UBE2E1C complex. A, TRIM21R–UBE2E1C complex crystallized in space group C2 where 15 protomers make up the crystallographic ASU. Eight protomers of TRIM21R form four homodimers (chains FD, EA, BC, and HG), whereas the remaining seven protomers comprise UBE2E1C (chains I, J, K, L, M, N, and O). Altogether, three complete TRIM21R:UBE2E1C 2:2 assemblies are present in the ASU comprising chains MBCO, IFDL, and NEAK, and a 1:2 complex is present comprising chains JHG. B, bead models representing the solution structure of free TRIM21R derived from the SAXS data using DAMMIF and assuming P2 symmetry (cyan) or no symmetry (blue dots) overlaid with the TRIM21R dimer crystal structure. C, cartoon representation of UBE2E1C (gray)–TRIM21R (green/cyan) complex crystal structure with Zn2+ shown as spheres (black); this coloring is maintained in Figs. 2–4. D, structure of UBE2E1C-TRIM21R direct interface (square in C; showing contacts in C). E, overview of TRIM21R-UBE2E1C contacts (lines), including hydrogen bonds or salt bridges and van der Waals interactions (black, solid); proposed interactions are in gray, dotted lines (44, 45). F, combined 1H and 15N CSPs of 15N-labeled UBE2E1C in the presence of 2.0 eq of unlabeled TRIM21R. Average CSP value is represented as the solid line; the dashed line is with one standard deviation added. Bars are colored according to reduced accessible surface area as determined by VADAR (73) from white (0%) to black (100%). Inset, cartoon representation of UBE2E1C. Residues with significant CSPs are shown as red spheres, smaller for CSPs above average, bigger for CSPs above 1 S.D. from average, and colored salmon if buried (>20%). The active-site region is indicated (yellow). G, in vitro autoubiquitination assays with UBE2E1 and UBE2D1 show extent of TRIM21R WT activity and loss of activity in TRIM21R mutants as annotated. IB, immunoblotting.
Table 1.
Crystallography data collection, phasing, and refinement statistics
aOne crystal was used for data collection and refinement.
bValues in parentheses are for highest-resolution shell.
cSpherical/Elliptical completeness, where the elliptical completeness is calculated by the Staraniso server.
The interface connecting UBE2E1C-H1, -L4, and -L7 with TRIM21R-L1, -H1, and -L2 (where H represents helix and L represents loop) is well-defined in the TRIM21R–UBE2E1C crystal structure (Fig. 1, D and E, and Table S1). Significant amide chemical shift perturbations (CSPs) were observed by NMR in the direct interface (Fig. 1F), and a Kd of 24 ± 11 μm was estimated for the TRIM21R–UBE2E1C interaction, based on CSPs in five titration points for nonbroadened residues (Fig. S4, A–C). Significant CSPs were also observed for residues in a contact network extending from the direct interface to the active-site region (Fig. 1F), in full agreement with previously proposed allosteric activation through the E2 core (39–41).
To functionally probe the interface, we used mutational mapping assayed by autoubiquitination (8) and E2–Ub hydrolysis assays of an oxyester-bonded UBE2E1–Ub complex (27, 34). By sequence homology, Arg55 in TRIM21 corresponds to the catalytic linchpin residue in E2-mediated Ub conjugation (24). In agreement, both autoubiquitination and E2–Ub hydrolysis assays were inhibited in TRIM21R-R55A (Fig. 1G). CSPs indicate that TRIM21-R55A interacts with UBE2E1 similarly as wildtype (WT) (Kd ≈ 50 μm; Fig. S4, D and E), supporting that both ubiquitination and E2–Ub hydrolysis depend on the presence of a catalytic element and not simply on complex formation (24, 34). In the direct contact interface, a TRIM21R-L20A mutation significantly reduces autoubiquitination (Fig. 1G), and very small NMR CSPs were observed for UBE2E1C with TRIM21-L20A (Fig. S4F), indicating disrupted binding. Similarly, in the SPA motif of UBE2E1C loop 7 (uL7), a UBE2E1C-A142D mutant entirely disrupts the complex formation as observed by NMR (Fig. S4G), in agreement with the SPA region being critical for TRIM21-catalyzed conjugation activity (25, 42).
UBE2D1 shows a similar but not complete loss of autoubiquitination with TRIM21R-R55K and -L20A mutants compared with UBE2E1 (Fig. 1G). The NMR CSP imprint of TRIM21R on UBE2D1 is highly similar but slightly less stringent compared with that of UBE2E1 (Fig. S1C). Jointly, these observations could indicate more promiscuous and thereby more permissive catalytic activation of UBE2D1 by TRIM21, in agreement with earlier findings (43). Previous studies have suggested UBE2E1-Glu105 (Asp in UBE2D1) and -Lys109 as hot-spot residues in UBE2E specificity (44, 45). We found small but distinct NMR CSPs for UBE2E1C-Lys109 (Fig. 1F) and for the corresponding Lys63 in UBE2D1 (Fig. S1C), supporting a possible role for this residue in TRIM21 recognition.
TRIM21R activity relies on a closed TRIM21R–UBE2E1C–Ub conformation
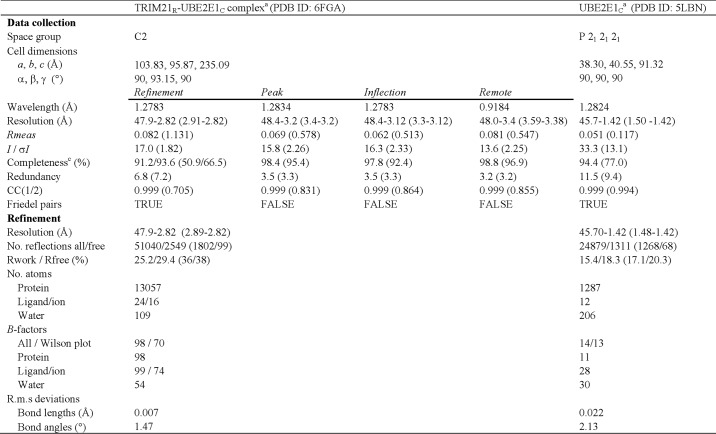
The ternary TRIM21R–UBE2E1C–Ub complex crystals resulted in low diffraction (>6 Å) and showed extensive line broadening by NMR experiments, suggesting dynamic properties. However, we could straightforwardly model the UBE2E1C–Ub–TRIM21R complex, supported by close E2–E3 structural similarity to a wealth of ternary E2–Ub–E3 complexes (24, 26–31) (Figs. 2A, S6, and S7A). In this model, TRIM21R residues Glu12, Glu13, Arg67′, and Asn71′ (′ symbol represents residues of the other RING protomer) hold positions that could stabilize a closed Ub conformation and thereby affect activity, as first shown for c-Cbl (46, 47) (Fig. 2A). Indeed, E2–Ub hydrolysis was severely compromised for TRIM21R mutants E12A, E13A, E13K, double mutant E12K/E13K, R67′A, and N71′A (Figs. 2B and S5) even if the E2–E3 interaction was retained as shown by NMR (Fig. S4H). The same TRIM21R mutants are also poorly active in autoubiquitination assays, where intrinsic UBE2E1 autoubiquitination instead becomes visible in reactions with no or poorly functioning E3 (Fig. 2C) (48). Interestingly, the single TRIM21R-E12K mutant is as active as WT TRIM21R in autoubiquitination assays but still shows greatly reduced activity in E2-Ub hydrolysis (Figs. 2B and S5). Indeed, a similar effect was observed for E12R in the related TRIM25, further supporting functional similarities between these TRIMs (30). Taken together, our results identify residues in TRIM21R helices flanking the core RING motif that significantly affect Ub transfer, presumably by stabilizing a “closed” Ub conformation in a ternary complex.
Figure 2.
Ubiquitin recognition and activation by TRIM21R. A, model showing the position of Ub (brown) in a conjugated, closed conformation relative to UBE2E1C and TRIM21R. The model was based on E2 structural superposition onto TRIM25–UBE2D1∼Ub (PDB code 5FER; Cα r.m.s.d., 0.54 Å). Inset, magnified view highlighting TRIM21R residues supporting the Ub closed state. B, reaction rates determined (mean ± S.D. of triplicates) for TRIM21-dependent hydrolysis of UBE2E1C (C131S)–O∼Ub (O represents oxyester linkage) by TRIM21 mutants as shown, color-coded corresponding to TRIM21R protomer chains. For complete gel source data, see Fig. S5C. Error bars represent S.D. C, in vitro autoubiquitination assays of TRIM21R and mutants with UBE2E1 as annotated. IB, immunoblotting.
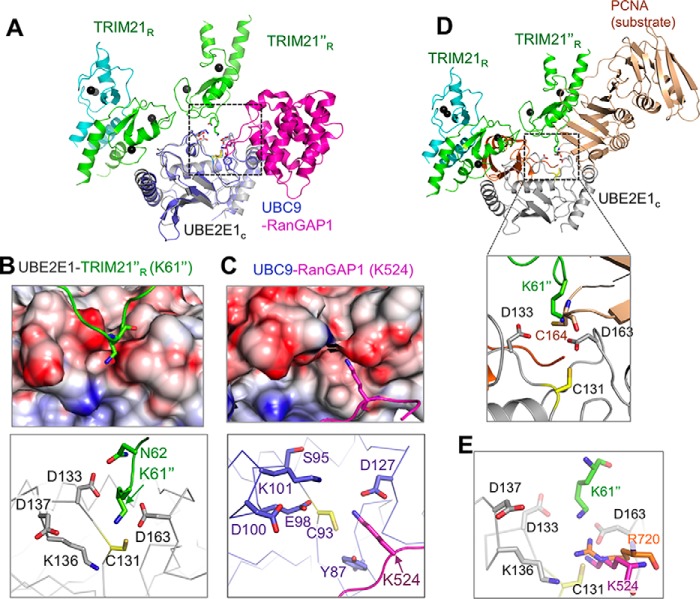
Crystal capture of a TRIM21 Lys61 acceptor lysine in the UBE2E1 active site
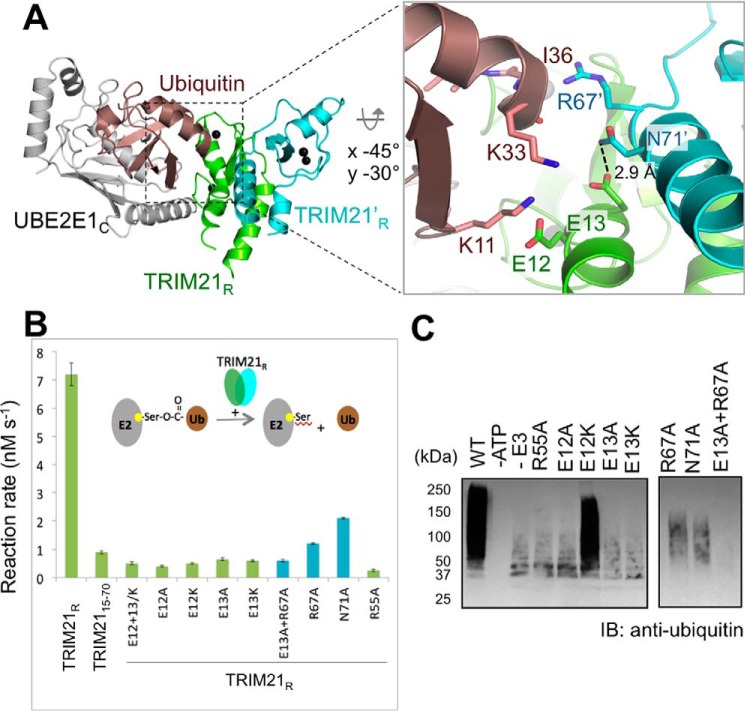
In the TRIM21R–UBE2E1C complex, we observed that the UBE2E1C active site of the NEA chain assembly contacts Lys61 in the TRIM21R F chain. This chain is adjacent to the NEA assembly in the asymmetric unit (ASU) and is here labeled TRIM21R″ (Figs. 1A and 3A). The Lys61″ side chain is well-accommodated in a pocket lined by UBE2E1C residues Asp133 and Asp163 (Fig. 3B). Within the resolution of the structure, the lysine ϵ acceptor group could easily form hydrogen bonds with Asp133 and Asp163 side-chain carboxylates and is within 5 Å of the active cysteine (Fig. 3B). Asp133 further bolsters the interaction by a hydrogen bond stabilizing the Asn62″ side-chain amide (Fig. 3B). In the complex, the side-chain orientations of Lys61″, Asp133, and Asp163 are all supported by well-defined electron densities (Figs. 3B and S7B). In contrast, the structure of free UBE2E1C shows very poor density for Asp163 despite the higher-resolution data, indicating a disordered orientation of this residue in the unbound state (Figs. 3C and S7C); similar disorder is observed also in substrate-free states of full-length UBE2E1 (PDB code 3BZH), UBE2D1 (PDB code 2C4P), and ubiquitin-conjugated UBE2D1 (PDB code 4AP4). Together, this suggests that lysine-coordinating residues are ordered on substrate lysine coordination in the active site.
Figure 3.
Structural positioning of the TRIM21R acceptor lysine in the UBE2E1 active site. A, cartoon representation of the UBE2E1C–TRIM21R complex (chains NEA) together with adjacent TRIM21R″ (chain F) in the asymmetric unit cell. Residues and regions highlighted in B–F are shown. B, 2Fo − Fc map (gray mesh; contoured at 1.2σ) for UBE2E1C active-site region (Cys131, Asp133, Lys136, Asp163, and the catalytic triad 123HPN125) together with Lys61″ and Asn62″ residues of TRIM21′R (PDB code 6FGA) with annotated amide distances to side-chain carboxyls in UBE2E1C. C, the 2Fo − Fc map (cyan mesh; contoured at 1.2σ) of the unbound UBE2E1C structure (PDB code 5LBN) with orientation and annotations as in B. D–F, magnified view of residues connecting Arg55(E3)–Asn138(E2)–Asp133(E2)–Lys61/Asn62(substrate) shown for free UBE2E1C (D), the UBE2E1C–TRIM21R complex (chains MBC) (E), and the TRIM21R″–UBE2E1C–TRIM21R (chains NEAF; similar to B) (F). G, in vitro autoubiquitination assay shows that the acidic residue Asp133 and Asp163 mutants of UBE2E1C are essential for substrate ubiquitination; remaining ubiquitination pertains to known E3-independent UBE2E1C autoubiquitination (48). H, E2–Ub oxyester hydrolysis assay of UBE2E1c shows that Asp163 does not affect Ub release, whereas D133S results in lost activity.
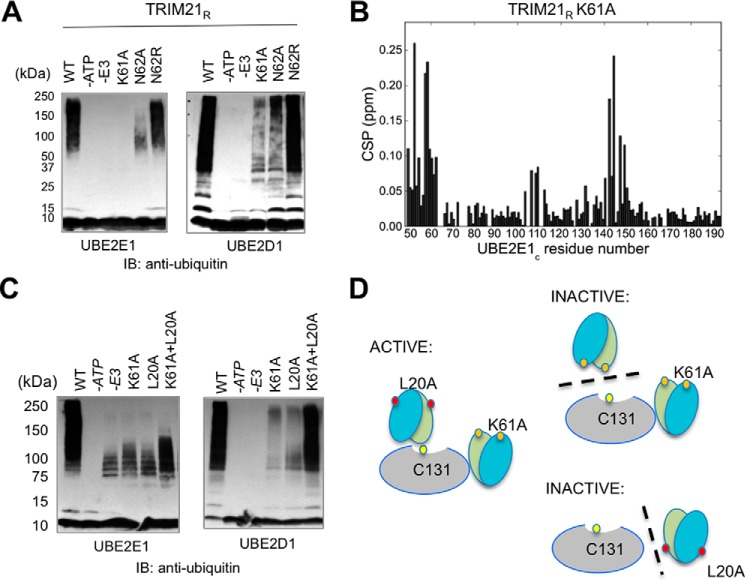
We probed the nature of Lys61″ as a possible target residue for autoubiquitination in several ways. First, a K61A mutation severely disrupts TRIM21R autoubiquitination with both UBE2E1 and UBE2D1 (Fig. 4A). A TRIM21R-N62A mutation similarly disrupts ubiquitination, whereas TRIM21R-N62R ubiquitination is close to WT, both by UBE2E1 and UBE2D1 (Fig. 4A), in agreement with a structural role of an adjacent side-chain amide in supporting ubiquitination (Fig. 4A). Ubiquitination at other sites in TRIM21R (Lys45 and Lys77) is very weak or absent as judged by the very low residual ubiquitination in TRIM21-K61A (Fig. 4A), suggesting Lys61 is the primary site for autoubiquitination in TRIM21R.
Figure 4.
Analysis of TRIM21R acceptor lysine Lys61 and reconstitutional in vitro autoubiquitination experiments using TRIM21R-K61A + TRIM21R-L20A mutants. A, in vitro autoubiquitination assays of TRIM21R and mutants with UBE2E1 and UBE2D1, respectively. B, CSPs of 15N-labeled UBE2E1CS in the presence of 2.0 eq of unlabeled TRIM21R-K61A (red) show binding similar to WT. C, autoubiquitination in a reconstitution experiment using substrate lysine-deficient mutant K61A and E2–E3 interaction–deficient mutant L20A. Ubiquitination is restored by an equimolar mixture of K61A and L20A mutants, in particular by UBE2D1 but also by UBE2E1. The outcome of the UBE2E1 experiment is slightly obscured by the known autoubiquitination of this E2 (48) as observed in the −E3 experiment. D, schematic overview of the interpretation of the reconstitution experiment: an active complex requires the presence of a substrate lysine (Lys61) and an uninterrupted E2–E3 interface (Leu20). Mutations L20A (red filled circles) and K61A (orange filled circles) are indicated. The schematic sketch here shows only the homodimer case for simplicity because only the mutation present in the protomer closest to E2 affects the outcome of experiment. IB, immunoblotting.
Because TRIM21 serves both as E3 and substrate in autoubiquitination, we critically interrogated whether our TRIM21R-K61A mutation might itself impair the E3 activity of TRIM21 by disrupting interactions or catalytic functions. First, NMR CSP analysis shows that the UBE2E1C–TRIM21R-K61A binding pattern is highly similar to that of WT TRIM21R (Fig. 4B). Second, in ubiquitin discharge assays, both K61A and N62A mutants are as active as TRIM21R, suggesting that these mutant E3s fully retain their ability to catalyze the release of Ub (Fig. S5). Third, to assay the capacity of TRIM21R-K61A in catalysis of Ub conjugation, we performed a reconstitution experiment with the non-E2–binding TRIM21R mutant L20A as a pseudosubstrate (Fig. 4, C and D). If deficient autoubiquitination in TRIM21R-K61A is only due to the lack of a target lysine and not to deficient catalysis, then TRIM21R-K61A should still be able to catalyze ubiquitination of the L20A mutant at its retained Lys61. In agreement with this, we found restored ubiquitination by an equimolar mixture of K61A and L20A mutants, in particular by UBE2D1 but also by UBE2E1 (Fig. 4C). Taken together, these experiments show that TRIM21R-K61A interacts with UBE2E1 similarly as WT and is catalytically active both in Ub discharge and conjugation, which together with the deficient autoubiquitination for K61A implies that Lys61 is indeed targeted in autoubiquitination by both UBE2E1 and UBE2D1.
Based on our structure, we then probed the roles of the Lys61″-coordinating residues Asp133 and Asp163 in catalysis and substrate recognition. A Ub-conjugated UBE2E1C-D163A mutant is hydrolyzed similarly as WT in the presence of TRIM21 (Fig. 3H), whereas the same mutation entirely abrogates TRIM21-mediated polyautoubiquitination (Fig. 3G), leaving only the known slow intramolecular UBE2E1-Lys136 ubiquitination (48) at a position close to the catalytic Cys131 (Fig. 3B). Thus, Asp163 appears to be primarily involved in substrate recognition. These results are in full agreement with corresponding D117A mutations in UBE2D1 (27, 49–51) and with the observation that serine phosphorylation in the corresponding position activates Ube2A for ubiquitination (for a review, see Ref. 57).
To assay the role of Asp133 in substrate recognition, we had to consider that this conserved aspartic acid anchors to the Ub C-terminal tail in the closed state in a range of Ub-conjugating E2s while employing the same rotamer as Asp133 in the free state (Fig. 3C) (26, 27, 30, 31, 35, 51). In SUMO-conjugating UBC9, a serine corresponding to Asp133 in UBE2E1 anchors identically to the SUMO C-terminal backbone (35, 36), suggesting that a D133S mutation in UBE2E1 could reveal a role in substrate recognition without distorting Ub anchoring. Indeed, as for D163A, we found that TRIM21-mediated substrate ubiquitination is interrupted by a D133S mutation, whereas UBE2E1 internal autoubiquitination to a lysine proximal to the active site can proceed (48) (Fig. 3G). However, in contrast to D163A, we found that that the D133S mutation also aborts TRIM21-mediated E2–Ub hydrolysis (Fig. 3H), suggesting an additional role for this residue in TRIM21-mediated catalysis.
Substrate-induced active-site remodeling extends to the RING linchpin
To investigate whether the substrate-induced reorientation of Asp133 could induce further structural changes, we compared our three structures of free, E3-bound, and E3 + substrate–bound UBE2E1 (Fig. 3, D–F). In the absence of E3 and substrate, the orientation of Asp133 is stabilized by an intramolecular side-chain hydrogen bond to Asn138, for which two side-chain rotamers were identified in the crystal structure (Fig. 3D). In the E2–E3 complex, the Asn138 side chain of UBE2E1 is constrained into a unique rotamer, supported by electrostatic interactions with TRIM21R-Arg55, but with the hydrogen bond to Asp133 maintained (Fig. 3E). Finally, in the substrate complex, the Asp133 side-chain carbonyl shows favorable interactions with Lys61″ and Asn62″ (Fig. 3, B and F), thereby releasing the Asn138 side chain to form a shorter hydrogen bond with the TRIM21R-Arg55 side chain. In this tentative Lys61/Asn62(substrate)–Asp133(E2)–Asn138(E2)–Arg55(E3) hydrogen-bonding network, a conservative TRIM21R-R55K mutation would interrupt Asn138 interactions. Indeed, an R55K mutation disables both UBE2E1-mediated ubiquitination (Fig. 1G) and UBE2E1–Ub hydrolysis (Fig. S5) even though CSPs suggest a maintained E2–E3 interaction (Fig. S4I). Taken together, an Arg55 linchpin-connected, hydrogen-bonding network may be critical for TRIM21-mediated catalysis of ubiquitination.
Residues in the UBE2E1 active-site entry path are conserved in Ub-conjugating E2s
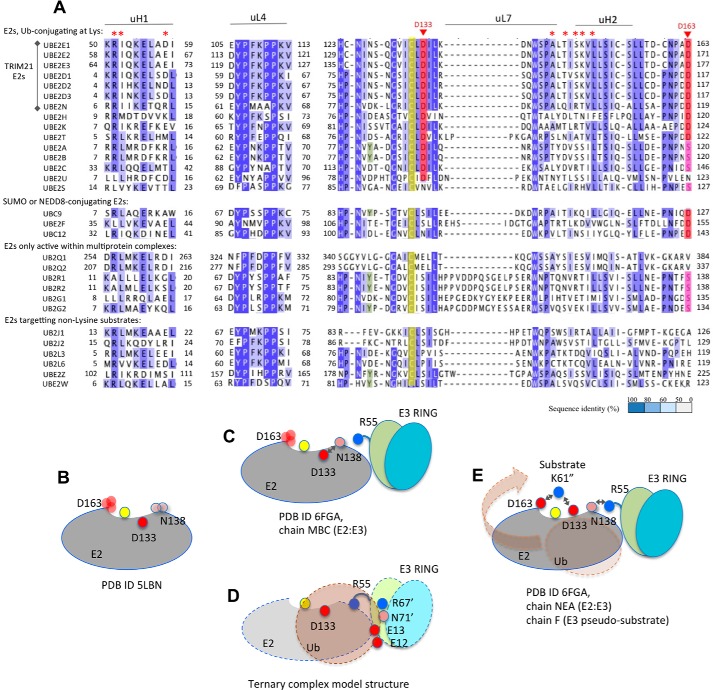
In the Ub-conjugating UBE2E1, the entry path of the targeted Lys61″ is guided by Asp133 and Asp163, which jointly line a negative crevice extending to the active-site cysteine (Fig. 5, A and B). Interestingly, ubiquitin-conjugating, lysine-targeting E2s either hold a conserved Asp133 (Asp/Glu), or a large, negatively charged L7 loop adjacent to the Asp133 position (UBE2E1 numbering; Fig. 6A). Similarly, at the Asp163 position, E2s active in lysine-anchored ubiquitination either hold a conserved Asp/Asn or a phosphorylatable serine (52, 53) (Fig. 6A). Conservation of this negative crevice at the active site suggests that the substrate entry path presented here for UBE2E1/TRIM21 could be accessible also to other Ub-conjugating E2s.
Figure 5.
Acceptor lysine positioning toward the E2 active site and the acidic residues of UBE2E1. A, E2-based superposition of TRIM21R″–UBE2E1C–TRIM21R (chains NEAF) with the human UBC9 (blue)–RanGAP1 (magenta) substrate complex (PDB code 2GRN). Electrostatic surface representation and side chains in the near vicinity of acceptor lysines are shown in insets, centered on TRIM21R″-Lys61 (B) and RanGAP1-Lys524 (C). D, structural superposition of E2s from the E2–SUMO–Siz1 E3–PCNA (dark gold) substrate complex (PDB code 5JNE; only PCNA is shown) onto TRIM21R″–UBE2E1C; only the acceptor residue analogs for PCNA (Cys164) and Lys6″ together with the Ub C-terminal tail (residues 72–75) from the TRIM25–UBE2D1∼Ub (orange; PDB code 5FER) ternary complex are shown. The inset shows the magnified view of the E2 active site, and residues are annotated. E, same as B, where the acceptor lysines Lys61″ from TRIM21R″ and Lys524 of RanGAP1 from UBC9–RanGAP1 substrate complex (PDB code 2GRN) and residue analog Arg720 of CUL1 from E2–NEDD8–RBL1–CUL1 (PDB code 4P5O) complex are shown.
Figure 6.
UBE2E1 residues involved in lysine anchoring and substrate-induced E2–E3 communication are conserved in lysine-targeting Ub-conjugating human E2s. A, human E2 sequence alignment, colored by degree of sequence identity and grouped by functional properties according to Ref. 53. Residues corresponding to UBE2E1-Asp133 and -Asp163 that anchor the substrate lysine in the present work are colored when conserved as Asp/Glu (red) or as Ser/Thr (pink; phosphorylatable). E2s active with TRIM21 are indicated (left), and UBE2E1 residues with significant TRIM21-induced CSPs in NMR spectra are annotated (on top, red asterisk). B–E, schematic showing how the Lys61/Asn62(substrate)–Asp133(E2)–Asn138(E2)–Arg55(E3) path, as structurally detailed in Fig. 3, D–F, relays communication during ubiquitination. UBE2E1 (E2), TRIM21 (E3 RING), and Ub domains are shown as simplified shapes, colored as in Figs. 1 and 3. On-path and Ub-interacting residues are shown as filled circles, labeled and colored according to charge; Cys131 is yellow and unlabeled throughout. Side-chain disorder is indicated by multiple, transparent circles, and hydrogen bonds are indicated as double arrows. Final transfer of Ub from E2 to substrate is indicated by a transparent brown arrow (E).
The targeted TRIM21-Lys61 in the current complex is well-positioned with respect to the active-cysteine compared with the substrate-containing structures obtained for SUMO- and NEDD8-conjugating E2s (Table S5). A SUMO substrate entry path similar to that employed in UBE2E1 was shown for yeast UBC9 sumoylation of human PCNA where the substrate is presented to E2 in a multimodular complex that steers the substrate into the E2 active site (Fig. 5D) (33). In contrast, in the human SUMO-conjugating UBC9 complex with the substrate RanGAP1, the targeted Lys524 enters at nearly right angles to TRIM21-Lys61″, similarly directed by UBC9-Asp127 (equivalent to UBE2E1-Asp163). Here, the substrate entry is critically bolstered by UBC9-Tyr87 (35, 36) (Fig. 5C), which only occurs in UBE2A and UBE2B among Ub-conjugating E2s (Fig. 5A). A UBE2E1-like acceptor lysine entry into UBC9 would be structurally hindered by a UBC9-Lys101-Asp127 ion pair gate (Fig. 5C). Reciprocally, a UBC9-like acceptor lysine entry into the UBE2E1 active site would be repelled by the equivalent of UBE2E1-Lys136 where a positive charge is conserved in Ub-conjugating E2s (Fig. 5C). NEDDylation relies on a complex but specific multimodular assembly that optimally positions the modules of E3, E2, and NEDD8 for catalysis, resulting in a lysine entry path similar to that in SUMO-conjugating UBC9 but does not hold the entire SUMO conserved pattern for substrate recognition of acceptor residue (Fig. 5D) (34).
Discussion
In this work, we present the crystal structure of a TRIM21–UBE2E1 complex comprising the TRIM21 RING domain. Although biochemical and mutational data for this complex consistently agree with observations for other Ub-conjugating E2s, our structure also presents the capture of a substrate lysine acceptor targeted for RING-mediated ubiquitination. The functional consistency between our UBE2E1–TRIM21 complex and other E2–RING complexes makes it plausible that also other Ub-conjugating E2s could conjugate their substrates in a similar manner.
A common denominator for both Ub- and SUMOylation is the critical functional role for the residue corresponding to UBE2E1-Asp163, which is conserved in both Ub- and SUMO/NEDDylating E2s (53) (Fig. 5A). This aspartic acid appears to coordinate the substrate lysine and prepare it for conjugation (35, 36) (Fig. 3). However, as shown here, the different substrate entry paths adopted by UBE2E1–TRIM21 and UBC9–human PCNA on one hand and UBC9–RanGAP1 and NEDD8–RBL1–CUL1 on the other jointly support the presence of varied substrate entry paths to E2 active sites. This suggests that the proposed general “gateway” role of the aspartic acid, UBE2E1-Asp163 (Fig. 5A), would primarily be to select and coordinate the acceptor lysine rather than to steer the substrate entry path. Taken together, this supports that, in addition to multimodular steering (33), E2 entry paths depend on small sequence variations, which may also guide substrate specificity.
Extending current views on substrate recognition, the acceptor lysine in the current structure is also coordinated by Asp133, which is uniquely conserved in Ub-conjugating E2s (Fig. 5A). At first glance, this could seem unexpected because, in the absence of substrate, residues equivalent to Asp133 in other Ub-conjugating E2s were shown to anchor to the C terminus of Ub in its closed state (26, 27, 30, 31, 35, 51). In the SUMO-conjugating UBC9, a serine in the position corresponding to Asp133 in UBE2E1 fulfils the same role in anchoring the highly similar SUMO C terminus but does not coordinate the substrate lysine (35, 36). If the sole and primary function of Asp133 in UBE2E1 is to stabilize a reactive, closed-state Ub, then a conservative D133S mutation in UBE2E1 should also support ubiquitination, which is opposed to our findings (Fig. 3G).
Our results suggest that, in ubiquitination, the conserved Asp133 might have dual roles in supporting the closed state of E2–Ub and in recognizing acceptor substrate lysines. Indeed, a structural overlay of our structure with the TRIM25–UBE2D1–Ub ternary complex (Fig. S7D) suggests that both UBE2E1-Asp133 and TRIM21-Arg55 would be prompted to release their Ub-stabilizing interactions in response to substrate binding. The release of Ub from its closed, E2-anchored state onto the substrate would then be triggered by the active-site coordination of the substrate lysine acceptor, by a chain of events affecting residues in the Lys61/Asn62(substrate)–Asp133(E2)–Asn138(E2)–Arg55(E3) contact chain (Fig. 6, B–E). By connecting the E2 active site with its corresponding E3, such a chain of activation would then imply that E3-catalyzed ubiquitination is jointly mediated by its substrate. The current structure, obtained in the absence of Ub, might then represent a model for a transition step where Ub is no longer anchored onto E2 in a closed conformation but is being released upon substrate conjugation (Fig. 6E).
The present structure together with mutational data suggests that TRIM21–UBE2E1 may hold substrate selectivity toward a K(N/R) pattern. Indeed, specific TRIM21-mediated monoubiquitination targets the 182KK pattern of the substrate GMP synthase (22). In DDX41, one of two Ub-Lys48–conjugated lysines holds an 8RKR motif (20). In IKKβ, a monoubiquitinated (21), TRIM21-mediated site at Lys163 and all three Ub-Lys48–conjugated sites hold motifs where Lys is flanked by an amide-containing side chain (162HK, 418KR, 555KQ, and 703KK), and monoubiquitination by UBE2E1 at histone H2A occurs at the 118PKKT motif (54). TRIM21 itself contains several additional K(N/R/K) motifs outside the TRIM21R domain that could be targeted by autoubiquitination. UBE2T in the Fanconi anemia pathway, which holds the conserved Asp133/163 pattern, spontaneously ubiquitinates FANCL at 522RKQ (55) (Fig. 6A). Finally, E2s specifically targeting hydroxyls, cysteines, lipids, or N termini do not hold the Asp133/163 conservation but instead show high variability in these active site–proximal positions (Fig. 6A), which may further support the importance of E2 active-site interplay with the substrate anchor site to fine-tune specificity in ubiquitination.
In a larger context, autoubiquitination of TRIM proteins has been observed as a mechanism for antiviral defense and correlates with inhibition of retroviral transcription (56, 57). In studies of TRIM5 assembly on capsids, a TRIM5–TRIM21 RING chimera spontaneously assembled into hexagonal two-dimensional arrays of TRIM dimers of antiparallel coiled coils, which resulted in the presentation of three RING domains at each hexagonal corner (58). It has been suggested that two of the RING domains could then dimerize and catalyze E2-mediated ubiquitination of the third RING (59). Our current structure supports this hypothesis by providing a detailed molecular mechanism for how such autoubiquitination occurs and a new structural scaffold for investigating how this could be facilitated in a trimeric arrangement. Further high-resolution structural analysis of TRIM substrate complexes with functionally complementary E2/E3/Ubl partners will be essential to map their structural and functional versatility and will advance the analysis of functional properties in multimodular ubiquitinating complexes.
Experimental procedures
Cloning of recombinant proteins
Human TRIM21 (UniProt accession number P19474) constructs were subcloned by ligation-independent cloning (67) into pET28-MHL expression vectors (TRIM21M1–R91) carrying an N-terminal, cleavable His6 tag. Full-length UBE2E1 (UniProt accession number P51965) and UBE2E1C (residues 36–193) were respectively subcloned into pET28b. Point mutations were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene). In addition, the UBE2E1C scaffold consistently included an S68R mutation to prevent noncovalent interactions between Ub and the backside of the E2's UBC domain, similar to UBE2D1-S22R (60). The Ube1/PET21d plasmid was a gift from Prof. Cynthia Wolberger (Addgene plasmid 34965) (61).
Recombinant protein expression and purification
TRIM21 constructs were expressed in Escherichia coli BL21(DE3) Rosetta-2 cells, induced with 0.2 mm isopropyl d-1-thiogalactopyranoside and 20 μm ZnCl2. After 16–18 h at 18 °C, the cells were lysed by sonication in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10% glycerol (v/v), 10 mm β-mercaptoethanol, 20 μm ZnCl2, and 5 units/ml DNase I (Roche Applied Science). The supernatant was purified on Ni2+-NTA-agarose resin (Qiagen) and eluted with 100–150 mm imidazole buffer. The His6 tag was cleaved off with tobacco etch virus recombinant protease (62) or thrombin as required. Cleaved protein was passed over Ni2+-NTA-agarose resin, and the flow-through was collected, concentrated, and subjected to Superdex 75 gel filtration (GE Healthcare) in 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% glycerol (v/v), 100 μm ZnCl2, and 10 mm β-mercaptoethanol. Buffer optimization was performed using static light-scattering StarGazer-384 (Harbinger), aiming for consistent high stability without signs of aggregation. Compared with our previous work (8), the stability of the TRIM21 RING was much improved by removal of the His tag, which in turn allowed for an increased ZnCl2 content without precipitation.
All UBE2E1 constructs were expressed in E. coli BL21(DE3) pLysS cells (Stratagene) at 37 °C and induced with 0.5 mm isopropyl d-1-thiogalactopyranoside for 20 h at 20 °C. Harvested cells were lysed by sonication in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10% glycerol (v/v), 10 mm β-mercaptoethanol, and 5 units/ml DNase I. The supernatant was applied to a 5-ml HisTrap column (GE Healthcare) and eluted with imidazole gradient. The His6 tag was cleaved by thrombin (25 °C, 4 h) followed by gel filtration (Superdex 200, GE Healthcare). Isotope-labeled proteins for NMR were expressed in M9 minimal medium supplemented with [13C]glucose and/or 15NH4Cl (Cambridge Isotopes). UBE2E1C mutants were prepared similarly as UBE2E1 WT. Preparation of recombinant Ube1 was carried out as described earlier (61).
E2–Ub oxyester hydrolysis assays
To generate the E2–Ub conjugate, UBE2E1C-S68R/C131S, denoted as UBE2E1SC (100 μm), His-tagged ubiquitin (120 μm), and His-tagged human Ube1 (5 μm) were incubated for 16–18 h at 30 °C in a reaction buffer containing 20 mm Tris-HCl, 200 mm NaCl, 5 mm ATP, 5 mm MgCl2, and 10 mm β-mercaptoethanol. The E2–o–Ub (o represents oxyester) conjugate was first purified by Ni2+-immobilized metal-affinity chromatography to separate the E2–o–Ub conjugate from unconjugated E2 followed by His tag cleavage and size-exclusion chromatography on a Superdex75 column. Purified E2–o–Ub (15 μm) was mixed with TRIM21R or TRIM21R mutants (10 μm) and incubated for 180 min at 27 °C with samples taken at several time points (5, 15, 30, 60, 90, and 180 min). Reactions were stopped by addition of SDS Laemmli buffer and analyzed by SDS-PAGE stained with Coomassie Blue R-250. The E2–o–Ub conjugate quantification on stained gels was performed using ImageQuant (GE Healthcare). Reactions were performed in triplicates, and rates are given as mean ± 1 S.D.
Autoubiquitination activity reaction
Autoubiquitination assays were performed in 20-μl reactions containing 0.50 μm TRIM21R or variants thereof, 100 ng of E1, 500 ng of UBE2E1, and 2.5 μg of ubiquitin in a buffer containing 50 mm Tris-HCl, 2.5 mm MgCl2, 0.5 mm DTT, and 2 mm ATP. Each reaction mixture was incubated for 2 h at room temperature and terminated by addition of 5 μl of 5× SDS-PAGE sample buffer containing 100 mm Tris-Cl, 10% (w/v) SDS, 0.5% (w/v) bromphenol blue, and 500 mm DTT followed by boiling. The total reaction mixture was loaded onto a 4–20% gradient gel, separated by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane for immunoblotting against ubiquitin.
Plasmids for localization experiments
pEGFP-TRIM21 (Ro52) and pEGFP-TRIM21β (Ro52β) were generated by subcloning TRIM21 and TRIM21β from pMyc-TRIM21 and pMyc-TRIM21β (12, 15), respectively, into pEGFP-C3 (Clontech) using the compatible EcoRI and SalI restriction sites and religating the plasmid retaining the correct reading frame. pJRed-UBE2D1 was generated by amplifying UBE2D1 mRNA by RT-PCR of human lymphocyte cDNA using the following primers: forward, CAACAAGTCGACATGGCGCTGAAGAGGATT; reverse, CAACAAGGATCCTTACATTGCATATTTCTGAGT. The PCR product was first subcloned to a pTOPO XL vector (Thermo Fisher Scientific). pTOPO XL-UBE2D1 was subsequently digested with BamHI and SalI and inserted into BamHI- and SalI-digested pJRed-C plasmid (Evrogen), retaining the correct reading frame, followed by religation of the plasmid.
Subcellular localization of UBE2E1, UBE2D1, TRIM21, and TRIM21β
HeLa cells were chosen for the localization experiments based on their morphology with a large thin cytoplasm when cultured on microscopic slides. Cells were cultured on Nunc Lab-Tek II chamber slides (Thermo Scientific). For transfection, 500 ng of plasmid (pJRed-UBE2D1, pEGFP-TRIM21, or pEGFP-TRIM21β) was used together with X-tremeGENE 9 reagent (Sigma-Aldrich, Merck). After 48 h, cells were washed with PBS before fixation with 4% paraformaldehyde for 10 min at 4 °C. For immunostaining, cells were fixed with 4% paraformaldehyde for 10 min at 4 °C and then permeabilized with 0.2% Triton X-100 followed by a blocking step with 5% fetal bovine serum in PBS for 30 min. 1 μg/ml rabbit anti-human UBE2E1 (ab36980, Abcam) was used as primary antibody and incubated for 60 min. Bound antibodies were detected by Alexa Fluor 594–conjugated donkey anti-rabbit antibodies in a 1:400 dilution (Molecular Probes).
Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Molecular Probes) in PBS for 2 min, and slides were mounted in Prolong Gold antifade mounting medium (Invitrogen) under a coverslip. Rinsing in PBS was performed two to four times between each step, and all steps but the fixation were performed at room temperature. A laser-scanning confocal microscope was used to assess and document the cells (63× magnification).
Analytical gel filtration
Tricorn Superdex 75 10/300 was used to perform the analytical gel filtration in an ÄKTA purifier using a standard low-molecular-weight calibration kit (GE Healthcare) to calculate void volume by blue dextran and molecular weight calibration curve from the standard proteins therein. TRIM21 and UBE2E1 constructs were analyzed in a concentration range of 50–400 μm with an injected volume of 100 μl and a flow rate of 0.5 ml/min.
NMR spectroscopy and data analysis
UBE2E1 NMR samples were prepared in buffer containing 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 0.02 mm NaN3, and 90% H20, 10% 2H20 (v/v). Triple-labeled [13C,2H,15N]UBE2E1 (600 μm) was prepared for the peptide backbone assignments. HNCA, HNCO, HNCOCA, HNCACO, HNCACB, and HNCOCACB triple-resonance experiments were collected at 30 °C on a Varian INOVA spectrometer operating at a proton frequency of 600 MHz (with cryoprobe). Due to poor stability at high concentrations for the UBE2E1C construct, UBE2E1CS (C131S) protein was prepared at a concentration of 400 μm, and HNCA, HNCO, and HNCACB resonance experiments were recorded to confirm assignments in the shorter construct. Data were processed using NMRPipe/NMRDraw (63), visualized and analyzed with the Sparky program (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). The backbone assignment was manually performed, assisted by the COMPASS software (64).
Titrations experiments were carried out in a buffer containing 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10 mm β-mercaptoethanol, 500 μm ZnCl2, and 90% H20, 10% 2H20 (v/v). CSP data were collected at 30 °C in 1H,15N heteronuclear single quantum coherence transverse relaxation optimized spectroscopy–based experiments on a Varian 500-MHz NMR spectrometer with uniformly 15N-labeled UBE2E1C (200 μm) apo constructs and addition of 0.25, 0.5, 0.75, 1, and 2 eq of unlabeled TRIM21R constructs. CSPs were calculated with the formula Δδ = [(Δδ1H)2 + (Δδ15N × 0.156)2]1/2 (65) where Δδ1H and Δδ15N are chemical shift perturbations (in ppm) with respect to the 1H and 15N chemical shifts and 0.156 is the normalization factor. To identify significant CSPs, a cutoff of two standard deviations from the trimmed mean was calculated in an iterative procedure as described (65).
Kd values were calculated by a nonlinear least-squares analysis using the following equation,
| (Eq. 1) |
where [P]T and [L]T are the total protein (NMR labeled) and ligand (unlabeled) concentrations at each aliquot, Δδ′ is the change in peak position with each aliquot, and Δδ′max is the change in shifts between apo and fully bound states of the protein, P. Kd values were only calculated for residues that show significant chemical shift perturbations upon TRIM21 binding and have signal intensities above the noise level. The dissociation constant of UBE2E1C–TRIM21R binding is an average over values obtained from fitting titrations on a per-residue basis for residues in UBE2E1C-H1, -L4, and -L7. Kd values obtained for residues in UBE2E1C-H2 were averaged separately, as CSPs observed for this region likely originate from allosteric effects.
Crystallization
Purified E2, UBE2E1C, and TRIM21R were mixed in 1:1.2 and 1:2 ratios, incubated overnight, and then concentrated to 35 mg ml−1. Initial crystal hits were optimized in both sitting-drop and hanging-drop vapor diffusion at 4 °C with a reservoir solution containing 100 mm Bicine, pH 9.0, and 5% (w/v) PEG 6000. Final crystals were obtained in the above-described reservoir conditions with 12.5% (v/v) glycerol and 5% (v/v) ethylene glycol and flash frozen in liquid nitrogen. Crystals belong to C2 space group with cell dimensions of a = 103.811 Å, b = 95.834 Å, c = 235.043 Å, α = γ = 90.0°, and β = 93.15° with a solvent content of 54%. For the free E2 structure, UBE2E1C was concentrated to 18 mg ml−1 and crystals were optimized in sitting-drop vapor diffusion at 4 °C. The initial crystals were obtained in 0.1 m sodium citrate, pH 6, and 8% (w/v) PEG 8000 at 4 °C. Final single crystals were obtained from the hanging-drop method in the same reservoir condition with added 10% (v/v) glycerol and flash frozen in liquid nitrogen.
Crystallography structure determination
Diffraction data for UBE2E1C–TRIM21R crystals were collected at BL14.1 beamline at BESSY Synchrotron (Berlin, Germany) and screened for TRIM21R presence by testing for diffraction at the Zn2+-absorption peak wavelength. The protein complex structure was solved by the three-wavelength multiple anomalous dispersion method using the anomalous signal from the two Zn2+ atoms in TRIM21R. The location of Zn2+ atoms and initial density modification were performed using SHELX (66) and its graphical user interface HKL2MAP (67) with a SHELXE-estimated mean figure of merit of 0.642 and pseudo-free correlation coefficient of 69%. For the structure refinement, we used the inflection point data set merged with “Fridel pairs = true” (2.82 Å) instead of “Fridel pairs = false” (3.1 Å) during the multiple anomalous dispersion phasing method. Our first model was built using the CCP4 (68) software Buccaneer (69) and completed by manual model building in Coot (70). The molecules in the asymmetric unit were initially refined with local noncrystallographic symmetry (NCS) restraints in BUSTER and later with Phenix_Rosetta (71) that does not use NCS but improved the local geometry as judged by MolProbity. For final refinement, we uploaded the unmerged XDS_ASCII.HKL inflection point data set with the STARANISO web server (http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi)7 (80) that performs an elliptical resolution cutoff for anisotropically diffracting crystals. Despite having a few diffraction spots to 2.57-Å resolution in the best-diffracting direction, we decided to remove the data in the 2.82–2.57-Å interval because spherical completeness was only 15% in that interval. After removing that interval, the spherical/elliptical completeness was 50.7/66.5% in the highest-resolution shell (2.91–2.82 Å), and overall the spherical/elliptical completeness was 91.2/93.6% in the 47.9–2.82-Å interval (Table 1). The final model was generated using local NCS restraints and jelly-body refinement in REFMAC5 (72) with 96.6/3.2/0.2% of the amino acids in the preferred/allowed/disallowed regions of the Ramachandran plot.
UBE2E1C crystals were produced from the same material as in the UBE2E1C–TRIM21R crystals, and data were recorded at the same beam time. Diffraction data were collected at BL14.1 beamline at BESSY Synchrotron. The structure was determined by molecular replacement in MOLREP using PDB code 3BZH (38) as a search model followed by manual model building in Coot and refinement in REFMAC5. All data collection and refinement statistics are summarized in Table 1.
Model building and structural presentations
The model of UBE2E1–Ub–TRIM21R was generated by superimposing UBE2E1C (module F) in the UBE2E1C–TRIM21R structure (PDB code 6FGA) onto the E2 module of the TRIM25–UBE2D1∼Ub ternary complex (PDB code 5FER; Cα r.m.s.d., 0.54 Å). The resulting UBE2E1–Ub conjugate in which Ub is in a closed conformation shows essentially no clashes with UBE2E1C or TRIM21 homodimers in the 6FGA structure, supporting its relevance in a ternary Ub–E2–E3–substrate complex. All figures were generated using PyMOL Molecular Graphics System, Version 1.2r3pre (Schrödinger LLC).
Structural interface analysis
The web server VADAR (Volume Area Dihedral Angle Reporter) (73) was used for structure evaluation, including hydrogen-bonding partners and accessible surface area for both TRIM21R (this study; PDB code 6FGA) and TRIM25 (PDB code 5FER) dimer analyses. Side chains were considered buried if their level of exposure was less than 20%.
SAXS sample preparation, data acquisition, analysis, and modeling
SAXS data were acquired for TRIM21R using the ESRF BM29 SAXS beamline (74, 75) with a robotic sample changer (76) and a Pilatus 1M detector (Dectris). SAXS data were also acquired using the Anton Paar SAXSess at Linköping University for TRIM21R. SAXS samples were prepared by extensive dialysis against their buffer: 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm β-mercaptoethanol, and 500 μm ZnCl2. For SAXSess measurements only, 10% glycerol was added to the buffer solution. Exact solvent blanks for all measurements were obtained from the last dialysis step. Table S3 provides the SAXS data acquisition parameters, sample parameters, and software used for data reduction to I(q) versus q, where I(q) = 4πsinθ/λ, 2θ is the angle between the incident and scattered X-rays, and λ is their wavelength), analysis, and interpretation. All data were placed on an absolute scale using the scattering from pure H2O (SAXSess) or incident beam flux (BM29).
I(q) versus q for the protein was obtained by subtraction of the solvent scattering from that of protein + solvent. For the BM29 data, solvent measurements taken immediately before and after the protein + solvent measurement were averaged to optimize solvent subtraction. As there was no discernible concentration dependence to I(q) for TRIM21R, SAXSess data from the highest concentration samples were averaged to improve signal to noise. Molecular weights for the proteins were estimated using the method of Orthaber and Glatter (77). Values for contrast and partial specific volumes were determined using the MULCh program (78) with the known chemical compositions of samples and solvent. SAXS data analysis and modeling were performed using the tools of the ATSAS program package (79). The online interface and software used are listed in Table S3. Default software parameters were employed unless otherwise specified. The ESRF and SAXSess data overlay well (Fig. S3D), and a Kratky (Fig. S3E) plot of the ESRF data shows the expected bell shape for a globular, mostly folded protein with a rising profile at high q-values, indicating some degree of flexibility in the structure. The Guinier results for the ESRF and SAXSess data were the same within error (Table S4), but the SAXSess data were measured to lower minimum q-values and hence are more reliable for dmax determination in P(r) calculations compared with the ESRF data. Indeed, the latter consistently showed a similar shape to that obtained with the SAXSess data with a weak tail to longer r values of indeterminate length that increased the apparent Rg values. Furthermore, the molecular weight values were slightly more consistent with full RING dimerisation for the higher concentration SAXSess data. Therefore, the DAMMIF models in Fig. 1B were obtained using the SAXSess data–derived P(r) (Fig. S3F) and represent the averaged and filtered models from 20 independent DAMMIF calculations. Normalized spatial discrepancy values were 0.735 (assuming P1, i.e. no specific symmetry) or 0.496 (assuming P2 symmetry), indicating similar structural solutions for all calculations.
Author contributions
M. A., M. W.-H., and M. S. conceptualization; M. A., N. C. K., V. C., A. A., A. R. R., J. T., M. M., M. W.-H., and M. S. data curation; M. A., N. C. K., V. C., A. W., M. M., M. W.-H., and M. S. formal analysis; M. A., N. C. K., V. C., A. A., J. T., M. M., M. W.-H., and M. S. validation; M. A., N. C. K., V. C., A. R. R., J. T., M. M., and M. S. investigation; M. A., N. C. K., V. C., J. T., M. M., and M. S. visualization; M. A., N. C. K., V. C., A. W., A. C. E., A. A., A. R. R., J. T., M. M., M. W.-H., and M. S. methodology; M. A., V. C., J. T., M. M., and M. S. writing-original draft; M. A., M. W.-H., and M. S. project administration; M. A., N. C. K., V. C., A. W., A. C. E., A. A., A. R. R., J. T., M. M., M. W.-H., and M. S. writing-review and editing; V. C., A. C. E., J. T., M. M., M. W.-H., and M. S. supervision; M. M. software; M. W.-H. and M. S. resources; M. W.-H. and M. S. funding acquisition.
Supplementary Material
Acknowledgments
We acknowledge staff, resources, and facility funding at beamlines BL14-1, BESSY, Berlin, Germany and BM29, ESRF, Grenoble, France and at core facilities ProLinC at Linköping University, Protein Science Facility at Karolinska Institute, and PReSTO compute platform at National Supercomputer Center (NSC), Linköping. We thank Dr. Patrik Lundström for expert NMR advice and Dr. Vivian Morad and M.Sc. students in the Sunnerhagen group for technical assistance.
This work was supported by funding from the Swedish Research Council (to M. S., J. T., and M. W.-H.); The Swedish Rheumatism Association, The Swedish Heart-Lung Foundation, The Stockholm County Council, the King Gustaf the Vth 80-year foundation, and Karolinska Institutet (to M. W.-H.); and the Swedish Foundation for International Cooperation in Research and Higher Education, the Swedish Child Cancer Foundation, the Swedish Cancer Foundation, the Carl Trygger Foundation, and Linköping University (to M. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7 and Tables S1–S5.
The atomic coordinates and structure factors (codes 5LBN and 6FGA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
NMR backbone chemical shifts for the UBE2E1 core domain have been deposited in the Biological Magnetic Resonance Data Bank under accession number 27587.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- TRIM
- tripartite motif
- UBE2E1
- ubiquitin-conjugating enzyme E2 E1
- PCNA
- proliferating cell nuclear antigen
- SUMO
- small ubiquitin-like modifier
- Ub
- ubiquitin
- Ubl
- ubiquitin-like
- IKKβ
- inhibitor of nuclear factor κ B kinase subunit β
- ESRF
- European Synchrotron Radiation Facility
- PDB
- Protein Data Bank
- SAXS
- small-angle X-ray scattering
- r.m.s.d.
- root mean square deviation
- CSP
- chemical shift perturbation
- ASU
- asymmetric unit
- UBC
- ubiquitin-conjugating
- NTA
- nitrilotriacetic acid
- H
- helix
- L
- loop
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- NCS
- noncrystallographic symmetry
- VADAR
- Volume Area Dihedral Angle Reporter.
References
- 1. Rajsbaum R., García-Sastre A., and Versteeg G. A. (2014) TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 426, 1265–1284 10.1016/j.jmb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hatakeyama S. (2011) TRIM proteins and cancer. Nat. Rev. Cancer 11, 792–804 10.1038/nrc3139 [DOI] [PubMed] [Google Scholar]
- 3. Metzger M. B., Pruneda J. N., Klevit R. E., and Weissman A. M. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 10.1016/j.bbamcr.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berndsen C. E., and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- 5. Ozato K., Shin D.-M., Chang T.-H., and Morse H. C. 3rd (2008) TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Napolitano L. M., Jaffray E. G., Hay R. T., and Meroni G. (2011) Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem. J. 434, 309–319 10.1042/BJ20101487 [DOI] [PubMed] [Google Scholar]
- 7. Oke V., and Wahren-Herlenius M. (2012) The immunobiology of Ro52 (TRIM21) in autoimmunity: a critical review. J. Autoimmun. 39, 77–82 10.1016/j.jaut.2012.01.014 [DOI] [PubMed] [Google Scholar]
- 8. Espinosa A., Hennig J., Ambrosi A., Anandapadmanaban M., Abelius M. S., Sheng Y., Nyberg F., Arrowsmith C. H., Sunnerhagen M., and Wahren-Herlenius M. (2011) Anti-Ro52 autoantibodies from patients with Sjögren's syndrome inhibit the Ro52 E3 ligase activity by blocking the E3/E2 interface. J. Biol. Chem. 286, 36478–36491 10.1074/jbc.M111.241786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qin Y., Liu Q., Tian S., Xie W., Cui J., and Wang R.-F. (2016) TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 26, 613–628 10.1038/cr.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hillje A.-L., Worlitzer M. M., Palm T., and Schwamborn J. C. (2011) Neural stem cells maintain their stemness through protein kinase C ζ-mediated inhibition of TRIM32. Stem Cells 29, 1437–1447 10.1002/stem.687 [DOI] [PubMed] [Google Scholar]
- 11. Plafker S. M., Plafker K. S., Weissman A. M., and Macara I. G. (2004) Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 167, 649–659 10.1083/jcb.200406001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Espinosa A., Oke V., Elfving A., Nyberg F., Covacu R., and Wahren-Herlenius M. (2008) The autoantigen Ro52 is an E3 ligase resident in the cytoplasm but enters the nucleus upon cellular exposure to nitric oxide. Exp. Cell Res. 314, 3605–3613 10.1016/j.yexcr.2008.09.011 [DOI] [PubMed] [Google Scholar]
- 13. Espinosa A., Zhou W., Ek M., Hedlund M., Brauner S., Popovic K., Horvath L., Wallerskog T., Oukka M., Nyberg F., Kuchroo V. K., and Wahren-Herlenius M. (2006) The Sjogren's syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J. Immunol. 176, 6277–6285 10.4049/jimmunol.176.10.6277 [DOI] [PubMed] [Google Scholar]
- 14. Wada K., and Kamitani T. (2006) Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 339, 415–421 10.1016/j.bbrc.2005.11.029 [DOI] [PubMed] [Google Scholar]
- 15. Strandberg L., Ambrosi A., Espinosa A., Ottosson L., Eloranta M.-L., Zhou W., Elfving A., Greenfield E., Kuchroo V. K., and Wahren-Herlenius M. (2008) Interferon-α induces up-regulation and nuclear translocation of the Ro52 autoantigen as detected by a panel of novel Ro52-specific monoclonal antibodies. J. Clin. Immunol. 28, 220–231 10.1007/s10875-007-9157-0 [DOI] [PubMed] [Google Scholar]
- 16. Kong H. J., Anderson D. E., Lee C. H., Jang M. K., Tamura T., Tailor P., Cho H. K., Cheong J., Xiong H., Morse H. C. 3rd, and Ozato K. (2007) Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J. Immunol. 179, 26–30 10.4049/jimmunol.179.1.26 [DOI] [PubMed] [Google Scholar]
- 17. Higgs R., Ní Gabhann J., Ben Larbi N., Breen E. P., Fitzgerald K. A., and Jefferies C. A. (2008) The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181, 1780–1786 10.4049/jimmunol.181.3.1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F. J., Sjöstrand M., Eloranta M.-L., Ní Gabhann J., Winqvist O., Sundelin B., Jefferies C. A., Rozell B., Kuchroo V. K., and Wahren-Herlenius M. (2009) Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206, 1661–1671 10.1084/jem.20090585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang K., Shi H.-X., Liu X.-Y., Shan Y.-F., Wei B., Chen S., and Wang C. (2009) TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J. Immunol. 182, 3782–3792 10.4049/jimmunol.0803126 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Z., Bao M., Lu N., Weng L., Yuan B., and Liu Y.-J. (2013) The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 14, 172–178 10.1038/ni.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wada K., Niida M., Tanaka M., and Kamitani T. (2009) Ro52-mediated monoubiquitination of IKKβ down-regulates NF-κB signalling. J. Biochem. 146, 821–832 10.1093/jb/mvp127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reddy B. A., van der Knaap J. A., Bot A. G., Mohd-Sarip A., Dekkers D. H., Timmermans M. A., Martens J. W., Demmers J. A., and Verrijzer C. P. (2014) Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol. Cell 53, 458–470 10.1016/j.molcel.2013.12.017 [DOI] [PubMed] [Google Scholar]
- 23. Fletcher A. J., Mallery D. L., Watkinson R. E., Dickson C. F., and James L. C. (2015) Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc. Natl. Acad. Sci. U.S.A. 112, 10014–10019 10.1073/pnas.1507534112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pruneda J. N., Littlefield P. J., Soss S. E., Nordquist K. A., Chazin W. J., Brzovic P. S., and Klevit R. E. (2012) Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 10.1016/j.molcel.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soss S. E., Yue Y., Dhe-Paganon S., and Chazin W. J. (2011) E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 286, 21277–21286 10.1074/jbc.M111.224006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dou H., Buetow L., Sibbet G. J., Cameron K., and Huang D. T. (2012) BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 10.1038/nsmb.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plechanovová A., Jaffray E. G., Tatham M. H., Naismith J. H., and Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 10.1038/nature11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dou H., Buetow L., Sibbet G. J., Cameron K., and Huang D. T. (2013) Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 20, 982–986 10.1038/nsmb.2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taherbhoy A. M., Huang O. W., and Cochran A. G. (2015) BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun. 6, 7621 10.1038/ncomms8621 [DOI] [PubMed] [Google Scholar]
- 30. Koliopoulos M. G., Esposito D., Christodoulou E., Taylor I. A., and Rittinger K. (2016) Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35, 1204–1218 10.15252/embj.201593741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanchez J. G., Chiang J. J., Sparrer K. M. J., Alam S. L., Chi M., Roganowicz M. D., Sankaran B., Gack M. U., and Pornillos O. (2016) Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 16, 1315–1325 10.1016/j.celrep.2016.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cappadocia L., Pichler A., and Lima C. D. (2015) Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 22, 968–975 10.1038/nsmb.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Streich F. C. Jr., and Lima C. D. (2016) Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 536, 304–308 10.1038/nature19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scott D. C., Sviderskiy V. O., Monda J. K., Lydeard J. R., Cho S. E., Harper J. W., and Schulman B. A. (2014) Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 10.1016/j.cell.2014.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reverter D., and Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 10.1038/nature03588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yunus A. A., and Lima C. D. (2006) Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 10.1038/nsmb1104 [DOI] [PubMed] [Google Scholar]
- 37. Yamaguchi M., VanderLinden R., Weissmann F., Qiao R., Dube P., Brown N. G., Haselbach D., Zhang W., Sidhu S. S., Peters J.-M., Stark H., and Schulman B. A. (2016) Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell 63, 593–607 10.1016/j.molcel.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sheng Y., Hong J. H., Doherty R., Srikumar T., Shloush J., Avvakumov G. V., Walker J. R., Xue S., Neculai D., Wan J. W., Kim S. K., Arrowsmith C. H., Raught B., and Dhe-Paganon S. (2012) A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell. Proteomics 11, 329–341 10.1074/mcp.O111.013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozkan E., Yu H., and Deisenhofer J. (2005) Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. U.S.A. 102, 18890–18895 10.1073/pnas.0509418102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benirschke R. C., Thompson J. R., Nominé Y., Wasielewski E., Juranić N., Macura S., Hatakeyama S., Nakayama K. I., Botuyan M. V., and Mer G. (2010) Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure 18, 955–965 10.1016/j.str.2010.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chakrabarti K. S., Li J., Das R., and Byrd R. A. (2017) Conformational dynamics and allostery in E2:E3 interactions drive ubiquitination: gp78 and Ube2g2. Structure 25, 794–805.e5 10.1016/j.str.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Christensen D. E., Brzovic P. S., and Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 10.1038/nsmb1295 [DOI] [PubMed] [Google Scholar]
- 43. Brzovic P. S., and Klevit R. E. (2006) Ubiquitin transfer from the E2 perspective: why is UbcH5 so promiscuous? Cell Cycle 5, 2867–2873 10.4161/cc.5.24.3592 [DOI] [PubMed] [Google Scholar]
- 44. Kar G., Keskin O., Nussinov R., and Gursoy A. (2012) Human proteome-scale structural modeling of E2-E3 interactions exploiting interface motifs. J. Proteome Res. 11, 1196–1207 10.1021/pr2009143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Wijk S. J., Melquiond A. S., de Vries S. J., Timmers H. T., and Bonvin A. M. (2012) Dynamic control of selectivity in the ubiquitination pathway revealed by an ASP to GLU substitution in an intra-molecular salt-bridge network. PLoS Comput. Biol. 8, e1002754 10.1371/journal.pcbi.1002754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dou H., Buetow L., Hock A., Sibbet G. J., Vousden K. H., and Huang D. T. (2012) Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19, 184–192 10.1038/nsmb.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Wu H., Wu W., Zhuo W., Liu W., Zhang Y., Cheng M., Chen Y.-G., Gao N., Yu H., Wang L., Li W., and Yang M. (2014) Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 24, 762–765 10.1038/cr.2014.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Banka P. A., Behera A. P., Sarkar S., and Datta A. B. (2015) RING E3-catalyzed E2 self-ubiquitination attenuates the activity of Ube2E ubiquitin-conjugating enzymes. J. Mol. Biol. 427, 2290–2304 10.1016/j.jmb.2015.04.011 [DOI] [PubMed] [Google Scholar]
- 49. Wenzel D. M., Stoll K. E., and Klevit R. E. (2011) E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42 10.1042/BJ20100985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Buetow L., Gabrielsen M., Anthony N. G., Dou H., Patel A., Aitkenhead H., Sibbet G. J., Smith B. O., and Huang D. T. (2015) Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol. Cell 58, 297–310 10.1016/j.molcel.2015.02.017 [DOI] [PubMed] [Google Scholar]
- 51. Wenzel D. M., Lissounov A., Brzovic P. S., and Klevit R. E. (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 10.1038/nature09966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valimberti I., Tiberti M., Lambrughi M., Sarcevic B., and Papaleo E. (2015) E2 superfamily of ubiquitin-conjugating enzymes: constitutively active or activated through phosphorylation in the catalytic cleft. Sci. Rep. 5, 14849 10.1038/srep14849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stewart M. D., Ritterhoff T., Klevit R. E., and Brzovic P. S. (2016) E2 enzymes: more than just middle men. Cell Res. 26, 423–440 10.1038/cr.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wheaton K., Sarkari F., Stanly Johns B., Davarinejad H., Egorova O., Kaustov L., Raught B., Saridakis V., and Sheng Y. (2017) UbE2E1/UBCH6 is a critical in vivo E2 for the PRC1 catalyzed ubiquitination of H2A at Lys-119. J. Biol. Chem. 292, 2893–2902 10.1074/jbc.M116.749564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Machida Y. J., Machida Y., Chen Y., Gurtan A. M., Kupfer G. M., D'Andrea A. D., and Dutta A. (2006) UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596 10.1016/j.molcel.2006.06.024 [DOI] [PubMed] [Google Scholar]
- 56. Campbell E. M., Weingart J., Sette P., Opp S., Sastri J., O'Connor S. K., Talley S., Diaz-Griffero F., Hirsch V., and Bouamr F. (2016) TRIM5α-mediated ubiquitin chain conjugation is required for inhibition of HIV-1 reverse transcription and capsid destabilization. J. Virol. 90, 1849–1857 10.1128/JVI.01948-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fletcher A. J., Christensen D. E., Nelson C., Tan C. P., Schaller T., Lehner P. J., Sundquist W. I., and Towers G. J. (2015) TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 34, 2078–2095 10.15252/embj.201490361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wagner J. M., Roganowicz M. D., Skorupka K., Alam S. L., Christensen D., Doss G., Wan Y., Frank G. A., Ganser-Pornillos B. K., Sundquist W. I., and Pornillos O. (2016) Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α. Elife 5, e16309 10.7554/eLife.16309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yudina Z., Roa A., Johnson R., Biris N., de Souza Aranha Vieira D. A., Tsiperson V., Reszka N., Taylor A. B., Hart P. J., Demeler B., Diaz-Griffero F., and Ivanov D. N. (2015) RING Dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep. 12, 788–797 10.1016/j.celrep.2015.06.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., and Klevit R. E. (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 10.1016/j.molcel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 61. Berndsen C. E., and Wolberger C. (2011) A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem. 418, 102–110 10.1016/j.ab.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van den Berg S., Löfdahl P.-A., Härd T., and Berglund H. (2006) Improved solubility of TEV protease by directed evolution. J. Biotechnol. 121, 291–298 10.1016/j.jbiotec.2005.08.006 [DOI] [PubMed] [Google Scholar]
- 63. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 64. Niklasson M., Ahlner A., Andresen C., Marsh J. A., and Lundström P. (2015) Fast and accurate resonance assignment of small-to-large proteins by combining automated and manual approaches. PLoS Comput. Biol. 11, e1004022 10.1371/journal.pcbi.1004022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schumann F. H., Riepl H., Maurer T., Gronwald W., Neidig K.-P., and Kalbitzer H. R. (2007) Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 39, 275–289 10.1007/s10858-007-9197-z [DOI] [PubMed] [Google Scholar]
- 66. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 10.1107/S0108767307043930 [DOI] [PubMed] [Google Scholar]
- 67. Pape T., and Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 10.1107/S0021889804018047 [DOI] [Google Scholar]
- 68. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- 70. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. DiMaio F., Echols N., Headd J. J., Terwilliger T. C., Adams P. D., and Baker D. (2013) Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat. Methods 10, 1102–1104 10.1038/nmeth.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nicholls R. A., Long F., and Murshudov G. N. (2012) Low-resolution refinement tools in REFMAC5. Acta Crystallogr. D Biol. Crystallogr. 68, 404–417 10.1107/S090744491105606X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R. F., Sykes B. D., and Wishart D. S. (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31, 3316–3319 10.1093/nar/gkg565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pernot P., Round A., Barrett R., De Maria Antolinos A., Gobbo A., Gordon E., Huet J., Kieffer J., Lentini M., Mattenet M., Morawe C., Mueller-Dieckmann C., Ohlsson S., Schmid W., Surr J., et al. (2013) Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J. Synchrotron Radiat. 20, 660–664 10.1107/S0909049513010431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brennich M. E., Kieffer J., Bonamis G., De Maria Antolinos A., Hutin S., Pernot P., and Round A. (2016) Online data analysis at the ESRF bioSAXS beamline, BM29. J. Appl Crystallogr. 49, 203–212 10.1107/S1600576715024462 [DOI] [Google Scholar]
- 76. Round A., Felisaz F., Fodinger L., Gobbo A., Huet J., Villard C., Blanchet C. E., Pernot P., McSweeney S., Roessle M., Svergun D. I., and Cipriani F. (2015) BioSAXS Sample Changer: a robotic sample changer for rapid and reliable high-throughput X-ray solution scattering experiments. Acta Crystallogr. D Biol. Crystallogr. 71, 67–75 10.1107/S1399004714026959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Orthaber D., and Glatter O. (2000) Synthetic phospholipid analogs: a structural investigation with scattering methods. Chem. Phys. Lipids 107, 179–189 10.1016/S0009-3084(00)00171-7 [DOI] [PubMed] [Google Scholar]
- 78. Whitten A. E., Cai S., and Trewhella J. (2008) MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. Appl. Crystallogr. 41, 222–226 10.1107/S0021889807055136 [DOI] [Google Scholar]
- 79. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tickle I.J., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., and Bricogne G. (2018) STARANISO, Global Phasing Ltd., Cambridge, UK [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.