Abstract
Aside from abasic sites and ribonucleotides, the DNA adduct N7-methyl deoxyguanosine (N7-CH3 dG) is one of the most abundant lesions in mammalian DNA. Because N7-CH3 dG is unstable, leading to deglycosylation and ring-opening, its miscoding potential is not well-understood. Here, we employed a 2′-fluoro isostere approach to synthesize an oligonucleotide containing an analog of this lesion (N7-CH3 2′-F dG) and examined its miscoding potential with four Y-family translesion synthesis DNA polymerases (pols): human pol (hpol) η, hpol κ, and hpol ι and Dpo4 from the archaeal thermophile Sulfolobus solfataricus. We found that hpol η and Dpo4 can bypass the N7-CH3 2′-F dG adduct, albeit with some stalling, but hpol κ is strongly blocked at this lesion site, whereas hpol ι showed no distinction with the lesion and the control templates. hpol η yielded the highest level of misincorporation opposite the adduct by inserting dATP or dTTP. Moreover, hpol η did not extend well past an N7-CH3 2′-F dG:dT mispair. MS-based sequence analysis confirmed that hpol η catalyzes mainly error-free incorporation of dC, with misincorporation of dA and dG in 5–10% of products. We conclude that N7-CH3 2′-F dG and, by inference, N7-CH3 dG have miscoding and mutagenic potential. The level of misincorporation arising from this abundant adduct can be considered as potentially mutagenic as a highly miscoding but rare lesion.
Keywords: DNA, DNA polymerase, DNA enzyme, DNA replication, DNA damage, DNA adduct, fidelity of DNA synthesis, miscoding, replication bypass, translesion synthesis
Introduction
DNA is constantly damaged by both endogenous (e.g. reactive oxygen species and SAM) and exogenous (e.g. polycyclic hydrocarbons and heterocyclic amines) sources (1). Examples of DNA damage include DNA adducts (e.g. alkylated and oxidized bases), single strand breaks, double strand breaks, DNA mismatches, abasic sites, and pyrimidine dimers (2). Such damage, if not repaired, can cause deleterious outcomes (e.g. stalled replication and miscoding events leading to cancer, teratogenesis, and cardiovascular disease) (3–6). Alkylating agents used in treatment of malignancies (such as cyclophosphamide, temozolomide, and melphalan) have been associated with causing cancers (e.g. lymphomas, malignant gliomas, and lung and ovarian cancers) (7–11).
The nitrogen and oxygen atoms of DNA bases are reactive toward several known alkylating agents, producing different types of DNA adducts (12, 13). Exposure of DNA to methylating agents forms several modified bases, including N3-methyl deoxyadenosine, N7-methyl deoxyguanosine (N7-CH3 dG),2 O6-methyl deoxyguanosine (O6-CH3 dG), and O4-methyl (deoxy)thymidine (13). O6-CH3 dG and O4-methyl (deoxy)thymidine are minor adducts but are highly cytotoxic and mutagenic; the mutagenicity of the abundant N7-CH3 dG and N3-methyl deoxyadenosine adducts is not known (14, 15).
The N7 atom of deoxyguanosine is the most nucleophilic site in DNA and is susceptible to alkylation, forming various N7-alkyl deoxyguanosine adducts (13, 16, 17). These adducts include N7-CH3 dG, N7-ethyl deoxyguanosine, and N7-benzyl deoxyguanosine (13, 18, 19). The deoxyguanosine adduct formed with the 8,9-exo-epoxide of the hepatocellular carcinogen aflatoxin B1 is highly mutagenic, causing GC to TA transversion mutations (6, 19).
N7-CH3 dG has been detected as the major DNA adduct formed by methylating agents and is the most abundant lesion in DNA aside from abasic sites (1, 20, 21) and ribonucleotides (22, 23), present in lymphocytes at levels of 14 adducts/107 normal nucleotides for nonsmokers and 25 adducts/107 nucleotides in smokers (24). Endogenous methylation of DNA, apparently from SAM, has been identified as the primary source of N7-CH3 dG adducts observed in the livers of untreated rats (25, 26).
The miscoding and mutagenic potentials of the resulting depurination (i.e. abasic sites) and ring-opened (i.e. N7-CH3 formamidopyrimidine (FAPY) dG) products of N7-CH3 dG have been extensively studied (27–31). The miscoding potential of N7-CH3 dG itself is not understood. Despite its abundance, N7-CH3 dG has been largely ignored in favor of other alkylated bases due to its instability to depurination and base-catalyzed ring-opening. It has been assumed that N7-CH3 dG is not miscoding because it should not alter the canonical Watson–Crick hydrogen-bonding pattern (32). However, the techniques that were used to reach this conclusion were not very sensitive compared with modern methods, and only a few model DNA polymerases were considered (21, 33).
In 1961, Lawley and Brookes (34) proposed that alkylation at the guanine N7 position might induce mispairing due to its lowering of the pKa of the N1 position from 9 to 7, favoring rare tautomers (17, 34–36). Even a low level of misincorporation across a very abundant lesion would be similar in risk to a highly miscoding but rare lesion.
Koag et al. (37) employed an isosteric fluorine transition-state destabilization approach to stabilize the glycosidic bond, to avoid depurination and mild deprotection conditions to prevent ring-opening to N7-CH3 FAPY dG. Although the lesion inhibited catalysis by pol β, replication was reported to be highly accurate (i.e. dCTP was inserted opposite N7-CH3 dG) (37). We used this 2′-fluoro analog, N7-CH3 2′-F dG, and analyzed its miscoding potential with several Y-family translesion synthesis polymerases (human pols (hpols) η, κ, and ι and Sulfolobus solfataricus Dpo4). N7-CH3 2′-F dG caused miscoding with hpol η and has mutagenic potential, which we infer is the case with N7-CH3 dG.
Results
Synthesis of 2′-F dG- and N7-CH3 2′-F dG–containing oligonucleotides
A fluorine analog of the lesion (N7-CH3 2′-F dG) was prepared by modifying the approach of Lee et al. (32) (Scheme S1 and Figs. S1–S4). The 23-mer oligonucleotides were characterized by LC-ESI-MS (Figs. S5B and S6A). The N7-CH3 2′-F dG–containing oligonucleotide was resistant to cleavage by FPG glycosylase, further confirming its identity as the intact lesion rather than the ring-opened N7-CH3 FAPY 2′-F dG oligonucleotide, which is a substrate for this glycosylase (Fig. S6B). Following alkaline treatment to form the FAPY lesion, FPG glycosylase cleaved the lesion (Fig. S6B). We conclude that the desired N7-CH3 2′-F dG lesion was present and that the two potential problems, depurination and ring-opening, had been avoided.
Primer extension past dG, 2′-F dG, and N7-CH3 2′-F dG by Y-family DNA polymerases
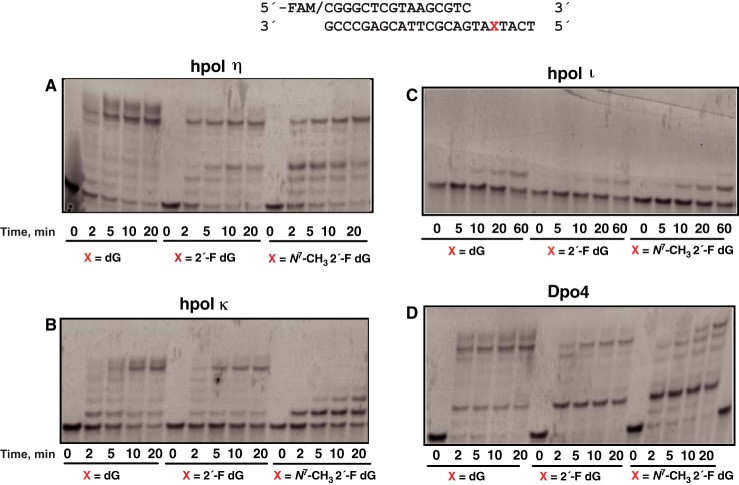
Four Y-family DNA polymerases were studied (hpol η, hpol κ, hpol ι, and Dpo4). hpol η and Dpo4 were the most effective of these in producing full-length extension products, although they both stalled following the P + 3 and P + 2 products, respectively (Fig. 1, A and D). In contrast, hpol κ stalled at the adduct (Fig. 1B), and hpol ι showed no distinction between the control templates and the adduct, stalling before the dG templates and the lesion (Fig. 1C).
Figure 1.
Bypass across and extension past dG, 2′-F dG, and N7-CH3 2′-F dG. A, hpol η; B, hpol κ; C, hpol ι; D, Dpo4. The sequences of the template and primer are shown at the top. Reactions contained 200 nm FAM-labeled primer–template oligonucleotide complex, 250 μm dNTPs, and 20 nm enzyme, except for pol ι (40 nm). Reactions were done at 37 °C for 2, 5, 10, 20, and 60 min.
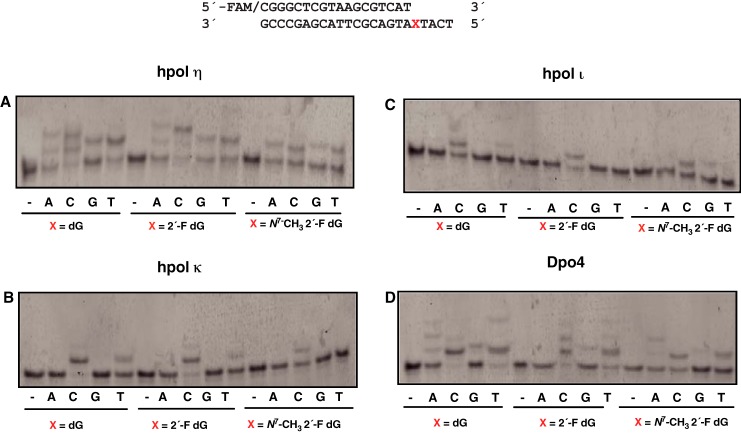
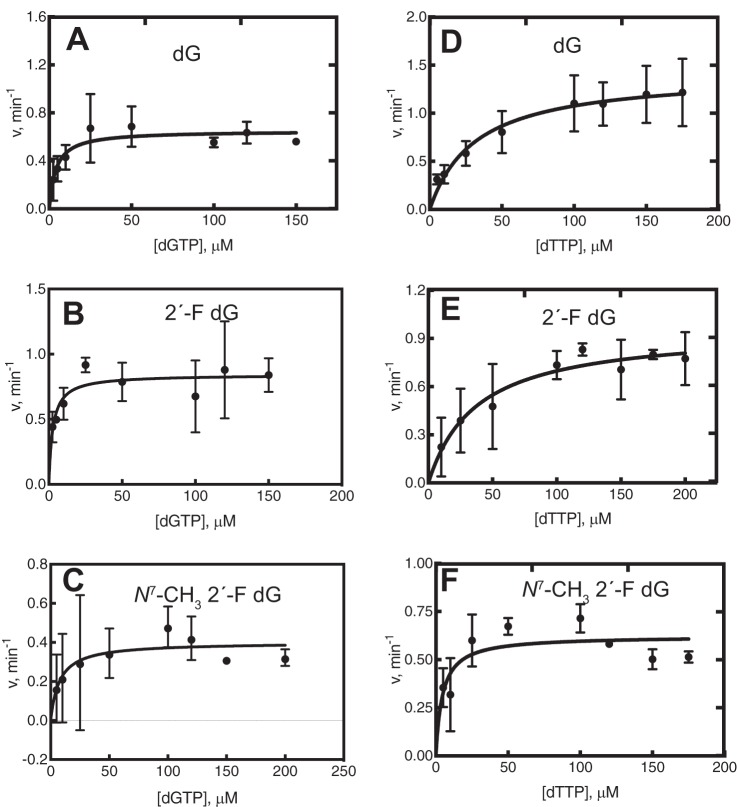
To determine the insertions across the adduct, primer extension experiments were done with individual dNTPs. hpol η and Dpo4 were highly error-prone with all three templates examined (Fig. 2, A and D). Dpo4 did not misincorporate dATP in the 2′-F dG control. In contrast with these two polymerases, hpol κ and hpol ι incorporated only the correct dCTP for all three templates under the same conditions (Fig. 2, B and C), and these two polymerases were not examined further. Rates for dCTP insertion by hpol κ were estimated (in single-nucleotide incorporation experiments) to be 1.9 min−1 for dG, 1.4 min−1 for 2′-F dG, and 0.9 min−1 for N7-CH3 2′-F dG (i.e. there was a ∼2-fold reduction in the rates when hpol κ encountered the lesion). For hpol ι, the rates of insertion of dCTP across the templates were 0.51 min−1 for dG, 0.56 min−1 for 2′-F dG, and 0.47 min−1 for N7-CH3 2′-F dG. Thus, the rates when hpol ι encountered the lesion were comparable with the control templates.
Figure 2.
Single-nucleotide incorporation opposite dG, 2′-F dG, and N7-CH3 2′-F dG. A, hpol η; B, hpol κ; C, hpol ι; D, Dpo4. The sequences of the template and primer are shown at the top. Reactions were conducted with 120 nm FAM-labeled primer–template oligonucleotide complex, 250 μm dNTPs, and 5 nm enzyme except for pol ι (10 nm). Reactions were conducted at 37 °C for 10 min.
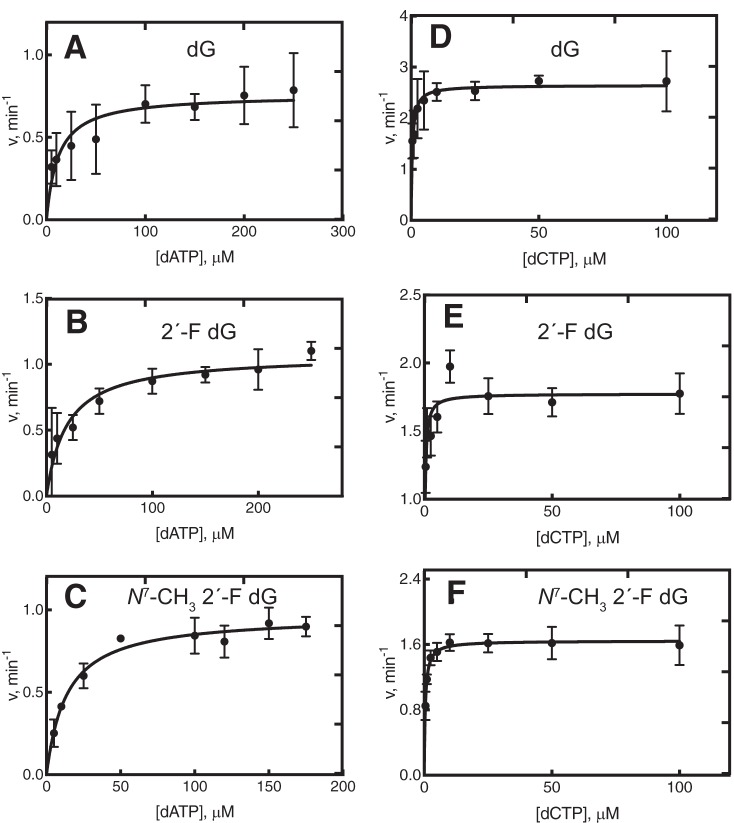
Steady-state kinetics of individual dNTP insertion opposite dG, 2′-F dG, and N7-CH3 2′-F dG by hpol η and Dpo4
Steady-state kinetic analysis was performed for hpol η and Dpo4 (Tables 1 and 2). The catalytic efficiencies and misincorporation frequencies for dG and 2′-F dG were comparable, as noted previously (38), suggesting that fluorine had little or no impact on polymerase recognition. With all three templates, hpol η preferred to insert dCTP relative to other dNTPs (Table 1 and Fig. 3). However, there was a 2-fold lower efficiency for incorporation of dCTP at the N7-CH3 2′-F dG lesion compared with dG and 2′-F dG. The efficiency for misinsertion of dATP was similar for all three templates, but the misinsertion frequency ∼2-fold higher with N7-CH3 2′-F dG (Table 1 and Figs. 3 and S7). The catalytic efficiency for dGTP misincorporation was ∼3-fold lower for the N7-CH3 2′-F dG lesion (Table 1 and Fig. 4). The efficiency for dTTP misincorporation was 3.5-fold greater for the N7-CH3 2′-F dG lesion compared with the control templates, and the misincorporation frequency was 5- and 11-fold higher relative to dG and 2′-F dG (Table 1 and Fig. 4). Thus, the misincorporation frequency for N7-CH3 2′-F dG was in the order dTTP > dATP > dGTP, ranging from 1 to 4% (Figs. S8 and S9).
Table 1.
Steady-state kinetics of single nucleotide insertion opposite dG, 2′-F dG, and N7-CH3 2′-F dG by hpol η
The oligonucleotides used were as follows,
5′-FAM-CGGGCTCGTAAGCGTCAT-3′ 3′-GCCCGAGCATTCGCAGTAXTACT-5′
where X represents dG, 2′-F dG, or N7-CH3 2′-F dG.
| Template base | dNTP | kcat | Km | kcat/Km | fa |
|---|---|---|---|---|---|
| min−1 | μm | μm−1 min−1 | |||
| dG | dCTP | 2.65 ± 0.11 | 0.45 ± 0.13 | 5.9 ± 1.7 | 1 |
| 2′-F dG | dCTP | 1.77 ± 0.05 | 0.23 ± 0.06 | 7.7 ± 2.0 | 1 |
| N7-CH3 2′-F dG | dCTP | 1.65 ± 0.04 | 0.43 ± 0.07 | 3.8 ± 0.6 | 1 |
| dG | dATP | 0.76 ± 0.06 | 12 ± 5 | 0.06 ± 0.03 | 0.01 |
| 2′-F dG | dATP | 1.1 ± 0.1 | 19 ± 6 | 0.06 ± 0.03 | 0.01 |
| N7-CH3 2′-F dG | dATP | 0.97 ± 0.04 | 14 ± 2 | 0.07 ± 0.01 | 0.018 |
| dG | dGTP | 0.65 ± 0.05 | 4.0 ± 1.7 | 0.16 ± 0.07 | 0.027 |
| 2′-F dG | dGTP | 0.84 ± 0.06 | 2.7 ± 1.2 | 0.31 ± 0.14 | 0.040 |
| N7-CH3 2′-F dG | dGTP | 0.40 ± 0.06 | 7.5 ± 6.3 | 0.05 ± 0.04 | 0.013 |
| dG | dTTP | 1.42 ± 0.14 | 32 ± 11 | 0.04 ± 0.01 | 0.0068 |
| 2′-F dG | dTTP | 0.96 ± 0.10 | 38 ± 15 | 0.03 ± 0.01 | 0.0033 |
| N7-CH3 2′-F dG | dTTP | 0.62 ± 0.05 | 4.6 ± 2.2 | 0.14 ± 0.07 | 0.037 |
a Misincorporation frequency f = (kcat/Km)incorrect/(kcat/Km)correct.
Table 2.
Steady-state kinetics of single nucleotide insertion opposite dG, 2′-F dG, and N7-CH3 2′-F dG by S. solfataricus Dpo4
The oligonucleotides used were as follows,
5′-FAM-CGGGCTCGTAAGCGTCAT-3′ 3′-GCCCGAGCATTCGCAGTAXTACT-5′
where X represents dG, 2′-F dG, and N7-CH3 2′-F dG.
| Template base | dNTP | kcat | Km | kcat/Km | fa |
|---|---|---|---|---|---|
| min−1 | μm | μm−1 min−1 | |||
| dG | dCTP | 158 ± 8 | 1.99 ± 0.41 | 79 ± 17 | 1 |
| 2′-F dG | dCTP | 115 ± 2 | 0.30 ± 0.03 | 383 ± 39 | 1 |
| N7-CH3 2′-F dG | dCTP | 2.88 ± 0.10 | 0.03 ± 0.01 | 96 ± 20 | 1 |
| dG | dATP | 0.36 ± 0.03 | 14 ± 6 | 0.03 ± 0.01 | 0.0004 |
| 2′-F dG | dATP | NDb | ND | ND | ND |
| N7-CH3 2′-F dG | dATP | 0.24 ± 0.02 | 9.8 ± 5.1 | 0.03 ± 0.02 | 0.0003 |
| dG | dGTP | 0.41 ± 0.04 | 29 ± 10 | 0.014 ± 0.005 | 0.0002 |
| 2′-F dG | dGTP | 0.40 ± 0.03 | 21 ± 7 | 0.019 ± 0.007 | 0.0002 |
| N7-CH3 2′-F dG | dGTP | 0.11 ± 0.01 | 24 ± 21 | 0.01 ± 0.01 | 0.0001 |
| dG | dTTP | 1.38 ± 0.36 | 76 ± 36 | 0.018 ± 0.010 | 0.0002 |
| 2′-F dG | dTTP | 0.93 ± 0.16 | 44 ± 16 | 0.021 ± 0.009 | 0.0001 |
| N7-CH3 2′-F dG | dTTP | 0.42 ± 0.04 | 16 ± 5 | 0.026 ± 0.009 | 0.0003 |
a Misincorporation frequency f = (kcat/Km)incorrect/(kcat/Km)correct.
b ND, not detected. DNA incorporation was below limits of quantitation (v < 0.002 min−1).
Figure 3.
Steady-state kinetic analysis of individual dATP and dCTP insertions by hpol η. Reactions contained templates dG (A and D), 2′-F dG (B and E), and N7-CH3 2′-F dG (C and F) at position X in the sequences 5′-CGGGCTCGTAAGCGTCAT-3′ and 3′-GCCCGAGCATTCGCAGTAXTACT-5′. Reactions were done at 37 °C for 5–10 min by incubating 120 nm primer–template oligonucleotide complex. For different panels, different hpol η concentrations were used as indicated. For A, B, C, and F, 5 nmol of enzyme was used, and the reaction was done for 5 min, with varying concentrations of dATP and dCTP. In the D and E, 2.5 nmol of enzyme was used, and the reaction was conducted for 5 min, with varying concentrations of dATP and dCTP. Fitting was to a hyperbolic equation in GraphPad Prism version 8.0, and kcat and Km values are presented in Table 1.
Figure 4.
Steady-state kinetic analysis of individual dGTP and dTTP insertions by hpol η. Reactions contained templates dG (A and D), 2′-F dG (B and E), and N7-CH3 2′-F dG (C and F) at position X in the sequences 5′-CGGGCTCGTAAGCGTCAT-3′ and 3′-GCCCGAGCATTCGCAGTAXTACT-5′. Reactions were done at 37 °C for 5–10 min by incubating 120 nm primer–template DNA complex with varying concentrations of hpol η. For A, B, and C, 8 nmol of hpol η was used, and the reaction was done for 10 min. In the case of D, we used 8 nmol of hpol η was used, and the reaction was done for 5 min; for E, 10 nmol of hpol η was used, and the reaction was conducted for 5 min; and for F, 5 nmol of hpol η was used, and the reaction was conducted for 5 min, varying concentrations of dGTP and dTTP. Fitting was to a hyperbolic equation in GraphPad Prism version 8.0, and kcat and Km values are presented in Table 1.
Dpo4 preferentially inserted dCTP opposite N7-CH3 2′-F dG (Table 2). There was a 4-fold lower efficiency for insertion across the lesion compared with 2′-F dG (Figs. S8 and S9). The efficiencies for incorporating other dNTPs were in the order dATP > dTTP > dGTP (Table 2).
Steady-state kinetics of post-lesion incorporation of individual dNTPs opposite 2′-F dG or N7-CH3 2′-F dG by hpol η
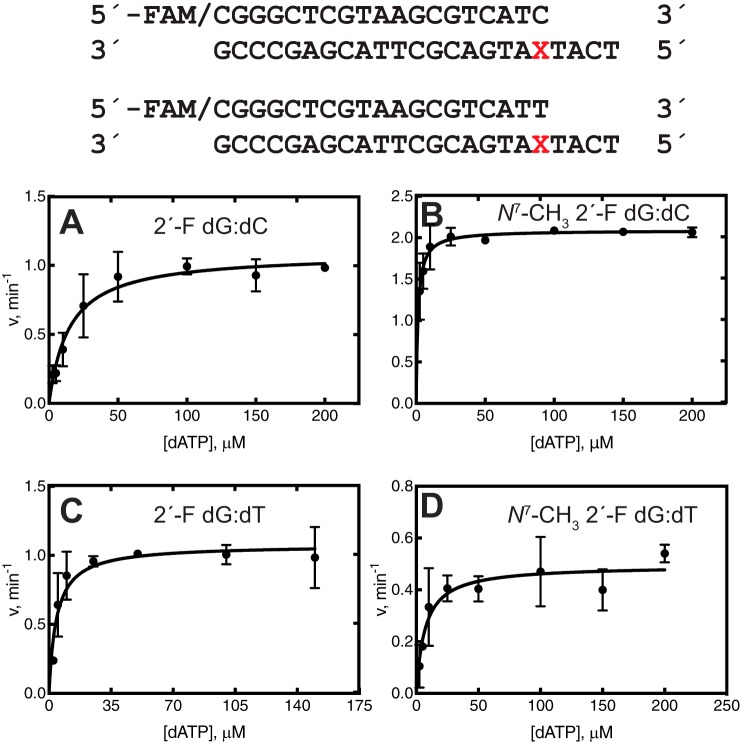
Steady-state insertion kinetics provides information on dNTP insertion across a lesion but does not provide information about extension past the lesion. Steady-state kinetics were done for further extension after the correct bp (N7-CH3 2′-F dG:dC) and a mispair (N7-CH3 2′-F dG:dT). dT was used as the misincorporated base opposite the lesion because it showed the greatest misincorporation frequency in the steady-state insertion kinetics with hpol η (Table 1). With the mispairs (2′-F dG:dT and N7-CH3 2′-F dG:dT), only dATP was incorporated opposite the next residue (dT) (Fig. S10). The efficiency of hpol η for incorporating dATP past the mispair was ∼4-fold lower for the lesion N7-CH3 2′-F dG than 2′-F dG, indicating some resistance to extension past the mispair (Table 3 and Fig. 5). On the other hand, the efficiency of hpol η for inserting dATP past the N7-CH3 2′-F dG:dC bp was 21-fold higher than the 2′-F dG:dC control, indicating that the correct pair was preferentially extended past the lesion. For the 2′-F dG control and lesion, dCTP had a similar efficiency of misincorporation past the correct pair. Finally, dTTP was misincorporated with a 6-fold higher efficiency for the lesion than the 2′-F dG control (Table 3 and Fig. 5).
Table 3.
Steady-state kinetics of single nucleotide extension past 2′-F dG:dC, 2′-F dG:dT and N7-CH3 2′-F dG:dC, and N7-CH3 2′-F dG:dT base pairs by hpol η
The oligonucleotides used were as follows,
5′-FAM-CGGGCTCGTAAGCGTCATC-3′ 3′-GCCCGAGCATTCGCAGTAXTACT-5′
5′-FAM-CGGGCTCGTAAGCGTCATT-3′ 3′-GCCCGAGCATTCGCAGTAXTACT-5′
where X represents 2′-F dG and N7-CH3 2′-F dG.
| Template base | Pairing | dNTP | kcat | Km | kcat/Km |
|---|---|---|---|---|---|
| min−1 | μm | μm−1 min−1 | |||
| 2′-F dG | Mispair | dATP | 1.1 ± 0.1 | 4.2 ± 1.2 | 0.26 ± 0.08 |
| N7-CH3 2′-F dG | dATP | 0.49 ± 0.04 | 7.1 ± 2.4 | 0.07 ± 0.02 | |
| 2′-F dG | Correct | dATP | 1.1 ± 0.1 | 15 ± 3 | 0.07 ± 0.02 |
| N7-CH3 2′-F dG | dATP | 2.1 ± 0.1 | 1.4 ± 0.3 | 1.5 ± 0.3 | |
| 2′-F dG | Correct | dCTP | 0.95 ± 0.06 | 5.5 ± 2.2 | 0.17 ± 0.07 |
| N7-CH3 2′-F dG | dCTP | 0.75 ± 0.03 | 4.6 ± 1.1 | 0.16 ± 0.04 | |
| 2′-F dG | Correct | dGTP | 2.1 ± 0.06 | 6.9 ± 1.0 | 0.3 ± 0.1 |
| N7-CH3 2′-F dG | dGTP | 0.97 ± 0.03 | 3.8 ± 0.7 | 0.26 ± 0.1 | |
| 2′-F dG | Correct | dTTP | 1.0 ± 0.1 | 3.4 ± 0.7 | 0.3 ± 0.10 |
| N7-CH3 2′-F dG | dTTP | 1.6 ± 0.1 | 0.96 ± 0.47 | 1.7 ± 0.8 |
Figure 5.
Steady-state kinetic analysis of dATP extension by hpol η. A, 2′-F dG:dC; B, N7-CH3 2′-F dG:dC; C, 2′-F dG:dT; D, N7-CH3 2′-F dG:dT bp. The sequences of the template and primer are shown at the top. Reactions were done at 37 °C for 5–10 min by incubating 120 nm primer–template oligonucleotide complex, 5–10 nm hpol η, and varying concentrations of dNTPs.
LC-MS/MS sequence analysis of extension products formed by hpol η and Dpo4
We introduced a dT:dU mismatch upstream of the site of dNTP addition to utilize uracil-DNA glycosylase (UDG) to cut the extension products for analysis by LC-MS/MS. Replication of the unmodified oligonucleotide gave only error-free products, as reported elsewhere (38). Replication across the lesion by Dpo4 also gave only error-free products, in support of the results of steady-state insertion kinetics (Table 4). hpol η replicated through the lesion in both an error-free and an error-prone manner, resulting in three main products (Table 5). The first product corresponded to error-free products (i.e. m/z 934.3: 5′-pTCATGA, m/z 1086.3: 5′-pTCATGAT, and m/z 613.2: 5′-pTCAT) Figs. S11 and S12. The second corresponded to misincorporation of dA (m/z 934.3: 5′-pTACTGA and m/z 1086.3: 5′-pTAGTCAT), and the third corresponded to misincorporation of dG (m/z 1086.3: 5′-pTGATCAT) (Figs. S9 and S10). The CID spectra of the products matched the predicted CID spectra of the sequences (Tables S1–S6).
Table 4.
LC-ESI-MS/MS analysis of full-length extension products across N7-CH3 2′-F dG by hpol η
The oligonucleotides used were as follows,
5′-FAM-CGGGCTCGTAAGCGTCUT-3′ 3′-GCCCGAGCATTCGCAGTAXTACT-5′
where X represents 2′-F dG and N7-CH3 2′-F dG. Products were cut at U, and the expected sequences began at the 3′ T of the primer.
| Product sequence | m/z, observed (charge) | m/z, theoretical (charge) | Relative peak area/tR (min) | % |
|---|---|---|---|---|
| 5′-pTCATGA | 934.27 (−2) | 1,870.22 (−1), 934.60 (−2) | 5,585.4/1.68 | 85 |
| 5′-pTCATGAT | 1,086.26 (−2), 724.07 (−3) | 1,086.70 (−2), 724.13 (−3) | 1,838.2/1.91 | |
| 5′-pTCAT | 613.22 (−2) | 1,227.80 (−1), 613.39 (−2) | 13,511/1.73 | |
| 5′-pTACTGA | 934.27 (−2) | 1,870.22 (−1), 934.60 (−2) | 620.6/1.68 | 10 |
| 5′-pTAGTCAT | 1,086.26 (−1), 724.07 (−2) | 1,086.70 (−1), 724.13 (−2) | 2,095.5/1.91 | |
| 5′-pTGATCAT | 1,086.26 (−1), 724.07 (−2) | 1,086.70 (−1), 724.13 (−2) | 1,397/1.91 | 5 |
Table 5.
LC-ESI-MS/MS analysis of full-length extension products across N7-CH3 2′-F dG by Dpo4
The oligonucleotides used were as follows,
5′-FAM-CGGGCTCGTAAGCGTCUT-3′ 3′-GCCCGAGCATTCGCAGTAXTACT-5′
where X represents 2′-F dG and N7-CH3 2′-F dG. Products were cut at U, and expected sequences began at 3′ T.
| Product sequence | m/z, observed (charge) | m/z, theoretical (charge) | Relative peak area/tR (min) | % |
|---|---|---|---|---|
| 5′-pTCATGA | 934.22 (−2) | 1,870.22 (−1), 934.60 (−2) | 6,417/ 1.67 | 100 |
| 5′-pTCATG | 777.56 (−2) | 1,557.01 (−1), 777.99 (−2) | 10,707/1.57 | |
| 5′-pTCAT | 613.09 (−2) | 1,227.80 (−1), 613.39 (−2) | 12,253/1.73 |
To confirm these assignments, mass spectra of commercial oligonucleotide standards with these sequences were compared with those of the observed products and were nearly identical. No products were observed containing the misincorporation of dT seen in the insertion kinetics experiments (Table 1). Relative areas were calculated for each product on the basis of the intensity of distinguishing CID ions (e.g. a3-B3 ions distinguish the error-free product from the product with misincorporation of dA). The yields of the observed products were estimated to be 85% for error-free bypass, 10% for misincorporation of dA, and 5% for misincorporation of dG (Table 4).
Discussion
Alkylation of DNA was first described in 1960 (20, 39), and the N7 atom of dG has long been known to be a major site of damage (34). The change in the pKa of the N1 atom (from 9 to 7) upon N7-methylation (34) was considered to be a potential reason for miscoding, evoking the original postulate of rare tautomer involvement in miscoding proposed by Watson and Crick (41). Due to this issue, one cannot consider an approach with 7-deaza dG for studying N7-alkyl dG miscoding, which would not reflect the electronic properties of the adduct. For discussion of the early studies on different alkylated bases and the development of a major role for O6-alkyl dG adducts in mutagenesis and carcinogenesis, see Lawley (39). Although O6-alkyl dG lesions are recognized to be important, the role of dG N7-alkylation has remained unclear. Some early studies concluded that N7-CH3 dG was not miscoding (39, 42), but the results of these studies are compromised by several issues, including the sensitivity of the assays in detecting miscoding, the lack of mammalian and microbial translesion DNA polymerases, and the lability of N7-CH3 dG. In 2009, Boysen et al. (21) concluded that there was no evidence for miscoding by N7-CH3 dG, although the authors suggested the 2′-F isostere approach we used here to address the issue. Lee and associates (37) used N7-CH3 2′-F dG with pol β and concluded that it was not miscoding but did not present limits of detection or utilize sensitive methods.
N7-Alkyl dG adducts are of particular interest because of their high endogenous levels and also high levels following exposure to alkylating agents (21, 39, 43, 44). N7-Alkyl dG adducts are found at the highest levels not only after exposure to methylating agents but with other alkylating agents as well (17, 39, 44, 45). Several examples of N7-alkyl dG adducts are found in laboratory animals and humans not knowingly exposed to exogenous agents, including N7-(2-hydroxy)ethyl dG, N7-(2-oxoethyl) dG, and N7-ethyl dG (44), but the origins of these adducts are not known. Although the levels of ribonucleotides and abasic sites have been reported to be higher than those of N7-CH3 dG, they are rapidly repaired by multiple pathways (22, 23), and the steady-state levels in cells are less than those of N7-CH3 dG (43).
The base-catalyzed imidazole ring opening of guanyl N7-alkyl adducts has been recognized for many years. As pointed out by Gates et al. (17), N7-CH3 dG is not unusually unstable, and at neutral pH, ring-opening is very slow; even at pH 8.9, the half-life is 9.8 h (46–49). Although there was original uncertainty about the multiple forms of N7-CH3 FAPY dG seen in chromatography, 15N NMR studies demonstrated that the site of the formyl group did not change (46) and that the adduct exists in slowly equilibrating rotomeric forms. Studies with rat liver and bladder DNA reported that levels of N7-CH3 dG decreased faster than those of the FAPY product, and levels of the two adducts were similar after 3–9 days (50, 51). However, Den Engelse et al. (49) reported only very low levels of the FAPY formed in rat liver following treatment with methylating agents. Some of the discrepancy may be due to the broadness of the N7-CH3 FAPY dG peaks, affecting both the resolution and the sensitivity (46, 49, 50, 52). In the report of Den Engelse et al. (49), no N7-CH3 FAPY dG adducts were detected in rat liver (<0.5% of N7-CH3 dG) up to 3 days after treatment with [14C]dimethylnitrosamine. Even in the report of Kadlubar et al. (51), the level of N7-CH3 FAPY dG did not reach the level of N7-CH3 dG (in the rat bladder epithelium) until 9 days after treatment with [14C]-methylnitrosourea. In considering all of this information, we conclude that the level of N7-CH3 dG is considerable and that any biological effects cannot be simply ascribed to abasic sites and N7-CH3 FAPY dG.
N7-CH3 dG is a substrate for several glycosylases, in addition to removal due to nonenzymatic depurination (53, 54), including 3-alkyladenine DNA glycosylase (AAG) in humans and the bacterial homologs 3-methyladenine glycosylase (AlkA), Bacillus cereus DNA glycosylase AlkD, and Streptomyces sahachiroi AlkZ (55–57). The chemical and biological half-lives of N7-CH3 dG have been estimated to be in the range of 69–192 h at 37 °C and neutral pH (chemical) (17) and 29–58 h (biphasic) in rat liver (presumably converting to an abasic site in the study cited, in that N7-CH3 dG was not detected (49). N7-CH3 FAPY dG is also a substrate for Escherichia coli FPG and other glycosylases (e.g. human OGG1, NTH1, and NEIL1) (58–61). The point made here is that N7-CH3 dG is persistent enough to be copied and miscoded, at least in tissues undergoing DNA replication.
In E. coli, N7-CH3 FAPY dG was not highly mutagenic when bypassed (G to T transversion mutation frequency of ≤2%) (62). When N7-CH3 FAPY dG was bypassed in a shuttle vector in simian kidney COS-7 cells, it readily produced G to T transversion mutations with 30% frequency (63). N7-CH3 FAPY dG was a strong block to replicative polymerases (e.g. pol α and pol δ/proliferating cell nuclear antigen), but hpol η, hpol κ, and the sequential action of hRev1/hpol ζ and Dpo4 were able to bypass N7-CH3 FAPY dG (29, 30). With hpol κ, N7-CH3 FAPY dG reduced the efficiency of dCTP insertion by an order of magnitude (29). Our previous work on the miscoding properties of N7-CH3 FAPY dG (29, 30) can be summarized and compared with the present work on N7-CH3 dG. Steady-state kinetic experiments on misinsertion showed only a low frequency of miscoding with S. solfataricus Dpo4 (0.01–0.04) but higher frequencies (0.28 and 0.29 for dT and dG insertion, respectively) with E. coli DNA polymerase I Klenow fragment. LC-MS analysis showed only misincorporation of dA for both polymerases examined with levels of misincorporation (2–35%) but considerable −1 frameshifts (11–17%) (30). In a later study with mammalian translesion DNA polymerases (29), we observed 2–5% misincorporation at N7-CH3 FAPY dG in steady-state kinetics and 11–29% misincorporation by LC-MS for extension products with hpol κ and η. Thus, the extents of misinsertion of hpol η (Tables 1 and 4) are similar in magnitude to those seen with N7-CH3 FAPY dG (29), although the oligonucleotide sequence is not the same.
Although Dpo4 and hpol κ are sometimes considered homologs (64, 65), they showed different abilities to replicate past N7-CH3 2′-F dG (Fig. 1, B and D), with hpol κ strongly blocked at the adduct site. hpol κ has been shown to bypass DNA adducts formed with methyl methanesulfonate more efficiently than hpols η and ι, and it also interacts directly with the ligase SHPRH to suppress methyl methanesulfonate–induced mutagenesis (66). hpol ι, which also inserted only dCTP, is also effective in inserting dNTPs across minor groove lesions, such as N3-methyl deoxyadenosine (67).
Koag et al. (37) evaluated the kinetics of insertion of dCTP and dTTP across N7-CH3 2′-F dG by pol β, a gap-filling X-family polymerase. The lesion decreased the rate of pol β catalysis by ∼300-fold, yet replication was accurate, and no misinsertion products were reported. The structures revealed Watson–Crick base pairing of N7-CH3 2′-F dG with an incoming dCTP, but the metal ion coordination was not optimal for catalysis. When N7-CH3 2′-F dG was crystallized with dTTP, an open conformation was found, with a staggered bp.
The relatively low but finite level of misincorporation at the N7-CH3 dG might seem unimportant. However, consideration needs to be given to the overall mutagenic load. In four different studies cited by Den Engelse et al. (49), the ratio of N7-CH3 dG to O6-CH3 dG adducts following treatment (of cells or rats) with dimethylnitrosamine or methylnitrosoureas was ∼10:1. In our own studies with hpol η (68), miscoding in the LC-MS assays was 77%, which may be compared with 15% here with N7-CH3 2′-F dG (Table 4). Multiplying the adduct level differences, 77 × 0.1 = 7.7 (O6-CH3 dG), which can be compared with 15 × 1 = 15 (N7-CH3 (2′-F) dG). Kunkel (43) has estimated a 200–3,000-fold difference in endogenous cellular levels of N7-CH3 dG over O6-CH3 dG. In a more recent study with cultured human lymphoblastoid cells, Sharma et al. (69) reported a 12-fold higher level of N7-CH3 dG adducts than O6-CH3 dG after treatment with methylnitrosourea and a 900-fold higher level of N7-CH3 dG in the untreated cells. Applying the difference in levels of miscoding to these levels of the adducts can therefore result in an even larger potential contribution of N7-CH3 dG to miscoding and mutagenesis.
In summary, we have shown that hpol η produces error-free bypass products in copying past N7-CH3 2′-F dG and also misinserts dA and dG, differing from the products seen for N7-CH3 FAPY dG, which inserted dT and produced a frameshift mutation (29). Our findings indicate that our results are not due to any contamination by the FAPY degradation product and also suggest N7-CH3 dG contribution to mutagenicity in cells. Caveats need to be considered about comparing miscoding frequencies in different sequence contexts, the potential roles of DNA polymerases that were not included here, rates of enzymatic repair in different cells, and possibly other issues. Inserting plasmid vectors containing N7-CH3 dG into cells to estimate mutation frequencies would be very problematic in terms of being sure that the lesion, even with the 2′-F group, was not modified before mutation occurred. In conclusion, the abundance of the adduct N7-CH3 dG, coupled with the evidence for miscoding, argues that this lesion should no longer be considered innocuous.
Experimental procedures
Materials
All chemicals and solvents were commercially available, of highest purity grade, and were used without additional purification. 9-(2-Deoxy-2-fluoro-β-d-arabinofuranosyl) guanine was purchased from Metkinen (Kuopio, Finland). Pyridine, N,N-dimethylformamide, dichloromethane, N,N-diisopropyethylamine, isobutyryl chloride, chlorotrimethylsilane, and 4,4′-dimethoxytrityl chloride were purchased from Sigma-Aldrich. Synthesis was monitored by TLC on Merck silica gel 60 F254 plates, with visualization at 254 nm and by spraying a solution of 5% concentrated H2SO4 in ethanol (v/v) and heating. Restriction endonucleases, UDG, FPG glycosylase, dNTPs, and T4 polynucleotide kinase were purchased from New England Biolabs (Ipswich, MA). Unmodified oligonucleotides and primers used for extension and steady-state kinetics were obtained from Integrated DNA Technologies (Coralville, IA) and were HPLC-purified. Primers used for LC-MS sequence analysis were also obtained from DNA Technologies (Coralville, IA) and were twice HPLC-purified. Human DNA polymerases hpol η (catalytic core residues 1–432), hpol ι (catalytic core residues 1–420), and hpol κ (catalytic core residues 19–526) and bacterial Dpo4 were expressed in E. coli and purified as described previously (70–73).
NMR spectroscopy and MS
1H and 13C NMR spectra were recorded on a 600-MHz Bruker NMR spectrometer; 31P NMR spectra were recorded on a 500-MHz Bruker NMR spectrometer. Mass spectrometry was performed at the Vanderbilt Mass Spectrometry Research Core Facility using both Thermo low-resolution (LTQ) and high-resolution (Orbitrap) spectrometers. Spectra of synthetic products (negative and positive ion modes) and modified oligonucleotides (negative ion mode) were obtained using a Waters Acquity UPLC instrument (Waters, Milford, MA) interfaced to a Thermo-Finnigan LTQ mass spectrometer (Thermo Scientific, San Jose, CA), also equipped with an electrospray source.
Synthesis of 9-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-1,9-dihydro-N2-isobutyrylguanosine (isobutyrylacetamido-6H-purin-6-one) (74)
Commercially available 9-(2-deoxy-2-fluoro-β-d-arabinofuranosyl) guanine (1) (10 mg, 1.05 mmol) was co-evaporated to dryness with anhydrous pyridine (3 × 10 ml) in vacuo. The residue was redissolved in anhydrous pyridine (10 ml) solution under an argon atmosphere, and chlorotrimethylsilane (334 μl, 7.88 mmol) was added. The mixture was stirred at room temperature for 2 h and then cooled to 0 °C. Isobutyryl chloride (110 μl, 3.15 mmol) was added in a dropwise manner over 20 min (74). The reaction mixture was allowed to warm to room temperature and further stirred for 3 h. The reaction mixture was then cooled to 0 °C, and water (10 ml) was added to quench the reaction. The reaction was stirred consecutively for 5 min at 0 °C and 5 min at room temperature, and then concentrated aqueous NH4OH (25 ml) was added, with more stirring for 30 min. H2O (170 ml) was added to dilute the reaction mixture, and the mixture was extracted with CH2Cl2 (50 ml). The aqueous phase was evaporated in vacuo to obtain a white solid, 9-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-N2-isobubutyrylguanosine (100 mg, 80%). 1H NMR (DMSO-d6): δ 8.10 (d, 1H, J = 2.0 Hz, H-8), 6.24 (dd, 1H, J = 4.2, 14.9 Hz, H-1′), 5.18 (dt, 1H, J = 4.2 Hz, 52 Hz, H-2′), 4.38 (dt, 1H, J = 4.2, 17.2 Hz, H-3′), 3.89 (dd, J = 4.9, 10.4 Hz, H-4′), 3.63 (m, 2H, J = 40.26 Hz, H-5′), 2.75 (m, 1H, J = 6.9 Hz, H-11), 1.08 (d, 6H, J = 6.62 Hz, H-12). 13C NMR (DMSO-d6): 180.8, 155.5, 148.8, 138.8, 120.1, 96.0, 94.8, 84.4, 82.3, 73.0, 60.9, 35.4, 19.5. MS: calculated for C14H18FN5O5 (M-H) 354.1; found 354.3.
Synthesis of 9-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-1,9-dihydro-N7-methyl-N2-isobutyrylacetamido-6H-purin-6-one (32)
To an anhydrous solution of N,N-dimethylformamide (5 ml) was added 9-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-1,9-dihydro-N2-isobutyrylguanosine (120 mg, 0.34 mmol) and methyl iodide (351 μl, 5.63 mmol) under an argon atmosphere. The reaction mixture was stirred at room temperature for 22 h and then poured into cold diethyl ether to precipitate the product, which was filtered and concentrated in vacuo to afford a white solid, 9-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-1,9-dihydro-N2- isobutyryacetamido-6H-purin-6-one (114 mg, 80%). 1H NMR (DMSO-d6): δ 9.69 (s, 1H, H-8), 6.28 (dd, 1H, J = 2.9, 13.8 Hz, H-1′), 5.88 (s, OH), 5.24 (d, 1H, J = 52 Hz, H-2′), 4.96 (s, OH), 4.40 (d, 1H, J = 17.2 Hz, H-3′) 4.08 (s, 3H, N7-CH3), 3.98 (s, H-4′), 3.61 (s, 2H, H-5′), 2.69 (t, 1H, J = 7.2 Hz, 13.6 Hz, H-11), 1.04 (d, 6H, J = 6.62 Hz, H-12). 13C NMR (DMSO-d6): 180.8, 155.5, 148.8, 138.8, 120.1, 96.0, 94.8, 84.4, 82.3, 73.0, 60.9, 35.4, 19.5. MS: calculated for C15H21FN5O5 (MH+) 370.2; found 370.2.
Synthesis of 9-[2-deoxy-5-O-(4,4′-dimethoxytrityl)-2-fluoro-β-d-arabinofuranosyl]-1,9-dihydro-N7-methyl-N2-isobutyrylacetamido-6H-purin-6-one
9-(2-Deoxy-2-fluoro-β-d-arabinofuranosyl)-1,9-dihydro-N7-methyl-N2- isobutyryacetamido-6H-purin-6-one (263 mg, 0.71 mmol), in anhydrous pyridine, and 4,4′-dimethoxytrityl chloride (721 mg, 2.1 mmol) were stirred at room temperature for 2 h under an argon atmosphere. The reaction mixture was diluted with CH2Cl2 (50 ml) and washed with saturated aqueous NaHCO3 and then brine (3 × 50 ml). The organic layer was dried over anhydrous Na2SO4 and filtered, and the solvent was evaporated. The crude residue was purified by silica gel column chromatography (3% CH3OH in CH2Cl2 plus 1% triethylamine, v/v) to afford 9-[2-deoxy-5-O-(4,4′-dimethoxytrityl)-2-fluoro-β-d-arabinofuranosyl]-1,9-dihydro-N7-methyl-N2-isobutyrylacetamido-6H-purin-6-one (290 mg, 60% yield). 1H NMR (600 MHz, CD2Cl2): δ 8.44 (1H, s, H-8), 6.93–7.52 (13H, m, aromatic H), 6.86 (1H, d, J = 7.7 Hz, H-1′), 5.34 (1H, t, 2.8, H-2′), 4.72 (1H, d, J = 17.22 Hz, H-3′), 4.45 (1H, m, H-4′), 3.97 (3H, s, N7-CH3), 3.79 (6H, s, OCH3, OCH3), 3.49–3.56 (2H, m, 7.27, 5.25 Hz, H-5 and H-5′), 2.72 (1H, m, H11), 1.109 (dd, 6H, J = 1.82, 11.62 Hz, H-12). MS: calculated for C36H39FN5O7 (MH+) 672.3; found 672.2.
Synthesis of 9-[2-deoxy-5-O-(4,4′-dimethoxytrityl)-2-fluoro-β-d-arabinofuranosyl]-1,9-dihydro-N7-methyl-N2-isobutyrylacetamido-6H-purin-6-one-3-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite
The dimethoxytrityl-protected nucleoside from the previous step (90 mg, 134 μmol) was dissolved in CH2Cl2 (2 ml), and N,N-diisopropyethylamine (55 μl, 0.33 mmol) was added. N,N-Diisopropylamino)chlorophosphine (45 μl, 0.2 mmol) was added, and then the reaction mixture was stirred at room temperature for 2 h under an argon atmosphere. The mixture was diluted with CH2Cl2 (50 ml) and washed with saturated aqueous NaHCO3 and then brine (3 × 50 ml), and the organic phase was dried over Na2SO4 and filtered. The solvent was evaporated in vacuo. The crude reaction mixture was purified by silica gel chromatography with 1% CH3OH in CH2Cl2 containing 1% trimethylamine (v/v) to afford 80 mg of 9-[2-deoxy-5-O-(4,4′-dimethoxytrityl)-2-fluoro-β-d-arabinofuranosyl]-1,9-dihydro-N7-methyl-N2-isobutyrylacetamido-6H-purin-6-one-3-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite, 68%. 31P NMR (500 MHz, CD2Cl2) δ 152.48, 152.30; MS: calculated for C36H39FN5O7 (MH+) 872.4; found 872.4.
Synthesis, purification, and characterization of 2′-F dG and N7-CH3 2′-F dG–containing DNA oligonucleotides
Modified oligonucleotides bearing 2′-fluorines were synthesized with Expedite reagents (Glen Research, Sterling, VA) on a 1-μmol scale utilizing a Perspective Biosystems model 8909 DNA synthesizer and a standard synthetic protocol (75). We chose the β-anomer for the 2′-fluoro analogs because this configuration has been shown not to alter sugar puckering in DNA; this is the typical configuration for the 2′-deoxynucleotides (76–78). The coupling of N7-CH3 2′-F dG phosphoramidite was performed off-line for 2 h. The remainder of the synthesis was done online using standard procedures. Modified oligonucleotides were cleaved from the solid support, and exocyclic groups were deprotected in a single step using anhydrous methanolic K2CO3 (50 mm), stirring at room temperature for 8 h. CH3OH was removed by sweeping with a stream of N2 gas. Oligonucleotides were purified by reversed-phase HPLC with a Phenominex Alumina RP octadecylsilane (C18) column (250 mm × 4.6 mm, 5 μm). The solvents used were aqueous 100 mm triethylammonium acetate (mobile phase A) and 100 mm triethylammonium acetate in H2O/CH3CN (1:1, v/v) (mobile phase B). The flow rate was 1.5 ml/min with the following gradient: initial 20% B, increased to 25% B over 5 min, held at 25% for 15 min, increased to 40% at 20 min, held for 5 min, then 100% at 25 min, and held until 30 min and 5% B at 31 min and re-equilibrated to 0% B for 5 min (all v/v). The UV detector was set at 240 nm. The collected fractions were lyophilized to dryness, redissolved in water, and desalted using ZipTip U-C18 columns prior to characterization.
Oligonucleotide 5′-TCAT(2′-F dG) ATGACGCTTACGAGCCCG-3′ was purified by HPLC, LC-ESI m/z calculated for [M-H]−, 7039.193; found 7043.000 (Fig. S5B).
Oligonucleotide 5′-TCAT(N7-CH3 2′-F dG)ATGACGCTTACGAGCCCG-3′ was purified by HPLC, LC-ESI m/z calculated for [M-H]−, 7054.216; found 7075.000 (Fig. S6A) (presumably sodium adduct).
The identity of the N7-CH3 2′-F dG–containing oligonucleotide was further confirmed by subjecting it to FPG glycosylase. The N7-CH3 2′-F dG–containing oligonucleotide was 32P-labeled at the 5′-end using T4 polynucleotide kinase (New England Biolabs) and annealed to its complementary strand by heating at 95 °C for 5 min and then allowing it to cool to room temperature overnight. A second portion of the N7-CH3 2′-F dG-oligonucleotide was treated with NaOH and stirred for 12 h at room temperature to create a hydrolyzed N7-CH3 FAPY-2′-F dG oligonucleotide. It was also 5′-end–labeled (32P-label and T4 polynucleotide kinase) and then annealed with its complementary strand. Both oligonucleotides were subjected to treatment with FPG glycosylase for 1 h at 37 °C. Reactions were quenched with 9 μl of quenching dye (20 mm EDTA, (pH 9.0) in 95% formamide, v/v) and the products were separated on a 20% acrylamide (w/v) electrophoresis gel. Results were visualized using a phosphorimaging system (Bio-Rad, Molecular Imager® FX) and analyzed by Quantity One software as described previously (38).
Primer annealing and extension assays
5′-FAM-labeled 16-mer, 18-mer, and 19-mer primers (5′-/FAM/CGGGCTCGTAAGCGTC-3′, 5′-/FAM/CGGGCTCGTAAGCGTCAT-3′, 5′-/FAM/CGGGCTCGTAAGCGTCATC-3′, and 5′-/FAM/CGGGCTCGTAAGCGTCATT-3′, respectively) were annealed to a 23-mer template (3′-GCCCGAGCATTCGCAGTAXTACT-5′, where X was dG, 2′-F dG, or N7-CH3 2′-F dG, in a 1:1 molar ratio at 95 °C for 5 min and slowly cooling to room temperature. For the full-length extension assays, WT hpol η (20 nm), hpol ι (40 nm), hpol κ (20 nm), and Dpo4 (20 nm) were incubated with the 16-mer primer–template DNA complex (200 nm) in 40 mm Tris-HCl buffer (pH 7.5) containing 5 mm MgCl2, 50 mm NaCl, 5% glycerol (v/v), 5 mm DTT, 50 μg/ml BSA, and 250 μm dNTPs. The reactions were done at 37 °C for 2, 5, 10, 20, and 60 min. For single-nucleotide incorporation experiments, an 18-mer primer–template DNA complex (120 nm) was used. Enzyme concentrations were as follows: hpol η (5 nm), hpol ι (10 nm), hpol κ (5 nm), and Dpo4 (5 nm). Reactions were done for 10 min. In the case of the single-nucleotide extension experiments, two primer–template DNA complexes (120 nm) were used with hpol η (5 nm) alone for 5 min. All other reaction conditions were the same as in the full-length extension experiments. Reactions were quenched as above, and products were separated on 18% denaturing acrylamide gels (w/v) and visualized with a Typhoon system (GE Healthcare).
Steady-state insertion and extension kinetics
Insertion reactions were done by incubating FAM-labeled 18-mer primer/23-mer template complexes (120 nm) with hpol η (2.5–10 nm) or Dpo4 (0.15–10 nm), and extension reactions were conducted by incubating two FAM-labeled 19-mer primer/23-mer template complexes (120 nm) with hpol η (5–10 nm). Both reactions were incubated at 37 °C for 5–10 min in 50 mm Tris-HCl buffer (pH 7.5) containing 5 mm MgCl2, 50 mm NaCl, 5% glycerol (v/v), 5 mm DTT, 50 μg/ml BSA, and varying concentrations of dNTPs. Reactions were quenched as described above, and products were separated on 18% denaturing acrylamide gels (w/v), visualized with a Typhoon system, and quantified utilizing ImageJ software (National Institutes of Health). Data obtained were fit to the hyperbolic Michaelis–Menten equation in GraphPad Prism software (version 8.0, La Jolla, CA).
LC-MS analysis of full-length extension products by hpol η and Dpo4
An 18-mer primer bearing a 2′-deoxyuridine (5′-FAM/CGGGCTCGTAAGCGTC(dU)T-3′) was annealed to the 23-mer oligomer used above, in a molar ratio of 1:1. Full-length extension reactions were done using similar conditions as in the steady-state experiments, with the exception of primer–template complex (2.5 μm), hpol η (150 nm), Dpo4 (300 nm), and dNTPs (500 μm). Reactions were incubated at 37 °C for 1 h. Reactions were quenched by spin column separation to remove Mg2+ and dNTPs, and the extension product was treated with 25 units of UDG at 37 °C for 4 h and then with 0.25 m piperidine, heating at 95 °C for 1 h. H2O was added to the reaction mixture, which was lyophilized and then redissolved in H2O (70). Products were analyzed by LC-MS/MS, performed using a Waters Acquity UPLC system linked to a Thermo-Finnigan LTQ mass spectrometer with electrospray ionization in the negative ion mode. Separation by chromatography was done using an Acquity UPLC system BEH octadecylsilane (C18) column (1.7 μm, 2.1 mm × 50 mm) with UPLC conditions as described previously (40).
Author contributions
F. P. G. conceived the studies; Y. S. expressed and purified hpol η; F. P. G. and O. J. N. designed the experiments, interpreted the data, and wrote the manuscript; O. J. N. carried out organic synthesis, characterization of the oligonucleotides, steady-state kinetics, and LC-MS sequence analysis.
Supplementary Material
Acknowledgments
We thank Drs. Carmelo J. Rizzo and Chanchal Malik for DNA synthesis and helpful discussions, Dr. Carl A. Sedgeman for helpful suggestions, and Dr. Plamen P. Christov for help with LC-MS sequencing analysis. Mass spectrometry was performed at the Vanderbilt Mass Spectrometry Research Core Facility. We also thank K. Trisler for assistance with preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES010546 (to F. P. G.) and T32 ES007028 (to F. P. G. and O. J. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Scheme S1, Tables S1–S6, and Figs. S1–S12.
- N7-CH3 dG
- N7-methyl deoxyguanosine
- N7-CH3 2′-F dG
- N7-methyl 2′-fluoro deoxyguanosine
- 2′-F dG
- 2′-fluoro deoxyguanosine
- FAM
- 6-carboxyfluorescein
- FAPY
- formamidopyrimidine
- FPG
- formamidopyrimidine DNA glycosylase
- pol
- DNA polymerase
- hpol
- human pol
- O6-CH3 dG
- O6-methyl deoxyguanosine
- UDG
- uracil-DNA glycosylase
- CID
- collision-induced dissociation
- ESI
- electrospray ionization.
References
- 1. Swenberg J. A., Lu K., Moeller B. C., Gao L., Upton P. B., Nakamura J., and Starr T. B. (2011) Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 120, S130–S145 10.1093/toxsci/kfq371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Helleday T., Eshtad S., and Nik-Zainal S. (2014) Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 10.1038/nrg3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weismann C. G., and Gelb B. D. (2007) The genetics of congenital heart disease: a review of recent developments. Curr. Opin. Cardiol. 22, 200–206 10.1097/HCO.0b013e3280f629c7 [DOI] [PubMed] [Google Scholar]
- 4. Shigenaga M. K., Hagen T. M., and Ames B. N. (1994) Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. U.S.A. 91, 10771–10778 10.1073/pnas.91.23.10771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sedgwick B. (2004) Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5, 148–157 10.1038/nrm1312 [DOI] [PubMed] [Google Scholar]
- 6. Erickson R. P. (2003) Somatic gene mutation and human disease other than cancer. Mut. Res. 543, 125–136 10.1016/S1383-5742(03)00010-3 [DOI] [PubMed] [Google Scholar]
- 7. Newell D., Gescher A., Harland S., Ross D., and Rutty C. (1987) N-Methyl antitumour agents: a distinct class of anticancer drugs? Cancer Chemother. Pharmacol. 19, 91–102 [DOI] [PubMed] [Google Scholar]
- 8. Friedman H. S., Kerby T., and Calvert H. (2000) Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 6, 2585–2597 [PubMed] [Google Scholar]
- 9. Lidor Y. J., Shpall E. J., Peters W. P., and Bast R. C. Jr. (1991) Synergistic cytotoxicity of different alkylating agents for epithelial ovarian cancer. Int. J. Cancer 49, 704–710 10.1002/ijc.2910490513 [DOI] [PubMed] [Google Scholar]
- 10. McGuire W. P. 3rd, and Markman M. (2003) Primary ovarian cancer chemotherapy: current standards of care. Br. J. Cancer 89, S3–S8 10.1038/sj.bjc.6601494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dolan M. E., McRae B. L., Ferries-Rowe E., Belanich M., van Seventer G. A., Guitart J., Pezen D., Kuzel T. M., and Yarosh D. B. (1999) O6-Alkylguanine-DNA alkyltransferase in cutaneous T-cell lymphoma: implications for treatment with alkylating agents. Clin. Cancer Res. 5, 2059–2064 [PubMed] [Google Scholar]
- 12. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- 13. Beranek D. T. (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mut. Res. 231, 11–30 10.1016/0027-5107(90)90173-2 [DOI] [PubMed] [Google Scholar]
- 14. Newbold R. F., Warren W., Medcalf A. S., and Amos J. (1980) Mutagenicity of carcinogenic methylating agents is associated with a specific DNA modification. Nature 283, 596–599 10.1038/283596a0 [DOI] [PubMed] [Google Scholar]
- 15. Mitra G., Pauly G. T., Kumar R., Pei G. K., Hughes S. H., Moschel R. C., and Barbacid M. (1989) Molecular analysis of O6-substituted guanine-induced mutagenesis of ras oncogenes. Proc. Natl. Acad. Sci. U.S.A. 86, 8650–8654 10.1073/pnas.86.22.8650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pullman A., and Pullman B. (1981) Molecular electrostatic potential of the nucleic acids. Q. Rev. Biophys. 14, 289–380 10.1017/S0033583500002341 [DOI] [PubMed] [Google Scholar]
- 17. Gates K. S., Nooner T., and Dutta S. (2004) Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol. 17, 839–856 10.1021/tx049965c [DOI] [PubMed] [Google Scholar]
- 18. Kou Y., Koag M. C., and Lee S. (2018) Structural and kinetic studies of the effect of guanine N7 alkylation and metal cofactors on DNA replication. Biochemistry 57, 5105–5116 10.1021/acs.biochem.8b00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bailey E. A., Iyer R. S., Stone M. P., Harris T. M., and Essigmann J. M. (1996) Mutational properties of the primary aflatoxin B1-DNA adduct. Proc. Natl. Acad. Sci. U.S.A. 93, 1535–1539 10.1073/pnas.93.4.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brookes P., and Lawley P. D. (1960) The reaction of mustard gas with nucleic acids in vitro and in vivo. Biochem. J. 77, 478–484 10.1042/bj0770478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boysen G., Pachkowski B. F., Nakamura J., and Swenberg J. A. (2009) The formation and biological significance of N7-guanine adducts. Mutat. Res. 678, 76–94 10.1016/j.mrgentox.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams J. S., and Kunkel T. A. (2014) Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst.) 19, 27–37 10.1016/j.dnarep.2014.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caldecott K. W. (2014) Molecular biology. Ribose–an internal threat to DNA. Science 343, 260–261 10.1126/science.1248234 [DOI] [PubMed] [Google Scholar]
- 24. Mustonen R., and Hemminki K. (1992) 7-Methylguanine levels in DNA of smokers' and non-smokers' total white blood cells, granulocytes and lymphocytes. Carcinogenesis 13, 1951–1955 10.1093/carcin/13.11.1951 [DOI] [PubMed] [Google Scholar]
- 25. Park J. W., and Ames B. N. (1988) 7-Methylguanine adducts in DNA are normally present at high levels and increase on aging: analysis by HPLC with electrochemical detection. Proc. Natl. Acad. Sci. U.S.A. 85, 7467–7470 10.1073/pnas.85.20.7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barrows L. R., and Magee P. N. (1982) Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis 3, 349–351 10.1093/carcin/3.3.349 [DOI] [PubMed] [Google Scholar]
- 27. Yu S. L., Lee S. K., Johnson R. E., Prakash L., and Prakash S. (2003) The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol. 23, 382–388 10.1128/MCB.23.1.382-388.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibbs P. E., and Lawrence C. W. (1995) Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol. 251, 229–236 10.1006/jmbi.1995.0430 [DOI] [PubMed] [Google Scholar]
- 29. Christov P. P., Yamanaka K., Choi J.-Y., Takata K., Wood R. D., Guengerich F. P., Lloyd R. S., and Rizzo C. J. (2012) Replication of the 2,6-diamino-4-hydroxy-N5-methyl-formamidopyrimidine (MeFapy-dGuo) adduct by eukaryotic DNA polymerases. Chem. Res. Toxicol. 25, 1652–1661 10.1021/tx300113e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christov P. P., Angel K. C., Guengerich F. P., and Rizzo C. J. (2009) Replication past the N5-methyl-formamidopyrimidine lesion of deoxyguanosine by DNA polymerases and an improved procedure for sequence analysis of in vitro bypass products by mass spectrometry. Chem. Res. Toxicol. 22, 1086–1095 10.1021/tx900047c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dizdaroglu M., Kirkali G., and Jaruga P. (2008) Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic. Biol. Med. 45, 1610–1621 10.1016/j.freeradbiomed.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 32. Lee S., Bowman B. R., Ueno Y., Wang S., and Verdine G. L. (2008) Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J. Am. Chem. Soc. 130, 11570–11571 10.1021/ja8025328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barbin A., Laib R. J., and Bartsch H. (1985) Lack of miscoding properties of 7-(2-oxoethyl)guanine, the major vinyl chloride-DNA adduct. Cancer Res. 45, 2440–2444 [PubMed] [Google Scholar]
- 34. Lawley P. D., and Brookes P. (1961) Acidic dissociation of 7:9-dialkylguanines and its possible relation to mutagenic properties of alkylating agents. Nature 192, 1081–1082 10.1038/1921081b0 [DOI] [PubMed] [Google Scholar]
- 35. Sowers L. C., Shaw B. R., Veigl M. L., and Sedwick W. D. (1987) DNA base modification: ionized base pairs and mutagenesis. Mutat. Res. 177, 201–218 10.1016/0027-5107(87)90003-0 [DOI] [PubMed] [Google Scholar]
- 36. Lawley P. D., and Brookes P. (1962) Ionization of DNA bases or base analogues as a possible explanation of mutagenesis, with special reference to 5-bromodeoxyuridine. J. Mol. Biol. 4, 216–219 10.1016/S0022-2836(62)80053-9 [DOI] [PubMed] [Google Scholar]
- 37. Koag M. C., Kou Y., Ouzon-Shubeita H., and Lee S. (2014) Transition-state destabilization reveals how human DNA polymerase β proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res. 42, 8755–8766 10.1093/nar/gku554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao L., Christov P. P., Kozekov I. D., Pence M. G., Pallan P. S., Rizzo C. J., Egli M., and Guengerich F. P. (2012) Replication of N2,3-ethenoguanine by DNA polymerases. Angew. Chem. Int. Ed. Engl. 51, 5466–5469 10.1002/anie.201109004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawley P. D. (1984) Carcinogenesis by alkylating agents. In Chemical Carcinogens, 2nd Ed. (Searle C. E., ed) pp. 324–484, American Chemical Society, Washington, D. C. [Google Scholar]
- 40. Sedgeman C. A., Su Y., and Guengerich F. P. (2017) Formation of S-[2-(N6-deoxyadenosinyl)ethyl]glutathione in DNA and replication past the adduct by translesion DNA polymerases. Chem. Res. Toxicol. 30, 1188–1196 10.1021/acs.chemrestox.7b00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watson J. D., and Crick F. H. C. (1953) Genetical implications of the structure of deoxyribonucleic acid. Nature 171, 964–967 10.1038/171964b0 [DOI] [PubMed] [Google Scholar]
- 42. Lawley P. D., and Orr D. J. (1970) Specific excision of methylation products from DNA of Escherichia coli treated with N-methyl-N′-nitro-N-nitrosoguanidine. Chem. Biol. Interact. 2, 154–157 10.1016/0009-2797(70)90047-5 [DOI] [PubMed] [Google Scholar]
- 43. Kunkel T. A. (1999) The high cost of living. American Association for Cancer Research Special Conference: Endogenous sources of mutations. Trends Genet. 15, 93–94 10.1016/S0168-9525(98)01664-3 [DOI] [PubMed] [Google Scholar]
- 44. De Bont R., and van Larebeke N. (2004) Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19, 169–185 10.1093/mutage/geh025 [DOI] [PubMed] [Google Scholar]
- 45. Inskeep P. B., Koga N., Cmarik J. L., and Guengerich F. P. (1986) Covalent binding of 1,2-dihaloalkanes to DNA and stability of the major DNA adduct, S-[2-(N7-guanyl)ethyl]glutathione. Cancer Res. 46, 2839–2844 [PubMed] [Google Scholar]
- 46. Humphreys W. G., and Guengerich F. P. (1991) Structure of formamidopyrimidine adducts as determined by NMR using specifically 15N-labeled guanosine. Chem. Res. Toxicol. 4, 632–636 10.1021/tx00024a005 [DOI] [PubMed] [Google Scholar]
- 47. Mao H., Deng Z., Wang F., Harris T. M., and Stone M. P. (1998) An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry 37, 4374–4387 10.1021/bi9718292 [DOI] [PubMed] [Google Scholar]
- 48. Hendler S., Fürer E., and Srinivasan P. R. (1970) Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial deoxyribonucleic acid or ribonucleic acid polymerase. Biochemistry 9, 4141–4153 10.1021/bi00823a017 [DOI] [PubMed] [Google Scholar]
- 49. Den Engelse L., Menkveld G. J., De Brij R. J., and Tates A. D. (1986) Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis 7, 393–403 10.1093/carcin/7.3.393 [DOI] [PubMed] [Google Scholar]
- 50. Beranek D. T., Weis C. C., Evans F. E., Chetsanga C. J., and Kadlubar F. F. (1983) Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem. Biophys. Res. Commun. 110, 625–631 10.1016/0006-291X(83)91195-6 [DOI] [PubMed] [Google Scholar]
- 51. Kadlubar F. F., Beranek D. T., Weis C. C., Evans F. E., Cox R., and Irving C. C. (1984) Characterization of the purine ring-opened 7-methylguanine and its persistence in rat bladder epithelial DNA after treatment with the carcinogen N-methylnitrosourea. Carcinogenesis 5, 587–592 10.1093/carcin/5.5.587 [DOI] [PubMed] [Google Scholar]
- 52. Oida T., Humphreys W. G., and Guengerich F. P. (1991) Preparation and characterization of oligonucleotides containing S-[2-(N7-guanyl)ethyl]glutathione. Biochemistry 30, 10513–10522 10.1021/bi00107a021 [DOI] [PubMed] [Google Scholar]
- 53. Xiao W., and Samson L. (1993) In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 90, 2117–2121 10.1073/pnas.90.6.2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barbado C., Córdoba-Canero D., Ariza R. R., and Roldán-Arjona T. (2018) Nonenzymatic release of N7-methylguanine channels repair of abasic sites into an AP endonuclease-independent pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E916–E924 10.1073/pnas.1719497115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Parsons Z. D., Bland J. M., Mullins E. A., and Eichman B. F. (2016) A catalytic role for C-H/π interactions in base excision repair by Bacillus cereus DNA glycosylase AlkD. J. Am. Chem. Soc. 138, 11485–11488 10.1021/jacs.6b07399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mullins E. A., Warren G. M., Bradley N. P., and Eichman B. F. (2017) Structure of a DNA glycosylase that unhooks interstrand cross-links. Proc. Natl. Acad. Sci. U.S.A. 114, 4400–4405 10.1073/pnas.1703066114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hollis T., Lau A., and Ellenberger T. (2000) Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat. Res. 460, 201–210 10.1016/S0921-8777(00)00027-6 [DOI] [PubMed] [Google Scholar]
- 58. Asagoshi K., Yamada T., Terato H., Ohyama Y., Monden Y., Arai T., Nishimura S., Aburatani H., Lindahl T., and Ide H. (2000) Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J. Biol. Chem. 275, 4956–4964 10.1074/jbc.275.7.4956 [DOI] [PubMed] [Google Scholar]
- 59. Dherin C., Radicella J. P., Dizdaroglu M., and Boiteux S. (1999) Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 27, 4001–4007 10.1093/nar/27.20.4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Katafuchi A., Nakano T., Masaoka A., Terato H., Iwai S., Hanaoka F., and Ide H. (2004) Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 279, 14464–14471 10.1074/jbc.M400393200 [DOI] [PubMed] [Google Scholar]
- 61. Asagoshi K., Yamada T., Okada Y., Terato H., Ohyama Y., Seki S., and Ide H. (2000) Recognition of formamidopyrimidine by Escherichia coli and mammalian thymine glycol glycosylases: distinctive paired base effects and biological and mechanistic implications. J. Biol. Chem. 275, 24781–24786 10.1074/jbc.M000576200 [DOI] [PubMed] [Google Scholar]
- 62. Patro J. N., Wiederholt C. J., Jiang Y. L., Delaney J. C., Essigmann J. M., and Greenberg M. M. (2007) Studies on the replication of the ring opened formamidopyrimidine, Fapy-dG in Escherichia coli. Biochemistry 46, 10202–10212 10.1021/bi700628c [DOI] [PubMed] [Google Scholar]
- 63. Kalam M. A., Haraguchi K., Chandani S., Loechler E. L., Moriya M., Greenberg M. M., and Basu A. K. (2006) Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 34, 2305–2315 10.1093/nar/gkl099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wolfle W. T., Washington M. T., Prakash L., and Prakash S. (2003) Human DNA polymerase κ uses template-primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. Genes Dev. 17, 2191–2199 10.1101/gad.1108603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ling H., Boudsocq F., Woodgate R., and Yang W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 10.1016/S0092-8674(01)00515-3 [DOI] [PubMed] [Google Scholar]
- 66. Lin J. R., Zeman M. K., Chen J. Y., Yee M. C., and Cimprich K. A. (2011) SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol. Cell 42, 237–249 10.1016/j.molcel.2011.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yoon J. H., Roy Choudhury J., Park J., Prakash S., and Prakash L. (2017) Translesion synthesis DNA polymerases promote error-free replication through the minor-groove DNA adduct 3-deaza-3-methyladenine. J. Biol. Chem. 292, 18682–18688 10.1074/jbc.M117.808659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Choi J.-Y., Chowdhury G., Zang H., Angel K. C., Vu C. C., Peterson L. A., and Guengerich F. P. (2006) Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 281, 38244–38256 10.1074/jbc.M608369200 [DOI] [PubMed] [Google Scholar]
- 69. Sharma V., Collins L. B., Clement J. M., Zhang Z., Nakamura J., and Swenberg J. A. (2014) Molecular dosimetry of endogenous and exogenous O6-methyl-dG and N7-methyl-G adducts following low dose D3-methylnitrosourea exposures in cultured human cells. Chem. Res. Toxicol. 27, 480–482 10.1021/tx5000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zang H., Goodenough A. K., Choi J.-Y., Irimia A., Loukachevitch L. V., Kozekov I. D., Angel K. C., Rizzo C. J., Egli M., and Guengerich F. P. (2005) DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 280, 29750–29764 10.1074/jbc.M504756200 [DOI] [PubMed] [Google Scholar]
- 71. Pence M. G., Choi J.-Y., Egli M., and Guengerich F. P. (2010) Structural basis for proficient incorporation of dTTP opposite O6-methylguanine by human DNA polymerase ι. J. Biol. Chem. 285, 40666–40672 10.1074/jbc.M110.183665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Patra A., Nagy L. D., Zhang Q., Su Y., Müller L., Guengerich F. P., and Egli M. (2014) Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase η. J. Biol. Chem. 289, 16867–16882 10.1074/jbc.M114.551820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Irimia A., Eoff R. L., Guengerich F. P., and Egli M. (2009) Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J. Biol. Chem. 284, 22467–22480 10.1074/jbc.M109.003905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilson T. J., Li N. S., Lu J., Frederiksen J. K., Piccirilli J. A., and Lilley D. M. (2010) Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl. Acad. Sci. U.S.A. 107, 11751–11756 10.1073/pnas.1004255107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Elmquist C. E., Stover J. S., Wang Z., and Rizzo C. J. (2004) Site-specific synthesis and properties of oligonucleotides containing C8-deoxyguanosine adducts of the dietary mutagen IQ. J. Am. Chem. Soc. 126, 11189–11201 10.1021/ja0487022 [DOI] [PubMed] [Google Scholar]
- 76. Marquez V. E., Tseng C. K.-H., Mitsuya H., Aoki S., Kelley J. A., Ford H. Jr., Roth J. S., Broder S., Johns D. G., and Driscoll J. S. (1990) Acid-stable 2′-fluoro purine dideoxynucleosides as active agents against HIV. J. Med. Chem. 33, 978–985 10.1021/jm00165a015 [DOI] [PubMed] [Google Scholar]
- 77. Schärer O. D., and Verdine G. L. (1995) A designed inhibitor of base-excision DNA repair. J. Am. Chem. Soc. 117, 10781–10782 10.1021/ja00148a036 [DOI] [Google Scholar]
- 78. Ikeda H., Fernandez R., Wilk A., Barchi J. J. Jr., Huang X., and Marquez V. E. (1998) The effect of two antipodal fluorine-induced sugar puckers on the conformation and stability of the Dickerson-Drew dodecamer duplex [d(CGCGAATTCGCG)]2. Nucleic Acids Res. 26, 2237–2244 10.1093/nar/26.9.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.