Abstract
CREB-target gene transcription during neuronal excitation is important for many aspects of neuronal development and function, including dendrite morphogenesis. However, the signaling events that regulate cAMP response element-binding protein (CREB)-mediated gene transcription during dendritic development are not well understood. Herein we report that the CREB coactivator TORC1 (transducer of regulated CREB 1) is required for activity-dependent CREB-target gene expression and dendrite growth in developing cortical neurons. Ca2+ influx via voltage-gated calcium channels induced TORC1 dephosphorylation and translocation into the nucleus in a calcineurin-dependent manner. Nuclear accumulation of TORC1 initiated the expression of CREB-target genes, including salt-inducible kinase 1 (SIK1). In response of persistent depolarization, de novo SIK1 protein in turn promoted TORC1 phosphorylation and consequent depletion of nucleus-localized TORC1. SIK1 induction thus appears to act as a negative feedback signal that prevents persistent CREB/TORC1-dependent transcription in the face of long-lasting neuronal activity. Overexpressing wild type TORC1 promoted basal as well as activity-induced dendritic growth, whereas expressing a dominant-negative form of TORC1 or downregulating TORC1 inhibited activity-dependent dendritic growth in vitro and in vivo. Together, these results suggest that neuronal activity-dependent dendritic growth in developing cortical neurons relies on transient TORC1-mediated CREB-target gene transcription.
Keywords: activity, CREB, dendritic arborization, gene, transcription, development
Introduction
Episodes of neuronal activity during development lead to Ca2+-dependent gene expression and long-lasting changes of dendrite morphology (Rajan and Cline, 1998; Cline, 2001; Wong and Ghosh, 2002; Parrish et al., 2007). The transcription factor cAMP-response element-binding protein (CREB) plays important roles in this process (Lonze et al., 2002; Redmond et al., 2002; Wayman et al., 2006). Phosphorylation of CREB at a key regulatory site, serine133 (Ser133) has been used a marker for the activation of CREB-mediated gene transcription (Bito et al., 1996; Radhakrishnan et al., 1997; Chawla et al., 1998; Shaywitz and Greenberg, 1999; Mayr and Montminy, 2001; West et al., 2001; Deisseroth and Tsien, 2002; Kornhauser et al., 2002). However, there is evidence showing that some extracellular stimuli that cause CREB phosphorylation at Ser133 fail to induce CREB-dependent transcription (Bonni et al., 1995; Thompson et al., 1995; Bito et al., 1996; Liu and Graybiel, 1996; Mayr et al., 2001). Moreover, it has been reported that depolarization of primary cortical neurons causes long-lasting phosphorylation of CREB at Ser133 but CREB-dependent gene transcription occurs only during a short time window (Tao et al., 1998). These findings suggest that there must exist additional CREB coactivators that control the kinetics of CREB-target gene expression (Bonni et al., 1995; Tao et al., 1998; Lonze and Ginty, 2002).
Efforts to identify novel CREB regulators led to the discovery of a conserved family of coactivators called transducers of regulated CREB activity (TORCs) (Conkright et al., 2003a,b; Bittinger et al., 2004). TORC family proteins bind with the bZIP domain of CREB, independent of the phosphorylation status of CREB at Ser133, and facilitate CREB-mediated gene transcription (Bittinger et al., 2004; Screaton et al., 2004; Koo et al., 2005). TORC1 is abundantly expressed in brain and is required for maintenance of the late phase long-term potentiation in the hippocampus (Zhou et al., 2006; Kovács et al., 2007).
Although there is extensive evidence for the critical role of Ca2+ in regulating dendrite development, how cytoplasmic Ca2+ signaling propagates to the nucleus for gene transcription is not clear. In the present study, we examined the role of TORC1 in regulating activity-dependent transcription of CREB-target genes and dendrite growth of developing cortical neurons. We found that TORC1 was highly expressed in developing cortical neurons, and membrane depolarization caused TORC1 dephosphorylation and nuclear accumulation in a time-dependent manner. TORC1 was required for the expression of CREB-target genes, such as salt-inducible kinase 1 (SIK1), in response to Ca2+ influx via VGCCs and activation of calcineurin. Interestingly, Ca2+-induced SIK1 was found to phosphorylate TORC1, driving TORC1 export from the nucleus and thus arresting TORC1/CREB-mediated transcription. Functionally, overexpressing TORC1 promoted dendritic growth, whereas expressing a dominant-negative form of TORC1 or downregulating TORC1 inhibited activity-dependent dendrite growth. Together this data supports a model wherein TORC1 and SIK1 act as ON-OFF switches regulating the kinetics of CREB-mediated gene transcription and dendrite growth.
Materials and Methods
Northern blotting.
Total cellular RNA was extracted from whole rat brain at different developmental stages using TRIzol (Invitrogen). RNA (20 μg) from each sample was loaded onto a 1.5% agarose/formaldehyde gel, transferred to a nylon membrane (GE Healthcare) after electrophoresis and hybridized with a 32P-labeled cDNA probe spanning nucleotides 48–647 of the TORC1 coding sequence (NM_001047115). 18S rRNA was used as loading controls.
In situ hybridization.
In situ hybridization was performed as previously described (Ding et al., 2003). Briefly, rats were deeply anesthetized and perfused with 4% paraformaldehyde (PFA) in 0.1 m phosphate buffer saline (DEPC-PBS; pH 7.4). Whole brains were removed and placed into 4%PFA in DEPC-PBS for 12–16 h at 4°C then dehydrated in a sucrose gradient (15%, 30%, dissolved in DEPC-PBS). Brains were cut into 20 μm coronal sections with a Cryostat CM1900 at −20°C (Leica) and mounted onto RNAase-free, saline-coated slides (stored at −20°C; Fisher Scientific). PCR products of rat TORC1 (forward primer: 5′-GCACAACCAGAAGCAGGC-3′; reverse primer: 5′-CAGGACTTGGGCCTGGAAC-3′) were ligated into pGEM-T Easy vector. Plasmids were then linearized for in vitro RNA transcription of antisense and sense DIG-labeled RNA probes using a SP6/T7 transcription kit (Roche). Hybridization was performed following standard protocols; anti-DIG-AP-Fab fragments, NBT/BCIP (Boehringer Mannheim) were used for detection.
Western blotting.
Cortices were dissected from embryonic (E16) and postnatal rats (P0, P3, P7, P14, P21 and adult) and homogenized with a dounce homogenizer in lysis buffer (137 mm NaCl, 20 mm Tris, 1% NP-40, 10% glycerol, 1 mm PMSF, 10 μg/ml aprotinin, 1 μg/ml pepstatin A and 1 μg/ml leupeptin; pH 8.0) as previously described (Redmond et al., 2000). Cultured neurons were lysed in RIPA buffer containing 1 mm Na3VO4 and 50 mm NaF. Protein concentrations were determined using the Lowry method. ∼10∼20 μg of each sample was loaded into each lane and size-fractionated by electrophoresis on 9% SDS-PAGE, followed by a 2 h, 400 mA electrotransfer onto nitrocellulose membranes. Membranes were blocked with 5% no-fat milk in 0.05% Tween 20 at room temperature for 1 h and probed with rabbit anti-TORC1 (1:2000; provided by Dr. H. Takemori) (Katoh et al., 2006; Zhou et al., 2006), HRP-coupled mouse anti-GAPDH (1:5000; Santa Cruz Biotechnology), rabbit anti-phospho-CREB (1:1000; Cell Signaling Technology), rabbit anti-CREB (1:1000; Cell Signaling Technology), rabbit anti-phospho-TORC1 (1:1500; provided by H. Takemori) (Screaton et al., 2004; Katoh et al., 2006), rabbit anti-SIK1 (1:1500; provided by H. Takemori) (Katoh et al., 2006) antibodies. The secondary antibody used was goat anti-rabbit IgG coupled to HRP (1:5000; GE Healthcare). Bands were visualized by enhanced chemiluminescence (ECL) and a Western blotting detection system (GE Healthcare). A ladder of prestained proteins (17–108 kDa; RYM-TECH) was used as weight markers. Band intensities were measured with ImageQuant 5.2 Software.
Electroporation.
Neurons suspended in transfection medium (0.1 ml) were mixed with 9 μg of plasmids, transferred into a 2.0 mm electroporation cuvette, and transfected by electroporation using the Amaxa Nucleofector apparatus (Amaxa) (Gärtner et al., 2006). Wt-TORC1 or TORC1 shRNA was cotransfected with pEGFP at a ratio of 3:1 (9 μg: 3 μg). Control and transfected neurons were plated onto coverslips or wells coated with 0.1 mg/ml poly-d-lysine. After plating for 4 h, culture medium was changed for neurobasal medium containing 10% FBS.
Expression constructs.
The pEF1-EGFP construct was a gift from H. Tabata (Institute of DNA Medicine, Tokyo, Japan). cDNAs encoding wild-type TORC1 and TORC1-N44-GFP (dn-TORC1, a GFP-tagged dominant-negative form of TORC1) were amplified by PCR and inserted into pEF1 expression vectors to produce wt-TORC1 and dn-TORC1 expression constructs (Bittinger et al., 2004; Zhou et al., 2006). SIK1-GFP, S577A SIK1-GFP, and DR-defective SIK1-GFP plasmids and were gifts from Dr. H. Takemori (Katoh et al., 2004) with modifications. The RNAi sequence targets were, TORC1: 5′-CGAACAATCCGCGGAAATTTA-3′; and TORC2: 5′-TCTCTGCCCAATGTTAACCA-3′ (Kovács et al., 2007). The knock-down efficiency of the SIK1 shRNA was reported previously (Koo et al., 2005).Sense and antisense shRNA were synthesized by Shanghai Genechem. A scrambled oligonucleotide (5′-TTCTCCGAACGTGTCACGT-3′) was used as a negative control in all knock-down experiments.
CRE-luciferase reporter assay.
In pharmacological experiments, dissociated cortical neurons were cotransfected before plating with pTAL-Cre firefly luciferase plasmid (Clontech), the Renilla luciferase expressing RL-SV40 vector (Promega) was used as an internal control. Neurons were stimulating with KCl (50 mm), KCl plus 25 μm nifedipine (Sigma-Aldrich) or KCl plus 5 μm FK506 (Sigma-Aldrich) for 24 h at DIV3. In transgenic experiments, TORC1 shRNA, dn-TORC1, wt-TORC, SIK1 shRNA, wt-SIK1, TORC1 shRNA plus SIK1 shRNA or empty vectors were cotransfected into cultured neurons with CRE-luciferase reporter vectors using a previously described calcium transfection protocol at DIV3 (Xia et al., 1996). Forty-eight hours after transfection, neurons were stimulated with KCl (50 mm) for 24 h. Luciferase assays were performed using a Dual Luciferase Assay kit (Promega) according to manufacturer's instructions.
Transfection, stimulation and immunostaining of primary neuronal cultures.
Cortical neuron cultures were prepared from E18 rat embryos as previously described (Redmond et al., 2002). For immunocytochemical analysis of endogenous TORC1 localization, DIV5 neurons were stimulated with 50 mm KCl for 15 min, 30 min, 60 min, 120 min, or 360 min. To assess the effects of pharmacological treatments on TORC1 translocation, nifedipine (25 μm), cycloheximide (0.1 mg/ml) (CHX; Sigma-Aldrich), or FK506 (5 μm) was added to the culture medium 15 min before the addition of KCl or glutamate (100 μm). To study depolarization-induced TORC1 translocation in cortical neurons, neurons were transfected at DIV4 and, 48 h later, were stimulated with 50 mm KCl for 60 min or 360 min. After stimulation, neurons were washed three times with PBS and fixed with 4% PFA in PBS at 4°C for 30 min. Fixed neurons were washed, incubated with 1% Triton X-100 in PBS for 5 min, and blocked with 5% FBS in PBS for 1 h at room temperature. Neurons were probed with the TORC1 antibody (1:1000) at 4°C overnight and washed three to six times with PBS. Neurons were then incubated with Alexa-488 or Alexta-546 conjugated goat anti-rabbit secondary antibody at 4°C for 1 h (1:2000; Invitrogen) at room temperature for 1 h and washed three to six times with PBS. All antibodies were diluted with PBS containing 5% FBS. Images were acquired with fluorescent confocal microscopes (LSM510 and Axiovert 200M, Zeiss) under 60×oil-immersion objectives with a numerical aperture of 2.0. Hoechst 33342 was used to validate the morphological identification of nuclei. The immunofluorescence intensity per unit area in the nucleus versus cytoplasm (N/C fluorescence ratio) was calculated as previously described (Zhou et al., 2006) and quantified with Image Pro Plus software (Media Cybernetics) (Dzeja et al., 2002).
Quantification of dendritic growth of cultured neurons.
After stimulation, cultured neurons were fixed and immunostained for EGFP according to established methods. Images were acquired with fluorescent microscopes (LSM510, Axiovert 200M, Zeiss) under a 40× oil-immersion objective with a numerical aperture of 0.7. To assess the effects of TORC1 on dendritic growth, dissociated neurons were transfected before plating and stimulated with or without 50 mm KCl at DIV3. After 24 h, neurons were fixed in 4% PFA in PBS and stained. At least 25 neurons per group were imaged. Reconstructions of dendritic arbors and quantification of dendrite length were determined using Neurolucida software. Image acquisition and data analysis were performed in a double-blind manner.
Virus vectors and packaging of virions.
The Semliki Forest Virus (SFV)-based vector and helper vector were kindly provided by Dr. Kenneth Lundstrom (Hoffmann-La Roche, Basel, Switzerland). Construction of pSFV (pd)-EGFP, and pSFV (pd)-dn-TORC1-EGFP vectors was described previously (Zhou et al., 2006). Briefly, a DNA fragment encoding EGFP (Clontech) was amplified by PCR and digested and inserted into pSFV (pd) vector with XhoI and SpeI restriction enzyme sites. The DNA fragment encoding full-length rat torc1 and 44 aa in the N termini of rat TORC1 were amplified by PCR and inserted into pSFV9 (pd)-EGFP vector and inserted into pSFV (pd)-EGFP vector to produce pSFV (pd) –TORC1-EGFP and pSFV (pd)-dn-TORC1-EGFP. Virus packaging was performed as previously described (Chen et al., 2007).
Semiquantitative reverse transcription-PCR analysis.
To assess activity-dependent sik1 transcription, cultures of DIV5 cortical neurons were stimulated with 50 mm KCl for 15, 30, 60, 120, or 360 min. To examine theeffects of pharmacological treatments on sik1 transcription, DIV5 cortical neurons were treated with KCl (50 mm), KCl plus FK506 (5 μm), KCl plus nifedipine (20 μm), KCl plus U0126 (inhibitor of MEK1/2, 40 μm), glutamate (100 μm), forskolin (25 μm), BDNF (25 ng/ml) (Sigma and PeproTech) for 6 h. To examine TORC1-dependent transcription of sik1, DIV 5 cortical neurons were transfected with activated virions (containing pSFV-GFP, pSFV-dn-TORC1-GFP or pSFV-wt-TORC1-GFP) for 9 h and stimulated with 50 mm KCl for 6 h; mRNA was then extracted and reverse transcribed for RT-PCR analysis. In knock-down experiments, dissociated cortical neurons were transfected with TORC1, TORC2 or SIK1 or scramble shRNA plasmids and mRNA was collected for reverse transcription (RT)-PCR analysis 3 d later. RNA (1 μg) was reverse transcribed and 1/20 of the RT products were used for PCR amplification. The primers and reaction conditions were listed below: TORC1 (forward primer: 5′-GCACAACCAGAAGCAGGC-3′; reverse primer: 5′-CAGGACTTGGGCCTGGAAC-3′). Cycling conditions were as follows: 95°C for 5 min; 31 cycles of 30 s at 94°C, 30 s at 56°C, then 30 s at 72°C; then 72°C for 7 min. Products were 600 bp. TORC2 (forward primer 5′-AGGTGATGATGGACATCGGCTCCA-3′; reverse primer: 5′-GTGGTACTGTTGCCCCCACT-3′). Cycling conditions were as follows: 95°C for 5 min; 36 cycles of 30 s at 94°C, 30 s at 56°C, then 40 s at 72°C; then 72°C for 7 min. Products were 799 bp. SIK1 (forward primer: 5′-AGTCAGATTGGCCTCACCCTTC-3′; reverse primer: 5′-TCAGCCCTTGAGTGAGAGACGT-3′). Cycling conditions were as follows: 95°C for 5 min; 31 cycles of 30 s at 94°C, 30 s at 56°C, then 15 s at 72°C; then 72°C for 7 min. Products were 146 bp. SIK2 (forward primer: 5′-TGGTGGAACGTCTGAAATCACA-3′; reverse primer: 5′-TCAGGTTCGGTGGATGCAA-3′). Cycling conditions were as follows: 95°C for 5 min; 36 cycles of 30 s at 94°C, 30 s at 56°C, then 15 s at 72°C; then 72°C for 7 min. Products were 150 bp. SIK3 (forward primer: 5′-CAGAGTTTGCCCAGCTCATCAA-3′; reverse primer: 5′-TAGGCTAACAGCATCCCCTTGG-3′). Cycling conditions were as follows: 95°C for 5 min; 38 cycles of 30 s at 94°C, 15 s at 56°C, then 56 s at 72°C; then 72°C for 7 min. Products were 128 bp. GAPDH (forward primer: 5′-ATGCCCCCATGTTTGTGATGG-3′; reverse primer: 5′-TGGTCATGAGCCCTTCCACGA-3′). Cycling conditions were as follows: 95°C for 5 min; 28 cycles of 30 s at 94°C, 30 s at 56°C, then 15 s at 72°C; then 72°C for 7 min. Products were 140 bp.
In utero electroporation.
In utero electroporation was performed as described previously (Saito and Nakatsuji, 2001). Briefly, uterine horns of pregnant rats were exposed at E16 and 1–2 μl of plasmid solution was injected into the lateral ventricle of the embryonic brain with a fine glass micropipette. Embryos were then clamped between 5 mm diameter tweezers-type disc electrodes (CUY650–5; Tokiwa Science) and, using an electroporator (ECM830; BTX), were given five 50 ms, 40 V electrical pulses at an interval of 950 ms. Uterine horns were then placed back into the abdominal cavity to allow the embryos to continue normal development.
Analysis of dendrite length in vivo.
Transfected P7 pups were deeply anesthetized and perfused with 4% paraformaldehyde in 0.1 m PBS, pH 7.4,. Whole brains were then removed and placed into 4% paraformaldehyde in 0.1 m PBS for 12 h and dehydrated in a sucrose gradient (15%, 30%, dissolved with PBS). Brains were cut into 60 μm coronal slices with a cryostat (CM1900, Leica) at −20°C followed by anti-GFP immunostaining. Confocal images (50 μm Z stacks) of isolated cells were acquired and three-dimensional (3D) reconstructions were made with Neurolucida software to trace dendritic arbors and measure total dendrite length.
Statistical analysis.
Statistical results are presented as the mean ± SEM. The significance of differences was determined by Student's t test compared with control group using Prism 4.0 software.
Results
Developmental expression profile of TORC1 in the rat cortex
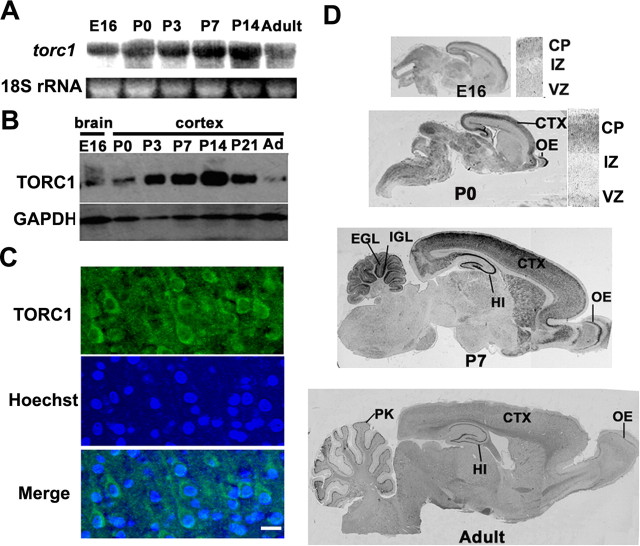
To determine the expression dynamics of torc1 in the rat brain, we performed Northern blot analysis at different stages. In total brain lysates, the expression of torc1 mRNA increased after birth (Fig. 1 A), peaked at P14, and decreased slightly in adulthood. In situ hybridization of rat brain sections with an antisense torc1 probe further revealed that torc1 mRNA was expressed in the forebrain, midbrain and hindbrain. Within the forebrain, torc1 was highly expressed in postmitotic cells of the cortical plate and expressed at low level in proliferating neuroblasts in the ventricular zone (Fig. 1 D). In the cortex, torc1 mRNA increased during the period of dendritic sprouting (Fig. 1 D). These hybridization signals were specific because no signal was detectable using a sense probe (data not shown). Using a polyclonal antibody specific to TORC1 (Zhou et al., 2006), we observed a dramatic increase of TORC1 protein in the cortex between P3 and P14 (Fig. 1 B). TORC1 protein distributed throughout the cytoplasm of the soma and dendrites of developing cortical neurons (Fig. 1 C). Together, our results indicate that both TORC1 mRNA and protein are highly expressed in developing cortical neurons and its expression peaks during the timing of dendrite development.
Figure 1.
Expression of torc1 mRNA and TORC1 protein during brain development. A, Northern blot of mRNA isolated from whole rat brains at different developmental stages hybridized with a torc1 probe. 18S rRNA was shown as RNA loading control. B, Western blot of protein extracts from rat cortex at indicated stages probed with anti-TORC1 antibody. Equivalent protein loading was confirmed by stripping and reprobing the same blots with a GAPDH antibody. C, Immunohistochemical staining of TORC1 in the layer II/II neuron of P7 rat cortex (top), using Hoechst 33324 as counterstaining (middle), the bottom shows the merge of both channels. Scale bar, 20 μm. D, In situ hybridization analysis of torc1 expression in the rat nervous system at indicated ages, presented as sagittal sections. VZ, Ventricular zone; IZ, intermediate zone; CP, cortical plate; EGL, external granular layer; IGL, internal granular layer; HI, hippocampus; CTX, neocortex; OE, olfactory epithelium; PK, Purkinje cell.
Ca2+-dependent translocation of TORC1 in developing cortical neurons
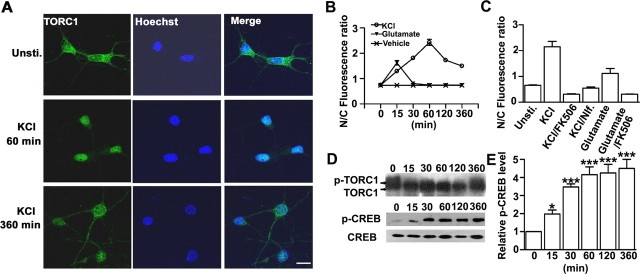
After culturing primary cortical neurons for 4 or 5 d (DIV4 or 5), we examined TORC1 expression and localization in response to neuronal activity. We examined whether membrane depolarization and glutamate stimulation had differential effects on TORC1 subcellular location by treating cortical neurons with 50 mm KCl or 100 μm glutamate. Under resting conditions, TORC1 protein was highly enriched in the cytoplasm, with only a relatively small amount localized in the nucleus. Depolarizing the cells with 50 mm KCl significantly increased nuclear accumulation of TORC1 in a time-dependent manner (Fig. 2 A), as quantified by the ratio of fluorescence intensity in the nucleus versus the cytoplasm (Fig. 2 B). Nuclear TORC1 increased markedly after 15 min, peaked at 60 min after KCl stimulation, and decreased to the resting level after 6 h. Application of either nifedipine, an L-type voltage-gated channel blocker, or the calcineurin inhibitor FK506 completely blocked KCl-induced nuclear translocation of TORC1 (Fig. 2 C). Interestingly, exposure to glutamate induced only a transient and less robust nuclear accumulation of TORC1, and this could also be blocked with FK506 (Fig. 2 C).
Figure 2.
Translocation of TORC1 in cultured cortical neurons in response to KCl stimulation. A, Representative image of TORC1 staining in neurons treated with KCl for the indicated time point (left), using Hoechst 33324 as counterstaining (middle), and the merge of both channels (right). Scale bar, 20 μm. B, Quantification analysis of ratio of nucleus to cytoplasm (N/C) TORC1 in neurons treated with KCl, glutamate, or vehicle control based on fluorescent signal. C, Quantitative analysis of TORC1 nuclear accumulation in neurons after treated with KCl for 60 min or glutamate for 15 min. nifedipine or FK506 are added for 15 min before KCl or glutamate stimulation. For each groups, >50 neurons were measured from at least three independent experiments (B, C). D, Western blot analysis of KCl-induced dephosphorylation of TORC1 and CREB phosphorylation for the indicated time point (minutes). Blots were probed with TORC1, phospho-CREB (p-CREB), or CREB antibodies sequentially. E, Normalization of phospho-CREB level at indicated time points with unstimulated control. Band intensities were quantified and averaged from six independent experiments. Error bars indicate SEM; *p < 0.05, **p < 0.01, ***p < 0.001, significant difference compared with the unstimulated control group.
Previous studies have shown that increased calcium and cAMP levels induce TORC2 dephosphorylation and subsequent transport to the nucleus (Screaton et al., 2004; Koo et al., 2005). We found that TORC1 was highly phosphorylated under basal conditions and membrane depolarization induced TORC1 dephosphorylation in a time-dependent manner (Fig. 2 D), correlating with the time course of TORC1 nuclear accumulation (Fig. 2 B). In contrast, phosphorylation of CREB at Ser133 was induced within 15 min of depolarization and maintained for at least 360 min in developing cortical neurons (Fig. 2 D,E). Together, our results suggest that membrane depolarization triggers TORC1 dephosphorylation and initiates TORC1 translocation to nucleus in a time-dependent manner, thus mirroring the previously established kinetics of KCl-induced CREB-target gene transcription (Tao et al., 1998; Kingsbury et al., 2007).
De novo TORC1-induced SIK1 phosphorylates TORC1 and is required for TORC1 nuclear export
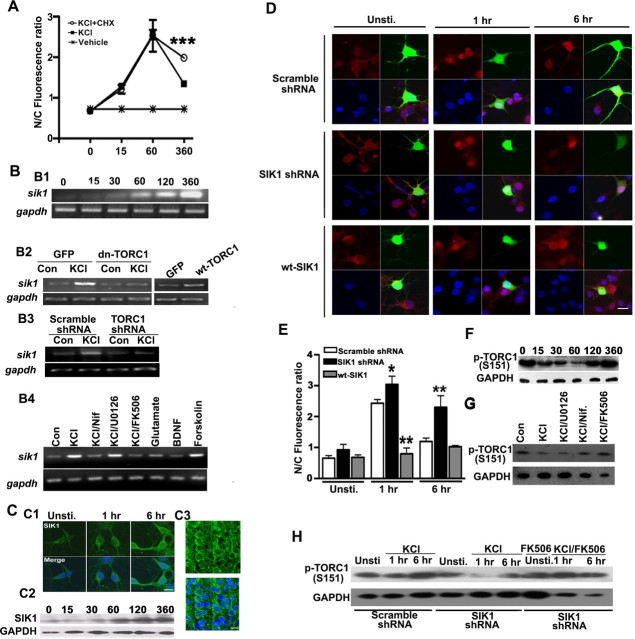
We next explored the factors that drive TORC1 phosphorylation and nuclear export. We first examined whether nuclear export was dependent on activity-dependent gene transcription and protein synthesis. Treatment of neurons with the protein synthesis inhibitor cycloheximide (CHX) in the presence of high KCl did not alter TORC1 nuclear accumulation over the first 60 min but blocked the subsequent TORC1 export (Fig. 3 A). This finding suggests that the TORC1 import-export cycle is controlled by activity-dependent protein synthesis.
Figure 3.
Effects of SIK1 on TORC1 translocation and phosphorylation. A, Quantification of the effects of cycloheximide (CHX) on KCl-stimulated TORC1 translocation for indicated time (min). CHX was added for 15 min before KCl stimulation. Error bars indicate SEM. ***p < 0.001; compared with the neurons treated with KCl. B, sik1 is upregulated by KCl stimulation. B1, RT-PCR analysis of sik1 mRNA from neurons stimulated with KCl as indicated time point (minutes). B2, Effects of wild-type (wt-TORC1) or dominant-negative TORC1 (dn-TORC1) constructs on basal and activity-induced expression of sik1 mRNA following stimulated with KCl for 6 h. B3, The effect of knocking down TORC1 on sik1 mRNA expression. Dissociated primary cortical neuronss were transfected with either scramble shRNA or TORC1 shRNA and, 3 d later, stimulated with KCl for 6 h. B4, Effects of KCl, BDNF, glutamate on sik1 transcription after stimulated for 6 h. FK506, nifedipine or U0126 (inhibitor of MEK1/2) was added for 15 min before KCl stimulation. GAPDH mRNA was used as RT-PCR control. C, SIK1 protein was upregulated after KCl stimulation in cortical neurons. C1, Immunostaining of SIK1 protein in neurons treated with KCl for indicated time (top), merged with Hoechst 33324 counterstaining (bottom). Scale bar, 20 μm. C2, SIK1 protein was induced by membrane depolarization at indicated time point (minutes). Equivalent protein loading was confirmed by probing the same blot with a GAPDH antibody. C3, Immunohistochemical analysis of SIK1 subcellular distribution in layer II/III neuons of P7 rat cortex (top), merged with Hoechst 33324 counterstaining (bottom). Scale bar, 20 μm. D, Immunostaining of TORC1 (red) in representative cortical neurons stimulated with KCl for the indicated time. DIV3 cortical neurons were transfected with indicated plasmids: EGFP plus scramble shRNA, EGFP plus SIK1 shRNA and SIK1-GFP, and neurons were stimulated with KCl for indicated time 48 h later. Scale bar, 20 μm. E, Quantification of (N/C) TORC1 ratio in transfected neurons. Error bars indicate SEM. *p < 0.05; **p < 0.01; compared with the scramble shRNA-transfected control group. F, Time course of KCl-induced dephosphorylation TORC1 at Ser 151. DIV5 cortical neurons were stimulated with KCl for indicated time (minutes). G, TORC1 phosphorylation (S151) after pharmacological treatments. DIV5 cortical neurons were stimulated with KCl for 1 h; FK506, nifedipine or U0126 were added for 15 min before KCl stimulation. H, KCl-stimulated dephosphorylation of TORC1 at Ser151 after knocking down SIK1. Dissociated cortical neurons were transfected with SIK1 shRNA and, 3 d later, stimulated with 50 mm KCl or KCl plus 5 μm FK506 for indicated time. Equivalent protein loading was confirmed by probing the same blot with a GAPDH antibody.
Salt-inducible kinases phosphorylate TORCs, triggering nuclear export and sequestration in the cytoplasm (Screaton et al., 2004; Koo et al., 2005; Katoh et al., 2006). We found that all of salt-inducible kinase isoforms (sik1, sik2 and sik3) were expressed at low level in developing cortical neurons. Interestingly, KCl treatment induced the upregulation of sik1, but not sik2 and sik3 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Using a polyclonal antibody specific to SIK1 (Katoh et al., 2006), we found SIK1 was mainly located in the cytoplasm both in vitro and in vivo and could be upregulated by membrane depolarization (Fig. 3 C). Expressing a dominant-negative form of TORC1 (dn-TORC1) or knocking down TORC1 with shRNA blocked KCl-induced sik1 transcription (Fig. 3 B). This indicates that activity-induced sik1 transcription is TORC1-dependent. Furthermore, overexpressing the wild type form of TORC1 in the absence of membrane depolarization was sufficient to increase sik1 expression (Fig. 3 B).
To test whether SIK1 expression influence the subcellular localization of TORC1, we manipulated SIK1 protein levels either by knocking down or overexpressing SIK1 in cultured cortical neurons. Knocking down SIK1 with a SIK1-specific shRNA (Koo et al., 2005) increased KCl-induced nuclear TORC1 at 6 h, whereas overexpressing wt-SIK1 inhibited KCl-induced nuclear accumulation of TORC1 (Fig. 3 D,E).
Previous reports suggest that SIK proteins phosphorylate TORC1 and TORC2 at Ser151 and Ser171, respectively. Dephosphorylation of TORC2 at Ser171 is required for nuclear import of TORC2 (Screaton et al., 2004; Koo et al., 2005) and the phosphorylation-defective form of TORC1 is not exported from the nucleus (Altarejos et al., 2008). Using an antibody that specifically recognizes Ser151-phospyorylated TORC1 (p-TORC1), we found that membrane depolarization induced time-dependent TORC1 dephosphorylation (Fig. 3 F), mirroring the kinetics of TORC1 nuclear export. In addition, we found that dephosphorylation of Ser-151 was dependent on calcium influx via VGCCs and calcineurin activity. Application of nifedipine or FK506 completely blocked the KCl-dependent TORC1 dephosphorylation (Fig. 3 G).
To further examine whether TORC1-dependent SIK1 upregulation triggers Ser151 phosphorylation in response to membrane depolarization, we examined p-TORC1 level after knocking down SIK1. We found that knocking down of SIK1 significantly inhibited TORC1 phosphorylation, an effect that was attenuated by FK506 (Fig. 3 H). Together, these results indicate that SIK1 mRNA and protein are upregulated by neuronal activity in a TORC1-dependent manner; SIK1 subsequently triggers TORC1 phosphorylation and nuclear export.
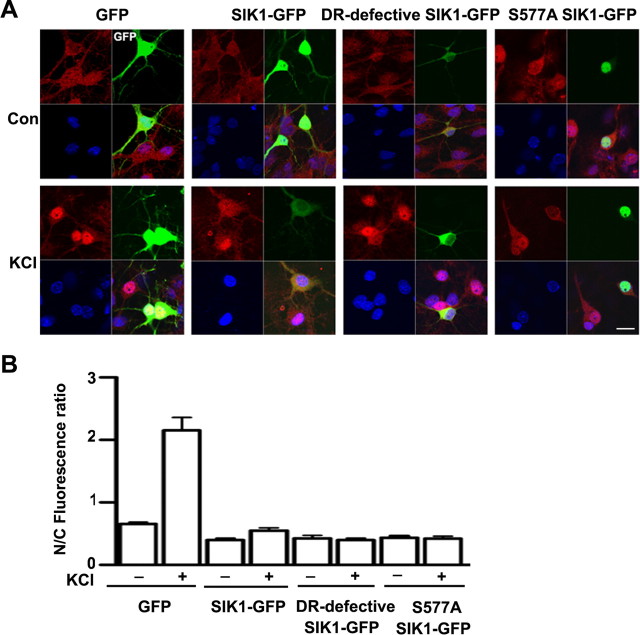
SIK1 was upregulated in response to membrane depolarization and was enriched, but not exclusively in the cytoplasm, whereas TORC1 shuttled between cytoplasm and nucleus depending on stimulation conditions. So we further asked whether SIK1 drives TORC1 export from the nucleus or prevents TORC1 from entering the nucleus. We transfected developing cortical neurons with wild-type and two mutant forms of SIK1 with GFP tag to examine TORC1 localization under basal and KCl stimulated conditions. The nuclear retention form of SIK1 (S577A SIK1-GFP), and the cytoplasmic retention form of SIK1 (DR-defective SIK1-GFP, ▵586–612) were constructed as previous reported (Katoh et al., 2004). As shown in Figure 4 A,S577A SIK1-GFP was restricted to the nucleus, whereas DR-defective SIK1-GFP was exclusively in the cytoplasm in cultured neurons. In response to KCl stimulation, both SIK1 mutant constructs inhibited TORC1 nuclear accumulation (Fig. 4 A,B), indicating that SIK1 inhibit nuclear accumulation of TORC1 by modulating either cytoplasmic- or nuclear-localized TORC1. In Y1 cells, it was reported that SIK1 phosphorylate TORC1 in both the nucleus and cytoplasm, thus inhibited CRE-mediated gene transcription (Katoh et al., 2004). Our results in cultured neurons also revealed that both cytoplasm and nuclear SIK1 inhibit activity-induced TORC1 nuclear accumulation through phosphorylating cytoplasmic or nuclear TORC1.
Figure 4.
Both cytoplasmic and nuclear SIK1 can inhibit activity-induced TORC1 nuclear accumulation. A, Cortical neurons were transfected with GFP, SIK1-GFP, DR-defective SIK1-GFP or S577A SIK1-GFP constructs at DIV4 and, 24 h later, were stimulated for 1 h with 50 mm KCl; Scale bar, 20 μm. B, The ratio of nuclear to cytoplasmic (N/C) TORC1 protein in neurons expressing the indicated constructs respectively; Error bars indicate mean ± SD. N = 20–30 neurons.
TORC1 is required for activity-induced CREB-target gene expression
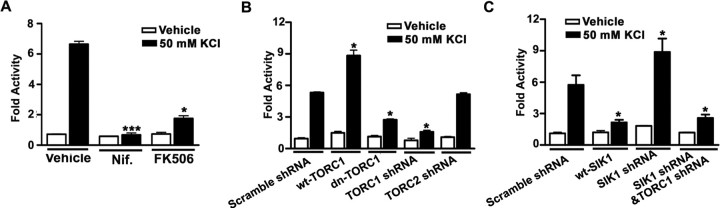
To determine whether TORC1 is required for CREB-target gene transcription, we transfected dissociated cortical neurons with a CRE-luciferase reporter to monitor CREB-target gene expression. We found that 50 mm KCl increased CRE-luciferase gene transcription in cultured cortical neurons (Fig. 5 A), which was blocked by either nifedipine or FK506 treatment (Fig. 5 A). Disrupting the functional interaction of TORC1 with CREB by expressing dn-TORC1 (Bittinger et al., 2004; Zhou et al., 2006) or knocking down TORC1 with shRNA blocked KCl-induce lucifease upregulation, whereas TORC2 shRNA had no effect (Fig. 5 B). Overexpression of wt-TORC1 increased both basal and KCl-induced luciferase expression (Fig. 5 B). Interestingly, overexpression of SIK1 inhibited, whereas knock-down of SIK1 promoted, activity-induced CRE-gene expression (Fig. 5 C). Thus, TORC1 is required for Ca2+-dependent CRE-activity, and SIK1, a CREB target-gene, may act as a negative feedback signal that arrests activity-dependent transcription of CREB-target genes.
Figure 5.
TORC1 is required for activity-induced CREB-target genes expression. A, CRE-luciferase activity in transfected neurons treated with KCl for 24 h in combination with inhibitors. B, Statistical analysis of CRE-luciferase activity cotransfected with either Scramble shRNA, wt-TORC1, dn-TORC1, TORC1 shRNA, or TORC2 shRNA vectors and stimulated with KCl for 24 h. C, Statistical analysis of CRE-luciferase activity cotransfected with either Scramble shRNA, wt-SIK1, SIK1 shRNA, or SIK1 shRNA plus TORC1 shRNA vectors and stimulated with KCl for 24 h. Results are presented as the mean ± SEM. *p < 0.05; **p < 0.01; compared with the scramble shRNA-transfected group. Luciferase activity from three independent experiments, four example wells were taken from each group.
TORC1 is required for dendritic growth of developing cortical neurons in vitro
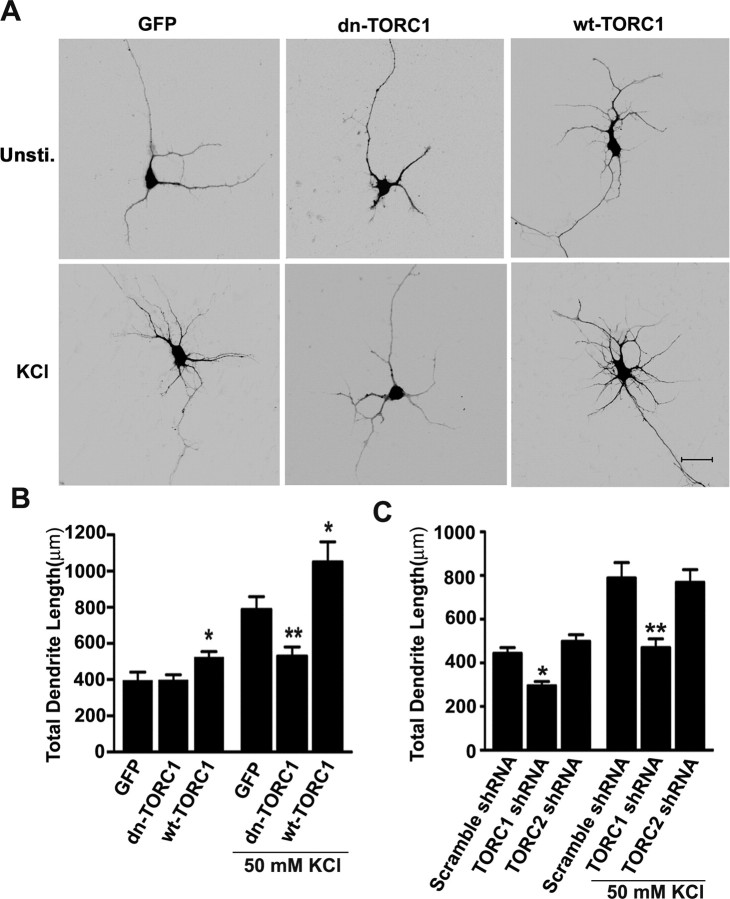
Ca2+-mediated gene expression is involved in activity-dependent regulation of dendritic development (Redmond et al., 2002; Wayman et al., 2006). To explore the role of TORC1 in Ca2+-induced dendrite outgrowth, EGFP-expressing cortical neurons prepared from E18 rats were cultured for 3 d before stimulating with 50 mm KCl for 24 h, a procedure previously designed to study the effects of CREB on dendrite arborization (Redmond et al., 2002). KCl stimulation promoted dendritic growth of cortical neurons (Fig. 6 A,B). However, activity-induced dendrite growth was significantly inhibited by expression of dn-TORC1 (Fig. 6 A,B). Furthermore, knocking down of TORC1, but not TORC2, inhibited both basal and activity-induced dendritic growth (Fig. 6 C). In contrast, overexpression of wt-TORC1 promoted both basal and activity-dependent dendritic growth (Fig. 6 A,B). These results indicate that TORC1 is required for the dendrite development in vitro.
Figure 6.
TORC1 is required for activity-induced dendrite arborization in vitro. A, Representative images of DIV4 cortical neurons transfected at DIV0 with EGFP plus empty vector, EGFP plus dn-TORC1 or EGFP plus wt-TORC1 construct, and stimulated with or without 50 mm KCl for 24 h. Scale bar, 25 μm. B, Effects of these treatments on total dendritic length per neuron quantified using Neurolucida software. For each groups, >25 neurons were measured from at least three independent experiments. Results are presented as the mean ± SEM. *p < 0.05; **p < 0.01 compared with the empty vector-transfected group. C, Effect of TORC1 shRNA or TORC2 shRNA on KCl-induced dendrite growth. For each groups, >25 neurons were measured from at least three independent experiments. Results are presented as the mean ± SEM. *p < 0.05; **p < 0.01; compared with the scramble shRNA-transfected group.
TORC1 is required for dendritic growth of developing cortical neurons
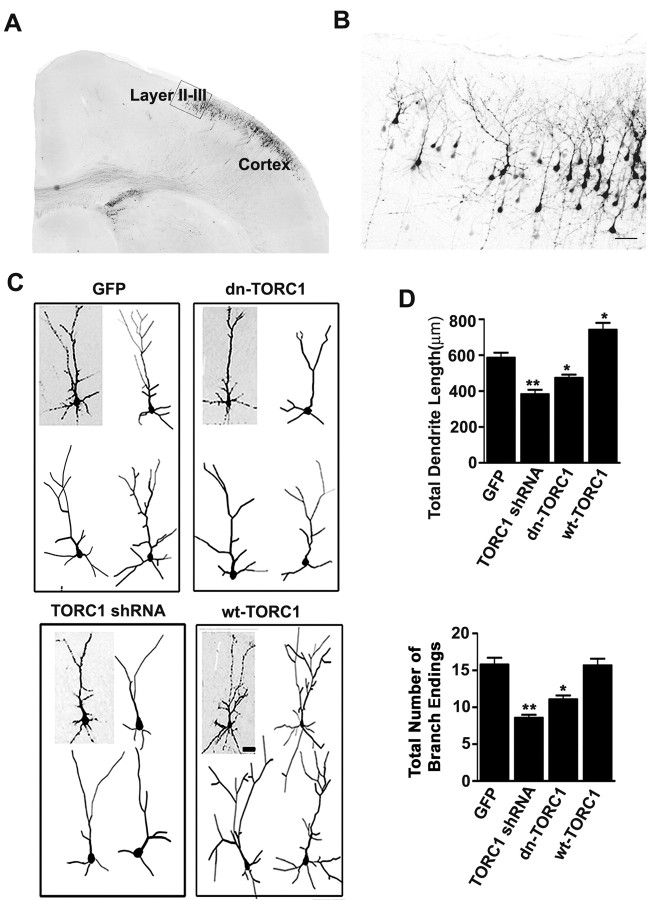
To examine whether TORC1 is involved in dendritic growth in vivo, we took advantage of the in utero electroporation technique to deliver expression constructs into rat cortical neurons (Saito and Nakatsuji, 2001). Control vectors, TORC1 shRNA, dn-TORC1, or wt-TORC1 expression vectors were injected into the lateral ventricle of E16 brains together with EGFP, then electroporated into a subpopulation of neuronal progenitor cells. At P7, TORC1 shRNA significantly decreased the level of endogenous TORC1 protein and wt-TORC1 significantly increased protein levels in expressing neurons (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). EGFP-positive neurons derived from transfected progenitor cells were mostly in layers II/III of the P7 somatosensory cortex (Fig. 7 A,B). Representative 3D reconstructions of transfected layer II/III neurons are shown in Figure 7 C. Compared with empty vector-transfected neurons, neurons expressing TORC1 shRNA or dn-TORC1 had significantly less mature dendritic arbors, as assessed by decreased total dendritic length and number of branches. In contrast, overexpressing wt-TORC1 significantly increased dendritic growth and branching (Fig. 7 C,D). Together these findings show that TORC1 is an important regulator of dendrite development both in vitro and in vivo.
Figure 7.
TORC1 is required for dendrite growth in vivo. A, Representative EGFP immunostaining of coronal section from somatosensory cortex of P7 rat brain. B, Representative EGFP-positive neurons in layer II/III cortex were selected for imaging and quantification. Scale bar: 100 μm. C, Representative 3D projections of confocal stacks from layer II/III EGFP-positive neurons in P7 cortices cotransfected with EGFP and the constructs indicated. In each example, one neuron is illustrated (gray background) along with 3D Neurolucida tracings of representative neurons (white background). Scale bar, 50 μm. D, Quantitative analysis of dendrite length and dendrite terminals measured from Neurolucida tracings. At least 25 neurons were examined in each group from at least three independent experiments. Results are presented as the mean ± SEM. *p < 0.05; **p < 0.01; compared with control group.
Discussion
In the present study, we examined the mechanism directing the nuclear-cytoplasmic shuttling of the CREB coactivator TORC1 in developing neurons in response to neuronal activity. We also uncovered the role of TORC1 in orchestrating the kinetics of CREB-mediated gene expression and in promoting dendrite growth in developing cortical neurons. The principal findings of this study are fivefold: (1) TORC1 mRNA and protein are enriched in developing cortical neurons, with expression peaking during the period of dendritic sprouting; (2) Ca2+ influx via VGCCs, but not glutamate receptors, regulates TORC1 dephosphorylation and nuclear accumulation in a time-dependent manner; (3) TORC1 is required for Ca2+-dependent CREB-target gene transcription, including sik1 transcription, in developing cortical neurons; (4) SIK1 is upregulated in response to membrane depolarization and sequesters TORC1 in the cytoplasm in a phosphorylation-dependent manner, thus arresting CREB-dependent transcription; (5) TORC1 is required for dendritic growth of developing cortical neurons.
Previous studies suggest that the phosphorylation site (Ser133) of CREB is a convergence point for multiple kinases and acts as a molecular switch for controlling gene activation kinetics (Bito et al., 1996; Liu and Graybiel, 1996; Shaywitz and Greenberg, 1999; Wu et al., 2001). Extracellular signals that cause influxes of Ca2+, like membrane depolarization, neurotransmitters and growth factors, activate intracellular kinases such as CaM kinase, cAMP/PKA and MAP kinase which, in turn, phosphorylate CREB at ser133 (Sheng et al., 1991; Bonni et al., 1995; Enslen et al., 1995; Shaywitz and Greenberg, 1999). However, not all stimuli that trigger Ser-133 phosphorylation provoke canonical CREB-dependent transcription (Bonni et al., 1995; Tao et al., 1998; Lonze and Ginty, 2002). For example, in cultured developing cortical neurons only membrane depolarization, but not glutamate, can effectively induce the expression of the CREB-target gene bdnf (Ghosh et al., 1994; West et al., 2001). The mechanism underlying the difference in CREB-target gene expression in response to KCl and glutamate in developing cortical neurons was unknown. In the current study, we found that KCl, but not glutamate, induces robust TORC1 nuclear translocation on a time course that correlates well with the kinetics of CREB-mediated gene transcription in response to KCl (Tao et al., 1998). Thus, phosphorylation of CREB at Ser133 is insufficient to drive CRE-gene activation in response to glutamate possibly because of the unresponsiveness of TORC1 to glutamate.
Both the effects of membrane depolarization-induced CREB-target gene expression and TORC1 nuclear accumulation are transient. This led us to examine the temporal relationship of CREB phosphorylation and TORC1 dephosphorylation. In developing cortical neurons, TORC1 was dephosphorylated and translocated to the nucleus within 1 h in response to KCl stimulation. However, TORC1 was then phosphorylated and shuttled back to the cytoplasm within 6 h, despite the persistent membrane depolarization and phosphorylation of CREB. Treatment with a protein synthesis inhibitor attenuated TORC1 export, suggesting the existence of a negative regulatory mechanism depending on de novo protein synthesis. Expression of SIK1, but not SIK2 or SIK3 is activity-dependent. Interestingly, activity-dependent expression of SIK1 is required for TORC1 phosphorylation and depletion of TORC1 from the nucleus. It seems that both cytoplasm and nuclear SIK1 could inhibit activity-induced TORC1 nuclear accumulation through phosphorylating TORC1. Although we found that the expression of SIK1 was regulated by neuronal activity, how the activity and subcellular location of SIK1 are regulated by neuronal activity is unknown.
Together, our results reveal a negative feedback loop involving TORC1 subcellular trafficking as means to control the kinetics of CRE-gene activation in response to neuronal activity. We used a CRE-reporter gene assay to examine the functional consequence of Ca2+-dependent TORC1 nuclear accumulation, and found that TORC1 was required for KCl-induced, CREB-mediated gene transcription in developing cortical neurons. Activity-induced CREB-target gene transcription has been implicated in a wide range of processes, from neuronal development and synaptic plasticity to disease conditions (Shaywitz and Greenberg, 1999; Lonze et al., 2002). Here we have shown that TORC1 is enriched in developing cortical neurons with levels peaking during the period of dendritic sprouting. In cultured cortical neurons, overexpression of TORC1 promotes basal and activity-dependent dendritic growth whereas arresting TORC1 function blocks activity-induced dendrite development. Interestingly, a previous study showed that new gene transcription and protein synthesis within 6 h after KCl stimulation is sufficient for activity-induced dendritic growth (Redmond et al., 2002), again correlating well with the dynamics of KCl-induced TORC1 nuclear accumulation. Our results suggest that TORC1 may control the dynamics of CRE-target gene transcription, and act as a critical molecular switch in controlling dendritic development of cortical neurons.
Footnotes
This work was supported by grants from the National Basic Research Program of China (2006CB806600), the Key State Research Program of China (2006CB943900), the Innovative Research Group of National Natural Science Foundation of China (30721004), and the Chinese Academy of Sciences (KSCX2-YW-R-099) to Z.-Q.X., and National Natural Science Foundation of China (30800318) to Y. Z.
References
- Altarejos JY, Goebel N, Conkright MD, Inoue H, Xie J, Arias CM, Sawchenko PE, Montminy M. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat Med. 2008;14:1112–1117. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Bonni A, Ginty DD, Dudek H, Greenberg ME. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- Chen Q, He S, Hu XL, Yu J, Zhou Y, Zheng J, Zhang S, Zhang C, Duan WH, Xiong ZQ. Differential roles of NR2A- and NR2B-containing NMDA receptors in activity-dependent brain-derived neurotrophic factor gene regulation and limbic epileptogenesis. J Neurosci. 2007;27:542–552. doi: 10.1523/JNEUROSCI.3607-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline HT. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 2001;11:118–126. doi: 10.1016/s0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Guzmán E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003a;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003b;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Tsien RW. Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron. 2002;34:179–182. doi: 10.1016/s0896-6273(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- Dzeja PP, Bortolon R, Perez-Terzic C, Holmuhamedov EL, Terzic A. Energetic communication between mitochondria and nucleus directed by catalyzed phosphotransfer. Proc Natl Acad Sci U S A. 2002;99:10156–10161. doi: 10.1073/pnas.152259999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enslen H, Tokumitsu H, Soderling TR. Phosphorylation of CREB by CaM-kinase IV activated by CaM-kinase IV kinase. Biochem Biophys Res Commun. 1995;207:1038–1043. doi: 10.1006/bbrc.1995.1289. [DOI] [PubMed] [Google Scholar]
- Gärtner A, Collin L, Lalli G. Nucleofection of primary neurons. Methods Enzymol. 2006;406:374–388. doi: 10.1016/S0076-6879(06)06027-7. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME. Requirement for BDNF in activity-dependent survival of cortical neurons. Science. 1994;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Min L, Muraoka M, Doi J, Horike N, Okamoto M. Salt-inducible kinase-1 represses cAMP response element-binding protein activity both in the nucleus and in the cytoplasm. Eur J Biochem. 2004;271:4307–4319. doi: 10.1111/j.1432-1033.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, Tsuchiya Y, Min L, Doi J, Miyauchi A, Witters LA, Nakamura H, Okamoto M. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. Febs J. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- Kingsbury TJ, Bambrick LL, Roby CD, Krueger BK. Calcineurin activity is required for depolarization-induced, CREB-dependent gene transcription in cortical neurons. J Neurochem. 2007;103:761–770. doi: 10.1111/j.1471-4159.2007.04801.x. [DOI] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Cowan CW, Shaywitz AJ, Dolmetsch RE, Griffith EC, Hu LS, Haddad C, Xia Z, Greenberg ME. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Kovács KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FC, Graybiel AM. Spatiotemporal dynamics of CREB phosphorylation: transient versus sustained phosphorylation in the developing striatum. Neuron. 1996;17:1133–1144. doi: 10.1016/s0896-6273(00)80245-7. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34:371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Mayr BM, Canettieri G, Montminy MR. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc Natl Acad Sci U S A. 2001;98:10936–10941. doi: 10.1073/pnas.191152098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I, Pérez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Rajan I, Cline HT. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo . J Neurosci. 1998;18:7836–7846. doi: 10.1523/JNEUROSCI.18-19-07836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, Okamoto M, Montminy M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Sheng M, Thompson MA, Greenberg ME. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ginty DD, Bonni A, Greenberg ME. L-type voltage-sensitive Ca2+ channel activation regulates c-fos transcription at multiple levels. J Biol Chem. 1995;270:4224–4235. doi: 10.1074/jbc.270.9.4224. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Wu GY, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS ONE. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]