Abstract
MicroRNAs are small non-coding RNA molecules which act as modulators of gene function, and have been identified as playing important roles in cancer as both tumor suppressors and oncogenes. The present study aimed to examine the role of miR-198 in prostate cancer aggression by analyzing how it influences several hallmarks of cancer. Abundance of miR-198 in prostate cancer and association with clinical characteristics was analyzed using a CPC-Gene prostate cancer dataset. Overexpression of miR-198 was performed using transient transfection of miR-198 mimic prior to assaying proliferation, cell cycle, and colony formation in LNCaP and DU145 cell lines using standard protocols. In vivo tumor formation in athymic nude mice was examined using LNCaP xenografts with stable overexpression conferred using lentiviral miR-198 transduction. Protein and mRNA abundance of MIB1 was determined using western blotting and RT-qPCR respectively, while miR-198 binding to MIB1 was validated using a luciferase reporter assay. miR-198 abundance was lower in high Gleason grade prostate cancer relative to intermediate and low-grade cancer. Overexpression of miR-198 diminished proliferation of prostate cancer cell lines, increased G0/G1 cell cycle arrest, and significantly impaired colony formation. Elevated miR-198 abundance was also demonstrated to impair tumor formation in vivo using LNCaP xenografts. Mindbomb E3 ubiquitin protein ligase 1 (MIB1) was demonstrated to be directly targeted by miR-198, and knockdown of MIB1 recapitulated the effects of miR-198 on proliferation and colony formation. The present evidence supports miR-198 as an important tumor suppressor in prostate cancer, and demonstrates for the first time that it acts by targeting MIB1. The present study reinforces the importance and complexity of miRNA in regulating prostate cancer aggression.
Keywords: microRNA, miR-198, prostate cancer, MIB1
Introduction
Patients with similar clinicopathological risk classification can display striking clinical heterogeneity in the progression of their prostate cancer (1). This limits clinical decision making for personalized management strategies. The molecular mechanisms underlying prostate cancer progression involve alterations in a variety of signaling pathways. Elucidating underlying mechanisms influencing prostate cancer progression can contribute to our understanding of how they drive different clinical phenotypes in prostate cancer (2), while providing potential candidates as novel biomarkers.
MicroRNAs (miRNAs) are a class of short non-protein coding RNA molecules which act within the RNA interference pathway. Each miRNA is a single-stranded RNA molecule approximately 22 nucleotides in length, and guides messenger RNA (mRNA) to post-transcriptional regulation machinery via binding to a sequence in the mRNA 3′ untranslated region (3′UTR). Through inhibition of targets which are tumor suppressors or oncogenes, miRNAs have been revealed to influence each of the classic hallmarks of cancer, and individual miRNAs often alter multiple hallmarks (3).
Evidence for the role of microRNA-198 (miR-198) as a tumor suppressor stems from its downregulation in numerous cancer types including multiple myeloma, gastric cancer, colorectal cancer, glioblastoma, osteosarcoma, and tumors of the breast, pancreas, liver and lungs (4–12). Low miR-198 levels have been revealed to predict poor survival in pancreatic and gastric cancer cohorts, and overexpression of miR-198 in vitro regulates several hallmarks of cancer (5,8). However, there is limited research on the role of miR-198 in prostate cancer.
The present findings demonstrated that miR-198 is reduced in high Gleason score tumors, and functions as a tumor suppressor in prostate cancer by inhibiting proliferation, anchorage-independent growth, and tumor growth, using in vitro and in vivo models. Mindbomb E3 ubiquitin protein ligase 1 (MIB1) was identified as a novel effector gene downstream of miR-198, which appears to function in a Notch pathway-independent manner. This research enhances our understanding of prostate cancer biology by identifying miR-198 as a novel tumor suppressor miRNA.
Materials and methods
Patient analyses
The Canadian Prostate Cancer Genome Network (CPC-GENE) dataset is comprised of 162 samples, of which 137 are intermediate risk Gleason 7 (3+4=99 and 4+3=38), 12 are low risk (Gleason 6), and 13 are high risk (Gleason >7). Differential expression between Gleason ≤7 and >7 was evaluated using two-sided Wilcoxon test, and all statistical analyses were completed using R statistical environment (v3.4.0). CPC-GENE is part of the international genome consortium (ICGC) and data access control is regulated via the ICGC-DACO (www.icgc.org). Informed consent, consistent with local Research Ethics Board (REB) and International Cancer Genome Consortium (ICGC) guidelines, was obtained at the time of clinical follow-up. Previously collected tumor tissues were used, following University Health Network REB-approved study protocols (UHN 06-0822-CE, UHN 11-0024-CE, CHUQ 2012-913:H12-03-192).
Cell culture
Human prostate carcinoma cell lines LNCaP (castrate-sensitive) and DU145 (castrate-resistant) were purchased from the American Type Culture Collection (ATCC). LNCaP cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) and 1% penicillin (100 U/ml)-streptomycin (100 µg/ml) (PS; Invitrogen; Thermo Fisher Scientific, Inc.). DU145 cells were cultured in Dulbecco's modified Eagle's medium containing 4.5 g/l D-glucose and GlutaMAX (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% PS. All cell lines were maintained in tissue-culture flasks within a humidified 37°C incubator with 5% CO2, and passaged when they reached 80% confluency. LS174T Notch-Luciferase cells were previously generated by our laboratory (13), and maintained in DMEM with 10% FBS and 1% PS. Cell lines were regularly confirmed to be free from mycoplasma contamination using MycoAlert Detection Kit (Lonza Group, Ltd.).
Transfection
Negative control and human miR-198 mimics (denoted as ‘Control’ and ‘miR-198’ respectively in figures; sequences in Table SI) were purchased from Shanghai GenePharma Co., Ltd. Three-sequence pooled siRNA for human MIB1 (sc-75781; sequence in Table SI) was obtained from Santa Cruz Biotechnology, Inc., along with a control siRNA (sc-37007; sequence in Table SI). MIB1 cDNA ORF in pcDNA3.1+ vector was purchased from GenScript (ID: OHu21837) and used in parallel with pcDNA3.1+ backbone plasmid as a control. All RNA products were stored at −80°C and exposed to a maximum of two freeze-thaw cycles. Sequences are listed in Table SI. Passaged cells were counted using a Countess automated cell counter (Invitrogen; Thermo Fisher Scientific, Inc.), and a total of 2.5×105 cells (DU145) or 4×105 cells (LNCaP) were seeded in 6-well plates, and transfected the following day using Lipofectamine 2000 as per the manufacturer's instructions (Thermo Fisher Scientific, Inc.). Cells were transfected for 24 h prior to being trypsinized, collected, and used in subsequent assays.
Quantitative real-time PCR (RT-qPCR)
For miRNA abundance, cells were lysed and total miRNA was extracted using mirVana miRNA isolation kit according to the manufacturer's instructions (Life Technologies; Thermo Fisher Scientific, Inc.). For gene expression, RNA was extracted using the RNeasy Mini Kit (Qiagen). RNA quality measured by absorbance at 260 nm was evaluated using a NanoDrop 2000 (Thermo Fisher Scientific). cDNA was synthesized using the miScript II RT kit for miRNA (Qiagen, Inc.) or Superscript VILO cDNA kit (Thermo Fisher Scientific, Inc.) for mRNA. Real-time quantitative PCR to assess gene expression was performed on a StepOnePlus Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the miScript SYBR Green PCR kit (Qiagen, Inc.) or SYBR Select Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) for mature miRNA or mRNA respectively. Primers for miRNA profiling were obtained from miScript Primer Assays for RNU6-2 and miR-198 (Qiagen, Inc.), whereas primers for mRNA abundance were designed using Primer-BLAST software (NCBI) and synthesized by Invitrogen; Thermo Fisher Scientific, Inc. (Table SII). Expression levels were calculated with the ΔΔCq method (14) using StepOne Software (Applied Biosystems; Thermo Fisher Scientific, Inc.), and relative abundance was normalized to RNU6-2 for miRNA or GADPH for mRNA.
Proliferation assay
Transfected cells were seeded in technical triplicates at 5×104 cells/well in a 6-well plate. After incubation for 4 days (LNCaP) or 5 days (DU145), cells were trypsinized and the total number of viable cells in each well was determined by mixing the cell suspension 1:1 (v/v) with 0.4% trypan blue solution, incubating for 1 min at room temperature, then measuring with a Countess automated cell counter. Results were graphed as the change in cell number from original number plated, relative to the amount of growth in the control condition.
Cell cycle analysis
LNCaP cells were seeded at 4×105 cells/well in a 6-well plate, and 24 h later transfected with miRNA mimics. The cells were incubated undisturbed for 48 h after transfection, at which time they were trypsinized, washed with phosphate-buffered saline (PBS), and fixed on ice for 30 min in cold 80% ethanol and Hank's balanced salt solution. Cells were pelleted by 5-min centrifugation at 200 × g, resuspended in RNAse A in HBSS (2 mg/ml), and stained with propidium iodide (0.1 mg/ml; Sigma-Aldrich; Merck KGaA) with 0.6% NP-40 (Thermo Fisher Scientific, Inc.). After 30 min of incubation in the dark, 50,000 events per sample were captured using a FACSCalibur flow cytometer (BD Biosciences), and the cell cycle profile was generated using FlowJo software (Version 10.0.4; FlowJo LLC).
Soft agar assay
Bottom soft agar layers were created in 24-well plates by adding a mixture containing 2X DMEM-F12 (Gibco; Thermo Fisher Scientific, Inc.), 10% FBS, 1% PS, and 0.8% w/v liquefied Agar A (Bio Basic, Inc.). Transfected cells were added to the upper agar layer mixture, which contained the same components as the base with the exception of the use of 0.5% liquefied Agar A, and plated in technical triplicates on top of the base layers at 700 cells/well. Approximately 30 days later, the plates were imaged using a Leica MZ FLIII stereomicroscope (Leica Microsystems GmbH), and the number of colonies were manually counted.
Generation of stable cell lines
Pre-miR-198 was cloned into a pBabe-puro vector, and transfected into Phoenix-AMPHO cells (a retroviral packaging cell line, ATCC), parallel to empty-pBabe vector as a matching control. After 24 h, the culture media containing pBabe-miR198 virus or pBabe-empty virus was collected. Viral media and polybrene (Sigma-Aldrich; Merck KGaA) were added onto adhered LNCaP cells in a 6-well plate, which were then spin transduced at 1,000 × g for 90 min in a 37°C centrifuge. Viral media was removed the following day, and puromycin (0.75 µg/ml) was added 3 days post spin-transduction for selection. After selection, the cells were maintained in puromycin (0.75 µg/ml) and miR-198 expression was determined using RT-qPCR.
In vivo tumor xenograft formation
All experiments involving animals were performed in accordance with the University of Toronto and Sunnybrook Research Institute Animal Care Committee guidelines using a peer-reviewed protocol (AUP #17-509). As per this AUP, humane endpoints included tumors >1.5 cm diameter, weight loss >20%, tumor ulceration, or abnormal posture. After reaching any of these endpoint criteria, mice were anaesthetized with isofluorane (4%) and sacrificed by cervical dislocation. Mice were housed and fed according to standard animal care policies, and welfare-related assessments were performed on a regular basis. Six to seven-week-old male athymic nude mice purchased from Charles River Laboratories were injected subcutaneously into the right flank with 1.5×106 LNCaP-ctrl or LNCaP-198 stable cell lines mixed in a 1:1 (v:v) ratio with Matrigel (Corning Incorporated). Prior to injection, the mice were weighed and distributed evenly into two experimental groups (LNCaP-ctrl or LNCaP-198) each containing four mice. Mice were monitored every 2–3 days and the tumor volume was measured using calipers and calculated using the modified ellipsoid formula: Volume=1/2 (length × width2). Tumor formation was graphed as the percent of mice tumor-free, defined as >60 mm3, and analyzed using log-rank test to compare survival curves.
Western blotting
Cells were rinsed with PBS and lysed in ice-cold radioimmunoassay precipitation lysis buffer containing Complete Mini protease inhibitor cocktail and PhosSTOP phosphatase inhibitor cocktail (Roche Diagnostics). Collected lysate was sonicated, and then centrifuged to pellet cell debris. Protein quantification was performed using the Bradford protein assay (Bio-Rad Laboratories, Inc.) with a Nanodrop spectrophotometer. Protein samples were combined with β-mercaptoethanol, and denatured by boiling. Protein lysate was run on a 4–20% polyacrimide gradient gel (Bio-Rad Laboratories, Inc.), wet-transferred to polyvinylidene difluoride membranes (Thermo Fisher Scientific, Inc.), and then blocked against non-specific binding for 1 h with gentle agitation in TBST containing either 5% non-fat dry milk or 5% bovine serum albumin (BSA). A primary antibody in appropriate solution (5% milk or 5% BSA) was added and incubated overnight at 4°C. The membranes were then washed three times with TBST, and then incubated with a horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (1:5,000; cat. no. 7074; Cell Signaling Technologies, Inc.) for 1 h at room temperature, and afterwards washed again three times in TBST. Protein detection was performed using incubation with an enhanced chemiluminescence solution (1.25 mM Luminol, 2 mM 4IPBA, 100 mM Tris-HCL pH 8.8), and imaged using a ChemiDoc Imaging System (Bio-Rad Laboratories, Inc.). The following antibodies from Cell Signaling Technologies, Inc. were used: β-actin (1:2,000; cat. no. 4967; anti-rabbit), MIB1 (1:1,000; cat. no. 4400; anti-rabbit).
Transcriptomic analysis
Total RNA from transfected cell lines was extracted using RNeasy Mini Kit (Qiagen, Inc.) as per the manufacturer's instructions. RNA quality was assessed using a spectrophotometer, and all samples were ethanol-precipitated if any sample was found to have an absorbance at 260 nm ≤1.8. Gene expression profiling was performed by the Centre for Applied Genomics (The Hospital for Sick Children, Toronto, Canada) using a GeneChip Human Gene 2.0 ST array (Affymetrix; Thermo Fisher Scientific, Inc.). Transcriptomic data was normalized using default parameters in Expression Console software (V.1.2; Affymetrix).
In silico analysis
miRNA target prediction was performed using miRWalk 2.0 atlas of predicted and published miRNA-gene interactions (15).
Notch-luciferase reporter assay
A Notch reporter cell line was previously generated and validated (13) by transducing a human colon adenocarcinoma line (LS174T) with lentivirus containing a minimal promoter with multiple recombination signal-binding protein 1 for J-Kappa (RBP-jκ) Notch response elements (5′-CGTGGGAA-3′) driving expression of the firefly luciferase gene (Qiagen, Inc.). The same lentivirus was used to transduce LNCaP cells to create a prostate cancer Notch reporter cell line. To determine Notch-luciferase activity, cells were seeded in a 24-well plate, transfected or treated with inhibitor for 24 h, then lysed and luciferase activity was assessed using a Luciferase Assay System (Promega Corporation) and a luminometer.
3′UTR luciferase assay
LNCaP cells were seeded in triplicate in a 96-well plate and co-transfected the following day with a luciferase reporter plasmid containing either wild-type MIB1 3′UTR or MIB1 3′UTR containing mutations to disrupt binding in the miR-198 predicted binding site (SwitchGear Genomics; Active Motif), and control or miR-198 mimic. After 24 h, luciferase activity was assayed according to the manufacturer's protocol using LightSwitch Luciferase Reagent (SwitchGear Genomics; Active Motif).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software), except for analyses conducted on patient samples which were performed in R. All in vitro experiments consisted of at least three experimental replicates, unless otherwise stated. Differences in means were compared using two-sided Student's t-tests, and graphed as the mean ± SEM. Statistical significance was defined as P<0.05, P<0.01 and P<0.001.
Results
miR-198 is downregulated in aggressive disease
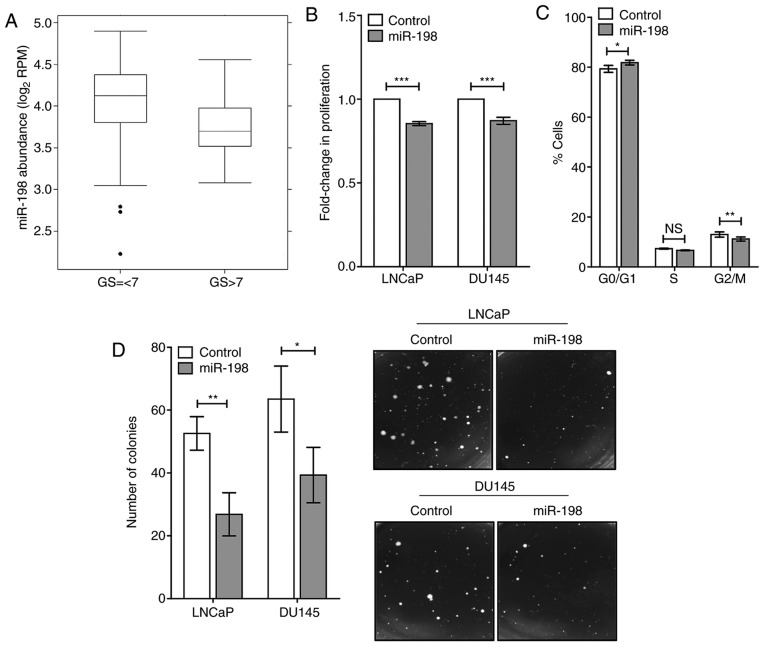
Given the role of miR-198 as a potential biomarker in other cancers, it was of interest in determining whether miR-198 abundance was altered in patient samples. We examined the Canadian Prostate Cancer Genome Network (CPC-GENE) sequencing project data, which contains miRNA abundance, and clinical information from 162 localized prostate cancer patients. An association with Gleason score was observed in the CPC-GENE dataset (Fig. 1A). In high Gleason score patients (Gleason >7), miR-198 abundance was significantly lower than in intermediate and low Gleason score patients (Gleason score ≤7; 1.2-fold decrease compared with GS ≤7, P<0.05). This analysis indicated a potential role for miR-198 in prostate cancer aggression, and warranted investigation into its function in prostate cancer.
Figure 1.
Low miR-198 tumor abundance is associated with high Gleason score and reduces the aggressive phenotype in vitro. (A) miR-198 abundance in prostate tumor samples from a CPC-GENE dataset with Gleason Score (GS) ≤7 (n=149) or GS >7 (n=13). The boxplot represents the median with IQR, and outliers are included in the analysis (two-sided Wilcoxon test, P=0.02) (B) Proliferation assays after transfection with control or miR-198 mimic in DU145 and LNCaP cells. Total viable cells were counted after 4 days (DU145) or 5 days (LNCaP), and normalized to the number of cells in the control mimic condition. (C) Cell cycle profile from flow cytometry of LNCaP cells 48 h after transfection with control or miR-198 mimic. (D) Soft agar assay results from DU145 and LNCaP cells with control/miR-198 mimic transfection. Number of colonies counted after 30 days incubation. Representative images from soft agar experiments. The mean ± SEM and statistical significance are denoted (*P<0.05, **P<0.01 and ***P<0.001).
Increased miR-198 suppresses aggressive phenotype
We assayed for important hallmarks of tumor aggression, including cellular proliferation, anchorage-independent growth, and invasive capacity in two human prostate cancer cell lines LNCaP and DU145. Transient transfection of miR-198 mimic in DU145 and LNCaP cells was performed, and overexpression was confirmed by RT-qPCR (Fig. S1A). This resulted in significantly reduced proliferation compared with the control mimic using viable cell counting (Fig. 1B). Analysis of cell cycle profiles was performed in LNCaP cells using flow cytometry and significant differences were revealed in the cell cycle distributions between miR-198 and control conditions (Fig. 1C). A greater percentage of miR-198 cells were observed in the G0/G1 phase (P<0.05), with less in the S phase (P=0.055) and G2/M phase (P<0.01) relative to the control, which was consistent with reduced cellular proliferation. To assay tumorigenic potential in vitro, anchorage-independent growth assays were performed and it was determined that miR-198 mimic-transfected DU145 and LNCaP cells had a significantly reduced ability to form colonies in soft agar (Fig. 1D).
miR-198 reduces tumorigenicity in vivo
In order to assay the influence of miR-198 on tumorigenicity in vivo, stably overexpressing cell lines were generated by transducing LNCaP cells with a lentivirus containing a miR-198 sequence (LNCaP-198) or a control sequence (LNCaP-ctrl). After antibiotic selection, surviving cells were pooled and assayed to confirm overexpression of miR-198 (Fig. S1B), and the ability to suppress colony formation in soft agar (Fig. 2A). Subsequently, male athymic nude mice were injected subcutaneously with either LNCaP-ctrl or LNCaP-198 cells, and tumor growth was monitored. At 35 days post-injection, only one LNCaP-198 mouse had an observable tumor, whereas all four LNCaP-ctrl mice developed tumors. Tumors ultimately formed in the entire LNCaP-198 group; however, the mean number of days required was 38.75 days vs. 28.25 in LNCaP-ctrl. Tumor-free survival analysis demonstrated miR-198 overexpression trended towards slower tumor formation in mice (Fig. 2B, log-rank P=0.0594).
Figure 2.
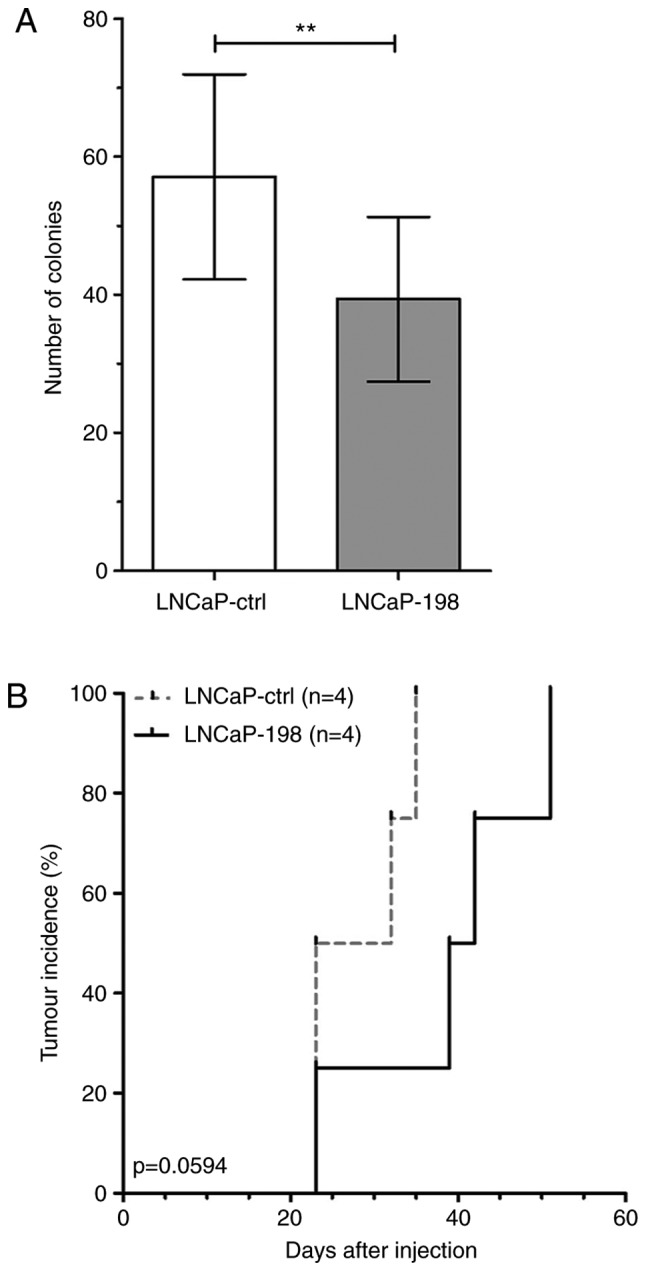
miR-198 impairs tumor formation in vivo. (A) Soft agar assays of stable control/miR-198 LNCaP cell lines. The mean ± SEM and statistical significance are denoted (**P<0.01). (B) Kaplan-Meier curves for an LNCaP xenograft experiment. Subjects were censored after the tumor volume reached ≥60 mm3. Log-rank tests were performed for statistical significance.
MIB1 is directly targeted by miR-198
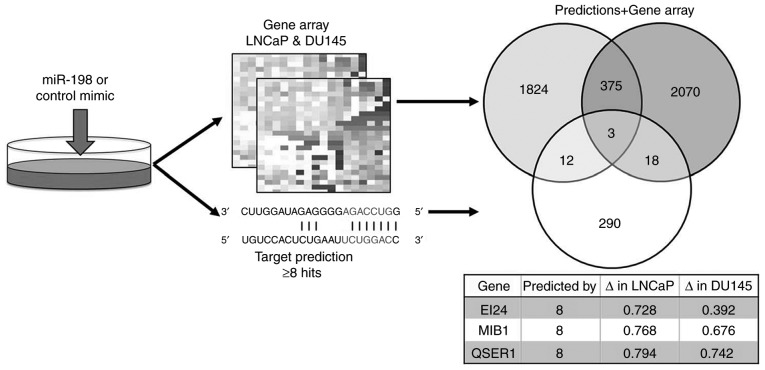
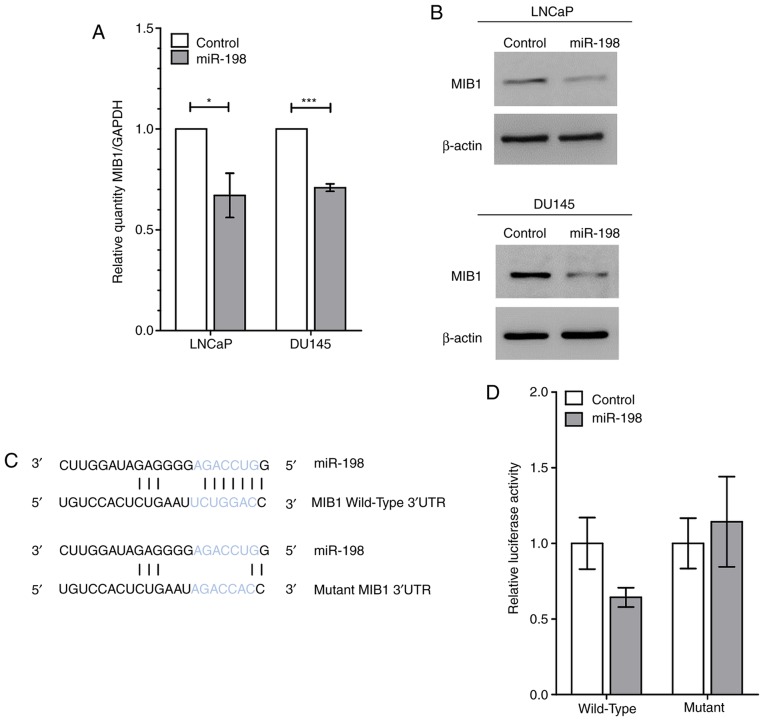
To identify targets of miR-198 which regulate tumorigenicity, transcriptomic analyses from DU145 and LNCaP cells transiently transfected with miR-198 or control mimics were performed, and these data were combined with in silico predicted targets from miRWalk 2.0 (Fig. 3). The three highest predicted targets were EI24 autophagy associated transmembrane protein (EI24), mindbomb E3 ubiquitin protein ligase 1 (MIB1), and glutamine and serine rich 1 (QSER1). QSER1 is a protein of unknown function, EI24 encodes a putative tumor suppressor (16), whereas MIB1 has a possible oncogenic function through promotion of Notch signaling and thus is a potential target of interest. In the transcriptomic analyses, miR-198 overexpression decreased MIB1 abundance by 32% in DU145 cells and 23% in LNCaP cells (Fig. 3). RT-qPCR performed after transient miR-198 transfection confirmed that MIB1 abundance was reduced by 29 and 33% in DU145 and LNCaP cells, respectively (Fig. 4A). In addition, western blot analysis of MIB1 confirmed a decrease at the protein level (Fig. 4B). Next, a luciferase reporter was designed and created, containing the predicted wild-type or a mutated miR-198 binding site from the MIB1 3′UTR in order to confirm direct binding of miR-198 to MIB1 (Fig. 4C). Luciferase activity was significantly decreased in LNCaP cells co-transfected with miR-198 mimic and wild-type MIB1 3′UTR compared to the control mimic, and exhibited no differences with miR-198 or control mimics with the mutant 3′UTR (Fig. 4D). These experiments established MIB1 as an authentic target of miR-198 in prostate cancer.
Figure 3.
Workflow for identification of miR-198 targets. Transcriptomic arrays were performed using RNA collected from LNCaP and DU145 cells transfected with either control or miR-198 mimic. miR-198 targets predicted by at least 8 out of 12 in silico prediction algorithms (n=323, white circle) were combined with putative targets decreased in the gene array analyses by at least 20% (LNCaP n=2214, light grey circle; DU145 n=2466, dark grey circle). Details of the three overlapping genes are presented in a table, including their fold change with miR-198 mimic compared with control mimic in each array. MIB1, mindbomb E3 ubiquitin protein ligase 1; EI24, EI24 autophagy associated transmembrane protein; QSER1, glutamine and serine rich 1.
Figure 4.
MIB1 is targeted by miR-198. (A) RT-qPCR results revealed lower MIB1 abundance in LNCaP and DU145 cells transfected with miR-198 mimic, relative to control mimic samples and normalized to GAPDH. (B) MIB1 protein in LNCaP and DU145 cells is reduced after miR-198 transfection compared with control mimic; representative blots are presented. β-actin was used as an endogenous control. (C) Predicted binding site of miR-198 within wild-type MIB1 3′UTR and mutated sequence used in luciferase assay. Location of miR-198 seed sequence denoted by blue text (AGACCUG), and sequence pairing represents complementarity predicted to influence binding efficacy. (D) Luciferase activity of MIB1 3′UTR wild-type reporter is reduced with miR-198 mimic while the mutant remains unchanged. Graph displaying a single representative experiment. The mean ± SEM and statistical significance are denoted (*P<0.05, ***P<0.001). MIB1, mindbomb E3 ubiquitin protein ligase 1.
MIB1 alteration recapitulates phenotype
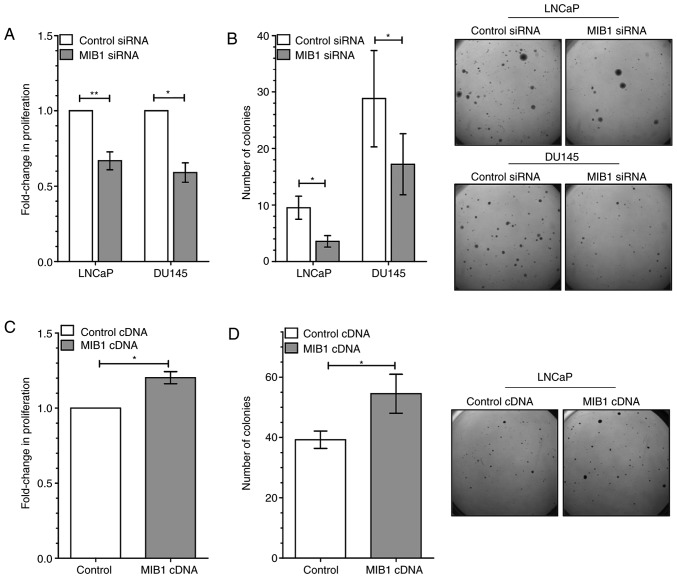
It was next determined whether MIB1 knockdown could result in a similar phenotype as miR-198. Knockdown of MIB1 was first confirmed at the mRNA and protein level, after siRNA transfection in DU145 and LNCaP cells using RT-qPCR and western blotting, respectively (Fig. S1C and D). Knockdown of MIB1 significantly reduced proliferation in both DU145 and LNCaP cells (Fig. 5A). Tumorigenic potential in soft agar was also decreased with MIB1 knockdown (Fig. 5B). In addition, MIB1 abundance was increased in LNCaP cells by transfecting with a vector containing MIB1 ORF cDNA, and subsequently assayed for phenotype. Cells with elevated MIB1 displayed significantly more proliferation (Fig. 5C) and colony formation ability (Fig. 5D) compared with the control vector. Thus, MIB1 knockdown effectively recapitulated the suppressive effects of miR-198 on cellular proliferation and tumorigenic potential, which was conversely promoted by elevation of MIB1.
Figure 5.
Knockdown of MIB1 recapitulates decreased proliferation and anchorage-independent growth. (A) Proliferation in both LNCaP and DU145 cells was reduced with MIB1 siRNA. (B) The number of colonies capable of anchorage-independent growth was lower with MIB1 siRNA in both cell lines. Representative images from soft agar colony formation with control or MIB1 siRNA presented on the right. (C) Proliferation and (D) colony formation of LNCaP cells is higher after MIB1 cDNA vector transfection. Representative images from soft agar colony formation assay presented on the right. The mean ± SEM and statistical significance are denoted (*P<0.05, **P<0.01). MIB1, mindbomb E3 ubiquitin protein ligase 1.
miR198/MIB1 effects on prostate cancer are likely Notch-independent
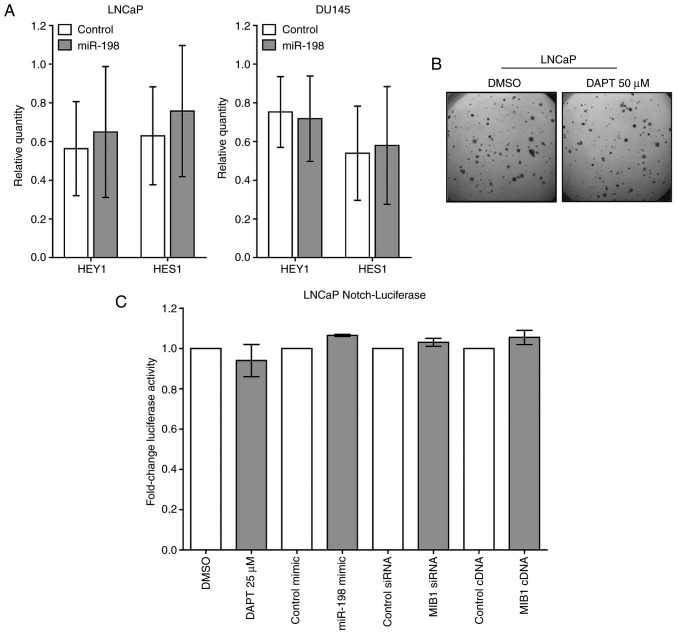
The most well-characterized role of MIB1 is as an E3 ubiquitin ligase which interacts with Delta to increase Notch signaling (17). To examine the effect of miR-198 on Notch signaling, the expression of the Notch transcription factors HEY1 and HES1 were examined, which are used as surrogate markers for Notch activation. RT-qPCR revealed no alterations in the expression of either mRNA with miR-198 transfection in either LNCaP or DU145 cells (Fig. 6A). Additionally, HEY1 and HES1 were unchanged by miR-198 transfection in both LNCaP and DU145 transcriptomic arrays (data not shown). To examine if Notch suppression could contribute to the miR-198 phenotype, soft agar colony formation assays were performed with DAPT, a Notch inhibitor, which revealed no difference in tumorigenic potential (Fig. 6B). Lastly, a LNCaP Notch-luciferase reporter cell line was generated to examine Notch activity, which exhibited no appreciable differences in Notch activity in LNCaP cells either after DAPT treatment, MIB1 siRNA, or miR-198 mimic transfection (Fig. 6C). To validate the Notch-luciferase assay, the effect of these manipulations in the previously validated LS174T cell line were examined, which confirmed reduced luciferase activity after treatment with the Notch inhibitor, but no effect from miR-198 or MIB1 transfections (Fig. S2). Collectively, our data strongly indicated that miR-198 and MIB1 exert their tumor suppressive effects independent of the Notch pathway.
Figure 6.
Effects on the Notch signaling pathway. (A) HEY1 and HES1 were not significantly altered by miR-198 transfection in LNCaP and DU145 cells. RT-qPCR normalized to GAPDH as an endogenous reference gene. (B) Treatment with the Notch inhibitor DAPT did not affect LNCaP colony formation in soft agar; representative soft agar images are presented. (C) Luciferase activity was not altered in Notch-luciferase LNCaP cells by either DAPT (Notch inhibitor) treatment, miR-198 mimic, MIB1 siRNA, or MIB1 cDNA vector. Fold change was normalized to the respective experimental control. MIB1, mindbomb E3 ubiquitin protein ligase 1; HES1, hairy and enhancer of split 1; HEY1, hes-related family bHLH transcription factor with YRPW motif 1.
Discussion
The present study revealed lower abundance of miR-198 in high Gleason grade tumors, highlighting its potential role as a tumor suppressor miRNA. In vitro experiments demonstrated that miR-198 overexpression reduced proliferation and anchorage-independent growth, and induced a G0/G1 cell cycle block. Tumor formation in vivo was reduced with elevated miR-198 abundance, confirming miR-198 functions as a tumor suppressive miRNA in prostate cancer. In prostate cancer, miR-198 is a low-abundance miRNA which may limit is utility as a biomarker; this contrasts to its higher abundance in other cancer types. For example, studies have reported miR-198 as a biomarker in other TCGA cancer cohorts, such as elevated abundance associated with increased overall survival in glioblastoma patients (9). However, miR-198 was undetectable (abundance level=zero, below detection threshold) in >90% of TCGA normal prostate and prostate cancer samples, which limited our ability to perform analyses in this dataset. CPC-GENE utilized a different platform to assess miRNA, which appears more sensitive to this low abundance miRNA, and thus was able to provide non-zero values for miR-198 abundance.
The previously identified targets of miR-198 vary widely in structure and function depending on the cancer type investigated. miR-198 has one previously described target in prostate cancer, Livin (or BIRC7) (18). However, no consistent decrease was observed in this mRNA after miR-198 transfection in both LNCaP and DU145 cells. In addition to Livin, previously identified targets from other cancer types were assayed, however, decreased abundance of these genes in LNCaP and DU145 cells (Table SIII) was not consistently observed. Combination of transcriptomic and in silico analyses identified MIB1 as a putative target of miR-198, as it contained a miR-198 binding site in its 3′UTR, and was consistently reduced by miR-198 overexpression in both cell line arrays. This effect was also observed using RT-qPCR and western blotting to analyze protein abundance. Subsequent MIB1 knockdown studies phenocopied miR-198 effects on reducing proliferation and tumorigenic potential, while a converse effect was observed with MIB1 elevation using an overexpressing vector.
MIB1 is described primarily as an integral positive regulator of Notch signaling, however, significant responsiveness in LNCaP cells was not observed. The role of Notch in prostate cancer is complex with clinical studies yielding conflicting results, demonstrating both up- and downregulation of components in prostate cancer samples and varying effects on deregulation in vitro (19,20). Several studies have concluded that aberrant activation of Notch signaling is consistently observed in metastatic samples, especially increases in NOTCH1 receptor and JAG1 ligand (19). In the present study, treatment of LNCaP cells with the Notch inhibitor DAPT did not reduce anchorage-independent growth. In addition, RT-qPCR analysis of downstream Notch reporter genes HEY1 and HES1 exhibited no change in their abundance after miR-198 transfection. Transfection with miR-198 mimic or treatment with a Notch inhibitor both failed to alter luciferase reporter activity in LNCaP cells, indicating that the key function of miR-198/MIB1 in prostate cancer may occur independently of the effects on Notch signaling. The present study is the first to report a miRNA targeting MIB1 in cancer, and therefore requires future exploration of pathways external to Notch signaling in which miR-198/MIB1 may exert a non-canonical function.
In summary, this research establishes a tumor suppressive role of miR-198 in prostate cancer. miR-198 displayed lower abundance in high Gleason grade tumors, and overexpression impaired tumor formation in mice. MIB1 was identified as a novel miR-198 target, through which miR-198 reduced proliferation and tumourigenicity of prostate cancer. This mechanism likely occurs in a Notch pathway-independent manner, which can be further explored to develop new therapeutic strategies. These study findings reinforce the importance and complexity of miRNA in regulating prostate cancer aggression.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- 3′UTR
3′ untranslated region
- CPC-GENE
Canadian Prostate Cancer Genome Network
- HES1
hairy and enhancer of split 1
- HEY1
hes-related family bHLH transcription factor with YRPW motif 1
- MIB1
mindbomb E3 ubiquitin protein ligase 1
- miRNA
microRNA
- miR-198
microRNA-198
- siRNA
small interfering RNA
Funding
SKL is a Movember Rising Star award recipient proudly funded by the Movember Foundation (grant. nos. RS2014-03, D2015-12 and D2017-1811), the Telus Motorcycle Ride For Dad (Huronia Branch), and a Ministry of Research and Innovation Early Researcher Award.
Availability of data and materials
All data generated or analyzed during the preset study are included in this published article.
Authors' contributions
JR designed and executed the majority of experiments, interpreted results, performed statistical analyses, and drafted the manuscript. SKL conceived the project, supervised direction, and revised the manuscript. CH, XH, MRD and ST assisted with the experiments and data interpretation. JJ, supervised by PCB, performed bioinformatic analyses of patient datasets. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
CPC-GENE is part of the international genome consortium (ICGC) and data access control is regulated via the ICGC-DACO (www.icgc.org). Informed consent, consistent with local Research Ethics Board (REB) and International Cancer Genome Consortium (ICGC) guidelines, was obtained at the time of clinical follow-up. Previously collected tumor tissues were used, following University Health Network REB-approved study protocols (UHN 06-0822-CE, UHN 11-0024-CE, CHUQ 2012-913:H12-03-192). All experiments involving animals were performed in accordance with the University of Toronto and Sunnybrook Research Institute Animal Care Committee guidelines using a peer-reviewed protocol (AUP #17-509).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 2.Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541:359–364. doi: 10.1038/nature20788. [DOI] [PubMed] [Google Scholar]
- 3.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi C, Chung TH, Huang G, Zhou J1, Yan J, Ahmann GJ, Fonseca R, Chng WJ. Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma. Oncotarget. 2015;6:26508–26518. doi: 10.18632/oncotarget.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Z, Zheng X, Kong D. Decreased miR-198 expression and its prognostic significance in human gastric cancer. World J Surg Oncol. 2016;14:33. doi: 10.1186/s12957-016-0784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Tang Z, Jiang B, Chen J, Fu Z. miR-198 functions as a tumor suppressor in breast cancer by targeting CUB domain-containing protein 1. Oncol Lett. 2017;13:1753–1760. doi: 10.3892/ol.2017.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang WT, Wang HL, Yang H, Ren FH, Luo YH, Huang CQ, Liang YY, Liang HW, Chen G, Dang YW. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: A clinicopathological and in silico study. Onco Targets Ther. 2016;9:5163–5180. doi: 10.2147/OTT.S108828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marin-Muller C, Li D, Bharadwaj U, Li M, Chen C, Hodges SE, Fisher WE, Mo Q, Hung MC, Yao Q. A tumorigenic factor interactome connected through tumor suppressor microRNA-198 in human pancreatic cancer. Clin Cancer Res. 2013;19:5901–5913. doi: 10.1158/1078-0432.CCR-12-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie E, Jin X, Wu W, Yu T, Zhou X, Shi Z, Zhang J, Liu N, You Y. MiR-198 enhances temozolomide sensitivity in glioblastoma by targeting MGMT. J Neurooncol. 2017;133:59–68. doi: 10.1007/s11060-017-2425-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Wang J, Kong X, Chen H, Wang Y, Qin M, Lin Y, Chen H, Xu J, Hong J, et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep. 2014;4:6145. doi: 10.1038/srep06145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S, Zhang G, Li P, Chen S, Zhang F, Li J, Jiang C, Chen X, Wang Y, Du Y, et al. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol. 2016;37:5193–5202. doi: 10.1007/s13277-015-4369-z. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Zhao Y, Wang L. MicroRNA-198 inhibited tumorous behaviors of human osteosarcoma through directly targeting ROCK1. Biochem Biophys Res Commun. 2016;472:557–565. doi: 10.1016/j.bbrc.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 13.Liu SK, Bham SA, Fokas E, Beech J, Im J, Cho S, Harris AL, Muschel RJ. Delta-like ligand 4-notch blockade and tumor radiation response. J Natl Cancer Inst. 2011;103:1778–1798. doi: 10.1093/jnci/djr419. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Mork CN, Faller DV, Spanjaard RA. Loss of putative tumor suppressor EI24/PIG8 confers resistance to etoposide. FEBS Lett. 2007;581:5440–5444. doi: 10.1016/j.febslet.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/S1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 18.Ye L, Li S, Ye D, Yang D, Yue F, Guo Y, Chen X, Chen F, Zhang J, Song X. Livin expression may be regulated by miR-198 in human prostate cancer cell lines. Eur J Cancer. 2013;49:734–740. doi: 10.1016/j.ejca.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Deng G, Ma L, Meng Q, Ju X, Jiang K, Jiang P, Yu Z. Notch signaling in the prostate: Critical roles during development and in the hallmarks of prostate cancer biology. J Cancer Res Clin Oncol. 2016;142:531–547. doi: 10.1007/s00432-015-1946-x. [DOI] [PubMed] [Google Scholar]
- 20.Su Q, Xin L. Notch signaling in prostate cancer: Refining a therapeutic opportunity. Histol Histopathol. 2016;31:149–157. doi: 10.14670/HH-11-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the preset study are included in this published article.