Abstract
N-linked glycosylation of proteins is the most common post-translational modification of proteins. The enzyme UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) catalyses the first step of N-glycosylation, and DPAGT1 knockout is embryonic lethal in mice. In this study, we identified the sole orthologue (algn-7) of the human DPAGT1 in the nematode C. elegans. The gene activity was disrupted by RNAi and deletion mutagenesis, which resulted in larval lethality, defects in oogenesis and oocyte-to-embryo transition. Endomitotic oocytes, abnormal fusion of pronuclei, abnormal AB cell rotation, disruption of permeation barriers of eggs, and abnormal expression of chitin and chitin synthase in oocytes and eggs were the typical phenotypes observed. The results indicate that N-glycosylation is indispensable for these processes. We further screened an N-glycosylated protein database of C. elegans, and identified 456 germline-expressed genes coding N-glycosylated proteins. By examining RNAi phenotypes, we identified five germline-expressed genes showing similar phenotypes to the algn-7 (RNAi) animals. They were ribo-1, stt-3, ptc-1, ptc-2, and vha-19. We identified known congenital disorders of glycosylation (CDG) genes (ribo-1 and stt-3) and a recently found CDG gene (vha-19). The results show that phenotype analyses using the nematode could be a powerful tool to detect new CDG candidate genes and their associated gene networks.
Keywords: ALG7, DPAGT1, gene knockout, N-glycosylated proteins, RNAi
Introduction
N-linked glycosylation of proteins is one of the post-translational modifications of proteins, and it is widely preserved in eukaryotes from archaea to humans. N-glycans are necessary for protein folding (Paulson 1989; Helenius 1994) and protein quality control (Helenius and Aebi 2004; Aebi et al. 2010). The biosynthesis of the N-glycan precursor, dolichol lipid-linked oligosaccharide (LLO), starts on the cytoplasmic side of the ER, and two N-acetylglucosamine (GlcNAc) and five mannose (Man) residues are added to the dolichol (Dol). Dol-PP-GlcNAc2Man5 is flipped in the lumen of the ER, where the complete LLO (Dol-PP-GlcNAc2Man9Glc3) is formed (Kornfeld and Kornfeld 1985; Helenius and Aebi 2004). The LLO is transferred onto an Asn residue of Asn-X-Ser/Thr (X can be any amino acid except proline) of the nascent polypeptide chain by an oligosaccharyltransferase (OST) complex (Gavel and von Heijne 1990). The N-glycans are processed in the ER, and undergo various modifications in the Golgi, resulting in matured glycoproteins.
The first step of the N-glycan synthesis (LLO synthesis) is catalyzed by the enzyme DPAGT1/ALG7, which is a UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase. DPAGT1 is the initiator in the biosynthesis of LLO of ER-localizing transmembrane proteins. The enzyme adds GlcNAc-phosphate to phospho-Dol at the first step of N-glycosylated protein synthesis which occurs on the cytoplasmic side of the ER. The gene encoding the enzyme (DPAGT1/ALG7) is reported as the most ancient and widely found gene of the ALG (asparagine-linked glycosylation) gene family (Samuelson et al. 2005). The ALG7 gene of Saccharomyces cerevisiae is an essential gene, and the null allele is inviable (Kukuruzinska and Robbins 1987). Disruption of DPAGT1/ALG7 in mice leads to lethality shortly after uterine implantation (Marek et al. 1999). In humans, the congenital disorders of glycosylation (CDG) type Ij (DPAGT1-CDG) and limb-girdle congenital myasthenic syndrome (CMS) with tubular aggregates are caused by mutation in the DPAGT1 gene (Wu et al. 2003; Belaya et al. 2012). The lethality observed in mice could be due to the deficiency of N-glycosylation of proteins expressed in gametogenesis and/or early embryogenesis, but it is generally difficult to study roles of N-glycosylation in germline development and early embryogenesis in mammalian systems.
The nematode C. elegans is a hermaphrodite, meaning that it is capable of self-fertilization. Due to the transparency of its body, it is possible to observe the processes of spermatogenesis, oogenesis, ovulation, fertilization and embryogenesis in vivo. In addition, C. elegans is an RNAi-friendly animal and it is easy to examine RNAi knockdown phenotypes of any gene. With a germline-specific RNAi technique, it is also easy to knockdown germline-specific gene functions involved in emergence of phenotypes (Sijen et al. 2001).
We have been studying C. elegans glycogenes which are human glycogene orthologues, and the results of our research are summarized in our C. elegans Glycogene Database (CGGDB). In the paper describing the database (Akiyoshi et al. 2015), we reported that inhibition of N-glycosylation pathway genes results in an ER stress response as judged by increased expression of the GFP-tagged hsp-4 (C. elegans BiP/HSP70 orthologue) transgene. Increased expression was observed in the following RNAi-treated animals: algn-7, algn-13, algn-14, algn-2 and algn-11 (these are cytoplasmic alg genes which are possible ALG gene orthologues in the nematode) and ostb-1, dad-1 and stt-3 (these are genes coding subunits of the oligosaccharyltransferase complex). Inhibition of ER alg genes (algn-3, -9, -12, -6, -8 and -10) does not affect hsp-4 expression (Akiyoshi et al. 2015). In our CGGDB database, we list the nematode algn-7 gene as a candidate human DPAGT1 orthologue. In previous studies by Struwe et al. (Struwe et al. 2009; Struwe and Warren 2010), tunicamycin, a widely used inhibitor of DPAGT1, was used to study functions of N-glycans in C. elegans. As discussed in our paper (Akiyoshi et al. 2015), slightly different phenotypes were observed between algn-7 RNAi-treated worms and tunicamycin-treated worms. In studies using other organisms, it was shown that tunicamycin affects various biochemical processes (Reiling et al. 2011) including glucose transport (Rojas et al. 2014), glycosphingolipid synthesis (Yusuf et al. 1983), and innate immunity without affecting N-glycosylation and ER-stress pathways (Kim et al. 2013). Thus, to resolve the discrepancy of phenotypes between the algn-7 RNAi-treated animals and the tunicamycin-treated animals, it is necessary to identify the true orthologue of human DPAGT1 in the worm genome, and examine its biochemical and biologic functions in the living organism.
In this study, we found that algn-7 was the sole orthologue of the human DPAGT1 in the C. elegans genome. Deletion of the gene resulted in larval-stage lethality (L2 lethality) and sterility. The lethality of the deletion allele was successfully rescued by introducing the human DPAGT1 gene. Our biochemical analysis showed that the algn-7 RNAi-treated wild type worms (N2) displayed a marked decrease in the nematode DPAGT1 enzymatic activity. algn-7 RNAi-treated animals laid osmotic integrity defective (OID) eggs, and the animals showed decreased expression of the chitin synthase CHS-1 in mature oocytes. The endomitotic oocyte (Emo) phenotype and abnormal oocyte-to-embryo transition phenotypes were observed in the algn-7 knockdown animals, indicating essential roles of algn-7 gene activities in chromosomal segregation in the oogenesis and oocyte-embryo-transition of C. elegans. Furthermore, we identified several N-glycosylated protein genes showing the same phenotypes observed in the algn-7 RNAi-treated worms. We identified 456 different genes encoding N-glycosylated proteins expressed in the C. elegans gonads. The RNAi phenotypes of these genes were examined, and we found five N-glycosylated protein-coding genes showing the same RNAi phenotypes as the algn-7 RNAi-treated worms. These results reveal roles of DPAGT1 and N-glycosylated proteins of the nematode from oogenesis to larval development. To our knowledge, this is the first direct study on the functions of the DPAGT1 gene of C. elegans, and is the first detailed study on the indispensable functions of the gene during oogenesis, oocyte-to-embryo transition and larval development.
Results
algn-7 is the sole orthologue of the human DPAGT1 in the C. elegans genome
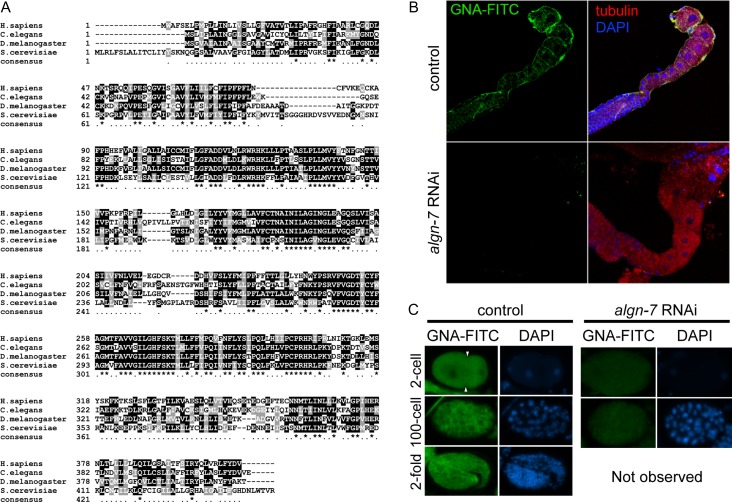
Using Position-Specific Iterated BLAST (PSI-BLAST) search with the human DPAGT1 sequence, we identified two candidate orthologue genes, algn-7 (Y60A3A.14) and T08D2.2. The genomic sequence of the latter contained multiple issues preventing a complete ORF and was classified as a pseudogene in the WormBase (WS261). Although RNA-Seq revealed that the gene was moderately expressed from germline to adults (WormBase, WS261), no phenotypes were observed by RNAi studies including ours. Thus, we identified the algn-7 gene as the sole orthologue of the human DPAGT1 (Figure 1A).
Fig. 1.
C. elegans DPAGT1(ALGN-7) is involved in N-glycosylation. (A) DPAGT1 sequence alignment. Human DPAGT1 (GenBank™ accession number, NP_009802.3) sequence aligned with C. elegans (NP_507859.2), D. melanogaster (NP_609608.1), and S. cerevisiae (NP_009802.3) using Clustal Omega. Strongly conserved residues are shaded black, and conserved residues are shaded dark gray. (B) GNA-FITC staining of C. elegans gonad. Oocytes were stained with FITC-labeled GNA-lectin (green) in the control (N2). algn-7 RNAi (rrf-1) resulted in loss of GNA-FITC staining. Tubulin (red) and DAPI (blue). (C) GNA-FITC staining in developing embryos. The fluorescence intensity in the control embryos (left) (N2; 2-cell, 100-cell, and 2-fold stages) decreased in the RNAi-treated animals (rrf-1) (right).
algn-7 encodes the nematode DPAGT1
To examine the expression level of N-glycosylated proteins in the C. elegans germline, we isolated gonads from the worm, and stained them with FITC-conjugated GNA (Galanthus nivalis) lectin. Gonads were stained brightly with lectin, indicating the expression of high mannose type (or complex type) N-glycans (Yamamoto 2014) in the germline. The staining disappeared in gonads isolated from the algn-7 RNAi-treated animals (Figure 1B). Eggs from the two-cell stage to the two-fold stage were also brightly stained with GNA-lectin, and the staining disappeared in the algn-7 RNAi-treated eggs (Figure 1C). These results strongly indicate that the algn-7 gene is responsible for N-glycan synthesis.
To examine the nematode DPAGT1 activity, we homogenized F1 adult worms and measured enzymatic activities by incubating the worm homogenates with P-dolichol and UDP-[H3] GlcNAc. We confirmed the DPAGT1 activity in the homogenate of the wild type worms, and the activity was significantly reduced when the algn-7 RNAi-treated worms were used for the experiment (Table I). From these experiments, we concluded that algn-7 encodes the nematode DPAGT1.
Table I.
DPAGT1 activity in mock RNAi- and algn-7 RNAi-treated animals
| dpm/200 μL | mean ± SE | dpm/min/mg protein | % activity | |||
|---|---|---|---|---|---|---|
| control 0 min | 55 | 55 | 66 | 59 ± 4 | ||
| mock (P-dol-) 10 min | 112 | 103 | 107 | 107 ± 3 | ||
| mock RNAi 10 min | 1220 | 1326 | 1412 | 1319 ± 55 | 10,080 ± 420 | 100% |
| algn-7 RNAi 10 min | 147 | 135 | 167 | 150 ± 9 | 801 ± 48 | 7.9% |
“control 0 min” indicates reaction mixture measured at time 0. “mock (P-dol-)” indicates reaction mixture without P-dolichol, and “mock RNAi” indicates RNAi with an empty vector.
Somatic phenotypes observed in the algn-7 knockout animals are rescued with the human DPAGT1 gene
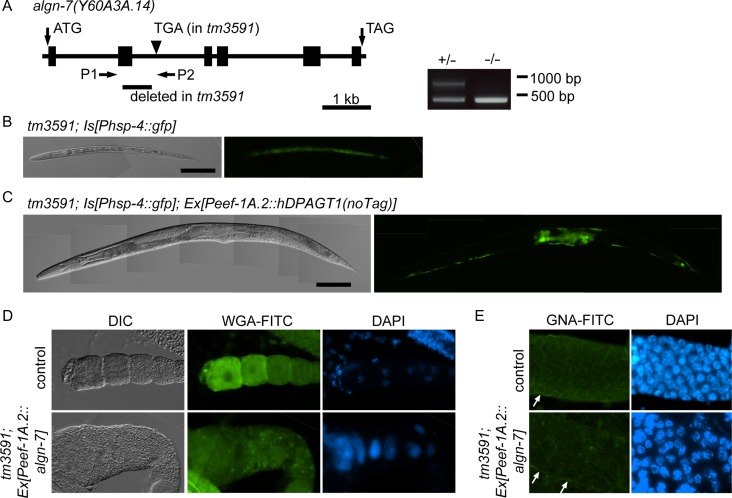
We isolated a deletion allele of the algn-7 gene by screening our TMP/UV-treated library of worms. One allele, tm3591, had a 590 bp deletion in the second exon and intron (Figure 2A), and the algn-7 (−/−) allele showed strong ER stress at the L2 stage (Figure 2B) and was lethal at the stage. Because the homozygote allele was L2 lethal, the animal was balanced with unc-51 (e369) and maintained as a heterozygote. algn-7 (−/−) animals born from the balanced heterozygotes were L2 lethal, and no adult animals were raised from the animals. The KO animals showed no germline phenotypes and embryonic lethal phenotypes that are described in the following sections of this paper. The lack of these phenotypes can be explained by maternal contribution of DPAGT1 activity from the heterozygous mothers. The L2-lethal phenotype and ER stress phenotypes were successfully rescued by introducing the human DPAGT1 gene driven by a ubiquitous promoter Peef-1A.2 (Wheeler et al. 2016) (Figure 2C) or C. elegans wild type algn-7 gene driven by the same promoter (not shown) by microinjection.
Fig. 2.
Isolation of algn-7 deletion allele, and its rescue by human DPAGT1 and wild-type algn-7. (A) Gene structure of algn-7 (N2) and the deletion in the algn-7 (tm3591) allele. tm3591 had a 590 bp deletion in the second exon and intron. Positions of the initiation and stop codons are indicated. The null allele was larval lethal, and the strain was balanced with unc-51(e369) and was maintained as hetero alleles. +/− and −/− alleles were distinguished by PCR using P1/P2 (see Materials and methods). (B) The algn-7 null allele (tm3591/tm3591) was L2-larval-stage lethal. The null allele carrying an ER-stress marker (Phsp-4::gfp) (tm3591; zcIs4) showed strong fluorescence in the whole body (L2 larvae), indicating a strong stress response in the null allele. Scale bar, 100 μm. (C) The L2-lethal phenotype was successfully rescued by injecting the human DPAGT1 gene, although the ER stress phenotype in the germline was not rescued by the extrachromosomal arrays. Scale bar, 100 μm. (D) Germline phenotypes of the null allele were not completely rescued by injection of the wild-type algn-7 gene. Bottom row (tm3591; Ex[Peef-1A.2::algn-7]). WGA-FITC staining was decreased in the rescued animal, and the DAPI staining showed the endomitotic oocyte (Emo) phenotype with abnormal nuclei. (E) GNA-FITC staining in the meiotic zone of wild-type and rescued animals. In the rescued animal (tm3591; Ex[Peef-1A.2::algn-7::venus]), a GNA-FITC signal was observed in the sheath cells of the somatic gonad (white arrows), while the staining in the meiotic zone of the gonad was decreased.
Germline abnormal phenotypes in the algn-7 knockout animals
Because algn-7 (−/−) animals were lethal at the L2 stage, we were unable to examine the germline of the KO animals. Introduction of the wild type algn-7 gene into the KO animals rescued the L2 lethal phenotype, and we were able to observe the gonads of the rescued animals. The functions of microinjected genes (transgene arrays) are silenced in the germline of the nematode (germline silencing) (Kelly et al. 1997). In the rescued animals, lectin staining with the WGA-FITC (Wheat Germ Agglutinin), which binds strongly to complex type and hybrid type glycans and weakly to high mannose type glycan (Yamamoto 2014), showed very weak fluorescence compared to the strong fluorescence in the wild type gonads (Figure 2D). Similar insufficient rescue of the fluorescent signal intensity was observed in the GNA-FITC stained gonads (Figure 2E). The insufficient rescue of the N-glycan expression observed in these experiments could be explained by the germline silencing of the injected wild type algn-7 gene, and it is expected that the abnormal phenotypes observed in the germline of the rescued animals resulted from deficiency of the algn-7 gene functions in the germline. In this respect, it is interesting to note that DAPI staining of the proximal gonads of the rescued animals (Figure 2D) showed oocytes with distended polyploid nuclei (endomitotic oocytes: Emo phenotype). This result strongly suggests that the functions of the algn-7 gene are necessary for the normal development of the oocytes.
Germline expression of algn-7 is essential for the normal development of oocytes
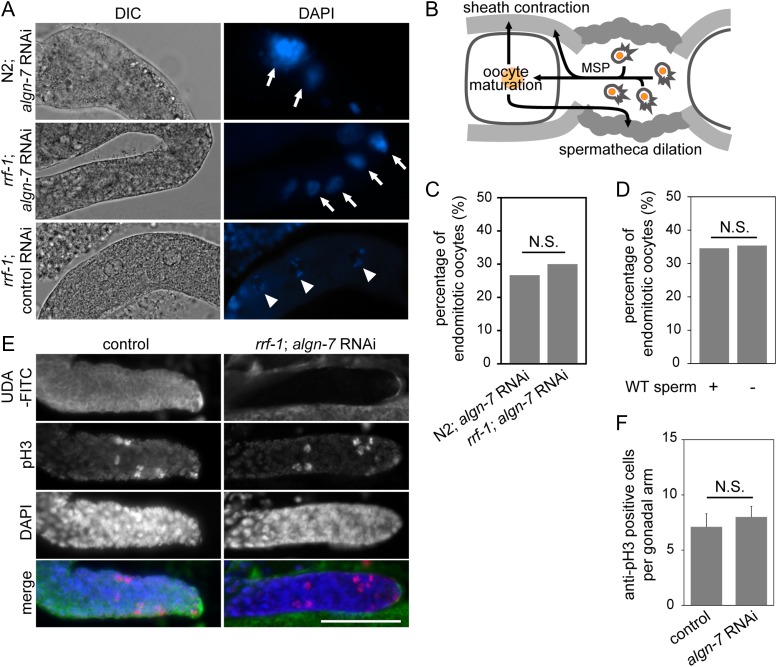
To investigate the roles of the algn-7 gene in the germline and in early embryos, the gene function was knocked down by feeding RNAi. As described above, algn-7 RNAi resulted in reduced DPAGT1 enzymatic activity in vivo, and it also reduced N-glycan expression in the germline (Table I and Figure 1B and C). Thus, we examined germline phenotypes in the algn-7 RNAi-treated animals. In the most severely affected cases, the RNAi-treated wild type animals (F1) were lethal at the L2 stage, but most of the animals were raised to adult animals. All the offspring born from the escapees showed germline abnormalities and they were 100% sterile. For instance, the RNAi-treated wild type animals (F1 adults at the 96 h time point) showed the Emo phenotype (Figure 3A) as found in the somatically rescued animals described above. This result indicates that the Emo phenotype resulted from deficiency of the algn-7 gene, suggesting the essential roles of the algn-7 gene in the proximal region of the gonad. Emo oocytes mature but are not properly ovulated or fertilized (Iwasaki et al. 1996). Ovulation follows increases in the contraction rate and strength of the gonadal sheath cell contraction, and dilation of the distal constriction of the spermatheca. These activations require oocyte maturation and sperm-derived major sperm protein (MSP) (McCarter et al. 1999; Miller et al. 2001, 2003) (Figure 3B). To examine whether the Emo phenotype in our algn-7 RNAi experiments was caused by defects of the somatic gonad (e.g., the distal tip cells, sheath cells, and spermathecal cells) or by defects of the germline, we used the rrf-1 (pk1417) mutant strain NL2098 in which RNAi is sensitive in the germline, but not in most of the somatic tissues including somatic gonad and muscle (Sijen et al. 2001; Kumsta and Hansen 2012). We found that algn-7 knockdown in the rrf-1 animals also resulted in the Emo phenotype (Figure 3A, middle row). The phenotype incidence was 30% (N = 30) and did not significantly change compared to N2; algn-7 RNAi (P = 1) (Figure 3C), indicating that the Emo observed in algn-7 RNAi was not due to defective motility of the gonadal sheath and spermatheca. The conclusion is also supported by our algn-7 RNAi experiment in a ppw-1 mutant background (NL3511). The NL3511 strain is a ppw-1(pk1425) mutant, which failed to produce a PAZ/PIWI domain-containing protein required for efficient germline RNAi (Tijsterman et al. 2002; Yoon et al. 2018). The ppw-1(pk1425) mutation prevents RNAi process in the germline, but not in the soma. Inhibition of algn-7 in this strain did not result in Emo or OID phenotype, while weak larval growth retardation was observed in the animals. These results strongly suggest that algn-7 deficiency caused abnormality in the ovulation signal transduction in the germline.
Fig. 3.
algn-7 in C. elegans oogenesis. (A) Gonads of algn-7 RNAi-treated hermaphrodite were dissected out and stained with DAPI. Emo phenotypes were observed in both N2 and rrf-1 strains. Wild-type chromosomes are marked by arrow heads and polyploid nuclei are marked by arrows. (B) Schematic representation of the ovulation system of C. elegans modified from the WormAtlas (Lints and Hall 2009). (C) The frequency of Emo in algn-7 RNAi-treated animals. The percentage of Emo for the germline-specific RNAi (rrf-1) did not significantly change compared to that for N2 RNAi (N > 30, P = 1, Fisher’s exact test). (D) Percentage of endomitotic oocytes in algn-7 RNAi-treated rrf-1 worms after mating with N2 males. Supply of WT sperm did not rescue Emo (N > 30, P = 1, Fisher’s exact test). (E) Effects of algn-7 RNAi at the distal tip of the gonad (rrf-1). While UDA-FITC lectin staining (green) was decreased in the region, the number of pH3-positive cells (red) did not change between the N2 and algn-7 RNAi-treated rrf-1 animals. DAPI (blue). Scale bar, 100 μm. (F) The number of germline stem cells in M phase was counted by pH3 staining at the distal tip region of gonads. The number of pH3-positive cells did not change between N2 (n = 10) and algn-7 RNAi-treated rrf-1 animals (n = 24) (Student’s t test).
We observed the Emo phenotype in the germline specific algn-7 RNAi-treated animals. From this result, we suspected that abnormality in either oogenesis or spermatogenesis (or both) resulted in the defective phenotype. To test which was essential, a mating experiment was performed. F1 progenies at the L4 stage of rrf-1 hermaphrodites that were fed with algn-7 dsRNAs were mated with N2 male worms fed with OP50 (control male). Sperm of N2 male were labeled with SYTO 17 to monitor mating (Figure 3D). SYTO 17 is a fluorescent nucleic acid-binding dye that labels mitochondria in sperm, but it does not affect sperm function (Hill and L’Hernault 2001). The Emo incidence in the algn-7 RNAi-treated rrf-1 worms supplied with the wild type (WT) sperm did not significantly change compared to the self-fertilized, non-mated worms (P = 1) (Figure 3D). In other experiments, we mated non-RNAi-treated N2 hermaphrodites with algn-7 RNAi-treated male worms. Normal male offspring were raised from the mating, and neither Emo nor embryonic lethal phenotype was observed in the experiments (three independent experiments, n > 300 each). These results indicate that the Emo phenotype observed in the rrf-1 algn-7 RNAi experiments resulted from the abnormal ovulation signal pathway in the oocyte. The results strongly suggest that N-glycosylation is required in oocytes rather than in sperm, and that the expression of algn-7 in the germline is essential for proper oogenesis and ovulation to occur.
N-glycosylation of proteins is dispensable in the germline stem cell niche of the nematode
Does N-glycosylation of proteins play a role in the production of germline stem cells in the nematode? In our previous study, we reported that synthesis of GPI-anchored proteins is essential for the maintenance of the germline stem cell niche of the nematode (Murata et al. 2012). At each distal end of the gonad, where a distal tip cell is covering the gonad, germline stem cells in the gonad divide and supply the oocyte precursors to the gonad. To examine whether algn-7 gene function is necessary for the germline stem cell niche maintenance, we isolated gonads from the algn-7 RNAi treated rrf-1 animals and counted the anti-phospho-histone H3 (pH3)-cross-reacting cells in the distal gonad. Although lectin staining with the UDA-FITC (Urtica dioica), which binds to high mannose type N-glycans (Itakura et al. 2017), was diminished in the RNAi-treated rrf-1 animals, the number of anti-pH3 positive cells did not significantly change in the algn-7 RNAi-treated rrf-1 animals (Figure 3E and F). Thus, the algn-7 gene activity in the nematode germline seems to be dispensable for the maintenance of the germline stem cell niche although there is a possibility that some residual DPAGT1 activity was sufficient to maintain the germline stem cell niche.
Abnormal phenotypes during oocyte-to-embryo transition
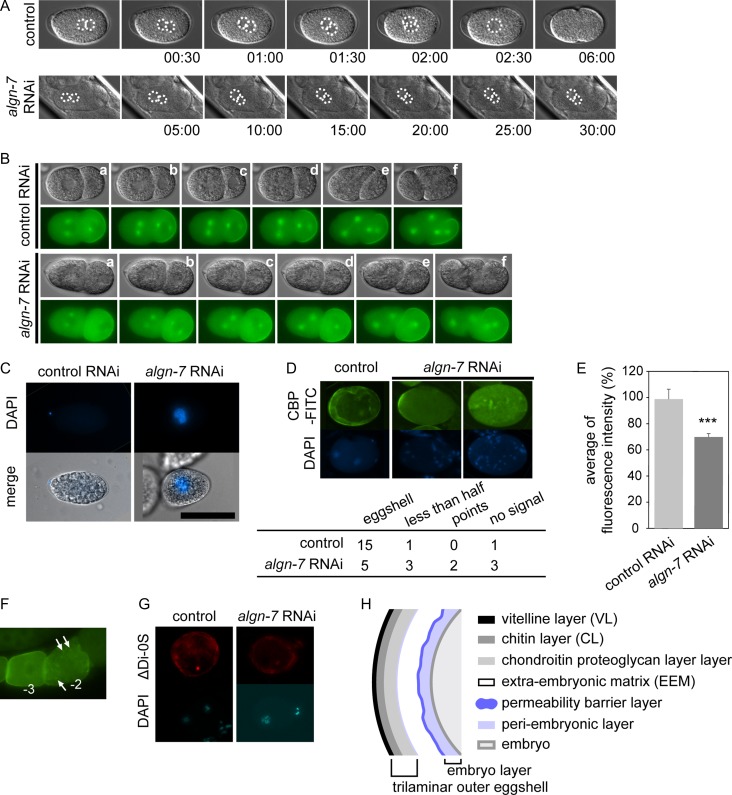
We found abnormal germline phenotypes in the algn-7 RNAi-treated animals. We next examined RNAi phenotypes of the algn-7 gene during oocyte-to-embryo transition (Robertson and Lin 2013). In some algn-7 RNAi-treated animals (5%>, n = 50), pronuclei stopped their movement just prior to the 90 degree rotation, and the fusion of pronuclei did not occur (Figure 4A). Misorientation of AB-cell in early embryonic cell division was also observed in some algn-7 RNAi-treated animals (48%, n = 27) (Figure 4B, see Supplemental Movie 1 and Movie 2). The affected embryos died.
Fig. 4.
Abnormal phenotypes in early embryogenesis and in eggshell formation. (A) Abnormal pronuclear fusion in an algn-7 RNAi-treated animal. Pronuclei stopped their movement at the timing of the 90-degree rotation, and fusion of pronuclei did not occur in the RNAi-treated animals. Normal pronuclear rotation and fusion of N2 are shown at the top (control) and a fertilized egg laid by an algn-7 RNAi-treated OD70 animal is shown at the bottom (algn-7 RNAi). Nuclei are indicated by dotted lines. Time is indicated as min:sec after fertilization. (B) Abnormal rotation of an AB cell in the algn-7 RNAi-treated animals. An egg from an RNAi-treated animal expressing Ppie-1::gfp::tbb-2; Ppie-1::par-2::gfp showed abnormal orientation of the AB-cell at the four-cell stage (bottom) compared to the control (non-RNAi treated animal). Figures are frames from the time-lapse movies (see Movie 1 and Movie 2). (C) A permeability test of the eggshell of an algn-7 RNAi-treated embryo. The algn-7 RNAi-treated egg was not oval, and its nucleus was stained with DAPI (scale bar: 50 μm). (D) FITC-CBP stained the eggshell of a control embryo (N2), while several algn-7-knockdown embryos lost fluorescent signals on the eggshells (freeze-cracked worms). In the table below the top photographs, the number of stained eggs observed (eggs with totally stained eggshells, eggs with less than half of the eggshell stained and eggs with dotted CBP-stained eggshells) and the number of eggs without FITC-CBP staining are shown. (E) Averages of fluorescence intensity of GFP::CHS-1 in the proximal oocytes (total fluorescence intensity of minus 1- to minus 3-oocytes) of AD265 strain (control vs algn-7 RNAi). algn-7 knockdown decreased the CHS-1 expression (N > 30, ***P < 0.001, Student’s t test, unpaired). (F) Typical abnormal distribution of CHS-1 in the algn-7 RNAi-treated AD265 animals. Comparison of the normal CHS-1 distribution in the minus-3 oocyte to the abnormal dotted distribution of CHS-1 in the minus-2 oocyte (arrows). (G) algn-7 RNAi- treatment did not alter the distribution of chondroitin (top right) in the eggshell compared to the control(top left). ΔDi-0S indicates non-sulphated chondroitin staining. (H) A diagram showing a C. elegans eggshell cross section. Modified from Stein and Golden (2015).
To test whether the permeability of the eggshell affected these phenotypes, we stained eggs with DAPI in isotonic solution (Zhang et al. 2005). Nuclei of eggs of the algn-7 RNAi-treated animals were stained, and the results indicated that the permeation barrier of the eggshell was defective. This was the typical osmotic integrity defective (OID) phenotype (Figure 4C). The shape of the OID embryos was not oval, like normal eggs from non-RNAi-treated animals, but was abnormally rounded. A meshwork of the chitin layer provides the embryo its ovoid shape (Stein and Golden 2015). The fifth layer from the outside of the eggshell functions as a permeability barrier layer (Figure 4H) and proper construction of this layer requires the chitin layer (Zhang et al. 2005; Stein and Golden 2015). Thus, we examined whether the eggshell of algn-7 RNAi-treated animals showed abnormal expression of chitin molecules. The presence of the chitin layer was examined by staining the eggshell with FITC-conjugated chitin-binding protein (FITC-CBP), which is a widely used chitin probe (Olson et al. 2012). The number of eggs labeled with FITC-CBP decreased compared to the control RNAi (Figure 4D). From these results, we concluded that algn-7 RNAi-treated eggs had deficient chitin layers, and this resulted in the OID phenotype. Localization of chitin synthase (CHS-1) on the mature oocyte membrane is required for successful construction of the intact chitin layer. To examine the variation in the localization and expression of CHS-1 in the algn-7 knockdown animals, the fluorescence intensity of AD265 (gfp::chs-1)(algn-7 RNAi) animals expressing GFP::CHS-1 was measured. Because the expression of CHS-1 varied gradually near the spermatheca, we measured the average of the fluorescence intensity in the three most proximal oocytes (Figure 4F). The average fluorescence intensity of GFP::CHS-1 in AD 265 (algn-7 RNAi) animals was approximately 20% lower than that for AD265 control RNAi (P < 0.001) (Figure 4E). These results show that algn-7 knockdown affected the expression of chitin synthase in the eggshell, strongly suggesting that inhibition of algn-7 induces down-regulation of chs-1, resulting in the OID phenotype. We also examined the expression of chondroitin, another essential component of the eggshell. By staining with anti-chondroitin stub antibody, we found that expression of chondroitin in the eggshell was not affected (Figure 4G). This result indicates that N-glycosylation of chondroitin proteoglycan does not affect its distribution in the eggshell.
Screening of the germline-expressed N-glycosylated proteins associated with abnormal eggshell formation and/or the Emo phenotype
We showed that inhibition of the algn-7 gene resulted in abnormalities in oocyte formation, oocyte-to-embryo transition and L2 larval lethality. These abnormalities could have resulted from malfunctions of N-glycosylated proteins and/or the unfolded protein response (UPR) induced by accumulated abnormal proteins. To examine the roles of N-glycosylated proteins and UPR in the development of the phenotype, we carried out RNAi screening of the genes encoding N-glycosylated proteins expressed in the germline. We chose this strategy because the phenotypes were mainly observed in the worm germline and in early embryos.
To our knowledge, no information on the N-glycosylated proteins showing the same RNAi phenotypes found in the algn-7-deficient animals has been reported. In the nematode, a list of experimentally verified N-glycosylated proteins is available (Kaji et al. 2007). The list was created by experiments employing lectin affinity purification of C. elegans proteins followed by confirmation of N-glycosylation using mass spectrometry. To determine which N-glycosylated proteins were associated with the OID/Emo phenotype of algn-7 RNAi-treated animals, we used this list in addition to the list of genes expressed in the gonadal transcriptome (RNA-Seq data) made by Ortiz et al. (2014). In the intersection of the two lists, we searched for the genes expressed in the gonads and showing the OID and/or Emo phenotype by searching the phenotype descriptions in WormBase (WS259) and in Textpresso (Müller et al. 2018).
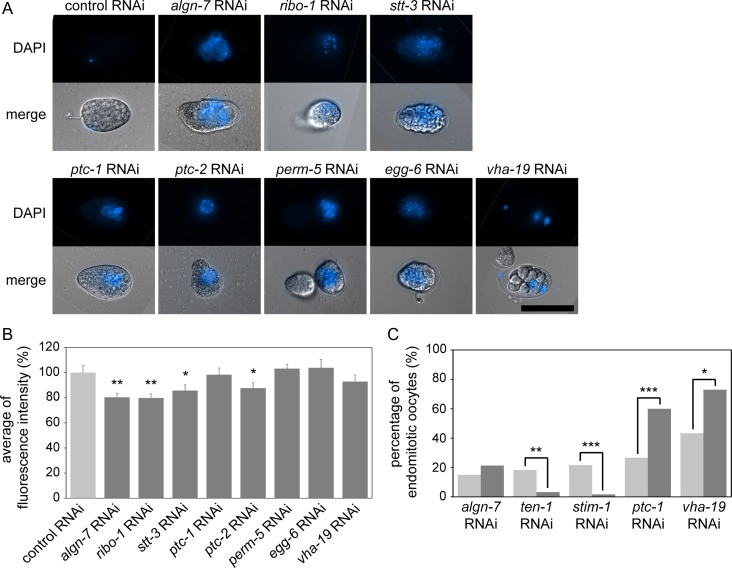
We found 456 genes encoding N-linked glycoproteins expressed in gonads (Supplemental Table S1) (Kaji et al. 2007; Ortiz et al. 2014), and our database search showed that seven of the 456 genes were associated with the OID phenotype: ribo-1, stt-3, ptc-1, ptc-2, egg-6, perm-5 and vha-19 (Table II and Supplemental Table S2). ptc-1/2 encode orthologues of human patched genes (PTCH1/2). RIBO-1 and STT-3 are orthologues of Rpn1/ribophorin I and STT3B, respectively, which encode subunits of the OST complex. VHA-19 is an orthologue of accessory subunit ATP6AP1 of the vacuolar (V)-ATPase proton pump (Lee et al. 2010). perm-5 is a taxonomically restricted gene (Verster et al. 2017) and encodes a protein of members of the human BPI (bactericidal permeability-increasing protein) fold-containing protein family. It is involved in the apoptotic process, embryo development and oogenesis in C. elegans (Green et al. 2011). perm-5 is expressed in gonadal sheath cells, vulval cells and in head neurons (Wang et al. 2006). EGG-6 is an extracellular leucine-rich-repeat-only protein and is required for proper organization of the apical extracellular matrix (Mancuso et al. 2012). We performed RNAi knockdown of the seven genes in the AD265 strain (see above) and measured the fluorescence intensity of GFP::CHS-1 in the proximal oocytes. The OID phenotype was confirmed for the seven genes in the RNAi-treated AD265 animals (Figure 5A). The average fluorescence intensity of the GFP-tagged CHS-1 in the ptc-2, ribo-1, stt-3 RNAi-treated animals (AD265) was significantly lower than that in AD265 control RNAi-treated animals (N > 20, P < 0.05) (Figure 5B). In the vha-19 (RNAi) animals, the distribution of CHS-1 was affected (Table II). RNAi knockdown of ptc-1, perm-5 or egg-6 did not affect the expression of GFP::CHS-1. The abnormal CHS-1 distribution phenotype was also detected in algn-7 (RNAi) animals (N2) at a low frequency (c.a., 2.5%, n = 40). These results suggest that the OID and chitin synthase phenotype under algn-7 RNAi can be ascribed to PTC-2/RIBO-1/STT-3 malfunctions.
Table II.
List of N-glycosylated proteins with Emo and/or OID phenotypes with algn-7 data for comparison
| worm genes | Emo/ OID | Emo in rrf-1; RNAi* | gfp::chs-1 | human orthologues | activity/function | human diseases |
|---|---|---|---|---|---|---|
| ribo-1 | − / ○ | 0.00% | down-regulation | Rpn1 | OST complex accessory subunit | NK (not known) |
| stt-3 | − / ○ | 57.6% | down-regulation | STT3B | OST complex catalytic subunit | STT3B-CDG |
| ptc-1 | ○ / ○ | 60.0% | WT | PTCH1/2 | protein patched homolog | Parkinson’s disease, nevoid vassal cell carcinoma syndrome, basal cell carcinoma, holoprosencephaly medulloblastoma |
| ptc-2 | − / ○ | 0.00% | down-regulation | ” | protein patched homolog | ” |
| egg-6 | − / ○ | 0.00% | WT | LRRC15 | extracellular leucine-rich repeat protein | NK |
| perm-5 | − / ○ | 0.00% | WT | BPI fold containing protein | NK | |
| vha-19 | ○ / ○ | 73.0% | localization variant | ATP6AP1 | subunit Ac45 of the membrane-bound (V0) domain of vacuolar proton translocating ATPase (V-ATPase) | Immunodeficiency 47 |
| stim-1 | ○ / − | 1.66% (n = 60) | − | STIM1 | orthologue of STIM1, a putative ER Ca2+ sensor | Immunodeficiency 10, myopathy, Stormorken syndrome |
| ten-1 | ○ / − | 3.27% | − | TENM1/2/3/4 | orthologue of fly Ten-m/Odz and vertebrate teneurins | Microphthalmia with coloboma-9 (MCOPCB9) |
| algn-7 | ○ / ○ | 21.3% | down regulation | DPAGT1 | DPAGT1 | DPAGT1-CDG (CDG-Ij) and limb-girdle congenital myasthenic syndrome (CMS) with tubular aggregates |
n = 30 except for stt-3 (n = 26) and stim-1 (n = 60).
“○” and “−” refer to presence and absence of the indicated phenotype, respectively.
Fig. 5.
RNAi phenotypes of the seven genes encoding different N-glycosylated proteins. (A) Permeability test in the AD265 strain. RNAi of seven genes encoding different N-glycosylated proteins induced the OID phenotype (scale bar: 50 μm). (B) Averages fluorescence intensities of GFP::CHS-1 in RNAi of seven genes coding N-glycosylated proteins in the AD265 strain (N > 20, *P < 0.05, **P < 0.01, Student’s t test, unpaired). (C) Comparison of Emo incidence in the RNAi experiments of the four N-glycosylated protein-coding genes. Results of the germline-specific RNAi (dark-gray bars) and the whole body RNAi (light-gray bars) are shown. The number of Emo worms for ten-1 and stim-1 RNAi-treated rrf-1 worms was less than that for N2 (N > 35, **P < 0.01, ***P < 0.001, Fisher’s exact test).
In the database search, we found four Emo genes encoding N-glycosylated proteins expressed in the germline; ten-1, stim-1, ptc-1 and vha-19 (Table II and Supplemental Table S2). As shown in Table II, we observed the Emo phenotype for the germline specific RNAi of stt-3, although no description of the Emo phenotype was found in our database search. TEN-1 is the C. elegans teneurin homolog and is required for proper formation of gonadal and pharyngeal basement membrane. It also has roles in hypodermal morphogenesis and proper elongation and placement of pharyngeal neurons (Trzebiatowska et al. 2008; Mörck et al. 2010). STIM-1 is an ER Ca2+ sensor that senses calcium ion depletion (Soboloff et al. 2012). PTC-1 and VHA-19, identified as OID genes, were also found to be Emo responsible genes. We examined the RNAi phenotypes of these genes in N2 and the somatic RNAi-defective strain rrf-1, and measured the incidence of the endomitotic oocytes. The percentage of Emo in the ten-1 RNAi-treated rrf-1 animals and in the stim-1 RNAi-treated rrf-1 animals decreased compared to the whole body RNAi (N > 35, P < 0.01) (Figure 5C). The results indicate that Emo in the ten-1- and stim-1-knockdown animals was due to ovulation abnormality in somatic gonads (e.g., sheath cells, spermatheca or the uterus). These results seemed to be consistent with the previous studies showing that TEN-1 is necessary for maintenance of the somatic gonadal sheath (Drabikowski et al. 2005; Trzebiatowska et al. 2008) and that STIM-1 has a role in calcium ion control in gonadal sheath in ovulation (Singaravelu and Singson 2013). The percentage of endomitotic oocytes in the rrf-1 animals treated with either ptc-1 or vha-19 RNAi increased to more than the percentage in the N2 animals (Figure 5C). For the whole-body vha-19-RNAi in N2 worms or in OD95 worms, a larval lethal phenotype and a sick phenotype are reported in addition to various oocyte phenotypes (Green et al. 2011) (WormBase, WS261). This is probably because the whole-body RNAi affects various somatic gonadal systems and other somatic cell types. The influence of ptc-1 and vha-19 RNAi in N2 was so severe that oocytes did not mature and the Emo incidence was lower than that in the germline-specific RNAi experiments. This possibility is supported by RNAi in the NL3511 strain. ptc-1 RNAi-treated NL3511 animals (F1) showed full penetrant sterility at 25°C, and vha-19 RNAi-treated NL3511 animals (F1) showed larval arrest at 25°C. The full penetrant sterility (ptc-1 RNAi) and the larval arrest (vha-19 RNAi) observed in the NL3511 animals indicate that ptc-1 and vha-19 are playing essential roles in somatic tissues as well. Because rrf-1 animals show deficiencies of RNAi processing in somatic gonads (Kumsta and Hansen 2012), we concluded that the Emo phenotype observed in the ptc-1 RNAi in a rrf-1 mutant background indicates that ptc-1 plays essential roles in the germline. Since the phenotype of ptc-1 and vha-19 deficiency in the germline was the same as that of algn-7, we identified these two genes as important factors associated with the glycosylation disorders found in this study.
Discussion
In this study, we identified the gene algn-7 as the sole C. elegans orthologue of the human DPAGT1 which catalyses the first step of N-glycosylated protein synthesis. Although previous studies on N-glycosylation pathways of C. elegans have focused on OST genes that function at the later step of N-glycan synthesis, no studies on the nematode DPAGT1 that functions at the first step of N-glycosylation have been published. As shown in this paper, inhibition of DPAGT1 activity of C. elegans by RNAi and gene knockout enabled us to study the role of DPAGT1 in vivo and in vitro with high reproducibility, and it was easy to examine the roles of the enzyme in gametogenesis and early embryonic development at a single-cell level.
Because inhibition of OST activity also results in inhibition of N-glycosylation of proteins, we compared the previously reported phenotypes with the algn-7 phenotypes. algn-7 (RNAi) animals displayed the Emo (endomitotic oocytes) phenotype and abnormal oocyte-to-embryo transition, indicating essential roles of DPAGT1 in cell cycle progression. In a previous paper, we reported that RNAi-mediated inhibition of OST genes (dad-1, ostb-1 or stt-3) results in an abnormal oocyte morphology phenotype in addition to the Sma (small body size) phenotype (Akiyoshi et al. 2015). This indicates possible roles of OST genes in cell cycle progression. The C. elegans emo-1 gene is of special interest regarding the cause of the Emo phenotype. As indicated in the name, inhibition of the emo-1 gene results in the Emo phenotype. The gene emo-1 is an orthologue of human SEC61 gene (Iwasaki et al. 1996), and the SEC61 and ribosome associate with the OST complex where nascent N-glycoproteins are synthesized (Harada et al. 2009; Pfeffer et al. 2014). In the present study, we discovered that inhibition of stt-3 also induced the Emo phenotype (Table II). Inhibition of ribo-1, another OST genes, results in abnormal chromosome segregation during oogenesis of the RNAi-treated animals in addition to the OID phenotype (Stevens and Spang 2013). Involvement of RIBO-1 and E-cadherin in polarity formation of early C. elegans embryos was also reported recently (Stevens and Spang 2017). The gene dad-1 (defender against cell death-1),which is the sole orthologue of human DAD1 (Roboti and High 2012), was originally identified as a suppressor of apoptosis in C. elegans (Sugimoto et al. 1995). The DAD1 gene encodes an essential component of the OST complex and plays essential roles in N-glycosylation of proteins (Kelleher and Gilmore 1997; Makishima et al. 1997). These analyses of algn-7 and OST genes indicate that synthesis of N-glycan is essential for cell cycle progression in oogenesis and in early embryonic development.
Inhibition of the algn-7 gene resulted in the OID phenotype as was reported for OST genes (ribo-1, ostd-1, stt-3 and dad-1) (Stevens and Spang 2013). In the algn-7 (RNAi) animals, the osmotic barrier of affected eggs was disrupted, and the expression of chitin synthase and its localization was disturbed in the eggs. Chitin is a polymer that does not require N-glycosylation directly. In Saccharomyces cerevisiae, Chs3, the catalytic subunit of chitin synthase III, is N-glycosylated (Sacristan et al. 2013). Although there is no evidence of N-glycosylation of the C. elegans CHS-1 protein, it could be N-glycosylated and underglycosylation of the CHS-1 might be involved in the eggshell malformation. Another possibility was suggested by our inspection of in vivo proteomics interaction data. C. elegans CHS-1 physically interacts with MBK-2, which is one of two C. elegans members of the DYRK (dual-specificity Yak1-related kinase) family of proteins. MBK-2 is expressed uniformly in the cortex of oocytes and newly fertilized eggs, and maternal MBK-2 is an important regulator of the oocyte-to-embryo transition (Chen et al. 2016). PTC-1 also interacts with MBK-2 (Chen et al. 2016), thus it is an intriguing possibility that PTC-1 and CHS-1 interact through MBK-2 and they are playing roles in eggshell formation and oocyte-to-embryo transition. The chitin layer is an indispensable component layer of the nematode eggshell (Olson et al. 2012), and normal synthesis and localization of chitin synthase (CHS-1) is indispensable for osmotic integrity and for inhibition of polyspermy (Johnston et al. 2006, 2010; Johnston and Dennis 2012). Inhibition of gna-2 (C. elegans glucosamine-6-phosphate N-acetyltransferase (GNPNAT1/GNA1)), which is essential for synthesis of uridine diphosphate-N-acetylhexosamine, results in the OID phenotype and a polarity defect phenotype. The gna-2 gene is in the hexosamine biosynthesis pathway, and is essential for N-glycan, O-glycan, proteoglycan and O-GlcNAc synthesis. Because chitin and either cpg-1 or cpg-2 (the two major core proteins of chondroitin proteoglycans that have chitin-binding motifs) are essential for osmotic/permeability barrier and normal polarization of fertilized eggs (Johnston et al. 2006), we examined the expression of chondroitin which is also a major component of C. elegans eggshell. Chondroitin is synthesized by chondroitin synthase SQV-5 and chondroitin polymerizing factor MIG-22. Inhibition of either sqv-5 or mig-22 results in a severe cytokinesis defect (Hwang et al. 2003; Mizuguchi et al. 2003), but we did not detect cytokinesis defects in all of the alg gene-/OST-gene knockdown animals (this study and (Akiyoshi et al. 2015)). Stevens and Spang reported that most ribo-1 (RNAi) one-cell stage embryos are able to divide into two cells, but subsequent cytokinesis events fails more often (Stevens and Spang 2013). The severe cytokinesis defects reported in their paper are different from the severe cytokinesis defects observed in chondroitin-defected embryos. This result strongly suggests that inhibition of N-glycan synthesis is not directly involved in the severe cytokinesis defects observed under inhibition of chondroitin synthesis although some of the chondroitin proteoglycan core proteins are N-glycosylated. This is also supported by our observation that the distribution of chondroitin was normal in the eggshell, indicating that inhibition of algn-7 resulted in disturbance of chitin synthase CHS-1, while it did not result in inhibition of SQV-5, MIG-22, or proteins involved in the cytokinesis directed by these proteins. In summary, N-glycan synthesis is indispensable for cell cycle progression during oogenesis and oocyte-to-embryonic transition and is dispensable for chondroitin-mediated embryonic cytokinesis.
Although we previously reported high expression of hsp-4::GFP in algn-7 RNAi-treated worms (Akiyoshi et al. 2015), the possibility that the phenotypes reported in this study were caused directly by the unfolded protein response (UPR) seems less likely because (1) involvement of the UPR in these phenotypes has not been reported before and (2) under ribo-1 RNAi-treatment, ER morphology is largely unaffected, while the phenotypes discussed above are observed (Stevens and Spang 2013). The UPR is also not active in fibroblasts derived from DPAGT1-CDG patients, unlike that seen in cells from patients with PMM2-CDG or DPM1-CDG (Yuste-Checa et al. 2017). Thus, the phenotypes reported in this study appear to be brought about by malfunctions in or deficiencies of a limited numbers of N-glycosylated proteins, including the ones identified in this study.
In the latter part of the present study, we screened 456 germline-expressed genes encoding N-glycosylated proteins for the algn-7 RNAi phenotypes. We found five germline-expressed genes (stt-3, ribo-1, ptc-1/2, and vha-19) showing the same algn-7 RNAi-phenotypes. As expected, two genes (stt-3 and ribo-1) were OST genes and they are well-known CDG genes. Inhibition of vha-19 resulted in the OID phenotype, and CHS-1 distribution was disturbed in the rrf-1 (vha-19 RNAi) animals indicating its function in the germline. A recent study using exome sequencing of human CDG patients showed that the human ATP6AP1, which is a vha-19 orthologue, is a gene responsible for a CDG with abnormal glycosylation of proteins and an immunodeficiency with hepatopathy (Jansen et al. 2016). ATP6AP1, an accessory subunit of the vacuolar (V)-ATPase proton pump, is required for luminal acidification of secretory vesicles, lysosomes and the Golgi (Toei et al. 2010). Inhibition of the vha-19 gene may cause an abnormality in vacuolar acidification (possibly the Golgi acidification) resulting in the abnormality of glycosylation as suggested in vertebrates (Hirata et al. 2010; Esmail et al. 2016; Pamarthy et al. 2018). Inhibition of the ptc-2 gene resulted in the OID and abnormal chitin synthase (chs-1) phenotype. In the supplemental table of a paper by Green et al. (2011), they reported that inhibition of ptc-2 causes abnormal condensation of pachytene chromatin of oocytes, resulting in multinucleated oocytes at the “medial” gonads of C. elegans. PTC-1 functions in the germline cytokinesis of the nematode (Kuwabara et al. 2000). Inhibition of ptc-1 resulted in the OID and Emo phenotype. Harris et al. reported that inhibition of ptc-1 results in MSP-independent MAPK activation, and the deletion of ptc-1 results in reorganization of meiotic microtubules of oocytes (Harris et al. 2006). In the paper, they also reported that deletion of emo-1, which is an orthologue of human SEC61 gene discussed above, results in the same phenotype. ptc-1 is a negative regulator of oocyte meiotic maturation (Govindan et al. 2006).
PTCH1 and PTCH2, which are human orthologues of ptc-1 and ptc-2, are not known to be CDG genes. PTCH1 is involved in BCC-1 (basal cell carcinoma: OMIM 605462), BCNS (basal cell nevus syndrome: OMIM 109400), HPE-7 (holoprosencephaly, type 7: OMIM 610828), KCOT (keratocystic odontogenic tumor), and ODA (ocular developmental anomalies such as anophthalmia/microphthalmia, anterior segment mesenchymal dysgenesis). PTCH2 is involved in BCC-1, BCNS, and MDB (medulloblastoma). Does N-glycosylation of PTCH1/2 affect pathogenesis of these diseases and CDGs? The chick patched gene PTCH1 is in the hedgehog signaling pathway, and N-glycosylation of PTCH1 is necessary for the interaction of PTCH1 with sonic hedgehog (SHH) (Marigo et al. 1996). It is also known that mammalian Smoothened protein (SMO) is N-glycosylated, and its glycosylation is essential for signal transduction (Marada et al. 2015). Thus, examining N-glycosylation patterns of PTCH1/2 in patients of CDGs and other diseases may help clarify molecular mechanisms of pathogenesis of these diseases. For example, PTCH1 is a dependence receptor of apoptosis and the lack of the ligand induces apoptosis (Fombonne et al. 2012). Underglycosylation of PTCH1 may result in dysregulation of apoptosis and might be involved in tumorigenesis in these diseases.
Although there are no obvious canonical orthologues of SHH and SMO (Bürglin 2008) in the C. elegans genome, there are various hedgehog related (Hh-related) genes in the genome (Roy et al. 2018). N-glycosylation of PTC-1 may be essential for interactions between PTC-1 and Hh-related protein ligands. Finding true interacting partners of PTC-1/2 in C. elegans may help understand the roles of PTCH1/2 gene functions in human diseases. The human orthologues of the ptc-1 and ptc-2 genes could be new CDG-related genes, and it will be intriguing to examine their roles in the CDG pathogenesis in CDG patients.
In conclusion, our screening of the germline-expressed N-glycoprotein genes associated with the OID and Emo phenotypes identified a recently identified CDG gene (vha-19), previously identified CDG genes (stt-3 and ribo-1), and two new candidate genes possibly involved in CDG pathogenesis (ptc-1 and ptc-2). Although CDG patients experience multiple symptoms, the molecular mechanisms underlying the symptoms have not been well understood. By using genetic and reverse genetic analyses of the identified CDG candidate genes in C. elegans, we can identify germline gene networks responsible for the osmoregulatory systems and cell-cycle regulatory systems associated with the OID and Emo phenotypes. It is interesting to note that a PMM2-CDG patient presented with infertility and premature ovarian failure (Vuillaumier-Barrot et al. 2012). Association of PMM2-CDG with primary ovarian failure is well known (Kristiansson et al. 1995). We expect that gene network analysis of the identified genes described in this study may shed light on the molecular mechanisms of the development of human CDG symptoms. In this respect, expression of PTC-1 and other N-glycosylated proteins with mutations of possible N-glycosylation sites (gene editing of the sequons) followed by examination of phenotypes will help understand the roles of N-glycan moieties and clarify the molecular mechanisms underlying the phenotypes. Studies in this direction are currently underway in our laboratory.
Materials and methods
Worm strains
The Bristol strain N2 was used as the wild-type animal (Brenner 1974). The algn-7 deletion allele tm3591 was obtained from the TMP/UV library as described previously (Gengyo-Ando and Mitani 2000) by using the following primers: P1: GTGTAATATGTGCAGCTGTC and P2: CAACGGATCCTGTAATGCTG. The animal was maintained by using a recessive balancer unc-51(e369). The tm3591 homozygous animals were chosen based on their phenotype (L2 lethal and/or increased expression of hsp-4::gfp).
The genotype of the somatically rescued animals with DPAGT1 was tm3591;zcIs4; Ex290[Peef-1A.2::hDPAGT1(noTag)], which were produced from tm3591/e369;zcIs4; Ex290 hermaphrodites. The following strains were obtained from the Caenorhabditis Genetics Center (CGC): N2, NL2098: rrf-1(pk1417) I, NL3511: ppw-1(pk1425) I, JH1473: [itIs153; ruIs57], AD265: nnIs2 [unc-119(+) Ppie-1::GFP::chs-1] (Maruyama et al. 2007), and SJ4005:hsp-4::gfp(zcIs4)V (Calfon et al. 2002). All worms were grown at 20°C on Nematode Growth Medium (NGM) agar plates with E. coli OP50 as a food source (Brenner 1974).
Immunofluorescence and fluorescent probe staining
The gonads and embryos were freeze-cracked and fixed in methanol at −20°C for 5 min. After being blocked with 1% BSA/TBST (Tris-buffered saline containing 0.2% Tween 20), samples were incubated with the GNA-FITC (1:40), WGA-FITC (1:50), UDA-FITC (1:1000) or CBP-FITC (1:250) at room temperature for 2 h or anti-phospho histone H3 (Ser10) antibody (anti-pH3 antibody, 1:1000) at 4°C overnight. For pH3 antibody staining, samples that were washed twice with TBST were further incubated with the Alexa Fluor 594 goat anti–rabbit IgG (1:200) at room temperature for 2 h in a dark, humid chamber. For chondroitin staining, freeze-cracked eggs were treated with 0.05 U/μL chondroitinase ABC for 1 h after blocking with TBS containing 0.1% Tween 80. The eggs were incubated with the anti-chondroitin stub antibody (Seikagaku Corporation, Tokyo) diluted 1:2000 with TBS at 20°C for 2–4 h. After being washed, the eggs were incubated with a rhodamine-conjugated goat anti-mouse IgG for 2 h at 20°C. The GNA-FITC (F-7401), WGA-FITC (F-2102) and UDA-FITC (F-8005) lectins were purchased from EY laboratories Inc. The CBP-FITC (P5211) was purchased from New England Biolabs. The anti-pH3 antibody was purchased from Upstate Biotechnology. The Alexa Fluor 594 goat anti–rabbit IgG was purchased from Invitrogen Molecular Probes.
Plasmid construction
For construction of Peef-1A.2::algn-7::Venus and Peef-1A.2::hDPAGT1, cDNAs were amplified by PCR from total cDNA of C. elegans and the LNCaP human cell line, respectively, using the primers listed below including restriction enzyme sites, followed by cloning into the NotI or NotI/BglII sites of the expression vector pFX_LVT-R03G5.1 (Kitagawa et al. 2007).
NotIY60A3A14Fwd1:
TGGCGGCCGCATGTCGCTGATATTTTTGGC
NotIY60A3A14Rev1:
TGGCGGCCGCATTCAACAACATCATAAAACAACG
hDPAGT1_NotIFwd1:
TGGGCGGCCGCATGTGGGCCTTCTCGGAATTG
hDPAGT1_BglIIRev2:
AGAGATCTTCAGACATCATAGAAGAGTC
RNAi interference
RNAi experiments in this study were based on the feeding method (Timmons et al. 2001). The cDNA clone for algn-7 was amplified by PCR from total cDNA of N2 worms. Primers for the algn-7 cDNA were 5′-TCGCTGATATTTTTGGCGAT-3′ and 5′-CGAGATGGTATGATGCCAGA-3′. The cDNA for T08D2.2 was also amplified by using the following primers: 5′GAGCCAACTTGGGCAGAAT-3′ and 5′-ACCCAAATCCGCTATTCTCC-3′. The fragments of the cDNAs were cloned into the EcoRV site of the L4440 (pPD129.36) vector by TA cloning (Borovkov and Rivkin 1997; Gengyo-Ando et al. 2006). The cloned plasmids were transformed into E. coli HT115 (DE3). All other bacterial feeding clones were obtained from the Ahringer RNAi clone library (Kamath and Ahringer 2003). Cultured bacterial strains were seeded onto NGM agar plates and incubated at 37°C overnight. To induce the expression of dsRNAs, the plates were treated with 2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated at 37°C for 6 h. L4-stage hermaphrodites were transferred onto the feeding plates and maintained at 25°C. The adult P0 worms and embryos were examined after 24 or 48 h. L3–L4 larvae of the F1 generation were moved to a new RNAi plate after 48 h, and we observed their phenotypes after 72–120 h.
Assay of C. elegans UDP-GlcNAc-1-phosphate:dolichyl phosphate GlcNAc-1-phosphotransferase (DPAGT1) activity
The DPAGT1 assay was performed using modification of the procedure of Eckert et al. (1998). Briefly, worms were homogenized in water in the presence of protease inhibitors (cOmplete ULTRA Tablets, Mini, EDTA-free, Roche Diagnostics) by sonication, and the samples were immediately frozen in liquid nitrogen. The worm homogenates containing 90–100 μg of protein were incubated for 10 min at 37°C in the presence of UDP-[3H]GlcNAc (37 kBq, PerkinElmer, MA), 10 mM MgCl2, 0.2% Triton X-100, 50 mM Tris-HCl (pH 7.4), and 10 μg of P-dolichol (Olbracht Serdary Research Lab., Canada) in a total volume of 50 μL. Incubations were terminated by addition of 6 mL of chloroform/methanol/water (1/1/1), vortexed and centrifuged. The aqueous phase was discarded, and the remaining organic phase was added to 5 mL of 50% methanol. After centrifugation, the aqueous phase was aspirated, and the radioactivity of the organic phase was measured by a liquid scintillation counter (Aloka LSC-6100, Tokyo, Japan).
DAPI staining of gonads
Adult hermaphrodites were mounted in M9 buffer containing 5 μg/mL DAPI (4′, 6′-diamidino-2-phenylindole hydrochloride) on poly-l-lysine-coated slide glass (Matsunami, Japan). Gonads were released by cutting just behind the pharynx under a dissecting microscope (Olympus SZX12), and incubated at room temperature for 30 min in the dark. After incubation, the samples were washed once in M9 buffer and mounted in 15 μL of 1, 4-diazabicyclo (2,2,2) octane (DABCO). Finally, samples were covered with an 18 × 18 mm cover glass (Matsunami, Japan), and the slide was sealed with nail polish.
Mating experiment
To examine whether the Emo phenotype in algn-7 RNAi-treated rrf-1 animals was rescued by supplying wild type sperm, we performed mating experiments according to the previously described method (Hill and L’Hernault 2001). L4 stage N2 male worms were transferred onto OP50 plates and incubated at 20°C overnight. The adult males were mounted in 100 μL of M9 buffer containing 10 μM SYTO17 red (Molecular Probes, Eugene, OR) on three-well printed slide glass (Matsunami, Japan). They were maintained at 25°C for 3 h in the dark. The stained males were moved onto an OP50 plate for 30 min. The same number of SYTO 17-labeled males and rrf-1 L4 hermaphrodites (F1) were fed with algn-7 dsRNAs, and they were both transferred onto a new OP50-mating plate. A plate on which only the hermaphrodites were placed was also prepared as a control. They were incubated at 25°C overnight in the dark and observed in the morning. In other experiments, we mated algn-7 (RNAi) males with N2 hermaphrodites. N2 hermaphrodites and males (10:2) were transferred onto algn-7 RNAi plates and incubated at 25°C for 72 h. L4 males (F1) from the algn-7 RNAi plates were mated with L4 hermaphrodites (N2) from separately prepared OP50 plates (10:2), and embryonic phenotypes of the offspring were monitored (three independent experiments, n > 300 each).
Eggshell permeability experiment
The eggshell permeability experiment was performed according to the previously described method (Zhang et al. 2005). Live embryos were dissected out from adult hermaphrodites fed with dsRNA-producing bacteria for 24 h in egg buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM HEPES, pH 7.3, 340 mOsm) containing 20 ng/mL DAPI on eight-well printed microscope slide glass (Matsunami Glass, Japan). The osmolality of egg buffer was measured with OSMOMAT 030-D Osmometer (Gonotec, Berlin, Germany). The embryos were covered with 24 × 50 mm cover glass and observed under a microscope.
Measurement of fluorescence intensity
To compare the expression level of chitin synthase CHS-1 in each RNAi knockdown animal, the fluorescence intensity of GFP::CHS-1 expressed in oocytes was measured. The oocytes were transferred from RNAi-treated adult hermaphrodites into M9 buffer on eight-well printed microscope slide glass (Matsunami Glass, Japan) and covered with 24 × 50 mm cover glass. The average fluorescence intensity in the three most proximal oocytes was measured using MetaMorph software (version 6.1r5, Molecular Devices Corporation).
Microscopy
Differential interference contrast (DIC) and fluorescence images were obtained using an Olympus BX51 fluorescence/DIC microscope (Olympus, Tokyo, Japan) equipped with a MAC5000 controller system (Ludl Electronic Products Ltd., NY) as previously described (Mizuguchi et al. 2008). Phenotypes were also monitored and scored by using a high resolution dissecting microscope (Olympus SZX12). The acquired images were processed by MetaMorph software (see above).
Bioinformatics and databases
Orthologous sequences were identified and aligned by using PSI-BLAST (five iterations) and Clustal Omega (Sievers et al. 2011). The list of the C. elegans N-glycosylated proteins was compiled from the supplemental materials of (Kaji et al. 2007) and GlycoProtDB (GPDB) [http://acgg.asia/gpdb2/; last accessed on November 28, 2018] at the Japan Consortium for Glycobiology and Glycotechnology DataBase. WormBase [http://www.wormbase.org/#012-34-5; last accessed on November 28, 2018] and Textpresso [https://textpressocentral.org/tpc; last accessed on November 28, 2018](Müller et al. 2018) were used to investigate genes with phenotypes similar to algn-7 RNAi. CGGDB [http://jcggdb.jp/cggdb/; last accessed on November 28, 2018] (Akiyoshi et al. 2015) was used to search the phenotype of RNAi knockdown of C. elegans glycogenes. We used the OMIM database [http://www.omim.org/; last accessed on November 28, 2018] when examining CDG and other diseases.
Statistical analysis
Statistical analyses, including Student’s t test and Fisher’s exact test were performed using the R statistical package (R, version 2.11.0) with R-Commander and its plugin package, EZR (Kanda 2013).
Supplementary Material
Acknowledgements
We acknowledge the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) for several strains used in this study. We thank Prof. Katsuko Yamashita and Dr. Keiko Fukushima for their help in the initial phase of our study.
Funding
This work was supported by JSPS KAKENHI Grant Number JP16K07298 to K.N. This work was also supported by grants from the Core Research for Evolutional Science and Technology (CREST) Program of the Japan Science and Technology Corporation (to K.N.) and by Grant 120140 from the Mizutani Foundation for Glycoscience (to K.N.).
Conflict of interest statement
None declared.
Abbreviations
CDG, congenital disorders of glycosylation; Dol, dolichol; DPAGT1, UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase; ER, endoplasmic reticulum; GFP, green fluorescent protein; GNA, Galanthus nivalis agglutinin; LLO, dolichol lipid-linked oligosaccharide; MSP, major sperm protein; OST, oligosaccharyltransferase; TMP, trimethylpsoralen; UDA, Urtica dioica agglutinin; UPR, unfolded protein response; WGA, wheat germ agglutinin.
References
- Aebi M, Bernasconi R, Clerc S, Molinari M. 2010. N-glycan structures: Recognition and processing in the ER. Trends Biochem Sci. 35(2):74–82, 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Akiyoshi S, Nomura KH, Dejima K, Murata D, Matsuda A, Kanaki N, Takaki T, Mihara H, Nagaishi T, Furukawa S et al. . 2015. RNAi screening of human glycogene orthologs in the nematode Caenorhabditis elegans and the construction of the C. elegans glycogene database. Glycobiology. 25(1):8–20, 10.1093/glycob/cwu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaya K, Finlayson S, Slater CR, Cossins J, Liu WW, Maxwell S, McGowan SJ, Maslau S, Twigg SRF, Walls TJ et al. . 2012. Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am J Hum Genet. 91(1):193–201, 10.1016/j.ajhg.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovkov AY, Rivkin MI. 1997. XcmI-containing vector for direct cloning of PCR products. Biotechniques. 22(5):812–814. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. 2008. The Hedgehog protein family. Genome Biol. 9(11):241, 10.1186/gb-2008-9-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415(6867):92–96, 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chen J-X, Cipriani PG, Mecenas D, Polanowska J, Piano F, Gunsalus KC, Selbach M. 2016. In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into P granule dynamics. Mol Cell Proteomics. 15(5):1642–1657, 10.1074/mcp.M115.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski K, Trzebiatowska A, Chiquet-Ehrismann R. 2005. ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev Biol. 282(1):27–38, 10.1016/j.ydbio.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Eckert V, Blank M, Mazhari-Tabrizi R, Mumberg D, Funk M, Schwarz RT. 1998. Cloning and functional expression of the human GlcNAc-1-P transferase, the enzyme for the committed step of the dolichol cycle, by heterologous complementation in Saccharomyces cerevisiae. Glycobiology. 8(1):77–85. [DOI] [PubMed] [Google Scholar]
- Esmail S, Yao Y, Kartner N, Li J, Reithmeier RAF, Manolson MF. 2016. N-linked glycosylation is required for vacuolar H+ -ATPase (V-ATPase) a4 subunit stability, assembly, and cell surface expression. J Cell Biochem. 117(12):2757–2768, 10.1002/jcb.25574. [DOI] [PubMed] [Google Scholar]
- Fombonne J, Bissey P-A, Guix C, Sadoul R, Thibert C, Mehlen P. 2012. Patched dependence receptor triggers apoptosis through ubiquitination of caspase-9. Proc Natl Acad Sci USA. 109(26):10510–10515, 10.1073/pnas.1200094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. 3(5):433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K, Mitani S. 2000. Characterization of Mutations Induced by Ethyl Methanesulfonate, UV, and Trimethylpsoralen in the Nematode Caenorhabditis elegans. [DOI] [PubMed]
- Gengyo-Ando K, Yoshina S, Inoue H, Mitani S. 2006. An efficient transgenic system by TA cloning vectors and RNAi for C. elegans. Biochem Biophys Res Commun. 349(4):1345–1350. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Cheng H, Harris JE, Greenstein D. 2006. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr Biol. 16(13):1257–1268, 10.1016/j.cub.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Green RA, Kao H-L, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K et al. . 2011. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 145(3):470–482, 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Li H, Li H, Lennarz WJ. 2009. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc Natl Acad Sci USA. 106(17):6945–6949, 10.1073/pnas.0812489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JE, Govindan JA, Yamamoto I, Schwartz J, Kaverina I, Greenstein D. 2006. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev Biol. 299(1):105–121, 10.1016/j.ydbio.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Helenius A. 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 5(3):253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 73:1019–1049, 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hill KL, L’Hernault SW. 2001. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev Biol. 232(1):105–114, 10.1006/dbio.2000.0136. [DOI] [PubMed] [Google Scholar]
- Hirata Y, Shimokawa N, Oh-hashi K, Yu Z-X, Kiuchi K. 2010. Acidification of the Golgi apparatus is indispensable for maturation but not for cell surface delivery of Ret. J Neurochem. 115(3):606–613, 10.1111/j.1471-4159.2010.06966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H-Y, Olson SK, Esko JD, Horvitz HR. 2003. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 423(6938):439–443, 10.1038/nature01634. [DOI] [PubMed] [Google Scholar]
- Itakura Y, Nakamura-Tsuruta S, Kominami J, Tateno H, Hirabayashi J. 2017. Sugar-binding profiles of chitin-binding lectins from the hevein family: A comprehensive study. Int J Mol Sci. 18(6):E1160, 10.3390/ijms18061160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, Francis R, Schedl T. 1996. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 134(3):699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen EJR, Timal S, Ryan M, Ashikov A, van Scherpenzeel M, Graham LA, Mandel H, Hoischen A, Iancu TC, Raymond K et al. . 2016. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat Commun. 7:11600, 10.1038/ncomms11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WL, Dennis JW. 2012. The eggshell in the C. elegans oocyte-to-embryo transition. Genesis. 50(4):333–349, 10.1002/dvg.20823. [DOI] [PubMed] [Google Scholar]
- Johnston WL, Krizus A, Dennis JW. 2006. The eggshell is required for meiotic fidelity, polar-body extrusion and polarization of the C. elegans embryo. BMC Biol. 4:35, 10.1186/1741-7007-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WL, Krizus A, Dennis JW. 2010. Eggshell chitin and chitin-interacting proteins prevent polyspermy in C. elegans. Curr Biol. 20(21):1932–1937, 10.1016/j.cub.2010.09.059. [DOI] [PubMed] [Google Scholar]
- Kaji H, Kamiie J-I, Kawakami H, Kido K, Yamauchi Y, Shinkawa T, Taoka M, Takahashi N, Isobe T. 2007. Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol Cell Proteomics. 6(12):2100–2109, 10.1074/mcp.M600392-MCP200. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. 2003. Genorne-wide RNAi screening in Caenorhabditis elegans. Methods. 30(4):313–321, 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kanda Y. 2013. Investigation of the freely available easy-to-use software ‘EZR’for medical statistics. Bone Marrow Transplant. 48(3):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. 1997. DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc Natl Acad Sci USA. 94(10):4994–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 146(1):227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-Y, Hwang J-S, Han I-O. 2013. Tunicamycin inhibits Toll-like receptor-activated inflammation in RAW264.7 cells by suppression of NF-κB and c-Jun activity via a mechanism that is independent of ER-stress and N-glycosylation. Eur J Pharmacol. 721(1–3):294–300, 10.1016/j.ejphar.2013.09.022. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Izumikawa T, Mizuguchi S, Dejima K, Nomura KH, Egusa N, Taniguchi F, Tamura J, Gengyo-Ando K, Mitani S et al. . 2007. Expression of rib-1, a Caenorhabditis elegans homolog of the human tumor suppressor EXT Genes, is indispensable for heparan sulfate synthesis and embryonic morphogenesis. J Biol Chem. 282(11):8533–8544, 10.1074/jbc.M611107200. [DOI] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 54:631–664, 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kristiansson B, Stibler H, Wide L. 1995. Gonadal function and glycoprotein hormones in the carbohydrate-deficient glycoprotein (CDG) syndrome. Acta Paediatr. 84(6):655–659. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska MA, Robbins PW. 1987. Protein glycosylation in yeast: Transcript heterogeneity of the ALG7 gene. Proc Natl Acad Sci USA. 84(8):2145–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. 2012. C. elegans rrf-1 mutations maintain RNAi efficiency in the Soma in addition to the germline. PLoS One. 7(5):e35428, 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Lee MH, Schedl T, Jefferis GS. 2000. A C. elegans patched gene, ptc-1, functions in germ-line cytokinesis. Genes Dev. 14(15):1933–1944. [PMC free article] [PubMed] [Google Scholar]
- Lee S-K, Li W, Ryu S-E, Rhim T, Ahnn J. 2010. Vacuolar (H+)-ATPases in Caenorhabditis elegans: What can we learn about giant H+ pumps from tiny worms? Biochim Biophys Acta. 1797(10):1687–1695, 10.1016/j.bbabio.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Lints R, Hall DH. 2009. Reproductive system, germ line. WormAtlas. doi: 10.3908/wormatlas.1.23 [accessed 2018 Apr 24]. http://www.wormatlas.org/hermaphrodite/germ%20line/Germframeset.html. [DOI]
- Makishima T, Nakashima T, Nagata-Kuno K, Fukushima K, Iida H, Sakaguchi M, Ikehara Y, Komiyama S, Nishimoto T. 1997. The highly conserved DAD1 protein involved in apoptosis is required for N-linked glycosylation. Genes Cells. 2(2):129–141. [DOI] [PubMed] [Google Scholar]
- Mancuso VP, Parry JM, Storer L, Poggioli C, Nguyen KCQ, Hall DH, Sundaram MV. 2012. Extracellular leucine-rich repeat proteins are required to organize the apical extracellular matrix and maintain epithelial junction integrity in C. elegans. Development. 139(5):979–990, 10.1242/dev.075135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marada S, Navarro G, Truong A, Stewart DP, Arensdorf AM, Nachtergaele S, Angelats E, Opferman JT, Rohatgi R, McCormick PJ et al. . 2015. Functional divergence in the role of N-linked glycosylation in smoothened signaling. PLoS Genet. 11(8):e1005473, 10.1371/journal.pgen.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek KW, Vijay IK, Marth JD. 1999. A recessive deletion in the GlcNAc-1-phosphotransferase gene results in peri-implantation embryonic lethality. Glycobiology. 9(11):1263–1271, 10.1093/glycob/9.11.1263. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. 1996. Biochemical evidence that patched is the Hedgehog receptor. Nature. 384(6605):176–179, 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Maruyama R, Velarde NV, Klancer R, Gordon S, Kadandale P, Parry JM, Hang JS, Rubin J, Stewart-Michaelis A, Schweinsberg P et al. . 2007. EGG-3 regulates cell-surface and cortex rearrangements during egg activation in Caenorhabditis elegans. Curr Biol. 17(18):1555–1560, 10.1016/j.cub.2007.08.011. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 205(1):111–128, 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 291(5511):2144–2147, 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. 2003. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 17(2):187–200, 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi S, Dejima K, Nomura KH, Murata D, Nomura K. 2008. Functional glycomics at the level of single cells: Studying roles of sugars in cell division, differentiation, and morphogenesis with 4d microscopy. Exp Glycosci. 290–294. [Google Scholar]
- Mizuguchi S, Uyama T, Kitagawa H, Nomura KH, Dejima K, Gengyo-Ando K, Mitani S, Sugahara K, Nomura K. 2003. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 423(6938):443–448. [DOI] [PubMed] [Google Scholar]
- Mörck C, Vivekanand V, Jafari G, Pilon M. 2010. C. elegans ten-1 is synthetic lethal with mutations in cytoskeleton regulators, and enhances many axon guidance defective mutants. BMC Dev Biol. 10:55, 10.1186/1471-213X-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H-M, Van Auken KM, Li Y, Sternberg PW. 2018. Textpresso central: A customizable platform for searching, text mining, viewing, and curating biomedical literature. BMC Bioinformatics. 19:94, 10.1186/s12859-018-2103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata D, Nomura KH, Dejima K, Mizuguchi S, Kawasaki N, Matsuishi-Nakajima Y, Ito S, Gengyo-Ando K, Kage-Nakadai E, Mitani S et al. . 2012. GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans. Mol Biol Cell. 23(6):982–995, 10.1091/mbc.E10-10-0855. [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson SK, Greenan G, Desai A, Müller-Reichert T, Oegema K. 2012. Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J Cell Biol. 198(4):731–748, 10.1083/jcb.201206008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz MA, Noble D, Sorokin EP, Kimble J. 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda). 4(9):1765–1772, 10.1534/g3.114.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamarthy S, Kulshrestha A, Katara GK, Beaman KD. 2018. The curious case of vacuolar ATPase: Regulation of signaling pathways. Mol Cancer. 17:41.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5815226/ 10.1186/s12943-018-0811-3. [accessed 2018 Jun 24]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JC. 1989. Glycoproteins: What are the sugar chains for? Trends Biochem Sci. 14(7):272–276. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Förster F. 2014. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun. 5:3072, 10.1038/ncomms4072. [DOI] [PubMed] [Google Scholar]
- Reiling JH, Clish CB, Carette JE, Varadarajan M, Brummelkamp TR, Sabatini DM. 2011. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc Natl Acad Sci USA. 108(29):11756–11765, 10.1073/pnas.1018098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S, Lin R. 2013. The oocyte-to-embryo transition. Adv Exp Med Biol. 757:351–372, 10.1007/978-1-4614-4015-4_12. [DOI] [PubMed] [Google Scholar]
- Roboti P, High S. 2012. The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J Cell Sci. 125(Pt 14):3474–3484, 10.1242/jcs.103952. [DOI] [PubMed] [Google Scholar]
- Rojas R, Segovia C, Trombert AN, Santander J, Manque P. 2014. The effect of tunicamycin on the glucose uptake, growth, and cellular adhesion in the protozoan parasite Crithidia fasciculata. Curr Microbiol. 69(4):541–548, 10.1007/s00284-014-0620-x. [DOI] [PubMed] [Google Scholar]
- Roy D, Kahler DJ, Yun C, Hubbard EJA. 2018. Functional interactions between rsks-1/S6K, glp-1/Notch, and regulators of Caenorhabditis elegans fertility and germline stem cell maintenance. G3 (Bethesda). 8(10):3293–3309, 10.1534/g3.118.200511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristan C, Manzano-Lopez J, Reyes A, Spang A, Muñiz M, Roncero C. 2013. Oligomerization of the chitin synthase Chs3 is monitored at the Golgi and affects its endocytic recycling. Mol Microbiol. 90(2):252–266, 10.1111/mmi.12360. [DOI] [PubMed] [Google Scholar]
- Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. 2005. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 102(5):1548–1553, 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539, 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RHA, Fire A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 107(4):465–476, 10.1016/S0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Singaravelu G, Singson A. 2013. Calcium signaling surrounding fertilization in the nematode Caenorhabditis elegans. Cell Calcium. 53(1):2–9, 10.1016/j.ceca.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. 2012. STIM proteins: Dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 13(9):549–565, 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KK, Golden A. 2015. The C. elegans eggshell. WormBook Online Rev C Elegans Biol. 1–35, 10.1895/wormbook.1.179.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Spang A. 2013. N-glycosylation is required for secretion and mitosis in C. elegans. PLoS One. 8(5):e63687, 10.1371/journal.pone.0063687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Spang A. 2017. Attenuation of N-glycosylation causes polarity and adhesion defects in the C. elegans embryo. J Cell Sci. 130(7):1224–1231, 10.1242/jcs.189316. [DOI] [PubMed] [Google Scholar]
- Struwe WB, Hughes BL, Osborn DW, Boudreau ED, Shaw KMD, Warren CE. 2009. Modeling a congenital disorder of glycosylation type I in C. elegans: A genome-wide RNAi screen for N-glycosylation-dependent loci. Glycobiology. 19(12):1554–1562, 10.1093/glycob/cwp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Warren CE. 2010. High-throughput RNAi screening for N-glycosylation dependent loci in Caenorhabditis elegans. Methods Enzymol. 480:477–493, 10.1016/S0076-6879(10)80021-7. [DOI] [PubMed] [Google Scholar]
- Sugimoto A, Hozak RR, Nakashima T, Nishimoto T, Rothman JH. 1995. dad-1, an endogenous programmed cell death suppressor in Caenorhabditis elegans and vertebrates. EMBO J. 14(18):4434–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Okihara KL, Thijssen K, Plasterk RHA. 2002. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr Biol. 12(17):1535–1540. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263(1):103–112. [DOI] [PubMed] [Google Scholar]
- Toei M, Saum R, Forgac M. 2010. Regulation and isoform function of the V-ATPases. Biochemistry. 49(23):4715–4723, 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, Chiquet-Ehrismann R. 2008. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell. 19(9):3898–3908, 10.1091/mbc.E08-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster AJ, Styles EB, Mateo A, Derry WB, Andrews BJ, Fraser AG. 2017. Taxonomically restricted genes with essential functions frequently play roles in chromosome segregation in Caenorhabditis elegans and Saccharomyces cerevisiae. G3 (bethesda). 7(10):3337–3347, 10.1534/g3.117.300193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillaumier-Barrot S, Isidor B, Dupré T, Le Bizec C, David A, Seta N. 2012. Expanding the spectrum of PMM2-CDG Phenotype. JIMD Rep. 5:123–125, 10.1007/8904_2011_114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao J, Corsi AK. 2006. Identification of novel target genes of CeTwist and CeE/DA. Dev Biol. 293(2):486–498, 10.1016/j.ydbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Wheeler BS, Anderson E, Frøkjær-Jensen C, Bian Q, Jorgensen E, Meyer BJ. 2016. Chromosome-wide mechanisms to decouple gene expression from gene dose during sex-chromosome evolution. eLife. 5, 10.7554/eLife.17365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rush JS, Karaoglu D, Krasnewich D, Lubinsky MS, Waechter CJ, Gilmore R, Freeze HH. 2003. Deficiency of UDP-GlcNAc:Dolichol phosphate N-acetylglucosamine-1 phosphate transferase (DPAGT1) causes a novel congenital disorder of Glycosylation Type Ij. Hum Mutat. 22(2):144–150, 10.1002/humu.10239. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. 2014. Carbohydrate-binding specificity of lectins using multiplexed glyco-bead array. Methods Mol Biol. 1200:319–326, 10.1007/978-1-4939-1292-6_27. [DOI] [PubMed] [Google Scholar]
- Yoon DS, Cha DS, Alfhili MA, Keiper BD, Lee M-H. 2018. Subunits of the DNA polymerase alpha-primase complex promote Notch-mediated proliferation with discrete and shared functions in C. elegans germline. FEBS J. 285(14):2590–2604, 10.1111/febs.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste-Checa P, Vega AI, Martín-Higueras C, Medrano C, Gámez A, Desviat LR, Ugarte M, Pérez-Cerdá C, Pérez B. 2017. DPAGT1-CDG: Functional analysis of disease-causing pathogenic mutations and role of endoplasmic reticulum stress. PLoS One. 12(6):e0179456, 10.1371/journal.pone.0179456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf HK, Pohlentz G, Sandhoff K. 1983. Tunicamycin inhibits ganglioside biosynthesis in rat liver Golgi apparatus by blocking sugar nucleotide transport across the membrane vesicles. Proc Natl Acad Sci USA. 80(23):7075–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Foster JM, Nelson LS, Ma D, Carlow CKS. 2005. The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Dev Biol. 285(2):330–339, 10.1016/j.ydbio.2005.06.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.