Abstract
Mitoviruses have small RNA(+) genomes, replicate in mitochondria, and have been shown to infect only fungi to date. For this report, sequences that appear to represent nearly complete plant mitovirus genomes were recovered from publicly available transcriptome data. Twenty of the refined sequences, 2684–2898 nt long and derived from 10 different species of land plants, appear to encompass the complete coding regions of contemporary plant mitoviruses, which furthermore constitute a monophyletic cluster within genus Mitovirus. Complete coding sequences of several of these viruses were recovered from multiple transcriptome (but not genome) studies of the same plant species and also from multiple plant tissues. Crop plants among implicated hosts include beet and hemp. Other new results suggest that such genuine plant mitoviruses were immediate ancestors to endogenized mitovirus elements now widespread in land plant genomes. Whether these mitoviruses are wholly cryptic with regard to plant health remains to be investigated.
Keywords: database mining, RNA virus, fungal virus, mitovirus, Narnaviridae, plant virus
Introduction
In current, ICTV-ratified taxonomy, family Narnaviridae comprises two genera, Narnavirus and Mitovirus, respectively containing two and five species of fungal viruses: Saccharomyces 20S RNA narnavirus and Saccharomyces 23S RNA narnavirus in genus Narnavirus; Cryphonectria mitovirus 1, Ophiostoma mitovirus 3a, Ophiostoma mitovirus 4, Ophiostoma mitovirus 5, and Ophiostoma mitovirus 6 in genus Mitovirus (Buck et al., 2005; Hong et al., 1998, 1999; Polashock and Hillman, 1994). Despite this small representation, many other apparent Mitovirus members have been reported to date from fungal hosts (Abdoulaye et al., 2017; Bartholomäus et al., 2016; Das et al., 2016; Heinze, 2012; Hillman and Cai, 2013; Khalifa and Pearson, 2014; Kitahara et al., 2014; Lakshman et al., 1998; Marzano et al., 2016; Vainio et al., 2015; Xie and Ghabrial, 2013; Xu et al., 2015; Zhang et al., 2015). Indeed, more than 90 accessions in the Nucleotide (nr/nt) database at GenBank appear to encompass the complete coding sequences of additional fungal mitoviruses. Recently, a number of apparent mitovirus sequences have been additionally identified in the transcriptomes of a large collection of invertebrates (Shi et al., 2016), but whether these viruses derived from the invertebrates themselves, or instead from associated organisms such as fungal symbionts, remains unclear.
Mitoviruses have small, nonsegmented RNA(+) genomes and are all thought to replicate in host mitochondria (Cole et al., 2000; Hillman and Cai, 2013; Hong et al., 1998, 1999; Polashock and Hillman, 1994; Rogers et al., 1987). Across a collection of 99 apparent fungal mitoviruses recently analyzed for another report (Nibert, 2017), genome lengths range between 2.1 and 4.4 kb. Each of the genome sequences encompasses a single long ORF, encoding a deduced protein that is 657 to 1137 aa long and includes conserved motifs of a viral RNA-dependent RNA polymerase (RdRp), presumably required for mitovirus replication. Indeed, the mitovirus RdRp is recognized to define a conserved protein domain family, pfam05919. Mitoviruses appear not to form virions that are released from cells and are instead thought to exist as strictly intracellular ribonucleoprotein complexes, which are transmitted without exposure to the extracellular environment during cell division and also during cell–cell fusion events (e.g., hyphal anastomosis) when mitochondrial exchange is known to occur (Giovannetti et al., 1999; Polashock et al., 1997). They are generally considered to be cryptic viruses, though effects on fungal growth and virulence for plants have been well demonstrated in some cases (Polashock et al., 1997; Wu et al., 2007, 2010; Xie and Ghabrial, 2013; Xu et al., 2015).
In addition to fungal mitoviruses, endogenized fragments of mitovirus genomes are found in many plant genomes. Such so-called nonretroviral endogenized RNA virus elements (NERVEs) are broadly distributed in eukaryotic genomes and derive from a wide variety of RNA viruses (Bruenn et al., 2015; Chiba et al., 2011; Crochu et al., 2004; Geuking et al., 2009; Horie et al., 2010; Katzourakis and Gifford, 2010; Liu et al., 2010; Tanne and Sela, 2005; Taylor and Bruenn, 2009). They consistently represent only portions of the original viral genomes, with subsequent mutations having accumulated that further reduce their coding capacities, making them readily distinguishable from intact viral genomes. In each case, their initial endogenization (copying and integration into a eukaryotic genome) seems to have required an exogenous reverse transcriptase, probably transposon encoded in most cases (Ballinger et al., 2012; Geuking et al., 2009). In fact, mitovirus NERVEs were among the first to be recognized (Hong et al., 1998; Marienfeld et al., 1997) and are now known to be widespread in the genomes of land plants, particularly in the mitochondrial genomes of flowering plants (Bruenn et al., 2015). Moreover, the sequences of many and perhaps all of these mitovirus NERVEs have been noted to form a monophyletic cluster, suggesting that they descend from a common ancestor (Bruenn et al., 2015; Xu et al., 2015). Their high prevalence in plant genomes seems fairly curious given the apparent absence of contemporary plant mitoviruses. However, several authors have proposed that mitovirus NERVEs in plants may have derived from a mitovirus originally infecting an endophytic or otherwise symbiotic fungus (Bruenn et al., 2015; Marienfeld et al., 1997; Xu et al., 2015), which transferred either its infected mitochondria or its mitovirus RNA into a plant, where the mitovirus sequences were then endogenized and subjected to ongoing divergence during subsequent plant evolution.
Based on such evidence and arguments, it seems at least plausible to consider that an ancient fungal mitovirus was common ancestor to most or all of the mitovirus NERVEs now widespread in land plant genomes. This consideration, however, does not rule out the possibility of genuine, replicating plant mitoviruses as intermediates in this chain of descent. For the current report, we therefore entertained the hypothesis that genuine plant mitoviruses are more likely to have been the immediate ancestors to plant mitovirus NERVEs. A corollary to this hypothesis is that replicating mitoviruses may remain extant in plants today, which the specific studies described below set out to investigate.
Results
Complete coding sequences of tentative plant mitoviruses
The deduced protein sequence of a mitovirus NERVE found in the mitochondrial genome of Arabidopsis thaliana (251 aa; GenBank P92543) was used as query in an initial tblastn search of the Transcriptome Shotgun Assembly (TSA) database for land plants (taxid 3193). The top hits included 60 with E-value scores ≤1e−30, indicative of strong sequence similarities. Among these 60 were 23 that derived from accessions between 2039 and 4223 nt long, approximating the genome lengths of fungal mitoviruses (see examples of the latter in Table 1, bottom) (Hillman and Cai, 2013). Moreover, in 13 of these 23, each apparent plus-strand sequence encompasses a single long ORF, encoding a deduced protein between 674 and 821 aa long and thus approximating the RdRp lengths of fungal mitoviruses (again see Table 1, bottom). Two of these 13 were derived from the same transcriptome project on Beta vulgaris (sugar beet strains) (BioProject PRJNA73561; Mutasa-Göttgens et al., 2012) and are 100% identical in their deduced protein sequences, and three others were derived from the same transcriptome project on Solanum chacoense (Chaco potato) (BioProject PRJNA299204) and are 100% identical in their deduced protein sequences. Thus, after excluding these replicates, we were left with 10 hits (E-values, 2e−95 to 1e−31) for further exploration as nearly complete genome sequences of tentative plant mitoviruses (Table 1, top).
Table 1.
Sequence features of newly identified plant mitoviruses
| ORFp length Virus name (aa) |
UTRs 5′ :3′ (nt)a |
Virus abbrev. | Host strain or source | GenBank accession or BioProject no. | Transcript length (nt): | |
|---|---|---|---|---|---|---|
| Original | Refined | |||||
| Ambrosia artemisiifolia mitovirus 1 | AmarMV1 | Pannonia | GEZL01037418 | 2341 | 2898 | |
| 821 | 317:115 | |||||
| Azolla filiculoides mitovirus 1 | AzfiMV1 | Stockholm1 | GBTV01009554 | 2858 | 2871 | |
| 795 | 336:145 | |||||
| Beta vulgaris mitovirus 1 | BevuMV1b | C600+Roberta | JP500572 | 2680 | 2680 | |
| 778 | 346:0 | |||||
| KWS2320 | PRJNA254489 | na | 2825 | |||
| 793 | 354:89 | |||||
| KWS2320 | PRJNA41497 | na | 2810 | |||
| 793 | 354:74 | |||||
| STR06A6001 | PRJNA41497 | na | 2786 | |||
| 793 | 340:64 | |||||
| STR06B6002 | PRJNA41497 | na | 2794 | |||
| 793 | 341:71 | |||||
| Cannabis sativa mitovirus 1 | CasaMV1c | Purple Kush | JP464487 | 2449 | 2857 | |
| 762 | 440:128 | |||||
| Finola | PRJNA73819 | na | 2805 | |||
| 762 | 424:92 | |||||
| MPC/MSU | PPJNA80055 | na | 2825 | |||
| 762 | 429:107 | |||||
| UC-COE | PRJNA178769 | na | 2824 | |||
| 762 | 430:105 | |||||
| Dahlia pinnata mitovirus 1 | DapiMV1d | Rio Riata | GBDN01010918 | 2684 | 2806 | |
| 782 | 375:82 | |||||
| UBC | PRJNA193277 | na | 2798 | |||
| 782 | 373:76 | |||||
| Erigeron breviscapus mitovirus 1 | ErbrMV1e | SMMU | GDQF01116002 | 2804 | 2804 | |
| 780 | 374:87 | |||||
| YAU | PRJNA277583 | na | 2829 | |||
| 780 | 377:109 | |||||
| YAU | PRJNA229196 | na | 2799 | |||
| 780 | 374:82 | |||||
| Humulus lupulus mitovirus 1 | HuluMV1f | Karahanasou | LA397818 | 2754 | 2795 | |
| 763 | 390:113 | |||||
| Oxybasis rubra mitovirus 1 | OxruMV1 | 374 | GEEQ01005055 | 2292 | 2734 | |
| 763 | 351:91 | |||||
| Petunia exserta mitovirus 1 | PeexMV1g | OPGC943 | GBRT01041798 | 2698 | 2701 | |
| 750 | 311:137 | |||||
| Bern | PRJNA300556 | na | 2684 | |||
| 750 | 302:129 | |||||
| Solanum chacoense mitovirus 1 | SochMV1h | G4 | GEDG01002811 | 2726 | 2773 | |
| 776 | 335:107 | |||||
| Cryphonectria [parasitica] mitovirus 1 | CpMV1 | NB631 | L31849 | 2728 | na | |
| 809 | 86:212 | |||||
| Ophiostoma [novo-ulmi] mitovirus 3a | OnuMV3a | Ld | AJ004930 | 2617 | na | |
| 718 | 268:192 | |||||
| Ophiostoma [novo-ulmi] mitovirus 4 | OnuMV4 | Ld | AJ132754 | 2599 | na | |
| 783 | 204:43 | |||||
| Ophiostoma [novo-ulmi] mitovirus 5 | OnuMV5 | Ld | AJ132755 | 2474 | na | |
| 729 | 227:57 | |||||
| Ophiostoma [novo-ulmi] mitovirus 6 | OnuMV6 | Ld | AJ132756 | 2343 | na | |
| 695 | 141:114 | |||||
3´ UTR value excludes stop codon
The two BevuMV1 nt sequences from B. vulgaris strain KWS2320 are 100% identical across their 2810-nt region of overlap and share a 1-nt insertion in their 5´ UTR relative to the other BevuMV1 strains. The five BevuMV1 nt sequences are ≥97.7% identical in pairwise comparisons across their 2664-nt region of overlap. BevuMV1-specific sequence reads are found in the transcriptome data of at least seven other B. vulgaris strains (DP02, KWS1P2, KWS230-DH1440, VDH66156, 0708724601, and 9877105301, plus an unnamed strain from Rothamsted Research; BioProjects PRJEB8344, PRJNA41497, PRJNA219421, PRJNA246329, and PRJNA254559), but too few to give rise to complete contigs for these additional strains of BevuMV1. BevuMV1-KWS2320 (BioProject PRJNA254489) is considered the reference strain for tentative species “Beta vulgaris mitovirus 1”.
The four CasaMV1 nt sequences are ≥95.3% identical in pairwise comparisons across their 2804-nt region of overlap. CasaMV1-Finola has a 1-nt insertion in its 5´ UTR relative to the other CasaMV1 strains. The deduced RdRp sequence of CasaMV1-MPC/MSU is 100% identical to that of CasaMV1-PurpleKush. CasaMV1-PurpleKush is considered the reference strain for tentative species “Cannabis sativa mitovirus 1”.
The two DapiMV1 nt sequences are 99.5% identical across their 2798-nt region of overlap. DapiMV1-RioRiata is considered the reference strain for tentative species “Dahlia pinnata mitovirus 1”.
The two ErbrMV1 nt sequences from Yunnan Agricultural University (YAU) are 100% identical across their 2799-nt region of overlap. The longer ErbrMV1 nt sequence from YAU and that from Second Military Medical University (SMMU) are 96.3% identical across their 2804-nt region of overlap. ErbrMV1-YAU (BioProject PRJNA277583) is considered the reference strain for tentative species “Erigeron breviscapus mitovirus 1”.
The original GenBank accession LA397818 indicates that it derived from samples of H. lupulus var. lupulus strain Shinsu Wase, but our analysis suggests that it derived instead from samples of H. lupulus var. cordifolius strain Karahanasou, which were part of the same project (BioProject PRJDB3233); there are HuluMV1-specific sequence reads in the Shinsu Wase transcriptome data, but too few to give rise to a complete contig.
The two PeexMV1 nt sequences are >99.9% identical (only 1 nt difference) across their 2684-nt region of overlap. PeexMV1- OPGC943 is considered the reference strain for tentative species “Petunia exserta mitovirus 1”.
Separate SRA data from S. chacoense strain G4 mutant frk1 was also analyzed (see Table S1) and gave rise to a 2799-nt contig for SochMV1, which was 100% identical to that from strain G4 across their 2761-nt region of overlap.
, not applicable
From previous experiences (Nibert et al, 2016; Pyle et al., 2017), we have learned that accessions in the TSA database are sometimes truncated at one or both termini. Before further analyzing the 10 tentative plant mitovirus sequences, we therefore accessed the raw sequence reads from which these transcript contigs had been assembled, as available for each in the Sequence Read Archive (SRA) database (Table S1). In this manner, we were able to generate refined assemblies that included 5´- and/or 3´-terminal sequence extensions for 8 of the 10 contigs (Table 1; other assembly details in Table S2).
During the course of accessing the SRA data sets for contig refinement, we also found that there are sequence reads available from other transcriptome projects on some of the same 10 plant species, involving other strains or sources of these plants, which were not represented in the TSA database. We therefore also searched the SRA data sets from these other projects for sequence reads highly similar to those of the original TSA hits from the respective plant species. Having identified such reads from several studies (Table S1), we were then able to assemble them into contigs that appear to represent nearly complete mitovirus genome sequences from three other sugar beet strains of Beta vulgaris (BioProjects PRJNA41497 and PRJNA254489; Dohm et al., 2014; Stracke et al., 2014), three other strains or sources of Cannabis sativa (hemp) (BioProjects PRJNA73819, PRJNA80055, and PRJNA178769; van Bakel et al., 2011), another source of Dahlia pinnata (a common ornamental) (BioProject PRJNA193277; Hodgins et al., 2014), another source of Erigeron breviscapus (a Chinese species of fleabane used in traditional medicine) (BioProjects PRJNA229196 and PRJNA277583; Zhang et al., 2015), and another source of Petunia exserta (a Brazilian species of increasing use as an ornamental) (BioProject PRJNA300556; Sheehan et al., 2016) (Table 1; other assembly details in Table S2).
Another finding of note is that the coding region of the originally identified, 2680-nt TSA sequence from a combination of Beta vulgaris strains C600 and Roberta (Mutasa-Göttgens et al., 2012) appears to remain truncated at its 3´ terminus, even after attempted refinement using the available SRA data. This deficiency is reflected by the fact that the single long ORF in this sequence extends all the way to its 3´ end, for a 3´ NTR length of 0 nt (Table 1). This deficiency is resolved, however, in the newly assembled contigs from three other Beta vulgaris sugar beet strains, which are 112–137 nt longer at their 3´ termini, including a 3´ NTR of 64–89 nt (Table 1). We additionally found that one of the newly assembled contigs from Cannabis sativa (source MPC/MSU) (BioProject PRJNA80055) encodes a deduced protein 100% identical to that encoded by the originally identified C. sativa transcript sequence from strain Purple Kush (BioProject PRJNA73819; van Bakel et al., 2011). Also, the two newly assembled contigs from Beta vulgaris strain KWS2320 (BioProjects PRJNA41497 and PRJNA254489) were 100% identical to one another across their region of overlap (2810 nt) including the coding sequences, and the two newly assembled contigs from Erigeron breviscapus source YAU (BioProjects PRJNA229196 and PRJNA277583) were 100% identical to one another across their region of overlap (2799 nt) including the coding sequences.
In summary, then, after the refinements and additions described in the preceding paragraphs, we were able to identify 20 nearly complete mitovirus genome sequences of apparent mitoviruses that (i) derive from samples of 10 different plant species; (ii) are between 2684 and 2898 nt long; and (iii) each encompasses a single long ORF that is bracketed by stop codons and encodes a deduced protein sequence between 750 and 821 aa long (Table 1, Fig. 1), for 17 of which the deduced protein sequences are unique. All 21 tentative plant mitovirus sequences listed in the body of Table 1 (including the 3´-truncated coding sequence from Beta vulgaris strains C600 and Roberta) have been submitted to GenBank as Third-Party Annotation (TPA) sequences with accession numbers BK010422–BK010442. The implicated hosts are all land plants and indeed all eudicot flowering plants, except for the fern Azolla filiculoides.
Fig. 1.
Scaled diagrams of apparent plant mitovirus genomes. Genome lengths are indicated at right. The genomic RNA plus strand of each virus is shown as a thick horizontal black line. The single long ORF, encoding the viral RdRp, is shown as a gray box above the line. The first and last nt positions in the ORF (including the stop codon) are labeled. The diagrams for the different viruses are aligned according to the position of the conserved GDD motif in each RdRp, which is also labeled. The diagrams for CasaMV1, BevuMV1, DapiMV1, ErbrMV1, and PeexMV1 are those for their reference strains (see Table 1). Color-coding: red, plant mitoviruses from flowering plants (Ambrosia artemisiifolia, Beta vulgaris, Cannabis sativa, Dahlia pinnata, Erigeron breviscapus, Humulus lupulus, Oxybasis rubra, Petunia exserta, and Solanum chacoense); orange, plant mitovirus from a fern (Azolla filiculoides).
Evidence for a monophyletic cluster of plant mitoviruses
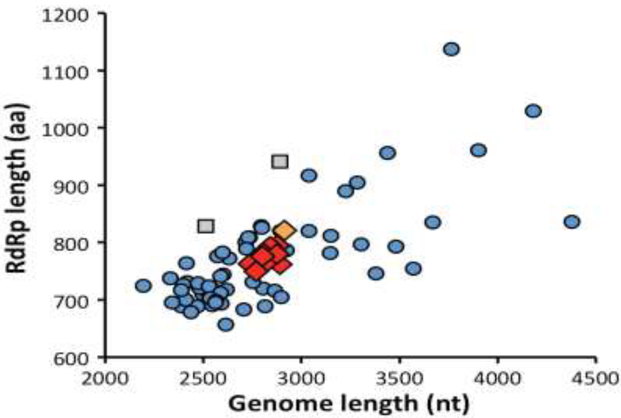
Do these 20 TSA- and/or SRA-derived complete coding sequences indeed represent a set of novel mitoviruses from land plants? As noted in the preceding paragraph, the RNA and protein lengths of these transcripts span relatively narrow ranges (median values, 2801 nt and 777 aa) and are appropriately sized to represent nearly complete mitovirus genomes (Table 1, Fig. 2). The lengths of their NTRs, though likely missing some terminal residues in most cases, are also relatively consistent (5´, 302–440 nt; 3´, 64–145 nt). Moreover, when we used each of the deduced protein sequences as queries in blastp searches of the NR database for viruses (NCBI taxid 10239), all were found to score top hits with the RdRp of a fungal mitovirus, with the top E-value scores between 4e–49 and 2e–40 (Table S3). Multiple sequence alignments of the new sequences indeed show strong conservation of the six motifs previously noted to be characteristic of mitovirus RdRps (Xie and Ghabrial, 2012) (Fig. S1). Global pairwise identity scores among the new RdRp sequences from different plant hosts range between 24% and 71%, with fern mitovirus AzfiMV1 consistently scoring near the bottom of this range (24–28%) (Fig. S2). The pairwise scores also show the close relationships (≥97.8% identity) among the RdRps of the different BevuMV1 strains, CasaMV1 strains, DapiMV1, ErbrMV1, and PeexMV1 strains, respectively (Fig. S2). It is also relevant to note that for these viruses for which complete coding sequences were obtained from multiple strains or sources of the plant host, the deduced RdRp length is consistent for each virus, e.g., 762 aa for all four strains or sources of CasaMV1 (Table 1).
Fig. 2.
Scatter plot of genome and RdRp lengths. Color-coding for the viruses new to this report (shown as diamonds) is the same as introduced in Fig. 1. Data from previously reported fungal mitoviruses and narnaviruses are shown as blue circles and gray squares, respectively.
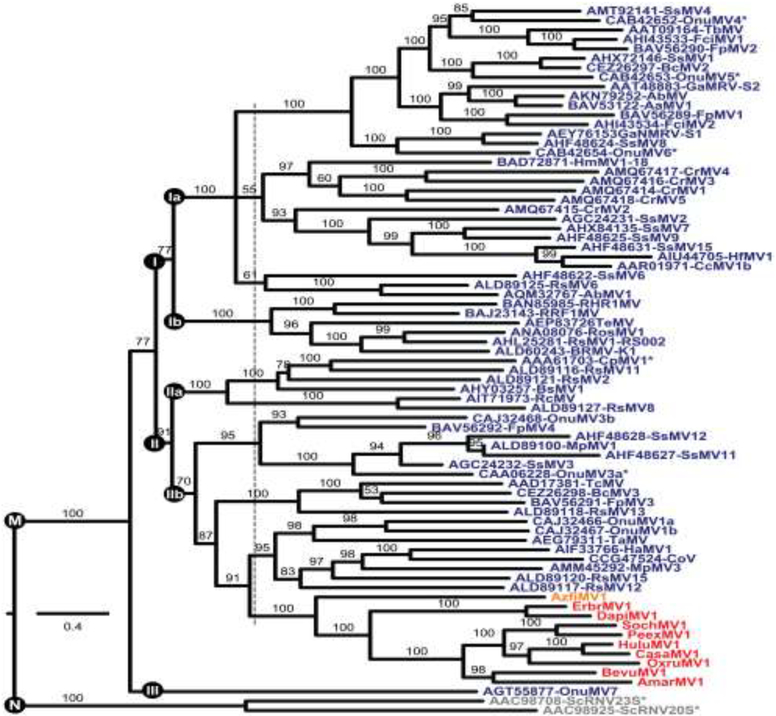
We next made use of phylogenetic methods to investigate the relationship between the RdRps of the tentative plant mitoviruses and those of a large collection of previously reported fungal mitoviruses (Table S4). The sequences of two viruses assigned to genus Narnavirus were included as a likely out-group. Results of these analyses show that the apparent plant mitoviruses are clearly embedded alongside fungal mitoviruses within the current bounds of genus Mitovirus (Fig. 3). Moreover, the apparent plant mitoviruses are specifically associated with one particular clade of fungal mitoviruses (designated Clade II/IIb in Fig. 3) and form a monophyletic cluster within that clade. The one virus from a more primitive plant host, fern mitovirus AzfiMV1, is the most basally branching member of this cluster. Potential implications for mitovirus evolution and taxonomy are considered in Discussion.
Fig. 3.
Phylogenetic tree of genus Mitovirus. Deduced RdRp sequences were aligned using MAFFT 7.310 (L-INS-i). The alignment was then analyzed using ModelFinder to determine the best-fit substitution model (VT+F+R6) and subjected to phylogenetic analysis using IQ-TREE and UFBoot as described in Materials and Methods. The site proportions and rates for the FreeRate model in this case were (0.0317,0.0275), (0.0531,0.1920), (0.1140,0.4564), (0.3158,0.8236), (0.3386,1.3151), and (0.1467,1.5775). The tree is displayed as a rectangular phylogram rooted on the branch to genus Narnavirus. Branch support values are shown in %, and branches with <50% support have been collapsed to the preceding node. Scale bar indicates average number of substitutions per alignment position. Color-coding is the same as in Fig. 2. CasaMV1, BevuMV1, DapiMV1, ErbrMV1, and PeexMV1 are represented by their reference strains. Viruses representing the 7 ratified species in genera Mitovirus (M) and Narnavirus (N) are highlighted by asterisks. Three or five apparent main clades within genus Mitovirus are labeled I–III. The dotted vertical line is explained in the main text. See Table S4 for a summary of abbreviations and GenBank numbers.
One important question is whether these novel mitovirus-like transcripts derived from the sampled plants or instead from fungal symbionts or contaminants associated with the respective plants at their times of sampling for transcriptome analyses. If the tentative plant mitoviruses instead derived from fungi, it seems unlikely that they would have been found to form a monophyletic cluster as seen in Fig. 3, but instead would have been dispersed among the other fungal mitoviruses within the phylogram. Thus, the monophyletic clustering of the tentative plant mitoviruses represents evidence that they derived from the sampled plants per se.
The RdRp ORF in most fungal mitoviruses is noteworthy for containing a number of internal UGA codons, since UGA encodes Trp in fungal mitochondria. In some fungi, however, UGA(Trp) is a rare mitochondrial codon (Hegedusova et al., 2014; Nibert, 2017), and the mitoviruses from those particular host species contain no, or only a few, UGA codons (Abdoulaye et al., 2017; Bartholomäus et al., 2016; Das et al., 2016; Heinze, 2012; Kitahara et al., 2014; Lakshman et al., 1998; Marzano et al., 2016; Nibert, 2017; Zhang et al., 2015). Thus, the presence of UGA(Trp) codons is not a definitive property of fungal mitoviruses. In plant mitochondrial transcripts, in contrast, UGA is a stop codon, as also in plant nuclear/cytosolic transcripts (Jukes and Osawa, 1990). Thus, plant mitoviruses would be expected to contain no UGA codons, except possibly as stop codons, and that is indeed the case for all of the apparent plant mitoviruses in Table 1. Also, if these viruses were instead derived from fungal symbionts or contaminants associated with the sampled plants, then those fungi would have all had to be ones in which UGA(Trp) is a rare mitochondrial codon. Because this again seems unlikely, the fact that all of the tentative plant mitoviruses in Table 1 contain no internal UGA codons represents further evidence that they are genuine plant mitoviruses.
Another important question is whether the newly identified sequences represent extant plant mitoviruses or instead mitovirus NERVEs. One argument for the former, as implied above, is that the newly identified sequences appear to represent similarly and appropriately sized intact mitovirus genomes, encoding similarly and appropriately sized full-length RdRps and terminated by similarly and appropriately sized 5´ and 3´ NTRs (Table 1, Fig. 1). Moreover, the newly identified sequences derive from RNA-based samples in the TSA or SRA database, not DNA-based samples in other databases. For the plant species from which the new mitoviruses derive, mitochondrial genome sequences are available in GenBank for Beta vulgaris (NC_002511 and NC_015099) and Cannabis sativa (NC_029855) and nuclear genome sequences are available for Beta vulgaris (BioProject PRJNA41497), Cannabis sativa (BioProject PRJNA350523), and Humulus lupulus (hop) (BioProject PRJDB3233). Megablast searches of these mitochondrial or nuclear genome sequences using the BevuMV1, CasaMV1, or HuluMV1 nt sequences as query identified no significant hits (E-values, >1), consistent with the conclusion that the BevuMV1, CasaMV1, and HuluMV1 sequences are unlikely to represent either mitochondrial or nuclear NERVEs.
Presence of apparent plant mitoviruses in different tissues
For some of the apparent plant mitoviruses originally identified in the TSA database, the samples that gave rise to their sequences derived from several different plant tissues, each of which had been separately subjected to library preparation and sequencing. The original TSA accessions had in each case then been assembled from the combination of sequence reads from the different tissues. However, by accessing the reads from each tissue, as separately deposited and annotated in the SRA database, we were able to assemble complete coding sequences in a tissue-specific manner for several of the viruses, including BevuMV1-KWS2320, CasaMV1-MPC/MSU, DapiMV1-RioRiata, PeexMV1-OPGC943, and SochMV1-G4 (Table 2). In each case, the nt sequences of the respective virus obtained from different tissues were >99.9% identical. Thus, as probably should be expected for genuine plant mitoviruses, essentially no tissue-specific sequence differences were observed for these five viruses. Finding the same virus in independent sets of transcriptome data obtained from different tissues of the same plant species provides further evidence that these viruses were derived from the plants per se.
Table 2.
Mitovirus sequences from different plant tissues
| Virus abbreviation and straina | Plant tissue | Virus-specific reads (fraction of total) reads × 1e5)b | Contig length (nt) | Mean coverage (per nt position)c |
|---|---|---|---|---|
| BevuMVl-KWS2320d | roots | ≥5.1 | 2704 | ≥454 |
| leaves | 1.0 | 2810 | 579 | |
| inflorescences | ≥9.2 | 2825 | ≥1416 | |
| seeds | ≥34 | 2799 | ≥357 | |
| seedlings | 4.9 | 2800 | 410 | |
| CasaMV1-MPC/MSUe | roots | 3.4 | 2818 | 137 |
| stems | 12 | 2824 | 450 | |
| leaves | 3.3 | 2805 | 177 | |
| flower buds | 9.8 | 2823 | 353 | |
| flowers | 10 | 2811 | 394 | |
| DapiMV1-RioRiataf | stem | 5.5 | 2796 | 93 |
| leaf | 4.9 | 2806 | 102 | |
| flower bud | 6.7 | 2791 | 106 | |
| PeexMV1-OPGC943g | apical shoot | 2.1 | 2659 | 43 |
| trichome | 2.3 | 2680 | 37 | |
| callus | 1.0 | 2667 | 14 | |
| flowers | ≥30 | 2700 | ≥740 | |
| seedling | 1.0 | 2664 | 17 | |
| SochMV1-G4h | leaves | 0.8 | 2750 | 84 |
| immature ovules | 5.5 | 2747 | 1326 | |
| mature ovules | ≥6.5 | 2754 | ≥1465 |
The viruses shown here are those from Table 1 for which a complete coding sequence could be assembled from each of the different sampled tissues.
Each fractional value has been multiplied by 100,000 for ease of comparison, The ≥ symbol reflects that in some cases a single SRA data set scored ≥20,000 hits (the limit for reporting by the online version of Blast at NCBI).
Positional coverage determined after using the Map Reads to Contigs function in CLC Genomics Workbench.
The five BevuMV1-KWS2320 nt sequences are 100% identical across a 2680-nt region of shared overlap. SRA data sets for young leaves (BioProject PRJNA254489) and old leaves (BioProject PRJNA41497) (Stracke et al., 2014) were combined for this analysis.
The five CasaMV1-MPC/MSU nt sequences are >99.9% identical (1 nt difference) across a 2804-nt region of shared overlap. SRA data sets for young, mature, and mixed leaves were combined for this analysis. SRA data sets for primary stem and stem–petioles were also combined for this analysis.
The three DapiMV1-RioRiata nt sequences are 100% identical across a 2791-nt region of shared overlap.
The five PeexMV1-OPGC943 nt sequences are 100% identical across a 2665-nt region of shared overlap.
The three SochMV1-G4 nt sequences are 100% identical across a 2732-nt region of shared overlap.
Another finding of possible interest from these analyses is that sequence reads respectively mapping to each of the five viruses in Table 2 are found at somewhat differing fractional frequencies (ranging between 0.8e−5 and 34e−5 overall) in the SRA data sets from different tissues. Of particular note is that the data sets containing higher frequencies of sequence reads for each virus are regularly those from flower-related samples, including seeds in the case of BevuMV1-KWS2320. The higher frequency of sequence reads in flowers is especially notable for PeexMV1-OPGC943 (more than 10-fold higher than in any of the other four sampled tissues). This seeming tendency for virus-specific reads to be found at higher frequencies in flower-related samples is intriguing and might suggest that mitovirus replication is somewhat enhanced in these tissues, possibly increasing the efficiency of vertical transmission to progeny plants.
Validation results for BevuMV1
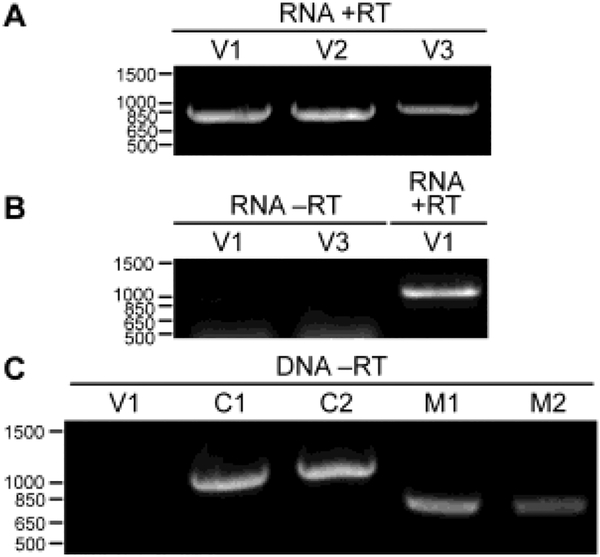
As indicated in the Table 1 legend, transcriptome data from Beta vulgaris sugar beet strain VDH66156 (BioProject PRJNA219421; Fugate et al., 2014) were also found to contain BevuMV1-specific reads, but not enough to assemble a complete genome sequence for this virus strain. We were nonetheless able to use leaves and seeds of this cultivar, obtained from the same source as for the transcriptome project, for validation testing of the virus. Following RNA extraction from either leaves or seeds, RT–PCR using different sets of BevuMV1-based primers yielded robust amplicons of expected sizes (Fig. 4A); in contrast, no BevuMV1-specific amplicons were obtained from leaf RNA or DNA using a PCR protocol lacking reverse transcriptase (Fig. 4B, C), consistent with the extrachromosomal, RNA-based origin of BevuMV1. Sanger sequencing of the RT–PCR amplicons then yielded 2693 nt of sequence representing the contiguous central region of the BevuMV1-VDH66156 genome and encompassing the complete coding region. Moreover, the 2693 nt of sequence obtained from either leaves or seeds are 100% identical to one another. The deduced RdRp sequence of BevuMV1-VDH66156 is ≥98.9% identical to that of the other BevuMV1 strains for which complete coding sequences were assembled (Fig. S2). The complete coding sequence of BevuMV1-VDH66156 has been deposited at GenBank as regular accession MG721540.
Fig. 4.
Validation results for BevuMV1-VDH66156. Positions of DNA markers are labeled at left (bp). RNA or DNA extracts were obtained from leaves of sugar beet strain VDH66156. (A) RNA extract was used for RT–PCR amplification using three different primer pairs specific for BevuMV1 (V1–V3). (B) RNA extract was used for PCR amplification (no RT step) using two different primer pairs specific for BevuMV1 (V1 and V3). At right is an RT-PCR control using one primer pair specific for BevuMV1 (V1). (C) DNA extract was used for PCR amplification (no RT step) using one primer pair specific for BevuMV1 (V1). At right are PCR controls using two primer pairs specific for B. vulgaris chloroplast DNA (C1 and C2) and two primer pairs specific for B. vulgaris mitochondrial DNA (M1 and M2).
Relationship of apparent plant mitoviruses to mitovirus NERVEs
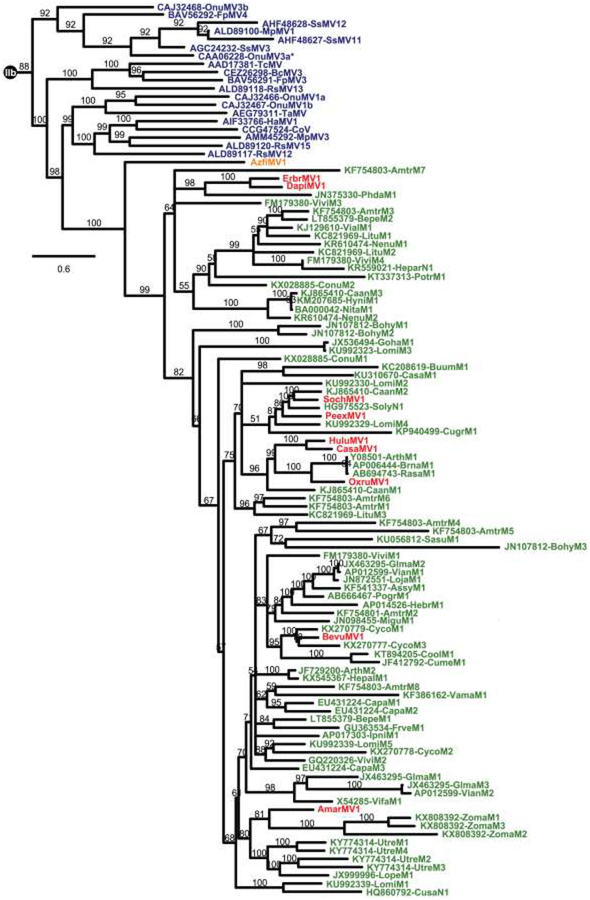
Previous phylogenetic analyses have provided evidence that the numerous mitovirus NERVEs in plant genomes are closely related to fungal mitoviruses (Bruenn et al., 2015; Marienfeld et al., 1997; Xu et al., 2015). To investigate how these elements may relate to the newly identified, apparent plant mitoviruses, additional phylogenetic analyses were performed. Although many mitovirus NERVEs have been previously identified, few are annotated as such in their GenBank accessions. Hence for this study, mitovirus NERVEs were re-identified by tblastn searches of the NR/NT database for green plants (taxid:33090), using the AzfiMV1 RdRp sequence or fungal mitovirus RdRp sequences as queries. To limit the number of sequences for analysis, several criteria were applied as described in Materials and Methods, ultimately yielding query-aligned regions of 79 mitovirus NERVEs (E-values, 1e−42 to 9e−5), all from flowering plants. These elements derive from mostly mitochondrial but also a few nuclear genomes and were translated into query-aligned protein sequences 121–522 aa long (Table S5) and encompassing RdRp motif C (Poch et al., 1989) with conserved sequence GDD (Fig. S3). Protein sequence-based phylogenetic analyses including these mitovirus NERVEs along with all of the viruses from Fig. 3 were then performed and strikingly showed that the NERVEs form a monophyletic cluster with the apparent plant mitoviruses, and apart from the fungal viruses (Fig. 5). In fact, the 79 NERVEs, all from flowering plants, are juxtaposed in the phylogram with the apparent plant mitoviruses from flowering plants, whereas the one fern mitovirus, AzfiMV1, branches next most basally to the flowering plant cluster, as also seen for the viruses in Fig. 3. These new phylogenetic results additionally concur with those in Fig. 3 in showing that a particular clade of fungal mitoviruses shares a proximate common ancestor with the apparent plant mitoviruses and, in Fig. 5, also the mitovirus NERVEs from plant genomes.
Fig. 5.
Phylogenetic tree including plant mitovirus NERVEs. Deduced protein sequences were aligned using MAFFT 7.310 (L-INS-i). The alignment was then analyzed using ModelFinder to determine the best-fit substitution model (VT+F+R6) and subjected to phylogenetic analysis using IQ-TREE and UFBoot as described in Materials and Methods. The site proportions and rates for the FreeRate model in this case were (0.0423,0.0776), (0.0552,0.2609), (0.1204,0.5237), (0.3069,0.8510), (0.3485,1.2689), and (0.1267,1.7035). The tree was displayed as a rectangular phylogram rooted on the branch to genus Narnavirus; the Clade IIb portion (see Fig. 3) was then excerpted for presentation here. Branch support values are shown in %, and branches with <50% support have been collapsed to the preceding node. Scale bar indicates average number of substitutions per alignment position. Color-coding for viruses is the same as in Fig. 2. CasaMV1, BevuMV1, DapiMV1, ErbrMV1, and PeexMV1 are represented by their reference strains. Mitovirus NERVEs are labeled in green; see Table S5 for a summary of their abbreviations and GenBank numbers.
Discussion
In this report, we provide strong evidence for contemporary (modern-day, extant) plant mitoviruses. The reported viruses derive from 10 different species of land plants, representing one family of ferns and four families of flowering plants. Among these plant hosts are several well known as either crops (Beta vulgaris, Cannabis sativa, and Humulus lupulus) or ornamentals (Dahlia pinnata and more recently Petunia exserta). Less well known but also useful are Azolla filiculoides (an Australian species of water fern used for water purification, nitrogen fixation, fertilizer, and livestock feed), Erigeron breviscapus (used for multiple purposes in traditional Chinese medicine), and Solanum chacoense (a South American species of wild potato foraged for its tubers and also used in breeding of cultivated potatoes). Reciprocally, both A. filiculoides and S. chacoense, in addition to Ambrosia artemisiifolia (common ragweed), are now invasive in many locations around the world, and the pollen of A. artemisiifolia is a well-known allergen. Also noteworthy is that evidence for several plant mitoviruses in this study has been found in multiple strains or sources of their respective hosts (B. vulgaris, C. sativa, D. pinnata, P. exserta, and E. breviscapus), suggesting that mitovirus infections of these plant species, and perhaps many others considering the wide distribution of mitovirus NERVEs in land plant genomes, are common.
Mitovirus evolution in plants
There are evolutionary congruencies between the plant hosts and their respective mitoviruses identified in this report. First, host Azolla filiculoides is the only representative of class Polypodiopsida (ferns), which diverged from Spermatophyta (seed plants, including flowering plants) ~400 million years ago (MYA) (time estimates from http://timetree.org/; Kumar et al., 2017). In accordance, fern mitovirus AzfiMV1 is more divergent than the other plant mitoviruses (see Figs. 3 and 5), which are all from flowering plants. Moreover, AzfiMV1 branches from the root of the flowering plant mitovirus cluster, suggesting that this monophyletic lineage of plant mitoviruses may have first entered plant hosts as early as the evolution of ferns. Indeed, Bruenn et al. (2015) have suggested an even earlier possible timing, as early as the evolution of clubmosses (~430 MYA). Mitovirus sequences from additional primitive plants (ferns, clubmosses, etc.) are needed for further clarifying these deeper evolutionary roots. It also remains possible that other, distinct lineages of plant mitoviruses have yet to be reported.
Second, among the flowering plant hosts and their respective mitoviruses identified in this report, three respective pairs appear most closely related (see Fig. 3): Dahlia pinnata (DapiMV1) and Erigeron breviscapus (ErbrMV1), which are members of family Asteraceae and diverged ~33 MYA; Petunia exserta (PeexMV1) and Solanum chacoense (SochMV1), which are members of family Solanaceae and diverged ~30 MYA; and Cannabis sativa (CasaMV1) and Humulus lupulus (HuluMV1), which are members of family Cannabaceae and diverged ~21 MYA. For these three pairs, it seems reasonable to conclude that an ancestral plant mitovirus was present before the divergence of each respective pair, explaining the phylogenetic correlations between these pairs of hosts and their viruses. AmarMV1, BevuMV1, and OxruMV1, on the other hand, do not appear to fit this simple pattern. Host Ambrosia artemisiifolia is another member of family Asteraceae, but its mitovirus AmarMV1 is more divergent from DapiMV1 and ErbrMV1 than might be expected from a simple co-descent model. Similarly, hosts Beta vulgaris and Oxybasis rubra (red goosefoot) are members of family Chenopodiaceae and diverged ~43 MYA, but their mitoviruses BevuMV1 and OxruMV1 are more divergent from one another than might be similarly expected. Overall, these findings for plant vs. mitovirus evolution are perhaps most consistent with a model in which mitoviruses have co-descended with their plant hosts but have occasionally been lost from a particular plant lineage, only to reenter that lineage at a later time, possibly in association with mitochondrial exchange of genetic material that is known to occur frequently among members of at least some plant taxa (Bruenn et al., 2015; Xi et al., 2013).
Further implications for mitovirus and mitovirus NERVE evolution
What other implications do the results in Figs. 3 and 5 suggest regarding the evolution of mitoviruses and mitovirus NERVEs? First, the monophyletic clustering and close juxtaposition of both the flowering plant mitoviruses and the flowering plant mitovirus NERVES in Fig. 5 suggests that plant mitoviruses are more likely than fungal mitoviruses to have been the immediate ancestors to most or all of the NERVEs examined here. This interpretation agrees with the hypothesis posited in the Introduction to this report but differs from thoughts of some previous authors, mostly because the existence of genuine plant mitoviruses had not been demonstrated until now.
Notably, the results in Figs. 3 and 5 do not exclude the possibility that a plant mitovirus may have in fact served as ancestor to the fungal mitoviruses. As noted by Shackelton and Holmes (2008), fungus-to-plant transmission of an ancestral fungal mitovirus would have necessitated traversal of a major host range barrier, in that the typical contemporary fungal mitovirus has a number of internal UGA codons (i.e., UGA(Trp) codons) in its RdRp ORF, which would function instead as stop codons in plant mitochondria, interfering with viral RdRp expression and genome replication upon fungus-to-plant transmission. In contrast, plant-to-fungus transmission of an ancestral plant mitovirus, lacking internal UGA codons because UGA is a stop codon in plant mitochondria, would not have encountered this barrier.
It is important to note, however, that a number of fungal mitoviruses lack internal UGA codons because UGA(Trp) is a rare mitochondrial codon in their particular fungal hosts, including a subset of basidiomycetes, possibly all glomeromycetes, and other primitive fungi (Nibert, 2017). Thus, if a fungal mitovirus with no internal UGA codons were to have been successfully transmitted into a plant, the host range barrier to RdRp expression and genome replication identified by Shackelton and Holmes (2008) would not have come into play. Fungus-to-plant transmission yielding an original plant mitovirus can therefore also be logically proposed to have occurred between an ancestral fungal mitovirus that lacked internal UGA codons and a primitive plant host. Since (i) UGA(Trp) is a rare mitochondrial codon in glomeromycetes (Nibert, 2017), (ii) the two previously reported glomeromycete mitoviruses wholly lack internal UGA codons (Kitahara et al., 2014), and (iii) glomeromycetes are routinely involved in endophytic interactions with many land plants, including more primitive ones (Brundrett, 2002; van der Heijden et al., 2015), an ancestral glomeromycete mitovirus might have been especially well suited for fungus-to-plant transmission followed by successful replication in plant mitochondria.
Are plant mitoviruses wholly cryptic with regard to plant health?
Some fungal mitoviruses have been directly implicated, by genetic studies involving viral gain or loss by particular fungal strains, in hypovirulence defects of their phytopathogenic fungal hosts (Polashock et al., 1997; Wu et al., 2007, 2010; Xie and Ghabrial, 2013; Xu et al., 2015). Moreover, in a few cases, both morphological abnormalities in mitochondria and in vitro growth defects have been demonstrated to accompany mitovirus-attributable fungal hypovirulence (Wu et al., 2007, 2010; Xu et al., 2015). Having provided strong evidence for contemporary plant mitoviruses in this report, important questions now arise as to whether these viruses may have any such noteworthy effects on their respective plant hosts, including several plants that serve as crops or ornamentals. Sequence database mining has also recently identified another group of putatively cryptic RNA viruses, plant amalgaviruses, in a number of newly ascribed hosts (Nibert et al., 2016), including common onion (Allium cepa), tea seed oil camellia (Camellia oleifera), pepper (Capsicum annuum), alfalfa (Medicago sativa), and rye (Secale cereale), as well as Erigeron breviscapus, which is identified as also a plant mitovirus host in this report. Additional studies on the effects of such presumably cryptic viruses on economically important plant hosts appear warranted. For plant mitoviruses in particular, their presumed association with mitochondria seems to make their potential for significant effects on plant hosts especially high.
Suggested changes to mitovirus classification
It appears from Fig. 3 that genus Mitovirus, as currently recognized, is phylogenetically broad and might be usefully subdivided, as also implied by previous authors (Hillman and Cai, 2013; Nibert, 2017; Xie and Ghabrial, 2013; Xu et al., 2015). A conservative proposal would be for current genus Mitovirus to be reclassified as subfamily “Mitovirinae” in family Narnaviridae. Within subfamily “Mitovirinae”, several new genera (possibly named “Alphamitovirus”, etc.) could then be recognized, possibly three genera corresponding to Clades I, II, and III, or five genera corresponding to Clades Ia, Ib, IIa, IIb, and III, as suggested in Fig. 3. The plant mitoviruses would thereby belong to the new genus corresponding to Clade II or IIb, which would include viruses from both plant and fungal hosts. The other new genera would contain viruses from only fungal hosts to date. There is a parallel to this circumstance in family Partitiviridae, where genera Alphapartitivirus and Betapartitivirus include viruses from both fungal and plant hosts to date, whereas Gammapartitivirus contains viruses from only fungal hosts and Deltapartitivirus contains viruses from only plant hosts to date (Nibert et al., 2014). One might wish to argue that the plant mitoviruses should instead warrant a separate genus, given their distinct host range; however, based on phylogenetic analyses such as that in Fig. 3, that viewpoint would seem to argue as well that the fungal mitoviruses should be divided among a much larger number of new genera (e.g., see dotted line in Fig. 3), which the current authors do not support at this time. Notably, Clades I and II suggested in Fig. 3 correspond to ones previously suggested by Hillman and Cai (2013), and Clades Ia and Ib correspond to ones previously suggested by Xu et al. (2015) and Nibert (2017), though numbered differently.
MATERIALS AND METHODS
Refining or newly generating transcript contigs from SRA data
For refining the termini of the 10 original TSA hits that were subjected to further study (Table 1), the 5´ and 3´-terminal sequences of each were used as queries to search the SRA data set(s) from which that transcript contig had been assembled (Table S1). The sequence reads identified by this search were then assembled into new contigs via CAP3 (Huang and Madan, 1999) or CLC Genomics Workbench 8.0 (QIAGEN). If these contigs extended either terminus of the original TSA accession, then the process was repeated, but including the new terminus in the query. This process was repeated until no additional, consistent extension could be obtained at either terminus. Terminal residues represented by single reads were trimmed from the final transcript contig. The original transcriptome studies not cited elsewhere in this report are those for Ambrosia artemisiifolia (BioProject PRJNA335689; Virág et al., 2016), Azolla filiculoides (BioProject PRJNA264391; de Vries et al., 2016), Dahlia pinnata (BioProject PRJNA189243; Lehnert and Walbot, 2014), Erigeron breviscapus (BioProject PRJNA293262; Chen et al., 2015), Humulus lupulus (BioProject PRJDB3233; Natsume et al., 2015), Oxybasis rubra (BioProject PRJNA305086), and Petunia exserta (BioProject PRJNA262142; Guo et al., 2015).
For newly generating transcript contigs, the available mitovirus sequence from the respective plant species was used as query to search the targeted SRA data set(s) from the same plant species The sequence reads identified by this search were then assembled into one or more contigs via CAP3 or CLC Genomics Workbench. As described in the preceding paragraph, the terminal sequences of these initial contigs were then reiteratively analyzed until no additional, consistent extension could be obtained at either terminus. In a few cases, two or more partial contigs assembled by CAP3 or CLC Genomics Workbench were found to share terminal overlaps, before or after further extensions had been made, and were then manually merged to generate the full-length transcript contig.
For each final transcript contig, the complete set of sequence reads that mapped to it were analyzed using CLC Genomics Workbench to determine the coverage values at each nt position in the contig, which were in turn used to calculate the mean coverage value (Tables S2 and 2). The number of matching sequence reads for each final transcript contig and the total sequence reads in the original SRA data set(s) were used to calculate the fraction of total (Tables S2 and 2).
Sequence and phylogenetic analyses
All database searches were performed with the indicated programs as implemented with defaults at http://blast.ncbi.nlm.nih.gov/Blast.cgi. Searches of the TSA or NR/NT database with protein sequence queries deduced from nucleotide sequences were performed using tblastn. Searches of SRA data sets with nucleotide sequence queries were performed using discontiguous megablast, or occasionally blastn if further efforts were needed to extend partial contigs. Searches of the NR database with protein sequence queries deduced from nucleotide sequences were performed using blastp.
ORFs in nucleotide sequences were identified and translated using ExPASy Translate as implemented at http://web.expasy.org/translate/ or SMS Translate as implemented at http://www.bioinformatics.org/sms2/. Multiple sequence alignments of protein sequences were performed using MAFFT 7.310 (L-INS-i) (Katoh and Standley, 2013) as implemented with defaults at http://mafft.cbrc.jp/alignment/server/. Global or local pairwise alignments of RNA sequences were performed using Needle, Needleall, or Water as implemented with defaults at http://www.bioinformatics.nl/emboss-explorer/. Codon frequencies were determined using SMS Codon Usage as implemented with defaults at http://www.bioinformatics.org/sms2/.
The best-fit substitution model for each multiple sequence alignment was identified according to the Bayesian information criterion using ModelFinder (Kalyaanamoorthy et al., 2017) as implemented with the “Find best and apply” option at https://www.hiv.lanl.gov/content/sequence/IQTREE/iqtree.html (Trifinopoulos et al., 2016). Phylogenetic analyses were then directly performed using IQ-TREE (Nguyen et al., 2015) and UFBoot (Minh et al., 2013) as implemented with defaults at that same website. The results in Newick format were submitted to TreeDyn 198.3 as implemented at http://www.phylogeny.fr/ for collapsing branches with lower support values. Tables S4 and S5 include full names, abbreviations, and GenBank accession numbers for any sequences in addition to those described in Table 1 that were used in generating Figs. 3, 5, and S3.
The following approach was used for identifying mitovirus NERVEs for phylogenetic analysis (E-values <1e−4 considered significant). Searches of the NR/NT database for green plants (taxid:33090), via tblastn using the RdRp sequence of fern mitovirus AzfiMV1 or fungal mitoviruses CpMV1, OnuMV4, OnuMV7, or TeMV (see Fig. 3) as queries, yielded no significant hits from non-flowering plants, but a merged total of 202 different significant hits from flowering plants (21 from non-eudicots, 181 from eudicots; E-values, 1e−42 to 7e−5). The query-aligned nt sequences from these hits were next downloaded and translated into aa sequences. To make the number of sequences for analysis more manageable, those with the following characteristics were discarded: sequences not clearly annotated as being genomic (mitochondrial or nuclear) in origin, translated sequences <120 aa long, and translated sequences not encompassing the conserved GDD motif or not aligning that motif as expected. In addition, a number of homologous sequences from plants of the same genus were noted (>65% identity in pairwise comparisons), and the shorter one of each of these replicate pairs was also discarded. Lastly, two NERVEs with large insertions relative to all the others were discarded because in preliminary trees they mapped on very long branches well within the plant mitovirus cluster.
Validation studies for BevuMV1
Leaves or seeds of B. vulgaris sugar beet strain VDH66156 were frozen in liquid nitrogen. Mortar and pestle were used to grind the frozen tissues into a fine powder and used directly for either RNA or DNA extraction. TRIzol Reagent (Invitrogen) was used to extract RNA according to manufacturer’s instructions. DNeasy Tissue Kit (Qiagen) was used to extract DNA according to manufacturer’s instructions. RNA extract was reverse-transcribed using SuperScript III Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions except that 4 primer pairs specific for different, overlapping regions of BevuMV1 were used for 4 separate reactions. One μL of RT reaction was then used directly per 20-μL total volume of PCR reaction. EconoTaq PLUS (Lucigen) was used for the PCR, according to manufacturer’s instructions. Separate PCR reactions were set up using the same 4 primer pairs with their respective RT reactions. For one set of controls, RNA extract was used directly for PCR with BevuMV1-specific primers. For another set of controls, DNA extract was used directly for PCR with BevuMV1-specific primers or with four other primer pairs specific for B. vulgaris chloroplast or mitochondrial DNA (Table S6). All PCR reactions used the following conditions: one cycle of 95°C for 30 s; 40 cycles of 95 °C for 30 s, 50°C for 30 s, 68°C for 1 min and 10 s; a final cycle of 68°C for 5 min and 4°C hold. For sequencing, RT–PCR amplicons were visualized by agarose gel electrophoresis using 1% agarose solution (SeaKem LE) and then purified from excised gel fragments using Monarch DNA Gel Extraction Kit (New England Biolabs) according to manufacturer’s instructions. Sanger sequencing was performed at the Dana-Farber/Harvard Cancer Center DNA Resource Core.
Supplementary Material
Highlights.
Mitoviruses are small RNA(+) viruses that replicate in mitochondria
They have been shown to infect only fungi to date
New evidence indicates that they also infect modern-day plants
These plant mitoviruses compose a monophyletic cluster in one Mitovirus clade
Mitovirus NERVEs in plant genomes have likely descended from plant mitoviruses
Acknowledgments
We thank Jesse Pyle for advice on RNA extraction from plant tissues. M. V. was supported in part by NIH Grant T32 GM007598 to the doctoral program in Molecules, Cells and Organisms at Harvard University. M.L.N. was supported in part by a subcontract from NIH grant 5R01GM033050. Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abdoulaye AH, Cheng J, Fu Y, Jiang D, Xie J. 2017. Complete genome sequence of a novel mitovirus from the phytopathogenic fungus Rhizoctonia oryzae-sativae. Arch Virol 162:1409–1412. [DOI] [PubMed] [Google Scholar]
- Ballinger MJ, Bruenn JA, Taylor DJ. 2012. Phylogeny, integration and expression of sigma virus-like genes in Drosophila. Mol Phylogenet Evol 65:251–258. [DOI] [PubMed] [Google Scholar]
- Bartholomäus A, Wibberg D, Winkler A, Pühler A, Schlüter A, Varrelmann M. 2016. Deep sequencing analysis reveals the mycoviral diversity of the virome of an avirulent isolate of Rhizoctonia solani AG-2–2 IV. PLoS One 11:e0165965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn JA, Warner BE, Yerramsetty P. 2015. Widespread mitovirus sequences in plant genomes. PeerJ. 3:e876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC. 2002. Coevolution of roots and mycorrhizas of land plants. New Phytol 154: 275–304. [DOI] [PubMed] [Google Scholar]
- Buck KW, Esteban R, Hillman BI. 2005. Family Narnaviridae In Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses (Eds, Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA), Elsevier Academic Press, San Diego, p. 751–756. [Google Scholar]
- Chen RB, Liu JH, Xiao Y, Zhang F, Chen JF, Ji Q, Tan HX, Huang X, Feng H, Huang BK, Chen WS, Zhang L. 2015. Deep sequencing reveals the effect of MeJA on scutellarin biosynthesis in Erigeron breviscapus. PLoS One 10:e0143881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. 2011. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog 7: e1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TE, Hong Y, Brasier CM, Buck KW. 2000. Detection of an RNA-dependent RNA polymerase in mitochondria from a mitovirus-infected isolate of the Dutch Elm disease fungus, Ophiostoma novoulmi. Virology 268:239–243. [DOI] [PubMed] [Google Scholar]
- Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, Belhouchet M, Lemasson JJ, deMicco P, de Lamballerie X. 2004. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol 85:1971–1980. [DOI] [PubMed] [Google Scholar]
- Das S, Falloon RE, Stewart A, Pitman AR. 2016. Novel mitoviruses in Rhizoctonia solani AG-3PT infecting potato. Fungal Biol 120:338–350. [DOI] [PubMed] [Google Scholar]
- de Vries J, Fischer AM, Roettger M, Rommel S, Schluepmann H, Bräutigam A, Carlsbecker A, Gould SB. 2016. Cytokinin-induced promotion of root meristem size in the fern Azolla supports a shoot-like origin of euphyllophyte roots. New Phytol 209:705–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depierreux D, Vong M, Nibert ML. 2016. Nucleotide sequence of Zygosaccharomyces bailii virus Z: evidence for +1 programmed ribosomal frameshifting and for assignment to family Amalgaviridae. Virus Res 217:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schulz B, Stadler PF, Schmidt T, Gabaldón T, Lehrach H, Weisshaar B, Himmelbauer H. 2014. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505:546–549. [DOI] [PubMed] [Google Scholar]
- Fugate KK, Fajardo D, Schlautman B, Ferrareze JP, Bolton MD, Campbell LG, Wiesman EC, Zalapa JE. 2014. Generation and characterization of a sugarbeet transcriptome and transcript-based SSR markers. The Plant Genome 7, doi: 10.3835/plantgenome2013.11.0038. [DOI] [Google Scholar]
- Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L. 2009. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323:393–396. [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Azzolini D, Citernesi AS. 1999. Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl Environ Microbiol 65:5571–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wiegert-Rininger KE, Vallejo VA, Barry CS, Warner RM. 2015. Transcriptome-enabled marker discovery and mapping of plastochron-related genes in Petunia spp. BMC Genomics 16:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedusova E, Brejova B, Tomaska L, Sipiczki M, Nosek J. 2014. Mitochondrial genome of the basidiomycetous yeast Jaminaea angkorensis. Curr Genet 60:49–59. [DOI] [PubMed] [Google Scholar]
- Heinze C 2012. A novel mycovirus from Clitocybe odora. Arch Virol 157:1831–1834. [DOI] [PubMed] [Google Scholar]
- Hillman BI, Cai G. 2013. The family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176. [DOI] [PubMed] [Google Scholar]
- Hintz WE, Carneiro JS, Kassatenko I, Varga A, James D. 2013. Two novel mitoviruses from a Canadian isolate of the Dutch elm pathogen Ophiostoma novo-ulmi (93–1224). Virol J 10:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins KA, Lai Z, Oliveira LO, Still DW, Scascitelli M, Barker MS, Kane NC, Dempewolf H, Kozik A, Kesseli RV, Burke JM, Michelmore RW, Rieseberg LH. 2014. Genomics of Compositae crops: reference transcriptome assemblies and evidence of hybridization with wild relatives. Mol Ecol Resour 14:166–177. [DOI] [PubMed] [Google Scholar]
- Hong Y, Cole TE, Brasier CM, Buck KW. 1998. Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 246:158–169. [DOI] [PubMed] [Google Scholar]
- Hong Y, Dover SL, Cole TE, Brasier CM, Buck KW. 1999. Multiple mitochondrial viruses in an isolate of the Dutch Elm disease fungus Ophiostoma novo-ulmi. Virology 258:118–127. [DOI] [PubMed] [Google Scholar]
- Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Osawa S. 1990. The genetic code in mitochondria and chloroplasts. Experientia 46:1117–1126. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genetics 6:e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa ME, Pearson MN. 2014. Molecular characterisation of novel mitoviruses associated with Sclerotinia sclerotiorum. Arch Virol 159:3157–3160. [DOI] [PubMed] [Google Scholar]
- Kitahara R, Ikeda Y, Shimura H, Masuta C, Ezawa T. 2014. A unique mitovirus from Glomeromycota, the phylum of arbuscular mycorrhizal fungi. Arch Virol 159:2157–2160. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: A resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- Lakshman DK, Jian J, Tavantzis SM. 1998. A double-stranded RNA element from a hypovirulent strain of Rhizoctonia solani occurs in DNA form and is genetically related to the pentafunctional AROM protein of the shikimate pathway. Proc Natl Acad Sci USA 95:6425–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert EM, Walbot V. 2014. Sequencing and de novo assembly of a Dahlia hybrid cultivar transcriptome. Front Plant Sci 5:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, Peng Y, Ghabrial SA, Yi X. 2010. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol 84:11876–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld JR, Unseld M, Brandt P, Brennicke A. 1997. Viral nucleic acid sequence transfer between fungi and plants. Trends Genet 13:260–261. [DOI] [PubMed] [Google Scholar]
- Marzano SY, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL. 2016. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Minh Anh Thi Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol, 30:1188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutasa-Göttgens ES, Joshi A, Holmes HF, Hedden P, Göttgens B. 2012. A new RNASeq-based reference transcriptome for sugar beet and its application in transcriptome-scale analysis of vernalization and gibberellin responses. BMC Genomics 13:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume S, Takagi H, Shiraishi A, Murata J, Toyonaga H, Patzak J, Takagi M, Yaegashi H, Uemura A, Mitsuoka C, Yoshida K, Krofta K, Satake H, Terauchi R, Ono E. 2015. The draft genome of hop (Humulus lupulus), an essence for brewing. Plant Cell Physiol 56:428–441. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML. 2017. Mitovirus UGA(Trp) codon usage parallels that of host mitochondria. Virology 507:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N. 2014. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188: 128–141. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Pyle JD, Firth AE. 2016. A +1 ribosomal frameshifting motif prevalent among plant amalgaviruses. Virology 498:201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polashock JJ, Bedker PJ, Hillman BI. 1997. Movement of a small mitochondrial double-stranded RNA element of Cryphonectria parasitica: ascospore inheritance and implications for mitochondrial recombination. Mol Gen Genet 256:566–571. [DOI] [PubMed] [Google Scholar]
- Polashock JJ, Hillman BI. 1994. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc Natl Acad Sci USA 91:8680–8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle JD, Keeling PJ, Nibert ML. 2017. Amalga-like virus infecting Antonospora locustae, a microsporidian pathogen of grasshoppers, plus related viruses associated with other arthropods. Virus Res 233:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ, Buck KW, Brasier CM. 1987. A mitochondrial target for the double-stranded RNAs in diseased isolates of the fungus that causes dutch elm disease. Nature 329:558–560. [Google Scholar]
- Shackelton LA, Holmes EC. 2008. The role of alternative genetic codes in viral evolution and emergence. J Theor Biol 254:128–134. [DOI] [PubMed] [Google Scholar]
- Sheehan H, Moser M, Klahre U, Esfeld K, Dell’Olivo A, Mandel T, Metzger S, Vandenbussche M, Freitas L, Kuhlemeier C. 2016. MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nat Genet 48:159–166. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu J, Holmes EC, Zhang YZ. 2016. Redefining the invertebrate RNA virosphere. Nature, doi: 10.1038/nature20167 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Stracke R, Holtgräwe D, Schneider J, Pucker B, Sörensen TR, Weisshaar B. 2014. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol 14:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne E, Sela I. 2005. Occurrence of a DNA sequence of a non-retro RNA virus in a host plant genome and its expression: evidence for recombination between viral and host RNAs. Virology 332:614–622. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Bruenn J. 2009. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol 7: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio EJ, Jurvansuu J, Streng J, Rajamäki ML, Hantula J, Valkonen JP. 2015. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J Gen Virol 96:714–725. [DOI] [PubMed] [Google Scholar]
- van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE. 2011. The draft genome and transcriptome of Cannabis sativa. Genome Biol 12:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MG, Martin FM, Selosse MA, Sanders IR. 2015. Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205:1406–1423. [DOI] [PubMed] [Google Scholar]
- Virág E, Hegedűs G, Barta E, Nagy E, Mátyás K, Kolics B, Taller J. 2016. Illumina sequencing of common (short) ragweed (Ambrosia artemisiifolia L.) reproductive organs and leaves. Front Plant Sci 7:1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Zhang L, Li G, Jiang D, Ghabrial SA. 2010. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406:117–126. [DOI] [PubMed] [Google Scholar]
- Wu MD, Zhang L, Li GQ, Jiang DH, Hou MS, Huang HC. 2007. Hypovirulence and double-stranded RNA in Botrytis cinerea. Phytopathology 97:1590–1599. [DOI] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley RK, Sugumaran M, Marx CJ, Rest JS, Davis CC. 2013. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genetics 9:e1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ghabrial SA. 2012. Molecular characterization of two mitoviruses co-infecting a hypovirulent isolate of the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 428:77–85. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wu S, Liu L, Cheng J, Fu Y, Jiang D, Xie J. 2015. A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res 197:127–136. [DOI] [PubMed] [Google Scholar]
- Zhang T, Li W, Chen H, Yu H. 2015. Full genome sequence of a putative novel mitovirus isolated from Rhizoctonia cerealis. Arch Virol 160:1815–1818. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wei X, Meng HL, Ma CH, Jiang NH, Zhang GH, Yang SC. 2015. Transcriptomic comparison of the self-pollinated and cross-pollinated flowers of Erigeron breviscapus to analyze candidate self-incompatibility-associated genes. BMC Plant Biol 15:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.