Dong et al. report gene rearrangement detection in circulating tumor cells by using covalent chemistry on nanosubstrates.
Abstract
Well-preserved mRNA in circulating tumor cells (CTCs) offers an ideal material for conducting molecular profiling of tumors, thereby providing a noninvasive diagnostic solution for guiding treatment intervention and monitoring disease progression. However, it is technically challenging to purify CTCs while retaining high-quality mRNA.Here, we demonstrate a covalent chemistry–based nanostructured silicon substrate (“Click Chip”) for CTC purification that leverages bioorthogonal ligation–mediated CTC capture and disulfide cleavage–driven CTC release. This platform is ideal for CTC mRNA assays because of its efficient, specific, and rapid purification of pooled CTCs, enabling downstream molecular quantification using reverse transcription Droplet Digital polymerase chain reaction. Rearrangements of ALK/ROS1 were quantified using CTC mRNA and matched with those identified in biopsy specimens from 12 patients with late-stage non–small cell lung cancer. Moreover, CTC counts and copy numbers of ALK/ROS1 rearrangements could be used together for evaluating treatment responses and disease progression.
INTRODUCTION
The use of circulating tumor cells (CTCs) (1) as a surrogate tumor source for molecular profiling of disease offers a potential noninvasive diagnostic solution for understanding underlying tumor biology, guiding treatment interventions, and monitoring disease progression. Unlike circulating tumor DNA or RNA (2), which are highly fragmented and compounded by substantial background, CTCs house intact genomic DNA and RNA, providing more genetic information about the tumors from which they originate. Recently, research focus has shifted away from the capture and enumeration of CTCs, and instead, toward approaches that enable in-depth mRNA profiling of both single and pooled CTCs (3–9). For example, transcriptome profiling was performed with full-length mRNA sequencing at the single CTC level with both prostate and melanoma CTCs, providing a glimpse into the in-depth biology of the diseases (4, 5). Compared with single-CTC mRNA assays, which are limited to relatively low quantities of RNA, enriched pools of CTCs that are coupled with sensitive downstream molecular profiling show greater translational potential in the clinic. So far, pooled CTCs have been used to detect hepatocellular carcinoma (HCC) (6), predict treatment responses and early dissemination in prostate cancer (7), and monitor early responses to immune checkpoint therapy in melanoma (8). We have also exploited pooled-CTC mRNA profiling for early detection of metastatic prostate cancer (9). However, to explore the diagnostic applications of pooled-CTC mRNA, technical challenges, including the low purity of enriched CTCs [from contamination by nonspecific binding of white blood cells (WBCs)] and the short lifetime of mRNA, must be addressed. Therefore, it is critical to develop a new platform that is optimized for pooled-CTC mRNA assays. Such a platform must be capable of (i) capturing and releasing pooled CTCs with minimal background WBC contamination and (ii) quickly and gently recovering pooled CTCs with well-preserved mRNA so that it can be seamlessly coupled with sensitive and reliable downstream molecular analysis.
In addition to CTC-based mRNA profiling, well-preserved mRNA in CTCs enables the detection and quantification of oncogenic gene rearrangements, which represent an unmet clinical need and remain unexploited by current CTC mRNA analyses. The presence of anaplastic lymphoma kinase (ALK) and ROS1 rearrangements define a subgroup of patients with non–small cell lung cancer (NSCLC) who are highly responsive to ALK/ROS1 tyrosine kinase inhibitors (TKIs), e.g., crizotinib (10, 11). Oncologists are increasingly receptive to noninvasive diagnostic solutions that enable both initial detection and continuous monitoring of NSCLC progression (12). Given that (i) mRNA has a relatively limited lifetime and (ii) there are multiple forms of rearranged transcripts (13–15), detection and precise quantification of ALK/ROS1 rearrangements in the blood of patients with NSCLC have proven challenging, further underscoring the need for an optimized CTC platform with the capacity to recover high-quality intact mRNA quickly for gene rearrangement analysis. Moreover, among the various downstream mRNA assays, reverse transcription Droplet Digital polymerase chain reaction (PCR) (RT-ddPCR) (16) is ideal for detecting gene rearrangements in CTCs. On the basis of a unique water-oil emulsion technology, RT-ddPCR offers a streamlined workflow for quantifying small amounts of CTC mRNA with single-molecule precision. Furthermore, RT-ddPCR can identify the partners of potential gene rearrangements through its PCR primer and reporter design.
In our previous studies, inspired by nanoscale features present in the tissue microenvironment (e.g., extracellular matrix and cell surface components), we pioneered (17, 18) the design and application of “NanoVelcro” cell-affinity substrates, in which capture agents (e.g., antibodies) can be grafted onto silicon nanowire substrates (SiNWS) (19) to capture cells of interest [e.g., CTCs and circulating fetal nucleated cells (20)]. The Velcro-like topographic interactions (17, 19) between the nanostructured substrates and nanoscale cellular surface components (i.e., microvilli) confer enhanced cell capture performance to the devices.
In this study, on the basis of our previous experience (9) isolating pooled CTCs, we exploited a new covalent chemistry–based CTC capture/release platform to address the unmet clinical need of quantifying gene rearrangements in CTCs. This platform was developed by integrating bioorthogonal ligation–mediated (21) CTC capture with disulfide cleavage–driven CTC release on nanostructured substrates to enable more efficient purification of pooled CTCs with well-preserved mRNA and less WBC contamination (Fig. 1). We used bioorthogonal click chemistry, i.e., inverse electron-demand Diels-Alder cycloaddition (22) between tetrazine (Tz) and trans-cyclooctene (TCO) with a rate constant (23) of 104 M−1 s−1, to mediate CTC capture and enumeration in SiNWS-embedded microfluidic devices and thus refer to them as “Click Chips.” Anti–epithelial cell adhesion molecule (EpCAM) has been widely used in many CTC enrichment platforms such as the only U.S. Food and Drug Administration–approved CellSearch (24). In contrast to previous anti-EpCAM–mediated CTC immobilization methods, a pair of highly reactive click chemistry motifs, i.e., Tz and TCO, were grafted onto cell capture substrates (via surface modification) and CTCs (via TCO–anti-EpCAM conjugation), respectively. The ligation between Tz-grafted SiNWS and TCO-grafted CTCs is facile, specific, irreversible, and insensitive to biomolecules, water, and oxygen (22), leading to specific, rapid, and irreversible immobilization of the CTCs with improved capture efficiency and reduced nonspecific immobilization of WBCs compared with that observed in previously reported anti-EpCAM–mediated approaches (19, 25). After CTC capture, exposure to a disulfide cleavage agent [i.e., 1,4-dithiothreitol (DTT)] (26) results in the prompt release of the CTCs from the SiNWS by cleaving the disulfide bond linking the Tz to the SiNWS. As WBCs are captured nonspecifically, they are not released by freeing Tz from the SiNWS, reducing WBC contamination in the released samples. In addition, mRNA stability is improved as a result of the (i) short CTC purification time (24 min) and (ii) mild processing conditions, enabling downstream mRNA analysis by RT-ddPCR (16). We demonstrate that Click Chips can be used for the detection and quantification of oncogenic gene rearrangements (i.e., ALK/ROS1 rearrangements) in CTCs isolated from patients with NSCLC, enabling serial monitoring of ALK/ROS1 rearrangements with single-molecule precision. Leveraging this capability, we correlate dynamic changes in both CTC count and quantity of CTC-derived ALK/ROS1 rearrangements with outcomes from radiographic imaging, the clinical gold standard for evaluating treatment responses of solid tumors.
Fig. 1. Schematic illustrating the combined use of bioorthogonal ligation (i.e., the reaction between Tz and TCO) and disulfide cleavage–driven by DTT to enable capture and release of CTCs using Click Chips.
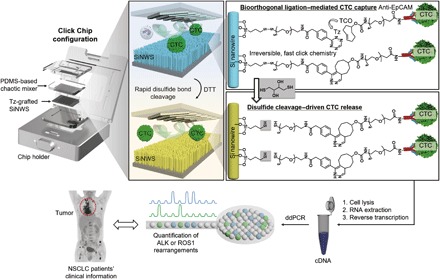
The purified CTCs can then be subjected to RT-ddPCR to detect ALK or ROS1 rearrangements in NSCLC samples with single-molecule precision. cDNA, complementary DNA.
RESULTS
Device fabrication and characterization
The SiNWS of Click Chips (Fig. 1) are composed of Tz-grafted silicon nanowires, which are fabricated via a combination of photolithographic patterning and Ag nanoparticle–templated wet etching (27). The resulting vertically aligned SiNWS (19) are cut into individual chips and then treated to tether Tz motifs onto the nanostructures. As illustrated in Fig. 2A, (3-mercaptopropyl)trimethoxysilane (MPS) is vapor-deposited onto the SiNWS. The terminal thiol groups presented by the silane-treated SiNWS are then reacted with ortho-pyridyl disulfide polyethylene glycol amine (OPSS-PEG-NH2) to incorporate a disulfide linker moiety terminated by an amine group. Last, a Tz-sulfo-NHS ester is coupled to the terminal amine to produce the Tz-grafted SiNWS.
Fig. 2. Surface modification, characterization, and verification of Tz-grafted SiNWS.
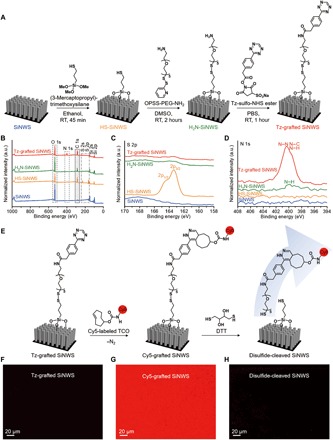
(A) Schematic summary of the stepwise functional group transformation developed for the preparation of Tz-grafted SiNWS. (B to D) X-ray photoelectron spectra of stepwise functionalized SiNWS: bare SiNWS (blue), SH-grafted SiNWS (orange), NH2-grafted SiNWS (green), and Tz-grafted SiNWS (red): (B) survey scans, (C) high-resolution XPS spectra in the energy range of the S 2p signals, and (D) high-resolution spectra in the energy range of the N 1s signals. (E) Scheme for verification of the successful preparation of Tz-grafted SiNWS via bioorthogonal ligation with Cy5-labeled TCO (2.6 mM) and disulfide cleavage by DTT (50 mM), respectively. Fluorescent images were captured through fluorescence microscopy (Nikon 90i, λex = 620/60) for (F) Tz-grafted SiNWS, (G) Cy5-grafted SiNWS, and (H) disulfide-cleaved SiNWS by DTT treatment. a.u., arbitrary units.
X-ray photoelectron spectroscopy (XPS) was used to characterize and confirm the stepwise modification of the SiNWS interface (Fig. 2, B to D, and fig. S1). Two new characteristic peaks of S 2s and 2p electrons from the thiol groups were observed for HS-SiNWS (Fig. 2, B and C), suggesting that the initial silanization reaction was successful. After treating the HS-SiNWS with the disulfide linker OPSS-PEG-NH2, a new N 1s electron peak was observed (Fig. 2, B and D), which confirms the presence of the linkers’ terminal amine group. When incubating the H2N-SiNWS with a Tz-sulfo-NHS ester, the intensity of the N 1s peak markedly increased, which is consistent with the nitrogen content of the Tz motif. Further analysis of deconvoluted x-ray photoelectron spectra at the C 1s and N 1s regions confirmed the bonding structure on the surface of SiNWS (see fig. S2).
In parallel, we tested and verified the preparation of Tz-grafted SiNWS via the bioorthogonal ligation between Tz on SiNWS and Cy5-labeled TCO, which was followed by disulfide cleavage using DTT (Fig. 2E and figs. S3 and S4). The Cy5 fluorophore was grafted onto the SiNWS through a TCO and Tz coupling reaction (Fig. 2F), leading to a strong fluorescent signal detected on treated Tz-grafted SiNWS (Fig. 2G and fig. S3A). In contrast, Tz-grafted flat Si substrates (without nanostructures) showed weak fluorescence after Cy5-labeled TCO treatment (fig. S3B), demonstrating fewer Tz motifs and less surface area than that of SiNWS. In addition, the lack of fluorescence on Cy5-labeled TCO-treated NH2-SiNWS (fig. S3C) confirms the specificity of the bioorthogonal click chemistry. Exposing the Cy5-grafted SiNWS to DTT for a series of incubation times induced disulfide cleavage and release of the fluorophore from the SiNWS, resulting in a rapid reduction in fluorescence (Fig. 2H) with 90% of Cy5 fluorophores detaching from the substrate within 12 min (fig. S4). These fluorescence data both confirm the successful preparation of Tz-grafted SiNWS and demonstrate the feasibility of CTC capture and release from the SiNWS.
CTC capture with Click Chips
A custom-designed microfluidic chip holder was used to integrate Tz-grafted SiNWS with an overlaid polydimethylsiloxane (PDMS) component consisting of a network of microchannels modified to induce chaotic mixing (25, 28) to form a Click Chip, as shown in Fig. 1. A complementary bioorthogonal motif (i.e., TCO) is covalently conjugated (29) onto anti-EpCAM. The resulting TCO–anti-EpCAM conjugate is then grafted onto target CTCs before capture. The PDMS chaotic mixer is integrated to facilitate direct physical contact between TCO-grafted CTCs and the Tz-grafted SiNWS. To optimize the operating parameters of Click Chips, we prepared artificial CTC samples by spiking 200 EpCAM-positive NSCLC cells into freshly purified human WBCs (5 × 106 cells ml−1) in a standard RPMI 1640 culture medium (200 μl). For convenience and precision during cell imaging and counting, these NSCLC cells were prestained with a DiO green fluorescent dye before spiking, while the WBCs were prestained with a DiD red fluorescent dye. We initially used a NSCLC cell line with a representative ROS1 rearrangement, i.e., HCC78 (SLC34A2-ROS1 rearrangement) to test and optimize the performance of the CTC capture and release (Fig. 3A). Before conducting CTC capture studies, the TCO–anti-EpCAM was incubated with the artificial NSCLC samples for 30 min at room temperature. After washing away excess antibody, the TCO-grafted model CTC samples were run through Click Chips. The SiNWS-immobilized cells or released/purified cell mixtures were then stained with 4′,6-diamidino-2-phenylindole (DAPI) and counted using a fluorescence microscope (Nikon 90i).
Fig. 3. Validation and optimization of Click Chips using artificial NSCLC samples.
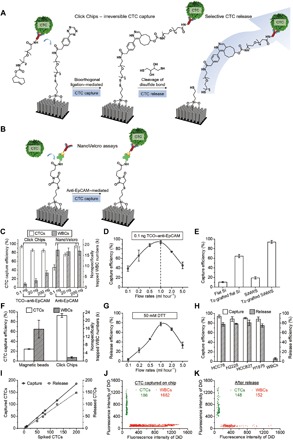
(A) Schematic representation of the mechanisms for bioorthogonal ligation–mediated capture and the disulfide cleavage–driven release of CTCs from Click Chips’ Tz-grafted SiNWS. (B) Schematic illustrating the conventional anti-EpCAM–mediated CTC capture process of NanoVelcro assays. (C) Comparison of CTC capture efficiency and specificity (i.e., nonspecifically captured WBCs) observed for Click Chips and NanoVelcro assays in the presence of 0.1, 20, and 200 ng of the respective anti-EpCAM capture agents (n = 3). (D) CTC capture efficiency of Click Chips was studied at flow rates of 0.1, 0.2, 0.5, 1.0, 2.0, and 5.0 ml hour−1 (n = 3). (E) CTC capture efficiency observed for the four different control groups: flat Si substrates without the Tz motif, Tz-grafted flat Si substrates, SiNWS without the Tz motif, and Click Chips based on Tz-grafted SiNWS (n = 3). (F) Comparison of capture efficiencies and specificity of bioorthogonal ligation–mediated CTC capture on Click Chips and magnetic beads. (G) CTC release efficiency was measured for Click Chips at flow rates of 0.1, 0.2, 0.5, 1.0, 2.0, and 5.0 ml hour−1 (n = 3). (H) General applicability of Click Chips for CTC capture and release was validated using artificial samples containing different NSCLC cell lines, i.e., HCC78, H2228, HCC827, H1975, and WBCs (n = 3). (I) Dynamic ranges observed for CTC capture and release using Click Chips. Spiked CTC numbers range from 5 to 200 cells ml−1. (J) A double parameter scatter plot showing the HCC78/WBC cell distribution observed for CTC capture in a Click Chip. (K) A double parameter scatter plot showing the cell distribution observed for CTC release, following the above experiment (J).
Figure 3C summarizes the CTC capture efficiencies and specificities observed for both bioorthogonal ligation–mediated CTC capture (Click Chips, Fig. 3A) and conventional anti-EpCAM–mediated CTC capture (NanoVelcro assays, Fig. 3B) (25) using flow rates of 1.0 ml hour−1. High CTC capture efficiencies (94 ± 3%) were demonstrated with Click Chips using a much smaller quantity of TCO–anti-EpCAM (as low as 0.1 ng) per capture study than used with previous NanoVelcro Chips. In comparison, anti-EpCAM–mediated NanoVelcro Chips had a lower capture efficiency (46 ± 6%) and higher WBC (17,547 ± 2757) contamination when using the same quantity of anti-EpCAM capture agent (0.1-ng biotinylated anti-EpCAM per capture study). Moreover, 200 ng of anti-EpCAM was needed for anti-EpCAM–mediated NanoVelcro Chips to achieve equivalent CTC capture efficiency. The incidence of nonspecifically captured WBCs (1684 ± 613 WBCs) on the Tz-grafted SiNWS was markedly reduced, with approximately 10 times fewer WBCs than anti-EpCAM–mediated NanoVelcro Chips.
Using 0.1 ng of TCO–anti-EpCAM, we evaluated the effect of different flow rates (0.1 to 5 ml hour−1) on the CTC capture efficiency of Click Chips. Our data suggest an optimum flow rate of 1.0 ml hour−1, which results in an average capture efficiency of 94 ± 3% (Fig. 3D). The effects of both nanoscale topography (19) and surface chemistry were evaluated by comparing the CTC capture performance of (i) flat Si substrates without the Tz motif, (ii) Tz-grafted flat Si substrates, (iii) SiNWS without the Tz motif, and (iv) Tz-grafted SiNWS. All studies were performed using a flow rate of 1.0 ml hour−1. The Tz-grafted SiNWS outperformed the three control groups, suggesting that the synergistic integration of the covalent bioorthogonal ligation–mediated (22) CTC capture and the topographic interactions (19) between cells and nanostructured substrates (i.e., SiNWS) are critical for optimal CTC capture efficiency (Fig. 3E). We then compared the performance of Click Chips with Tz-grafted magnetic beads (fig. S5) using the same quantity of TCO–anti-EpCAM (0.1 ng per capture study). The magnetic bead–based sorting method exhibited a capture efficiency of 24 ± 1% with a high number (15,781 ± 4362) of nonspecifically trapped WBCs (Fig. 3F). These results demonstrate the superiority of Click Chip, which combines the advantages of bioorthogonal ligation, nanostructured substrates, and herringbone-patterned PDMS together into a single device.
CTC release from Click Chips
The capability to release CTCs from the Tz-grafted SiNWS was studied using the optimized CTC capture conditions identified above (i.e., 0.1-ng TCO–anti-EpCAM, capture flow rate = 1.0 ml hour−1). The captured cells were released from the nanostructured surface by injecting 200 μl of DTT (50 mM) into Click Chip devices. We examined the effect of flow rate (0.1 to 5 ml hour−1) on the CTC release efficiency and found that a rate of 1.0 ml hour−1 resulted in the most robust release and collection of cells (78 ± 4%) (Fig. 3G). Higher flow rates (>1.0 ml hour−1) resulted in damage to cells as the result of the higher shear forces and mechanical collision in the devices. Operating Click Chips at optimal conditions enables rapid CTC purification (capture and release) in 24 min. The general applicability of Click Chips was tested and validated using four artificial CTC samples composed of NSCLC cell lines, i.e., HCC78 (SLC34A2-ROS1), H2228 (EML4-ALK), HCC827 (EGFR exon19del E746-A750), or NCI-H1975 (EGFR L858R/T790M) mixed with populations of WBCs. In these studies, Click Chips can capture the NSCLC cells with average efficiencies between 88 and 94% and release the cells with comparable average efficiencies ranging between 75 and 81% (Fig. 3H). The dynamic range of Click Chips was evaluated using artificial CTC samples containing 5 to 200 HCC78 cells spiked into a population of WBCs (Fig. 3I). Click Chips exhibit consistent capture (y = 0.93x − 1.10, R2 = 0.999) and recovery rates (y = 0.75x − 1.84, R2 = 0.993) that are sufficient to detect CTCs in clinical samples. A representative study using an artificial CTC sample containing 200 HCC78 cells (Fig. 3, J and K) demonstrated efficient CTC capture (186 of 200) and release (148 of 186) from Click Chips with minimal background (1682 WBC capture and 152 WBC release). More comprehensive experimental data are compiled in table S1. These data suggest that the disulfide cleavage–driven release mechanism used here will further improve the purity of recovered CTC populations (ca. one order of magnitude reduction in WBC contamination), ensuring the collection of robust and reproducible samples for downstream molecular characterization.
Quantification of rearrangements using CTC-derived mRNA
We first demonstrated the feasibility and dynamic ranges of detecting ALK/ROS1 rearrangements via RT-ddPCR with CTCs purified from artificial NSCLC cell line samples (figs. S6 and S7). Furthermore, we showed that disulfide cleavage–driven CTC release improved signal-to-noise ratio compared with on-chip cell lysis in detecting ROS1 rearrangement (fig. S8). Click Chip–mediated capture and release of CTCs were then validated using blood samples collected from patients with NSCLC (Fig. 4A). As shown in Table 1, blood was sampled from 12 late-stage patients with NSCLC who were treated with crizotinib and 6 healthy donors. Among the 12 patients with NSCLC, 7 patients harbored ALK rearrangements and 5 patients harbored ROS1 rearrangements. Blood was drawn before and after initiation of crizotinib therapy for some patients. For each patient with NSCLC and each healthy donor, two 2.0-ml blood samples were separately subjected to (i) CTC capture in a Click Chip, followed by immunostaining and CTC enumeration (30), and (ii) CTC purification in a Click Chip, followed by RT-ddPCR analysis to quantify the copy numbers of rearranged ALK or ROS1 transcripts. The CTC enumeration counts and copy numbers of rearranged ALK or ROS1 transcripts for individual samples are summarized in Table 1. The number of CTCs ranged from 0 to 36, and there are 2780 to 3196 WBCs in the Click Chips for each 2-ml blood sample. We detected positive ALK or ROS1 rearrangements in the CTCs collected from all 12 patients with NSCLC that were consistent with biopsies obtained at diagnosis. All the six healthy donors were negative for CTCs and rearranged ALK/ROS1 transcripts.
Fig. 4. Click Chips combined with RT-ddPCR analysis can be used to monitor dynamic changes in CTC count and CTC-derived ALK/ROS1 rearrangements in patients with NSCLC over the course of crizotinib (ALK/ROS1-TKI) treatments.
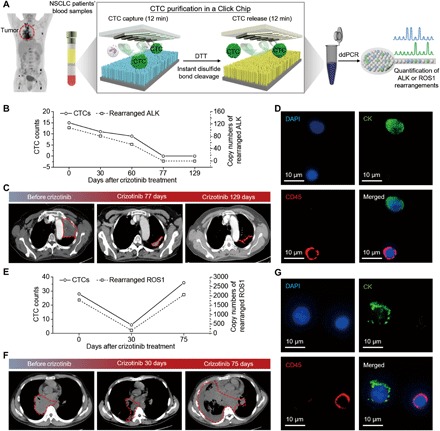
(A) Schematic illustrating the general workflow developed for conducting CTC enumeration and CTC-based ALK/ROS1 rearrangement quantification. (B) The dynamic changes (0 to 129 days) of CTC counts and rearranged ALK transcripts (per 2 ml of blood) observed for a patient with NSCLC harboring the EML4-ALK rearrangement (A07) before and after crizotinib treatment. (C) CT images of patient A07 taken at days 0, 77, and 129, after crizotinib treatment. (D) Representative fluorescent micrographs of CTCs (DAPI+/CK+/CD45–) captured from patient A07’s blood samples using Click Chips. (E) Dynamic change (0 to 75 days) of CTC counts and rearranged ROS1 transcripts (per 2 ml of blood) observed for a patient with NSCLC with the CD74-ROS1 rearrangement (R05) before and after crizotinib treatment. (F) Chest CT scans of patient R05 taken at days 0, 30, and 75 after crizotinib treatment. (G) Representative fluorescent micrographs of the CTC populations obtained from patient R05.
Table 1. Clinical characteristics of patients with NSCLC (adenocarcinoma) and healthy donors (HD) enrolled in our study.
N/A, not applicable.
|
Patient number |
Gender | Age (years) |
Smoking history (years) |
Tumor grade |
Clinical stage |
CTC counts* |
ALK/ ROS1 status (tissue) |
ALK/ ROS1 status (CTCs) |
Copy numbers of ALK/ROS1 rearrangements in CTCs* |
| A01 | Male | 53 | None | 3 | IV | 18 | EML4-ALK | EML4-ALK | 333 |
| A02 | Male | 38 | None | 3 | IV | 15 | EML4-ALK | EML4-ALK | 603 |
| A03 | Male | 65 | None | 3 | IV | 32 | EML4-ALK | EML4-ALK | 198 |
| A04 | Female | 65 | None | 3 | IV | 27 | EML4-ALK | EML4-ALK | 1728 |
| A05 | Male | 53 | None | 3 | IV | 16 | EML4-ALK | EML4-ALK | 252 |
| A06 | Male | 58 | None | 3 | IV | 26 | EML4-ALK | EML4-ALK | 207 |
| A07-1 | Male | 39 | None | 2 | IV | 15 | EML4-ALK | EML4-ALK | 108 |
| A07-2 | Male | 39 | None | 2 | IV | 11 | EML4-ALK | EML4-ALK | 81 |
| A07-3 | Male | 39 | None | 2 | IV | 9 | EML4-ALK | EML4-ALK | 54 |
| A07-4 | Male | 39 | None | 2 | IV | 0 | EML4-ALK | EML4-ALK | 0 |
| A07-5 | Male | 39 | None | 2 | IV | 0 | EML4-ALK | EML4-ALK | 0 |
| R01 | Female | 62 | None | 3 | IIIB | 28 | CD74-ROS1 | CD74-ROS1 | 504 |
| R02 | Male | 41 | None | 2 | IIIB | 22 | CD74-ROS1 | CD74-ROS1 | 594 |
| R03 | Male | 61 | 35 | 2 | IV | 17 | CD74-ROS1 | CD74-ROS1 | 684 |
| R04 | Male | 34 | None | 3 | IV | 27 | CD74-ROS1 | CD74-ROS1 | 963 |
| R05-1 | Male | 32 | None | 3 | IV | 28 | CD74-ROS1 | CD74-ROS1 | 1773 |
| R05-2 | Male | 32 | None | 3 | IV | 6 | CD74-ROS1 | CD74-ROS1 | 162 |
| R05-3 | Male | 32 | None | 3 | IV | 36 | CD74-ROS1 | CD74-ROS1 | 2061 |
| HD01 | Male | 30 | None | N/A | N/A | 0 | N/A | N/A | 0 |
| HD02 | Male | 26 | None | N/A | N/A | 0 | N/A | N/A | 0 |
| HD03 | Male | 29 | None | N/A | N/A | 0 | N/A | N/A | 0 |
| HD04 | Male | 46 | None | N/A | N/A | 0 | N/A | N/A | 0 |
| HD05 | Female | 36 | None | N/A | N/A | 0 | N/A | N/A | 0 |
| HD06 | Male | 32 | None | N/A | N/A | 0 | N/A | N/A | 0 |
*Per 2 ml of blood.
Dynamic monitoring of CTCs and ALK/ROS1 rearrangements during treatment
In Fig. 4 (B to D), we track the treatment of patient A07 over time. Blood was collected serially before initiating crizotinib and then 30, 60, 77, and 129 days after treatment. The dynamic changes are plotted for both the CTC count and the copy numbers of rearranged ALK transcripts detected in Fig. 4B. Computed tomography (CT) images of the patient showed that before treatment, the patient exhibited a heavy tumor burden in his left lung. Following the initiation of crizotinib, the patient’s tumor burden was reduced progressively over the course of 129 days (Fig. 4C). These radiographic observations are consistent with the reductions observed in both CTC count and ALK rearrangement transcripts detected. At the time of data cutoff, the patient’s treatment response with crizotinib was durable. Figure 4D shows a typical micrograph of CTCs from patient A07 captured using the Click Chip system.
Serial blood draws were obtained before and after initiation of crizotinib therapy from a patient with NSCLC (R05) harboring ROS1 rearrangement. The CTC count and copy numbers of rearranged ROS1 transcripts are plotted in Fig. 4E. Corresponding CT images from the patient show lesions throughout the patient’s chest before starting crizotinib. The patient was found to have a partial response after 30 days of treatment but unfortunately relapsed after 75 days (Fig. 4F). The patient died of intractable tumor burden 78 days after entering the study. Dynamic changes in both the number of CTCs detected and the copy number of rearranged ROS1 transcripts were consistent with CT images and clinical outcomes. A typical micrograph of the CTCs captured by Click Chips from this patient with NSCLC is shown in Fig. 4G. These data serve as a powerful proof of concept for the capability to use CTCs collected via Click Chips to serve as a diagnostic tool for monitoring disease status in critically ill patients with cancer.
DISCUSSION
In this study, we have successfully developed and validated a new covalent chemistry–based CTC purification device (Click Chip) that enables efficient purification of pooled CTCs with well-preserved mRNA and low levels of WBC contamination, making it an excellent option for a CTC mRNA assay. We demonstrated the clinical utilities of Click Chips in NSCLC for (i) enumerating CTCs and (ii) quantifying ALK/ROS1 rearrangements in pooled CTCs with well-preserved mRNA by coupling with RT-ddPCR. Our experimental data unveil that Click Chips exhibit markedly improved CTC capture efficiency and specificity compared with the controlled studies, paving the way for precise quantification of gene rearrangements in CTCs. The improved CTC capture efficiency and specificity observed for Click Chips are attributed to the synergistic integration of the covalent bioorthogonal ligation–mediated (22) CTC capture and nanostructured substrates (i.e., SiNWS) (19). The bioorthogonal ligation–mediated CTC capture is used to overcome the limitations associated with the conventional antibody–mediated CTC capture methods. A pair of highly reactive click chemistry motifs (i.e., TCO and TZ) were grafted onto CTCs and SiNWS via antibody conjugation and surface modification, respectively. When a blood sample is run through a Click Chip, the PDMS chaotic mixer facilitates physical contact between the TCO-grafted CTCs and the Tz-grafted SiNWS. These interactions trigger inverse electron-demand Diels-Alder reactions between TCO on the CTCs and Tz on the SiNWS, instantly and irreversibly immobilizing the CTCs. Previously, we demonstrated that the incorporation of nanostructured substrates (19, 25) into cell-affinity substrates resulted in enhanced cell capture affinities compared with those observed for nonstructured (i.e., flat) substrates. The results summarized in this study also suggest that incorporating SiNWS into the click chemistry–mediated CTC capture platform resulted in a markedly improved CTC capture efficiency (94 ± 3%) and specificity (with a 10-fold reduction in WBC contamination) using a much smaller quantity of TCO–anti-EpCAM (as low as 0.1 ng) per capture study. Our present experiential data suggest that the use of a single capture agent (i.e., TCO–anti-EpCAM) is sufficient to achieve the desired CTC capture performance in NSCLC. We envision that the proposed Click Chips can be synergistically coupled with a multimarker capture cocktail to capture and purify CTCs in the cancer types (e.g., HCC) that require multiple capture agents (31).
In addition to the improved CTC capture efficiency and specificity observed for Click Chips, incorporating a disulfide bond into the surface linker that tethers Tz motifs onto the SiNWS allows for disulfide cleavage–driven CTC release. Upon exposure to a mild disulfide cleavage agent (i.e., DTT) (26), the surface linkers anchoring the CTCs onto the SiNWS are cleaved, releasing the CTCs and leaving the nonspecifically trapped WBCs behind. We note that disulfide cleavage–driven CTC release confers a second layer of specificity, further improving CTC purification performance. Moreover, the well-preserved mRNA of purified pooled CTCs paves the way for seamless coupling with detection and quantification of oncologic gene rearrangements (i.e., ALK and ROS1). Compared with several reported release strategies such as dual-mode responsive gelatin (32), ligand competition (9), DNA displacement (33), aptamer/enzyme digestion (34), and light exposure (35), our method could retrieve CTCs under milder conditions with enhanced purity and reduced processing time, thereby providing better preserved CTC-derived mRNA. Besides, our experimental data have demonstrated the advantages of post-release lysis of purified CTCs over on-chip lysis in the ROS1 rearrangement detection using artificial HCC78 CTC samples. We attribute the higher copy number of ROS1 rearrangement detected using the mRNA extracted from the post-release lysis to (i) the high yield of CTC capture/release, (ii) the short CTC purification time (24 min), (iii) the mild processing conditions, and (iv) the protection of CTC membranes for their RNA contents. In comparison, mRNA exposed by on-chip lysis of CTCs is contaminated by mRNA from a relatively large number of WBCs, which leads to the targetable gene rearrangement being buried in irrelevant background. Moreover, it is difficult to flush CTC-RNA from the densely arranged Si nanowires, further increasing the value of the CTC releasing process in our platform.
In patients with NSCLC, the presence of ALK/ROS1 rearrangements defines a distinct molecular subgroup that may benefit from the treatment of ALK/ROS1-TKIs (i.e., crizotinib) (36). Despite the effectiveness of crizotinib in ALK/ROS1-positive NSCLC, most patients eventually develop acquired resistance to crizotinib, typically within 1 year (37, 38). Therefore, quantifying ALK/ROS1 rearrangement by RT-ddPCR in CTCs may serve as a complementary method providing early prediction of clinical treatment responses, offering an invaluable glimpse into the longitudinal evolution of ALK/ROS1 rearranged NSCLC during ALK/ROS1-TKI treatment. Although fluorescence in situ hybridization (FISH) tests have been used to detect gene rearrangements in CTCs, adopting these CTC-based FISH tests in clinical settings is challenging (39). The rare FISH events need to be visually identified with high-resolution fluorescent microscopy after confirming the CTCs’ positions via immunofluorescence staining. These laborious procedures would introduce significant burden to laboratory personnel and pathologists. These CTC-based FISH tests are unable to provide quantitative readouts or to identify the fusion gene partners (13). We demonstrate that Click Chips can be applied for both CTC enumeration and purification using blood samples collected from patients with NSCLC. The purified CTCs can then be quantified in terms of ALK/ROS1 rearrangements by RT-ddPCR with single-molecule precision. We applied this capability to correlate dynamic changes in both CTC count and in the quantification of CTC-derived ALK/ROS1 rearrangements to outcomes from radiographic imaging, the clinical gold standard for evaluating responses of solid tumors. Compared with CTC counts, quantification of CTC-derived ALK/ROS1 rearrangements yields a broader dynamic range, enabling more precise monitoring of treatment response and/or disease progression in patients with NSCLC. These results can empower oncologists to make more informed and precise therapeutic decisions as they direct the care of their patients.
MATERIALS AND METHODS
Fabrication of Tz-grafted SiNWS
Vertically aligned SiNWS were prepared via a combination of photolithographic patterning and silver (Ag) nanoparticle–templated wet etching (27) according to the procedures described in our previous publication (25). Briefly, a thin-film photoresist (AZ 5214, AZ Electronic Materials USA Corp.) was spin-coated onto a (100) p-type silicon (Si) wafer (Silicon Quest International) with resistivity approximately 10 to 20 ohm·cm. After being exposed to ultraviolet (UV) light, the Si wafer was immersed into the etching solution containing hydrofluoric acid (4.6 M; Sigma-Aldrich), silver nitrate (AgNO3, 0.2 M; Sigma-Aldrich), and deionized water. After that, the Si wafer was immersed in boiling aqua regia [hydrochloric acid (HCl)/nitric acid (HNO3), 3:1 (v/v); Sigma-Aldrich] for 15 min to remove the silver film. The resulting Si nanowires had a length of approximately 10 μm. The SiNWS were rinsed with acetone (≥99.5%; Sigma-Aldrich) and then ethanol (anhydrous; Sigma-Aldrich) several times to remove the patterned photoresist. A three-step chemical modification was developed to introduce a disulfide linker coupled to the Tz motif onto the chip surfaces. (i) For surface silanization, the SiNWS were placed into a Teflon frame in a glass beaker and incubated with a piranha solution [sulfuric acid (H2SO4)/hydrogen peroxide (H2O2), 2:1 (v/v); Sigma-Aldrich] for 1 hour. After rinsing successively with deionized water and ethanol three times, the SiNWS were then dried with nitrogen gas and then sealed in a vacuum desiccator and exposed to silane vapor coming from MPS (211.4 mg, 200 μl; Sigma-Aldrich) for 45 min to introduce thiol groups onto the SiNWS. (ii) Freshly prepared HS-SiNWS were incubated with OPSS-PEG-NH2 (0.30 mg, 3.8 mM; Nanocs Inc.) in dimethyl sulfoxide (200 μl) solution for 2 hours to incorporate disulfide linkers with terminal amine groups. The obtained amine-terminated silicon nanowire substrates (H2N-SiNWS) were rinsed with ethanol three times. (iii) To graft Tz motifs onto SiNWS, the H2N-SiNWS were reacted with Tz-sulfo-NHS ester (0.32 mg, 3.8 mM; Click Chemistry Tools Bioconjugate Technology Company) in phosphate-buffered saline (PBS) buffer (200 μl) for 1 hour. The functionalized Tz-grafted SiNWS were rinsed with PBS three times before use for CTC capture and release experiments.
Preparation of microfluidic chaotic mixer components
Microfluidic chaotic mixer substrates were fabricated from a photolithographically prepared master wafer prepared by inductively coupled plasma-reactive ion etching (25, 40). The master wafer was fabricated by spin-coating a 75-μm-thick layer of negative photoresist (MicroChem Corp.) onto a 76-mm silicon wafer. The silicon wafer was then exposed to UV light through a photomask composed of a serpentine rectangular microfluidic channel (22-cm length and 2.0-mm width). Next, a layer of a second negative photoresist was spin-coated onto the wafer with a thickness of 40 μm. A second photomask composed of herringbone ridges was aligned between the previously exposed pattern and the pattern to be imprinted using a Mask Aligner (Karl Suss America Inc.). The tops of the resulting microchannels have ridges that were used to promote chaotic mixing within the microfluidic network. Before replica molding, the Si master was pretreated by exposure to trimethylchlorosilane vapor for 1 min. The silane-treated master was transferred to a petri dish. The petri dish was filled with the well-mixed PDMS precursor (RTV 615 A and B in a 10:1 ratio; GE Silicones), degassed, and then incubated in an oven at 80°C for 48 hours to make a 5-mm-thick chip. The PDMS microfluidic chaotic mixer was peeled off from the silicon master wafer/mold, and two through-holes were punched at the ends of the channel for insertion of tubing.
Assembling Click Chips in chip holders
A chip holder was custom-designed to establish a secure seal between the Tz-grafted SiNWS and the PDMS component. Multiple alignment markers were designed on the substrates to enable rapid assembly of Click Chip devices (within 1 min). A fully automated digital fluidic handling system composed of two PSD4 syringe pumps (Hamilton Robotics), rotary valves, and a controller program with graphical user interface was used to control the loading of reagents and test samples as well as automatically coordinate CTC capture and release.
X-ray photoelectron spectroscopic characterization
The analysis was carried out in a PHI 5000 VersaProbe system (ULVAC-PHI; Chigasaki) using a microfocus (100 μm, 25 W) Al Kα x-ray source with photoelectrons collected from a takeoff angle of 45°. Functionalized SiNWS were vacuum-dried overnight before being transferred into the system for analysis. During spectral acquisition, a dual-beam charge neutralizer (7-V Ar+ and 1-V flooding electron beam) was used to compensate for any charging effects. Survey spectra over a binding energy range of 0 to 1000 were measured for SiNWS, HS-SiNWS, H2N-SiNWS, and Tz-grafted SiNWS. High-resolution XPS spectra of S 2p, N 1s, C 1s, and O 1s were also collected. The C─C peak corresponding to adventitious carbon (C 1s = 284.6 eV) was used to calibrate binding energies.
Fluorescence characterization of Tz-grafted SiNWS
To demonstrate the feasibility and specificity of bioorthogonal ligation between Tz and TCO motifs on Tz-grafted SiNWS, Cy5-labeled TCO reagent (2.6 mM, Click Chemistry Tools Bioconjugate Technology Company) in PBS (200 μl) was used to treat Tz-grafted SiNWS as well as H2N-SiNWS (as a control) for 30 min. Unbound Cy5-labeled TCO was rinsed off with deionized water five times before characterization. Fluorescence images were collected with a fluorescence microscope (Nikon 90i; λex = 590 to 650 nm, exposure time = 500 ms). To investigate disulfide cleavage after bioorthogonal ligation, the Cy5-grafted SiNWS were treated with DTT (3.8 mM; Sigma-Aldrich) in PBS (200 μl) for 0, 15, 30, 45, and 60 min and DTT (50 mM) in PBS (200 μl) for 0, 3, 6, 9, 12, 15, 20, 30, 45, and 60 min, respectively. After being rinsed by deionized water five times, the fluorescence images were collected using a fluorescence microscope. Average fluorescence intensity of Cy5 on substrates was calculated using the NIS-Elements AR 4.20 software package.
Preparation of TCO–anti-EpCAM conjugate
TCO–anti-EpCAM conjugate was prepared by incubating TCO-PEG4-NHS ester (0.5 mM; Click Chemistry Tools Bioconjugate Technology Company) with human EpCAM/TROP-1 antibody [goat immunoglobulin G (IgG), 0.5 μg μl−1; R&D Systems Inc.] in PBS at room temperature for 30 min. The TCO–anti-EpCAM conjugate was diluted to 0.005 μg μl−1 and stored at −20°C.
NSCLC cell lines
Four NSCLC cell lines including HCC78, H2228, HCC827, and H1985 were purchased from American Type Culture Collection and cultured using RPMI 1640 1640 growth medium with 10% fetal bovine serum, 1% GlutaMAX-I, and penicillin-streptomycin (100 U ml−1) (Thermo Fisher Scientific) in a humidified incubator with 5% CO2.
Artificial CTC sample studies
To enable convenient cell imaging and counting, cultured EpCAM-positive NSCLC cells (1 × 106 ml−1) were prestained with a Vybrant DiO green fluorescent dye (Invitrogen) in serum-free culture medium at 37°C for 1 hour. EpCAM-negative WBCs were isolated from blood samples of the healthy donors with approval from the University of California, Los Angeles Institutional Review Board (no. 00000173) and then prestained with a Vybrant DiD red fluorescent dye (Invitrogen). Excess cell-labeling dye was removed by centrifuging the labeled suspension at 1500 rpm for 5 min and washed with PBS twice. Typical artificial CTC samples were prepared by spiking 200 prestained NSCLC cells into the prestained WBCs (5 × 106 cells ml−1) in 200 μl of RPMI 1640. The artificial CTC sample was incubated with TCO–anti-EpCAM (0.1 ng) in RPMI 1640 (200 μl) at room temperature for 30 min and then centrifuged at 300g for 10 min to remove the excess TCO–anti-EpCAM and nonreactive TCO-PEG4-NHS ester. The resulting samples containing TCO-grafted CTCs were then purified using Click Chips. For CTC enumeration, the cells captured on Tz-grafted SiNWS were stained with DAPI and imaged under a fluorescence microscope (Nikon 90i, W1, 325 to 375 nm; W2, 465 to 495 nm; W3, 590 to 650 nm).
CTC capture and release from Click Chips
For CTC capture, PBS (200 μl) was first introduced into a Click Chip via a digital fluidic handler at a flow rate of 1 ml hour−1 to confirm that an appropriate seal was made between the patterned PDMS chaotic mixer and the Tz-grafted SiNWS. The TCO-grafted artificial CTC samples and peripheral blood mononuclear cell (PBMC) samples (200 μl) collected from patients with NSCLC were infused into Click Chip microfluidic devices at the optimum flow rate of 1 ml hour−1. For CTC enumeration, the CTCs captured in Click Chip were fixed with 2.0% formaldehyde (Electron Microscopy Sciences) in PBS (200 μl). CTCs captured on the chips were enumerated. To release CTCs, 200 μl of DTT (50 mM) was injected into the Click Chip system at a flow rate of 1 ml hour−1, and then 100 μl of PBS was injected at a flow rate of 1 ml hour−1 to collect the released CTCs. The released CTCs were collected into either a 96-well plate for fluorescence imaging or 1.5-ml ribonuclease (RNase)–free Eppendorf tubes for subsequent molecular analysis using RT-ddPCR.
Patients with NSCLC and treatments
We consecutively enrolled 12 patients with treatment-naïve advanced NSCLC (stages III and IV) from October 2016 to June 2017, including 7 patients who harbored the ALK rearrangement (13, 14) and 5 patients with known ROS1 rearrangement. Patients who had other uncontrolled malignant tumors, uncontrolled infection or Mycobacterium tuberculosis, or severe mental disease were excluded. Physicians and patients made treatment decisions together, and all the 12 enrolled patients received crizotinib as a first-line therapy. All the enrolled patients underwent follow-up imaging examinations every 2 to 3 months, and their clinical responses were evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST 1.1). This study was approved by the Ethics Committee of Guangdong Provincial Hospital of Traditional Chinese Medicine, and all patients provided written informed consent for this study.
Pathological examination of NSCLC tissue sections
Pathological examination including hematoxylin and eosin (H&E) staining, immunohistochemistry (IHC), and ALK/ROS1 rearrangement analysis of the biopsies obtained from the 12 enrolled patients was performed independently by experienced pathologists from Guangdong Provincial Hospital of Traditional Chinese Medicine. All the tumor samples were fixed in 10% neutral formalin for 24 to 48 hours and embedded in paraffin. The H&E staining was performed following Clinical Laboratory Improvement Amendments–compliant methods and equipment. Serial 4-μm-thick tissue sections from formalin-fixed paraffin-embedded (FFPE) blocks were cut and mounted on poly-l-lysine–coated glass slides. Standard IHC staining on 4-μm-thick tissue sections was performed on Ventana BenchMark ULTRA Slide Stainer according to a protocol optimized for each antibody as determined by conventional IHC. The IHC analyses for P63, CK5/6, CK7, thyroid transcription factor-1 (TTF-1), Napsin A, CD56, synaptophysin (SYN), and chromogranin A (CgA) were performed to differentiate between NSCLC (adenocarcinoma and squamous cell carcinoma) and small cell lung cancer in the poorly differentiated cases. Positive CK7, Napsin A, and/or TTF-1 staining combined with negative P63, CK5/6, CD56, SYN, and CgA staining confirmed the diagnosis of lung adenocarcinoma enrolled in this study.
ALK/ROS1 rearrangement analysis of NSCLC tissue
ALK/ROS1 rearrangement analysis of tissue was performed according to the standard protocols in the Molecular Pathology Department of Guangdong Provincial Hospital of Traditional Chinese Medicine.The ALK/ROS1 rearrangements were detected by RT real-time PCR using a fusion gene detection kit (Amoy, Xiamen, China, China Food and Drug Administration–approved), which covers 26 ALK gene rearrangement subtypes and 14 ROS1 gene rearrangement subtypes. Total RNA was extracted from FFPE tissue sections using Qiagen (Dusseldorf, Germany) RNeasy FFPE kit. Complementary DNA (cDNA) was synthesized under the following conditions: 42°C, 1 hour; 95°C, 5 min. The conditions used for real-time PCR were as follows: an initial denaturation at 95°C for 5 min, followed by 15 cycles of 95°C for 25 s, 64°C for 20 s, and 72°C for 20 s and 31 cycles of 93°C for 25 s, 60°C for 35 s, and 72°C for 20 s.
Collection and treatment of blood samples from patients with NSCLC
Peripheral venous blood samples were collected serially before and after initiation of crizotinib therapy with the written informed consent of these patients. Each 5.0-ml blood sample was collected in a BD Vacutainer glass tube (BD Medical, Thermo Fisher Scientific catalog no. 02-684-26) with acid citrate dextrose. For each sample of the patient with NSCLC, two 2.0-ml vials of blood were separately centrifuged to collect PBMCs. The isolated PBMCs were then incubated with TCO–anti-EpCAM (0.1 ng) in PBS (200 μl) at room temperature for 30 min before being loaded into Click Chip devices. One 2.0-ml blood sample was used for CTC capture and enumeration. In parallel, the other 2.0-ml blood sample was used for CTC release and ALK/ROS1 rearrangement quantification by RT-ddPCR.
Immunofluorescence characterization of CTCs captured from blood samples
A three-color immunocytochemistry analysis (30) was adopted for immunofluorescence characterization of CTCs captured from blood samples collected from patients with NSCLC. The cells captured on Tz-grafted SiNWS were incubated with 0.05% Triton X-100 [in PBS (200 μl)] for 10 min. The captured cells were incubated overnight with a mixture of primary antibodies including Pan-CK antibody [rabbit, polyclonal, 1:100 (v/v); Dako] and anti-CD45 antibody [F10-89-4] [mouse, monoclonal, 1:400 (v/v); Abcam] in a PBS solution (200 μl) containing 2% normal donkey serum (Jackson ImmunoResearch) at 4°C. After washing with PBS three times, the captured cells were further incubated at room temperature for 1 hour with a mixture of secondary antibodies including donkey anti-rabbit IgG (H+L) [Alexa Fluor 488, 1:500 (v/v); Invitrogen] and donkey anti-mouse IgG (H+L) [Alexa Fluor 647, 1:500 (v/v); Invitrogen] in a PBS solution (200 μl) containing 2% donkey serum. After washing with PBS three times, the cells were treated with ProLong Gold Antifade Mountant with DAPI (Invitrogen). The substrates were then imaged with a fluorescence microscope (Nikon 90i). The captured CTCs were differentiated from background WBCs according to their unique staining pattern (CK+/CD45−/DAPI+) and intact nuclear morphology (WBCs were stained CK−/CD45+/DAPI+).
RNA extraction and RT-ddPCR detection of ALK/ROS1 rearrangement in CTCs
The CTCs released from Click Chips were collected in a 1.5-ml RNase-free Eppendorf tube and lysed by TRI Reagent [1:3, (v/v), Zymo Research Corp.]. The collected RNA was purified using a Direct-zol RNA MicroPrep Kit (Zymo Research Corp.) according to the manufacturer’s protocol. The purified RNA was reverse-transcribed to cDNA using a Thermo Scientific Maxima H Minus Reverse Transcriptase Kit according to the manufacturer’s instructions. Samples of cDNA were detected with a PrimePCR ddPCR Expert Design Assay Kit from Bio-Rad that covers 26 ALK gene rearrangement subtypes and 14 ROS1 gene rearrangement subtypes. Data were analyzed using the QuantaSoft software package to calculate the corresponding copy numbers of ALK or ROS1 rearrangements detected from individual samples.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Institutes of Health (R33CA174562, R44CA180482, R01CA218356, U01CA198900, and R21CA235340). J.D. gratefully acknowledges financial support from the China Scholarship Council (201706010064). Author contributions: Y.Z., H.-R.T., and J.D. designed the research and analyzed the data. J.D., M.M., and X.T. performed the device modification, CTC capture and CTC release experiments, and CTC enumeration. Y.J.J., J.C., and R.Y.Z. performed the RT-ddPCR experiments. L.B. and J.D. performed the cell culture and artificial CTC sample preparation. T.-Y.H., J.-J.S., and H.-h.Y. performed XPS characterization. Y.Z., D.Z., X.C., Y.L., H.Z., and G.Z. performed the detection of CTCs and ALK/ROS1 rearrangement from blood samples or tissues, as well as clinical information collection. P.-J.C. and A.Z. helped with CTC enumeration. J.D., H.-R.T., and Y.Z. prepared the manuscript. M.Z., M.L., P.S.W., and S.J.J. revised the manuscript. P.T., M.S., V.G.A., and E.M.P. commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaav9186/DC1
Fig. S1. High-resolution XPS characterization.
Fig. S2. Deconvolution of x-ray photoelectron spectra of stepwise functionalized SiNWS.
Fig. S3. Fluorescence imaging for exploring bioorthogonal ligation.
Fig. S4. Dynamic changes of Cy5 fluorescence intensity upon disulfide cleavage by DTT.
Fig. S5. Scheme of CTC capture on Tz-grafted magnetic beads.
Fig. S6. Scheme of RT-ddPCR analysis of ALK/ROS1 rearrangements in CTCs.
Fig. S7. Dynamic ranges for RT-ddPCR quantification of ALK/ROS1 rearrangements.
Fig. S8. Comparison of cell lysis on-chip with cell lysis of DTT-released cells.
Table S1. CTC purity during capture and release processes.
REFERENCES AND NOTES
- 1.Krebs M. G., Metcalf R. L., Carter L., Brady G., Blackhall F. H., Dive C., Molecular analysis of circulating tumour cells—Biology and biomarkers. Nat. Rev. Clin. Oncol. 11, 129–144 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Schwarzenbach H., Hoon D. S. B., Pantel K., Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Jack R. M., Grafton M. M., Rodrigues D., Giraldez M. D., Griffith C., Cieslak R., Zeinali M., Kumar Sinha C., Azizi E., Wicha M., Tewari M., Simeone D. M., Nagrath S., Ultra-specific isolation of circulating tumor cells enables rare-cell RNA profiling. Adv. Sci. 3, 1600063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramskold D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O. R., Daniels G. A., Khrebtukova I., Loring J. F., Laurent L. C., Schroth G. P., Sandberg R., Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto D. T., Zheng Y., Wittner B. S., Lee R. J., Zhu H., Broderick K. T., Desai R., Fox D. B., Brannigan B. W., Trautwein J., Arora K. S., Desai N., Dahl D. M., Sequist L. V., Smith M. R., Kapur R., Wu C. L., Shioda T., Ramaswamy S., Ting D. T., Toner M., Maheswaran S., Haber D. A., RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalinich M., Bhan I., Kwan T. T., Miyamoto D. T., Javaid S., LiCausi J. A., Milner J. D., Hong X., Goyal L., Sil S., Choz M., Ho U., Kapur R., Muzikansky A., Zhang H., Weitz D. A., Sequist L. V., Ryan D. P., Chung R. T., Zhu A. X., Isselbacher K. J., Ting D. T., Toner M., Maheswaran S., Haber D. A., An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 114, 1123–1128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto D. T., Lee R. J., Kalinich M., LiCausi J. A., Zheng Y., Chen T., Milner J. D., Emmons E., Ho U., Broderick K., Silva E., Javaid S., Kwan T. T., Hong X., Dahl D. M., McGovern F. J., Efstathiou J. A., Smith M. R., Sequist L. V., Kapur R., Wu C.-L., Stott S. L., Ting D. T., Giobbie-Hurder A., Toner M., Maheswaran S., Haber D. A., An RNA-based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer. Cancer Discov. 8, 288–303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong X., Sullivan R. J., Kalinich M., Kwan T. T., Giobbie-Hurder A., Pan S., LiCausi J. A., Milner J. D., Nieman L. T., Wittner B. S., Ho U., Chen T., Kapur R., Lawrence D. P., Flaherty K. T., Sequist L. V., Ramaswamy S., Miyamoto D. T., Lawrence M., Toner M., Isselbacher K. J., Maheswaran S., Haber D. A., Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl. Acad. Sci. U.S.A. 115, 2467–2472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen M. Y., Chen J. F., Luo C. H., Lee S., Li C. H., Yang Y. L., Tsai Y. H., Ho B. C., Bao L. R., Lee T. J., Jan Y. J., Zhu Y. Z., Cheng S., Feng F. Y., Chen P., Hou S., Agopian V., Hsiao Y. S., Tseng H. R., Posadas E. M., Yu H. H., Glycan stimulation enables purification of prostate cancer circulating tumor cells on PEDOT NanoVelcro chips for RNA biomarker detection. Adv. Healthc. Mater. 7, 1700701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solomon B. J., Mok T., Kim D. W., Wu Y. L., Nakagawa K., Mekhail T., Felip E., Cappuzzo F., Paolini J., Usari T., Iyer S., Reisman A., Wilner K. D., Tursi J., Blackhall F., First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 373, 1582 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Shaw A. T., Ou S. H. I., Bang Y.-J., Camidge D. R., Solomon B. J., Salgia R., Riely G. J., Varella-Garcia M., Shapiro G. I., Costa D. B., Doebele R. C., le L. P., Zheng Z., Tan W., Stephenson P., Shreeve S. M., Tye L. M., Christensen J. G., Wilner K. D., Clark J. W., Iafrate A. J., Crizotinib in ROS1-rearranged non–small-cell lung cancer. N. Engl. J. Med. 371, 1963–1971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A., Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 10, 472–484 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Rimkunas V. M., Crosby K. E., Li D., Hu Y., Kelly M. E., Gu T. L., Mack J. S., Silver M. R., Zhou X., Haack H., Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: Identification of a FIG-ROS1 fusion. Clin. Cancer Res. 18, 4449–4457 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Katayama R., Lovly C. M., Shaw A. T., Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: A paradigm for precision cancer medicine. Clin. Cancer Res. 21, 2227–2235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao W. J., Choi Y. L., Song J. Y., Zhu Y., Xu Q., Zhang F., Jiang L., Cheng J., Zheng G., Mao M., ALK, ROS1 and RET rearrangements in lung squamous cell carcinoma are very rare. Lung Cancer 94, 22–27 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Taylor S. C., Laperriere G., Germain H., Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 7, 2409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin M., Chen J.-F., Lu Y.-T., Zhang Y., Song J., Hou S., Ke Z., Tseng H.-R., Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc. Chem. Res. 47, 2941–2950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jan Y. J., Chen J.-F., Zhu Y., Lu Y.-T., Chen S. H., Chung H., Smalley M., Huang Y.-W., Dong J., Chen L.-C., Yu H.-H., Tomlinson J. S., Hou S., Agopian V. G., Posadas E. M., Tseng H.-R., NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Adv. Drug Deliv. Rev. 125, 78–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Wang H., Jiao J., Chen K. J., Owens G. E., Kamei K. I., Sun J., Sherman D. J., Behrenbruch C. P., Wu H., Tseng H. R., Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew. Chem. Int. Ed. 48, 8970–8973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou S., Chen J.-F., Song M., Zhu Y., Jan Y. J., Chen S. H., Weng T.-H., Ling D.-A., Chen S.-F., Ro T., Liang A.-J., Lee T., Jin H., Li M., Liu L., Hsiao Y. S., Chen P., Yu H.-H., Tsai M.-S., Pisarska M. D., Chen A., Chen L.-C., Tseng H.-R., Imprinted NanoVelcro microchips for isolation and characterization of circulating fetal trophoblasts: Toward noninvasive prenatal diagnostics. ACS Nano 11, 8167–8177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devaraj N. K., The future of bioorthogonal chemistry. ACS Cent. Sci. 4, 952–959 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackman M. L., Royzen M., Fox J. M., Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels−Alder reactivity. J. Am. Chem. Soc. 130, 13518–13519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karver M. R., Weissleder R., Hilderbrand S. A., Synthesis and evaluation of a series of 1,2,4,5-tetrazines for bioorthogonal conjugation. Bioconjug. Chem. 22, 2263–2270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riethdorf S., O’Flaherty L., Hille C., Pantel K., Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 125, 102–121 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Wang S. T., Liu K., Liu J., Yu Z. T.-F., Xu X., Zhao L., Lee T., Lee E. K., Reiss J., Lee Y.-K., Chung L. W. K., Huang J., Rettig M., Seligson D., Duraiswamy K. N., Shen C. K.-F., Tseng H.-R., Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem. Int. Ed. 50, 3084–3088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith E. A., Wanat M. J., Cheng Y., Barreira S. V. P., Frutos A. G., Corn R. M., Formation, spectroscopic characterization, and application of sulfhydryl-terminated alkanethiol monolayers for the chemical attachment of DNA onto gold surfaces. Langmuir 17, 2502–2507 (2001). [Google Scholar]
- 27.Peng K.-Q., Yan Y.-J., Gao S.-P., Zhu J., Synthesis of large-area silicon nanowire arrays via self-assembling nanoelectrochemistry. Adv. Mater. 14, 1164–1167 (2002). [Google Scholar]
- 28.Stroock A. D., Dertinger S. K. W., Ajdari A., Mezić I., Stone H. A., Whitesides G. M., Chaotic mixer for microchannels. Science 295, 647–651 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Haun J. B., Devaraj N. K., Hilderbrand S. A., Lee H., Weissleder R., Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat. Nanotechnol. 5, 660–665 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y.-T., Zhao L., Shen Q., Garcia M. A., Wu D., Hou S., Song M., Xu X., OuYang W.-H., OuYang W. W.-L., Lichterman J., Luo Z., Xuan X., Huang J., Chung L. W. K., Rettig M., Tseng H.-R., Shao C., Posadas E. M., NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods 64, 144–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Court C. M., Hou S., Winograd P., Segel N. H., Li Q. W., Zhu Y., Sadeghi S., Finn R. S., Ganapathy E., Song M., French S. W., Naini B. V., Sho S., Kaldas F. M., Busuttil R. W., Tomlinson J. S., Tseng H.-R., Agopian V. G., A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 24, 946–960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reátegui E., Aceto N., Lim E. J., Sullivan J. P., Jensen A. E., Zeinali M., Martel J. M., Aranyosi A. J., Li W., Castleberry S., Bardia A., Sequist L. V., Haber D. A., Maheswaran S., Hammond P. T., Toner M., Stott S. L., Tunable nanostructured coating for the capture and selective release of viable circulating tumor cells. Adv. Mater. 27, 1593–1599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Chen N., Li S., Battig M. R., Wang Y., Programmable hydrogels for controlled cell catch and release using hybridized aptamers and complementary sequences. J. Am. Chem. Soc. 134, 15716–15719 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Shen Q., Xu L., Zhao L., Wu D., Fan Y., Zhou Y., OuYang W.-H., Xu X., Zhang Z., Song M., Lee T., Garcia M. A., Xiong B., Hou S., Tseng H.-R., Fang X., Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv. Mater. 25, 2368–2373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L., Lu Y.-T., Li F., Wu K., Hou S., Yu J., Shen Q., Wu D., Song M., OuYang W.-H., Luo Z., Lee T., Fang X., Shao C., Xu X., Garcia M. A., Chung L. W. K., Rettig M., Tseng H.-R., Posadas E. M., High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv. Mater. 25, 2897–2902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reck M., Rabe K. F., Precision diagnosis and treatment for advanced non–small-cell lung cancer. N. Engl. J. Med. 377, 849–861 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Katayama R., Shaw A. T., Khan T. M., Mino-Kenudson M., Solomon B. J., Halmos B., Jessop N. A., Wain J. C., Yeo A. T., Benes C., Drew L., Saeh J. C., Crosby K., Sequist L. V., Iafrate A. J., Engelman J. A., Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci. Transl. Med. 4, 120ra17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gainor J. F., Tseng D., Yoda S., Dagogo-Jack I., Friboulet L., Lin J. J., Hubbeling H. G., Dardaei L., Farago A. F., Schultz K. R., Ferris L. A., Piotrowska Z., Hardwick J., Huang D., Mino-Kenudson M., Iafrate A. J., Hata A. N., Yeap B. Y., Shaw A. T., Patterns of metastatic spread and mechanisms of resistance to crizotinib in ROS1-positive non–small-cell lung cancer. JCO Precis. Oncol. 2017, 10.1200/PO.17.00063, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiley C. T., le Quesne J., Santis G., Sharpe R., de Castro D. G., Middleton G., Swanton C., Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet 388, 1002–1011 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Sheng W., Ogunwobi O. O., Chen T., Zhang J., George T. J., Liu C., Fan Z. H., Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 14, 89–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaav9186/DC1
Fig. S1. High-resolution XPS characterization.
Fig. S2. Deconvolution of x-ray photoelectron spectra of stepwise functionalized SiNWS.
Fig. S3. Fluorescence imaging for exploring bioorthogonal ligation.
Fig. S4. Dynamic changes of Cy5 fluorescence intensity upon disulfide cleavage by DTT.
Fig. S5. Scheme of CTC capture on Tz-grafted magnetic beads.
Fig. S6. Scheme of RT-ddPCR analysis of ALK/ROS1 rearrangements in CTCs.
Fig. S7. Dynamic ranges for RT-ddPCR quantification of ALK/ROS1 rearrangements.
Fig. S8. Comparison of cell lysis on-chip with cell lysis of DTT-released cells.
Table S1. CTC purity during capture and release processes.


