Zic family proteins regulate GLUT1 gene expression in a β-catenin/Tcf7l2–dependent manner in colorectal cancer cells.
Abstract
The zinc finger of the cerebellum (ZIC) proteins has been implicated to function in normal tissue development. Recent studies have described the critical functions of Zic proteins in cancers and the potential tumor-suppressive functions in colon cancer development and progression. To elucidate the functional roles of Zic proteins in colorectal cancer, we knocked out the Zic5 gene and analyzed the chromatin localization pattern and transcriptional regulation of target gene expression. We found that Zic5 regulates glucose metabolism, and Zic5 knockout is accompanied by an increased glycolytic state and tolerance to a low-glucose condition. Furthermore, loss of β-catenin or TCF7l2 diminishes the chromatin binding of Zic5 globally. Our studies suggest that the Wnt/β-catenin signaling pathway has a strong influence on the function of Zic proteins and glucose metabolism in colorectal cancers through GLUT1. Interfering Wnt/-catenin–Zic5 axis–regulated aerobic glycolysis represents a potentially effective strategy to selectively target colon cancer cells.
INTRODUCTION
The zinc finger of the cerebellum (ZIC) protein family members contain Cys2His2 zinc finger (C2H2-ZF) domains with strong sequence conservation, and they act as transcription activators or repressors by directly binding to DNA (1). The human Zic gene family is composed of five members Zic1 to Zic5, and they play pleiotropic roles in developmental stages and diseases, including cell specification, stimulation of cell proliferation, and delay of cell differentiation (1–3). Although their roles have been extensively studied during development and pathogenesis of neuronal disorders when the genes are mutated or deleted, there is a lack of knowledge of how they may mechanistically influence cancer pathogenesis.
A recent study revealed that Zic1 and Zic2 participate in the polycomb complex PRC1-controlled self-renewal of intestinal stem cells (4). Zic1 and Zic2 inhibit the canonical Wnt/β-catenin signaling in SW480 colon cancer cells (4). Zic gene expression is suppressed by PRC1 activity in the intestinal stem cells; therefore, Wnt/β-catenin signaling is sustained under normal and pathological conditions (4). This study links the potential tumor-suppressive functions of Zic proteins in colon cancer development and progression. However, previous studies have presented contradictory roles of Zic proteins in Wnt/β-catenin signaling: In one study, Zic2 is a repressor (5), while Zic1 is an activator (6) of Wnt/β-catenin signaling; conversely, in another study, Wnt/β-catenin signaling could also regulate Zic gene expression (7, 8). Whether or not different members of Zic protein family have differential roles in colon cancer and what the direct target genes are still remain unknown.
To define the roles of Zic proteins in cancer, we used the HCT116 colon cancer cell line that only expresses Zic2 and Zic5 transcripts [as detected by RNA sequencing (RNA-seq)]. Specifically, we focused on Zic5 because the function and the regulation of this protein are underexplored, especially at the genomics level (9–11). We found that genes up-regulated in Zic5 knockout (KO) cells correlated with the increased glycolytic state. Both glucose transporter 1 (GLUT1/SLC2A1) and hexokinase 2 (HK II), the crucial players involved in glucose uptake and metabolism, were up-regulated when Zic5 was depleted, which resulted in the metabolic switch toward aerobic glycolysis and enabled the cells to tolerate low glucose availability. Moreover, the Wnt/β-catenin pathway may play an essential role on glucose metabolism through the Zic proteins in colorectal cancer cells. Our results provide a mechanistic rationale for exploring the therapeutic against Wnt/β-catenin–Zic axis for colorectal cancers with Zic5 expression loss or Zic2 expression gain.
RESULTS
Repression of gene expression by Zic5 correlates with the increased glycolytic state
A potential prognostic value of Zic5 was found in colon cancers because high ZIC5 transcript levels (1552938_at) were correlated with better overall survival using the PrognoScan database–based Kaplan-Meier analysis of the overall survival of 177 and 55 patients with colorectal cancer in two independent cohorts (fig. S1, A and B) (12, 13). We further explored PRECOG (PREdiction of Clinical Outcomes from Genomic Profiles) database and also found similar results (fig. S1, C and D) (14, 15). In other types of cancer such as breast cancer and ovarian cancer, high levels of Zic5 expression are also correlated with better survival outcome on the basis of the Kaplan-Meier plotter analysis (fig. S1, E and F) (16, 17). To define the detailed functions of Zic proteins in colon cancer, we used the HCT116 cell line that only expresses detectable ZIC2 and ZIC5 transcripts by RNA-seq. We first used CRISPR-Cas9 technology to knock out the ZIC5 gene in HCT116 cells (Fig. 1, A and B, clones C16 and C18) and examined the morphological changes with F-actin staining (Fig. 1C). To identify the gene target for Zic5, we profiled the gene expression changes in Zic5 KO cells compared to Zic5 wild-type (WT) cells with adjusted P < 0.01 (Fig. 1D). We identified 246 down-regulated and 475 up-regulated genes in Zic5 KO clones, suggesting the general transcriptional repression function of Zic5. This is consistent with the previous studies suggesting that Zic1 and Zic2 potentially inhibit β-catenin transcriptional activity and Zic family proteins act as transcriptional repressors (4). The gene ontology (GO) analysis of the up-regulated genes with Zic5 depletion demonstrated enrichment in the cellular metabolic process. When we correlated the gene expression profile in Zic5 WT and KO cells with the genes induced by (i) genotoxic stress upon 5-fluorouracil treatment (12 hours), (ii) glucose deprivation (24 hours), or (iii) hypoxia (1% O2, 24 hours) (18), we found that the genes whose expression is up-regulated in Zic5 KO cells were significantly correlated with the glucose deprivation state, indicating elevated glycolysis (Fig. 1E). This triggered us to further investigate the role of Zic5 in the glucose metabolism pathway in colon cancers.
Fig. 1. Genes up-regulated in Zic5 KO cells correlate with the increased glycolytic state.
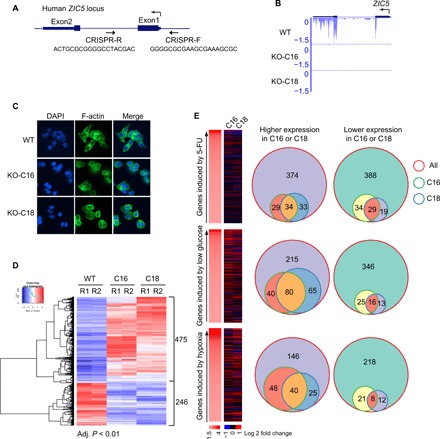
(A) Generation of Zic5 KO HCT116 cells by CRISPR-Cas9. (B) RNA-seq track to show the complete KO of Zic5 in C16 and C18 clones. (C) Immunofluorescence showing the cellular morphology of Zic5 WT and KO HCT116 cells by F-actin staining. (D) Hierarchical clustering analysis and corresponding heat map of differentially expressed genes in HCT116 Zic5 WT and KO cells. The color scale bar indicates the fold change ranges for the comparisons (ratios) indicated on the top of the heat map. (E) Effects of Zic5 KO on gene expression induced by three different conditions. 5-FU, 5-fluorouracil; DAPI, 4′,6-diamidino-2-phenylindole.
Depletion of Zic5 renders cells more resistant to low glucose–induced apoptosis
We noted that when HCT116 cells were subject to glucose withdrawal, apoptosis occurred within 48 hours. Therefore, we challenged the Zic5 WT and KO cells to high (25 mM), normal (5.5 mM), or low (0.5 mM) glucose–containing media for 2 days and monitored cell viability. Zic5 KO cells were more resistant to low glucose–induced apoptosis compared to the WT counterpart, indicated by the lower levels of cleaved poly(adenosine 5′-diphosphate–ribose) polymerase (PARP) when the glucose level dropped (Fig. 2A). Additional clones were also selected and confirmed with a reduction in apoptosis compared to the WT cells when challenged to low glucose–containing media. Further, we treated the cells with 2-deoxy-d-glucose (2-DG), a glycolysis inhibitor that cannot undergo further glycolysis, mimicking the low-glucose condition. Similarly, Zic5 KO cells were relatively more resistant to 2-DG treatment, as shown by the cleaved PARP and cleaved caspase3 levels (Fig. 2B). Conversely, cells were more prone to low glucose–induced apoptosis when Zic5 was ectopically expressed (Fig. 2, C and D). To exclude the effect of clonal selection and assess the requirement of DNA binding ability coupled with glucose sensitivity, full length or N terminus of Zic5 (Zic5-FL or Zic5-N, respectively) lacking the DNA binding domain at the C terminus of the protein was restored in the Zic5 KO cells, and the cell death was monitored when cells were cultured in low-glucose media or with 2-DG treatment. Zic5-FL, but not Zic5, lacking DNA binding domain could rescue the sensitivity to low-glucose or 2-DG treatment, excluding the clonal effect and suggesting the requirement of DNA binding (Fig. 2, E and F). We also expressed either Zic5-FL or Zic5-N in WT HCT116 cells and found that actually only Zic5-FL could enhance the apoptosis but not Zic5-N, suggesting that the DNA binding domain of Zic5 was required to induce apoptosis (Fig. 2G).
Fig. 2. Depletion of Zic5 renders the cells more resistant to low glucose–induced apoptosis.
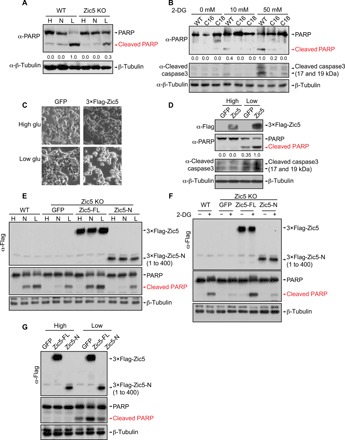
(A) Western blot showing the cleaved PARP levels of Zic5 WT and KO cells cultured in the Dulbecco’s modified Eagle’s medium (DMEM) medium with high (H, 25 mM), normal (N, 5.5 mM), or low glucose (L, 0.5 mM). (B) Western blot showing the cleaved PARP and cleaved caspase3 levels in Zic5 WT and KO cells treated with an increasing concentration of 2-DG. (C) Bright-field cellular morphology and (D) Western blot showing the cleaved PARP and cleaved caspase3 levels of HCT116 cells expressing GFP or 3×Flag-Zic5 cultured in the DMEM medium with high or low glucose. (E) Western Blot showing the cleaved PARP levels of Zic5 WT and KO cells and KO cells restored with Zic5-FL or Zic5-N cultured in the DMEM medium with high (H, 25 mM), normal (N, 5.5 mM), or low glucose (L, 0.5 mM). (F) Western blot showing the cleaved PARP levels in Zic5 WT and KO cells, and KO cells restored with Zic5-FL or Zic5-N treated with or without 50 mM 2-DG. (G) Western blot showing the cleaved PARP levels of HCT116 cells expressing green fluorescent protein (GFP), 3×Flag-Zic5-FL, or 3×-Flag-Zic5-N cultured in the DMEM medium with high or low glucose. Zic5-FL and Zic5-N were detected by antibody against Flag. Cleaved PARP signals were quantified with ImageJ.
Zic5 acts as a transcriptional repressor for the glycolysis-related genes
To understand the role of Zic5 function on the regulation of the glucose metabolism, we compared global genome-wide binding locations using Zic5 WT and KO cells. To analyze the chromatin localization pattern for the gene targets of Zic5, we performed chromatin immunoprecipitation sequencing (ChIP-seq) using our homemade Zic5 antibodies (fig. S2). We identified 7040 peaks specific to Zic5 (fig. S2B). Forty-five percent of the binding regions occupied the transcription start site (TSS), whereas the rest of the peaks localized in the non-TSS (fig. S3A). The GO analysis for Zic5-occupied loci showed the enrichment of biological processes in various metabolic pathways and glycolysis regulation (fig. S3B), which echoed the RNA-seq results in glucose metabolism–related gene expression regulated by Zic5 (Fig. 1, D and E). We also performed the Homer de novo motif analysis and found that the sequence of the Zic5 binding motif (CCCCCTGCTG[ATG]G) was highly overlapped with the Zic2 motif reported in previous studies (fig. S3C) (19). To further analyze Zic5-specific peaks in Homer de novo motif analysis, we searched the best matches of the top two motifs of Zic5 (fig. S3, D and E). Motif 1 (P = 1e-2646) and Motif 2 (P = 1e-310) are both significant among all motifs from Homer results. As expected, the best matches of motif 1 are enriched in the Zic family members (fig. S3D). When we ranked the overall Zic5 binding intensity, the heat map revealed a strong correlation with polymerase II (Pol II) and H3K4me3 occupancies (Fig. 3A). About 75% of the Zic5-bound TSS regions were also co-bound by RNA Pol II and H3K4me3, as shown in the Venn diagrams (Fig. 3B) and the average plot (Fig. 3C). Using K-means clustering to partition genes on the basis of the differential expression in the Zic5 WT and KO cells, our analysis revealed cluster 3 as a distinct set of genes that are derepressed with increased Pol II occupancy when Zic5 was deleted (Fig. 3D). Cluster 3 was significantly enriched for the regulation of various cellular metabolic processes (Fig. 3E), further highlighting the role of Zic5 in controlling the cellular metabolic pathways.
Fig. 3. Zic5 acts as a transcriptional repressor of glycolysis-related genes.
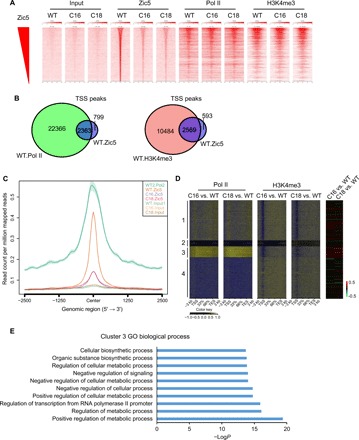
(A) Binding intensity heat maps showing the genomic occupancy distribution of Zic5 and RNA Pol II and H3K4me3 modifications from 2-kb downstream to 2-kb upstream from the Zic5 peak summits (base pair showing the highest Zic5 read pileup) of Zic5-occupied genes in Zic5 WT and KO cells. The depicted regions were ranked by decreasing Zic5 binding strength, as measured by the normalized fold enrichment of reads under Zic5 peaks to the respective input regions. (B) Venn diagram showing the overlap between RNA Pol II or H3K4me3 and Zic5 occupancy at the TSS regions in the WT HCT116 cells. (C) Average binding profiles of Zic5 and RNA Pol II around the TSS of Zic5-occupied genes in HCT116 cells. The profiles were calculated on the basis of average normalized coverage (reads per base pair). (D) The metagene analysis showing the Pol II and H3K4me3 binding peaks at the gene body and the gene expression change in the Zic5 KO cells. TES, transcription end site. (E) GO term analysis of cluster 3 in the metagene analysis.
Loss of Zic5 derepresses GLUT1/SLC2A1 gene expression with an increase in GLUT1 protein levels and glucose uptake
Reprogramming energy metabolism is one of the major hallmarks of cancer cells, and they tend to use glucose for aerobic glycolysis (20, 21). This metabolic switch will further lead to oncogene activation and a hypoxic state within the tumors. First, we scanned through a series of genes involved in the glycolysis pathway and that are induced by a low-glucose condition and found that both GLUT1/SLC2A1 and HK II, the crucial players involved in glucose uptake and metabolism, were up-regulated when Zic5 was depleted. This resulted in the metabolic switch toward aerobic glycolysis and enabled the cells tolerant to low-glucose availability (Fig. 4A). Consistently, we found the GLUT1 gene promoter bound by Zic5 in WT cells and GFP-tagged Zic5 using GFP antibody, but not Zic5 KO cells (Fig. 4C and fig. S4), which suggested that the change in GLUT1/SLC2A1 gene expression was a direct effect (Fig. 4B). When Zic5 was depleted, Pol II occupancy was concomitantly increased in the GLUT1 gene, suggesting the repressive function of Zic5 on GLUT1 gene expression regulation (Fig. 4C). Overexpression of Zic5-FL but not Zic5-N lacking the DNA binding domain decreased GLUT1 gene expression (Fig. 4D). We also knocked out Zic5 in 293T cells, measured the mRNA levels of GLUT1, and found a consistent up-regulation among different clones (Fig. 4E). The gene expression regulation in HCT116 colon cancer cells and 293T cells is consistent, and further study in other cell types is warranted to see whether this is a general phenomenon. The mRNA level increase of the GLUT1 gene also correlated with the protein level increase measured by the GLUT1 protein staining with flow cytometry (Fig. 4F), GLUT1 protein levels in Zic5 WT and KO cells with Western blot (Fig. 4G and fig. S4B), and increase in glucose uptake measured by 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (2-NBDG) uptake into the cells (Fig. 4H). We also depleted GLUT1 in Zic5 WT and KO cells and performed the colony formation assay to check the dependence of GLUT1 when Zic5 was lost (Fig. 4I). Knockdown of GLUT1 in each cell line was verified by Western blot (fig. S4C), and it markedly decreased the cell proliferation and colony formation in both WT and Zic5 KO cells, with a more profound effect in Zic5 KO cells, suggesting greater reliance of the GLUT1 when Zic5 is lost in the cells (Fig. 4I). We believe that Zic5 may act as a transcription factor or co-factor that directly regulates GLUT1 gene expression and glucose metabolism.
Fig. 4. Loss of Zic5 derepresses GLUT1/SLC2A1 gene expression.
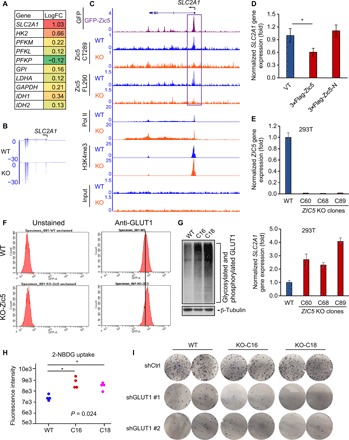
(A) Glycolysis-related gene expression change in Zic5 WT and KO cells are shown in the color-coded format. The ratio indicates the log fold change (logFC). (B) Loss of Zic5 increased GLUT1/SLC2A1 mRNA level as shown by RNA-seq. (C) Zic5 binding at the promoter of GLUT1/SLC2A1 gene is lost in Zic5 KO clones. (D) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) showing that overexpression of Zic5 but not Zic5-N decreases GLUT1/SLC2A1 mRNA level. (E) qRT-PCR showing that the KO of Zic5 in 293T cells increases the GLUT1/SLC2A1 mRNA level. (F) GLUT1 protein levels in Zic5 WT and KO HCT116 cells are measured by flow cytometry. (G) Western blot showing GLUT1 expression levels in Zic5 WT and KO cells. (H) Glucose uptake in Zic5 WT and KO HCT116 cells is measured by the amount of 2-NBDG taken up by the cells. The fluorescent intensity is quantified by the plate reader (n = 4). The pairwise Wilcoxon rank sum tests were used to calculate the significance of C16 and C18 cells glucose uptake compared with WT cells. *P < 0.05. (I) Colony formation assay showing the effect of GLUT1 depletion in Zic5 WT and KO cells.
β-Catenin/TCF7l2 complex as a central hub for Zic5 recruitment and transcriptional regulation
To gain more insight into the molecular mechanism of how Zic5 controls transcriptional regulation of target genes, we sought to identify Zic5-interacting proteins in HCT116 cells. We generated a stable HCT116 line expressing either the Zic2 or Zic5 protein with 3×Flag tag. 3×Flag-Zic2 or 3×Flag-Zic5 and GFP vector control purifications from nuclear extracts were analyzed by Multidimensional Protein Identification Technology (MudPIT), which resulted in the identification of the β-catenin/Tcf7l2 complex, the crucial effectors of the Wnt signaling pathway (Fig. 5A and table S1) (22). The co-purification of these two factors was also validated by Western blotting when cells were transfected with 3×Flag-Zic2 or Zic5 and immunoprecipitated against Flag to observe the interaction with the endogenous Tcf7l2 or β-catenin in vivo. Consistent with our mass spectrometry (MS) data, Zic5 could pull down both Tcf7l2 and β-catenin, although β-catenin pulldown was weaker compared with Tcf7l2 (Fig. 5B); however, with Zic2, we could only detect Tcf7l2, but not β-catenin, likely because of the sensitivity of the antibody (Fig. 5C). We were also able to detect the endogenous interaction between Zic2 and Tcf7l2 when Zic2 was immunoprecipitated (fig. S5A). To further validate the physical interaction between Zic5/Zic2 and Tcf7l2/β-catenin complex, we performed in vitro binding assay using purified recombinant His-tag TCF7l2 and His-tag β-catenin and in vitro–translated Flag-Zic2 and Flag-Zic5. The use of His-tag pulldown was not feasible because Zic2 protein has intrinsic His-tag in its protein sequence. For that reason, we used M2 beads to pull down Flag-Zic2 or Flag-Zic5 and checked the binding with the recombinant TCF7l2 or β-catenin proteins. As revealed by the binding assay, His-tag Tcf7l2 has a direct physical interaction with both Zic2 and Zic5, while His-tag β-catenin does not in the in vitro binding assay (Fig. 5D). Zic5 has zinc finger domain at the C terminus of the protein (485 to 575 amino acids). Therefore, for domain-mapping experiments, we transfected 293T cells with FL (1 to 664 amino acids), N terminus (1 to 400 amino acids), and C terminus (401 to 664 amino acids) of Zic5 and immunoprecipitated against flag to see the interaction with Tcf7l2/β-catenin complex in vivo (Fig. 5E). Both Zic5-FL and Zic5-C interacted with Tcf7l2/β-catenin complex in vivo, but not Zic5-N, suggesting that the region containing the DNA binding domain may be required for the direct interaction with Tcf7l2 (Fig. 5E).
Fig. 5. β-Catenin/Tcf7l2 complex recruits Zic5 to regulate gene expression.
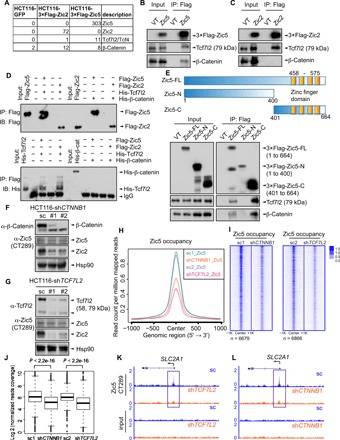
(A) MudPIT analysis of the purification of 3×Flag-Zic5 and 3×Flag-Zic2 in HCT116 cells from nuclear extracts. (B and C) Validation of Tcf7l2 and β-catenin co-immunoprecipitation with Zic5 or Zic2 by Western blot. VT, vector control. (D) Tcf7l2 but not β-catenin directly binds to Zic5 and Zic2 in the in vitro binding assay. IB, immunoblotting. (E) C terminus of Zic5 is required to be directly associated with the β-catenin/Tcf7l2 complex. (F and G) CTNNB1 or TCF7L2 knockdown in HCT116 cells by distinct short hairpin RNAs (shRNAs). The protein levels of β-catenin, Tcf7l2, Zic2, and Zic5 were examined by Western Blot. (H) Average plot and (I) heat maps showing the global binding of Zic5 in shControl cells and shCTNNB1 or shTCF7L2 cells from −1 to +1 kb centered on Zic5 peaks in shControl cells. (J) Boxplot depicts the normalized reads coverage for Zic5 peaks in shControl cells and shCTNNB1 or shTCF7L2 cells. The P values were calculated with the Wilcoxon signed-rank test. (K and L) Track examples showing the Zic5 binding at the GLUT1 gene promoter region with Tcf7l2 or β-catenin knockdown. IgG, immunoglobulin G.
We also found transcription factor (TCF) binding motif ranking as top 3 with a score of 0.57 in the best matches of Zic5 motif 2 (fig. S3, C and E). The interaction between Zic5 and the β-catenin/Tcf7l2 complex and the possible co-localization of Zic5 and TCF on the chromatin predicted by Homer de novo motif analysis triggered us to further investigate the chromatin binding and recruitment process for each of these factors. First, β-catenin or Tcf7l2 was depleted by short hairpin RNAs (shRNAs) in WT HCT116 cells, and Zic5 protein levels were not altered by β-catenin or Tcf7l2 knockdown (Fig. 5, F and G). Tcf7l2/β-catenin complex knockdown led to the significant down-regulation of Zic2, indicating a positive feedback loop in the Zic2-Tcf7l2/β-catenin signaling axis (Fig. 5, F and G). We found a marked decrease of Zic5 binding in the cells depleted with Tcf7l2 or β-catenin genome wide (Fig. 5, H to J) and at the promoter regions of GLUT1/SLC2A1 as well (Fig. 5, K and L), indicating that the regulation of GLUT1 gene expression by Zic5 was mediated by the β-catenin/Tcf7l2 complex and the Wnt signaling pathway. Reciprocally, Tcf7l2 binding was affected to a much lesser extent when Zic5 was depleted (fig. S5B), suggesting that Zic5 was recruited by the β-catenin/Tcf7l2 complex to regulate the metabolic gene expression, especially at the GLUT1 gene locus.
We also analyzed the Zic5 binding using cluster 3 genes from Fig. 3D that are derepressed with increased Pol II occupancy when Zic5 was deleted and found the consistent decrease in Zic5 occupancy at the TSS regions of these genes when β-catenin or Tcf7l2 was knocked down (fig. S5, C and D), indicating the β-catenin/Tcf7l2–dependent regulation of metabolic gene transcriptional control by Zic5.
Because both Zic2 and Zic5 were co-purified with β-catenin/Tcf7l2 complex (Fig. 5, A to C), it was intriguing to investigate whether Zic2 and/or Zic5 has competitive or cooperative functions in regulating glucose metabolism. First, we further knocked down Zic2 in the Zic5 KO cells (Fig. 6A), determined GLUT1/SLC2A1 gene expression changes, and found a consistent decrease with three distinct shRNAs against Zic2 (Fig. 6B). Endogenous Zic2 levels are sensitive to acute glucose withdrawal, and an additional depletion of Zic2 restored cellular sensitivity to glucose deprivation, as demonstrated by PARP cleavage (Fig. 6C). Glucose uptake was also decreased in the Zic2-depleted cells, partially rescued by the effect of Zic5 KO (Fig. 6D). About 58 genes that were activated by Zic5 KO were rescued in Zic2 knockdown cells, including the SLC2A1 gene, suggesting the reciprocal function of Zic2 and Zic5 in regulating glucose metabolism (Fig. 6E). Actually, 418 genes were found to be down-regulated and only 84 genes were up-regulated in Zic2 knockdown cells, indicating the transcriptional activation function of Zic2 in HCT116 cells, further supporting the antagonistic roles of Zic2 (transcriptional activator) and Zic5 (transcriptional repressor). To support our findings with gene expression regulation by Zic2 and Zic5, we generated Zic2 KO cells by CRISPR-Cas9 (fig. 6A) and performed ChIP-seq analysis of Zic2 in WT and KO cells. The depletion of Zic2 was verified by Western blot and RNA-seq (fig. S6, B and C). We further annotated Zic2-specific peaks by subtracting the peaks found in KO cells from WT cells (fig. S6D). Motif analysis showed that Zic2 binding motif 1 was highly similar compared with Zic5 motif 1 (figs. S6, E and F, and S3C), suggesting the potential agonistic or antagonistic effects between Zic2 and Zic5. About 50% of Zic2 and Zic5 peaks overlapped with each other, as shown in the Venn diagram (fig. S6G), and Zic2 was also found to bind at the promoter region of GLUT1 (fig. S6H). Last, the genome-wide competitive binding of Zic5/Zic2 on the overlap peaks of Zic2 and Zic5 was shown in fig. S6I. The average ratio of Zic5 to Zic2 was significantly higher than 1, suggesting the competitive binding of Zic5 and Zic2. The reciprocal ChIP-seq of Zic2 and Zic5 in WT, Zic5 KO, or Zic2 KO cells showed that the ratio of Zic2 to Zic5 increases at the GLUT1 promoter region and genome wide in Zic5 KO cells and that the ratio of Zic5 to Zic2 increases in Zic2 KO cells, further indicating the antagonistic effects of Zic2 and Zic5 in regulating gene expression. Figure 6F summarizes our studies in HCT116 WT and Zic5 KO cells: During the normal state, GLUT1 gene expression was maintained by the balanced controls between Zic2 and Zic5 recruitment by β-catenin/Tcf7l2 and normal glucose uptake. When cells were depleted of Zic5, GLUT1 gene expression activated abnormally and glucose uptake increased, leading to the increased glycolytic state.
Fig. 6. Zic5 regulates glucose metabolism in a β-catenin/Tcf7l2–dependent manner in HCT116 colon cancer cells.
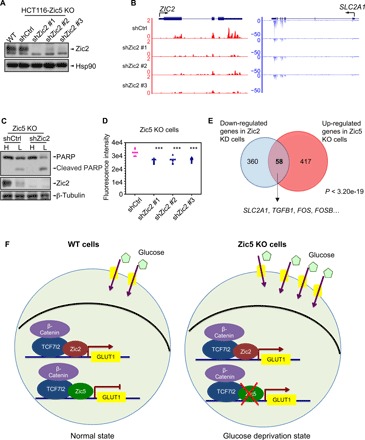
(A) Zic5 KO cells (C18) were infected with three independent shRNAs against Zic2. Western blot showed the knockdown efficiency of Zic2 knockdown after puromycin selection. HCT116 parental cells were included as positive control. (B) RNA-seq tracks showed the GLUT1/SLC2A1 gene expression decrease with Zic2 knockdown. (C) Western blot showing the cleaved PARP levels of Zic2 knockdown in Zic5 KO cells cultured in the DMEM medium with high (H, 25 mM) or low glucose (L, 0.5 mM). (D) Glucose uptake in Zic5 KO cells with shCtrl of shZic2 is measured by 2-NBDG uptake. The fluorescence intensity is quantified by the plate reader (n = 8). ***P < 0.001. (E) Venn diagram analysis identifies 58 common genes by Zic2 knockdown and Zic5 KO. The P value was determined with the hypergeometric test. (F) Zic5 acts as a transcriptional repressor in regulating SLC2A1/GLUT1 gene expression. Loss of Zic5 derepresses GLUT1 gene expression and further increases the protein level of GLUT1. Glucose uptake is increased and thus leads to the more glycolytic features of the cells. This may link to the tumor progression in the tumor microenvironment.
DISCUSSION
Although the Zic family of proteins have a highly conserved C2H2-ZF domain toward the C-terminal region, N-terminal and C-terminal coding regions outside of the zinc finger domains are unique for each family member (1). This raises the possibility of a division of labor among the five family members of this class of transcription factors in regulating gene expression. Zic5 is the largest Zic protein in this family and with the most divergent sequence similarity compared with Zic1 to Zic4. In the current study, we particularly explored the function of Zic5 in colorectal cancer cells and characterized the transcriptional regulation of Zic5 on GLUT1, the glucose transporter 1, the most critical gene involved in glucose uptake and glycolysis in human cancers. Together, our results revealed that Zic5 negatively regulates glucose metabolism in HCT116 colon cancer cells and uncover a previously unidentified link between Zic protein–regulated transcription and the metabolic signaling pathways in colon cancer cells.
Studies of the Zic family proteins in human cancers have been investigated in the previous reports mainly focusing on Zic1 and Zic2. Zic1 has been shown to exert an oncogenic function in medulloblastoma (23), meningioma (24), and liposarcoma (25) and tumor-suppressive roles in gastric cancer (26, 27), colorectal cancer (28), and thyroid cancer (29). In contrast, studies on Zic2 revealed its oncogenic functions in pancreatic cancer (30), epithelial ovarian cancer (31), oral squamous cell carcinoma (32), cervical cancer (33), and hepatocellular carcinoma (34). However, the role of Zic5 is underexplored, probably because of its low expression and restricted expression pattern in tumors (9–11). Unexpectedly, we noted that the function of Zic2 was not dispensable, but rather, it may antagonize the function of Zic5 in glucose metabolism (Fig. 6, A to D and fig. S6, F to I). Our laboratory has previously demonstrated that Zic2 is a transcriptional repressor in regulating the embryonic stem cell specification (35). Our results also revealed Zic5 as a transcriptional repressor in regulating glucose metabolism in colon cancer cells. It remains unknown whether human Zic proteins function similarly or has distinct binding partners on chromatin to regulate transcription in a cell type–dependent manner or cell type–independent manner. In addition, it will be important to examine the collaborative or competitive actions of different Zic proteins in regulating glucose metabolism in other types of tumor cells.
The Wnt signaling pathway has been shown to participate in the reprogramming of cancer cell metabolism (36), the activation of which enhances the glucose metabolism through glycolysis (37). We found that the effectors of Wnt signaling β-catenin and Tcf7l2 in collaboration with Zic proteins directly regulate GLUT1 gene expression and, consequently, glucose uptake and metabolism. Overexpression of GLUT1 has been considered as the prognostic indicator for cancer. The Zic2 and Zic5 protein levels enable the fine tuning of the homeostasis of glucose metabolism in normal and disease states by regulating GLUT1 gene expression. In Zic5 low-expressing cells, interfering with the glucose metabolism pathways remains promising for colon cancer treatment in the future. Because of the low glucose availability in the tumors, inactivating the genes induced by low glucose or restoring the genes repressed by a low-glucose condition warranted further investigation. Overall, we have identified a key node regulating GLUT1 gene expression by the Wnt/β-catenin and Zic2/Zic5 signaling pathway. Selective restoration of Zic5 function or elimination of Zic2 might serve as a new strategy for cancer therapy.
MATERIALS AND METHODS
Cell culture
DLD1 and RKO GLUT1+/+ and GLUT1−/− cells were provided by B. Vogelstein’s laboratory. All the other cell lines were purchased from the American Type Culture Collection (Rockville, MD). The 293T and HCT116 cell lines were maintained in Dulbecco’s modified Eagle’s medium (Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Gibco) at 37° with 5% CO2. For the low-glucose condition, cells were plated in high glucose–containing media (25 mM) for 24 hours before switching to low glucose–containing (0.5 mM) or normal glucose–containing (5.5 mM) media.
ZIC5 and ZIC2 KOs by CRISPR-Cas9–targeted genome editing
Cells were co-transfected with a pair of guide RNAs and pX330-U6-Chimeric_BB-CBh-hSpCas9 (38) and another vector with puromycin resistance using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) and selected with puromycin (Thermo Fisher Scientific) at 2 μg/ml for 3 days. Cells were seeded in 96-well plates and selected for single clones after 2 to 3 weeks. Genomic DNA was isolated using genomic lysis buffer [10 mM tris-HCl (pH 7.5), 10 mM EDTA, 10 mM NaCl, and 0.5% sarcosyl] at 60°C for 2 hours and precipitation buffer (150 mM NaCl in 100% EtOH) at room temperature for 30 min. Polymerase chain reaction screening was performed using the primers as follows and further confirmed by Western blot and RNA-seq: Zic5 WT, TGAATCACGTCACGGTGGAG (forward) and GGCCTGTGTGTGCTATCGAA (reverse); Zic5 KO, CGTCTACTGTCTATGCGCCC (forward) and TGAAGAGTGTGTCACGGTGG (reverse); Zic2 WT, CCACCAACAACGCTTGTGAA (forward) and TGAGGCGTACCCAGGTCAAT (reverse); and Zic2 KO, CCCAGCAGCCTGTGTGATTT (forward) and GAGACCTTCTGCCAAACGGG (reverse). pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from F. Zhang (Addgene plasmid no. 42230).
Virus packaging, infection, and stable cell line generation
Lentiviruses for knocking down β-catenin, Tcf7l2, or Zic2, were packaged as previously described (39). shRNA sequences are listed as follows: shCTNNB1 #1, ccggatctgtctgctctagtaataactcgagttattactagagcagacagattttttg (forward) and aattcaaaaaatctgtctgctctagtaataactcgagttattactagagcagacagat (reverse); shCTNNB1 #2, ccgggggagtggtttaggctatttgctcgagcaaatagcctaaaccactccctttttg (forward) and aattcaaaaagggagtggtttaggctatttgctcgagcaaatagcctaaaccactccc (reverse); shTCF7L2 #1 ccgggcgccaacgacgaactgatttctcgagaaatcagttcgtcgttggcgctttttg (forward) and aattcaaaaagcgccaacgacgaactgatttctcgagaaatcagttcgtcgttggcgc (reverse); shTCF7L2 #2, ccggccgaaagtttccgagacaaatctcgagatttgtctcggaaactttcggtttttg (forward) and aattcaaaaaccgaaagtttccgagacaaatctcgagatttgtctcggaaactttcgg (reverse); shZic2 #1, ccggcggaagcacatgaaggtccaactcgagttggaccttcatgtgcttccgtttttg (forward) and aattcaaaaacggaagcacatgaaggtccaactcgagttggaccttcatgtgcttccg (reverse); shZic2 #2, ccgggcaactgagcaatcccaagaactcgagttcttgggattgctcagttgctttttg (forward) and aattcaaaaagcaactgagcaatcccaagaactcgagttcttgggattgctcagttgc (reverse); and shZic2 #1, ccggccggagtctttgaagctgaaactcgagtttcagcttcaaagactccggtttttg (forward) and aattcaaaaaccggagtctttgaagctgaaactcgagtttcagcttcaaagactccgg (reverse). GLUT1 shRNAs were purchased from Sigma-Aldrich (TRCN0000043583 and TRCN0000043584). Cells were selected with puromycin (2 μg/ml) for 3 days before the Western blot and ChIP-seq experiments. Retroviruses for overexpressing 3×Flag-Zic5 were packaged, as previously described (39). Cells were selected with G418 (800 μg/ml) for 1 week before Western blot and MudPIT analysis.
Quantitative reverse transcription polymerase chain reaction
Quantitative reverse transcription polymerase chain reaction was performed as previously described (40). Primer sequences used for the reactions are listed as follows: Zic5, TCAAGATCCACAAGCGTACTC (forward) and AGCCTCGAATCTTGCAGTAG (reverse); SLC2A1/GLUT1, GGACAGGCTCAAAGAGGTTATG (forward) and AGGAGGTGGGTGGAGTTAAT (reverse).
F-actin staining
F-actin staining was performed as previously described (41). Alexa Fluor 488 phalloidin was purchased from Cell Signaling Technology (Danvers, MA). VECTASHIELD Antifade Mounting Medium with 4′,6-diamidino-2-phenylindole was purchased from Vector Laboratories (Burlingame, CA).
Two-dimensional colony formation assay
Cells were seeded at 500 cells per well in six-well plates, and culture medium was replaced every 4 days for 2 weeks. Cells were then fixed with 3.7% paraformaldehyde and stained with 0.05% crystal violet.
Immunoprecipitation
HCT116 cells were lysed in Triton X-100 lysis buffer [50 mM tris (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM dithiothreitol (DTT), protease inhibitors, and Benzonase]. After centrifugation at 13,000g for 10 min, the supernatants (1 mg of total protein) were collected and incubated with anti-Flag M2 affinity gel at 4°C for 2 hours with rotation. Samples were washed with lysis buffer four times and competed with 3×Flag peptides for 15 min with vigorous agitation. Proteins were resuspended in 5× SDS sample loading buffer, heated to 95°C for 5 min, and subjected to SDS–polyacrylamide gel electrophoresis (PAGE).
In vitro translation of Zci2 and Zic5 constructs and in vitro binding assay
In vitro binding assay was performed, as previously described (42). β-Catenin and Tcf7l2 were cloned into pET28a(+) vector and expressed in BL21(DE3) cells. His-tagged recombinant proteins were purified with Ni-NTA agarose resin (Qiagen) following the manufacturer’s instruction. In vitro translation of Flag-Zic2 or Flag-Zic5 was carried out using T7 Quick Coupled Translation/Transcription system (Promega) following the manufacture’s instruction. The FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2 agarose beads (Sigma-Aldrich), followed by incubation with bacterially expressed His-tag proteins in binding buffer (1× phosphate-buffered saline, 0.1% NP40, 0.5 mM DTT, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride) for 4 hours at 4°C. The beads were washed for four times using ice-cold wash buffer and eluted with excess 3×Flag peptides (APExBio). Samples were boiled with SDS loading buffer before loading for SDS-PAGE.
Western blot analysis
Western blot analysis was performed, as previously described (40). Anti-Flag antibody was purchased from Sigma-Aldrich (St. Louis, MO). Anti–β-tubulin antibody was purchased from The Developmental Studies Hybridoma Bank. Anti-PARP, anti-caspase3, anti-Tcf7l2, and anti–β-catenin antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-His antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-GLUT1 and anti-Zic2 antibodies were purchased from Abcam (Cambridge, MA).
Cell viability assay
Cell viability was measured by cell counting using the Vi-CELL Cell Counter (Beckman Coulter Life Sciences, Indianapolis, IN).
Flow cytometry
Cells were fixed, permeabilized, and immunostained with antibody against GLUT1 (Cell Signaling Technology) and subjected to flow cytometry analysis using the BD FACSAria Special Order Research Product (BD Biosciences, San Jose, CA).
2-NBDG glucose uptake assay
The 2-NBDG was purchased from Cayman Chemical (Ann Arbor, MI). Ten thousand cells were seeded in black 96-well flat plates. Media were replaced to glucose-free media the next day, and cells were cultured for 20 hours. The 2-NBDG (50 μg/ml) was added for 7 hours and immediately subjected to glucose uptake assay using excitation and emission wavelengths of 485 and 535 nm.
Next-generation sequencing data processing
RNA-seq and ChIP-seq samples were sequenced with the Illumina NextSeq technology, and output data were processed with the bcl2fastq software tool. Sequence quality was assessed using FastQC v0.11.2 (43), and quality trimming was done using the FASTX toolkit. RNA-seq and ChIP-seq reads were aligned to the hg19 genome using TopHat v2.0.9 (44) and Bowtie v0.12.9 (45), and only uniquely mapped reads with a two-mismatch threshold were considered for downstream analysis. Gene annotations from Ensembl 72 were used. Output BAM files were converted into bigwig track files to display coverage throughout the genome (in reads per million) using the GenomicRanges package (46) and other standard Bioconductor R packages.
RNA-seq analysis
Gene count tables were constructed using HTseq (47) with Ensembl gene annotations and used as input for edgeR 3.0.8 (48). Genes with Benjamini-Hochburg–adjusted P < 0.01 were considered to be differentially expressed. GO enrichment analysis was evaluated by the MetaScape online software suite (49).
ChIP-seq analysis
Cells (5 × 107) were used for each ChIP assay, as performed as previously described (50). Peaks were called with MACS v1.4.2 (51) using default parameters and were annotated by the HOMER software (52). Metaplots were generated using ngsplot (53). Bedtools was used to determine the raw counts at these merged peaks (54). Using in-house Perl scripts, raw counts at each peak were converted to reads per kilobase per million mapped reads values with total library counts, and log fold change values between conditions were then computed with these normalized values. Differential occupancy was evaluated by edgeR, and peaks with Benjamini-Hochburg–adjusted P < 0.05 were considered to be differentially occupied.
Mass spectrometry
We followed the protocol described previously (55). For Orbitrap Fusion Tribrid MS analysis, the tryptic peptides were purified with Pierce C18 spin columns (Thermo Fisher Scientific). Three micrograms of each fraction was autosampler loaded with a Thermo EASY nLC 1000 UPLC pump onto a vented Acclaim PepMap 100 (75 μm by 2 cm) nanoViper trap column coupled to a nanoViper analytical column (3 μm, 100 Å, C18, 0.075 mm, 500 mm; 164570, Thermo Fisher Scientific) with a stainless steel emitter tip assembled on the Nanospray Flex Ion Source with a spray voltage of 2000 V. Buffer A contained 94.785% H2O with 5% acetonitrile (ACN) and 0.125% formic acid (FA), and buffer B contained 99.875% ACN with 0.125% FA. The chromatographic run was 2 hours in total, with the following profiles: 0 to 7% for 3 min, 10% for 3 min, 25% for 80 min, 33% for 20 min, 50% for 3 min, 95% for 3 min, and again 95% for 8 min. Additional MS parameters include ion transfer tube temperature = 300°C, EASY-IC internal mass calibration, default charge state = 2, and cycle time = 3 s. Detector type was set to Orbitrap with a resolution of 60 K and a wide quad isolation [mass range = normal, scan range = 300 to 1500 mass/charge ratio (m/z), maximum injection time = 50 ms, automatic gain control (AGC) target = 200,000, microscans = 1, S-lens radio frequency level = 60, without source fragmentation, and data type = centroid]. Monoisotopic precursor selection was set as on, including charge states of 2 to 6 (reject unassigned). Dynamic exclusion was enabled with n = 1 for 30- and 45-s exclusion duration at 10 parts per million (ppm) for high and low (precursor selection decision = most intense, top 20, isolation window = 1.6, scan range = auto normal, first mass = 110, collision energy = 30%, collision-induced dissociation, detector type = ion trap, OT resolution = 30 K, IT scan rate = rapid, maximum injection time = 75 ms, AGC target = 10,000, Q = 0.25, and injected ions for all available parallelizable time).
Tandem mass spectra analysis
Spectrum raw files from samples were extracted into ms1 and ms2 files using in-house program RawXtractor or RawConverter (http://fields.scripps.edu/downloads.php) (56), and the tandem mass spectra were searched against UniProt human protein database (downloaded on 25 March 2014) (57) and matched to sequences using the ProLuCID/SEQUEST algorithm (ProLuCID version 3.1) (58, 59), with 50-ppm peptide mass tolerance for precursor ions and 600 ppm for fragment ions. The search space included all fully and half-tryptic peptide candidates that fell within the mass tolerance window with no miscleavage constraint, assembled and filtered with DTASelect2 (version 2.1.3) (60, 61) through Integrated Proteomics Pipeline (IP2 version 3, Integrated Proteomics Applications Inc., CA, USA; www.integratedproteomics.com). To accurately estimate peptide probabilities and false discovery rates (FDRs), we used a target/decoy database containing the reversed sequences of all the proteins appended to the target database (62). Each protein identified was required to have a minimum of one peptide of minimum length of six amino acid residues and to be within 10 ppm of the expected m/z. However, this peptide had to be an excellent match with an FDR less than 0.001 and needed to have at least one excellent peptide match. After the peptide/spectrum matches were filtered, we estimated that the protein FDRs were ≤1% for each sample analysis.
Supplementary Material
Acknowledgments
We thank C. Collings for initial data analysis, K. Cozzolino for initial MS analysis, and A. Whelan for technical assistance. Z.Z. was supported, in part, by the NIH/NCI training grant T32 CA070085 and Alex’s Lemonade Stand Foundation (ALSF) Young Investigator Award by Northwestern Mutual. L.W. was supported by the Training Program in Signal Transduction and Cancer (T32 CA070085-19). E.B. was supported by the Research Specialist Award R50-CA221848 from the National Cancer Institute. N.C. was supported by R35CA197532 (NSC). This study was supported, in part, by the Outstanding Investigator Award R35CA197569 from the National Cancer Institute to A.S. Author contributions: Z.Z. and A. Shilatifard designed the study. Z.Z., L.W., C.R., and A. Shilati performed most of the experiments and part of the analyses. Z.Z. wrote the first draft of the manuscript. E.R. and S.M. generated and sequenced the next-generation sequencing libraries. J.S. performed the MS experiments and analyzed the results. E.B. performed the bioinformatics analyses. Z.Z., N.C., and A. Shilatifard revised the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Next-generation sequencing datasets have been deposited at Gene Expression Omnibus with the accession number GSE127960. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaax0698/DC1
Fig. S1. Potential prognostic value of Zic5 in colon cancer.
Fig. S2. Generation of our homemade antibodies against Zic5 for ChIP-seq analysis.
Fig. S3. Comparative ChIP-seq analysis of ZIC5 in HCT116 ZIC5 WT and KO cells.
Fig. S4. Loss of Zic5 derepresses GLUT1/SCL2A1 gene expression.
Fig. S5. β-Catenin/Tcf7l2 complex recruits Zic5 to regulate gene expression.
Fig. S6. Comparative ChIP-seq analysis of ZIC2 in HCT116 ZIC2 WT and KO cells.
Table S1. Mass spectrometry data.
REFERENCES AND NOTES
- 1.Grinberg I., Millen K. J., The ZIC gene family in development and disease. Clin. Genet. 67, 290–296 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Merzdorf C. S., Emerging roles for zic genes in early development. Dev. Dyn. 236, 922–940 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Watabe Y., Baba Y., Nakauchi H., Mizota A., Watanabe S., The role of Zic family zinc finger transcription factors in the proliferation and differentiation of retinal progenitor cells. Biochem. Biophys. Res. Commun. 415, 42–47 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Chiacchiera F., Rossi A., Jammula S., Piunti A., Scelfo A., Ordóñez-Morán P., Huelsken J., Koseki H., Pasini D., Polycomb complex PRC1 preserves intestinal stem cell identity by sustaining Wnt/β-catenin transcriptional activity. Cell Stem Cell 18, 91–103 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Pourebrahim R., Houtmeyers R., Ghogomu S., Janssens S., Thelie A., Tran H. T., Langenberg T., Vleminckx K., Bellefroid E., Cassiman J.-J., Tejpar S., Transcription factor Zic2 inhibits Wnt/β-catenin protein signaling. J. Biol. Chem. 286, 37732–37740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merzdorf C. S., Sive H. L., The zic1 gene is an activator of Wnt signaling. Int. J. Dev. Biol. 50, 611–617 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Murgan S., Kari W., Rothbächer U., Iché-Torres M., Mélénec P., Hobert O., Bertrand V., Atypical transcriptional activation by TCF via a Zic transcription factor in C. elegans neuronal precursors. Dev. Cell 33, 737–745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nyholm M. K., Wu S.-F., Dorsky R. I., Grinblat Y., The zebrafish zic2a-zic5 gene pair acts downstream of canonical Wnt signaling to control cell proliferation in the developing tectum. Development 134, 735–746 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Satow R., Inagaki S., Kato C., Shimozawa M., Fukami K., Identification of zinc finger protein of the cerebellum 5 as a survival factor of prostate and colorectal cancer cells. Cancer Sci. 108, 2405–2412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satow R., Nakamura T., Kato C., Endo M., Tamura M., Batori R., Tomura S., Murayama Y., Fukami K., ZIC5 drives melanoma aggressiveness by PDGFD-mediated activation of FAK and STAT3. Cancer Res. 77, 366–377 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Sun Q., Shi R., Wang X., Li D., Wu H., Ren B., Overexpression of ZIC5 promotes proliferation in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 479, 502–509 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Mizuno H., Kitada K., Nakai K., Sarai A., PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2, 18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J. J., Deane N. G., Wu F., Merchant N. B., Zhang B., Jiang A., Lu P., Johnson J. C., Schmidt C., Bailey C. E., Eschrich S., Kis C., Levy S., Washington M. K., Heslin M. J., Coffey R. J., Yeatman T. J., Shyr Y., Beauchamp R. D., Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, 958–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentles A. J., Newman A. M., Liu C. L., Bratman S. V., Feng W., Kim D., Nair V. S., Xu Y., Khuong A., Hoang C. D., Diehn M., West R. B., Plevritis S. K., Alizadeh A. A., The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid J. F., Gariboldi M., Sokolova V., Capobianco P., Lampis A., Perrone F., Signoroni S., Costa A., Leo E., Pilotti S., Pierotti M. A., Integrative approach for prioritizing cancer genes in sporadic colon cancer. Genes Chromosomes Cancer 48, 953–962 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., Szallasi Z., An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Győrffy B., Lánczky A., Szállási Z., Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 19, 197–208 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Galbraith M. D., Allen M. A., Bensard C. L., Wang X., Schwinn M. K., Qin B., Long H. W., Daniels D. L., Hahn W. C., Dowell R. D., Espinosa J. M., HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Najafabadi H. S., Mnaimneh S., Schmitges F. W., Garton M., Lam K. N., Yang A., Albu M., Weirauch M. T., Radovani E., Kim P. M., Greenblatt J., Frey B. J., Hughes T. R., C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Annibaldi A., Widmann C., Glucose metabolism in cancer cells. Curr. Opin. Clin. Nutr. Metab. Care 13, 466–470 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Brantjes H., Barker N., van Es J., Clevers H., TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 383, 255–261 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Yokota N., Aruga J., Takai S., Yamada K., Hamazaki M., Iwase T., Sugimura H., Mikoshiba K., Predominant expression of human Zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 56, 377–383 (1996). [PubMed] [Google Scholar]
- 24.Aruga J., Nozaki Y., Hatayama M., Odaka Y. S., Yokota N., Expression of ZIC family genes in meningiomas and other brain tumors. BMC Cancer 10, 79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brill E., Gobble R., Angeles C., Lagos-Quintana M., Crago A., Laxa B., DeCarolis P., Zhang L., Antonescu C., Socci N. D., Taylor B. S., Sander C., Koff A., Singer S., ZIC1 overexpression is oncogenic in liposarcoma. Cancer Res. 70, 6891–6901 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong J., Chen S., Xue M., Du Q., Cai J., Jin H., Si J., Wang L., ZIC1 modulates cell-cycle distributions and cell migration through regulation of sonic hedgehog, PI3K and MAPK signaling pathways in gastric cancer. BMC Cancer 12, 290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L. J., Jin H. C., Wang X., Lam E. K. Y., Zhang J. B., Liu X., Chan F. K. L., Si J. M., Sung J. J. Y., ZIC1 is downregulated through promoter hypermethylation in gastric cancer. Biochem. Biophys. Res. Commun. 379, 959–963 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Gan L. H., Chen S., Zhong J., Wang X., Lam E. K. Y., Liu X., Zhang J., Zhou T., Yu J., Si J., Wang L., Jin H., ZIC1 is downregulated through promoter hypermethylation, and functions as a tumor suppressor gene in colorectal cancer. PLOS ONE 6, e16916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang W., Zhao Y., Yang Q., Liu W., Guan H., Lv S., Ji M., Shi B., Hou P., ZIC1 is a putative tumor suppressor in thyroid cancer by modulating major signaling pathways and transcription factor FOXO3a. J. Clin. Endocrinol. Metab. 99, E1163–E1172 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Inaguma S., Ito H., Riku M., Ikeda H., Kasai K., Addiction of pancreatic cancer cells to zinc-finger transcription factor ZIC2. Oncotarget 6, 28257–28268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini S., Poynor E., Barakat R. R., Clivio L., Cinquini M., Fruscio R., Porcu L., Bussani C., D'Incalci M., Erba E., Romano M., Cattoretti G., Katsaros D., Koff A., Luzzatto L., The zinc finger gene ZIC2 has features of an oncogene and its overexpression correlates strongly with the clinical course of epithelial ovarian cancer. Clin. Cancer Res. 18, 4313–4324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuma K., Kasamatsu A., Yamatoji M., Yamano Y., Fushimi K., Iyoda M., Ogoshi K., Shinozuka K., Ogawara K., Shiiba M., Tanzawa H., Uzawa K., Expression status of Zic family member 2 as a prognostic marker for oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 136, 553–559 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Chan D. W., Liu V. W. S., Leung L. Y., Yao K. M., Chan K. K. L., Cheung A. N. Y., Ngan H. Y. S., Zic2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J. Pathol. 225, 525–534 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Zhu P., Wang Y., He L., Huang G., Du Y., Zhang G., Yan X., Xia P., Ye B., Wang S., Hao L., Wu J., Fan Z., ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J. Clin. Invest. 125, 3795–3808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo Z., Gao X., Lin C., Smith E. R., Marshall S. A., Swanson S. K., Florens L., Washburn M. P., Shilatifard A., Zic2 is an enhancer-binding factor required for embryonic stem cell specification. Mol. Cell 57, 685–694 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherwood V., WNT signaling: An emerging mediator of cancer cell metabolism? Mol. Cell. Biol. 35, 2–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cisternas P., Salazar P., Silva-Álvarez C., Barros L. F., Inestrosa N. C., Activation of Wnt signaling in cortical neurons enhances glucose utilization through glycolysis. J. Biol. Chem. 291, 25950–25964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cong L., Zhang F., Genome engineering using CRISPR-Cas9 system. Methods Mol. Biol. 1239, 197–217 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z., Wang L., Xu W., IL-13Rα2 mediates PNR-induced migration and metastasis in ERα-negative breast cancer. Oncogene 34, 1596–1607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Z., Wang L., Wen Z., Ayaz-guner S., Wang Y., Ahlquist P., Xu W., Systematic analyses of the cytotoxic effects of compound 11a, a putative synthetic agonist of photoreceptor-specific nuclear receptor (PNR), in cancer cell lines. PLOS ONE 8, e75198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Collings C. K., Zhao Z., Cozzolino K. A., Ma Q., Liang K., Marshall S. A., Sze C. C., Hashizume R., Savas J. N., Shilatifard A., A cytoplasmic COMPASS is necessary for cell survival and triple-negative breast cancer pathogenesis by regulating metabolism. Genes Dev. 31, 2056–2066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Zhao Z., Meyer M. B., Saha S., Yu M., Guo A., Wisinski K. B., Huang W., Cai W., Pike J. W., Yuan M., Ahlquist P., Xu W., CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.S. Andrew, FastQC: A quality control tool for high throughput sequence data (2010); https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 44.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T., Carey V. J., Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tripathi S., Pohl M. O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D. A., Moulton H. M., DeJesus P., Che J., Mulder L. C. F., Yángüez E., Andenmatten D., Pache L., Manicassamy B., Albrecht R. A., Gonzalez M. G., Nguyen Q., Brass A., Elledge S., White M., Shapira S., Hacohen N., Karlas A., Meyer T. F., Shales M., Gatorano A., Johnson J. R., Jang G., Johnson T., Verschueren E., Sanders D., Krogan N., Shaw M., König R., Stertz S., García-Sastre A., Chanda S. K., Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen F. X., Woodfin A. R., Gardini A., Rickels R. A., Marshall S. A., Smith E. R., Shiekhattar R., Shilatifard A., PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 162, 1003–1015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen L., Shao N., Liu X., Nestler E., ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hickox A. E., Wong A. C. Y., Pak K., Strojny C., Ramirez M., Yates J. R. III, Ryan A. F., Savas J. N., Global analysis of protein expression of inner ear hair cells. J. Neurosci. 37, 1320–1339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He L., Diedrich J., Chu Y.-Y., Yates J. R. III, Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal. Chem. 87, 11361–11367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.UniProt Consortium , UniProt: A hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu T., Park S. K., Venable J. D., Wohlschlegel J. A., Diedrich J. K., Cociorva D., Lu B., Liao L., Hewel J., Han X., Wong C. C. L., Fonslow B., Delahunty C., Gao Y., Shah H., Yates J. R. III, ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eng J. K., McCormack A. L., Yates J. R. III, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Tabb D. L., McDonald W. H., Yates J. R. III, DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cociorva D., Tabb D. L., Yates J. R., Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.14 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Elias J. E., Gygi S. P., Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaax0698/DC1
Fig. S1. Potential prognostic value of Zic5 in colon cancer.
Fig. S2. Generation of our homemade antibodies against Zic5 for ChIP-seq analysis.
Fig. S3. Comparative ChIP-seq analysis of ZIC5 in HCT116 ZIC5 WT and KO cells.
Fig. S4. Loss of Zic5 derepresses GLUT1/SCL2A1 gene expression.
Fig. S5. β-Catenin/Tcf7l2 complex recruits Zic5 to regulate gene expression.
Fig. S6. Comparative ChIP-seq analysis of ZIC2 in HCT116 ZIC2 WT and KO cells.
Table S1. Mass spectrometry data.


