ABSTRACT
Background
Little is known about the contribution of genetic variation to food timing, and breakfast has been determined to exhibit the most heritable meal timing. As breakfast timing and skipping are not routinely measured in large cohort studies, alternative approaches include analyses of correlated traits.
Objectives
The aim of this study was to elucidate breakfast skipping genetic variants through a proxy-phenotype genome-wide association study (GWAS) for breakfast cereal skipping, a commonly assessed correlated trait.
Methods
We leveraged the statistical power of the UK Biobank (n = 193,860) to identify genetic variants related to breakfast cereal skipping as a proxy-phenotype for breakfast skipping and applied several in silico approaches to investigate mechanistic functions and links to traits/diseases. Next, we attempted validation of our approach in smaller breakfast skipping GWAS from the TwinUK (n = 2,006) and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium (n = 11,963).
Results
In the UK Biobank, we identified 6 independent GWAS variants, including those implicated for caffeine (ARID3B/CYP1A1), carbohydrate metabolism (FGF21), schizophrenia (ZNF804A), and encoding enzymes important for N6-methyladenosine RNA transmethylation (METTL4, YWHAB, and YTHDF3), which regulates the pace of the circadian clock. Expression of identified genes was enriched in the cerebellum. Genome-wide correlation analyses indicated positive correlations with anthropometric traits. Through Mendelian randomization (MR), we observed causal links between genetically determined breakfast skipping and higher body mass index, more depressive symptoms, and smoking. In bidirectional MR, we demonstrated a causal link between being an evening person and skipping breakfast, but not vice versa. We observed association of our signals in an independent breakfast skipping GWAS in another British cohort (P = 0.032), TwinUK, but not in a meta-analysis of non-British cohorts from the CHARGE consortium (P = 0.095).
Conclusions
Our proxy-phenotype GWAS identified 6 genetic variants for breakfast skipping, linking clock regulation with food timing and suggesting a possible beneficial role of regular breakfast intake as part of a healthy lifestyle.
Keywords: food timing, breakfast, GWAS, chronobiology, circadian clock
Introduction
The timing of food intake is a modifiable risk factor for weight management and chronic disease prevention (1, 2). Although food timing choices are influenced by physiologic, behavioral, and environmental factors, little is known about the contribution of genetic variation (3). The estimated heritability of food timing has been investigated in twin studies and ranges from 18% to 56%, with breakfast timing being the most heritable among meals (3, 4). Shared heritability has also been observed between food timing and chronotype (4) (i.e., morning or evening preference), offering preliminary insight into the genetics of food timing. Elucidating variants and genes may disclose underlying regulatory biological processes and inform the development of personalized nutrition recommendations based on genetic preference for food timing.
The discovery of genetic variants regulating food timing is hindered by the lack of its routine assessment in population-based cohort studies together with limited integrated genetic data. Breakfast skipping, more commonly assessed in some larger epidemiologic cohorts, is shown to be correlated with the timing of other meals (5–7), such as an earlier lunch among breakfast skippers (8); therefore, genetic findings from breakfast skipping may extend to food timing in general. Breakfast skipping is itself also a heritable trait with an estimated heritability of 41–66% (9), and has also been linked to higher risk of type 2 diabetes (10–12), subclinical atherosclerosis (13), and detrimental cardiometabolic health independent of dietary quality (14). In addition, morning anorexia, a clinical feature of 2 eating disorders, night eating syndrome and sleep-related eating syndrome (15), results in skipped breakfast, and thus skipping breakfast is often considered a subclinical eating disorder (16). Therefore, unraveling the genetic architecture of breakfast skipping may further have clinical implications and reveal genetic instruments to assess causal links with cardiometabolic traits (17, 18).
An earlier genome-wide association study (GWAS) for breakfast skipping in the TwinUK study found no significant association signals; however, the null findings in that study may be due to incomplete genetic coverage and a modest sample size (n = 2,006) (19). Breakfast cereal is commonly measured in dietary assessment tools, and in the United Kingdom, breakfast cereal is the most commonly consumed breakfast food (>50% of the population) (20, 21) and is primarily consumed in the mornings (22). To leverage the statistical power of the UK Biobank (23) [n = 193,860; almost 100-fold that of the prior study (19)] to identify genetic variants related to breakfast skipping, our primary outcome for this research, we conducted a proxy-phenotype GWAS (24) based on the consumption of breakfast cereal, a correlated trait, as has been conducted for other traits measured in limited datasets (24–26). We applied an array of in silico approaches to investigate mechanistic functions and links to other traits and diseases. Next, we sought replication of our findings in the published TwinUK breakfast skipping GWAS (19), and a breakfast skipping GWAS meta-analysis in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium (n = 11,963).
Methods
Study populations
The present GWAS included 193,860 participants of European ancestry with genetic information and breakfast cereal–skipping data from 24-h recalls (27) from the UK Biobank, a prospective population-based cohort study (23). In addition, replication was conducted in 2,006 female participants from the previously published TwinUK breakfast skipping GWAS (19), and 11,963 participants of European ancestry with genetic information and breakfast-skipping data from 5 adult epidemiologic population-based cohorts from the CHARGE consortium nutrition working group: the Bogalusa Heart Study (BHS), the Coronary Artery Risk Development in Young Adults (CARDIA) Study, the Cardiovascular Health Study (CHS), The Netherlands Epidemiology of Obesity (NEO) Study, and the Women's Health Initiative (WHI) (Supplemental Table 1). Analysis was limited to participants of European ancestry to maximize statistical power and limit heterogeneity that may obscure signal detection. All participants provided written informed consent, and each cohort's study protocol was reviewed and approved by their respective institutional review board.
UK Biobank: assessment of breakfast skipping, genotyping, and GWAS
In the UK Biobank, dietary data were collected from 211,036 participants through the use of the Oxford WebQ, a web-based 24-h diet recall that asks participants to self-report on the frequency of intake of ∼200 commonly consumed foods and drinks in the preceding 24 h (28, 29). Breakfast cereal skipping was estimated with the use of data from up to 5 web-based 24-h diet recalls. Participants were asked, “Did you eat any breakfast cereal yesterday? This could be at any time of the day. Please include hot cereals, but not cereal bars”, with responses “Yes” or “No”. Breakfast cereals encompassed a range of hot and cold cereal including the following: bran cereal, muesli, oat crunch, other cereal, plain cereal, porridge, sweetened cereal, and whole-wheat cereal. Responses from all completed recalls (≤5) were considered for each participant (n = 193,860). Responses were categorized as “breakfast skipping” if the participant always responded “No”, “sometimes breakfast skipping” if the participant sometimes responded “Yes”, and “breakfast consumers” if the participant always responded “Yes”.
In the UK Biobank, genotyping was performed centrally by the biobank, and genotyping, quality control, and imputation procedures have been described in detail previously (23). In brief, blood, saliva, and urine were collected from participants, and DNA was extracted from the samples. Participant DNA was genotyped on 2 arrays, UK BiLEVE and UK Biobank Axiom, with >95% common content, and genotypes for ∼800,000 autosomal single nucleotide polymorphisms (SNPs) were imputed to 2 reference panels. Genotypes were called with the use of the Affymetrix Power Tools software. Detailed information on sample and SNP quality control, population structure by principal component analysis, and imputation have been described previously (23, 30, 31).
GWAS was performed for breakfast skipping in related participants of European ancestry (n = 193,860) through the use of the BOLT-LMM (32) linear mixed models (3-category breakfast skipping variable treated continuously) and an additive genetic model adjusted for age, sex, 10 principal components of ancestry, genotyping array, and accounting for relatedness with the use of a genetic correlation matrix with a maximum per-SNP missingness of 10% and per-sample missingness of 40%. We used a genome-wide significance threshold of 5 × 10−8. We used a SNP imputation quality threshold of 0.80, and a minimum minor allele frequency (MAF) threshold of 0.001. Trait heritability was calculated as the proportion of trait variance due to additive genetic factors measured in this study through the use of BOLT-REML (32), to leverage the power of raw genotype data together with low-frequency variants (MAF ≥0.001). In the UK Biobank, we had >80% power to detect an allele with frequency 0.01 and effect size of β >0.01 for breakfast skipping at the genome-wide significance level (Quanto version 1.2.4).
A polygenic risk score (PRS) for breakfast skipping was tested for association with 10 other commonly consumed noncereal breakfast items through the use of a weighted PRS calculated by summing the products of the breakfast-skipping risk allele count for all 6 SNPs multiplied by the scaled effect from the primary UK Biobank GWAS; this calculation was performed with the GTX package in R (33). PRS associations were tested for the following 10 noncereal breakfast items: bread (UK Biobank Field Identifier (FID):1438), butter/spreadable butter (FID:1428_1), coffee (FID:1498), eggs (FID:102,930), fruits (FID:1309), milk added (FID:100,890), orange juice (FID:100,190), processed meat (FID:1349), tea intake (FID:1488), yogurt/ice-cream (FID:102,080). All GWAS summary statistics were retrieved from publicly available sources (34) and significance was considered at the Bonferroni-corrected threshold [P < 0.005 (= 0.05/10)].
Gene and pathway enrichment analyses, genetic correlation analyses, and Mendelian randomization
Pathway analysis was conducted with the use of MAGMA (35) gene-set analysis in FUMA (36), which uses the full distribution of SNP P values and is performed for curated gene sets and GO terms obtained from MsigDB (total of 10,891 pathways). A significance threshold was set after Bonferroni correction accounting for all pathways tested (P < 0.05/10,891). Tissue enrichment analysis was also conducted with FUMA (36) for 53 tissue types from the Genotype-Tissue Expression (GTEx) project, and a significance threshold was set following Bonferroni correction accounting for all tested tissues (P < 0.05/53).
Genome-wide genetic correlation analysis of linkage disequilibrium score regression (37–39) with LDHub was conducted for the breakfast skipping GWAS and publicly available data from 224 published non-UK Biobank GWAS and other lifestyle UK Biobank GWASs for chronotype (40), sleep duration (30), and macronutrient intake. Linkage disequilibrium score regression estimates the genetic correlation between 2 traits from summary statistics (ranging from −1 to 1) based on the fact that the GWAS effect-size estimate for each SNP incorporates effects of all SNPs in linkage disequilibrium with that SNP, and a similar relation is observed when single-study test statistics are replaced with the product of z scores from 2 studies of traits with some correlation. Furthermore, genetic correlation is possible between case-control studies and quantitative traits, as well as within these trait types. A Bonferroni-corrected significance threshold was selected accounting for all tests performed [P < 0.00022 (=0.05/227 tests)].
Mendelian randomization (MR) analysis was carried out with MR-Base (41), with the inverse variance weighted (IVW) approach serving as our main analysis method (42), and MR-Egger (43) and weighted median estimation (44) as sensitivity analyses. MR results may be biased by horizontal pleiotropy—i.e., where the genetic variants that are robustly related to the exposure of interest (here sleep duration) independently influence levels of a causal risk factor for the outcome. IVW assumes that there is either no horizontal pleiotropy, or that, across all SNPs, horizontal pleiotropy is 1) uncorrelated with SNP-risk factor associations and 2) has an average value of zero. MR-Egger assumes the first of these conditions but relaxes the second by explicitly estimating the nonzero mean pleiotropy and adjusting the causal estimate accordingly. Estimation of the pleiotropy parameter means that the MR-Egger estimate is generally far less precise than the IVW estimate. The weighted median approach is valid if <50% of the weight is pleiotropic (i.e., no single SNP that contributes 50% of the weight or a number of SNPs that together contribute 50% should be invalid because of horizontal pleiotropy). Given these different assumptions, if all 3 methods are broadly consistent, this strengthens our causal inference. For our 2- and 1-sample MR analyses, we used all identified signals from the UK Biobank GWAS and looked for the per-allele difference in odds (binary outcomes) or means (continuous) with correlated outcomes from publicly available summary data in the MR-Base platform or UK Biobank published summary statistics [for chronotype (40) and insomnia (31)]. Results are a likelihood measure of “more breakfast skipping”; sample 1 is our UK Biobank GWAS and sample 2 is a different GWAS consortia (or UK Biobank in 1-sample analysis) investigating the outcomes we explored. The exact number of SNPs used in the MR analysis depends on the SNPs available in the outcome GWAS. We estimate we have >95% power with 193,860 participants in the UK Biobank to detect causal links between breakfast skipping and outcomes (https://sb452.shinyapps.io/power/).
To further interrogate observed correlations between breakfast skipping and chronotype reported earlier (4), we examined causality of chronotype on breakfast skipping in bidirectional MR. For all 351 GWAS hits identified for chronotype (40), we assessed the per-allele difference in means with the breakfast skipping GWAS from the UK Biobank. Sample 1 is the chronotype GWAS (40) (23&Me participants only) and sample 2 is our UK Biobank GWAS. Results are therefore a measure of “more evening chronotype.”
TwinUK: in silico replication
Based on reported summary statistics from the previously published TwinUK breakfast skipping GWAS (19), we tested for independent replication of the UK Biobank breakfast skipping GWAS signals. In the published TwinUK study, breakfast skipping was assessed by the following question: “How often do you eat breakfast (e.g., bread, toast, milk) at the start of the day?” and responses (every morning, 5–6 d/wk, 3–4 d/wk, 1–2 d/wk and <1 d/wk) were dichotomized into breakfast skippers (skip breakfast ≥1 time/wk), or breakfast consumers (consume breakfast every day). A total of 283,744 directly typed SNPs were tested for association with breakfast skipping in 2,006 female participants (403 skippers/1,603 consumers). In that published study, all analyses were adjusted for family relatedness, but not age. Analysis details can be found in Supplemental Material from that publication (19). Furthermore, a PRS for breakfast skipping, as previously described, was tested for replication in the TwinUK with the use of all available SNPs of the 6 UK Biobank GWAS signals in the TwinUK. If the lead SNP from the UK Biobank signal was unavailable, due to absence of imputation in the TwinUK study, an appropriate proxy SNP in high linkage disequilibrium in the 1KGP3 reference panel (r2 > 0.8) was used instead.
CHARGE consortium: assessment of breakfast skipping, genotyping, GWAS, and meta-analysis
In cohorts from the CHARGE consortium, breakfast skipping was assessed in each cohort through the use of a diet or lifestyle questionnaire designed to capture the habits of the population investigated. Typically, participants were asked to indicate how often, on average, they had consumed breakfast according to multiple frequency categories (e.g., never, 2–3 times/wk, 6–7 times/wk) (see Supplemental Table 2 for cohort-specific questions). Breakfast intake frequency was dichotomized into breakfast skippers and breakfast consumers for the purpose of attaining homogeneity across cohorts. Because most participants had breakfast every day, those who reported skipping breakfast most of the days of the week (i.e., skipped breakfast ≥4 times/wk) were considered breakfast skippers, as has been conducted previously (19), except for the Dutch study, NEO, which had the lowest prevalence of breakfast skipping, where breakfast skipped ≥2 times/wk was considered breakfast skipping (details in Supplemental Table 2).
In the CHARGE consortium, genome-wide genotyping was conducted in each cohort through the use of Affymetrix or Illumina platforms. Each study performed quality control for genotyped SNPs based on MAF, call-rate, and departure from Hardy-Weinberg equilibrium (Supplemental Table 3). Phased haplotypes from 1000 G were used to impute ∼38 million autosomal variants with the use of a hidden Markov model algorithm implemented in BEAGLE (45), MACH/minimac (46, 47), or SHAPEIT/IMPUTE (48, 49). Study-specific GWAS was conducted with the use of genotyped and imputed SNP dosages assuming an additive genetic model. Breakfast skipping was evaluated as the dependent variable through the use of logistic regression, adjusted for age, sex, study-specific centers, or population stratification principal components, where applicable. Variants with low MAC (<20) and low imputation quality (BEAGLE and MACH: r2 < 0.3; or IMPUTE: proper info <0.4) were removed. Quality control for cohort-level GWAS results was performed to ensure correct specification of the minor allele and agreement in frequencies with the reference population, and examination of QQ plots assessed any large inflation of test statistics.
Results across studies were combined through the use of fixed-effect meta-analysis with inverse variance weights with the use of METAL software (University of Michigan, Center for Statistical Genetics) (50). The final GWAS meta-analysis included 5 adult cohorts (n = 1,487 skippers; n = 10,476 consumers). To account for population stratification, the association results from individual studies were adjusted for genomic control before meta-analysis with METAL by estimating the inflation of the test statistic by comparing the median test statistic to that expected by chance, and then applying genomic control corrections to the standard error. Heterogeneity across studies was tested with Cochran's Q statistic and quantified through the use of the heterogeneity statistic, I2, and presented as percentages. Genome-wide significance was considered at P < 5 × 10−8. Summary statistics are available in dbGaP. In the meta-analysis, we had 80% power to detect an allele with frequency 0.15 and effect of OR = 1.4 for breakfast skipping at the genome-wide significance level (Quanto version 1.2.4).
Similar to the TwinUK study PRS analysis described above, a PRS for breakfast skipping based on the UK Biobank lead signals was further tested for replication in the CHARGE consortium with the use of GWAS meta-analyzed results and the GTX package in R (33). In sensitivity subgroup meta-analysis, meta-analysis was restricted to only younger cohorts (average cohort age <50 y) with a higher prevalence of breakfast skipping (i.e., BHS and CARDIA; n = 1989; % skippers = 28.5%) or older cohorts (average cohort age >50 y) of similar demographic to the UK Biobank (i.e., CHS, NEO, and WHI; n = 9974; % skippers = 9.2%), and PRS association analyses were repeated.
Results
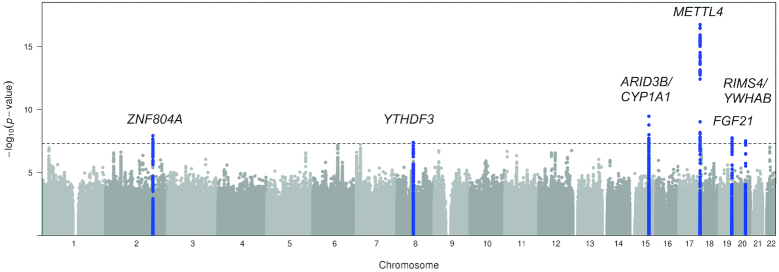
Figure 1 represents a schematic of the study design and the main findings. Among UK Biobank participants of European ancestry with 24-h recall data on breakfast cereal (n = 193,860), 25%, 20%, and 55% were classified as always, sometimes, and never breakfast skippers (Supplemental Table 4). Participants classified as always skippers were, on average, younger, had a higher BMI, were more likely to be evening chronotype, had shorter sleep duration, had lower total energy intake, and lower relative intakes of carbohydrate and protein, but higher intake of fat, as compared with sometimes or never skippers (Supplemental Table 4). The UK Biobank GWAS for breakfast skipping identified 6 independent genome-wide significant (P < 5 × 10−08) loci (Figures 2A, and 3, Table 1), and genome-wide SNP-based heritability was estimated at 7.0% (SE = 0.2%). Of the 6 genetic variants, the ARID3B/CYP1A1 locus has previously been implicated in caffeine metabolite concentrations (51), with the higher caffeine metabolite A allele associated with higher breakfast skipping. Genes nearest to the remaining identified variants have been associated with carbohydrate metabolism (FGF21) (52), smoking (METTL4) (53), and schizophrenia (ZNF804A) (54). A combined weighted PRS of the 6 breakfast skipping signals further associated with other commonly consumed noncereal breakfast items, including bread, coffee, fruits, milk added, processed meats, and tea (all P < 0.005; Figure 4). Pathway analysis of these genes with the use of MAGMA (35) did not identify enrichment of specific biological pathways reaching the multiple-testing threshold of significance; however, tissue enrichment analysis of gene expression from GTEx tissues identified enrichment of associated genes in the cerebellum (P = 5.43 × 10−5; Supplemental Figure 1).
FIGURE 1.
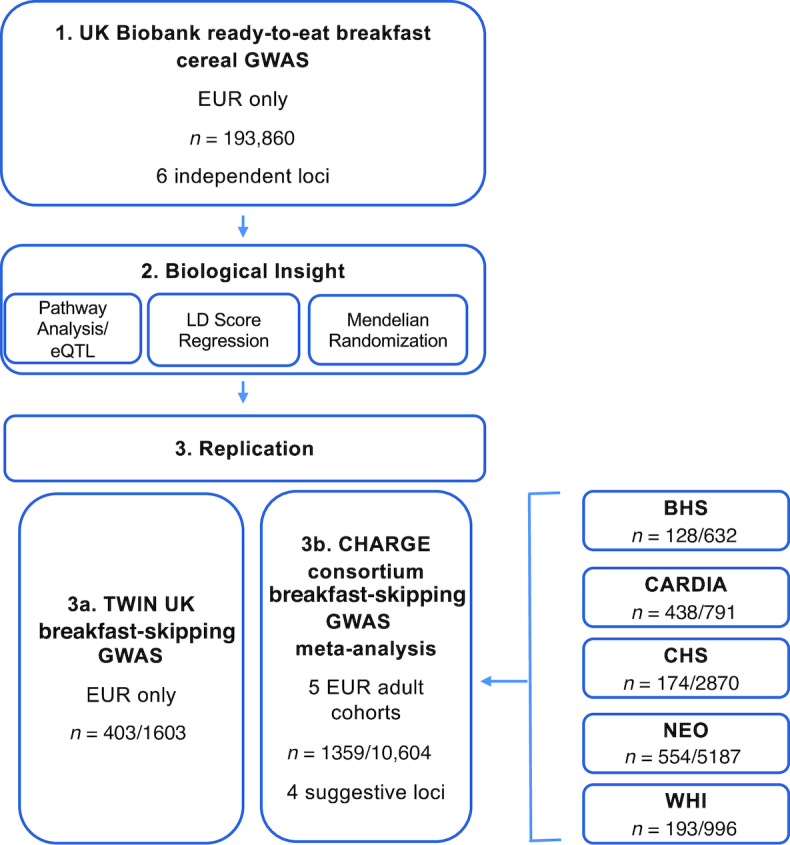
Schematic of the study design and main findings. Sample size (n) is total n or n skippers/n consumers. BHS, Bogalusa Heart Study; CARDIA, Coronary Artery Risk Development in Young Adults; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; CHS, Cardiovascular Health Study; EUR, European; GWAS, genome-wide association study; LD, linkage disequilibrium; eQTL, expression quantitative trait locus; NEO, The Netherlands Epidemiology of Obesity; WHI, Women's Health Initiative.
FIGURE 2.
Manhattan plot for GWAS of breakfast skipping in participants of European ancestry from the UK Biobank (n = 193,860). Novel loci are highlighted in blue and nearest gene(s) are annotated. Manhattan plot shows the –log10P values (y-axis) for all genotyped and imputed SNPs passing quality control, plotted by chromosome (x-axis). Horizontal dashed line denotes genome-wide significance (5 × 10−8). GWAS, genome-wide association study; SNP, single nucleotide polymorphism.
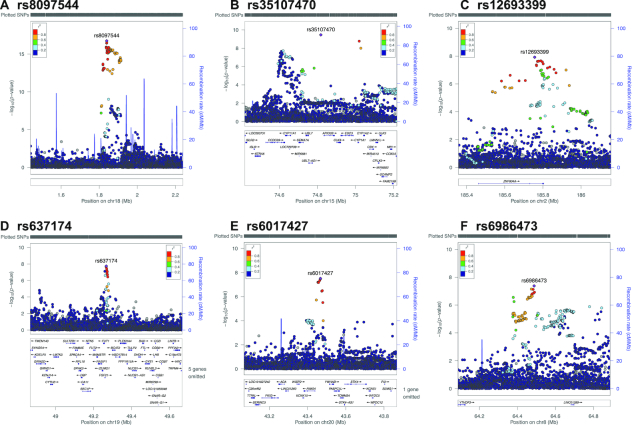
FIGURE 3.
(A–F) Regional association plots for lead signals from the GWAS of breakfast skipping in the UK Biobank (n = 193,860). The panels show –log10P values for lead SNPs. The SNPs shown are those within 400 kb of the lead SNP. LD is indicated in color scale in relation to the highlighted marker (purple). The scheme is red for strong LD (r2 ≥ 0.8), orange, green and blue for lower LD, and navy blue for no LD. chr, chromosome; cM, centimorgan; GWAS, genome-wide association study; LD, linkage disequilibrium; Mb, mega base pair; SNP, single-nucleotide polymorphism.
TABLE 1.
Top signals (P < 5 × 10−8) from the UK Biobank GWAS of breakfast skipping in participants of European ancestry (n = 193,860)1
| SNP | Chr:position | Nearest gene(s) | Alleles (E/A) | EAF | Info | β | SE | P value |
|---|---|---|---|---|---|---|---|---|
| rs8097544 | 18:1,839,564 | METTL4 | G/A | 0.146 | 0.98 | 0.0326 | 0.0038 | 1.80 × 10–17 |
| rs35107470 | 15:74,817,689 | ARID3B | G/A | 0.324 | 0.94 | 0.0185 | 0.0030 | 3.50 × 10–10 |
| rs12693399 | 2:185,757,011 | ZNF804A | A/T | 0.218 | 1.00 | 0.0186 | 0.0033 | 1.20 × 10–8 |
| rs637174 | 19:49,266,936 | FGF21 | A/G | 0.341 | 0.98 | 0.0161 | 0.0029 | 1.80 × 10–8 |
| rs6017427 | 20:43,467,380 | RIMS4/YWHAB | A/G | 0.142 | 0.99 | 0.0214 | 0.0039 | 3.10 × 10–8 |
| rs6986473 | 8:64,487,672 | YTHDF3 | T/C | 0.773 | 1.00 | 0.0175 | 0.0032 | 4.20 × 10–8 |
The GWAS was performed in related participants of European ancestry with the use of BOLT-LMM linear mixed models and an additive genetic model adjusted for age, sex, 10 principal components of ancestry, genotyping array and genetic correlation matrix. Nearest genes are within the locus of interest. β (SE) estimates are per each additional effect allele. Positive β reflects higher breakfast skipping. Chr, chromosome; E/A, effect/alternative alleles; EAF, effect allele frequency; GWAS, genome-wide association study; Info, imputation quality score; position, base pair coordinate hg19; SNP, single nucleotide polymorphism.
FIGURE 4.
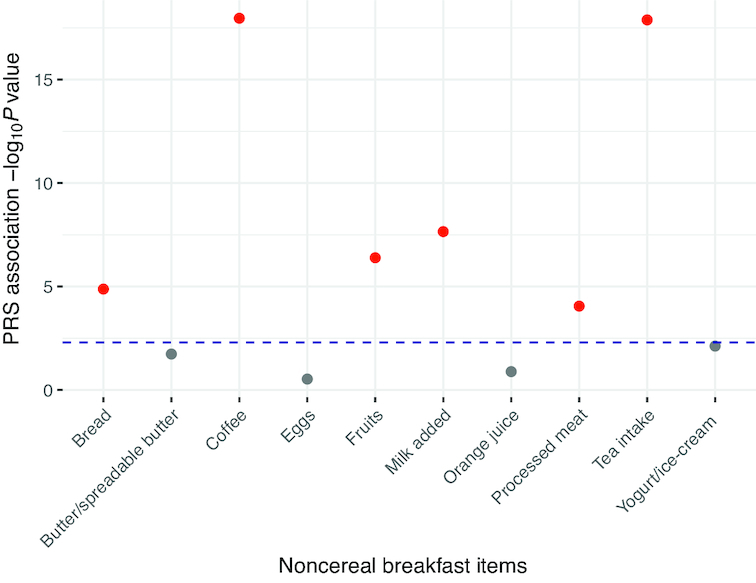
Breakfast skipping polygenic risk score association with other commonly consumed noncereal breakfast items in the UK Biobank. Weighted PRS for breakfast skipping comprised 6 breakfast-skipping signals from the UK Biobank tested for association with 10 other commonly consumed noncereal breakfast items with the use of the GTX package in R (33). All GWAS summary statistics were retrieved from publicly available resources (34) and significance was considered at the Bonferroni-corrected threshold [P < 0.005 (= 0.05/10)], represented by the horizontal dashed blue line. The plot shows the –log10P values (y-axis) for the PRS association with the 10 noncereal breakfast items. Red denotes significant association and grey denotes nonsignificant association. GWAS, genome-wide association study; PRS, polygenic risk score.
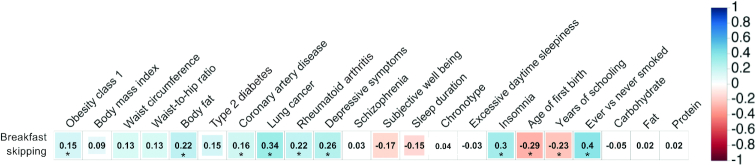
Genome-wide genetic correlations indicated shared biological links between breakfast skipping and anthropometric, psychiatric, and physical disease traits (P < 2.14 × 10−4; Figure 5, Supplemental Table 5). Positive genome-wide correlations with breakfast skipping were observed for obesity, body fat, coronary artery disease, lung cancer, depressive symptoms, insomnia, and smoking, and negative genome-wide correlations were observed for age of first birth and years of schooling. In addition, from other UK Biobank GWASs, we observed a trend towards genome-wide genetic correlation between breakfast skipping and evening chronotype [rg (SE) = −0.040 (0.020); P = 0.046] but no correlations with carbohydrate, fat, or protein intake (all P > 0.01). To assess for causal links between breakfast skipping and related traits, in 2-sample MR analyses, we observed causal associations between genetically defined breakfast skipping and higher BMI [IVW: 0.304 (0.153); P = 4.62 × 10−2], more depressive symptoms [IVW: 0.289 (0.146); P = 4.86 × 10−2], and smoking [IVW: 0.974 (0.311); P = 1.75 × 10−3], all with consistent effect direction in sensitivity analyses across other MR methods (Table 2). To further interrogate previously observed correlations between chronotype and breakfast skipping, we conducted bidirectional MR and observed that genetically defined breakfast skipping was not causal of evening chronotype (P = 0.67); however, genetically defined evening chronotype was causally associated with breakfast skipping [IVW: 0.019 (0.006); P = 9.36 × 10−4] (Table 2).
FIGURE 5.
Genetic architecture shared between breakfast skipping and selected behavioral and disease traits. LD score regression estimates of genetic correlation (rg) were obtained by comparing summary statistics from the UK Biobank breakfast skipping GWAS with summary statistics from publicly available GWASs for psychiatric and metabolic disorders, immune diseases, and other traits of natural variation. Traits in this figure are selected based on previously observed epidemiologic findings; full genetic correlations with other traits can be found in Supplemental Table 5. Blue, positive genetic correlation; red, negative genetic correlation; rg values are displayed for significant correlations. Larger squares correspond to more significant P values. Genetic correlations significantly different from zero after Bonferroni correction (P < 0.00022) are labeled with an asterisk. GWAS, genome-wide association study, LD, linkage disequilibrium.
TABLE 2.
Causal links between breakfast skipping and correlated traits analyzed through the use 1- and 2-sample MR1
| IVW | MR Egger | Weighted median | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Outcome | Study | PubMed ID | SNPs | β | SE | P value | β | SE | P value | β | SE | P value |
| Body fat, % | N/A | 26833246 | 5 | 0.261 | 0.167 | 0.12 | −0.154 | 0.702 | 0.84 | 0.104 | 0.211 | 0.62 |
| BMI, kg/m2 | GIANT | 25673413 | 5 | 0.304 | 0.153 | 0.046 | 1.151 | 0.589 | 0.15 | 0.439 | 0.131 | 0.001 |
| Chronotype, evening | UK Biobank | N/A | 6 | 0.074 | 0.177 | 0.67 | −1.082 | 0.563 | 0.13 | 0.076 | 0.157 | 0.63 |
| Coronary heart disease, log OR | C4D | 21378988 | 3 | −0.387 | 0.726 | 0.59 | 10.590 | 11.880 | 0.54 | −0.049 | 0.834 | 0.95 |
| Depressive symptoms, log OR | SSGAC | 27089181 | 6 | 0.289 | 0.146 | 0.049 | 0.642 | 0.706 | 0.42 | 0.367 | 0.126 | 0.004 |
| Insomnia, log OR | UK Biobank | N/A | 6 | 0.052 | 0.042 | 0.22 | −0.082 | 0.180 | 0.67 | 0.028 | 0.054 | 0.61 |
| Lung cancer, log OR | ILCCO | 24880342 | 5 | 0.155 | 0.713 | 0.83 | −1.423 | 3.169 | 0.68 | 0.260 | 0.634 | 0.68 |
| Rheumatoid arthritis, log OR | N/A | 24390342 | 2 | −1.271 | 1.609 | 0.43 | N/A | N/A | N/A | N/A | N/A | N/A |
| Smoking (ever vs never), log OR | TAG | 20418890 | 5 | 0.974 | 0.311 | 0.002 | 0.626 | 1.380 | 0.68 | 0.914 | 0.397 | 0.02 |
| Type 2 diabetes, log OR | DIAGRAM | 24509480 | 4 | 0.431 | 0.790 | 0.59 | 2.722 | 3.473 | 0.52 | 0.696 | 0.680 | 0.31 |
| Schooling, y | SSGAC | 27225129 | 6 | −0.118 | 0.103 | 0.25 | −0.482 | 0.392 | 0.29 | −0.170 | 0.080 | 0.03 |
| Breakfast skipping [exposure: chronotype (23&Me participants only)] | UK Biobank | Present | 338 | 0.019 | 0.006 | 9.36 × 10–4 | 0.018 | 0.014 | 0.20 | 0.035 | 0.016 | 0.02 |
β is the estimated effect of change on outcome in units indicated or log OR for case-control traits using the SNPs associated with breakfast skipping (or evening chronotype) and effect estimates for breakfast skipping from the UK Biobank GWAS of breakfast skipping. Number of SNPs used in each analysis is in the SNPs column. Each outcome was obtained from publicly available GWAS summary data, except for chronotype and insomnia. The number of SNPs used varies for each outcome because of not being able to find some of the 6 SNPs (or proxies) in the summary data. Three MR methods were used. Two-sample MR was used for all outcomes, except for chronotype and insomnia. C4D, Coronary Artery Disease Genetics Consortium; DIAGRAM, DIAbetes Genetics Replication And Meta-analysis consortium; GIANT, Genetic Investigation of ANthropometric Traits; GWAS, genome-wide association study; ILCCO, International Lung Cancer Consortium; MR, Mendelian randomization; N/A, not applicable/available; SNP, single nucleotide polymorphism; SSGAC, Social Science Association Consortium; TAG, Tobacco and Genetics Consortium; IVW, inverse variance weighted.
Replication of the UK Biobank signals in the independent breakfast-skipping GWAS reported from the TwinUK study (19) (20.1% breakfast skippers; n = 403 skippers/1,603 consumers) provided further support for our findings (Supplemental Table 6), where a combined PRS of the 6 UK Biobank breakfast skipping signals showed significant association with breakfast skipping [β (SE) = 0.014 (0.007) per allele; P value = 0.032]. The 5 cohorts from the CHARGE consortium were from the United States and the Netherlands and included a total of 11,963 participants of whom 11.4% (n = 1359) were classified as breakfast skippers. Across participating cohorts, breakfast skipping ranged from 5.7% to 35.6% (Supplemental Table 7), and in general, younger cohorts (i.e., <40 y BHS and CARDIA) had more than double the prevalence of breakfast skipping than older cohorts, with the exception of WHI. A combined weighted PRS of the UK Biobank breakfast skipping signals did not show significant association with breakfast skipping in the CHARGE consortium GWAS meta-analysis [β (SE) = 0.048 (0.029) per allele; P value = 0.095] (Supplemental Table 8). In sensitivity meta-analysis restricted to either younger or older cohorts, stronger evidence of PRS association was evident among the younger cohorts (P = 0.06) compared with the older cohorts (P = 0.70) (Supplemental Table 9). Furthermore, in the CHARGE GWAS meta-analysis, no genome-wide significant associations were observed (Supplemental Figures 2–4), and a total of 4 variants showed suggestive associations of P < 1 × 10−05 (Table 3).
TABLE 3.
Top signals (P < 1 × 10−5) from CHARGE consortium genome-wide association meta-analysis of breakfast skipping (n = 11,963)1
| SNP | Chr:position | Nearest gene | Alleles (E/A) | EAF | OR (95% CI) | P value | Study association direction | I 2 |
|---|---|---|---|---|---|---|---|---|
| rs76211599 | 21:20,129,400 | LOC101927797 | A/G | 0.111 | 1.54 (1.29, 1.84) | 1.83 × 10–6 | – – – + – | 12 |
| rs2732520 | 11:35,056,846 | PDHX | A/G | 0.785 | 1.40 (1.22, 1.60) | 2.26 × 10–6 | – – – – | 0 |
| rs144181848 | 4:128,263,712 | INTU | C/G | 0.025 | 2.15 (1.55, 2.99) | 5.16 × 10–6 | – – + + – – | 61 |
| rs9838428 | 3:66,842,859 | KBTBD8 | G/T | 0.131 | 1.41 (1.21, 1.64) | 7.99 × 10–6 | + + + – + | 66 |
A GWAS was performed in each cohort in participants of European ancestry with the use of logistic regression, adjusted for age, sex, study-specific centers, and population stratification principal components, where applicable, then meta-analyzed using fixed-effect meta-analysis with inverse variance weights in METAL software. OR (95% CI) estimates are per each additional effect allele. ORs >1.00 reflect greater odds of breakfast skipping. Order of study in the Study association direction column: BOGALUSA, CARDIA, CHS, NEO, and WHI. I2 represents the heterogeneity statistic, presented as a percentage. BHS, Bogalusa Heart Study; CARDIA, Coronary Artery Risk Development in Young Adults; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; Chr, chromosome; CHS, Cardiovascular Health Study; E/A, effect/alternative alleles; EAF, effect allele frequency; GWAS, genome-wide association study; NEO, The Netherlands Epidemiology of Obesity; position, base pair coordinate hg19; SNP, single nucleotide polymorphism; WHI, Women's Health Initiative.
Discussion
Our multiple-stage proxy-phenotype GWAS, including the UK Biobank, TwinUK study, and CHARGE consortium, supports the role of 6 genetic variants in breakfast skipping, and links clock regulation with food timing. Expression of identified genes is enriched in the cerebellum, a region previously implicated in lifestyle traits (30). Among the identified signals is an ARID3B/CYP1A1 genetic variant related to caffeine metabolism, suggesting shared association signals for commonly consumed morning food and beverage items, which is further supported by our observed association of a breakfast PRS with other commonly consumed noncereal breakfast items. In addition, FGF21, which previously associated with carbohydrate intake (52), exhibits circadian rhythmicity and is considered an important metabolic regulator integrating the circadian clock with energy homeostasis (55). Interestingly, 3 of the 6 loci harbor genes (METTL4, YWHAB, and YTHDF3) that encode homologs of enzymes important for N6-methyladenosine (m6A) RNA transmethylation, a prevalent posttranscriptional modification that regulates mRNA decay, triages RNAs to stress granules (56), and regulates the pace of the circadian clock (57), among other functions (58). In addition, the rs6017427 allele A at RIMS4/YWHAB associated with higher breakfast skipping is an expression quantitative trait locus for reduced YWHAB expression in blood. Our findings suggest one intriguing possibility that the breakfast skipping variants lead to lower m6A mRNA methylation, resulting in lengthening of the circadian period, contributing to evening preference, breakfast skipping, and later food timing. Consistently, variants in this pathway have been identified as chronotype-related variants [rs2580160 near METTL4 and rs34054660 near YTHDF3 (40)], supporting this hypothesis. Additional replication and future functional studies will be required to test the hypothesis that m6A RNA methylation plays a role in breakfast skipping and later food timing, and extend previously observed phenotypic links between the circadian clock and food timing (59, 60).
Through genome-wide correlation analyses, we were able to agnostically interrogate correlations between breakfast skipping and hundreds of publicly available traits. As expected, genetic correlations were consistent with previously observed epidemiologic and experimental findings (1), such as correlations between breakfast skipping and obesity (61) and depressive symptoms (62). Genetic correlations with insomnia and rheumatoid arthritis, however, are novel. As causality cannot be inferred from correlations, we subsequently performed MR and observed that genetically defined breakfast skipping is causal of higher BMI, more depressive symptoms, and smoking. Interestingly, the findings related to breakfast skipping and higher BMI are evident despite the small contribution of breakfast to total energy intake.
Our results provide further support of the link between breakfast skipping and chronotype (59, 60) beyond individual loci. In bidirectional MR, we observed a causal link between being an evening person and skipping breakfast, but not vice versa. The identified causal links may implicate breakfast skipping and late food timing as a mediator of certain epidemiologic associations reported between evening chronotype and cardiometabolic diseases and mortality (63, 64); however, confirmatory analyses and larger-scale studies of gene-behavior mismatch are necessary.
Replication of our UK Biobank breakfast-skipping signals in the TwinUK, a second British cohort, supports our rationale for using breakfast cereal in the UK Biobank as a proxy-phenotype for breakfast. The CHARGE consortium facilitated collaboration across 5 adult cohort studies, tripling the number of breakfast skipping cases compared with the published TwinUK efforts (19). However, the lack of verification in the CHARGE consortium breakfast skipping meta-analysis, which comprises a US and a Dutch cohort, may suggest differences in the phenotype. The stronger PRS association observed in the subgroup meta-analysis restricted to younger cohorts may possibly be as a result of the higher prevalence of breakfast skipping in that subgroup (younger = 28.5% skipping compared with older = 9.2% skipping). In addition, none of the tested GWAS variants achieved the genome-wide significance threshold at P < 5 × 10−08, likely due to the overall low prevalence of breakfast skipping (11.4%) and continued insufficient statistical power.
Various strengths in our investigation are worth noting. Our study employs a proxy-phenotype approach to leverage the statistical power of a large biobank, the UK Biobank, that focuses on dietary composition to assess dietary pattern. A similar proxy-phenotype approach (24) has been successfully implemented in earlier GWASs when the phenotype of interest is difficult to ascertain, available in a limited dataset, or unmeasured (25). Thus, we proposed the use of breakfast cereal, a commonly eaten breakfast food in the United Kingdom (20, 21) temporally consumed during the morning (22), as a proxy-phenotype to advance our understanding of the role of breakfast. We validated our approach by demonstrating associations of our PRS with other noncereal breakfast items and in an independent UK cohort with breakfast skipping data. Via this approach, other foods that exhibit distinct temporal peaks (i.e., based on time of day or season) may possibly also be used as proxies to advance studies on food timing and patterns. The lack of PRS association with butter, eggs, orange juice, and yogurt may be a result of weak breakfast temporal peaks for these foods. In addition, we utilized genome-wide genetic correlation analyses to support previously identified associations observed from cross-sectional epidemiologic analyses, which are prone to various limitations, and extend findings to indicate causality through the use of MR. Lastly, the CHARGE consortium cohorts in the present meta-analysis used comparable dietary assessment tools that were appropriate for the population under study, providing the highest-quality data that can be reasonably collected across epidemiologic studies.
A major limitation of the current GWAS is the reliance on existing large data not designed to capture dietary intake beyond composition. Thus, further studies with improved phenotyping are necessary to verify our findings. A limitation of taking breakfast cereal as a proxy-phenotype for breakfast intake is exposure misclassification. Considering that 94% of people in the UK are breakfast consumers (65), we may be misclassifying an important number of participants (∼48.8%) in the UK Biobank, a bias that will persist despite our large sample size. However, misclassification resulting from this will likely reduce our statistical power to detect genetic variants and bias our results towards the null. Thus, it is plausible that additional signals exist beyond the 6 we reported. In addition, we did not factor in the dietary quality, macronutrient composition, and added-sugar content of cereals in the analysis, which may influence satiety and weight regulation, particularly when considering the range of cereals included in the cereal definition. Furthermore, we may have missed other important environmental factors that could potentially confound or interact with genetic factors to influence our results. It is worth noting that there was no significant (P = 0.22) genome-wide genetic correlation observed between breakfast cereal, a carbohydrate-rich food, and carbohydrate intake, which indicates that the identified signals are not entirely driven by the carbohydrate content. Ordinal scaling of breakfast skipping in the UK Biobank hinders our ability to appropriately interpret the magnitude of findings, including causal links. In addition, power was a limitation in our CHARGE consortium GWAS, given the modest number of studies with available breakfast data and the low prevalence of breakfast skipping at a population level (8). The low prevalence of breakfast skipping, ranging from 5.7% to 35.6% in the included cohorts, necessitated the use of weekly frequency intake (i.e., >3 or <4 times/wk) to dichotomize the trait. Lastly, the present analysis was limited to participants of European ancestry, and further investigation in other ethnicities is necessary for generalizability of our findings.
Recommendations to consume a nutrient-dense breakfast introduced in the 2010 Dietary Guidelines for Americans (66) have been removed from the 2015 iteration (67), and should be critically re-evaluated in light of increasing prevalence of breakfast skipping (11% in 1970 to 18% in 2002) (68) and later times of food intake in the United States (8, 68). Our comprehensive and complementary UK Biobank, TwinUK study, and CHARGE consortium proxy-phenotype GWAS approach identified 6 genetic variants for food timing, linking the role of the circadian clock in regulating food timing. Overall, our observed genetic correlation and MR results are in keeping with a potentially healthy role of regular breakfast intake, consistent with recommendations from the American Heart Association (1). Furthermore, we recommend that future studies, including large biobanks such as the US-based AllOfUs initiative (69), collect data on dietary pattern, including timing, to further interrogate their genetic and physiologic effects on human health.
Supplementary Material
Acknowledgments
This research has been conducted with the use of the UK Biobank Resource (UK Biobank application number 6818) and CHARGE consortium. We thank the researchers from the UK Biobank and CHARGE consortium who contributed or collected data. CHARGE cohort-specific sources of support and acknowledgments are presented in Supplemental Table 1.
The authors’ contributions were as follows—HSD, JM, FAJLS, MG, and RS: designed the study; HSD, JM, JMM, YS, DS, TMB, RL-G, VBP, and HN: conducted research and contributed to statistical analyses; HSD, JM, CES, TT, NMM, MLN, FAJLS, MKR, MG, and RS: interpreted data; HSD, JM, JML, CES, TT, NMM, CT, and RS wrote the manuscript; and all authors: read and approved the final version of the manuscript. HSD and RS are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. BMP serves on the Data and Safety Monitoring Board of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. FAJLS received speaker fees from Bayer Healthcare, Sentara Healthcare, Philips, Kellogg Company, Vanda Pharmaceuticals, and Pfizer Pharmaceuticals. NMM is a scientific advisor on the Whole Grains Council. HSD, JM, JML, YS, CES, TT, CT, DS, TMB, RL-G, HN, SR, RNL, TMA, RdM, LB, LQ, KLK, DOM-K, VBP, MLN, MKR, MG, and RS have no conflicts of interest.
Notes
HSD was supported by NIH grant R01DK107859. JM was supported by a postdoctoral fellowship funded by the European Commission Horizon 2020 program: Marie Skłodowska-Curie Actions (H2020-MSCA-IF- 2015-703787). FAJLS was supported in part by NIH grants R01HL094806, R01HL118601, R01DK099512, R01DK102696, and R01DK105072. NMM is supported in part by USDA agreement #58-1950-4-003 and funding from the General Mills Bell Institute of Health and Nutrition. MG was supported by the Spanish Government of Investigation, Development, and Innovation (SAF2017-84135-R) including FEDER co-funding, and NIDDK R01DK105072. RS was supported by NIH grants R01DK107859, R01HL113338, and R01DK105072, and the Phyllis and Jerome Lyle Rappaport Massachusetts General Hospital Research Scholar Award. The Infrastructure for the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium is supported in part by National Heart, Lung, and Blood Institute (NHLBI) grant HL105756. Funding sources for individual CHARGE cohort studies appear in Supplemental Table 1. General Mills Bell Institute of Health and Nutrition was not involved in the design, implementation, analysis, or interpretation of the data.
Supplemental Tables 1–9 and Supplemental Figures 1–4 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: BHS, Bogalusa Heart Study; CARDIA, Coronary Artery Risk Development in Young Adults; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; CHS, Cardiovascular Health Study; FID, UK Biobank Field Identifier; GWAS, genome-wide association study; IVW, inverse variance weighted/weighting; m6A, N6-methyladenosine; MAF, minor allele frequency; MR, Mendelian randomization; NEO, The Netherlands Epidemiology of Obesity; PRS, polygenic risk score; SNP, single nucleotide polymorphism; WHI, Women's Health Initiative.
References
- 1. St-Onge M-P, Ard J, Baskin ML, Chiuve SE, Johnson HM, Kris-Etherton P, Varady K, American Heart Association Obesity Committee of the Council on Lifestyle and Cardiometabolic Health; Council on Cardiovascular Disease in the Young; Council on Clinical Cardiology; and Stroke Council . Meal timing and frequency: implications for cardiovascular disease prevention: a scientific statement from the American Heart Association. Circulation. 2017;135:e96–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mattson MP, Allison DB, Fontana L, Harvie M, Longo VD, Malaisse WJ, Mosley M, Notterpek L, Ravussin E, Scheer FAJL et al.. Meal frequency and timing in health and disease. Proc Natl Acad Sci USA. 2014;111:16647–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Castro JM. Heritability of diurnal changes in food intake in free-living humans. Nutrition. 2001;17:713–20. [DOI] [PubMed] [Google Scholar]
- 4. Lopez-Minguez J, Dashti HS, Madrid-Valero JJ, Madrid JA, Saxena R, Scheer FA, Ordoñana JR, Garaulet M. Heritability of the timing of food intake. Clin Nutr. 2019;38(2):767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Castro JM. The time of day of food intake influences overall intake in humans. J Nutr. 2004;134:104–11. [DOI] [PubMed] [Google Scholar]
- 6. Jakubowicz D, Froy O, Wainstein J, Boaz M. Meal timing and composition influence ghrelin levels, appetite scores and weight loss maintenance in overweight and obese adults. Steroids. 2012;77:323–31. [DOI] [PubMed] [Google Scholar]
- 7. Gill S, Panda S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 2015;22:789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kant AK, Graubard BI. Within-person comparison of eating behaviors, time of eating, and dietary intake on days with and without breakfast: NHANES 2005–2010. Am J Clin Nutr. 2015;102:661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keski-Rahkonen A, Viken RJ, Kaprio J, Rissanen A, Rose RJ. Genetic and environmental factors in breakfast eating patterns. Behav Genet. 2004;34:503–14. [DOI] [PubMed] [Google Scholar]
- 10. Mekary RA, Giovannucci E, Willett WC, van Dam RM, Hu FB. Eating patterns and type 2 diabetes risk in men: breakfast omission, eating frequency, and snacking. Am J Clin Nutr. 2012;95:1182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mekary RA, Giovannucci E, Cahill L, Willett WC, van Dam RM, Hu FB. Eating patterns and type 2 diabetes risk in older women: breakfast consumption and eating frequency. Am J Clin Nutr. 2013;98:436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reutrakul S, Hood MM, Crowley SJ, Morgan MK, Teodori M, Knutson KL. The relationship between breakfast skipping, chronotype, and glycemic control in type 2 diabetes. Chronobiol Int. 2014;31:64–71. [DOI] [PubMed] [Google Scholar]
- 13. Uzhova I, Fuster V, Fernández-Ortiz A, Ordovás JM, Sanz J, Fernández-Friera L, López-Melgar B, Mendiguren JM, Ibáñez B, Bueno H et al.. the importance of breakfast in atherosclerosis disease. J Am Coll Cardiol. 2017;70:1833–42. [DOI] [PubMed] [Google Scholar]
- 14. Smith KJ, Gall SL, McNaughton SA, Blizzard L, Dwyer T, Venn AJ. Skipping breakfast: longitudinal associations with cardiometabolic risk factors in the Childhood Determinants of Adult Health Study. Am J Clin Nutr. 2010;92:1316–25. [DOI] [PubMed] [Google Scholar]
- 15. O'Reardon JP, Peshek A, Allison KC. Night eating syndrome: diagnosis, epidemiology and management. CNS Drugs. 2005;19:997–1008. [DOI] [PubMed] [Google Scholar]
- 16. Melve KK, Baerheim A. Signs of subclinical eating disorders in teenage girls. Scand J Prim Health Care. 1994;12:197–203. [DOI] [PubMed] [Google Scholar]
- 17. Li X, Meng X, Spiliopoulou A, Timofeeva M, Wei W-Q, Gifford A, Shen X, He Y, Varley T, McKeigue P et al.. MR-PheWAS: exploring the causal effect of SUA level on multiple disease outcomes by using genetic instruments in UK Biobank. Ann Rheum Dis. 2018;77(7):1039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–90. [DOI] [PubMed] [Google Scholar]
- 19. Boraska V, Davis OSP, Cherkas LF, Helder SG, Harris J, Krug I, Pei-Chi Liao T, Treasure J, Ntalla I, Karhunen L et al.. Genome-wide association analysis of eating disorder-related symptoms, behaviors, and personality traits. Am J Med Genet Part B Neuropsychiatr Genet. 2012;159B:803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gibson SA, Gunn P. What's for breakfast? Nutritional implications of breakfast habits: insights from the NDNS dietary records. Nutr Bull. 2011;36:78–86. [Google Scholar]
- 21. Mckevith B, Jarzebowska A. The role of breakfast cereals in the UK diet: headline results from the National Diet and Nutrition Survey (NDNS) year 1. Nutr Bull. 2010;35:314–19. [Google Scholar]
- 22. Burke SJ, McCarthy SN, O'Dwyer NA, Gibney MJ. Analysis of the temporal intake of cereal and dairy products in Irish adults: implications for developing food-based dietary guidelines. Public Health Nutr. 2005;8:238–48. [DOI] [PubMed] [Google Scholar]
- 23. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J et al.. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, Chabris CF, Emilsson V, Johnson AD, Lee JJ et al.. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111:13790–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hormozdiari F, Kang EY, Bilow M, Ben-David E, Vulpe C, McLachlan S, Lusis AJ, Han B, Eskin E. Imputing phenotypes for genome-wide association studies. Am J Hum Genet. 2016;99:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen G-B, Emilsson V, Meddens SFW et al.. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galante J, Adamska L, Young A, Young H, Littlejohns TJ, Gallacher J, Allen N. The acceptability of repeat Internet-based hybrid diet assessment of previous 24-h dietary intake: administration of the Oxford WebQ in UK Biobank. Br J Nutr. 2016;115:681–6. [DOI] [PubMed] [Google Scholar]
- 28. Liu B, Young H, Crowe FL, Benson VS, Spencer EA, Key TJ, Appleby PN, Beral V. Development and evaluation of the Oxford WebQ, a low-cost, web-based method for assessment of previous 24 h dietary intakes in large-scale prospective studies. Public Health Nutr. 2011;14:1998–2005. [DOI] [PubMed] [Google Scholar]
- 29. Bradbury KE, Young HJ, Guo W, Key TJ. Dietary assessment in UK Biobank: an evaluation of the performance of the touchscreen dietary questionnaire. J Nutr Sci. 2018;7:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dashti HS, Jones SE, Wood AR, Lane JM, van Hees VT, Wang H, Rhodes JA, Song Y, Patel K, Anderson SG et al.. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10:1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lane JM, Jones SE, Dashti HS, Wood AR, Aragam KG, van Hees VT, Strand LB, Winsvold BS, Wang H, Bowden J et al.. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019;51:387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B et al.. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC et al.. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. UK Biobank – Neale lab [Internet]. Available from: www.nealelab.is/uk-biobank [Accessed February 1, 2019]. [Google Scholar]
- 35. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K et al.. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium et al.. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium, Duncan L et al.. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, Jeffries AR, Dashti HS, Hillsdon M, Ruth KS et al.. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R et al.. The MR-Base platform supports systematic causal inference across the human phenome. [Internet]. Elife. 2018;7:e34408 Available from: https://elifesciences.org/articles/34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am J Hum Genet. 2016;98:116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delaneau O, Marchini J, 1000 Genomes Project Consortium, Lunter G, Marchini JL, Myers S, Gupta-Hinch A, Iqbal Z, Mathieson I et al.. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cornelis MC, Kacprowski T, Menni C, Gustafsson S, Pivin E, Adamski J, Artati A, Eap CB, Ehret G, Friedrich N et al.. Genome-wide association study of caffeine metabolites provides new insights to caffeine metabolism and dietary caffeine-consumption behavior. Hum Mol Genet. 2016;25:5472–82. [DOI] [PubMed] [Google Scholar]
- 52. Frayling TM, Beaumont RN, Jones SE, Yaghootkar H, Tuke MA, Ruth KS, Casanova F, West B, Locke J, Sharp S et al.. A common allele in FGF21 associated with sugar intake is associated with body shape, lower total body-fat percentage, and higher blood pressure. Cell Rep. 2018;23:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park SL, Carmella SG, Chen M, Patel Y, Stram DO, Haiman CA, Le Marchand L, Hecht SS. Mercapturic acids derived from the toxicants acrolein and crotonaldehyde in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. PLoS One. 2015;10:e0124841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang H, Xiao X, Li M. The schizophrenia risk gene ZNF804A: clinical associations, biological mechanisms and neuronal functions. Mol Psychiatry. 2017;22:944–53. [DOI] [PubMed] [Google Scholar]
- 55. Andersen B, Beck-Nielsen H, Højlund K. Plasma FGF21 displays a circadian rhythm during a 72-h fast in healthy female volunteers. Clin Endocrinol (Oxf). 2011;75:514–9. [DOI] [PubMed] [Google Scholar]
- 56. Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian S-B, Ignatova Z. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci Alliance. 2018;1:e201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fustin J-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. [DOI] [PubMed] [Google Scholar]
- 58. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. [DOI] [PubMed] [Google Scholar]
- 59. Muñoz JSG, Cañavate R, Hernández CM, Cara-Salmerón V, Morante JJH. The association among chronotype, timing of food intake and food preferences depends on body mass status. Eur J Clin Nutr. 2017;71:736–42. [DOI] [PubMed] [Google Scholar]
- 60. Maukonen M, Kanerva N, Partonen T, Kronholm E, Tapanainen H, Kontto J, Männistö S. Chronotype differences in timing of energy and macronutrient intakes: a population-based study in adults. Obesity (Silver Spring). 2017;25:608–15. [DOI] [PubMed] [Google Scholar]
- 61. Deshmukh-Taskar P, Nicklas TA, Radcliffe JD, O'Neil CE, Liu Y. The relationship of breakfast skipping and type of breakfast consumed with overweight/obesity, abdominal obesity, other cardiometabolic risk factors and the metabolic syndrome in young adults. The National Health and Nutrition Examination Survey (NHANES): 1999–2006. Public Health Nutr. 2013;16:2073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee SA, Park E-C, Ju YJ, Lee TH, Han E, Kim TH. Breakfast consumption and depressive mood: a focus on socioeconomic status. Appetite. 2017;114:313–9. [DOI] [PubMed] [Google Scholar]
- 63. Knutson KL, von Schantz M. Associations between chronotype, morbidity and mortality in the UK Biobank cohort. Chronobiol Int. 2018;35(8):1045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vera B, Dashti HS, Gómez-Abellán P, Hernández-Martínez AM, Esteban A, Scheer FAJL, Saxena R, Garaulet M. Modifiable lifestyle behaviors, but not a genetic risk score, associate with metabolic syndrome in evening chronotypes. Sci Rep. 2018;8:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gaal S, Kerr MA, Ward M, McNulty H, Livingstone MBE. Breakfast consumption in the UK: patterns, nutrient intake and diet quality. A study from the International Breakfast Research Initiative Group. Nutrients. 2018;10:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McGuire S. US Department of Agriculture and US Department of Health and Human Services , Dietary Guidelines for Americans, 2010 7th edition, Washington (DC): US Government Printing Office, January 2011. Adv Nutr2011;2:293–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. DeSalvo KB, Olson R, Casavale KO. Dietary Guidelines for Americans. JAMA. 2016;315:457. [DOI] [PubMed] [Google Scholar]
- 68. Kant AK, Graubard BI. 40-year trends in meal and snack eating behaviors of American adults. J Acad Nutr Diet. 2015;115:50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jaffe S. Planning for US Precision Medicine Initiative underway. Lancet. 2015;385:2448–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.