Abstract
Background
Parasitic plants engage in a complex molecular dialog with potential host plants to identify a host and overcome host defenses to initiate development of the parasitic feeding organ, the haustorium, invade host tissues, and withdraw water and nutrients. While one of two critical signaling events in the parasitic plant life cycle (germination via stimulant chemicals) has been relatively well-studied, the signaling event that triggers haustorium formation remains elusive. Elucidation of this poorly understood molecular dialogue will shed light on plant-plant communication, parasitic plant physiology, and the evolution of parasitism in plants.
Results
Here we present an experimental framework that develops easily quantifiable contrasts for the facultative generalist parasitic plant, Triphysaria, as it feeds across a broad range of diverse flowering plants. The contrasts, including variable parasite growth form and mortality when grown with different hosts, suggest a dynamic and host-dependent molecular dialogue between the parasite and host. Finally, by comparing transcriptome datasets from attached versus unattached parasites we gain insight into some of the physiological processes that are altered during parasitic behavior including shifts in photosynthesis-related and stress response genes.
Conclusions
This work sheds light on Triphysaria’s parasitic life habit and is an important step towards understanding the mechanisms of haustorium initiation factor perception, a unique form of plant-plant communication.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1856-1) contains supplementary material, which is available to authorized users.
Background
Triphysaria versicolor is a model parasitic plant in the family Orobanchaceae [1, 2], a family that represents one of a likely 12 independent origins of parasitism in flowering plants [3, 4]. T. versicolor is a facultative parasite, and a generalist that can parasitize a wide range of monocot and eudicot hosts, both in nature [5], and in the laboratory [6]. Other members of this family are a primary constraint to African agriculture [7], infesting 40% of all cereal crops in sub-Saharan Africa [8], and causing an estimated $US 10 billion in crop damage annually [9, 10]. The Orobanchaceae also provide unique opportunities to study parasitism as it is the only plant family with the full range of parasitic lifestyles [11], plus a fully autotrophic sister lineage, Lindenbergia [12]. In addition to their usefulness for understanding the evolution of parasitism (and thus novel traits, [13, 14]), these plants display extremes of physiology and development that can help us understand many facets of plant biology. For example, strigolactones, long known as germination stimulants [15] for parasitic members of Orobanchaceae, were discovered in 2008 to be important plant hormones [16, 17], the likely receptors for which have been recently identified [18].
Strigolactones are also important signaling molecules perceived by arbuscular mycorrhizal (AM) fungi during symbiosis [19], suggesting that parasitic plants have evolved to eavesdrop on the molecular dialogue between potential hosts and symbiotic fungi [20]. Interruption of this dialogue has been identified as one of the potential control points for weedy parasitic Orobanchaceae [21–23]. However, the impact of altering strigolactone levels in the plant and in the rhizosphere as part of an effort to control parasitic weeds is still being explored. This is complicated by recent work reporting protective effects of AM fungi against Striga hermonthica in Sorghum [24]. Another potential point of control is post attachment physiology of the parasite [10]. Post attachment resistance traits are usually polygenic and breeding programs have targeted these modes of resistance, though only partial and short-term resistance has been achieved [10]. A third point of control is the mechanism by which parasitic plants initiate haustorium formation [1], including the perception of haustorium inducing factors (HIFs). Raw host root exudates contain active HIFs including various quinones, hydroquinones, phenolic acids and flavonoids [25]. It is likely that the considerable redundancy in host derived HIFs contributes to the broad host range of parasitic Orobanchaceae [25]. It also presents the possibility that a complex HIF profile conveys host quality information, providing a point at which the parasite can evaluate its host in preparation for attachment [25]. The mechanism of this process is largely unknown, save the following observations: 1) structurally diverse active HIFs all have a narrow window of redox potentials [25], 2) the quinone reductase TvQR1 is important for haustorium initiation in Triphysaria and acts very early in HIF perception [25, 26], and 3) that TvPirin is necessary for haustorium formation [27]. Interestingly, TvQR1 has a much greater allelic diversity than TvPirin, with the highest diversity in a protein domain that determines substrate specificity [28]. This diversity may help explain Triphysaria’s ability to respond to a wide variety of host root exudates and hence feed across a broad host range.
In obligate parasites like Striga the commitment to haustorium formation (i.e. haustoriogenesis) is made at germination, because even though separate signaling events must occur to initiate haustoriogenesis, seed resources are quickly exhausted and provide only a few days of resources to effect successful attachment to a host root, without which the seedling dies [29]. Therefore, it is critical to coordinate haustorium formation with radicle growth and with regard to the proximity and orientation to the potential host root via the perception of HIFs. In contrast, the commitment to haustoriogenesis in facultative hemiparasites, like Triphysaria, occurs via HIF perception by roots of ostensibly free-living plants. The facultative generalist parasite must also evaluate potential hosts during the free-living phase of growth to identify high quality versus low quality hosts. The general lack of self-haustorium formation, plus the reduced rate of haustorium formation on congeneric plants (i.e. T. eriantha), compared to Arabidopsis thaliana, shows that Triphysaria has the ability to evaluate host quality [30]. Because Triphysaria does not require a host-derived germination simulant, host evaluation is uncoupled from germination, making the facultative generalist a useful model for characterization of HIF perception processes in parasitic plants. Importantly, the host range of Triphysaria overlaps with that of the weedy Orobanchaceae and provides a framework for discovery of host recognition and evaluation machinery that is shared family-wide. Previous work has shown that another facultative parasitic Orobanchaceae, Castilleja densiflora (syn. Orthocarpus densiflora) displays host dependent floral phenotypes [31] as well as host dependent survivorship [32]. Furthermore, phenotypic transitions to more vigorous growth, thought to occur after successful attachment, have been noted [32, our unpublished field observations]. Therefore, we hypothesized that Triphysaria would display host dependent phenotypes during interactions with various hosts that we could magnify by growing the parasite on distantly related plants that span the parasite’s host range.
We selected a group of experimentally tractable host plant genera, based in part upon the survey by Thurman [5], that includes three eudicots (Arabidopsis, Medicago, and Solanum) and three monocots (Zea, Oryza, and Juncus). Here we describe experiments that reveal clines of easily quantifiable parasite phenotypes displayed by Triphysaria while it fed across its host range. These phenotypes suggest that the generalist parasite may have the ability to evaluate host quality, and our framework provides a means to evaluate parasite success nondestructively throughout the parasite’s life cycle. Surprisingly, we found that phenotypes that indicate enhanced parasite vigor were strongly correlated with low parasite survivorship. We also show that direct parasite-host contact, not just host root exudate, is necessary for the development of a distinct growth phenotype. Finally, we developed image-based analytics that recapitulate destructive measurements that will allow us to capture phenotypic transitions during host-parasite interactions with non-destructive time course measurements.
Results
Co-culture across Triphysaria’s host range
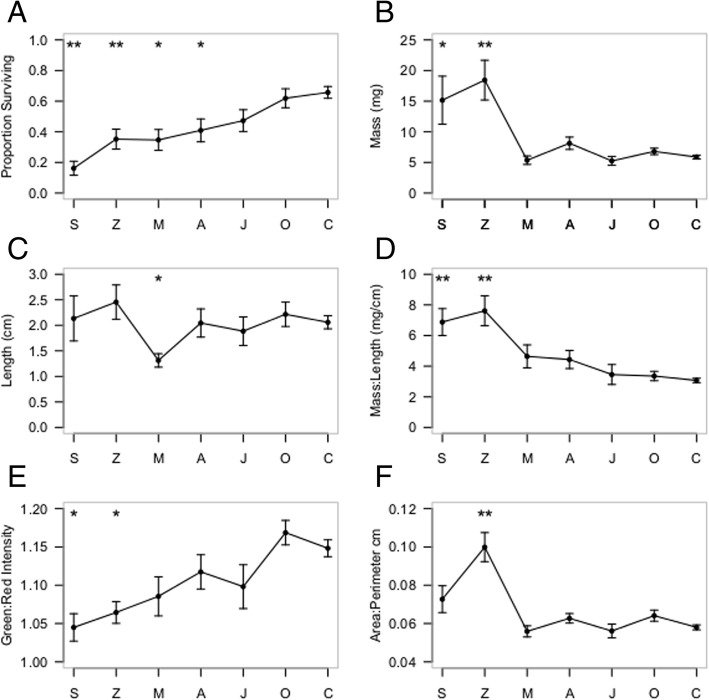
The host range co-culture experiment was monitored daily and survivorship of Triphysaria was recorded weekly by counting surviving individuals. By the 5th week of greenhouse co-culture, parasites in the Solanum (p > 0.001), Zea (p = 0.001), Medicago (p = 0.003), and Arabidposis (p = 0.034) pots showed significantly fewer surviving individuals than the control pots (Fig. 1a.) Because the trend appeared to be toward very low parasite survivorship in the experimental treatments, the greenhouse experiment was ended and several measurements, some destructive, were made to discover host dependent parasite phenotypes. Furthermore, because some of these patterns were very surprising, we employed very conservative statistics to avoid false positives. Simple parasite growth parameters were significantly different than the control with hosts that induced the lowest survivorship. This included higher dry mass (Fig. 1b: for Zea p < 0.001 & Solanum p = 0.026) indicating that even though the parasites were less likely to survive with Solanum and Zea hosts, survivors accrued more tissue than free-living individuals.
Fig. 1.
The characteristics of Triphysaria grown across its host range are significantly different from host-free plants and are often highly correlated. ANOVA (Dunnett-Hsu correction) statistical significance compared to the control *p < 0.05 and **p < 0.01. S=Solanum, Z = Zea, M = Medicago, A = Arabidopsis, J = Juncus, O=Oryza, C = host-free control. Pearson’s R2 for A vs. E = 0.86, D vs. E = 0.69
Compared to the gracile control plants, hosts which induced the highest parasite mortality also induced a novel phenotype – the survivors were “pale and plump” (Fig. 2: e.g. Solanum and Zea compared to the control), apparently due to shortened internodes and altered leaf morphology. We attempted to quantify the “plump” phenotype by integrating aspects of the growth parameter data. We integrated the dry mass (Fig. 1b) and plant height measurements (Fig. 1c) to produce a ratio to quantify the “plump” phenotype (Fig. 1d). Compared to the control, there were significant differences for Triphysaria grown with Zea (p = < 0.001) and Solanum (p = 0.0083). We hypothesized that the paleness of the plants was due to reduced chlorophyll content, therefore we attempted to quantify the “pale” phenotype by estimating the red:green ratio of plants (as described in [33] because this method was useful to estimate changes in chlorophyll content in senescing wheat). We analyzed photographs of all surviving individuals (See Fig. 2 for representative images from each treatment). The red:green ratio was significantly different than the host-free control for Triphysaria grown with Solanum (p = 0.0247) and Zea (p = 0.0162) (Fig. 1e). The gradation of paleness was strongly correlated (R2 = 0.86) with survivorship rates (Fig. 1a) and moderately so with the mass length ratio (Fig. 1e, R2 = 0.69) suggesting that these phenotypes may be related. Considering all evidence together, the trend for Triphysaria grown with known hosts was fewer surviving individuals that were hardier and ostensibly less autotrophic.
Fig. 2.

Triphysaria displays host dependent phenotypes. Representative images of Triphysaria showing the average number of surviving plants in each treatment, plus controls. The host genus is listed above each set of parasites. The control plants were grown in identical conditions, in an identical circular arrangement, but without hosts
Because analyzing the plant images allowed us to quantify the “greenness” of plants, thereby confirming visual observations and showing significant experimental contrasts, we attempted to recapitulate the “plumpness” (e.g. mass/height ratio) of plants as well. By analyzing each photograph to outline each plant, we generated perimeter to area ratios. This approach recapitulated our plant mass measurements (R2 = 0.87; see Fig. 1b vs. 1f) and showed a significant difference of parasites grown with Zea versus the host-free control. The correlation with the mass/height ratio was good, but lower (R2 = 0.72) indicating further refinement of the method is needed to accurately capture the experimental contrasts, namely a way to estimate plant height in a high throughput manner. Importantly, these image-based analytics showed us significant phenotypic differences between at least partly heterotrophic parasites and the autotrophic parasite controls.
Co-culture with Solanum and subirrigation of Triphysaria
Observations in the first co-culture experiment led to hypotheses about the cause of the host-dependent phenotypes. We sought to separate stimuli that induced the growth phenotypes, so we designed an experiment to isolate signaling cues that involved top watering larger co-culture pots as in the multi host experiment, but instead collecting the flow through and then using it to sub-irrigate smaller pots containing Triphysaria only (Fig. 3a). We hypothesized that the “pale and plump” phenotype was a result of successful parasite attachment. Therefore, we predicted that this phenotype would be absent from the sub-irrigated pots which lacked direct host contact, yet these plants would be exposed to water soluble host exudates that have been shown to induce haustoria in Triphysaria [34]. Indeed the only parasites in the experiment that showed the phenotype were the Triphysaria that were grown in direct contact with the tomato host (Fig. 3b SlTv vs. others: see 3c & 3d for representative plant images).
Fig. 3.
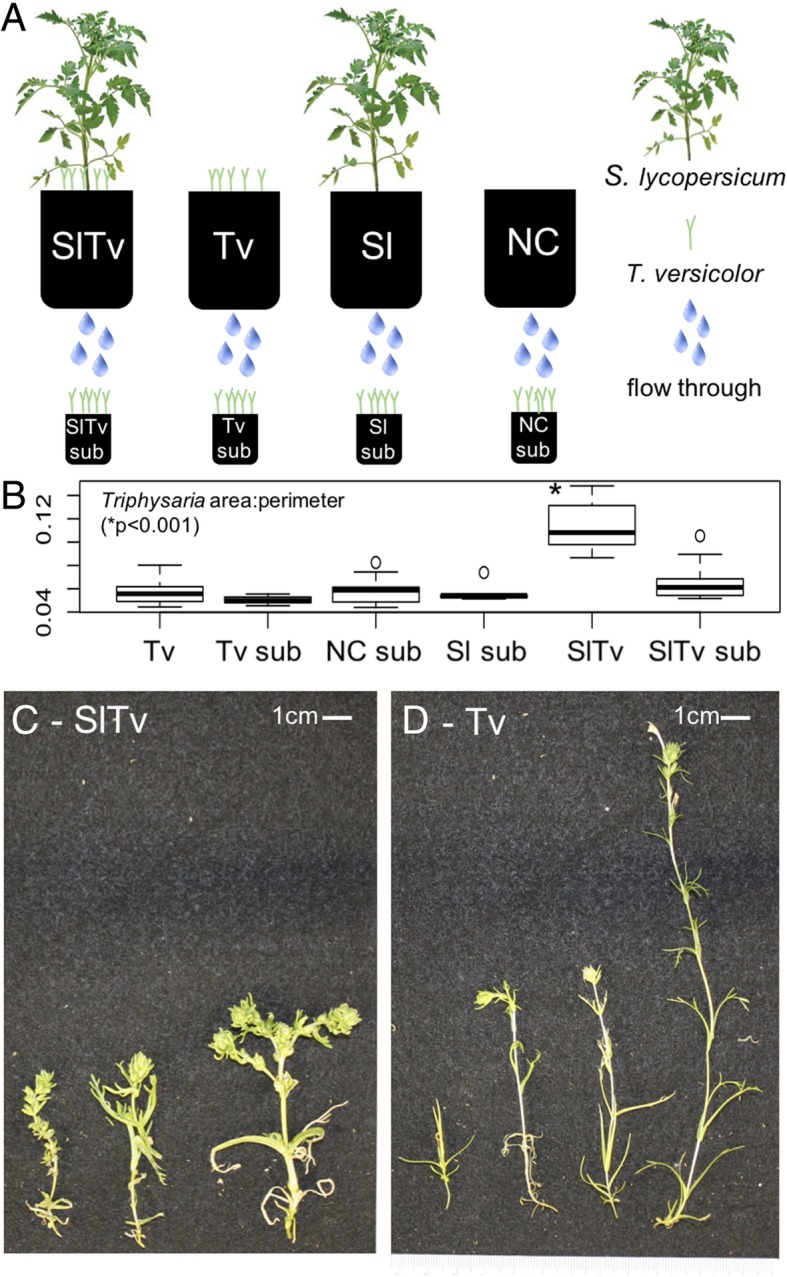
The “plump” phenotype is dependent on direct host contact. a experimental design, Sl = S. lycopersicum, Tv = T. versicolor, NC = negative control; b box plot of area:perimeter ratios for all parasites in the experiment; c & d example Triphysaria images from pot SlTv (parasite + host) and Tv (parasite only) showing the “plump” phenotype that parasites display when grown with S. lycopersicum hosts. ANOVA (Tukey-Kramer correction) *p < 0.001
There was a very weak pattern (paired t-test p = 0.04) that suggested parasite survivorship was lower in SlTv Sub (27 ± 9% surviving Triphysaria when watered with flow through from tomato only pots) than NC Sub (50 ± 6% surviving Triphysaria when watered with flow through from soil only pots), yet when we corrected for multiple comparisons the result was not significant. Thus, the survivorship of parasites in this experiment was not significantly different, possibly for one or more of three reasons: 1) growing conditions were cooler, hence more favorable causing more co-cultured parasites to establish, 2) host-free plants, which, although they were more likely to survive without hosts, still showed a trend of decreasing survivorship and thus had more time to die (8 vs. 5 weeks), and 3) lack of statistical power – the control group was roughly 1/5 of the size of the multi-host experiment, which we designed with a very large control group (n = 45) based upon a power analysis from preliminary experiments (data not shown). Importantly, parasites in the sub-irrigated pots did not display the plump phenotype, supporting our hypothesis that host contact was required for this distinct phenotype.
Differentially expressed genes in autotrophic vs. heterotrophic Triphysaria
The Parasitic Plant Genome Project (PPGP; [35]; http://ppgp.huck.psu.edu) hosts a publicly available compendium of life stage specific transcriptomes for species including Phelipanche (syn. Orobanche) aegyptiaca, Striga hermonthica and Triphysaria versicolor (see Yang et al., 2015). The observations we have made with Triphysaria feeding across its host range suggest that the parasite’s physiology is substantially altered in a host-dependent fashion. The PPGP transcriptome database includes data for Triphysaria grown with and without a host (M. truncatula) for flowers/reproductive structures, shoots, and roots. We compared these previously analyzed digital gene expression datasets [13] to find gene activity that differed significantly between the autotrophic vs. heterotrophic modes of the facultative parasite (Table 1).
Table 1.
GO enrichment of differentially expressed genes in the feeding parasite support the observed host dependent phenotypes. Bold numbers indicate P < 0.05, adapted from [13]
| GOSlim Term | DE genes | p-value | |
|---|---|---|---|
| Vegetative Shoots | upreg | downreg | |
| ATPase activity | 28 | 4 | 6.26E-06 |
| peptidase activity | 19 | 1 | 2.22E-05 |
| carbohydrate metabolic process | 13 | 3 | 1.78E-02 |
| response to stress | 13 | 2 | 5.98E-03 |
| nucleus | 12 | 2 | 1.09E-02 |
| translation | 3 | 15 | 2.76E-03 |
| intracellular | 3 | 16 | 1.50E-03 |
| thylakoid | 2 | 33 | 1.24E-09 |
| protein complex | 2 | 42 | 6.07E-13 |
| photosynthesis | 1 | 28 | 6.88E-09 |
| structural constituent of ribosome | 1 | 15 | 1.43E-04 |
| ribosome | 1 | 15 | 1.43E-04 |
| Reproductive Shoots | |||
| peptidase activity | 20 | 4 | 9.56E-04 |
| ion binding | 18 | 51 | 1.21E-06 |
| oxidoreductase activity | 6 | 41 | 3.16E-09 |
| transmembrane transport | 3 | 14 | 5.03E-03 |
| transport | 3 | 19 | 2.22E-04 |
| transmembrane transporter activity | 3 | 16 | 1.50E-03 |
| translation | 2 | 12 | 5.18E-03 |
| response to stress | 1 | 8 | 1.74E-02 |
| structural constituent of ribosome | 1 | 8 | 1.74E-02 |
| ribosome | 1 | 8 | 1.74E-02 |
| Roots | |||
| ion binding | 46 | 16 | 3.37E-05 |
| peptidase activity | 37 | 2 | 2.34E-10 |
| response to stress | 23 | 1 | 1.21E-06 |
| cellular protein modification process | 22 | 2 | 1.70E-05 |
| kinase activity | 20 | 2 | 6.66E-05 |
| biosynthetic process | 19 | 8 | 4.13E-02 |
| DNA metabolic process | 17 | 6 | 2.76E-02 |
| DNA binding | 16 | 6 | 4.35E-02 |
| hydrolase activity, acting on glycosyl bonds | 11 | 2 | 1.95E-02 |
| lipid metabolic process | 9 | 1 | 1.93E-02 |
| translation | 1 | 11 | 2.36E-03 |
| intracellular | 1 | 10 | 4.65E-03 |
Not surprisingly, genes related to photosynthesis with the GO Biological Process term “photosynthesis” and Cellular Component term “thylakoid” are under-represented in Triphysaria when it feeds on Medicago compared to the autotrophic (host-free) mode of growth. Consistent with our previous work examining all parasite life stages [13], the Molecular Function GO term “peptidase activity” was overrepresented in the feeding parasite’s root tissue and the Biological Process GO term “translation” was underrepresented among differentially expressed (DE) genes. The GO Biological Process terms “biosynthetic process” and “carbohydrate metabolic process” are notably higher, respectively in root and shoot, in the feeding parasite compared to fully autotrophic Triphysaria. Consistent with elevated mortality rates in our experiment that suggest increased parasite stress, both in the roots and shoots of feeding parasites, “response to stress” category genes were strongly upregulated. These gene expression signals are correlated with altered growth patterns and provide candidate genes and processes to examine in future experiments.
Discussion
Interpreting the responses of a generalist parasitic plant to a range of hosts
Previous work has suggested that other facultative generalists in Orobanchaceae may show host preference or selectivity [36–38], an observation widely made of parasitic angiosperms [39, 40]. Therefore, in order to gain insight into the host evaluation process, we set out to establish a framework to observe phenotypic clines and transitions associated with host exposure across the parasite’s confirmed host range. Our observations of Triphysaria shoots display a spectrum of phenotypic characteristics along the host range of the parasite.
Typically, parasitic plant success is defined as a successful connection to a suitable host [41]. Because we (unpublished field and lab observations) and others [31] have noted transitions in parasite growth patterns that are thought to occur after successful attachment of parasites to host roots (via haustoria), we reasoned that similar obvious transitions in our experiment could be used as a proxy for successful attachment of Triphysaria to a suitable host.
Triphysaria plants show a range of a “pale and plump” phenotype that is more pronounced on certain hosts than others. This parasite phenotype resulted from shortened internodes and stubbier, fleshier, and more pale leaves. Follow-up analyses of the parasite’s anatomy may provide some additional insight in the processes that drive these growth patterns. Of particular interest would be changes in leaf anatomy, as it is known that the related obligate parasites Striga gesnerioides and Alectra orobanchoides display diminished leaf morphology compared to free living relatives [42–45]. While the overall height and dry mass of the heterotrophic individuals showed no clear trend, when used to calculate a mass:length ratio, it revealed a clear gradation that might be useful as a proxy for success of the parasite. This is because parasites that displayed the most dramatic phenotypes (grown on Zea and Solanum, see Fig. 2) accrued more biomass compared to the more gracile individuals grown on other hosts. Indeed, our observations are consistent with previous work in a closely related facultative hemi-parasite [31, 32] as Triphysaria also displayed host dependent survivorship as well as host-dependent growth characteristics. Additionally, the distinct paleness of the “plump” individuals is concordant with significant under-representation of genes related to photosynthesis in the feeding parasite, suggesting increased heterotrophy compared to fully autotrophic Triphysaria. Together these data show that the hemi-parasite Triphysaria displays clear host-dependent phenotypes that are suggestive of variable parasite success, or perhaps even host selectivity, though more work on this question is needed. In fact, Atsatt noted that facultative members of Orobanchaceae (syn. Scrophulariaceae) were ideal candidates to characterize host-specific parasite responses, in part because of the frequently observed enhanced vigor and more rapid growth after a presumed attachment to a host plant [31].
The host dependent phenotypes suggest increased parasite vigor, even though reduced overall survivorship for these heterotrophs was observed compared to more gracile host-free controls in our 5-week co-culture experiment. These observations are consistent with work by Atsatt, who found the closely related facultative parasite, Castilleja densiflora (syn. Orthocarpus densiflora) has initially low survivorship when grown with hosts versus without, but after 2 months the parasites with hosts were more likely to survive [32]. Therefore, the initial low survivorship may reflect a gamble that pays off for successful parasites late in their life cycle when water stress increases late in the season in the northern California part of the parasite’s native range [32]. A larger (necessarily due to high parasite mortality) and longer controlled study, perhaps in the field, would help determine if the same long term trends are seen with Triphysaria when feeding on various hosts.
The low survivorship of discernably more successful individuals seems to indicate that, like recent work in pea suggests [46], these plants are engaging in risky behavior possibly by allocating resources away from autotrophic modes of growth. This risky behavior may be buffered or canalized by host plants for successfully attached parasites – like the increased survivorship of inbred albino Orthocarpus purpurascens (syn. Castileja exserta) when grown with a host versus without [47]. It is therefore possible that the parasites die when attempting to transition to heterotrophic modes of growth. In this way Triphysaria may be engaging in risk in a similar way as a forager on a negative energy budget [48] – when resources are so limited that survival is unlikely, risk prone behavior in the form of a gamble for a big payoff (in this case a successful union with a host plant root) may be the only viable strategy.
These data shed light on a long-held hypothesis for which relevant data has been very limited – in fact Heide-Jørgensen makes the argument that the distinction between facultative and obligate parasitism is irrelevant because definitive evidence for facultative parasitism does not yet exist and is very hard to obtain [41]. He argues that a fully autotrophic mode of growth in a host-free system may be an artifact of highly favorable growth conditions in the lab that do not reflect growth conditions for facultative parasitic plants in nature. We contend that if autotrophy were a viable strategy, the plants would likely not risk costly haustorium formation on hosts that significantly increase mortality. Conversely, we have observed what seem to be autotrophic Triphysaria growing and flowering ~ 1 m away from any potential host plant in the field (unpublished). So while these data suggesting risky behavior by Triphysaria when grown with known hosts does support the hypothesis that Triphysaria is functionally an obligate parasite [41], definitive support for this or the alternative hypothesis remains elusive.
We did attempt to excavate pots from the multi-host experiment to survey haustorium formation, but the very dense, wet, and sandy soil made this extremely difficult. While we frequently observed haustoria, we were unable to attribute connections to certain individuals, or even accurately count the tiny ~ 1 mm haustoria. Development of a co-culture system that would induce the phenotypes we observed and that also allows researchers to monitor haustorium development with ease is needed.
Previous observations have shown that root exudates are sufficient to induce haustorium formation in Triphysaria [30]. We therefore attempted to determine if the plump phenotype was due to exudates, induced tomato defenses, or direct host contact. We confirmed that the “plump” phenotype was dependent upon direct contact with a known host, supporting the hypothesis that the phenotype co-occurs with a switch to heterotrophy, not simply exposure to host exudates or allelopathic compounds. Although the signal was very weak that Solanum exudate is sufficient to reduce survivorship, this hints that survivorship is linked to haustorium formation and can be induced without direct host contact. Using our image-based analytics, it should be possible to longitudinally monitor plant growth, and select individuals to excavate to search for haustoria when a significant contrast appears in the non-destructive analysis.
Conclusions
Characterization of the elusive molecular dialogue between parasitic plants and their host plants requires a tractable framework. To that end, we have created a framework that includes non-destructive methods for longitudinal studies and demonstrated that significant differences in easily quantifiable growth patterns are obtainable. Furthermore, these patterns of parasite growth and survival do shed light on long held questions about whether true facultative parasites exist in nature – our data suggest that Triphysaria is engaging in risky behavior to potentially parasitize certain hosts more than others. Our framework was optimized with hosts that have resources for molecular genomics work; this will facilitate next steps to explore mechanisms of host choice or evaluation by parasitic plants, as well as the nature of the unique plant-plant molecular dialogue between parasitic Orobanchaceae and their hosts.
Methods
Co-culture across Triphysaria’s host range
Host plants selection
Putative hosts were selected from surveys by Thurman [5] and were further refined to include plants with publicly available genome or transcriptome sequence data resources anticipating molecular studies that leverage this experimental framework. Host plants selected for this project were Arabidopsis thaliana (Col-0), Medicago truncatula (A17), Solanum lycopersicum (Heinz 57), Zea mays (B73), Oryza sativa subspecies Japonica cv. Nipponbare, and Juncus effuses. Haustorium formation resulting from host contact was confirmed for each host (Additional file 1: Figure S1).
Seed germination
Triphysaria versicolor, Medicago truncatula (A17), and Zea mays (B73) seeds were obtained and germinated as described by [49]. Solanum lycopersicum (Heinz 57) seed were produced in the Penn State Biology Greenhouse. S. lycopersicum seeds were surface sterilized using a 50% bleach (5.25% hypochlorite) + 0.01% Triton X-100 (Sigma) solution for 30 min, then washed 10x with sterile distilled water and germinated on Triphysaria co-culture medium (1/4x Hoagland’s basal salt and nutrient mix, 7.5 g/L sucrose, 6 g/L plant tissue culture grade agarose, pH of 6.1). Oryza sativa subspecies Japonica cv. Nipponbare seeds were incubated in sterile distilled water at 28–37 °C until germination then transferred to Triphysaria co-culture medium. Arabidopsis thaliana (Col-0) seeds were surface sterilized using 70% ethanol + 0.01% Triton X-100 for 8 min, then washed with 100% ethanol, air-dried for ~ 15 min, and germinated on Triphysaria co-culture medium. Juncus effusus seed was obtained from a commercial vendor (http://www.everwilde.com/). J. effusus seed was washed 2–3 times with sterile distilled water, washed with 70% ethanol for 3–5 min, washed with undiluted commercial bleach (5.25% hypochlorite) for 40 min, washed with sterile molecular biology grade water 3–5 times, and germinated on Triphysaria co-culture medium.
Experiment layout and watering
The host range co-culture experiment was conducted in the College of Agricultural Sciences Greenhouse #111 at Penn State University from March 3 to May 19, 2014. The experiment was set up in a complete randomized block design (randomizer.org) with 27 pots in each of 5 blocks. Each block contained 6 hosts × 3 replicates plus 9 control [no host] pots each with 7 parasites. A custom drip irrigation system was made using watering timers (Orbit Digital 2-Outlet timer Model #: 27133), ¾” (~ 19 mm) irrigation tubing and various couplers and fittings widely available at hardware stores (Additional file 2: Figure S2). Weighted irrigation drippers were fitted into the ¾” irrigation tubing and standard garden hose ball-check valves were calibrated to flow 100 mL/minute of water to each drip line outlet. Soil media was Sunshine mix #1 and sand (Quickcrete Medium - Lowes) mixed 1:1 (by volume) and measured volumetrically into pots with a triple layer of newspaper in the bottom of each pot to prevent media loss. Solanum, Maize and Oryza were planted in large pots (2.5 gal, ~ 9.5 L) and received 200 mL of water twice per day, while Triphysaria (controls), Arabidopsis, Medicago and Juncus were planted in small pots (1 gal, ~ 3.8 L) and received 100 mL of water twice per day. The watering regime was calibrated to through-water pots with excess water to maintain high levels of soil moisture. 5 mL granular Osmocote Plus (15–9-12) was added to each irrigated pot and sticky cards were used to control insect pests during the experiment.
Planting timeline
After germination (time on plates: Juncus 17d, Medicago and Maize 8d, Arabidopsis 7d, Solanum and Oryza - 6d) host plants were transplanted into soil media (see above) 2 weeks prior to co-culture and grown in a growth chamber (21 °C, 16/8 light/dark cycle) for 1 week (flats with domes for 3 days, with “cracked” domes for 4 days). 1 week prior to transplant to experimental pots, host plants were hardened off in the greenhouse where the experiment took place (3 days with “cracked” domes, 4 days without domes). ~ 6000 Triphysaria were germinated and grown in tandem batches at 16 °C ~ 25–35 days prior (Triphysaria germination is not highly synchronous) to co-culture. 1 day after host plants were added to experimental pots, germinated Triphysaria were transplanted serially (i.e. a researcher transplanted a suitable parasitic plant (defined as ≥1 cm root and first true leaves open) in one pot, then moved to the next pot, until the usable parasitic plants were exhausted). Seven Triphysaria plants were added to each host pot ~ 25 cm from the host stem, in a circle, at intervals of roughly 50° (~ 2.5 cm spacing) and Triphysaria only pots were planted in the same configuration sans host in small pots (see above). After initial planting the watering regime was supplemented by extensive hand misting with DI water (as needed during the first week ~ 4–5 times daily) to help all plants establish in the greenhouse. Survivorship at week 2 was very high and not significantly different for any treatment, indicating successful establishment.
Data collection
The experiment was monitored daily. A survey of survivorship was taken at 1 week intervals. When Triphysaria die, they rapidly oxidize and wilt, so our definition of “survivor” was “upright Triphysaria with some green leaf tissue”. This definition could include dying individuals, though once senescence had begun, the plant was always counted as dead at the next week interval. On May 16, 2014, 5 weeks after the beginning of co-culture, all parasitic plants were harvested by cutting Triphysaria at the soil surface. All plants were photographed (Nikon D80) and then numbered and bagged for drying. Plants were dried for 48 h at 65 °C, at which time each Triphysaria was weighed, replaced into the bag, and sealed in an airtight storage container. Plant height was calculated using ImageJ (http://imagej.nih.gov/). Each photograph was analyzed manually by measuring the length, in pixels, of 1 cm on the reference scale bar. These values were averaged, and this value was used to calibrate (86.05 ± 0.12 pixels/cm) the measurement function in ImageJ. The calibrated measurement tool was then used to measure the length from the cut plant (at soil surface) to the apex of each Triphysaria plant. To describe the “plump” phenotype observed in the experiment, we normalized the dry mass (in mg) of each Triphysaria by the height (in mm).
Data analysis
Statistical analysis was done using SAS PROC MIXED [50] as a randomized complete block design with block as a random effect. Dunnett’s test [51] for differences between the treatments (hosts) and the control (no host) was done for each response to control for multiple testing. The unequal allocation of replicates was done to optimize the power of Dunnett’s test. Each of the response variables was analyzed separately.
Co-culture with Solanum lycopersicum and sub-irrigation of Triphysaria
Seed germination
Solanum lycopersicum (Heinz 57) and Triphysaria versicolor seeds were obtained and germinated as described above.
Experiment layout and watering
A sub-irrigation co-culture experiment was conducted in the College of Agricultural Sciences Greenhouse #85 at Penn State University in from September 16, 2014 to December 14, 2014. Treatments (10 each of Solanum, Solanum + Triphysaria, Triphysaria only, soil only control, for a total of 40 pots) were randomly arranged (randomizer.org) and placed in two columns of 20. A drip irrigation system like that described above was used to water each large pot (2.5 gal, 4 L soil media) except that only one timer was used to regulate water to the whole experiment and calibrated to deliver 250 mL of water twice daily to each pot. These irrigated pots were placed at the high end of slightly inclined trays. One small pot (300 ml) with 7 Triphysaria only were placed one each at the low end of the inclined trays (that also contained the large pot) so as to be sub-irrigated with the flow-through of the larger pot. A small drainage port in each tray allowed for excess flow-through water to be drained away avoiding water logging of these smaller pots.
Planting timeline
Solanum and Triphysaria were germinated and grown prior to co-culture as described above. Co-culture and control pots were set up as described above. During the first week, watering was calibrated up (from 200 to 250 mL) to deliver sufficient water to sub-irrigated pots. During this optimization phase, dead Triphysaria were replaced as needed. Supplemental lighting from overhead sodium vapor lamps was supplied from 8 am to 8 pm if light intensity dropped below 200μE. After the watering regime was established, 5 mL granular Osmocote Plus (15–9-12) was added to each top irrigated pot.
Data collection
The experiment was monitored daily and data was collected as described above with the following exception: the duration of the experiment was ~ 8 weeks (59 days) instead of 5 weeks.
Data analysis
Statistical analysis was done using SAS PROC MIXED [50] taking into account the pairing of the large and small pots. Tukey’s test for all pairwise comparisons was applied for each response to control for multiple testing.
Non-destructive, image-based analysis
Image acquisition
Images were taken with a D80 Nikon camera on a tripod at a fixed distance from the imaging stage, with fixed focus. Auto exposure was used to compensate for changing light conditions throughout the day in the greenhouse. The plants were processed iteratively through each block. Because each block was randomized the effect of dynamic lighting conditions would affect treatments randomly. The imaging stage had a dark felt background and metric/SAE rulers were fixed to the stage on both X and Y axes.
Image segmentation and processing to estimate perimeter:area ratio
All image segmentation was performed using ImageJ (https://imagej.nih.gov/ij/). Prior to segmentation the RGB images were either broken into separate channels or transformed into other color spaces for easier analysis: HSV, HSI, or CMYK. Once a particular channel was chosen the image was thresholded to capture the plant ROIs. The ROI could then be cleaned using an additional thresholding to capture the background within and outside the plant; this created a more clearly defined edge. From the refined ROI, the Particle Analyzer function was used to measure the area and the perimeter of the plant. The raw RGB data from the ROI was also obtained for the purpose of normalization against a white reference. The raw RGB data from the white reference was sampled and normalized using the grey world method [52].
Color normalization was performed using both the white reference (ruler) and plant ROIs by converting raw RGB values to XYZ, where the white reference was applied to normalize ROI values for chromatic adaptation using Bradford matrices [53–55]. Conversion back to RGB color space provides the corrected values needed for estimation of chlorophyll content through Green/Red ratio [33]. For the ImageJ macro, see Additional file 3.
Data analysis
Statistical analysis was done using SAS PROC MIXED [50] using the mean values for the surviving plants in each pot. To account for the different numbers of survivors, weighted analysis was done using the number of surviving plants as weights. Block was included as a random effect. Dunnett’s test [51] for multiple comparisons with a control was done to determine differences between the pots with hosts and the control. Unequal allocation of replicates was used to optimize the power of Dunnett’s test.
Gene expression analysis and function enrichment test of Triphysaria
Transcriptome analysis of Triphysaria versicolor (functional annotation and identification of differentially expressed (DE) genes in the parasite during free-living versus parasitic modes of growth) was reported previously [13]. The DE genes were subject to a Fisher’s exact test using GO slim terms for upregulated and down-regulated DE genes as the input table (fisher.test function in R), which identified enriched GO slim terms associated with upregulated or downregulated features for shoots, flowers, and roots; alpha was set to 0.05.
Additional files
Figure S1. All hosts were verified by direct observation of haustorium formation by Triphysaria. A) Arabidopsis, B) Juncus, C) Medicago, D) Oryza, E) Solanum, F) Zea. (TIFF 4874 kb)
Figure S2. Images of experimental apparati and planting scheme. A) Seven Triphysaria were planted around each host equidistant from each other and the host plant. For control pots, the arrangement was identical, except without host plants. B) the watering control system. (TIFF 6785 kb)
Archive containing the ImageJ macro. (IJM 2 kb)
Acknowledgements
We thank Scott DiLoretto for use of the Horticulture greenhouse for these experiments and Anthony Omeis for additional support in the Biology Greenhouses at Penn State University. We also thank the Tomato Genetic Resource Center at The University of California, Davis, for Heinz 57 seeds, Charles Anderson for seeds of Arabidopsis thaliana Col-0, and Emily Helliwell and Yinong Yang for Oryza sativa seeds.
Abbreviations
- HIF
Haustorium inducing factor
- PPGP
Parasitic Plant Genome Project
- ROI
Region of interest
Authors’ contributions
Conception and design of study LAH, CWD, NSA, JIY; greenhouse work, experimental setup, plant cultivation LAH, SJ, NF, WK, KV, HZ, JIY; data analysis and presentation LAH, SJ, NF, WK, KV, HZ, NSA, CWD; wrote manuscript LAH, CWD. All authors read and approved the final manuscript.
Funding
This research was supported by awards DBI-0701748 and IOS-1238057 to C.W.D. and J.H.Y., and by The United States Department of Agriculture's Agricultural Research Service.
Availability of data and materials
Transcriptome data is available at http://ppgp.huck.psu.edu.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Loren A. Honaas, Phone: (509)664-2280, Email: loren.honaas@ars.usda.gov
Sam Jones, Email: samueljones84@yahoo.com.
Nina Farrell, Email: nfarrell@broadinstitute.org.
William Kamerow, Email: wkamerow@gmail.com.
Huiting Zhang, Email: zhanght120@gmail.com.
Kathryn Vescio, Email: kvescio@umass.edu.
Naomi S. Altman, Email: nsa1@psu.edu
John I. Yoder, Email: jiyoder@ucdavis.edu
Claude W. dePamphilis, Email: cwd3@psu.edu
References
- 1.Bandaranayake Pradeepa C. G., Yoder John I. Parasitic Orobanchaceae. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. Haustorium Initiation and Early Development; pp. 61–74. [Google Scholar]
- 2.Goldwasser Y, Westwood J, Yoder J. The use of Arabidopsis to study interactions between parasitic angiosperms and their plant hosts. Arabidopsis Book. 2002;1:e0035. doi: 10.1199/tab.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkman Todd J, McNeal Joel R, Lim Seok-Hong, Coat Gwen, Croom Henrietta B, Young Nelson D, dePamphilis Claude W. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evolutionary Biology. 2007;7(1):248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellot S, Cusimano N, Luo SX, Sun GL, Zarre S, Groger A, Temsch E, Renner SS. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biol Evol. 2016;8(7):2214–2230. doi: 10.1093/gbe/evw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thurman LD. Genecological studies in Orthocarpus subgenus Triphysaria. Berkeley: University of California; 1966. [Google Scholar]
- 6.Estabrook E, Yoder J. Plant-plant communications: rhizosphere signaling between parasitic angiosperms and their hosts. Plant Physiol. 1998;116(1):1–7. doi: 10.1104/pp.116.1.1. [DOI] [Google Scholar]
- 7.Vurro M, Bonciani B, Vannacci G. Emerging infectious diseases of crop plants in developing countries: impact on agriculture and socio-economic consequences. Food Security. 2010;2(2):113–132. doi: 10.1007/s12571-010-0062-7. [DOI] [Google Scholar]
- 8.Rodenburg J, Riches CR, Kayeke JM. Addressing current and future problems of parasitic weeds in rice. Crop Prot. 2010;29(3):210–221. doi: 10.1016/j.cropro.2009.10.015. [DOI] [Google Scholar]
- 9.Ejeta G. Integrating new Technologies for Striga Control: towards ending the witch-hunt. Singapore: World Scientific Publishing Co; 2007. The Striga scourge in Africa: a growing pandemic. [Google Scholar]
- 10.Scholes JD, Press MC. Striga infestation of cereal crops - an unsolved problem in resource limited agriculture. Curr Opin Plant Biol. 2008;11(2):180–186. doi: 10.1016/j.pbi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15(4):227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Olmstead RG, DePamphilis CW, Wolfe AD, Young ND, Elisons WJ, Reeves PA. Disintegration of the Scrophulariaceae. Am J Bot. 2001;88(2):348–361. doi: 10.2307/2657024. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PC, Wu B, Der JP, et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol. 2015;32(3):767–790. doi: 10.1093/molbev/msu343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida S, Cui S, Ichihashi Y, Shirasu K. The Haustorium, a specialized invasive organ in parasitic plants. Annu Rev Plant Biol. 2016;67:643–667. doi: 10.1146/annurev-arplant-043015-111702. [DOI] [PubMed] [Google Scholar]
- 15.Cook C, Whichard LP, Wall M, Egley GH, Coggon P, Luhan PA, McPhail A. Germination stimulants. II. Structure of strigol, a potent seed germination stimulant for witchweed (Striga lutea) J Am Chem Soc. 1972;94(17):6198–6199. doi: 10.1021/ja00772a048. [DOI] [Google Scholar]
- 16.Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 17.Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- 18.Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. 2015;349(6247):540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- 19.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 20.Bouwmeester HJ, Roux C, Lopez-Raez JA, Becard G. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 2007;12(5):224–230. doi: 10.1016/j.tplants.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Yoder JI, Scholes JD. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr Opin Plant Biol. 2010;13(4):478–484. doi: 10.1016/j.pbi.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Irving LJ, Cameron DD. Advances in Botanical Research, Vol 50. Vol. 50. 2009. You are what you eat: interactions between root parasitic plants and their hosts; pp. 87–138. [Google Scholar]
- 23.Spallek T, Mutuku M, Shirasu K. The genus Striga: a witch profile. Mol Plant Pathol. 2013;14(9):861–869. doi: 10.1111/mpp.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lendzemo VW, Kuyper TW, Matusova R, Bouwmeester HJ, Van Ast A. Colonization by arbuscular mycorrhizal Fungi of Sorghum leads to reduced germination and subsequent attachment and emergence of Striga hermonthica. Plant Signal Behav. 2007;2(1):58–62. doi: 10.4161/psb.2.1.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoder JI. Host-plant recognition by parasitic Scrophulariaceae. Curr Opin Plant Biol. 2001;4(4):359–365. doi: 10.1016/S1369-5266(00)00185-0. [DOI] [PubMed] [Google Scholar]
- 26.Bandaranayake PC, Filappova T, Tomilov A, Tomilova NB, Jamison-McClung D, Ngo Q, Inoue K, Yoder JI. A single-electron reducing quinone oxidoreductase is necessary to induce haustorium development in the root parasitic plant Triphysaria. Plant Cell. 2010;22(4):1404–1419. doi: 10.1105/tpc.110.074831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandaranayake PCG, Tomilov A, Tomilova NB, Ngo QA, Wickett N, dePamphilis CW, Yoder JI. The TvPirin gene is necessary for Haustorium development in the parasitic plant Triphysaria versicolor. Plant Physiol. 2012;158(2):1046–1053. doi: 10.1104/pp.111.186858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo QA, Albrecht H, Tsuchimatsu T, Grossniklaus U. The differentially regulated genes TvQR1 and TvPirin of the parasitic plant Triphysaria exhibit distinctive natural allelic diversity. BMC Plant Biol. 2013;13:28. doi: 10.1186/1471-2229-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matusova R, Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 2005;139(2):920–934. doi: 10.1104/pp.105.061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoder JI. A species-specific recognition system directs haustorium development in the parasitic plant Triphysaria (Scrophulariaceae) Planta. 1997;202(4):407–413. doi: 10.1007/s004250050144. [DOI] [PubMed] [Google Scholar]
- 31.Atsatt PR, Guldberg LD. Host influence on floral variability in Orthocarpus densiflorus (Scrophulariaceae) Plant Syst Evol. 1978;129(3):167–176. doi: 10.1007/BF00990758. [DOI] [Google Scholar]
- 32.Atsatt PR. The population biology of closely related species of Orthocarpus. PhD thesis. Los Angeles: University of California; 1966.
- 33.Adamsen FJ, Pinter PJ, Barnes EM, LaMorte RL, Wall GW, Leavitt SW, Kimball BA. Measuring wheat senescence with a digital camera. Crop Sci. 1999;39(3):719–724. doi: 10.2135/cropsci1999.0011183X003900030019x. [DOI] [Google Scholar]
- 34.Jamison DS, Yoder JI. Heritable variation in quinone-induced haustorium development in the parasitic plant Triphysaria. Plant Physiol. 2001;125(4):1870–1879. doi: 10.1104/pp.125.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westwood JH, dePamphilis CW, Das M, Fernández-Aparicio M, Honaas LA, Timko MP, Wafula EK, Wickett NJ, Yoder JI. The parasitic plant genome project: new tools for understanding the biology of Orobanche and Striga. Weed Sci. 2012;60(2):295–306. doi: 10.1614/WS-D-11-00113.1. [DOI] [Google Scholar]
- 36.Atsatt PR, Strong DR. The population biology of annual grassland Hemiparasites. I The Host Environment. Evolution. 1970;24(2):278–291. doi: 10.1111/j.1558-5646.1970.tb01761.x. [DOI] [PubMed] [Google Scholar]
- 37.Werth Charles R., Riopel James L. A Study of the Host Range of Aureolaria pedicularia (L.) Raf. (Scrophulariaceae) American Midland Naturalist. 1979;102(2):300. doi: 10.2307/2424657. [DOI] [Google Scholar]
- 38.Gibson CC, Watkinson AR. Host selectivity and the mediation of competition by the root hemiparasite Rhinanthus minor. Oecologia. 1991;86(1):81–87. doi: 10.1007/BF00317393. [DOI] [PubMed] [Google Scholar]
- 39.Shen H, Ye W, Hong L, Huang H, Wang Z, Deng X, Yang Q, Xu Z. Progress in parasitic plant biology: host selection and nutrient transfer. Plant Biol (Stuttg) 2006;8(2):175–185. doi: 10.1055/s-2006-923796. [DOI] [PubMed] [Google Scholar]
- 40.Runyon JB, Mescher MC, De Moraes CM. Volatile chemical cues guide host location and host selection by parasitic plants. Science (New York, NY) 2006;313(5795):1964–1967. doi: 10.1126/science.1131371. [DOI] [PubMed] [Google Scholar]
- 41.Henning S. Heide-Jørgensen: introduction: the parasitic syndrome in higher plants. In: Joel DM, Gressel J, Musselman LJ, editors. Parasitic Orobanchaceae: parasitic mechanisms and control strategies. Berlin: Springer; 2013. pp. 1–14. [Google Scholar]
- 42.Press MC, Smith S, Stewart GR. Carbon acquisition and assimilation in parasitic plants. Funct Ecol. 1991;5(2):278–283. doi: 10.2307/2389265. [DOI] [Google Scholar]
- 43.Tuohy J, Smith EA, Stewart GR: The parasitic habit: trends in morphological and ultrastructural reductionism. In Proceedings of Biology and Control of Orobanche: 13–17 January 1986; Wageningen, Netherlands. Edited by S. J. ter Borg; pp. 86–95.
- 44.Shah N, Smirnoff N, Stewart GR. Photosynthesis and stomatal characteristics of Striga hermonthica in relation to its parasitic habit. Physiol Plantarum. 1987;69(4):699–703. doi: 10.1111/j.1399-3054.1987.tb01987.x. [DOI] [Google Scholar]
- 45.Press MC, Tuohy JM, Stewart GR. Gas exchange characteristics of the sorghum-striga host-parasite association. Plant Physiol. 1987;84(3):814–819. doi: 10.1104/pp.84.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dener E, Kacelnik A, Shemesh H. Pea plants show risk sensitivity. Curr Biol. 2016;26(13):1763–1767. doi: 10.1016/j.cub.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Atsatt PR. Hemiparasitic flowering plants: phenotypic canalization by hosts. Nature. 1970;225(5238):1161–1163. doi: 10.1038/2251161b0. [DOI] [PubMed] [Google Scholar]
- 48.Kacelnik A, Teson M. Risky theories—the effects of variance on foraging decisions. Integr Comp Biol. 1996;36(4):402–434. [Google Scholar]
- 49.Honaas LA, Wafula EK, Yang Z, Der JP, Wickett NJ, Altman NS, Taylor CG, Yoder JI, Timko MP, Westwood JH, et al. Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 2013;13(9):9. doi: 10.1186/1471-2229-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Institute S . SAS/STAT 9.1 User's Guide the Reg Procedure:(Book Excerpt): SAS Institute. 2008. [Google Scholar]
- 51.Dunnett CW. New tables for multiple comparisons with a control. Biometrics. 1964;20(3):482–491. doi: 10.2307/2528490. [DOI] [Google Scholar]
- 52.Finlayson Graham D., Schiele Bernt, Crowley James L. Computer Vision — ECCV'98. Berlin, Heidelberg: Springer Berlin Heidelberg; 1998. Comprehensive colour image normalization; pp. 475–490. [Google Scholar]
- 53.Hunt R. Chromatic adaptation transforms and a colour inconstancy index. The Reproduction of Colour. 6th ed. 2004; p. 587–95.
- 54.Tooms MS. Colour reproduction in electronic imaging systems: photography, television, cinematography. Chichester: Wiley; 2016.
- 55.Luo M, Hunt R. A chromatic adaptation transform and a colour inconstancy index. Color Res Appl. 1998;23(3):154–158. doi: 10.1002/(SICI)1520-6378(199806)23:3<154::AID-COL7>3.0.CO;2-P. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. All hosts were verified by direct observation of haustorium formation by Triphysaria. A) Arabidopsis, B) Juncus, C) Medicago, D) Oryza, E) Solanum, F) Zea. (TIFF 4874 kb)
Figure S2. Images of experimental apparati and planting scheme. A) Seven Triphysaria were planted around each host equidistant from each other and the host plant. For control pots, the arrangement was identical, except without host plants. B) the watering control system. (TIFF 6785 kb)
Archive containing the ImageJ macro. (IJM 2 kb)
Data Availability Statement
Transcriptome data is available at http://ppgp.huck.psu.edu.