Abstract
Background:
Na+/K+ ATPase enzyme is essential for nerve cell membrane integrity, and reduction in its activity, probably due to ATP1A1 gene polymorphisms, is related to diabetic neuropathy progression. Therefore, the goal of the existent study is to evaluate the Na+/K+ ATPase activity in type 2 diabetes mellitus (T2DM) Egyptian patients with or without neuropathy, search for polymorphism(s) in the highly polymorphic region of ATP1A1 gene, exon 2, and study its (their) associations with T2DM with and without neuropathy.
Materials and Methods:
A total number of 150 individuals were subclassified into healthy controls (n = 30), T2DM without complications (n = 60), and T2DM with neuropathy (n = 60).
Results:
The biochemical results exhibited a significant reduction in fasting C-Peptide and activity of Na+/K+ ATPase in T2DM patients with neuropathy followed by T2DM without complication in comparison with healthy controls. ATP1A1 exon2 was amplified by polymerase chain reaction (PCR) then digested by the PstI restriction enzyme, and the obtained data from restriction fragment length polymorphism-PCR and sequencing revealed the existence of a novel synonymous G94A single nucleotide polymorphism (SNP) at nucleotide 27 in exon 2 of ATP1A1 gene (rs1060366). Diabetic groups had only allele A, while the control group had G allele. Interestingly, individuals carrying AA genotype had a significantly lower Na+/K+ ATPase, C-peptide, and higher glycosylated hemoglobin (HBA1c %) than those having GG genotype, suggesting a possible association for this SNP, and this developed phenomenon of not only T2DM but also diabetic neuropathy.
Conclusion:
Thus, allele A of G94A SNP (rs1060366) could be a risk allele for diabetes susceptibility among Egyptian patients.
Keywords: C-peptide, diabetes mellitus, diabetic neuropathies, polymorphism, sodium-potassium-exchanging ATPase
INTRODUCTION
Type 2 diabetes mellitus (T2DM), as a most prevalent form of diabetes which accounts for about 90%–95%, is associated with abnormalities in carbohydrate, lipid, and protein metabolism. Chronic exposure to hyperglycemia can result in dysfunction and defeat of various organs chiefly the eyes (retinopathy), kidneys (nephropathy), nerves (neuropathy), and heart and blood vessels (macrovascular diseases).[1,2] Diabetic peripheral neuropathy is the main cause of disability in patients due to diffuse or focal damage to peripheral somatic or autonomic nervous system.[3] This complication is mainly caused due to the disrupted effect of DM on the membrane structure of protein, organization, and enzymatic activities. Among these membrane enzymes, Na+/K+ ATPase (EC 3.6.3.9) stimulates the Na+ and K+ transport through the cell membrane using adenosine triphosphate (ATP) as an energy source[4] that showed the most noticeable distorted effects and impaired function.[5] Although excessive oxygen-free radicals’ production, glycated proteins’ formation, and disruption in the metabolism of nerve growth factor can reduce the activity of Na+/K+ ATPase, the specific mechanism by which DM inhibits the enzyme activity remains unclear.
The human ATPase gene contains several α subunits and their related genes. Each α subunits is 20–25 kb in length. α1, α2, and α4 are encoded by ATP1A1, ATP1A2, and ATP1AL2 genes, respectively that localized on chromosome 1. However, α3 is encoded by ATP1A3 gene that localized on chromosome 19.[6] Among these genes, ATP1A1 gene is robustly expressed in peripheral nerves and red blood cells, and its polymorphisms were reported to be associated with T1DM patients with neuropathy.[7,8] The single nucleotide polymorphism (SNP) was detected in intron 1 of ATP1A1 by restriction fragment length polymorphism (RFLP) using Bgl II enzyme[7] in Caucasian patients with >10 years of T1DM.[8] In T2DM patients, a PstI SNP was detected in exon 2 of ATP1A1 in Chinese,[9] and a Bgl II SNP was discovered in intron 1 of ATP1A1 in Turkish population.[10] Of these two SNPs, only PstI SNP was related with the lower level of the activity of Na+/K+ ATPase in T2DM neuropathy. Other than Chinese, no other population was checked for the SNP presence or not and its association with neuropathy. Therefore, the purpose of this study was to assess the Na+/K+ ATPase activity in T2DM Egyptian patients with or without neuropathy, search for polymorphism(s) in exon 2 of ATP1A1 gene, and study its (their) associations with T2DM.
MATERIALS AND METHODS
Subjects
This randomized study included 150 individuals who were subclassified into 30 healthy controls without DM (normal fasting blood glucose [FBG] and postprandial blood glucose [PBG]), 60 T2DM without complications, and 60 T2DM with neuropathy. The selected T2DM patients from Diabetes and Endocrinology Unit, Department of Internal Medicine, Hospital of Tanta University, Egypt were conducted and approved by the Local Ethical Committee (FWA00006444). T2DM was diagnosed according to the criteria of American Diabetes Association.[11] The criteria of neuropathic patients were detected by the Diabetes Control and Complications Trial Research Group.[12] The neuropathic T2DM patients with at least two of the following three criteria were subjected in the study: abnormal motor and sensorial system findings; symptoms of peripheral neuropathy; and abolished deep nerve reflexes. Patients with following symptoms as uncontrolled hypertension, congestive heart failure, thyroid and parathyroid disorders, liver or kidney diseases, hematological diseases, neuromuscular diseases, exposure to neurotoxins, and lumbar or cervical discopathy were all excluded from the study. The full history of patients was an emphasis on the duration of diabetes, smoking habits, body mass index (BMI), and history of any other associated disease and therapeutic history.
Biochemical parameters
Venous blood was drawn from each individual in the morning after an overnight fast; 0.5 ml blood was collected in the test tube containing sodium fluoride for the determination of FBG and 2 ml blood were collected in a dry centrifuge tubes, allowed to clot then centrifuged, and the obtained serum was utilized for estimation of lipid profile and fasting C-peptide level. A volume of 2 ml blood was collected in EDTA tube for determination of glycosylated hemoglobin (HbA1c) and ATP1A1 gene polymorphism. A volume of 5 ml blood was collected in sodium-citrated tube for determination of Na+/K+ ATPase activity. A volume of 0.5 ml blood was collected 2 h after breakfast in sodium fluoride tube for determination of PBG.
FBG and PBG were estimated using commercial kit that was supplied by Spinreact, Egypt. Glycosylated hemoglobin in whole blood was assayed using the NycoCard READER® supplied by Axis-Shield, Oslo, Norway. Lipid profile was determined using commercial kit supplied by Human® company, Egypt. Fasting C-peptide was measured using human C-peptide enzyme-linked immunosorbent assay kit (immunospec, Cat# E29-071) with detection limit 0.03 ng/ml, and inter- and intra-assay coefficient of variations were <4.75% and <8.72%, respectively.
Extraction of erythrocyte membranes
The hemoglobin free ghost membranes were obtained according to the method of Dodge et al.[13] All samples were centrifuged at 5500 g for 6 min. After the separation of buffy coat, the rest of the samples were washed twice with isotonic NaCl. After each washing, the supernatant was extracted. Final cells were lysed by the 5 mmol/L, ice-cold, hypotonic phosphate buffer, pH 8.0, and centrifugated at 4°C for 40 min with 16000 g till white pinky precipitate. Finally, the extracted ghost membranes were suspended in tris buffer and kept at −80°C until the time of analysis. Then protein content was measured as previously described by Lowry et al.[14] The erythrocyte membrane protein content suspensions were expressed as mg protein/ml.
The erythrocyte membrane Na+/K+ ATPase activity was measured according to the method described by Kitao and Hattori.[15] The membranes were incubated at 37°C with buffer pH 7.7. ATPase reaction was started by the addition of 3 mmol/L Na2ATP and after 20 min stopped with 1 mL of 15% TCA. After 5 min of centrifuging at 6000 g, supernatant was extracted, and 3 mL 0.1 N Na acetate, 0.4 mL molybdate/H2SO4 mixture (1:1), and 0.4 mL of 1% ascorbic acid were added; after 20 min, absorbance was measured at 800 nm. All samples were duplicated, with and without 1 mmol/L ouabain. The difference between inorganic phosphates in two calculations was defined as the Na+/K+ ATPase activity (nmole Pi/mg protein/h).
Restriction fragment length polymorphism-polymerase chain reaction (RFLP-PCR)
(RFLP PCR) was performed as the following steps: DNA extraction, PCR, and digestion by restriction enzyme and gel electrophoresis.[16] The genomic DNA was extracted using Gene JET genomic DNA extraction kit (Fermentas, #K0721) depends on digestion of the blood samples with proteinase K in either the supplied digestion or lysis solution. The lysate was then mixed with ethanol and loaded on the purification column where the DNA binds to the silica membrane. Impurities were effectively removed by washing the column with the prepared wash buffers. Genomic DNA was then eluted under low ionic strength conditions with the elution buffer. Concentration and purity of DNA were measured by Nanodrop (ultraviolet–visible spectrophotometer Q5000/USA). The ATP1A1 gene was amplified using a forward primer 5’ TCCAGAATTTTCAGTTTCAG 3’ and a reverse primer 5’ AGATGAGATCTGTAC AGCTG 3’ which was designed by Zhang et al.[9] and confirmed by Primer3 software on the published human sequence in GenBank databases. To ensure the sequencing primer is unique for the template sequence, we checked up for the similarity to other known sequences with basic local alignment search tool (BLAST) (www.ncbi.nlm.nih.gov/blast/Blast.cgi). PCR was obtained in 50 μl-containing genomic DNA (5–20 ng/μl), primers (0.1–0.5 μM), PCR Master Mix, and nuclease-free water. The final reaction mixture was placed in a Techne thermal cycler (TC-3000, USA). The PCR was programmed under the following conditions: initial denaturation at 95°C for 5 min followed by 35 cycles of 95°C for 45 s for DNA denaturation, annealing temperatures (Ta) 55°C for 45 s, extension at 72°C for 1 min, and a final extension at 72°C for 7 min. The amplified DNA segment of the ATP1A1 gene was digested with PstI restriction enzyme (ThermoScientific) at 37 C° for 16 h, and the cleaved fragments were detected by agarose gel electrophoresis, and then, the visualization of fragment patterns was obtained under UV in gel documentation system.
Gene sequencing
The purified products of PCR were performed to MacroGen Company (South Korea) to be sequenced in both directions using ABI 3730XL-DNA sequencer (Applied Biosystem, USA). The sequences were analyzed using the Chromas Lite 2.1 program (http://technelysium.com.au/?page_id=13), and the identity of the sequenced PCR product was examined using Blast search against GenBank database of Homo sapiens ATP1A1 (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The alignments, annotations, and assembly of the sequences were applied by using Geneious 4.8.4 software http://www.geneious.com/web/geneious/home.
Data analysis
All values resulted as a mean ± standard error of the mean and then compared by GraphPad Prism version 7.00 for Windows, GraphPad Software, La Jolla California USA. The differences between the two groups were analyzed with Student's t-test, and the differences between the means of all the groups were analyzed with one-way ANOVA test. Levels of P < 0.05 were expressed as significant. The correlations between Na+/K+ ATPase activity and other were calculated by Pearson's correlation coefficient (r). Genotype frequencies were determined using the Chi-squared test.
RESULTS
A total of 150 participants were included in the current study. Males and females represented 42.66% and 57.33%, respectively, in the study. Our results exhibited a significant (P < 0.001) increase in diabetes duration, BMI, FBG, PBG, HbA1c %, total cholesterol level, triacylglycerol, and low-density lipoprotein cholesterol (LDL-C) level and a significant decrease in high-density lipoprotein cholesterol (HDL-C) level in diabetic groups, with remarkable changes in patients with neuropathy as compared to healthy control groups [Table 1]. On the other hand, fasting C-Peptide and Na+/K+ ATPase were significant decrease (P < 0.001) in diabetic neuropathic patients followed by T2DM without complication as compared to control groups [Table 1].
Table 1.
Demographic, clinical data, and biochemical parameters of the studied groups
| Group 1 Control (n=30) | Group 2 Diabetic without complication (n=60) | P | Group 3 Patients with diabetic neuropathy (n=60) | P* | |
|---|---|---|---|---|---|
| Age (years) | 42±0.83 | 50±1.06 | <0.0001 | 52±1.1 | 0.3379 |
| Sex (male/female) | 14/16 | 26/34 | NS | 24/36 | NS |
| Diabetes duration (years) | 5.5±0.3 | - | 7.2±0.35 | <0.001 | |
| BMI (kg/m2) | 23.54±0.28 | 32.8±0.35 | <0.0001 | 33±0.31 | 0.8910 |
| Smoking habits, n (%) | 6 (20) | 16 (26.7) | - | 18 (30) | - |
| FBG (mg/dl) | 88.6±1.1 | 163±1.13 | <0.0001 | 211±1.39 | <0.0001 |
| PBG (mg/dl) | 108.8±1.65 | 249±1.44 | <0.0001 | 299±1.48 | <0.0001 |
| HbA1c % | 5.6±0.14 | 7.0±0.1 | <0.0001 | 7.6±0.15 | 0.0020 |
| TC (mg/dl) | 170.7±4.2 | 211.5±3.3 | <0.0001 | 231±2.8 | <0.0001 |
| TGs (mg/dl) | 120.1±6 | 184.3±10.05 | 0.0002 | 203.8±9.5 | 0.2760 |
| HDL-C (mg/dl) | 67.2±1.3 | 52.2±0.64 | <0.0001 | 50.8±0.93 | 0.4539 |
| LDL-C (mg/dl) | 81.6±1.4 | 118.7±0.68 | <0.0001 | 138±0.89 | <0.0001 |
| Fasting C-peptide (ng/ml) | 3.2±0.17 | 2.2±0.06 | <0.0001 | 1.3±0.08 | <0.0001 |
| Na+/K+ ATPase activity (nmol Pi/mg protein/h) | 423.2±10.6 | 318.8±3.5 | <0.0001 | 266.8±4.06 | <0.0001 |
Values are expressed as mean±SE. P value versus Group 1. *P value versus Group 2. BMI=Body mass index; FBG=Fasting blood glucose; PBG=Postprandial blood glucose; HbA1c=Glycosylated hemoglobin; HDL-C=High density lipoprotein-cholesterol; LDL-C=Low density lipoprotein-cholesterol; TC=Total cholesterol; TG=Triacylglycerol; NS=Not significant; SE=Standard error
Na+/K+ ATPase activity revealed a significant positive correlation with fasting C-peptide (r = 0.379, P < 0.001), HDL-C with r = 0.371 and P < 0.001, respectively, and a significant negative correlation with FBG (r = −0.209, P < 0.05), PBG (r = −0.276, P < 0.01), HbA1c (r = −0.333, P < 0.01), total cholesterol (r = −0.328, P < 0.01), triacylglycerol (r = −0.256, P < 0.05), and LDL-C (r = −0.388, P < 0.001). However, Na+/K+ ATPase activity exhibited an insignificantly negative (P > 0.05) correlation with Diabetes duration (r = −0.037, P > 0.05) and age with r = −0.174 and P > 0.05, respectively [Table 2].
Table 2.
Correlation between Na+/K+ ATPase activity and biochemical variables
| Parameters | Na+/K+ ATPase activity (nmol Pi/mg protein/h) in diabetic patients (n=120) | |
|---|---|---|
| Correlation coefficient (r) | P | |
| Age (years) | −0.174 | 0.102 |
| BMI (kg/m2) | −0.099 | 0.353 |
| Diabetes duration (years) | −0.037 | 0.725 |
| FBG (mg/dl) | −0.209 | <0.05 |
| PBG (mg/dl) | −0.276 | <0.01 |
| HbA1c (%) | −0.333 | <0.01 |
| Fasting C-peptide (ng/ml) | 0.379 | <0.001 |
| TC (mg/dl) | −0.328 | <0.01 |
| TG (mg/dl) | −0.256 | <0.05 |
| HDL-C (mg/dl) | 0.371 | <0.001 |
| LDL-C (mg/dl) | −0.388 | <0.001 |
BMI=Body mass index; FBG=Fasting blood glucose; PBG=Postprandial blood glucose; HbA1c=Glycosylated hemoglobin; HDL-C=High density lipoprotein-cholesterol; LDL-C=Low density lipoprotein-cholesterol; TC=Total cholesterol; TG=Triacylglycerol
A fragment of ATP1A1 gene (311 bp) was digested with PstI restriction enzyme into 217 and 94 bp fragments in all groups except control group [Figure 1a and b]. Lack of digestion in this group may indicate the presence of a novel SNP at the restriction site of PstI. To confirm this result, gene sequencing was applied, and the obtained results were submitted to GenBank database with accession number MG053109. After analysis of this SNP based on Genome Reference Consortium Human Build 38 patch release 12 (GRCh38.p12), reference SNP is rs1060366. Analysis of these sequences revealed the presence of a synonymous G94A SNP at nucleotide 27 in exon 2 [equivalent to nucleotide 94 of PCR product, Figure 1c]. Diabetic groups had only A allele, while control had G allele of this SNP, odds ratio = 14701, 95% confidence interval 285.63–756,649, P < 0.0001. This means that A is the risk allele for diabetes and that of G is the protective allele.
Figure 1.
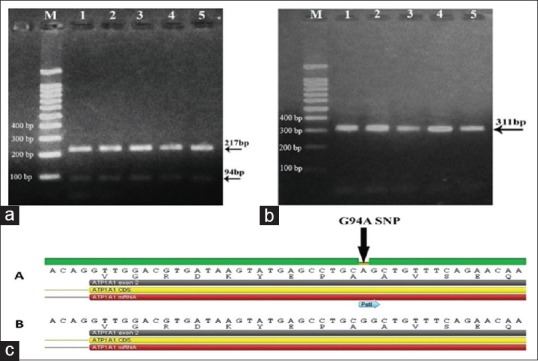
(a) PstI digestion of 5 ATP1A1 polymerase chain reaction products in diabetic groups shows digestion in all samples. M represents 1kb ladder. Lane 1 and 2; Group II (diabetic patients without complications). Lane 3, 4 and 5; Group III (patients with diabetic neuropathy). (b) PstI digestion of five ATP1A1 polymerase chain reaction products in control group shows no digestion. M represents 1kb ladder. (c) Gene sequencing for amplified ATP1A1 gene (311 bp) after purification. Synonymous G94A single nucleotide polymorphism. (A) diabetic groups and (B) control group (MG053109)
This SNP resulted in only two genotypes, AA and GG with frequencies of 100% in diabetic groups and 100% in control group, respectively [Table 3]. In addition, Chi-square (χ2) of genotype frequencies revealed a significant difference between the two diabetic groups and the control group (χ2 = 150, P < 0.0001).
Table 3.
Genotype frequencies and distribution in the studied groups
| Control (n=30) | Diabetic without complication (n=60) | Patients with diabetic neuropathy (n=60) | |
|---|---|---|---|
| Presence of risk allele (AA) | 0 | 60 | 60 |
| Protective allele (GG) | 30 | 0 | 0 |
χ2=150; P<0.0001
Table 4 shows the relation between the genotype distribution of G94A ATP1A1 gene and Na+/K+ ATPase, C-peptide, and HBA1c %. Individuals carrying the risk alleles AA genotype showed significant decline (P < 0.001) in Na+/K+ ATPase activity and C-peptide levels but elevating HBA1c % in comparison with individuals carrying GG genotype.
Table 4.
Biochemical parameters among studied subjects according to their genotypes
| GG genotype 30/150 | AA genotype 120/150 | P | |
|---|---|---|---|
| Na+/K+ ATPase (nmol Pi/mg protein/h) | 423.2±10.6 | 319.94±1.9 | <0.001 |
| Fasting C-peptide (ng/ml) | 3.2±0.17 | 1.38±0.03 | <0.001 |
| HBA1c % | 5.6±0.14 | 7.76±0.08 | <0.001 |
HbA1c=Glycosylated hemoglobin
DISCUSSION
The study detected the presence of a significant decrease in fasting C-peptide, the activity of Na+/K+ ATPase in diabetic patients with or without neuropathy as compared to healthy controls. We detected a novel PstI SNP in ATP1A1 gene of Egyptian individuals differ from that detected in Chinese individuals and studied its relation with diabetes susceptibility.
In the current study, Na+/K+ ATPase activity was significantly decreased in T2DM groups than control (P < 0.001) as we described in previous study.[17] Parallel results were reported by Koc et al.[18] and Zhang et al.[9] In contrast, Vague et al.[6] and Humayoun et al.[5] showed no significant change in Na+/K+ ATPase activity between T2DM patients and healthy controls. Among the two diabetic groups, diabetic neuropathy group showed the lower activity of Na+/K+ ATPase than diabetic without neuropathy (P < 0.001). These results are consistent with Koc et al.[18] and Zhang et al.[9] As while the inhibition mechanism of DM to the activity of these two enzymes have not been elucidated yet, our data along with reports published by others speculate that hyperglycemia causes the increment levels of nervous intracellular glucose that leading to normal glycolytic pathway saturation.[19] The shunted glucose level that entered into the polyol pathway and converted to sorbitol and fructose occurred by aldose reductase and sorbitol dehydrogenase enzymes. Meanwhile, aldose reductase enzyme requires NADPH. Reduction of the NADPH decreases the nitric oxide synthase amount so that the levels of nitric oxide are decreased, diminishing blood flow.[20] Decreased NADPH also reduces the level of myo-inositol. Moreover, the accumulated sorbitol and fructose reduce nerve myo-inositol. Finally, the reduction of myo-inositol levels result in decreased Na+/K+ ATPase activity.[18] Another possible cause for reduction of Na+/K+ ATPase activity is the oxidative stress triggered by DM. Increased oxidative stress causes peroxidation of lipid in the cell membrane, and subsequently, Na+/K+ ATPase activity declines.[21] Taken together decreased Na+/K+ ATPase activity either due to lower levels of myo-inositol or higher oxidative stress, induces accumulation of sodium ions in the axons, thereby hindering the depolarization of the neural membrane leading to a decrease in nerve conduction velocity.[9,18]
Moreover, C-peptide and insulin are secreted together into the blood circulation due to absorption of intestinal glucose; insulin insufficiency in diabetic patients is related to a low C-peptide level. Several studies shed light on physiologic insulin and C-peptide that are necessary for restore activity of Na+/K+ ATPase.[22] It was revealed that C-peptide stimulates directly Na+/K+ ATPase activity in renal tubule cells.[23] Several arguments highlighted the biological effects of C-peptide.[6] Furthermore, C-peptide administration improves parameters that altered in diabetic complications. In addition, C-peptide increases oxygen uptake, blood flow, stimulates glucose transport in skeletal muscle, improves vascular dysfunction and autonomic or sensory nerve dysfunction.[24] In the existent study, fasting C-Peptide was significantly decreasing in diabetics as compared with controls (P < 0.001) and reached the lowest level in diabetic neuropathic patients. In addition, fasting C-peptide showed a positive significant correlation with Na+/K+ ATPase. These results are in consistent with De La Tour et al.[22] and Vague et al.[6] who observed a positively significant correlation between fasting C-Peptide with Na+/K+ ATPase activity among T2DM patients. Moreover, Djemli-Shipkolye et al.[25] suggested that physiological infusion of C-peptide could be useful for the prohibition of diabetic complications. In contrast, Koc et al.[18] observed no correlation between activity of Na+/K+ ATPase and fasting C-Peptide, BMI, HbA1c, serum glucose, LDL cholesterol, HDL cholesterol, or triglycerides in the diabetic patients’ groups.
Interestingly, we shed light on the key role of Na+/K+ ATPase activity in prevention, and treatment of diabetic neuropathy by restoring its activity in agreement with Iannello et al.[26] point out the beneficial effect of C-peptide infusion with insulin for prevention of diabetic neuropathy. Moreover, Akbulut et al.[27] suggested that erythropoietin therapy will restore neuropathy due to increase in Na+/K+ ATPase activity. Furthermore, oral supplementation of curcumin increases Na+/K+ ATPase activity and it may help to explain some of the biological effects of curcumin in prevention of diabetic neuropathy.[28] Recently prevention role of coenzyme Q10 in DM may be involving in prevention of diabetic neuropathy by reduction of oxidative stress leading to increasing Na+/K+ ATPase activity.[29]
The majority of DM patients have reduced Na+/K+ ATPase activity (mainly due to metabolic change in myo-inositol or higher oxidative stress); however, not all DM have diabetic neuropathy. Therefore, gene mutation, rather than metabolic disorder, may be the key to the occurrence of diabetic neuropathy (genetic susceptibility). This prompts us to check the association between ATP1A1 SNP and Na+/K+ ATPase activity and occurrence of diabetic neuropathy. To achieve this goal, we first search for the presence of the previously discovered PstI SNP among Egyptian population. Indeed, we found a synonymous G94A SNP at nucleotide 27 in exon 2 (equivalent to nucleotide 94 of PCR product) as revealed by PstI digestion which gave 217 and 94 bp fragments in diabetic groups only. The diabetic groups had only A (risk/restrictive) allele with AA genotype, while the control group had G (protective) allele with GG genotype. T2DM patients with or without neuropathy (with AA genotypes) were significantly different from controls (with GG genotypes) in genotype distribution (χ2 = 150, P < 0.0001). We did not find any significant association between G94A SNP and diabetic neuropathy as the two diabetic groups showed the same genotypic distribution. In contrast, Zhang et al.[9] found a lower incidence of A allele and AA genotype in T2DM with neuropathy than T2DM without neuropathy. Interestingly, in the present study, T2DM patients carrying A allele and AA genotype had a significantly lower Na+/K+ ATPase than those having G allele and GG genotype, suggesting a possible association for A allele and development of not only T2DM but also diabetic neuropathy. This means that A allele is a risk factor for T2DM patients in the studied Egyptian population. In contrast, Zhang et al.[9] found an association between A allele and increased Na+/K+ ATPase activity, indicating its role as a protective allele for diabetic neuropathy in a Chinese population. This difference in the association of A allele may be attributed to different ethnic groups (Egyptian vs. Chinese). In addition to Na+/k+ ATPase, we also found a similar association between this SNP and decreased C-Peptide level, and increased HBA1c %. The presence of a synonymous SNP, which does not change amino acids, does not indicate that the mutation has no effect on gene function because this SNP can modify mRNA stability, gene expression level, and protein conformation.[30] In the agreement, our results and those reported by Zhang et al.[9] demonstrated a significant association between the synonymous G94A SNP and T2DM.
CONCLUSION
In the present study, we found a significant decrease in C-peptide level and activity of Na+/K+ ATPase in diabetic patients without complications and those with neuropathy as compared to healthy controls, suggesting a role for ATPase in the development of T2DM and neuropathy. We also find an association between the risk allele A of G94A ATP1A1 gene (rs1060366) and T2DM. However, further confirmatory investigations on a large number population using genome-wide association studies and quantitative trait loci are required to validate whether this SNP is a positional marker for T2DM.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank to all patients that participated in the study.
REFERENCES
- 1.Zadhoush F, Sadeghi M, Pourfarzam M. Biochemical changes in blood of type 2 diabetes with and without metabolic syndrome and their association with metabolic syndrome components. J Res Med Sci. 2015;20:763–70. doi: 10.4103/1735-1995.168383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasom S, Komanasin N, Settasatian N, Settasatian C, Kukongviriyapan U, Intharapetch P, et al. Association of a disintegrin and metalloproteinase with a thrombospondin type 1 motif member 13 polymorphisms with severity of coronary stenosis in type 2 diabetes mellitus. J Res Med Sci. 2018;23:59. doi: 10.4103/jrms.JRMS_518_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Kishore L, Kaur N. Diabetic peripheral neuropathy: Current perspective and future directions. Pharmacol Res. 2014;80:21–35. doi: 10.1016/j.phrs.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Clausen MV, Hilbers F, Poulsen H. The structure and function of the Na, K-ATPase isoforms in health and disease. Front Physiol. 2017;8:371. doi: 10.3389/fphys.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humayoun M, Khalid A, Nizami NA. Association of decrease Na+/K+-ATPase activity with diabetes mellitus type 2. Pak J Med Health Sci. 2016;10:507–9. [Google Scholar]
- 6.Vague P, Coste TC, Jannot MF, Raccah D, Tsimaratos M. C-peptide, Na+, K(+)-ATPase, and diabetes. Exp Diabesity Res. 2004;5:37–50. doi: 10.1080/15438600490424514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shull MM, Pugh DG, Lingrel JB. The human Na, K-ATPase CWI gene: Characterization of the 5 ‘ -flanking region and identification of a restriction fragment length polymorphism. Genomics. 1990;460:451–60. doi: 10.1016/0888-7543(90)90475-a. [DOI] [PubMed] [Google Scholar]
- 8.Vague P, Dufayet D, Coste T, Moriscot C, Jannot MF, Raccah D. Association of diabetic neuropathy with Na/K ATPase gene polymorphism. Diabetologia. 1997;40:506–11. doi: 10.1007/s001250050708. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Bai R, Du J. Study on the association of Na/K ATPase activity and ATP1A1 gene polymorphism with type 2 diabetic peripheral neuropathy. Chin J Diabetes. 2007;4:10. [Google Scholar]
- 10.Topcu C, Gurbilek M, Akoz M, Cora T. The investigation of relationship between Na+/K+ATPase enzyme activity, ATP1A1 gene polymorphism and C-peptide in type 2 diabetic patients with neuropathy. Turk J Biochem. 2009;34:154–9. [Google Scholar]
- 11.American Diabetes Association 2. Classification and diagnosis of diabetes. Diabetes Care. 2016;39 Suppl 1:S13–22. doi: 10.2337/dc16-S005. [DOI] [PubMed] [Google Scholar]
- 12.The effect of intensive diabetes therapy on the development and progression of neuropathy. The diabetes control and complications trial research group. Ann Intern Med. 1995;122:561–8. doi: 10.7326/0003-4819-122-8-199504150-00001. [DOI] [PubMed] [Google Scholar]
- 13.Dodge JT, Mitchell C, Hanahan DJ. The preparation and chemical characteristics of hemoglobin-free ghosts of human erythrocytes. Arch Biochem Biophys. 1963;100:119–30. doi: 10.1016/0003-9861(63)90042-0. [DOI] [PubMed] [Google Scholar]
- 14.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 15.Kitao T, Hattori K. Inhibition of erythrocyte ATPase activity by aclacinomycin and reverse effects of ascorbate on ATPase activity. Experientia. 1983;39:1362–4. doi: 10.1007/BF01990105. [DOI] [PubMed] [Google Scholar]
- 16.Maleki F, Haghani K, Shokouhi S, Mahmoodi K, Sayehmiri K, Mahdieh N, et al. A case-control study on the association of common variants of CAPN10 gene and the risk of type 2 diabetes in an Iranian population. Clin Lab. 2014;60:663–70. doi: 10.7754/clin.lab.2013.130630. [DOI] [PubMed] [Google Scholar]
- 17.Abosheasha MA, Zahran F, Bessa SS, Mohamed TM. Na + /K + -ATPase activity as a potential biomarker for Type 2 Diabetes mellitus. Res J Pharm, Biol Chem Sci. 2018;9:1227–31. [Google Scholar]
- 18.Koc B, Erten V, Yilmaz MI, Sonmez A, Kocar IH. The relationship between red blood cell Na/K-ATPase activities and diabetic complications in patients with type 2 diabetes mellitus. Endocrine. 2003;21:273–8. doi: 10.1385/ENDO:21:3:273. [DOI] [PubMed] [Google Scholar]
- 19.Nagilla B, Reddy P. Neuroprotective and antinociceptive effect of curcumin in diabetic neuropathy in rats. Int J Pharm Pharm Sci. 2014;6:131–8. [Google Scholar]
- 20.Suzen S, Buyukbingol E. Recent studies of aldose reductase enzyme inhibition for diabetic complications. Curr Med Chem. 2003;10:1329–52. doi: 10.2174/0929867033457377. [DOI] [PubMed] [Google Scholar]
- 21.Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De La Tour DD, Raccah D, Jannot MF, Coste T, Rougerie C, Vague P. Erythrocyte Na/K ATPase activity and diabetes: Relationship with C-peptide level. Diabetologia. 1998;41:1080–4. doi: 10.1007/s001250051033. [DOI] [PubMed] [Google Scholar]
- 23.Tsimaratos M, Roger F, Chabardès D, Mordasini D, Hasler U, Doucet A, et al. C-peptide stimulates Na+, K+-ATPase activity via PKC alpha in rat medullary thick ascending limb. Diabetologia. 2003;46:124–31. doi: 10.1007/s00125-002-0996-1. [DOI] [PubMed] [Google Scholar]
- 24.Jannot MF, Raccah D, De La Tour DD, Coste T, Vague P. Genetic and environmental regulation of Na/K adenosine triphosphatase activity in diabetic patients. Metabolism. 2002;51:284–91. doi: 10.1053/meta.2002.29009. [DOI] [PubMed] [Google Scholar]
- 25.Djemli-Shipkolye A, Gallice P, Coste T, Jannot MF, Tsimaratos M, Raccah D, et al. The effects ex vivo and in vitro of insulin and C-peptide on Na/K adenosine triphosphatase activity in red blood cell membranes of type 1 diabetic patients. Metabolism. 2000;49:868–72. doi: 10.1053/meta.2000.6753. [DOI] [PubMed] [Google Scholar]
- 26.Iannello S, Milazzo P, Belfiore F. Animal and human tissue Na, K-ATPase in normal and insulin-resistant states: Regulation, behaviour and interpretative hypothesis on NEFA effects. Obes Rev. 2007;8:231–51. doi: 10.1111/j.1467-789X.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 27.Akbulut S, Gurbilek M, Kiyici A, Akoz M, Altintepe L, Karakuscu A, Topcu C. The effect of erythropoietin application on erythrocyte Na, K- ATPase activities in patients with diabetic polyneuropathy. Turk J Biochem. 2013;38:13–7. [Google Scholar]
- 28.Singh P, Kesharwani RK, Misra K, Rizvi SI. The modulation of erythrocyte na(+)/K(+)-ATPase activity by curcumin. J Adv Res. 2015;6:1023–30. doi: 10.1016/j.jare.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarei P, Rezvanfar MR, Ansarihadipour H, Delavar M, Abdollahi M, Khosrowbeygi A. Effects of coenzyme Q10 supplementation on the serum levels of amylase, adenosine deaminase, catalase, and total antioxidant capacity in women with type 2 diabetes mellitus: A randomized, double-blind placebo-controlled trial. J Res Med Sci. 2018;23:91. doi: 10.4103/jrms.JRMS_970_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Magd MA, Saleh AA, Nafeaa AA, El-Komy SM, Afifi MA. Polymorphisms of the IGF1 gene and their association with growth traits, serum concentration and expression rate of IGF1 and IGF1R in buffalo. J Zhejiang Univ Sci B. 2017;18:1064–74. doi: 10.1631/jzus.B1600573. [DOI] [PMC free article] [PubMed] [Google Scholar]


