Abstract
Understanding the cell–cell interactions that control CNS development and function has long been limited by the lack of methods to cleanly separate neural cell types. Here we describe methods for the prospective isolation and purification of astrocytes, neurons, and oligodendrocytes from developing and mature mouse forebrain. We used FACS (fluorescent-activated cell sorting) to isolate astrocytes from transgenic mice that express enhanced green fluorescent protein (EGFP) under the control of an S100β promoter. Using Affymetrix GeneChip Arrays, we then created a transcriptome database of the expression levels of >20,000 genes by gene profiling these three main CNS neural cell types at various postnatal ages between postnatal day 1 (P1) and P30. This database provides a detailed global characterization and comparison of the genes expressed by acutely isolated astrocytes, neurons, and oligodendrocytes. We found that Aldh1L1 is a highly specific antigenic marker for astrocytes with a substantially broader pattern of astrocyte expression than the traditional astrocyte marker GFAP. Astrocytes were enriched in specific metabolic and lipid synthetic pathways, as well as the draper/Megf10 and Mertk/integrin αvβ5 phagocytic pathways suggesting that astrocytes are professional phagocytes. Our findings call into question the concept of a “glial” cell class as the gene profiles of astrocytes and oligodendrocytes are as dissimilar to each other as they are to neurons. This transcriptome database of acutely isolated purified astrocytes, neurons, and oligodendrocytes provides a resource to the neuroscience community by providing improved cell-type-specific markers and for better understanding of neural development, function, and disease.
Keywords: astrocyte, neuron, oligodendrocyte, GeneChip, Aldh1L1, culture, gene profiling, microarray, transcriptome, phagocytosis, astroglia, Megf10, Mertk, Draper, Mfge8
Introduction
The nature and role of neuron–glia interactions in controlling the development, function, and pathology of our brains remain among the greatest unsolved mysteries in neurobiology today. In particular, the development and function of astrocytes, a cell type that constitutes approximately one-third of mouse brain cells and nearly half of human brain cells, remain essentially uncharacterized. A central limitation in advancing our understanding of mature astrocyte development and function has been the lack of procedures that allow for their prospective purification. Primary cultures of astroglia, which can only be prepared from neonatal rodent brain, have been useful for gene profiling and other studies (McCarthy and de Vellis, 1980; Bachoo et al., 2004). These astroglial cultures, however, are apparently derived from a small, unidentified population of proliferating glial precursor cells that express several astrocyte markers, but which appear to have an immature or reactive phenotype. Their properties are thus of unclear relationship to those of mature astrocytes in postnatal and adult brain.
Genome-wide transcriptional profiling has recently emerged as a powerful tool for better understanding the development and function of defined cell types. These studies have, for instance, revealed a detailed taxonomic tree and cell-type-specific gene expression patterns for 12 different neuronal subtypes (Sugino et al., 2006), characterized the specification and development of cortical projection neurons (Arlotta et al., 2005), and profiled subtypes of striatal projection neurons (Lobo et al., 2006). In addition, purified rat oligodendrocyte progenitor cells (OPCs) and premyelinating, postmitotic oligodendrocytes (OLs) have recently been profiled to reveal developmental gene expression changes during OL specification and differentiation (Dugas et al., 2006; Nielsen et al., 2006). These studies have established gene expression profiles for mouse neurons and rat OLs, but a global direct comparison between the main CNS neural cell types has not yet been possible because of the lack of methods to purify postnatal astrocytes and myelinating OLs, both highly fragile cell types.
In this study, we report the development of methods to highly purify astrocytes, OLs, and neurons from the postnatal mouse forebrain. We acutely purified mouse astrocytes from early postnatal ages [postnatal day 1 (P1)] to later postnatal ages (P30), when astrocyte differentiation is morphologically complete (Bushong et al., 2004), and acutely purified mouse OL-lineage cells from stages ranging from OPCs to newly differentiated OLs to myelinating OLs. We extracted RNA from each of these highly purified, acutely isolated cell types and used GeneChip Arrays to determine the expression levels of >20,000 genes and construct a comprehensive database of cell-type-specific gene expression in the mouse forebrain. Analysis of this database confirms cell-type- specific expression of many well characterized and functionally important genes. In addition, we have identified thousands of new cell-type-enriched genes, thereby providing important new information about astrocyte, OL, and neuron interactions, metabolism, development, and function. This database provides a comparison of the genome-wide transcriptional profiles of the main CNS cell types and is a resource to the neuroscience community for better understanding the development, physiology, and pathology of the CNS.
Materials and Methods
These purification procedures are based on previously described dissociation (Segal et al., 1998) and immunopanning purification protocols (Barres et al., 1992). All important aspects of the fluorescent-activated cell sorting (FACS) and panning purification procedures are summarized below. Additional experimental details are in supplemental material (available at www.jneurosci.org).
FACS purification of astrocytes.
Dissociated forebrains from S100β-EGFP mice were resuspended in panning buffer [Dulbecco's PBS (DBPS) containing 0.02% BSA and 12.5 U/ml DNase] and sequentially incubated on the following panning plates: secondary antibody only plate to deplete microglia, O4 plate to deplete OLs, PDGFRα plate to deplete OPCs, and a second O4 plate to deplete any remaining OLs. This procedure was sufficient to deplete all OL-lineage cells from animals P8 and younger, however, in older animals that had begun to myelinate, additional depletion of OLs and myelin debris was accomplished as follows. The nonadherent cells from the last O4 dish were harvested by centrifugation, and the cells were resuspended in panning buffer containing GalC, MOG, and O1 supernatant and incubated for 15 min at room temperature. The cell suspension was washed and then resuspended in panning buffer containing 20 μg of donkey anti-mouse allophycocyanin for 15 min. The cells were washed and resuspended in panning buffer containing propidium iodide (PI). EGFP+ astrocytes were then purified by FACS. Dead cells were gated out using high PI staining and forward light scatter. Astrocytes were identified based on high EGFP fluorescence and negative APC fluorescence from indirect immunostaining for OL markers GalC, MOG, and O1. Cells were sorted twice and routinely yielded >99.5% purity based on reanalysis of double sorted cells.
FACS purification of neurons.
EGFP− cells were the remaining forebrain cells after microglia, OLs, and astrocytes had been removed, and were primarily composed of neurons, and to a lesser extent, endothelial cells (we estimate <4% endothelial cells at P7 and <20% endothelial cells at P17). EGFP− cells from S100β-EGFP dissociated forebrain were FACS purified in parallel with astrocyte purification and were sorted based on their negative EGFP immunofluorescence. Cells were sorted twice and routinely yielded >99.9% purity. In independent preparations, the EGFP− cell population was additionally depleted of endothelial cells and pericytes by sequentially labeling with biotin-BSL1 lectin and streptavidin-APC while also labeling for OL markers as described above. Cells were sorted twice and routinely yielded >99.9% purity.
Panning purification of oligodendrocyte lineage cells.
Dissociated mouse forebrains were resuspended in panning buffer. To deplete microglia, the single-cell suspension was sequentially panned on four BSL1 panning plates. The cell suspension was then sequentially incubated on two PDGFRα plates (to purify and deplete OPCs), one A2B5 plate (to deplete any remaining OPCs), two MOG plates (to purify and deplete myelinating OLs), and one GalC plate (to purify the remaining PDGFRα−, MOG−, OLs). The adherent cells on the first PDGFRα, MOG, and GalC plates were washed to remove all antigen-negative nonadherent cells. The cells were then lysed while still attached to the panning plate with Qiagen RLT lysis buffer, and total RNA was purified. Purified OPCs were >95% NG2 positive and 0% MOG positive. Purified Myelin OLs were 100% MOG positive, >95% MBP positive, and 0% NG2 positive. Purified GalC OLs depleted of OPCs and Myelin OLs were <10% MOG positive and ∼50% weakly NG2 positive, a reflection of their recent development as early OLs.
Data normalization and analysis.
Raw image files were processed using Affymetrix GCOS and the Microarray Suite (MAS) 5.0 algorithm. Intensity data were normalized per chip to a target intensity value of 500, and expression data and absent/present calls for individual probe sets were determined. Gene expression values were also calculated by normalization and modeling across arrays using the dChip software package with invariant-set normalization and a PM model (www.dchip.org) (Li and Wong, 2001). Using these described methods, we have reported a PCR validation rate of >90% (Dugas et al., 2006). The 29 samples were grouped into 9 sample types: Astros P7-P8, Astros P17, Astros P17-gray matter (P17g), Neurons P7, Neurons P17, Neurons-endothelial cell depleted (P7n, P17n), OPCs, GalC-OLs, and MOG-OLs. Gene filtering was performed to select probe sets that were consistently expressed in at least one cell type, in which consistently expressed was defined as being called present and having a MAS 5.0 intensity level of >200 in at least two-thirds of the samples in the cell type. We identified 20,932 of the 45,037 probe sets that were consistently expressed in at least one of the nine cell types. The Significance Analysis of Microarrays (SAM) method (Tusher et al., 2001) was used to determine genes that were significantly differentially expressed between different cell types (for SAM cell type groupings, see supplemental Table S2, available at www.jneurosci.org as supplemental material). Clustering was performed using the hclust method with complete linkage in R. Expression values were transformed for clustering by computing a mean expression value for the gene using those samples in the corresponding SAM statistical analysis, and then subtracting the mean from expression intensities. To preserve the log2 scale of the data, unless otherwise indicated, no normalization by variance was performed. Plots were created using the gplots package in R. The Bioconductor software package (Gentleman et al., 2004) was used throughout the expression analyses. Functional analyses were performed through the use of Ingenuity Pathways Analysis (IPA) (Redwood City, CA).
The GeneChip microarray data has been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE9566.
Immunohistochemistry.
Mice or rats were perfused with DPBS followed by 4% PFA and then postfixed in 4% PFA overnight at 4°C. Brains were cryoprotected overnight at 4°C in 15% sucrose and frozen in 1 part Tissue-Tek O.C.T. and 2 parts 30% sucrose. Sections were blocked in 50% goat serum with 0.4% Triton X-100 in PBS for 45 min at room temperature. Rabbit serum against rat Aldh1L1 (Krupenko and Oleinik, 2002) used at 1:1000 was added over two nights at 4°C. Sections were incubated with Alexa Fluor 594 or 488 goat anti-rabbit highly crossed adsorbed secondary antibodies at 1:500 for 2 h at room temperature.
Results
Purification of CNS cell types
To purify astrocytes, we took advantage of a transgenic line of mice that expresses EGFP under the control of an S100β promoter (Zuo et al., 2004), a well established glial cell marker (Ludwin et al., 1976). We selected the S100β-EGFP Kosmos line that showed strong EGFP fluorescence in astrocytes. Because we found this transgenic line also expresses EGFP in OL-lineage cells, our purification strategy (Fig. 1 A) was to first deplete the EGFP-positive (EGFP+) OLs and OPCs by immunopanning. This procedure was sufficient to deplete all OL lineage cells from animals P8 and younger, however, in older animals that had begun to myelinate, additional depletion of OLs and myelin debris was necessary using immunolabeling with GalC, MOG, and O1 and an APC fluorescence secondary antibody to remove any remaining OLs during FACS sorting. After depleting OL-lineage cells, we then FACS purified the EGFP+ astrocytes by sequential double sorting to yield >99% pure astrocytes (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). We purified forebrain astrocytes at both early developmental stages, when astrocytes are newly generated and immature (P1–P8), and at later stages, when astrocyte development is nearly complete and the astrocytes have matured (P17–P30) (Bushong et al., 2004). In some experiments, we also purified astrocytes specifically from P17 cerebral cortical gray matter to purify protoplasmic astrocytes. Based on these experiments, and the fragility of mature fibrous astrocytes, we suspect that our purified populations of forebrain astrocytes are highly enriched for protoplasmic astrocytes. The high purity of astrocytes was confirmed by examining well described markers of CNS cell types: astrocyte markers were only expressed by FACS-purified EGFP+ astrocytes and neuron, OL, microglia, and endothelial cell markers were at background levels (Fig. 1 C–E) (supplemental Table S1, available at www.jneurosci.org as supplemental material).
Figure 1.
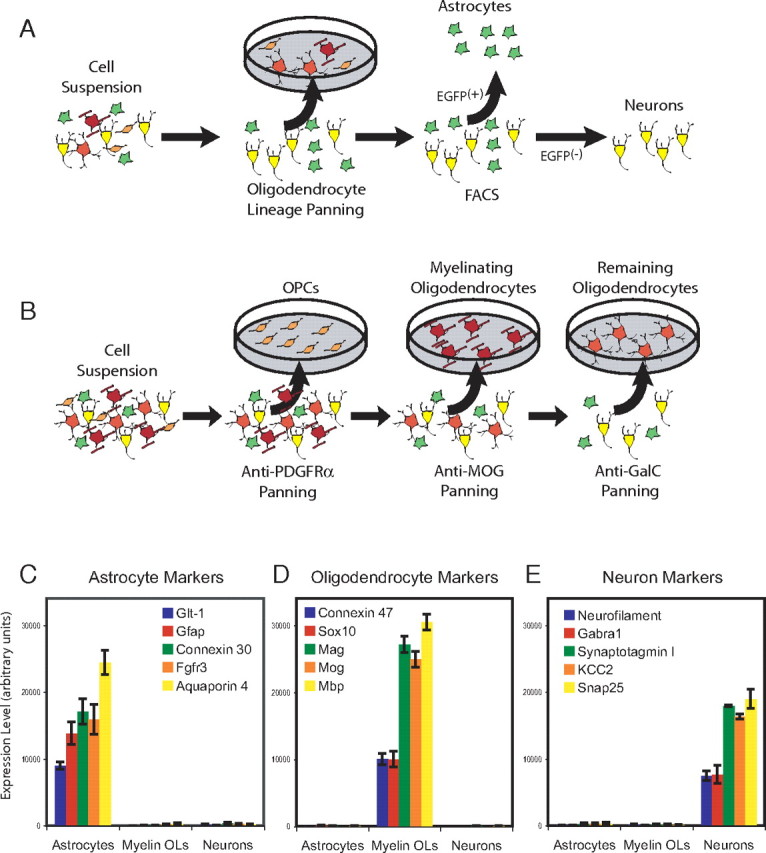
Purification of astrocytes, neurons, and oligodendrocytes. A, Purification of astrocytes and neurons. A cell suspension of mouse forebrain was depleted of OPCs and OLs using immunopanning with OL lineage-specific antibodies. Astrocytes were then isolated from the remaining cells by FACS purifying the EGFP-positive astrocytes. The FACS-purified EGFP-negative cells were isolated and represent an enriched neuron population. B, Purification of OLs. A cell suspension of mouse forebrain was first depleted of microglia by panning on BSL1 lectin panning plates, and the remaining cells were then incubated on PDGFRα panning plates to purify and deplete OPCs, MOG panning plates to purify and deplete myelinating OLs, and a GalC panning plate to purify the remaining OLs. C–E, Expression levels of well described markers for astrocytes (C), OLs (D), and neurons (E) demonstrates high purity of each cell type. The y-axis represents the level of gene expression determined by MAS 5.0. Error bars represent ±SEM.
Neurons were collected from the population of EGFP-negative (EGFP−) cells that remained after depletion of microglia, OLs, and astrocytes (Fig. 1 A). These EGFP− cells are primarily composed of neurons and to a lesser extent endothelial cells. In some preparations, the endothelial cells were depleted from the EGFP− population by labeling with BSL1 lectin. The resulting purified neuronal population was negative for markers of astrocytes, OLs, microglia, and endothelial cells (Fig. 1 C–E) (supplemental Table S1, available at www.jneurosci.org as supplemental material).
To purify mouse OPCs, OLs, and myelinating OLs (Myelin OLs), we used immunopanning techniques and stage-specific antibodies to purify cells from P16 mouse forebrain (Fig. 1 B). Our strategy relied on the stage-specific OL progenitor marker platelet-derived growth factor receptor α (PDGFRα), which is expressed by OPCs (Hall et al., 1996), and the stage-specific OL marker myelin oligodendrocyte glycoprotein (MOG), whose expression in vivo is restricted to mature OLs coinciding with the onset of myelination by these cells (Solly et al., 1996). After purifying and depleting OPCs and Myelin OLs from the cell suspension, we then purified newly differentiated OLs using the pan-OL monoclonal antibody GalC to yield a population of newly differentiated OLs that are depleted of OPCs and Myelin OLs. The high purity of OPCs, OLs, and Myelin OLs was confirmed by examining well described markers of CNS cell types: OL markers were only expressed by immunopurified OPCs, OLs, and Myelin OLs, and astrocyte, neuron, microglia, and endothelial cell markers were at background levels (Fig. 1 C–E, supplemental Table S1, available at www.jneurosci.org as supplemental material).
These purification methods for astrocytes, neurons, and OLs rely on known cell-type-specific markers. To confirm our purified cells were representative of the major cell types in the mouse forebrain, we purified mRNA from whole, undissociated forebrain. We then examined the genes expressed by whole forebrain to confirm that they could be accounted for by our purified populations of astrocytes, neurons, and OLs, and crude populations of microglia and endothelial cells. We found that essentially all genes expressed by whole forebrain were represented by at least one of these cell populations suggesting that our purified cell populations together constitute the major cell classes in the mouse forebrain.
A complete description of all sample ages and purification methods appears in supplemental Table S2 and supplemental Experimental Procedures (available at www.jneurosci.org as supplemental material).
Gene expression profiling of CNS cell types
Total RNA prepared from purified cell types was used to generate labeled cRNA using a two-step linear amplification protocol with poly(A) primers that amplify the 3′ end of the mRNA. This labeled cRNA was hybridized to Affymetrix Mouse 430 2.0 Arrays containing oligonucleotide probe sets complementary to 3′-ends of the mRNA transcripts (3′-arrays); each array contained 45,037 oligonucleotide probe sets representing 20,832 unique genes. In addition, labeled cDNA was generated with a one-step linear amplification protocol using random primers to amplify all RNA. This labeled cDNA was hybridized to Affymetrix Mouse Exon 1.0 ST Arrays with oligonucleotide probe sets complementary to regions along the whole mRNA molecule (exon-arrays), representing 17,213 genes that have defined transcript level probe selection and expression indexes (Xing et al., 2006). We performed our main analyses with the data generated from the 3′-arrays using the MAS 5.0 algorithm to generate expression values and absent/present (A/P) calls, and dChip, SAM, and Bioconductor software were used for statistical analysis and clustering.
Samples representing biological replicates of the same purified cell type and developmental stage were grouped and we then created a master list of probe sets that were consistently expressed in at least one cell population (see Materials and Methods). These 20,932 probe sets represent 12,416 unique genes that are expressed at significant levels in at least one CNS cell type. A master data table of the expression values for all 20,932 expressed probe sets appears in supplemental Table S3 (available at www.jneurosci.org as supplemental material). Our statistical analysis and clustering proceeded with this filtered list of expressed genes.
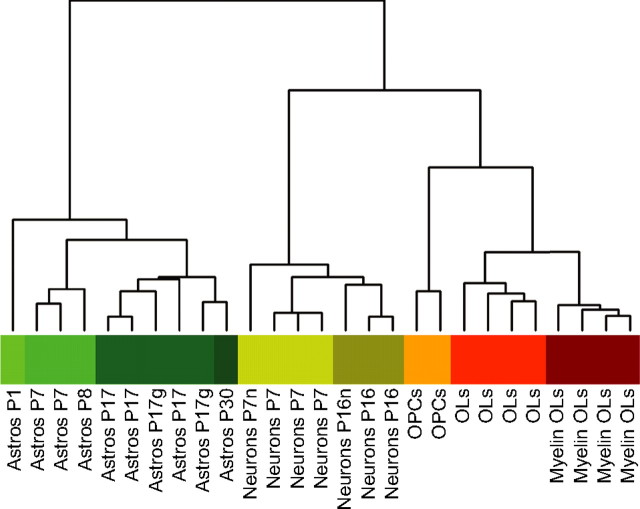
Unsupervised hierarchical clustering of purified cell type samples revealed that astrocytes, neurons, and OLs each have a distinct gene expression pattern. The dendrogram in Figure 2 shows three main branches representing astrocytes, neurons, and OLs. Within each cell type, samples cluster according to their developmental stage. The clustering shows high reproducibility between biological replicates and strong similarity within one cell type. The similarity within cell types is in contrast to the large differences found between astrocytes, neurons, and OLs. Interestingly, the correlation between pairwise comparisons of astrocytes and OLs is not any stronger than the pairwise correlation of either of these cell types with neurons. Considering the highly specialized functions of each of these mature CNS cell types, it should not be unexpected that their gene expression profiles are highly divergent and that each represents an equally unique cell type.
Figure 2.
Dendrogram and sample clustering of purified CNS cell types. Hierarchical clustering of highly purified CNS cell type samples from different developmental stages reveals three distinct clusters representing astrocytes, neurons, and oligodendrocytes. The similarity of gene expression between different samples is represented by the vertical distances on each branch of the dendrogram. Biological replicates show the highest degree of correlation within samples, represented by short vertical distances. Within each cell population, gene expression is more highly correlated between maturing and mature samples (Astros P7, Astros P17, OLs, Myelin OLs) than between immature and maturing samples (Astros P1, Astros P7, OPCs, OLs). Color bar and sample labels describe each individual sample type (green, astrocytes; yellow, neurons; orange–red, OL lineage cells; P, postnatal day, represented by different color shades; g, cerebral cortical gray matter astrocytes; n, neuron samples depleted of residual endothelial cells).
The heat map generated from unsupervised hierarchical gene clustering of all 12,416 expressed genes shows cell type gene expression patterns at the gene level (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Nearly all genes show cell type clustering between astrocytes, neurons, and OLs and many also cluster based on the cells' developmental stage. These gene clusters appear equally distributed among all cell types, reinforcing our finding that astrocytes and OLs are not more similar to each other than they are to neurons. Astrocytes and OLs have long been classified together under the designation “glia” to signify that they may share common features and functions. Although our data does not address the question of a common lineage of astrocytes and OLs, these data indicate that mature astrocytes and OLs do not share a large cohort of common “glial” genes, and suggests that the notion of a molecularly defined glial cell type is largely misleading because all three mature cell types differ vastly based on the genes that they express.
Identification and validation of neural cell-type-specific genes
Quantitative comparison of astrocyte, neuron, and OL gene expression allowed for identification of all genes that are differentially overexpressed in each cell type. All genes enriched by at least 1.5-fold and statistically different by significance analysis of microarrays (SAM) with a false discovery rate (FDR) threshold of <1% (Tusher et al., 2001) are listed in supplemental Tables S4–S6 (available at www.jneurosci.org as supplemental material). These include 2618 astrocyte-enriched genes, 2036 neuron-enriched genes, and 2228 OL-enriched genes. Based on the enrichment levels of previously well established cell type markers (supplemental Table S1, available at www.jneurosci.org as supplemental material), genes enriched >20-fold are considered to be cell-type-specific genes. The top 40 astrocyte, neuron, and OL cell-type-specific genes are shown in the heat map in Figure 3 and the top 250 genes appear in supplemental Figure S3 (available at www.jneurosci.org as supplemental material). As expected, the top 40 cell-type-specific genes include well established cell type specific markers, verifying our isolation procedure. However, many of these genes have not previously been characterized as cell type specific in the CNS and their functional roles are not yet known.
Figure 3.
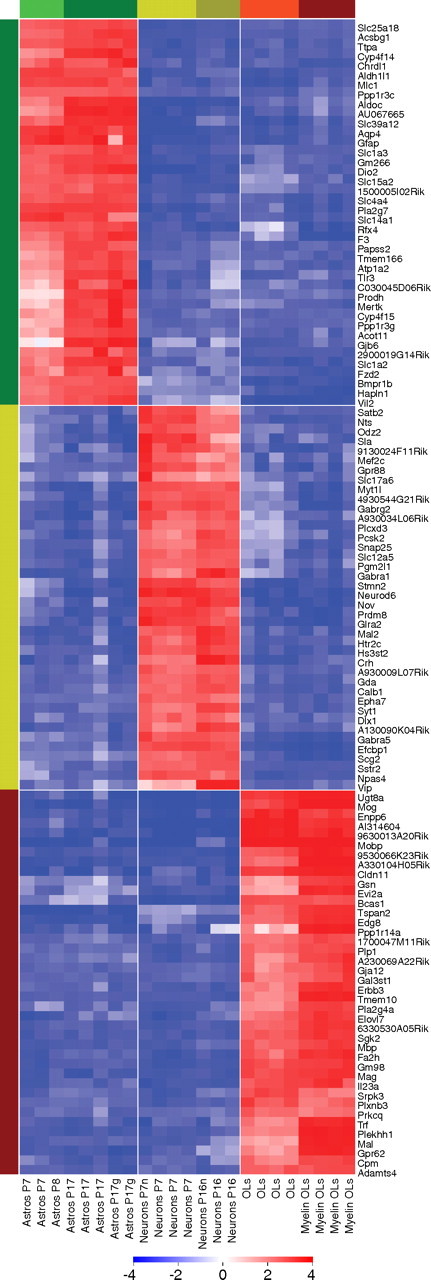
Expression of astrocyte, oligodendrocyte, and neuron-specific genes. The top 40 cell-type-specific genes expressed by astrocytes (green bar), neurons (yellow bar), and oligodendrocytes (red bar) are depicted. Each individual gene expression level was normalized (see Materials and Methods) and plotted on a log2 color scale, with blue representing low expression and red representing high expression. The fold enrichment can be estimated from the log2 color bar scale, for example, the change from medium blue (−2) to red (3) represents a 32-fold difference in expression level.
We compared our lists of the top 40 cell-type-specific genes based on 3′-array data with our exon-array data to confirm the cell type specificity of these genes. We first mapped the top 40 astrocyte, neuron, and OL-specific genes from the 3′-array probe sets (a total of 120 probe sets) to mouse exon-array transcript clusters using a mapping file provided by Affymetrix and our manual query of NetAffx. Of the 120 3′-array probe sets, 101 were mapped unambiguously to unique exon-array gene identifiers that were represented by core probe sets. Of these 101 genes, 92 (91%) were at least 15-fold enriched in their respective cell type according to the exon-array expression indexes, 100 (99%) were at least fivefold enriched, and the minimal exon-array fold change was 3.9 (supplemental Table S7, available at www.jneurosci.org as supplemental material). In addition to providing validation of the 3′-array results, the exon-array transcript level data algorithm is optimized to provide overall gene expression levels independent of different mRNA splice forms (Xing et al., 2006). Thus, when a gene is confirmed by the exon array data, it suggests the gene is cell type specific regardless of gene splicing. A full list of the exon-array expression values for 17,213 genes independent of splicing appears in supplemental Table S8 (available at www.jneurosci.org as supplemental material). Additional analysis using the exon-array data to identify cell-type-specific gene splicing is ongoing. The exon-array results, which use different labeling methods and probe selection regions, provide strong independent evidence for CNS cell-type-specific genes discovered from the 3′-arrays.
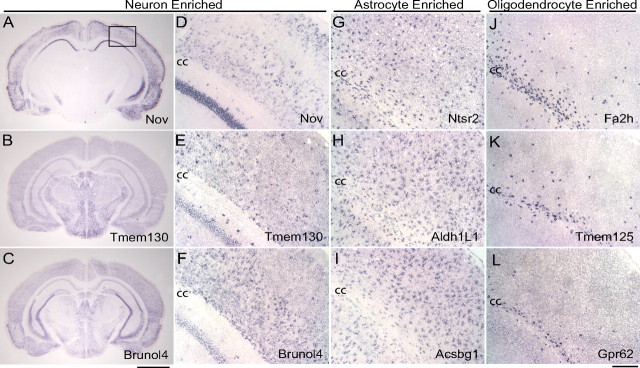
We next performed in situ hybridization (ISH) to confirm some of the new cell-type-specific genes in the CNS (Fig. 4). ISH for several arbitrarily selected genes identified as being highly enriched in neurons (Fig. 4 A–F, Nov, Tmem130, and Brunol4) confirmed in all three cases that the genes had an expression pattern consistent with neuron-specific expression, although the distribution between neuronal subtypes varied between genes, with Nov displaying a considerably more restricted cortical expression than Tmem130 or Brunol4. Similarly, ISH for genes identified as being highly enriched in astrocytes (Fig. 4 G–I, Ntsr2, Aldh1L1 and Acsbg1) showed a pattern consistent with astrocyte-specific expression, with staining showing labeling of many small cell bodies and processes broadly distributed throughout the cerebral cortex. Genes identified as being highly enriched in OLs (Fig. 4 J–L, Fah2, Tmem125/6330530A05Rik, and Gpr62) gave a staining pattern consistent with OL-specific expression, with staining predominantly restricted to the white matter and scattered cells within the cortex. For genes we identified as being cell type specific that gave above background ISH, our analyses indicated excellent concordance between the cell type specificity predicted using our array data and the regional distribution seen using ISH.
Figure 4.
Validation of gene expression data by in situ hybridizations. A–C, Coronal brain sections showing ISH for genes identified by array data as having specific neuronal expression: A, Nov; B, Tmem130; C, Brunol4. D–L, Higher-magnification images corresponding to outlined region in A showing hippocampus, corpus callosum (cc), and the overlying cortex. D–F, Genes identified by array data as having specific neuronal expression, displaying expression in the hippocampus and cortex: D, Nov; E, Tmem130; F, Brunol4. G–I, Genes identified by array data as having astrocyte- enriched expression, showing fibrous, positive cells throughout the white and gray matter: G, Ntsr2; H, Aldh1L1; I, Acsbg1. J–L, ISH for genes identified by array data as having specific OL expression, showing white matter expression in the corpus callosum and the occasional positive cell in the overlying cortex: J, Fa2h; K, Tmem125/6330530A05Rik; L, Gpr62. All ISH performed on P17 mouse brains. Scale bars: A–C, 2 mm; D–L, 200 μm.
Because our database lists many more cell-type-enriched genes than we can examine by ISH ourselves, we next compared our findings with those of the Allen Brain Atlas (ABA) project, which is a comprehensive ISH database for identifying the anatomical expression patterns of ∼20,000 genes in the adult mouse brain. The ABA informatics filter identifies genes with similar spatial distribution patterns. Training these filters with the ISH pattern for established cell-type-specific markers was used to predict new cell-type-enriched genes (Lein et al., 2007). We compared the ABA list of 44 astrocyte-enriched genes, 78 OL-enriched genes, and 69 neuron-enriched genes to our quantitative 3′-array determination of the mRNA levels in each cell type. A heat map of astrocyte, OL, and neuron gene expression values for each gene identified by the ABA appears in supplemental Figures S4–S6 (available at www.jneurosci.org as supplemental material). Of the 191 ABA cell-type-specific genes, 178 (93%) were expressed in purified CNS cells in our 3′-array analysis. Of these expressed genes, 141 (79%) were >1.5-fold cell-type-enriched, 106 (60%) were >4-fold cell-type-enriched, and 30 (17%) were >20-fold enriched (cell type specific) in the same cell type as identified by the ABA. This shows overall concordance between the ABA filters and our data analysis, although our analysis suggests that 23 (13%) of the genes identified by the ABA are in fact enriched in another cell type.
This comparison between our two datasets shows a high degree of agreement and also how the ABA represents a resource that complements our database of genome-wide gene expression values for each of the main CNS cell types. It also shows the power of quantitative genome-wide gene expression analysis. Although the ABA was able to identify the correct cell-type-enriched gene expression patterns in almost 80% of their selected genes, few of these genes represent the most cell-type-specific genes. Furthermore, our database also identifies many more genes as cell type enriched and includes quantitative gene enrichment levels in different cell types and at different developmental stages.
Aldh1L1 is a new astrocyte-specific marker
To characterize new astrocyte-specific markers, we examined our gene expression database to identify astrocyte-specific genes with the highest levels of mRNA expression, widest patterns of ISH labeling throughout the brain, and proteins expected to be expressed throughout the whole cell (i.e., not restricted to cellular subcompartments). One such gene that fit these criteria is the aldehyde dehydrogenase 1 family, member L1 (Aldh1L1) gene, also known as 10-formyltetrahydrofolate dehydrogenase (FDH) (Cook et al., 1991; Anthony and Heintz, 2007). ISH for this gene indicated that its mRNA is expressed throughout the CNS in a pattern consistent with pan-astrocyte expression (Fig. 4 H), and it has been previously shown that Aldh1L1 protein is expressed in adult rat astrocytes (Neymeyer et al., 1997). Immunohistochemical staining with an Aldh1L1-specific polyclonal antibody revealed the highly branched astrocyte morphology, including the astrocyte cell body and its extensive processes. In contrast, GFAP antibodies primarily labeled the thick main processes of some astrocytes (Fig. 5 A–C). All cells positive for GFAP are also labeled with Aldh1L1, whereas Aldh1L1 strongly labels many more astrocytes. Furthermore, Aldh1L1 mRNA is more widely expressed throughout the brain, whereas GFAP show more predominant expression in white matter (see Allen Brain Atlas) (supplemental Fig. S7, available at www.jneurosci.org as supplemental material). Double labeling with Aldh1L1 and markers to neurons (tubulin), OLs (CC1 and MBP), and OPCs (NG2) indicate that Aldh1L1 does not label other brain cell types (Fig. 5 D–F).Together, these findings identify Aldh1L1 as a highly, broadly, and specifically expressed astrocyte gene and indicate that it is a useful astrocyte-specific marker.
Figure 5.
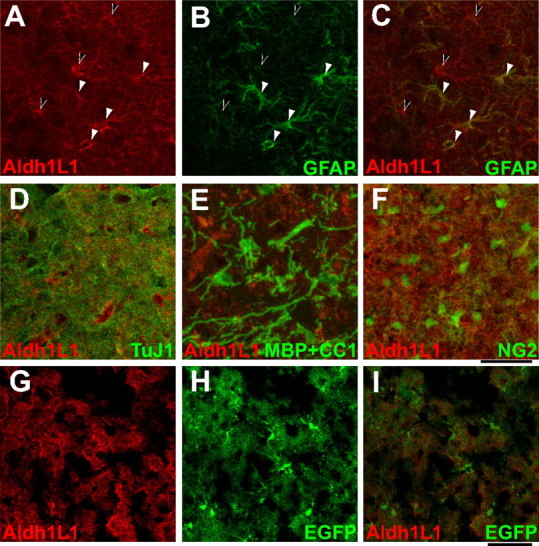
Aldh1L1 is a specific pan-astrocyte marker. A–F, Immunohistochemical staining of P15 rat cortex shows Aldh1L1 is a cell-type-specific pan-astrocyte marker. A–C, Costaining of rat cortex for Aldh1L1 (red) and GFAP (green) staining. A, Aldh1L1 labels both the cell bodies and extensive processes of astrocytes in the cortex. B, GFAP labels the astrocyte intermediate filament cytoskeleton but not the finer processes that Aldh1L1 is capable of labeling. C, GFAP labels a subset of astrocytes: cells labeled by GFAP are also Aldh1L1 positive (white arrowheads), whereas Aldh1L1 labels many astrocytes not labeled by GFAP (black arrowheads). D–F, Aldh1L1 does not label neurons (D, Tuj1), OLs (E, MBP + CC1), or OPCs (F, NG2). G–I, The merge (I) of Aldh1L1 immunostaining (G) and strong BAC Aldh1L1-EGFP fluorescence (H) seen in the Aldh1L1-EGFP transgenic mouse cortex shows that all cells expressing the EGFP transgene also express the endogenous Aldh1L1 protein. Scale bars: A–F, 40 μm; G–I, 60 μm.
A BAC Aldh1L1-EGFP mouse has already been generated by the GENSAT project (Heintz, 2004). The diffuse expression pattern of Aldh1L1 throughout the CNS of this mouse is consistent with pan-astrocyte expression (supplemental Fig. S7, available at www.jneurosci.org as supplemental material). We analyzed brain cryosections from this mouse line to both further verify the expression pattern of Aldh1L1 and to determine whether this mouse faithfully reports EGFP specifically in astrocytes. As shown in Figure 5 G–I, we found that throughout the brain Aldh1L1 immunohistochemical staining colocalized with EGFP, confirming that the BAC-Aldh1L1-EGFP transgene drives EGFP expression in the same astrocyte-specific pattern as the endogenous Aldh1L1 protein. In addition, EGFP diffused into the nucleus, making it particularly easy to identify astrocyte cell bodies in these mice. We then prepared a cell suspension from the Aldh1L1-EGFP mice, immunostained with O4 monoclonal antibody to OLs, and FACS analyzed to determine whether the Aldh1L1-EGFP mice will be useful for FACS purifying astrocytes. Supplemental Figure S8 (available at www.jneurosci.org as supplemental material) shows a good separation between the EGFP-positive astrocytes and remaining EGFP-negative cells. In addition, the O4-positive OLs are EGFP negative, indicating that the Aldh1L1-EGFP mouse is better mouse for FACS purifying astrocytes compared with the S100β mouse. These data demonstrate that the Aldh1L1 protein is expressed specifically by astrocytes, but not by OLs or neurons, and thus that Aldh1L1 is a better immunohistochemical astrocyte marker than GFAP because it better labels both the astrocyte cell body and processes throughout both gray and white matter.
The most specific neural cell type markers that we have identified have been provided to the Neuromab Project (http://www.neuromab.org), NINDS GENSAT Project, and the NIH Neuromouse Cre Project so that low-cost, high-quality antibodies and mouse lines should soon be available to the neuroscience community.
Analysis of canonical pathways enriched in the main CNS cell types
The large number of genes preferentially expressed by each neural cell type suggests that astrocytes, neurons, and OLs may differ in many fundamental signaling and metabolic pathways. To more systematically determine canonical signaling and metabolic pathways enriched in each CNS cell type, we used the IPA tool from Ingenuity Systems. Given a list of genes, IPA performs a statistical test for enrichment of these genes in its hand-curated canonical pathway database. Each individual IPA signaling pathway includes extracellular signaling components, cell membrane receptors, downstream effectors, and transcription factors that have been described to interact the published scientific literature, with each pathways often including from 30 to 100 or more individual genes. The IPA metabolic pathways are derived from the KEGG metabolic pathways (Kanehisa et al., 2006). These tools allow us to determine the top canonical signaling and metabolic pathways represented among the genes that are significantly enriched in each given cell type (supplemental Tables S4–S6, available at www.jneurosci.org as supplemental material). Table 1 lists signaling pathways (A–C) and metabolic pathways (D–F) enriched in each cell type at p < 0.05. These enriched signaling pathways are likely influenced by cell–cell interactions, perturbed by disease processes, and affected by drugs to yield physiological effects. We observed 12 significantly enriched metabolic pathways in astrocytes, but only three in OLs and neurons, indicating significant roles for astrocytes in energy metabolite and amino acid production in the CNS (see Discussion).
Table 1.
Ingenuity pathway analysis identified cell-type-enriched pathways
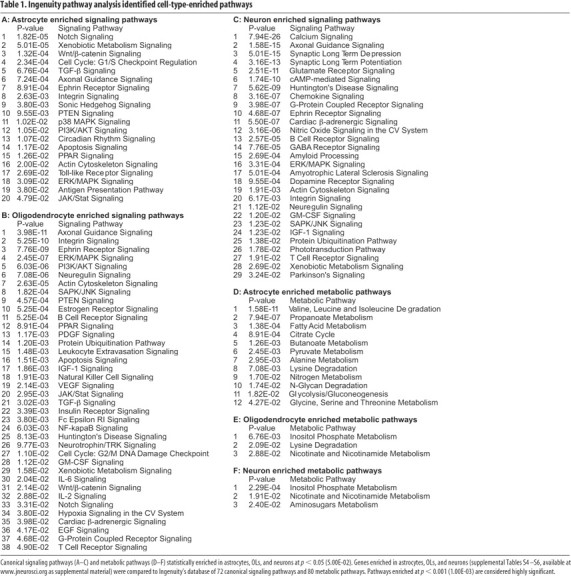
Canonical signaling pathways (A–C) and metabolic pathways (D–F) statistically enriched in astrocytes, OLs, and neurons at p < 0.05 (5.00E-02). Genes enriched in astrocytes, OLs, and neurons (supplemental Tables S4–S6, available at www.jneurosci.org as supplemental material) were compared to Ingenuity's database of 72 canonical signaling pathways and 80 metabolic pathways. Pathways enriched at p < 0.001 (1.00E-03) are considered highly significant.
Analysis of genes expressed by astrocytes
By analyzing acutely isolated, highly purified astrocytes, our genome-wide analysis of their mRNA expression provides the first truly comprehensive look at the genes expressed by astrocytes in the developing and mature mouse CNS, providing new insight into astrocyte specification, development, function, and signaling interactions with blood vessels and synapses. Our analysis identified many transcription factors, signaling transmembrane receptors, and secreted proteins specifically and highly expressed by astrocytes (supplemental Tables S9–S13, available at www.jneurosci.org as supplemental material). The development of astrocytes is less well understood than that of neurons and OLs. We identified DNA binding proteins that are highly enriched in astrocytes, some at very high levels (supplemental Table S10, available at www.jneurosci.org as supplemental material). These genes include both transcriptional activators and repressors, and are likely to encompass many of the key genes that specify astrocyte gene expression. As there are presently no known astrocyte-specific transcription factors in the mammalian brain [although one has been described in a restricted spinal cord domain (Muroyama et al., 2005)], these astrocyte-enriched genes represent the first astrocyte-specific transcription factors. Many of the genes are astrocyte-enriched, although only three (Rfx4, Pbxip1, and Gli3) meet our criteria for being astrocyte-specific (at least 20-fold astrocyte-enriched). Many other transcription factors such as Sox2, Pax6, Id1, and Id3, thought to be enriched in progenitor cells (Ramalho-Santos et al., 2002), were expressed at surprisingly high levels in astrocytes. These findings raise the persistent question of whether the majority of adult astrocytes are terminally differentiated or may, under the right circumstances, be able to divide or even revert to stem-cell-like behavior (Doetsch et al., 1999).
A number of receptors already reported to be important for early astrocyte development such as Bmpr1a, Bmpr1b, Bmpr2, Notch1/2/3, and Fgfr3 (Song and Ghosh, 2004) are highly expressed and enriched in our profiles of developing and mature astrocytes (supplemental Table S11, available at www.jneurosci.org as supplemental material). The Sonic Hedgehog receptor patched (Ptch1) and the Shh signal transducer smoothened (Smo) are both enriched in astrocytes along with their downstream transcriptional mediators Gli2 and Gli3 (Ruiz i Altaba et al., 2002; Palma et al., 2005). Frizzled homolog 2 (Fzd2), a Wnt receptor, is highly expressed and is one of the most astrocyte specific genes. The signaling pathways characterized by these receptors, such as TGF-β (BMP), Notch, Sonic Hedgehog, and Wnt/β-catenin signaling pathways, are determined to be all significantly enriched in astrocytes by IPA (Table 1 A). Interestingly, although downstream Notch target genes turned off as OPCs matured into myelinating OLs, Notch target genes remain high, and sometimes increase, as astrocytes mature. Although Notch signaling has been implicated in inducing neural progenitor cells to develop into astrocytes (Tanigaki et al., 2001), these findings suggest a more sustained role of Notch signaling in maintaining astrocyte fate.
We identified specific high-level expression by astrocytes of several highly conserved, complete molecular pathways for engulfment and phagocytosis (Table 2). One was the cell death abnormality (ced) pathway first identified as having defective apoptotic cell phagocytosis in C. elegans and also shown to function in axon pruning in Drosophila (Zhou et al., 2004; Awasaki et al., 2006). The ced genes form two parallel pathways leading to ced-10 (Rac1) activation and engulfment. The first pathway contains the genes ced-1 (Megf10, Drosophila draper), ced-7 (Abca1), and ced-6 (Gulp1), all of which are enriched in astrocytes (Table 2). Another homolog of ced-1 is Lrp1 (cd91), which is also highly expressed and enriched in astrocytes and is a receptor for ApoE, a gene linked to familial Alzheimer's disease (Corder et al., 1993). The second parallel ced pathway is the ced-2 (Crk), ced-5 (Dock1), ced-12 (Elmo) complex, and all of these genes are also expressed at high levels in astrocytes, although not as highly enriched as the ced-7/ced-6/ced-1 pathway (Table 2). The molecular components for other well defined phagocytosis pathways, the receptor tyrosine kinase (Mertk and Axl) and αvβ5 integrin pathway (Finnemann and Nandrot, 2006), were also found to be expressed by astrocytes. These are all recognition receptors (see Discussion) that are engaged to engulf debris coated by specific opsonins. We also identified their cognate ligands, including Mfge8 and Gas6, as enriched in astrocytes. Mfge8 was one of the few most highly expressed astrocyte genes encoding a secreted protein, as was ApoE, both of which function, at least in part, as opsonins. Together, these findings strongly suggest an important role for developing and mature astrocytes in phagocytosis.
Table 2.
Genes in defined phagocytosis and engulfment pathways
| Probe set ID | Astrocytes (P7) | Astrocytes (P17) | Neurons (P16) | Myelin oligos | Gene symbol | Gene title |
|---|---|---|---|---|---|---|
| 1451086_s_at | 12835 | 17817 | 9937 | 19272 | Rac1 (ced-10) | RAS-related C3 botulinum substrate 1 |
| 1421840_at | 13229 | 11313 | 443 | 239 | Abca1 (ced-7) | ATP-binding cassette, subfamily A (ABC1), member 1 |
| 1453771_at | 840 | 770 | 105 | 69 | Gulp1 (ced-6) | GULP, engulfment adaptor PTB domain containing 1 |
| 1429841_at | 4746 | 8268 | 149 | 2340 | Megf10 (ced-1) | Multiple EGF-like-domains 10 |
| 1448655_at | 4671 | 5922 | 1693 | 86 | Lrp1 (ced-1) | Low-density lipoprotein receptor-related protein 1 |
| 1448248_at | 5831 | 6609 | 4578 | 5183 | Crk (ced-2) | v-crk sarcoma virus CT10 oncogene homolog (avian) |
| 1452220_at | 4040 | 3898 | 175 | 1574 | Dock1 (ced-5) | Dedicator of cyto-kinesis 1 |
| 1456098_a_at | 1232 | 4115 | 1553 | 801 | Elmo2 (ced-12) | Engulfment and cell motility 2, ced-12 homolog (C. elegans) |
| 1422869_at | 1948 | 7370 | 59 | 89 | Mertk | c-mer proto-oncogene tyrosine kinase |
| 1423586_at | 2145 | 2598 | 52 | 43 | Axl | AXL receptor tyrosine kinase |
| 1417399_at | 503 | 1991 | 1485 | 268 | Gas6 | Growth arrest specific 6 |
| 1452784_at | 9018 | 10595 | 1757 | 4162 | Itgav | Integrin α V |
| 1417533_a_at | 8569 | 6783 | 193 | 231 | Itgb5 | Integrin β 5 |
| 1420911_a_at | 25011 | 31089 | 617 | 754 | Mfge8 | Milk fat globule-EGF factor 8 protein |
| 1417876_at | 36 | 50 | 11 | 36 | Fcgr1 | Fc receptor, IgG, high-affinity I |
| 1435477_s_at | 48 | 21 | 32 | 54 | Fcgr2b | Fc receptor, IgG, low-affinity IIb |
| 1448620_at | 24 | 33 | 34 | 72 | Fcgr3 | Fc receptor, IgG, low-affinity III |
| 1418340_at | 21 | 55 | 22 | 80 | Fcer1g | Fc receptor, IgE, high-affinity I, gamma polypeptide |
| 1450678_at | 55 | 33 | 79 | 79 | Cd11/Itgb2 | Integrin β 2 |
| Reference cell-type-enriched genes | ||||||
| 1440142_s_at | 9742 | 13827 | 31 | 77 | Gfap | Glial fibrillary acidic protein |
| 1448768_at | 7 | 14 | 6 | 24964 | Mog | Myelin oligodendrocyte glycoprotein |
| 1433884_at | 326 | 339 | 18197 | 239 | Syt1 | Synaptotagmin I |
Molecular components of phagocytosis and engulfment pathways are enriched and expressed at high levels in astrocytes. See Results and Discussion for a complete description of the different pathways.
Analysis of genes expressed by oligodendrocytes
The list of genes specific to OLs indicates that, in contrast to the list of astrocyte- and neuron-enriched genes, many of the cell-type-specific genes in OLs have been previously characterized. Fifty cell-type-specific genes (>20-fold enrichment) were identified. Of these genes, 16 are previously well established OL genes such as myelin-associated oligodendrocytic basic protein (Mobp), Transferrin (Trf), and Claudin 11 (Cldn11). More than half of these 50 OL-specific genes are associated with the term “membrane” in the DAVID database (http://david.abcc.ncifcrf.gov/), perhaps not surprisingly given the highly specialized function of OLs in myelin membrane formation. These included previously described markers of the OL lineage such as MOG and myelin and lymphocyte protein (Mal), enzymes required for lipid metabolism and synthesis such as galactose-3-o-sulfotransferase 1 (Gal3st1) and UDP galactosyltransferase 8a (Ugt8a), and also novel transmembrane proteins of unknown function such as transmembrane protein 10 (Tmem10/opalin), Tmem125/6330530A05Rik, and ecotropic viral integration site 2a (Evi2a) (supplemental Table S5, available at www.jneurosci.org as supplemental material). Gene model 98, a likely transcription factor (B. Emery and B. A. Barres, unpublished observations), was identified as one of the most broadly and specifically expressed genes by OLs, which was confirmed by ISH in the ABA. Surprisingly, APP, APLP1, APLP2, and the genes involved in their processing (with the exception of Tace) were highly expressed in OLs, and nearly all were more highly expressed in OLs than in neurons. For example, β-site APP-cleaving enzyme 2 (Bace2) showed an enrichment of 12-fold in the OLs, and was strongly upregulated as OLs differentiated (Fig. 6. We observed that the entire Wnt planar polarity pathway became highly activated as OLs matured, suggesting an important role for this pathway in node of Ranvier and paranodal organization. We also identified many OL-enriched transmembrane receptors, transcription factors, and secreted proteins whose specific roles in OL biology are not yet known. Metabolic differences include the preferential expression of enzymes for the synthesis of creatine (Gamt and Gatm) and carnitine (supplemental Table S14, available at www.jneurosci.org as supplemental material) in OLs.
Figure 6.
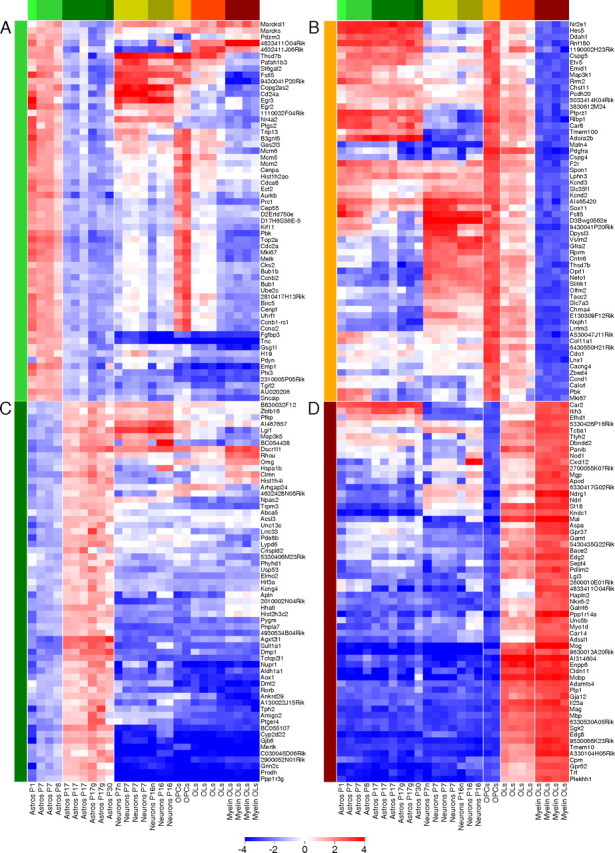
Genes upregulated and downregulated during astrocyte and oligodendrocyte development. The top 60 genes most downregulated during astrocyte development (A, light green bar) and OL development (B, orange bar), and the top 60 genes most upregulated during astrocyte development (C, dark green bar) and OL development (D, red bar). The genes are plotted on a heat map to illustrate gene expression patterns in all CNS cell types at different developmental stages. The individual gene expression level for each cell type is normalized to the age averaged astrocyte expression (A, C) and the age averaged OL expression (B, D). The normalized values are plotted on a log2 color scale, with blue representing low expression and red representing high expression. The fold enrichment can be estimated from the log2 color bar scale. For example, the change from light blue (−1) to medium red (2) represents an eightfold difference in expression level. Note that, although few genes strongly downregulated during development (A, B) are expressed in a cell-type-specific pattern, the majority of genes strongly upregulated during development (C, D) are expressed in a cell-type-specific pattern.
Analysis of genes expressed by neurons
Identification of pan-neuronal markers has long been problematic because of the great degree of regional and subtype neuronal heterogeneity. Our findings, combined with the ISH patterns on the ABA, demonstrate that widely used neuronal markers such as Map2, Tau, HuC/Elav3, Gap43, Prpc, and voltage-dependent sodium channels are not specifically expressed by neurons, and that many specific neuronal genes such as Eno3/NSE, Camk2, and neurofilament chains L, M, and H are only expressed by subsets of neurons. As confirmed by our profiles, many synaptic vesicle proteins such as synaptotagmin I (Syt1) are specifically and broadly expressed by neurons, but, because of their synapse accumulation, are not useful for identification of neuronal cell bodies in cryosections. The neuronal gene that we identified as most highly, specifically, and broadly expressed in our profiles was stathmin-like-2 (see ABA), also known as Scg10. We also identified Brunol4, Brunol5, and Brunol6, members of the Bruno family of RNA binding proteins (Good et al., 2000), as being highly neuron-enriched (although they are by no means pan-neuronal). This gene family has been implicated in developmentally regulated alternative splicing (Ladd et al., 2001), and Brunol6 has been previously shown by ISH to be neuron-specific (McKee et al., 2005). The strong neuronal enrichment of three members of the Bruno family suggests that these genes are important factors for neuron-specific gene splicing. Last, we identified several highly expressed neuronal secreted proteins including neuregulin 3 and Nov/Ccn3. The latter is a member of the connective tissue growth factor family, with strong expression in the cerebral cortex (Fig. 4 A). Nov is a cysteine-rich secreted protein thought to function as a growth factor (Liu et al., 1999) One proposed receptor for Nov is Ddr1 (Das et al., 2006), which our data indicate is highly expressed by developing, but not mature, astrocytes and OLs.
Gene expression changes during astrocyte and oligodendrocyte development
We next identified the genes significantly upregulated and downregulated during astrocyte and OL development (by >1.5-fold and at a FDR <1%) by comparing developing (P7–P8) with mature (P17) astrocytes and by comparing OPCs with myelinating OLs (MOG+ OLs) (supplemental Tables S15–S18, available at www.jneurosci.org as supplemental material). The top 60 genes most upregulated and downregulated during astrocyte and OL development are shown in Figure 6 and the top 250 are shown in supplemental Figure S9 (available at www.jneurosci.org as supplemental material). Global gene expression in P17 and P30 astrocytes was quite similar, and genes upregulated during astrocyte development did not continue to change from P17 to P30 (supplemental Figs. S2 and S9, P30 astrocyte column, available at www.jneurosci.org as supplemental material). This suggests that just as astrocytes acquire their mature morphology at approximately postnatal day 14–21 (Bushong et al., 2004) that by P17 they also express their mature gene expression profile and astrocyte development is essentially complete.
We found that nearly all of the genes that are strongly downregulated during astrocyte and OL development (Fig. 6 A,B) are expressed by another cell type in the CNS, only 11 of 60 (18%) are astrocyte-enriched and only 3 of 60 (5%) are enriched within the OL lineage. This suggests that the most downregulated genes represent those generally required for proliferation and CNS development rather than genes involved in specific cell type developmental processes. In contrast, of those genes most strongly upregulated during astrocyte development, 46 of 60 (77%) are astrocyte-enriched, and nearly all genes upregulated during OL development, 58 of 60 (97%), are OL-enriched.
More than half of the genes most downregulated during astrocyte development are also downregulated during OL development, and many of these genes are involved in the cell cycle such as mitogen signaling pathways, cell division cycle regulating proteins, centromere proteins, and genome replication proteins. One typical gene downregulated in both astrocytes and OPCs is maternal embryonic leucine zipper kinase (Melk), a marker for self-renewing neural progenitor cells that promotes the cell cycle in proliferating progenitors (Nakano et al., 2005). Melk was tenfold downregulated from P7 to P17 and was called absent in all P17 astrocyte and Myelin OL samples. A closer look at the top 50 genes downregulated in astrocytes shows a similar pattern, with 48 of 50 (96%) below the expression threshold in P17 cerebral cortical gray matter astrocytes, strongly suggesting complete senescence of cell division in the vast majority of cortical astrocytes by P17, as has previously been documented. One example of a well characterized astrocyte progenitor cell marker that is not expressed by OPCs is nestin, a marker expressed by radial glia and by astrocytes during development and after injury, but not by normal mature astrocytes. Nestin is downregulated threefold from P1 to P7 astrocytes, downregulated another fourfold from P7 to P17 astrocytes, and called absent in all P17 astrocyte samples. Two of the most downregulated genes during astrocyte development are uncharacterized DNA binding proteins (Tcfcp2l1 and Riken gene 2310005P05 with a helix-hairpin-helix DNA-binding, class 1 protein domain). They are both astrocyte-enriched (13-fold and fivefold respectively) and not expressed by OPCs, suggesting a specific role in astrocyte lineage development.
One of the genes most strongly upregulated during astrocyte development is BC055107, the homolog in humans known as Drr1/Tu3a/Fam107a. Among the CNS neural cell types, Drr1 is astrocyte-specific (21-fold enriched in astrocytes), and one of the most highly expressed genes. Drr1, a 144 amino acid protein with a nuclear localization signal, was identified as a human tumor suppressor gene deleted in tumors from multiple tissues (Wang et al., 2000). The strong upregulation of Drr1 during mouse astrocyte development, combined with its characterization as a tumor suppressor, strongly suggest a functional role of Drr1 in maintaining cells in a quiescent, nonproliferating state. This gene deserves attention in future studies to determine whether polymorphisms or mutations predispose to glioblastoma susceptibility.
The finding that genes downregulated during OPC differentiation are generally expressed by other CNS cell types (Fig. 6 B) suggests a paucity of OPC-specific markers relative to markers for differentiated OLs. The exceptions to this trend included the two well established OPC markers, PDGFRα and NG2/Cspg4, but also some less well characterized genes such as Matrilin-4 (Matn4), Ligand-of-Numb protein X (Lnx1), protein kinase cGMP-dependent (Prkg2), and zinc finger BED domain containing 4 (Zbed4). Zbed4 is a member of the human hAT family that self-associates and acts as a transcriptional activator of cell cycle genes (Yamashita et al., 2007). G-protein-coupled receptor 17 (Gpr17) is strongly and specifically expressed by developing OLs and by adulthood appears specific to adult OPCs.
As OPCs differentiate into OLs and then Myelin OLs (supplemental Tables S18, S19, available at www.jneurosci.org as supplemental material), concurrent with the upregulation of myelin genes, their gene profiles reveal a dramatic downregulation of cytoskeletal and matrix genes that might sustain the cytoskeleton, and a strong upregulation of a large number of genes whose function appears to be to depolymerize the cytoskeleton (such as gelsolin) or to collapse the cytoskeleton (such as proteins in the ephrin and semaphorin signaling pathways), all of which may potentially be required for myelin compaction. Genes upregulated concurrently with MOG and in vivo myelination include genes associated with the paranode, and genes required for normal node of Ranvier formation, such as MAL, hyaluronan and proteoglycan link protein 2 (Hapln2), Contactin 3 (Cntn3), Cntn2, Nkx6.2, Stathmin1 (Stmn1), and RIKA330104H05 (Ermin). These observations strongly suggest that the genes underlying myelination may be regulated in distinct waves (Dugas et al., 2006), the first being involved in the ensheathment of the nerve fibers and the second in establishing the intercellular junctions involved in node of Ranvier formation and maintenance.
Comparison of in vivo astrocytes with in vitro cultured astroglia
Primary cultures of astroglia (McCarthy and de Vellis, 1980) have long served as an in vitro proxy for studying in vivo astrocytes, however, the relationship between preparations of cultured astroglia to normally functioning astrocytes is poorly understood. Unsupervised hierarchical clustering of in vitro cultured astroglia samples along with in vivo purified brain cell types showed that cultured astroglia cluster with the astrocytes, but they branch off at a level similar to the level at which OPC branch away from OLs (supplemental Fig. S10, available at www.jneurosci.org as supplemental material). Based on their gene expression profiles, this data suggests that cultured astroglia and in vivo astrocytes are as different from each other as OPCs are from OLs. The heat map in supplemental Figure S11 (available at www.jneurosci.org as supplemental material) shows the global gene comparison of all 12,416 CNS expressed genes and indicates that cultured astroglia express many of the genes expressed by astrocytes and generally do not express those genes enriched in neurons or OLs. Although their expression is similar, it is clear that some astrocyte genes are not expressed by cultured astroglia, whereas some genes expressed by cultured astroglia are not expressed by astrocytes in vivo. This is in contrast to in vitro cultures of other CNS cell types such as OPCs, OLs, and retinal ganglion cells, in which the genes expressed by cultured cells closely match the genes expressed in vivo (Dugas et al., 2006; Wang et al., 2007). This could not be explained by the presence of serum in the astroglia culture medium, as these differences persisted when we instead cultured the astroglia in serum-free medium (data not shown). We compared cultured astroglia and in vivo astrocytes and identified 2103 cultured astroglia-enriched genes and 2819 in vivo astrocyte-enriched genes (supplemental Tables S20, S21, available at www.jneurosci.org as supplemental material).
These results clearly show that cultured astroglia do not represent the same cell type as in vivo astrocytes, but instead represent an astrocyte-like cell type. One hypothesis is that cultured astroglia represent an immature stage of the astrocyte lineage, such as a glial precursor cell; an alternative hypothesis is that cultured astroglia reflect a reactive astrocyte phenotype. In support of the first hypothesis, the Notch signaling pathway is the most significantly enriched signaling pathway in astrocytes in vivo (Table 1 A) (supplemental Table S22, available at www.jneurosci.org as supplemental material) and we found that Notch target genes remained high, and sometimes increased, as astrocytes in vivo mature. Our profiles also indicated that the Notch pathway is not activated in cultured astroglia. Although the Notch1 and Notch2 ligands are expressed by both in vitro astroglia and in vivo astrocytes, the downstream Notch effecter Hes5 is not expressed in vitro, and Hes1 is expressed at much lower levels in vitro. Interestingly, Notch ligands delta-like 4 (Dll4), Jagged 1 (Jag1), and Jag2 are highly expressed by endothelial cells (R. Daneman and B. A. Barres, unpublished data), which ubiquitously contact astrocytes in vivo, suggesting the possibility that active Notch signaling in the mature CNS is required to maintain a mature astrocyte fate. In support of the alternative hypothesis, the presence of immune system genes (such as complement protein C3) at high levels in cultured astroglia, but not in astrocytes in vivo, provided some evidence that astroglia in culture are more similar to reactive astrocytes. Future studies using similar methods to purify astrocyte progenitor cells and reactive astrocytes, and using the databases we have presented here, should be able to more fully describe the nature of cultured astroglia.
Discussion
A database of transcriptional profiles for CNS neural cell types
In this study, we have described the development of new methods to highly purify acutely isolated astrocytes, neurons, and OLs from developing to mature (P1 to P30) mouse forebrain and used these techniques to produce a transcriptome database of genome-wide gene expression values for the main neural cell types in the CNS. This database provides a resource to the neuroscience community to better understand the development and function of the brain. These same methods can also in principle now be used to study how gene expression changes in astrocytes, neurons, and OLs during brain pathology.
This database of cell-type-specific gene expression complements existing qualitative databases that describe regional gene expression patterns such as the ABA and GENSAT. Comparing our lists of genes displaying CNS cell type specificity with the ABA revealed a high degree of correspondence; however, the coverage of the genome enabled by GeneChip Arrays enabled us to identify a much greater number of differentially expressed genes in a more quantitative manner. Moreover, as we were able to isolate cells from a variety of postnatal ages, our data allows for analysis of developmental profiles of gene expression. This data, in conjunction with the information on regional expression provided by the ABA and mouse lines available from GENSAT, provides neuroscientists with powerful tools to dissect temporal, spatial, and cell-type-specific CNS gene expression.
Identification of new cell-type-specific markers
There is a need for improved neural cell type markers in the CNS. Identification of pan-neuronal, astrocyte, and OL markers has long been problematic because of the great degree of regional and subtype neuronal heterogeneity. In particular, current markers for astrocytes are either not uniformly expressed in all astrocytes, or do not fully label both the astrocyte cell body and all processes. For example, GFAP, the most widely used astrocyte marker, is preferentially expressed in white matter over gray matter astrocytes and does not label all processes (Bushong et al., 2002). Similarly, aquaporin 4, although highly astrocyte specific, is localized to astrocyte endfeet. Connexin 43 only labels some astrocytes and is also expressed by endothelial cells, and although S100β labels both gray matter and white matter astrocytes, it also labels OPCs and OLs.
Here, we identified Aldh1L1 as a highly useful new astrocyte-specific marker. We also showed that the GENSAT Aldh1L1-EGFP BAC mouse has fluorescence labeling of astrocytes that faithfully follows the astrocyte-specific Aldh1L1 expression pattern and therefore provides a superior tool for visualization of astrocytes in the developing and adult mouse brain. Given that Aldh1L1 appears to be expressed by nearly all astrocytes, but not other CNS cell types, the Aldh1L1 promoter will likely be a useful tool to drive Cre expression specifically in astrocytes. By their specific expression patterns, Pla2g7 and Ascbg1 are also likely to be useful astrocyte-specific markers.
In a similar manner, our dataset allows for the identification of many suitable markers for neurons and OLs. For OLs, there are many myelin genes such as MBP that have long served as useful markers, but as these are highly localized to myelin, they have not been useful for quantitative studies involving staining of OL cell bodies. We have identified several highly OL specific genes including Fa2h, Gpr62, Tmem125/6330530A05Rik, gene model 98, and Plekhh1 that have great potential to be improved markers for this lineage. Although many genes whose expression was highly specific to neurons were identified, the vast majority of these genes displayed regional specificity when analyzed by ISH or cross referenced with the ABA. Stathmin-like-2 (Scg10) was the most specific and broadly expressed gene we could identify, suggesting its suitability for a pan-neuronal marker.
The astrocyte transcriptome provides many new clues to astrocyte development and function
The astrocyte transcriptome that we have identified provides new clues to the dramatic nature of the division of metabolic labor between the major cell types in the brain. The preferential storage of glycogen and glycoylsis in astrocytes has long been known (Hertz, 2004). Astrocytes are thought to play a key role in degrading glutamate to glutamine and in coupling synaptic activity and glucose utilization (Magistretti, 2006). Our findings confirm the significant astrocyte enrichment of these metabolic pathways, such as glycolysis and the Krebs cycle, but also reveal a large number of other metabolic pathways that are enriched in astrocytes (Table 1 D). For instance, there is a significant enrichment of pathways involved in amino acid synthesis and degradation, such as a preferential presence of the pathways for synthesis of glycine, serine, and cysteine. Certain lipid synthetic pathways are also highly enriched in astrocytes, as shown, for instance, by their preferential expression of Acsbg1/bubblegum/lipidosin, a protein with very long-chain acyl-CoA synthetase activity (Fraisl et al., 2004). Although astrocytes have long been thought to play a primary role in glutamate degradation, our metabolic pathway analysis indicated that astrocytes are also highly enriched for most or all of the major pathways for glutamate production or the 4 carbon backbone of glutamate, suggesting that an important role of astrocytes may actually be to produce neurotransmitters such as glutamate and then send them to neurons in the form of neuroactive immediate precursors such as glutamine. The astrocyte transcriptome, however, does not include mRNAs for vesicular glutamate transporters or synaptic proteins including Vglut1, Vglut2, synapsin 1, and synaptotagmin found in neurons, and thus does not support the idea that astrocytes conduct regulated secretion of glutamate in vivo.
The astrocyte transcriptome that we have elucidated also points to many hypotheses about astrocyte function. Arguably, knowledge of the most highly expressed specific genes within a cell type should provide important clues as to the likely functions of that cell type. For OLs, the most highly expressed specific genes are primarily myelin genes, consistent with their most important role in myelination. For neurons, highly expressed synaptic genes correlate with their main role of neurotransmission. For astrocytes, an examination of the top most expressed specific genes reveals such secreted proteins as ApoE, ApoJ/clusterin, Pla2g7, Sparc, hevin/Sparcl1, and Mfge8, genes about whose function in astrocytes we know very little. Some of these astrocyte-enriched genes might be important for astrocyte-synapse or astrocyte-endothelial cell interactions. Many of these genes are strongly upregulated in astrocytes from P1 to P7 to P17, suggesting that the proteins they encode play important roles later in CNS development around and after the time most synapses are formed. Such a possibility is consistent with evidence of the ability of astrocytes to regulate synaptic formation (Christopherson et al., 2005). We found many other astrocyte-enriched genes that are implicated in psychiatric diseases such as schizophrenia and bipolar disorder, including Npas3 (Pieper et al., 2005), Mlc1 [found in distal astrocyte processes (Boor et al., 2005)], Lgi1/4 (Gu et al., 2004; Schulte et al., 2006), and Gpr56 (Mochida, 2005), providing clues to the potential ways that astrocytes may contribute to synaptic regulation and neurological and psychiatric diseases.
Several evolutionarily conserved phagocytic pathways are highly enriched in astrocytes
In culture, astroglia are highly phagocytic cells (Roldan et al., 1997; Chang et al., 2000). Although the extent to which mammalian astrocytes can phagocytose in vivo has not been clear, CNS glial cells in Drosophila brain that are similar to mammalian astrocytes have been shown to mediate clearance of apoptotic cells and participate in axon pruning (Freeman et al., 2003; MacDonald et al., 2006). Our studies identified the same evolutionarily conserved phagocytic pathways used by fly glia and C. Elegans cells as being specifically and highly expressed by mouse astrocytes in vivo. These include the Ced-1/Draper/Megf10 (and Lrp1/CD91), Ced-7, Ced-6/Gulp1 and ced-2/Crk, ced-5/Dock1, ced-12/Elmo pathways. In addition, we also identified the Mertk/Axl, αvβ5 integrin pathway previously shown to mediate phagocytosis of shedding rod outer segments by retinal pigment epithelial cells (Feng et al., 2002; Duncan et al., 2003; Finnemann and Nandrot, 2006; Nandrot et al., 2007), as highly enriched in astrocytes in vivo. Ligands for each of these pathways, ApoE and Mfge8, are among the most highly expressed astrocyte genes, emphasizing their likely importance to the function of astrocytes in vivo. It is unclear whether these pathways synergize or independently mediate clearance of specific types of debris dependent on receptor specificity. Astrocytes, OLs, and neurons do not express Fc receptor and Cd11/CD18b/integrin β2 phagocytic pathways to mediate clearance of antibody or complement opsonized debris, although these two pathways are highly expressed by microglia.
What are the potential targets of astrocyte phagocytosis? During development, apoptotic cells must be cleared by phagocytosis, and some evidence suggests that mammalian astrocytes in vivo may participate in this process (Krueger et al., 1995). In Drosophila, it has been shown that draper (Megf10), Dock1, and Rac1 are required for cell death attributable to growth differences, and the same neighboring cell receptors that promote death are also those responsible for engulfment (Li and Baker, 2007). A functional role of glial cells in phagocytosis in vivo in the peripheral nervous system has been shown for Schwann cell engulfment of axon tips (axosomes) during the process of synapse elimination (Bishop et al., 2004). Similarly astrocytes have been found to phagocytose CNS synapses (Steward and Messenheimer, 1978). Both Megf10 and Mertk are highly expressed by Schwann cells, and thus are excellent candidates to help mediate this process, suggesting the possibility that the role of these phagocytic pathways in astrocytes may be to help mediate synapse elimination. The phagocytic pathways that we have identified are also excellent candidates to help mediate the clearance of amyloid. Astroglia in vitro, and when cultured on brain slices, robustly clear amyloid deposits (Wyss-Coray et al., 2003), and both ApoE and Mfeg8 help mediate amyloid clearance (Boddaert et al., 2007). Cumulatively, these data suggest that the glial phagocytic pathways we have identified are important new candidates to participate in axon pruning, CNS and PNS synapse elimination, amyloid clearance within the brain, and that disruption of these pathways could lead to amyloid plaque accumulation in Alzheimer's disease.
Footnotes
This work was supported by National Institute of Neurological Disease Grant R01 NS045621 (B.A.B.), National Eye Institute Grant R01 EY10257 (B.A.B.), Medical Scientist Training Program Grant MSTP GM07365 (J.D.C., A.K.), Australian National Health and Medical Research Council CJ Martin Fellowship 400438 (B.E.), a Singapore Agency of Science, Technology and Research graduate scholarship (L.C.F.), National Eye Institute National Research Service Award Postdoctoral Fellowship Grant EY07033 (K.S.C.), and National Institutes of Heath Grant DK54388 and CA095030 (S.A.K.). We extend special thanks to Dr. Lubert Stryer for advice, encouragement, and support in doing the Affymetrix gene profiling and members of the Barres laboratory for discussions and critical readings of this manuscript.
References
- Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol. 2007;500:368–383. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, DePinho RA. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci USA. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, Raff MC. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–661. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Boddaert J, Kinugawa K, Lambert JC, Boukhtouche F, Zoll J, Merval R, Blanc-Brude O, Mann D, Berr C, Vilar J, Garabedian B, Journiac N, Charue D, Silvestre JS, Duyckaerts C, Amouyel P, Mariani J, Tedgui A, Mallat Z. Evidence of a role for lactadherin in Alzheimer's disease. Am J Pathol. 2007;170:921–929. doi: 10.2353/ajpath.2007.060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boor PK, de Groot K, Waisfisz Q, Kamphorst W, Oudejans CB, Powers JM, Pronk JC, Scheper GC, van der Knaap MS. MLC1: a novel protein in distal astroglial processes. J Neuropathol Exp Neurol. 2005;64:412–419. doi: 10.1093/jnen/64.5.412. [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci. 2002;22:183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Ellisman MH. Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int J Dev Neurosci. 2004;22:73–86. doi: 10.1016/j.ijdevneu.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Chang GH, Barbaro NM, Pieper RO. Phosphatidylserine-dependent phagocytosis of apoptotic glioma cells by normal human microglia, astrocytes, and glioma cells. Neuro Oncol. 2000;2:174–183. doi: 10.1093/neuonc/2.3.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Cook RJ, Lloyd RS, Wagner C. Isolation and characterization of cDNA clones for rat liver 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1991;266:4965–4973. [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Das S, Ongusaha PP, Yang YS, Park JM, Aaronson SA, Lee SW. Discoidin domain receptor 1 receptor tyrosine kinase induces cyclooxygenase-2 and promotes chemoresistance through nuclear factor-κB pathway activation. Cancer Res. 2006;66:8123–8130. doi: 10.1158/0008-5472.CAN-06-1215. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JL, LaVail MM, Yasumura D, Matthes MT, Yang H, Trautmann N, Chappelow AV, Feng W, Earp HS, Matsushima GK, Vollrath D. An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest Ophthalmol Vis Sci. 2003;44:826–838. doi: 10.1167/iovs.02-0438. [DOI] [PubMed] [Google Scholar]
- Feng W, Yasumura D, Matthes MT, LaVail MM, Vollrath D. Mertk triggers uptake of photoreceptor outer segments during phagocytosis by cultured retinal pigment epithelial cells. J Biol Chem. 2002;277:17016–17022. doi: 10.1074/jbc.M107876200. [DOI] [PubMed] [Google Scholar]
- Finnemann SC, Nandrot EF. MerTK activation during RPE phagocytosis in vivo requires alphaVbeta5 integrin. Adv Exp Med Biol. 2006;572:499–503. doi: 10.1007/0-387-32442-9_69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisl P, Forss-Petter S, Zigman M, Berger J. Murine bubblegum orthologue is a microsomal very long-chain acyl-CoA synthetase. Biochem J. 2004;377:85–93. doi: 10.1042/BJ20031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PJ, Chen Q, Warner SJ, Herring DC. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- Gu W, Sander T, Becker T, Steinlein OK. Genotypic association of exonic LGI4 polymorphisms and childhood absence epilepsy. Neurogenetics. 2004;5:41–44. doi: 10.1007/s10048-003-0158-8. [DOI] [PubMed] [Google Scholar]
- Hall A, Giese NA, Richardson WD. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development. 1996;122:4085–4094. doi: 10.1242/dev.122.12.4085. [DOI] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Hertz L. Intercellular metabolic compartmentation in the brain: past, present and future. Neurochem Int. 2004;45:285–296. doi: 10.1016/j.neuint.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BK, Burne JF, Raff MC. Evidence for large-scale astrocyte death in the developing cerebellum. J Neurosci. 1995;15:3366–3374. doi: 10.1523/JNEUROSCI.15-05-03366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13:227–236. [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. research0032.1–0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Baker NE. Engulfment is required for cell competition. Cell. 2007;129:1215–1225. doi: 10.1016/j.cell.2007.03.054. [DOI] [PubMed] [Google Scholar]
- Liu C, Liu XJ, Crowe PD, Kelner GS, Fan J, Barry G, Manu F, Ling N, De Souza EB, Maki RA. Nephroblastoma overexpressed gene (NOV) codes for a growth factor that induces protein tyrosine phosphorylation. Gene. 1999;238:471–478. doi: 10.1016/s0378-1119(99)00364-9. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Ludwin SK, Kosek JC, Eng LF. The topographical distribution of S-100 and GFA proteins in the adult rat brain: an immunohistochemical study using horseradish peroxidase-labelled antibodies. J Comp Neurol. 1976;165:197–207. doi: 10.1002/cne.901650206. [DOI] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Magistretti PJ. Neuron-glia metabolic coupling and plasticity. J Exp Biol. 2006;209:2304–2311. doi: 10.1242/jeb.02208. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH. Cortical malformation and pediatric epilepsy: a molecular genetic approach. J Child Neurol. 2005;20:300–303. doi: 10.1177/08830738050200040501. [DOI] [PubMed] [Google Scholar]
- Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438:360–363. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- Nakano I, Paucar AA, Bajpai R, Dougherty JD, Zewail A, Kelly TK, Kim KJ, Ou J, Groszer M, Imura T, Freije WA, Nelson SF, Sofroniew MV, Wu H, Liu X, Terskikh AV, Geschwind DH, Kornblum HI. Maternal embryonic leucine zipper kinase (MELK) regulates multipotent neural progenitor proliferation. J Cell Biol. 2005;170:413–427. doi: 10.1083/jcb.200412115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC. Essential role for MFG-E8 as ligand for αvβ5 integrin in diurnal retinal phagocytosis. Proc Natl Acad Sci USA. 2007;104:12005–12010. doi: 10.1073/pnas.0704756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neymeyer V, Tephly TR, Miller MW. Folate and 10-formyltetrahydrofolate dehydrogenase (FDH) expression in the central nervous system of the mature rat. Brain Res. 1997;766:195–204. doi: 10.1016/s0006-8993(97)00528-3. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci. 2006;26:9881–9891. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma V, Lim DA, Dahmane N, Sanchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Ruiz i Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper AA, Wu X, Han TW, Estill SJ, Dang Q, Wu LC, Reece-Fincanon S, Dudley CA, Richardson JA, Brat DJ, McKnight SL. The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc Natl Acad Sci USA. 2005;102:14052–14057. doi: 10.1073/pnas.0506713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Roldan A, Gogg S, Ferrini M, Schillaci R, De Nicola AF. Glucocorticoid regulation of in vitro astrocyte phagocytosis. Biocell. 1997;21:83–89. [PubMed] [Google Scholar]
- Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- Schulte U, Thumfart JO, Klocker N, Sailer CA, Bildl W, Biniossek M, Dehn D, Deller T, Eble S, Abbass K, Wangler T, Knaus HG, Fakler B. The epilepsy-linked lgi1 protein assembles into presynaptic kv1 channels and inhibits inactivation by kvbeta1. Neuron. 2006;49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Segal MM, Baughman RW, Jones KA, Huettner JE. Mass cultures and microislands of neurons from postnatal rat brain. In: Banker G, Goslin K, editors. Culturing nerve cells. Ed 2. Cambridge, MA: MIT; 1998. pp. 309–338. [Google Scholar]
- Solly SK, Thomas JL, Monge M, Demerens C, Lubetzki C, Gardinier MV, Matthieu JM, Zalc B. Myelin/oligodendrocyte glycoprotein (MOG) expression is associated with myelin deposition. Glia. 1996;18:39–48. doi: 10.1002/(SICI)1098-1136(199609)18:1<39::AID-GLIA4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Steward O, Messenheimer JA. Histochemical evidence for a post-lesion reorganization of cholinergic afferents in the hippocampal formation of the mature cat. J Comp Neurol. 1978;178:697–709. doi: 10.1002/cne.901780407. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001;29:45–55. doi: 10.1016/s0896-6273(01)00179-9. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci. 2007;27:8593–8603. doi: 10.1523/JNEUROSCI.4488-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Darling J, Zhang JS, Liu W, Qian J, Bostwick D, Hartmann L, Jenkins R, Bardenhauer W, Schutte J, Opalka B, Smith DI. Loss of expression of the DRR 1 gene at chromosomal segment 3p21.1 in renal cell carcinoma. Genes Chromosomes Cancer. 2000;27:1–10. doi: 10.1002/(sici)1098-2264(200001)27:1<1::aid-gcc1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, Silverstein SC, Husemann J. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med. 2003;9:453–457. doi: 10.1038/nm838. [DOI] [PubMed] [Google Scholar]
- Xing Y, Kapur K, Wong WH. Probe selection and expression index computation of affymetrix exon arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita D, Komori H, Higuchi Y, Yamaguchi T, Osumi T, Hirose F. Human DNA replication-related element binding factor (hDREF) self-association via hATC domain is necessary for its nuclear accumulation and DNA binding. J Biol Chem. 2007;282:7563–7575. doi: 10.1074/jbc.M607180200. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Mangahas PM, Yu X. The genetics of hiding the corpse: engulfment and degradation of apoptotic cells in C. elegans and D. melanogaster . Curr Top Dev Biol. 2004;63:91–143. doi: 10.1016/S0070-2153(04)63004-3. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Lubischer JL, Kang H, Tian L, Mikesh M, Marks A, Scofield VL, Maika S, Newman C, Krieg P, Thompson WJ. Fluorescent proteins expressed in mouse transgenic lines mark subsets of glia, neurons, macrophages, and dendritic cells for vital examination. J Neurosci. 2004;24:10999–11009. doi: 10.1523/JNEUROSCI.3934-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]