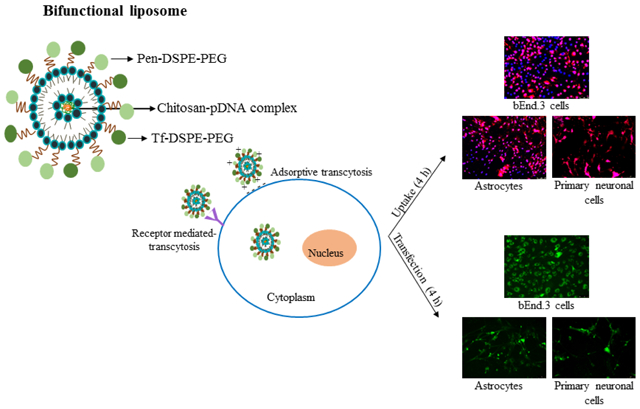
Abstract
Liposome based delivery systems provide a promising strategy for treatment of neurodegenerative diseases. A rational design of brain-targeted liposomes can support the development of more efficient treatments with drugs and gene materials. Here, we characterized surface modified liposomes with transferrin (Tf) protein and penetratin (Pen), a cell-penetrating peptide, for efficient and targeted gene delivery to brain cells. PenTf-liposomes efficiently encapsulated plasmid DNA, protected them against enzymatic degradation and exhibited a sustained in vitro release kinetics. The formulation demonstrated low cytotoxicity and was non-hemolytic. Liposomes were internalized into cells mainly through energy-dependent pathways especially clathrin-mediated endocytosis. Reporter gene transfection and consequent protein expression in different cell lines were significantly higher using PenTf-liposomes compared to unmodified liposomes. The ability of these liposome to escape from endosomes can be an important factor which may have likely contributed to the high transfection efficiency observed. Rationally designed bifunctional targeted-liposomes provide an efficient tool for improving the targetability and efficacy of synthesized delivery systems. This investigation of liposomal properties attempted to address cell differences, as well as, vector differences, in gene transfectability. The findings indicate that PenTf-liposomes can be a safe and non-invasive approach to transfect neuronal cells through multiple endocytosis pathways.
Keywords: bifunctional liposome, transferrin, penetratin, gene delivery, brain targeting
Graphical abstract
1. Introduction
The introduction of nanoparticles in pharmaceutical drug delivery has led to tremendous advancement in the field by opening avenues for transport and target specific delivery of therapeutic molecules, especially those with unfavorable physicochemical properties. Liposomes are widely used nanoparticle based delivery systems with different applications due to their versatility in modifying physicochemical and biophysical properties of drugs, thus modulating the biological outcome (Bulbake et al., 2017; Sercombe et al., 2015; Zylberberg and Matosevic, 2016). They are vesicles consisting of one or more concentric bilayers of phospholipids with an aqueous core to enable loading of both hydrophilic and hydrophobic molecules. The dynamic and adaptable nature of liposomes allow prolonged systemic circulation as well as delivery and accumulation in specific tissues (Panahi et al., 2017; Zylberberg et al., 2017). Liposomes confer additional advantages including increased drug stability (Zhang and Anchordoquy, 2004), bioavailability (Çağdaş et al., 2004), controlled release (Sercombe et al., 2015), biocompatibility (Knudsen et al., 2015) and capacity to carry molecules with a wide range of size (Balazs and Godbey, 2011).
Currently, neurodegenerative disorders represent a growing burden to the public health. Scarcity of efficient treatments for these diseases not only lead to suffering of patients but also present financial challenges (Kirson et al., 2016; Prince et al., 2013). Since the current treatments have largely failed in reversing the disease progression and restoring cellular functions, many efforts have been made with the intention to discover innovative and definitive treatments for these types of disorders. Among the therapies, gene therapy appears promising and offers to counter or replace the malfunctioning/causative gene (Kumar et al., 2016; Naldini, 2015). The biological barriers remain the main obstacles in the delivery of therapeutic molecules to brain cells that is particularly pertinent for DNA molecules owing to their high molecular weights and anionic nature. Liposomes can be designed to breach the blood brain barrier (BBB) and deliver drugs specifically to the brain cells. Additionally, they are suitable gene carrier that can not only carry plasmid DNA (pDNA), but also protect them against enzymatic degradation(Ma et al., 2007; Safari and Zarnegar, 2014). Efficient gene delivery capacity has been demonstrated by cationic liposomes and therefore they have been extensively utilized as gene carriers (Li et al., 2011; Michel et al., 2017; Shim et al., 2013; Yang et al., 2014). The electrostatic interactions between positively charged liposomes and negatively charged cell membrane usually aid liposome internalization into cells and consequently support pDNA delivery to the nucleus (Obata et al., 2009; Wiethoff et al., 2002). However, the cationic surfaces of liposomes also have the propensity to interact electrostatically with blood and components related to the immune system. Hydrophilic polymer such as poly(ethylene glycol) (PEG) has demonstrated the ability to reduce protein binding interaction to liposomes and prolong the half-life of particles in blood (Kolate et al., 2014; Ogris et al., 1999).
Effective delivery of liposomes to target cells can be achieved by decorating liposomal surface with ligands of specific receptors that are present on the surface of target tissues, which enables development of strategic targeted delivery systems (Zylberberg et al., 2017). Transferrin receptor (TfR), a transmembrane protein overexpressed on brain endothelial cells, has been extensively explored for delivery of therapeutics to the brain. However, the mechanism involved in the receptor-mediated transcytosis of Tf-modified liposomes across brain endothelial cells is still unclear (Bien-Ly et al., 2014; Johnsen and Moos, 2016). Nevertheless, targeting via TfR allows a high degree of internalization of carriers, but receptor saturation can be a drawback (Xiao and Gan, 2013). The capacity of cell-penetrating peptide (CPP) in translocating a variety of cargoes into the cell in a non-invasive manner without the use of receptors might be an additional strategy to enhance carrier internalization. CPPs have been successfully applied in drug delivery amongst which penetratin (Pen), a CPP derived from Antennapedia homeodomain, has demonstrated capability to penetrate neurons and accumulate in the nucleus (Ramsey and Flynn, 2015). The cationic-amphiphilic character of Pen is involved in interaction with lipid components of cellular membrane and subsequent internalization into the cell (Bashyal et al., 2016; Zhang et al., 2016). Numerous studies have demonstrated the enhanced drug delivery abilities of Pen-modified liposomes (Chikh et al., 2001; Marty et al., 2004). However, the combination of multiple strategies including receptor targeting and enhanced cell penetration, has been found to deliver genes across the BBB more efficiently (Balducci et al., 2014; Bana et al., 2014; Chen et al., 2016; Sharma et al., 2013).
In this study, we designed liposomes for efficient gene delivery to neuronal cells by modifying the surface of liposomes with Tf protein and Pen. Two plasmids (plasmid green fluorescent protein- pGFP and plasmid βgalactosidase- pβgal) were used as models for transfection. To achieve the best transfection efficiency, we complexed DNA with chitosan and loaded them into liposomes, thereby taking advantage of the unique gene delivery properties of chitosan such as DNA condensation, protection against enzymatic degradation and enhancement in transfection efficiency (Cifani et al., 2015; Mao et al., 2010). The binding affinity of chitosan to pDNA as well as the capacity of the nanoparticles to protect pDNA against enzymatic degradation were evaluated. Hemolytic activity and cytotoxicity profile of the formulations were also evaluated to determine the in vivo biocompatibility of liposomes. Cellular uptake mechanisms and transfection efficiency of liposomal formulations were examined in bEnd.3 cells, astrocytes and primary neuronal cells. Finally, the contribution of endosomal escape in improving transfection efficiency in bEnd.3 cells was also investigated.
2. Material and methods
2.1. Materials
The phospholipids, dioleoyl-3-trimethylammonium-propane chloride (DOTAP), dioleoyl-snglycero-3-phosphoethanolamine (DOPE), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) were purchased from Avanti Polar Lipids (Birmingham, AL, USA). The phospholipid DSPE–PEG2000–NHS was purchased from Biochempeg Scientific Inc (Watertown, MA, USA). Holo-transferrin bovine, cholesterol, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), Ethylenediaminetetraacetic acid (EDTA), 1,1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (DiI), Hoechst 33342, Ethidium bromide (EtBr), Sodium azide, Amiloride and Triton™ X-100 were obtained from Sigma–Aldrich (St. Louis, MO, USA). Chlorpromazine and Colchicine were obtained from Enzo Life Sciences (Farmingdale, NY, USA). Chitosan (MW 30 kDa) was purchased from Glentham Life Sciences (Corsham, UK). Plasmid DNA encoding β-galactosidase (gWiz-βGal) and plasmid DNA encoding Green Fluorescent Protein (gWiz-GFP) were purchased from Aldevron LLC (Fargo, ND, USA). Dulbecco’s Modified Eagle Medium (DMEM), and phosphate buffered saline (PBS) were purchased from Corning Incorporated (Corning, NY, USA). Fetal bovine serum (FBS) was purchased from JR Scientific Inc. (Woodland, CA, USA). β-galactosidase enzyme assay kit with reporter lysis buffer was supplied by Promega (Madison, WI, USA).
2.2. Conjugation of Pen to DSPE-PEG2000-NHS and Tf to DSPE-PEG2000-NHS
Pen and Tf were conjugated to terminal NHS-activated DSPE-PEG2000 phospholipid, separately. Pen and DSPE-PEG2000-NHS were dissolved in anhydrous DMF at 1:5 molar ratio, after adjusting the pH to 8.0-8.5 with triethylamine. The reaction was allowed to continue for 120 h at room temperature with gentle stirring. The resultant reaction mixture was dialyzed (molecular weight cut-off of 3500 Da) in deionized water for 48 h to remove uncoupled Pen. The dialysate was lyophilized and stored at −20 °C until use.
For conjugation of Tf to DSPE-PEG2000-NHS, 125 μg Tf/μM phospholipid were dissolved in anhydrous DMF, after adjusting pH to 8.0-8.5 with triethylamine. The reaction was continued for 24 h at room temperature under moderate stirring. Unbound protein was removed using Sephadex G-100 column. Coupling efficiencies of both reactions were determined using BCA protein assay (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Preparation and characterization of liposomes
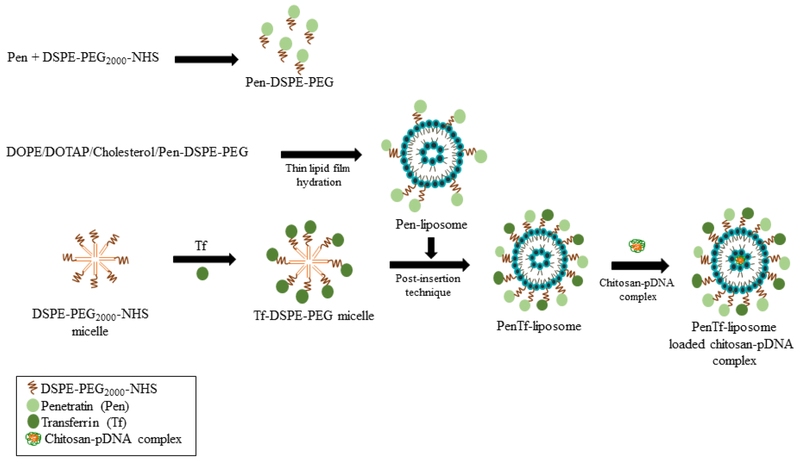
Liposomes were prepared in a three-step process using thin lipid film hydration method and were made bi-functional through incorporation of Tf-micelles into Pen-liposomes by post-insertion technique(Sharma et al., 2012). First, DOPE/DOTAP/Cholesterol/Pen-PEG-lipid (45:45:2:4 mol %) were dissolved in chloroform:methanol (2:1, v/v) and dried to form a thin lipid film, which was thereafter hydrated using HEPES buffer (pH 7.4). DiI or Lisamime rhodamine B (0.5 mol %) were incorporated into the liposomes to prepare fluorescence labeled-liposomes. Second, Pen-liposomes were stirred overnight with Tf-micelles to form PenTf-liposomes. Free Tf was removed from the PenTf-liposomes using Sephadex G-100 column and the incorporation percentage of Tf in Pen-liposomes surface to form PenTf-liposomes was determined using micro BCA assay. At last, chitosan-pGFP or chitosan-pβgal polyplexes were added to hydration buffer for incorporation into liposomes (Figure 1). Hydrodynamic size and zeta potential of the formulations were determined by dynamic light scattering using Zetasizer Nano ZS 90 (Malvern Instruments, Malvern, UK) at 25 °C.
Fig. 1.
Preparation of liposomes: Schematic represents formation of brain targeted dual modified liposomes encapsulating pDNA/chitosan complex.
2.4. Chitosan-pDNA binding ability
The optimal chitosan-pDNA N/P ratio (molar ratios of the amine groups in chitosan and phosphate groups in DNA) was monitored by EtBr exclusion assay and agarose gel electrophoresis(Layek and Singh, 2013a). Naked pDNA was used as a positive control. The complexes containing 1 μg of pDNA at different chitosan weight ratios were stained with EtBr (0.5 μg) for 5 min and the fluorescence intensity was measured using a spectrophotometer (excitation/emission wavelengths: 260/600 nm, respectively). Relative fluorescence intensity of EtBr solution in the presence of free plasmid corresponded to 0% condensation while fluorescence intensity without plasmid corresponded to 100% condensation. For agarose gel electrophoresis, the complexes (1 μg pDNA at different chitosan weight ratios) were loaded in 0.8% w/v agarose gel stained with EtBr (0.5 μg/mL) and electrophoresed at 80 V in 0.5X Tris-acetate-EDTA (TAE, Bio-Rad, CA, USA) buffer for 80 min. The pDNA migration was thereafter recorded.
2.5. pDNA encapsulation efficiency
Encapsulation efficiencies of liposomal formulations containing pGFP or pβgal were calculated using DNA intercalating dye Hoechst 33342 (0.15 μg/mL)(Zhang et al., 2007). Fluorescence was measured using Spectramax M5 microplate spectrophotometer (Molecular Devices, Sunnyvale, CA) at 354 nm excitation and 458 nm emission wavelengths. Percent encapsulation was calculated considering the absorbance in presence of 0.5% v/v Triton X-100 as 100%.
2.6. DNase protection assay
The ability of liposomal formulations to protect pDNA against enzymatic degradation was examined by DNase I protection assay(Layek et al., 2014). Formulations containing 1 μg pDNA were incubated for 60 min at 37 °C with 1-unit DNase I. Naked pDNA incubated with DNase I was used as a positive control. After incubation, the reaction was stopped by adding 5 μl of EDTA (100 mM). Subsequently, 20 μL of heparin (5 mg/mL) was added and the mixture incubated for 2 h at room temperature to release the pDNA from the complex. The released pDNA samples were subjected to agarose gel electrophoresis 0.8% (w/v) at 80 V for 80 min.
2.7. In vitro release
Liposomal formulations containing 50 μg of pDNA were dispersed in 10 mL of PBS (pH 7.4) and incubated at 37 °C under constant shaking at 50 rpm. At predetermined time intervals, 300 μL of the suspension was withdrawn and centrifuged at 5,000 rpm, 4 °C, for 30 min. After staining with Hoechst 33342 (0.15 μg/mL), the content of pDNA was determined using fluorescence spectrophotometer (excitation/emission wavelengths: 354/458 nm, respectively). The cumulative amount of released pDNA from the liposomes was then calculated.
2.8. Blood compatibility study
Hemolytic activity of liposomal formulation was assessed in vitro to estimate compatibility of liposomes with blood. Erythrocytes were harvested from freshly withdrawn blood of Sprague-Dawley rats followed by centrifugation at 1,500 rpm for 10 min and washed three times with PBS, pH 7.4, 10 mM CaCl2. The erythrocyte solution (2% v/v) was incubated with negative control (PBS), positive control (Triton X-100 1% v/v) or liposomal formulations (31.25-1000 μM) for 1 h at 37 °C, 5% CO2. Thereafter, cell suspension was centrifuged at 1,500 rpm for 10 min. The supernatants were then removed and the absorbance of released hemoglobin was measured using a spectrophotometer at 540 nm. Percent hemolysis was calculated considering the absorbance in the presence of Triton X-100 as 100%.
2.9. Cell culture
Primary astrocytes and primary neuronal cells were isolated from 1-day old rats (Sumners and Fregly, 1989). Briefly, the meninges were removed from the dissected brains. Minced brains were incubated with DMEM containing 0.25% trypsin and DNase I (8μg/mL) at 37 °C. Cells were isolated and cultured in DMEM with 10% v/v fetal bovine serum (FBS) and 1% v/v penicillin streptomycin fungizone (Psf). Primary neuronal cells were obtained after treatment with 10 μM cytosine arabinoside on day 3. The purity of cell cultures was tested by respectively). immunostaining for glial fibrillary acidic protein (GFAP) and anti-MAP2 antibody and the cultures were considered ideal when they consisted of >80% astrocytes or primary neuronal cells. bEnd.3 cells obtained from ATCC (Manassas, VA, USA) were cultured in DMEM 10% v/v FBS and 1% v/v Psf. Cells were incubated in atmosphere of 5% CO2 at 37 °C.
2.10. Cytotoxicity assay
Cytotoxic potential of liposomal formulations (Plain-, Tf-, Pen-, or PenTf-liposomes) were evaluated in bEnd.3 cells, astrocytes and primary neuronal cells using MTT assay (Layek and Singh, 2013b; Sharma et al., 2012). All cell lines were plated in 96-well plates (1×104 cells/well), 24 h prior to the assay. Cells were exposed to liposomal formulations at different phospholipid concentrations (100, 200, 400 and 600 nM) in serum free media during 4 h. After incubation, the media was replaced and cells incubated for a total of 48 h. Cell viability was evaluated by adding 10 μL MTT (5 mg/mL) to each well. After 3 h of incubation, MTT solution was removed and the formazan crystals were solubilized in dimethyl sulfoxide. The absorbance was measured at 570 nm. Control group consisted of untreated cells.
2.11. Cellular uptake study
Cellular uptake of DiI-labeled liposomes was evaluated in bEnd.3, astrocytes and primary neuronal cells. About 5×105 cells/well were seeded in 24-well plates, 24 h prior to uptake analysis. Cells were incubated with liposomal formulation (100 nM) and the uptake was investigated at different time intervals. Following liposomal uptake, cells were rinsed with PBS (pH 7.4). Cellular uptake was analyzed using fluorescence microscope (Leica DMi8, IL, USA) and quantitative estimation of liposomal uptake was performed by lysis of cells in Triton-X 100 (0.5% v/v) followed by fluorescent dye extraction in methanol. Fluorescence intensity was measured using spectrophotometer (excitation/emission wavelength: 553/570 nm, respectively).
2.12. Uptake mechanism
In order to elucidate the mechanisms associated with cellular uptake, bEnd.3, astrocytes and primary neuronal cells were seeded in 24-well plates (5×105 cells/well), 24 h prior to the experiment. Cells were treated with well-known endocytosis inhibitors such as sodium azide (10 mM) to inhibit all energy-dependent endocytosis, chlorpromazine (10 μg/mL) to block clathrin-mediated endocytosis, colchicine (100 μg/mL) to prevent caveaolae formation or amiloride (50 μg/mL) to inhibit macropinocytosis for 30 min at 37 °C before application of DiI-labeled liposomes (100 nM)(Layek et al., 2014). Cellular uptake was analyzed after 4 h using fluorescence microscope and fluorescence intensity was measured using spectrophotometer.
2.13. In vitro transfection efficiency
The transfection potentials of liposomal formulations loaded with pGFP or pβgal were evaluated in bEnd.3, astrocytes and primary neuronal cells. About 1x106 cells/well were seeded in 6-well plates, 24 h prior to transfection analysis. Liposomal formulations (100 nM) containing either chitosan-pGFP, or chitosan-pβgal complexes (1μg), were added to the cells in serum-free medium. After 4 h, the media was replaced and cells incubated for a total of 48 h. Cellular GFP expression was analyzed using fluorescence microscope, and quantitative evaluation was performed using FACS analysis-Accuri C6 flow cytometer (Ann Arbor, MI, USA) at excitation wavelength 488 nm and emission wavelength using optical filter FL1 533/30 nm. β-galactosidase activity was quantified using βgal assay kit (Promega, Madison, WI, USA). Cells were lysed using βgal assay buffer and incubated with substrate for 60 min at 37 °C. Addition of sodium carbonate stopped the reaction and the absorbance was measured at 420 nm.
2.14. Endosomal escape
Enhancement in transfection efficiency due to endosomal escape was evaluated in bEnd.3 cells. About 5×105 cells/well were seeded in 24 well-plates, 24 h prior to the assay. Thereafter, cells were treated with 50 mM sucrose (a lysosomotropic agent) and liposomal formulations containing 1 μg pGFP (Ciftci and Levy, 2001). After a 4 h incubation, the media was replaced and cells were incubated for a total of 48 h. Cellular GFP expression was analyzed using fluorescence microscope and flow cytometer.
2.15. Data analysis
Four independent experiments were performed, and all data expressed as mean ± standard deviation (S.D.). Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey multiple comparison post-hoc test (GraphPad Prism version 5.0). Differences were considered statistically significant for p values lower than 0.05.
3. Results
3.1. Characterization of liposomes
Coupling efficiencies of Pen with PEG-lipid and Tf with PEG-lipid were found to be 90.1±9.4% and 71.7±3.9%, respectively. On the other hand, incorporation of Tf into Pen-liposomes surface to form PenTf-liposomes was found to be 94.9±5.4%. Plain-, Tf-, Pen- and PenTf-liposomes loaded with pGFP or pβgal had similar average diameter (155-165 nm), with no significant difference in size between all formulations, as illustrated in Table 1. Furthermore, the formulations had small size distribution (PDI < 0.2), demonstrating tendency for monodispersity and lack of aggregation, suggesting stability. The preparation of nanocarriers with homogeneous particle size and low size distribution is essential for development of efficient and stable delivery systems (Danaei et al., 2018).
Table 1:
Characteristics of Liposome-pDNA: particle size, zeta potential, PDI
| Liposomes | Particle size (nm) | PDIa | Zeta potential (mV) |
|---|---|---|---|
| Plain-liposome | |||
| pGFP | 158.1±1.55 | 0.084±0,04 | 17.8±1.93 |
| Pβgal | 157.7±2.93 | 0.118±0,03 | 31.9±1.36 |
| Tf-liposome | |||
| pGFP | 156.2±1.91 | 0.113±0.04 | 10.6±1.20 |
| Pβgal | 164.2±5.84 | 0.137±0.01 | 9.41±0.18 |
| Pen-liposome | |||
| pGFP | 157.7±2.12 | 0.075±0.03 | 24.7±2.81 |
| Pβgal | 156.9±1.97 | 0.126±0.04 | 23.3±0.93 |
| PenTf-liposome | |||
| pGFP | 155.9±5.84 | 0.038±0.02 | 32.5±1.25 |
| Pβgal | 156.2±1.84 | 0.085±0.06 | 19.5±1.22 |
Data are presented as mean ± SD from four different preparations.
PDI: polydispersity index
Despite being positively charged, the liposomal formulations exhibited differences in zeta potential which indicate that surface modification influence the charge of nanoparticles. In these formulations, the cationic lipid DOTAP may have contributed to positive zeta potential of Plain-liposome, Pen-liposome and PenTf-liposome. Conversely, the lower zeta potential of Tf-liposomes could be attributed to the presence of negatively charged protein on the surface of liposomes. The overall charge of Pen conjugated liposomes was positive. The balance between the positive charge of Pen and the negative charge of Tf probably led to overall positive zeta potential of PenTf-liposome.
3.2. Plasmid DNA affinity test
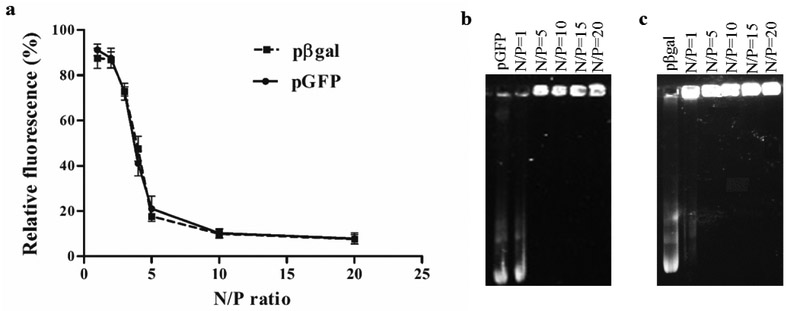
The formation of chitosan-pDNA complexes was evaluated using EtBr exclusion assay and confirmed by agarose gel retardation assay. Naked pGFP and naked pβgal were used as positive controls. EtBr intercalates with plasmid base pairs resulting in significant increase in fluorescence intensity. When the plasmid complexes with chitosan, the EtBr-plasmid intercalation is prevented, reducing the fluorescence intensity(Madeira et al., 2011). Therefore, a decrease in fluorescence intensity indicates chitosan plasmid interaction. The polyplexes were prepared at different N/P ratios to investigate the optimal concentration of chitosan required to complex completely with pDNA while still being able to dissociate from the chitosan inside the cells. High ratios of chitosan-plasmid can result in very stable complexes, which will not dissociate easily inside the cells, compromising transfection efficiency. As shown in the Figure 2a, EtBr fluorescence decreased as chitosan-plasmid N/P ratio increased and a significant reduction of up to 20% fluorescence was observed at N/P=5, for both chitosan-pGFP and -pβgal complexes. The reduction of EtBr fluorescence at N/P=10 and 20 were not significantly different compared to N/P=5. This pattern of fluorescence reduction was similar for both plasmids. Complex formation was confirmed using gel electrophoresis, where light bands of pGFP (Figure 2b) and pβgal (Figure 2c) were observed at N/P= 1, but no bands were observed at higher N/P ratios, for both plasmids. The electrostatic interaction of chitosan and plasmids lead to formation of polyplexes which migrate slower than uncomplexed free DNA during gel electrophoresis. Based on this data, a N/P=5 was chosen for preparation of chitosan-pDNA complexes.
Fig. 2.
Binding affinity of plasmid DNA to chitosan. (a) Relative fluorescence of chitosan-pGFP and chitosan-pβgal complexes in different N/P ratios (1, 2, 3, 4, 5, 10 and 20). Data expressed as mean ± SD (n=4). (b) Agarose gel electrophoresis of chitosan-pGFP and (c) chitosan-pβgal complexes in different N/P ratios (1, 5, 10, 15 and 20). Naked pGFP and naked pβgal were used as controls.
3.3. Encapsulation efficiency and DNase I protection assay
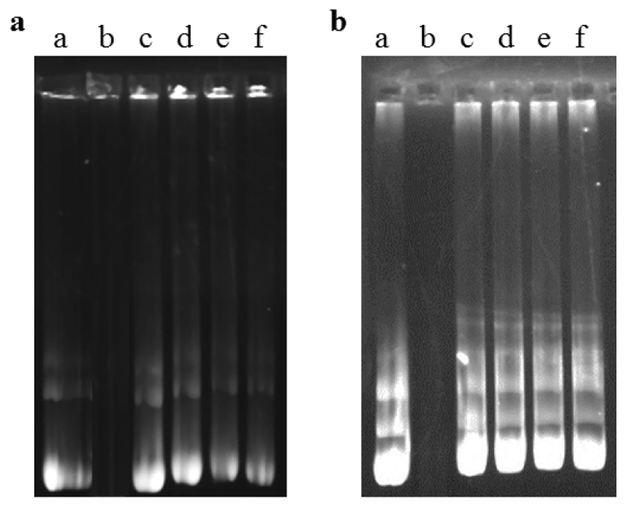
Encapsulation efficiencies above 80 % was obtained for Plain-, Tf-, Pen- and PenTf-liposomes loaded with pGFP or βgal and they were not affected significantly by liposome surface modifications, as shown in Table 2. A suitable gene delivery system should not only transport the therapeutic genes to the target cells, but also protect them against enzymatic degradation. Free DNA is vulnerable to DNase degradation and nanocarriers can potentially protect the encapsulated therapeutic molecules(Zylberberg et al., 2017). The ability of liposomal formulations containing chitosan-pGFP or chitosan-pβgal complexes to protect pGFP and pβgal against enzymatic degradation was compared to free pDNA. Naked pGFP (Figure 3a, lane b) and pβgal (Figure 3b, lane b) were fully degraded upon incubation with DNase I, while pDNA encapsulated in liposomal formulations remained intact as shown by presence of bright bands, suggesting that all liposomal formulations could effectively protect the pDNA from enzymatic degradation.
Table 2:
Liposome-pDNA encapsulation efficiency
| Encapsulation Efficiency | ||||
|---|---|---|---|---|
| Plain-liposome | Tf-liposome | Pen-liposome | PenTf-liposome | |
| pGFP | 87.1±3.17% | 84.2±8.03% | 85.3±8.71% | 86.4±3.56% |
| pβgal | 87.9±3.23% | 94.9±0.86% | 87.2±1.89% | 90.7±6.82% |
Fig. 3.
Protective effect of liposomal formulation containing pDNA against nuclease degradation. (a) Liposomal formulations containing chitosan-GFP (N/P 5). Lane a, naked pGFP; lane b, naked pGFP+DNase I; lanes c-f, Plain-, Tf-, Pen-, and PenTf- chitosan-GFP liposomes, respectively + DNase I. (b) Liposomal formulations containing chitosan-βgal (N/P 5). Lane a, naked pβgal, lane b naked pβgal+DNase I, lane c-f, Plain-, Tf-, Pen-, PenTf- chitosan-βgal liposomes, respectively, + DNase I.
3.4. In vitro release of pDNA
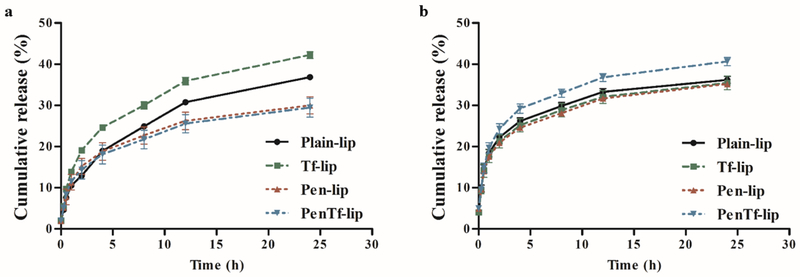
The cumulative release profile of pGFP (Figure 4a) and pβgal (Figure 4b) from the liposomal formulations were monitored at 37 °C for 24 h in phosphate buffer containing 5% FBS. Plasmid GFP was continually released in the first 8 h from all formulations that amounted to 22% cumulative release from Plain-, Pen- and PenTf-liposomes and 29% from Tf-liposomes. On the other hand, approximately 28% of pβgal was released from Plain-, Tf- and Pen-liposomes in 8 h and 32% from PenTf-liposomes. After 8 h, the liposomal formulations exhibited a relatively slow release profile. The release rates of pGFP from Plain-, Tf-, Pen- and PenTf-liposomes were 37.3±0.4%, 42.2±0.8%, 29.9±2.7%, 29.4±2.3%, respectively after 24 h while that for pβgal from Plain, Tf-, Pen and PenTf-liposomes were 36.2±0.8%, 35.4±1.6%, 35.2±0.5% and 40.6±0.9%, respectively.
Fig. 4.
Cumulative release of pDNA from liposomes at N/P ratio 5 for up to 24 h containing (a) pGFP-chitosan and (b) pβgal-chitosan. Data expressed as mean ± SD (n=4).
3.5. Blood compatibility
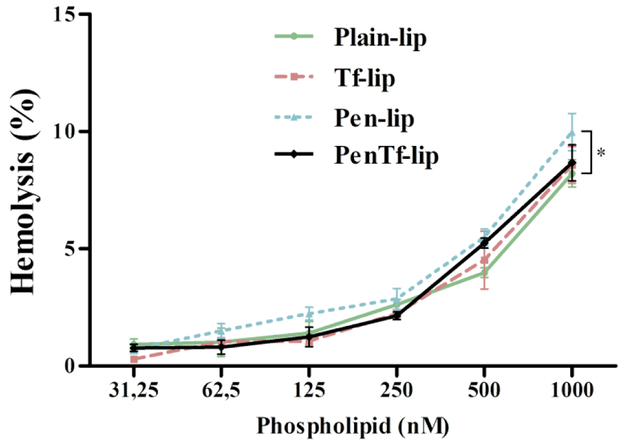
Evaluation of interaction of liposomal formulations with blood cells can help examine possible undesirable effects after systemic administration in vivo. The interaction of cationic liposomes with negatively charged membrane of blood cells can cause erythrocyte lysis and release of hemoglobin(Sharma et al., 2013). Determination of hemolytic activity of liposomal formulations demonstrated a low cytotoxic potential with no significant negative impact on erythrocytes at lipid concentrations up to 500 nM (Figure 5). However, the hemoglobin release was concentration dependent and at highest phospholipid concentration of 1,000 nM the Plain-, Tf-, Pen- and PenTf-liposomes caused 8.2±0.5%, 8.6±0.8%, 9.9±0.8%, 8.7±0.8% hemolysis, respectively. Particularly, Pen-liposomes exhibited significantly (p<0.05) higher hemolysis percentage compared to Plain-liposomes at 1,000 nM. In general, formulations with in vitro hemolysis percentage less than 5% is considered within the critical safe hemolytic level for biomaterials according to ISO/TR 7406(Li et al., 2012). Hemolysis percentage of 5% was observed at 500 nM phospholipid concentration for Pen- and PenTf-liposomes, while lower values (4.5% and 4%) were observed for Tf- and Plain-liposomes, suggesting good compatibility of the liposomes with blood at phospholipid concentrations ≤500 nM.
Fig. 5.
Hemolytic activity of Plain-, Tf-, Pen- and PenTf-liposomes at different phospholipid concentrations (31.25-1000 nM) in erythrocyte solution (2% v/v) after 1 h of incubation at 37 °C. Hemolytic activity of 1% v/v Triton X-100 was considered as 100% hemolysis. Data expressed as mean ± SD (n=4). Significantly higher hemolysis was observed with Pen-liposomes compared to Plain-, Tf- and PenTf-liposomes (***p<0.001).
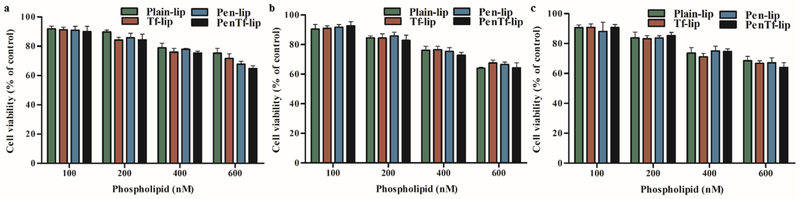
3.6. Cytotoxicity
Cellular viability after treatment for 4 h with different liposomal concentrations (100, 200, 400 and 600 nM) was studied in bEnd.3 (Figure 6a), astrocytes (Figure 6b) and primary neuronal cells (Figure 5c). The cytotoxicity was concentration-dependent in all cell lines studied. Cell viability did not significantly decrease at 100 nM phospholipid concentration and more than 90% cells survived till the end of study at 4 h. However, at 600 nM phospholipid concentration, cell survival decreased significantly to approximately ~68% in all three cell lines. These results demonstrate the low cytotoxic potential of liposomal formulations at 100 nM phospholipid concentration, which was chosen for subsequent experiments.
Fig. 6.
In vitro cytotoxicity of Plain-, Tf-, Pen- and PenTf-liposomes at different phospholipid concentrations (100, 200, 400 and 600 nM) in (a) bEnd.3 cells, (b) astrocytes and (c) primary neuronal cells after 4 h of incubation. Cell viability was determined by MTT assay. Data expressed as mean ± SD (n=4).
3.7. Cellular uptake and its mechanism
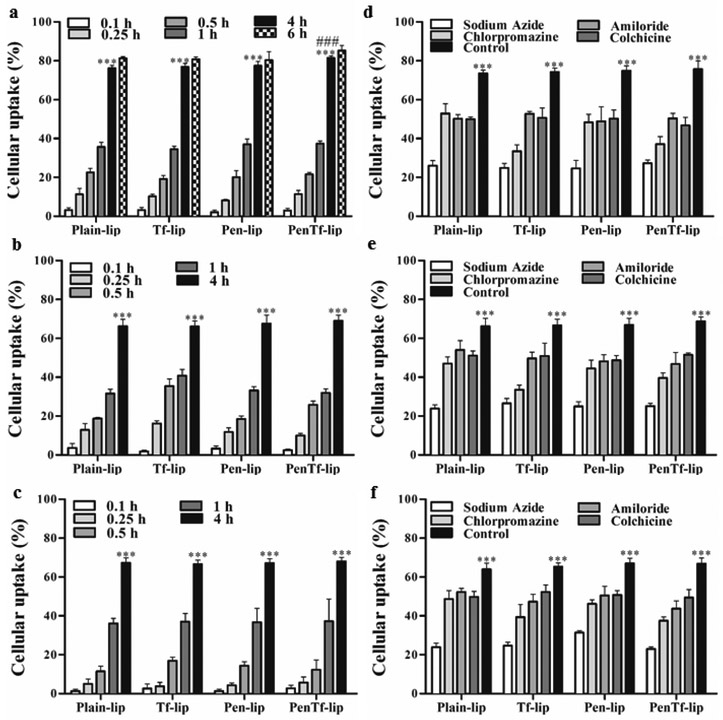
Cellular uptake of liposomes was investigated in bEnd.3, astrocytes and primary neuronal cells at different time intervals. Maximum liposome uptake was observed in bEnd.3 cells in 4 h (Figure 7a) and an increase in incubation time to 6 h led to no significant difference in the uptake. For this cell line, the average uptake of Plain-, Tf-, Pen- and PenTf-liposomes after 4 h was ~78%, while that after 6 h was not of significantly different at 81%. Based on this information, the uptake in other cells lines was subsequently investigated for up to 4 h. As shown in the Figure 7a (bEnd.3), 7b (astrocytes) and 7c (primary neuronal cells), uptake amounts gradually increased from 0.1 h and reached optimal levels at 4 h, which was significantly higher for all formulations compared to other time points (p<0.001) in all the three cell lines tested. Particularly, the internalization of PenTf-liposomes was significantly higher (p<0.001) compared to Plain-, Tf- and Pen-liposome in bEnd.3 cells at 4 h. Furthermore, the percent uptake of Plain, Tf-, Pen- and PenTf-liposomes in bEnd.3 cells at 4 h was significantly greater (p<0.001) than respective liposomal formulations in astrocytes and primary neuronal cells.
Fig. 7.
Cellular uptake in (a) bEnd.3, (b) astrocytes and (c) primary neuronal cells treated with Plain-, Tf-, Pen- and PenTf-liposomes for 0.1, 0.25, 1 and 4 h. Data expressed as mean ± SD (n=4). Significantly higher uptake of liposomes in all cells was observed at 4 h (*** p<0.001) compared to 0.1, 0.25, 0.5 and 1 h. Significantly higher uptake of PenTf-liposomes in bEnd.3 cells was observed at 4 h (### p<0.001) compared to uptake of Plain-, Tf- and Pen-liposome at 4 h. Effect of chemical inhibitors on uptake of Plain-, Tf-, Pen- and PenTf-liposomes in (d) bEnd.3, (e) astrocytes and (f) primary neuronal cells at 4 h. Data expressed as mean ± SD (n=4). Statistically significant uptake (p<0.001) compared to sodium azide treatment is represented as ***.
However, carrier internalization can involve different active and passive uptake mechanisms. To investigate this, various endocytosis inhibitors were used that selectively block different active uptake endocytosis routes. Figures 7d (bEnd.3), 7e (astrocytes), and 7f (primary neuronal cells) depict the relative liposomal uptake percentages after pre-treatment with endocytosis inhibitors (sodium azide, chlorpromazine, amiloride and colchicine). The depletion of intracellular ATP by pre-incubation of cells with sodium azide led to a substantial reduction in cellular internalization of liposomal formulations. Liposomal uptake of bEnd.3, astrocytes and primary neuronal cells were ~35.5%, corresponding to about 50% suppression in uptake as compared to liposomal uptake of untreated (control) cells at 4 h.
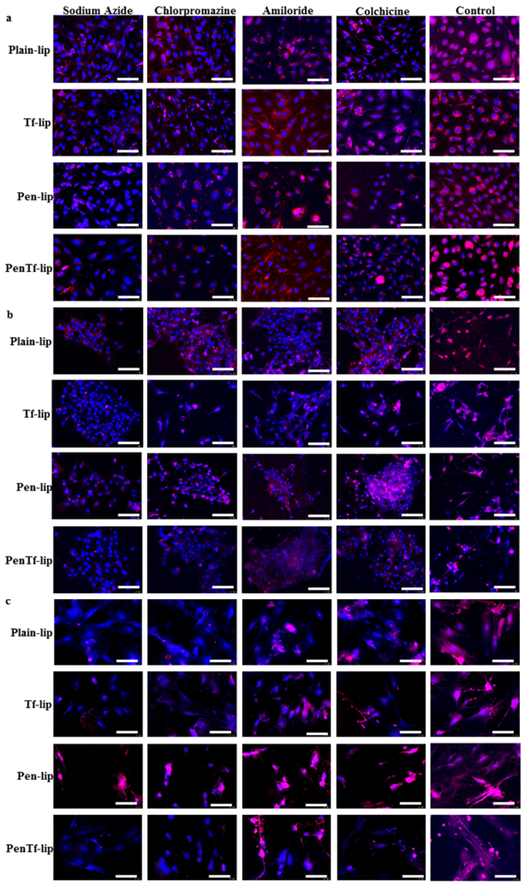
In order to determine whether this energy-dependent uptake mechanism involved any specific endocytic pathway, the cell lines were pretreated with other specific inhibitors as well. Chlorpromazine is a well-known inhibitor of clathrin-mediated endocytosis. Pre-treatment of cells with this drug caused significant reduction in liposomal uptake in all cell lines, however different extent of inhibition was observed amongst different liposomal formulations. The uptake of liposomes without surface modifications (Plain-liposomes) was reduced by ~27% in all cell lines, while the reduction of uptake of Tf-, Pen- and PenTf-liposomes were ~47%, ~33% and ~47%, respectively compared to control cells. Macropinocytosis blockage was induced by pretreating cells with amiloride, which significantly reduced liposomal uptake by 32% in bEnd.3 cells, compared to uptake of non-treated cells. Amiloride pre-treatment also caused specific reduction in uptake of each liposomal formulation in astrocytes and primary neuronal cells. The reduction observed for Plain-, Tf, Pen and PenTf-liposomes in both cells were about 18%, 27%, 26% and 33%, respectively, compared to control cells. Pre-treatment with colchicine, which is a caveolae-endocytosis inhibitor, decreased the internalization of liposomal formulations by 33% in bEnd.3 cells, 24% in astrocytes and 23% in primary neuronal cells, compared to control cells. Fluorescence microscopy image analysis of the effect of endocytosis inhibitors on uptake of liposomal formulations in bEnd.3 (Figure 8a), astrocytes (Figure 8b) and primary neuronal cells (Figure 8c) confirmed the quantitative analysis.
Fig. 8.
Fluorescence images of the effect of chemical inhibitors (sodium azide, chlorpromazine, amiloride and colchicine) on uptake of Plain-, Tf-, Pen- and PenTf-liposomes in (a) bEnd.3, (b) astrocytes and (c) primary neuronal cells at 4 h (Scale bar: 100 μm).
3.8. Transfection efficiency
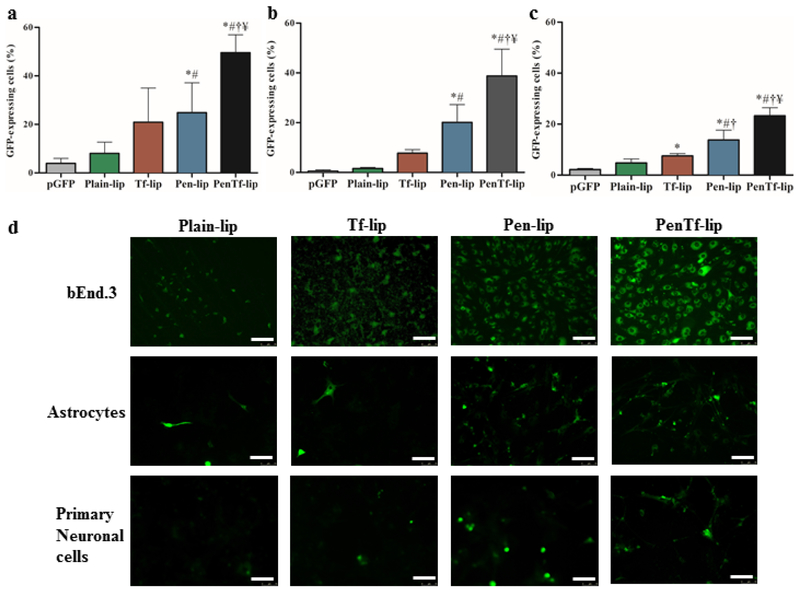
The efficiency of liposomal formulations as a gene delivery vector was evaluated in bEnd.3, astrocytes and primary neuronal cells using pGFP and pβgal as reporter genes. The cell lines used in these experiments comprise the BBB and therefore they can be used as a model to evaluate the transfection in the targeted biological barrier. For this purpose, the transfection efficiencies of various liposomal formulations were evaluated in the aforementioned cells using a GFP reporter gene delivered either alone (no vector) or encapsulated in the liposomal formulations. As shown in the Figure 9a, PenTf-liposomes possessed higher gene transfection capability in bEnd.3 cells yielding 48.7% cells as GFP positive, which was significantly higher than pGFP, Plain-, Tf- and Pen-liposomes (p<0.005). Similar pattern was observed in astrocytes (Figure 9b) and primary neuronal cells (Figure 9c). In astrocytes and primary neuronal cells, the highest level of transfection efficiency was demonstrated by PenTf-liposome at 38.8% and 23.2% respectively, which were significantly higher than pGFP, Plain-, Tf- and Pen-liposomes (p<0.05). Fluorescence microscopy images of cells treated with liposomal formulations confirmed the higher fluorescence observed with PenTf-liposomes (Figure 9d).
Fig. 9.
GFP expression levels 48 h after transfection in (a) bEnd.3 cells, (b) astrocytes and (c) primary neuronal cells treated with Plain-, Tf-, Pen- and PenTf-liposomes containing pGFP (1μg). Data expressed as mean ± SD (n=4). Statistically significant (p<0.05) differences are shown as (*) with pGFP, (#) with Plain-lip, (†****) with Tf-lip and (¥) with Pen-lip. (d) Fluorescence microscopy images of GFP expression in bEnd.3, astrocytes and primary neuronal cells treated with Plain-, Tf-, Pen- and PenTf-liposomes containing pGFP (Scale bar: 100 μm).
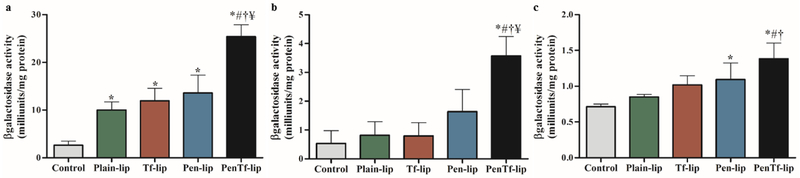
βgal reporter gene was also used to determine transfection efficiencies in the aforementioned cells. The transfection efficiency of liposomes loaded with pβgal followed similar pattern as that observed with pGFP. PenTf-liposomes containing pβgal induced a 10-fold enhancement in gene expression in bEnd.3 cells (Figure 10a) compared to cellular basal levels and it was significantly higher compared to the other liposomal formulations (p<0.001). On the other hand, βgal expression in astrocytes treated with PenTf-liposome was 6-fold enhanced compared to cellular basal levels, and it was also significantly (p<0.005) higher than the other liposomal formulations, as shown in Figure 10b. In primary neuronal cells, PenTf-liposomes induced a 2-fold enhancement in βgal expression compared to cellular basal levels (Figure 10c), and it was significantly higher than Tf- and Plain-liposomes (p<0.05).
Fig. 10.
βgalactosidase expression levels 48 h after transfection in (a) bEnd.3 cells, (b) astrocytes and (c) primary neuronal cells treated with Plain-, Tf-, Pen- and PenTf-liposomes containing pβgal (1μg). Data expressed as mean ± SD (n=4). Statistically significant (p<0.05) differences are shown as (*) with control, (#) with Plain-lip, (†****) with Tf-lip and (¥) with Pen-lip.
3.9. Endosomal escape
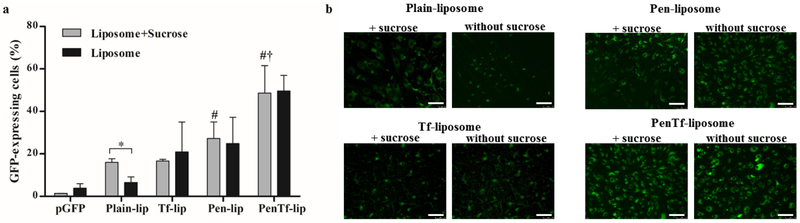
Endosomal escape of formulations in bEnd.3 cells was investigated by treating the cells with sucrose, a lysosomotropic agent that causes osmotic inflow of water into lysosomes, thereby reducing nuclease-mediated degradation(Ciftci and Levy, 2001). Enhancement in transfection efficiency by sucrose was quantified using a GFP reporter gene. As shown in Figure 11a, transfection efficiencies in bEnd.3 cells treated with sucrose and pGFP, Plain-, Tf-, Pen- or PenTf-liposomes were 1.3±0.1%, 16.0±1.6%, 16.6±0.8%, 27.2±7.8% and 48.6±12.8%, respectively. Significant enhancement in transfection in bEnd.3 cells treated with Plain-liposomes + sucrose was observed compared to Plain-liposomes without sucrose, as evident by the increased fluorescent signal produced in the cells. In contrast, other treatment groups (pGFP, Tf-, Pen and PenTf-liposomes + sucrose) did not enhance the transfection efficiency in bEnd.3 cells significantly compared to the respective treatments without sucrose. As earlier observed, PenTf-liposomes showed higher capacity to transfect bEnd.3 cells irrespective of presence of sucrose compared to the other liposomal formulations. Fluorescence microscopy images of cells treated with liposomal formulation expressing GFP confirmed the quantitative analysis (Figure 11b).
Fig. 11.
Assessment of ability of liposomes to escape endosome. (a) GFP expression levels 48 h after transfection in bEnd.3 cells treated with or without 50 mM sucrose and Plain-, Tf-, Pen- or PenTf-liposomes containing pGFP (1μg). Data expressed as mean ± SD (n=4). Statistically significant (p<0.05) differences are shown as (*) with Plain-lip, (#) with pGFP+sucrose and (†) with Plain-lip+sucrose, Tf-lip+sucrose and Pen-lip+sucrose. (b) Fluorescence microscopy images of GFP expression in bEnd.3 cells treated with or without 50 mM sucrose and Plain-, Tf-, Pen- or PenTf-liposomes containing pGFP (Scale bar: 100 μm).
4. Discussion
The need for more efficient approaches to treat different diseases, especially neurodegenerative diseases has led to significant improvements in drug delivery technology. Liposomes can be structurally modified to adjust their properties according to the desired function (Safari and Zarnegar, 2014). Effective carriers for treatment of neurodegenerative diseases should be able to cross the BBB, the major obstacle for delivering therapeutic genes, and reach the nucleus of neuronal cells (Bergen et al., 2008). Based on the abundant expression of transferrin receptors on BBB, these receptors have been extensively studied as a potential brain drug delivery target. Several reports have focused on Tf receptors as a brain targeting moiety and have demonstrated effective delivery to the brain using Tf-targeted nanoparticles, which rely on transcytosis of the complexes and encapsulated cargo (Girão da Cruz et al., 2005; Girão Da Cruz et al., 2004; Johnsen et al., 2017; Lee et al., 2000; Liu et al., 2017; Zheng et al., 2015). Nonetheless, additional challenges exist in the development of brain delivery systems that includes receptor saturation, inefficient cellular uptake, inability to escape the endosome and formulation toxicity. To this end, we rationally designed liposomal nanoparticles capable of not only targeting BBB and efficiently transfecting neuronal cells but also protecting the plasmid DNA and assisting in its endosomal escape. Surface modification with Tf protein was used for targeting the brain endothelial cells, which comprise the BBB, while conjugation to Pen was intended to improve liposome internalization into cells, in the event of Tf receptor saturation. Stealth properties were instilled by incorporation of DSPE-PEG phospholipid into liposomal surface and improvement in gene transfection through endosomal escape was achieved by complexation of pDNA to low molecular weight chitosan.
Chitosan is a polymer widely used in drug delivery for its ability to protect molecules against enzymatic degradation and promote transfection. It has been shown that transfection properties of chitosan depends on its high buffering capacity in the endosomal pH range (4.5-6.5), which mediate endosomal escape by the proton-sponge effect (Mansouri et al., 2004; Saikia and Gogoi, 2015). Different chitosan-plasmid N/P ratios can affect the stability of chitosan-DNA complex, which can influence plasmid delivery into cells and consequently, cellular transfection. Determination of optimal chitosan-plasmid ratio is therefore critical for protecting plasmids against enzymatic degradation while allowing its release in the intracellular space for subsequent cell transfection. The strong electrostatic interaction between chitosan and DNA, drives gene packing and protection (Lavertu et al., 2006). Depending on nitrogen to phosphate (N/P) charge ratio, chitosan can condense DNA to sizes compatible for cellular uptake and the steric protection afforded thereof, can prevent nucleases from accessing the DNA (Mao et al., 2010). In the present study, EtBr assay together with agarose gel electrophoresis suggested that chitosan-plasmid interactions increased upon increasing N/P ratio and N/P of 5 was the optimal ratio for this study.
Design of stable liposomal carriers with targeting ligands requires a broader understanding of the target site in order to maximize therapeutic efficacy (Zylberberg et al., 2017). Physicochemical properties are critical determinants of the abilities of delivery systems to overcome the biological barriers. Therefore, a proper characterization of liposomes not only enables determination of their delivery potentials, but also their safety profiles and transfection properties (Balbino et al., 2012). The liposomal formulations were synthesized by thin-lipid film and post-insertion methods generating consistent particle size, positive zeta potential and high pDNA encapsulation efficiency, which were in accordance with other studies utilizing multifunctionalized liposome-based delivery systems (Chen et al., 2016; Michel et al., 2017; Sharma et al., 2012). Despite the limited permeability of BBB, functionalization of liposomes have shown to be essential for efficient penetration of nanoparticles with size below 200 nm into brain (Chen et al., 2016; dos Santos Rodrigues et al., 2018; Ordóñez-Gutiérrez et al., 2016). Additionally, cationic liposomes may facilitate electrostatic interaction to cell membrane and consequent cellular uptake (Fröhlich, 2012).
The induction of specific gene modification requires stable gene delivery system, which enables high cellular uptake, protection from enzymatic degradation as well as pDNA release from the endosome and transport to cell nucleus (Ibraheem et al., 2014; Zylberberg et al., 2017). In vitro experiments demonstrated that the liposomal formulations could not only encapsulate pDNA efficiently but also thwart enzymatic degradation of gene load. Furthermore, they exhibited a sustained release kinetics in vitro that likely contributed to enhanced transfection efficiency by providing time for the carriers to reach neuronal cells and release pDNA.
However, effectiveness of nanocarriers is contingent upon its interactions with other cells and proteins in biological systems that dictate their distribution, elimination fate as well as biocompatibility (Düzgüneş and Nir, 1999). Despite synthesis using biocompatible phospholipids, liposomes can cause in vitro and in vivo toxicity. Cationic carriers can interact electrostatically with the negatively charged membrane of red blood cells causing their lysis (Ju et al., 2016; Qi et al., 2016; Roursgaard et al., 2016). Therefore, hemocompatibility studies were performed to obtain information about the systemic safety of the synthesized liposomes. Liposomal formulations were found to be non-hemolytic even at high phospholipid concentrations. Pen-liposomes exhibited a more cytotoxic profile and induced higher levels of hemolysis compared to the other liposomal formulations. This could be attributed to the cationic character of Pen that may have enhanced Pen-liposome interactions with erythrocytes and, consequently induced their lysis. In general, the low hemolytic potential of liposomes demonstrated good biocompatibility of the formulations. These results were in accordance with findings obtained from other hemolysis studies using modified liposomes, which suggest that phospholipid composition and concentration influences their hemolytic potential (Michel et al., 2017; Mourtas et al., 2009; Sharma et al., 2013). Cytotoxicity studies confirmed that liposomal formulations do not affect cell viability in low phospholipid concentrations, as has been previously reported by other researchers using bEnd.3, astrocytes and PC-12 cells treated with multifunctionalized nanoparticles (Sánchez-López et al., 2016; Zheng et al., 2016). It is to be noted that low cytotoxic profile is a highly desired property for gene carriers.
Cellular internalization of cationic liposomes occurs mainly due to electrostatic interactions with cell membranes and multiple strategies have been used to further enhance liposome cellular uptake (Obata et al., 2009; Sharma et al., 2012; Shim et al., 2013). In this study, liposomal formulations followed a time-dependent uptake reaching significant amounts at 4 h. Quantification of percent uptake demonstrated that liposome internalization is slower in astrocytes and primary neuronal cells compared to bEnd.3 cells. PenTf-liposomes exhibited significantly higher internalization in bEnd.3 cells at 4 h compared to the other liposomal formulations; however this was not observed in astrocytes and primary neuronal cells. One possible reason for the differences in uptake amongst the cells may be due to individual cellular characteristics (Mailä Nder and Landfester, n.d.). Expression of specific genes by primary cells and cell lines can generate different cellular phenotypes and functions, which may dictate individual uptake properties for instance (Alge et al., 2006; Maurisse et al., 2010; Pan et al., 2009). Secondly, the incubation period used in the experiment may have allowed time for non-specific liposome-cell interactions and liposome internalization. Our findings support previous reports showing time dependent cellular uptake of liposomal formulations (Kang et al., 2017). Liposome surface modification, specially CPP and Tf targeted-liposomes, have demonstrated to be key factors in enhancing cellular uptake (Girão Da Cruz et al., 2004; Zheng et al., 2015).
Many reports on cellular uptake have suggested that non-viral gene carriers are preferentially internalized via endocytosis. Different pathways are involved in carrier uptake, which depends on the cell type and carrier characteristics including particle size, surface charge and surface modifications (Kang et al., 2017; Mailä Nder and Landfester, n.d.; Salatin and Yari Khosroushahi, 2017; Yameen et al., 2014). Therefore, assessment of endocytic pathway in liposomal uptake can help elucidate intracellular processing and subsequent transfection efficiency. Due to the cationic nature of liposomal formulation and the abundance of negatively charged moieties on cell surface, high levels of electrostatic interactions were expected. Our results suggested that internalization of liposomal formulations occurred through multiple mechanisms especially energy-dependent pathways such as macropinocytosis, clathrin- and caveolae-mediated endocytosis. ATP depletion induced in the cell lines by sodium azide demonstrated that liposomal uptake involved endocytosis, however, incomplete uptake suppression suggested that passive transport might be occurring simultaneously. Participation of caveolae and macropinocytosis in the uptake process of liposomal formulations in bEnd.3, astrocytes and primary neuronal cells were also observed by pretreating cells with colchicine and amiloride, respectively. On the other hand, inhibition of clathrin-mediated endocytosis by chlorpromazine resulted in differential extent of uptake in each formulation. Clathrin-coated vesicles based internalization was observed to be the main route of uptake of PenTf-liposomes. These observations are in accordance with the known endocytosis pathway of Tf-TfR complex and with studies that investigated the uptake of Tf-conjugated liposomes in various cell lines (Anabousi et al., 2006; Johnsen et al., 2017; Liu et al., 2017; Voinea et al., 2002), thereby emphasizing the role of Tf ligand in liposomal uptake. Penetratin conjugation also assisted liposomal-cell interaction and subsequent internalization, therefore contributing to the liposome uptake through multiple pathways. The involvement of different routes (Kang et al., 2017) and participation of surface modification such as Pen in uptake of multifunctionalized liposomes have been commonly reported (Console et al., 2003; Tseng et al., 2002). Indeed, cell type and nanoparticle characteristics can together influence cellular uptake and the pathways of uptake.
The respective transfection efficiencies of bEnd.3, astrocytes and primary neuronal cells were established using GFP and βgal as reporter genes delivered either alone or encapsulated in liposomes. The liposomal formulations demonstrated the ability to efficiently deliver genes and transfect bEnd.3 cells, astrocytes and primary neuronal cells. A comparison of the percent transfection of DNA alone or encapsulated in different liposomal formulations showed higher transfection of both plasmids in bEnd.3 cells compared to astrocytes or primary neuronal cells. This suggests that bEnd.3 cells may be internalizing liposomes, trafficking them to nucleus and transcribing the DNA more efficiently than astrocytes and primary neuronal cells. This was also observed in uptake studies, where bEnd.3 cells demonstrated greater ability to take up liposomal formulations.
Transfection efficiency in neuronal cells has been reported to be low but it can be significantly improved using dual-modified targeted liposomes compared to unmodified liposomes (Girão Da Cruz et al., 2004). In line with published literature, our findings demonstrated more efficient transfection of cells using bifunctional liposomes (PenTf-liposomes) compared to Plain-, Tf- and Pen-liposomes, in all BBB cells studied. Higher gene delivery efficiency exhibited by PenTf-liposomes may be attributed to higher uptake of liposomal formulations in 4 h, ability of DNA to escape the endosome and get shuttled to cell nucleus. We believe that Pen and Tf conjugation to liposomes as well as chitosan complexation to pDNA may have collectively worked to improve transfection efficiency observed with PenTf-liposomes.
At the BBB, liposomes can be subjected to either direct transcytosis across brain endothelial cells, followed by subsequent release in abluminal side or they could be taken up in endosomes, leading to consequent enzymatic degradation and release in cell cytoplasm (Kim et al., 2015; Yang et al., 2013). For efficient transfection, the carriers need to escape the endosome to reach the cytoplasm and get trafficked to the nucleus. Here, evaluation of endosomal escape in bEnd.3 cells indicated significant effect on transfection efficiency only with Plain-liposomes and sucrose. These results suggest that liposome surface modification influences endosomal uptake and escape mechanisms, which contributes to improved transfection efficiency observed with surface modified liposomes. This observation is in agreement with cellular internalization and lysosomal escape studies that were conducted using liposomes dually modified with Tf ligand and a CPP in glioma C6 cells (Zheng et al., 2015). Varkouhi et. al. also suggested that CPPs including Pen are involved in liposome endosomal escape, highlighting their importance as endosomal escape agents (Varkouhi et al., 2011). Therefore, we believe that the enhanced transfection in bEnd.3 cells treated with PenTf-liposomes could have also resulted due to participation of Pen in endosomal escape. Moreover, chitosan-pDNA polyplexes might be synergistically involved in endosomal escape process and provide additional pDNA protection against endosomal degradation through proton sponge effect(Huang et al., 2005). These results cannot directly predict the in vivo performance of the liposomes and animal studies are necessary to determine efficacy of the formulation, which are being planned. However, through these studies, we demonstrate that bifunctional PenTf-liposomes with chitosan are ideal gene delivery vectors that provide protection to pDNA against enzymatic degradation, while possessing low cytotoxicity and high transfection efficiencies.
Actively targeting transferrin receptors using transferrin functionalized liposomes has been shown as an efficient strategy for gene delivery to the brain. However, the efficiency of transferrin-liposomes can be limited due to saturation of transferrin receptor (Qian, 2002), which was overcome by attaching penetratin on liposome surface. This system showed efficient cellular internalization and endosomal escape properties that resulted in transfection enhancement. Therefore, dual-targeting liposome gene delivery system can potentially provide better brain targeting ability and therapeutic efficacy for treatment of neurodegenerative diseases.
5. Conclusions
The designed PenTf-liposomes were found to be efficient in transfecting different cell lines, especially primary neuronal cells. The mechanism that mediated liposome internalization can be characterized mainly as an energy-dependent process involving different pathways with an emphasis on clathrin-mediated endocytosis probably relating to TfR. The ability of liposomal formulations to escape the endosome can play an important role in transfection efficiency. Overall, the studies indicate that PenTf-liposomes hold great promise as an efficient gene carrier to the brain.
Acknowledgment
B.S.R. is supported by a fellowship from Sciences without Borders Program (Conselho Nacional de Desenvolvimento Cientifico e Technológico – CNPq, Brazil).
Funding
This research was supported by National Institutes of Health (Grant R01AG051574).
Footnotes
Conflict of interest
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alge CS, Hauck SM, Priglinger SG, Kampik A, Ueffing M, 2006. Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival. J. Proteome Res 5, 862–878. 10.1021/pr050420t [DOI] [PubMed] [Google Scholar]
- Anabousi S, Bakowsky U, Schneider M, Huwer H, Lehr CM, Ehrhardt C, 2006. In vitro assessment of transferrin-conjugated liposomes as drug delivery systems for inhalation therapy of lung cancer. Eur. J. Pharm. Sci 29, 367–374. 10.1016/j.ejps.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Balazs DA, Godbey WT, 2011. Liposomes for Use in Gene Delivery 2011. 10.1155/2011/326497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbino TA, Gasperini AAM, Oliveira CLP, Azzoni AR, Cavalcanti LP, De La Torre G, 2012. Correlation of the physicochemical and structural properties of pDNA/cationic liposome complexes with their in vitro transfection. Langmuir 28, 11535–11545. 10.1021/la302608g [DOI] [PubMed] [Google Scholar]
- Balducci C, Mancini S, Minniti S, La Vitola P, Zotti M, Sancini G, Mauri M, Cagnotto A, Colombo L, Fiordaliso F, Grigoli E, Salmona M, Snellman A, Haaparanta-Solin M, Forloni G, Masserini M, Re F, 2014. Multifunctional Liposomes Reduce Brain - Amyloid Burden and Ameliorate Memory Impairment in Alzheimer’s Disease Mouse Models. J. Neurosci 34, 14022–14031. 10.1523/JNEUROSCI.0284-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bana L, Minniti S, Salvati E, Sesana S, Zambelli V, Cagnotto A, Orlando A, Cazzaniga E, Zwart R, Scheper W, Masserini M, Re F, 2014. Liposomes bi-functionalized with phosphatidic acid and an ApoE-derived peptide affect Aβ aggregation features and cross the blood-brain-barrier: Implications for therapy of Alzheimer disease. Nanomedicine Nanotechnology, Biol. Med 10, 1583–1590. 10.1016/j.nano.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Bashyal S, Noh G, Keum T, Choi YW, Lee S, 2016. Cell penetrating peptides as an innovative approach for drug delivery; then, present and the future. J. Pharm. Investig 46, 205–220. 10.1007/s40005-016-0253-0 [DOI] [Google Scholar]
- Bergen JM, Park IK, Horner PJ, Pun SH, 2008. Nonviral approaches for neuronal delivery of nucleic acids. Pharm. Res 25, 983–998. 10.1007/s11095-007-9439-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Ly N, Yu YJ, Bumbaca D, Elstrott J, Boswell CA, Zhang Y, Luk W, Lu Y, Dennis MS, Weimer RM, Chung I, Watts RJ, 2014. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. J. Exp. Med 211, 233–44. 10.1084/jem.20131660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulbake U, Doppalapudi S, Kommineni N, Khan W, 2017. Liposomal formulations in clinical use: An updated review. Pharmaceutics 9, 1–33. 10.3390/pharmaceutics9020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ğağdaş M, Sezer AD, Bucak S, 2004. Liposomes as Potential Drug Carrier Systems for Drug Delivery. Appl. Nanotechnol. Drug Deliv 1–50. 10.5772/58459 [DOI] [Google Scholar]
- Chen ZL, Huang M, Wang XR, Fu J, Han M, Shen YQ, Xia Z, Gao JQ, 2016. Transferrin-modified liposome promotes α-mangostin to penetrate the blood-brain barrier. Nanomedicine Nanotechnology, Biol. Med 12, 421–430. 10.1016/j.nano.2015.10.021 [DOI] [PubMed] [Google Scholar]
- Chikh GG, Kong S, Bally MB, Meunier JC, Schutze-Redelmeier MP, 2001. Efficient delivery of Antennapedia homeodomain fused to CTL epitope with liposomes into dendritic cells results in the activation of CD8+ T cells. J. Immunol 167, 6462–70. 10.4049/jimmunol.167.11.6462 [DOI] [PubMed] [Google Scholar]
- Cifani N, Chronopoulou L, Pompili B, Di Martino A, Bordi F, Sennato S, Di Domenico EG, Palocci C, Ascenzioni F, 2015. Improved stability and efficacy of chitosan/pDNA complexes for gene delivery. Biotechnol. Lett 37, 557–565. 10.1007/s10529-014-1727-7 [DOI] [PubMed] [Google Scholar]
- Ciftci K, Levy RJ, 2001. Enhanced plasmid DNA transfection with lysosomotropic agents in cultured fibroblasts. Int. J. Pharm 218, 81–92. 10.1016/S0378-5173(01)00623-8 [DOI] [PubMed] [Google Scholar]
- Console S, Marty C, García-Echeverría C, Schwendener R, Ballmer-Hofer K, 2003. Antennapedia and HIV transactivator of transcription (TAT) “protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J. Biol. Chem 278, 35109–35114. 10.1074/jbc.M301726200 [DOI] [PubMed] [Google Scholar]
- Danaei M, Dehghankhold M, Ataei S, Hasanzadeh Davarani F, Javanmard R, Dokhani A, Khorasani S, Mozafari MR, 2018. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 10, 1–17. 10.3390/pharmaceutics10020057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Rodrigues B, Oue H, Banerjee A, Kanekiyo T, Singh J, 2018. Dual functionalized liposome-mediated gene delivery across triple co-culture blood brain barrier model and specific in vivo neuronal transfection. J. Control. Release 286, 264–278. 10.1016/j.jconrel.2018.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düzgüneş N, Nir S, 1999. Mechanisms and kinetics of liposome-cell interactions. Adv. Drug Deliv. Rev 40, 3–18. 10.1016/S0169-409X(99)00037-X [DOI] [PubMed] [Google Scholar]
- Fröhlich E, 2012. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomedicine 7, 5577–5591. 10.2147/IJN.S36111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girão da Cruz MT, Cardoso ALC, de Almeida LP, Simões S, Pedroso de Lima MC, 2005. Tf-lipoplex-mediated NGF gene transfer to the CNS: Neuronal protection and recovery in an excitotoxic model of brain injury. Gene Ther. 12, 1242–1252. 10.1038/sj.gt.3302516 [DOI] [PubMed] [Google Scholar]
- Girão Da Cruz MT, Simões S, Pedroso De Lima MC, 2004. Improving lipoplex-mediated gene transfer into C6 glioma cells and primary neurons. Exp. Neurol 187, 65–75. 10.1016/j.expneurol.2003.12.013 [DOI] [PubMed] [Google Scholar]
- Huang M, Fong CW, Khor E, Lim LY, 2005. Transfection efficiency of chitosan vectors: Effect of polymer molecular weight and degree of deacetylation. J. Control. Release 106, 391–406. 10.1016/j.jconrel.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Ibraheem D, Elaissari A, Fessi H, 2014. Gene therapy and DNA delivery systems. Int. J. Pharm 459, 70–83. 10.1016/j.ijpharm.2013.11.041 [DOI] [PubMed] [Google Scholar]
- Johnsen KB, Burkhart A, Melander F, Kempen PJ, Vejlebo JB, Siupka P, Nielsen MS, Andresen TL, Moos T, 2017. Targeting transferrin receptors at the blood-brain barrier improves the uptake of immunoliposomes and subsequent cargo transport into the brain parenchyma. Sci. Rep 7, 1–13. 10.1038/s41598-017-11220-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen KB, Moos T, 2016. Revisiting nanoparticle technology for blood-brain barrier transport: Unfolding at the endothelial gate improves the fate of transferrin receptor-targeted liposomes. J. Control. Release 222, 32–46. 10.1016/j.jconrel.2015.11.032 [DOI] [PubMed] [Google Scholar]
- Ju J, Huan ML, Wan N, Hou YL, Ma XX, Jia YY, Li C, Zhou SY, Zhang B. Le, 2016. Cholesterol derived cationic lipids as potential non-viral gene delivery vectors and their serum compatibility. Bioorganic Med. Chem. Lett 26, 2401–2407. 10.1016/j.bmcl.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Kang JH, Jang WY, Ko YT, 2017. The Effect of Surface Charges on the Cellular Uptake of Liposomes Investigated by Live Cell Imaging. Pharm. Res 34, 704–717. 10.1007/s11095-017-2097-3 [DOI] [PubMed] [Google Scholar]
- Kim BK, Hwang GB, Seu YB, Choi JS, Jin KS, Doh KO, 2015. DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes. Biochim. Biophys. Acta - Biomembr 1848, 1996–2001. 10.1016/j.bbamem.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Kirson NY, Desai U, Ristovska L, Cummings AKG, Birnbaum HG, Ye W, Andrews JS, Ball D, Kahle-Wrobleski K, 2016. Assessing the economic burden of Alzheimer’s disease patients first diagnosed by specialists. BMC Geriatr. 16, 1–8. 10.1186/s12877-016-0303-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KB, Northeved H, Pramod Kumar EK, Permin A, Gjetting T, Andresen TL, Larsen S, Wegener KM, Lykkesfeldt J, Jantzen K, Loft S, Møller P, Roursgaard M, 2015. In vivo toxicity of cationic micelles and liposomes. Nanomedicine Nanotechnology, Biol. Med 11, 467–477. 10.1016/j.nano.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Kolate A, Baradia D, Patil S, Vhora I, Kore G, Misra A, 2014. PEG - A versatile conjugating ligand for drugs and drug delivery systems. J. Control. Release 192, 67–81. 10.1016/j.jconrel.2014.06.046 [DOI] [PubMed] [Google Scholar]
- Kumar SR, Markusic DM, Biswas M, High KA, Herzog RW, 2016. Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. - Methods Clin. Dev 3, 16034 10.1038/mtm.2016.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavertu M, Méthot S, Tran-Khanh N, Buschmann MD, 2006. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 27, 4815–24. 10.1016/j.biomaterials.2006.04.029 [DOI] [PubMed] [Google Scholar]
- Layek B, Haldar MK, Sharma G, Lipp L, Mallik S, Singh J, 2014. Hexanoic acid and polyethylene glycol double grafted amphiphilic chitosan for enhanced gene delivery: Influence of hydrophobic and hydrophilic substitution degree. Mol. Pharm 11, 982–994. 10.1021/mp400633r [DOI] [PubMed] [Google Scholar]
- Layek B, Singh J, 2013a. Amino acid grafted chitosan for high performance gene delivery: Comparison of amino acid hydrophobicity on vector and polyplex characteristics. Biomacromolecules. 10.1021/bm301720g [DOI] [PubMed] [Google Scholar]
- Layek B, Singh J, 2013b. Amino Acid Grafted Chitosan for High Performance Gene Delivery: Comparison of Amino Acid Hydrophobicity on Vector and Polyplex Characteristics. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Engelhardt B, Lesley J, Bickel U, Pardridge WM, 2000. Targeting rat anti-mouse transferrin receptor monoclonal antibodies through blood-brain barrier in mouse. J. Pharmacol. Exp. Ther 292, 1048–1052. [PubMed] [Google Scholar]
- Li P, Liu D, Sun X, Liu C, Liu Y, Zhang N, 2011. A novel cationic liposome formulation for efficient gene delivery via a pulmonary route. Nanotechnology 22 10.1088/0957-4484/22/24/245104 [DOI] [PubMed] [Google Scholar]
- Li X, Wang L, Fan Y, Feng Q, Cui FZ, 2012. Biocompatibility and toxicity of nanoparticles and nanotubes. J. Nanomater 2012 10.1155/2012/548389 [DOI] [Google Scholar]
- Liu C, Liu XN, Wang GL, Hei Y, Meng S, Yang LF, Yuan L, Xie Y, 2017. A dual-mediated liposomal drug delivery system targeting the brain: Rational construction, integrity evaluation across the blood–brain barrier, and the transporting mechanism to glioma cells. Int. J. Nanomedicine 12, 2407–2425. 10.2147/IJN.S131367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Zhang S, Jiang H, Zhao B, Lv H, 2007. Lipoplex morphologies and their influences on transfection efficiency in gene delivery. J. Control. Release 123, 184–194. 10.1016/j.jconrel.2007.08.022 [DOI] [PubMed] [Google Scholar]
- Madeira C, Loura LMS, Aires-Barros MR, Prieto M, 2011. Fluorescence methods for lipoplex characterization. Biochim. Biophys. Acta - Biomembr 1808, 2694–2705. 10.1016/j.bbamem.2011.07.020 [DOI] [PubMed] [Google Scholar]
- Mailä Nder V, Landfester K, n.d. Interaction of Nanoparticles with Cells. 10.1021/bm900266r [DOI] [PubMed] [Google Scholar]
- Mansouri S, Lavigne P, Corsi K, Benderdour M, Beaumont E, Fernandes JC, 2004. Chitosan-DNA nanoparticles as non-viral vectors in gene therapy: Strategies to improve transfection efficacy. Eur. J. Pharm. Biopharm 57, 1–8. 10.1016/S0939-6411(03)00155-3 [DOI] [PubMed] [Google Scholar]
- Mao S, Sun W, Kissel T, 2010. Chitosan-based formulations for delivery of DNA and siRNA. Adv. Drug Deliv. Rev 62, 12–27. 10.1016/j.addr.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Marty C, Meylan C, Schott H, Ballmer-Hofer K, Schwendener RA, 2004. Enhanced heparan sulfate proteoglycan-mediated uptake of cell-penetrating peptide-modified liposomes. C. Cell. Mol. Life Sci 61, 1785–1794. 10.1007/s00018-004-4166-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurisse R, De Semir D, Emamekhoo H, Bedayat B, Abdolmohammadi A, Parsi H, Gruenert DC, 2010. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol. 10, 1–9. 10.1186/1472-6750-10-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel T, Luft D, Abraham MK, Reinhardt S, Salinas Medina ML, Kurz J, Schaller M, Avci-Adali M, Schlensak C, Peter K, Wendel HP, Wang X, Krajewski S, 2017. Cationic Nanoliposomes Meet mRNA: Efficient Delivery of Modified mRNA Using Hemocompatible and Stable Vectors for Therapeutic Applications. Mol. Ther. - Nucleic Acids 8, 459–468. 10.1016/j.omtn.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtas S, Michanetzis GPAK, Missirlis YF, Antimisiaris SG, 2009. Haemolytic activity of liposomes: Effect of vesicle size, lipid concentration and polyethylene glycol-lipid or arsonolipid incorporation. J. Biomed. Nanotechnol 5, 409–415. 10.1166/jbn.2009.1050 [DOI] [PubMed] [Google Scholar]
- Naldini L, 2015. Gene therapy returns to centre stage. Nature 526, 351–360. 10.1038/nature15818 [DOI] [PubMed] [Google Scholar]
- Obata Y, Saito S, Takeda N, Takeoka S, 2009. Plasmid DNA-encapsulating liposomes: Effect of a spacer between the cationic head group and hydrophobic moieties of the lipids on gene expression efficiency. Biochim. Biophys. Acta - Biomembr 1788, 1148–1158. 10.1016/j.bbamem.2009.02.014 [DOI] [PubMed] [Google Scholar]
- Ogris M, Brunner S, Schüller S, Kircheis R, Wagner E, 1999. PEGylated DNA/transferrin-PEI complexes: Reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 6, 595–605. 10.1038/sj.gt.3300900 [DOI] [PubMed] [Google Scholar]
- Ordóñez-Gutiérrez L, Posado-Fernández A, Ahmadvand D, Lettiero B, Wu L, Antón M, Flores O, Moghimi SM, Wandosell F, 2016. ImmunoPEGliposome-mediated reduction of blood and brain amyloid levels in a mouse model of Alzheimer’s disease is restricted to aged animals. Biomaterials 112 [DOI] [PubMed] [Google Scholar]
- Pan C, Kumar C, Bohl S, Klingmueller U, Mann M, 2009. Comparative Proteomic Phenotyping of Cell Lines and Primary Cells to Assess Preservation of Cell Type-specific Functions. Mol. Cell. Proteomics 8, 443–450. 10.1074/mcp.M800258-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahi Y, Farshbaf M, Mohammadhosseini M, Mirahadi M, Khalilov R, Saghfi S, Akbarzadeh A, 2017. Recent advances on liposomal nanoparticles:synthesis, characterization and biomedical applications. Artif. Cells, Nanomedicine Biotechnol 45, 788–799. 10.1080/21691401.2017.1282496 [DOI] [PubMed] [Google Scholar]
- Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP, 2013. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimer’s Dement. 9, 63–75. 10.1016/j.jalz.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Qi P, Cao M, Song L, Chen C, Liu M, Li N, Wu D, Peng J, Hu G, Zhao J, 2016. The biological activity of cationic liposomes in drug delivery and toxicity test in animal models. Environ. Toxicol. Pharmacol. 47, 159–164. 10.1016/j.etap.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Qian ZM, 2002. Targeted Drug Delivery via the Transferrin Receptor-Mediated Endocytosis Pathway. Pharmacol. Rev 54, 561–587. 10.1124/pr.54.4.561 [DOI] [PubMed] [Google Scholar]
- Ramsey JD, Flynn NH, 2015. Cell-penetrating peptides transport therapeutics into cells. Pharmacol. Ther 154, 78–86. 10.1016/j.pharmthera.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Roursgaard M, Knudsen KB, Northeved H, Persson M, Christensen T, Kumar PEK, Permin A, Andresen TL, Gjetting T, Lykkesfeldt J, Vesterdal LK, Loft S, Møller P, 2016. In vitro toxicity of cationic micelles and liposomes in cultured human hepatocyte (HepG2) and lung epithelial (A549) cell lines. Toxicol. Vitr 36, 164–171. 10.1016/j.tiv.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Safari J, Zarnegar Z, 2014. Advanced drug delivery systems: Nanotechnology of health design A review. J. Saudi Chem. Soc 18, 85–99. 10.1016/j.jscs.2012.12.009 [DOI] [Google Scholar]
- Saikia C, Gogoi P, 2015. Chitosan: A Promising Biopolymer in Drug Delivery Applications. J. Mol. Genet. Med s4, 1–10. 10.4172/1747-0862.S4-006 [DOI] [Google Scholar]
- Salatin S, Yari Khosroushahi A, 2017. Overviews on the cellular uptake mechanism of polysaccharide colloidal nanoparticles. J. Cell. Mol. Med 21, 1668–1686. 10.1111/jcmm.13110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-López E, Ettcheto M, Egea MA, Espina M, Calpena AC, Folch J, Camins A, García ML, 2016. New Potential Strategies for Alzheimer’s Disease Prevention: Pegylated Biodegradable Dexibuprofen Nanospheres Administration to APPswe/PS1dE9. Nanomedicine Nanotechnology, Biol. Med 10.1016/j.nano.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S, 2015. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol 6, 1–13. 10.3389/fphar.2015.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G, Modgil A, Layek B, Arora K, Sun C, Law B, Singh J, 2013. Cell penetrating peptide tethered bi-ligand liposomes for delivery to brain in vivo: Biodistribution and transfection. J. Control. Release 167, 1–10. 10.1016/j.jconrel.2013.01.016 [DOI] [PubMed] [Google Scholar]
- Sharma G, Modgil A, Sun C, Singh J, 2012. Grafting of cell-penetrating peptide to receptor-targeted liposomes improves their transfection efficiency and transport across blood-brain barrier model. J. Pharm. Sci 101, 2468–78. 10.1002/jps.23152 [DOI] [PubMed] [Google Scholar]
- Shim G, Kim MG, Park JY, Oh YK, 2013. Application of cationic liposomes for delivery of nucleic acids. Asian J. Pharm. Sci 8, 120–128. 10.1016/j.ajps.2013.07.009 [DOI] [Google Scholar]
- Sumners C, Fregly MJ, 1989. Modulation of angiotensin II binding sites in neuronal cultures by mineralocorticoids. Am. J. Physiol 256, C121–C129. [DOI] [PubMed] [Google Scholar]
- Tseng Y-L, Liu J-J, Hong R-L, 2002. Translocation of liposomes into cancer cells by cell-penetrating peptides penetratin and tat: a kinetic and efficacy study. Mol. Pharmacol 62, 864–72. 10.1124/mol.62.4.864 [DOI] [PubMed] [Google Scholar]
- Varkouhi AK, Scholte M, Storm G, Haisma HJ, 2011. Endosomal escape pathways for delivery of biologicals. 10.1016/j.jconrel.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Voinea M, Dragomir E, Manduteanu I, Simionescu M, 2002. Binding and uptake of transferrin-bound liposomes targeted to transferrin receptors of endothelial cells. Vascul. Pharmacol 39, 13–20. 10.1016/S1537-1891(02)00165-9 [DOI] [PubMed] [Google Scholar]
- Wiethoff CM, Gill ML, Koe GS, Koe JG, Middaugh CR, 2002. The structural organization of cationic lipid-DNA complexes. J. Biol. Chem 277, 44980–44987. 10.1074/jbc.M207758200 [DOI] [PubMed] [Google Scholar]
- Xiao G, Gan L-S, 2013. Receptor-mediated endocytosis and brain delivery of therapeutic biologics. Int. J. Cell Biol 2013, 703545 10.1155/2013/703545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yameen B, Choi W Il, Vilos C, Swami A, Shi J, Farokhzad OC, 2014. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release 190, 485–499. 10.1016/j.jconrel.2014.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. ye, Zheng Y, Chen J. yin, Zhang Q. yang, Zhao D, Han D. en, Chen X. jing, 2013. Comprehensive study of cationic liposomes composed of DC-Chol and cholesterol with different mole ratios for gene transfection. Colloids Surfaces B Biointerfaces 101, 6–13. 10.1016/j.colsurfb.2012.05.032 [DOI] [PubMed] [Google Scholar]
- Yang Z, Li J, Wang Z, Dong D, Qi X, 2014. Biomaterials Tumor-targeting dual peptides-modified cationic liposomes for delivery of siRNA and docetaxel to gliomas. Biomaterials 35, 5226–5239. 10.1016/j.biomaterials.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Zhang D, Wang J, Xu D, 2016. Cell-penetrating peptides as noninvasive transmembrane vectors for the development of novel multifunctional drug-delivery systems. J. Control. Release 229, 130–139. 10.1016/j.jconrel.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Zhang HW, Zhang L, Sun X, Zhang ZR, 2007. Successful transfection of hepatoma cells after encapsulation of plasmid DNA into negatively charged liposomes. Biotechnol. Bioeng 10.1002/bit.21146 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Anchordoquy TJ, 2004. The role of lipid charge density in the serum stability of cationic lipid/DNA complexes. Biochim. Biophys. Acta - Biomembr 1663, 143–157. 10.1016/j.bbamem.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Zheng C, Ma C, Bai E, Yang K, Xu R, 2015. Transferrin and cell-penetrating peptide dual-functioned liposome for targeted drug delivery to glioma. Int. J. Clin. Exp. Med 8, 1658–1668. [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Pang X, Yang P, Wan X, Wei Y, Guo Q, Zhang Q, Jiang X, 2016. A hybrid siRNA delivery complex for enhanced brain penetration and precise amyloid plaque targeting in Alzheimer’s disease mice. Acta Biomater. 10.1016/j.actbio.2016.11.029 [DOI] [PubMed] [Google Scholar]
- Zylberberg C, Gaskill K, Pasley S, Matosevic S, 2017. Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther. 24, 441–452. 10.1038/gt.2017.41 [DOI] [PubMed] [Google Scholar]
- Zylberberg C, Matosevic S, 2016. Pharmaceutical liposomal drug delivery: a review of new delivery systems and a look at the regulatory landscape. Drug Deliv 7544, 1–11. 10.1080/10717544.2016.1177136 [DOI] [PubMed] [Google Scholar]