Abstract
Misfolded proteins compromise cellular homeostasis. This is especially problematic in the endoplasmic reticulum (ER), which is a high-capacity protein-folding compartment and whose function requires stringent protein quality-control systems. Multiprotein complexes in the ER are able to identify, remove, ubiquitinate, and deliver misfolded proteins to the 26S proteasome for degradation in the cytosol, and these events are collectively termed ER-associated degradation, or ERAD. Several steps in the ERAD pathway are facilitated by molecular chaperone networks, and the importance of ERAD is highlighted by the fact that this pathway is linked to numerous protein conformational diseases. In this review, we discuss the factors that constitute the ERAD machinery and detail how each step in the pathway occurs. We then highlight the underlying pathophysiology of protein conformational diseases associated with ERAD.
PROTEIN FOLDING, MOLECULAR CHAPERONES, AND PROTEOSTASIS
The birth of a protein begins in the cytoplasm with the emergence of a polypeptide chain from the ribosome exit tunnel. For a protein to attain its native conformation, it must navigate a complex folding landscape and endure the relentless surveillance of quality-control pathways. The information for a protein's tertiary structure is contained within its primary amino acid sequence (Anfinsen 1973), yet the growing polypeptide does not exit the ribosome in isolation. Rather, it emerges into a crowded environment filled with other proteins, lipid membranes, and cellular metabolites, which can alter the folding pathway. In addition, exposed hydrophobic side chains in soluble proteins must be shielded from solvent, whereas integral membrane proteins and proteins that enter the secretory pathway are directed to the endoplasmic reticulum (ER). Moreover, translated regions may need to interact with more carboxy-terminal regions before the native structure is attained, leaving unfolded polypeptide chains to linger in solution (van den Berg et al. 1999; Frydman 2001; Braselmann et al. 2013). Consequently, proteins are initially deposited in an energetically unfavorable environment, and there is a strong propensity to reach an unproductive or deleterious energy minimum that can give rise to toxic protein aggregates (Hartl et al. 2011).
To overcome these challenges, a conserved and extensive network of proteins, known as molecular chaperones, are dedicated to ushering newly translated polypeptides through the folding and maturation process. Molecular chaperones also aid in the identification and elimination of polypeptides that are unable to fold (Klaips et al. 2018). Collectively, the group of diverse factors that maintain these protein homeostatic systems is known as the proteostasis network (Sala et al. 2017). Although there is an enormous investment of cellular resources dedicated to maintaining proteostasis, some protein-folding errors are unrecognized or the machinery that comprises the proteostasis network malfunctions. In turn, some misfolded proteins are prematurely captured and destroyed before they can fold. These phenomena lie at the heart of several protein conformational diseases in humans and have been implicated in cellular and organismal aging (Labbadia and Morimoto 2015; Higuchi-Sanabria et al. 2018).
In this review, we will first briefly discuss the biogenesis of proteins in the ER and the major chaperone classes that engage nascent polypeptides. We will then review the individual steps in the ER-associated degradation (ERAD) pathway and highlight how chaperones and other factors facilitate these steps. We conclude with a discussion of select diseases associated with this pathway.
MOLECULAR CHAPERONES PLAY CRITICAL ROLES DURING PROTEIN BIOGENESIS IN THE ER
The ER is the entry point to the secretory pathway and assembles integral membrane proteins in eukaryotes (Fig. 1). Soluble ER-directed proteins contain a signal sequence at the amino terminus. During signal sequence emergence from the ribosome, this short peptide is recognized and bound by the signal recognition particle (SRP), a ribonucleoprotein complex, which temporarily interrupts translation and directs the ribosome to the ER membrane after docking to the SRP receptor (Saraogi and Shan 2011). The SRP–SRP receptor interaction positions the polypeptide at the translocation channel, which is composed of the trimeric Sec61 protein complex along with several regulatory subunits (Rapoport 2007). After SRP dissociates, translation continues, and Sec61 allows passage of the polypeptide into the ER or, for membrane proteins, into the ER bilayer by virtue of a lateral gate in Sec61 (Osborne et al. 2005). The signal sequence is cleaved inside the ER by signal peptidase. In contrast, the first transmembrane helix (TMH) in integral membrane proteins often serves as a signal sequence and in most cases is not cleaved (Shao and Hegde 2011).
Figure 1.
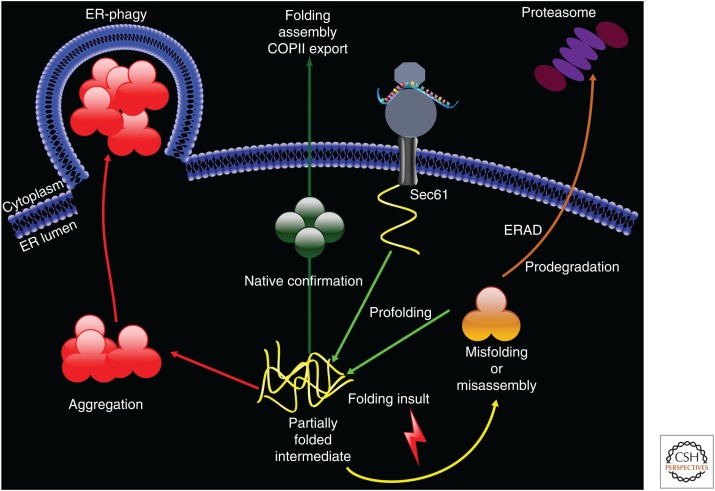
Nascent polypeptide biogenesis in the endoplasmic reticulum (ER). After docking at the ER membrane on the Sec61 translocation channel, the ribosome cotranslationally translocates a polypeptide (in yellow) into the ER lumen. Chaperones, including Hsp70s, Hsp40s, NEFs, lectins, disulfide isomerases, and peptidyl proline isomerase, act as “profolding” factors (see text for details). If the polypeptide acquires its native conformation and—as shown in one example, oligomerizes (in green)—it traffics from the ER in COPII vesicles. Genetic mutations, errors in transcription or translation, defects in the acquisition of posttranslational modifications, and/or ER stress can result in misfolding or a misassembled intermediate (in orange). If the substrate is unable to fold, “prodegradation” chaperones facilitate ER-associated degradation (ERAD). In contrast, partially folded intermediates can aggregate (in red) and are instead cleared by ER-phagy. One route for this pathway leads to the formation of an “omegasome,” as depicted (Simonsen and Stenmark 2008). For simplicity, only a soluble ER lumenal protein is presented.
Proteins that enter the ER progress through several maturation steps that require the addition, formation, and/or modification of sugar moieties, cis–trans prolyl isomers, disulfide bonds and—for integral proteins—the adoption of complex transmembrane topologies (Braakman and Bulleid 2011). An Hsp70 molecular chaperone, known as BiP (Gething 1999), is a central player in ER homeostasis. Hsp70s have two allosterically linked functional domains, a substrate-binding domain that interacts with short stretches of hydrophobic residues, and an ATP-binding domain that drives a conformational change in the protein via ATP hydrolysis to regulate substrate-binding affinity (Hageman et al. 2011; Zhuravleva et al. 2012; Zuiderweg et al. 2013). By associating with exposed hydrophobic patches, Hsp70s, like BiP, shield a polypeptide from aggregating until productive intramolecular interactions occur (Mayer and Bukau 2005; Otero et al. 2010; Young 2010; Mayer 2013). This is especially important given the vectorial nature of protein translocation. In fact, BiP has also been shown to augment protein translocation (Brodsky et al. 1995; Matlack et al. 1999) as well as ERAD, as discussed below. More recently, the reversible oligomerization of BiP was found to be associated with the unfolded protein load in the ER (Preissler et al. 2015), and oxidation of a key cysteine in the chaperone regulates its activity in response to oxidative stress (Wang et al. 2014). These data indicate that the activity of this chaperone also responds to the ER environment.
The peptide binding and release cycle of Hsp70 is intrinsically slow but is stimulated by Hsp40 cochaperones (Kampinga and Craig 2010; Kityk et al. 2018) and by nucleotide exchange factors (NEFs) (Bracher and Verghese 2015), both of which also reside in the ER and facilitate protein folding and quality control. Hsp40s can also bind and deliver substrates to Hsp70. Recent data indicate that Hsp40s in the ER aid in the assembly of oligomeric membrane proteins (Li et al. 2017), regulate protein translocation (Schorr et al. 2015), and differentially recognize unique motifs in nascent polypeptides, some of which are aggregation-prone (Behnke et al. 2016).
As noted above, the activity of Hsp70 chaperones is also enhanced by NEFs, and in the ER several members of this family are important during early steps in protein biogenesis. NEFs in the Hsp110/170 family resemble Hsp70 in that they contain an ATP-binding domain as well as a substrate-binding domain, but they also harbor an extended carboxy-terminal motif and acidic insertions in the substrate-binding domain (Liu and Hendrickson 2007). Like Hsp70s and Hsp40s, members of this family bind protein substrates and prevent aggregation (Park et al. 2003; Goeckeler et al. 2008; Behnke and Hendershot 2014), but the importance of this activity—at least for the cytosolic homologs—is unclear (Garcia et al. 2017). Nevertheless, conserved ER NEFs liberate bound ADP from BiP (Steel et al. 2004; Andréasson et al. 2010; Hale et al. 2010). This event drives substrate release not only because the exchange of bound ADP with ATP lowers peptide affinity, but because some NEFs possess a peptide-like motif that displaces the BiP-associated substrate (Rosam et al. 2018). Another recent finding is that the Sil1 NEF in yeast acts as a BiP reductant, thereby altering its substrate-binding properties after an oxidative stress has been resolved (Siegenthaler et al. 2017).
Nearly all secreted proteins contain disulfide bonds, which are formed in the more oxidizing environment of the ER and stabilize native conformations (Frand et al. 2000; Tu and Weissman 2004). To this end, protein disulfide isomerases (PDIs) catalyze the formation of disulfide bonds in nascent polypeptides (Feige and Hendershot 2011) and can associate with other chaperones, such as BiP, which deliver substrates to the PDIs (Jessop et al. 2009). One PDI family member, ERdj5, is both an Hsp40 and a PDI family member. Unlike other PDIs, ERdj5 is a reductase and helps unfold proteins before ERAD (see below) (Ushioda et al. 2008).
Most secreted and membrane proteins contain glycans, which are added cotranslationally by the action of the ER oligosaccharyltransferase (Aebi et al. 2010; Cherepanova et al. 2016). Glycosylation occurs on an asparagine in the N-X-(S/T) consensus site and serves several purposes, such as stabilizing the protein and preventing aggregation. As discussed in the next paragraph, glycans also engage ER chaperone-like lectins, which monitor protein folding (Caramelo and Parodi 2007; Pearse and Hebert 2010). Finally, glycans mediate cell–cell contact and signaling cascades when positioned on cell surface proteins.
Soon after the addition of the core glycan, which is composed of Glc3Man9(GlcNAc)2, two of the three outer glucose residues are trimmed by glucosidases (Deprez et al. 2005). The monoglucosylated glycan is then recognized by two calcium-binding lectins, calnexin (CNX), an integral ER membrane protein associated with Sec61, and calreticulin (CRT), an ER lumenal protein (Schrag et al. 2001). Both CNX and CRT act as chaperones, stabilizing ER proteins, promoting folding, and recruiting other folding components, such as PDIs. Removal of the final glucose by another glucosidase results in release from CNX/CRT and, potentially, transport of a folded protein from the ER. However, the folding status of the released substrate is assessed by the UDP glucose glycosyltransferase (UGGT) (Sousa and Parodi 1995; Trombetta and Helenius 2000; Taylor et al. 2004; Ritter et al. 2005). If the protein has not yet folded, UGGT adds back a single glucose to the appended glycan so CNX/CRT can rebind, allowing for additional cycles of protein folding.
Another rate-limiting step during protein folding is cis–trans isomerization of prolines (Reimer et al. 1998), which is catalyzed by the peptidyl-prolyl isomerases (PPIs) (Schmid et al. 1993; Gothel and Marahiel 1999). Spontaneous isomerization of proline is too slow to support protein folding, and specific prolines may need to isomerize multiple times before a native conformation is achieved. Consistent with their role in protein folding, one PPI associates with both a PDI as well as a chaperone-like lectin that facilitates protein folding in the ER (Kozlov et al. 2017).
Finally, the ER in most eukaryotes contains an Hsp90 homolog, known as Grp94. Cytosolic and nuclear Hsp90s are best known for their role in the maturation of transcription factors, protein kinases, and cell surface receptors (Pratt et al. 2008; Taipale et al. 2012), which has spurred interest in the development of Hsp90 inhibitors in cancer. In the cytoplasm, Hsp70 and Hsp90 interact through a cofactor known as Hop that also regulates their activity (Johnson et al. 1998; Alvira et al. 2014). Surprisingly, however, the ER lacks a Hop homolog, and there is a relatively small number of known Grp94 substrates (Ansa-Addo et al. 2016).
Overall, molecular chaperones assemble into large multifunctional complexes, which augment protein folding and other processes (Freilich et al. 2018; Joshi et al. 2018). This is especially critical in the ER, which receives, sorts, and posttranslationally modifies one-third of the eukaryotic proteome. However, the efficiency of the ER-folding machinery is not foolproof, and when errors in posttranslational modification occur or mutations compromise protein folding, aberrant species must be removed or cellular homeostasis will fail. Somewhat surprisingly, some of the same chaperones dedicated to the folding of nascent polypeptides in the ER also become essential recognition components of the degradation machinery (Brodsky 2007). However, by associating with proteins during folding, chaperones are in a prime position to identify when a protein has failed to reach its native state. The switch from a profolding to a prodegradative function may be based on interactions with different motifs in the protein or by differential association with cofactors. The mechanism of how this switch is made is not understood, but, as discussed above, glycosylation can act as a switch (Roth and Zuber 2017). Notably, slow-acting mannosidases interrupt CNX/CRT binding and impart a time frame for folding (Liu et al. 1999; Kanehara et al. 2007). Specifically, after the initial removal of mannose, an unfolded protein becomes a substrate for the ER degradation-enhancing mannosidase (EDEM) (Molinari et al. 2003; Oda et al. 2003; Clerc et al. 2009). EDEM binding results in further glycan processing and ERdj5 recruitment (see above), which breaks disulfide bonds in preparation for ERAD (Ushioda et al. 2008; Ninagawa et al. 2014). At this point, the commitment to degradation has been made and EDEM hands a substrate to the OS9 or XTP3-B lectin, which escorts it to the ERAD machinery (Christianson et al. 2008; van der Goot et al. 2018). There is also evidence of spatial separation of the mannosidases, which allow folding attempts to occur before mannose trimming (Pan et al. 2013; Benyair et al. 2015). Nevertheless, some proteins in the ER are not glycosylated. In some of these cases, the lectins still function as ER quality-control gatekeepers, and in other cases alternate mechanisms of substrate recognition exist or recognition is mediated by BiP (Brodsky et al. 1999; Okuda-Shimizu and Hendershot 2007; Ushioda et al. 2013).
ENDOPLASMIC RETICULUM QUALITY CONTROL AND ERAD
The clearance or repair of misfolded proteins in the ER depends on ERAD as well as the unfolded protein response (UPR) and ER-phagy (Fig. 1). The UPR has been reviewed in detail elsewhere (see, for example, Karagöz et al. 2018; Preissler and Ron 2018), but in brief this transcriptional response is activated by the accumulation of misfolded proteins in the ER and triggered by integral ER membrane sensors. The UPR mitigates stress by slowing general protein translation, thus reducing the protein load in the ER while simultaneously up-regulating factors that facilitate the folding or clearance of misfolded species, including ERAD-requiring factors (Travers et al. 2000). However, continued ER stress shifts the UPR from a protective to a proapoptotic pathway, which underlies the pathology of numerous diseases. In contrast, ER-phagy selectively removes portions of the ER after the UPR has been resolved (Schuck et al. 2014) or more directly targets substrates via recently identified ER-phagy receptors (Smith and Wilkinson 2017). Select ERAD substrates become substrates for ER-phagy when overexpressed (Kruse et al. 2006b; Ishida et al. 2009), but to date it is unclear whether these substrates are targeted for autophagy after retrotranslocation and aggregation in the cytoplasm or through encapsulation of ER fragments (Fig. 1).
ERAD takes place in distinct yet coordinated steps (Fig. 2). First, misfolded substrates are recognized, which is followed by substrate tagging via the conjugation of a polyubiquitin motif. Acquisition of this motif serves as a signal for the energy-dependent extraction of the substrate from the ER as well as its subsequent delivery to the proteasome (Meusser et al. 2005; Vembar and Brodsky 2008; Araki and Nagata 2011; Smith et al. 2011; Ruggiano et al. 2014; Pisoni and Molinari 2016; Berner et al. 2018).
Figure 2.
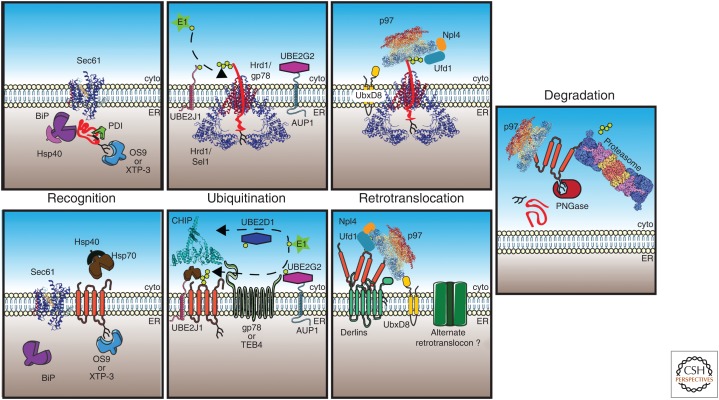
Endoplasmic reticulum-associated degradation (ERAD) of lumenal and integral membrane substrates. (Top) During translocation through Sec61 (RCSB PDB 3JC2), a soluble ERAD substrate (in red) encounters BiP, which aids in polypeptide folding and translocation across the membrane. BiP activity is enhanced by ER lumenal Hsp40s. The polypeptide can be modified by the formation of disulfide bonds by protein disulfide isomerases (PDIs) and by addition of N-linked glycans, which are bound by lectins such as OS9 or XTP-3 (in mammals). If the polypeptide fails to fold, it is transferred to Hrd1. The E3 ligase/RING domain of Hrd1 is located in the cytoplasm, so lumenal substrates must be partially retrotranslocated for ubiquitin conjugation, which occurs via an enzymatic cascade that includes an E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme such as UBE2G2/AUP1 or UBE2J1), and E3 (ubiquitin ligase such as Hrd1 or gp78), which build a polyubiquitin chain (lime hexagons) on the substrate. AUP1 tethers the E2 to the ER membrane and functions analogously to Cue1 in yeast. Following ubiquitination, the substrate is bound by the p97 (RCSB PDB 5C1A) complex, which is recruited to the membrane by UbxD8 and interacts with the substrate via ubiquitin-binding partners Ufd1 and Npl1. (Bottom) Integral membrane ERAD substrates (in orange) may also interact with BiP, but also have access to cytoplasmic chaperones such as Hsp70-Hsp40 that recognize folding lesions. Cytoplasmic lysines are then polyubiquitinated by E3 ligases such as TEB4, gp78, and/or CHIP (cyan, RCSB PDB 2C2L) in mammals. The p97–Ufd1–Npl1 heterotrimer is recruited by UbxD8 and, in some cases, Derlin family members, which facilitate retrotranslocation. The nature of the retrotranslocon channel (green) for membrane proteins is not established. Finally, the retrotranslocated protein can be deglycosylated by PNGase, followed by delivery to the 26S proteasome (RCSB PDB 4CR2), deubiquitinated by components in the 19S particle (purple/blue), and proteolyzed in the 20S core particle (yellow/salmon).
ERAD substrates are recognized by molecular chaperones and the chaperone-like lectins discussed above. Chaperone-based recognition takes advantage of surface-exposed, hydrophobic polypeptides that should be sequestered within the protein interior. Early results implicated BiP as a partner of misfolded or orphaned proteins in mammalian cells (Bole et al. 1986; Knittler et al. 1995), and later genetic studies in yeast directly implicated BiP in ERAD substrate selection (Plemper et al. 1997; Brodsky et al. 1999; Kabani et al. 2003). The contributions of Hsp40 partners (Nishikawa et al. 2001) and NEFs (Buck et al. 2013; Williams et al. 2015) during BiP-mediated selection and the maintenance of substrate solubility underscores the importance of chaperone-based complexes in promoting both folding and ERAD.
In the second step, the identified substrate is ubiquitinated. Ubiquitin is a 76-amino-acid protein that forms isopeptide-linked polyubiquitin chains, which serve as a signal for proteasome-mediated degradation during ERAD (Preston and Brodsky 2017). Ubiquitin is primed for addition to misfolded proteins by an enzymatic cascade starting with a ubiquitin-activating enzyme (E1). Next, ubiquitin becomes covalently linked to a ubiquitin-conjugating enzyme (E2) before a ubiquitin ligase (E3) catalyzes the final transfer of ubiquitin to exposed lysine residues in the targeted substrate (Deshaies and Joazeiro 2009; Finley 2009). In yeast, where ERAD was first defined and has been extensively studied, there are two ER-localized E3 ligases associated with this pathway, Hrd1 (Wilhovsky et al. 2000) and Doa10 (Swanson et al. 2001), but the contributions of other E3s were uncovered more recently (Kohlmann et al. 2008; Stolz et al. 2013; Foresti et al. 2014). As anticipated, based on the greater number of potential ERAD substrates, the list of ERAD-contributing E3s in mammals is significantly longer (Claessen et al. 2012; Olzmann et al. 2013a).
Based on the specific site of the misfolded lesion within a protein, different branches of the ERAD pathway have been described (Huyer et al. 2004; Vashist and Ng 2004; Carvalho et al. 2006; Denic et al. 2006). These distinctions are most clearly defined in yeast but are somewhat blurred in mammalian cells (Bernasconi et al. 2010). Nevertheless, the term ERAD-L has been used to designate a soluble protein within the ER lumen or an ER membrane–associated protein with a misfolded domain facing the lumen. Hrd1 is the E3 ubiquitin ligase used for ERAD-L. Other factors that recognize and transfer substrates from lumenal chaperones to Hrd1 or stabilize the Hrd1 complex include the Derlins (Knop et al. 1996), Usa1 (Horn et al. 2009; Carroll and Hampton 2010) or HERP in mammals (Okuda-Shimizu and Hendershot 2007), and Hrd3 (or Sel1 in mammals) (Gardner et al. 2000; Mueller et al. 2008; Vashistha et al. 2016). Hrd3 interacts with BiP as well as an Hsp40 partner and even shows chaperone-like properties (Mehnert et al. 2015), which may help retain a misfolded protein in solution before retrotranslocation.
An ER membrane protein with a misfolded cytoplasmic-facing domain is inaccessible to ER resident chaperones. In this case, recognition requires cytoplasmic chaperones, and a different E3 ligase, Doa10, is used along with a distinct set of E2-conjugating enzymes and cofactors in yeast (Vashist and Ng 2004; Carvalho et al. 2006; Ravid et al. 2006). This route is known as ERAD-C, and the recognition and ubiquitination of ERAD-C substrates depends on Hsp70 and Hsp40 partners (Youker et al. 2004; Han et al. 2007; Nakatsukasa et al. 2008). More recently, the yeast cytosolic protein disaggregase, Hsp104, was shown to enhance the degradation of an aggregation-prone ERAD-C substrate (Preston et al. 2018). Based on the diversity of potential ERAD-C substrates (i.e., membrane proteins with cytoplasmic domains), it is likely that other recognition factors exist.
The identification of integral membrane proteins with folding lesions within the lipid bilayer is especially problematic as no classical chaperones have been identified that possess this activity. However, a single-pass transmembrane protein with low hydrophobicity was transiently displaced into the ER lumen in which BiP could bind and facilitate ERAD (Feige and Hendershot 2013). For more firmly anchored membrane proteins, the Hrd1 ubiquitin ligase, along with the associated Hrd3 cofactor, appears to bind and then ubiquitinate these ERAD-M substrates (Sato et al. 2009). Nevertheless, based on the complex nature of membrane proteins, which deposit domains in three unique chemical environments (i.e., the ER, the lipid bilayer, and the cytosol), it is likely that most misfolded integral membrane proteins will follow more than one of these three ERAD pathways (see, for example, Buck et al. 2010).
Once selected, ERAD substrates are transported or retrotranslocated from the ER lumen or membrane back to the cytosol where they are delivered to and degraded by the 26S proteasome. Initially, it was assumed that an ER resident protease handled misfolded ER proteins (Needham and Brodsky 2013), but it subsequently became evident that aberrant soluble and even membrane proteins are retrotranslocated to the cytosol for disposal (Hiller et al. 1996; McCracken and Brodsky 1996; Wiertz et al. 1996; Plemper et al. 1997; Nakatsukasa et al. 2008; Garza et al. 2009). Initially, the Sec61 translocation channel was the best candidate for a retrotranslocation channel (Pilon et al. 1997; Romisch 1999), and more recently an ER metalloprotease was shown to help clear substrates that become trapped in Sec61 caused by the premature folding of a cytoplasmic domain (Ast et al. 2016). Hrd1 is involved in the destruction of similar substrates (Rubenstein et al. 2012). Other work suggested that the Derlins might be the retrotranslocation channel, or at least act as a major contributor to retrotranslocation (Lilley and Ploegh 2004; Ye et al. 2004; Wahlman et al. 2007; Greenblatt et al. 2011; Mehnert et al. 2014). However, more recent data are consistent with Hrd1 acting as both a ligase and a retrotranslocation channel, at least for soluble substrates (Baldridge and Rapoport 2016; Schoebel et al. 2017). It also remains possible that integral membrane proteins are extracted directly from the lipid bilayer or are somehow solubilized within the bilayer and retrotranslocated without the aid of a channel (Avci and Lemberg 2018).
The force to pull substrates from the ER is provided by the AAA-ATPase Cdc48 in yeast or the homologous p97 protein in mammals (Bays et al. 2001; Rabinovich et al. 2002; Ye et al. 2004). Hexameric, chaperone-like AAA-ATPase ring structures form components of multichaperone complexes and proteolytic machines. In general, this class of proteins act as force-generating engines in numerous energy-dependent processes, such as protein degradation and disaggregation and DNA and lipid remodeling (Mogk et al. 2008; Sauer and Baker 2011; van den Boom and Meyer 2018). Cdc48/p97 functions in a complex with Ufd1 and Npl4, which recognize polyubiquitinated substrates and bind several components at the ER membrane (Stolz et al. 2010; Buchberger et al. 2015; Neal et al. 2018). The appended polyubiquitin chain not only marks the substrate for proteasome-dependent degradation, but Cdc48 uses this motif as a handle (and the ER membrane as an anchor) for substrate retrotranslocation by coupling ATP hydrolysis to drive the motor. Only a few substrates can be extracted in a ubiquitin-independent manner, and in this case AAA-ATPases in the proteasome are sufficient (Lee et al. 2004). A recent study also reported that more hydrophobic transmembrane domains were less efficiently retrotranslocated, suggesting that degradation at the ER, perhaps via clipping by integral membrane proteases (Loureiro et al. 2006; Fleig et al. 2012), contributes to retrotranslocation/degradation (Guerriero et al. 2017). Other recent data indicate that polyubiquitinated substrates are threaded through the Cdc48 core after partial deubiquitination (Bodnar and Rapoport 2017). Whether species with shorter polyubiquitin chains interact efficiently with the proteasome or whether another round of polyubiquitination is needed is unknown.
After a polyubiquitinated substrate is removed from the ER, it is delivered to the proteasome with the help of the Rad23 and Dsk2 shuttling factors (Medicherla et al. 2004). In some cases, appended glycans are removed by a protein N-glycanase (PNGase) before proteasome delivery (Hirayama et al. 2015). Rad23 also interacts with this enzyme (Kim et al. 2006), suggesting that delivery is coupled with glycan removal, which might otherwise impede proteasome entry. The proteasome has a barrel-like catalytic core housing two copies of three unique proteases (Voges et al. 1999; Bard et al. 2018), and entry into a narrow aperture at the end of the barrel is regulated by the 19S “cap,” which itself recognizes ubiquitinated substrates and possesses deubiqutinating enzymes as a substrate is fed into the proteasome core for degradation (Finley 2009; Liu and Ye 2012).
REGULATION OF PROTEIN ABUNDANCE BY ERAD
The ER serves as a portal for residents of the ER, Golgi, lysosome, and plasma membrane, as well as secreted proteins. Included among this diverse group of substrates are hormones, enzymes, cell surface receptors, and nutrient and ion transporters and channels. This positions the ER as a potential regulator of downstream pathways (Halperin et al. 2014). Indeed, ERAD is implicated not only in quality control but in quantity control (Printsev et al. 2017). For example, HMG-CoA reductase catalyzes the rate-limiting step in cholesterol biosynthesis and is the target of cholesterol- lowering statins (Goldstein and Brown 1990). The appearance of sterol end products or biosynthetic intermediates leads directly to the acquisition of a structurally disordered form of the enzyme, which triggers ubiquitination and ERAD (Jo and Debose-Boyd 2010; Wangeline et al. 2017). Other enzymes in the sterol and lipid droplet biosynthetic pathway are also regulated by ERAD (Foresti et al. 2013; Olzmann et al. 2013b; Ruggiano et al. 2016). Another example of regulated degradation occurs when the ERAD pathway is hijacked by a viral pathogen. The human cytomegalovirus (HCMV) genome encodes several proteins that reduce surface expression of major histocompatibility class I (MHC-1), thus disabling of the host immune response. MHC-1 degradation is induced via an ERAD-like pathway that requires select HCMV gene products. A similar situation exists after HIV infection (Morito and Nagata 2015; van den Boomen and Lehner 2015). Interestingly, none of these processes require molecular chaperones, consistent with the idea that ERAD-mediated quality and quantity control can proceed via distinct routes.
In contrast, the metabolic regulation of apolipoprotein B (ApoB), a protein component of chylomicrons, very low-density lipoproteins (VLDLs), and LDLs (Fisher and Ginsberg 2002), is chaperone-dependent. When lipid availability is limited, the cotranslational translocation of ApoB through Sec61 is halted and the protein is instead routed for ERAD in an Hsp70, Hsp90, and PDI-dependent manner (Fisher et al. 1997; Gusarova et al. 2001; Grubb et al. 2012); the PDIs also play a role in ApoB lipidation (Wang et al. 2015). Interestingly, the Hsp110 chaperone, which shows substrate-binding activity and functions as an NEF (see above), protects ApoB, potentially acting as a holdase as the protein translocates into the ER (Hrizo et al. 2007).
ERAD SUBSTRATE RECOGNITION
As outlined above, ERAD substrate recognition requires chaperones and chaperone-like lectins, but the biochemical and biophysical features of a misfolded protein or an orphaned subunit that are sufficient for ERAD targeting are poorly defined. Simply exposing hydrophobic motifs favors chaperone binding, but whether the chaperone now plays a profolding or prodegradative role clearly requires other events, which were discussed in the previous section. Nevertheless, the presence of an aggregation-prone domain was recently shown to support ERAD-C targeting (Preston et al. 2018), and studies on a related substrate indicated that aggregation propensity influences whether a substrate is delivered to the ERAD or lysosomal/vacuolar degradation pathway (Sun and Brodsky 2018). However, aggregation propensity for many ERAD-L substrates may be less relevant as the ER has an unusually robust capacity to prevent protein aggregation compared with the cytosol (Rousseau et al. 2004; Vincenz-Donnelly et al. 2018). Thus, the ER provides a protective environment in which most aggregation-prone proteins remain soluble for recognition by chaperones and lectins. Nevertheless, several proteins can still aggregate or illegitimately oligomerize in the ER and undergo ER-phagy (Kruse et al. 2006a,b; Ishida et al. 2009).
PROTEIN CONFORMATIONAL DISEASES AND ERAD
In principle, proteins can adopt a stunning array of conformations during folding—estimated to be as high as 10300 (Levinthal 1969; Karplus 1997)—and given the diverse conformations of secreted proteins and the preponderance of genetic mutations it is not unreasonable to imagine that many of the ∼6700 secretory proteins in humans may be a harbinger of disease. To date, 64 diseases in which the conformationally defective protein is destroyed by ERAD are known (Table 1). For inclusion, a mutant protein associated with a human disease must be retained in the ER and stabilized by treatment with a proteasome inhibitor and/or is polyubiquitinated. In contrast to a more extensive previous list (Guerriero and Brodsky 2012), we excluded toxins that masquerade as ERAD substrates and diseases associated with the ERAD-like action of viral pathogens.
Table 1.
Resource list in which disease-associated proteins are destroyed ERAD
| Chromosomal locus | Gene | Protein | Disease | References |
|---|---|---|---|---|
| Receptors | ||||
| 11p15.1 | ABCC8 | Sulfonylurea receptor 1 (SUR1) | Persistent hyperinsulinemic hypoglycemia of infancy (PHHI) | Taschenberger et al. 2002; Yan et al. 2005, 2010 |
| 4q21.21 | ANTXR2 | Anthrax toxin receptor 2 | Hyaline fibromatosis syndrome | Deuquet et al. 2009, 2011 |
| Xq28 | AVPR2 | V2 vasopressin receptor 2 (V2R) | Nephrogenic diabetes insipidus (NDI) | Schwieger et al. 2008 |
| 3q13 | CASR | Calcium-sensing receptor (CasR) | Familial hypocalciuric hypercalcemia (FHH) in heterozygotes | Huang et al. 2006; Pidasheva et al. 2006; Huang and Breitwieser 2007 |
| 3q13 | CASR | Calcium-sensing receptor (CasR) | Neonatal severe hyperparathyroidism (NSHPT) in homozygotes | Huang et al. 2006; Pidasheva et al. 2006; Huang and Breitwieser 2007 |
| 5q34 | GABRA1 | γ-amino butyric acid receptor (GABAR) α1 | Juvenile myoclonic epilepsy | Gallagher et al. 2007; Bradley et al. 2008 |
| 5q34 | GABRA1 | γ-amino butyric acid receptor (GABAR) α1 | Childhood absence epilepsy (CAE) | Kang et al. 2009b |
| 5q34 | GABRG2 | γ-amino butyric acid receptor (GABAR) γ2 | Generalized epilepsy with febrile seizures plus (GEFS+) | Kang et al. 2009a |
| 4q13.2 | GNRHR | Gonadotropin-releasing hormone (GnRH) receptor | Hypogonadotropic hypogonadism | Brothers et al. 2004; Houck et al. 2014 |
| 19p13.3-13.2 | INSR | Insulin receptor | Insulin resistance syndrome (type 2 diabetes) | Kadowaki et al. 1991; Accili et al. 1992; Imamura et al. 1998; Ramos et al. 2007 |
| 19p13.3 | LDLR | Low-density lipoprotein receptor (LDLR) | Familial hypercholesterolemia | Lehrman et al. 1987; Jensen et al. 1997; Li et al. 2004 |
| 3p22-21.1 | PTH1R | Parathyroid hormone receptor (PTHR) | Parathyroid hormone resistance | Alonso et al. 2011 |
| 10q11.2 | RET | RET receptor tyrosine kinase | Hirschprung disease | Kjaer and Ibáñez 2003; Kjaer et al. 2010 |
| 3q21-24 | RHO | Rhodopsin | Retinitis pigmentosa | Illing et al. 2002; Griciuc et al. 2010a,b |
| Transporters | ||||
| 7q21.1 | ABCB4 | ATP-binding cassette, subfamily B, member 4 (MDR3) | Progressive familial intrahepatic cholestasis type 3 (PFIC3) | Delaunay et al. 2009; Gautherot et al. 2012; Gordo-Gilart et al. 2016; Park et al. 2016 |
| 2q24 | ABCB11 | Bile salt export pump (BSEP) | Progressive familial intrahepatic cholestasis type II (PFIC II) | Hayashi et al. 2005; Mochizuki et al. 2007; Wang et al. 2008 |
| 16p13.1 | ABCC1 | P-glycoprotein | Implications for cancer treatment | Loo and Clarke 1998 |
| 10q24 | ABCC2 | Multidrug resistance protein 2 (MRP2) | Dubin–Johnson syndrome | Hashimoto et al. 2002; Keitel et al. 2003 |
| 4q22 | ABCG2 | Breast cancer resistance protein | Implications for cancer treatment | Nakagawa et al. 2008, 2009; Furukawa et al. 2009; Sugiyama et al. 2011 |
| 13q14.3 | ATP7B | Copper transporting P-type ATPase | Wilson disease | Harada et al. 2001; Huster et al. 2003; de Bie et al. 2007; van der Velden et al. 2010 |
| 1p36 | ATP13A2 | ATPase type 13A2 cation transporter | Kufor–Rakeb syndrome (KRS)/Parkinson's disease 9 | Ugolino et al. 2011 |
| Channels | ||||
| 12q12-13 | AQP2 | Aquaporin 2 | Nephrogenic diabetes insipidus (NDI) | Tamarappoo et al. 1999; Hirano et al. 2003 |
| 7q31.2 | CFTR | Cystic fibrosis transmembrane conductance regulator (CFTR) | Cystic fibrosis (CF) | Jensen et al. 1995; Ward et al. 1995 |
| 7q36.1 | KCNH2 | Human ether-a-go-go-related gene (HERG) voltage-gated potassium channel | Congenital long QT syndrome | Zhou et al. 1998, 1999; Furutani et al. 1999; Gong et al. 2005; Walker et al. 2010 |
| 11q24.3 | KCNJ1 | Renal outer medullary potassium channel (ROMK) | Bartter syndrome | O'Donnell et al. 2017a,b |
| 11p15.5-15.4 | KCNQ1 | Potassium voltage-gated channel subfamily Q member 1 | Long QT syndrome | Gouas et al. 2004; Dahimene et al. 2006; Peroz et al. 2009; Huang et al. 2018 |
| 4q21.1 | PKD2 | Polycystin 2 | Autosomal dominant polycystic kidney disease type 2 (ADPKD) | Liang et al. 2008; Gao et al. 2010 |
| 3p22.2 | SCN5A | Cardiac sodium channel (SCN5A) | Brugada syndrome | Baroudi et al. 2002, 2004; Lin et al. 2008; Shao et al. 2009; Clatot et al. 2012 |
| 17q21-22 | SLC4A1 | Kidney chloride/bicarbonate anion exchanger (kAE1) | Autosomal recessive renal tubular acidosis | Kittanakom et al. 2004; Patterson and Reithmeier 2010; Chu et al. 2014 |
| Secreted/other | ||||
| 2p24-23 | APOB | Apolipoprotein B (Apo B) | A-β-lipoproteinemia | Yeung et al. 1996; Liao et al. 1998; Liang et al. 2003; Burnett et al. 2007 |
| 20p13 | AVP | Vasopressin precursor protein | Autosomal-dominant neurohypophyseal diabetes insipidus (ADNDI) | Ito and Jameson 1997; Nijenhuis et al. 1999; Friberg et al. 2004 |
| 11q13 | BSCL2 | Seipin | Silver's syndrome/distal hereditary motor neuropathy type V | Ito and Suzuki 2007 |
| 17q21.33 | COL1A1 | Type I procollagen pro-α1 chain | Osteogenesis imperfecta | Lamandé et al. 1995; Fitzgerald et al. 1999; Ishida et al. 2009 |
| 2p13.3 | DYSF | Dysferlin | Limb girdle muscular dystrophy type 2B/Miyoshi myopathy (LBMD2B/MM) | Fujita et al. 2007 |
| 19p13.3 | Elane | Neutrophil elastase | Severe congenital neutropenia (SCN) | Kollner et al. 2006; Grenda et al. 2007; Xia and Link 2008; Nanua et al. 2011; Nustede et al. 2016 |
| 2p12 | EIF2AK3 | Eukaryotic translation initiation factor 2α kinase 3 (PERK) | Walcott–Rallison syndrome (WRS) | Gupta et al. 2010 |
| 15q21.1 | FBN1 | Fibrillin 1 | Marfan syndrome | Whiteman and Handford 2003; Chao et al. 2010 |
| 4q28 | FGB | Fibrinogen | Hereditary hypofibrinogenemia | Xia and Redman 1999, 2001; Brennan et al. 2000; Kruse et al. 2006b |
| 1q21 | GBA | Glucocerebrosidase (GC) | Gaucher disease | Ron and Horowitz 2005; Mu et al. 2008; Bendikov-Bar et al. 2011 |
| Xq13.1 | GJB1 | Connexin 32 (Cx32) | X-linked Charcot–Marie–Tooth disease (CMTX) | VanSlyke et al. 2000; Kleopa et al. 2002; Sakaguchi et al. 2011 |
| Xq22 | GLA | α-Galactosidase A (α-Gal A) | Fabry disease | Yam et al. 2006; Hamanaka et al. 2008 |
| 15q24.1 | HEXA | β-Hexosaminidase α subunit | Tay-Sachs | Lau and Neufeld 1989; Maegawa et al. 2007; Mu et al. 2008 |
| 6p21.3 | HFE | HFE protein | Hereditary hemochromatosis | Waheed et al. 1997 |
| 11p15.5 | INS | Proinsulin | Neonatal diabetes (type 1) | Allen et al. 2004; Hartley et al. 2010; Park et al. 2010 |
| 17q23.1 | MPO | Myeloperoxidase (MPO) | Hereditary myeloperoxidase deficiency | DeLeo et al. 1998 |
| 18q11-12 | NPC1 | Niemann–Pick type C1 (NPC1), sterol transporting protein | Niemann–Pick type C disease | Gelsthorpe et al. 2008; Wang et al. 2011 |
| 19q13.1 | NPHS1 | Nephrin mutations unfold and ER stress | Congenital nephrotic syndrome of the Finnish type | Liu et al. 2001, 2004; Drozdova et al. 2013 |
| 1q25.2 | NPHS2 | Podocin mutations unfold and ER stress | Focal segmental glomeulonecrosis (FCGS) | Roselli et al. 2004; Fan et al. 2009; Serrano-Perez et al. 2018 |
| Xq22 | PLP1 | Proteolipid protein (PLP) DM20 (splice isoform) | Pelizaeus–Merzbacher disease (PMD)/spastic paraplegia type 2 (SPG-2) | Gow et al. 1998; Krämer-Albers et al. 2006 |
| 17p12 | PMP22 | Peripheral myelin protein 22 (PMP22) | Charcot–Marie–Tooth disease type 1A | Ryan et al. 2002; Fortun et al. 2005, 2007 |
| 2q13-14 | PROC | Protein C | Protein C deficiency | Katsumi et al. 1996, 1998; Tokunaga et al. 1996; Nishio et al. 2008 |
| 3q11.2 | PROS1 | Protein S | Protein S deficiency | Tsuda et al. 2006 |
| 14q32.1 | SERPINA1 | α-1-Antitrypsin (AT) | α-1-Aantitrypsin deficiency | Qu et al. 1996; Teckman et al. 2001 |
| Xq22.2 | SERPINA7 | Thyroxine-binding globulin (TBG) | Thyroxine-binding globulin deficiency | Miura et al. 1994a,b; Soheilipour et al. 2016 |
| 1q23-25.1 | SERPINC1 | Antithrombin | Type I antithrombin deficiency | Tokunaga et al. 1997, 2003 |
| 17p13 | SERPINF2 | α-2-Antiplasmin (A2AP) | α-2-Plasmin inhibitor deficiency | Chung et al. 2000; Nishio et al. 2008 |
| 11q13.5 | SERPINH1 | Heat shock protein 47 (Hsp47) | Osteogenesis imperfecta | Christiansen et al. 2010 |
| 3q26.1 | SERPINI1 | Neuroserpin | Familial encephalopathy with neuroserpin inclusion bodies (FENIB) | Kroeger et al. 2009; Ying et al. 2011 |
| 8q21 | SFTPC | Surfactant protein C (SP-C) | Interstitial lung disease | Dong et al. 2008; Wang et al. 2009 |
| 9q34 | TOR1A | Torsin A | Early onset torsion dystonia | Giles et al. 2008; Li et al. 2014; Zacchi et al. 2014, 2017 |
| 18q12.1 | TTR | Transthyretin (TTR) | Familial amyloid polyneuropathy | Sekijima et al. 2005; Sato et al. 2007; Susuki et al. 2009 |
| 8q24.22 | TG | Thyroglobulin | Congenital hypothyroid goiter | Guaraldi et al. 1991; Medeiros-Neto et al. 1996; Hishinuma et al. 1999; Kim et al. 2008 |
| 11q14-21 | TYR | Tyrosinase (TYR) | Oculocutaneous albinism | Halaban et al. 2000; Svedine et al. 2004; Watabe et al. 2004; Ballar et al. 2011 |
| 16p12.3 | UMOD | Uromodulin | Medullary cyst kid disease/familial juvenile hyperuricemic nephropathy | Rampoldi et al. 2003; Bernascone et al. 2006; Williams et al. 2009; Schaeffer et al. 2017 |
| 12p13.3 | VWF | von Willebrand factor (vWF) | von Willebrand's disease type IIA | Lyons et al. 1992; Allen et al. 2000, 2001; Bodó et al. 2001 |
The manifestation of a protein conformational diseases depends on how a mutation alters the structure and function of the gene product. Loss-of-function mutations that lead to conformational diseases usually result in protein misfolding and/or defects in multiprotein complex assembly. For proteins that enter the ER, the most common outcome is ERAD.
A mutant form of the cystic fibrosis transmembrane conductance regulator (CFTR) was the first disease-causing/loss-of-function protein linked to ERAD (Cheng et al. 1990; Jensen et al. 1995; Ward et al. 1995). CFTR is a member of the ATP-binding cassette (ABC) family, which are made-up of two membrane-spanning domains, each with six TMHs and two cytoplasmic nucleotide-binding domains (NBDs). CFTR is unique among ABC transporters in that it is a chloride channel and contains a regulatory domain between NBD1 and MSD2. The topological complexity of CFTR combined with extensive co- and posttranslational folding result in a substantial amount of the wild-type protein and essentially all of the F508del disease-causing mutant allele being targeted for ERAD (Jensen et al. 1995; Ward et al. 1995; Kleizen et al. 2005; Thibodeau et al. 2005, 2010; Du and Lukacs 2009; Khushoo et al. 2011). In addition to the common F508del variant, ∼120 other disease-causing mutations have been identified in CFTR (Sosnay et al. 2013), many of which also affect protein folding (Veit et al. 2016). F508del CFTR is recognized by Hsp70, Hsp40, small HSPs, and PPIs, and is then polyubiquitinated and delivered to the proteasome (Meacham et al. 1999, 2001; Youker et al. 2004; Grove et al. 2011; Hutt et al. 2012).
Cystic fibrosis patients are faced with pathologies in multiple organ systems, including the respiratory and gastrointestinal tracts and the reproductive and endocrine systems, which is caused by increased viscosity of lumenal secretions (Rowe et al. 2005; Cutting 2015). In the 29 years since the CFTR gene was identified (Kerem et al. 1989; Riordan et al. 1989; Rommens et al. 1989), small molecules that facilitate F508del CFTR biogenesis have been actively sought. Early work focused on nonspecific chemical chaperones that enhance the cellular folding environment (Brown et al. 1996; Sato et al. 1996; Howard et al. 2003). In addition, simply subjecting cells to low-temperature increased F508del CFTR maturation and cell surface activity, presumably by decreasing kinetic barriers during folding (Denning et al. 1992). More recently, combinations of a folding “corrector” and a channel potentiator significantly increased F508del CFTR surface expression and activity (Van Goor et al. 2009, 2011). This drug combination was approved by the Food and Drug Administration (FDA) in 2015, and even with some positive outcomes the effects in patients have shown limited efficacy and give rise to unwanted side effects (Horsley and Barry 2017). Improved corrector–potentiator combinations will undoubtedly be forthcoming. In theory, the addition of an ERAD inhibitor might further improve the efficacy of these therapeutics (Chung et al. 2016).
Under some circumstances, misfolded conformers may overwhelm or even escape the detection of quality-control pathways, resulting in a toxic gain-of-function phenotype. For example, transthyretin (TTR) is secreted into the vitreous humor by retinal pigment epithelial cells, the cerebrospinal fluid by the choroid plexus, and the bloodstream by the liver (Hamilton and Benson 2001). More than 135 mutations in TTR have been identified, which vary in their amyloidogenicity and, in turn, the age of onset and disease presentation (Connors et al. 2003; Hammarström et al. 2003; Sekijima et al. 2005). Some mutant TTR variants escape ERAD and form amyloids in the heart and peripheral nerves, which presents as TTR cardiomyopathy, familial amyloid polyneuropathy, or central nervous system amyloidosis. Mutations that destabilize the TTR tetramer are more prone to oligomerize (Hammarström et al. 2002). Interestingly, the D18G variant is one of the most highly aggregation-prone TTR forms in vitro, but gives rise to a mild disease phenotype, likely because of the protective effect of ERAD (Hammarström et al. 2003; Sekijima et al. 2003, 2005). Another TTR mutant, A25T is secreted more efficiently, and recent data indicate that an ER lumenal Hsp40 chaperone, ERdj3, escorts the mutant protein through the secretory pathway, potentially suppressing ERAD and extracellular aggregation (Genereux et al. 2015). Although a drug that stabilizes the TTR tetramer shows efficacy for some mutant alleles (Klabunde et al. 2000; Bulawa et al. 2012; Rappley et al. 2014), these data highlight a more general role for chaperones as therapeutic targets (Lindquist and Kelly 2011; Brandvold and Morimoto 2015; Li et al. 2016).
CONCLUDING REMARKS
In this review, we introduced ER quality-control pathways, highlighted the molecular chaperones and other factors required for protein folding in the ER as well as ERAD, and provided a resource list in which disease-associated proteins are destroyed by ERAD (Table 1). Given the magnitude of coding variations in human genome databases (Telenti et al. 2016)—and the astronomical number of possible mutations that might compromise protein folding—it is possible that any protein that enters the ER might ultimately be included in this list.
One category of disease-causing substrates we predict will increasingly be linked to ERAD is ion channels. Ion channels present cells with multiple folding challenges as they are large, show complex topologies, form hetero- or homo-oligomers, and/or contain charged residues within their membrane-spanning domains, which allows for ion permeation or voltage sensing (Green 1999). For example, the epithelial sodium channel, ENaC forms a heterotrimer of homologous α, β, and γ subunits (Buck and Brodsky 2018). In some cells, the β and γ subunits are constitutively produced and are targeted for ERAD, whereas the α subunit is only synthesized after aldosterone secretion. When expressed individually, each subunit is degraded in an Hsp40-dependent manner (Buck et al. 2010). In contrast, only the orphaned α subunit is degraded in an Lhs1/Grp170-dependent manner (Buck et al. 2013). Mutations in ENaC that alter ERAD and lead to salt-wasting have not yet been identified, but based on these complex regulatory and folding pathways—and because the assembled channel can still be degraded by ERAD (Staub et al. 1997; Valentijn et al. 1998; Malik et al. 2001)—we anticipate that syndrome-associated ENaC polymorphisms will be identified. Similarly, loss-of-function mutations in the thiazide-sensitive NaCl cotransporter, NCC, cause the salt-wasting disorder Gitelman syndrome (Simon et al. 1996). One Gitelman-causing NCC mutation truncates the cytoplasmic tail of the protein, which binds more readily to Hsp70 and Hsp40 (Donnelly et al. 2013). Future work will be required to determine whether this mutant and any of the other ∼40 disease-causing mutants is more efficiently targeted for ERAD, as these data suggest.
Although drugs have been identified to treat both loss-of-function (e.g., CFTR) and gain-of-function (e.g., TTR) diseases, these examples represent the exception rather than the rule. Thus, another important future goal is to find chemical—or better, pharmacological—chaperones that facilitate the folding of mutant proteins and prevent ERAD. Proteostasis modulators such as 4-phenylbutyric acid, celastrol, verapamil, and suberoylanilide hydroxamic acid (SAHA) have been used with some success (see Kelly 2018), and in one study the combined administration of SAHA and an ERAD inhibitor led to synergistic, corrective effects on a mutant ion channel (Han et al. 2015). One hopes that other examples of this approach will soon follow.
ACKNOWLEDGMENTS
We thank Jen Goeckeler-Fried for critical reading of the manuscript and helpful suggestions. Work on ERAD in the Brodsky laboratory is supported by Grants GM75061 and DK79307 to J.L.B and by Grant DK101584 to C.J.G. from the National Institutes of Health.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Accili D, Kadowaki T, Kadowaki H, Mosthaf L, Ullrich A, Taylor SI. 1992. Immunoglobulin heavy chain-binding protein binds to misfolded mutant insulin receptors with mutations in the extracellular domain. J Biol Chem 267: 586–590. [PubMed] [Google Scholar]
- Aebi M, Bernasconi R, Clerc S, Molinari M. 2010. N-glycan structures: Recognition and processing in the ER. Trends Biochem Sci 35: 74–82. 10.1016/j.tibs.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Allen S, Abuzenadah AM, Hinks J, Blagg JL, Gursel T, Ingerslev J, Goodeve AC, Peake IR, Daly ME. 2000. A novel von Willebrand disease–causing mutation (Arg273Trp) in the von Willebrand factor propeptide that results in defective multimerization and secretion. Blood 96: 560–568. [PubMed] [Google Scholar]
- Allen S, Goodeve AC, Peake IR, Daly ME. 2001. Endoplasmic reticulum retention and prolonged association of a von Willebrand's disease-causing von Willebrand factor variant with ERp57 and calnexin. Biochem Biophys Res Commun 280: 448–453. 10.1006/bbrc.2000.4139 [DOI] [PubMed] [Google Scholar]
- Allen JR, Nguyen LX, Sargent KEG, Lipson KL, Hackett A, Urano F. 2004. High ER stress in β-cells stimulates intracellular degradation of misfolded insulin. Biochem Biophys Res Commun 324: 166–170. 10.1016/j.bbrc.2004.09.035 [DOI] [PubMed] [Google Scholar]
- Alonso V, Ardura JA, Wang B, Sneddon WB, Friedman PA. 2011. A naturally occurring isoform inhibits parathyroid hormone receptor trafficking and signaling. J Bone Miner Res 26: 143–155. 10.1002/jbmr.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvira S, Cuéllar J, Röhl A, Yamamoto S, Itoh H, Alfonso C, Rivas G, Buchner J, Valpuesta JM. 2014. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat Commun 5: 5484 10.1038/ncomms6484 [DOI] [PubMed] [Google Scholar]
- Andréasson C, Rampelt H, Fiaux J, Druffel-Augustin S, Bukau B. 2010. The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110. J Biol Chem 285: 12445–12453. 10.1074/jbc.M109.096735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. 1973. Principles that govern the folding of protein chains. Science 181: 223–230. 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- Ansa-Addo EA, Thaxton J, Hong F, Wu BX, Zhang Y, Fugle CW, Metelli A, Riesenberg B, Williams K, Gewirth DT, et al. 2016. Clients and oncogenic roles of molecular chaperone gp96/grp94. Curr Top Med Chem 16: 2765–2778. 10.2174/1568026616666160413141613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Nagata K. 2011. Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol 3: a007526 10.1101/cshperspect.a007526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T, Michaelis S, Schuldiner M. 2016. The protease Ste24 clears clogged translocons. Cell 164: 103–114. 10.1016/j.cell.2015.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci D, Lemberg MK. 2018. Membrane protein dislocation by the rhomboid pseudoprotease Dfm1: No pore needed? Mol Cell 69: 161–162. 10.1016/j.molcel.2017.12.031 [DOI] [PubMed] [Google Scholar]
- Baldridge RD, Rapoport TA. 2016. Autoubiquitination of the Hrd1 ligase triggers protein retrotranslocation in ERAD. Cell 166: 394–407. 10.1016/j.cell.2016.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballar P, Pabuccuoglu A, Kose FA. 2011. Different p97/VCP complexes function in retrotranslocation step of mammalian Er-associated degradation (ERAD). Int J Biochem Cell Biol 43: 613–621. 10.1016/j.biocel.2010.12.021 [DOI] [PubMed] [Google Scholar]
- Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. 2018. Structure and function of the 26S proteasome. Annu Rev Biochem 87: 697–724. 10.1146/annurev-biochem-062917-011931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudi G, Acharfi S, Larouche C, Chahine M. 2002. Expression and intracellular localization of an SCN5A double mutant R1232W/T1620M implicated in Brugada syndrome. Circ Res 90: E11–E16. 10.1161/hh0102.102977 [DOI] [PubMed] [Google Scholar]
- Baroudi G, Napolitano C, Priori SG, Del Bufalo A, Chahine M. 2004. Loss of function associated with novel mutations of the SCN5A gene in patients with Brugada syndrome. Can J Cardiol 20: 425–430. [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. 2001. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12: 4114–4128. 10.1091/mbc.12.12.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Hendershot LM. 2014. The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s. J Biol Chem 289: 2899–2907. 10.1074/jbc.M113.507491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Mann MJ, Scruggs FL, Feige MJ, Hendershot LM. 2016. Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol Cell 63: 739–752. 10.1016/j.molcel.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendikov-Bar I, Ron I, Filocamo M, Horowitz M. 2011. Characterization of the ERAD process of the L444P mutant glucocerebrosidase variant. Blood Cells Mol Dis 46: 4–10. [DOI] [PubMed] [Google Scholar]
- Benyair R, Ogen-Shtern N, Lederkremer GZ. 2015. Glycan regulation of ER-associated degradation through compartmentalization. Semin Cell Dev Biol 41: 99–109. 10.1016/j.semcdb.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Bernascone I, Vavassori S, Di Pentima A, Santambrogio S, Lamorte G, Amoroso A, Scolari F, Ghiggeri GM, Casari G, Polishchuk R, et al. 2006. Defective intracellular trafficking of uromodulin mutant isoforms. Traffic 7: 1567–1579. 10.1111/j.1600-0854.2006.00481.x [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. 2010. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol 188: 223–235. 10.1083/jcb.200910042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner N, Reutter KR, Wolf DH. 2018. Protein quality control of the endoplasmic reticulum and ubiquitin–proteasome-triggered degradation of aberrant proteins: Yeast pioneers the path. Annu Rev Biochem 87: 751–782. 10.1146/annurev-biochem-062917-012749 [DOI] [PubMed] [Google Scholar]
- Bodnar NO, Rapoport TA. 2017. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169: 722–735.e9. 10.1016/j.cell.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodó I, Katsumi A, Tuley EA, Eikenboom JCJ, Dong Z, Sadler JE. 2001. Type 1 von Willebrand disease mutation Cys1149Arg causes intracellular retention and degradation of heterodimers: A possible general mechanism for dominant mutations of oligomeric proteins. Blood 98: 2973–2979. 10.1182/blood.V98.10.2973 [DOI] [PubMed] [Google Scholar]
- Bole DG, Hendershot LM, Kearney JF. 1986. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol 102: 1558–1566. 10.1083/jcb.102.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. 2011. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem 80: 71–99. 10.1146/annurev-biochem-062209-093836 [DOI] [PubMed] [Google Scholar]
- Bracher A, Verghese J. 2015. GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: Nucleotide exchange factors for Hsp70 molecular chaperones. Subcell Biochem 78: 1–33. 10.1007/978-3-319-11731-7_1 [DOI] [PubMed] [Google Scholar]
- Bradley CA, Taghibiglou C, Collingridge GL, Wang YT. 2008. Mechanisms involved in the reduction of GABAA receptor α1-subunit expression caused by the epilepsy mutation A322D in the trafficking-competent receptor. J Biol Chem 283: 22043–22050. 10.1074/jbc.M801708200 [DOI] [PubMed] [Google Scholar]
- Brandvold KR, Morimoto RI. 2015. The chemical biology of molecular chaperones—Implications for modulation of proteostasis. J Mol Biol 427: 2931–2947. 10.1016/j.jmb.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselmann E, Chaney JL, Clark PL. 2013. Folding the proteome. Trends Biochem Sci 38: 337–344. 10.1016/j.tibs.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan SO, Wyatt J, Medicina D, Callea F, George PM. 2000. Fibrinogen brescia: Hepatic endoplasmic reticulum storage and hypofibrinogenemia because of a γ284 Gly→Arg mutation. Am J Pathol 157: 189–196. 10.1016/S0002-9440(10)64530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. 2007. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation). Biochem J 404: 353–363. 10.1042/BJ20061890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci 92: 9643–9646. 10.1073/pnas.92.21.9643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. 1999. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem 274: 3453–3460. 10.1074/jbc.274.6.3453 [DOI] [PubMed] [Google Scholar]
- Brothers SP, Cornea A, Janovick JA, Conn PM. 2004. Human loss-of-function gonadotropin-releasing hormone receptor mutants retain wild-type receptors in the endoplasmic reticulum: molecular basis of the dominant-negative effect. Mol Endocrinol 18: 1787–1797. 10.1210/me.2004-0091 [DOI] [PubMed] [Google Scholar]
- Brown CR, Hong-Brown LQ, Biwersi J, Verkman AS, Welch WJ. 1996. Chemical chaperones correct the mutant phenotype of the ΔF508 cystic fibrosis transmembrane conductance regulator protein. Cell Stress Chaperones 1: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Schindelin H, Hänzelmann P. 2015. Control of p97 function by cofactor binding. FEBS Lett 589: 2578–2589. 10.1016/j.febslet.2015.08.028 [DOI] [PubMed] [Google Scholar]
- Buck TM, Brodsky JL. 2018. Epithelial sodium channel biogenesis and quality control in the early secretory pathway. Curr Opin Nephrol Hypertens 27: 364–372. 10.1097/MNH.0000000000000438 [DOI] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. 2010. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell 21: 1047–1058. 10.1091/mbc.e09-11-0944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Plavchak L, Roy A, Donnelly BF, Kashlan OB, Kleyman TR, Subramanya AR, Brodsky JL. 2013. The Lhs1/GRP170 chaperones facilitate the endoplasmic reticulum-associated degradation of the epithelial sodium channel. J Biol Chem 288: 18366–18380. 10.1074/jbc.M113.469882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa CE, Connelly S, DeVit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, et al. 2012. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci 109: 9629–9634. 10.1073/pnas.1121005109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JR, Zhong S, Jiang ZG, Hooper AJ, Fisher EA, McLeod RS, Zhao Y, Barrett PHR, Hegele RA, van Bockxmeer FM, et al. 2007. Missense mutations in APOB within the βα1 domain of human APOB-100 result in impaired secretion of ApoB and ApoB-containing lipoproteins in familial hypobetalipoproteinemia. J Biol Chem 282: 24270–24283. 10.1074/jbc.M702442200 [DOI] [PubMed] [Google Scholar]
- Caramelo JJ, Parodi AJ. 2007. How sugars convey information on protein conformation in the endoplasmic reticulum. Semin Cell Dev Biol 18: 732–742. 10.1016/j.semcdb.2007.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Hampton RY. 2010. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem 285: 5146–5156. 10.1074/jbc.M109.067876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373. 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Chao SC, Chen JS, Tsai CH, Lin JM, Lin YJ, Sun HS. 2010. Novel exon nucleotide substitution at the splice junction causes a neonatal Marfan syndrome. Clin Genet 77: 453–463. 10.1111/j.1399-0004.2009.01337.x [DOI] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. 1990. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63: 827–834. 10.1016/0092-8674(90)90148-8 [DOI] [PubMed] [Google Scholar]
- Cherepanova N, Shrimal S, Gilmore R. 2016. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr Opin Cell Biol 41: 57–65. 10.1016/j.ceb.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen HE, Schwarze U, Pyott SM, AlSwaid A, Al Balwi M, Alrasheed S, Pepin MG, Weis MA, Eyre DR, Byers PH. 2010. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet 86: 389–398. 10.1016/j.ajhg.2010.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. 2008. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 10: 272–282. 10.1038/ncb1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, King J, Berrini M, Rumley AC, Apaja PM, Lukacs GL, Alexander RT, Cordat E. 2014. Degradation mechanism of a Golgi-retained distal renal tubular acidosis mutant of the kidney anion exchanger 1 in renal cells. Am J Physiol Cell Physiol 307: C296–C307. 10.1152/ajpcell.00310.2013 [DOI] [PubMed] [Google Scholar]
- Chung DH, Ohashi K, Watanabe M, Miyasaka N, Hirosawa S. 2000. Mannose trimming targets mutant α2-plasmin inhibitor for degradation by the proteasome. J Biol Chem 275: 4981–4987. 10.1074/jbc.275.7.4981 [DOI] [PubMed] [Google Scholar]
- Chung WJ, Goeckeler-Fried JL, Havasi V, Chiang A, Rowe SM, Plyler ZE, Hong JS, Mazur M, Piazza GA, Keeton AB, et al. 2016. Increasing the endoplasmic reticulum pool of the F508del allele of the cystic fibrosis transmembrane conductance regulator leads to greater folding correction by small molecule therapeutics. PLoS ONE 11: e0163615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Kundrat L, Ploegh HL. 2012. Protein quality control in the ER: Balancing the ubiquitin checkbook. Trends Cell Biol 22: 22–32. 10.1016/j.tcb.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatot J, Ziyadeh-Isleem A, Maugenre S, Denjoy I, Liu H, Dilanian G, Hatem SN, Deschenes I, Coulombe A, Guicheney P, et al. 2012. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Nav1.5 α-subunits. Cardiovasc Res 96: 53–63. 10.1093/cvr/cvs211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. 2009. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol 184: 159–172. 10.1083/jcb.200809198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors LH, Lim A, Prokaeva T, Roskens VA, Costello CE. 2003. Tabulation of human transthyretin (TTR) variants, 2003. Amyloid 10: 160–184. 10.3109/13506120308998998 [DOI] [PubMed] [Google Scholar]
- Cutting GR. 2015. Cystic fibrosis genetics: From molecular understanding to clinical application. Nat Rev Genet 16: 45–56. 10.1038/nrg3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahimene S, Alcolea S, Naud P, Jourdon P, Escande D, Brasseur R, Thomas A, Baro I, Merot J. 2006. The N-terminal juxtamembranous domain of KCNQ1 is critical for channel surface expression: Implications in the Romano-Ward LQT1 syndrome. Circ Res 99: 1076–1083. 10.1161/01.RES.0000250262.12219.95 [DOI] [PubMed] [Google Scholar]
- de Bie P, van de Sluis B, Burstein E, van de Berghe PVE, Muller P, Berger R, Gitlin JD, Wijmenga C, Klomp LWJ. 2007. Distinct Wilson's disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology 133: 1316–1326. 10.1053/j.gastro.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay JL, Durand-Schneider AM, Delautier D, Rada A, Gautherot J, Jacquemin E, Aït-Slimane T, Maurice M. 2009. A missense mutation in ABCB4 gene involved in progressive familial intrahepatic cholestasis type 3 leads to a folding defect that can be rescued by low temperature. Hepatology 49: 1218–1227. 10.1002/hep.22775 [DOI] [PubMed] [Google Scholar]
- DeLeo FR, Goedken M, McCormick SJ, Nauseef WM. 1998. A novel form of hereditary myeloperoxidase deficiency linked to endoplasmic reticulum/proteasome degradation. J Clin Invest 101: 2900–2909. 10.1172/JCI2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359. 10.1016/j.cell.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. 1992. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 358: 761–764. 10.1038/358761a0 [DOI] [PubMed] [Google Scholar]
- Deprez P, Gautschi M, Helenius A. 2005. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell 19: 183–195. 10.1016/j.molcel.2005.05.029 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Deuquet J, Abrami L, Difeo A, Ramirez MC, Martignetti JA, van der Goot FG. 2009. Systemic hyalinosis mutations in the CMG2 ectodomain leading to loss of function through retention in the endoplasmic reticulum. Hum Mutat 30: 583–589. 10.1002/humu.20872 [DOI] [PubMed] [Google Scholar]
- Deuquet J, Lausch E, Guex N, Abrami L, Salvi S, Lakkaraju A, Ramirez MCM, Martignetti JA, Rokicki D, Bonafe L, et al. 2011. Hyaline fibromatosis syndrome inducing mutations in the ectodomain of anthrax toxin receptor 2 can be rescued by proteasome inhibitors. EMBO Mol Med 3: 208–221. 10.1002/emmm.201100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. 2008. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell 19: 2620–2630. 10.1091/mbc.e07-07-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly BF, Needham PG, Snyder AC, Roy A, Khadem S, Brodsky JL, Subramanya AR. 2013. Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J Biol Chem 288: 13124–13135. 10.1074/jbc.M113.455394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdova T, Papillon J, Cybulsky AV. 2013. Nephrin missense mutations: Induction of endoplasmic reticulum stress and cell surface rescue by reduction in chaperone interactions. Physiol Rep 1: e00086 10.1002/phy2.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Lukacs GL. 2009. Cooperative assembly and misfolding of CFTR domains in vivo. Mol Biol Cell 20: 1903–1915. 10.1091/mbc.e08-09-0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Zhang H, Ding J, Liu S, Miao J, Xing Y, Yu Z, Guan N. 2009. R168H and V165X mutant podocin might induce different degrees of podocyte injury via different molecular mechanisms. Genes Cells 14: 1079–1090. 10.1111/j.1365-2443.2009.01336.x [DOI] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM. 2011. Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol 23: 167–175. 10.1016/j.ceb.2010.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM. 2013. Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol Cell 51: 297–309. 10.1016/j.molcel.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513. 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher EA, Ginsberg HN. 2002. Complexity in the secretory pathway: The assembly and secretion of apolipoprotein B-containing lipoproteins. J Biol Chem 277: 17377–17380. 10.1074/jbc.R100068200 [DOI] [PubMed] [Google Scholar]
- Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. 1997. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem 272: 20427–20434. 10.1074/jbc.272.33.20427 [DOI] [PubMed] [Google Scholar]
- Fitzgerald J, Lamandé SR, Bateman JF. 1999. Proteasomal degradation of unassembled mutant type I collagen pro-α1(I) chains. J Biol Chem 274: 27392–27398. 10.1074/jbc.274.39.27392 [DOI] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK. 2012. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell 47: 558–569. 10.1016/j.molcel.2012.06.008 [DOI] [PubMed] [Google Scholar]
- Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. 2013. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife 2: e00953 10.7554/eLife.00953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. 2014. Quality control of inner nuclear membrane proteins by the Asi complex. Science 346: 751–755. 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- Fortun J, Li J, Go J, Fenstermaker A, Fletcher BS, Notterpek L. 2005. Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. J Neurochem 92: 1531–1541. 10.1111/j.1471-4159.2004.02987.x [DOI] [PubMed] [Google Scholar]
- Fortun J, Verrier JD, Go JC, Madorsky I, Dunn WA, Notterpek L. 2007. The formation of peripheral myelin protein 22 aggregates is hindered by the enhancement of autophagy and expression of cytoplasmic chaperones. Neurobiol Dis 25: 252–265. 10.1016/j.nbd.2006.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Cuozzo JW, Kaiser CA. 2000. Pathways for protein disulphide bond formation. Trends Cell Biol 10: 203–210. 10.1016/S0962-8924(00)01745-1 [DOI] [PubMed] [Google Scholar]
- Freilich R, Arhar T, Abrams JL, Gestwicki JE. 2018. Protein–protein interactions in the molecular chaperone network. Acc Chem Res 51: 940–949. 10.1021/acs.accounts.8b00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg MA, Spiess M, Rutishauser J. 2004. Degradation of wild-type vasopressin precursor and pathogenic mutants by the proteasome. J Biol Chem 279: 19441–19447. 10.1074/jbc.M310249200 [DOI] [PubMed] [Google Scholar]
- Frydman J. 2001. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem 70: 603–647. 10.1146/annurev.biochem.70.1.603 [DOI] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. 2007. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: Ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum Mol Genet 16: 618–629. 10.1093/hmg/ddm002 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Wakabayashi K, Tamura A, Nakagawa H, Morishima Y, Osawa Y, Ishikawa T. 2009. Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm Res 26: 469–479. 10.1007/s11095-008-9752-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Trudeau MC, Hagiwara N, Seki A, Gong Q, Zhou Z, Imamura S-i, Nagashima H, Kasanuki H, Takao A, et al. 1999. Novel mechanism associated with an inherited cardiac arrhythmia: Defective protein trafficking by the mutant HERG (G601S) potassium channel. Circulation 99: 2290–2294. 10.1161/01.CIR.99.17.2290 [DOI] [PubMed] [Google Scholar]
- Gallagher MJ, Ding L, Maheshwari A, Macdonald RL. 2007. The GABAA receptor α1 subunit epilepsy mutation A322D inhibits transmembrane helix formation and causes proteasomal degradation. Proc Natl Acad Sci 104: 12999–13004. 10.1073/pnas.0700163104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang Y, Wegierski T, Skouloudaki K, Pütz M, Fu X, Engel C, Boehlke C, Peng H, Kuehn EW, et al. 2010. PRKCSH/80K-H, the protein mutated in polycystic liver disease, protects polycystin-2/TRPP2 against HERP-mediated degradation. Hum Mol Genet 19: 16–24. 10.1093/hmg/ddp463 [DOI] [PubMed] [Google Scholar]
- Garcia VM, Nillegoda NB, Bukau B, Morano KA. 2017. Substrate binding by the yeast Hsp110 nucleotide exchange factor and molecular chaperone Sse1 is not obligate for its biological activities. Mol Biol Cell 28: 2066–2075. 10.1091/mbc.e17-01-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol 151: 69–82. 10.1083/jcb.151.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Sato BK, Hampton RY. 2009. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem 284: 14710–14722. 10.1074/jbc.M809607200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautherot J, Durand-Schneider AM, Delautier D, Delaunay JL, Rada A, Gabillet J, Housset C, Maurice M, Aït-Slimane T. 2012. Effects of cellular, chemical, and pharmacological chaperones on the rescue of a trafficking-defective mutant of the ATP-binding cassette transporter proteins ABCB1/ABCB4. J Biol Chem 287: 5070–5078. 10.1074/jbc.M111.275438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsthorpe ME, Baumann N, Millard E, Gale SE, Langmade SJ, Schaffer JE, Ory DS. 2008. Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem 283: 8229–8236. 10.1074/jbc.M708735200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genereux JC, Qu S, Zhou M, Ryno LM, Wang S, Shoulders MD, Kaufman RJ, Lasmézas CI, Kelly JW, Wiseman RL. 2015. Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J 34: 4–19. 10.15252/embj.201488896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ. 1999. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol 10: 465–472. 10.1006/scdb.1999.0318 [DOI] [PubMed] [Google Scholar]
- Giles LM, Chen J, Li L, Chin L-S. 2008. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum Mol Genet 17: 2712–2722. 10.1093/hmg/ddn173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler JL, Petruso AP, Aguirre J, Clement CC, Chiosis G, Brodsky JL. 2008. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett 582: 2393–2396. 10.1016/j.febslet.2008.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. 1990. Regulation of the mevalonate pathway. Nature 343: 425–430. 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- Gong Q, Keeney DR, Molinari M, Zhou Z. 2005. Degradation of trafficking-defective long QT syndrome type II mutant channels by the ubiquitin-proteasome pathway. J Biol Chem 280: 19419–19425. 10.1074/jbc.M502327200 [DOI] [PubMed] [Google Scholar]
- Gordo-Gilart R, Andueza S, Hierro L, Jara P, Alvarez L. 2016. Functional rescue of trafficking-impaired ABCB4 mutants by chemical chaperones. PLoS ONE 11: e0150098 10.1371/journal.pone.0150098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothel SF, Marahiel MA. 1999. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci 55: 423–436. 10.1007/s000180050299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouas L, Bellocq C, Berthet M, Potet F, Demolombe S, Forhan A, Lescasse R, Simon F, Balkau B, Denjoy I, et al. 2004. New KCNQ1 mutations leading to haploinsufficiency in a general population; Defective trafficking of a KvLQT1 mutant. Cardiovasc Res 63: 60–68. 10.1016/j.cardiores.2004.02.011 [DOI] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Lazzarini RA. 1998. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J Cell Biol 140: 925–934. 10.1083/jcb.140.4.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green WN. 1999. Ion channel assembly: Creating structures that function. J Gen Physiol 113: 163–170. 10.1085/jgp.113.2.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR. 2011. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol 18: 1147–1152. 10.1038/nsmb.2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenda DS, Murakami M, Ghatak J, Xia J, Boxer LA, Dale D, Dinauer MC, Link DC. 2007. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood 110: 4179–4187. 10.1182/blood-2006-11-057299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Aron L, Piccoli G, Ueffing M. 2010a. Clearance of RhodopsinP23H aggregates requires the ERAD effector VCP. Biochim Biophys Acta 1803: 424–434. 10.1016/j.bbamcr.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Griciuc A, Aron L, Roux MJ, Klein R, Giangrande A, Ueffing M. 2010b. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded Rhodopsin in Drosophila. PLoS Genet 6: e1001075 10.1371/journal.pgen.1001075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove DE, Fan CY, Ren HY, Cyr DM. 2011. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRΔF508. Mol Biol Cell 22: 301–314. 10.1091/mbc.e10-09-0760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb S, Guo L, Fisher EA, Brodsky JL. 2012. Protein disulfide isomerases contribute differentially to the endoplasmic reticulum-associated degradation of apolipoprotein B and other substrates. Mol Biol Cell 23: 520–532. 10.1091/mbc.e11-08-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaraldi M, Martoni A, Tononi A, Occhiuzzi L, Caliceti U, Dall'Olio D, Laudadio P, Pannuti F. 1991. 5-Fluorouracil + folinic acid with cisplatinum and bleomycin in the treatment of advanced head and neck squamous cell carcinoma. Ann Oncol 2: 379–381. 10.1093/oxfordjournals.annonc.a057961 [DOI] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. 2012. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev 92: 537–576. 10.1152/physrev.00027.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Reutter KR, Augustine AA, Preston GM, Weiberth KF, Mackie TD, Cleveland-Rubeor HC, Bethel NP, Callenberg KM, Nakatsukasa K, et al. 2017. Transmembrane helix hydrophobicity is an energetic barrier during the retrotranslocation of integral membrane ERAD substrates. Mol Biol Cell 28: 2076–2090. 10.1091/mbc.e17-03-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, McGrath B, Cavener DR. 2010. PERK (EIF2AK3) regulates proinsulin trafficking and quality control in the secretory pathway. Diabetes 59: 1937–1947. 10.2337/db09-1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. 2001. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J Biol Chem 276: 24891–24900. 10.1074/jbc.M100633200 [DOI] [PubMed] [Google Scholar]
- Hageman J, van Waarde MA, Zylicz A, Walerych D, Kampinga HH. 2011. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem J 435: 127–142. 10.1042/BJ20101247 [DOI] [PubMed] [Google Scholar]
- Halaban R, Svedine S, Cheng E, Smicun Y, Aron R, Hebert DN. 2000. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc Natl Acad Sci 97: 5889–5894. 10.1073/pnas.97.11.5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale SJ, Lovell SC, de Keyzer J, Stirling CJ. 2010. Interactions between Kar2p and its nucleotide exchange factors Sil1p and Lhs1p are mechanistically distinct. J Biol Chem 285: 21600–21606. 10.1074/jbc.M110.111211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin L, Jung J, Michalak M. 2014. The many functions of the endoplasmic reticulum chaperones and folding enzymes. IUBMB Life 66: 318–326. 10.1002/iub.1272 [DOI] [PubMed] [Google Scholar]
- Hamanaka R, Shinohara T, Yano S, Nakamura M, Yasuda A, Yokoyama S, Fan JQ, Kawasaki K, Watanabe M, Ishii S. 2008. Rescue of mutant α-galactosidase A in the endoplasmic reticulum by 1-deoxygalactonojirimycin leads to trafficking to lysosomes. Biochim Biophys Acta 1782: 408–413. 10.1016/j.bbadis.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Hamilton JA, Benson MD. 2001. Transthyretin: A review from a structural perspective. Cell Mol Life Sci 58: 1491–1521. 10.1007/PL00000791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström P, Jiang X, Hurshman AR, Powers ET, Kelly JW. 2002. Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity. Proc Natl Acad Sci 99: 16427–16432. 10.1073/pnas.202495199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström P, Sekijima Y, White JT, Wiseman RL, Lim A, Costello CE, Altland K, Garzuly F, Budka H, Kelly JW. 2003. D18G transthyretin is monomeric, aggregation prone, and not detectable in plasma and cerebrospinal fluid: A prescription for central nervous system amyloidosis? Biochemistry 42: 6656–6663. 10.1021/bi027319b [DOI] [PubMed] [Google Scholar]
- Han S, Liu Y, Chang A. 2007. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem 282: 26140–26149. 10.1074/jbc.M701969200 [DOI] [PubMed] [Google Scholar]
- Han DY, Di XJ, Fu YL, Mu TW. 2015. Combining valosin-containing protein (VCP) inhibition and suberanilohydroxamic acid (SAHA) treatment additively enhances the folding, trafficking, and function of epilepsy-associated γ-aminobutyric acid, type A (GABAA) receptors. J Biol Chem 290: 325–337. 10.1074/jbc.M114.580324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Sakisaka S, Terada K, Kimura R, Kawaguchi T, Koga H, Kim M, Taniguchi E, Hanada S, Suganuma T, et al. 2001. A mutation of the Wilson disease protein, ATP7B, is degraded in the proteasomes and forms protein aggregates. Gastroenterology 120: 967–974. 10.1053/gast.2001.22543 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hartley T, Siva M, Lai E, Teodoro T, Zhang L, Volchuk A. 2010. Endoplasmic reticulum stress response in an INS-1 pancreatic β-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol 11: 59 10.1186/1471-2121-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Uchiumi T, Konno T, Ebihara T, Nakamura T, Wada M, Sakisaka S, Maniwa F, Amachi T, Ueda K, et al. 2002. Trafficking and functional defects by mutations of the ATP-binding domains in MRP2 in patients with Dubin–Johnson syndrome. Hepatology 36: 1236–1245. 10.1053/jhep.2002.36368 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Takada T, Suzuki H, Akita H, Sugiyama Y. 2005. Two common PFIC2 mutations are associated with the impaired membrane trafficking of BSEP/ABCB11. Hepatology 41: 916–924. 10.1002/hep.20627 [DOI] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Frankino PA, Paul JW III, Tronnes SU, Dillin A. 2018. A futile battle? Protein quality control and the stress of aging. Dev Cell 44: 139–163. 10.1016/j.devcel.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science 273: 1725–1728. 10.1126/science.273.5282.1725 [DOI] [PubMed] [Google Scholar]
- Hirano K, Zuber C, Roth J, Ziak M. 2003. The proteasome is involved in the degradation of different aquaporin-2 mutants causing nephrogenic diabetes insipidus. Am J Pathol 163: 111–120. 10.1016/S0002-9440(10)63635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama H, Hosomi A, Suzuki T. 2015. Physiological and molecular functions of the cytosolic peptide: N-glycanase. Semin Cell Dev Biol 41: 110–120. 10.1016/j.semcdb.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Hishinuma A, Takamatsu J, Ohyama Y, Yokozawa T, Kanno Y, Kuma K, Yoshida S, Matsuura N, Ieiri T. 1999. Two novel cysteine substitutions (C1263R and C1995S) of thyroglobulin cause a defect in intracellular transport of thyroglobulin in patients with congenital goiter and the variant type of adenomatous goiter. J Clin Endocrinol Metab 84: 1438–1444. [DOI] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schütz A, Heinemann U, Sommer T, Jarosch E. 2009. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell 36: 782–793. 10.1016/j.molcel.2009.10.015 [DOI] [PubMed] [Google Scholar]
- Horsley A, Barry P. 2017. Orkambi in patients with severe disease—Bumps in the road to CFTR modulation. J Cyst Fibros 16: 311–312. 10.1016/j.jcf.2017.04.008 [DOI] [PubMed] [Google Scholar]
- Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, Janovick JA, Conn PM, Cyr DM. 2014. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell 54: 166–179. 10.1016/j.molcel.2014.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Fischer H, Roux J, Santos BC, Gullans SR, Yancey PH, Welch WJ. 2003. Mammalian osmolytes and S-nitrosoglutathione promote ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) protein maturation and function. J Biol Chem 278: 35159–35167. 10.1074/jbc.M301924200 [DOI] [PubMed] [Google Scholar]
- Hrizo SL, Gusarova V, Habiel DM, Goeckeler JL, Fisher EA, Brodsky JL. 2007. The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic reticulum-associated degradation (ERAD). J Biol Chem 282: 32665–32675. 10.1074/jbc.M705216200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Breitwieser GE. 2007. Rescue of calcium-sensing receptor mutants by allosteric modulators reveals a conformational checkpoint in receptor biogenesis. J Biol Chem 282: 9517–9525. 10.1074/jbc.M609045200 [DOI] [PubMed] [Google Scholar]
- Huang Y, Niwa Ji, Sobue G, Breitwieser GE. 2006. Calcium-sensing receptor ubiquitination and degradation mediated by the E3 ubiquitin ligase dorfin. J Biol Chem 281: 11610–11617. 10.1074/jbc.M513552200 [DOI] [PubMed] [Google Scholar]
- Huang H, Kuenze G, Smith JA, Taylor KC, Duran AM, Hadziselimovic A, Meiler J, Vanoye CG, George AL Jr, Sanders CR. 2018. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci Adv 4: eaar2631 10.1126/sciadv.aar2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster D, Hoppert M, Lutsenko S, Zinke J, Lehmann C, Mössner J, Berr F, Caca K. 2003. Defective cellular localization of mutant ATP7B in Wilson's disease patients and hepatoma cell lines. Gastroenterology 124: 335–345. 10.1053/gast.2003.50066 [DOI] [PubMed] [Google Scholar]
- Hutt DM, Roth DM, Chalfant MA, Youker RT, Matteson J, Brodsky JL, Balch WE. 2012. FK506 binding protein 8 peptidylprolyl isomerase activity manages a late stage of cystic fibrosis transmembrane conductance regulator (CFTR) folding and stability. J Biol Chem 287: 21914–21925. 10.1074/jbc.M112.339788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem 279: 38369–38378. 10.1074/jbc.M402468200 [DOI] [PubMed] [Google Scholar]
- Illing ME, Rajan RS, Bence NF, Kopito RR. 2002. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem 277: 34150–34160. 10.1074/jbc.M204955200 [DOI] [PubMed] [Google Scholar]
- Imamura T, Haruta T, Takata Y, Usui I, Iwata M, Ishihara H, Ishiki M, Ishibashi O, Ueno E, Sasaoka T, et al. 1998. Involvement of heat shock protein 90 in the degradation of mutant insulin receptors by the proteasome. J Biol Chem 273: 11183–11188. 10.1074/jbc.273.18.11183 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Yamamoto A, Kitamura A, Lamande SR, Yoshimori T, Bateman JF, Kubota H, Nagata K. 2009. Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell 20: 2744–2754. 10.1091/mbc.e08-11-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Jameson JL. 1997. Molecular basis of autosomal dominant neurohypophyseal diabetes insipidus. Cellular toxicity caused by the accumulation of mutant vasopressin precursors within the endoplasmic reticulum. J Clin Invest 99: 1897–1905. 10.1172/JCI119357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Suzuki N. 2007. Molecular pathogenesis of seipin/BSCL2-related motor neuron diseases. Ann Neurol 61: 237–250. 10.1002/ana.21070 [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. 1995. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83: 129–135. 10.1016/0092-8674(95)90241-4 [DOI] [PubMed] [Google Scholar]
- Jensen HK, Holst H, Jensen LG, Jørgensen MM, Andreasen PH, Jensen TG, Andresen BS, Heath F, Hansen PS, Neve S, et al. 1997. A common W556S mutation in the LDL receptor gene of Danish patients with familial hypercholesterolemia encodes a transport-defective protein. Atherosclerosis 131: 67–72. 10.1016/S0021-9150(96)06059-5 [DOI] [PubMed] [Google Scholar]
- Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. 2009. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci 122: 4287–4295. 10.1242/jcs.059154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y, Debose-Boyd RA. 2010. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol 45: 185–198. 10.3109/10409238.2010.485605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BD, Schumacher RJ, Ross ED, Toft DO. 1998. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem 273: 3679–3686. 10.1074/jbc.273.6.3679 [DOI] [PubMed] [Google Scholar]
- Joshi S, Wang T, Araujo TLS, Sharma S, Brodsky JL, Chiosis G. 2018. Adapting to stress—Chaperome networks in cancer. Nat Rev Cancer 18: 562–575. 10.1038/s41568-018-0020-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL. 2003. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell 14: 3437–3448. 10.1091/mbc.e02-12-0847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Kadowaki H, Accili D, Yazaki Y, Taylor SI. 1991. Substitution of arginine for histidine at position 209 in the α-subunit of the human insulin receptor. A mutation that impairs receptor dimerization and transport of receptors to the cell surface. J Biol Chem 266: 21224–21231. [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Kawaguchi S, Ng DT. 2007. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol 18: 743–750. 10.1016/j.semcdb.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Kang JQ, Shen W, Macdonald RL. 2009a. The GABRG2 mutation, Q351X, associated with generalized epilepsy with febrile seizures plus, has both loss of function and dominant-negative suppression. J Neurosci 29: 2845–2856. 10.1523/jneurosci.4772-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Shen W, Macdonald RL. 2009b. Two molecular pathways (NMD and ERAD) contribute to a genetic epilepsy associated with the GABAA receptor GABRA1 PTC mutation, 975delC, S326fs328X. J Neurosci 29: 2833–2844. 10.1523/jneurosci.4512-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Karagöz GE, Acosta-Alvear D, Walter P. 2018. The unfolded protein response: Detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M. 1997. The Levinthal paradox: Yesterday and today. Fold Des 2: S69–S75. 10.1016/S1359-0278(97)00067-9 [DOI] [PubMed] [Google Scholar]
- Katsumi A, Senda T, Yamashita Y, Yamazaki T, Hamaguchi M, Kojima T, Kobayashi S, Saito H. 1996. Protein C Nagoya, an elongated mutant of protein C, is retained within the endoplasmic reticulum and is associated with GRP78 and GRP94. Blood 87: 4164–4175. [PubMed] [Google Scholar]
- Katsumi A, Kojima T, Senda T, Yamazaki T, Tsukamoto H, Sugiura I, Kobayashi S, Miyata T, Umeyama H, Saito H. 1998. The carboxyl-terminal region of protein C is essential for its secretion. Blood 91: 3784–3791. [PubMed] [Google Scholar]
- Keitel V, Nies AT, Brom M, Hummel-Eisenbeiss J, Spring H, Keppler D. 2003. A common Dubin-Johnson syndrome mutation impairs protein maturation and transport activity of MRP2 (ABCC2). Am J Physiol Gastrointest Liver Physiol 284: G165–G174. 10.1152/ajpgi.00362.2002 [DOI] [PubMed] [Google Scholar]
- *.Kelly JW. 2018. Restoring cellular function by proteostasis regulators. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a034108 [DOI] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. 1989. Identification of the cystic fibrosis gene: Genetic analysis. Science 245: 1073–1080. 10.1126/science.2570460 [DOI] [PubMed] [Google Scholar]
- Khushoo A, Yang Z, Johnson Arthur E, Skach William R. 2011. Ligand-driven vectorial folding of ribosome-bound human CFTR NBD1. Mol Cell 41: 682–692. 10.1016/j.molcel.2011.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Ahn J, Liu C, Tanabe K, Apodaca J, Suzuki T, Rao H. 2006. The Png1–Rad23 complex regulates glycoprotein turnover. J Cell Biol 172: 211–219. 10.1083/jcb.200507149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PS, Lee J, Jongsamak P, Menon S, Li B, Hossain SA, Bae JH, Panijpan B, Arvan P. 2008. Defective protein folding and intracellular retention of thyroglobulin-R19K mutant as a cause of human congenital goiter. Mol Endocrinol 22: 477–484. 10.1210/me.2007-0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittanakom S, Cordat E, Akkarapatumwong V, Yenchitsomanus P, Reithmeier RAF. 2004. Trafficking defects of a novel autosomal recessive distal renal tubular acidosis mutant (S773P) of the human kidney anion exchanger (kAE1). J Biol Chem 279: 40960–40971. 10.1074/jbc.M405356200 [DOI] [PubMed] [Google Scholar]
- Kityk R, Kopp J, Mayer MP. 2018. Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol Cell 69: 227–237.e4. 10.1016/j.molcel.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Kjaer S, Ibáñez CF. 2003. Intrinsic susceptibility to misfolding of a hot-spot for Hirschsprung disease mutations in the ectodomain of RET. Hum Mol Genet 12: 2133–2144. 10.1093/hmg/ddg227 [DOI] [PubMed] [Google Scholar]
- Kjaer S, Hanrahan S, Totty N, McDonald NQ. 2010. Mammal-restricted elements predispose human RET to folding impairment by HSCR mutations. Nat Struct Mol Biol 17: 726–731. 10.1038/nsmb.1808 [DOI] [PubMed] [Google Scholar]
- Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. 2000. Rational design of potent human transthyretin amyloid disease inhibitors. Nat Struct Biol 7: 312–321. 10.1038/74082 [DOI] [PubMed] [Google Scholar]
- Klaips CL, Jayaraj GG, Hartl FU. 2018. Pathways of cellular proteostasis in aging and disease. J Cell Biol 217: 51–63. 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. 2005. Folding of CFTR is predominantly cotranslational. Mol Cell 20: 277–287. 10.1016/j.molcel.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Kleopa KA, Yum SW, Scherer SS. 2002. Cellular mechanisms of connexin32 mutations associated with CNS manifestations. J Neurosci Res 68: 522–534. 10.1002/jnr.10255 [DOI] [PubMed] [Google Scholar]
- Knittler MR, Dirks S, Haas IG. 1995. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: Quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci 92: 1764–1768. 10.1073/pnas.92.5.1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J 15: 753–763. 10.1002/j.1460-2075.1996.tb00411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmann S, Schäfer A, Wolf DH. 2008. Ubiquitin ligase Hul5 is required for fragment-specific substrate degradation in endoplasmic reticulum-associated degradation. J Biol Chem 283: 16374–16383. 10.1074/jbc.M801702200 [DOI] [PubMed] [Google Scholar]
- Kollner I, Sodeik B, Schreek S, Heyn H, von Neuhoff N, Germeshausen M, Zeidler C, Kruger M, Schlegelberger B, Welte K, et al. 2006. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood 108: 493–500. 10.1182/blood-2005-11-4689 [DOI] [PubMed] [Google Scholar]
- Kozlov G, Muñoz-Escobar J, Castro K, Gehring K. 2017. Mapping the ER interactome: The P domains of calnexin and calreticulin as plurivalent adapters for foldases and chaperones. Structure 25: 1415–1422.e3. 10.1016/j.str.2017.07.010 [DOI] [PubMed] [Google Scholar]
- Krämer-Albers EM, Gehrig-Burger K, Thiele C, Trotter J, Nave KA. 2006. Perturbed interactions of mutant proteolipid protein/DM20 with cholesterol and lipid rafts in oligodendroglia: Implications for dysmyelination in spastic paraplegia. J Neurosci 26: 11743–11752. 10.1523/jneurosci.3581-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger H, Miranda E, MacLeod I, Pérez J, Crowther DC, Marciniak SJ, Lomas DA. 2009. Endoplasmic reticulum-associated degradation (ERAD) and autophagy cooperate to degrade polymerogenic mutant serpins. J Biol Chem 284: 22793–22802. 10.1074/jbc.M109.027102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA. 2006a. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: One for soluble Z variant of human α-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell 17: 203–212. 10.1091/mbc.e04-09-0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse KB, Dear A, Kaltenbrun ER, Crum BE, George PM, Brennan SO, McCracken AA. 2006b. Mutant fibrinogen cleared from the endoplasmic reticulum via endoplasmic reticulum-associated protein degradation and autophagy: An explanation for liver disease. Am J Pathol 168: 1299–1308. 10.2353/ajpath.2006.051097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. 2015. The biology of proteostasis in aging and disease. Annu Rev Biochem 84: 435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamandé SR, Chessler SD, Golub SB, Byers PH, Chan C, Cole WG, Sillence DO, Bateman JF. 1995. Endoplasmic reticulum-mediated quality control of type i collagen production by cells from osteogenesis imperfecta patients with mutations in the proα1(I) chain carboxyl-terminal propeptide which impair subunit assembly. J Biol Chem 270: 8642–8649. 10.1074/jbc.270.15.8642 [DOI] [PubMed] [Google Scholar]
- Lau MM, Neufeld EF. 1989. A frameshift mutation in a patient with Tay–Sachs disease causes premature termination and defective intracellular transport of the α-subunit of β-hexosaminidase. J Biol Chem 264: 21376–21380. [PubMed] [Google Scholar]
- Lee RJ, Liu CW, Harty C, McCracken AA, Latterich M, Römisch K, DeMartino GN, Thomas PJ, Brodsky JL. 2004. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J 23: 2206–2215. 10.1038/sj.emboj.7600232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman MA, Schneider WJ, Brown MS, Davis CG, Elhammer A, Russell DW, Goldstein JL. 1987. The Lebanese allele at the low density lipoprotein receptor locus. Nonsense mutation produces truncated receptor that is retained in endoplasmic reticulum. J Biol Chem 262: 401–410. [PubMed] [Google Scholar]
- Levinthal C. 1969. How to fold graciously. In Mossbauer spectroscopy in biological systems, Proceedings of meeting held at Allerton House, Monicello, Illinois, pp. 22–24. University of Illinois Press, Urbana, IL. [Google Scholar]
- Li Y, Lu W, Schwartz AL, Bu G. 2004. Degradation of the LDL receptor class 2 mutants is mediated by a proteasome-dependent pathway. J Lipid Res 45: 1084–1091. 10.1194/jlr.M300482-JLR200 [DOI] [PubMed] [Google Scholar]
- Li H, Wu HC, Liu Z, Zacchi LF, Brodsky JL, Zolkiewski M. 2014. Intracellular complexes of the early-onset torsion dystonia-associated AAA+ ATPase TorsinA. SpringerPlus 3: 743 10.1186/2193-1801-3-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shao H, Taylor IR, Gestwicki JE. 2016. Targeting allosteric control mechanisms in heat shock protein 70 (Hsp70). Curr Top Med Chem 16: 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Jiang Q, Bai X, Yang YF, Ruan MY, Cai SQ. 2017. Tetrameric assembly of K+ channels requires ER-located chaperone proteins. Mol Cell 65: 52–65. 10.1016/j.molcel.2016.10.027 [DOI] [PubMed] [Google Scholar]
- Liang J, Kim T, Fang S, Yamaguchi J, Weissman AM, Fisher EA, Ginsberg HN. 2003. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J Biol Chem 278: 23984–23988. 10.1074/jbc.M302683200 [DOI] [PubMed] [Google Scholar]
- Liang G, Li Q, Tang Y, Kokame K, Kikuchi T, Wu G, Chen XZ. 2008. Polycystin-2 is regulated by endoplasmic reticulum-associated degradation. Hum Mol Genet 17: 1109–1119. 10.1093/hmg/ddm383 [DOI] [PubMed] [Google Scholar]
- Liao W, Yeung SCJ, Chan L. 1998. Proteasome-mediated degradation of apolipoprotein B targets both nascent peptides cotranslationally before translocation and full-length apolipoprotein B after translocation into the endoplasmic reticulum. J Biol Chem 273: 27225–27230. 10.1074/jbc.273.42.27225 [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840. 10.1038/nature02592 [DOI] [PubMed] [Google Scholar]
- Lin MT, Wu MH, Chang CC, Chiu SN, Theriault O, Huang H, Christe G, Ficker E, Chahine M. 2008. In utero onset of long QT syndrome with atrioventricular block and spontaneous or lidocaine-induced ventricular tachycardia: Compound effects of hERG pore region mutation and SCN5A N-terminus variant. Heart Rhythm 5: 1567–1574. 10.1016/j.hrthm.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Lindquist SL, Kelly JW. 2011. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: Progress and prognosis. Cold Spring Harb Perspect Biol 3: a004507 10.1101/cshperspect.a004507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA. 2007. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131: 106–120. 10.1016/j.cell.2007.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye Y. 2012. Roles of p97-associated deubiquitinases in protein quality control at the endoplasmic reticulum. Curr Protein Pept Sci 13: 436–446. 10.2174/138920312802430608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. 1999. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem 274: 5861–5867. 10.1074/jbc.274.9.5861 [DOI] [PubMed] [Google Scholar]
- Liu L, Done SC, Khoshnoodi J, Bertorello A, Wartiovaara J, Berggren PO, Tryggvason K. 2001. Defective nephrin trafficking caused by missense mutations in the NPHS1 gene: Insight into the mechanisms of congenital nephrotic syndrome. Hum Mol Genet 10: 2637–2644. 10.1093/hmg/10.23.2637 [DOI] [PubMed] [Google Scholar]
- Liu XL, Done SC, Yan K, Kilpelainen P, Pikkarainen T, Tryggvason K. 2004. Defective trafficking of nephrin missense mutants rescued by a chemical chaperone. J Am Soc Nephrol 15: 1731–1738. 10.1097/01.ASN.0000129826.28932.FD [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. 1998. Quality control by proteases in the endoplasmic reticulum. J Biol Chem 273: 32373–32376. 10.1074/jbc.273.49.32373 [DOI] [PubMed] [Google Scholar]
- Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL. 2006. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 441: 894–897. 10.1038/nature04830 [DOI] [PubMed] [Google Scholar]
- Lyons SE, Bruck ME, Bowie EJ, Ginsburg D. 1992. Impaired intracellular transport produced by a subset of type IIA von Willebrand disease mutations. J Biol Chem 267: 4424–4430. [PubMed] [Google Scholar]
- Maegawa GHB, Tropak M, Buttner J, Stockley T, Kok F, Clarke JTR, Mahuran DJ. 2007. Pyrimethamine as a potential pharmacological chaperone for late-onset forms of GM2 gangliosidosis. J Biol Chem 282: 9150–9161. 10.1074/jbc.M609304200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik B, Schlanger L, Al-Khalili O, Bao HF, Yue G, Price SR, Mitch WE, Eaton DC. 2001. Enac degradation in A6 cells by the ubiquitin-proteosome proteolytic pathway. J Biol Chem 276: 12903–12910. 10.1074/jbc.M010626200 [DOI] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA. 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97: 553–564. 10.1016/S0092-8674(00)80767-9 [DOI] [PubMed] [Google Scholar]
- Mayer MP. 2013. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci 38: 507–514. 10.1016/j.tibs.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. 2005. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684. 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. 1996. Assembly of ER-associated protein degradation in vitro: Dependence on cytosol, calnexin, and ATP. J Cell Biol 132: 291–298. 10.1083/jcb.132.3.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. 1999. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J 18: 1492–1505. 10.1093/emboj/18.6.1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3: 100–105. 10.1038/35050509 [DOI] [PubMed] [Google Scholar]
- Medeiros-Neto G, Kim PS, Yoo SE, Vono J, Targovnik HM, Camargo R, Hossain SA, Arvan P. 1996. Congenital hypothyroid goiter with deficient thyroglobulin. Identification of an endoplasmic reticulum storage disease with induction of molecular chaperones. J Clin Invest 98: 2838–2844. 10.1172/JCI119112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH. 2004. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep 5: 692–697. 10.1038/sj.embor.7400164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert M, Sommer T, Jarosch E. 2014. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol 16: 77–86. 10.1038/ncb2882 [DOI] [PubMed] [Google Scholar]
- Mehnert M, Sommermeyer F, Berger M, Kumar Lakshmipathy S, Gauss R, Aebi M, Jarosch E, Sommer T. 2015. The interplay of Hrd3 and the molecular chaperone system ensures efficient degradation of malfolded secretory proteins. Mol Biol Cell 26: 185–194. 10.1091/mbc.E14-07-1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. 2005. ERAD: The long road to destruction. Nat Cell Biol 7: 766–772. 10.1038/ncb0805-766 [DOI] [PubMed] [Google Scholar]
- Miura Y, Kambe F, Yamamori I, Mori Y, Tani Y, Murata Y, Oiso Y, Seo H. 1994a. A truncated thyroxine-binding globulin due to a frameshift mutation is retained within the rough endoplasmic reticulum: A possible mechanism of complete thyroxine-binding globulin deficiency in Japanese. J Clin Endocrinol Metab 78: 283–287. [DOI] [PubMed] [Google Scholar]
- Miura Y, Mori Y, Kambe F, Tani Y, Oiso Y, Seo H. 1994b. Impaired intracellular transport contributes to partial thyroxine-binding globulin deficiency in a Japanese family. J Clin Endocrinol Metab 79: 740–744. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Kagawa T, Numari A, Harris MJ, Itoh J, Watanabe N, Mine T, Arias IM. 2007. Two N-linked glycans are required to maintain the transport activity of the bile salt export pump (ABCB11) in MDCK II cells. Am J Physiol Gastrointest Liver Physiol 292: G818–G828. 10.1152/ajpgi.00415.2006 [DOI] [PubMed] [Google Scholar]
- Mogk A, Haslberger T, Tessarz P, Bukau B. 2008. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans 36: 120–125. 10.1042/BST0360120 [DOI] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299: 1397–1400. 10.1126/science.1079474 [DOI] [PubMed] [Google Scholar]
- Morito D, Nagata K. 2015. Pathogenic hijacking of ER-associated degradation: Is ERAD flexible? Mol Cell 59: 335–344. 10.1016/j.molcel.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Mu TW, Ong DST, Wang YJ, Balch WE, Yates JR III, Segatori L, Kelly JW. 2008. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell 134: 769–781. 10.1016/j.cell.2008.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci 105: 12325–12330. 10.1073/pnas.0805371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Tamura A, Wakabayashi K, Hoshijima K, Komada M, Yoshida T, Kometani S, Matsubara T, Mikuriya K, Ishikawa T. 2008. Ubiquitin-mediated proteasomal degradation of non-synonymous SNP variants of human ABC transporter ABCG2. Biochem J 411: 623–631. 10.1042/BJ20071229 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Wakabayashi-Nakao K, Tamura A, Toyoda Y, Koshiba S, Ishikawa T. 2009. Disruption of N-linked glycosylation enhances ubiquitin-mediated proteasomal degradation of the human ATP-binding cassette transporter ABCG2. FEBS J 276: 7237–7252. 10.1111/j.1742-4658.2009.07423.x [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132: 101–112. 10.1016/j.cell.2007.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanua S, Murakami M, Xia J, Grenda DS, Woloszynek J, Strand M, Link DC. 2011. Activation of the unfolded protein response is associated with impaired granulopoiesis in transgenic mice expressing mutant Elane. Blood 117: 3539–3547. 10.1182/blood-2010-2010-311704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal S, Jaeger PA, Duttke SH, Benner C, Glass CK, Ideker T, Hampton RY. 2018. The Dfm1 Derlin is required for ERAD retrotranslocation of integral membrane proteins. Mol Cell 69: 915 10.1016/j.molcel.2018.02.014 [DOI] [PubMed] [Google Scholar]
- Needham PG, Brodsky JL. 2013. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: The early history of ERAD. Biochim Biophys Acta 1833: 2447–2457. 10.1016/j.bbamcr.2013.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis M, Zalm R, Burbach JPH. 1999. Mutations in the vasopressin prohormone involved in diabetes insipidus impair endoplasmic reticulum export but not sorting. J Biol Chem 274: 21200–21208. 10.1074/jbc.274.30.21200 [DOI] [PubMed] [Google Scholar]
- Ninagawa S, Okada T, Sumitomo Y, Kamiya Y, Kato K, Horimoto S, Ishikawa T, Takeda S, Sakuma T, Yamamoto T, et al. 2014. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J Cell Biol 206: 347–356. 10.1083/jcb.201404075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol 153: 1061–1070. 10.1083/jcb.153.5.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Koyama T, Nakahara M, Egawa N, Hirosawa S. 2008. Proteasome degradation of protein C and plasmin inhibitor mutants. Thromb Haemost 100: 405–412. 10.1160/TH08-04-0252 [DOI] [PubMed] [Google Scholar]
- Nustede R, Klimiankou M, Klimenkova O, Kuznetsova I, Zeidler C, Welte K, Skokowa J. 2016. ELANE mutant-specific activation of different UPR pathways in congenital neutropenia. Br J Haematol 172: 219–227. 10.1111/bjh.13823 [DOI] [PubMed] [Google Scholar]
- Oda Y, Hosokawa N, Wada I, Nagata K. 2003. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299: 1394–1397. 10.1126/science.1079181 [DOI] [PubMed] [Google Scholar]
- O'Donnell BM, Mackie TD, Brodsky JL. 2017a. Linking chanelopathies with endoplasmic reticulum associated degradation. Channels (Austin) 11: 499–501. 10.1080/19336950.2017.1357944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell BM, Mackie TD, Subramanya AR, Brodsky JL. 2017b. Endoplasmic reticulum-associated degradation of the renal potassium channel, ROMK, leads to type II Bartter syndrome. J Biol Chem 292: 12813–12827. 10.1074/jbc.M117.786376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. 2007. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell 28: 544–554. 10.1016/j.molcel.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC. 2013a. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol 5: a013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Richter CM, Kopito RR. 2013b. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci 110: 1345–1350. 10.1073/pnas.1213738110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. 2005. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol 21: 529–550. 10.1146/annurev.cellbio.21.012704.133214 [DOI] [PubMed] [Google Scholar]
- Otero JH, Lizák B, Hendershot LM. 2010. Life and death of a BiP substrate. Semin Cell Dev Biol 21: 472–478. 10.1016/j.semcdb.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Cheng X, Sifers RN. 2013. Golgi-situated endoplasmic reticulum α-1, 2-mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with γ-COP. Mol Biol Cell 24: 1111–1121. 10.1091/mbc.e12-12-0886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Easton DP, Chen X, MacDonald IJ, Wang XY, Subjeck JR. 2003. The chaperoning properties of mouse grp170, a member of the third family of hsp70 related proteins. Biochemistry 42: 14893–14902. 10.1021/bi030122e [DOI] [PubMed] [Google Scholar]
- Park SY, Ye H, Steiner DF, Bell GI. 2010. Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted. Biochem Biophys Res Commun 391: 1449–1454. 10.1016/j.bbrc.2009.12.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kim TH, Kim SW, Noh SH, Cho KJ, Choi C, Kwon EY, Choi YJ, Gee HY, Choi JH. 2016. Functional characterization of ABCB4 mutations found in progressive familial intrahepatic cholestasis type 3. Sci Rep 6: 26872 10.1038/srep26872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson ST, Reithmeier RA. 2010. Cell surface rescue of kidney anion exchanger 1 mutants by disruption of chaperone interactions. J Biol Chem 285: 33423–33434. 10.1074/jbc.M110.144261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BR, Hebert DN. 2010. Lectin chaperones help direct the maturation of glycoproteins in the endoplasmic reticulum. Biochim Biophys Acta 1803: 684–693. 10.1016/j.bbamcr.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peroz D, Dahimene S, Baro I, Loussouarn G, Merot J. 2009. LQT1-associated mutations increase KCNQ1 proteasomal degradation independently of Derlin-1. J Biol Chem 284: 5250–5256. 10.1074/jbc.M806459200 [DOI] [PubMed] [Google Scholar]
- Pidasheva S, Grant M, Canaff L, Ercan O, Kumar U, Hendy GN. 2006. Calcium-sensing receptor dimerizes in the endoplasmic reticulum: Biochemical and biophysical characterization of CASR mutants retained intracellularly. Hum Mol Genet 15: 2200–2209. 10.1093/hmg/ddl145 [DOI] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K. 1997. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J 16: 4540–4548. 10.1093/emboj/16.15.4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisoni GB, Molinari M. 2016. Five questions (with their answers) on ER-associated degradation. Traffic 17: 341–350. 10.1111/tra.12373 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Böhmler S, Bordallo J, Sommer T, Wolf DH. 1997. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388: 891–895. 10.1038/42276 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y. 2008. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem 283: 22885–22889. 10.1074/jbc.R800023200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Preissler S, Ron D. 2018. Early events in the endoplasmic reticulum unfolded protein response (UPR). Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Chambers JE, Crespillo-Casado A, Avezov E, Miranda E, Perez J, Hendershot LM, Harding HP, Ron D. 2015. Physiological modulation of BiP activity by trans-protomer engagement of the interdomain linker. eLife 4: e08961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Brodsky JL. 2017. The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem J 474: 445–469. 10.1042/BCJ20160582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Guerriero CJ, Metzger MB, Michaelis S, Brodsky JL. 2018. Substrate insolubility dictates Hsp104-dependent endoplasmic-reticulum-associated degradation. Mol Cell 70: 242–253.e6. 10.1016/j.molcel.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printsev I, Curiel D, Carraway KL III. 2017. Membrane protein quantity control at the endoplasmic reticulum. J Membr Biol 250: 379–392. 10.1007/s00232-016-9931-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D, Teckman JH, Omura S, Perlmutter DH. 1996. Degradation of a mutant secretory protein, α1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem 271: 22791–22795. 10.1074/jbc.271.37.22791 [DOI] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22: 626–634. 10.1128/MCB.22.2.626-634.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos RR, Swanson AJ, Bass J. 2007. Calreticulin and Hsp90 stabilize the human insulin receptor and promote its mobility in the endoplasmic reticulum. Proc Natl Acad Sci 104: 10470–10475. 10.1073/pnas.0701114104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi L, Caridi G, Santon D, Boaretto F, Bernascone I, Lamorte G, Tardanico R, Dagnino M, Colussi G, Scolari F, et al. 2003. Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet 12: 3369–3384. 10.1093/hmg/ddg353 [DOI] [PubMed] [Google Scholar]
- Rapoport TA. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669. 10.1038/nature06384 [DOI] [PubMed] [Google Scholar]
- Rappley I, Monteiro C, Novais M, Baranczak A, Solis G, Wiseman RL, Helmke S, Maurer MS, Coelho T, Powers ET, et al. 2014. Quantification of transthyretin kinetic stability in human plasma using subunit exchange. Biochemistry 53: 1993–2006. 10.1021/bi500171j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J 25: 533–543. 10.1038/sj.emboj.7600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. 1998. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J Mol Biol 279: 449–460. 10.1006/jmbi.1998.1770 [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. 1989. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245: 1066–1073. 10.1126/science.2475911 [DOI] [PubMed] [Google Scholar]
- Ritter C, Quirin K, Kowarik M, Helenius A. 2005. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J 24: 1730–1738. 10.1038/sj.emboj.7600645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K. 1999. Surfing the Sec61 channel: Bidirectional protein translocation across the ER membrane. J Cell Sci 112: 4185–4191. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. 1989. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science 245: 1059–1065. 10.1126/science.2772657 [DOI] [PubMed] [Google Scholar]
- Ron I, Horowitz M. 2005. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum Mol Genet 14: 2387–2398. [DOI] [PubMed] [Google Scholar]
- Rosam M, Krader D, Nickels C, Hochmair J, Back KC, Agam G, Barth A, Zeymer C, Hendrix J, Schneider M, et al. 2018. Bap (Sil1) regulates the molecular chaperone BiP by coupling release of nucleotide and substrate. Nat Struct Mol Biol 25: 90–100. 10.1038/s41594-017-0012-6 [DOI] [PubMed] [Google Scholar]
- Roselli S, Moutkine I, Gribouval O, Benmerah A, Antignac C. 2004. Plasma membrane targeting of podocin through the classical exocytic pathway: Effect of NPHS2 mutations. Traffic 5: 37–44. 10.1046/j.1600-0854.2003.00148.x [DOI] [PubMed] [Google Scholar]
- Roth J, Zuber C. 2017. Quality control of glycoprotein folding and ERAD: The role of N-glycan handling, EDEM1 and OS-9. Histochem Cell Biol 147: 269–284. 10.1007/s00418-016-1513-9 [DOI] [PubMed] [Google Scholar]
- Rousseau E, Dehay B, Ben-Haiem L, Trottier Y, Morange M, Bertolotti A. 2004. Targeting expression of expanded polyglutamine proteins to the endoplasmic reticulum or mitochondria prevents their aggregation. Proc Natl Acad Sci 101: 9648–9653. 10.1073/pnas.0403015101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe SM, Miller S, Sorscher EJ. 2005. Cystic fibrosis. New Engl J Med 352: 1992–2001. 10.1056/NEJMra043184 [DOI] [PubMed] [Google Scholar]
- Rubenstein EM, Kreft SG, Greenblatt W, Swanson R, Hochstrasser M. 2012. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J Cell Biol 197: 761–773. 10.1083/jcb.201203061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. 2014. Quality control: ER-associated degradation: Protein quality control and beyond. J Cell Biol 204: 869–879. 10.1083/jcb.201312042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Mora G, Buxó L, Carvalho P. 2016. Spatial control of lipid droplet proteins by the ERAD ubiquitin ligase Doa10. EMBO J 35: 1644–1655. 10.15252/embj.201593106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MC, Shooter EM, Notterpek L. 2002. Aggresome formation in neuropathy models based on peripheral myelin protein 22 mutations. Neurobiol Dis 10: 109–118. 10.1006/nbdi.2002.0500 [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Yamashita S, Miura A, Hirahara T, Kimura E, Maeda Y, Terasaki T, Hirano T, Uchino M. 2011. A novel GJB1 frameshift mutation produces a transient CNS symptom of X-linked Charcot–Marie–Tooth disease. J Neurol 258: 284–290. 10.1007/s00415-010-5752-8 [DOI] [PubMed] [Google Scholar]
- Sala AJ, Bott LC, Morimoto RI. 2017. Shaping proteostasis at the cellular, tissue, and organismal level. J Cell Biol 216: 1231–1241. 10.1083/jcb.201612111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraogi I, Shan SO. 2011. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic 12: 535–542. 10.1111/j.1600-0854.2011.01171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Ward CL, Krouse ME, Wine JJ, Kopito RR. 1996. Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J Biol Chem 271: 635–638. 10.1074/jbc.271.2.635 [DOI] [PubMed] [Google Scholar]
- Sato T, Susuki S, Suico MA, Miyata M, Ando Y, Mizuguchi M, Takeuchi M, Dobashi M, Shuto T, Kai H. 2007. Endoplasmic reticulum quality control regulates the fate of transthyretin variants in the cell. EMBO J 26: 2501–2512. 10.1038/sj.emboj.7601685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. 2009. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell 34: 212–222. 10.1016/j.molcel.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. 2011. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80: 587–612. 10.1146/annurev-biochem-060408-172623 [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Merella S, Pasqualetto E, Lazarevic D, Rampoldi L. 2017. Mutant uromodulin expression leads to altered homeostasis of the endoplasmic reticulum and activates the unfolded protein response. PLoS ONE 12: e0175970 10.1371/journal.pone.0175970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid FX, Mayr LM, Mucke M, Schonbrunner ER. 1993. Prolyl isomerases: Role in protein folding. Adv Protein Chem 44: 25–66. 10.1016/S0065-3233(08)60563-X [DOI] [PubMed] [Google Scholar]
- Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D, et al. 2017. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 548: 352–355. 10.1038/nature23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr S, Klein MC, Gamayun I, Melnyk A, Jung M, Schauble N, Wang Q, Hemmis B, Bochen F, Greiner M, et al. 2015. Co-chaperone specificity in gating of the polypeptide conducting channel in the membrane of the human endoplasmic reticulum. J Biol Chem 290: 18621–18635. 10.1074/jbc.M115.636639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. 2001. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell 8: 633–644. 10.1016/S1097-2765(01)00318-5 [DOI] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 127: 4078–4088. 10.1242/jcs.154716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieger I, Lautz K, Krause E, Rosenthal W, Wiesner B, Hermosilla R. 2008. Derlin-1 and p97/Valosin-containing protein mediate the endoplasmic reticulum-associated degradation of human v2 vasopressin receptors. Mol Pharmacol 73: 697–708. 10.1124/mol.107.040931 [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Hammarström P, Matsumura M, Shimizu Y, Iwata M, Tokuda T, Ikeda S, Kelly JW. 2003. Energetic characteristics of the new transthyretin variant A25T may explain its atypical central nervous system pathology. Lab Invest 83: 409–417. 10.1097/01.LAB.0000059937.11023.1F [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarström P, Miller SR, Sawkar AR, Balch WE, Kelly JW. 2005. The biological and chemical basis for tissue-selective amyloid disease. Cell 121: 73–85. 10.1016/j.cell.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Serrano-Perez MC, Tilley FC, Nevo F, Arrondel C, Sbissa S, Martin G, Tory K, Antignac C, Mollet G. 2018. Endoplasmic reticulum-retained podocin mutants are massively degraded by the proteasome. J Biol Chem 293: 4122–4133. 10.1074/jbc.RA117.001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Hegde RS. 2011. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol 27: 25–56. 10.1146/annurev-cellbio-092910-154125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D, Okuse K, Djamgoz MB. 2009. Protein-protein interactions involving voltage-gated sodium channels: Post-translational regulation, intracellular trafficking and functional expression. Int J Biochem Cell Biol 41: 1471–1481. 10.1016/j.biocel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Siegenthaler KD, Pareja KA, Wang J, Sevier CS. 2017. An unexpected role for the yeast nucleotide exchange factor Sil1 as a reductant acting on the molecular chaperone BiP. eLife 6: e24141 10.7554/eLife.24141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DB, Nelson-Williams C, Johnson Bia M, Ellison D, Karet FE, Morey Molina A, Vaara I, Iwata F, Cushner HM, Koolen M, et al. 1996. Gitelman's variant of Barter's syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet 12: 24–30. 10.1038/ng0196-24 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Stenmark H. 2008. Self-eating from an ER-associated cup. J Cell Biol 182: 621–622. 10.1083/jcb.200807061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Wilkinson S. 2017. ER homeostasis and autophagy. Essays Biochem 61: 625–635. 10.1042/EBC20170092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. 2011. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090. 10.1126/science.1209235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soheilipour F, Fazilaty H, Jesmi F, Gahl WA, Behnam B. 2016. First report of inherited thyroxine-binding globulin deficiency in Iran caused by a known de novo mutation in SERPINA7. Mol Genet Metab Rep 8: 13–16. 10.1016/j.ymgmr.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnay PR, Siklosi KR, Van Goor F, Kaniecki K, Yu H, Sharma N, Ramalho AS, Amaral MD, Dorfman R, Zielenski J, et al. 2013. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet 45: 1160–1167. 10.1038/ng.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M, Parodi AJ. 1995. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J 14: 4196–4203. 10.1002/j.1460-2075.1995.tb00093.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. 1997. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J 16: 6325–6336. 10.1093/emboj/16.21.6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. 2004. Coordinated activation of Hsp70 chaperones. Science 303: 98–101. 10.1126/science.1092287 [DOI] [PubMed] [Google Scholar]
- Stolz A, Schweizer RS, Schäfer A, Wolf DH. 2010. Dfm1 forms distinct complexes with Cdc48 and the ER ubiquitin ligases and is required for ERAD. Traffic 11: 1363–1369. 10.1111/j.1600-0854.2010.01093.x [DOI] [PubMed] [Google Scholar]
- Stolz A, Besser S, Hottmann H, Wolf DH. 2013. Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc Natl Acad Sci 110: 15271–15276. 10.1073/pnas.1304928110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Shuto T, Suzuki S, Sato T, Koga T, Suico MA, Kusuhara H, Sugiyama Y, Cyr DM, Kai H. 2011. Posttranslational negative regulation of glycosylated and non-glycosylated BCRP expression by Derlin-1. Biochem Biophys Res Commun 404: 853–858. 10.1016/j.bbrc.2010.12.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Brodsky JL. 2018. The degradation pathway of a model misfolded protein is determined by aggregation propensity. Mol Biol Cell 29: 1422–1434. 10.1091/mbc.E18-02-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susuki S, Sato T, Miyata M, Momohara M, Suico MA, Shuto T, Ando Y, Kai H. 2009. The endoplasmic reticulum-associated degradation of transthyretin variants is negatively regulated by BiP in mammalian cells. J Biol Chem 284: 8312–8321. 10.1074/jbc.M809354200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedine S, Wang T, Halaban R, Hebert DN. 2004. Carbohydrates act as sorting determinants in ER-associated degradation of tyrosinase. J Cell Sci 117: 2937–2949. 10.1242/jcs.01154 [DOI] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev 15: 2660–2674. 10.1101/gad.933301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. 2012. Quantitative analysis of HSP90–client interactions reveals principles of substrate recognition. Cell 150: 987–1001. 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarappoo BK, Yang B, Verkman AS. 1999. Misfolding of mutant aquaporin-2 water channels in nephrogenic diabetes insipidus. J Biol Chem 274: 34825–34831. 10.1074/jbc.274.49.34825 [DOI] [PubMed] [Google Scholar]
- Taschenberger G, Mougey A, Shen S, Lester LB, LaFranchi S, Shyng S-L. 2002. Identification of a familial hyperinsulinism-causing mutation in the sulfonylurea receptor 1 that prevents normal trafficking and function of KATP channels. J Biol Chem 277: 17139–17146. 10.1074/jbc.M200363200 [DOI] [PubMed] [Google Scholar]
- Taylor SC, Ferguson AD, Bergeron JJ, Thomas DY. 2004. The ER protein folding sensor UDP-glucose glycoprotein–glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat Struct Mol Biol 11: 128–134. 10.1038/nsmb715 [DOI] [PubMed] [Google Scholar]
- Teckman JH, Burrows J, Hidvegi T, Schmidt B, Hale PD, Perlmutter DH. 2001. The proteasome participates in degradation of mutant α1-antitrypsin Z in the endoplasmic reticulum of hepatoma-derived hepatocytes. J Biol Chem 276: 44865–44872. 10.1074/jbc.M103703200 [DOI] [PubMed] [Google Scholar]
- Telenti A, Pierce LC, Biggs WH, di Iulio J, Wong EH, Fabani MM, Kirkness EF, Moustafa A, Shah N, Xie C, et al. 2016. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci 113: 11901–11906. 10.1073/pnas.1613365113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. 2005. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol 12: 10–16. 10.1038/nsmb881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau PH, Richardson JM, Wang W, Millen L, Watson J, Mendoza JL, Du K, Fischman S, Senderowitz H, Lukacs GL, et al. 2010. The cystic fibrosis-causing mutation ΔF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem 285: 35825–35835. 10.1074/jbc.M110.131623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Tsukamoto T, Koide T. 1996. Cellular basis for protein c deficiency caused by a single amino acid substitution at Argl5 in the γ-carboxyglutamic acid domain. J Biochem 120: 360–368. 10.1093/oxfordjournals.jbchem.a021421 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Shirotani H, Hara K, Kozuki D, Omura S, Koide T. 1997. Intracellular degradation of secretion defect-type mutants of antithrombin is inhibited by proteasomal inhibitors. FEBS Lett 412: 65–69. 10.1016/S0014-5793(97)00745-X [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Hara K, Koide T. 2003. N-Linked oligosaccharide processing, but not association with calnexin/calreticulin is highly correlated with endoplasmic reticulum-associated degradation of antithrombin Glu313-deleted mutant. Arch Biochem Biophys 411: 235–242. 10.1016/S0003-9861(02)00717-8 [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258. 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Helenius A. 2000. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J Cell Biol 148: 1123–1130. 10.1083/jcb.148.6.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Tokunaga F, Nagamitsu H, Koide T. 2006. Characterization of endoplasmic reticulum-associated degradation of a protein S mutant identified in a family of quantitative protein S deficiency. Thromb Res 117: 323–331. 10.1016/j.thromres.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. 2004. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J Cell Biol 164: 341–346. 10.1083/jcb.200311055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolino J, Fang S, Kubisch C, Monteiro MJ. 2011. Mutant Atp13a2 proteins involved in parkinsonism are degraded by ER-associated degradation and sensitize cells to ER-stress induced cell death. Hum Mol Genet 20: 3565–3577. 10.1093/hmg/ddr274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. 2008. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321: 569–572. 10.1126/science.1159293 [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Nagata K. 2013. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell 24: 3155–3163. 10.1091/mbc.e13-03-0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn JA, Fyfe GK, Canessa CM. 1998. Biosynthesis and processing of epithelial sodium channels in Xenopus oocytes. J Biol Chem 273: 30344–30351. 10.1074/jbc.273.46.30344 [DOI] [PubMed] [Google Scholar]
- van den Berg B, Ellis RJ, Dobson CM. 1999. Effects of macromolecular crowding on protein folding and aggregation. EMBO J 18: 6927–6933. 10.1093/emboj/18.24.6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom J, Meyer H. 2018. VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling. Mol Cell 69: 182–194. 10.1016/j.molcel.2017.10.028 [DOI] [PubMed] [Google Scholar]
- van den Boomen DJ, Lehner PJ. 2015. Identifying the ERAD ubiquitin E3 ligases for viral and cellular targeting of MHC class I. Mol Immunol 68: 106–111. 10.1016/j.molimm.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot AT, Pearce MMP, Leto DE, Shaler TA, Kopito RR. 2018. Redundant and antagonistic roles of XTP3B and OS9 in decoding glycan and non-glycan degrons in ER-associated degradation. Mol Cell 70: 516–530.e6. 10.1016/j.molcel.2018.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden LM, Stapelbroek JM, Krieger E, van den Berghe PVE, Berger R, Verhulst PM, Holthuis JCM, Houwen RHJ, Klomp LWJ, van de Graaf SFJ. 2010. Folding defects in P-type ATP 8B1 associated with hereditary cholestasis are ameliorated by 4-phenylbutyrate. Hepatology 51: 286–296. 10.1002/hep.23268 [DOI] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PDJ, Burton B, Cao D, Neuberger T, Turnbull A, Singh A, Joubran J, Hazlewood A, et al. 2009. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci 106: 18825–18830. 10.1073/pnas.0904709106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J, Olson ER, et al. 2011. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci 108: 18843–18848. 10.1073/pnas.1105787108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanSlyke JK, Deschenes SM, Musil LS. 2000. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol Biol Cell 11: 1933–1946. 10.1091/mbc.11.6.1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Ng DT. 2004. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol 165: 41–52. 10.1083/jcb.200309132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashistha N, Neal SE, Singh A, Carroll SM, Hampton RY. 2016. Direct and essential function for Hrd3 in ER-associated degradation. Proc Natl Acad Sci 113: 5934–5939. 10.1073/pnas.1603079113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit G, Avramescu RG, Chiang AN, Houck SA, Cai Z, Peters KW, Hong JS, Pollard HB, Guggino WB, Balch WE, et al. 2016. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell 27: 424–433. 10.1091/mbc.e14-04-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. 2008. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957. 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz-Donnelly L, Holthusen H, Korner R, Hansen EC, Presto J, Johansson J, Sawarkar R, Hartl FU, Hipp MS. 2018. High capacity of the endoplasmic reticulum to prevent secretion and aggregation of amyloidogenic proteins. EMBO J 37: 337–350. 10.15252/embj.201695841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. 1999. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068. 10.1146/annurev.biochem.68.1.1015 [DOI] [PubMed] [Google Scholar]
- Waheed A, Parkkila S, Zhou XY, Tomatsu S, Tsuchihashi Z, Feder JN, Schatzman RC, Britton RS, Bacon BR, Sly WS. 1997. Hereditary hemochromatosis: Effects of C282Y and H63D mutations on association with β2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proc Natl Acad Sci 94: 12384–12389. 10.1073/pnas.94.23.12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. 2007. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129: 943–955. 10.1016/j.cell.2007.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker VE, Wong MJH, Atanasiu R, Hantouche C, Young JC, Shrier A. 2010. Hsp40 chaperones promote degradation of the hERG potassium channel. J Biol Chem 285: 3319–3329. 10.1074/jbc.M109.024000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dong H, Soroka CJ, Wei N, Boyer JL, Hochstrasser M. 2008. Degradation of the bile salt export pump at endoplasmic reticulum in progressive familial intrahepatic cholestasis type II. Hepatology 48: 1558–1569. 10.1002/hep.22499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJHJ, Hansen TH. 2009. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol 187: 655–668. 10.1083/jcb.200908036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang J, Li N, Ge L, Li B, Song B. 2011. Molecular characterization of the NPC1L1 variants identified from cholesterol low absorbers. J Biol Chem 286: 7397–7408. 10.1074/jbc.M110.178368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pareja KA, Kaiser CA, Sevier CS. 2014. Redox signaling via the molecular chaperone BiP protects cells against endoplasmic reticulum-derived oxidative stress. eLife 3: e03496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Park S, Kodali VK, Han J, Yip T, Chen Z, Davidson NO, Kaufman RJ. 2015. Identification of protein disulfide isomerase 1 as a key isomerase for disulfide bond formation in apolipoprotein B100. Mol Biol Cell 26: 594–604. 10.1091/mbc.E14-08-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangeline MA, Vashistha N, Hampton RY. 2017. Proteostatic tactics in the strategy of sterol regulation. Annu Rev Cell Dev Biol 33: 467–489. 10.1146/annurev-cellbio-111315-125036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83: 121–127. 10.1016/0092-8674(95)90240-6 [DOI] [PubMed] [Google Scholar]
- Watabe H, Valencia JC, Yasumoto K, Kushimoto T, Ando H, Muller J, Vieira WD, Mizoguchi M, Appella E, Hearing VJ. 2004. Regulation of tyrosinase processing and trafficking by organellar pH and by proteasome activity. J Biol Chem 279: 7971–7981. 10.1074/jbc.M309714200 [DOI] [PubMed] [Google Scholar]
- Whiteman P, Handford PA. 2003. Defective secretion of recombinant fragments of fibrillin-1: Implications of protein misfolding for the pathogenesis of Marfan syndrome and related disorders. Hum Mol Genet 12: 727–737. 10.1093/hmg/ddg081 [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84: 769–779. 10.1016/S0092-8674(00)81054-5 [DOI] [PubMed] [Google Scholar]
- Wilhovsky S, Gardner R, Hampton R. 2000. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell 11: 1697–1708. 10.1091/mbc.11.5.1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Reed AA, Galvanovskis J, Antignac C, Goodship T, Karet FE, Kotanko P, Lhotta K, Moriniere V, Williams P, et al. 2009. Uromodulin mutations causing familial juvenile hyperuricaemic nephropathy lead to protein maturation defects and retention in the endoplasmic reticulum. Hum Mol Genet 18: 2963–2974. 10.1093/hmg/ddp235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Chen G, Tsai B. 2015. The nucleotide exchange factors Grp170 and Sil1 induce cholera toxin release from BiP to enable retrotranslocation. Mol Biol Cell 26: 2181–2189. 10.1091/mbc.E15-01-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Link DC. 2008. Severe congenital neutropenia and the unfolded protein response. Curr Opin Hematol 15: 1–7. 10.1097/MOH.0b013e3282f13cd2 [DOI] [PubMed] [Google Scholar]
- Xia H, Redman C. 1999. The degradation of nascent fibrinogen chains is mediated by the ubiquitin proteasome pathway. Biochem Biophys Res Commun 261: 590–597. 10.1006/bbrc.1999.1081 [DOI] [PubMed] [Google Scholar]
- Xia H, Redman CM. 2001. Differential degradation of the three fibrinogen chains by proteasomes: Involvement of Sec61p and cytosolic Hsp70. Arch Biochem Biophys 390: 137–145. 10.1006/abbi.2001.2374 [DOI] [PubMed] [Google Scholar]
- Yam GHF, Bosshard N, Zuber C, Steinmann B, Roth J. 2006. Pharmacological chaperone corrects lysosomal storage in Fabry disease caused by trafficking-incompetent variants. Am J Physiol Cell Physiol 290: C1076–C1082. 10.1152/ajpcell.00426.2005 [DOI] [PubMed] [Google Scholar]
- Yan FF, Lin CW, Cartier EA, Shyng SL. 2005. Role of ubiquitin-proteasome degradation pathway in biogenesis efficiency of β-cell ATP-sensitive potassium channels. Am J Physiol Cell Physiol 289: C1351–C1359. 10.1152/ajpcell.00240.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan FF, Pratt EB, Chen PC, Wang F, Skach WR, David LL, Shyng SL. 2010. Role of Hsp90 in biogenesis of the β-cell ATP-sensitive potassium channel complex. Mol Biol Cell 21: 1945–1954. 10.1091/mbc.e10-02-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429: 841–847. 10.1038/nature02656 [DOI] [PubMed] [Google Scholar]
- Yeung SJ, Chen SH, Chan L. 1996. Ubiquitin–proteasome pathway mediates intracellular degradation of apolipoprotein B. Biochemistry 35: 13843–13848. 10.1021/bi9618777 [DOI] [PubMed] [Google Scholar]
- Ying Z, Wang H, Fan H, Wang G. 2011. The endoplasmic reticulum (ER)-associated degradation system regulates aggregation and degradation of mutant neuroserpin. J Biol Chem 286: 20835–20844. 10.1074/jbc.M110.200808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell 15: 4787–4797. 10.1091/mbc.e04-07-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC. 2010. Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol 88: 291–300. 10.1139/O09-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Wu HC, Bell SL, Millen L, Paton AW, Paton JC, Thomas PJ, Zolkiewski M, Brodsky JL. 2014. The BiP molecular chaperone plays multiple roles during the biogenesis of torsinA, an AAA+ ATPase associated with the neurological disease early-onset torsion dystonia. J Biol Chem 289: 12727–12747. 10.1074/jbc.M113.529123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Dittmar JC, Mihalevic MJ, Shewan AM, Schulz BL, Brodsky JL, Bernstein KA. 2017. Early-onset torsion dystonia: A novel high-throughput yeast genetic screen for factors modifying protein levels of torsinAΔE. Dis Model Mech 10: 1129–1140. 10.1242/dmm.029926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Gong Q, Epstein ML, January CT. 1998. HERG channel dysfunction in human long QT syndrome. J Biol Chem 273: 21061–21066. 10.1074/jbc.273.33.21061 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Gong Q, January CT. 1999. Correction of defective protein trafficking of a mutant HERG potassium channel in human long QT syndrome. J Biol Chem 274: 31123–31126. 10.1074/jbc.274.44.31123 [DOI] [PubMed] [Google Scholar]
- Zhuravleva A, Clerico EM, Gierasch LM. 2012. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151: 1296–1307. 10.1016/j.cell.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuiderweg ER, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. 2013. Allostery in the Hsp70 chaperone proteins. Top Curr Chem 328: 99–153. 10.1007/128_2012_323 [DOI] [PMC free article] [PubMed] [Google Scholar]