Abstract
Drosophila Dscam isoforms are derived from two alternative transmembrane/juxtamembrane domains (TMs) in addition to thousands of ectodomain variants. Using a microRNA-based RNA interference technology, we selectively knocked down different subsets of Dscams containing either the exon 17.1- or exon 17.2-encoding TM. Eliminating Dscam[TM1] reduced Dscam expression but minimally affected postembryonic axonal morphogenesis. In contrast, depleting Dscam[TM2] blocked axon arborization. Further removal of Dscam[TM1] enhanced the loss-of-Dscam[TM2] axonal phenotypes. However, Dscam[TM1] primarily regulates dendritic development, as evidenced by the observations that removing Dscam[TM1] alone impeded elaboration of dendrites and that transgenic Dscam[TM1], but not Dscam[TM2], effectively rescued Dscam mutant dendritic phenotypes in mosaic organisms. These distinct Dscam functions can be attributed to the juxtamembrane regions of TMs that govern dendritic versus axonal targeting of Dscam as well. Together, we suggest that specific Drosophila Dscam juxtamembrane variants control dendritic elaboration and axonal arborization.
Keywords: protein targeting, axon, dendrite, down syndrome, Drosophila, RNA interference
Introduction
The insect Dscam gene encodes thousands of distinct immunoglobulin/fibronectin-type cell adhesion molecules that mainly differ in their extracellular domain and can carry one of the two well-conserved transmembrane/juxtamembrane segments (TMs) (Schmucker et al., 2000). The variations in the ectodomain have drastic impacts on the inter-Dscam binding in vitro (Wojtowicz et al., 2004), whereas distinct TMs may target Dscam to different subcellular compartments (Wang et al., 2004; Zhan et al., 2004). Such a huge repertoire of diverse homophilic cell adhesion molecules may help govern how a complex nervous system is specifically wired.
In Drosophila, Dscam is widely required for proper neuronal morphogenesis, especially the bifurcation/arborization of neurites. Interestingly, loss of Dscam function affects neurite trajectories selectively at the loci where bifurcation normally occurs (Wang et al., 2002; Chen et al., 2006). Some Dscam mutant neurites stall with clumps of possibly numerous short branches at the ends, resulting in truncation of axonal trees at their first points of bifurcation. These characteristic phenotypes are probably derived from excessive repetitive bifurcations of mutant growth cones, because others that have fully extended frequently exhibit evidence for additional bifurcations at the normal branching points. In addition, proper guidance of individual growth cones at the points of ramification, especially with respect to their sister growth cones, requires Dscam. It appears that Dscam controls neurite arborization by preventing comigration of sister growth cones, thus preventing the number of sister growth cones from exceeding the number of available fascicles. Dynamic stochastic expression of distinct Dscam ectodomains (Neves et al., 2004) would permit self recognition in such “like-kill-like” morphogenetic processes. Consistent with this model, most Dscam ectodomain exon alternatives are not conserved through evolution, arguing that the overall diversity is more critical than the identities of individual variants (Graveley et al., 2004).
In contrast, the two exon alternatives that encode the TM of Dscam are well conserved (Graveley et al., 2004). Interestingly, ectopic Dscam can be preferentially localized to dendrites by selective utilization of exon 17.1 (encoding TM1) or localized to axons by using exon 17.2 (encoding TM2) (Wang et al., 2004). To determine the roles of Dscam[TM1] versus Dscam[TM2], we previously created Dscams lacking either exon 17.1 or exon 17.2. Deletion of either exon 17 led to skipping of the other exon 17 in many Dscam transcripts (our unpublished results). Manipulating genomic Dscam can be problematic for other reasons (see Discussion). We, thus, resorted to a microRNA(miRNA)-based RNA interference (RNAi) technology (Chen et al., 2007) for specifically silencing exon 17.1- or exon 17.2-containing Dscam transcripts. We found that endogenous Dscam[TM1] and Dscam[TM2] primarily act to mediate dendritic elaboration and axonal arborization, respectively. Consistent results were obtained with transgenic Dscam alleviating dendrite versus axon phenotypes in Dscam mutant clones depending on the nature of its TM. In addition, the cytoplasmic juxtamembrane regions of the TMs are sufficient to control Dscam localization as well as its differential roles in dendrites versus axons. Together, we suggest that Dscams with distinct TMs control dendritic elaboration versus axonal arborization.
Materials and Methods
Transgenes.
Standard molecular biological techniques were used to generate UAS-17.1 miRNA, UAS-17.2 miRNA, and UAS-18 miRNA, which encode microRNAs carrying unique Dscam sequences derived from exon 17.1, exon 17.2, and exon 18, respectively. The complete nucleotide sequences of the microRNA constructs are available on request. In addition, the details about how the two UAS-Dscams with chimeric TMs were constructed are available on request.
Flies.
Transgenic flies carrying various UAS-miRNA and UAS-Dscam with chimeric TM were obtained by P element-mediated germ line transformation with technical support from Genetic Services.
Tissue-specific induction of RNAi and phenotypic analysis by immunohistochemistry.
Targeted induction of UAS-miRNA involved use of various tissue-specific galactosidase-4 (GAL4) drivers, and their phenotypic analysis mainly involved coexpression of various upstream activation sequence (UAS)-reporter genes. Both flip-out-marked clones of ellipsoid body (EB) neurons and MARCM (mosaic analysis with a repressible cell marker)-labeled clones of projection neurons (PNs) were induced in newly hatched larvae by heat shock for 40 min at 37°C. Whole fly brains were prepared for immunostaining as described previously (Lee et al., 1999). Detection of endogenous Dscam proteins involved a mouse monoclonal antibody (mAb) that was raised against the peptide ATLDKRRPDLRDELG. The anti-Dscam mAb, 1D4 mAb, and anti-mCD8 mAb were used at 1:20, 1:50, and 1:100, respectively. Fluorescence signals were captured with confocal microscopy and processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
Results
microRNA-based RNA interference permits selective depletion of Dscam[TM1] versus Dscam[TM2]
Transgenic Drosophila Dscams with distinct TMs are enriched in dendrites or axons (Wang et al., 2004) and, consistent with such differential protein targeting phenomena, potently affect different aspects of neuronal morphogenesis (Wang et al., 2004; Zhan et al., 2004). To determine whether endogenous Dscam with TM1 versus TM2 indeed helps govern different neuronal morphogenetic processes, we examined whether and how targeted depletion of Dscam[TM1] or Dscam[TM2] perturbs distinct aspects of neuronal morphogenesis in intact Drosophila brains. We first explored whether one can effectively knock down Dscam[TM1] versus Dscam[TM2] by specifically silencing the Dscam transcripts that carry exon 17.1 or exon 17.2 using a miRNA-based RNAi technology (Chen et al., 2007). We engineered three UAS-miRNA constructs, UAS-17.1 miRNA, UAS-17.2 miRNA, and UAS-18 miRNA, to specifically target the Dscam exon 17.1, exon 17.2, and exon 18 (a common Dscam exon), respectively. For each UAS-miRNA transgene, we identified the most potent transgenic line from multiple independent transformants by individually assaying its ability to antagonize GAL4-induced coexpression of UAS-Dscam::GFP (green fluorescent protein). We resorted to transgenic Dscam::GFP, because direct visualization of Dscam[TM1] versus Dscam[TM2] was not possible without antibodies against different TMs. Pairing transgenic miRNA with Dscam::GFP that carries the miRNA target sequences consistently led to a drastic reduction in the level of Dscam-GFP expression (Fig. S1C,F, available at www.jneurosci.org as supplemental material). In contrast, even with the most potent transgenic line, UAS-17.1 miRNA and UAS-17.2 miRNA exerted no detectable effect on the alternative isoform (Fig. S1D,E, available at www.jneurosci.org as supplemental material). It assures no cross-reactivity, also eliminating the trivial explanation that suppression of Dscam transgenes might result from the presence of multiple UAS transgenes. These observations provide us with the opportunity to use transgenic miRNA to knock down specific subsets of Dscam isoforms in intact fly brains.
We then examined how a broad induction of various Dscam-targeted miRNAs affects the endogenous Dscam expression in the developing Drosophila CNS. We could normally detect abundant Dscam proteins in the larval neuropils by immunostaining with a peptide antibody against some Dscam common motif(s) (Wang et al., 2004) (Fig. 1A). Interestingly, such Dscam immunoreactivity was differentially abolished after pan-neuronal induction of various miRNA transgenes. First, we were encouraged by the lack of detectable residual Dscam after induction of UAS-18 miRNA (Fig. 1B) or coinduction of UAS-17.1 miRNA and UAS-17.2 miRNA (Fig. 1C), which are both expected to silence all Dscam splice variants. Second, induction of UAS-17.1 miRNA or UAS-17.2 miRNA alone, even in multiple copies, left its overall pattern of expression essentially unchanged (Fig. 1D,E). Depleting Dscam[TM1] versus Dscam[TM2] should provide insight into the spatial/temporal patterns of TM1 versus TM2 expression. Interestingly, close inspection revealed that Dscam[TM1] apparently exists more abundantly than Dscam[TM2] in the larval CNS, especially within the abdominal ganglion (Fig. 1, compare D with E). Together, these observations demonstrate the general feasibility of knocking down Dscam[TM1] versus Dscam[TM2] using UAS-17.1 miRNA or UAS-17.2 miRNA. Additionally, both UAS-18 miRNA alone and UAS-17.1 miRNA plus UAS-17.2 miRNA should allow us to determine the Dscam “null” phenotypes one can obtain with various GAL4 drivers.
Figure 1.
Silencing of endogenous Dscam expression by various transgenic miRNAs. Composite confocal images of wandering larvae CNS showing endogenous Dscam expression (magenta; as revealed by immunostaining with an anti-Dscam exon 18 peptide mAb), in wild-type (WT) and after asense-GAL4/GAL4-C155-dependent induction of various anti-Dscam miRNAs (B–E). A, Wild-type control. Note that UAS-18 miRNA alone (B) or only UAS-17.1 miRNA plus UAS-17.2 miRNA (C) could effectively eliminate the entire Dscam expression. In addition, regardless of the levels of Dscam, major neural structures, as revealed by coinduction of UAS-mCD8::GFP (green), remained comparable (insets). Scale bar: (here and in all figures) 20 μm.
Dscam[TM2], but not Dscam[TM1], plays an essential role in governing axon arborization, probably because of differential protein targeting
To determine the roles of Dscam[TM1] versus Dscam[TM2] in Dscam-dependent axonal morphogenesis, we first examined how expression of specific Dscam-targeted miRNAs affects the morphogenesis of mushroom body (MB) axons. Transgenic miRNA was induced through MB development using GAL4-OK107. We selectively focused on its effects on the orthogonal α and β lobes, because they are normally derived via Dscam-governed axon bifurcation, and one can readily identify the MB α/β axons based on their strong immunoreactivity with the 1D4 monoclonal antibody (Wang et al., 2002). Interestingly, induction of UAS-18 miRNA or UAS-17.2 miRNA alone, but not UAS-17.1 miRNA, drastically disrupted the formation of α/β lobes (Fig. 2). Abnormal α/β lobes were often misshapen and variably truncated (Fig. 2B–E), reminiscent of the deformed MBs in Dscam mutant organisms (Wang et al., 2004). This suggests involvement of Dscam[TM2], but not Dscam[TM1], in MB axonal morphogenesis. We further classified the anomalies based on the presence or absence of any α/β lobe residue and its degree of extension (severe, no α/β neurite extended beyond the peduncle terminus; strong, no neurite reached the tips of α/β lobes; medium, small subsets of α/β neurites were fully extended; weak, grossly intact lobes failed to segregate). Quantitative analysis of the above phenotypes (n > 100, each) revealed that UAS-17.2 miRNA alone did not cause as much of a defect as UAS-18 miRNA (Fig. 2I) (5.0 ± 2.0 vs 52.7 ± 4.5% for the severe phenotype). This is apparently a result of the possibility that endogenous Dscam[TM1] could partially compensate for the loss of Dscam[TM2] in the Dscam-governed bifurcation of MB axons, because doubling the dosage of UAS-17.2 miRNA did not enhance the phenotypes (8.7 ± 2.5%), but coinduction of UAS-17.1 miRNA and UAS-17.2 miRNA fully recapitulated the 18 miRNA-derived null phenotypes (59.0 ± 6.0%) (Fig. 2I). Together, these results provide direct evidence for differential involvement of Dscam[TM1] and Dscam[TM2] in supporting neuronal morphogenesis.
Figure 2.
Dscam[TM2], but not Dscam[TM1], plays an essential role in MB axonal morphogenesis. A–E, Adult MB lobes visualized by 1D4 mAb. Compared with the wild type (A), induction of certain anti-Dscam miRNAs (see I) disrupted the formation of MB α/β lobes to various extents (B–E). F–H, Derivation of two chimeric Dscam TMs (F) and their effects on Dscam::GFP (green) protein targeting. After binary induction with GAL4–201Y, Dscam[17.1/17.2]::GFP, like Dscam[TM2]::GFP, is uniformly distributed in the larval MBs (G). In contrast, Dscam[17.2/17.1]::GFP, like Dscam[TM1]::GFP, is enriched in dendrites (H). Additionally, both Dscam[17.1/17.2]::GFP and Dscam[TM2]::GFP are preferentially targeted to axons after suppression of the induction by RNAi (data not shown) (similar to Fig. S1F,H, available at www.jneurosci.org as supplemental material). I, Quantitative analysis of MB lobe phenotypes, based on the above classification (A–E), after GAL4-OK107-dependent induction of various anti-Dscam miRNAs and in the absence or presence of distinct transgenic Dscam::GFP.
Preferential usage of exon 17.2 over exon 17.1 might explain why Dscam[TM2] plays a dominant role in this particular developmental event. To rule out this possibility, we attempted to rescue the 18 miRNA-derived null phenotypes by supplementing transgenic Dscam[TM1] or Dscam[TM2]. We reasoned that both Dscam transgenes carry wild-type exon 18 and should be suppressed by 18 miRNA to an analogous degree, if they were comparably induced. Additionally, their ability to antagonize silencing of endogenous Dscam by 18 miRNA should be identical. Thus, any difference in their rescue of 18 miRNA-derived loss-of-Dscam phenotypes can be ascribed to the presence of TM1 or TM2 in the comparably residual transgenic Dscam. We previously identified UAS-Dscam::GFPs that exhibit similar levels of induction (Wang et al., 2004). Using these lines, we obtained substantially better rescue after coinduction of UAS-18 miRNA with UAS-Dscam[exon 17.2]::GFP than with UAS-Dscam[exon 17.1]::GFP (Fig. 2I) (38.0 ± 1.8 vs 6.9 ± 0.4% for the weak plus normal phenotypes). This supports the notion that Dscam[TM2] and Dscam[TM1], which differ only in the TMs, are functionally distinct. To locate the structural basis for such a functional distinction between Dscam[TM1] and Dscam[TM2], we conducted structural-functional analysis and generated two chimeric TMs by swapping the cytoplasmic juxtamembrane portions between TM1 and TM2 (Fig. 2F). Interestingly, this exchange reversed the functional distinction (Fig. 2I) (rescued to 69.0 ± 3.0% with 17.1/17.2 vs 7.3 ± 2.4% with 17.2/17.1) as well as their differential protein targeting between Dscam[TM1] and Dscam[TM2] (Fig. 2G,H). These results suggest that the cytoplasmic juxtamembrane domains of Dscam govern its TM-dependent differential subcellular localization and further imply that, probably because of differential targeting, TM2-containing Dscam isoforms function primarily in axons, whereas TM1-containing Dscams possibly act in dendrites within neurons.
To examine whether Dscam[TM2], but not Dscam[TM1], is broadly used to mediate diverse axonal morphogenesis, we further investigated whether and how knocking down Dscam[TM1] or Dscam[TM2] levels affects the morphogenesis of EB neurons. We reported previously that Dscam-dependent proper arborization of axons is also required for full elaboration of EB neurites in the centrally placed EB neuropil (Wang et al., 2002). Interestingly, after separate induction of UAS-17.1 miRNA and UAS-17.2 miRNA using asense-GAL4 (Zhu et al., 2006b) plus GAL4-EB1 (Wang et al., 2002), only UAS-17.2 miRNA could phenocopy the morphogenetic defects characteristic of Dscam mutant EB neurons (Fig. S2A–D, available at www.jneurosci.org as supplemental material) (100%; n = 10, each). This observation again supports the notion that Dscam[TM2], but not Dscam[TM1], plays an essential role in governing diverse axonal morphogenetic processes, and, as addressed above, this difference in function is probably derived from differential protein targeting.
Dscam[TM1] is involved primarily in dendritic elaboration
Transgenic Dscam[TM1]::GFP is selectively enriched in dendrites in diverse model neurons, including the antennal lobe (AL) PNs (Wang et al., 2004). To determine whether Dscam[TM1] is preferentially involved in dendritic morphogenesis, we resorted to PNs for additional analysis of the TM-dependent distinct Dscam morphogenetic functions. In contrast with MB and EB neurons where Dscam appears dispensable for dendritic morphogenesis (Wang et al., 2002), the PNs, especially the ventral lineage-derived multiglomeruli-targeting PNs (in the vPN clones), require Dscam for full elaboration of their dendrites in the ALs in addition to proper arborization of their axons in the lateral horns (LHs) (Zhu et al., 2006a).
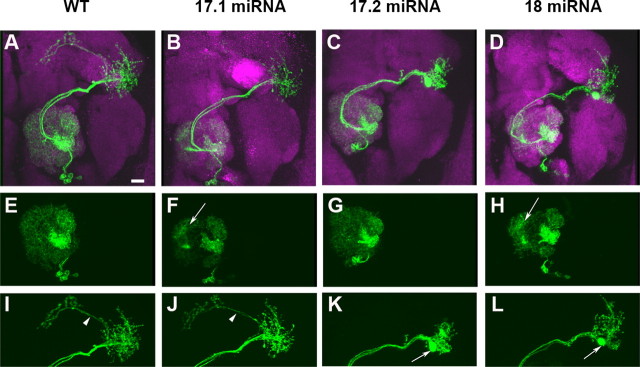
We first demonstrated that GAL4-GH146-dependent induction of UAS-18 miRNA in the otherwise wild-type vPN neuroblast (Nb) clones potently suppressed the elaboration of MARCM-labeled dendrites in the ALs and disrupted their axonal arborization in the LHs (Fig. 3D,H,L) (100%; n = 10). The dendrites of multiglomeruli-targeting PNs became aberrantly restricted to the AL medial upper portion where the axon passage resides (Fig. 3H, arrow). In the LHs, their axonal branches failed to extend away from one another and often stalled with abnormal aggregates (Fig. 3L, arrow). In addition, the projection out of the LH (Fig. 3I,J, arrowheads) is essentially missing. These morphogenetic defects are analogous to the previously documented Dscam loss-of-function PN phenotypes (Zhu et al., 2006a). We then wondered whether depleting Dscam[TM1] versus Dscam[TM2] in the same neurons might differentially affect the morphogenesis of their dendrites versus axons. UAS-17.1 miRNA and UAS-17.2 miRNA were separately induced by GAL4-GH146 in the vPN Nb clones. Remarkably, the loss-of-Dscam dendritic and axonal phenotypes were uncoupled and well correlated with the protein targeting phenomena. Induction of UAS-17.1 miRNA alone effectively suppressed PN dendritic elaboration but minimally affected their axonal arborization (Fig. 3B,F,J) (100%; n = 9). In contrast, induction of UAS-17.2 miRNA alone selectively disrupted the arborization of PN axons in the LHs (Fig. 3C,G,K) (100%; n = 15). We further quantified dendrite phenotypes by counting the glomeruli that became not innervated by GAL4-GH146-positive PNs in the above vPN Nb clones. Given the presence of several GAL4-GH146-positive pan-glomerular PNs in the vPN lineage, there is normally no glomerulus where we could not detect any vPN-lineage-derived GAL4-GH146-labeled neurites. Such vPN dendrites were also fully elaborated after depletion of endogenous Dscam[TM2], as revealed by no GAL4-GH146-unlabeled glomerulus despite induction of 17.2 miRNA. In contrast, depleting endogenous Dscam[TM1] with 17.1 miRNA made a significant and comparable number of AL glomeruli devoid of MARCM-labeled vPN dendrites as knocking down all Dscam isoforms by 18 miRNA (17.1, 10.86 ± 0.69 vs 18, 11.00 ± 0.82). These results collectively suggest that differential protein targeting of Dscam[TM1] and Dscam[TM2], respectively, controls dendritic elaboration and axonal arborization.
Figure 3.
Dscam[TM1] and Dscam[TM2] primarily govern dendritic and axonal morphogenesis, respectively. MARCM-labeled adult vPN Nb clones. Compared with the wild-type clone (A, green), induction of 17.1 miRNA, 17.2 miRNA, and 18 miRNA in vPN Nb clones (B–D, green) specifically disrupted dendritic elaboration (F, arrow), axonal arborization (K, arrow), and both (H, L; arrows), respectively. Adult fly brains were counterstained with nc82 mAb (magenta). The cropped images selectively show dendritic elaboration (E–H) or axonal arborization (I–L) of the clones.
Consistent with this notion, single-ectodomain transgenic Dscam significantly rescued certain Dscam mutant phenotypes in dendrites versus axons, depending on the presence of TM1 or TM2. Briefly, using two distinct Dscam transgenes that differ only in their exon 17, we had shown previously that transgenic Dscam[TM2], but not Dscam[TM1], could effectively rescue various axonal morphogenetic defects in single-cell clones of Dscam mutant MB neurons (Wang et al., 2004). Interestingly, when the same pair of Dscam transgenes were respectively examined for its effects on Dscam mutant DL-1 PNs, we found that PN dendrite defects were rescued only by transgenic Dscam[TM1], whereas only transgenic Dscam[TM2] partially rescued PN bouton formation in the MB calyces (Fig. 4). This provides RNAi-independent evidence for involvement of Dscam[TM1] and Dscam[TM2] in the morphogenesis of dendrites and axons, respectively.
Figure 4.
Rescue of Dscam mutant PN morphogenesis by transgenic Dscam with TM1 versus TM2. A–H, Adult single-cell clones of DL-1 PNs (green) of which the dendrite elaboration in the DL-1 glomeruli (as revealed by nc82 immunostaining; magenta) and axon arborization in the MB calyces and the LHs are, respectively, shown in A–D and E–H. A, E, Wild-type clones. B, F, Dscam mutant clones. C, G, Rescue of mutant clones with pDscam-Dscam[3.36.25.1-genomic 18–24]. D, H, Rescue with pDscam-Dscam[3.36.25.2-genomic 18–24]. Note partial coverage of DL-1 glomeruli by the green PN dendrites in B and D and the absence of bouton-like structures (arrowheads) in F and G. I–J, Quantitative analysis of the coverage of DL-1 glomeruli by single-cell PN clones (I) and the numbers of PN-derived bouton-like structures in the MB calyces (J). n = 25 in every condition.
Discussion
Our use of an miRNA-based RNAi technology permits depletion of distinct subsets of Dscam isoforms based on their exon compositions. Reducing TM1-containing (encoded by exon 17.1) versus TM2-containing (encoded by exon 17.2) Dscams during morphogenesis of various model neurons allowed us to demonstrate that Dscam[TM1] and Dscam[TM2] are preferentially involved in dendritic elaboration and axonal arborization, respectively. This differential involvement of distinct Dscams is likely a result of differential protein targeting, because they are specified by similar cytoplasmic juxtamembrane portions of the Dscam exon 17-encoding TMs. However, after strong binary induction, transgenic Dscam[TM2] can exist abundantly in both dendrites and axons, whereas Dscam[TM1] remains primarily restricted to dendrites, although Dscam[TM1] exhibits more broad function than Dscam[TM2]. Knocking down both further impeded MB axons (Fig. 2I), whereas eliminating exon 17.1-containing Dscam was equally potent as silencing all Dscam transcripts in the inhibition of PN dendrites (Fig. 3). These discrepancies suggest targeting-independent functional distinction between Dscam[TM1] and Dscam[TM2].
Despite some recently raised concerns on the specificity of RNAi-mediated gene silencing (Ma et al., 2006), we are confident in this study for several reasons. First, it should be more straightforward to obviate potential off-target effects with the microRNA-based RNAi constructs, because each of them only yields two 22-nucleotide-long double-stranded RNAs. Second, we generated an independent miRNA-based RNAi construct against a common Dscam exon in addition to the ones targeting exon 17.1 versus exon 17.2 and have obtained the expected results after analogous induction of RNAi against these discrete Dscam sequences. Third, the specificity and effectiveness of our RNAi was also confirmed by direct visualization of specific Dscam proteins in vivo. Fourth, all of our RNAi-induced phenotypes were comparable both qualitatively and quantitatively to previously known Dscam loss-of-function phenotypes. Fifth, we could also rescue the RNAi-mediated silencing of Dscam using appropriate Dscam transgenes. In addition, our demonstration that endogenous Dscam mediates dendritic versus axonal morphogenesis depending on the presence of TM1 or TM2 has been implicated by multiple independent lines of research. In particular, whereas transgenic Dscam with TM2 consistently rescues of Dscam mutant axons, Dscam[TM1] is much more potent than Dscam[TM2] in the rescue of Dscam mutant dendrite elaboration.
Alternative methods for knocking out subsets of isoforms mainly involve deletion of specific exon alternatives at the genomic level. In theory, by gene targeting (Gong and Golic, 2003), one can modify a genomic Dscam to carry only exon 17.1 or exon 17.2 in Drosophila. But such genomic manipulation is potentially problematic. Assuming simple gene targeting does not alter the levels of Dscam expression, elimination of subsets of isoforms could inevitably lead to expression of other isoforms in much broader patterns and/or at higher levels than the normal unperturbed conditions. This may attenuate the defects resulting from the loss of specific isoforms and/or elicit additional phenotypes because of ectopic expression of the residual isoforms, further complicating interpretation. In contrast, knocking down various subsets of Dscam isoforms at the level of translation (e.g., by microRNA) should deplete the isoforms of interest without affecting the expression of others. This way, one can potentially map the endogenous patterns of expression for various specific subsets of Dscam isoforms and, more importantly, unequivocally determine the contributions made by given subsets of Dscam isoforms to various neural developments. In the case of Dscam[TM1] versus Dscam[TM2], we found that, although the relative abundance might differ at various developmental stages, they are apparently coexpressed in most cells. The coexpression with differential requirements implies that they are functionally distinct. Dscam[TM1] selectively concentrates in dendritic growth cones, whereas Dscam[TM2] is preferentially located to axonal growth cones. Nevertheless, we could not observe their differential targeting after depletion of one versus the other, because most neuropils are made up of both dendrites and axons. We also recently learned that retrograde transport plays an essential role in the dendritic enrichment of Dscam[TM1] (our unpublished results), potentially explaining why both Dscam[TM1] and Dscam[TM2] could be detected in the cores of larval MB peduncles where newly derived MB axons are selectively fasciculated (our unpublished observation). Finally, compared with the constitutive changes associated with gene targeting, GAL4/UAS-mediated targeted induction of RNAi permits more varied control over when and/or where to knock down the isoforms of interest and should better illustrate the stage- as well as tissue-specific isoform-characteristic functions.
Together, our results indicate Dscams with distinct TMs are differentially involved in morphogenesis of dendrites versus axons, most likely because of differential protein targeting. What might be the advantages for having two exon alternatives to specify where a Dscam protein should be located? One possible advantage is the ability to independently control the Dscam repertoire at dendrite and axon within a single neuron. For instance, coupling different ectodomains with TM1 would permit selective enrichment of Dscams with distinct homophilic binding specificities in the dendrites but not axons. Additionally, for a given ectodomain, varying the ratio of TM1/TM2 could simultaneously alter the amounts of dendritic and axonal Dscams. Given that dendrites and axons of the same neurons often acquire different morphological characteristic features, it would be interesting to determine whether the TM1/TM2-governed differential distribution of Dscam directly underlies some aspects of the differential morphogenesis of dendrites versus axons.
Footnotes
This work was supported by the National Institutes of Health and the March of Dimes Birth Defects Foundation. We thank B. Leung for critical reading of this manuscript and members of the Lee laboratory for helpful discussions throughout the entire project.
References
- Chen BE, Kondo M, Garnier A, Watson FL, Puettmann-Holgado R, Lamar DR, Schmucker D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Kaur A, Gunning D, Zipursky SL, Rowen L, Clemens JC. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA. 2004;10:1499–1506. doi: 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee C-HJ, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee C-HJ, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006a;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006b;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]