Abstract
Protoplasmic astrocytes are critically important to energy metabolism in the CNS. Our current understanding of the metabolic interactions between neurons and glia is based on studies using cultured cells, from which mainly inferential conclusions have been drawn as to the relative roles of neurons and glia in brain metabolism. In this study, we used functional genomics to establish the relative compartmentalization of neuronal and astrocytic metabolic pathways in the adult brain. To this end, fluorescence-activated cell sorting was used to directly isolate neurons and protoplasmic astrocytes from the cortex of adult mice. Microarray analysis showed that astrocytes and neurons each express transcripts predicting individual self-sufficiency in both glycolysis and oxidative metabolism. Surprisingly, most enzymes in the tricarboxylic acid (TCA) cycle were expressed at higher relative levels in astrocytes than in neurons. Mass spectrometric analysis of the TCA cycle intermediates confirmed that freshly isolated adult astrocytes maintained an active TCA cycle, whereas immuno-electron microscopy revealed that fine astrocytic processes encompassing synapses contained a higher density of mitochondria than surrounding cells. These observations indicate that astrocytes exhibit robust oxidative metabolism in the intact adult brain and suggest a prominent contribution of astrocytic metabolism to functional brain imaging, including BOLD (blood-oxygen level-dependent) functional magnetic resonance imaging signals.
Keywords: FACS, microarray, glutamate, lactate, glycolysis, TCA cycle, mass spectrometry
Introduction
It has long been recognized that astrocytes play essential roles in brain metabolism (Hertz et al., 2007). In situ, protoplasmic astrocytes are highly polarized cells, with distinct sets of processes that project to contact either synapses or vascular walls (Simard et al., 2003; Volterra and Meldolesi, 2005). This unique morphology permits astrocytes to concurrently assess and coregulate synaptic activity with the vascular supply of metabolic substrates, thereby effecting neurovascular coupling (Takano et al., 2006). However, the structural complexity of intact brain tissue has been a serious impediment for dissecting the metabolic roles of astrocytes in situ. Currently, there are no techniques available with the necessary spatial and temporal resolution to study metabolic processes at the single-cell level (Hertz et al., 2007). Therefore, concepts as important as the lactate shuttle and the glutamate–glutamine cycle are based on data from cultures with only indirect evidence obtained from studies using intact tissue. Extending in vitro observations to intact brain is difficult, because the phenotypic characteristics of cultured astrocytes differ significantly from their in situ counterparts. For instance, the morphological distinctiveness of astrocytes in situ is lost as they are reduced in vitro to nonpolarized, epitheloid-like cells lacking vascular processes and synaptic contact. In addition, many cultured cells derive from progenitors that differentiate in culture with induction of genes not necessarily expressed in vivo (Wilhelm et al., 2004).
In this study, we asked whether astrocytes and neurons differed in their expression of genes associated with both anaerobic and oxidative glycolytic metabolism. We focused on genes encoding metabolic enzymes, with a special emphasis on identifying pathways that might exhibit functional complementation between neurons and astrocytes. To this end, we used fluorescence-activated cell sorting (FACS) to purify neurons and two distinct populations of astrocytes from dissociated adult mouse neocortex, using methods that were previously established for use with parenchymal progenitors cells (Wang et al., 1998; Keyoung et al., 2001; Nunes et al., 2003). Astrocytes were separated into two pools, defined as GLT1+/GFAP+ and GLT1+/GFAP−, based on their expression of the glutamate transporter 1 (GLT1), or expression of green fluorescent protein (GFP) in the transgenic Gfap-driven GFP mouse (Zhuo et al., 1997). We then harvested RNA for subsequent genomic expression profiling (Lobo et al., 2006; Sim et al., 2006). We found that GLT1-positive astrocytes only exhibited subtle gene expression differences associated with the glial fibrillary acidic protein (GFAP) phenotype and shared the expression profile of key metabolic enzymes. Using a similar approach, neurons were isolated based on expression of yellow fluorescent protein (YFP) under the Thy1 promoter (Feng et al., 2000). Surprisingly, we found that most enzymes in the tricarboxylic acid (TCA) cycle, including Cs (citrate synthase) and Mdh1 (malate dehydrogenase), were overexpressed by astrocytes relative to neurons but did not appear compartmentalized. Both astrocytes and neurons manifested a full repertoire of enzymes mediating aerobic metabolism and were hence independently competent in that regard. In contrast, many pathways entering or exiting glycolysis or the TCA cycle, such as those for lactate and glutamate metabolism, were clearly compartmentalized, with some reactions catalyzed by neurons and others catalyzed by astrocytes. These expression-based observations were confirmed by direct metabolic measurements, by exposing acutely isolated GLT1+ astrocytes to [U-13C]glucose and analyzing the labeled intermediates by mass spectrometry (MS) (McKenna et al., 2005; Bak et al., 2006; Olstad et al., 2007). TCA cycle intermediates and derivates, such as citrate, malate, and glutamate, contained 13C label from [U-13C]glucose, confirming that cortical astrocytes have a robust capacity for oxidative metabolism. This conclusion found support in an ultrastructural analysis of the GFAP-driven, GFP-immunolabeled cortex, which confirmed that astrocytes contain a large number of mitochondria in their fine processes that contact synapses.
Thus, astrocytes and neurons share the principal aerobic pathways of glucose oxidation. As such, astrocytes must contribute to functional brain imaging based on surrogate metabolic indicators of neuronal activation, such as blood-oxygen level-dependent signal changes.
Materials and Methods
Dissociation of cortex.
Brain tissue was from 10- to 12-week-old FVB/N-Tg(GFAPGFP)14Mes/J or B6.Cg-Tg(Thy1-YFPH)2Jrs/J mice (The Jackson Laboratory, Bar Harbor, ME). Mice were anesthetized with pentobarbital (50 mg kg−1, i.p.), perfused with cold Hanks buffer (Invitrogen, Carlsbad, CA), and decapitated. The brain was immediately removed to cold Hanks buffer containing glutamate receptor antagonists, 3 μm DNQX and 100 μm APV (Tocris, Ellisville, MO). The cortex was dissected free of white matter, cut into small pieces, and digested with 8 U/ml papain (Worthington, Lakewood, NJ) in Ca2+/Mg2+-free PIPES/cysteine buffer, pH 7.4, for 1 h at 37°C/5% CO2. After one wash, the tissue was then further digested with 40 U/ml DNase I (Sigma, St. Louis, MO) in Mg2+-containing MEM (Invitrogen) with 1% bovine serum albumin (BSA) (Invitrogen) for 15 min at 37°C/5% CO2. The tissue was then carefully triturated in cold MEM with 1% BSA, centrifuged over a 90% Percoll gradient (GE Healthcare, Piscataway, NJ) to collect all cells below and including the lipid layer, which then was further diluted five times in MEM with 1% BSA and centrifuged to collect the pellet, which normally included five million viable cells per brain used. The cells were then resuspended in 2% normal donkey serum (Jackson ImmunoResearch, West Grove, PA) before labeling with rabbit GLT1 antibody (1:150, custom made; Invitrogen) and secondary donkey anti-rabbit R-phycoerythrin (R-PE) (1:200; Jackson ImmunoResearch). Cells were resuspended in cold MEM with 1% BSA and 4 μg ml−1 propidium iodide (PI) (Sigma) and immediately sorted by FACS. Usually 150,000 astrocytes (GLT1+/Gfap-GFP+) were recovered after FACS.
FACS.
Cells were sorted using either the BD FACSVantage Cell Sorting System (13 psi sheath pressure, Cell Quest software) or the BD FACSAria Cell Sorting System (35 psi sheath pressure, FACSDiva software; BD Biosciences, San Jose, CA). GFP/YFP, R-PE, and PI were all excited by a 488 nm laser, and emissions were collected by 530/30 nm, 575/26 nm, and 675/20 nm discrimination filters, respectively. The signals were manually compensated, and cells were sorted into cold MEM with 1% BSA. Data were analyzed by FlowJo (Ashland, OR).
RNA processing, microarray, and quantitative PCR.
After FACS, cells were immediately extracted for total RNA and DNase treated using the RNAqueous Micro kit (Ambion, Austin, TX). RNA quantity was assessed using the NanoDrop-1000 (NanoDrop Technologies, Wilmington, DE), and RNA integrity was assessed using the 2100-Bioanalyzer (Agilent Technologies). For microarray, 20 ng of total RNA was amplified and labeled with biotin using the Ovation kit (NuGEN, San Carlos, CA) according to the manufacturer's instruction and hybridized to the Affymetrix (Santa Clara, CA) GeneChip Mouse Genome 430 2.0 Array. For quantitative PCR (qPCR), total RNA was reverse transcribed using oligo random hexamer primers and Taqman Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). Relative quantity of transcripts was assessed using Taqman Assays on Demand and the 7000 Sequence Detection System (Applied Biosystems). 18S served as an internal control that all samples were normalized to before calculating relative expression.
Metabolic labeling studies.
Ten- to 12-week-old C57 black mice (Charles River, Wilmington, MA) were anesthetized using Forane (Baxter, Deerfield, IL) and perfused with Hanks buffer, and cortices were dissociated and labeled with rabbit GLT1 (rbGLT1) antibody as described above. Cells were labeled with secondary rat anti-rabbit magnetic bead-conjugated antibody and purified on magnetic-activated cell sorting (MACS) columns according to the manufacture's instructions (Miltenyi Biotec, Auburn, CA). MACS-separated cells were plated onto tissue culture plates in freshly prepared and CO2-equilibrated D3050 MEM (Sigma) supplemented with 44 mm Na2CO3 and 2 mm U-13C6-d-glucose, pH 7.4 (Cambridge Isotopes, Andover, MA). Cells were incubated at 37°C/5%CO2 for 1 h. Then, the medium was collected, and metabolic substrates were extracted from the cells in 70% ethanol.
Biochemical analysis.
Lyophilized cell extracts and media were dissolved in HCl, adjusted to pH <2, and dried under atmospheric air. The metabolites were extracted into an organic phase of ethanol and benzene and dried again under atmospheric air before derivatization with MTBSTFA [N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide] plus 1% t-BDMS-Cl (tert-butyldimethylchlorosilane) (Regis Technologies, Morton Grove, IL) (Mawhinney et al., 1986a,b). The samples were analyzed on a Hewlett Packard (Palo Alto, CA) 5890 Series II gas chromatograph linked to a Hewlett Packard 5972 Series mass spectrometer. Peaks from gas chromatography (GS)/MS spectra were integrated, and atom percentage excess values were calculated by comparison with unlabeled standard solutions. Atom percentage excess (13C) of aspartate, malate, and citrate was determined after allowing for naturally abundant 13C and silicon (from silyl groups) as described by Biemann (1962).
Immunochemistry.
Dissociated cells plated on glass coverslips were fixed with 4% paraformaldehyde, labeled with mouse anti-Aquaporin 4 (AQP4; 1:400; Santa Cruz Biotechnology, Santa Cruz, CA) and/or guinea pig anti-GLT1 (1:4000; Chemicon, Temecula, CA), and then labeled with Cy3-, Cy5-, or FITC-conjugated donkey secondary antibodies (1:250; Jackson Immunoresearch). For Thy1-YFP+-sorted populations, cells were labeled for microtubule-associated protein 2 (MAP2) instead using mouse anti-MAP2 (1:500; Chemicon). For immunohistochemistry, either coronal 14-μm-thick cryostat sections or 100 μm vibratome sections were prepared from 10- to 12-week-old mice as described previously (Simard et al., 2003). Antibodies against rabbit anti-GLT1 (1:500, custom made), chicken anti-GFAP (1:500; Chemicon), and guinea pig anti-GLT1 (1:5000; Chemicon) were used. Secondary antibodies were from donkey and conjugated with Cy3, Cy5, or FITC (Jackson Immunoresearch). Sections were visualized using either a fluorescence microscope (IX70; Olympus, Tokyo, Japan) or a confocal microscope (FV500; Olympus). For immuno-electron microscopy, sections were prepared from 8- to 12-week-old Gfap-GFP mice as described previously (Goldman and Nedergaard, 1992). Images were collected with Hitachi (Tokyo, Japan) 7100 electron microscope and analyzed with ImageJ.
Western blotting.
For Western blotting, 5 μg protein samples from homogenized adult mouse cortex were run on a 10% acrylamide/bisacrylamide gel, transferred to a polyvinylidene difluoride membrane, blocked in 5% milk, and incubated with rabbit anti-GLT1 (1:250, custom made) and secondary anti-rabbit HRP-conjugated (1:20,000; GE Healthcare) antibodies. Blots were visualized using the ECL Chemiluminescence System (GE Healthcare).
Microarray data mining and statistical analysis.
Microarray data were analyzed using the Arrayassist 5.0.0 software package (Stratagene, La Jolla, CA). We normalized the data using the MAS5 algorithm, followed by log2 transformation and filtering out any probe set that had only one present call across all samples or a maximum intensity value <100 across all samples that reduced the initial data set of 45,101 probe sets to 19,369 probe sets. To assure reproducibility among independent biological replicates in the genomic data set, we compared the correlation coefficient within groups of sorted cells (19,369 probe sets). The degree of similarity across all samples was assessed by hierarchical clustering using Euclidean average distances. All statistical comparisons between groups used the nonpaired parametric Student's t test using the Benjamini-Hochberg false discovery rate (FDR) correction algorithm of 5%. Only corrected p values are shown in this study. For selection of “astrocyte-enriched genes,” we subtracted the profiles of GLT1+/Gfap-GFP+ cells from that of GLT1−/Gfap-GFP− and Thy1-YFP+, respectively [corrected, p < 0.05; fold change (FC) > 2]. Then, the intersection (924 probe sets) of these two independent subtractions was extracted and compared with the GLT1+/Gfap-GFP− group, which did not differ significantly. The 924 probe sets were designated astrocyte-enriched genes. Because of many incorrect annotations of the Affymetrix probe sets, we manually annotated all probe sets used in Results and have only included probe sets that bind at the same chromosomal location as the target gene as well as being annotated as the correct gene in the Ensembl database (www.ensembl.org). Metabolic pathways were mapped using the KEGG encyclopedia (www.genome.jp/KEGG). Statistical testing of other data than microarray was performed using Excel. The specific test is indicated in figure legend.
Results
GLT1 identifies two populations of astrocytes based on Gfap-GFP expression
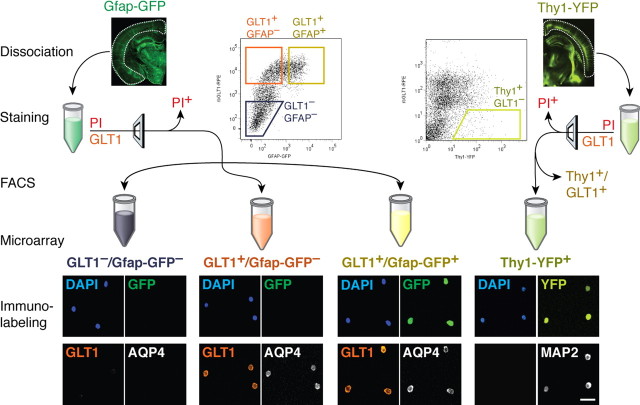
Astrocytes are traditionally recognized by their expression of GFAP. However, GFAP is expressed to a variable degree in the adult mouse brain (Wang and Walz, 2003) diminishing its utility as a marker to identify all astrocytes in an unbiased manner. To identify astrocytes in situ to assess their gene expression, we thus used the glial-specific glutamate transporter GLT1 (Regan et al., 2007) as a marker in addition to GFAP (Fig. 1). To label cells expressing GLT1, we generated an antibody directed against its extracellular loop (Fig. 2a–d), whereas labeling of cells expressing GFAP was achieved using the transgenic Gfap-driven GFP mouse (Zhuo et al., 1997) (Fig. 2f–m). By using these overlapping markers of the astrocytic phenotype, we applied FACS immediately after dissociation of adult mouse cortices, to isolate a population of cortical astrocytes, defined as GLT1+/Gfap-GFP+ (Fig. 1). We also identified a rather large pool of GLT1+ but Gfap-GFP− cells, defined as GLT1+/Gfap-GFP−, which potentially could be a subpopulation of astrocytes with low or absent GFAP expression (Fig. 1). Additionally, we collected a population of cells negative for both GLT1 and Gfap-GFP, defined as GLT1−/Gfap-GFP−, which contained mostly endothelial cells, neurons, oligodendrocytes, smooth muscle cells, and microglia.
Figure 1.
Schematic outline of FACS procedure and microarray analysis. Left, Gfap-GFP-positive cortices (left image) were dissociated, immunolabeled with rbGLT1-PE, and purified by FACS into three populations: GLT1+/Gfap-GFP+, GLT1+/Gfap-GFP−, and GLT1−/Gfap-GFP− based on sequentially gating of size (FSC-SSC plot), viability (PI), and Gfap-GFP and rbGLT1-PE fluorescence. Right, Thy1-YFP-positive cortices (right image) were dissociated and immunolabeled with rbGLT1-PE, and GLT1−/Thy-YFP+ neurons were purified by FACS using a similar approach as above. After FACS, RNA was extracted from all the purified populations for microarray analysis. Bottom, Small aliquots of the FACS-purified populations were assayed for purity and specificity by immunocytochemistry. Cells were immunolabeled with guinea pig anti-GLT1 (red) and mouse anti-AQP4 (white) to confirm the astrocytic phenotype, whereas all nuclei labeled with DAPI (4′,6-diamidino-2-phenylindole) (blue). Only the GLT1+/Gfap-GFP+ population was GFP+ (green). The GLT1+/Gfap-GFP+ population had 97.3 ± 0.3% GLT1+ cells, the GLT1+/Gfap-GFP− population had 96.0 ± 1.6% GLT1+ cells, and the GLT1−/Gfap-GFP− population had 1.0 ± 0.4% GLT1+ cells (n = 3 biological replicates; mean ± SEM). GLT1−/Thy1-YFP+-purified cells were immunolabeled with mouse anti-MAP2 (white) and 97 ± 4% labeled with MAP2, confirming the neuronal phenotype.
Figure 2.
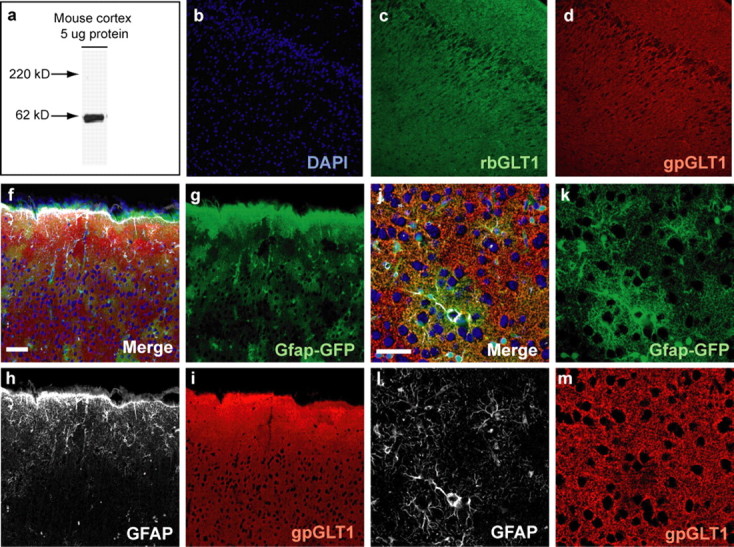
Validation of custom-made rabbit anti-GLT1 antibody. A, Western blot analysis of the rabbit anti-GLT1 antibody (1:250) was performed on 5 μg of protein extracted from adult mouse cortex. Only one band of the expected size of ∼62 kDa was observed in the lane. B–D, Custom-made rabbit anti-GLT1 antibody stained in an identical manner as commercial guinea pig anti-GLT1 antibody. Cryosections of perfusion-fixed adult mouse brain were used for immunohistochemistry. Sections were incubated with rabbit anti-GLT1and guinea pig anti-GLT1 for double immunolabeling, followed by donkey anti-rabbit FITC and anti-guinea pig Cy3 labeling. Sections were visualized using a fluorescence microscope (IX70; Olympus). F–I, GFAP, Gfap-GFP, and GLT1 coexpression patterns; all GFAP-defined astrocytes were GFP and GLT1 positive, whereas only some GLT1-positive astrocytes were GFAP or GFP positive. Vibratome sections from perfusion-fixed adult mouse GFAP–GFP brain was used for immunohistochemistry. Sections were incubated with mouse anti-GFAP and guinea pig anti-GLT1 followed by anti-guinea pig Cy3 and anti-mouse Cy5 labeling. Sections were visualized using a confocal microscope (FV500; Olympus). Scale bars: F, 50 μm; J, 40 μm. DAPI, 4′,6-diamidino-2-phenylindole.
Some reports have suggested that a minor fraction of Gfap-GFP+ cells are NG2+ (Cspg4) progenitor cells (Lin et al., 2005; Paukert and Bergles, 2006). We thus tested for the presence of progenitor cells using a NG2 antibody on dissociated cells from adult Gfap-GFP+ cortices. We found with flow cytometry that 0.03% of the Gfap-GFP+ cells colabeled with NG2. Similarly, immunolabeling of postsorted, unstained Gfap-GFP+ cells demonstrated that 0.4% labeled for NG2. These data eliminate a possible contamination of NG2+ progenitor cells in the astrocytic pool and demonstrate the major population of NG2+ cells was Gfap-GFP− (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
To validate the purity of the sorted populations, we immunolabeled both for GLT1, using a second antibody that targeted the intracellular portion of GLT1, and for the prototypic astrocyte protein, AQP4 (Nielsen et al., 1997) (Fig. 1). The initial unsorted dissociate had 45 ± 5% GLT1+ and 47 ± 6% AQP4+ cells (n = 3 biological replicates; mean ± SEM) in which >99% of the GLT1+ cells colocalized with AQP4. When we applied the same procedure to the sorted populations, we found that the GLT1+/Gfap-GFP+ population contained 97.3 ± 0.3% GLT1+ cells, whereas the GLT1+/Gfap-GFP− population had 96.0 ± 1.6% GLT1+ cells (n = 3 biological replicates; mean ± SEM) demonstrating that we efficiently had enriched two subpopulations of astrocytes. In contrast, the GLT1+-depleted cell population (GLT1−/Gfap-GFP−) retained only 1.0 ± 0.4% GLT1+ cells (n = 3 biological replicates; mean ± SEM).
To purify neuronal RNA for genomic comparison, we isolated Thy1-driven, YFP-expressing neurons from adult mouse cortex (Feng et al., 2000) using FACS (Fig. 1). Because astrocytes also express Thy1 at both the protein and transcript level, albeit at low levels, we eliminated all GLT1+ astrocytes from the neuronal Thy1+ pool. To validate the neuronal phenotype of the GLT1−/Thy1-YFP+ purified cells, we immunolabeled postsorted cells with MAP2 antibody, a neuronal marker, and found that 97 ± 4% expressed MAP2.
After isolation, we immediately extracted total RNA from the enriched pools, which we used for genomic expression profiling using Affymetrix mouse genome expression array and for validation of selected probes using qPCR. To demonstrate enrichment of astrocytic markers at the transcript level in the postsorted populations, we validated the expression of Aqp4 using qPCR. As expected, we found Aqp4 expression significantly enriched 364-fold in the GLT1+/Gfap-GFP+ astrocytes compared with the GLT1−/Gfap-GFP− cells (p = 0.0011). In contrast, Aqp4 was depleted from the Thy1+ neuronal pool 236-fold compared with the GLT1+/Gfap-GFP+ astrocytes (p < 0.0001) (Fig. 3) (n = 3 biological replicates; paired two-sided Student's t test). However, Aqp4 was only enriched twofold in GLT1+/Gfap-GFP+ astrocytes compared with GLT1+/Gfap-GFP− (p = 0.069), demonstrating that these cells also express high levels of astrocyte markers at the transcript level. By this means, we used FACS to enrich a population of GLT1+/Gfap-GFP+ astrocytes, a population of GLT1+/Gfap-GFP− astrocytes, a pool of Thy1+-defined neurons, and an astrocyte-depleted GLT1−/Gfap-GFP− population.
Figure 3.
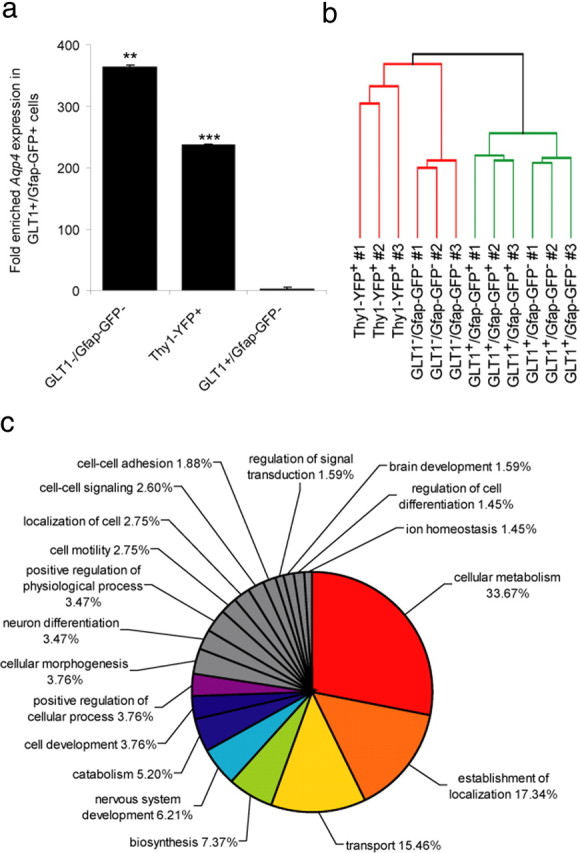
Generation of an astrocyte-enriched gene expression set. a, Reverse transcription qPCR on purified RNA showed that Aqp4 expression was enriched 364-fold in the GLT1+/Gfap-GFP+ group compared with the GLT1−/Gfap-GFP− group (**p = 0.0011) and 236-fold compared with the Thy1+ neuron group (***p < 0.0001); however, Aqp4 was only twofold higher in GLT1+/Gfap-GFP+ astrocytes compared with GLT1+/Gfap-GFP− astrocytes (p = 0.069, paired Student's t test; mean ± SEM). b, Similarity of groups was compared using MAS5 normalized and filtered data (19,369 probe sets) by hierarchical clustering of the Euclidean average distances (n = 3 biological samples per group). The GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− groups were the two most similar. c, Categorizing GO terms of the biological functions associated with the astrocyte-enriched genes (924 probe sets) were analyzed in DAVID 2007 (http://david.abcc.ncifcrf.gov/), and the relative proportions were presented in a pie chart (functional annotation threshold; count >10; p < 0.05; FDR cutoff 5%). Three genes from each of the five major and least similar GO term groups are presented in Table 2.
The GFAP phenotype is associated with subtle differences in astrocytic expression profile
We next examined the gene expression of the GFAP+ and GLT1+ enriched populations using microarray. To assess the degree of similarity between samples, we used hierarchical clustering (Eisen et al., 1998), which confirmed that each sample clustered to its anticipated group (Fig. 3). The GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− groups were the two most similar groups, reflecting the molecular similarity of GLT1+/GFAP+ and GLT1+/GFAP− astrocytic subpopulations. Each astrocytic population differed substantially from the astrocyte-depleted (GLT1−/Gfap-GFP−) and neuronal (Thy1+)-enriched populations (Fig. 3) (Euclidean average distance). Given the strong similarity between the two astrocytic subgroups, we next asked how many genes differed in their expression patterns between the four groups.
From a normalized and filtered set of genes (19,369 probe sets), only 0.96% of the genes were differentially expressed between the GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− populations (Table 1). A similar analysis of the GLT1−/Gfap-GFP− and Thy1+ groups showed they differed from the GLT1+/Gfap-GFP+ group by 31 and 14%, respectively (Table 1). The high degree of similarity between the GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− groups could not have been attributable to a large variance within one or both groups because the correlation coefficient within groups was similar among all groups (Table 1). Finally, we also expressed the expression of cell death and stress response-related genes using Gene Ontology (Lobo et al., 2006), and found that “cell death”- and “stress response”-categorized genes contributed to <1% of the total number of expressed genes in all cell types (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). In addition, hypoxia-related genes did not reach significance of overrepresentation in a Gene Ontology functional annotation analysis.
Table 1.
Microarray comparison of FACS-purified groups
| Cell type | GLT1+/Gfap-GFP+ | GLT1+/Gfap-GFP− | GLT1−/Gfap-GFP− | Thy1-YFP+ |
|---|---|---|---|---|
| Percentage of significant genesa compared with GLT1+/Gfap-GFP+ | 0.96% (186/19,369) | 31% (6000/19,369) | 14% (2670/19,369) | |
| Correlation within groups (n = 3)b | 0.98 | 0.98 | 0.99 | 0.94 |
| Percentage of genes expressed within each groupc | 48.8% (9452/19,369) | 48.1% (9313/19,369) | 50.4% (9759/19,369) | 32.7% (6340/19,369) |
| Percentage of significant metabolism-related genesd compared with GLT1+/Gfap-GFP+ | 0% (0/87) | 58.0% (51/87) | 43.2% (38/87) |
aStudent's t test, Benjamini-Hochberg FDR correction of 5%, and fold difference > 2.
bThe correlation coefficient within the indicated group.
cPercentage of genes with present calls in all samples within the same group.
dPercentage of 87 metabolism-related genes presented in this study that were significantly different between the GLT1+/Gfap-GFP+ group and the indicated group.
We next generated a list of astrocyte-enriched genes by subtracting the profiles of both Thy1-defined neurons (n = 3) and the GLT1−/Gfap-GFP− cells (n = 3) from those of GLT1+/Gfap-GFP+ astrocytes (n = 3), additionally excluding their intersection of 924 probe sets. The 924 probe sets did not significantly differ between the GLT1+/Gfap-GFP− and GLT1+/Gfap-GFP+ populations (Student's t test; Benjamini-Hochberg 5% FDR correction; FC > 2; p < 0.05; n = 3 samples per group), suggesting that the 186 differentially expressed genes (186 probe sets of 19,369; 0.96%) found between the GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− populations are not astrocyte specific. We therefore pooled the GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− populations, defined as GLT1+ astrocytes, to study astrocyte-enriched genes, because they had shown similarity in all aspects as well as expressing a similar profile with respect to cell-type markers (supplemental Table 1, available at www.jneurosci.org as supplemental material). On this list, we found many genes already known to be astrocyte specific (Table 2), such as AQP4 (Aqp4), Connexin 30 (Gja1), Connexin 43 (Gjb6), and GLT1 (Slc1a2) (Nielsen et al., 1997; Regan et al., 2007).
Table 2.
GLT1+ astrocyte-enriched genes
| Gene symbol | Gene title | Entrez gene | FD | Corr. p value |
|---|---|---|---|---|
| Known astrocyte-specific genes | ||||
| Aqp4 | Aquaporin 4 | 11829 | 72 | 0.00006 |
| Gja1 | Gap junction membrane channel protein α1 (Connexin 30) | 14609 | 9 | 0.00009 |
| Gjb6 | Gap junction membrane channel protein β6 (Connexin 43) | 14623 | 22 | 0.00002 |
| S100b | S100 protein, β polypeptide, neural | 20203 | 32 | 0.00001 |
| Slc1a2 | Solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 20511 | 159 | 0.00022 |
| Cellular metabolism | ||||
| Cbs | Cystathionine β-synthase | 12411 | 361 | 0.00001 |
| Ppp1r3c | Protein phosphatase 1, regulatory subunit 3C (PTG) | 53412 | 9 | 0.00076 |
| Plcd4 | Phospholipase C, δ4 | 18802 | 84 | 0.00018 |
| Transport | ||||
| Slc14a1 | Solute carrier family 14 (urea transporter), member 1 | 108052 | 39 | 0.00036 |
| Slc4a4 | Solute carrier family 4 (anion exchanger), member 4 | 54403 | 88 | 0.00003 |
| Slc6a11 | Solute carrier family 6 (GABA transporter), member 11 (GAT4) | 243616 | 19 | 0.00007 |
| Establishment of localization | ||||
| Mlc1 | Megalencephalic leukoencephalopathy 1 | 170790 | 75 | 0.00012 |
| Ttpa | Tocopherol (α) transfer protein | 50500 | 65 | 0.00011 |
| Ednrb | Endothelin receptor type B | 13618 | 29 | 0.00002 |
| Nervous system development | ||||
| Hes5 | Hairy and enhancer of split 5 | 15208 | 19 | 0.00009 |
| Fgfr3 | Fibroblast growth factor receptor 3 | 14184 | 23 | 0.00002 |
| Tagln3 | Transgelin 3 | 56370 | 7 | 0.00001 |
| Positive regulation of cellular process | ||||
| Sox9 | SRY-box containing gene 9 | 20682 | 17 | 0.00003 |
| Tlr3 | Toll-like receptor 3 | 142980 | 3 | 0.00337 |
| Egfr | Epidermal growth factor receptor | 13649 | 52 | 0.00004 |
From an initial set of genes (19,369 probe sets), the GLT1+/Gfap-GFP+ astrocytic profile was compared with the Thy1-YFP+ neuronal profile and the GLT1−/Gfap-GFP− cell profile, respectively. The intersection (924 probe sets) of these two independent comparisons was extracted and called astrocyte-enriched genes. There was no statistical significant difference in any of the astrocyte-enriched genes between the GLT1+/Gfap-GFP+ and the GLT1+/Gfap-GFP− astrocytic profiles (nonpaired Student's t test; Benjamini-Hochberg FDR correction of 5%). FD, Fold difference between GLT1+/Gfap-GFP−/+ astrocytes (n = 6) versus GLT1−/Gfap-GFP− cells (n = 3); Corr. p value, Nonpaired Student's t test, Benjamini-Hochberg FDR correction of 5%.
Astrocytes express genes with biological functions in metabolism, development, and transport
To better understand the overall function of the astrocyte-enriched genes, we used Gene Ontology to categorize the role of each differentially expressed gene. We narrowed the list of 924 probe sets to show only the three most interesting or important genes from five major nonoverlapping categories to represent a summary of the overall data (Fig. 3, Table 2). We found that 33% of the differentially expressed genes were involved in cellular metabolism, 7.8% in biosynthesis, and 5.2% in catabolism, indicating that astrocytes are active participants in energy homeostasis (Pellerin and Magistretti, 2004; Koehler et al., 2006; Hertz et al., 2007). Many of these metabolism genes were involved in glucose metabolism (e.g., Ppp1r3c), but astrocytes also expressed genes involved in amino acid and fatty acid metabolism (e.g., Cbs). Another large proportion of genes were involved in patterning and localization (17.3%), supporting the notion that astrocytes are important for establishing a regionally specified domain within the brain. There was also a large proportion of genes related to transport (15.5%), consistent with the role of astrocytes in blood–brain barrier function. Many additional genes associated with phenotypic development and cell-fate commitment. Together, these findings add new insight into the functional roles of astrocytes in the cellular homeostasis of the adult CNS.
Because one major functional role of astrocytes is participation in energy homeostasis, we wanted to more specifically study the compartmentalization of metabolic pathways between astrocytes and neurons in situ. Using the astrocytic and neuronal expression profiles, we first asked what proportion of metabolic genes of interest were differentially expressed between the GLT1+/Gfap-GFP+ group and any of the other groups. The Thy1+ and GLT1− groups had 58.0 and 43.2% significantly expressed metabolism-related genes, respectively (Table 1). Interestingly, the GLT1+/Gfap-GFP+ and GLT1+/Gfap-GFP− groups exhibited no significant difference in any metabolism-related genes, suggesting their metabolism is identical. To simplify the study, we therefore studied metabolism between GLT1+ astrocytes and Thy1+ neurons.
Most glycolytic enzymes are enriched in astrocytes
Glucose is the main energy substrate of the adult brain. It can be metabolized either via glycolysis to pyruvate for entry into the TCA cycle, via the pentose phosphate pathway to generate sugars for nucleotides and reducing equivalents, or stored as glycogen (Berg et al., 2002). Yet, although it is well established that astrocytes have glycolytic and glycogenic metabolism (Dringen et al., 1993; Wender et al., 2000), the existence and extent of oxidative metabolism in astrocytes has remained controversial (Aubert et al., 2007; Hertz et al., 2007). To assess the infrastructural competence for oxidative metabolism in astrocytes, we assessed the relative expression between astrocytes and neurons of all genes within the metabolic pathways for glucose. In particular, we examined the major pathways of glucose oxidation including glycogen and lactate metabolism, anaplerotic pathways, and the glutamate–glutamine cycle. For all comparisons of gene expression between groups, we used three biological replicates, analyzed by unpaired two-sided Student's t tests with a Benjamini-Hochberg correction (α = 5%).
We found that, relative to neurons, astrocytes expressed enriched levels of all glycogen-metabolizing enzymatic steps, confirming previous findings and supporting the efficient separation of these phenotypes (Pfeiffer et al., 1992; Wender et al., 2000; Brown et al., 2003) (Fig. 4a and supplemental Table 2, available at www.jneurosci.org as supplemental material). We next examined the glycolytic pathway and found that both astrocytes and neurons expressed all the necessary enzymes to perform glycolysis (Fig. 4b and supplemental Table 2, available at www.jneurosci.org as supplemental material). The transcript for neuronal-specific Enolase 2, Eno2, was significantly enriched in neurons (2.2-fold; p = 0.011), whereas Eno1 was significantly enriched in astrocytes (50.2-fold; p = 0.011) (Schmechel et al., 1978). When we compared the relative expression of all glycolytic enzymes from glucose phosphorylation to pyruvate synthesis, 7 of the 10 enzymatic steps exhibited significantly enriched expression in astrocytes relative to neurons, consistent with the high capacity of astrocytes to metabolize glucose.
Figure 4.
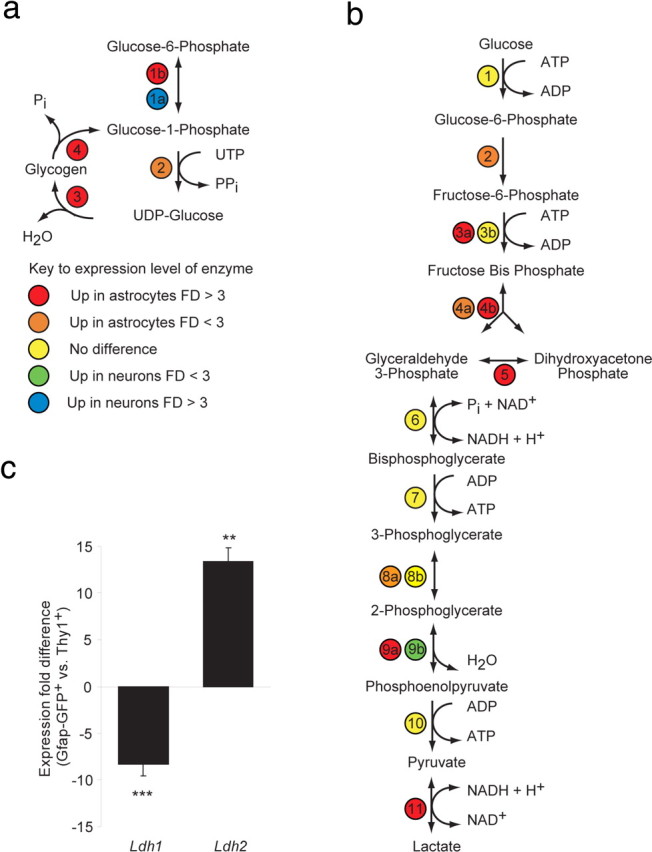
Expression of glycogenic and glycolytic enzymes is enhanced in astrocytes. a, b, Microarray analysis of the expression fold difference in glycogenic and glycolytic enzymes between GLT1+/Gfap-GFP−/+ astrocytes (n = 6) and Thy1+ neurons (n = 3). The number- and color-coded circles indicate an enzyme (number) and the expression level (color) of that specific enzyme. The coding is indicated in the key; enzymes labeled “No difference” were not significantly different between astrocytes and neurons. “Up in astrocytes” and “Up in neurons” indicate a significantly increased fold difference (FD) in the gene expression in astrocytes relative to neurons and in neurons relative to astrocytes, respectively (unpaired two-sided Student's t test, corrected p < 0.05). a, Key to enzymes in the glycogen synthesis and breakdown pathways: 1a, Pgm2l1 and Pgm5; 1b, Pgm2; 2, Ugp2; 3, Gys1; 4, Pygb, Pygl, and Pygm. b, Key to enzymes in the glycolysis pathway: 1, Hk1 and Hk2; 2, Gpi1; 3a, Pfkm and Pfkp; 3b, Pfkl; 4a, Aldoa; 4b, Aldoc; 5, Tpi; 6, G6pdx; 7, Pgk1; 8a, Pgam1; 8b, Bpgm; 9a, Eno1; 9b, Eno2; 10, Pkm2; 11, Ldhb. c, The histograms show the expression fold difference of Ldha and Ldhb assessed by qPCR between Gfap-GFP+ astrocytes and Thy1+ neurons. **p < 0.01; ***p < 0.001. n = 3 biological independent, unpaired two-sided Student's t test; mean ± SEM.
Astrocytes produce lactate directly from glycolysis and from TCA cycle intermediates
Pyruvate is the end-product of glycolysis, at which point it is either converted to lactate in the cytosol or enters the mitochondria for catabolism via the TCA cycle to either acetyl CoA or oxaloacetate. Whereas the conversion of pyruvate to lactate is catalyzed principally by lactate dehydrogenase b (Ldhb), the reverse reaction is mediated principally by Ldha. These two reactions play a major role in the astrocyte–neuron–lactate shuttle hypothesis by Magistretti et al., (1999), which proposes that astrocytic and neuronal lactate metabolism is compartmentalized by the complementary expression of Ldha and Ldhb in neurons and astrocytes, respectively (O'Brien et al., 2007). We therefore assessed the relative expression patterns of Ldha and Ldhb by astrocytes and neurons. We first noted that Ldhb was enriched >13-fold in astrocytes (p < 0.001) relative to neurons. qPCR was next used to address this question, because Ldha is not uniquely represented on the Affymetrix 430 2.0 chip. qPCR revealed that Ldha was enriched 8.3 ± 1.2-fold in neurons relative to astrocytes (p < 0.001), whereas Ldhb was enriched 13.4 ± 1.4-fold in astrocytes relative to neurons (p = 0.003) (Fig. 4c). These data suggest that astrocytes preferentially synthesize lactate from pyruvate, whereas neurons preferentially generate pyruvate from lactate, suggesting a compartmentalized relationship of lactate metabolism between astrocytes and neurons.
To validate lactate production of and provision by astrocytes, we exposed acutely MACS-isolated neocortical GLT1+ astrocytes to [U-13C]glucose, so as to monitor the appearance of glucose-derived metabolites in both the cells and their media, using MS. Because these studies require a large number of cells to generate enough material for MS analysis, we were limited to study astrocytes. The viability (90 ± 3% viable cells) and purity (94 ± 3% GLT1+ cells) of each astrocytic isolate was established by flow cytometry. Mass spectroscopy confirmed the generation of 13C-labeled lactate, consistent with their enriched levels of Ldhb (Table 3). Lactate was found primarily in the cell media. Here, 61.1 ± 3.4% of all lactate was M+3 labeled, indicating that lactate is produced directly from glycolysis and immediately released from astrocytes (M + X indicates the molecular weight of the metabolite in which X atoms are 13C atoms). Surprisingly, we also found M+1- and M+2-labeled lactate species, which could only be derived from TCA cycle precursors (supplemental Fig. 3c,d, available at www.jneurosci.org as supplemental material). These results suggest that astrocytes do not metabolize all glucose exclusively to lactate but, in addition, also exhibit oxidative metabolism of glucose. To confirm this observation, we next asked whether TCA cycle enzymes were expressed and functional in astrocytes.
Table 3.
Percentage enrichment in cell extracts and medium from MACS purified GLT1+ astrocytes incubated with d-[U-13C]glucose
| M+1 | M+2 | M+3 | |
|---|---|---|---|
| Medium | |||
| Lactate | 0.3 ± 0.15 | 2.7 ± 0.3 | 61.1 ± 3.4a |
| Cell extract | |||
| Glutamate | 8.8 ± 1.0 | 5.1 ± 1.6 | 6.5 ± 0.5 |
| Aspartate | 16 ± 4 | 1.1 ± 0.5 | NE |
| Malate | NE | 2.5 ± 0.8 | 1.3 ± 0.8 |
| Citrate | NE | NE | 2.4 ± 1.1 |
Medium and cell extracts were analyzed for GC/MS, except for glutamate, which was analyzed by liquid chromatography/MS because of peak overlap in the GC/MS for this metabolite. M+x is the molecular weight of the metabolite plus x number of 13C atoms incorporated. NE, Not enriched. n = 5 for lactate, malate, citrate, and aspartate; n = 3 for glutamate; mean ± SEM.
aDirectly from glycolysis.
Astrocytes have oxidative metabolism of glucose and active TCA cycling
Besides lactate synthesis, the main route of pyruvate metabolism is oxidation in the mitochondria via the TCA cycle. Entry of pyruvate into the TCA cycle can start with oxidative decarboxylation to form acetyl-CoA, which reacts with oxaloacetate to form citrate (Fig. 5a). Through one turn of the TCA cycle, citrate is oxidized to oxaloacetate eliminating two carbon atoms as CO2. When acetyl-CoA contains 13C, labeled glutamate and aspartate can be derived by the exit of labeled TCA cycle intermediates (supplemental Table 3a, available at www.jneurosci.org as supplemental material). However, if the labeled TCA cycle intermediates do not exit the cycle, labeled oxaloacetate can participate in additional turns incorporating more labeled acetyl-CoA and contributing to distinct labeling patterns of TCA cycle intermediates and amino acid derivatives. An active TCA cycle has been demonstrated repeatedly in cultured astrocytes and neurons, but it is still debated whether this is the case in astrocytes in vivo (Pellerin and Magistretti, 2004; Schurr, 2006). Thus, to confirm the existence of oxidative metabolism in astrocytes in situ, we next asked to what extent astrocytes have TCA cycle enzymes relative to neurons. Astrocytes express all the necessary enzymes for a functioning TCA cycle (Fig. 5a and supplemental Table 2, available at www.jneurosci.org as supplemental material). Surprisingly, in all TCA cycle enzymatic steps but one, the relative expression was significantly higher in astrocytes than neurons.
Figure 5.
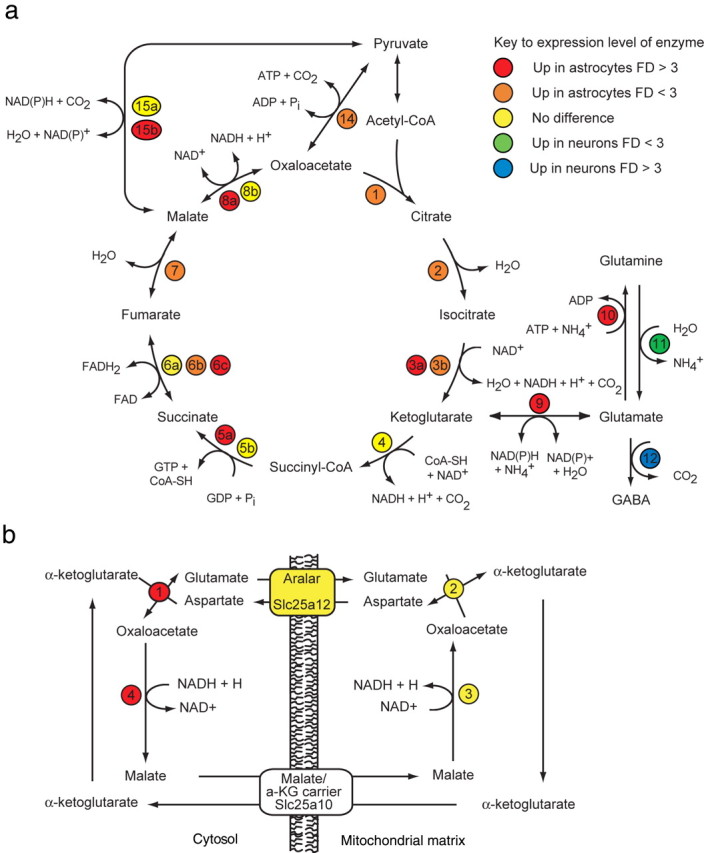
Microarray analysis of the expression fold difference in TCA cycle and aspartate–malate shuttle enzymes between GLT1+/Gfap-GFP−/+ astrocytes (n = 6) and Thy1+ neurons (n = 3). The number- and color-coded circles indicate an enzyme (number) and the expression level (color) of that specific enzyme. The coding is indicated in the key; enzymes labeled “No difference” were not significantly different between astrocytes and neurons. “Up in astrocytes” and “Up in neurons” indicate a significantly increased fold difference (FD) in the gene expression in astrocytes relative to neurons and in neurons relative to astrocytes, respectively (unpaired two-sided Student's t test, corrected p < 0.05). a, Key to TCA cycle enzymes: 1, Cs; 2, Aco1 and Aco2; 3a, Idh3b and Idh3g; 3b, Idh3a; 4, Ogdh and Dlst; 5a, Suclg1 and Suclg2; 5b, Sucla2; 6a, Sdha and Sdhb; 6b, Sdhc; 6c, Sdhd; 7, Fh1; 8a, Mdh1; 8b, Mdh2; 9, Glud1; 10, Glul; 11, Gls; 12, Gad1; 14, Pcx; 15a, Me2; 15b, Mod1. b, Key to NADH shuttle pathway: 1, Got1; 2, Got2; 3, Mdh1; 4, Mdh2.
To further confirm the existence of oxidative metabolism in astrocytes, we next asked whether TCA cycle activities could be observed by incubating acutely isolated astrocytes in medium containing [U-13C]glucose and tracing the labeled intermediates as described above. We found that TCA cycle pathways were functionally active because labeling from [U-13C]glucose could be found in TCA cycle intermediates, such as citrate and malate, as well as the amino acids glutamate and aspartate (Table 3). More specifically, the labeling pattern of citrate, malate, aspartate, and glutamate indicated that their precursors had cycled multiple turns in the TCA cycle (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). This observation clearly demonstrates oxidative metabolism of glucose in astrocytes.
As noted, we found M+1- and M+2-labeled lactate (Table 3), indicating pyruvate recycling of cycling TCA intermediates (supplemental Fig. 3c,d, available at www.jneurosci.org as supplemental material), which is supported by specific expression of cytosolic malic enzyme (Mod1) in astrocytes (Fig. 5a and supplemental Table 2, available at www.jneurosci.org as supplemental material). Anaplerosis is an alternative entry into the TCA cycle that involves carboxylation of pyruvate to oxaloacetate catalyzed by pyruvate carboxylase (Pcx). In the brain, anaplerosis is only performed by Pcx, which is specific to astrocytes (Yu et al., 1983; Shank et al., 1985) (Fig. 5a and supplemental Table 2, available at www.jneurosci.org as supplemental material (n = 3 biological replicates). We confirmed that Pcx was specifically expressed by astrocytes, in addition to detection of M+3-labeled citrate that indicates carboxylation of pyruvate (supplemental Fig. 3b, available at www.jneurosci.org as supplemental material) (Waagepetersen et al., 2001).
The labeling pattern of glutamate provides critical information about the degree of cycling and hence the activity of the TCA cycle. Glutamate synthesis from α-ketoglutarate is catalyzed by an aminotransferase or glutamate dehydrogenase; the latter encoded by Glud1 was previously found in astrocytes (Aoki et al., 1987). In line with this, we found Glud1 expression 4.3-fold higher (p = 0.0023) in astrocytes than neurons, suggesting higher astrocytic capacity to metabolize glutamate. When we incubated the acutely isolated GLT1+ astrocytes in medium containing [U-13C]glucose, we found an even enrichment of the M+1-, M+2-, and M+3-labeled glutamate, demonstrating oxidative metabolism of pyruvate with an active TCA cycle in addition to confirming active glutamate synthesis with participation of pyruvate carboxylase (Fig. 5a and supplemental Table 2, available at www.jneurosci.org as supplemental material). As expected, the glutamate–glutamine–GABA cycle involving glutamine synthetase (Glul), glutamate decarboxylase (Gad1), and glutaminase (Gls) appeared highly compartmentalized between astrocytes and neurons, confirming previous studies (Fig. 5a and supplemental Table 2, available at www.jneurosci.org as supplemental material).
Astrocytes express transcripts for NADH/NAD+ shuttle systems
The metabolic fate of pyruvate is directly linked to cellular NADH/NAD+ homeostasis. After glycolysis, cytosolic NADH must be converted back to NAD+ for glycolysis to continue. Cytosolic NADH can be oxidized to NAD+ by the conversion of pyruvate to lactate by Ldhb. However, if pyruvate enters the mitochondria for oxidation an equimolar amount of cytosolic NADH must be reoxidized for glycolysis to continue. Because NADH is impermeable to the mitochondrial membrane, shuttle systems transfer reducing equivalents into the mitochondria without physically moving them over the membrane by using membrane permeable substrates, such as glutamate and malate, as energy carriers. Classically there are described two such shuttle systems, the malate–aspartate shuttle (MAS) and the glycerol–phosphate shuttle (Fitzpatrick et al., 1983; Cheeseman and Clark, 1988; Lai et al., 1989; Berg et al., 2002). The malate–aspartate shuttle has been reported to be active in neurons, whereas the glycerol–phosphate shuttle has been reported as less active in the brain. Studies from cultured neocortical and cerebellar astrocytes have clearly demonstrated that the MAS operated in glia (Waagepetersen et al., 2001).
We therefore examined the expression of components of both the malate–aspartate shuttle (Slc25a12, Slc25a10, Got1, and Got2) and the glycerol–phosphate shuttle (Gpd1 and Gpd2) in astrocytes and neurons (Fig. 5b and supplemental Table 2, available at www.jneurosci.org as supplemental material). Interestingly, Gpd1 and Gpd2 were expressed by both astrocytes and neurons, suggesting that the glycerol–phosphate shuttle is operative in both cell types. Similarly, Got2 and Slc25a12 were expressed by all cell types. In contrast, Got1 was enriched 3.2-fold in astrocytes (p = 0.017). We found Slc25a10 expression under the detection limit by both microarray and qPCR, suggesting that Slc25a10 in adult cortex is scarce. Thus, although the abundance of glycerol–phosphate shuttle components is consistent with astrocytic oxidative metabolism, the different expression levels of components of the malate–aspartate shuttle suggests a difference in neuronal and astrocytic utilization of the malate carrier, especially in regard to the malate-α-ketoglutarate carrier (slc25a10).
Fine astrocytic processes contain an abundance of mitochondria
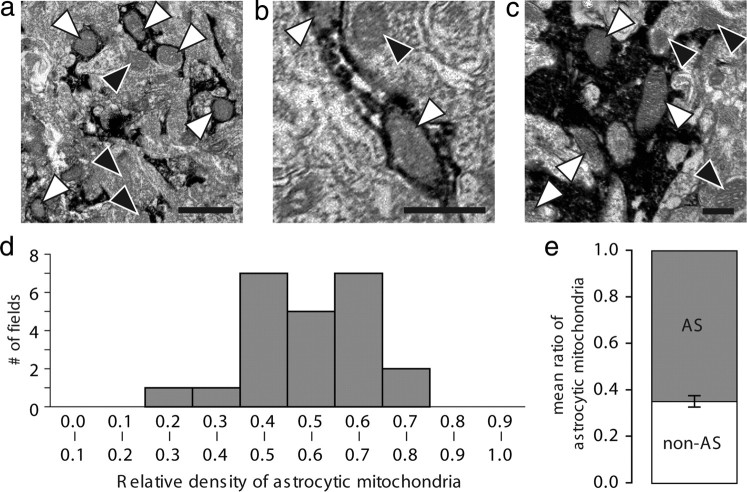
If oxidative metabolism is high in astrocytes, one might expect that astrocytes contain a large number of mitochondria. Several past EM studies have quantified the number of mitochondria in astrocytic and neuronal cell bodies (Pysh and Khan, 1972). However, these studies have not been able to identify the fine processes in astrocytes because of their lack of GFAP fibers or other astrocyte-specific features. To overcome this problem, we used sections prepared from the Gfap-GFP mice and immunostained them against GFP, which distributes throughout the cytosol. The relative number of mitochondria in GFP-positive processes, including fine processes, was compared with the relative number of mitochondria in the remaining part of the field, which would include neuronal processes, synapses, glial, or vascular cells (Fig. 6a–c). Surprisingly, the relative density of mitochondria in GFP-positive astrocytic processes was significantly higher than in the surrounding neuropil (65.5 ± 5.5% mitochondria in GFP-positive processes normalized to relative astrocyte area; n = 18, 3 animals) (Fig. 6d,e). Thus, astrocytic processes are enriched with mitochondria (p < 0.0001, Student's t test). Combined with previous EM studies, this analysis adds strong morphological support to the concept that oxidative metabolism is a prominent part of energy metabolism in protoplasmic astrocytes.
Figure 6.
Electron microscopy images of mitochondria in astrocytic processes. Cortical sections were prepared from GFAP–GFP mice and immunolabeled against GFP. GFP-positive astrocytes throughout the cortical layers were evaluated. a–c, Representative EM images of different sizes in astrocytic processes immunolabeled against GFP. GFP+ astrocytic mitochondria are indicated with white arrowheads, and nonastrocytic mitochondria are indicated with black arrowheads. Note a relative higher number of astrocytes in astrocytic processes compared with surrounding neuropil. Scale bars: a, 1 μm; b, 0.5 μm; c, 0.5 μm. d, Plot of the relative density of astrocytic mitochondria compared the density of mitochondria in the surrounding neuropil. The relative density of mitochondria was defined as number of mitochondria in astrocytic processes (defined by the staining for GFP) versus the relative number of mitochondria in rest of the section (not stained for GFP). The relative density of mitochondria was quantified in a total of 23 sections from three animals. e, Comparison of the mean of the density of mitochondria in and outside GFP-positive astrocytic (AS) processes in the same sections plotted in d (Student's t test).
Discussion
This study used a novel approach to analyze compartmentalization of energy metabolism in adult brain. Acutely dissociated astrocytes and neurons from adult cortical brain tissue were isolated based on promoter-driven GFP or YFP expression, or immunolabeling against cell-specific surface antigens. RNA extracted from the purified pools of cells was subsequently used for genomic expression profiling (Wang et al., 1998; Keyoung et al., 2001; Nunes et al., 2003; Lobo et al., 2006). In this way, metabolic pathways could be analyzed with cell-type specificity not readily feasible in situ. The analysis showed that both astrocytes and neurons expressed transcripts for oxidative metabolism of glucose and, surprisingly, that the expression level of the majority of enzymes in the TCA cycle was higher in astrocytes than in neurons. To obtain a functional validation of the transcript analysis, we incubated the acutely isolated astrocytes with [U-13C]glucose, followed by MS, to determine the activity of metabolic pathways. We found 13C enrichment of TCA cycle intermediates and derivatives, supporting the functional competence and robust activity of aerobic metabolism in astrocytes. This direct observation speaks to a number of inferential reports to the contrary, which have suggested that astrocytic metabolism is either predominantly (Schurr, 2006) or exclusively (Pellerin and Magistretti, 1994; Sibson et al., 1998; Magistretti et al., 1999; Attwell and Laughlin, 2001) glycolytic.
In addition, we noted by electron microscopy that the relative number of mitochondria in astrocytes is higher than in the surrounding neuropil in mice expressing GFP under the Gfap promoter. To identify astrocytic processes, we took advantage of GFP being a small cytosolic protein present in even the very fine GFAP-negative astrocytic processes. Previous EM studies using staining of GFAP could not with certainty identify these fine structures of astrocytes, because GFAP is present only in larger main processes. The fine processes of astrocytes are in direct contact with synapses and play an important role in several energy-demanding processes, most notably glutamate uptake. The finding that astrocytes contain a large number of mitochondria provided additional support for an active oxidative metabolism in cortical astrocytes, in addition of being the first study of mitochondria in fine astrocytic processes covering synapses.
Active glycolysis inevitably raises the question by what mechanism cytosolic NADH is reoxidized. In neurons, the malate–aspartate shuttle has been demonstrated to account for maintaining homeostatic levels of NAD+/NADH, whereas in astrocytes, the presence of the malate–aspartate shuttle is debated (Ramos et al., 2003). However, studies using cultured astrocytes suggested the presence of a shuttle of NADH that is consistent with the expression of aralar1 in cultured astrocytes (Waagepetersen et al., 2001). In this study, we identified expression of all components of the glycerol–phosphate shuttle. All but one component of the malate–aspartate shuttle was expressed in astrocytes. The low expression of the malate–aspartate shuttle component, Slc25a10, was supported by the Allen Brain Atlas. This observation suggests that other mechanisms may exist as, for instance, the proposed transcellular redox coupling mechanism in which astrocytes shuttle lactate to neurons, for neuronal pyruvate synthesis and shuttling of neuronal pyruvate back to astrocytes (Cerdan et al., 2006).
Previous cell culture studies have suggested that lactate metabolism is compartmentalized (Itoh et al., 2003). Our analysis supported this concept, because astrocytes at the transcript level had a high capacity for lactate synthesis and after acute isolation produced large quantities of lactate. The majority of lactate produced by astrocytes was found in the medium, and not in the cell extract, suggesting that lactate is released as soon as it is synthesized. Not all released lactate derived from glycolysis, but some also derived from the TCA cycle. In contrast to the astrocytic lactate production, the genomic analysis showed that neurons exhibited a high capacity for lactate consumption. Because of the limited number of Thy1-positive cells isolated from one brain (∼10,000), it was not feasible to analyze active neuronal metabolism by incubating in [U-13C]glucose. Nevertheless, the observation that lactate metabolism is compartmentalized between astrocytes and neurons support the astrocyte–neuron–lactate shuttle hypothesis originally proposed by Magistretti et al., (1999). The high capacity for oxidative metabolism in astrocytes, as well as the labeling of TCA cycle intermediates and amino acids derived from these, provide, on the other hand, clear evidence that lactate only in part constitutes the metabolic end product of glucose metabolism. The observation of oxidative metabolism in astrocytes is supported by a recent study (Takano et al., 2007). Two-photon imaging of NADH in live animals indicated that astrocytic vascular endfeet are oxidatively active and may consume O2 at the expense of tissue located more distant to the vasculature during metabolic stress. The changes in NADH fluorescence signal suggested that glucose oxidation is compartmentalized according to access to O2 rather than in a cell-specific pattern, thus supporting the concept that both astrocytes and neurons are active O2 consumers.
Several entry or exit pathways related to glycolysis or TCA cycle were clearly compartmentalized. For example, we found enriched levels of glycogen-metabolizing enzymes in astrocytes supporting the notion that astrocytes are the compartment of glycogen metabolism in CNS (Ibrahim, 1975). Glutamate metabolism was also compartmentalized as reported previously. Glutamate can be metabolized either by Glud1 or aminotransferases. We found specific expression of Glud1 by astrocytes that also was confirmed at the functional level by 13C labeling of glutamate. Importantly, we found M+1-, M+2-, and M+3-labeled glutamate indicating active TCA cycling, which further supports oxidative metabolism. In addition, glutamate can be regenerated in the glutamate–glutamine cycle, which was clearly compartmentalized between astrocytes and neurons, as expected. Astrocytes expressed Glul supporting glutamine synthesis from glutamate, and in contrast, neurons expressed Gls and Gad1, for glutamate and GABA synthesis from glutamine. Finally, pyruvate recycling was also prevalent in astrocytes. At the genomic level, astrocytes expressed Mod1 specifically. At the functional level, malic enzyme activity was demonstrated by detection of M+1- and M+2-labeled lactate species. Astrocytes specifically expressed Pcx, which carboxylates pyruvate to form oxaloacetate, which was confirmed by identification of M+3-labeled citrate and glutamate in astrocyte extracts.
By directly isolating astrocytes from the adult brain, using FACS based on either a GFAP promoter-driven GFP reporter or GLT1 surface expression, we have identified those genes differentially expressed in live, unperturbed neocortical astrocytes. We identified and validated transcriptional patterns strongly suggestive of active oxidative metabolism in protoplasmic astrocytes, a point that has proven controversial in recent studies of cell–cell interactions in cerebral metabolism. More broadly though, these data comprise the first systematic genomic analysis of adult astrocytes. By comparing the gene expression patterns of freshly isolated astrocytes to those of adult glial progenitors (Sim et al., 2006), we may now define the molecular hallmarks of astrocytic development as well as metabolism. As such, the genomic analysis of adult astrocytes provide a wealth of molecular targets for modulating astrocytic fate and physiology, as well as an important baseline against which to assess those pathways differentially perturbed in the setting of disease. This strategy provides both an operational model and genomics database by which to assess the involvement of astrocytes in murine models of CNS disease. In this regard, recent observations show that reactive changes in astrocytes play a prominent role in the degeneration of motor neurons in the SOD1 mutant model of amyotrophic lateral sclerosis (Clement et al., 2003; Nagai et al., 2007). The investigation of astrocytic gene expression patterns in such mice, and the direct comparison of these to the normal patterns of expression reported here, should permit the efficient and algorithmic identification of those pathways associated with disease pathogenesis.
Footnotes
This work was supported by the Mathers Charitable Foundation; National Institutes of Health Grants NS30007, NS38073, NS39559, and NS050315; the Adelson Program in Neural Repair and Regeneration; the CNS Foundation; and the Ataxia-Telangiectasia Children's Program. H.S.W. and A.S. were supported by Danish State Medical Research Council Grants 22-04-0314 and 22-03-0250. We thank Lars Evje for excellent technical help.
References
- Aoki C, Milner TA, Sheu KF, Blass JP, Pickel VM. Regional distribution of astrocytes with intense immunoreactivity for glutamate dehydrogenase in rat brain: implications for neuron–glia interactions in glutamate transmission. J Neurosci. 1987;7:2214–2231. doi: 10.1523/JNEUROSCI.07-07-02214.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- Aubert A, Pellerin L, Magistretti PJ, Costalat R. A coherent neurobiological framework for functional neuroimaging provided by a model integrating compartmentalized energy metabolism. Proc Natl Acad Sci USA. 2007;104:4188–4193. doi: 10.1073/pnas.0605864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak LK, Schousboe A, Sonnewald U, Waagepetersen HS. Glucose is necessary to maintain neurotransmitter homeostasis during synaptic activity in cultured glutamatergic neurons. J Cereb Blood Flow Metab. 2006;26:1285–1297. doi: 10.1038/sj.jcbfm.9600281. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. Ed 5. New York: W. H. Freeman; 2002. [Google Scholar]
- Biemann K. Mass spectroscopy, organic chemistry applications. New York: McGraw; 1962. [Google Scholar]
- Brown AM, Tekkok SB, Ransom BR. Glycogen regulation and functional role in mouse white matter. J Physiol (Lond) 2003;549:501–512. doi: 10.1113/jphysiol.2003.042416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdan S, Rodrigues TB, Sierra A, Benito M, Fonseca LL, Fonseca CP, Garcia-Martin ML. The redox switch/redox coupling hypothesis. Neurochem Int. 2006;48:523–530. doi: 10.1016/j.neuint.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Cheeseman AJ, Clark JB. Influence of the malate-aspartate shuttle on oxidative metabolism in synaptosomes. J Neurochem. 1988;50:1559–1565. doi: 10.1111/j.1471-4159.1988.tb03044.x. [DOI] [PubMed] [Google Scholar]
- Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillee S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, Brown RH, Jr, Julien JP, Goldstein LS, Cleveland DW. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- Dringen R, Gebhardt R, Hamprecht B. Glycogen in astrocytes: possible function as lactate supply for neighboring cells. Brain Res. 1993;623:208–214. doi: 10.1016/0006-8993(93)91429-v. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick SM, Cooper AJ, Duffy TE. Use of beta-methylene-D,L-aspartate to assess the role of aspartate aminotransferase in cerebral oxidative metabolism. J Neurochem. 1983;41:1370–1383. doi: 10.1111/j.1471-4159.1983.tb00835.x. [DOI] [PubMed] [Google Scholar]
- Goldman SA, Nedergaard M. Newly generated neurons of the adult songbird brain become functionally active in long-term culture. Brain Res Dev Brain Res. 1992;68:217–223. doi: 10.1016/0165-3806(92)90063-3. [DOI] [PubMed] [Google Scholar]
- Hertz L, Peng L, Dienel GA. Energy metabolism in astrocytes: high rate of oxidative metabolism and spatiotemporal dependence on glycolysis/glycogenolysis. J Cereb Blood Flow Metab. 2007;27:219–249. doi: 10.1038/sj.jcbfm.9600343. [DOI] [PubMed] [Google Scholar]
- Ibrahim MZ. Glycogen and its related enzymes of metabolism in the central nervous system. Adv Anat Embryol Cell Biol. 1975;52:3–89. doi: 10.1007/978-3-642-86875-7. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Esaki T, Shimoji K, Cook M, Law MJ, Kaufman E, Sokoloff L. Dichloroacetate effects on glucose and lactate oxidation by neurons and astroglia in vitro and on glucose utilization by brain in vivo. Proc Natl Acad Sci USA. 2003;100:4879–4884. doi: 10.1073/pnas.0831078100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyoung HM, Roy NS, Benraiss A, Louissaint A, Jr, Suzuki A, Hashimoto M, Rashbaum WK, Okano H, Goldman SA. High-yield selection and extraction of two promoter-defined phenotypes of neural stem cells from the fetal human brain. Nat Biotechnol. 2001;19:843–850. doi: 10.1038/nbt0901-843. [DOI] [PubMed] [Google Scholar]
- Koehler RC, Gebremedhin D, Harder DR. Role of astrocytes in cerebrovascular regulation. J Appl Physiol. 2006;100:307–317. doi: 10.1152/japplphysiol.00938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JC, Murthy CR, Cooper AJ, Hertz E, Hertz L. Differential effects of ammonia and beta-methylene-DL-aspartate on metabolism of glutamate and related amino acids by astrocytes and neurons in primary culture. Neurochem Res. 1989;14:377–389. doi: 10.1007/BF01000042. [DOI] [PubMed] [Google Scholar]
- Lin SC, Huck JH, Roberts JD, Macklin WB, Somogyi P, Bergles DE. Climbing fiber innervation of NG2-expressing glia in the mammalian cerebellum. Neuron. 2005;46:773–785. doi: 10.1016/j.neuron.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Magistretti PJ, Pellerin L, Rothman DL, Shulman RG. Energy on demand. Science. 1999;283:496–497. doi: 10.1126/science.283.5401.496. [DOI] [PubMed] [Google Scholar]
- Mawhinney TP, Robinett RS, Atalay A, Madson MA. Analysis of amino acids as their tert.-butyldimethylsilyl derivatives by gas-liquid chromatography and mass spectrometry. J Chromatogr. 1986a;358:231–242. doi: 10.1016/s0021-9673(01)90333-4. [DOI] [PubMed] [Google Scholar]
- Mawhinney TP, Robinett RS, Atalay A, Madson MA. Gas-liquid chromatography and mass spectral analysis of mono-, di- and tricarboxylates as their tert.-butyldimethylsilyl derivatives. J Chromatogr. 1986b;361:117–130. doi: 10.1016/s0021-9673(01)86899-0. [DOI] [PubMed] [Google Scholar]
- McKenna MC, Waagepetersen H, Gruetter R, Sonnewald U, Schousboe A. Basic neurochemistry. Ed 7. London: Elsevier; 2005. Cerebral metabolism and blood flow. [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S, Nagelhus EA, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen OP. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MC, Roy NS, Keyoung HM, Goodman RR, McKhann G, II, Jiang L, Kang J, Nedergaard M, Goldman SA. Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat Med. 2003;9:439–447. doi: 10.1038/nm837. [DOI] [PubMed] [Google Scholar]
- O'Brien J, Kla KM, Hopkins IB, Malecki EA, McKenna MC. Kinetic parameters and lactate dehydrogenase isozyme activities support possible lactate utilization by neurons. Neurochem Res. 2007;32:597–607. doi: 10.1007/s11064-006-9132-9. [DOI] [PubMed] [Google Scholar]
- Olstad E, Qu H, Sonnewald U. Glutamate is preferred over glutamine for intermediary metabolism in cultured cerebellar neurons. J Cereb Blood Flow Metab. 2007;27:811–820. doi: 10.1038/sj.jcbfm.9600400. [DOI] [PubMed] [Google Scholar]
- Paukert M, Bergles DE. Synaptic communication between neurons and NG2+ cells. Curr Opin Neurobiol. 2006;16:515–521. doi: 10.1016/j.conb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci USA. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Neuroenergetics: calling upon astrocytes to satisfy hungry neurons. Neuroscientist. 2004;10:53–62. doi: 10.1177/1073858403260159. [DOI] [PubMed] [Google Scholar]
- Pfeiffer B, Meyermann R, Hamprecht B. Immunohistochemical co-localization of glycogen phosphorylase with the astroglial markers glial fibrillary acidic protein and S-100 protein in rat brain sections. Histochemistry. 1992;97:405–412. doi: 10.1007/BF00270387. [DOI] [PubMed] [Google Scholar]
- Pysh JJ, Khan T. Variations in mitochondrial structure and content of neurons and neuroglia of rat brain: an electron microscopoy study. Brain Res. 1972;36:1–18. doi: 10.1016/0006-8993(72)90762-7. [DOI] [PubMed] [Google Scholar]
- Ramos M, del Arco A, Pardo B, Martinez-Serrano A, Martinez-Morales JR, Kobayashi K, Yasuda T, Bogonez E, Bovolenta P, Saheki T, Satrustegui J. Developmental changes in the Ca2+-regulated mitochondrial aspartate-glutamate carrier aralar1 in brain and prominent expression in the spinal cord. Brain Res Dev Brain Res. 2003;143:33–46. doi: 10.1016/s0165-3806(03)00097-x. [DOI] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel D, Marangos PJ, Zis AP, Brightman M, Goodwin FK. Brain endolases as specific markers of neuronal and glial cells. Science. 1978;199:313–315. doi: 10.1126/science.339349. [DOI] [PubMed] [Google Scholar]
- Schurr A. Lactate: the ultimate cerebral oxidative energy substrate? J Cereb Blood Flow Metab. 2006;26:142–152. doi: 10.1038/sj.jcbfm.9600174. [DOI] [PubMed] [Google Scholar]
- Shank RP, Bennett GS, Freytag SO, Campbell GL. Pyruvate carboxylase: an astrocyte-specific enzyme implicated in the replenishment of amino acid neurotransmitter pools. Brain Res. 1985;329:364–367. doi: 10.1016/0006-8993(85)90552-9. [DOI] [PubMed] [Google Scholar]
- Sibson NR, Dhankhar A, Mason GF, Rothman DL, Behar KL, Shulman RG. Stoichiometric coupling of brain glucose metabolism and glutamatergic neuronal activity. Proc Natl Acad Sci USA. 1998;95:316–321. doi: 10.1073/pnas.95.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim F, Lang J, Waldau B, Roy N, Schwartz T, Pilcher W, Chandross K, Natesan S, Merrill J, Goldman S. Complementary patterns of gene expression by adult human oligodendrocyte progenitor cells and their white matter environment. Ann Neurol. 2006;59:763–779. doi: 10.1002/ana.20812. [DOI] [PubMed] [Google Scholar]
- Simard M, Arcuino G, Takano T, Liu QS, Nedergaard M. Signaling at the gliovascular interface. J Neurosci. 2003;23:9254–9262. doi: 10.1523/JNEUROSCI.23-27-09254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Tian GF, Peng W, Lou N, Libionka W, Han X, Nedergaard M. Astrocyte-mediated control of cerebral blood flow. Nat Neurosci. 2006;9:260–267. doi: 10.1038/nn1623. [DOI] [PubMed] [Google Scholar]
- Takano T, Tian GF, Peng W, Lou N, Lovatt D, Hansen AJ, Kasischke KA, Nedergaard M. Cortical spreading depression causes and coincides with tissue hypoxia. Nat Neurosci. 2007;10:754–762. doi: 10.1038/nn1902. [DOI] [PubMed] [Google Scholar]
- Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- Waagepetersen HS, Qu H, Schousboe A, Sonnewald U. Elucidation of the quantitative significance of pyruvate carboxylation in cultured cerebellar neurons and astrocytes. J Neurosci Res. 2001;66:763–770. doi: 10.1002/jnr.10061. [DOI] [PubMed] [Google Scholar]
- Wang K, Walz W. Unusual topographical pattern of proximal astrogliosis around a cortical devascularizing lesion. J Neurosci Res. 2003;73:497–506. doi: 10.1002/jnr.10683. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu H, Jiang J, Delohery TM, Isdell F, Goldman SA. Isolation of neuronal precursors by sorting embryonic forebrain transfected with GFP regulated by the T alpha 1 tubulin promoter. Nat Biotechnol. 1998;16:196–201. doi: 10.1038/nbt0298-196. [DOI] [PubMed] [Google Scholar]
- Wender R, Brown AM, Fern R, Swanson RA, Farrell K, Ransom BR. Astrocytic glycogen influences axon function and survival during glucose deprivation in central white matter. J Neurosci. 2000;20:6804–6810. doi: 10.1523/JNEUROSCI.20-18-06804.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A, Volknandt W, Langer D, Nolte C, Kettenmann H, Zimmermann H. Localization of SNARE proteins and secretory organelle proteins in astrocytes in vitro and in situ. Neurosci Res. 2004;48:249–257. doi: 10.1016/j.neures.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Yu AC, Drejer J, Hertz L, Schousboe A. Pyruvate carboxylase activity in primary cultures of astrocytes and neurons. J Neurochem. 1983;41:1484–1487. doi: 10.1111/j.1471-4159.1983.tb00849.x. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Sun B, Zhang CL, Fine A, Chiu SY, Messing A. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol. 1997;187:36–42. doi: 10.1006/dbio.1997.8601. [DOI] [PubMed] [Google Scholar]