Abstract
Eph receptors play critical roles in the establishment and remodeling of neuronal connections, but the signaling pathways involved are not fully understood. We have identified a novel interaction between the C terminus of the EphA4 receptor and the PDZ domain of the GTPase-activating protein spine-associated RapGAP (SPAR). In neuronal cells, this binding mediates EphA4-dependent inactivation of the closely related GTPases Rap1 and Rap2, which have recently been implicated in the regulation of dendritic spine morphology and synaptic plasticity. We show that SPAR-mediated inactivation of Rap1, but not Rap2, is critical for ephrin-A-dependent growth cone collapse in hippocampal neurons and decreased integrin-mediated adhesion in neuronal cells. Distinctive effects of constitutively active Rap1 and Rap2 on the morphology of growth cones and dendritic spines support the idea that these two GTPases have different functions in neurons. Together, our data implicate SPAR as an important signaling intermediate that links the EphA4 receptor with Rap GTPase function in the regulation of neuronal morphology.
Keywords: growth cone collapse, dendritic spine, hippocampus, ephrin, integrin, tyrosine kinase
Introduction
Eph receptor tyrosine kinases have a pivotal role in axon pathfinding during neural development (O'Leary and Wilkinson, 1999; Pasquale, 2005; Egea and Klein, 2007). These receptors are expressed in growth cones and are activated by glycosylphosphatidylinositol-linked ephrin-A ligands or transmembrane ephrin-B ligands on neighboring cells. EphA receptor signaling after ephrin stimulation typically causes growth cone collapse and neurite retraction in cultured neurons. In vivo, axons with high EphA expression avoid regions with high ephrin expression, which is critical for topographic organization of neuronal connections. In particular, knock-out of the EphA4 receptor results in disrupted midline repulsion, generation of ectopic connections in the spinal cord, and defects in limb motor axon pathfinding (Helmbacher et al., 2000; Kullander et al., 2001, 2003). EphA4 also regulates the structural plasticity of synaptic connections in the mature brain (Murai et al., 2003). Dendritic spines (the small dendritic protrusions that are postsynaptic to excitatory terminals) are elongated and more numerous in mice lacking EphA4. Furthermore, stimulation of EphA4 in hippocampal slices with soluble ephrin-A ligands causes shortening and loss of spines.
The signaling mechanisms responsible for EphA-dependent morphological effects in neurons and other cell types have not been fully elucidated, but it is becoming increasingly evident that small GTPases of the Rho and Ras families play a critical role (Noren and Pasquale, 2004). These GTPases regulate cell morphology primarily through reorganization of the actin cytoskeleton and by modulating cell adhesion. They switch between an inactive GDP-bound conformation and an active GTP-bound conformation that allows interaction with downstream effectors. Guanine nucleotide exchange factors promote GTPase activation by accelerating the replacement of GDP with GTP, and several exchange factors for Rho family GTPases have been shown to function downstream of EphA receptors (Cowan et al., 2005; Sahin et al., 2005). In contrast, GTPase-activating proteins (GAPs) accelerate the hydrolysis of GTP to GDP. α2-Chimaerin, a GTPase-activating protein for Rac, and p120RasGAP, a GTPase-activating protein for H-Ras and R-Ras, have been recently implicated in EphA-dependent repulsive responses (Dail et al., 2006; Beg et al., 2007; Iwasato et al., 2007; Wegmeyer et al., 2007).
Here we show that EphA4 regulates the Ras family proteins, Rap1 and Rap2, through a novel interaction with spine-associated RapGAP (SPAR) (Pak et al., 2001). Ephrin stimulation promotes association of the EphA4 C terminus with the SPAR PDZ domain, and this interaction is important for inactivation of Rap1 and Rap2, two GTPases involved in the regulation of synaptic plasticity and dendritic spine morphology (Pak et al., 2001; Zhu et al., 2002, 2005; Xie et al., 2005; Fu et al., 2007b). Furthermore, we found that inactivation of Rap1 by SPAR is necessary for ephrin-A-dependent growth cone collapse. Interestingly, Rap1 and Rap2 have different effects on neuronal morphology and on ephrin-A-dependent reduction of integrin-mediated adhesion, suggesting that these closely related GTPases have distinct functions in developing neurons. These results provide new insight into EphA signaling mechanisms that control neuronal morphology through regulation of GTPases of the Ras family.
Materials and Methods
DNA constructs.
The cDNAs encoding the PDZ domain of mouse SPAR (amino acids 992–1056, GenBank accession AF026504), human FAP1 (Fas-associated phosphatase 1; amino acids 1597–1678 corresponding to PDZ domain 4, GenBank accession D021211), mouse protein interacting with C kinase 1 (PICK1; amino acids 22–105, GenBank accession Z46720), rat proline-rich synapse-associated protein 1/cortactin binding protein 1 (CBP1; amino acids 37–131, GenBank accession AJ131899) and human ERBB2-interacting protein (Erbin; amino acids 1280–1369, GenBank accession NM_018695) and the cDNAs encoding the C-terminal tails of mouse EphA4 (amino acids 892–986, GenBank accession NM_007938), EphA5 (amino acids 801–876, GenBank accession NM_007937), EphA6 (amino acids 934–1035, GenBank accession NM_007938), and EphA7 (amino acids 890–995, GenBank accession BC026153) were obtained by PCR amplification from cDNA libraries using specific primers. The cDNAs were then cloned into the pGEX-4T1 bacterial expression vector (GE Healthcare, Piscataway, NJ), and the constructs were verified by DNA sequencing.
cDNAs encoding full-length chicken EphA4, EphB2, and EphB5 and human EphA2 and EphB4 were cloned into the mammalian expression vector pcDNA3 (Invitrogen, Carlsbad, CA); the chicken EphA3 cDNA was cloned into a noncommercial vector under the control of the CMV promoter. N-terminal myc-tagged constructs of wild-type and mutant rat SPAR were cloned into the pGW1 vector have been described (Pak et al., 2001). Chicken EphA4 in pcDNA3 was used as a template to amplify EphA4 lacking the C-terminal MVPV sequence (EphA4ΔMVPV) or to generate the kinase-dead EphA4K653R construct. Mouse Rap2A was cloned from an EST clone into pcDNA3 fused to an N-terminal myc tag. The Rap2 S17N and Rap2 Q63E mutants were generated by PCR mutagenesis. Hemagglutinin (HA)-tagged wild-type and mutant human Rap1A (GenBank accession X12533) were purchased from University of Missouri-Rolla cDNA Resource Center (http://www.cdna.org/). The yellow fluorescent protein (YFP)-actin plasmid was purchased from BD Biosciences (San Jose, CA). The N-terminally FLAG-tagged Rap1GAP cDNA (GenBank accession NM_002885) in pcDNA3 has been described by Rubinfeld et al. (1991).
Antibodies.
Rabbit SPAR polyclonal antibodies were generated using an N-terminal SPAR peptide (amino acids 4–22) for immunization and affinity purification. Affinity-purified EphA4 polyclonal antibodies were either directed against a C-terminal peptide (Soans et al., 1994) or against amino acids 925–986 of chicken EphA4 fused to glutathione S-transferase (GST). Anti-GST antibodies were absorbed on a GST affinity column. Monoclonal EphB2 (1A6H12) and EphA4 (4C8H5) antibodies and polyclonal EphA2 (34-7400), EphA3 (Z30125), EphB4 (35-2900), and EphB5 (Z30134) antibodies were from Zymed/Invitrogen; a monoclonal antibody to Rap2 was from BD Transduction Laboratories; a monoclonal anti-HA antibody was from Covance (Princeton, NJ); a monoclonal anti-α-tubulin antibody and polyclonal anti-Rap1 and Rap2 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); monoclonal anti-myc, synaptophysin, and β-actin antibodies were from Sigma (St. Louis, MO); polyclonal anti-myc antibodies and monoclonal anti-phosphotyrosine (4G10) and PSD-95 were from Millipore (Billerica, MA).
Transfection of HEK 293T and HT22 cells.
Human embryonic kidney (HEK) 293T cells were transiently transfected with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. HT22 cells were transfected with Lipofectamine 2000 (Invitrogen) using 12.5 μg of DNA and 32.5 μl of Lipofectamine complexed for 20 min in 1.5 ml of OptiMEM medium and then diluted in a final volume of 5.5 ml of OptiMEM to transfect a 10 cm plate. Alternatively, to prepare HT22 cells for the adhesion assays 10 μg of DNA and 20 μl of Lipofectamine were complexed for 20 min in 0.6 ml of OptiMEM and then diluted in a final volume of 2.1 ml of OptiMEM to transfect a 6 cm plate. Cells were incubated with DNA/Lipofectamine complexes for 6 h at 37°C, washed with PBS, and grown for ∼24 h in DMEM containing 10% FCS.
For SPAR siRNA knock down, pSuper vector (Oligoengine, Seattle, WA) containing the target sequence 5′-GAA GTT CAC AGA GTG GGT C-3′ of mouse SPAR/Sipa1l1 (signal-induced proliferation-associated 1-like 1, nucleotides 5907–5925, GenBank accession number NM_172579) was transfected in HT22 cells. The SPAR siRNA pSuper vector or a control pSuper vector containing a luciferase siRNA target sequence were cotransfected with pcDNA3 encoding HA-tagged human Rap1.
GST pull-down assays.
Forty-eight hours after transient transfection, HEK 293T cells were lysed in Brij buffer [1 mm EDTA, 1% Brij in PBS (CellGro), pH 7.4, with 1 mm PMSF, 1 mm sodium orthovanadate, 1 mm NaF, and protease inhibitors (Sigma)]. Lysates were precleared with glutathione Sepharose 4B beads (Pharmacia) for 2 h at 4°C and incubated with glutathione beads loaded with GST-fusion proteins purified from bacterial extracts and blocked with 6% BSA. The beads were washed once with high-salt Brij buffer (1 mm EDTA, 0.25% Brij in PBS with a total of 300 mm NaCl) followed by three washes with 350 mm NaCl and a wash with high-salt Brij buffer before elution by boiling in SDS-PAGE sample buffer.
For pull-down assays of Rap-GTP with GST fused to the Rap-binding domain of RalGDS (GST-RalGDS RBD), fresh hippocampal slices were prepared from P7 mice and stimulated with 10 μg/ml ephrin-A1 Fc or control Fc (R & D Systems) for 60 min at room temperature. In cultured cells, the activities of endogenous Rap GTPases or exogenous HA-tagged Rap1 and myc-tagged Rap2 were analyzed after stimulation at 37°C with 2 μg/ml ephrin-A1 Fc or control Fc preclustered by incubation with a 1/10 concentration of goat anti-human IgG antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Tissue or cells were lysed in GAP activity buffer (25 mm HEPES, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 1% v/v NP-40, 0.25% w/v Na deoxycholate, 1 mm PMSF, 1 mm sodium orthovanadate, 1 mm NaF, and protease inhibitors (Sigma)]. GST-RalGDS RBD beads were incubated with lysates for 45 min at 4°C and washed with lysis buffer before elution by boiling in SDS-PAGE sample buffer.
Immunoprecipitations.
HEK 293T cells were lysed 48 h after transfection in modified radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% NP-40, and 0.25% Na-deoxycholate) containing 1 mm PMSF, 1 mm sodium orthovanadate, 1 mm NaF, and protease inhibitors. Hippocampal slices were lysed in modified RIPA buffer, and supernatants were used after centrifugation for 30 min at 20,000 × g. Lysates were precleared by incubation with GammaBind Sepharose (GE Healthcare) and then incubated for 4 h at 4°C with 2.5 μg of polyclonal EphA4 antibodies or control nonimmune IgGs coupled to 20 μl of GammaBind Sepharose or with 10 μl of anti-myc antibodies conjugated to agarose beads (Santa Cruz Biotechnology). EphA4 coimmunoprecipitated with myc-SPAR was detected by immunoblotting with rabbit polyclonal EphA4 antibodies, whereas EphA4 immunoprecipitated with rabbit polyclonal EphA4 antibodies was detected using mouse monoclonal anti-EphA4 antibodies.
Cell attachment assays.
HT22 cells were transfected with Rap constructs together with EGFP to identify the transfected cells. Twenty-four hours after transfection, cells were starved for 5 h in DMEM with 1% BSA, detached, and kept in suspension for 40 min at 37°C in DMEM-1% BSA. Cells (7 × 104) were then allowed to attach to coverslips coated with 10 μg/ml fibronectin (Millipore) or 20 μg/ml poly-l-lysine (Sigma) for 10 min at 37°C in the presence of 10 μg/ml Fc or 10 μg/ml ephrin-A3 Fc. Attached cells were fixed in 4% paraformaldehyde and stained with 4′,6′-diamidino-2-phenylindole (Invitrogen, Carlsbad, CA). Attached green fluorescent (transfected) cells were counted blind, and their number was corrected for transfection efficiency. Seven 10× microscopic fields were counted for each coverslip, and three coverslips per condition were analyzed in each of three experiments, except for the Rap1 G12V-transfected cells plated on poly-l-lysine, for which only two coverslips were used in two of the four experiments.
Synaptosomes.
The preparation of synaptosomal fractions was adapted from Rogers and Lemaire (1991) and is described in more detail by Bourgin et al. (2007).
Neuronal and glial culture, transfection, and electroporation.
Primary hippocampal neurons and glial cells were prepared from embryonic day 17 rat embryos and cultured according to Zafra et al. (1990) with minor modifications. For growth cone collapse assays, dissociated primary hippocampal cells were resuspended in DMEM supplemented with 10% FCS without antibiotics and counted. Approximately 4.5–5.5 × 106 cells were transfected by electroporation according to the manufacturer's instructions (Amaxa Biosystems, Gaithersburg, MD). Cells were centrifuged and resuspended in 100 μl of Nucleofector solution, mixed with a total of 3 μg of DNA (1:1 ration of EGFP and Rap1, Rap2, or SPAR DNA) and electroporated using program G-13. Transfected cells were plated at low density (1.5 × 104 cells/cm2) on poly-d-lysine- (50 μg/ml) and laminin- (2.5 μg/ml) coated coverslips and grown for 2 d in Neurobasal medium (Invitrogen) supplemented with B27, l-glutamine, and penicillin/streptomycin. Growth cone collapse was induced by stimulating the neurons for 20 min with 2 μg/ml preclustered ephrin-A1 Fc or Fc as a control. The neurons were then fixed and immunostained as described below.
For long-term cultures and immunoblotting, hippocampal cells were resuspended in DMEM and 10% FCS, and glial cells were removed by attachment to uncoated culture plates for 2 h. Neurons were recovered as nonadherent cells, plated at high density (4 × 104 cells/cm2) in Neurobasal medium and cultured in the presence of 2 μm AraC to avoid glial cell contamination. The hippocampal glial cells were cultured in DMEM, 10% FCS with penicillin/streptomycin and l-glutamine. Neuronal and glial cultures were collected after 14 d in culture for immunoblotting. Neuronal cultures were also collected at different time periods in culture for the time course experiment shown in supplemental Figure 1 (available at www.jneurosci.org as supplemental material). For dendritic spine analysis, transfection of hippocampal neurons was performed after 12 d in culture using the calcium phosphate precipitation method (Ethell et al., 2001). Nine days after transfection, neurons were fixed and immunostained.
Immunostaining.
Dissociated hippocampal neurons in culture were fixed with 4% paraformaldehyde, 4% sucrose in 0.1 m PBS, pH 7.4, blocked with 5% goat serum, 0.1% Triton X-100 in TBS for 1 h at room temperature. For analysis of growth cone collapse, neurons were double labeled with rhodamine–phalloidin (Invitrogen) and anti-tubulin antibodies (Santa Cruz Biotechnology) followed by goat anti-mouse antibodies conjugated to Alexa 635 (Invitrogen). For immunolocalization of Rap1, Rap2, SPAR, and EphA4, neurons transfected with YFP-actin were incubated sequentially with monoclonal EphA4 antibody (0.5 μg/ml, Zymed) overnight at 4°C, goat anti-mouse Alexa 635-conjugated IgG (1/300, Invitrogen) for 2 h at room temperature, polyclonal antibodies against Rap1 (1 μg/ml, Santa Cruz Biotechnology), Rap2 (1 μg/ml, Santa Cruz Biotechnology), or SPAR (0.75 μg/ml) overnight at 4°C and goat anti-rabbit Alexa 594-coupled IgG (1:300, Invitrogen) for 2 h at room temperature. Antibodies were diluted in 5% goat serum, 0.1% Triton X-100 in TBS. Between antibody incubations, coverslips were washed three times with 0.1% Triton X-100 in TBS for 10 min at room temperature.
Measurement of growth cones and dendritic spines.
For growth cone analysis, 50–70 EGFP-positive (transfected) neurons per condition were photographed under a fluorescence microscope in each experiment (two independent experiments were carried out). Images showing actin and tubulin staining were merged, and the growth cones at the tip of the longest process, which typically corresponds to the axon, were analyzed using NIH ImageJ. Only neurons with at least three neurites, and one neurite at least twice as long as the others, were analyzed. Only the growth cone of the longest neurite, the axon, was scored. The area of the growth cones was measured by framing the actin-positive extensions using the freehand modus. Axonal length was determined by measuring the longest process from the soma of the neuron to the distal tip of the growth cone. For growth cone collapse analysis, axon tips with filopodia extensions and visible lamellipodia were considered as noncollapsed. Axon tips without lamellipodia and with ≤3 retraction fibers were counted as collapsed.
For dendritic spine analysis, images of dissociated neurons were acquired using a Bio-Rad 1024 confocal laser scanning microscope. Z-series encompassing the entire dendritic segment were stacked and analyzed using NIH ImageJ. Photographs were taken of three to five neurons per experiment (three independent repeat experiments), and 700–1000 spines were counted per condition (365 spines for Rap2Q63E-transfected neurons).
Results
EphA4 associates with the Rap-specific GTPase-activating protein SPAR
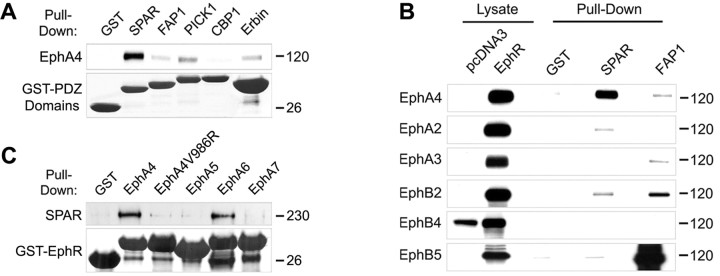
Eph receptors use several cytoplasmic regions to interact with signaling proteins and transduce intracellular signals (Pasquale, 2005; Egea and Klein, 2007). We were interested in identifying novel signaling interactions that may be important in the regulation of neuronal morphology downstream of EphA4. A potential site of interaction that has not been well characterized for EphA4 is its C-terminal PDZ-binding motif (Torres et al., 1998; Buchert et al., 1999). To identify PDZ domain proteins that may interact with EphA4, we performed GST pull-down assays using the PDZ domains of several candidate proteins known to be expressed in neurons. In pull-down assays using lysates from HEK 293T cells transiently transfected with EphA4, we detected a strong binding preference of EphA4 for the PDZ domain of the Rap-specific GTPase-activating protein SPAR (Fig. 1A). Further experiments showed that the SPAR PDZ domain preferentially binds to EphA4 among a number of EphA and EphB receptors examined (Fig. 1B). We also performed the reverse pull-down assays using GST fusion proteins containing the C-terminal tails of EphA receptors known to be expressed in neurons and lysates of HEK 293T cells transiently transfected with SPAR. These experiments revealed that the C termini of EphA4 and EphA6, but not those of EphA5 and EphA7, interact strongly with SPAR. Furthermore, inactivation of the EphA4 PDZ-binding motif by mutation of the C-terminal hydrophobic valine (Sheng and Sala, 2001) prevented efficient binding of SPAR (Fig. 1C). These results suggest that EphA4 can bind SPAR through a PDZ domain-mediated interaction.
Figure 1.
The C-terminal sequences of EphA4 and EphA6 bind the PDZ domain of the GTPase-activating protein SPAR. A, The PDZ domains of the indicated proteins, fused to GST, were used for pull-down assays with lysates of EphA4-transfected HEK 293T cells. Bound EphA4 was detected by immunoblotting and the GST-PDZ domain fusion proteins were detected by amido black staining. A lane between the CBP1 and Erbin lanes was digitally removed. B, Lysates from HEK 293T cells transfected with the indicated Eph receptors were used for pull-down assays with the PDZ domain of SPAR, and of FAP1 as a control, fused to GST. Bound Eph receptors were detected by immunoblotting. A lane between the lysate and pull-down lanes was digitally removed. C, The C-terminal tails of the indicated receptors, fused to GST, were used for pull-down assays with lysates of HEK 293T cells transfected with Myc-SPAR. SPAR bound to the GST fusion proteins was detected by immunoblotting with anti-Myc antibodies, and the GST fusion proteins were detected by amido black staining. In EphA4V986R, the C-terminal valine was mutated to arginine to inactivate the PDZ domain-binding motif.
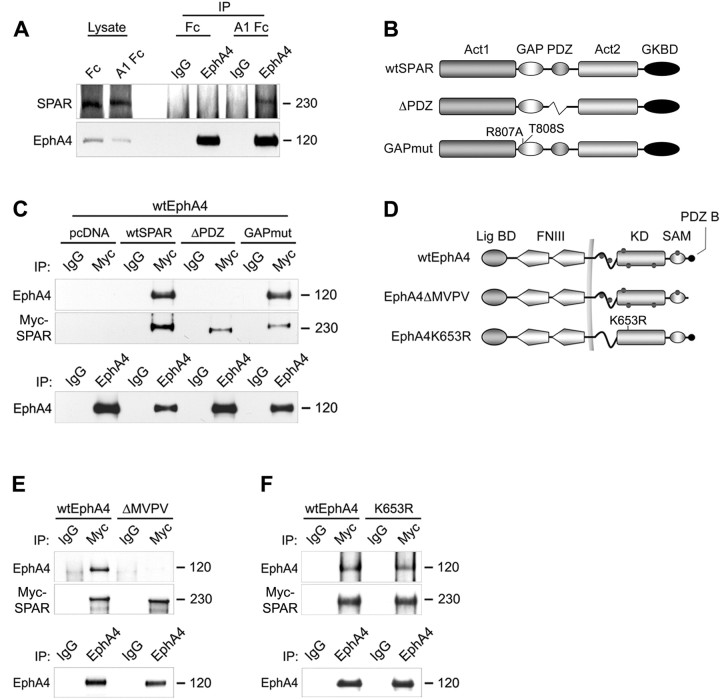
We further investigated this interaction and found that endogenous EphA4 and SPAR coimmunoprecipitated from freshly prepared hippocampal slices treated with ephrin-A1 Fc (Fig. 2A). The specificity of the SPAR antibodies was verified as shown in supplemental Figure 1A (available at www.jneurosci.org as supplemental material). Furthermore, EphA4 and SPAR also coimmunoprecipitated from cotransfected HEK 293T cells (Fig. 2C). Consistent with the GST pull-down assays showing an interaction of EphA4 with the PDZ domain of SPAR, EphA4 failed to coimmunoprecipitate with a SPAR mutant lacking the PDZ domain (ΔPDZ) (Fig. 2B,C). Furthermore, an EphA4 mutant lacking the PDZ domain-binding site (ΔMVPV) did not coimmunoprecipitate with wild-type SPAR (wtSPAR) (Fig. 2D,E). In contrast, mutations inactivating the GTPase-activating domain of SPAR (GAPmut) (Fig. 2B,C) or the kinase domain of EphA4 (K653R) (Fig. 2D,F) did not affect the interaction between the two cotransfected proteins. Thus, EphA4-SPAR association does not require EphA4 phosphorylation. Whether ephrin-A1 Fc-dependent EphA4 dimerization or conformational changes promote interaction with endogenously expressed SPAR remains to be determined. These data demonstrate that EphA4 associates with SPAR, that the interaction is mediated by the C-terminal PDZ-binding motif of EphA4 and the PDZ domain of SPAR, and that ephrin stimulation enhances the interaction between endogenous EphA4 and SPAR.
Figure 2.
EphA4 binds SPAR through a PDZ domain-mediated interaction. A, Freshly prepared hippocampal slices from 3-week-old mice were stimulated with soluble ephrin-A1 Fc (A1 Fc), or Fc as a control, and EphA4 was immunoprecipitated (IP). EphA4 and coimmunoprecipitated SPAR were detected by immunoblotting with the respective antibodies. B, D, Schematic representation of SPAR and EphA4 wild-type and mutant forms. B, GKBD, Guanylate kinase-binding domain. D, Lig BD, Ligand-binding domain; FNIII, fibronectin type III repeat; KD, kinase domain; SAM, sterile-α motif domain; PDZ B, PDZ-domain binding motif; wtEphA4, wild-type EphA4; EphA4K653R, kinase-inactive EphA4; dots indicate tyrosine phosphorylation sites, and the plasma membrane is indicated by a vertical line. C, E, F, The indicated EphA4 constructs and Myc-tagged SPAR constructs were cotransfected in HEK 293T cells. SPAR was immunoprecipitated and detected with anti-Myc antibodies. Coimmunoprecipitated EphA4 was detected by immunoblotting with anti-EphA4 antibodies. Bottom, Similar amounts of EphA4 could be immunoprecipitated from the transfected cells. Lanes between wild-type and mutated EphA4 were digitally removed.
Ephrin-A stimulation causes inactivation of Rap1 and Rap2 in neuronal cells
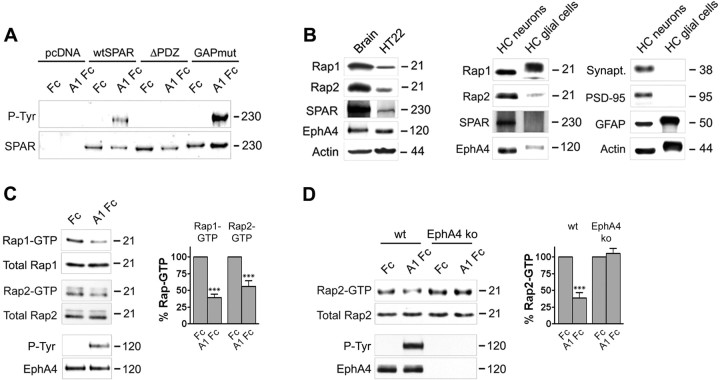
Consistent with a functional connection between EphA4 and SPAR, ephrin-A1 Fc treatment of HT22 cells, which are derived from immortalized hippocampal neurons (Li et al., 1997), resulted in tyrosine phosphorylation of SPAR (Fig. 3A). The phosphorylation was dependent on the presence of the PDZ domain of SPAR (Fig. 3A), suggesting that a physical association with EphA4 is required for SPAR phosphorylation. Because SPAR is a GTPase-activating protein for Rap1 and Rap2 (Pak et al., 2001), we examined whether EphA4 stimulation by ephrin affects activation of these Rap GTPases in HT22 cells, which express endogenous SPAR (Fig. 3B). Pull-down assays with a GST fusion protein of the Rap-binding domain of RalGDS to isolate active, GTP-bound Rap1 and Rap2 (Franke et al., 1997) demonstrated that ephrin-A1 Fc treatment decreases activation of both GTPases (to 36% of Fc control levels for Rap1 and to 57% of control levels for Rap2) (Fig. 3C). Rap1 inactivation appeared to be more sensitive to ephrin-A1 stimulation because a decrease in Rap1-GTP was observed even with ephrin concentrations as low as 0.25 μg/ml, whereas we did not detect Rap2 inactivation with ephrin concentrations lower than 1.5 μg/ml (data not shown).
Figure 3.
EphA4 stimulation by ephrin decreases Rap1 and Rap2 activation. A, HT22 cells were transfected with the indicated SPAR constructs and stimulated with preclustered ephrin-A1 Fc (A1 Fc) or Fc control for 8 min. Immunoprecipitated SPAR was probed with anti-phosphotyrosine antibodies (P-Tyr) followed by reprobing with anti-Myc antibodies (SPAR). B, Lysates from 1-month-old mouse brain, HT22 cells, and primary hippocampal neurons or glial cells cultured for 14 d were probed by immunoblotting with the indicated antibodies. The synaptic proteins synaptophysin (Synapt.) and PSD-95 and glial fibrillary acidic protein (GFAP) were detected as controls; actin detection shows equal loading. C, HT22 neuronal cells were stimulated with preclustered ephrin-A1 Fc (A1 Fc) or Fc control for 7.5 min. GTP-bound Rap1 or Rap2 were isolated by pull-down with GST-RalGDS RBD and detected by immunoblotting. To verify EphA4 activation by ephrin, EphA4 immunoprecipitates were probed with anti-phosphotyrosine antibodies (P-Tyr) and reprobed for EphA4. For quantification, the levels of Rap1-GTP or Rap2-GTP (determined as the optical density of scanned autoradiographs) were normalized to the Rap protein levels detected in the lysates (Total Rap). The histogram shows the average levels of Rap1-GTP and Rap2-GTP from four experiments ± SE, expressed as a percentage of the levels after Fc treatment. ***p < 0.001 for the comparison of ephrin-A1 Fc-treated with Fc-treated by two-way ANOVA and Bonferroni posttest. D, Freshly prepared hippocampal slices from wild-type mice and EphA4 knock-out littermates were stimulated with ephrin-A1 Fc or Fc control for 60 min and GTP-bound Rap2 was isolated by pull-down and quantified as described in C. The histogram shows the average levels of Rap2-GTP from four independent experiments ± SE. ***p < 0.001 by two-way ANOVA and Bonferroni posttest.
We also found that ephrin-A1 Fc stimulation substantially decreased Rap2 activation (to 43% of Fc control levels) in hippocampal slices, concomitant with increased EphA4 tyrosine phosphorylation (Fig. 3D). Similar results were obtained with ephrin-A3 Fc stimulation (data not shown). Importantly, ephrin-A1 Fc did not cause Rap2 inactivation in hippocampal slices obtained from EphA4 knock-out mice (Fig. 3D) indicating that EphA4 is the main EphA receptor that downregulates Rap2 activation in the hippocampus. Although Rap1 is well expressed in the hippocampus, we could not detect sufficient levels of active Rap1 in control or ephrin-A1 Fc-treated slices under the conditions of our assay (data not shown), suggesting that overall basal Rap1 activation is low. This likely represents the level of Rap1 activation in glial cells, which are much more numerous than neurons and also express Rap1 (Fig. 3B). These results show that ephrin-A stimulation results in the inactivation of Rap GTPases in cultured neuronal cell lines and in the hippocampus.
SPAR mediates Rap1 and Rap2 inactivation downstream of EphA4
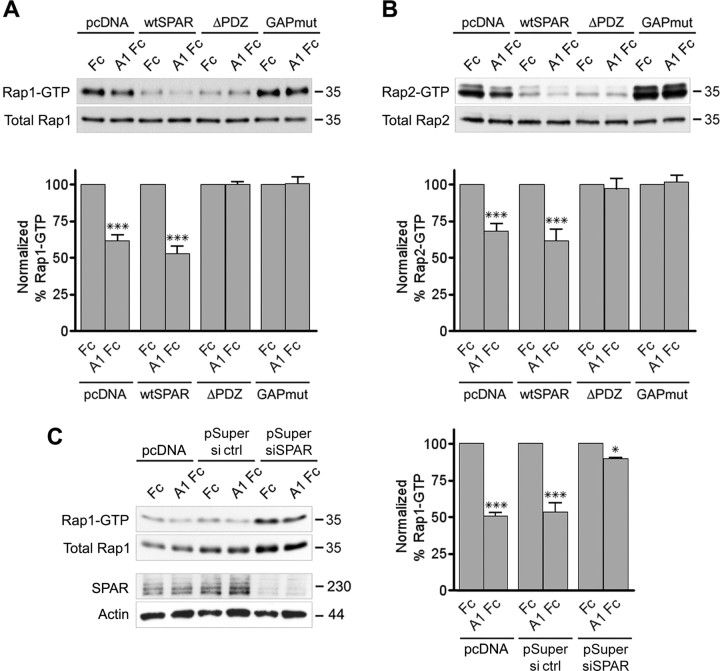
To investigate the involvement of SPAR in Rap GTPase inactivation downstream of EphA4, we transfected HT22 cells with wild-type or mutant SPAR. Given the low transfection efficiencies in these cells, we also cotransfected HA-tagged Rap1 or Myc-tagged Rap2 to allow analysis of Rap activity preferentially in the cells also expressing transfected SPAR. Overexpression of SPAR containing a functional GAP domain caused substantial inactivation of cotransfected Rap1 and Rap2 (blots in Fig. 4A,B, wtSPAR and ΔPDZ, Fc treatment), as previously shown for other Rap GTPase-activating proteins (Tsukamoto et al., 1999; Shimonaka et al., 2003; McLeod et al., 2004). Nevertheless, ephrin-A1 Fc stimulation still significantly decreased Rap1 and Rap2 activation in cells expressing wild-type SPAR (Fig. 4A,B, histograms), whereas Rap inactivation was blocked in cells expressing SPAR with an active GAP domain but lacking the PDZ domain (Fig. 4A,B, histogram). On the other hand, expression of SPAR with an inactive GAP domain increased the levels of GTP-bound Rap1 and Rap2, suggesting that this dominant-negative SPAR mutant (Pak et al., 2001) inhibits the basal activity of endogenous SPAR (blots in Fig. 4A,B, GAPmut, Fc treatment). The SPAR GAP mutant also blocked Rap inactivation after ephrin-A1 Fc stimulation (Fig. 4A,B, histograms). SPAR siRNA-mediated knock down also increased the basal level of Rap1 activation and inhibited Rap1 inactivation by ephrin-A1 Fc (Fig. 4C). These data indicate that SPAR mediates Rap1 and Rap2 inactivation downstream of EphA4 and that ephrin-dependent upregulation of SPAR GAP activity requires a physical interaction between EphA4 and SPAR.
Figure 4.
SPAR mediates ephrin-A-dependent inactivation of Rap1 and Rap2. A–C, HT22 cells were cotransfected with HA-Rap1 (A) or Myc-Rap2 (B) and the indicated SPAR constructs (see Fig. 2B) or HA-Rap1 and pSuper siRNA vector targeting SPAR (pSuper siRNA) or luciferase as a control (pSuper ctrl) (C). The transfected cells were stimulated with preclustered ephrin-A1 Fc (A1 Fc) or Fc control for 8 min. Active Rap1 and Rap2 were isolated by pull-down with GST-RalGDS RBD and detected by immunoblotting with antibodies against the HA or Myc tag. Total HA-Rap1, Myc-Rap2, endogenous SPAR, and β-actin were detected by immunoblotting cell lysates. The levels of each GTP-bound Rap protein were normalized to the corresponding levels of total Rap protein in the lysates. The histograms show the average levels of HA-Rap1-GTP from three experiments and of Myc-Rap2-GTP from five experiments ± SE expressed as a percentage of the levels after Fc treatment of similarly transfected cells. Statistical significance was determined by a two-way ANOVA and Bonferroni posttest. *p < 0.05; ***p < 0.001 for the comparison between ephrin-A1 Fc-treated and Fc-treated.
SPAR mediates ephrin-A-dependent growth cone collapse
Rap1, Rap2, SPAR, and EphA4 were all found to be expressed in cultured hippocampal neurons (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material), consistent with previous reports (Pak et al., 2001; Fu et al., 2007b). Interestingly, SPAR became detectable only 8 h after plating, suggesting that its expression is upregulated concomitant with the extension of neuronal processes (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). EphA4, SPAR, and Rap immunoreactivities were also all detectable in the growth cones of hippocampal neurons (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). To determine whether SPAR plays a role in the growth cone collapse that occurs when cultured hippocampal neurons are stimulated with ephrin-A1 Fc, we cotransfected SPAR constructs together with EGFP to mark the transfected neurons. After 2 d in culture, the neurons were stimulated with ephrin-A1 Fc and immunostained for α-tubulin and F-actin to visualize growth cones. The transfected dominant-negative SPAR GAP mutant but not wild-type SPAR inhibited ephrin-A1 Fc-induced growth cone collapse (collapsed growth cones: in control-transfected neurons 25.8 ± 3.0% for Fc vs 65.0 ± 2.7% for ephrin-A1 Fc, p < 0.001; in wtSPAR-transfected neurons 21.7 ± 1.0% for Fc vs 50.8 ± 1.6% for ephrin-A1 Fc, p < 0.001; in SPAR GAPmut-transfected neurons 13.1 ± 1.6% for Fc vs 19.6 ± 0.03% for ephrin-A1 Fc, p > 0.05) (Fig. 5A,B). This result suggests that SPAR GAP activity is required for ephrin-A-dependent growth cone collapse.
Figure 5.
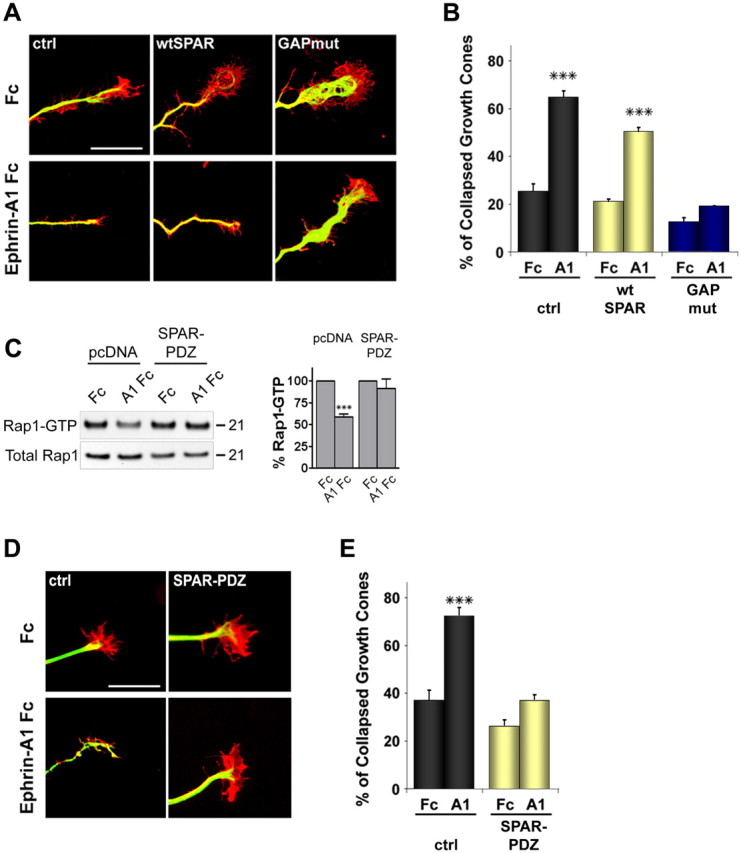
SPAR mediates ephrin-A-dependent growth cone collapse. A, Primary hippocampal neurons were cotransfected before plating with wtSPAR, GAPmut, or pcDNA3-Myc as a control (ctrl) together with EGFP. Neurons were stimulated with ephrin-A1 Fc or Fc as a control, fixed, and stained for tubulin (Alexa-635 digitally converted to green) and F-actin (red). Scale bar, 10 μm. B, The histogram shows average percentages of collapsed growth cones ± SE. ***p < 0.001 by two-way ANOVA and Bonferroni posttest. C, HT22 cells were cotransfected with HA-Rap1 and the SPAR PDZ domain or pcDNA3 as a control. The transfected cells were stimulated with preclustered ephrin-A1 Fc (A1 Fc) or Fc control for 8 min. Active Rap1 was isolated by pull-down with GST-RalGDS RBD and detected by immunoblotting with antibodies against the HA tag. Total Rap1 was detected by immunoblotting cell lysates. The levels of GTP-bound Rap1 were normalized to the corresponding levels of total Rap protein in the lysates. For each transfection, the histogram shows the average levels of Rap1-GTP ± SE from three experiments expressed as a percentage of the levels after Fc treatment. Statistical significance was determined by a two-way ANOVA and Bonferroni posttest. ***p < 0.001 for the comparison between ephrin-A1 Fc-treated and Fc-treated. D, Neurons were cotransfected with a construct encoding the SPAR PDZ domain or pcDNA3-Myc as a control (ctrl) together with EGFP. Neurons were stimulated with ephrin-A1 Fc or Fc as a control, fixed, and stained as in A. Scale bar, 10 μm. E, The histogram shows average percentages of collapsed growth cones ± SE from experiments that also included the transfections shown in Figure 6D. **p < 0.01 by two-way ANOVA and Bonferroni posttest.
We also transfected the isolated SPAR PDZ domain to uncouple EphA4 from SPAR. This inhibited Rap1 inactivation in HT22 cells (Fig. 5C) and growth cone collapse in neurons treated with ephrin-A1 Fc (collapsed growth cones: in control-transfected neurons 37.0 ± 4.3% for Fc versus 72.5 ± 3.5% for ephrin-A1 Fc, p < 0.001; in neurons transfected with the SPAR PDZ domain 21.4 ± 1.9% for Fc vs 30.3 ± 1.6% for ephrin-A1 Fc, p > 0.05) (Fig. 5D,E), supporting the importance of a physical interaction between EphA4 and SPAR for downstream Rap inactivation and growth cone collapse.
Rap1 but not Rap2 inactivation is required for ephrin-A-dependent growth cone collapse
The finding that SPAR mutants inhibit Rap inactivation and growth cone collapse in response to ephrin-A1 suggested a role for Rap1 and/or Rap2 inactivation in growth cone collapse downstream of EphA4. We were interested in discriminating the roles of these two GTPases in growth cone collapse. However, dominant-negative S17N mutants of the individual GTPases were not useful because transfection of each of these mutants caused inactivation of endogenous Rap1 as well as Rap2 (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). RNAi knock-down was not attempted because there are two Rap1 and three Rap2 genes. The GTPase-activating protein Rap1GAP has been shown to preferentially inactivate Rap1 in in vitro assays (Rubinfeld et al., 1991). However, in our hands transfected Rap1GAP caused extensive inactivation of both Rap1 and Rap2 (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material). To overcome these difficulties, we used constitutively active forms of the two proteins, Rap1 G12V and Rap2 Q63E (supplemental Fig. 2C, available at www.jneurosci.org as supplemental material) (Kitayama et al., 1990; Kelley et al., 2004), to separately examine the effects of Rap1 and Rap2.
Transfection of hippocampal neurons showed that constitutively active Rap1 abrogates ephrin-A1 Fc-dependent growth cone collapse (collapsed growth cones: in control-transfected neurons 25.6 ± 5.5% for Fc versus 79.5 ± 10.9% for ephrin-A1 Fc, p < 0.001; in RapG12V-transfected neurons 18.2 ± 1.8% for Fc versus 31.3 ± 0.9% for ephrin-A1 Fc, p > 0.05) (Fig. 6A,C). In contrast, growth cones transfected with wild-type Rap1, which can be inactivated by GTPase-activating proteins, underwent significant collapse in response to ephrin-A1 Fc (collapsed growth cones: in wtRap-transfected neurons 33.8 ± 3.4% for Fc vs 57.8 ± 2.6% for ephrin-A1 Fc, p < 0.05). Growth cone collapse still occurred in neurons transfected with constitutively active Rap2 Q63E (collapsed growth cones: in control-transfected neurons 37.0 ± 4.3% for Fc vs 72.0 ± 3.5% for ephrin-A1 Fc, p < 0.001; in wtRap2-transfected neurons 36.6 ± 1.6% for Fc vs 70.6 ± 1.6% for ephrin-A1 Fc, p < 0.001; in Rap2 Q63E-transfected neurons 54.6 ± 0.9% for Fc versus 88.4 ± 5.7% for ephrin-A1 Fc, p < 0.001) (Fig. 6B,D). These data indicate that inactivation of Rap1, but not Rap2, is required for growth cone collapse caused by ephrin-A stimulation, suggesting that Rap1 and Rap2 have different functions in axon guidance.
Figure 6.
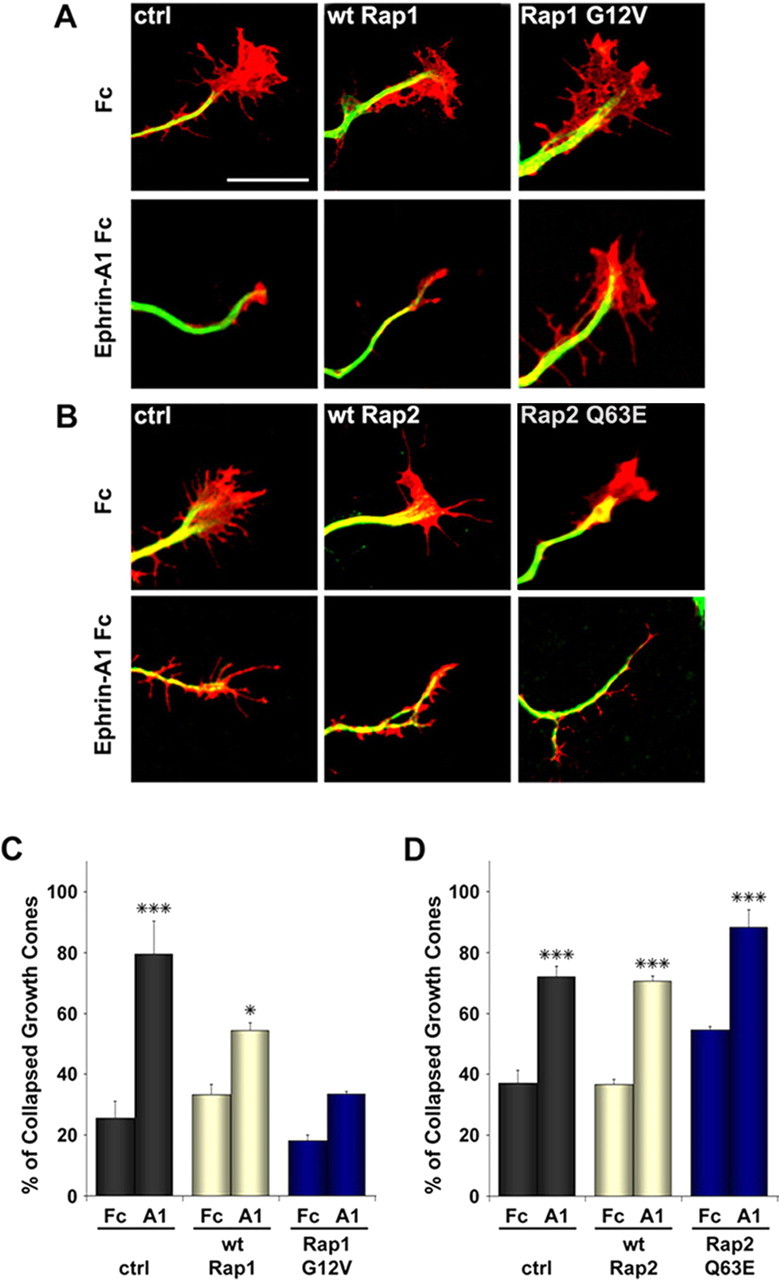
Constitutively active Rap1 but not Rap2 inhibits ephrin-A-dependent growth cone collapse in hippocampal neurons. A, B, Primary hippocampal neurons were cotransfected before plating with the indicated HA-tagged Rap1 or Myc-tagged Rap2 constructs or pcDNA3-Myc as a control (ctrl) together with EGFP, cultured for 2 d, and then stimulated with ephrin-A1 Fc or Fc as a control. Neurons were then fixed and stained for α-tubulin (Alexa-647 digitally converted to green) and F-actin (red). Scale bar, 10 μm. C, D, The histograms show the mean percentages of collapsed growth cones ± SE. *p < 0.05; **p < 0.01; ***p < 0.001 by two-way ANOVA and Bonferroni posttest.
Rap1 and Rap2 have different effects on neuronal morphology
To further investigate possible differences in the biological activities of Rap1 and Rap2 in neurons, we examined the effects of constitutively active Rap constructs on growth cone size and dendritic spine morphology. Interestingly, Rap1 G12V caused a pronounced increase in the area of axonal growth cones when compared with wild-type Rap1 and vector control (growth cone area: 49.8 ± 3.7 μm2 for control; 38.6 ± 2.6 μm2 for wtRap1; 70.7 ± 4.4 μm2 for Rap1 G12V) (Fig. 7). On the other hand, growth cones from neurons transfected with Rap2 Q63E were smaller than control-transfected growth cones (growth cone area: 45.0 ± 2.6 μm2 for control; 37.5 ± 2.9 μm2 for wtRap2; 26.2 ± 1.9 μm2 for Rap2 Q63E) (Fig. 7). In contrast to these opposite effects on growth cone size (enlargement by active Rap1 and reduced size by active Rap2), axon length was not significantly changed by either constitutively active Rap1 or Rap2 (data not shown).
Figure 7.
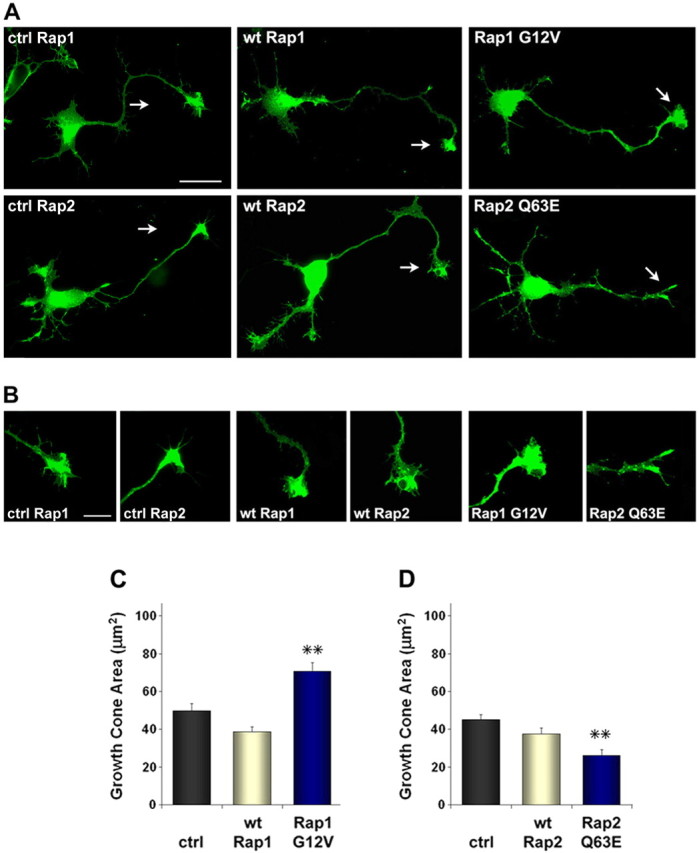
Active Rap1 increases and active Rap2 decreases growth cone size in hippocampal neurons. A, Primary hippocampal neurons were cotransfected before plating with the indicated HA-tagged Rap1 or Myc-tagged Rap2 constructs or pcDNA3-Myc as a control (ctrl) together with EGFP. Neurons were fixed and photographed after 2 d in culture. Scale bar, 25 μm. B, Digitally enlarged images of growth cones. Scale bar, 10 μm. C, D, The histograms show the average size of growth cones ± SE in neurons transfected as in A and B and, in addition, control neurons transfected with pcDNA3-Myc (ctrl). **p < 0.01 compared with control by one-way ANOVA and Dunnett's posttest.
Rap1, Rap2, SPAR, and EphA4 are all present in brain synaptosomal preparations, including the postsynaptic density fraction (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material) (Husi et al., 2000; Pak et al., 2001; Grunwald et al., 2004; Xie et al., 2005), suggesting that the EphA4-SPAR signaling pathway could also function in dendritic spines. Because overexpression of constitutively active Rap1 Q63E was previously shown to induce spine elongation without affecting spine density (Xie et al., 2005), we examined the effects of constitutively active Rap2 Q63E on dendritic spines. Primary hippocampal neurons cultured for 10 d were transfected with Rap2 constructs together with EGFP. At 21 d in culture, neurons expressing wild-type Rap2 and constitutively active Rap2 had significantly shorter spines than neurons transfected with control empty vector (spine length: 1.15 ± 0.02 μm for control; 0.99 ± 0.01 μm for wtRap2; 0.72 ± 0.02 μm for Rap2 Q63E) (supplemental Fig. 3B,C, available at www.jneurosci.org as supplemental material). The width of dendritic spines transfected with the Rap2 constructs was slightly decreased when compared with control-transfected neurons (spine width: 0.57 ± 0.01 μm for control; 0.50 ± 0.01 μm for wtRap2; 0.47 ± 0.01 μm for Rap2 Q63E). Constitutively active Rap2, but not wild-type Rap2, also decreased spine density (6.3 ± 0.37 spines/10 μm for control; 5.7 ± 0.33 spines/10 μm for wtRap2; 3.2 ± 0.25 spines/10 μm for Rap2 Q63E) (supplemental Fig. 3B,C, available at www.jneurosci.org as supplemental material) as previously reported (Fu et al., 2007b). These effects of Rap2 suggest that Rap1 and Rap2 have distinct effects not only on growth cone morphology but also on dendritic spine morphology and density.
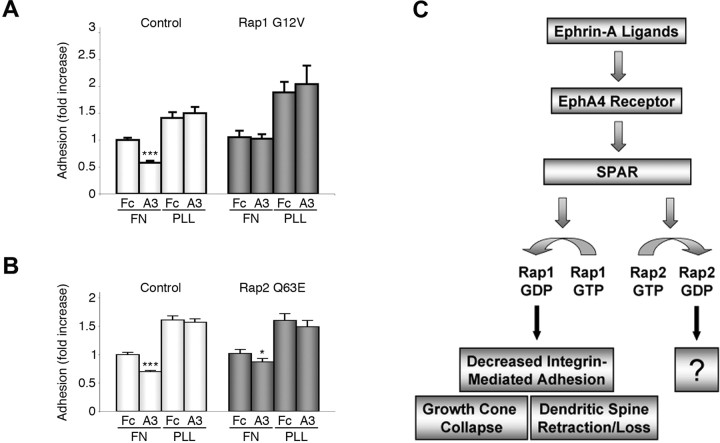
Rap1 but not Rap2 inactivation is required for ephrin-A-dependent downregulation of integrin-mediated adhesion in neuronal cells
The effects of Rap1 and Rap2 on growth cone size and ephrin-induced growth cone collapse suggest that these GTPases may differentially affect integrin-mediated cell adhesion of hippocampal neurons. We recently reported that ephrin-A treatment inhibits β1 integrin activity in HT22 cells and that inactivation of β1 integrins is required for reduction of dendritic spine length and density downstream of EphA4 in hippocampal neurons (Bourgin et al., 2007). Because GTP-bound Rap1 is well known to promote integrin-mediated adhesion in non-neuronal cells (Caron, 2003; Stork, 2003; Bos, 2005), Rap1 inactivation by SPAR may play a role in the downregulation of integrin function downstream of EphA4. Indeed, we found that transfection of constitutively active Rap1 G12V blocked the negative effects of ephrin-A3 Fc on HT22 cell attachment to the integrin ligand fibronectin (Fig. 8A). In contrast, a significant decrease in cell attachment after ephrin-A3 Fc stimulation was still observed in cells transfected with constitutively active Rap2 Q63E, although the decrease in cell attachment was less pronounced compared with control-transfected cells (Fig. 8B). Together, our data indicate that inactivation of Rap1 and not Rap2 is required for EphA4-dependent decrease in integrin-mediated adhesion in neurons.
Figure 8.
Constitutively active Rap1 but not Rap2 blocks ephrin-A3-dependent inhibition of integrin-mediated cell attachment. A, B, HT22 cells were transfected with the constitutively active Rap1 G12V (A), Rap2 Q63E (B), or pcDNA3 (control) together with EGFP and plated for 10 min on fibronectin (FN) or poly-l-lysine (PLL). The histograms shows the average percentage of EGFP-positive adherent cells ± SE from four experiments, corrected for transfection efficiency and normalized to control-transfected cells plated on fibronectin and treated with control Fc. *p < 0.05 and ***p < 0.001 for the comparison of ephrin-A1 Fc-treated cells with the corresponding Fc-treated cells by two-way ANOVA. C, Schematic illustration of the signaling pathway linking EphA4 receptor stimulation by ephrins in neurons to SPAR-mediated inactivation of Rap GTPases and morphological changes. Rap1 inactivation contributes to ephrin-induced growth cone collapse and loss of integrin-mediated attachment, whereas the effects of Rap2 inactivation downstream of EphA4 remain to be determined.
Discussion
We have identified a novel EphA receptor signaling pathway that couples EphA4 with the regulation of the small GTPases, Rap1 and Rap2 (Fig. 8C). This signaling pathway depends on an interaction of the C terminus of EphA4 with the PDZ domain of the Rap GTPase-activating protein, SPAR. We found that ephrin stimulation plays a key role in SPAR-dependent Rap1 inactivation and that this signaling is important for growth cone collapse and downregulating integrin-mediated adhesion. We also demonstrated that Rap1 and Rap2 have differential effects on neuronal morphology and signaling downstream of EphA4. These results provide novel insight into signaling mechanisms that may be critical for neural circuitry formation and maintenance.
SPAR, also known as E6TP1 (E6-targeted protein 1) and SPA1L1 (SpaI-like protein 1) (Deguchi et al., 1998; Gao et al., 1999; Roy et al., 2002), is the first protein found to interact with the PDZ domain-binding motifs of EphA4 and EphA6. Although other EphA receptors have been reported to associate with PDZ domain-containing proteins, the functional significance of these interactions is not yet known (Torres et al., 1998; Buchert et al., 1999). Associations mediated by the PDZ domain-binding motifs of EphB receptors (the other class of Eph receptors) have been shown to regulate Eph receptor trafficking and clustering in dendrites, dendrite morphogenesis, and glutamatergic synaptogenesis (Torres et al., 1998; Hoogenraad et al., 2005; Kayser et al., 2006). It will be interesting to examine whether SPAR may link EphA4 to actin filaments through its actin-binding domain 1 (Act1) and Act2 actin-binding domains and to PSD-95 through its guanylate kinase-binding domain (Pak et al., 2001), in addition to promoting Rap inactivation.
Our data show that EphA4 is a physiological upstream regulator of SPAR. Interaction with ligand-activated EphA4 promotes SPAR-dependent inactivation of Rap1 and Rap2 concomitant with elevation of SPAR tyrosine phosphorylation. Phosphorylation may regulate SPAR activity, because changes in SPAR phosphorylation have also been observed after NMDA receptor activation (Roy et al., 2002). Furthermore, tyrosine phosphorylation is known to modulate the function of other GTPase-activating proteins (Park and Jove, 1993; Roof et al., 2000). The similarities in the morphological effects of EphA4 and SPAR on dendritic spines are consistent with a function of these two proteins in a common signaling pathway. For example, hippocampal pyramidal neurons transfected with an EphA4 kinase inactive mutant and pyramidal neurons from EphA4 knock-out mice have elongated dendritic spines (Murai et al., 2003). Similarly, SPAR degradation induced by SNK (serum-inducible kinase) and SPAR inactivation achieved by transfection of the dominant-negative GAP domain mutant result in spine elongation (Pak et al., 2001; Pak and Sheng, 2003). Furthermore, activation of EphA4 and transfection of active SPAR lacking the Act2 actin-binding domain both cause spine shortening. Thus, both EphA4 kinase activity and SPAR GAP activity suppress dendritic spine elongation. EphA4 signaling and SPAR transfection also decrease spine density. Furthermore, we found that transfection of dominant-negative SPAR with an inactive GAP domain or uncoupling of EphA4 and SPAR by the overexpressed SPAR PDZ domain abrogate ephrin-A-induced growth cone collapse in hippocampal neurons, suggesting a role for this pathway in axon guidance.
SPAR-mediated inactivation of Rap1 likely contributes to the morphological changes induced by EphA4 signaling in neurons, as indicated by our results showing that expression of constitutively active Rap1 G12V blocks the growth cone collapsing and integrin inactivating effects of ephrin-A ligands. Consistent with a role for Rap1 in mediating the morphological effects of Eph receptors, stimulation of EphB2 with ephrin-B1 Fc has also been recently shown to inactivate Rap1 in colon carcinoma cell lines, leading to retraction of the cell periphery and cell detachment (Riedl et al., 2005). These morphological changes were abrogated by transfection of constitutively active Rap1 G12V, but the mechanism of Rap1 inactivation was not defined. It will be interesting to determine whether SPAR and the two other GTPase-activating proteins of the same family, SPAL2 and SPAL3 (Roy et al., 2002), may link different EphA and EphB receptors to Rap1 and Rap2 inactivation in different cell types. In brain neuroepithelial cells and aortic endothelial cells, Eph receptors have instead been reported to activate Rap1 leading to neuronal differentiation and cell spreading, respectively (Nagashima et al., 2002; Aoki et al., 2004). Thus, Eph receptors might also regulate Rap1 activity through different mechanisms depending on the cellular context and the Eph receptor involved.
Rap1 is well known to promote neuronal differentiation and neurite outgrowth (York et al., 1998; Yamada et al., 2005) and has been recently implicated in the depolarization-induced growth of cortical dendrites (Chen et al., 2005) and activity-dependent dendritic spine remodeling (Xie et al., 2005). Furthermore, Rap1B has been shown to play a critical role in axon specification in hippocampal neurons (Schwamborn and Puschel, 2004). A role for Rap1 in growth cone spreading has not been previously reported, but is consistent with the known role of Rap1 in promoting integrin-mediated adhesion, actin polymerization, and cell spreading (Ohba et al., 2001; Stork, 2003; Enserink et al., 2004; Bos, 2005). Our data suggest that Rap1 inactivation decreases integrin-mediated adhesion downstream of EphA4 in neuronal cells. We also recently reported that EphA4 regulates dendritic spine morphology by influencing integrin activity (Bourgin et al., 2007). That growth cones and dendritic spines use some of the same mechanisms for remodeling is not surprising, given the importance of cell adhesion and the actin cytoskeleton for the architecture of both structures. Thus, we propose a model where decreased integrin activity through SPAR-mediated Rap1 inactivation contributes to ephrin-A-dependent growth cone collapse and decreased dendritic spine length and density (Fig. 8C). However, inactivation of Rap1 (and Rap2) may not be sufficient for the effects of EphA4 on spine structural plasticity because transfection of Rap1GAP did not cause dendritic spine shortening and loss, at least in cortical neurons (Xie et al., 2005). Similarly, transfection of Rap1GAP was not sufficient to mimic the cell retraction and detachment effects of ephrin-B1 in colon cancer cells (Riedl et al., 2005).
Our results suggest that Rap2 inactivation does not have as critical a role as Rap1 in ephrin-A-dependent growth cone collapse and inhibition of integrin activity in neuronal cells. Furthermore, we found that constitutively active Rap2 decreases growth cone size and dendritic spine length and density, whereas constitutively active Rap1 increases growth cone size (Fig. 7) and spine length (Xie et al., 2005). The dissimilar morphological effects of Rap1 and Rap2 on growth cones and dendritic spines are surprising because these two GTPases are closely related (∼60% amino acid identity), share a number of the same regulators, and have some analogous functions (Ohba et al., 2000, 2001; Zhu et al., 2002, 2005; McLeod et al., 2004; Fu et al., 2007b). However, one of the amino acids in the effector domain is different between Rap1 and Rap2 (Caron, 2003), and some effectors may selectively mediate Rap1 or Rap2 signals. For example, Rap1-dependent spine elongation in hippocampal neurons requires the scaffolding protein AF6, which preferentially interacts with Rap1 over Rap2 and may selectively link Rap1 to regulators of the actin cytoskeleton (Boettner et al., 2000; Xie et al., 2005). Active Rap1 has also been shown to influence the subcellular localization of Rac and Rho regulatory proteins, leading to localized cell spreading and the formation of cell protrusions (Arthur et al., 2004; Yamada et al., 2005), consistent with positive effects of Rap1 on growth cone and dendritic spine size. On the other hand, Rap2 but not Rap1 interacts with TNIK (Traf2- and Nck-interacting kinase), a kinase expressed in the brain (Machida et al., 2004; Taira et al., 2004; Zhu et al., 2005). Binding of activated Rap2 enhances the ability of TNIK to decrease F-actin levels and inhibit cell spreading. Such effects are consistent with the decreased size of growth cones and dendritic spines that we have observed in neurons transfected with constitutively active Rap2. Whether the direct association of Rap2 with actin filaments (Torti et al., 1999) may also influence its effects on neuronal morphology remains to be determined.
In addition to affecting neuronal morphology, inactivation of Rap1 and Rap2 downstream of EphA4 and SPAR would be expected to enhance synaptic transmission, because Rap1 and Rap2 have been shown to drive the removal of synaptic AMPA receptors that underlies long-term depression (LTD) and depotentiation, respectively (Zhu et al., 2002, 2005; Fu et al., 2007b). It will be interesting to determine whether EphA4-dependent Rap inactivation is responsible for the long-term potentiation-like effects observed after stimulation of hippocampal slices with an ephrin-A Fc ligand (Gao et al., 1998).
The novel EphA4–SPAR–Rap1 pathway that we have identified likely acts in concert with other Ras and Rho pathways that are also activated downstream of EphA receptors to sculpt neuronal morphology and control the establishment and remodeling of neuronal connections (Cowan et al., 2005; Sahin et al., 2005; Beg et al., 2007; Fu et al., 2007a; Iwasato et al., 2007; Wegmeyer et al., 2007). Because SPAR is a ubiquitously expressed protein and part of a family of three related proteins, the Eph-SPAR signaling connection may operate widely to regulate adhesion and morphology of many cell types through control over the different activities of Rap1 and Rap2.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants NS454092 and HD025938 (E.B.P.), Deutsche Forschungsgemeinschaft postdoctoral fellowship RI 1208/1-2 (M.R.), a postdoctoral fellowship from the Fondation pour la Recherche Medicale (C.B.), and NIH postdoctoral fellowship NS43029 (K.K.M.). We are grateful to P. Stork for providing the Rap1GAP construct; F. Irie for providing the EphA4 K653R construct; J. Yu for the pSuper luciferase siRNA vector; L. N. Nguyen, F. Valencia, and D. Melendez for excellent technical assistance; J. Meerloo and E. Monosov for assistance with microscopy; and N. K. Noren for helpful comments on this manuscript. M.R. performed all the experiments shown in this manuscript, except those in Figure 8, and wrote this manuscript together with E.B.P. K.K.M. found that Rap2 is inactivated by EphA4, performed initial Rap2 overexpression experiments in hippocampal slices, and contributed to plasmid construction and editing this manuscript. C.B. performed the cell attachment assays shown in Figure 8 and helped with editing this manuscript. D.T.P. provided the SPAR constructs and siRNA vector and assisted with editing this manuscript. E.B.P. oversaw all aspects of the project and wrote this manuscript with M.R.
References
- Aoki M, Yamashita T, Tohyama M. EphA receptors direct the differentiation of mammalian neural precursor cells through a mitogen-activated protein kinase-dependent pathway. J Biol Chem. 2004;279:32643–32650. doi: 10.1074/jbc.M313247200. [DOI] [PubMed] [Google Scholar]
- Arthur WT, Quilliam LA, Cooper JA. Rap1 promotes cell spreading by localizing Rac guanine nucleotide exchange factors. J Cell Biol. 2004;167:111–122. doi: 10.1083/jcb.200404068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AA, Sommer JE, Martin JH, Scheiffele P. alpha2-Chimaerin is an essential EphA4 effector in the assembly of neuronal locomotor circuits. Neuron. 2007;55:768–778. doi: 10.1016/j.neuron.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc Natl Acad Sci USA. 2000;97:9064–9069. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Bourgin C, Murai KK, Richter M, Pasquale EB. The EphA4 receptor regulates dendritic spine remodeling by affecting beta1 integrin signaling pathways. J Cell Biol. 2007;178:1295–1307. doi: 10.1083/jcb.200610139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert M, Schneider S, Meskenaite V, Adams MT, Canaani E, Baechi T, Moelling K, Hovens CM. The junction-associated protein AF-6 interacts and clusters with specific Eph receptor tyrosine kinases at specialized sites of cell-cell contact in the brain. J Cell Biol. 1999;144:361–371. doi: 10.1083/jcb.144.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J Cell Sci. 2003;116:435–440. doi: 10.1242/jcs.00238. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang PY, Ghosh A. Regulation of cortical dendrite development by Rap1 signaling. Mol Cell Neurosci. 2005;28:215–228. doi: 10.1016/j.mcn.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Vav family GEFs link activated Ephs to endocytosis and axon guidance. Neuron. 2005;46:205–217. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Dail M, Richter M, Godement P, Pasquale EB. Eph receptors inactivate R-Ras through different mechanisms to achieve cell repulsion. J Cell Sci. 2006;119:1244–1254. doi: 10.1242/jcs.02842. [DOI] [PubMed] [Google Scholar]
- Deguchi M, Hata Y, Takeuchi M, Ide N, Hirao K, Yao I, Irie M, Toyoda A, Takai Y. BEGAIN (brain-enriched guanylate kinase-associated protein), a novel neuronal PSD-95/SAP90-binding protein. J Biol Chem. 1998;273:26269–26272. doi: 10.1074/jbc.273.41.26269. [DOI] [PubMed] [Google Scholar]
- Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007;17:230–238. doi: 10.1016/j.tcb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Enserink JM, Price LS, Methi T, Mahic M, Sonnenberg A, Bos JL, Tasken K. The cAMP-Epac-Rap1 pathway regulates cell spreading and cell adhesion to laminin-5 through the alpha3beta1 integrin but not the alpha6beta4 integrin. J Biol Chem. 2004;279:44889–44896. doi: 10.1074/jbc.M404599200. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Irie F, Kalo MS, Couchman JR, Pasquale EB, Yamaguchi Y. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Franke B, Akkerman JW, Bos JL. Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci. 2007a;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- Fu Z, Lee SH, Simonetta A, Hansen J, Sheng M, Pak DT. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J Neurochem. 2007b;100:118–131. doi: 10.1111/j.1471-4159.2006.04195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Srinivasan S, Boyer SN, Wazer DE, Band V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao WQ, Shinsky N, Armanini MP, Moran P, Zheng JL, Mendoza-Ramirez JL, Phillips HS, Winslow JW, Caras IW. Regulation of hippocampal synaptic plasticity by the tyrosine kinase receptor, REK7/EphA5, and its ligand, AL-1/Ephrin-A5. Mol Cell Neurosci. 1998;11:247–259. doi: 10.1006/mcne.1998.0696. [DOI] [PubMed] [Google Scholar]
- Grunwald IC, Korte M, Adelmann G, Plueck A, Kullander K, Adams RH, Frotscher M, Bonhoeffer T, Klein R. Hippocampal plasticity requires postsynaptic ephrinBs. Nat Neurosci. 2004;7:33–40. doi: 10.1038/nn1164. [DOI] [PubMed] [Google Scholar]
- Helmbacher F, Schneider-Maunoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development. 2000;127:3313–3324. doi: 10.1242/dev.127.15.3313. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Iwasato T, Katoh H, Nishimaru H, Ishikawa Y, Inoue H, Saito YM, Ando R, Iwama M, Takahashi R, Negishi M, Itohara S. Rac-GAP alpha-chimerin regulates motor-circuit formation as a key mediator of EphrinB3/EphA4 forward signaling. Cell. 2007;130:742–753. doi: 10.1016/j.cell.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Kayser MS, McClelland AC, Hughes EG, Dalva MB. Intracellular and trans-synaptic regulation of glutamatergic synaptogenesis by EphB receptors. J Neurosci. 2006;26:12152–12164. doi: 10.1523/JNEUROSCI.3072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley GG, Reks SE, Smrcka AV. Hormonal regulation of phospholipase Cepsilon through distinct and overlapping pathways involving G12 and Ras family G-proteins. Biochem J. 2004;378:129–139. doi: 10.1042/BJ20031370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama H, Matsuzaki T, Ikawa Y, Noda M. Genetic analysis of the Kirsten-ras-revertant 1 gene: potentiation of its tumor suppressor activity by specific point mutations. Proc Natl Acad Sci USA. 1990;87:4284–4288. doi: 10.1073/pnas.87.11.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullander K, Mather NK, Diella F, Dottori M, Boyd AW, Klein R. Kinase-dependent and kinase-independent functions of EphA4 receptors in major axon tract formation in vivo. Neuron. 2001;29:73–84. doi: 10.1016/s0896-6273(01)00181-7. [DOI] [PubMed] [Google Scholar]
- Kullander K, Butt SJ, Lebret JM, Lundfald L, Restrepo CE, Rydstrom A, Klein R, Kiehn O. Role of EphA4 and EphrinB3 in local neuronal circuits that control walking. Science. 2003;299:1889–1892. doi: 10.1126/science.1079641. [DOI] [PubMed] [Google Scholar]
- Li Y, Maher P, Schubert D. A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron. 1997;19:453–463. doi: 10.1016/s0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- Machida N, Umikawa M, Takei K, Sakima N, Myagmar BE, Taira K, Uezato H, Ogawa Y, Kariya K. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem. 2004;279:15711–15714. doi: 10.1074/jbc.C300542200. [DOI] [PubMed] [Google Scholar]
- McLeod SJ, Shum AJ, Lee RL, Takei F, Gold MR. The Rap GTPases regulate integrin-mediated adhesion, cell spreading, actin polymerization, and Pyk2 tyrosine phosphorylation in B lymphocytes. J Biol Chem. 2004;279:12009–12019. doi: 10.1074/jbc.M313098200. [DOI] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat Neurosci. 2003;6:153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- Nagashima K, Endo A, Ogita H, Kawana A, Yamagishi A, Kitabatake A, Matsuda M, Mochizuki N. Adaptor protein Crk is required for Ephrin-B1-induced membrane ruffling and focal complex assembly of human aortic endothelial cells. Mol Biol Cell. 2002;13:4231–4242. doi: 10.1091/mbc.E02-04-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Pasquale EB. Eph receptor-ephrin bidirectional signals that target Ras and Rho proteins. Cell Signal. 2004;16:655–666. doi: 10.1016/j.cellsig.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Ohba Y, Mochizuki N, Matsuo K, Yamashita S, Nakaya M, Hashimoto Y, Hamaguchi M, Kurata T, Nagashima K, Matsuda M. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol Cell Biol. 2000;20:6074–6083. doi: 10.1128/mcb.20.16.6074-6083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y, Ikuta K, Ogura A, Matsuda J, Mochizuki N, Nagashima K, Kurokawa K, Mayer BJ, Maki K, Miyazaki J, Matsuda M. Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis. EMBO J. 2001;20:3333–3341. doi: 10.1093/emboj/20.13.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Wilkinson DG. Eph receptors and ephrins in neural development. Curr Opin Neurobiol. 1999;9:65–73. doi: 10.1016/s0959-4388(99)80008-7. [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- Park S, Jove R. Tyrosine phosphorylation of Ras GTPase-activating protein stabilizes its association with p62 at membranes of v-Src transformed cells. J Biol Chem. 1993;268:25728–25734. [PubMed] [Google Scholar]
- Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- Riedl JA, Brandt DT, Batlle E, Price LS, Clevers H, Bos JL. Down-regulation of Rap1 activity is involved in ephrinB1-induced cell contraction. Biochem J. 2005;389:465–469. doi: 10.1042/BJ20050048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Lemaire S. Role of the sigma receptor in the inhibition of [3H]-noradrenaline uptake in brain synaptosomes and adrenal chromaffin cells. Br J Pharmacol. 1991;103:1917–1922. doi: 10.1111/j.1476-5381.1991.tb12352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof RW, Dukes BD, Chang JH, Parsons SJ. Phosphorylation of the p190 RhoGAP N-terminal domain by c-Src results in a loss of GTP binding activity. FEBS Lett. 2000;472:117–121. doi: 10.1016/s0014-5793(00)01439-3. [DOI] [PubMed] [Google Scholar]
- Roy BC, Kohu K, Matsuura K, Yanai H, Akiyama T. SPAL, a Rap-specific GTPase activating protein, is present in the NMDA receptor-PSD-95 complex in the hippocampus. Genes Cells. 2002;7:607–617. doi: 10.1046/j.1365-2443.2002.00546.x. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Munemitsu S, Clark R, Conroy L, Watt K, Crosier WJ, McCormick F, Polakis P. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–1042. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- Sahin M, Greer PL, Lin MZ, Poucher H, Eberhart J, Schmidt S, Wright TM, Shamah SM, O'Connell S, Cowan CW, Hu L, Goldberg JL, Debant A, Corfas G, Krull CE, Greenberg ME. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 2005;46:191–204. doi: 10.1016/j.neuron.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Puschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Shimonaka M, Katagiri K, Nakayama T, Fujita N, Tsuruo T, Yoshie O, Kinashi T. Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J Cell Biol. 2003;161:417–427. doi: 10.1083/jcb.200301133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soans C, Holash JA, Pasquale EB. Characterization of the expression of the Cek8 receptor-type tyrosine kinase during development and in tumor cell lines. Oncogene. 1994;9:3353–3361. [PubMed] [Google Scholar]
- Stork PJ. Does Rap1 deserve a bad Rap? Trends Biochem Sci. 2003;28:267–275. doi: 10.1016/S0968-0004(03)00087-2. [DOI] [PubMed] [Google Scholar]
- Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem. 2004;279:49488–49496. doi: 10.1074/jbc.M406370200. [DOI] [PubMed] [Google Scholar]
- Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- Torti M, Bertoni A, Canobbio I, Sinigaglia F, Lapetina EG, Balduini C. Interaction of the low-molecular-weight GTP-binding protein rap2 with the platelet cytoskeleton is mediated by direct binding to the actin filaments. J Cell Biochem. 1999;75:675–685. doi: 10.1002/(sici)1097-4644(19991215)75:4<675::aid-jcb13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Tsukamoto N, Hattori M, Yang H, Bos JL, Minato N. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J Biol Chem. 1999;274:18463–18469. doi: 10.1074/jbc.274.26.18463. [DOI] [PubMed] [Google Scholar]
- Wegmeyer H, Egea J, Rabe N, Gezelius H, Filosa A, Enjin A, Varoqueaux F, Deininger K, Schnutgen F, Brose N, Klein R, Kullander K, Betz A. EphA4-dependent axon guidance is mediated by the RacGAP alpha2-chimaerin. Neuron. 2007;55:756–767. doi: 10.1016/j.neuron.2007.07.038. [DOI] [PubMed] [Google Scholar]
- Xie Z, Huganir RL, Penzes P. Activity-dependent dendritic spine structural plasticity is regulated by small GTPase Rap1 and its target AF-6. Neuron. 2005;48:605–618. doi: 10.1016/j.neuron.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Yamada T, Sakisaka T, Hisata S, Baba T, Takai Y. RA-RhoGAP, Rap-activated Rho GTPase-activating protein implicated in neurite outgrowth through Rho. J Biol Chem. 2005;280:33026–33034. doi: 10.1074/jbc.M504587200. [DOI] [PubMed] [Google Scholar]
- York RD, Yao H, Dillon T, Ellig CL, Eckert SP, McCleskey EW, Stork PJ. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- Zafra F, Hengerer B, Leibrock J, Thoenen H, Lindholm D. Activity dependent regulation of BDNF and NGF mRNAs in the rat hippocampus is mediated by non-NMDA glutamate receptors. EMBO J. 1990;9:3545–3550. doi: 10.1002/j.1460-2075.1990.tb07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, Baumgart JP, Velamoor V, Auberson YP, Osten P, van Aelst L, Sheng M, Zhu JJ. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 2005;46:905–916. doi: 10.1016/j.neuron.2005.04.037. [DOI] [PubMed] [Google Scholar]