Summary:
Over two decades ago, it was discovered that the human T-cell repertoire contains T cells that do not recognize peptide antigens in the context of MHC molecules but instead respond to lipid antigens presented by CD1 antigen-presenting molecules. The ability of T cells to ‘see’ lipid antigens bound to CD1 enables these lymphocytes to sense changes in the lipid composition of cells and tissues as a result of infections, inflammation, or malignancies. Although foreign lipid antigens have been shown to function as antigens for CD1-restricted T cells, many CD1-restricted T cells do not require foreign antigens for activation but instead can be activated by self-lipids presented by CD1. This review highlights recent developments in the field, including the identification of common mammalian lipids that function as autoantigens for αβ and γδ T cells, a novel mode of T-cell activation whereby CD1a itself rather than lipids serves as the autoantigen, and various mechanisms by which the activation of CD1-autoreactive T cells is regulated. As CD1 can induce T-cell effector functions in the absence of foreign antigens, multiple mechanisms are in place to regulate this self-reactivity, and stimulatory CD1-lipid complexes appear to be tightly controlled in space and time.
Keywords: CD1 antigen-presenting molecules, lipid antigens, T cells
CD1 as a lipid antigen-presenting molecule
Since the discovery of the interactions between peptide-loaded major histocompatibility complex (MHC) complexes and αβ T-cell receptors (TCRs) (1–4), additional MHC class I homologs have been added to the list of TCR targets, including CD1, MR1, EPCR, and HFE in humans (reviewed in 5). These MHC class I like molecules all possess an antigen-binding groove, formed by α1 and α2 helices, but their grooves differ significantly in size and physicochemical properties, which in turn affect the nature of the ligands that they bind. To date, besides MHC, only CD1 and MR1 have been shown to present antigens to T cells. These antigens include lipid-based antigens presented by CD1 molecules (6) and recently described intermediates of the riboflavin pathway presented by MR1 (7, 8).
The finding that lipids could function as antigens for T cells (6) established a new paradigm for antigen recognition by T cells, which was no longer limited to protein-derived antigens. Thus, T cells can sense and respond to changes in lipid repertoire that result from infection, inflammation, and even malignancies. Since the identification of the first lipid antigen, C80 mycolic acid (6), a component of the mycobacterial cell wall, additional classes of lipid antigens have been identified, including glycolipids (9–13), phospholipids (14, 15), lysophospholipids (16, 17), gangliosides (18, 19), lipopeptides (20), amphipathic small molecules (21), and even oils (22). The first crystal structure of murine CD1 (23) revealed deep antigen-binding pockets or tunnels, lined with hydrophobic aminoacids, and the first crystal structure of CD1 bound to lipid showed how these hydrophobic tunnels interacted with the alkyl chains of lipid antigens, anchoring them inside the protein. The hydrophilic head-group of the antigen protruded from a portal on the surface of CD1b protein, where it was exposed to the aqueous environment (24). Subsequently, trimolecular structures of CD1-antigen and TCR showed that the TCR interacts with the surface of the CD1 molecule and the exposed hydrophilic part of the antigen (25).
Humans express five CD1 proteins, four of which are surface expressed antigen-presenting molecules (CD1a, CD1b, CD1c, and CD1d) (26, 27). Each of the four CD1 isoforms differ in size of the antigen-binding grooves (24, 28–30), intracellular trafficking patterns (31), lipid ligand repertoire (32), and tissue distribution of expression (33). Together with the observation that multiple CD1 isoforms have been maintained throughout mammalian evolution (34), this argues that each CD1 isoform plays a non-redundant role in the immune system.
CD1-restricted T cells in the human T-cell repertoire
αβ T cells
Genomes of muroid rodents, including common laboratory strains of mice, harbor only two CD1 genes, both encoding CD1d (35), and lack genes encoding other CD1 isoforms, presumably through a break in chromosome 1 that occurred during mammalian evolution (34). Therefore, studies of CD1 and CD1-restricted T-cell function in mice are likely to provide an incomplete view of the functions of lipid-specific T cells in humans. The most extensively studied subset of CD1-restricted T cells is a subset of CD1d-restricted T cells, present in both mice and humans, referred to as invariant natural killer T cells (iNKT cells) based on their expression of an invariant TCRα chain paired with a TCRβ chain with limited Vβ usage (TRAV11 TRAJ18 paired with TRBV13–2, TRBV29, TRBV1 in mice, and TRAV10 TRAJ18 paired with TRBV25 in humans) (36, 37). The development and in vivo functions of iNKT cells have been addressed in numerous reviews (38–48) and are therefore not be extensively discussed in this review.
Although in mice iNKT cells comprise a significant fraction of T cells [approximately 1% spleen T cells, and up to 30% of all lymphocytes in liver (49)], in humans their representation in the T-cell repertoire is significantly smaller [median 0.03% of the peripheral blood T cells, range <0.010–2.3% (50), 0.5% of liver T cells (51)], and it is unclear whether the role of iNKT cells in the human immune system is of equal importance as it is in that of mice. Studies of the polyclonal CD1-restricted T-cell repertoire and ex vivo frequency analysis of CD1-reactive T-cell clones in humans showed that CD1a- and CD1c-autoreactive T cells were most frequently detected, with frequencies that were estimated to be up to 10% of peripheral blood T cells (52–54). This autoreactivity, which is defined as reactivity to CD1-expressing antigen-presenting cells in the absence of exogenously added antigen, likely represents the recognition of endogenous lipids, and is a common feature of CD1-restricted T cells (55). Overall, the presence of CD1a- and CD1c-autoreactive T cells at significant frequencies in non-diseased individuals (52), supports the notion that these autoreactive T cells fulfill a physiological role in the immune system, yet this role remains incompletely understood. Recently, certain CD1c-restricted T cells were shown to respond to a class of lipids identified as methyl-lysophosphatidic acids (mPLA), which accumulated in leukemia cells (56). These mPLA-specifc T cells efficiently killed acute leukemia cells, but not non-transformed CD1c-expressing cells, and a potential role for these T cells in immune surveillance against hematological malignancies was proposed.
CD1a-autoreactive T cells have been suggested to play a role in skin immunity and homeostasis (53). CD1a has a restricted expression pattern in humans, where it is expressed on thymocytes (57), and on certain tissue resident dendritic cells subsets at variable levels (33, 58). However, the only cells that constitutively express very high levels of CD1a are Langerhans cells. These resident antigen-presenting cells of the epidermis express CD1a on their cell surface at higher levels even than MHC class II (59), and they form a contiguous network lipid antigen-presenting cells in the skin, and are also found in squamous epithelia of the mouth, esophagus, and genital tract (60–62). In line with the abundance of CD1a in the skin, the majority of CD1a-autoreactive T cells in the peripheral blood of healthy individuals was detected in a CD4+ T-cell subset that expressed cutaneous lymphocyte antigen (CLA), and other skin homing chemokine receptors, such as CCR4 and CCR10, suggesting that these T cells home to the skin (63, 64). Indeed, CD1a-autoreactive T cells were detected in normal dermis ex vivo and were activated by CD1a expressing epidermal Langerhans cells in vitro (53). Cytokine profiles of CD1a-autoreactive T cells showed a predominant upregulation of Interleukin-22 (IL-22), a cytokine that acts directly on keratinocytes resulting in production of anti-microbial peptides (e.g. β-defensin-2), increased proliferation and decreased differentiation (65–67). IL-22 plays a role in epithelial anti-microbial immunity, tissue remodeling and wound healing, and the expression of IL-22 by CD1a-autoreactive T cells suggests that lipid-specific T cells may contribute to these functions in the skin. In addition, IL-22 producing T cells have been implicated in T cell-mediated skin diseases such as psoriasis and atopic dermatitis (68–73), pointing to a potential role for lipid-specific T cells in these skin pathologies.
CD1a, CD1b, and CD1c tetramers
The development of tetramers, fluorescently labeled tetrameric complexes of MHC molecules bound to peptides (74), revolutionized the analysis of antigen-specific T cells and made it possible to quantify and phenotypically characterize specific T cells without the need for in vitro activation. Similarly, CD1d tetramers loaded with the iNKT cell agonist α-galactosylceramide have been used for many years to quantify and isolate both human and murine iNKT cells (75–77). Recently, newly developed CD1a, CD1b, CD1c tetramers, and dextramers loaded with known mycobacterial lipid antigens have proven ternary interactions between CD1, mycobacterial lipid and TCR, and identified populations of specific T cells against the mycobacterial antigens dideoxymycobactin (DDM), glucose monomycolate (GMM), and phosphomycoketide (PM) in M.tb-exposed individuals (78–80). Interestingly, these studies also revealed that in humans, iNKT cells and mucosa-associated invariant T (MAIT) cells (TRAV1–2 TRAJ33) (81) are not the only two T-cell subsets expressing a semi-invariant TCR. A subset of CD1b-restricted T cells also harbors a nearly invariant α-chain and β-chains with restricted variable gene usage (TRAV1–2 TRAJ9 with no N-region additions, paired with TRBV6–2). This T-cell subset has a high affinity for GMM bound to CD1b and is called germline-encoded mycolyl-reactive T cells (GEM T cells) (82). Although detected at very low levels in some healthy individuals, the frequency of these mycobacterial lipid-specific T cells is increased upon exposure to M.tb. In addition to GEM T cells, the repertoire contains another subset with restricted variable region usage, namely LDN5-like T cells, which have intermediate affinity for GMM bound to CD1b (TRAV17 paired with TRBV4) (83). The existence of T-cell populations with (semi) invariant TCRs among lipid-specific CD1b-restricted T cells likely derives from the non-polymorphic nature of CD1b molecules. As these T cells expand in response to M.tb exposure, the detection and quantification of GEM and LDN5-like T cells has the potential to be a diagnostic marker of M.tb. exposure, measured either through tetramer staining or high throughput TCR sequencing (84).
γδ and δ/αβ hybrid TCRs
The initial report of CD1 as an antigen-presenting molecule for T cells described two T cells clones, one with an αβ TCR restricted by CD1a, and one with a γδ TCR restricted by CD1c (85). In the years since this initial finding, sporadic reports have shown links between CD1 and γδ TCRs (14, 15, 86–88); however, the vast majority of reported CD1-restricted T-cell clones and semi-invariant polyclonal T-cell populations (e.g. iNKT cells, GEM T cells, LDN5-like T cells) express αβ TCRs. Renewed interest in the CD1 and γδ TCR connection came from studies using CD1d tetramers loaded with sulfatide, which demonstrated that in humans the majority of T cells binding to sulfatide tetramers were Vδ1 γδ T cells (89). In normal human gut, Vδ1 cells are abundant, and recently it was shown that an estimated 5% of the Vδ1 cells among intraepithelial lymphocytes (IELs) of the ileum responded to CD1d and sulfatide, and an even larger fraction (15%) was estimated to be reactive to CD1d with other endogenous lipid ligands (90). This is consistent with earlier reports of γδ CD1d-autoreactive T-cell clones among gut IELs (15, 91) and clearly puts lipid-reactive γδ T cells on the map as an important intestinal T-cell population. Another study showed CD1d-autoreactive Vδ1 T cells among in vitro expanded and CD1d-α-GalCer enriched PBMCs, some of which also recognized sulfatide (92). Both studies described ternary complexes of γδ TCRs with lipid-loaded CD1d, one with sulfatide and one with α-GalCer, and were the first to describe a γδ TCR bound to an antigen-presenting molecule plus ligand. Surprisingly, the γδ TCR bound to CD1d-sulfatide showed that interaction occurred solely between the δ chain and the CD1d-lipid complex, with the germline encoding regions interacting with CD1d, and the CDR3δ interacting with both sulfatide headgroup and CD1d α1-helix (90). A γδ TCR complexed with CD1d α-GalCer showed an orthogonal docking over the A′ pocket, and the interaction with CD1d was also dominated by the germline-encoded Vδ1 chain, whereas in this structure the CDR3γ was responsible for the main interactions with α-GalCer (92).
Not only Vδ1 T cells but also Vδ3 cells have been shown to be CD1d-restricted, and these T cells displayed Th1, Th2, and Th17 cytokine profiles (93). Given the fact that γδ T cells make up a large fraction of the human T-cell population, especially in the intestine, it is conceivable that among lipid-reactive T cells in humans, γδ T cells outnumber αβ T cells (90). A surprising finding following the identification of Vδ1 T cells that responded to CD1d loaded with α-GalCer, was that approximately 50% of all Vδ1 cells in humans expresses a TCR α chain comprised of δ1 variable region rearranged with an α chain joining (J) and constant (C) region, paired with a diverse array of β chains (94). Although sporadic reports of unusual recombination events in TCRs have appeared over the years (95–98), this study showed that among Vδ1 cells, these δ/αβ hybrid TCRs are as common as γδ TCRs. The δ/αβ TCR adopted a very similar mode of CD1d α-GalCer recognition to the γδ TCR, with an orthogonal docking over the A′ pocket and the germline-encoded Vδ1 chain dominating the interaction with CD1d (94). The γδ and the δ/αβ TCR binding to CD1d αGalCer differs substantially from the parallel docking mode over the F′ pocket that is characteristic for all αβ iNKT cell TCRs (99). Overall, the new findings show a clear contribution of Vδ1 cells to the population of CD1-restricted T cells in humans. Expansions of Vδ1 T cells have been reported in several pathological situations, including in celiac disease (100–102), and certain viral infections (e.g. CMV) (103, 104), yet the involvement of CD1d and/or lipid presentation in these particular conditions remains unclear.
Novel mammalian lipids identified as antigens for CD1-restricted T cells
Analogous to the MHC–peptide antigen-presenting system, it was initially thought that CD1-restricted T cells would primarily be activated by lipid antigens of foreign origin. However, it has since become clear that many CD1-restricted T cells display overt in vitro reactivity to CD1-presenting endogenous lipids and perform in vivo functions independent of foreign lipids.
Endogenous lipids involved in NKT cell selection and activation
In recent years, several classes of self-lipids have been identified as antigens for CD1d-restricted invariant (iNKT cells) and diverse NKT cells (dNKT cells). These include phospholipids (105), lysophospholipids (16), plasmalogen lysophospholipids (106), and isoglobotriosylceramide (lc I) (iGb3, mouse) (13). Peroxisome-derived ether lipids (e.g. plasmalogen lysophospholipids) were recently suggested to be important in the selection and activation of iNKT cells in vivo, since mice lacking the enzyme glyceronephosphate O-transferase (GNPAT, a peroxisomal enzyme essential for the synthesis of ether lipids), had a reduction in the number of iNKT cells both in thymus and peripheral organs (106). Synthetic lysoplasmalogens (mono alkyl glycero phospholipids that contain an ether bond at the sn-1 position, instead of an ester bond) were shown to be strong antigens for murine iNKT cells, however, only weak antigens for human iNKT cells (106). A different study provided insight in another class of endogenous iNKT cell antigens, namely α-glycosylceramides. As glycosylceramide synthases in mammals are β-transferases, it was long thought that mammalian glycosylceramides are exclusively present in the form of β-anomers. This led to the assumption that endogenous monoglycosylceramide ligands similar in structure to the prototypical NKT cell antigen, α-galactosylceramide (derived from a marine sponge), would not exist in mammals. However, recently it was shown that murine immune cells constitutively produce small quantities of α-glycosylceramides and that the amounts of their glycolipids are regulated by catabolic enzymes of the ceramide and glycolipid pathway (107). It remains unknown, however, what mechanisms lead to the production of α-anomers of glycolipids. It is possible that minute amounts of α-anomers arise during glycosphingolipid synthesis, or they are potentially generated from β-glycosphingolipids by anomerase activity of specific isomerases (108). In line with this was another study, in which a minor constituent of the glucosylceramide fraction of various mammalian sources was shown to be a strong activator of iNKT cells (109). Although the exact identity of the activating ligand could not be determined, it retained activity after the treatment with β glucosidase, and it was speculated that trace amounts of naturally occurring α-linked glycolipids (α-GalCer or α-GlcCer) potentially accounted for the strong reactivity. Both of these studies support the notion that anomeric a-linked glycolipids, which can function as strong antigens for iNKT cells, are not limited to marine sponges and certain microbes (9, 110) but are also broadly detected in mammalian sources.
Skin oils as antigens for CD1a-restricted T cells
The general model of lipid antigen recognition by CD1-restricted T cells involves the essential role of the hydrophilic head group in mediating contacts with the TCR. However, this notion was recently challenged by the identification of several highly hydrophobic oils extracted from human epidermis as antigens for CD1a-autoreactive T cells (22, 53). Using plate-bound recombinant CD1a protein and tissues extracted with chloroform and methanol, the highest responses from CD1a-autoreactive T-cell lines (BC2 and Bgp) were seen with lipids from human epidermis. The response was stronger when pure chloroform extracts were used, which contained only the most hydrophobic lipids, suggesting that the stimulating lipids were very hydrophobic compounds. Indeed, fractionation of epidermal lipids identified several components of human skin, specifically sebaceous oils and waxes, as CD1a antigens, including waxesters, triacylglycerides, free fatty acids, and squalene. A common feature of these antigens is the absence of a hydrophilic headgroup and specifically in the case of squalene, a C30 terpenoid, the complete absence of any polar functional group (Fig. 1). As it was unclear how a molecule lacking chemical basis for hydrogen bonding or charge–charge interactions could interact with a TCR, it raised the question of how these newly identified skin oils functioned as antigens. In addition, previously identified CD1 antigens have been shown to be recognized based on their protrusion from the groove, yet almost all of the identified CD1a antigens were significantly smaller (C16–C30) than those (approximately C42) matching the CD1a groove volume (1350 A3) and are therefore not expected to protrude to the outer surface of CD1a. Based on this, it was hypothesized that these ‘headless’ hydrophobic lipids fully nest in the CD1a molecule, thereby allowing direct interaction between CD1a and TCR (Fig. 1). In support of this model was the observation that analogs of the lipid antigens showed an inverse correlation between headgroup size and polarity and T-cell activation (22). Analysis by mass spectrometry of lipids eluted from CD1a molecules, showed that most cellular lipids are headgroup-containing lipids, whereas antigenic hydrophobic oils are less common. This suggests that under normal circumstances the CD1a bound ligand repertoire on antigen-presenting cells consist of ubiquitous headgroup-containing and therefore non-activating lipids (e.g. phospholipids, sphingomyelin), which need to be replaced by hydrophobic headless lipids for T-cell activation to occur.
Fig. 1. Proposed model for the activation of CD1a-autoreactive T cells.
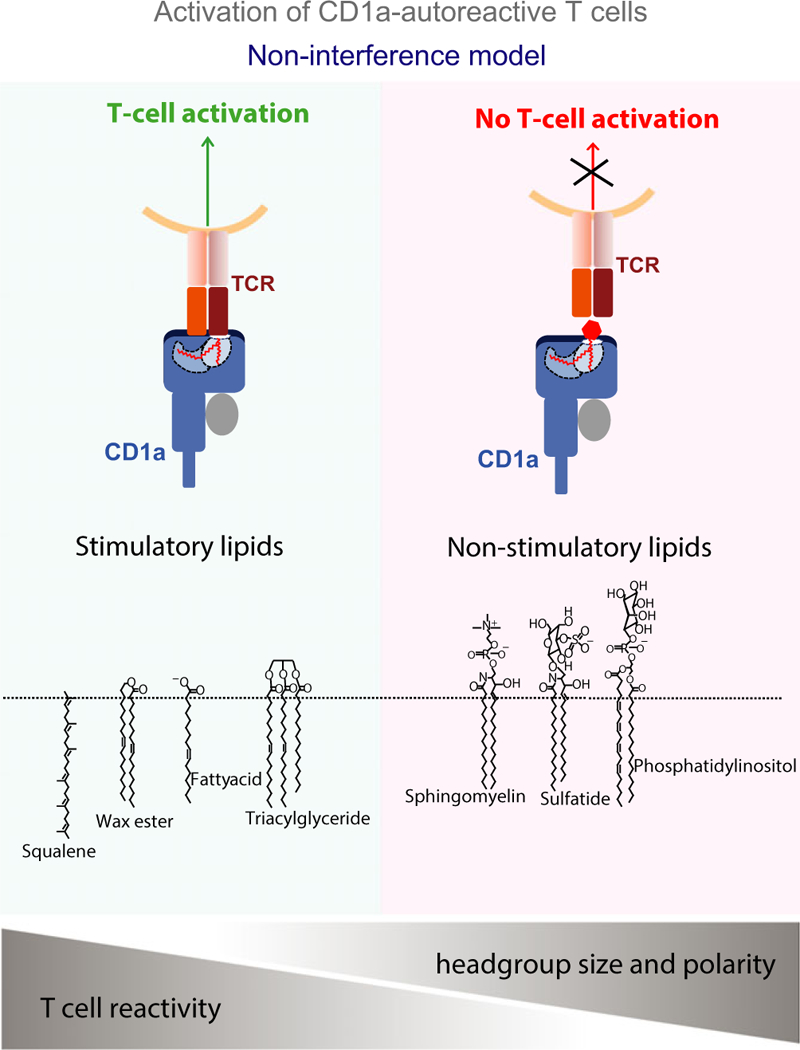
Small apolar lipids without headgroups (left panel) nest in the CD1a groove and allow interactions between the T-cell receptor (TCR) and CD1a. Lipids with large polar headgroups (right panel), which protrude from the CD1a groove, inhibit interactions between TCR and CD1a.
Overall, the identification of components of human sebum as T-cell antigens shows that skin lipids do not solely act as a protective barrier but also function as T-cell antigens. This leads to the novel idea that human skin lipids may act as an antigen reservoir that can actively influence T-cell activation. It further implies that shifts in lipid composition, as is observed in certain autoimmune skin diseases (e.g. atopic dermatitis, psoriasis) (111–116) and skin conditions (e.g. acne, rosacea) (117, 118) may affect the activation of skin resident CD1a-restricted T cells. In addition, the identification of hydrophobic oils and in particular squalene as antigens for CD1a-restricted T cells may also provide further insight in the immunological mechanisms that underlie the adjuvanticity of squalene- and mineral oil-based adjuvants. MF59 and A S03 are widely used squalene-based adjuvants used in influenza vaccines in Europe (119), and it is conceivable that CD1a-restricted T cells contribute to their adjuvant properties. Furthermore, many small hydrophobic molecules function as skin allergens. These include certain fragrances (e.g. geraniol and fatty aldehydes) (120), which are not known to act through haptenation of MHC or peptides, but are potential candidates for functioning as headless antigens for CD1a-autoreactive T cells.
CD1a-lipid-TCR ternary structure
Consistent with certain aspects of the proposed non-interference model of CD1a-autoreactive T-cell activation (Fig. 1), the crystal structure of CD1a protein bound to an autoreactive TCR was solved (121). The CD1a-autoreactive T-cell clone used in this study, BK6, was the first described CD1a-reactive T-cell clone (85). BK6 was shown to bind unloaded CD1a tetramers, suggesting that an endogenous lipid from the cell line in which CD1a protein was expressed, could function as an antigen for the T-cell line. However, mass spectrometry analysis of the lipids eluting from recombinant CD1a proteins, showed a diverse array of hundreds of lipids ranging from highly hydrophobic to polar phosphoglycolipids. As it would be highly unlikely that a single antigen would be present in multiple CD1a molecules in the tetramer, it was thought that polymeric binding of CD1a to the TCR was the result of multiple antigenic lipids bound to CD1a. Indeed, using size-exclusion chromatography to purify the ternary complexes of lipid-loaded CD1a bound to BK6 TCR and compare the eluted lipids to those eluted from unbound CD1a, multiple lipid ligands that were permissive for CD1a-TCR binding were identified by mass spectrometry. Among permissive ligands were phospholipids, lysophosphatidylcholine, and fatty acids, whereas sphingomyelin and sulfatide were identified as non-permissive ligands. The ternary structure revealed that the BK6 TCR docked orthogonally across the CD1a antigen-binding cleft (Fig. 2A) with the non-germline encoded CDR3α and CDR3β dominating the interactions with CD1a (Fig. 2B). The CDR3α was positioned at the edge of the A′ roof, which is a structure found on all CD1 molecules and forms platform that shields the hydrophobic lipid. The CDR3β was located centrally above the A′ roof. Both CDR3 regions formed contacts with both the a1 and a2 helix of CD1a, but did not contact the lipid antigen in the groove. The ability of certain lipids, such as sulfatide, to prevent TCR binding, was explained by the fact that the galactosyl sulfate head group disrupted interactions between residues from α1 and α2 helices (Arg76 and Glu154) and thereby disrupting the docking site of the TCR on the A′ roof of CD1a (121).
Fig. 2. Crystal structure of CD1a-autoreactive T-cell receptor (TCR) (BK6) bound to CD1a-lysophosphatidyl choline (LPC) complex.
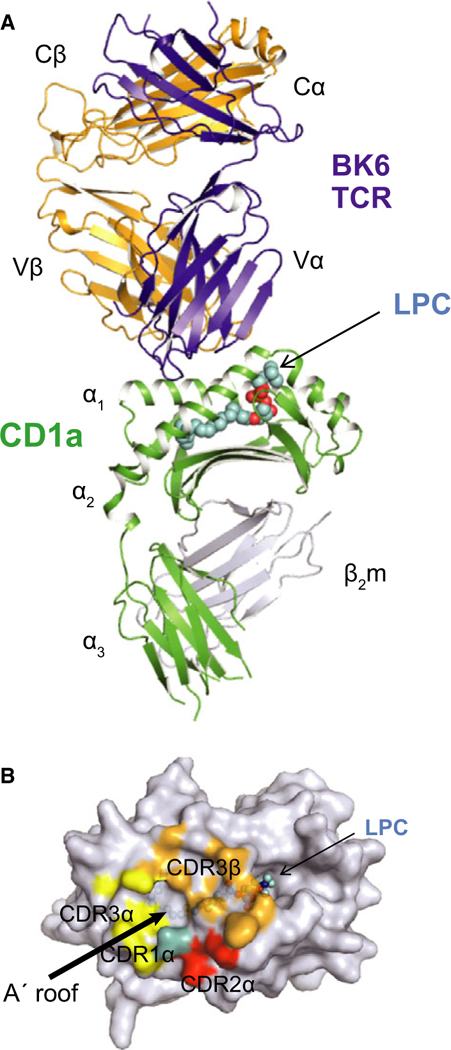
Overall docking mode (A) and the structural footprint of the TCR on the A′ roof surface of CD1a. (B) Modified from Birkinshaw et al. (121).
This study provided structural evidence for a novel mode of T-cell activation by antigen-presenting molecules. Although most CD1-restricted TCRs studied to date have shown a strong specificity for structural features of the polar headgroup of the antigen with which they interact, this TCR shows no interaction with lipid that is bound in the antigen-binding groove. As an antigen is considered to be the defining structure on the composite surface of the antigen-presenting molecule-antigen complex with which the TCR interacts, it could be argued that the CD1a self-ligands that allowed binding of the BK6 TCR to CD1a are not antigens. Rather, they are a group of permissive ligands that allow TCR interaction with CD1a without disrupting the contact surface. Although the prevalence of this type of TCR-CD1a interaction among all CD1a-restricted T cells remains to be determined, it is likely not a rare phenomenon, since other CD1a-autoreactive TCRs were determined to have sequence similarities in the TCR α and β chains (121). The lipid reactivity pattern of the T-cell clones that were activated by skin lipids (BC2 and Bgp) differed slightly from that of BK6, but overall the ternary structure is consistent with the non-interference model (Fig. 1), in that small ‘headless’ hydrophobic lipids allow interaction between the TCR and CD1a, whereas headgroup-containing lipids (including sphingomyelin) inhibit the interaction (22). It remains to be determined through structural and mutational studies if the TCRs of BC2 and Bgp also only interact with CD1a protein or that the TCR also forms hydrophobic interactions with the non-polar lipids in the groove.
Modes of CD1-lipid recognition by TCRs
The interaction between the BK6 autoreactive TCR and CD1a without contacting the lipid (Fig. 2) can be considered one end of the antigen recognition spectrum. As mentioned, it is technically speaking not antigen recognition but rather permissive ligand-dependent CD1a binding. As this mode of recognition does not require specific TCR–lipid interactions, there is likely a wide array of lipids that allows for T-cell activation, as long as they do not disrupt the contact surface on CD1a. Furthermore, it suggests that T-cell activation is determined by the relative amounts of permissive ligands and non-permissive ligands, rather than the presence of a specific antigenic lipid. This allows T cells to broadly respond to shifts in the lipid environment without the need for precise antigen recognition. At the other end of the spectrum are CD1-restricted TCRs that show exclusive specificity for the polar headgroup of the lipid antigen, and minor structural changes in the antigen will result in loss of recognition. This has mainly been shown for T cells specific for foreign lipid antigens. For instance, the activation of CD1b-restricted GMM-specific T-cell line was completely abrogated when the glucose was replaced by galactose or mannose (122), and CD1c-restricted mannosyl-β PM specific T cells were highly specific for the β-linked anomer of the mycobacterial antigen (123). In these cases, it is the absence or presence of the antigens, and not relative amounts of activating and non-activating lipids that dictates T-cell activation. Beside these two extreme modes of CD1-lipid recognition, there are many CD1-restricted TCRs that reside somewhere in the middle, showing a lower degree of specificity, as their TCR can recognize multiple lipid antigens. This is the case for iNKT cells, which all recognize α-GalCer, and subsets also recognize structurally distinct antigens, such as isoglobotrihexosylceramide (iGb3, mouse), β-GalCer, phospholipids, and lysophosphatidylcholine. Structural evidence provided explanation for the recognition of these distinct antigens, showing for iGb3 and β-GalCer, that the iNKT TCR molds these antigens to resemble the conformation of foreign a-linked ligands (induced fit molecular mimicry) (124, 125), and showing an alternative docking for the recognition of lysophosphatidylcholine by iNKT cell TCRs (126). In another study, the ability of structurally distinct self-lipids to be recognized by iNKT cell TCRs was systematically addressed and, it was proposed that activation is dictated by relative amounts of lipids that are either permissive or non-permissive to CD1d–TCR interactions (127).
Regulation of autoreactive CD1-restricted T-cell activation
The fact that CD1-restricted T cells, unlike MHC-restricted T cells, show overt reactivity to endogenous lipids implies that there must be mechanisms in place that prevent unwanted activation of lipid-specific T cells. Several distinct mechanisms have been described that can regulate CD1-restricted T-cell activation, and act to ensure that activation only occurs when needed, such as in response to pathogens, inflammation, or tissue damage. These mechanisms are depicted in Fig. 3 and described in the following paragraphs.
Fig. 3. Regulation of autoreactive CD1-restricted T-cell activation.
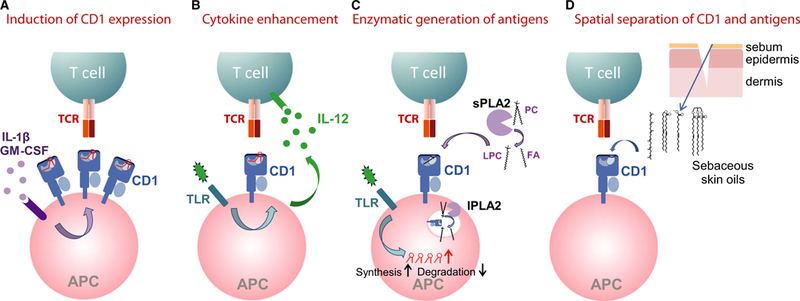
The activation of CD1-autoreactive T cells has been described to be regulated by distinct mechanisms, including the regulation of surface CD1 expression levels (A), the enhancement of T cell activation through cytokines (B), the enzymatic regulation of antigen levels (C), and the spatial separation of CD1 and antigens (D).
Regulation of CD1 surface expression levels (Fig. 3A)
Cell surface levels of MHC molecules are one of the factors determining the magnitude of T-cell activation. Both MHC class I and class II are constitutively expressed on the surface of antigen-presenting cells, but their levels increase significantly upon antigen-presenting cell (APC) activation (128). MHC class II, though generally not expressed on non-hematopoietic cells, is often expressed on certain epithelial cells in response to infection or inflammation (down-stream of IFN-γ) (129). Of CD1 molecules, CD1d is the only isoform that is constitutively expressed on all antigen-presenting cells, including monocytes, macrophages, B cells, and dendritic cells (33), and is expressed on non-hematopoietic cells, such as intestinal epithelial cells, hepatocytes, and keratinocytes, in response to IFN-c (17, 130–132). By contrast, the expression of CD1a, CD1b, and CD1c is highly variable and is likely one of the mechanisms by which activation of CD1-restricted T cells is controlled (Fig. 3A). CD1a, CD1b, and CD1c are upregulated on monocytes during differentiation into dendritic cells in vitro in response to GM-CSF/IL-4 (133, 134). In addition, mycobacterial and borrelial lipids induce the upregulation the transcription and surface expression levels of CD1a, CD1b, and CD1c on monocytes, in a Toll-like receptor 2 (TLR2) and IL-1β-dependent mechanism (135, 136). The abundant expression of CD1a, CD1b, and CD1c on dendritic cells at the site of tuberculoid leprosy lesions suggest that similarly in vivo, mycobacterial lipids induce CD1 upregulation (137). Conversely, human serum contains factors that prevent the upregulation of CD1a, CD1b, and CD1c on monocytes differentiating into dendritic cells. Lysophosphatidic acid and cardiolipin were shown to inhibit CD1 expression at the level of gene transcription in a peroxisome proliferator activated receptor (PPAR)-dependent mechanism (138). In addition, binding of IgG to FcγRIIa on dendritic cells inhibited the expression of CD1a, CD1b, and CD1c (139). These data suggest that group 1 CD1 expression levels on monocytes/dendritic cells is tightly controlled in the peripheral blood, and that upregulation only occurs when the APCs enter the tissues where inhibitory serum components are absent. Downregulation of surface CD1d (and CD1a) through several distinct mechanisms in HIV and Herpes simplex infection of antigen-presenting cells has been proposed as an immune evasion mechanism (140–145), further emphasizing the role of surface CD1 expression levels on T-cell activation.
Although CD1a, CD1b, and CD1c levels on monocyte-derived and tissue resident dendritic cells appear to be tightly controlled, some other antigen-presenting cells are not dependent on inflammation and/or infection-derived signals for CD1 upregulation but instead constitutively express high CD1 levels. For instance, the constitutive high expression of CD1a on epidermal Langerhans cells is not significantly affected by TLR signaling or other stimuli. This suggests that for local immune response in the skin mechanisms other than CD1 upregulation are in place to regulate the activation of CD1a-autoreactive T cells.
Enhancement of T-cell activation through cytokines (Fig. 3B)
It was long thought that recognition of foreign lipids antigens was the main driver for CD1d-restricted iNKT cell activation in infection, and many studies have identified microbial glycolipids that can function as antigens for iNKT cells (10, 12, 146–148). However, both in vitro and in vivo, the release of IFN-γ by iNKT cells in response to diverse bacteria (including Sphingomonas yanoikuyae and Streptococcus pneumoniae, which both harbor iNKT cell antigens) was shown to strongly depend on TLR-driven IL-12 production by the APC rather than on the cognate recognition of foreign lipid antigens (149) (Fig. 3B). This cytokine-driven activation was explained by the constitutive expression of high levels of IL-12 receptor by iNKT cells, enabling instant IL-12-induced STAT4 activation (149, 150). Importantly, the presence of CD1d with endogenous lipid was still required for activation, yet without the IL-12 production, the self-reactivity did not elicit IFN-γ release by iNKT cells in vivo. The foreign lipid independent mechanism is not limited to bacterial infections, as also in fungal infections caused by Aspergillus,Candida, Histoplasma, and Alternaria, fungal lipids were not required for activation. Rather, the binding of β−1,3 glucans (fungal cell wall polysaccharides) to Dectin-1 triggered IL-12 production, which in combination with CD1d plus endogenous lipids, resulted in iNKT cell activation (151). Overall, the dependence of IFN-γ production by iNKT cells on the combination of endogenous lipid presented by CD1d and IL-12, can be considered a mode of regulation that limits pro-inflammatory cytokine release by these T cells to situations where IL-12 is induced, which include bacterial, fungal infections, and inflammatory conditions.
Enzymatic regulation of antigens levels (Fig. 3C)
Enzymes involved in the synthesis and degradation of glycolipids have been implicated in the activation of iNKT and other CD1-restricted T cells, and act through regulating the levels of endogenous lipid antigens (Fig. 3C). The treatment of human antigen-presenting cells with Toll-like receptor ligands (LPS, R848) resulted in the upregulation of several enzymes in biosynthesis of lipids of the globo, isoglobo, and ganglioside series, and increased iNKT cell activation. Effects from soluble factors like IL-12 were separated from effects on endogenous antigen levels by fixing the APC after TLR activation (152). Fluorescently labelled tetrameric complexes of iNKT TCRs showed increased binding to APC upon TLR activation, despite similar levels of CD1d, suggesting that innate stimuli modulate the endogenous lipid repertoire, resulting in increased stimulatory CD1d-lipid complexes. Dendritic cells deficient in the lysosomal enzyme α-galactosidase A showed increased activation of iNKT cells, presumably resulting from an accumulation of endogenous lipid antigen. Lysosomal α-galactosidase A is the enzyme that is deficient in Fabry’s disease, resulting in the accumulation of iGb3, Gb3, and possibly other glycolipids (153). MyD88-dependent TLR stimulation was shown to result in the temporary inhibition of α-galactosidase A, again supporting a mechanistic link between pathogen recognition and self-lipid antigen accumulation for NKT cells (153). For CD1a- and CD1b-restricted T-cell clones specific for sulfatide and monosialotetrahexosylganglioside (GM1), respectively, a similar mechanism of TLR-dependent changes in endogenous antigen levels was described. Lipopolysaccharide (LPS) induced upregulation of glycosphingolipid synthesis in APCs and subsequently increased T-cell activation (154). Overall, several studies support the notion that activation of antigenpresenting cells, specifically through TLR engagement, can result in increased levels of endogenous lipid antigens through increased biosynthesis or decreased degradation (Fig. 3C).
Fatty acids and lysophosphatidyl choline have been identified as antigens/permissive ligands for CD1a-autoreactive T cells (22, 121), and these two compounds can be generated by the degradation of the ubiquitous membrane phospholipid, phosphatidylcholine, upon the catalytic hydrolyzation of the sn-2 acyl bond by phospholipase A2 (PLA2) (Fig. 3C). A recent study showed that bee and wasp venom were able to activate human T-cell lines, polyclonal T-cell cultures, and ex vivo T cells in a PLA2 dependent and CD1a-dependent manner (155). PLA2, which is a component of bee and wasp venom, indeed activated the CD1a-restricted T cells through generation of the degradation products (neoantigens) of phosphodiacylglycerols, namely fatty acids and lysophospholipids. The analysis of fluid of suction blisters, harvested after intradermal injection of bee venom into healthy skin, showed a decrease in phosphatidylcholines and an increase in lysophospholipids as a result of phospholipid activity in the dermis in vivo. This showed that injection of bee venom altered the local lipid environment to generate more of the CD1a antigens fatty acids and lysophospholipids, as would be predicted from PLA2 activity. Therefore, exogenous sources of PLA2 can generate CD1a antigens in vivo, and similarly, endogenous-secreted PLA2, which is expressed in inflammation and infection, is expected to increase CD1a autoreactivity. Previously, several studies have shown that both lysophospholipids and ether lysophospholipids can function as antigens for NKT cells (16, 17, 106, 156), and endogenous phospholipase A2 activity has been shown to increase CD1d autoreactivity. In a mouse model of hepatitis B infection, the virus induced upregulation of endogenous-secreted phospholipase A2 (Pla2g2c) in hepatocytes, and a subsequent increase in iNKT cell activation (17). In a different study, lysosomal phospholipase A2 was shown to be involved in the generation of activating CD1d lipid complexes in the thymus, as Lpla2ȡ/− mice showed reduced numbers of iNKT cells (157) (Fig. 3C). Overall, these studies show that PLA2 does not only generate precursors of inflammatory lipid mediators generated from arachidonic acid release but also releases CD1a and CD1d antigens, which links phospholipase activity to T-cell activation (158).
In human skin, these findings can have implications for skin barrier sensing by T cells and mechanisms underlying inflammatory skin disease in which phospholipase activity is altered. For instance, in psoriasis, the expression of several secreted PLA2 enzymes (sPLA2-X, -IIA, -IID, and -IB) were upregulated in the dermis (159). This suggests that levels of CD1a antigens/permissive ligands increase in the dermis, possibly resulting in increased T-cell activation. In addition, Filaggrin (FLG) mutations, which are strongly associated with atopic dermatitis, are generally thought to increase susceptibility to disease through decreased skin barrier function. However, studies using FLG knockdown 3D skin constructs showed a significant upregulation of secreted PLA2 (IIA) in the FLG deficient tissue and a twofold higher level of free fatty acids, possibly implicating a link between commonly mutated protein and skin fatty acid levels (160). Furthermore, human skin is colonized by large number of bacteria and fungi, many of which express different phospholipases and lipases. For instance, Malassezia species represent fungi that are abundantly found on human skin and are dependent on exogenous fatty acids for their growth (161). The genomes of Malassezia species contain a number of genes encoding lipases [e.g. Malassezia globosa encodes 14 lipases and 9 phospholipases (162)], and Malassezia has been shown to contribute to increased fatty acids levels in the skin due to their cleavage from mammalian triacylglycerides and phospholipids (163). Other skin resident fungi (e.g. Candida species), and bacteria found on human skin (e.g. Staphylococcus) also express secreted (phospho)lipases (162, 164). Therefore, in addition to endogenous phospholipase activity, our skin microbiome likely contributes to release of free fatty acids and lysophospholipids, and potentially influences the activation of CD1a-restricted T cells in the skin. This is a largely unexplored area, which could provide novel insights in the regulation of skin immunity.
Spatial separation of CD1 and antigens (Fig. 3D)
The repertoire of lipids presented by the different human CD1 isoforms is dictated by various characteristics, including the size and properties of the antigen-binding grooves, and the intracellular trafficking pathways. After assembly and maturation of CD1 proteins in the endoplasmic reticulum (165), CD1 associates with β2-m and traffics through the Golgi network to the cell surface (166). Here, the paths of the four surface-expressed human CD1 isoforms diverge. CD1b, CD1c, and CD1d traffic extensively through the late endosomal and lysosomal compartments (reviewed in 167), where the loading and unloading of lipids is regulated through pH and saposins (168–171). Therefore, for these CD1 isoforms, the endosomal system can be considered a site for regulating the repertoire of bound and presented lipid antigens. In contrast, CD1a is predominantly detected at the cell surface and in the early endosomal recycling compartments (31, 172), since it lacks an endosomal localization motif. Based on its shallow antigen-binding groove (28), limited intracellular trafficking (173), relative stability in the absence of lipid ligand (174), and capacity for surface lipid exchange (175), it is conceivable that the repertoire of CD1a presented lipids is dictated more by the extracellular milieu, than by factors involving intracellular lipid exchange, processing and selection. As a consequence, compared with other CD1 isoforms, the CD1a bound lipid repertoire is likely to most reflect the external lipid milieu.
The mechanisms described thus far for the regulation of CD1-autoreactive T-cell activation do not explain how the activation of skin resident human CD1a-autoreactive T cells is controlled. All the components required to induce a T-cell response are present under normal conditions in human skin, namely CD1ahigh Langerhans cells, CD1a antigens (in sebum), and CD1a-autoreactive T cells. Yet, in steady state conditions, continuous T-cell activation is not predicted to occur; one of the speculations being that the Langerhans cells, T cells, and antigens are physically separated in healthy skin. Langerhans cells reside primarily in the suprabasal spinous layer of the epidermis, whereas CD1a-autoreactive T cells were detected in the dermis (53); therefore, activation may be restricted to situations when the Langerhans cells migrate out of the epidermis through the dermis. Furthermore, the ‘headless’ hydrophobic skin lipids (including squalene, fatty acids, and waxesters) (22) that allow for T-cell activation are the main constituents of human sebum, which is not in direct contact with Langerhans cells. Sebum is sequestered in sebaceous glands, and exits the glands via the hair follicles to coat the outer layer of the epidermis. However, upon barrier breach due to infection or injury, it was postulated that the sebum contents permeate the epidermis and become accessible to CD1a expressing Langerhans cells, resulting in T-cell activation (22, 176) (Fig. 3D). This would further imply that in situations where the barrier is chronically disrupted, such as atopic dermatitis, continuous accessibility to stimulatory skin lipids is possibly a factor in chronic T-cell activation.
In summary, whereas MHC class I functions as a nearly universally expressed sentinel to capture and display any foreign peptide antigen that is generated inside a cell, stimulatory CD1-self lipid complexes appear to be more tightly regulated. In response to triggers (e.g. viral infection, TLR stimulation, tissue damage, venom), enzymatic or physical release of self-lipid antigens and/or upregulated CD1 expression levels result in a temporary increase in stimulatory CD1-lipid complexes, enabling T-cell activation. Overall, multiple mechanisms are in place to ensure localized and timely activation of CD1-autoreactive T cells.
Concluding thoughts
Our knowledge of human CD1-restricted T cells and their lipid specificities has increased significantly over the past 5 years. Yet there are still many unknowns about the diversity and frequencies of the T cells restricted by the different CD1 isoforms as well as the dynamics of these T-cell populations in response to infections and inflammatory conditions. Tools such as newly developed lipid-loaded CD1 tetramers and dextramers (78–80), as well as human CD1 transgenic mice (177) and humanized mouse models (178) will help further studies aimed at identifying the roles of CD1-restricted T cells in the human immune system. The structural insights in the novel mode of CD1-lipid complex recognition by CD1a-autoreactive T cells has paved the way for studies focused on defining permissive and non-permissive lipid ligands, which could be used to modulate immune responses.
The fact that several CD1-autoreactive T-cell populations, such as CD1d-restricted Vδ1 T cells in the gut and CD1a-restricted T cells in the skin, are in close contact with our gut and skin microbiome raises questions about the interplay between commensals, self and foreign lipids, and the CD1 system at our epithelia. In mice, commensal gut microbiota have been shown to regulate NKT cells and NKT cell-dependent inflammatory conditions at the mucosa (179–182). In humans, it is unknown if and how skin and gut microbiome shape and regulate CD1-restricted responses, but the ability of bacteria and fungi to alter the CD1 presented self-lipid repertoire, supports an active role for the microbiome in regulating CD1-autoreactive T cells at the epithelia, where they may act as general sentinels of tissue damage or infection.
Acknowledgements
This study was supported by (NIAMS K01AR068475), and a Research Career Development Award from the Dermatology Foundation. The author thanks Richard Birkinshaw and Jamie Rossjohn for providing the figure of the CD1a-lipid-TCR crystal structure, and Branch Moody and Ildiko van Rhijn for critical reading of the manuscript.
Footnotes
The author has no conflicts of interest.
References
- 1.Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 1974;248:701–702. [DOI] [PubMed] [Google Scholar]
- 2.Zinkernagel RM, Doherty PC. The discovery of MHC restriction. Immunol Today 1997;18:14–17. [DOI] [PubMed] [Google Scholar]
- 3.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 1996;384:134–141. [DOI] [PubMed] [Google Scholar]
- 4.Garcia KC, et al. An αβ T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 1996;274:209–219. [PubMed] [Google Scholar]
- 5.Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol 2013;31:529–561. [DOI] [PubMed] [Google Scholar]
- 6.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature 1994;372:691–694. [DOI] [PubMed] [Google Scholar]
- 7.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012;491:717–723. [DOI] [PubMed] [Google Scholar]
- 8.Eckle SB, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med 2014;211:1585–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 1997;278:1626–1629. [DOI] [PubMed] [Google Scholar]
- 10.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 2005;434:520–525. [DOI] [PubMed] [Google Scholar]
- 11.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol 2006;7:978–986. [DOI] [PubMed] [Google Scholar]
- 12.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol 2011;12:966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science 2004;306:1786–1789. [DOI] [PubMed] [Google Scholar]
- 14.Russano AM, et al. Recognition of pollen-derived phosphatidyl-ethanolamine by human CD1d-restricted gamma delta T cells. J Allergy Clin Immunol 2006;117:1178–1184. [DOI] [PubMed] [Google Scholar]
- 15.Russano AM, et al. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. J Immunol 2007;178:3620–3626. [DOI] [PubMed] [Google Scholar]
- 16.Fox LM, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol 2009;7:e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeissig S, et al. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med 2012;18:1060–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shamshiev A, et al. The alphabeta T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity 2000;13:255–264. [DOI] [PubMed] [Google Scholar]
- 19.Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Self glycolipids as T-cell autoantigens. Eur J Immunol 1999;29:1667–1675. [DOI] [PubMed] [Google Scholar]
- 20.Moody DB, et al. T cell activation by lipopeptide antigens. Science 2004;303:527–531. [DOI] [PubMed] [Google Scholar]
- 21.Van Rhijn I, et al. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci U S A 2004;101:13578–13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong A, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol 2014;15:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science 1997;277:339–345. [DOI] [PubMed] [Google Scholar]
- 24.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol 2002;3:721–726. [DOI] [PubMed] [Google Scholar]
- 25.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 2007;448:44–49. [DOI] [PubMed] [Google Scholar]
- 26.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol 1989;19:285–292. [DOI] [PubMed] [Google Scholar]
- 27.Balk SP, Bleicher PA, Terhorst C. Isolation and characterization of a cDNA and gene coding for a fourth CD1 molecule. Proc Natl Acad Sci U S A 1989;86:252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2 15 A. Nat Immunol 2003;4:808–815. [DOI] [PubMed] [Google Scholar]
- 29.Koch M, et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol 2005;6:819–826. [DOI] [PubMed] [Google Scholar]
- 30.Scharf L, et al. The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 2010;33:853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugita M, et al. Separate pathways for antigen presentation by CD1 molecules. Immunity 1999;11:743–752. [DOI] [PubMed] [Google Scholar]
- 32.Huang S, et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci U S A 2011;108:19335–19340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol 2007;314:113–141. [DOI] [PubMed] [Google Scholar]
- 34.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol 2007;314:3–26. [DOI] [PubMed] [Google Scholar]
- 35.Balk SP, Bleicher PA, Terhorst C. Isolation and expression of cDNA encoding the murine homologues of CD1. J Immunol 1991;146:768–774. [PubMed] [Google Scholar]
- 36.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med 1994;180:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8-alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med 1993;178:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol 2007;25:297–336. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol 2008;20:358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol 2009;9:28–38. [DOI] [PubMed] [Google Scholar]
- 41.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol 2010;11:197–206. [DOI] [PubMed] [Google Scholar]
- 42.Hegde S, Fox L, Wang X, Gumperz JE. Autoreactive natural killer T cells: promoting immune protection and immune tolerance through varied interactions with myeloid antigen-presenting cells. Immunology 2010;130:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L, Van Kaer L. Natural killer T cells in health and disease. Front Biosci (Schol Ed) 2011;3:236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engel I, Kronenberg M. Making memory at birth: understanding the differentiation of natural killer T cells. Curr Opin Immunol 2012;24:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol 2013;25:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol 2013;13:101–117. [DOI] [PubMed] [Google Scholar]
- 47.Gapin L, Godfrey DI, Rossjohn J. Natural Killer T cell obsession with self-antigens. Curr Opin Immunol 2013;25:168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghazarian L, Simoni Y, Magalhaes I, Lehuen A. Invariant NKT cell development: focus on NOD mice. Curr Opin Immunol 2014;27:83–88. [DOI] [PubMed] [Google Scholar]
- 49.Hammond KJ, et al. CD1d-restricted NKT cells: an interstrain comparison. J Immunol 2001;167:1164–1173. [DOI] [PubMed] [Google Scholar]
- 50.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med 2002;195:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenna T, et al. NKT cells from normal and tumor-bearing human livers are phenotypically and functionally distinct from murine NKT cells. J Immunol 2003;171:1775–1779. [DOI] [PubMed] [Google Scholar]
- 52.de Lalla C, et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol 2011;41:602–610. [DOI] [PubMed] [Google Scholar]
- 53.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol 2010;11:1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young MH, Gapin L. Group 1 CD1-restricted T cells take center stage. Eur J Immunol 2011;41:592–594. [DOI] [PubMed] [Google Scholar]
- 55.Vincent MS, Gumperz JE, Brenner MB. Understanding the function of CD1-restricted T cells. Nat Immunol 2003;4:517–523. [DOI] [PubMed] [Google Scholar]
- 56.Lepore M, et al. A novel self-lipid antigen targets human T cells against CD1c(+) leukemias. J Exp Med 2014;211:1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin LH, Calabi F, Lefebvre FA, Bilsland CA, Milstein C. Structure and expression of the human thymocyte antigens CD1a, CD1b, and CD1c. Proc Natl Acad Sci U S A 1987;84:9189–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moody DB. TLR gateways to CD1 function. Nat Immunol 2006;7:811–817. [DOI] [PubMed] [Google Scholar]
- 59.Meunier L, et al. Quantification of CD1a, HLA-DR, and HLA class I expression on viable human Langerhans cells and keratinocytes. Cytometry 1996;26:260–264. [DOI] [PubMed] [Google Scholar]
- 60.Yu RC, Abrams DC, Alaibac M, Chu AC. Morphological and quantitative analyses of normal epidermal Langerhans cells using confocal scanning laser microscopy. Br J Dermatol 1994;131:843–848. [DOI] [PubMed] [Google Scholar]
- 61.de Fraissinette A, Schmitt D, Thivolet J. Langerhans cells of human mucosa. J Dermatol 1989;16:255–262. [DOI] [PubMed] [Google Scholar]
- 62.van Loon LA, Krieg SR, Davidson CL, Bos JD. Quantification and distribution of lymphocyte subsets and Langerhans cells in normal human oral mucosa and skin. J Oral Pathol Med 1989;18:197–201. [DOI] [PubMed] [Google Scholar]
- 63.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol 2009;10:857–863. [DOI] [PubMed] [Google Scholar]
- 64.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T (H)1 and T(H)2 cells. Nat Immunol 2009;10:864–871. [DOI] [PubMed] [Google Scholar]
- 65.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity 2004;21:241–254. [DOI] [PubMed] [Google Scholar]
- 66.Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol 2005;174:3695–3702. [DOI] [PubMed] [Google Scholar]
- 67.Wolk K, et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol 2006;36:1309–1323. [DOI] [PubMed] [Google Scholar]
- 68.Boniface K, et al. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol 2007;150:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma HL, et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest 2008;118:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nograles KE, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol 2009;123:1244–1252 e1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eyerich S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest 2009;119:3573–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kagami S, Rizzo HL, Lee JJ, Koguchi Y, Blauvelt A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J Invest Dermatol 2010;130:1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang N, Pan HF, Ye DQ. Th22 in inflammatory and autoimmune disease: prospects for therapeutic intervention. Mol Cell Biochem 2011;353:41–46. [DOI] [PubMed] [Google Scholar]
- 74.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996;274:94–96. [PubMed] [Google Scholar]
- 75.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med 2000;192:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karadimitris A, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci U S A 2001;98:3294–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med 2000;191:1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kasmar AG, et al. CD1b tetramers bind alphabeta T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med 2011;208:1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kasmar AG, et al. Cutting edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol 2013;21:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ly D, et al. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med 2013;210:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 2003;422:164–169. [DOI] [PubMed] [Google Scholar]
- 82.Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol 2013;14:706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Rhijn I, et al. TCR bias and affinity define two compartments of the CD1b-glycolipid-specific T Cell repertoire. J Immunol 2014;192:4054–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Rhijn I, Moody DB. CD1 and mycobacterial lipids activate human T cells. Immunol Rev 2015;264:138–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature 1989;341:447–450. [DOI] [PubMed] [Google Scholar]
- 86.Spada FM, et al. Self-recognition of CD1 by gamma/delta T cells: implications for innate immunity. J Exp Med 2000;191:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med 2005;202:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dieude M, et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted gammadelta T cells in the normal murine repertoire. J Immunol 2011;186:4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bai L, et al. The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. Eur J Immunol 2012;42:2505–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luoma AM, et al. Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity 2013;39:1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Balk SP, et al. Oligoclonal expansion and CD1 recognition by human intestinal intraepithelial lymphocytes. Science 1991;253:1411–1415. [DOI] [PubMed] [Google Scholar]
- 92.Uldrich AP, et al. CD1d-lipid antigen recognition by the gammadelta TCR. Nat Immunol 2013;14:1137–1145. [DOI] [PubMed] [Google Scholar]
- 93.Mangan BA, et al. Cutting edge: CD1d restriction and Th1/Th2/Th17 cytokine secretion by human Vdelta3 T cells. J Immunol 2013;191:30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pellicci DG, et al. The molecular bases of delta/ alphabeta T cell-mediated antigen recognition. J Exp Med 2014;211:2599–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hochstenbach F, Brenner MB. T-cell receptor delta-chain can substitute for alpha to form a beta delta heterodimer. Nature 1989;340:562–565. [DOI] [PubMed] [Google Scholar]
- 96.Peyrat MA, et al. Repertoire analysis of human peripheral blood lymphocytes using a human V delta 3 region-specific monoclonal antibody. Characterization of dual T cell receptor (TCR) delta-chain expressors and alpha beta T cells expressing V delta 3J alpha C alpha-encoded TCR chains. J Immunol 1995;155: 3060–3067. [PubMed] [Google Scholar]
- 97.Miossec C, et al. Further analysis of the T cell receptor gamma/delta+ peripheral lymphocyte subset. The V delta 1 gene segment is expressed with either C alpha or C delta. J Exp Med 1990;171:1171–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bowen S, Sun P, Livak F, Sharrow S, Hodes RJ. A novel T cell subset with trans-rearranged Vgamma-Cbeta TCRs shows Vbeta expression is dispensable for lineage choice and MHC restriction. J Immunol 2014;192:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol 2012;12:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halstensen TS, Scott H, Brandtzaeg P. Intraepithelial T cells of the TcR gamma/delta+ CD8- and V delta 1/J delta 1+ phenotypes are increased in coeliac disease. Scand J Immunol 1989;30:665–672. [DOI] [PubMed] [Google Scholar]
- 101.Spencer J, Isaacson PG, MacDonald TT, Thomas AJ, Walker-Smith JA. Gamma/delta T cells and the diagnosis of coeliac disease. Clin Exp Immunol 1991;85:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dunne MR, et al. Persistent changes in circulating and intestinal gammadelta T cell subsets, invariant natural killer T cells and mucosal-associated invariant T cells in children and adults with coeliac disease. PLoS ONE 2013;8:e76008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dechanet J, et al. Implication of gammadelta T cells in the human immune response to cytomegalovirus. J Clin Invest 1999;103:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Knight A, et al. The role of Vdelta2-negative gammadelta T cells during cytomegalovirus reactivation in recipients of allogeneic stem cell transplantation. Blood 2010;116:2164–2172. [DOI] [PubMed] [Google Scholar]
- 105.Tatituri RV, et al. Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proc Natl Acad Sci U S A 2013;110:1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Facciotti F, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol 2012;13:474–480. [DOI] [PubMed] [Google Scholar]
- 107.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity 2014;41:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim J, Kim JH, Winau F. Thinking inside the box: endogenous alpha-anomeric lipid antigens. Immunity 2014;41:505–506. [DOI] [PubMed] [Google Scholar]
- 109.Brennan PJ, et al. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci U S A 2014;111:13433–13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol 2013;11: e1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamamoto A, Serizawa S, Ito M, Sato Y. Stratum corneum lipid abnormalities in atopic dermatitis. Arch Dermatol Res 1991;283:219–223. [DOI] [PubMed] [Google Scholar]
- 112.Imokawa G, Abe A, Jin K, Higaki Y, Kawashima M, Hidano A. Decreased level of ceramides in stratum corneum of atopic dermatitis: an etiologic factor in atopic dry skin? J Invest Dermatol 1991;96:523–526. [DOI] [PubMed] [Google Scholar]
- 113.Janssens M, et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J Lipid Res 2012;53:2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Motta S, Monti M, Sesana S, Caputo R, Carelli S, Ghidoni R. Ceramide composition of the psoriatic scale. Biochim Biophys Acta 1993;1182:147–151. [DOI] [PubMed] [Google Scholar]
- 115.Motta S, Monti M, Sesana S, Mellesi L, Ghidoni R, Caputo R. Abnormality of water barrier function in psoriasis. Role of ceramide fractions. Arch Dermatol 1994;130:452–456. [PubMed] [Google Scholar]
- 116.van Smeden J, et al. The importance of free fatty acid chain length for the skin barrier function in atopic eczema patients. Exp Dermatol 2014;23:45–52. [DOI] [PubMed] [Google Scholar]
- 117.Ni Raghallaigh S, Bender K, Lacey N, Brennan L, Powell FC. The fatty acid profile of the skin surface lipid layer in papulopustular rosacea. Br J Dermatol 2012;166:279–287. [DOI] [PubMed] [Google Scholar]
- 118.Pappas A, Johnsen S, Liu JC, Eisinger M. Sebum analysis of individuals with and without acne. Dermatoendocrinol 2009;1:157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Atmar RL, Keitel WA. Adjuvants for pandemic influenza vaccines. Curr Top Microbiol Immunol 2009;333:323–344. [DOI] [PubMed] [Google Scholar]
- 120.Barany E, Loden M. Content of fragrance mix ingredients and customer complaints of cosmetic products. Am J Contact Dermat 2000;11:74–79. [DOI] [PubMed] [Google Scholar]
- 121.Birkinshaw RW, et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol 2015;16:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moody DB, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science 1997;278:283–286. [DOI] [PubMed] [Google Scholar]
- 123.de Jong A, et al. CD1c presentation of synthetic glycolipid antigens with foreign alkyl branching motifs. Chem Biol 2007;14:1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y, et al. The Valpha14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med 2010;207:2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pellicci DG, et al. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol 2011;12:827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lopez-Sagaseta J, Sibener LV, Kung JE, Gumperz J, Adams EJ. Lysophospholipid presentation by CD1d and recognition by a human Natural Killer T-cell receptor. EMBO J 2012;31:2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 2011;34:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Turley SJ, et al. Transport of peptide-MHC class II complexes in developing dendritic cells. Science 2000;288:522–527. [DOI] [PubMed] [Google Scholar]
- 129.Kambayashi T, Laufer TM. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat Rev Immunol 2014;14:719–730. [DOI] [PubMed] [Google Scholar]
- 130.Bonish B, et al. Overexpression of CD1d by keratinocytes in psoriasis and CD1d-dependent IFN-gamma production by NK-T cells. J Immunol 2000;165:4076–4085. [DOI] [PubMed] [Google Scholar]
- 131.Colgan SP, Morales VM, Madara JL, Polischuk JE, Balk SP, Blumberg RS. IFN-gamma modulates CD1d surface expression on intestinal epithelia. Am J Physiol 1996;271:C276–C283. [DOI] [PubMed] [Google Scholar]
- 132.Agrati C, et al. CD1d expression by hepatocytes is a main restriction element for intrahepatic T-cell recognition. J Biol Regul Homeost Agents 2005;19:41–48. [PubMed] [Google Scholar]
- 133.Pickl WF, et al. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J Immunol 1996;157:3850–3859. [PubMed] [Google Scholar]
- 134.Zhou LJ, Tedder TF. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc Natl Acad Sci U S A 1996;93:2588–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roura-Mir C, et al. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J Immunol 2005;175:1758–1766. [DOI] [PubMed] [Google Scholar]
- 136.Yakimchuk K, et al. Borrelia burgdorferi infection regulates CD1 expression in human cells and tissues via IL1-beta. Eur J Immunol 2011;41:694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sieling PA, et al. CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol 1999;162:1851–1858. [PubMed] [Google Scholar]
- 138.Leslie DS, et al. Serum lipids regulate dendritic cell CD1 expression and function. Immunology 2008;125:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smed-Sorensen A, et al. IgG regulates the CD1 expression profile and lipid antigen-presenting function in human dendritic cells via FcgammaRIIa. Blood 2008;111:5037–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yuan W, Dasgupta A, Cresswell P. Herpes simplex virus evades natural killer T cell recognition by suppressing CD1d recycling. Nat Immunol 2006;7:835–842. [DOI] [PubMed] [Google Scholar]
- 141.Shinya E, et al. Endogenously expressed HIV-1 nef down-regulates antigen-presenting molecules, not only class I MHC but also CD1a, in immature dendritic cells. Virology 2004;326:79–89. [DOI] [PubMed] [Google Scholar]
- 142.Cho S, et al. Impaired cell surface expression of human CD1d by the formation of an HIV-1 Nef/ CD1d complex. Virology 2005;337:242–252. [DOI] [PubMed] [Google Scholar]
- 143.Chen N, et al. HIV-1 down-regulates the expression of CD1d via Nef. Eur J Immunol 2006;36:278–286. [DOI] [PubMed] [Google Scholar]
- 144.Moll M, Andersson SK, Smed-Sorensen A, Sandberg JK. Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood 2010;116:1876–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kelly H, Mandraju R, Coelho-dos-Reis JG, Tsuji M. Effects of HIV-1-induced CD1c and CD1d modulation and endogenous lipid presentation on CD1c-restricted T-cell activation. BMC Immunol 2013;14:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 2005;434: 525–529. [DOI] [PubMed] [Google Scholar]
- 147.Albacker LA, et al. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med 2013;19:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol 2005;35:1692–1701. [DOI] [PubMed] [Google Scholar]
- 149.Brigl M, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med 2011;208:1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol 2007;178:2706–2713. [DOI] [PubMed] [Google Scholar]
- 151.Cohen NR, et al. Innate recognition of cell wall beta-glucans drives invariant natural killer T cell responses against fungi. Cell Host Microbe 2011;10:437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Salio M, et al. Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc Natl Acad Sci U S A 2007;104:20490–20495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Darmoise A, et al. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity 2010;33:216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.De Libero G, et al. Bacterial infections promote T cell recognition of self-glycolipids. Immunity 2005;22:763–772. [DOI] [PubMed] [Google Scholar]
- 155.Bourgeois EA, et al. Bee venom processes human skin lipids for presentation by CD1a. J Exp Med 2015;212:149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cox D, et al. Determination of cellular lipids bound to human CD1d molecules. PLoS ONE 2009;4:e5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Paduraru C, et al. Role for lysosomal phospholipase A2 in iNKT cell-mediated CD1d recognition. Proc Natl Acad Sci U S A 2013;110:5097–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Salio M, Cerundolo V. Linking inflammation to natural killer T cell activation. PLoS Biol 2009;7: e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Haas U, et al. Characterization and differentiation-dependent regulation of secreted phospholipases A in human keratinocytes and in healthy and psoriatic human skin. J Invest Dermatol 2005;124:204–211. [DOI] [PubMed] [Google Scholar]
- 160.Vavrova K, et al. Filaggrin deficiency leads to impaired lipid profile and altered acidification pathways in a 3D skin construct. J Invest Dermatol 2014;134:746–753. [DOI] [PubMed] [Google Scholar]
- 161.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013;498:367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Park M, Do E, Jung WH. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 2013;41:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Harada K, Saito M, Sugita T, Tsuboi R. Malassezia species and their associated skin diseases. J Dermatol 2015;42:250–257. [DOI] [PubMed] [Google Scholar]
- 164.Rosenstein R, Gotz F. Staphylococcal lipases: biochemical and molecular characterization. Biochimie 2000;82:1005–1014. [DOI] [PubMed] [Google Scholar]
- 165.Kang SJ, Cresswell P. Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J Biol Chem 2002;277:44838–44844. [DOI] [PubMed] [Google Scholar]
- 166.Odyniec AN, Barral DC, Garg S, Tatituri RV, Besra GS, Brenner MB. Regulation of CD1 antigen-presenting complex stability. J Biol Chem 2010;285:11937–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol 2003;3:11–22. [DOI] [PubMed] [Google Scholar]
- 168.Relloso M, et al. pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity 2008;28:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol 2004;5:169–174. [DOI] [PubMed] [Google Scholar]
- 170.Yuan W, et al. Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc Natl Acad Sci U S A 2007;104:5551–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leon L, et al. Saposins utilize two strategies for lipid transfer and CD1 antigen presentation. Proc Natl Acad Sci U S A 2012;109:4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cernadas M, Cavallari M, Watts G, Mori L, De Libero G, Brenner MB. Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J Immunol 2010;184:1235–1241. [DOI] [PubMed] [Google Scholar]
- 173.Barral DC, et al. CD1a and MHC class I follow a similar endocytic recycling pathway. Traffic 2008;9:1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Garzon D, Anselmi C, Bond PJ, Faraldo-Gomez JD. Dynamics of the antigen-binding grooves in CD1 proteins: reversible hydrophobic collapse in the lipid-free state. J Biol Chem 2013;288: 19528–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Manolova V, et al. Functional CD1a is stabilized by exogenous lipids. Eur J Immunol 2006;36:1083–1092. [DOI] [PubMed] [Google Scholar]
- 176.Kronenberg M, Havran WL. Immunology: oiling the wheels of autoimmunity. Nature 2014;506:42–43. [DOI] [PubMed] [Google Scholar]
- 177.Felio K, et al. CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. J Exp Med 2009;206:2497–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lockridge JL, et al. Analysis of the CD1 antigen presenting system in humanized SCID mice. PLoS ONE 2011;6:e21701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nieuwenhuis EE, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest 2009;119:1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wei B, et al. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J Immunol 2010;184:1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012;336:489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wingender G, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology 2012;143:418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]


