Abstract
Eukaryotic gene expression is often tightly regulated by interactions between transcription factors (TFs) and their DNA cis targets. Yeast one-hybrid (Y1H) is one of the most extensively used methods to discover these interactions. We developed a high-throughput meiosis-directed yeast one-hybrid system using the Magic Markers of the synthetic genetic array analysis. The system has a transcription factor–DNA interaction discovery rate twice as high as the conventional diploid-mating approach and a processing time nearly one-tenth of the haploid-transformation method. The system also offers the highest accuracy in identifying TF–DNA interactions that can be authenticated in vivo by chromatin immunoprecipitation. With these unique features, this meiosis-directed Y1H system is particularly suited for constructing novel and comprehensive genome-scale gene regulatory networks for various organisms.
Growth and development in prokaryotic and eukaryotic organisms are modulated by gene expression through transcriptional regulation (Chan et al. 2010). Messenger RNA transcription is controlled by physical interactions between TFs and their cis targets on the chromosomes. Several methods have been developed to enumerate such TF–DNA interactions, for example, yeast one-hybrid (Y1H) (Li and Herskowitz 1993), chromatin immunoprecipitation (ChIP) (Solomon et al. 1988), ChIP-seq, and DNA affinity purification (DAP) (O'Malley et al. 2016). These approaches have been used to construct gene regulatory networks (GRNs) for system-biology analysis. GRNs are highly valuable in providing crucial clues to underlying genetic regulatory mechanisms, leading to deep understanding of developmental processes, causes of human diseases (Fuxman Bass et al. 2015), and novel methods for breeding better crops (Lin et al. 2013; Yang et al. 2017).
Among all the methods developed for detecting TF–DNA interactions, Y1H remains one of the most popular choices owing to its straightforward and low-cost nature. Y1H has been used extensively to construct GRNs in Drosophila melanogaster, Caenorhabditis elegans, Homo sapiens, and Arabidopsis thaliana (Gaudinier et al. 2011, 2018; Hens et al. 2011; Reece-Hoyes et al. 2011; Fuxman Bass et al. 2015; Taylor-Teeples et al. 2015). Y1H typically relies on introducing a DNA bait fused to a reporter gene and a separate TF prey into the same yeast cell to screen for prey and bait (TF–DNA) binding through yeast viability selection or colorimetric assays induced by reporter gene activation (Reece-Hoyes and Marian Walhout 2012). Introduction of the bait and prey can be achieved by (1) a “diploid-mating” system that involves mating two haploid strains, one strain with the TF prey (Fig. 1A,B) and the other with the DNA bait (Fig. 1D,E), into a diploid strain (Fig. 1C; Gaudinier et al. 2011, 2018; Reece-Hoyes et al. 2011; Taylor-Teeples et al. 2015); or (2) a “haploid-transformation” system, involving two rounds of consecutive transformation of a haploid strain (Fig. 1D–F; Deplancke et al. 2006a; Hens et al. 2011; Li et al. 2012; Yang et al. 2017; Petzold et al. 2018). Using the viability selection, the haploid-transformation system was found to yield approximately twofold higher discovery rate of TF–DNA interactions than that of the diploid format (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011; Taylor-Teeples et al. 2015). However, the haploid-transformation system, needing two rounds of yeast transformation, is more laborious than the diploid-mating system and so requires a much longer processing time (Hens et al. 2011). This long processing time substantially limited its usage in large-scale analysis at the whole-genome level. Therefore, nearly all the present large-scale GRNs were constructed using the diploid-mating system, which unfortunately suffers from limited resolution power owing to its inherent lower discovery rate.
Figure 1.
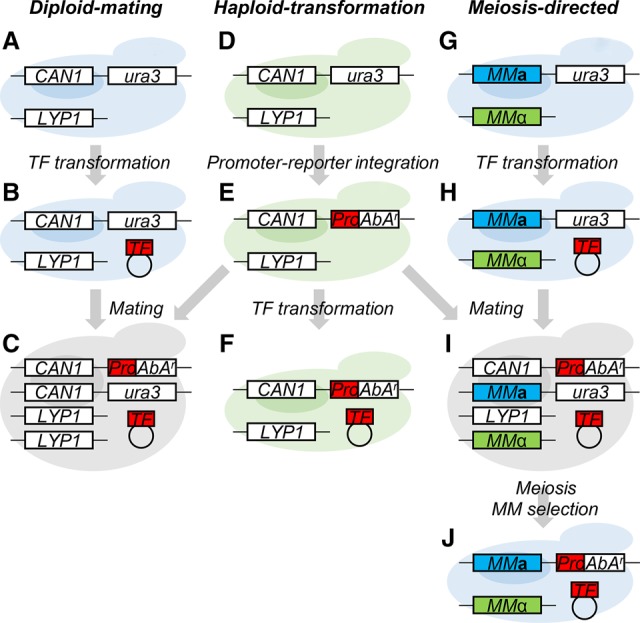
Three types of Y1H systems. (A–E) Diploid-mating system. A haploid MATa strain (light blue for MATa mating type) (A) was transformed with a plasmid carrying the transcription factor gene (TF) (B). An antibiotic Aureobasidin A (AbA) selection marker (AbAr) driven by a promoter (Pro) of a secondary cell wall biosynthesis gene was chromosomally integrated into a haploid MATα strain (light green for MATα mating type) (D) at the ura3 locus to yield the MATα_Pro strain (E). The TF-transformed strain (B) was then mated with the MATα_Pro (E) to produce a diploid strain (C). (D–F) Haploid-transformation system. Two-round transformation of a haploid MATα strain (D) were carried out for promoter integration (E) and TF plasmid introduction (F). (D,E, G–J) Meiosis-directed system. MMa and MMα (Magic Marker; in blue and green) were used to replace CAN1 and LYP1, resulting in the Y1HGold-MM strain (can1Δ/lyp1Δ) (G). A TF plasmid was transformed into the Y1HGold-MM strain (H) followed by mating with the MATα_Pro strain (E), resulting in a heterologous diploid strain (CAN1/can1Δ/LYP1/lyp1Δ) (I). This diploid strain then underwent meiosis, and the MATa cells (J) were selected via MMa and MMα.
In yeast genetics, the synthetic genetic array (SGA) analysis was developed to identify positive association of two genes or genetic traits (Tong et al. 2001; Tong and Boone 2007; Yeh et al. 2017). For example, two single-mutant strains are tested, with one mutant carrying a knockout gene A (or a genetic trait) and a wild-type gene B (i.e., aB) and the other mutant a gene B knockout, wild-type gene A, and two Magic Markers (MMs) (Tong and Boone 2006, 2007) (i.e., Ab_MMs). These two single mutants are mated to incorporate the two traits (a and b) to yield a heterozygous diploid strain (AaBb_MMs). This diploid strain is induced for meiosis where MMs convert AaBb_MMs to a haploid double mutant (ab_MMs). The inviable haploid double mutant would then allow identification of functional interaction, that is, a “synthetic lethal” interaction, between genes A and B.
In this study, we exploited this SGA system for use in Y1H screening, in which cell viability, in the presence of antibiotics, serves as a read-out for reporting interaction between DNA bait and TF prey. We called our SGA-based screening system “meiosis-directed Y1H” (Fig. 1G–J) because of the adaptation of the unique meiosis step. This system combines the advantages of the two current Y1H systems. It has (1) a mating step to incorporate both TF preys and DNA baits to achieve short processing time and (2) a meiosis step after mating to generate haploid cells for viability selection to yield high discovery rate.
Results
Construction of the Y1H strain with Magic Markers
To develop this meiosis-directed Y1H system, we first constructed a MATa haploid strain containing two MMs (Tong and Boone 2006, 2007), MMa and MMα, (Fig. 1G; for details, see Supplemental Fig. S1D) based on the commercially available Y1HGold strain (MATα, Clontech) (Supplemental Fig. S1A). MMa was PCR-amplified from the previously reported Y8205 strain (Tong and Boone 2007; Yeh et al. 2017) and chromosomally recombined into the Y1HGold at the CAN1 (arginine permease) locus. The resulting strain (Supplemental Fig. S1B) was then used for the transformation to introduce the amplified MMα at the LYP1 (lysine permease) locus (Supplemental Fig. S1C). To switch the mating type of the transformants, the entire MATa locus was amplified from strain BY4741 (Brachmann et al. 1998) and used to replace the MATα locus. We named the final strain as Y1HGold-MM (Fig. 1G; Supplemental Fig. S1D).
Selection and construction of TF preys and DNA baits for Y1H
To select the TF-prey candidates, we carried out transcriptomic analysis to identify the specifically or highly expressed TFs in wood-forming tissue and cells. In the tissue-type level, we previously identified stem differentiating xylem (SDX)-specific TFs through RNA-seq analysis on four types of tissue, that is, SDX, phloem, leaf, and young shoot, of the model woody plant, Populus trichocarpa (Lin et al. 2017). In the cell-type level, laser capture microdissection was used to isolate two types of wood-forming cells, fibers and vessels, from SDX (Fig. 2) for RNA-seq analysis (Lin et al. 2017; Shi et al. 2017) that led to the identification of 20 fiber-specific and 37 vessel-specific TFs (FDR < 0.05 and transcript abundance ratio >1.5 [fiber vs. vessel]) (Supplemental Table S3). We combined the SDX-, fiber-, and vessel-specific TFs with those TFs highly expressed (>3 CPM, counts per million) in SDX, fibers, and vessels, resulting in a final list of 1301 TFs. We then defined these 1301 TFs as wood-forming-related TFs (Supplemental Table S3). Ninety-two TFs were randomly selected and cloned as the TF-prey candidates. We selected the 2-kb promoter regions of seven genes as the DNA baits including five lignin biosynthesis genes (PtrCCoAOMT1, PtrCCoAOMT2, PtrHCT1, PtrCCR2, and PtrC3H3) and two cellulose biosynthesis genes (PtrCesA7 and PtrCesA8) involved in wood formation (Suzuki et al. 2006; Wang et al. 2014; Yan et al. 2019). The TF preys and DNA baits were then cloned into pGADT7 and pAbAi vectors (Clontech), respectively, for the Y1H screening (Supplemental Table S3).
Figure 2.
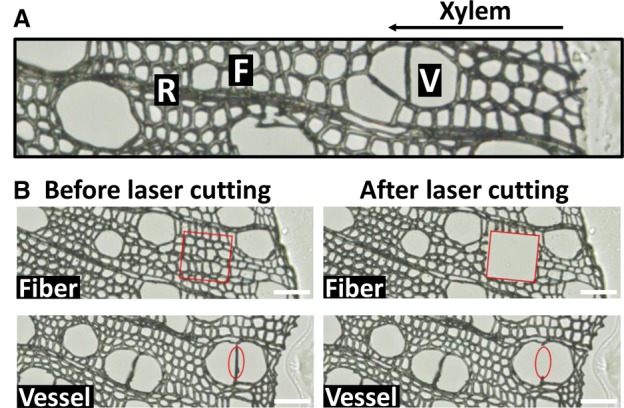
Laser capture microdissection (LCM) for collecting different cell types. (A) A cross section of stem differentiating xylem containing fiber (F), vessel (V), and ray cells (R). (B) Fiber and vessel cells collected by LCM. Scale bars, 50 µm.
The meiosis-directed Y1H system
We individually introduced TF-prey-containing plasmids into the Y1HGold-MM strain. The resulting strain (Fig. 1H) was then mated with the MATα strain containing the integrated DNA bait (Fig. 1E) to generate a diploid strain containing the TF prey and the DNA bait (Fig. 1I). Meiosis of this diploid strain yielded a mixture of unsporulated diploids (diploid without meiosis) and sporulated haploid segregants. MMs in the system then allowed the elimination of the unsporulated diploid cells and selection for specific mating-type haploid cells containing both TF prey and DNA bait (Fig. 1J), thus permitting the viability selection in the haploid format for positive TF–DNA interaction. The MMs-mediated diploid elimination and haploid selection are the two most critical steps in the new Y1H system, requiring specifically designed yeast strains to function.
To achieve diploid elimination, MMa and MMα were used to replace the yeast chromosomal CAN1 and LYP1, two permease genes that can mediate uptake of toxic arginine and lysine analogs, respectively, resulting in a can1Δ and lyp1Δ haploid strain (Fig. 1GH) that would be incapable of up-taking these analogs. Upon mating of DNA-bait (CAN1/ LYP1) (Fig. 1E) and TF-prey strains (can1Δ /lyp1Δ) (Fig. 1H), the heterozygous diploid cells (CAN1/can1Δ/LYP1/lyp1Δ) (Fig. 1I) would then regain the ability to uptake the toxic arginine and lysine analogs to kill the unsporulated diploid cells to only retain the MATa and MATα haploid segregants (can1Δ/lyp1Δ) (Fig. 1J). We then selected the MATa haploid segregants through MMa in the cells. MMa is a STE2 promoter-driven his5 gene (see structure in Supplemental Fig. S1D) that can only be turned on in MATa background to enable the selection of the MATa haploid segregants through the his5+ phenotype. This allows yeast viability selection for TF–DNA interactions in a haploid format with specific mating type. In this way, the new Y1H system would combine the advantages of diploid-mating for short processing time and haploid-transformation for high discovery rate of TF–DNA interactions.
Y1H screening format and yeast colony image processing
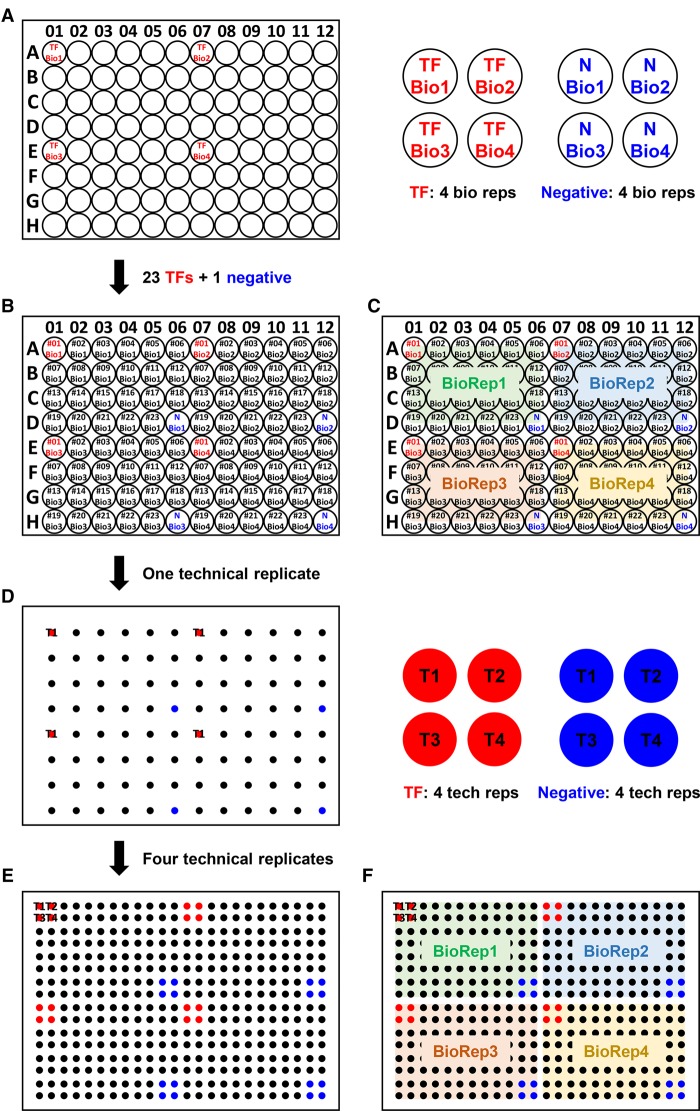
In all of our screening experiments, the HDA robotic yeast arraying platform (Yeh et al. 2017) was used to carry out a high-throughput 384-colony format, in which a single plate contained four biological replicates (“reps”) of a set of 23 TF preys and one negative control (empty vector) with each set being further replicated four times [(23 + 1) × 4 biological reps. × 4 technical reps. = 384] (Fig. 3). The DNA bait was introduced into the yeast in ways specific for each type of Y1H system tested (Methods). Therefore, we performed 16 replicates for each DNA-bait and TF-prey interaction experiment. We then used PhenoBooth (Singer) to capture images of the colonies grown on the Aureobasidin A (AbA)-containing selection plates (Supplemental Figs. S2–S5). The size of yeast colonies was quantified using a subtraction threshold and a circularity range by the PhenoBooth software.
Figure 3.
Experimental arrangement of yeast strains on 96- and 384-format plates. (A) A 96-well plate was divided into four sectors for housing four biological replicates. TF Bio 1–4 (red) are the four biological replicates of the same strain. (B,C) Full views after final assignments. Negative controls are in blue. (D) Appearance of the plate after first pinning of the 96 strains from B. (E,F) Full views of the 384-format after pinning four technical replicated from B.
Comparison of meiosis-directed Y1H to two traditional Y1H systems
We then tested this meiosis-directed Y1H system for interactions between 7 DNA baits and 92 TF preys and compared it with the two most commonly used Y1H methods, the diploid-mating and haploid-transformation systems. Two types of haploid-transformation systems, “smart-pooling” and “array-screens,” were described in the previous study (Reece-Hoyes and Marian Walhout 2012). Because the array-screens method has higher discovery rate and requires less effort for the library maintenance, we selected the array-screens method as the haploid-transformation system in this study. For each Y1H system, we tested 644 (92 TFs × 7 DNA) TF–DNA combinations, representing one of the few most comprehensive and simultaneous comparisons of multiple high-throughput Y1H systems.
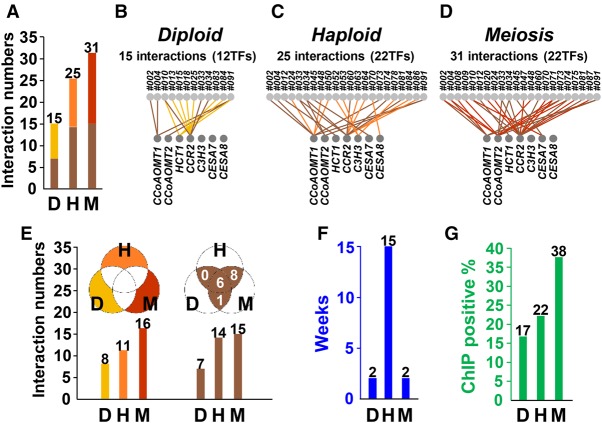
The comparative analysis demonstrated that among the three Y1H systems, the meiosis-directed screening identified the largest number of TF–DNA interactions (Fig. 4A) and was highly reproducible (Supplemental Fig. S6). Consistent with previous studies (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011), the haploid-transformation system yielded more interactions than did the diploid-mating system (Fig. 4A). The numbers of the TFs found to interact with the seven baits in the meiosis-directed system (22/92) (Fig. 4D) were the same to that in the haploid-transformation system (22/92) (Fig. 4C), whereas only 12 TFs could be scored in the diploid-mating system (Fig. 4B). The results of same numbers of identified TFs but more TF–DNA interactions suggest that the meiosis-directed Y1H may discover novel interactions undetected by the traditional Y1H systems.
Figure 4.
Comparison of experimental data obtained by three Y1H systems and in vivo validation. (A) Numbers of TF–DNA interactions identified from diploid-mating (D), haploid-transformation (H), and meiosis-directed (M) systems. (B–D) TF–DNA interactions identified in each of the three Y1H systems are graphically shown onto the GRN. The light gray circles on the top panel represent the TFs with interactions against the seven promotors (bottom panel; gray circles). (E) Numbers of TF–DNA interactions that appeared only in each system (yellow, orange, and red), and in more than two systems (brown). The color scheme matches that of A–D. (F) Blue bars represent the time of yeast strain production for the screening of 92 TFs against seven targeted promotors. (G) Percentages of TF–DNA interactions that could be in vivo validated by ChIP are shown in each Y1H system.
The analysis also showed that each of the three systems identified a unique set of interactions, with the meiosis-directed screening covering the greatest number of the unique interactions (Fig. 4A,E, yellow bars/parts). The meiosis-directed and the haploid-transformation systems, both using a haploid format for viability selection, identified a set of identical interactions (Fig. 4E, eight interactions in the brown part), of which none could be detected by the diploid-mating system. These results confirm a ploidy effect (haploid vs. diploid) on the viability selection suggested by previous studies (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011). Of all TF–DNA interactions identified, six were commonly detected by all three systems (Fig. 4A,E, brown bars/parts).
As designed, the meiosis-directed Y1H has a much reduced processing time (2 wk) in yeast strain production, the most time-consuming step in Y1H, compared to the haploid-transformation system (15 wk) (Fig. 4F) that has been thought to have the highest discovery rate of TF–DNA interactions (Gaudinier et al. 2011; Reece-Hoyes et al. 2011; Taylor-Teeples et al. 2015). An exponential increase in this time difference, for example, decades versus months, is expected as the scale of screening increases (Supplemental Table S1). In addition, it should be noted that our meiosis-directed system by design also permits scoring TF–DNA interactions at the diploid state (Fig. 1I) prior to meiosis that, when performed for scoring and then combined with the intended haploid-format viability selection (Fig. 1J), would integrate the features of both systems.
In vivo validation of the TF–DNA interactions
We next used chromatin immunoprecipitation (ChIP) to analyze in vivo the Y1H-identified TF–DNA interactions to determine false positives and validate the authenticity and biological relevance of the interactions. Because the TF–DNA interactions identified here are potential regulators for wood formation, we performed ChIP in a P. trichocarpa stem differentiating xylem (SDX) protoplast system, which we have previously demonstrated to reliably and effectively represent wood formation in planta (Lin et al. 2013, 2014). We overexpressed the identified TFs individually in the form of TF-GFP fusion in SDX protoplasts, which were analyzed by ChIP using anti-GFP antibody following our previously established procedure specifically optimized for woody plants (Lin et al. 2013; Li et al. 2014). For ChIP analysis of each interaction, three to four biological replicates were performed and only the interactions that could be validated three or more times were considered genuine interactions in vivo (Methods; Supplemental Fig. S7). Y1H screening is known to generate false positives in high frequencies. Previous Y1H studies demonstrated that only about 10%–30% of the detected TF–DNA interactions are authentic in vivo, validated by ChIP (Hens et al. 2011; Fuxman Bass et al. 2015). We selected 70%–80% of the interactions identified by each of the three Y1H systems for ChIP validation. The positive rates of the three systems were 17% (two positives of 12 tested interactions) for the diploid-mating, 22% (four of 18) for the haploid-transformation, and 38% (nine of 24) for the meiosis-directed systems (Fig. 4G; Supplemental Fig. S8). We also performed statistical analysis (Mann–Whitney U test) to investigate the ChIP enrichment of all ChIP-validated samples from three Y1H systems (Methods; Supplemental Figs. S9–S11). The relative ChIP-enrichment values from the ChIP-positives (Supplemental Fig. S9A) and ChIP-negatives (Supplemental Fig. S9B) were both used for statistical analysis. In ChIP-positives, only the enrichment values from the positive regions were used (Supplemental Fig. S9A). In ChIP-negatives, the enrichment values from each of the three regions were used (Supplemental Fig. S9B). The enrichment values of ChIP-positives were grouped with that of ChIP-negatives in each region (Supplemental Fig. S9C–H), resulting in three combinations of enrichment values (Supplemental Fig. S9F–H). Take one combination for example (ChIP-positive with ChIP-negative −1 to −650 region) (Supplemental Fig. S10), the enrichment values of meiosis-directed (Supplemental Fig. S10C) were statistically compared to that of diploid-mating (Supplemental Fig. S10A) and haploid-transformation (Supplemental Fig. S10B). Our statistical results of all three combinations demonstrated that meiosis-directed Y1H showed significantly higher ChIP enrichment than diploid-mating Y1H (P = 0.020, 0.012, 0.027) and slightly higher enrichment than haploid-transformation Y1H (P = 0.097, 0.115, 0.162) (Supplemental Fig. S11). Therefore, the meiosis-directed screening appears to be a more powerful system for identifying authentic in vivo TF–DNA interactions.
The nine ChIP-validated TF–DNA interactions uncovered in this study represent six TFs binding directly to three lignin biosynthesis genes (Supplemental Fig. S8). Among these six TFs, we found that Potri.005G001600 (also known as PtrMYB170) could bind to the promoters of PtrCCoAOMT1 and PtrCCoAOMT2 (Supplemental Fig. S8A). The PtrMYB170 homolog in another poplar, Populus tomentosa (PtoMYB170), was reported as an activator for inducing many monolignol biosynthesis genes including CCoAOMT1. The transgenic poplar with PtoMYB170 overexpression showed increased lignin deposition (Xu et al. 2017). Similarly, the knockout mutant atmyb61, the PtrMYB170 homolog in Arabidopsis, exhibited decreased lignin contents (Romano et al. 2012). With these previous studies, our Y1H and ChIP results suggested that PtrMYB170/PtoMYB170/AtMYB61 all bind directly to the promoters of lignin biosynthesis genes for the regulation of lignin contents. We also found that the protein products of Potri.005G231300 and Potri.004G158200 bind to the promoters of PtrCCoAOMT2 or PtrCCR2 (Supplemental Fig. S8D,E), and the mutant of their homolog AtbZIP44 in Arabidopsis also exhibits the effects on the micropylar endosperm cell walls (Iglesias-Fernández et al. 2013). Our results of the direct binding of three TFs (PtrMYB170, and the protein products of Potri.005G231300 and Potri.004G158200) to monolignol biosynthesis genes are consistent with previous studies (Romano et al. 2012; Iglesias-Fernández et al. 2013; Xu et al. 2017). We also identified three TFs (Potri.004G174400, Potri.009G134000, and Potri.015G082700) with unknown functions (Supplemental Fig. S8A). Further characterization of their regulatory roles using reporter assays, TF knockdown, or TF mutants may generate novel knowledge of the regulation in lignin biosynthesis during wood formation.
Discussion
Y1H screening is one of the most popular methods to build GRNs and may also be modified to identify potential regulatory TF complexes (Yang et al. 2017). The two most critical parameters for Y1H are the processing time and the discovery rate of TF–DNA interactions. Each of the two traditional Y1H systems possesses a severe drawback, namely, very long processing time for the haploid-transformation system and low discovery rate for the diploid-mating system. In this study we introduced SGA into the conventional Y1H screening and developed the meiosis-directed system. This novel system uses mating to introduce the TF preys and DNA baits into yeast strains, thus affording short processing time. Subsequent viability selection for the haploid segregants provides the high discovery rate. Although the present study was done in the 384 format, preliminary study showed that it can be easily expanded into the 1536 format for even higher throughput. In addition to the short processing time and high discovery rate, the meiosis-directed system has the highest in vivo validation rate using ChIP analysis, showcasing that the uncovered increased interaction events indeed occur in planta, rather than being false positives. Previous large-scale studies were often involved in screening through thousands of TFs in combination with hundreds of DNAs (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011; Fuxman Bass et al. 2015; Taylor-Teeples et al. 2015). At the moment, our study appears to be on a smaller scale. Further large-scale investigation should provide more information regarding the discovery rate.
Although the meiosis-directed system needs an extra haploid selection step, the time spent on this operation is minimal. Our system by design also produces diploid strains as intermediate products containing TF preys and DNA baits (Fig. 1I). This feature offers a unique advantage to perform viability selection using both the intermediate products (i.e., diploid strains) (Fig. 1I) and the final products (i.e., haploid strains) (Fig. 1J), allowing an additive and therefore a much higher discovery rate. For example, in this study, we uncovered 31 TF–DNA interactions using the final products (Fig. 4A) and the other eight interactions specifically identified at the diploid stage (Fig. 4A), for a total of 39 interactions, thereby offering a 26% (8/31) increase in the discovery rate with limited extra processing time.
One critical element for the success of this new Y1H system is the development of the Y1HGold-MM strain (Fig. 1G; Supplemental Fig. S1D). Whether the discovery rate of our system is superior to other Y1H systems, which use a variety of vectors and strains, needs further systematic investigation. However, we wish to point out that our Y1HGold-MM strain offers a unique flexibility that takes advantage of all existing yeast strain libraries with a variety of TF-prey or DNA-bait vectors used in the previous Y1H systems. These yeast library strains were produced for constructing the GRNs from different species, including Drosophila melanogaster, Caenorhabditis elegans, Homo sapiens, and Arabidopsis thaliana, for the haploid-transformation or diploid-mating systems. The most time-consuming step in Y1H screening is the yeast transformation step in which TF preys and DNA baits are introduced into the same yeast strains. Using our Y1HGold-MM strain, the existing yeast strains from other systems can be used to perform the meiosis-directed system. For example, if the existing TF preys were prepared in MATa strains, then the DNA baits can be transformed into the MATα Y1HGold-MM strain, and vice versa. Our Y1HGold-MM strain thus provides a convenient way to expand the current GRNs using the existing yeast strains.
During viability selection, the ploidy effect results in significantly different discovery rates between haploid-transformation and diploid-mating systems (Vermeirssen et al. 2007; Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011). In a previous study involved in screening of 384 TFs against one REV promoter (Gaudinier et al. 2011), the low discovery rate of the diploid-mating system was partially rectified using high-copy vectors to express TF preys. In this study, we also used high-copy vectors for screening 644 TF–DNA combinations to test the ploidy effect. We also found that the high-copy vectors indeed increased the discovery rate of the diploid-mating system. We demonstrated a ratio of 0.6 (15 by diploid-mating and 25 by haploid-transformation system) (Fig. 4A) of the discovery rates, whereas a ratio of 0.36 was reported previously using low-copy vectors (Vermeirssen et al. 2007). Yet this improvement remained somewhat unsatisfactory. In other words, using haploid format for viability selection appears to be the best strategy to discover TF–DNA interactions. Now, our meiosis-directed system essentially overcome this challenge through the deployment of the meiosis step, yielding a haploid format for the viability selection to obtain the highest discovery rate.
Many high-throughput screening systems, including Y1H, use robotics to array yeast or bacterial colonies on solid agar plates, but the colony sizes are strongly affected by the spatial locations on the plates (Typas et al. 2008; Mangat et al. 2014; French et al. 2016). Such effect appears to be more severe when the colonies are growing at plate edges, also known as the “edge effect” (Lundholt et al. 2003; French et al. 2016). The colonies on plate edges tend to grow faster, leading to inaccurate comparison of the colonies at different locations on a plate. In the Y1H high-throughput screening, the edge effect would become more serious if the potential TF–DNA interactions and the negative control were tested by only one biological sample (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011). In this study, we applied 16 replicates, including four biological replicates each with four technical replicates, to examine the TF–DNA interactions with the negative control. These four biological replicates were spatially separated from each other on the plates (Fig. 3), thus minimizing the edge effect and ensuring robust comparison among the three Y1H systems in this study. To reduce the false positives, previous studies used two reporters for yeast viability selection or colorimetric assays to increase the scoring stringency (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011; Fuxman Bass et al. 2015). In our system, we used the 16 yeast replicates to enhance the robustness of the scoring and to effectively reduce the potential false positives. This is supported by our observation that the in vivo validation rates among the three Y1H systems compared in this study is consistent with studies reported to date (∼10%– 30%), suggesting that using one reporter but with more extensive biological and technical repetitions generates similar false positive rates as Y1H systems using two reporters. However, we acknowledge that a single reporter system may not be able to completely eliminate systematic artifacts by just increasing the number of replicates. Should there be a need for higher stringency, our system can be easily converted to a two-reporter system.
Although the Y1H screening is well recognized as a powerful approach for delineating gene-expression regulation, its deployment to the genome- or large-scale analysis, in which tens of thousands of interactions are involved, remains a daunting task. For such scales, speedy processing time, technical robustness, and output precision will have to be simultaneously fulfilled. In this regard, the meiosis-directed system we developed would be a method of choice for more comprehensive understanding of gene transregulation in complex biological processes in plants and other organisms.
Methods
Construction of the Y1HGold-MM strain
To convert the commercially available Y1HGold strain (MATα, Clontech) into a strain harboring two Magic Markers, MMa (can1Δ::STE2pr-his5) and MMα (lyp1Δ::STE3pr-LEU2) (Tong and Boone 2007), but in the context of MATa (designated as Y1HGold-MM strain), the following steps were taken. MMa was PCR-amplified from the Y8205 strain (Tong and Boone 2007) and homologously recombined into the Y1HGold (Supplemental Fig. S1A) at the CAN1 (arginine permease) locus. Successful replacement was identified by first growing transformants on YPD plates for 2 d for diluting away the endogenous Can1p, followed by replica plating onto solid SD medium (Tong and Boone 2006) with arginine dropout, but with addition of canavanine (100 mg/L) (Supplemental Fig. S1B). The resulting strain was then used for introducing PCR-amplified MMα at the LYP1 (lysine permease) locus using the preceding procedure, except that the selection plate was without lysine but containing thialysine (100 mg/L) (Supplemental Fig. S1C). To switch the mating type, the entire MATa locus was amplified from strain BY4741 (Brachmann et al. 1998) and used to replace the MATα locus in the constructed strain. Successful MATa-switched transformants were then selected on the histidine dropout medium (Supplemental Fig. S1D). All steps of genetic manipulations were confirmed by PCR analysis, and the genotype of the final Y1HGold-MM strain was sequentially validated through testing on specific media plates. Primers used are described (Supplemental Table S2). The Y1HGold-MM strain is freely available on request.
Construction of TF-prey and DNA-bait plasmids
Total RNA was extracted from 6-mo-old P. trichocarpa SDX using RNeasy Plant Mini kit (Qiagen). Reverse transcription was performed using PrimeScript RT reagent kit with gDNA Eraser (Takara). TF coding regions, including their respective upstream and downstream UTRs, were amplified and cloned into pENTR vector (Invitrogen) and validated by DNA sequencing (Supplemental Table S4). Subsequent subcloning of the TF coding regions into pGADT7-AD prey vector (Clontech) yielded the final TF-prey plasmids. Genomic DNA was extracted from 2-mo-old P. trichocarpa young leaves. The 2-kb regions upstream of the translational start sites were amplified and cloned into the URA3-marked pABAi vector (Clontech), which contains an Aureobasidin A resistance (AbAr) reporter, resulting in DNA-bait plasmids and validated by DNA sequencing (Supplemental Table S5). Primers used are described (Supplemental Table S2).
Chromosomal integration of DNA baits and reporter assay optimization
DNA-bait plasmids were linearized by BstBI or BbsI and chromosomally integrated into the Y1HGold strain (Clontech). Screening of the transformants with low reporter background was done as described (Deplancke et al. 2006b) with the following modifications. For each DNA bait, at least 24 transformants were tested for AbA resistance ranging from 100 to 1000 ng/mL. The determined optimal concentrations were 300 ng/mL (PtrCCoAOMT2), 500 ng/mL (PtrCCoAOMT1, PtrC3H3, PtrHCT1, PtrCesA8), 700 ng/mL (PtrCesA7), and 800 ng/mL (PtrCCR2). All selected strains were further validated for the correct integrations by PCR (Matchmaker Insert Check PCR Mix I, Clontech) and used as the DNA-bait strains in this study. Primers used are described (Supplemental Table S2).
Strain preparation for three Y1H systems
For the haploid-transformation system, the TF-prey plasmids and an empty pGADT7-AD were individually transformed into each of the DNA-bait strains, and the transformants were selected using SD-Ura-Leu medium. Six transformants from each experiment were picked and restreaked for single colonies. Four colonies with similar growth rates (four biological replicates) were transferred separately into SD-Leu liquid medium containing 25% glycerol in a 96-well format (Fig. 3A–C) and pinned onto SD-Leu plates four times (four technical replicates) by ROTOR HDA (Singer) (Supplemental Fig. S2A), which yielded a 384 format (Fig. 3D–F). Strains in the 96-well plates were stored at −80°C for future use. The 92 TF preys were divided into four batches (23 TFs/batch) for all Y1H screens (Supplemental Fig. S3).
For the diploid-mating system, TF-prey plasmids and an empty pGADT7-AD were individually transformed into the Y1HGold-MM strain (Fig. 1G; Supplemental Fig. S1D), and the transformants were selected using SD-Leu medium (Fig. 1E). Four colonies with similar growth rates (four biological replicates) were transferred separately into SD-Leu liquid medium containing 25% glycerol in a 96-well format (Fig. 3A–C) and pinned onto SD-Leu plate four times (four technical replicates) by ROTOR HDA (Singer), which yielded a 384 format (Fig. 3D–F). Strains in the 96-well plates were stored at −80°C for future use. To generate diploid strains for the diploid-mating system, each DNA-bait strain was cultured in 3 mL YPDA medium overnight. The next day, 1 mL of cells were pelleted, resuspended in 200 µL sterile water, evenly spread on a YPDA plate, and grown for 12–16 h. This lawn of cells was then transferred onto another blank YPDA plate using 384-format short pins and further cultured for 24 h. Mating was conducted on a separate blank YPDA plate through sequential pinning a set of TF-prey strains (MATa) and each of the DNA-bait strains (MATα) to generate 384 mating spots. After 24-h incubation, the bait-and-prey-containing diploids (Fig. 1I) were selected twice in sequence on the SD-Leu-Ura plates.
For the meiosis-directed system, the diploid strains generated from the diploid-mating system were sporulated on the sporulation medium containing 2% potassium acetate (w/v) for 3–5 d at 22°C. The haploid strains (MATa) containing preys and baits (Fig. 1J) were selected on the SD-Leu-Ura-His-Lys-Arg + canavanine (100 mg/L) + thialysine (100 mg/L) medium twice in sequence. Canavanine and thialysine select for meiotic segregants harboring can1Δ and lyp1Δ markers, respectively (Tong and Boone 2006, 2007). Histidine dropout was used for selecting MATa segregants.
AbA reporter selection and image processing
AbA selection was used to score bait–prey interactions. For all three Y1H systems, yeast strains containing both prey and bait were first refreshed on SD-Leu-Ura plates and incubated at 30°C for 2 d before AbA selection. The yeast strains were then pinned onto SD-Leu-Ura “master” plates. Two subsequent and sequential pinnings were done, both using fresh pins, with the first from master plates to SD-Leu-Ura + AbA (“AbA-1”) plates and the second from “AbA-1” to “AbA-2” plates. The purpose of sequential pinnings using fresh pins was to reduce cell numbers on AbA-2 plates, thereby tightening the AbA selection. Only master plates and AbA-2 plates were used for image capturing (Supplemental Fig. S4A,B). Image capturing and quantification of the colony sizes were done by using PhenoBooth Colony Counter (Singer) (Supplemental Fig. S2B) on days 0, 4, 5, 6, and 7 (for all raw and processed images, see Data access). The subtraction threshold and the circularity range were set at 245 and 0.05–1, respectively. The day zero values were treated as background signals.
Scoring TF–DNA interactions
To qualify for scoring, a TF–DNA interaction, which is represented by 16 colonies (i.e., four biological and four technical replicates) in each of the three Y1H screens, must grow on the master plate (i.e., 48 colonies in total) to ensure the presence of both prey and bait plasmids (Supplemental Fig. S4A). For those TF–DNA interactions that did not fulfill this criterion, they were eliminated for further consideration in all three screens. We first ranked the colony-size values of the negative control (i.e., empty vector) on the AbA-2 plates (Supplemental Fig. S4B, blue colonies) and used the averaged value from the 5th to the 12th ranks (Supplemental Fig. S4C) as the threshold value. A twofold threshold value was then taken as a cutoff to judge whether or not other colonies (Supplemental Fig. S4B, red colonies) on the same plate grew (Supplemental Fig. S4D). We defined a scored TF–DNA interaction as one for which at least half of the total number of the colonies must grow on the selection plates as previously described (Gaudinier et al. 2011; Hens et al. 2011; Reece-Hoyes et al. 2011; Fuxman Bass et al. 2015). In our case, this would represent at least eight of the 16 colonies on the AbA-2 plates, which is equivalent to the success of at least two biological replicates with eight technical replicates (Supplemental Fig. S5). All interactions that were scored positively in the course of imaging analysis from the 4th to the 7th d were pooled and documented in the final list (Supplemental Table S6).
SDX protoplast chromatin immunoprecipitation (ChIP)
The P. trichocarpa SDX protoplast isolation and transfection were done as described (Lin et al. 2013, 2014) with minor modifications. Six 8-cm debarked stem segments of 6-mo-old P. trichocarpa were immersed in 40 mL cell wall digestion enzyme solution in a 50-mL centrifuge tube for 3 h at room temperature (RT). The digested debarked stem segments were removed and transferred into the 30 mL MMG solution in another 50-mL centrifuge tube. The protoplasts were released by gentle shaking for 30 sec, filtered by the 75-μm nylon membrane, and centrifuged at 500g for 3 min at RT. The pelleted protoplasts were resuspended in the MMG solution, and the cell density was adjusted to 5 × 105 cells/mL. Twenty mL of the protoplasts were used for transfection with 2 mL plasmid DNA (2 mg/mL) and 22 mL PEG solution. The protoplast-DNA-PEG mixture was fully mixed in a 250-mL bottle and incubated for 10 min at RT. Transfection was terminated by adding 88-mL WI solution, followed by centrifugation at 500g for 10 min at RT. The pelleted protoplasts were resuspended in 50-mL WI solution and incubated at RT in a petri dish coated with 1% (w/v) BSA for 7 h in the dark. The transfected protoplasts were collected by centrifugation at 700g for 10 min at RT and resuspended in the 20 mL WI solution.
ChIP was performed as described (Lin et al. 2013; Li et al. 2014) with minor modifications. Crosslinking was done by adding 540 μL formaldehyde (37%) followed by gentle mixing and incubation at RT for 10 min. Reaction was stopped by adding 1.375 mL of 2 M glycine and incubated for 5 min at RT. Treated protoplasts were centrifuged at 700g for 10 min at 4°C, washed twice by 1.8 mL WI solution (4°C) in a 2-mL microfuge tube, pelleted by centrifugation at 700g for 5 min at 4°C, and resuspended in 1.8 mL Buffer 2 (0.25 M sucrose, 10 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 5 mM ß-mercaptoethanol, 1% Triton X-100, protease inhibitors). To ensure removing residual formaldehyde, the protoplast suspension was transferred to a new 2-mL microfuge tube and recentrifuged at 16,000g for 10 min at 4°C. The pelleted protoplasts were resuspended in 500 μL Buffer 3 (1.7 M sucrose, 10 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 5 mM ß-mercaptoethanol, 0.15% Triton X-100, protease inhibitors). The mixture was carefully laid on top of 500 μL Buffer 3 in a 1.5-mL microfuge tube, centrifuged at 16,000g for 1 h at 4°C. The pelleted nuclei were resuspended in 300 μL lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS, protease inhibitors) and transferred to a TPX 1.5-mL tube (Diagenode C30010010) for sonication, which was done in Bioruptor (Diagenode) for three rounds. Each round consists of five cycles of 30 sec “On” and 30 sec “Off” set at High Power. Between each round, the tube was removed and hand-flickered to ensure even sonication in the next round. After sonication, the mixture was transferred to a 1.5-mL microfuge tube, centrifuged at 16,000g for 10 min at 4°C, from which 250 μL supernatant was recovered. A fraction of the supernatant (50 μL) was saved at −80°C as “Input” control. The remaining 200 µL was transferred to a 2-mL microfuge tube, in which 1.8 mL dilution buffer (16.7 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.1% Triton X-100, 1.2 mM EDTA [pH 8.0], protease inhibitors without PMSF) and anti-GFP antibody (6–10 μg) (Abcam, ab290) were added. After rotating at 10 rpm for 12–16 h at 4°C, Dynabeads protein G (Thermo) (40–50 μL) was added, and the mixture was further rotated for 2–4 h at 4°C. The beads were sequentially washed by 1 mL high-salt buffer (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 2 mM EDTA [pH 8.0], 0.25% Triton X-100), 1 mL LiCl buffer (10 mM Tris-HCl [pH 8.0], 25 mM LiCl, 1 mM EDTA [pH 8.0], 0.5% NP-40, 0.25% sodium deoxycholate), and twice with 1 mL TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, pH 8.0). A magnetic stand was used to collect the beads during each wash, which was done at 4°C for 5 min on a rotator (10 rpm). The immunoprecipitated protein-DNA was eluted with 250 μL prewarmed (65°C) elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at 65°C. The elution step was performed twice. To the combined eluted sample (“IP” sample; 500 μL) as well as the “Input” control sample (brought to 500 µL by adding 450 μL prewarmed elution buffer), 20 μL of 5 M NaCl was added to revert protein-DNA crosslinking at 65°C for 6–12 h. Finally, 32 μL of protease/RNase buffer (150 mM EDTA [pH 8.0], 615 mM Tris-HCl [pH 6.5], 0.65 mg/mL Proteinase K, 0.31 mg/mL RNase A) was added for incubation at 45°C for 1 h, and the DNA was purified by Qiagen MinElute kit.
ChIP-qPCR was performed using Roche SYBR Green Master Mix. The DNA from “Input” was diluted 50 times, and from “IP” was diluted 10 times as the templates. PtrActin was used as the negative control. All the primer sequences used for qPCR were listed in Supplemental Table S2.
Statistical analyses of the relative ChIP-enrichment values
Statistical analyses were performed to examine the relative ChIP-enrichment values of the TF–DNA interactions identified by three Y1H systems. The relative ChIP-enrichment values from all ChIP-validated samples were analyzed, including all three biological replicates of the ChIP-positives and ChIP-negatives. ChIP validations were performed for the diploid-mating (12 presumed TF–DNA interactions), haploid-transformation (18), and meiosis-directed (24) Y1H systems. Therefore, 36 (12 × 3 biological reps. for diploid-mating), 54 (18 × 3 for haploid-transformation), and 72 (24 × 3 for meiosis-directed) ChIP-enrichment values were used. Because the mean of the enrichment values in meiosis-directed is higher than diploid-mating and haploid-transformation (Supplemental Fig. S11), one-tailed Mann–Whitney U test (nonparametric test) was applied for the statistical analysis.
Data access
The raw and processed plate images generated in this study have been uploaded to the Google Drive (https://drive.google.com/open?id=156WjEuY3wcfPjKVXkstFDuvxbXw6l5JT).
Supplementary Material
Acknowledgments
W.L. is supported by the National Natural Science Foundation of China (NSFC 31522014) and China 1000-Talents Plan for young researchers. T.-H.C. is supported by the Taiwan Ministry of Science and Technology (MOST 105-2311-B-001-059), Academia Sinica (Taiwan) and MOST (106-0210-01-15-02 and 107-0210-01-19-01), and Academia Sinica Thematic Project (AS-103-TP-B12). Y.-C.J.L. is supported by Taiwan MOST (106-2311-B-002-001-MY2 and 107-2636-B-002-003) and MOST Young Scholar Fellowship Columbus Program. C.-S.Y. is supported by an Academia Sinica Postdoctoral Fellowship. V.L.C. is supported by the NSFC Grant (31430093), U.S. National Science Foundation, Plant Genome Research Program Grant DBI-0922391, U.S. Office of Science (Biological and Environmental Research), Department of Energy Grant DE-SC000691, North Carolina State University Jordan Family Distinguished Professor Endowment, and North Carolina State University Forest Biotechnology Industrial Research Consortium.
Author contributions: T.-H.C., Y.-C.J.L., C.-S.Y., W.L., and V.L.C. designed the experiments. T.-H.C., Y.-C.J.L., W.L., and V.L.C. supervised the project. C.-S.Y., F.M., C.-T.K., T.-S.H., J.-H.Y., E.-T.H., N.-C.T., and Y.-C.J.L. performed the Y1H experiments. Z.W. and H.M. performed the ChIP assays. Z.W., H.M., C.Z., G.-Z.Q., J.J., G.L., J.P.W., and Y.-C.J.L. contributed to cloning of the 92 TFs. C.-S.Y., Z.W., F.M., C.-T.K., T.-S.H., N.-C.T., and Y.-C.J.L. analyzed the data. T.-H.C., Y.-C.J.L., C.-S.Y., C.-Y.K., C.-C.L., W.L., and V.L.C. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.245951.118.
Freely available online through the Genome Research Open Access option.
References
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. 10.1126/science.1182213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, et al. 2006a. A gene-centered C. elegans protein-DNA interaction network. Cell 125: 1193–1205. 10.1016/j.cell.2006.04.038 [DOI] [PubMed] [Google Scholar]
- Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJ. 2006b. Gateway-compatible yeast one-hybrid screens. CSH Protoc 2006 10.1101/pdb.prot4590 [DOI] [PubMed] [Google Scholar]
- French S, Mangat C, Bharat A, Côté JP, Mori H, Brown ED. 2016. A robust platform for chemical genomics in bacterial systems. Mol Biol Cell 27: 1015–1025. 10.1091/mbc.E15-08-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Sahni N, Shrestha S, Garcia-Gonzalez A, Mori A, Bhat N, Yi S, Hill DE, Vidal M, Walhout AJM. 2015. Human gene-centered transcription factor networks for enhancers and disease variants. Cell 161: 661–673. 10.1016/j.cell.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, Liu Z, Breton G, Pruneda-Paz JL, Kim D, Kay SA, et al. 2011. Enhanced Y1H assays for Arabidopsis. Nat Methods 8: 1053–1055. 10.1038/nmeth.1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudinier A, Rodriguez-Medina J, Zhang L, Olson A, Liseron-Monfils C, Bagman AM, Foret J, Abbitt S, Tang M, Li B, et al. 2018. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563: 259–264. 10.1038/s41586-018-0656-3 [DOI] [PubMed] [Google Scholar]
- Hens K, Feuz JD, Isakova A, Iagovitina A, Massouras A, Bryois J, Callaerts P, Celniker SE, Deplancke B. 2011. Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat Methods 8: 1065–1070. 10.1038/nmeth.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Barrero-Sicilia C, Carrillo-Barral N, Oñate-Sánchez L, Carbonero P. 2013. Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J 74: 767–780. 10.1111/tpj.12162 [DOI] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. 1993. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262: 1870–1874. 10.1126/science.8266075 [DOI] [PubMed] [Google Scholar]
- Li Q, Lin YC, Sun YH, Song J, Chen H, Zhang XH, Sederoff RR, Chiang VL. 2012. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc Natl Acad Sci 109: 14699–14704. 10.1073/pnas.1212977109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lin YC, Li Q, Shi R, Lin CY, Chen H, Chuang L, Qu GZ, Sederoff RR, Chiang VL. 2014. A robust chromatin immunoprecipitation protocol for studying transcription factor–DNA interactions and histone modifications in wood-forming tissue. Nat Protoc 9: 2180–2193. 10.1038/nprot.2014.146 [DOI] [PubMed] [Google Scholar]
- Lin YC, Li W, Sun YH, Kumari S, Wei H, Li Q, Tunlaya-Anukit S, Sederoff RR, Chiang VL. 2013. SND1 transcription factor–directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell 25: 4324–4341. 10.1105/tpc.113.117697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Li W, Chen H, Li Q, Sun YH, Shi R, Lin CY, Wang JP, Chen HC, Chuang L, et al. 2014. A simple improved-throughput xylem protoplast system for studying wood formation. Nat Protoc 9: 2194–2205. 10.1038/nprot.2014.147 [DOI] [PubMed] [Google Scholar]
- Lin YJ, Chen H, Li Q, Li W, Wang JP, Shi R, Tunlaya-Anukit S, Shuai P, Wang Z, Ma H, et al. 2017. Reciprocal cross-regulation of VND and SND multigene TF families for wood formation in Populus trichocarpa. Proc Natl Acad Sci 114: E9722–E9729. 10.1073/pnas.1714422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundholt BK, Scudder KM, Pagliaro L. 2003. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen 8: 566–570. 10.1177/1087057103256465 [DOI] [PubMed] [Google Scholar]
- Mangat CS, Bharat A, Gehrke SS, Brown ED. 2014. Rank ordering plate data facilitates data visualization and normalization in high-throughput screening. J Biomol Screen 19: 1314–1320. 10.1177/1087057114534298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598 10.1016/j.cell.2016.08.063 [DOI] [PubMed] [Google Scholar]
- Petzold HE, Rigoulot SB, Zhao C, Chanda B, Sheng X, Zhao M, Jia X, Dickerman AW, Beers EP, Brunner AM. 2018. Identification of new protein–protein and protein–DNA interactions linked with wood formation in Populus trichocarpa. Tree Physiol 38: 362–377. 10.1093/treephys/tpx121 [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Marian Walhout AJ. 2012. Yeast one-hybrid assays: a historical and technical perspective. Methods 57: 441–447. 10.1016/j.ymeth.2012.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, Walhout AJ. 2011. Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat Methods 8: 1059–1064. 10.1038/nmeth.1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JM, Dubos C, Prouse MB, Wilkins O, Hong H, Poole M, Kang KY, Li E, Douglas CJ, Western TL, et al. 2012. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol 195: 774–786. 10.1111/j.1469-8137.2012.04201.x [DOI] [PubMed] [Google Scholar]
- Shi R, Wang JP, Lin YC, Li Q, Sun YH, Chen H, Sederoff RR, Chiang VL. 2017. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 245: 927–938. 10.1007/s00425-016-2640-1 [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. 1988. Mapping protein–DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53: 937–947. 10.1016/S0092-8674(88)90469-2 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun YH, Chiang VL. 2006. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142: 1233–1245. 10.1104/pp.106.086678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, et al. 2015. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol 313: 171–192. [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C. 2007. High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae. In Methods in microbiology (ed. Stansfield I, Stark MJR), Vol. 36, pp. 369–386, 706–707. Academic Press, London: 10.1016/S0580-9517(06)36016-3 [DOI] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, et al. 2008. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods 5: 781–787. 10.1038/nmeth.1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Deplancke B, Barrasa MI, Reece-Hoyes JS, Arda HE, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Brent MR, et al. 2007. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods 4: 659–664. 10.1038/nmeth1063 [DOI] [PubMed] [Google Scholar]
- Wang JP, Naik PP, Chen HC, Shi R, Lin CY, Liu J, Shuford CM, Li Q, Sun YH, Tunlaya-Anukit S, et al. 2014. Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26: 894–914. 10.1105/tpc.113.120881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fu X, Liu R, Guo L, Ran L, Li C, Tian Q, Jiao B, Wang B, Luo K. 2017. PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol 37: 1713–1726. 10.1093/treephys/tpx093 [DOI] [PubMed] [Google Scholar]
- Yan X, Liu J, Kim H, Liu B, Huang X, Yang Z, Lin YJ, Chen H, Yang C, Wang JP, et al. 2019. CAD1 and CCR2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytol 222: 244–260. 10.1111/nph.15505 [DOI] [PubMed] [Google Scholar]
- Yang F, Li W, Jiang N, Yu H, Morohashi K, Ouma WZ, Morales-Mantilla DE, Gomez-Cano FA, Mukundi E, Prada-Salcedo LD, et al. 2017. A maize gene regulatory network for phenolic metabolism. Mol Plant 10: 498–515. 10.1016/j.molp.2016.10.020 [DOI] [PubMed] [Google Scholar]
- Yeh CS, Chang SL, Chen JH, Wang HK, Chou YC, Wang CH, Huang SH, Larson A, Pleiss JA, Chang WH, et al. 2017. The conserved AU dinucleotide at the 5′ end of nascent U1 snRNA is optimized for the interaction with nuclear cap-binding-complex. Nucleic Acids Res 45: 9679–9693. 10.1093/nar/gkx608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.