Abstract
Genome editing tools have simplified the generation of knock-in gene fusions, yet the prevalent use of gene-specific homology-directed repair (HDR) templates still hinders scalability. Consequently, realization of large-scale gene tagging requires further development of approaches to generate knock-in protein fusions via generic donors that do not require locus-specific homology sequences. Here, we combine intron-based protein trapping with homology-independent repair-based integration of a generic donor and demonstrate precise, scalable, and efficient gene tagging. Because editing is performed in introns using a synthetic exon, this approach tolerates mutations in the unedited allele, indels at the integration site, and the addition of resistance genes that do not disrupt the target gene coding sequence, resulting in easy and flexible gene tagging.
Fusing endogenous proteins with fluorescence or epitope tags is a widely used and essential approach for studying proteins within their natural regulatory context. The advent of CRISPR/Cas tools for modifying the genome (Ran et al. 2013; Doudna and Charpentier 2014; Hsu et al. 2014) has made this easier and even more accessible, yet scalability is still very limited. The need for a gene-specific homology-directed repair (HDR) template requires costly synthesis or labor-intensive molecular cloning, and because precise targeting must be achieved in frame and within the coding sequence, it necessitates careful design of reagents and screening of clonal cell lines to avoid disruptive editing at the nontagged allele. The development of split fluorescent proteins has simplified the generation of fluorescent fusions, because only a minimal tag is required for genomic knock-in (Cabantous et al. 2005; Kamiyama et al. 2016; Leonetti et al. 2016; Feng et al. 2017). Nevertheless, these endogenous tagging methods still require synthesizing individual HDR donors. Several approaches to develop generic exon-tagging methods have been demonstrated (Lackner et al. 2015; Schmid-Burgk et al. 2016), but because these require precise tagging at the coding sequence, they are limited in design flexibility and are prone to disruptive mutations at the nontagged allele as well as to indels within the tagged allele that can lead to frameshifts. Derivative strategies have been developed to increase the efficiency of homology-independent repair-dependent tagging methods but at the cost of no longer utilizing a generic donor (Suzuki et al. 2016).
An alternative approach for generating endogenous fusions is by random integration of synthetic exons delivered by transposons or retroviral particles (Trinh le and Fraser 2013). This approach, known as “protein trapping” or “CD-tagging” (Jarvik et al. 1996), is not restricted to small donors and has been used in both model organisms (Clyne et al. 2003; Buszczak et al. 2007; Trinh le et al. 2011) and mammalian cells (Sigal et al. 2007; Cohen et al. 2008). Although protein trapping is inexpensive and scalable, the random nature of tag integration precludes its use for the generation of curated libraries of fusion cell lines.
Here, by utilizing a combination of protein trapping and gene targeting, we demonstrate a novel strategy to tag genes. This approach targets introns and is efficient, easy to implement, and does not limit the size of the donor. Furthermore, in contrast to generic exon tagging, generic intron tagging allows for especially flexible donor design owing to the splice acceptor and donor sites: Any incorporated vector sequence external to those sites has no effect on the coding sequence of the tagged protein. This property not only enables protein tagging with precisely defined tags, but also allows for the addition of genomic elements, such as those encoding a resistance gene, that do not disrupt the target gene coding sequence. We use this approach to introduce antibiotic selection markers in a nondisruptive way to obtain high proportions of positively tagged cells. Generic intron tagging also uniquely tolerates mutations in the nontagged allele (because those are intronic and typically nondisruptive) as well as indels that flank the inserted donor as a result of editing that could lead to frameshifts in an exonic setting. Because the donor is generic, the generation of more fusion cell lines only requires the cloning of additional intron-targeting sgRNAs. Furthermore, choosing sgRNAs with few off-target effects is easy because introns provide a wide range of protospacer options. The high efficiency and overall high flexibility of this system would thus potentially be extremely useful for large-scale tagging experiments, as well as for quickly screening many sites for protein tagging.
Results
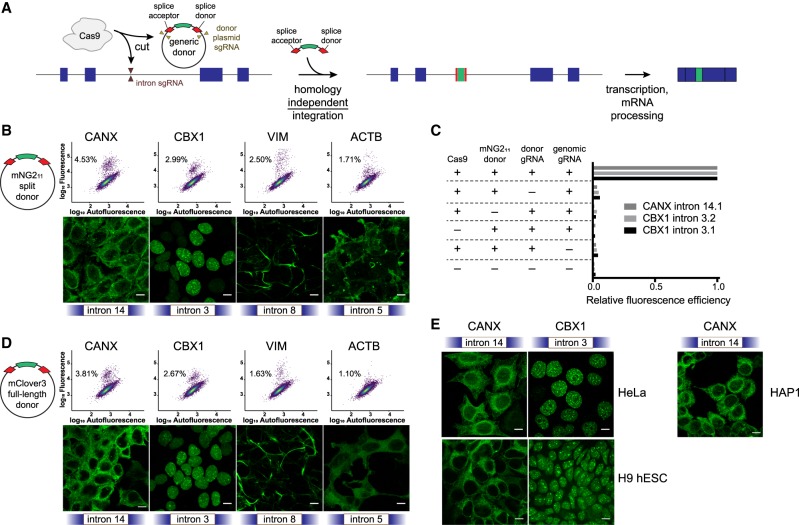
We combined homology-independent repair-based editing with the use of a generic synthetic exon donor containing a fluorescent tag to perform targeted protein trapping at intronic locations (Fig. 1A). Our synthetic donor contained the mNG211 tag, part of a previously published split fluorophore system (Feng et al. 2017), flanked by linker sequences and splice acceptor (SA) and splice donor (SD) sites (Supplemental Table S1). We embedded this sequence between two identical sgRNA target sites chosen to have minimal off-target activity in the human genome, such that cutting of the plasmid in cells generates a linear DNA donor molecule. Plasmids encoding SpCas9, sgRNAs against the donor plasmid, intron-targeting sgRNAs, and the donor plasmid itself were transfected into HEK293 cells stably expressing mNG21–10. Multiple introns for each gene were chosen semirandomly, because the generic nature of the approach allowed for the interrogation of multiple sites at once with minimal additional effort or cost. Intron “frame” was the only criterion that made intron selection nonrandom: We targeted introns that lay precisely in between would-be codons in the adjacent exons, because the donor used in this study is compatible with frame 1. However, it would be trivial to target introns that bisect would-be codons by using a donor containing the appropriate frameshift mutations.
Figure 1.
Homology-independent generic intron tagging enables efficient and easy generation of endogenous fusions. (A) Illustration of the tagging approach: Double-strand breaks are generated in the intron and donor resulting in the addition of a synthetic exon and fusion of the tag to the coding sequence. (B) Using a small donor composed of the mNG211 epitope flanked by splice acceptor and donor sites results in efficient tagging of CANX, CBX1, VIM, and ACTB at the indicated introns (all sgRNA “target 1”), as observed by flow cytometry (upper panels, colored by density) and by confocal microscopy (lower panels). Percentages in the dot plots represent the green population as a subset of the total. (C) All transfection mix components are required for tagging of CANX intron 14, sgRNA target 1 (14.1), and of CBX1 intron 3, targets 1 and 2 (3.1 and 3.2). The table indicates which component was removed, and bar plots represent the relative percentage of fluorescence-positive cells compared to the full mix. (D) Tagging using a full-length mClover3 fluorophore as a donor. (E) Tagging of CANX and CBX1 in HeLa cells, H9 human embryonic stem cells (hESC), and HAP1 cells. All images are maximum projections of Z-stacks, and scale bars correspond to 10 µm.
Proteins successfully tagged with mNG211 emit a fluorescence signal upon binding of mNG21–10. Using this approach, we were able to tag four tested genes with well-established localization patterns (CANX, CBX1, VIM, and ACTB) at a frequency that enabled easy isolation of both clonal and polyclonal tagged populations of cells (Fig. 1B). To test that our tagging approach was mediated by double-strand breaks in both the genomic sequence and the donor plasmid, we removed each individual component of the transfection mix and found that efficient tagging indeed required all components (Fig. 1C). We then tested the feasibility of integrating larger donors by replacing the mNG211 small tag (∼4.15 kDa) with a full-length mClover3 fluorescent protein (FP) (∼28.9 kDa) and found comparable integration efficiencies (Fig. 1D). In the specific case of intron 5 of ACTB, integrating a full-length FP resulted in a lower expression level and a diffuse localization pattern, consistent with the production of nonfunctional protein (rightmost panels in Fig. 1B–D). Tagging with a full-length FP versus a split FP is likely to affect the folding dynamics of the targeted protein differently at certain sites, potentially explaining the difference seen with ACTB. Last, to verify that the activity we observed was not specific to HEK293 cells, we also successfully tagged HeLa cells, H9 human embryonic stem cells, and HAP1 cells (Fig. 1E). All of these cell types exhibited tagging efficiencies <0.5% for either CANX or CBX1 at the conditions tested.
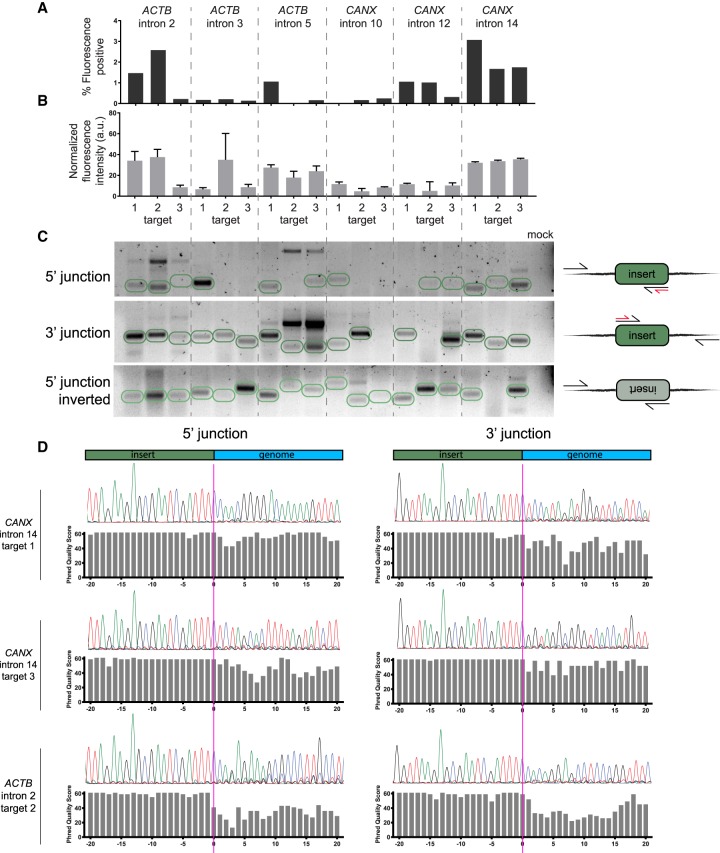
Unsuccessful tagging can be a result of, but not limited to, inefficient genomic DNA cutting, low donor integration, or a fusion location that detrimentally affects protein folding. To investigate these possibilities, we chose two genes, ACTB and CANX, and designed nine sgRNAs for each that spanned three introns. We then measured tagging efficiency and the protein expression levels in pre-enriched, polyclonal tagged cells at each of these locations (Fig. 2A,B). We found that efficient integration associated with high expression levels of the protein typically coincided within the same intron, indicating that the location of the fusion within the protein is a more critical parameter than the choice of the sgRNA within an intron. Integration of the donor construct appeared to occur for all locations whether or not successful tagging was observed, as analyzed by PCR using genomic templates from the total transfected cell populations and primers that were designed to amplify the genomic DNA-to-donor junction on both sides of the donor (Fig. 2C). We observed little discernable directional preference for donor integration, and tandem insertions were also observed, as evidenced by the upper bands corresponding to twice and sometimes three times the expected molecular weight of a single insertion (Fig. 2C). We also used Sanger sequencing on some of the amplified junctions and found accurate integration sometimes flanked by junction indels (Fig. 2D), further emphasizing the advantages of flanking donors with splice acceptor and donor sites.
Figure 2.
Successful tagging is mostly determined by the choice of intron. (A) Tagging with mNG211 across introns in ACTB and CANX. Bar plots represent the percent of fluorescence-positive cells for each sgRNA position. (B) Expression mean and standard error for positive cells in each location. Sample sizes are proportional to the bar plots in A. (C) Gel image showing the amplification of donor-to-genomic DNA junctions, as illustrated in the right-hand diagrams. Expected band sizes for insertion of a single copy of donor are circled in green. In the diagrams, black arrows represent primer sites for amplification and red arrows represent primer sites for sequencing in D. The last lane corresponds to a PCR reaction with primers for CANX intron 14, target 1, but without a template. (D) Sanger sequencing of donor-to-genomic DNA junctions shows dephasing at the donor and genomic DNA junction, which indicates indels at the integration site.
Picking three introns at random for CANX resulted in the identification of two feasible fusion locations that do not disrupt protein localization: at intron 14 (Fig. 1B) and at intron 12 (Supplemental Fig. S1), emphasizing the ease with which fusion locations can be identified using this approach. However, not all fusions that resulted in high tagging efficiency and fluorescence intensity indicated a successful fusion, because tagging ACTB at intron 2 disrupted proper localization (Supplemental Fig. S1). Therefore, it is recommended that novel fusion locations would be validated by additional methods.
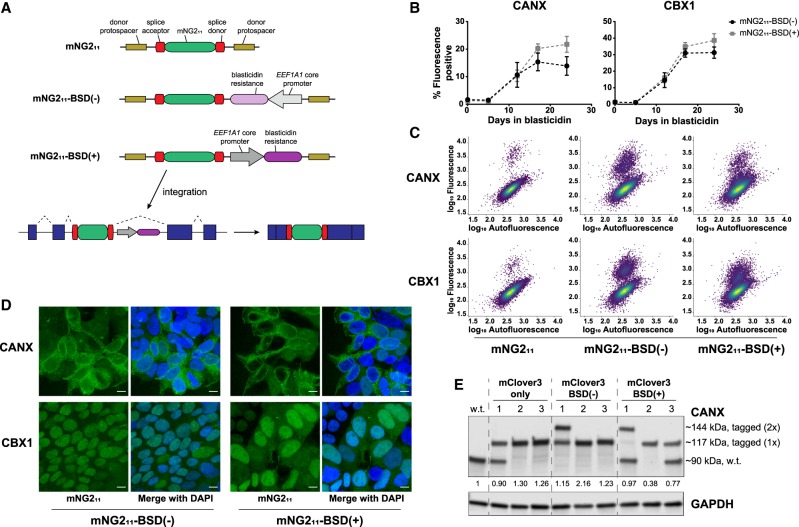
We took additional advantage of the use of splicing for the generation of protein fusions and added a blasticidin resistance gene to the donor cassette outside of the splice acceptor and donor sites but still between the donor protospacer sequences such that integration events can be selected without fusing the resistance cassette to the target protein-coding sequence (Fig. 3A). Because the blasticidin resistance gene would be expressed whether or not the donor construct is integrated in the proper orientation, the theoretical maximum percentage of positively tagged cells is 50% after blasticidin selection. This degree of enrichment would be immensely beneficial when tagging efficiency is very low and when isolation of clones without sorting is required (e.g., for nonfluorescent tags). In addition, in cases for which clonal isolation is not possible, increasing the number of tagged cells can facilitate analysis of a polyclonal population.
Figure 3.
A modified donor allows for easy selection of tagged cells. (A) Schematic of donor constructs without and with a blasticidin resistance (BSD) gene. (B) Enrichment of fluorescence-positive HEK293 mNG21–10 cells tagged with mNG211-BSD(−/+) at CANX intron 14 and CBX1 intron 3 after blasticidin treatment. Data represent mean ± SEM (n = 3). (C) Dot plots of total HEK293 cell populations tagged with mNG211 or with mNG211-BSD(−/+) and selected for 12 d. Plots are colored by density. (D) Confocal microscopy of total cell populations as in C. Images are maximum projections of Z-stacks, and scale bars correspond to 10 µm. (E) Western blot of clonal HAP1 lysates tagged with mClover3 only or mClover3-BSD(−/+) at CANX intron 14, target 1. The values below the anti-CANX blot indicate total levels of the major CANX band (tagged and untagged) relative to levels in wild-type (w.t.) cells.
Because the resistance gene is close to the splice donor and also contains an active promoter, we anticipated a potential effect on splicing efficiency and thus tested a donor cassette with the resistance gene inverted (mNG211-BSD(−)) and in parallel (mNG211-BSD(+)) relative to the splice donor site (Supplemental Table S2). Tagging of CANX and CBX1 with mNG211-BSD(−/+) revealed a large increase in the percent of positively tagged cells after 2–3 wk selection with blasticidin (Fig. 3B). CBX1 seemed to benefit more greatly from blasticidin selection than CANX in terms of fold change, potentially owing to locus-specific effects. Although there was no significant difference between mNG211-BSD(−) and mNG211-BSD(+) in terms of the percent of positively tagged cells over time, tagging with mNG211-BSD(−) appeared to result in a fluorescent cell population with an overall higher fluorescence intensity compared to the nonfluorescent population (Fig. 3C). This effect could result from the promoter of the BSD gene interfering more strongly with splicing machinery in the mNG211-BSD(+) cassette, or attributable to other effects on protein expression. Imaging of cells after blasticidin selection but before sorting confirmed a high efficiency as well as the anticipated protein localization patterns (Fig. 3D), supporting the notion that the BSD gene does not affect the targeted protein function more so than only introducing the fluorescence tag.
To more thoroughly evaluate donor splicing and protein stability after integration of a large tag and a resistance gene, we generated a BSD-containing mClover3 generic donor (mClover3-BSD(−) and mClover3-BSD(+)) (Supplemental Table S2). All mClover3-based generic donors were transfected into predominantly haploid populations of HAP1 cells targeting CANX, which after several passages can transform into diploid cells with homozygous tagged alleles (Olbrich et al. 2017). Clonal cell lines were obtained by cell sorting, expanded, and modified CANX protein was analyzed by western blotting. The primary band of most clones corresponded to a single size of CANX, indicating that the splicing efficiency of all donors is virtually 100% (Fig. 3E). Clones with multiple sizes of CANX typically corresponded to cells with heterozygous tag integration as assessed by genomic PCR (Supplemental Fig. S2A), and none of the tagging led to unexpected protein sizes (Supplemental Fig. S2B). CANX levels as assessed by the sum of the bands were largely unchanged by tagging, except possibly in the case of mClover3-BSD(+), where protein levels generally appeared lower (Fig. 3E). This was confirmed by flow cytometry analysis of the clones (Supplemental Fig. S2C) and is consistent with the BSD(+) population appearing dimmer than the BSD(−) population in Figure 3C. Taken together with the imaging data, internally tagging endogenous genes by intron-targeted protein trapping can be performed without necessarily disrupting protein localization or stability, even in the presence of a proximal resistance gene.
To summarize, we showed that generating endogenous fusions by generic intron tagging is efficient and easy to implement. Testing a small number of sgRNAs spanning multiple introns is sufficient to identify a successful tagging site, and because these do not require a locus-specific donor, costs are minimal.
Discussion
Proteins are commonly fused to either fluorescence or epitope tags to study their function, localization, and interactions within living cells. Although exogenous delivery of fused proteins using either plasmid or viral vectors is easy and widely used, results from such experiments are confounded by many factors including overexpression artifacts and the lack of endogenous regulatory context. The advent of easy-to-use genome editing tools has made endogenous tagging much more prevalent but still not common practice. The dependence on HDR limits efficiency and requires costly synthesis of gene-specific HDR templates, which also limits scalability. This motivates the development of additional tagging methods, especially those that use generic donors that are better suited for large-scale applications. Here, we demonstrated a tagging strategy that relies on a generic synthetic exon donor. We were able to successfully tag a variety of targets in multiple different cell lines, indicating the general application of this system. Importantly, intronic tagging seemed largely insensitive to donor size, because we were able to incorporate tags as small as mNG211 and as large as full-length mClover3. This particular quality of our system stands in contrast to other potentially scalable tagging systems, such as those that are only feasible with small tags and still require donor synthesis (Leonetti et al. 2016).
Methods that use generic donors have been previously demonstrated for N- and C-terminal tagging (Lackner et al. 2015; Schmid-Burgk et al. 2016), yet because tagging is performed directly in the coding sequence, these tools are limited in design flexibility and are prone to disruptive indel mutations both in the tagged and nontagged allele. Compared to the more restricted N- and C-terminal tagging method, intron tagging as described here expands the possible locations for tag integration within the coding region. It is likely that for many proteins, N- or C-terminal tagging would be disruptive, whereas an exposed, nonterminal location would result in a viable fusion. Indeed, internal tagging has borne out various protein- and gene-trap libraries constructed by random intronic integration (Sigal et al. 2006; Buszczak et al. 2007; Bürckstümmer et al. 2013). We found that successful tagging was largely determined by the relative position of the tag site within the protein-coding region, further emphasizing the importance of tag location within the protein. More information will be needed to better understand how to integrate tags within proteins in the least disruptive way, and the scalability of the intron tagging approach described here will enable systematic tagging experiments to better understand those rules.
The realization of large-scale tagging experiments will depend on the ability to achieve efficient tagging for each gene. Intron tagging increases the number of available protospacer sequences, enabling the selection of the most efficient sgRNAs. To further increase apparent tagging efficiency, we took advantage of the intronic location of the integrated tags and added a blasticidin resistance marker that will not be fused to the mature tagged protein (Fig. 3A). This enabled antibiotic selection for tag sites within the protein-coding sequence and not just those at the C-terminus (Schmid-Burgk et al. 2016). This approach can increase the apparent tagging efficiency to as high as 50% (it is limited by the random orientation of tag integration). This will enable more challenging applications such as tagging at low efficiency sites, isolation of cells without sorting, and direct analysis of polyclonal tagged cells, which are not amenable to clonal isolation.
Genomic integration of tags to study protein function at the endogenous context will continue to be vital in cell biology research. As more tools to simplify tagging become available, it will become a common practice to avoid artifacts associated with exogenous overexpression. Generic tagging methods are especially attractive because they enable large-scale tagging at minimal cost. We present an easy, flexible, scalable and robust method for gene tagging that we hope will help open the door toward the interrogation of proteome dynamics at scale both in arrayed and pooled formats.
Methods
Cloning
The mNG211 donor tag (Feng et al. 2017) flanked by flexible 15 amino acid linkers was synthesized as two complementary oligos from IDT and annealed. This template was amplified by primers to add splice donor and acceptor sites, sgRNA target sequences external to the splice sites, and 25-nt overhangs into the pMC.BESPX-MCS2 parental vector (System Biosciences). pMC.BESPX-MCS2 was digested with EcoRI and ApaI and combined with the mNG211 amplicon by Gibson assembly (New England Biolabs), generating the pMC-mNG211 donor plasmid (Supplemental Table S1). The pMC-mClover3 donor plasmid was generated by replacing the mNG211 sequence from the pMC-mNG211 plasmid with the sequence of mClover3 (Addgene 74257) by Gibson assembly (Supplemental Table S1). The BSD(−)- and BSD(+)-containing plasmids were generated by inserting DNA encoding the EEF1A1 core promoter, a blasticidin resistance gene, and an SV40 poly(A) sequence in the reverse and forward orientations, respectively, into the pMC-mNG211 or pMC-mClover3 plasmids by Gibson assembly (Supplemental Table S2).
To generate HEK293 cells stably expressing mNG21–10, mNG21–10 (Feng et al. 2017) fused to the self-cleaving 2A peptide and tdTomato (Addgene 37347) was cloned into the lenti dCAS-VP64_Blast (Addgene 61425) backbone in place of dCas9-VP64 by 3-piece Gibson assembly. sgRNA-expressing plasmids (Supplemental Table S3) were generated by digesting a lentiGuide-Puro plasmid (Addgene 52963) with Esp3I and ligating an annealed sgRNA oligo duplex as described previously (Ran et al. 2013).
Cell culture and transfection
Experiments were performed in HEK293 (ATCC CRL-1573), HeLa cells (ATCC CCL-2), H9 hESCs (WiCell), and HAP1 cells (Horizon). The HEK293 cells were generated to constitutively express mNG21–10 and tdTomato from a stably integrated lentiviral cassette. Individual clones were sorted based on the tdTomato signal, and a line with stable expression over time was selected for experiments. HEK293 and HeLa cells were cultured in DMEM (Thermo Fisher Scientific) + 10% fetal bovine serum (FBS; VWR) + antibiotic-antimycotic (Thermo Fisher Scientific). HAP1 cells were cultured in IMDM (Thermo Fisher Scientific) + 10% fetal bovine serum (FBS; VWR) + pen-strep (Thermo Fisher Scientific). H9 cell lines were cultured in a feeder-free system on plates coated with hESC-qualified Matrigel (Corning 354277) and were maintained in mTeSR1 media (STEMCELL Technologies 85850). H9 cells were dissociated using StemPro Accutase (Gibco) and 2 × 105 cells were replated per well of a 12-well plate in mTeSR1 supplemented with 10 µM ROCK inhibitor (Stemolecule Y-27632, Stemgent) for 24 h. Blasticidin selection of HEK293 and HAP1 cells was performed with 5 µg/mL blasticidin (Thermo Fisher Scientific).
For transfection experiments, cells were plated across a 12-well plate such that they would be ∼60% confluent on the day of transfection. The donor plasmid was delivered at 5× the molar ratio of lentiCas9-Blast plasmid (Addgene 53962) and the two lentiGuide-Puro plasmids (Addgene 52963) encoding (1) the donor-cutting sgRNA, and (2) the genomic locus-targeting sgRNA (Supplemental Table S3). In total, each well received ∼1.4 µg of DNA. For HEK293 and HeLa cells, DNA was delivered in 100 µL Opti-MEM (Thermo Fisher Scientific) with 4.3 µL of 1 g/L PEI (Polysciences 24765). For HAP1 cells, DNA was delivered in 100 µL Opti-MEM with 4.3 µL TurboFectin 8.0 (OriGene). For H9 cells, DNA was delivered in 50 µL Opti-MEM with 3 µL Lipofectamine Stem reagent (Thermo Fisher Scientific), along with equal amounts relative to the Cas9- and sgRNA-expressing plasmids of the episomal vector expressing TP53 inhibitor (Addgene 41856). After 6 days, all cells were harvested, analyzed, and sorted by flow cytometry.
Flow cytometry and cell sorting
Cultured cells were trypsinized, resuspended in the appropriate media to ∼1 × 106 cells/mL, and filtered through a cell strainer. Cellular fluorescence was measured on a BD FACSAria Fusion (BD Biosciences). mClover3 and mNG2 fluorescence were detected by the 488 nm laser and filters 502LP and 530/30. Autofluorescence was detected by the 405 nm laser and the 450/50 filter. Polyclonal fluorescent cell populations were acquired by isolating 1000 cells by sorting. Data were analyzed using Flowing Software 2 ver. 2.5.1 (http://flowingsoftware.btk.fi/index.php).
Confocal microscopy and image processing
For imaging experiments, cells were grown on coverslips and directly fixed in 4% formaldehyde (Electron Microscopy Sciences) in PBS (Thermo Fisher Scientific). Fixed cells were washed in PBS, and coverslips were mounted on microscopy slides in VECTASHIELD mounting medium (Vector Laboratories). Images were acquired on a Leica TCS SP8 confocal microscope. Z-stacks (0.6-µm slices) spanning the entire volume of the cells were recorded with oil-immersion 63× Plan-Apochromat lenses, 1.4 NA. Images were processed using Fiji (Schindelin et al. 2012).
Western blotting
Cultured cells were pelleted, washed with PBS, and resuspended in RIPA lysis buffer (Cell Signaling 9806) with 1× protease inhibitor cocktail (MilliporeSigma P8340). Samples were normalized by bicinchoninic acid (BCA) assay (Cell Signaling 7780), and loaded on a precast SDS–PAGE gel (Bio-Rad 4561086). Western blotting followed using standard protocols. Imaging of blots was performed on a LI-COR Odyssey (LI-COR). The following antibodies were used: α-CANX (Novus Biologicals, NBP2-53352, 1:1000), α-GAPDH (Cell Signaling 2118, 1:2000), IRDye 680LT Goat anti-Rabbit (LI-COR 926-68021, 1:10,000), and IRDye 800CW Goat anti-Mouse (LI-COR 926-32210, 1:10,000).
PCR analysis of genomic regions
Roughly 2 to 3 × 106 cells were harvested for genomic DNA extraction in 100 µL of QuickExtract (Epicentre) according to the manufacturer's protocol. Amplification of edited genomic regions was performed with the EmeraldAmp MAX PCR Master Mix (Takara Bio USA). For analysis of polyclonal cell populations (Fig. 2), primers were designed using the default parameters of Primer3 (http://primer3.ut.ee/) to produce amplicons 250–300 nt in length at the 5′ and 3′ junctions of each targeted site. Amplification reactions included a genomic primer upstream of the target integration site paired with a reverse primer hybridizing to the 3′ end of the tag, or a genomic primer downstream from the target integration site with a forward primer hybridizing to the 5′ end of the tag (Supplemental Table S4). The amplicons were imaged alongside a 100-bp DNA ladder (New England Biolabs) and extracted from a 2% agarose gel using the Monarch Gel Extraction kit (New England Biolabs), and analyzed by Sanger sequencing (GENEWIZ) using the tag-hybridizing primers from the amplification reaction. For analysis of monoclonal cell populations tagged with a longer DNA insert (Fig. 3E; Supplemental Fig. S2), primers were again designed using Primer3 to produce an amplicon 1921 bp in wild-type cells (Supplemental Table S4). After amplification, PCR products were run alongside a 1-kb DNA ladder (New England Biolabs).
Supplementary Material
Acknowledgments
This research was supported by funds from the Children's Hospital of Philadelphia and the Center for Cellular and Molecular Therapeutics. Y.V.S. is supported by F32CA239499 fellowship from the NIH National Cancer Institute. S.E.S. is partially supported by NIH T32GM008216 genetics training grant.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.246413.118.
Freely available online through the Genome Research Open Access option.
References
- Bürckstümmer T, Banning C, Hainzl P, Schobesberger R, Kerzendorfer C, Pauler FM, Chen D, Them N, Schischlik F, Rebsamen M, et al. 2013. A reversible gene trap collection empowers haploid genetics in human cells. Nat Methods 10: 965–971. 10.1038/nmeth.2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. 2007. The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–1531. 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous S, Terwilliger TC, Waldo GS. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol 23: 102–107. 10.1038/nbt1044 [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Brotman JS, Sweeney ST, Davis G. 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics 165: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, et al. 2008. Dynamic proteomics of individual cancer cells in response to a drug. Science 322: 1511–1516. 10.1126/science.1160165 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Feng S, Sekine S, Pessino V, Li H, Leonetti MD, Huang B. 2017. Improved split fluorescent proteins for endogenous protein labeling. Nat Commun 8: 370 10.1038/s41467-017-00494-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik JW, Adler SA, Telmer CA, Subramaniam V, Lopez AJ. 1996. CD-tagging: a new approach to gene and protein discovery and analysis. BioTechniques 20: 896–904. 10.2144/96205rr03 [DOI] [PubMed] [Google Scholar]
- Kamiyama D, Sekine S, Barsi-Rhyne B, Hu J, Chen B, Gilbert LA, Ishikawa H, Leonetti MD, Marshall WF, Weissman JS, et al. 2016. Versatile protein tagging in cells with split fluorescent protein. Nat Commun 7: 11046 10.1038/ncomms11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Carré A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SM, Brummelkamp TR, et al. 2015. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun 6: 10237 10.1038/ncomms10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. 2016. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci 113: E3501–E3508. 10.1073/pnas.1606731113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich T, Mayor-Ruiz C, Vega-Sendino M, Gomez C, Ortega S, Ruiz S, Fernandez-Capetillo O. 2017. A p53-dependent response limits the viability of mammalian haploid cells. Proc Natl Acad Sci 114: 9367–9372. 10.1073/pnas.1705133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Höning K, Ebert TS, Hornung V. 2016. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun 7: 12338 10.1038/ncomms12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Alaluf I, Swerdlin N, Perzov N, Danon T, Liron Y, et al. 2006. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat Methods 3: 525–531. 10.1038/nmeth892 [DOI] [PubMed] [Google Scholar]
- Sigal A, Danon T, Cohen A, Milo R, Geva-Zatorsky N, Lustig G, Liron Y, Alon U, Perzov N. 2007. Generation of a fluorescently labeled endogenous protein library in living human cells. Nat Protoc 2: 1515–1527. 10.1038/nprot.2007.197 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. 2016. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540: 144–149. 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh le A, Fraser SE. 2013. Enhancer and gene traps for molecular imaging and genetic analysis in zebrafish. Dev Growth Differ 55: 434–445. 10.1111/dgd.12055 [DOI] [PubMed] [Google Scholar]
- Trinh le A, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, et al. 2011. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev 25: 2306–2320. 10.1101/gad.174037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.