Abstract
The cellular and molecular mechanisms underlying the development and maintenance of dendritic spines are not fully understood. ADP-ribosylation factor 6 (ARF6) is a small GTPase known to regulate actin remodeling and membrane traffic. Here, we report involvement of ARF6 and exchange factor for ARF6 (EFA6A) in the regulation of spine development and maintenance. An active form of ARF6 promotes the formation of dendritic spines at the expense of filopodia. EFA6A promotes spine formation in an ARF6 activation-dependent manner. Knockdown of ARF6 and EFA6A by small interfering RNA decreases spine formation. Live imaging indicates that ARF6 knockdown decreases the conversion of filopodia to spines and the stability of early spines. The spine-promoting effect of ARF6 is partially blocked by Rac1. ARF6 and EFA6A protect mature spines from inactivity-induced destabilization. These results suggest that ARF6 and EFA6A may regulate the conversion of filopodia to spines and the stability of both early and mature spines.
Keywords: spine, filopodia, ARF6, EFA6A, Rac1, synapse
Introduction
Dendritic spines are small actin-rich protrusions that receive most of the excitatory synaptic inputs. These spines are associated with synapse formation, synaptic function and plasticity, as well as brain dysfunctions such as mental retardation (Hering and Sheng, 2001; Fiala et al., 2002; Nimchinsky et al., 2002; Yuste and Bonhoeffer, 2004; Carlisle and Kennedy, 2005; Ethell and Pasquale, 2005; Hayashi and Majewska, 2005; Matus, 2005; Segal, 2005). Spine morphogenesis is thought to involve several cellular events, including the early extension of filopodia from dendritic shafts, disappearance of filopodia, emergence of early spines and their maturation, and maintenance. However, the relationships between these cell biological events have yet to be clarified.
At the molecular level, a key regulator of spine morphogenesis is the Rac1 small GTPase. Rac1 promotes the formation and maturation of dendritic spines (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Tashiro and Yuste, 2004). The effect of Rac1 is assisted by diverse upstream and downstream molecules, including the EphB receptor kalirin-7 and PAK (Penzes et al., 2003). However, our understanding of the molecular organization of the Rac1 signaling pathway contributing to spine morphogenesis is poor.
ADP ribosylation factor (ARF) is a Ras-related family of small GTPases that regulate the actin cytoskeleton and membrane traffic (Chavrier and Goud, 1999). Among the six known ARF isoforms, ARF6 has received increasing attention because of its unique localization at the plasma membrane and the regulation of actin cytoskeleton, receptor endocytosis, and endosome recycling (Donaldson, 2003). ARF activity is regulated by diverse guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Among them, exchange factor for ARF6 (EFA6A) is an ARF6-specific GEF that is mainly expressed in brain (Perletti et al., 1997; Franco et al., 1999) and regulates the actin cytoskeleton and endosomal trafficking in non-neural cells (Franco et al., 1999; Derrien et al., 2002).
In the nervous system, ARF6 has been shown to suppress the branching of dendrites and axons in hippocampal neurons (Hernandez-Deviez et al., 2002; Hernandez-Deviez et al., 2004), promote neurotransmitter release at the Xenopus neuromuscular junction (Ashery et al., 1999), and stimulate clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol 4-phosphate 5-kinase (PIP5-kinase) type Iγ (Krauss et al., 2003). ARF6-dependent regulation of neurite branching suggests involvement in early neuronal morphogenesis. However, the following findings indicate the involvement of ARF6 in later morphogenesis, in particular, spine development and maintenance. ARF6 transcripts are highly expressed at later developmental stages (Suzuki et al., 2001). Recent proteomic analyses have identified various ARF GEFs and GAPs as components of the postsynaptic density (PSD) (Jordan et al., 2004; Peng et al., 2004). GIT1, a multidomain PSD protein that regulates spine morphogenesis (Zhang et al., 2003), contains an ARF GAP domain. These findings, along with the well known actin-regulatory effect of ARF6, suggest that this protein is involved in the regulation of dendritic spines.
In the present study, we investigated the involvement of ARF6 and EFA6A spine morphogenesis. Our data suggest that ARF6 and EFA6A are important regulators of spine development and maintenance.
Materials and Methods
Antibodies.
Rabbit polyclonal ARF6 (1654) and guinea pig polyclonal EFA6A (1626) antibodies were generated using glutathione S-transferase-ARF6 (amino acids 90–175; rat) and hexahistidine (H6)-EFA6A (full length; human) as immunogens. ARF6 antibody specificities were confirmed using ARF-Myc constructs and immunoblot analysis (data not shown). Guinea pig polyclonal enhanced green fluorescent protein (EGFP) (1432) antibody was generated using H6-EGFP (amino acids 1–240) as an immunogen. The following antibodies were purchased: PSD-95 (Affinity BioReagents, Golden, CO), synaptophysin, α-tubulin, and FLAG (Sigma, St. Louis, MO), and HA (hemagglutinin) and Myc rabbit polyclonal (Santa Cruz Biotechnology, Santa Cruz, CA).
Expression and small interfering RNA constructs.
pXS-ARF6-HA (wild-type and T27N; human) have been described previously (Peters et al., 1995). Full-length EFA6A (rat and human), ARF1, ARF3, and ARF5 (human), and ARF6 (rat) were amplified by PCR from human and rat brain cDNAs (BD Biosciences, San Jose, CA) and subcloned into pFLAG-CMV2 (human EFA6A; Sigma), GW1 (human and rat EFA6A; British Biotechnology, Oxford, UK), and pcDNA3.1/Myc-HisA (ARF1, ARF3, ARF5, ARF6; Invitrogen, San Diego, CA). Human EFA6A pleckstrin homology plus the C-terminal region (PH+C) (amino acids 349–645) and Rac1N17 were subcloned into pFLAG-CMV2. Full-length KIAA2011 was inserted into GW1. Point mutants (pXS-ARF6-HA T157A and pFLAG-EFA6A E242K) were generated using the Quickchange kit (Stratagene, La Jolla, CA). The following RNA interference sequences were used: 5′-ATTGGTGGCTGGCGAGTAT-3′ of rat EFA6A mRNA (GenBank accession number NM_134370) and 5′-AGCTGCACCGCATTATCAA-3′ of rat ARF6 mRNA (GenBank accession number NM_024152). Complementary small interfering RNA (siRNA) oligonucleotides were annealed and inserted into pSUPER.gfp/neo (Oligoengine Platform, Seattle, WA). Human EFA6A was used as a rescue construct because it differs from rat EFA6A at three nucleotides in the siRNA target sequence (5′-ACTGGTGGCTGGGGAGTAC-3′; different nucleotides are underlined). An ARF6 rescue construct that has two different nucleotides in the siRNA target sequence was generated using the Quickchange kit (5′-AGCTACATCGCATTATCAA-3′; changed nucleotides are underlined).
Hippocampal neuron culture, transfection, and immunocytochemistry.
Cultured hippocampal neurons were prepared from embryonic day 18 rat brain. Cultures were plated on coverslips coated with poly-l-lysine (1 mg/ml) and grown in Neurobasal medium supplemented with B27 (Invitrogen), 0.5 mm glutamine, and 12.5 μm glutamate in a 10% CO2 incubator. Transfection of expression and siRNA constructs was performed using the calcium phosphate kit (Invitrogen). Cultured neurons at 17 d in vitro (DIV) were treated with 2 μm tetrodotoxin (TTX) for 3 d. Hippocampal neurons were fixed in PBS containing 4% paraformaldehyde and 4% sucrose, permeabilized in PBS containing 0.2% Triton X-100, and incubated with primary antibodies, followed by cyanine 3 (Cy3)-, Cy5-, or FITC-conjugated secondary antibodies. Immunostaining was performed using mouse PSD-95 (1:500; Affinity BioReagents), mouse FLAG (2 μg/ml), rabbit HA (1 μg/ml), and guinea pig and rabbit EGFP (1:500) antibodies. Secondary antibodies used for immunostaining includes the following: FITC-conjugated goat anti-rabbit and guinea pig IgG (1:200; Jackson ImmunoResearch, West Grove, PA), Cy3-conjugated goat anti-mouse, Cy3-conjugated goat anti-mouse and rabbit IgG (1:500; Jackson ImmunoResearch), and Cy5-conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch).
Microscopy and time-lapse imaging.
Digital images of neurons were captured with an LSM 510 or PASCAL confocal microscope (Zeiss, Nussloch, Germany) using a C-Apochromat 63×, 1.20 numerical aperture (NA) water-immersion objective at room temperature. For time-lapse imaging, live cells in culture medium were mounted in a chamber at 37°C through which a continuous flow of 10% CO2 was passed (Zeiss). Images were acquired with an LSM510 confocal microscope (Zeiss) using a C-Apochromat 63×, 1.20 NA water-immersion objective and an additional electronic zooming factor of 3×, resulting in a nominal spatial resolution of 20 pixels per micrometer. The laser power was attenuated to 3% to avoid neuronal toxicity, and images were taken every 5 min for 2 h.
Image analysis and quantification.
Morphometric measurements of Z-series images were performed using MetaMorph image analysis software (Universal Imaging Corporation, West Chester, CA). Neurons were selected randomly, and aspiny interneurons were excluded from the analysis. One or two of the thickest dendrites in each neuron were chosen for analysis. To determine spine and filopodia density, spines were defined as dendritic protrusions of 0.4–3 μm length (with or without a head) and filopodia as dendritic protrusions of 3–10 μm in length. For the analysis of protrusion morphology, protrusions located in dendritic segments 100 μm distal from the first branching point of primary dendrites were selected, and the maximum length and width of each protrusion were measured manually. The density and dimensions of protrusions from a single dendrite were grouped and averaged; means from multiple individual neurons were averaged to obtain a population mean. To quantitate net filopodia length change in time-lapse imaging, we subtracted the length of filopodia at the beginning from that at the end of the time course. For the quantitation of mean filopodia length change, we measured individual length changes every 5 min over a 120 min period and averaged the values. Data were compared using one-way ANOVA with Tukey’s multiple comparison test or Student’s t test, and histograms depicting means ± SEM were generated.
Results
Expression patterns of EFA6A and ARF6 in brain
Because expression patterns of EFA6A and ARF6 proteins are yet to be extensively characterized (Hernandez-Deviez et al., 2002; Krauss et al., 2003), we initially determined their spatiotemporal expression patterns in rat brain using specific antibodies generated in this study (1626 for EFA6A and 1654 for ARF6). EFA6A was mainly expressed in brain (Fig. 1A), in agreement with the Northern blot patterns (Perletti et al., 1997; Derrien et al., 2002). The EFA6A antibody reacted with a major band of ∼140 kDa that matched the size of KIAA2011 proteins (lanes 3, 5) but not the smaller splice variant of EFA6A (rEFA6A and hEFA6A cDNA; lanes 1, 2). Both EFA6A and ARF6 were widely expressed in brain; expression levels of EFA6A were higher in the cortex and hippocampus, whereas ARF6 was higher in cortex and cerebellum (Fig. 1B), in accordance with previous in situ hybridization data (Suzuki et al., 2001, 2002; Sakagami et al., 2004). During postnatal rat brain development, EFA6A expression peaked at approximately postnatal day 7 (P7) followed by a slow decrease to the adult level, whereas ARF6 expression reached a peak at approximately P21 and was steadily maintained thereafter (Fig. 1C). In brain subcellular fractions, EFA6A and ARF6 were mainly detected in the crude synaptosome (P2) and synaptosomal membrane (LP1) fractions (Fig. 1D). EFA6A was highly enriched in PSD fractions, but ARF6 was not (Fig. 1E), indicating that the former is a novel PSD protein. Our results show that EFA6A and ARF6 have overlapping, yet partially distinct, expression patterns.
Figure 1.
Expression patterns of EFA6A and ARF6 proteins in rat brain. A, Tissue distribution of EFA6A. Two splice variants of EFA6A were used as controls (KIAA2011 and rEFA6A/hEFA6A; indicated by an arrow and arrowhead, respectively). B, Expression in different rat brain regions. Ctx, Cortex; Hc, hippocampus; Cb, cerebellum; R, other regions. C, Expression during rat brain development. wk, Weeks. D, Distribution in rat brain fractions. H, Homogenates; P1, crude nuclear fraction; P2, crude synaptosomes; S2, supernatant after P2 precipitation; S3, cytosol; P3, light membranes; LP1, synaptosomal membranes; LS2, synaptosomal cytosol; LP2, synaptic vesicle-enriched fraction; SynPhy, synaptophysin. E, Enrichment in PSD fractions.
ARF6 promotes spine formation
To determine the role of ARF6 in spine morphogenesis, we tested the effects of ARF6 overexpression (wild-type and mutants) in cultured neurons (DIV 9–11). The ARF6 mutants used included dominant-negative ARF6 T27N, which is defective in GTP binding, and fast cycling T157A, which is spontaneously more active than the wild-type protein (Santy, 2002). The latter mutant binds and releases GTP more quickly and induces phenotypes characteristic of ARF6 activation without disrupting the normal functions of the protein (Donaldson, 2003). When expressed in hippocampal neurons, ARF6 T157A showed the most prominent effect. ARF6 T157A increased the linear density of dendritic spines (defined as dendritic protrusions of 0.4–3 μm in length with or without a head), with a concomitant decrease in the density of filopodia (defined as dendritic protrusions of 3–10 μm in length) and hence no change in the density of total dendritic protrusions (spine plus filopodia) (Fig. 2A–D). ARF6 T157A also decreased the length of dendritic protrusions (Fig. 2E,H) and increased the localization of PSD-95 in dendritic spines (Fig. 2G). Protrusion width was not affected (Fig. 2F,I). These results suggest that ARF6 has an ability to promote the formation of dendritic spines at the expense of dendritic filopodia, which have been suggested as precursors of spines (Dailey and Smith, 1996; Ziv and Smith, 1996; Fiala et al., 1998; Maletic-Savatic et al., 1999; Marrs et al., 2001; Okabe et al., 2001).
Figure 2.

ARF6 promotes spine formation. A, Cultured hippocampal neurons were doubly transfected at DIV 9 with ARF6-HA [wild type (WT), T27N, and T157A] plus EGFP (farnesylated EGFP; used for an efficient visualization of membrane structures), or EGFP alone, and double labeled at DIV 11 for EGFP, HA, and PSD-95. Scale bar, 5 μm. B–D, Quantitation of the effect of ARF6 on the linear densities of spines, filopodia, and total protrusions (spines plus filopodia). The followings are statistical values: EGFP alone (protrusion density, 48.46 ± 2.24; spine density, 35.40 ± 1.85; filopodia density, 13.06 ± 1.06; length, 2.39 ± 0.08; width, 0.94 ± 0.02); ARF6 wild type (protrusion density, 47.18 ± 2.18; spine density, 36.72 ± 2.29; filopodia density, 10.46 ± 1.62; length, 2.15 ± 0.13; width, 1.02 ± 0.03); ARF6 T27N (protrusion density, 45.33 ± 1.85; spine density, 32.57 ± 1.69; filopodia density, 12.76 ± 1.15; length, 2.42 ± 0.08; width, 0.91 ± 0.02); and ARF6 T157A (protrusion density, 51.09 ± 1.82; spine density, 44.76 ± 1.63; filopodia density, 6.33 ± 0.87; length, 1.82 ± 0.05; width, 1.01 ± 0.02). E–I, Quantitation of the effect of ARF6 on the lengths and widths of protrusions and spine localization of PSD-95. E, F, Histogram of mean values. H, I, Cumulative frequency distribution. EGFP alone (32.17 ± 5.21); ARF6 wild type (47.26 ± 3.45); ARF6 T27N (29.93 ± 5.19); ARF6 T157A (51.94 ± 3.51). Data were collected from 400–1100 protrusions on 11–29 dendrites of 8–19 neurons for each group in the three independent experiments. **p < 0.01; ***p < 0.001 (one-way ANOVA).
EFA6A promotes spine formation through ARF6 activation
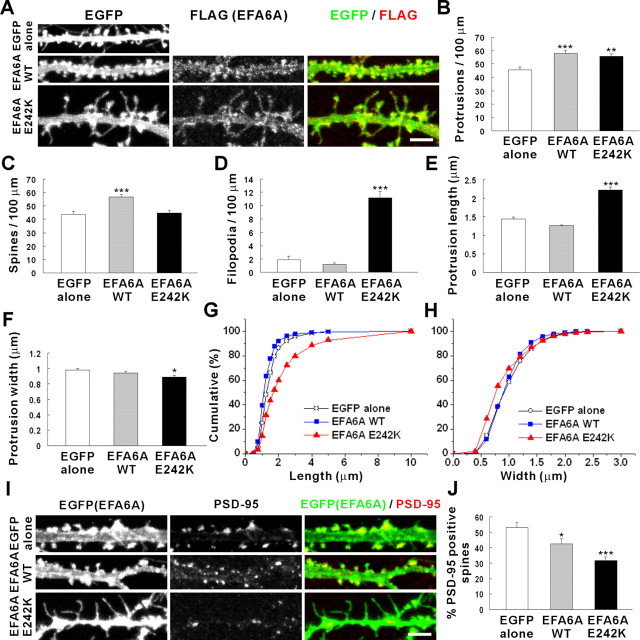
Next, we examined the effects of overexpression of EFA6A, an ARF6 exchange factor, in cultured hippocampal neurons (DIV 12–19). EFA6A increased the linear density of spines (Fig. 3A–D), consistent with its role in ARF6 activation. In contrast, an EFA6A variant with a point mutation in the Sec7 domain that eliminates exchange activity (EFA6A E242K) markedly increased the density of filopodia (Fig. 3A–D), probably because of the lack of the ARF6-dependent conversion of filopodia to spines. Consistently, EFA6A E242K caused an increase in the length of dendritic protrusions and decreases in protrusion width and PSD-95 localization in spines (Fig. 3E–J). These results suggest that EFA6A promotes spine formation in an ARF6-dependent manner.
Figure 3.
EFA6A promotes spine formation through ARF6 activation. A, Cultured hippocampal neurons were transfected at DIV 12 with FLAG-EFA6A [wild type (WT) or E242K] plus EGFP, or EGFP alone, and double labeled at DIV 19 for EGFP and FLAG. Scale bar, 5 μm. B–D, Quantitation of the effects of EFA6A (wild type and E242K) expression on the linear densities of spines, filopodia, and total protrusions. EGFP alone (protrusion density, 45.59 ± 2.09; spine density, 43.71 ± 2.02; filopodia density, 1.88 ± 0.51; length, 1.44 ± 0.05; width, 0.98 ± 0.02); EFA6A wild type (protrusion density, 58.05 ± 1.88; spine density, 56.86 ± 1.78; filopodia density, 1.19 ± 0.21; length, 1.26 ± 0.03; width, 0.94 ± 0.02); EFA6A E242K (protrusion density, 55.68 ± 1.99; spine density, 44.53 ± 2.18; filopodia density, 11.16 ± 0.98; length, 2.23 ± 0.08; width, 0.89 ± 0.02). E–H, Quantitation of the effects of EFA6A on the lengths and widths of dendritic protrusions. I, Cultured neurons expressing FLAG-EFA6A (wild type and E242K) were double labeled for EGFP or EFA6A and PSD-95. Scale bar, 5 μm. J, Quantitation of the effects of EFA6A expression on spine localization of PSD-95. EGFP alone (53.31 ± 2.94); EFA6A wild type (42.45 ± 3.47); EFA6A E242K (31.68 ± 2.4). Data were collected from 700–1900 protrusions on 17–38 dendrites of 13–23 neurons for each group in the three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (one-way ANOVA).
EFA6A promotes filopodia formation through an ARF6-independent mechanism
In addition to the Sec7 domain in the N-terminal half, EFA6A contains the pleckstrin homology (PH) domain in the middle of the protein, which may regulate the membrane association of EFA6A, and a C-terminal region, which modulates the actin cytoskeleton (Franco et al., 1999; Derrien et al., 2002). Overexpression of an EFA6A variant containing PH+C induced an increase in the density of spines, but not filopodia, with minimal effects on protrusion morphology (length and width) and PSD-95 localization in spines (Fig. 4A–J). This could be either caused by a direct induction of spine formation from dendrites or a consequence of initial filopodia proliferation followed by their conversion to spines by endogenous ARF6. In support of the latter possibility, overexpression EFA6A PH+C at earlier stages (DIV 6–9), in which expression levels of endogenous ARF6 are relatively low (Fig. 1C), caused an increase in the density of filopodia but not spines (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). These results suggest that EFA6A may promote filopodia formation through the ARF6-independent C-terminal region.
Figure 4.
The C-terminal (PH+C) region of EFA6A leads to an increase in spine density. A, Cultured neurons were transfected at DIV 12 with FLAG-EFA6A PH+C and EGFP, or EGFP alone, and labeled at DIV 19 for EGFP and FLAG. Scale bar, 5 μm. B–D, Quantitation of the effect of EFA6A PH+C expression on the linear density of spines, filopodia, and total protrusions. EGFP alone (protrusion density, 44.3 ± 1.19; spine density, 42.05 ± 1.24; filopodia density, 2.25 ± 0.69; length, 1.58 ± 0.05; width, 1.04 ± 0.02); EFA6A PH+C (protrusion density, 54.85 ± 1.40; spine density, 51.93 ± 1.35; filopodia density, 3.22 ± 0.34; length, 1.59 ± 0.03; width, 0.97 ± 0.02). E–H, Quantitation of the effect of EFA6A expression on the lengths and widths of protrusions. I, Cultured neurons expressing FLAG-EFA6A PH+C were double labeled for EGFP or EFA6A and PSD-95. Scale bar, 5 μm. J, Quantitation of the effect of EFA6A PH+C expression on spine localization of PSD-95. EGFP alone (53.31 ± 2.94); EFA6A PH+C (50.55 ± 2.36). Data were collected from 800–1500 protrusions on 20–27 dendrites of 16–20 neurons for each group in the three independent experiments. *p < 0.05; ***p < 0.001 (Student’s t test).
EFA6A and ARF6 are required for spine formation
The roles of EFA6A and ARF6 in spine morphogenesis were further explored by siRNA knockdown. EFA6A and ARF6 siRNA constructs markedly suppressed the levels of target proteins in both HEK293T cells and cultured hippocampal neurons (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Knockdown of ARF6 in cultured neurons (DIV 13–16) led to a decrease in spine density, with a concomitant increase in filopodia density (Fig. 5A–D), probably because of a reduced filopodia-to-spine conversion. Consistently, ARF6 siRNA caused an increase in protrusion length and decreases in both the width of protrusions and spine localization of PSD-95 (Fig. 5E–J). Knockdown of EFA6A also resulted in similar changes, although to a lesser extent in some parameters, compared with ARF6 knockdown (Fig. 5A–J). The effects of ARF6 and EFA6A knockdown were reversed by coexpression of siRNA-resistant rescue constructs (Fig. 5A–H) (supplemental Fig. 2A–C,E, available at www.jneurosci.org as supplemental material). These results suggest that ARF6 and EFA6A may be required for spine formation.
Figure 5.
Knockdown of EFA6A and ARF6 by siRNA results in a decrease in spine formation. A, Cultured neurons were transfected at DIV 13 singly with pSUPER.gfp/neo EFA6A (si-EFA6A), pSUPER.gfp/neo ARF6 (si-ARF6), or pSUPER.gfp/neo alone (si-vec), or doubly with pSUPER constructs plus rescue FLAG-EFA6A or ARF6-HA expression constructs (Rescue), were immunostained at DIV 16 with the antibodies indicated. Scale bar, 5 μm. B–D, Quantitation of the effects of siRNA knockdown of EFA6A and ARF6 on the linear densities of spines, filopodia, and total protrusions. si-vec (protrusion density, 45.51 ± 1.347; spine density, 40.49 ± 1.36; filopodia density, 5.03 ± 0.44; length, 1.91 ± 0.04; width, 1.21 ± 0.02); si-EFA6A (protrusion density, 37.73 ± 1.24; spine density, 25.27 ± 1.03; filopodia density, 12.45 ± 0.84; length, 2.66 ± 0.07; width, 1.07 ± 0.02); si-EFA6A + Rescue (protrusion density, 45.68 ± 1.46; spine density, 42.19 ± 1.49; filopodia density, 3.48 ± 0.48; length, 1.70 ± 0.04; width, 1.19 ± 0.02); si-ARF6 (protrusion density, 38.92 ± 1.16; spine density, 21.92 ± 0.94; filopodia density, 17.0 ± 0.69; length, 3.13 ± 0.06; width, 0.99 ± 0.01); si-ARF6 + Rescue (protrusion density, 46.06 ± 1.83; spine density, 39.84 ± 1.74; filopodia density, 6.23 ± 0.67; length, 2.03 ± 0.04; width, 1.21 ± 0.02). E–H, Quantitation of the effects of siRNA knockdown of EFA6A and ARF6 on the lengths and widths of protrusions. I, Cultured neurons transfected with si-EFA6A, si-ARF6, or si-vec were doubly labeled for EGFP and PSD-95. Scale bar, 5 μm. J, Quantitation of the effects of siRNA knockdown of EFA6A and ARF6 on the localization of PSD-95 in spines. si-vec (47.48 ± 1.99); si-EFA6A (28.42 ± 2.80); si-ARF6 (30.98 ± 3.46). Data were collected from 1200–1800 protrusions on 30–39 dendrites of 22–37 neurons for each group in the three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (one-way ANOVA).
Because EFA6A expression peaks at approximately P7, we also tested the effects of EFA6A knockdown at the early stages (DIV 6–9). Intriguingly, EFA6A knockdown did not induce any changes in protrusion density, although coexpression of EFA6A siRNA with EFA6A PH+C resulted in an increase in protrusion density (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). These results suggest that the C-terminal region of EFA6A may be sufficient to promote but not required for filopodia proliferation at the early stages.
ARF6 is required for the conversion of filopodia to spines and stabilization of early spines
The ARF6-dependent promotion of spine formation during development may be mediated by enhanced conversion of filopodia to spines and/or inhibition of spine destabilization. To address this issue, cultured neurons expressing ARF6 siRNA (DIV 10–12, a stage of active filopodia-to-spine conversion) were subjected to time-lapse imaging (Fig. 6A–C). We analyzed the fate of every filopodium (Fig. 6D) or spine (Fig. 6E) at the end of the 2 h recording session. ARF6 knockdown markedly decreased the percentage of filopodia converted to spines (Fig. 6D), indicating that ARF6 is required for filopodia-to-spine conversion. When spines were monitored, ARF6 knockdown increased the percentage of spines disappeared but did not affect that converted to filopodia (Fig. 6E), indicating that ARF6 is required for the stabilization of early spines, in addition to filopodia-to-spine conversion. Most of the disappeared spines (96%; n = 49 spines) in ARF6 siRNA-expressing neurons directly disappeared without, for instance, being converted to filopodia first before they disappear. Consistent with these results, the length of filopodia in ARF6 siRNA-expressing neurons (net change) remained unchanged but decreased in control neurons (Fig. 6F). The length of spines was not significantly changed by ARF6 siRNA (Fig. 6F). ARF6 siRNA did not affect protrusive motility of filopodia (Fig. 6G). Based on these findings, we suggest that ARF6 is required for the conversion of filopodia to spines and stabilization of early spines.
Figure 6.

ARF6 is required for the conversion of filopodia to spines and stabilization of early spines. A, Cultured neurons were transfected at DIV 10 with si-ARF6 or si-vec (control), and time-lapse images of living neurons were captured at DIV 12 for 2 h. Lowercase letters indicate examples of filopodia plotted in b and c. Arrowheads indicate that spines disappeared at the end of recording (120 min). Scale bar, 3 μm. B, C, Changes in the lengths of individual filopodia. The lengths of individual filopodia over the 2 h recording period (5 min interval) are plotted. Note the difference in the final filopodial length between the two groups at the end of the recording. D, Fate of filopodia during the 2 h recording period. Percentages of filopodia converted to spines, disappeared, or remained as filopodia are indicated. si-vec (filopodia to spines, 47.72 ± 8.02; filopodia to filopodia, 46.35 ± 9.75; filopodia disappeared, 5.94 ± 4.29, n = 4 neurons); si-ARF6 (filopodia to spines, 10.97 ± 4.75; filopodia to filopodia, 85.76 ± 6.42; filopodia disappeared, 3.27 ± 2.03; n = 4 neurons). E, Fate of spines during the 2 h recording period. Percentages of spines converted to filopodia, disappeared, or remained as spines are indicated. si-vec (spines to spines, 89.63 ± 3.45; spines to filopodia, 3.41 ± 2.09; spines disappeared, 6.97 ± 1.74; n = 4 neurons); si-ARF6 (spines to spines, 79.26 ± 3.90, p = 0.0023; spines to filopodia, 3.05 ± 1.07; spines disappeared, 17.69 ± 3.67; n = 4 neurons). F, Net changes in the lengths of protrusions during the 2 h recording. (n = 48 for filopodia and 301 for spines in control neurons; n = 84 for filopodia and 295 for spines in ARF6 siRNA-expressing neurons). si-vec (filopodia, −0.75 ± 0.19; spines, 0.01 ± 0.03, n = 48 for filopodia and 301 for spines); si-ARF6 (filopodia, −0.02 ± 0.12; spines, 0.10 ± 0.03, n = 84 for filopodia and 295 for spines). G, Quantitation of the protrusive motility of filopodia. Mean length changes in filopodia between two consecutive time points (5 min intervals) were measured. si-vec (0.32 ± 0.02, n = 48); si-ARF6 (0.34 ± 0.02, n = 84). Data were collected from four independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (Student’s t test).
The effect of ARF6 on spine development is partially blocked by a dominant-negative Rac1
Previous studies have shown that ARF6 signaling is mediated by Rac1 small GTPase to a certain extent (Donaldson, 2003). To determine whether the spine-promoting effect of ARF6 is mediated by Rac1, we cotransfected cultured neurons with ARF6 T157A and Rac1 N17, a dominant-negative mutant. Rac1 N17 partially blocked ARF6 T157A-induced changes in spine formation and protrusion morphology (length and width) (Fig. 7A–H). These results suggest that Rac1 is an important downstream effector of ARF6. The incomplete blockade of ARF6 T157A effects may be attributable to the presence of additional effectors downstream of ARF6.
Figure 7.
Effect of ARF6 on spine development is partly blocked by dominant-negative Rac1. A, Cultured neurons were transfected at DIV 9 with ARF6-HA T157A plus EGFP, ARF6-HA T157A plus EGFP plus FLAG-Rac1 N17, or EGFP alone, and double-labeled at DIV 11 for EGFP, HA, and FLAG. Scale bar, 5 μm. B–D, Quantitation of the Rac1 N17-dependent inhibition of the ARF6 T157A effects on the linear densities of spines, filopodia, and total protrusions. EGFP alone (protrusion density, 54.44 ± 2.19; spine density, 35.72 ± 2.30; filopodia density, 18.72 ± 1.02; length, 2.73 ± 0.10; width, 0.80 ± 0.02); ARF6 T157A (protrusion density, 55.95 ± 1.69; spine density, 48.77 ± 1.82; filopodia density, 7.18 ± 0.84; length, 1.85 ± 0.07; width, 1.00 ± 0.02); ARF6 T157A + Rac1 N17 (protrusion density, 64.89 ± 2.58; spine density, 44.15 ± 2.12; filopodia density, 20.74 ± 1.56; length, 2.60 ± 0.07; width, 0.89 ± 0.03). E–H, Quantitation of the Rac1 N17-dependent blockade of the ARF6 T157A effects on the lengths and widths of protrusions. Data were collected from 800–100 protrusions on 18–26 dendrites of 14–21 neurons for each group in the two independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (one-way ANOVA).
ARF6 and EFA6A protect mature spines from destabilization
Interestingly, both ARF6 and EFA6A are highly expressed at the late stages (Fig. 1C), suggesting that they may play an additional role in the stabilization of mature spines. TTX, which inhibits neuronal activity by blocking sodium channels, converts spines into long filopodia-like protrusions in cultured hippocampal neurons (Papa and Segal, 1996). We examined the effects of ARF6 and EFA6A on TTX-induced spine destabilization. TTX treatment resulted in increased length of dendritic protrusions (Fig. 8A–F), consistent with previous findings (Papa and Segal, 1996). However, neurons expressing ARF6 T157A or EFA6A (DIV 17–20) were resistant to TTX-induced elongation of spines, with complete protection seen in ARF6 T157A (Fig. 8A-F). These results suggest that ARF6 and EFA6A may be involved in the stabilization of mature spines at the later stages.
Figure 8.
ARF6 and EFA6A protect mature spines from destabilization. A, Cultured neurons were transfected at DIV 17 with FLAG-EFA6A plus EGFP, ARF6-HA plus EGFP, or EGFP alone, treated with TTX for 3 d, and labeled at DIV 20 for EGFP, HA, and FLAG. Scale bar, 5 μm. B, C, Quantitation of protection from TTX-induced spine elongation by EFA6A and ARF6. EGFP alone (1.78 ± 0.06); EGFP + TTX (2.25 ± 0.14); EFA6A (1.35 ± 0.03); EFA6A + TTX (1.51 ± 0.07); ARF6 T157A (1.56 ± 0.08); ARF6 T157A + TTX (1.53 ± 0.06). Data were collected from 400–2000 protrusions on 9–36 dendrites of 6–28 neurons for each group in the three independent experiments. *p < 0.05; **p < 0.01 (one-way ANOVA).
Discussion
Role of ARF6 in spine development and maintenance
Our data suggest that ARF6 is widely involved in distinct steps of spine development and maintenance. First, ARF6 promotes the conversion of dendritic filopodia to spines and stabilization of early spines. In support of this, overexpression of an active form of ARF6 (T157A) promotes spine formation even at an early stage (DIV 11), whereas siRNA knockdown of ARF6 suppresses it. Critically, our live imaging results demonstrate that ARF6 is required for the conversion of filopodia to spines and stabilization of early spines. Active ARF6 (T157A) also causes a decrease in protrusion length and increases in protrusion width and spine localization of PSD-95, all of which may reflect a morphological maturation accompanying filopodia-to-spine conversion. Conversely, siRNA knockdown of ARF6 results in increased protrusion length and decreased protrusion width and spine localization of PSD-95. Last, ARF6 protects mature spines from TTX-induced destabilization.
What are the molecular mechanisms underlying the effects of ARF6 on dendritic spines? Our results indicate that the effects of ARF6 T157A on spine development are partially blocked by dominant-negative Rac1, suggesting that Rac1 is an important downstream effecter of ARF6. Consistent with this, the effects of Rac1 that have been reported are similar to those of ARF6. Overexpression of constitutively active Rac1 in various neuronal preparations increases spine density (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Pilpel and Segal, 2004) and maturation (Tashiro and Yuste, 2004). Conversely, dominant-negative Rac1 reduces spine density (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Pilpel and Segal, 2004), maturation (Tashiro and Yuste, 2004), and stability (Tashiro and Yuste, 2004).
Rac1-dependent regulation of actin rearrangement and spine dimensions has been closely associated with the modulation of synaptic strength and plasticity (Matsuzaki et al., 2001; Yuste and Bonhoeffer, 2001; Hayashi and Majewska, 2005; Matus, 2005; Segal, 2005). Thus, an intriguing possibility is that ARF6 may be linked, through Rac1, to the regulation of synaptic strength and plasticity. In this context, it should be noted that ARF6 regulates membrane traffic, including receptor endocytosis and endosomal recycling, in addition to the actin cytoskeleton.
Another possible downstream effector of ARF6 is PIP5-kinase (Honda et al., 1999), which catalyzes the production of phosphatidylinositol (4,5)-bisphosphate, a major phosphoinositide in the plasma membrane involved in membrane traffic and actin rearrangements (Czech, 2003; Yin and Janmey, 2003). Alternatively, ARF6 may act on phospholipase D, which can also stimulate PIP5-kianse through the production of phosphatidic acid (Donaldson, 2003). Whether these molecules mediate the effects of ARF6 on dendritic spines remains to be determined.
Role of EFA6A in spine development and maintenance
Our data suggest the EFA6A promotes spine formation by acting as an exchange factor for ARF6. In support of this, overexpression of EFA6A promotes spine formation. In contrast, EFA6A E242K that lacks ARF6 activation causes an increase in filopodia density. Consistently, siRNA knockdown of EFA6A decreases spine density while increasing filopodia density at the same time. Of note, the effects of EFA6A siRNA knockdown were similar to those of ARF6 knockdown, suggesting that EFA6A may be a major upstream activator of ARF6.
However, it should be pointed out that some effects of EFA6A knockdown were slightly smaller than those of ARF6 knockdown. A possible explanation for this quantitative difference is that ARF6 GEFs other than EFA6A may function as upstream activators of ARF6. The EFA6 family of ARF6 GEFs, which is conserved in lower species, including Caenorhabditis elegans and Drosophila, has additional isoforms (EFA6B, EFA6C, and EFA6D) (Derrien et al., 2002). Northern blot results indicate that EFA6C is mainly expressed in brain, similar to EFA6A, whereas EFA6B shows a widespread tissue distribution (Derrien et al., 2002). EFA6C is detected in spines at the electron microscopic level and enriched in PSD fractions (Matsuya et al., 2005). Our results (Fig. 1B) and previous reports indicate that EFA6C is highly expressed in cerebellar Purkinje cells (Matsuya et al., 2005), in contrast to the preferential expression of EFA6A in forebrain areas (Suzuki et al., 2002) and widespread expression of ARF6 (Suzuki et al., 2001), suggesting that EFA6 family isoforms may differentially interact with ARF6 in brain regions. In addition to EFA6 family proteins, brain expresses other families of ARF GEFs, including ARF GEP100 (ARF6 specific) (Someya et al., 2001) and ARNO/GRP/cytohesin (ARF1 and ARF6 specific) (Nie et al., 2003). Msec7-1, a rat homolog of human cytohesin-1, has been implicated in the regulation of neurite branching and presynaptic function, respectively (Ashery et al., 1999; Hernandez-Deviez et al., 2002; Hernandez-Deviez et al., 2004). Thus, it remains to be determined whether additional EFA6 isoforms (EFA6B and EFA6C) and other ARF6 GEFs (cytohesins and ARF-GEP100) are also involved, through their ARF6 activation, in the regulation of spine development.
EFA6A contains two distinct functional domains: the ARF6-activating Sec7 domain and the C-terminal (PH+C) region. Our data indicate that the C-terminal region of EFA6A has an ability to induce filopodia formation at an early stage (DIV 6–9), independent of ARF6 activation (Fig. 4) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). However, siRNA knockdown of EFA6A at an early stage (DIV 6–9) did not significantly reduce the density of dendritic protrusions. These results suggest that the C-terminal region of EFA6A may be sufficient but not required for filopodia production. In this context, it should be noted that the EFA6A expression peaks at approximately P7, in which ARF6 expression is relatively low compared with the later stages. This suggests that early EFA6A may have additional functions other than ARF6 activation.
Dendritic filopodia as precursors of spines
Previous studies support the hypothesis that dendritic filopodia serve as precursors of spines (Dailey and Smith, 1996; Ziv and Smith, 1996; Fiala et al., 1998; Maletic-Savatic et al., 1999; Marrs et al., 2001; Okabe et al., 2001), although there are evidence against this hypothesis (Yuste and Bonhoeffer, 2004) and suggestions for alternative functions of filopodia such as involvement in synapse formation (Parnass et al., 2000; Portera-Cailliau et al., 2003). This model, however, can be supported by testing whether the blockade of filopodia-to-spine conversion reduces the final number of spines or, conversely, whether an enhanced production of filopodia leads to an increase in final spine number.
Indeed, our data indicate that the blockade of filopodia-to-spine conversion by ARF6 siRNA knockdown (DIV 13–16) results in a decrease in spine density with a concomitant increase in filopodia density. In addition, overexpression of the C-terminal region of EFA6A (DIV 12–19), which promotes filopodia proliferation at the ARF6-limiting early stages (DIV 6–9), leads to an increase in the final density of spines. Along with our live imaging data that directly demonstrate the involvement of ARF6 in filopodia-to-spine conversion, these results support the hypothesis that dendritic filopodia may function as precursors of spines.
Footnotes
This work was supported by the National Creative Research Initiative Program of the Korean Ministry of Science and Technology (E.K.) and Korea Health 21 Research and Development Project, Ministry of Health and Welfare, Republic of Korea Grant 02-PJ1-PG1-CH06-0001 (H.K.). S.C. is supported in part by the Bojeong Kim fellowship. We thank Dr. Julie G. Donaldson (National Institutes of Health) for pXS-ARF6-HA cDNAs (wild type and T27N) and the Kazusa DNA Research Institute for KIAA2011 cDNA.
References
- Ashery U, Koch H, Scheuss V, Brose N, Rettig J (1999). A presynaptic role for the ADP ribosylation factor (ARF)-specific GDP/GTP exchange factor msec7–1. Proc Natl Acad Sci USA 96:1094–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB (2005). Spine architecture and synaptic plasticity. Trends Neurosci 28:182–187. [DOI] [PubMed] [Google Scholar]
- Chavrier P, Goud B (1999). The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol 11:466–475. [DOI] [PubMed] [Google Scholar]
- Czech MP (2003). Dynamics of phosphoinositides in membrane retrieval and insertion. Annu Rev Physiol 65:791–815. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Smith SJ (1996). The dynamics of dendritic structure in developing hippocampal slices. J Neurosci 16:2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien V, Couillault C, Franco M, Martineau S, Montcourrier P, Houlgatte R, Chavrier P (2002). A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J Cell Sci 115:2867–2879. [DOI] [PubMed] [Google Scholar]
- Donaldson JG (2003). Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem 278:41573–41576. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB (2005). Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol 75:161–205. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM (1998). Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci 18:8900–8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM (2002). Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev 39:29–54. [DOI] [PubMed] [Google Scholar]
- Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, Chavrier P (1999). EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J 18:1480–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Majewska AK (2005). Dendritic spine geometry: functional implication and regulation. Neuron 46:529–532. [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M (2001). Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci 2:880–888. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Casanova JE, Wilson JM (2002). Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci 5:623–624. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Roth MG, Casanova JE, Wilson JM (2004). ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell 15:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y (1999). Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99:521–532. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA (2004). Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics 3:857–871. [DOI] [PubMed] [Google Scholar]
- Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V (2003). ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol 162:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN (1996). Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379:837–840. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K (1999). Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science 283:1923–1927. [DOI] [PubMed] [Google Scholar]
- Marrs GS, Green SH, Dailey ME (2001). Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat Neurosci 4:1006–1013. [DOI] [PubMed] [Google Scholar]
- Matsuya S, Sakagami H, Tohgo A, Owada Y, Shin HW, Takeshima H, Nakayama K, Kokubun S, Kondo H (2005). Cellular and subcellular localization of EFA6C, a third member of the EFA6 family, in adult mouse Purkinje cells. J Neurochem 93:674–685. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H (2001). Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci 4:1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A (2005). Growth of dendritic spines: a continuing story. Curr Opin Neurobiol 15:67–72. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L (2000). Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci 20:5329–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hirsch DS, Randazzo PA (2003). Arf and its many interactors. Curr Opin Cell Biol 15:396–404. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K (2002). Structure and function of dendritic spines. Annu Rev Physiol 64:313–353. [DOI] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H (2001). Spine formation and correlated assembly of presynaptic and postsynaptic molecules. J Neurosci 21:6105–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa M, Segal M (1996). Morphological plasticity in dendritic spines of cultured hippocampal neurons. Neuroscience 71:1005–1011. [DOI] [PubMed] [Google Scholar]
- Parnass Z, Tashiro A, Yuste R (2000). Analysis of spine morphological plasticity in developing hippocampal pyramidal neurons. Hippocampus 10:561–568. [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M (2004). Semi-quantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem 279:21003–21011. [DOI] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL (2003). Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron 37:263–274. [DOI] [PubMed] [Google Scholar]
- Perletti L, Talarico D, Trecca D, Ronchetti D, Fracchiolla NS, Maiolo AT, Neri A (1997). Identification of a novel gene, PSD, adjacent to NFKB2/lyt-10, which contains Sec7 and pleckstrin-homology domains. Genomics 46:251–259. [DOI] [PubMed] [Google Scholar]
- Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD (1995). Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol 128:1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Segal M (2004). Activation of PKC induces rapid morphological plasticity in dendrites of hippocampal neurons via Rac and Rho-dependent mechanisms. Eur J Neurosci 19:3151–3164. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Pan DT, Yuste R (2003). Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. J Neurosci 23:7129–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami H, Matsuya S, Nishimura H, Suzuki R, Kondo H (2004). Somatodendritic localization of the mRNA for EFA6A, a guanine nucleotide exchange protein for ARF6, in rat hippocampus and its involvement in dendritic formation. Eur J Neurosci 19:863–870. [DOI] [PubMed] [Google Scholar]
- Santy LC (2002). Characterization of a fast cycling ADP-ribosylation factor 6 mutant. J Biol Chem 277:40185–40188. [DOI] [PubMed] [Google Scholar]
- Segal M (2005). Dendritic spines and long-term plasticity. Nat Rev Neurosci 6:277–284. [DOI] [PubMed] [Google Scholar]
- Someya A, Sata M, Takeda K, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M (2001). ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc Natl Acad Sci USA 98:2413–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Owada Y, Suzuki R, Yoshimoto T, Kondo H (2001). Localization of mRNAs for six ARFs (ADP-ribosylation factors) in the brain of developing and adult rats and changes in the expression in the hypoglossal nucleus after its axotomy. Brain Res Mol Brain Res 88:124–134. [DOI] [PubMed] [Google Scholar]
- Suzuki I, Owada Y, Suzuki R, Yoshimoto T, Kondo H (2002). Localization of mRNAs for subfamily of guanine nucleotide-exchange proteins (GEP) for ARFs (ADP-ribosylation factors) in the brain of developing and mature rats under normal and postaxotomy conditions. Brain Res Mol Brain Res 98:41–50. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Yuste R (2004). Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol Cell Neurosci 26:429–440. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R (2000). Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex 10:927–938. [DOI] [PubMed] [Google Scholar]
- Yin HL, Janmey PA (2003). Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65:761–789. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2001). Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci 24:1071–1089. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2004). Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5:24–34. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Horwitz AF (2003). Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol 161:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ (1996). Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17:91–102. [DOI] [PubMed] [Google Scholar]