Abstract
Neural development involves the expression of ensembles of regulatory genes that control the coordinate and region-specific expression of a host of other genes, resulting in the unique structure, connectivity, and function of each brain region. Although the role of some specific genes in neural development has been studied in detail, we have no global view of the orchestration of spatial and temporal aspects of gene expression across multiple regions of the developing brain. To this end, we used transcriptional profiling to examine expression levels of 9955 genes in the hypothalamus, hippocampus, and frontal cortex across seven stages of postnatal development and up to four stages of prenatal development in individual male rats (six per group). The results reveal dramatic changes across development in >97% of the neurally expressed genes. They also uncover a surprising degree of regional differentiation occurring after birth and through the first 2 weeks of life. Cluster analysis identifies 20 clusters of transcripts enriched in genes related to particular functions, such as DNA metabolism, nuclear function, synaptic vesicle transport, myelination, and neuropeptide hormone activity. Thus, groups of genes with related functions change in the brain at specific times, possibly marking critical periods for each function. These findings can broadly serve as a backdrop for studying the role of individual genes in neural development. They also underscore the importance of early postnatal life in the rat, which corresponds to late gestation in the human, as a critical late phase of neural organization and differentiation, even in subcortical regions.
Keywords: brain development, microarray, cortex, hippocampus, hypothalamus, developmental disorders
Introduction
Neural development involves the unfolding of a highly orchestrated biological program of spatial and temporal gene expression to control the birth, trajectory, final location, and connectivity of neurons and circuits (Tessier-Lavigne and Goodman, 1996; Jessell and Sanes, 2000; Marin and Rubenstein, 2003). This structural elaboration determines all aspects of brain function. A wealth of evidence indicates that genetic and/or environmentally induced perturbations arising during brain development can lead to a range of pathologies in later life. This is not only true for developmental and childhood disorders such as autism (Rubenstein and Merzenich, 2003; Insel and Fernald, 2004), but for adult onset disorders such as schizophrenia that exhibit disorganization in the morphology and pattern of gene expression in key brain regions (Akbarian et al., 1993; Akil and Lewis, 1997). Even subtler disorders, such as major depression, are thought to result from genetic predisposition coupled with developmental events that lead to vulnerability to stress later in life (Caspi et al., 2003). An example of the combined genetic and developmental basis of affective disorders is the finding that the hippocampal volume of an individual can predict vulnerability to post-traumatic stress disorder in his identical twin who is exposed to trauma (Gilbertson et al., 2002).
Recently, advances in whole-genome expression analyses have allowed systematic studies examining global changes in gene expression associated with disease in humans (Mirnics et al., 2001; Evans et al., 2004). Interestingly, a broad unbiased survey in human postmortem tissue showed that the most striking change in major depression is in growth factor gene expression (Evans et al., 2004), consistent with candidate studies of specific growth factors (Dwivedi et al., 2003) and reinforcing the view that changes in neural development and plasticity play a role in the pathophysiology of these disorders.
Although human studies are critical in shedding light on the biological basis of neural disorders, they can have significant intrinsic limitations (Li et al., 2004). Thus, animal models are required to define the normal developmental trajectory of the brain, and to identify critical periods in organization that might represent points of great vulnerability to environmental perturbation. Here we use the rat as an animal model because of its broad use in neurobiological and behavioral research. We rely on gene expression profiling (Affymetrix GeneChips) to describe the unfolding of global gene expression throughout the development of three separate regions of the male rat brain each implicated in stress responsiveness and in cognitive and affective brain disorders: the hippocampus, hypothalamus, and frontal cortex.
Our results demonstrate dramatic changes in expression (both increases and decreases) over time within a given region, as well as a significant increase in differentiation between regions, especially after birth. Thus, the first 2 weeks of life appear critical in completing the neural differentiation in gene expression that eventually endows each brain region with its distinct tissue specificity.
The full dataset, plus raw data from all analyses presented are available at www.jneurosci.org as supplemental material.
Materials and Methods
Subjects
Sprague Dawley rats from Charles River (Wilmington, MA) were bred in-house, with date of conception determined by detection of sperm plugs. After successful mating, females were initially housed two per cage, and then housed singly on the 18th day of gestation. After birth, litters were culled to 12 (six males and six females where possible) on postnatal day 1 (P1). We define the day of birth as P0, which generally corresponds to embryonic day 22 (E22). Pups were weaned at P21, with males and females separated on P30. Animals were killed by decapitation, and the brains were immediately removed. Frontal cortex, hippocampus, and hypothalamus were rapidly dissected on ice, fast-frozen at –40°C, and stored at –80°C before processing. Residual brain tissue was collected from embryos for sex determination. For each developmental stage, brains were collected from three entire litters, with tissue from two males per litter selected for analysis based on the quality of dissection. In total, 11 developmental stages were analyzed: E16, E17, E18, and E20, as well as P1, P4, P7, P14, P21, P30, and P90. Because of technical difficulties in the accurate and reliable dissection of some brain regions from fresh embryonic tissue, we omitted analysis of E16 and E17 hypothalamus, plus hippocampus at all embryonic ages.
All experiments were conducted in accordance with the guidelines of the animal ethics committee at the University of Michigan following the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996).
Sex determination
To avoid the potential confound of sexual dimorphism and the effects of estrous state, tissue was only analyzed from male rats. Sex of postnatal animals was determined by anogenital distance and presence of a scrotal sac. Sex determination in embryos was achieved by multiplex PCR amplification of DNA extracted from residual brain tissue using primers specific to a 490 bp region intronic to the Y chromosomal Tspy gene (5′-GATGTGGTGAACCCTGTGCTA, 3′-CATTTCACATGTAAGCAGCTTTTA) plus primers to amplify a 182 bp amplicon intronic to the autosomal Agrp gene as a positive amplification control (5′-AACTCCTTAGGGAAAGGGATAAA, 3′-ACCTGCGTTCGTAAGGGAGTA).
GeneChip hybridization
We used a total of 162 Affymetrix (Santa Clara, CA) RAE230A Gene-Chips to analyze cRNA derived from frontal cortex, hypothalamus, and hippocampus from 11, 9, and 7 different developmental time points, respectively. For each combination of time point and developmental stage, cRNA derived from six animals was individually hybridized to six different arrays. No pooling of samples was necessary.
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the instructions of the manufacturer, followed by an additional clean-up step using RNeasy RNA purification columns (Qiagen, Valencia, CA). RNA quality and concentration were both determined by absorbance at 260 and 280 nm and by analysis using an Agilent (Palo Alto, CA) bioanalyzer. First- and second-strand cDNA synthesis were performed as detailed in the Affymetrix Expression Analysis Technical Manual, version 3, using 6 μg of total RNA. In vitro transcription (IVT) of cDNA to biotinylated cRNA was performed with the Ambion (Austin, TX) MEGAscript T7 High Yield Transcription kit. Products of second-strand cDNA synthesis and IVT were purified with the Affymetrix GeneChip Sample Cleanup Module. IVT products were prepared for hybridization to Affymetrix RAE230A GeneChips per Affymetrix instructions. Arrays were hybridized for 18 h at 45°C, washed, and stained on an Affymetrix fluidics station using the standard EukGE-WS2v4_450 protocol and scanned with an Affymetrix GeneChip scanner.
GeneChip data description and normalization
Design of the Affymetrix RAE230A GeneChip was originally based on information within the UniGene build 99, dating from June 2002. To account for recent advances in rat genomics, GeneChip signal intensity data were interpreted by use of a custom chip description file (cdf) (filename RN230A_RN_UG_4) based on UniGene build 139 (released January 2005). Design of the custom cdf will be described in detail previously (Dai et al., 2005). Briefly, the custom cdf was generated by individually analyzing the sequence of every probe within each probe set on the RAE230A GeneChip and reassigning individual probes to new probe sets or removing probes entirely from analysis, to ensure that all probes within a probe set detect sequences within a single UniGene cluster from build 139. Custom cdf files are available at http://brainarray.mhri.med.umich.edu/brainarray/. Using the UniGene custom cdf file, a probe set is defined as all probes within a specific UniGene cluster, which serves as the probe set identification name. In contrast to the default Affymetrix cdf that contains 15,866 probe sets, the custom cdf file contains 9955 probe sets.
Cell intensity files from the Affymetrix GeneChip arrays were normalized by the RMA (robust multiarray average) algorithm (Irizarry et al., 2003) available at www.bioconductor.org. The presence or absence of gene expression was determined by the ratio of signals from perfect match versus mismatch probes on the GeneChips. Any gene that was not detected as being present in at least four of the six biological replicates, from at least one combination of brain region and developmental stage, was removed from additional analysis. Using these criteria, 6653 of the 9955 genes were expressed at detectable levels. All additional statistical analyses were performed on log2 transformed data using Partek Pro 6.0 (Partek, St. Charles, MO).
Statistical analyses of gene expression profiles
Principal components analysis. Principal components analysis (PCA) was performed on data from all 162 GeneChips after exclusion of probes that were not detectably expressed at any time point or brain region. Data were analyzed using the correlation method, which adjusts the mean of the data to zero and the SD to 1. This is a conservative method and is appropriate for variables that differ in their variance.
Hierarchical clustering. Unsupervised hierarchical clustering was performed to determine the relative similarities of tissues derived from different developmental stages and brain regions. Analyses were performed using data from the same probe sets as those described for PCA. Before clustering, mean expression levels were determined for each probe set across the six biological replicates of each combination of specific developmental stage and brain region. Clustering was performed on the log2 values of these means. Interpoint distances were calculated using the coefficient of shape difference, which is a function of the average Euclidean distance between samples but discounts additive differences; thus, it is appropriate for analyzing changes in the ratios of expression, independent of absolute expression levels. Distance between clusters was determined from the average distance between all pairs of objects in the two different clusters. The topology of the dendrogram was highly robust and varied little based on changes in the methodology for calculating relationships between samples.
Partitional clustering of gene expression profiles. Partitional clustering was used to subdivide probe sets into groups based on similarities in expression profiles between developmental stages and brain regions. Probe sets were filtered to exclude genes that were either undetected in all samples or failed to differ significantly between either brain regions of developmental stages. Furthermore, to focus on only the genes with the most informative expression profiles, genes were only included if they showed at least a twofold difference in expression between any two samples, reducing the data to 3159 probe sets. Similarities between expression profiles from different probe sets were determined using the coefficient of shape applied to log2-transformed mean expression values from each combination of brain region and developmental stage, as described for our hierarchical cluster analyses. To determine the optimal number of clusters, K means clustering was applied to every number of clusters from 3 to 50, and the Davies Bouldin measure was calculated for each iteration. This measure reflects cluster validity by determining the compactness of each cluster divided by the distance between clusters, normalized by the total number of clusters. The optimum number of clusters was thus determined at 26 clusters. Six clusters were each composed of a single gene (Rn.37881, hemoglobin, ϵ 1; Rn.1404, transthyretin; Rn.9864, Forkhead box G1; Rn.11400, prostaglandin D2 synthase; Rn.35223, Src-like adaptor; Rn.45546, myeloid ecotropic viral integration site 1 homolog 2). Gene expression profiles for the remaining 20 clusters were visualized using GeneSpring 6.0 (Silicon Genetics, Redwood City, CA). Finally, gene ontology (GO) analyses used the “function mapper” tool available at http://brainarray.mhri.med.umich.edu/brainarray/, which employs a hypergeometric distribution based on the number of genes within a gene list and within a GeneChip to calculate significance levels for enrichment of specific ontological terms.
Real-time PCR confirmation of microarray expression data.
Real-time (RT)-PCR was used to validate expression profiles of 33 genes, all of which had been implicated previously in susceptibility to either schizophrenia or bipolar disorder. Selection of genes for confirmation was independent of expression profile determined by microarray. The list of genes for analysis was generated by mining the Online Mendelian Inheritance in Man (OMIM) database, with seven additional genes identified from the literature. Primers specific to each gene of interest were designed to generate amplicons of 70–110 bp using Eprimer3 software within the Jemboss bioinformatics suite (Carver and Bleasby, 2003). To minimize the impact of any contamination with genomic DNA, most primers were designed to flank intronic sequences. RT-PCR primer sequences are listed in Table S1 (available at www.jneurosci.org as supplemental material).
RT-PCR was performed on a subset of samples used in microarray analysis. Between three and six biological replicates were analyzed from all samples of frontal cortex and hippocampus between P1 and P30. Template cDNA was synthesized from 1 μg of total RNA (as used for array experiments) using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and quantified using the Quant-IT PicoGreen dsDNA kit (Invitrogen, Eugene, OR). RT-PCR was performed using template synthesized as above and the iQ SYBR green Supermix qRT PCR kit (Bio-Rad) with each sample represented once. Samples from the same brain region samples were always amplified on the same PCR plate for each gene being assayed. PCRs were amplified using an iCycler RT-PCR system (Bio-Rad) for 40 cycles using parameters of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. Before performing PCRs, each primer pair was tested using a fivefold dilution series of pooled cDNA template to ensure that the efficiency of amplifications was linear and that the primers specifically produced a single amplified product. Cycle threshold (CT) was defined as the cycle required to reach a fluorescence value of 10 times the mean plus SD of the first 10 cycles. Melt curve analysis was also performed for each reaction to ensure that a single product contributed to the fluorescent signal. We required a primary peak to comprise at least 90% of the total area of all peaks.
Given the major expression changes reported here for all detectable genes, no housekeeping genes could be used to normalize expression data. Therefore, CTs for each gene were normalized individually to the median CT value obtained across all 33 genes analyzed by RT-PCR. After exclusion of outliers, correlations were determined between RT-PCR and microarray data separately for each brain region using Pearson's product moment correlation coefficient.
Results
Profiling rat brain development
We analyzed changes in the global patterns of gene expression across seven stages of postnatal development in three regions of the male rat brain: frontal cortex, hypothalamus, and hippocampus. In addition to P1, P4, P7, P14, P21, P30, and P90, analyses were extended back to E16, E17, E18, and E20 for frontal cortex, and E18 and E20 (1–2 d before birth) for the hypothalamus. For each brain region and developmental stage, six biological replicates were analyzed independently on Affymetrix RAE230A GeneChip arrays. To account for recent advances in both annotation and accuracy of rat genome bioinformatics, GeneChip data were interpreted using a custom cdf, which groups individual probes from the GeneChip into newly defined probe sets based on a very recent version of UniGene (build 139, January 2005). This approach apportioned all probes on the RAE230A array into a total of 9955 probe sets, each corresponding to a single cluster of sequences within UniGene build 139. Complete gene expression data are provided in Table S2 (available at www.jneurosci.org as supplemental material).
The presence or absence of gene expression was determined by comparing signal intensity between probes that perfectly match their target sequences against probes containing mismatches. If expression was detected in at least 4 of 6 replicates for any combination of brain region and developmental stage, it was considered as being detectably expressed. From the 9955 probe sets defined from the GeneChip data, significant levels of expression were detected for 6653 probe sets (66.8%). To identify genes for which expression differed significantly between either different brain regions or developmental stages, we applied two-way ANOVA to the 6653 probe sets. We set a significance threshold of p < 0.05, after false discovery rate correction (Benjamini and Hochberg, 1995). This correction means that 5% of all significantly different genes will be false positives, in contrast to uncorrected significance thresholds in which 5% of all genes analyzed would be false positive. A total of 5844 genes (87.8%) showed significant differences in expression between brain regions, with 6470 genes (97.2%) changing significantly over time. Remarkably, of the 6653 probe sets for which gene expression was detectable, only 33 (0.5%) failed to reach statistical significance for differences between either different brain regions or different developmental stages. Indeed, the majority of probe sets detected highly significant and large changes in expression between either time points or brain regions (median significance level across all 6653 probe sets was p = 10–28; median fold change was 1.9-fold) (Fig. 1).
Figure 1.
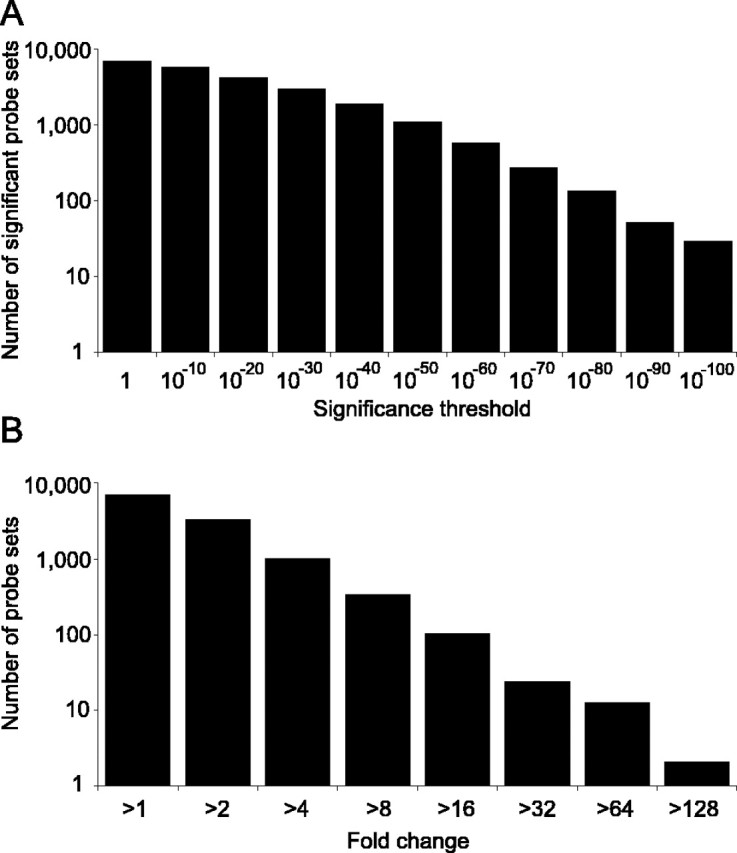
Distribution of significance levels and fold changes. Gene expression was detectably present for 6653 probe sets from the 9955 probe sets on the Affymetrix GeneChip. Significance levels for these 6653 probe sets after Benjamini–Hochberg false discovery rate correction are shown, based on the lesser of the two significance levels determined for differences between either developmental stage or brain region (A), as is the distribution of maximum fold change between any two combinations of brain region and time point (B).
Global comparison between samples
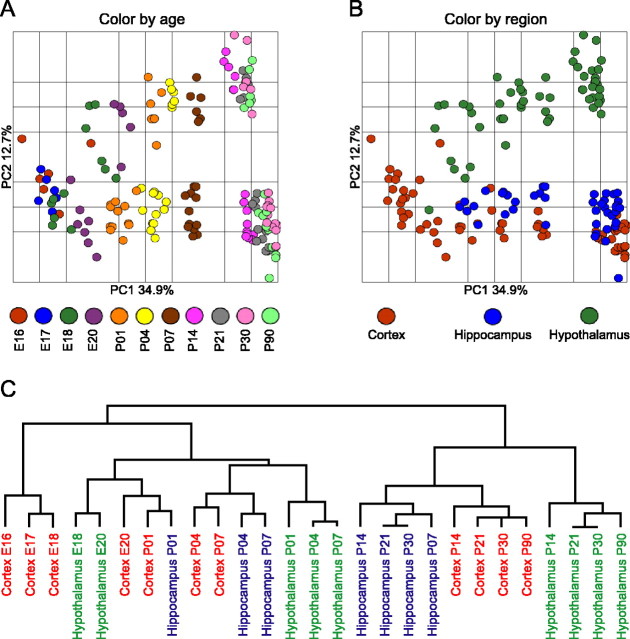
PCA is a data reduction technique that apportions the major components of variance within a dataset into a limited number of dimensions, thus facilitating visualization of the global similarities and differences between samples. The dimension that accounts for the single greatest portion of the total variance is termed principal component 1 (PC1). Application of PCA to our data clearly shows that the single greatest component of the variance (PC1) corresponds to the developmental stage of the brain (Fig. 2A). Inspection of the distribution of developmental stages across PC1 reveals dramatic changes in overall gene expression from E16 continuing through to P14, 2 weeks after birth. In contrast, between P14 and adulthood, additional changes in gene expression are relatively minor, as demonstrated by the tight clustering of all samples from P14 to P90 along PC1. In addition to the major impact of developmental stage on gene expression profiles, differences between the three brain regions are clearly revealed by inspection along PC 2 (Fig. 2B). Hippocampus and frontal cortex show relatively similar expression profiles, with hypothalamus highly diverged from both brain regions. Combining the distribution of variance across principal components 1 and 2 (Fig. 2A,B) shows that divergence in expression profiles between all three brain regions increases with age, with most of this region-specific differentiation being established by 2 weeks postpartum.
Figure 2.
Relationships between developmental ages and brain regions. PCA of gene expression profiles from each of the 162 GeneChips shows the major component of the variance (PC1) to separate samples by developmental age (A), with PC2 accounting for differences in expression between brain regions (B). PC1 and PC2 account for 34.9 and 12.7% of the total variance in the data, respectively. Hierarchical clustering (C) similarly details relationships between all samples. The vertical distance connecting two samples with in the dendrogram reflects similarity between the samples; for example, the two samples with the greatest similarity to each other are the hypothalamus at postnatal days 21 and 30.
Relationships between brain regions and time points are further clarified by hierarchical clustering of samples (Fig. 2C), which shows a dramatic difference between the brain before and after P14. Indeed, expression profiles of the hypothalamus at P7 appear more closely related to those of embryonic cortex than to the hypothalamus at P14. This clustering indicates that, after P14, samples sharing the greatest similarity are from within the same brain regions as opposed to the same developmental stages. In contrast, before P14, samples tend to cluster by a combination of both brain region and developmental age. Interestingly, both PCA and hierarchical analysis indicate that development of the hypothalamus occurs in advance of both frontal cortex and hippocampus, consistent with previous observations that medial regions of the telencephalon and diencephalon mature earlier than more lateral structures (for review, see Rice and Barone, 2000). For example, alignment of samples along PC1 (Fig. 2A) shows hypothalamus at E18 to be more similar to cortex and hippocampus at E20 than at E18. Similarly, hypothalamus at P1 more closely resembles cortex and hippocampus at P4. These trends are further confirmed by cluster analysis (Fig. 2C).
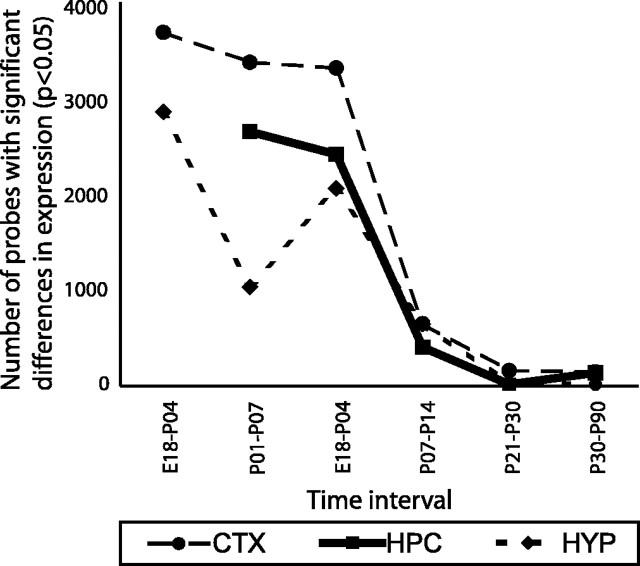
The dramatic reduction in the rate of change in gene expression profiles after P14 is further illustrated by analysis of the number of genes that differ significantly between pairs of developmental time points (Fig. 3). Comparison of brains from animals separated by 6–8 d in age generally shows 2000–4000 genes being significantly differentially expressed between time points for each brain region, up to and including P14. However, after P14, the rate of change in expression slows dramatically, with a maximum of 649 genes differing between P14 and P21 and only 147 changing over the 60 d period from P30 to P90. Although there is an apparent reduction in the rate of change in the hypothalamus between P1 and P7 (Fig. 3), extending PCA to incorporate principal components 3 and 4 indicates that variation among P1 hypothalamic samples is unusually high (data not shown), perhaps as a result of inconsistencies during dissection. Therefore, the apparent reduction in the rate of gene expression change at that point should be interpreted cautiously.
Figure 3.
Number of probe sets that differ between developmental stages. Data were analyzed for the number of significant differences in gene expression levels between pairs of time points. Given the unequal distribution of data collection over developmental time, developmental stages separated by 6–8 d were compared for ages less than P14. Probe sets for which expression was not detectable were excluded. The remaining 6653 probe sets were analyzed by one-way ANOVA for significant differences in expression, with a Benjamini–Hochberg false discovery rate (FDR) correction applied post hoc (Benjamini and Hochberg, 1995). The number of genes that were significantly different between time points (p < 0.05) is presented. CTX, Cortex; HPC, hippocampus; HYP, hypothalamus.
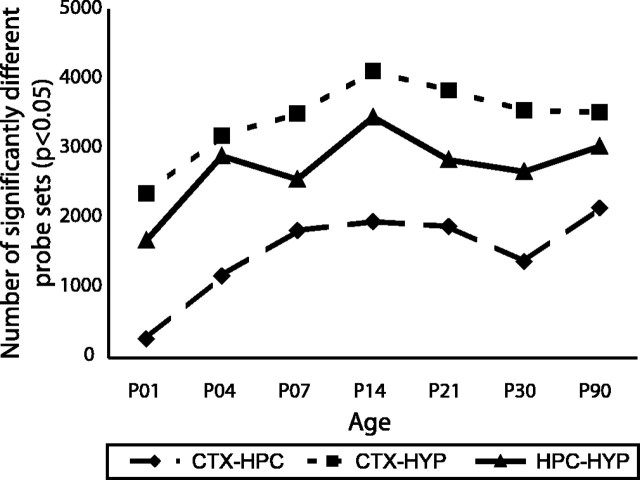
As indicated by principal components analysis (Fig. 2), divergence between brain regions increases as a function of maturity. A similar picture emerges from a statistical comparison of the number of probe sets that differ significantly between pairs of brain regions (Fig. 4). Pairwise comparisons between brain regions again show greatest similarity in expression profiles between the frontal cortex and the hippocampus, with greatest divergence between the frontal cortex and hypothalamus. This rank order of pairwise similarities is maintained throughout the entire postnatal period. Remarkably, very few differences in expression are detected between hippocampal and cortical samples from P1 neonates, with only ∼300 probe sets showing significant differences between these samples (p < 0.05). This compares with ∼2000 probe sets that differ in expression between the adult hippocampus versus frontal cortex.
Figure 4.
Number of probe sets that differ between brain regions. Probe sets that differed significantly between brain regions at each developmental stage were identified by one-way ANOVA (p < 0.05 following Benjamini–Hochberg false discovery rate correction).
Global analysis of gene expression profiles
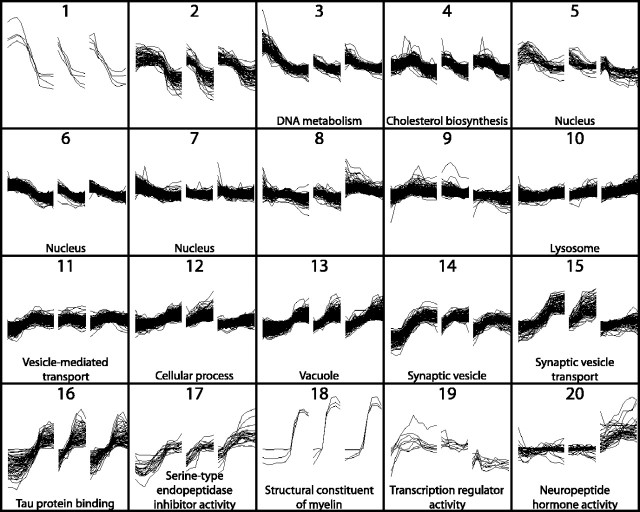
One of the major challenges in current genetics is to assign functionality to the host of relatively uncharacterized genes identified through genome sequencing projects. Although simplistic, it may be assumed that some genes that share similarities in their expression profiles may share similar or related functions. Partitional clustering was therefore applied to group genes into clusters based on similarity of expression profiles (see Materials and Methods). Probe sets were clustered into 26 groups, 6 of which contained single genes. Expression patterns of genes within the remaining 20 clusters are shown in Figure 5. Gene ontology analyses revealed significant enrichment (p < 0.05) for genes with common functions for 16 of these 20 clusters. Identities of all genes within these 26 clusters are available in supplemental Table S3 (available at www.jneurosci.org as supplemental material).
Figure 5.
Partitional clustering of gene expression profiles. Partitional clustering of 3159 genes (showing significant variation over time or between brain regions, plus a minimum twofold difference in expression between samples) generated 20 clusters containing more than one gene. Expression profiles of each cluster are shown as the ratio of signal intensity to the median signal intensity across all samples. The y-axis indicates fold change on a log scale from 0.01 to 100. Changes in expression with increasing age are shown from left to right for frontal cortex (left), hippocampus (center), and hypothalamus (right). Time points are shown with equidistant spacing, despite marked differences in the intervals between consecutive time points. GO analysis of transcripts within each cluster found significant enrichment for specific GO terms for 16 of 20 clusters (p < 0.05 after false discovery rate correction). The most significantly enriched GO terms are shown.
RT-PCR validation of microarray expression data
We used RT-PCR to validate our microarray data. Given the evidence that perturbations during brain development can predispose to pathologies in later life, we focused on genes that had been implicated previously in susceptibility to either schizophrenia or bipolar disorder. Using a combination of literature searches and inspection of the OMIM database, 55 genes were identified that had been implicated in disease and that had corresponding rat homologs present on our microarray. Expression profiles of these genes are shown in Figure 6. Thirty-three of these genes were further analyzed by RT-PCR. Correlations between results obtained by RT-PCR and those obtained by microarray are summarized in Table 1. Overall, expression patterns of genes that had been shown by microarray to vary by more than twofold during development were well replicated by RT-PCR (mean correlation coefficient between platforms, r = 0.60), whereas genes previously shown to vary by lesser amounts generally replicated poorly (mean correlation, r = 0.16). Where a lack of correlation was observed between platforms, this was generally attributable to high levels of sample-to-sample variation within the RT-PCR data (data not shown) preventing the more subtle changes in expression from being reliably detected.
Figure 6.
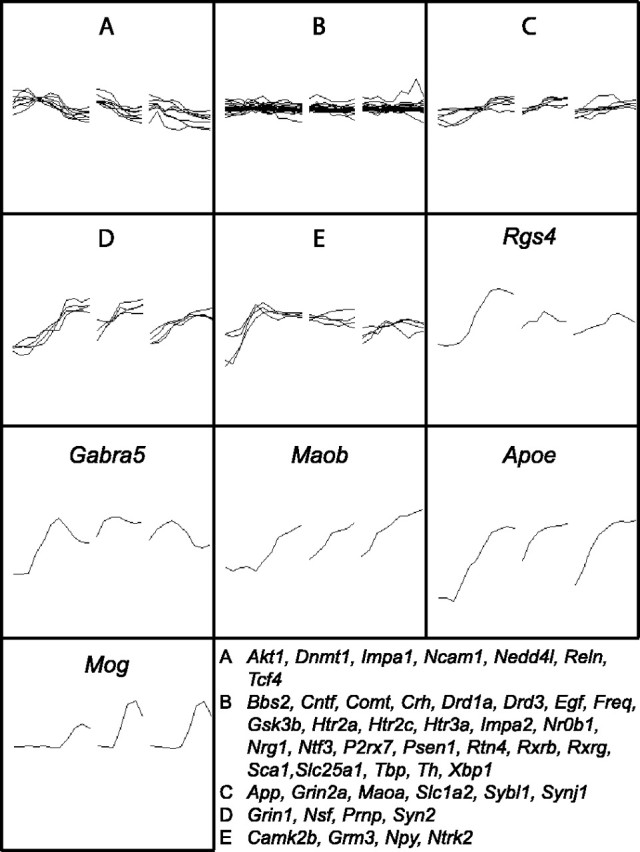
Expression profiles of susceptibility loci for schizophrenia and bipolar disorder. Inspection of the OMIM database combined with literature searches identified 127 genes that have been implicated in susceptibility to either schizophrenia or bipolar disorder. Fifty-five of these loci had identifiable rat homologs that were detected as being expressed in this study. To facilitate presentation, partitional clustering was applied to these 55 genes resulting in the generation of 10 clusters, expression profiles of which are shown. A–E, Five clusters were composed of more than one gene. The loci within each of these five clusters are listed. Rgs4, Regulator of G-protein signalling 4; Gabra4, GABAA receptor α 4; Maob, monoamine oxidase B; Apoe, apolipoprotein E; Mog, myelin oligodendrocyte glycoprotein.
Table 1.
Comparison of expression data determined by microarray and RT-PCR
|
|
Frontal cortex |
Hippocampus |
||||
|---|---|---|---|---|---|---|
| Gene |
r
|
Fold |
r
|
Fold |
||
| Adcy9 | 0.60** | (3.4) | 0.85** | (4.6) | ||
| Akt1 | 0.77** | (2.7) | 0.41* | (2.1) | ||
| Apoe | 0.81** | (6.6) | 0.83** | (4.9) | ||
| App | -0.17 | (1.8) | 0.29 | (1.5) | ||
| Bbs2 | -0.19 | (1.1) | 0.03 | (1.1) | ||
| Camk2b | 0.18 | (1.4) | 0.38* | (1.7) | ||
| Comt | -0.08 | (1.4) | -0.09 | (1.6) | ||
| Drd1a | -0.38 | (1.3) | 0.08 | (1.1) | ||
| Freq | -0.26 | (1.1) | 0.37* | (1.2) | ||
| Gria3 | 0.61** | (2.7) | 0.46** | (1.5) | ||
| Grin2a | 0.73** | (1.4) | 0.01 | (1.5) | ||
| Gsk3b | 0.33 | (1.3) | 0.46* | (1.2) | ||
| Htr2a | 0.13 | (1.1) | -0.10 | (1.1) | ||
| Htr2c | 0.51** | (1.2) | -0.11 | (1.2) | ||
| Impa2 | -0.17 | (1.3) | -0.03 | (1.1) | ||
| Maoa | 0.27 | (1.7) | -0.16 | (1.6) | ||
| Maob | 0.74** | (6.7) | 0.47** | (4.2) | ||
| Nedd41 | 0.02 | (1.3) | -0.10 | (1.6) | ||
| Npy | 0.07 | (2.1) | -0.17 | (1.6) | ||
| Nsf | 0.04 | (4.3) | 0.73** | (4.5) | ||
| Prnp | 0.59** | (2.7) | 0.64** | (1.9) | ||
| Psen1 | 0.27 | (1.2) | 0.53** | (1.2) | ||
| Reln | 0.66** | (1.9) | 0.82** | (3.5) | ||
| Rgs4 | 0.78** | (8.0) | 0.25 | (2.1) | ||
| Rtn4 | 0.11 | (1.5) | 0.16 | (1.2) | ||
| Slc1a2 | 0.71** | (1.9) | 0.75** | (2.0) | ||
| Sybl1 | 0.38* | (1.6) | 0.65** | (2.0) | ||
| Syn2 | 0.66** | (8.0) | 0.63** | (6.0) | ||
| Synj1 | 0.42* | (1.7) | 0.62** | (1.8) | ||
| Tbp | 0.10 | (1.3) | 0.29 | (1.2) | ||
| Tcf4 | 0.01 | (1.9) | 0.40* | (1.9) | ||
| Th | -0.08 | (1.1) | 0.09 | (1.1) | ||
|
Xbp1
|
-0.04 |
(1.4) |
-0.06 |
(1.5) |
||
Correlation coefficients between data generated by RT-PCR and by microarray are presented separately for each of the 33 genes analyzed by RT-PCR. Maximum fold changes over developmental time, as determined by microarray analyses, are shown in parentheses. Significant correlations: *p < 0.05 and **p < 0.01. Adcy9, Adenylate cyclase 9; Akt1, thymoma viral proto-oncogene 1; Apoe, apolipoprotein E; App, amyloid β precursor protein; Bbs2, Bardet—Biedl syndrome 2; Camk2b, calcium/calmodulin-dependent protein kinase II β; Comt, catechol-O-methyl-transferase; Drd1a, dopamine receptor 1A; Freq, frequenin homolog; Gria3, glutamate receptor, ionotropic, AMPA3; Grin2a, glutamate receptor, ionotropic, NMDA 2A; Gsk3b, glycogen synthase kinase 3β; Htr2a, 5-hydroxytryptamine receptor 2A; Htr2c, 5-hydroxytryptamine receptor 2C; Impa2, inositol(myo)-1(or 4)-monophosphatase 2; Maoa, monoamine oxidase A; Maob, monoamine oxidase B; Nedd41, neural precursor cell expressed, developmentally downregulated 4-like; Npy, neuropeptide Y; Nsf, N-ethylmaleimide-sensitive factor protein; Prnp, prion protein; Psen1, presenilin 1; Reln1, reelin; Rgs4, regulator of G-protein signalling 4; Rtn4, reticulon 4; Slc1a2, solute carrier family 1 (glial high-affinity glutamate transporter), member 2; Sybl1, synaptobrevin-like 1; Syn2, synapsin II; Synj1, synaptojanin 1; Tbp, TATA-box-binding protein; Tcf4, transcription factor 4; Th, tyrosine hydroxylase; Xbp1, X-box-binding protein 1.
Discussion
Transcriptional profiling of the hypothalamus, hippocampus, and frontal cortex across development shows the following: (1) more than two-thirds of probed genes are expressed in brain postnatally; (2) >95% of expressed genes show highly significant changes during development; (3) >85% of them show significant differences in expression between regions. The magnitude of these spatiotemporal changes is very large: nearly one-half of expressed genes exceed twofold, with more than one-half being significant at the level of p < 10–20 (Fig. 1); (4) Dramatic changes in gene expression occur early in postnatal life (1–2 weeks) and plateau thereafter; (5) differences between regions increase significantly during that time period. For example, <300 genes differentiate hippocampus from frontal cortex at P1, whereas >2000 genes differentiate them in adult brains; (6) clustering of brain regions based on transcriptional profiles shows a clear break at 2 weeks postpartum. From 14 d of age onwards, a given region resembles itself more than any other region. Before then, transcriptional profiles are simultaneously determined by both region and age. For example, E20 hypothalamus resembles E20 cortex more closely than it resembles a P1 hypothalamus; (7) clustering of transcripts as a function of temporal and regional expression profiles reveals 20 distinctive clusters, 80% of which are enriched in particular functions.
This study represents the first global characterization of gene expression across multiple brain regions and at various stages of prenatal and postnatal development in rodent brain. A pioneering study (Mody et al., 2001) profiled mouse hippocampus until P30, detecting several gene families related to waves of functional reorganization of the hippocampus. Our findings are generally consistent with their interpretations, despite species differences. In addition, contrasting multiple areas provides a molecular basis for understanding the nature and timing of regional differentiation.
Although some of the findings could have been broadly hypothesized based on knowledge of developmental neurobiology, they can now be instantiated with unique molecular profiles. For example, we would have predicted that adult hippocampus and cortex would resemble each other more than they resemble the hypothalamus and that hippocampus would lie between the other two structures. However, the present data now provide the broad molecular signatures that support these ideas.
Other findings were less expected, especially in their magnitude. We would have predicted that the first 2 weeks postpartum would be critical for continued neural development, because rat pups display rapid behavioral changes during that time. This includes increased motor coordination and locomotion, as well as maturation of sensory systems such as taste, sound, and vision (Smart and Dobbing, 1971; Altman and Sudarshan, 1975; Routtenberg et al., 1978; Uziel et al., 1981; Hill and Mistretta, 1990; Westerga and Gramsbergen, 1990; Vorhees et al., 1994), as well as increased communication and social behavior (Bolles and Woods, 1964; Hofer et al., 1998). However, it was surprising to discover the magnitude and breadth of reorganization of gene expression postpartum, with a given brain region not recognizable as “itself” before 2 weeks of life. This sheds new light on the role of maternal behavior and other early environmental factors in shaping brain function into adulthood (Meaney, 2001).
Our findings implicate several ensembles of functionally related genes, some known and some novel, in critical periods of brain development. Thus, some clusters of functionally related genes exhibit maximum activity likely relevant to early organizational events in brain. Cell proliferation within the ventricular and subventricular zones of the rat neocortex peaks between E13 and E18, declining toward birth (Rakic and Caviness, 1995). This pattern may be driven in part by cluster 3 transcripts enriched for genes involved in DNA metabolism, including five genes involved in DNA replication and chromosome cycling. Similarly, expression levels in clusters 5, 6, and 7 are highest in frontal cortex at E17–E18, declining in all three brain regions with increasing age. The 123 genes in these three clusters are enriched for genes involved in nuclear function and include 48 genes involved in cell proliferation. Their interactions and coordinate regulation are likely critical in initial phases of development.
Functional clusters also identify actors that may mediate later developmental events. For example, all three genes within gene cluster 18 (Fig. 5) relate to myelination: myelin basic protein, myelin-associated oligodendritic basic protein, and proteolipid protein. Their expression rises from P4–P7 to P21, resulting in a 10-fold to 100-fold increase in mRNA levels, likely reflecting brain myelination. Expression profiles of genes within cluster 14 are highly enriched for components of synaptic vesicles, whereas cluster 15 is enriched for synaptic vesicle transport transcripts. Both clusters display relatively stable expression in forebrain before approximately P4 and after P21. However, expression of these genes increases by ∼10-fold between P4 and P21. Thus, these clusters provide a wealth of novel candidates mediating the peak of synaptogenesis that occurs between P11 and P20 in rat cortex (Sutor and Luhmann, 1995).
Major differences in expression are also apparent between different regions of the developing rat brain (Figs. 2, 5). Cluster analyses reveal this most dramatically for genes enriched for neuropeptide hormone activity (cluster 20), which increases markedly beginning at E20 and continuing to adulthood, specifically in the hypothalamus. Examples include arginine vasopressin, cocaine and amphetamine-regulated transcript, galanin, hypocretin, oxytocin, preproenkephalin-related sequence, pro-melanin-concentrating hormone, and thyrotropin releasing hormone, which provide a unique signature for that region.
The increasing prevalence of whole-genome profiling (Chesler et al., 2005; Hubner et al., 2005) requires improved functional characterization of the genome. Although this will ultimately depend on gene-by-gene analyses, characteristics of known genes within a cluster can lead to hypotheses about functions of unknown genes within that ensemble. For example, cluster 19 is composed of eight transcripts, five of which are unknown. Interestingly, the three known genes are all transcription factors, namely human immunodeficiency virus type 1 enhancer binding protein (Hivep2), POU domain, class 3, transcription factor 1 (Pou3f1), and zinc finger protein 238 (Znf238). Expression of this cluster is greatest during the first postnatal week, and its members are implicated in the regulation of cell differentiation, cytoskeletal development, and cell death. Thus, Pou3f1 regulates the transition of promyelin cells into myelinating cells (Jaegle et al., 1996), and its expression peaks at P4, just before the dramatic induction of myelination genes. Znf238 is a transcriptional repressor, with putative targets including the plasma ion exchanger S1c9a1 (which regulates the cortical cytoskeleton) and protein C (which regulates apoptosis) (Aoki et al., 1998). We therefore suggest that the five previously uncharacterized transcripts within cluster 19 may be important in transcriptional regulation during the first week of life.
This approach can also suggest novel functions for previously characterized genes. Thus, cluster 20 is highly enriched in neuropeptide signaling genes. However, many of its members had not been previously implicated in hypothalamic functions. For instance, Abelson helper integration site 1 (Ahi1) is a signal transduction adaptor molecule recently implicated in Joubert Syndrome (Dixon-Salazar et al., 2004), which is characterized by brainstem malformations, neonatal hypotonia, ataxia, developmental delay, and mental retardation (Louie and Gleeson, 2005). Ahi1 was known to be expressed in embryonic brain, but the marked increase in expression within the developing hypothalamus demonstrated here was not previously reported. This finding suggests some hitherto unknown functions for this gene in adult hypothalamus and may lead to insights into the pathology of Joubert Syndrome. Similarly, hippocalcin-like 1 (Hpcal1) displays high homology to hippocalcin, a neuron-specific calcium-binding protein highly expressed in hippocampal pyramidal cells, which interacts with neuronal apoptosis inhibitory protein to promote neuronal survival (Mercer et al., 2000). Its marked increase in expression in the developing hypothalamus suggests that Hpcal1 may promote neuronal survival in this region.
Other discoveries emerge from comparing expression patterns within a biological family. For instance, a member of the fibroblast growth factor (FGF) family, FGF13, had been shown to be highly expressed in hippocampus, (Smallwood et al., 1996). Our arrays reveal that its expression levels are an order of magnitude higher than any other member of the FGF family and changes the most dramatically across development, with a distinct peak in all three brain regions at P4–P7 revealed for this first time by our analysis. FGF13 is expressed at the nodes of Ranvier of sensory axons and modulates the functionality of voltage-gated sodium channels (Wittmack et al., 2004). Interestingly, this gene is X linked and implicated in Börjeson-Forssman-Lehmann syndrome characterized by mental retardation (Gecz et al., 1999). Given the intense and highly dynamic expression pattern of FGF13 during development, its central functions and role in cognitive development deserve study.
Our results offer a useful backdrop for neurobiologists studying the expression of any candidate gene across development or the impact of behavioral or pharmacological manipulations at a particular developmental stage. They can also identify genes that serve as intermediaries between early development and the final adult phenotype, and whose modification can alter subsequent neural function. Finally, we note that the period of early postnatal development in rat corresponds to late gestation in humans (Morgane et al., 2002). The substantial reorganization in gene expression during this stage points to the importance of late gestation in determining vulnerability to human brain disorders.
Footnotes
This work was funded by United States Office of Naval Research Grant N00014-02-1-0879, National Institute of Mental Health Grant PO1 MH42251, and National Institute on Drug Abuse Grant RO1 DA 13386. We are grateful to Jim Stewart and Mary Hoversten for technical assistance.
Correspondence should be addressed to Dr. John Stead, Department of Psychology, Institute for Neuroscience, Carleton University, Ottawa, Ontario, Canada K1S 5B6. E-mail: John_Stead@Carleton.ca.
DOI:10.1523/JNEUROSCI.2755-05.2006
Copyright © 2006 Society for Neuroscience 0270-6474/06/260345-09$15.00/0
References
- Akbarian S, Vinuela A, Kim JJ, Potkin SG, Bunney Jr WE, Jones EG (1993) Distorted distribution of nicotinamide-adenine dinucleotide phosphate-diaphorase neurons in temporal lobe of schizophrenics implies anomalous cortical development. Arch Gen Psychiatry 50: 178–187. [DOI] [PubMed] [Google Scholar]
- Akil M, Lewis DA (1997) Cytoarchitecture of the entorhinal cortex in schizophrenia. Am J Psychiatry 154: 1010–1012. [DOI] [PubMed] [Google Scholar]
- Altman J, Sudarshan K (1975) Postnatal development of locomotion in the laboratory rat. Anim Behav 23: 896–920. [DOI] [PubMed] [Google Scholar]
- Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M (1998) RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem 273: 26698–26704. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery ratea practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Bolles RC, Woods PJ (1964) The ontogeny of behavior in the albino rat. Anim Behav 12: 427–441. [Google Scholar]
- Carver T, Bleasby A (2003) The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics 19: 1837–1843. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R (2003) Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301: 386–389. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW (2005) Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37: 233–242. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar T, Silhavy JL, Marsh SE, Louie CM, Scott LC, Gururaj A, Al-Gazali L, Al-Tawari AA, Kayserili H, Sztriha L, Gleeson JG (2004) Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet 75: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN (2003) Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry 60: 804–815. [DOI] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, Lopez JF, Thompson RC, Meng F, Stead JD, Walsh DM, Myers RM, Bunney WE, Watson SJ, Jones EG, Akil H (2004) Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA 101: 15506–15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gecz J, Baker E, Donnelly A, Ming JE, McDonald-McGinn DM, Spinner NB, Zackai EH, Sutherland GR, Mulley JC (1999) Fibroblast growth factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum Genet 104: 56–63. [DOI] [PubMed] [Google Scholar]
- Gilbertson MW, Shenton ME, Ciszewski A, Kasai K, Lasko NB, Orr SP, Pitman RK (2002) Smaller hippocampal volume predicts pathologic vulnerability to psychological trauma. Nat Neurosci 5: 1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DL, Mistretta CM (1990) Developmental neurobiology of salt taste sensation. Trends Neurosci 13: 188–195. [DOI] [PubMed] [Google Scholar]
- Hofer MA, Masmela JR, Brunelli SA, Shair HN (1998) The ontogeny of maternal potentiation of the infant rats' isolation call. Dev Psychobiol 33: 189–201. [DOI] [PubMed] [Google Scholar]
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Muller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ (2005) Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 37: 243–253. [DOI] [PubMed] [Google Scholar]
- Insel TR, Fernald RD (2004) How the brain processes social information: searching for the social brain. Annu Rev Neurosci 27: 697–722. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D (1996) The POU factor Oct-6 and Schwann cell differentiation. Science 273: 507–510. [DOI] [PubMed] [Google Scholar]
- Jessell TM, Sanes JR (2000) Development. The decade of the developing brain. Curr Opin Neurobiol 10: 599–611. [DOI] [PubMed] [Google Scholar]
- Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, Lopez JF, Avelar A, Shokoohi V, Chung T, Mesarwi O, Jones EG, Watson SJ, Akil H, Bunney Jr WE, Myers RM (2004) Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet 13: 609–616. [DOI] [PubMed] [Google Scholar]
- Louie CM, Gleeson JG (2005) Genetic basis of Joubert syndrome and related disorders of cerebellar development. Hum Mol Genet 14 [Suppl 2]: R235–R242. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL (2003) Cell migration in the forebrain. Annu Rev Neurosci 26: 441–483. [DOI] [PubMed] [Google Scholar]
- Meaney MJ (2001) Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci 24: 1161–1192. [DOI] [PubMed] [Google Scholar]
- Mercer EA, Korhonen L, Skoglosa Y, Olsson PA, Kukkonen JP, Lindholm D (2000) NAIP interacts with hippocalcin and protects neurons against calcium-induced cell death through caspase-3-dependent and -independent pathways. EMBO J 19: 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Lewis DA, Levitt P (2001) Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci 24: 479–486. [DOI] [PubMed] [Google Scholar]
- Mody M, Cao Y, Cui Z, Tay KY, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien JZ (2001) Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci USA 98: 8862–8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgane PJ, Mokler DJ, Galler JR (2002) Effects of prenatal protein malnutrition on the hippocampal formation. Neurosci Biobehav Rev 26: 471–483. [DOI] [PubMed] [Google Scholar]
- Rakic P, Caviness Jr VS (1995) Cortical development: view from neurological mutants two decades later. Neuron 14: 1101–1104. [DOI] [PubMed] [Google Scholar]
- Rice D, Barone Jr S (2000) Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect 108 [Suppl 3]: 511–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtenberg A, Strop M, Jerdan J (1978) Response of the infant rat to light prior to eyelid opening: mediation by the superior colliculus. Dev Psychobiol 11: 469–478. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM (2003) Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J (1996) Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci USA 93: 9850–9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart JL, Dobbing J (1971) Vulnerability of developing brain. VI. Relative effects of fetal and early postnatal undernutrition on reflex ontogeny and development of behavior in the rat. Brain Res 33: 303–314. [DOI] [PubMed] [Google Scholar]
- Sutor B, Luhmann HJ (1995) Development of excitatory and inhibitory postsynaptic potentials in the rat neocortex. Perspect Dev Neurobiol 2: 409–419. [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS (1996) The molecular biology of axon guidance. Science 274: 1123–1133. [DOI] [PubMed] [Google Scholar]
- Uziel A, Romand R, Marot M (1981) Development of cochlear potentials in rats. Audiology 20: 89–100. [DOI] [PubMed] [Google Scholar]
- Vorhees CV, Acuff-Smith KD, Moran MS, Minck DR (1994) A new method for evaluating air-righting reflex ontogeny in rats using prenatal exposure to phenytoin to demonstrate delayed development. Neurotoxicol Teratol 16: 563–573. [DOI] [PubMed] [Google Scholar]
- Westerga J, Gramsbergen A (1990) The development of locomotion in the rat. Brain Res Dev Brain Res 57: 163–174. [DOI] [PubMed] [Google Scholar]
- Wittmack EK, Rush AM, Craner MJ, Goldfarb M, Waxman SG, Dib-Hajj SD (2004) Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J Neurosci 24: 6765–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]