Abstract
Homer is a crucial postsynaptic scaffolding protein involved in both maintenance and activity-induced plasticity of the synapse. However, its quaternary structure has yet to be determined. We conducted a series of biophysical experiments that provide the first evidence that Homer forms a tetramer via its coiled-coil domain, in which all subunits are aligned in parallel orientation. To test the importance of the tetrameric structure for functionality, we engineered dimeric and tetrameric Homer by deleting a part of coiled-coil domain or replacing it with artificially engineered dimeric or tetrameric coiled-coil domain from a yeast protein. The structure–activity relationship was determined by assaying cocluster formation with its ligand in heterologous cells, distribution in dendritic spines, and turnover rate of protein exist in dendritic spines. Our results provide the first insight into the structure of native Homer protein as a tetramer and the functional significance conferred by that structure.
Keywords: Homer, metabotropic glutamate receptor, coiled-coil, quaternary structure, synapse, scaffolding protein
Introduction
Homer is a synaptic scaffolding protein with an N-terminal EVH1 and a C-terminal coiled-coil domain (Xiao et al., 2000; Thomas, 2002). Its family is composed of three members, and each of them is alternatively spliced to long and short forms (Fig. 1). The long forms contain both EVH1 and coiled-coil domains and are constitutively expressed, whereas the short forms contain only the EVH1 domain and are expressed in an activity-dependent manner (Brakeman et al., 1997). The binding of its EVH1 domain to various other scaffolding and signal transduction molecules, including metabotropic glutamate receptors (mGluRs), IP3 receptors (IP3Rs) (Tu et al., 1998), Shank (Tu et al., 1999), and canonical transient receptor potential (TRPC) family channels (Yuan et al., 2003), allows Homer to serve as a postsynaptic scaffold that crosslinks and regulates the functionality of these ligands. For example, long Homer forms a link between mGluR1/5 and downstream effector IP3R (Tu et al., 1998) or works as an inverse agonist of mGluR1/5 (Ango et al., 2001). For this functional coupling of postsynaptic proteins, the coiled-coil domain of long Homer plays an important role through its capacity to oligomerize. The short Homer lacking the coiled-coil domain acts as an endogenous dominant negative to block the function of long Homer, consequently reducing the amount of synaptic proteins, the size of dendritic spines, and the amplitude of synaptic current (Sala et al., 2003).
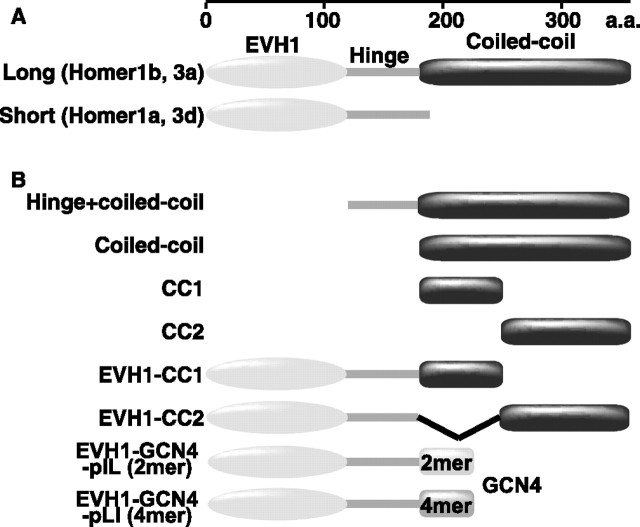
Figure 1.
General scheme of Homer proteins. A, Schematic drawing of domain organization of alternatively spliced forms of Homers. Both long and short forms have an N-terminal EVH1 domain. C-terminal coiled-coil domain is unique for long forms. B, Deletion mutants used in this study. The exact numbers of residues in each deletion mutants are as follows. Hinge+coiled-coil, 138–361 of Homer3a; Coiled-coil, 184–361 of Homer3a; CC1, 181–246 of Homer1b and 184–275 of Homer3a; CC2, 247–354 of Homer1b and 253–361 of Homer3a; EVH1-CC1, 1–246 of Homer1b and 1–275 of Homer3a; EVH1-CC2, 1–176 + 247–354 of Homer1b.
Although a number of studies point to the functional importance of oligomerization of long Homer, there has been no consensus on its quaternary structure. Long Homer has been depicted as a dimer, tetramer, or hexamer, formed via the coiled-coil domain, in both parallel and antiparallel configurations (Xiao et al., 2000; Fagni et al., 2002; Thomas, 2002). In contrast, the EVH1 domain has been crystallized and an atomic-resolution structure has been solved (Beneken et al., 2000; Barzik et al., 2001). To understand how Homer regulates the function of various binding partners through its EVH1 domain, structural information about the coiled-coil domain is indispensable.
Here we describe a series of biophysical and cell biological experiments to elucidate the quaternary structure of Homer and its functional significance. First, we show that long Homer forms a tetramer through the coiled-coil domain, the individual monomers of which are aligned in parallel formation. To test the significance of this structure, we then generated Homer constructs in which the coiled-coil domain was replaced with a dimeric or tetrameric coiled-coil domain artificially engineered from a yeast protein. The cell biological experiments using these constructs indicate that the tetramer formation is both sufficient and necessary for cocluster formation with its ligand mGluR1α, whereas monomeric and dimeric Homer did not have such capacity. The localization and turnover rate at the dendritic spines were affected by multimerization status of Homer as well but our results also suggest the presence of additional factor(s), conferred by full-length coiled-coil domain, which affect these parameters. Our results provide the first account that the long Homer protein can serve as a tetravalent hub, capable of interacting with four ligand proteins at the same time.
Materials and Methods
Purification of Homer proteins from bacteria.
pET15b and pET28a plasmids (EMD Biosciences, Madison, WI) with LacI sequence removed were used for expression in Escherichia coli. Expression vectors for rat Homer1, human Homer3, and their deletion mutants were constructed using PCR-based methods. Point mutations or deletions were introduced with QuikChange Site-Directed Mutagenesis System (Stratagene, La Jolla, CA). BL21(DE3) was used as a host bacteria for Homer1 coiled-coil domain constructs, and BL21(DE3)pLacI was used for all others. Expression was induced by adding final 1 mm isopropyl-β-d-thiogalactopyranoside to the culture medium at OD600 = 0.8 at 25°C. Cells were harvested and washed with 20 mm Tris-Cl, pH 8.0, resuspended in 20 mm Tris-Cl, 30 mm imidazole-Cl, 300 mm NaCl, pH 8.0, and stored at −80°C until use. Cells were lysed by sonication and centrifuged, and the resultant supernatant was loaded onto 5 ml of Ni-nitrilotriacetic acid (NTA) agarose column (EMD Biosciences) pre-equilibrated with the loading buffer. After wash, full-length Homers were eluted by a 6 ml linear gradient of 30–500 mm imidazole-Cl and 1 m NaCl, pH 8.0. The pooled fractions were loaded onto HiPrep 16/60 Sephacryl S-300 HR gel filtration column (GE Healthcare, Piscataway, NJ) equilibrated by 20 mm potassium phosphate buffer, 1 m NaCl, and 0.1% 2-mercaptoethanol, pH 7.4. For other deletion mutants, NaCl was not used for elution from Ni-NTA agarose, and PBS and 0.1% 2-mercaptoethanol, pH 7.4, was used for gel filtration.
To identify a junction region within the coiled-coil domain, Homer1b was partially digested by trypsin (10 μg/ml) at room temperature for 20 min. In some preparations of Homer3, we observed degradation of the protein during prolonged storage. The N-terminal sequences for these cleavage sites were analyzed by Edman degradation in the biopolymer facility at the Massachusetts Institute of Technology Cancer Research Center and Tufts University Core Facility (Boston, MA).
Analytical gel filtration.
Analytical gel filtration was performed on a 24 ml Superose 6 10/300 GL column (GE Healthcare), equilibrated with 20 mm potassium phosphate buffer, 1 mm EDTA, 1 m NaCl, and 0.1% 2-mercaptoethanol, pH 7.4 (for full-length Homers), or PBS and 0.1% 2-mercaptoethanol (for others) and run at a flow rate of 0.25 ml/min. The elution profile was analyzed by absorption at 280 nm or by measurement of protein concentration with Coomassie plus protein assay reagent (Perbio Science, Rockford, IL).
Preparation of brain tissue sample.
Cerebrum of postnatal day 7 (P7) and adult rats was homogenized in PBS, pH 7.4, with 0.1% 2-mercaptoethanol in a glass–Teflon homogenizer. The soluble fraction was obtained as a supernatant of 50,000 × g centrifuge and loaded onto a Superose 6 10/300 GL column as above. Each fraction was analyzed by Western blotting with monoclonal anti-Vesl-L/Homer1b/c antibody (Cell Signaling Technology, Danvers, MA).
Circular dichroism spectroscopy.
Circular dichroism spectroscopy was performed on a CD spectrometer 60DS (AVIV Biomedical, Lakewood, NJ) using a cuvette with 1 cm path length at a protein concentration of 0.1–0.2 mg/ml in 5 mm potassium phosphate buffer, pH 7.4. Hexahistidine tags were removed by thrombin digestion of fragments of the Homer coiled-coil domain.
Analytical ultracentrifugation.
Beckman Coulter (Fullerton, CA) Optima XL-A analytical ultracentrifuge was used with an An-60 Ti rotor for sedimentation equilibrium. Homer1b was dialyzed in 20 mm potassium phosphate buffer, 2 m NaCl, and 0.1% 2-mercaptoethanol, pH 7.4. Homer3a was prepared similarly to Homer1b, except with 1 m NaCl. All of the other samples were dialyzed in PBS. The absorption measurements were taken at 245 nm for the C-terminal half of the coiled-coil domain (CC2) and 280 nm for all the others. Experiments were performed at 20°C for Homer1b and Homer3a and at 4°C for all the other samples. Partial specific volume and sample density were calculated using the program SEDNTERP (John Philo, Thousand Oaks, CA) (Laue et al., 1992). Molecular weights were determined by WinNonlin (University of Connecticut, http://spin6.mcb.uconn.edu/winnonlin/winnonln.html) (Johnson et al., 1981) using global analysis of sedimentation data at different speeds and concentrations.
For sedimentation velocity experiments, starting sample concentrations were 0.5–1.0 mg/ml in 20 mm potassium phosphate buffer, 1 m NaCl, and 0.1% 2-mercaptoethanol, pH 7.4. Experiments were performed at 4°C with the rotor speed of 40,000 rpm. The absorbance profiles were analyzed using SEDFIT (National Institutes of Health, http://www.analyticalultracentrifugation.com/default.htm) (Schuck, 2000) to determine the sedimentation coefficient distributions.
Chemical crosslink of coiled-coil region of Homer.
C-terminal fragment of Homer1b (290–354) was expressed and purified as described above. The purified sample was crosslinked by incubation with 0.2% glutaraldehyde for 20 min at room temperature. The reaction was stopped by addition of 0.1 m Tris-HCl, pH 6.8. The crosslinked sample was separated in 14% SDS-PAGE gel and stained with Coomassie brilliant blue R-250.
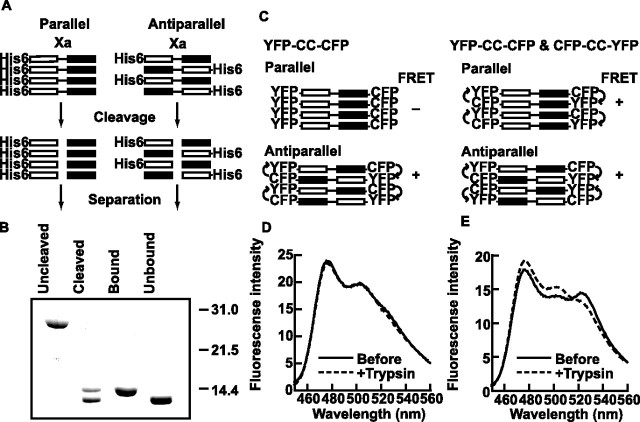
Cleavage of coiled-coil domain at an inserted Factor Xa recognition sequence.
A Factor Xa recognition site was introduced to the Homer3a 184–361 fragment by mutation of the 252GQGQ255 sequence in Homer3a to IDGR. Factor Xa (EMD Biosciences) was added to the purified protein at a final concentration of 10 U/ml and incubated at room temperature overnight. The cleavage product was loaded onto a 1 ml Histrap column (GE Healthcare) to separate the N- and C-terminal halves and eluted using a 30–500 mm gradient of imidazole, pH 8.0.
Fluorescence resonance energy transfer.
pECFP-EYFP or pEYFP-ECFP vectors were constructed from pECFP-C1 and pEYFP-N1 or pECFP-N1 and pEYFP-C1 (Clontech, Mountain View, CA). Rat Homer1b cDNA was amplified by PCR to obtain a fragment coding the hinge and the coiled coil 137–354 and inserted into pECFP-EYFP and pEYFP-ECFP. These plasmids were transfected to human embryonic kidney 293 (HEK293)-T cells using Genejuice (EMD Biosciences). The cells were harvested 4 d after transfection, washed with PBS, and homogenized in 20 mm Na-HEPES, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100, pH 7.5. Supernatant obtained after centrifugation at 16,300 × g for 20 min was used for fluorospectrometry on RF-5301PC (Shimazu, Kyoto, Japan). Specific CFP excitation at 433 nm was used for excitation to assess fluorescence resonance energy transfer (FRET).
Mammalian expression vectors.
cDNAs for mGluR1α, Homer1a-green fluorescent protein (GFP), and Homer1b-GFP were kindly provided by Dr. Yoshiaki Tagawa (Kyoto University, Kyoto, Japan) and Dr. Shigetada Nakanishi (Osaka Bioscience Institute, Osaka, Japan). pGW-myc-Homer1a and pGW-myc-Homer1b were described previously (Sala et al., 2001, 2003). EVH1-GCN4-pLI was constructed by connecting synthesized linkers to code RMKQIEDKLEEILSKLYHIENELARIKKLLGER, and EVH1-GCN4-pIL by connecting RMKQLEQKIEELLSKIYHLENEIARLKKLIGER after the 176th residue of Homer1b.
mGluR1α clustering assay.
COS-7 cells were cotransfected with myc-tagged Homer and mGluR1α using Genejuice (EMD Biosciences). The cells were fixed, permeabilized, and immunostained 4 d after transfection. Anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-mGluR1α (Upstate, Charlottesville, VA) antibodies were used for immunostaining. Coclusters were defined as intense stains observed in the perinuclear area in both channels and visually assessed blind to the constructs used.
Neuronal imaging.
Hippocampal organotypic slice culture was prepared from P6 or P7 rats and transfected at 5–6 d in vitro (DIV) with various Homer constructs tagged with GFP and pDsRed2-C1 as described previously (Okamoto et al., 2004). Images were taken on 7–8 DIV with a custom-built two-photon microscope with an excitation at 890 nm under constant perfusion with artificial CSF and analyzed off-line on MetaMorph (Molecular Devices, Downingtown, PA). The fluorescence profiles of well separated spines and adjacent dendritic shafts were measured at both channels, and the fluorescence peak of both spine and dendrite were obtained. Synapse targeting was determined as follows:
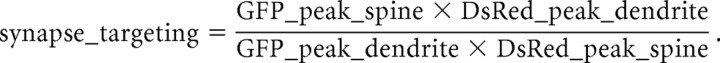 |
For fluorescence recovery after photobleaching (FRAP) assay, the slice culture was transfected with GFP-tagged Homer constructs alone. Images of a single spine were taken with an Olympus (Tokyo, Japan) FV 300/IX70 inverted laser-scanning confocal microscope with excitation at 488 nm and 6% laser output intensity. For photobleaching, the spine was scanned 10 times with 2 s intervals at the same wavelength, with 100% laser output intensity. Thereafter, the imaging was continued at the original laser intensity and sampling interval. The fluorescent intensity exist in the dendritic spine was analyzed off-line with MetaMorph and plotted. Recovery was fitted to a single exponential curve (Okamoto et al., 2004).
Results
Long forms of Homer proteins behave as a multimeric complex
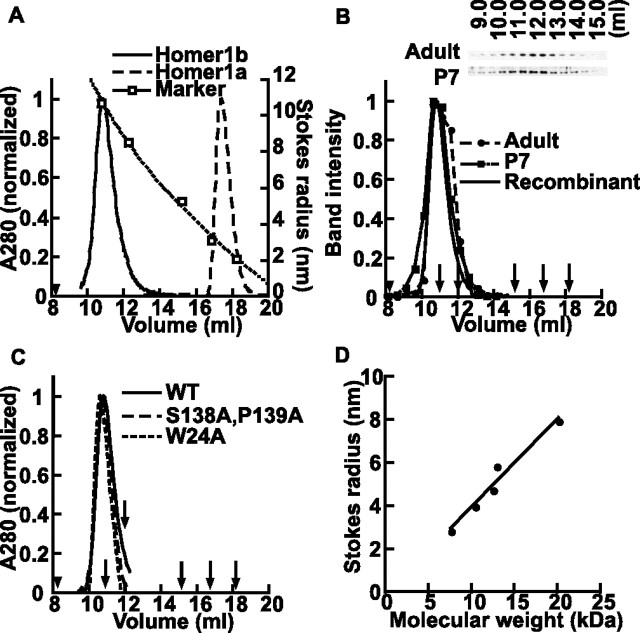
To estimate the native protein size of Homer proteins, we first used the analytical gel filtration of bacterially expressed and purified proteins. A short Homer, Homer1a eluted between 44 and 17 kDa markers (Fig. 2A), consistent with globular monomeric proteins with calculated molecular weight of 23 kDa. However, the long form of Homer, Homer1b, eluted in much earlier fractions than expected (Fig. 2A). Although the calculated molecular weight of Homer1b as a monomer is 42 kDa, it was larger than the largest globular molecular weight marker, thyroglobulin (670 kDa), and was similar to that of fibrinogen, an elongated molecule with a Stokes radius of 10.7 nm (van Holde, 1985). Homer3a and 3d, long and short forms of Homer3, showed elution profiles similar to those of Homer1b and 1a (Table 1). The gel filtration pattern resembles a normal Gaussian distribution without skew, indicating that the protein is rather homogenous. This unexpected result prompted us to further study the native structure of Homer proteins.
Figure 2.
Analytical gel filtration of Homer. A, Elution profiles of purified Homer1a and Homer1b from Superose 6 gel filtration column. The molecular weights and Stokes radii of markers indicated with white squares are as follows: human plasma fibrinogen, 341 kDa, 10.7 nm; thyroglobulin, 670 kDa, 8.5 nm; bovine γ-globulin, 158 kDa, 5.2 nm; chicken ovalbumin, 44 kDa, 3.1 nm; equine myoglobin, 17 kDa, 2.1 nm, from left to right. The void volume was 8.2 ml and is indicated by an arrowhead. B, Elution profile of endogenous Homer1b/c from P7 and adult rat, detected by Western blotting. The scanned band intensity was normalized and plotted in the graph and overlaid with the elution profile of recombinant Homer1b. Positions of molecular weight markers are indicated by arrows. C, Elution profile of Homer P-motif mutant (S138A, P139A) and EVH1 domain ligand-binding site mutant (W24A). WT, Wild type. D, Relationships between Stokes radius and molecular weight of coiled-coil fragments.
Table 1.
Stokes radius and oligomerization status of Homer deletion mutants
| Homer1 |
Homer3 |
|||||
|---|---|---|---|---|---|---|
| Calculated MW | Stokes radius | X-mer | Calculated MW | Stokes radius | X-mer | |
| Long | 42.3 | 10.5 | 3.3 | 42.0 | 9.6 | 3.5 |
| Short | 23.0 | 2.8 | 1.0 | 15.8 | 1.2 | 1.0 |
| Hinge + coiled-coil | N.D. | N.D. | N.D. | 26.8 | 8.8 | 4.1 |
| Coiled-coil | N.D. | N.D. | N.D. | 20.2 | 7.9 | 4.0 |
| CC1 | 7.7 | 2.8 | 3.4 | 10.5 | 3.9 | 1.9 |
| CC2 | 13.0 | 5.8 | 3.4 | 12.6 | 4.7 | 3.3 |
| EVH1 + CC1 | 29.6 | 5.5 | 2.2 | 32.4 | 5.7 | 2.4 |
| EVH1 + CC2 | 34.6 | 9.4 | 3.9 | N.D. | N.D. | N.D. |
Molecular weight (MW) calculated from amino acid composition (in kDa), Stokes radii determined by gel filtration (in nm), and average number of monomers in an oligomer determined by analytical ultracentrifugation, are shown. Molecular weight includes His6 except for coiled-coil, CC1, and CC2 of both Homers. For Homer1, the long form is Homer1b, and the short form is Homer1a. For Homer3, the long form is Homer3a, and the short form is Homer3d. Other constructs are listed in Figure 1B. N.D., Not determined.
We first confirmed this finding with the endogenous Homer protein by performing gel filtration of crude rat brain cytosolic fraction and monitored the elution pattern by Western blot using anti-Homer1b/c antibody. The native Homer, from both adult and P7 rats, was eluted as a single peak at the same position as bacterially expressed Homer1b (Fig. 2B). This indicates that native Homer protein also forms an apparently large complex in gel filtration. In addition, the size of the Homer complex is independent of synaptic development status.
A crystallographic study of the EVH1 domain of Homer shows that there is an internal ligand of the EVH1 domain, named the P-motif, within the hinge region between the EVH1 and coiled-coil domains (Irie et al., 2002). Intermolecular interaction between the EVH1 domain and P-motif is especially intriguing because it can potentially form large head-to-head concatemers with no theoretical length limit, potentially explaining the large observed size. To test whether such an interaction mediates oligomer formation, we introduced a mutation to the P-motif (S138A, P139A) or to the ligand-binding site of the EVH1 domain (W24A) and determined their gel filtration elution profiles (Fig. 2C). The elution profiles of the mutants were identical to those of the wild type, showing that the interaction between the EVH1 domain and P-motif does not play a major role in Homer oligomer formation. Although it remains possible that such an interaction takes place in intact cells, it does not contribute to complex formation in extracted and purified protein.
Homer is an elongated molecule
The elution volume from the gel filtration is a reflection of the Stokes radius of the protein, an effective radius of a molecule as it tumbles freely in solution. For a spherical molecule, it is proportional to the cubic root of the molecular weight of the molecule. Globular proteins follow the rule and thus gel filtration is widely used to estimate a molecular weight of proteins. However, a nonglobular protein does not strictly follow such a relationship, and for the same molecular weight, the Stokes radius is larger for proteins with an elongated shape. Homers have a leucine-rich stretch of 180 aa residues at the C-terminal half that has been implicated as a coiled-coil domain (Fig. 1A). The existence of such a domain in Homer molecules may explain their large Stokes radii. Therefore, in the following sections, we focus on a characterization of the coiled-coil domain of Homer proteins.
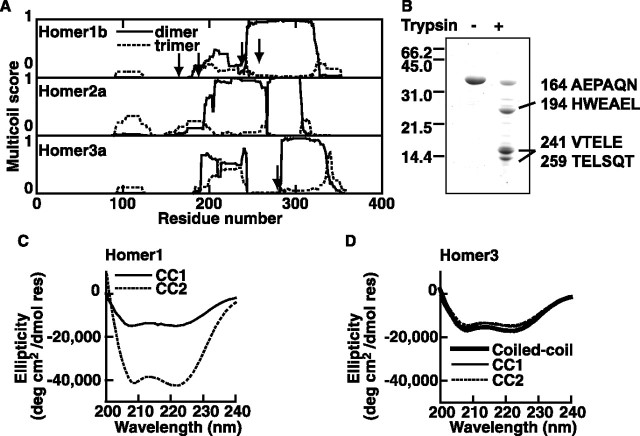
We first used Multicoil, a prediction program for amino acid sequences (Wolf et al., 1997), to assess the likelihood of dimeric and trimeric coiled-coil formation in a protein. All subtypes of Homers exhibit high scores for dimeric, but not trimeric, coiled-coil (Fig. 3A). To experimentally test whether the C-terminal region of Homer forms α-helix, a prerequisite for the formation of a coiled-coil, we subjected it to circular dichroism analysis (Fig. 3C,D). The coiled-coil domain of Homer3 showed two minima at 208 and 222 nm, typical for proteins with α-helices. Circular dichroism spectrum analysis of Homer1 coiled-coil domain was not possible because the fragment was insoluble.
Figure 3.
Homer C terminus forms coiled-coil structure. A, Results of computer-based prediction (Multicoils) of coiled-coil of Homer1b, 2a, and 3a. Scores as a dimer and as a trimer are shown. The program cannot predict coiled-coil larger than trimers. Arrows indicate protease degradation sites. B, Partial digestion of Homer1b hinge + coiled coil fragment (110–354) with trypsin. The N-terminal sequences of the digestion products determined by Edman degradation methods are shown on the right. The number indicates the amino acid position. They both turned out to be mixtures of two proteins. The bottom two bands resulted in the same sequences. C, D, Circular dichroism spectra of Homer1b (C) and Homer3a (D) coiled-coil domains.
When analyzing the coiled-coil prediction score (Fig. 3A), we noticed that in all forms of Homer, there is a decrease in coiled-coil probability at the middle of the coiled-coil domain, indicating that there may be a discontinuity of coiled-coils. Consistent with this observation, limited treatment of Homer1 with trypsin produced a cleavage site around this region (Fig. 3A; B, arrows). Also, the degradation of purified Homer3 proteins during storage occurred at this region. We tentatively named the N- and C-terminal halves of the coiled-coil as CC1 and CC2 (Fig. 1B). Both CC1 and CC2 were expressed and purified separately. In all cases, the circular dichroism results are consistent with α-helix folding patterns (Fig. 3C,D).
We next determined the Stokes radii of CC1 and CC2 by analytical gel filtration and plotted all available radii versus calculated molecular weight (Fig. 2D). This showed a linear relationship, as expected from linear molecule. It is, therefore, likely that the C-terminal region makes a linear coiled-coil domain and contributes to a large Stokes radius.
This view was further supported by the frictional ratio of the molecule in solution determined from the Stokes radius and sedimentation coefficient of long Homer proteins (Dam and Schuck, 2004). It was 2.4 for Homer1b and 2.3 for Homer3a, supporting the view that Homer is an elongated molecule.
Homer exists as a tetrameric molecule
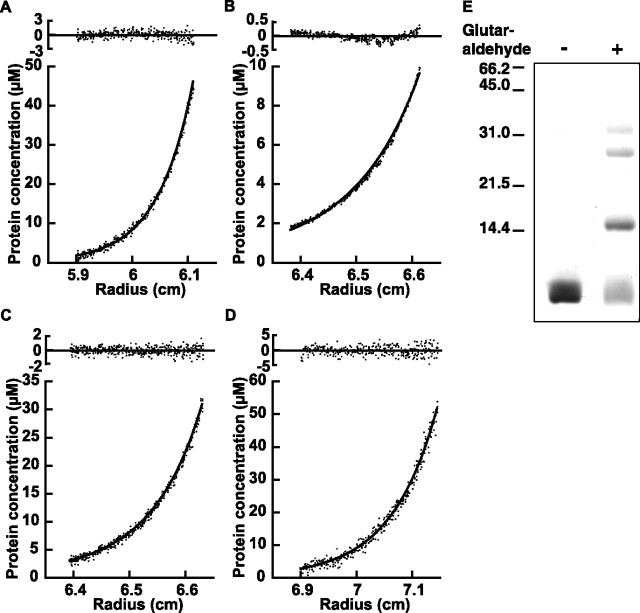
Another contributing factor to the large Stokes radius is oligomerization. To determine molecular weight of the complex, we performed a sedimentation equilibrium assay on an analytical ultracentrifuge. The ratio of weight, average molecular weight, and calculated size of monomer gives the average number of monomers existing in an oligomer (Table 1 and Fig. 4). Sedimentation equilibrium of short Homers, Homer1a and 3d, showed that they exist as monomers. In contrast, the long forms of Homer1b and 3a were 3.3 mer and 3.5 mer on average. This may represent tetramerization with partial dissociation because of the high concentration of salt added to avoid precipitation.
Figure 4.
Homer forms a tetramer. A–D, Sedimentation equilibrium analysis of Homer proteins. A, Representative run of Homer1a in PBS at a centrifugation speed of 30,000 rpm. The sample was also centrifuged at 12,000, 16,000, and 20,000 rpm and fitted to a global single-species model. The residuals are shown at the top. B, Homer1b at 9000 rpm. This sample was also centrifuged at 6000, 7500, and 12,000 rpm and globally fitted to dimer-tetramer equilibration model. C, Homer3a at 10,000 rpm. This sample was also centrifuged at 5000, 6000, and 8000 rpm, and the plot was globally fitted to dimer-tetramer equilibrium model. D, Homer3d, run as described in A. E, Chemical crosslink of Homer1b CC2 fragment with glutaraldehyde. The purified fragments before and after the crosslink reaction are shown with the positions of molecular weight markers.
To increase solubility of the protein and more accurately determine oligomeric status at physiological salt concentrations, we tested two Homer3a deletion mutants, one lacking the EVH1 domain (hinge + coiled-coil), and another lacking both EVH1 domain and hinge region (coiled-coil region only) (Fig. 1B and Table 1). Both were soluble in PBS. The coiled-coil domain construct of Homer3, determined under this condition, was 4.0 mer; the hinge + coiled-coil construct was 4.1 mer, indicating that Homer forms a tetramer at physiological salt concentration.
In addition, we chemically crosslinked purified CC2 fragment of Homer1 with glutaraldehyde (Fig. 4E). The crosslinked fragment showed four bands on SDS-PAGE, with molecular masses estimated to be 8.5, 15.2, 27.4, and 32.8 kDa, respectively, which can be explained as a monomer, dimer, trimer, or tetramer of 8.0 kDa CC2 fragments. This result also supports the formation of a tetramer, excluding the possibility that long Homer exists as a mixture of smaller and larger oligomers in equilibrium. In sum, the long form of Homer exists as tetramer, which, in conjunction with the linear shape of the molecules, contributes to the large Stokes radius observed in long Homer proteins.
Homer makes a parallel coiled-coil
Coiled-coil domains, in general, can either be in parallel configuration, in which all monomers are aligned in the same direction, or in antiparallel configuration, in which monomers are aligned in opposing directions. We therefore asked whether long Homer makes a parallel or an antiparallel tetramer. For this purpose, we individually expressed CC1 and CC2 (Fig. 1B) to determine whether they could still form tetramers by themselves. If Homer forms a parallel tetramer, CC1 and CC2 would each form tetramers. In contrast, if it forms an antiparallel tetramer, the individually expressed CC1 or CC2 will fail to form tetramers. The result was that the coiled-coil domain composed only of CC1 or CC2 was still 3.4 mer for Homer1b. The CC2 region of Homer3a was 3.3 mer, whereas CC1 falls apart to be 1.9 mer (Table 1). This supports a view that Homer coiled-coil domain forms a parallel tetramer.
However, it is still possible that individually expressed CC1 and CC2 are forming abnormal tetramers in bacteria. Therefore, we first expressed a full-length coiled-coil domain of Homer3a, cleaved at the junction between CC1 and CC2, and tested whether we could separate the two fragments. The expectation from this experiment was that, if the coiled-coil domain makes a parallel tetramer, we would be able to separate CC1 and CC2 (Fig. 5A, left). In contrast, if it makes an antiparallel tetramer, CC1 and CC2 will be inseparable even after cleavage (Fig. 5A, right). For this purpose, we generated a coiled-coil domain of Homer3a with a Factor Xa cleavage site inserted between CC1 and CC2. A hexahistidine tag (His6) was attached at the N terminus for Histrap column purification (His6-CC1-Xa-CC2). Practically all His6-CC1-Xa-CC2 was cleaved by Factor Xa without leaving any intact population, as indicated by two separate bands on SDS-PAGE (Fig. 5B). We then loaded it onto a Histrap column, and after the separation of bound and unbound proteins, the CC2 came exclusively to the unbound fraction, whereas the bound protein was exclusively His6-CC1. This suggests that Homer coiled-coil domain makes a parallel tetramer. We confirmed that the coiled-coil domain before cleavage formed a 4.0 mer on analytical ultracentrifugation. The CC2 domain obtained after the cleavage was 3.2 mer on average, indicating that it was eluted from the column as tetramer, although cleaved CC1 became 2.0 mer.
Figure 5.
Homer forms a parallel oligomer. A, Experimental design of Factor Xa cleavage test. See Results for details. B, SDS-PAGE of samples at each step of separation stained with Coomassie brilliant blue. From left, purified protein before and after cleavage with Factor Xa, cleaved protein bound and unbound to the Histrap column. The result was consistent with parallel oligomer formation. C, Experimental paradigm of FRET experiments. See Results for details. D, Fluorescence emission spectrum of YFP-CC-CFP at CFP-specific excitation at 433 nm. No peak at 527 nm corresponding with YFP was observed. Trypsin digestion did not have an effect on the fluorescence profile. E, Fluorescence spectrum of CFP-CC-YFP and YFP-CC-CFP coexpressed. A peak at 527 nm, which corresponds to YFP, was observed. Also note a decrease in this peak and a concomitant increase in CFP peak by trypsin digestion. Fluorescence intensities are expressed by arbitrary units.
FRET experiments show consistency with parallel oligomerization of Homer
To further confirm that long Homer forms a parallel linear tetramer, we used FRET technique (Heyduk, 2002). We first constructed Homer1b coiled-coil domain with CFP and YFP at the N and C termini (YFP-CC-CFP). If Homer is a parallel linear molecule, YFP-CC-CFP should not show FRET, because all YFP and CFP moieties will be separated by coiled-coil domain. The length of the coiled-coil domain is estimated to be 26 nm from the crystal structure of soluble N-ethylmaleimide-sensitive factor attached protein receptor (SNARE) complex, a tetrameric coiled-coil (Sutton et al., 1998), which is beyond the efficient range of FRET (<10 nm) (Fig. 5C). In contrast, if Homer is an antiparallel linear molecule, it should show FRET because the N and C termini come closer.
The construct expressed in HEK293-T cell did not show any evidence for FRET, supporting the view that Homer is a parallel linear tetramer (Fig. 5D). As a positive control of FRET, we coexpressed YFP-CC-CFP with CFP-CC-YFP. This combination should allow FRET in both parallel and antiparallel configurations. As expected, the HEK293-T cell homogenate showed a YFP peak after CFP-specific excitation at 433 nm (Fig. 5E). This peak was eliminated by limited digestion with trypsin, with a concomitant increase of the CFP peak, confirming FRET. Our data show that Homer exists as a tetramer with all monomers aligned in parallel. It also confirms that the coiled-coil domain is linear and that there is no turn in the middle bringing the two ends closer.
Tetramerization of Homer is necessary and sufficient for coclustering with mGluR1α in COS-7 cells
It has been demonstrated that long Homer has an ability to form a cocluster with mGluR1α at the perinuclear region, likely in endoplasmitic reticulum or Golgi apparatus in heterologous cells (Roche et al., 1999; Tadokoro et al., 1999). Such a localization of mGluR to perinuclear region is also reported in neurons and is hypothesized to detect glutamate pumped into luminal side of the organella by glutamate transporter, thereby involved in signal transduction from synapse to the nucleus (O’Malley et al., 2003; Jong et al., 2005). In contrast to Homer1b, Homer1a, although it can bind to mGluR1α through the common EVH1 domain, does not form cocluster. Therefore, the coclustering reflects both binding with mGluR1α through the N-terminal EVH1 domain and the function of the coiled-coil domain of Homer. We thus used this assay to test the functionality of the coiled-coil domain in living cells. We constructed two deletion mutants of Homer1b; one contained the EVH1 domain and the CC1 domain (EVH1-CC1), and the other contained the EVH1 and the CC2 domain (EVH1-CC2) (Fig. 1B). Analytical ultracentrifugation showed that, although the EVH1-CC2 construct remained 3.9 mer, the EVH1-CC1 construct was reduced to 2.2 mer (Table 1). We first used these two constructs and wild-type proteins to compare the multimeric status of Homer and its effect on function.
When mGluR1α was coexpressed with Homer1b, they clearly colocalized at the perinuclear region but failed to localize when individually expressed (Fig. 6). The monomeric Homer1a did not show coclustering, confirming that multimerization is important for this function. In contrast, tetrameric EVH1-CC2 clearly indicated coclustering, whereas dimeric EVH1-CC1 did not. Therefore, the ability to form a tetramer is crucial for efficient cocluster formation with mGluR1α.
Figure 6.
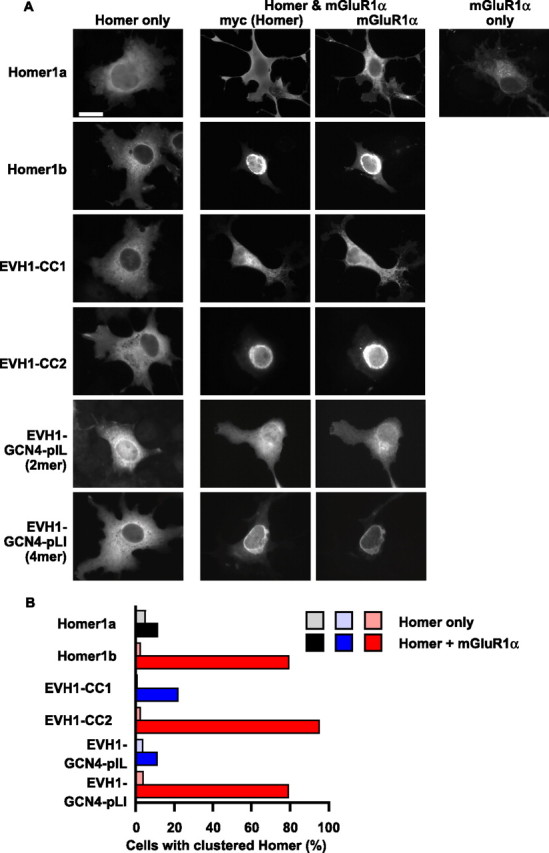
Cocluster formation of Homer deletion mutants with mGluR1α in COS-7 cells. A, Images of COS-7 cells expressing myc-tagged Homer or its mutants and mGluR1α. Cells were immunostained with anti-myc and anti-mGluR1α antibodies and imaged. Scale bar, 20 μm. B, Quantification of cluster formation of various Homer constructs in the presence and absence of mGluR1α. One hundred to two hundred cells were used for quantification in each group. Color codes are as follows: black, monomer in biophysical assay (see Table 1); blue, dimer; red, tetramer. Similar color codes are used in Figs. 7–9.
However, it is still possible that this simply indicates a requirement of the CC2 region but not of the CC1 region. For example, the CC2 region may be required for binding with some additional proteins necessary for coclustering. We therefore fused the EVH1 domain with unrelated coiled-coil peptides, a dimeric GCN4-pIL or a tetrameric GCN4-pLI, both artificially engineered from the dimeric leucine zipper DNA-binding domain of yeast transcription factor (Harbury et al., 1993) (Fig. 1B). Coiled-coil domains in general have heptad repeats with nonpolar residues at the first a and fourth d positions, which are located at the interface between α-helices of the coiled-coil. The dimeric GCN4-pIL peptide has isoleucines at a positions and leucines at d positions, whereas the tetrameric GCN4-pLI peptide has leucines at a positions and isoleucines at d positions; otherwise, they are identical. We considered that such constructs allow pure evaluation of the role of oligomerization. Tetrameric EVH1-GCN4-pLI fully restored the ability of the EVH1 domain to cocluster with mGluR1α, whereas dimeric EVH1-GCN4-pIL did not (Fig. 6). We confirmed that EVH1-GCN4-pIL and EVH1-GCN4-pLI form dimers and tetramers, respectively, using analytical ultracentrifuge of purified proteins (data not shown).
Together, coclustering activity was only associated with Homer1b, EVH1-CC2, or EVH1-GCN4-pLI, all of which were confirmed to be tetramer in biophysical analyses of the purified proteins. This good correlation between the multimeric status deduced from biophysical analyses and clustering activity is most conceivably explained by the following: (1) Homer1b forms tetramer in mammalian cells as well, not dimer or less; (2) the tetramerization of Homer is necessary and sufficient for cocluster formation in COS-7 cells.
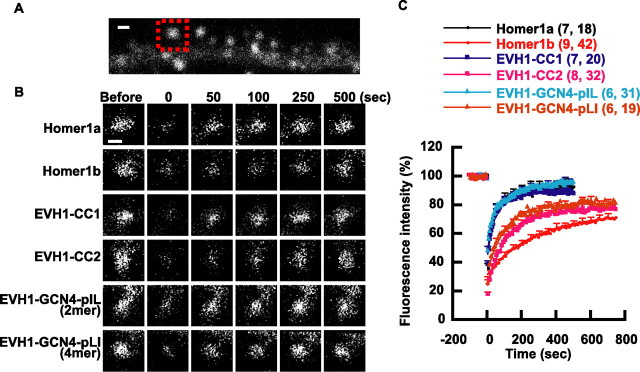
FRAP assay indicates that tetramerization of Homer1b contributes to slower turnover of the protein
Next, we used FRAP assay to estimate the rate of turnover of molecules between spines and dendritic shaft (Fig. 7). The rate of FRAP is affected by several parameters of the molecule such as its hydrodynamic properties in aqueous solution, the stability of its interaction with the binding site, and its active transport through interaction with motor proteins. Homer–GFP fusion proteins are expressed in CA1 pyramidal neurons in organotypically cultured hippocampus. Fluorescence in a single dendritic spine was photobleached, and then the FRAP was monitored with confocal microscopy. The recovery curves show the recovery of Homer1a in ∼3 min, whereas Homer1b takes >12 min. Dimeric mutants EVH1-CC2 and EVH1-GCN4-pIL did not show any significant difference from Homer1a in their recovery times. In contrast, the turnover of tetrameric constructs EVH1-CC2 and EVH1-GCN4-pLI was significantly slower than Homer1a, although it did not reach the extent of Homer1b. The most plausible explanation of the results is that the four EVH1 domains allow Homer1b to more efficiently bind with the ligand proteins that anchor Homer in dendritic spines.
Figure 7.
Tetramerization of Homer contributes to the slower turnover rate. A, Experimental scheme of spine FRAP assay. A single spine separated well from a dendritic shaft such as the one marked by a red square was selected as a target of photobleaching. The image shown is a neuron expressing GFP-Homer1b. B, Representative FRAP images are shown for each construct. Images are noisy because the laser intensity was minimized to avoid photobleaching during the experiment. C, Averaged FRAP time course of multiple experiments. Average fluorescence intensity of 10 images before photobleaching was taken as 100%. Error bars show SEM for every 50 s. Numbers of cells/spines used are in parentheses. Scale bar, 1 μm.
A full-length coiled-coil domain is necessary for efficient synaptic targeting of Homer proteins
Homer is highly concentrated at dendritic spines, where it is involved in assembling various postsynaptic proteins (Brakeman et al., 1997; Xiao et al., 1998). We therefore tested the relationship between multimerization and spine targeting by expressing the Homer constructs used above in CA1 pyramidal neurons in organotypically cultured hippocampus. The constructs were fused with GFP and coexpressed with red fluorescent protein (DsRed2) and visualized using two-photon microscopy (Fig. 8A). The accumulation of Homer–GFP at a spine head versus dendritic shaft was calculated from a fluorescent profile across a line drawn through the spine head and the nearby dendritic shaft. By including DsRed2 as volume filler, we were able to account for variation in the size of dendritic spines. Homer1b accumulated in spines with an average spine/dendrite ratio of 3.50 ± 0.06 (mean ± SEM), whereas Homer1a much less accumulated and almost diffuse, giving an average ratio of 1.51 ± 0.02 (Fig. 8B). Also, the spines were smaller than those of the cells expressing Homer1b, as estimated by DsRed2 fluorescence intensity (p < 0.001 by Kolmogorov–Smirnov test) (data not shown), confirming our previous results (Sala et al., 2003).
Figure 8.
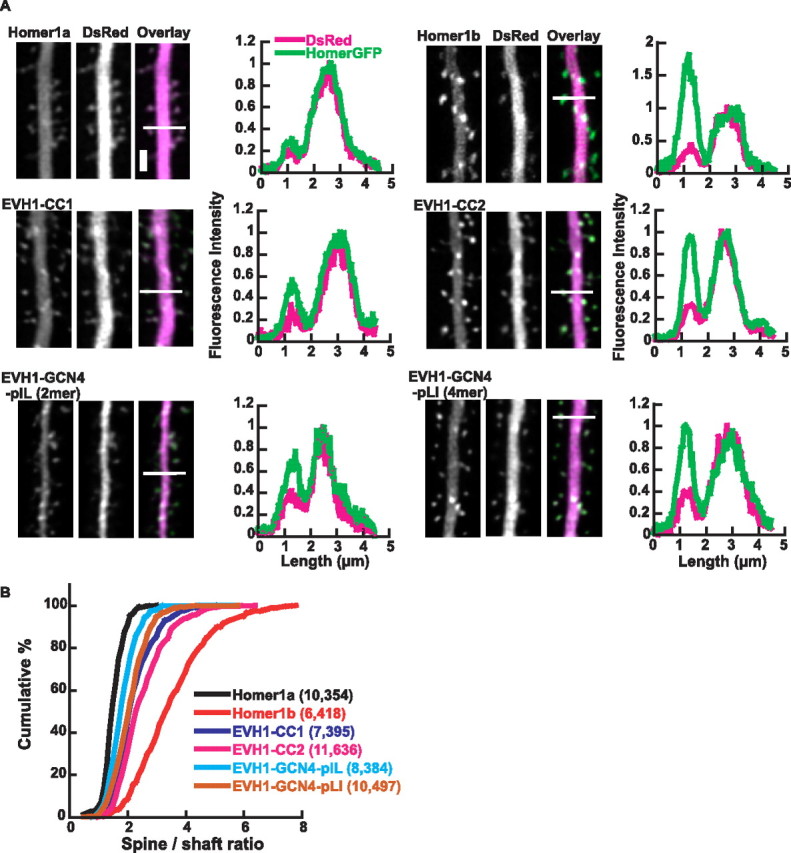
Synaptic delivery of Homer requires intact coiled-coil domain. The CA1 pyramidal neurons were transfected with GFP-tagged Homer or its mutants. DsRed2 was cotransfected as a volume filler. The fluorescence profile was taken, and the synaptic accumulation of GFP signal versus dendritic shaft was calculated. A, Representative images of Homer1a, Homer1b, EVH1-CC1, EVH1-CC2, EVH1-GCN4-pIL, and EVH1-GCN4-pLI (green) with cotransfected DsRed2 (magenta). The images were deconvoluted using Huygens (Scientific Volume Imaging, Hilversum, The Netherlands). The fluorescence profiles across the line in the images are shown in right. The peaks in the middle of the plot correspond with the dendritic shaft, and those on the left are spine heads. The intensity is adjusted so that the peaks in both channels at dendritic shaft are 1. Scale bar, 2 μm. B, The summary of synapse targeting of different constructs in cumulative plot. Numbers of cells/spines analyzed are in parentheses. The statistical significances between all samples were p < 0.0005 by Kolmogorov–Smirnov test.
Deletion mutants of Homer1b, EVH1-CC1, and EVH1-CC2 were targeted to spines more efficiently than those of monomeric Homer1a, indicating that dimer or tetramer formation contributes to protein delivery to the spines. Similarly, dimeric and tetrameric EVH1, EVH1-GCN4-pIL, and EVH1-GCN4-pLI were also targeted to spines more robustly than monomeric Homer1a. These mutants, however, did not localize to spines as strongly as Homer1b, suggesting that tetramerization contributes to the spine localization to a certain degree but by itself is not sufficient and that the coiled-coil domain of Homer may have an additional feature contributing to efficient spine targeting.
Discussion
We analyzed the quaternary structure of long Homer and found it to form a linear tetramer, in which all individual monomers are aligned in parallel (Fig. 9). During development, there is no change in the complex, indicating that oligomerization status of long Homer is not developmentally regulated.
Figure 9.
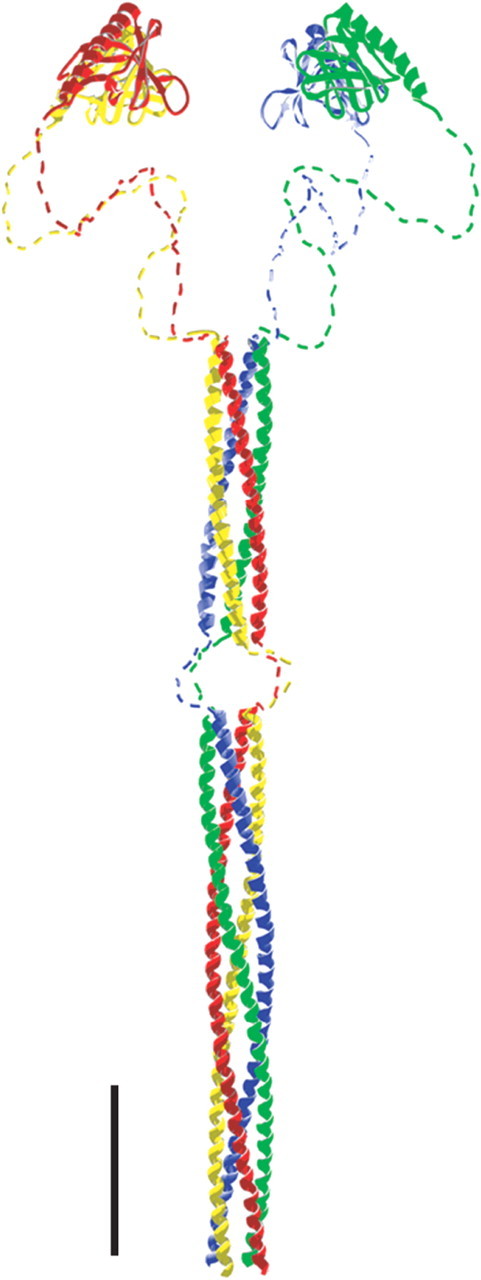
An illustration of Homer long form. EVH1 domain structure is based on crystallographic data of PDB ID 1I2H (Irie et al., 2002). The structure of the coiled-coil domain is based on the data of tetrameric coiled-coil GCN4-pLI (Protein Data Bank 1GCL) (Harbury et al., 1993) adjusted to the length of Homer coiled-coil domain. The hinge region is assumed to take a flexible structure and is drawn arbitrarily. Swiss-PDBViewer (Guex and Peitsch, 1997) and POV-Ray were used for model construction. Scale bar, 5 nm.
One previous report used protein crosslinker on purified Homer and suggested that Homer forms a multimer even larger than a tetramer through the C-terminal half of CC2 region (Tadokoro et al., 1999). Under their conditions, the crosslinked product formed a complex unresolvable on SDS-PAGE and did not even enter the gel, indicating that multiple monomers are in the complex, and they could not show how many monomers are actually involved in the complex. We are aware that Homer forms aggregate, especially under low salt concentration, and therefore their observation, made at 50 mm NaCl, may represent such an aggregate, which explains why the complex was so large and unresolvable on the SDS-PAGE. In our study, we chemically crosslinked soluble CC2 fragment and observed the formation of a tetramer but no larger complex. Although we cannot rule out that such a multimerization represents certain forms of Homer in situ, we unequivocally show that the tetramer is the minimum stable building block of naturally existing Homer.
We are aware that deletion mutants, particularly those without the CC2 regions of both Homer1 and Homer3, tend to form dimers rather than completely dissociating into monomers (Table 1). Dimerization was also observed in the experiment in which tetramer was formed initially and then cleaved at the middle (Fig. 5). This may indicate that Homer tetramer is actually a dimer of dimers, rather than a fourfold symmetric tetramer.
The tetramer formation allows Homer to serve as a hub for two- or three-dimensional complexes, as opposed to a dimer, which can only form linear complexes. Single long Homer can bind to four different ligands to make a functionally coupled complex for effective coupling of different components of signal transduction system.
Our finding that Homer takes a parallel configuration indicates that EVH1 domain ligands attached to the long Homer are all more-or-less equivalent in terms of the distance between them, within the length of EVH1 domain and flexible hinge region. If the long Homer were antiparallel, two of the ligands would be close to each other, and two would be distant, by the length of coiled-coil domain of 180 aa residues, which is translated to ∼26 nm. By comparison, the size of a tetrameric IP3R complex, a 1252 kDa membrane protein, is 17.5 nm in diameter (Sato et al., 2004). Also, by analogy with a known structure of KvAP, a Homer ligand TRPC can be estimated to be a tetramer with a diameter of ∼10 nm (Jiang et al., 2003). A difference of 26 nm would be significant in terms of dimension of these molecules and thus likely affect the coupling efficiency of signaling. It is also possible that all EVH1 domains of a tetrameric long Homer bind to these tetramers simultaneously. Such a tetramer–tetramer interaction can potentially regulate the channel activity of the channel complexes through the conformational regulation.
In addition to coupling signal transduction molecules, Homer in concert with Shank determines the size of dendritic spines. Recently, it was described that Shank can form an antiparallel homodimer through an interaction between two PDZ domains (Im et al., 2003). The tetrameric Homer and this dimeric Shank can form a three-dimensional lattice with no theoretical size limit. Other postsynaptic ligands for Homers could be embedded within such a lattice, causing local interruption in the lattice but without breaking its continuity. In fact, rotary shadow electron microscopic observation of isolated postsynaptic density reveals a mesh-like structure, in which Shank immunostaining has been found (Petersen et al., 2003). Although we currently have little experimental evidence for this view, it is consistent with previous study that a coexpression of Shank and Homer1b generates a synergistic effect to enlarge the width of dendritic spine, greater than a simple additive effect of individual expression of the two proteins (Sala et al., 2001). Also, disruption of such putative mesh by the overexpression of monomeric Homer1a reduced the synaptic structure and distribution of various postsynaptic proteins (Sala et al., 2003).
In neurons, tetramerization contributes to the slow turnover rate and localization at the dendritic spines; both EVH1-CC2 and EVH1-GCN4-pLI partially restored the effects. However, the full-length coiled-coil domain was necessary for the most efficient effect (Figs. 7, 8). This indicates that not only tetramerization, but also additional features conferred by the full-length coiled-coil domain, contributes to slow turnover rate and efficient spine localization. The length of Homer coiled-coil domain is much longer than necessary for formation of a stable coiled-coil. For example, VASP, another tetrameric EVH1 domain protein, forms a tetramer just with 45 aa, and GCN4-pLI with 34 aa. This extra length of coiled-coil domain in Homer increases the complexity of the protein and affects the hydrodynamic nature of the protein. Also, the coiled-coil domain has been shown to interact with Rho family small G-proteins (Shiraishi et al., 1999), a SNARE family protein syntaxin-13 (Minakami et al., 2000), and a ubiquitin pathway protein 2B28 (Ishibashi et al., 2005); such interactions might also contribute to our observation. Additional multimerization of the tetramer may also be a potential mechanism for spine localization. As an alternative possibility, a specific three-dimensional configuration of four EVH1 domains may be important. Another rescue construct with a right-handed coiled-coil domain from VASP (EVH1-CCVASP) was not targeted to the synapse (data not shown), in contrast to a partial targeting in EVH1-GCN4-pLI, which is left-handed coiled-coil (Fig. 8), although both equally rescued the coclustering with mGluR1α (Fig. 6) (data not shown). These and other possible features conferred by the full-length coiled-coil together determine the efficiency of targeting to the synapse.
Footnotes
H. M. Ames’ present address: University of Michigan Medical Scientist Training Program, University of Michigan Medical School, Ann Arbor, MI 48109.
This work was supported by RIKEN and The Ellison Medical Foundation. We thank Jun-wha Rhee, Christine Fanchiang, Michael Churchill, Drs. Ken-ichi Okamoto, Randall Burton, Christina Marie Taylor, Christopher C. Liu, Robert T. Sauer, Rui-Ming Xu, Yoshiaki Tagawa, Shigetada Nakanishi, Terunaga Nakagawa, Carlo Sala, and Morgan Sheng for valuable advice and sharing of resources, and Travis Emery for editing.
References
- Ango F, Prézeau L, Muller T, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L (2001). Agonist-independent activation of metabotropic glutamate receptors by the intracellular protein Homer. Nature 411:962–965. [DOI] [PubMed] [Google Scholar]
- Barzik M, Carl UD, Schubert WD, Frank R, Wehland J, Heinz DW (2001). The N-terminal domain of Homer/Vesl is a new class II EVH1 domain. J Mol Biol 309:155–169. [DOI] [PubMed] [Google Scholar]
- Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF, Leahy DJ (2000). Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron 26:143–154. [DOI] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF (1997). Homer: a protein that selectively binds metabotropic glutamate receptors. Nature 386:284–288. [DOI] [PubMed] [Google Scholar]
- Dam J, Schuck P (2004). Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol 384:185–212. [DOI] [PubMed] [Google Scholar]
- Fagni L, Worley PF, Ango F (2002). Homer as both a scaffold and transduction molecule. Sci STKE 2002:RE8. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993). A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401–1407. [DOI] [PubMed] [Google Scholar]
- Heyduk T (2002). Measuring protein conformational changes by FRET/LRET. Curr Opin Biotechnol 13:292–296. [DOI] [PubMed] [Google Scholar]
- Im YJ, Lee JH, Park SH, Park SJ, Rho SH, Kang GB, Kim E, Eom SH (2003). Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization. J Biol Chem 278:48099–48104. [DOI] [PubMed] [Google Scholar]
- Irie K, Nakatsu T, Mitsuoka K, Miyazawa A, Sobue K, Hiroaki Y, Doi T, Fujiyoshi Y, Kato H (2002). Crystal structure of the Homer 1 family conserved region reveals the interaction between the EVH1 domain and own proline-rich motif. J Mol Biol 318:1117–1126. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Ogawa S, Hashiguchi Y, Inoue Y, Udo H, Ohzono H, Kato A, Minakami R, Sugiyama H (2005). A novel protein specifically interacting with Homer2 regulates ubiquitin-proteasome systems. J Biochem (Tokyo) 137:617–623. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R (2003). X-ray structure of a voltage-dependent K+ channel. Nature 423:33–41. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Correia JJ, Yphantis DA, Halvorson HR (1981). Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J 36:575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong YJ, Kumar V, Kingston AE, Romano C, O’Malley KL (2005). Functional metabotropic glutamate receptors on nuclei from brain and primary cultured striatal neurons. Role of transporters in delivering ligand. J Biol Chem 280:30469–30480. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL (1992). Computer-aided interpretation of analytical sedimentation data for proteins. In: Analytical ultracentrifugation in biochemistry and polymer science (Harding SE, Rowe AJ, Horton JC, eds) Cambridge, UK: Royal Society of Chemistry.
- Minakami R, Kato A, Sugiyama H (2000). Interaction of Vesl-1L/Homer 1c with syntaxin 13. Biochem Biophys Res Commun 272:466–471. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Nagai T, Miyawaki A, Hayashi Y (2004). Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci 7:1104–1112. [DOI] [PubMed] [Google Scholar]
- O’Malley KL, Jong YJ, Gonchar Y, Burkhalter A, Romano C (2003). Activation of metabotropic glutamate receptor mGlu5 on nuclear membranes mediates intranuclear Ca2+ changes in heterologous cell types and neurons. J Biol Chem 278:28210–28219. [DOI] [PubMed] [Google Scholar]
- Petersen JD, Chen X, Vinade L, Dosemeci A, Lisman JE, Reese TS (2003). Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J Neurosci 23:11270–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche KW, Tu JC, Petralia RS, Xiao B, Wenthold RJ, Worley PF (1999). Homer 1b regulates the trafficking of group I metabotropic glutamate receptors. J Biol Chem 274:25953–25957. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M (2001). Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31:115–130. [DOI] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M (2003). Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci 23:6327–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Hamada K, Ogura T, Miyazawa A, Iwasaki K, Hiroaki Y, Tani K, Terauchi A, Fujiyoshi Y, Mikoshiba K (2004). Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. J Mol Biol 336:155–164. [DOI] [PubMed] [Google Scholar]
- Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Mizutani A, Bito H, Fujisawa K, Narumiya S, Mikoshiba K, Furuichi T (1999). Cupidin, an isoform of Homer/Vesl, interacts with the actin cytoskeleton and activated rho family small GTPases and is expressed in developing mouse cerebellar granule cells. J Neurosci 19:8389–8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395:347–353. [DOI] [PubMed] [Google Scholar]
- Tadokoro S, Tachibana T, Imanaka T, Nishida W, Sobue K (1999). Involvement of unique leucine-zipper motif of PSD-Zip45 (Homer 1c/vesl-1L) in group 1 metabotropic glutamate receptor clustering. Proc Natl Acad Sci USA 96:13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U (2002). Modulation of synaptic signalling complexes by Homer proteins. J Neurochem 81:407–413. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF (1998). Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron 21:717–726. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF (1999). Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23:583–592. [DOI] [PubMed] [Google Scholar]
- van Holde KE (1985). In: Physical biochemistry Englewood Cliffs, NJ: Prentice-Hall.
- Wolf E, Kim PS, Berger B (1997). MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci 6:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF (1998). Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron 21:707–716. [DOI] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Worley PF (2000). Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol 10:370–374. [DOI] [PubMed] [Google Scholar]
- Yuan JP, Kiselyov K, Shin DM, Chen J, Shcheynikov N, Kang SH, Dehoff MH, Schwarz MK, Seeburg PH, Muallem S, Worley PF (2003). Homer binds TRPC family channels and is required for gating of TRPC1 by IP3 receptors. Cell 114:777–789. [DOI] [PubMed] [Google Scholar]