Abstract
RNA interference (RNAi) offers the potential to selectively silence disease-related genes in defined cell subsets. Translation into the clinical routine is, however, still hampered by the lack of efficient carrier systems for therapeutic siRNA, endosomal entrapment presenting a major hurdle. A promising siRNA delivery system has previously been developed on the base of polyethylenimine (PEI) and the targeting ligand transferrin (Tf) to specifically reach activated T cells in the lung. In the present work, the focus is on optimizing Tf-PEI polyplexes for gene knockdown in primary activated T cells by improving their endosomal escape properties. Blending of the conjugate with membrane lytic melittin significantly enhanced endosomal release and thereby cytoplasmic delivery, while maintaining selective T cell targeting abilities and overall cell tolerability. The gathered data furthermore demonstrate that melittin addition also distinctly improves several other essential particle characteristics, such as siRNA encapsulation efficiency and stability in lung lining fluids. In conclusion, this results in a novel upgraded siRNA delivery system that is not only able to specifically deliver its payload to the desired target cells via receptor-mediated endocytosis, but also shows enhanced release from endosomal vesicles in order to initiate RNAi in the cytoplasm.
Keywords: endosomal escape, melittin, siRNA delivery, T cell targeting, transferrin receptors
1. Introduction
Small interfering RNA (siRNA) has the capability to target and functionally inhibit any chosen gene via RNA interference (RNAi).[1] This specific altering of gene expression is not only of great interest for functional analysis, but can moreover be exploited therapeutically to precisely influence pathological processes. While siRNA has already emerged to be a standard research tool, its translation into therapeutic usage in the clinical routine has been slow, with overcoming extra- and intracellular barriers being a major impediment. Due to unfavorable characteristics such as relatively high molecular weight and strong charge, naked siRNA is hardly able to cross hydrophobic cell membranes and is furthermore very susceptible to degradation.[2] Therefore, it is necessary to employ a suitable delivery system that efficiently protects the siRNA, facilitates cellular delivery and aids cytoplasmic transport to achieve passage to the target location. Cationic lipids and polymers can spontaneously condense siRNA and form nanoscale complexes, showing high gene delivery efficiencies and eluding immune responses often occurring with viral vectors.[3] However, these carrier systems usually have two major drawbacks: high toxicity due to their positive charge and abundant endosomal entrapment of their payload. Endosomal escape is generally important to avoid degradation of sensitive material in the acidic and enzymatic conditions of the endolysosomal compartment.[4] For siRNA, it is yet particularly relevant, as it has to reach the RNA-induced silencing complex, the RNAi machinery located in the cytoplasm, in order to bring about its effect.[5]
A promising approach to facilitate efficient delivery of siRNA to the desired target region is to use local administration routes, circumventing systemic degradation, and first pass metabolism of the sensitive cargo while at the same time reducing required doses and minimizing side effects. By combining this strategy with specific targeting of diseased cells it is feasible to reduce the dose even further as delivery becomes as exact and efficient as possible. In various inflammatory and immune-related diseases, T cells play a key role as they mediate a majority of cellular immune responses. Via production of Th2 cytokines, activated T cells orchestrate several pathologic cascades underlying chronic inflammatory diseases, such as asthma.[6] Here, blockage of these cytokines or preferably even their transcription factors can early on undermine disease-triggering processes and bears the potential to treat even severe forms in patients whose symptoms are currently not adequately controlled. Unfortunately, T cells have proven to be difficult to transfect as they are resistant to common non-viral vector based transfection methods. In contrast to most other cell types, T cells are devoid of caveolae as they do not express caveolin,[7] meaning that their active endocytosis of nanoparticles is limited. While other techniques such as viral delivery systems or electroporation as a physical method may be effective in vitro, but are not well suited for therapeutic in vivo application,[8] a more sophisticated way has to be found to reach the T cells.
Previously, our group developed a conjugate consisting of transferrin (Tf) and polyethylenimine (PEI), namely Tf-PEI, that can efficiently and selectively deliver siRNA to activated T cells in the lung,[9] exploiting the fact that Tf receptor (TfR) expression is upregulated in activated T cells, while naïve T cells only show negligible expression.[10] The differential expression of this activation marker concomitantly being an internalizing receptor allows the targeting of specific cell subsets while leaving naïve T cells unaffected, thereby avoiding influence on the general immune system. Besides exhibiting optimal physicochemical properties, Tf-PEI showed significant gene silencing efficiencies in vitro and preferentially delivered siRNA to activated T cells in vivo.[9] However, despite promising results, one aspect that still left room for optimization and potentially prevented the conjugate from achieving therapeutically relevant in vivo knockdown rates[11] was sufficient endosomal release of delivered siRNA. Recent studies have identified escape of the endosomal pathway as the rate-determining step in delivering therapeutic agents, being the only way to circumvent lysosomal entrapment and subsequent degradation.[12] We therefore hypothesized that the achieved gene silencing effect could possibly be enhanced by including an endosomolytic component. The peptide melittin (Mel) exhibits an inherent capacity for membrane disruption and was shown to be able to lyse red blood cells and model membranes.[13,14] As this effect is not impaired by acidic conditions, melittin exhibits optimal features as an endosomolytic agent and was proven to be a useful tool to release delivered therapeutics from the endosomal compartment.[15] Besides enhancing transfection efficiencies of both polymeric[16–18] and lipid-based[19] gene delivery vectors, it can also be used directly as an oligonucleotide transfection agent.[20] The evoked pore-formation in membranes can, however, cause general toxicity if this effect is not limited to the endosomes.[15] For this reason, a pH-sensitive shielding of melittin was applied, yielding a responsive conjugate with minimized membrane-effects at extracellular neutral conditions, but precise lytic activity upon endolysosomal acidification.
The aim of this study was therefore to refine and optimize our Tf-PEI conjugate by incorporating a bioresponsive endosomolytic domain while maintaining the specific T cell targeting ability in order to develop a versatile siRNA delivery system combining the two essential features for successful directed delivery. The resulting Tf-Mel-PEI blend was fully characterized and compared to its single components Tf-PEI and Mel-PEI as well as unmodified PEI polymer and demonstrated superior competences in terms of siRNA encapsulation and protection, Tf-related cellular uptake and transfection efficiency, sequence specific gene, and protein knockdown and, moreover, endosomal release properties in various cell models, including primary human T cells. Based on the findings presented here, we therefore conclude that the blending of Tf-PEI and Mel-PEI results in an efficient siRNA delivery system with optimal properties to preferentially reach and treat activated T cells in the lung.
2. Results
2.1. Conjugate Synthesis
Tf-PEI conjugates were prepared as described before,[9] resulting in a molecular ratio of Tf to PEI of approximately 1.5:1. Low molecular weight (LMW, 5k) PEI was successfully coupled to 2,3-Dimethyl-maleic anhydride (DMMAn) modified melittin (Mel) to yield the Mel-PEI conjugate with pH-responsive lytic ability. The reversible masking of lysine residues and N-terminal amino groups with DMMAn resulted in bioresponsive membrane-disruptive properties only emerging in acidic environments, leading to a distinct reduction of toxic side effects in the extracellular area. As a first step, PEI was coupled to the SPDP-PEG4 linker and melittin was reacted with DMMAn after activation with dithiothreitol (DTT). DMMAn-Mel was then conjugated with PDP-modified PEI via formation of a disulfide bond in the presence of 1 m guanidine hydrochloride to prevent aggregation of positively charged modified PEI and negatively charged modified melittin. Acidic cleavage of DMMAn was in turn avoided by conducting the proceeding purification at pH 8.
2.2. Particle Characterization
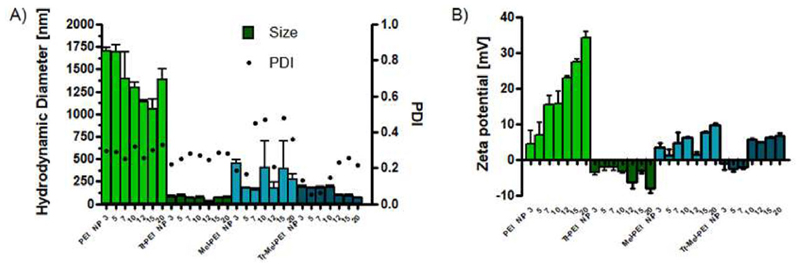
Hydrodynamic diameter, polydispersity index (PDI), and zeta potential of prepared polyplexes were determined by dynamic light scattering and laser Doppler anemometry (LDA), respectively. As Figure 1A shows, PEI/siRNA polyplexes demonstrated agglomeration with sizes above 1000 nm in physiologic salt conditions for all tested N/P ratios (nitrogen to phosphate ratio, see experimental section) with an average PDI of 0.29, while Tf-PEI formulations resulted in significantly smaller particle sizes ranging from 37 to 106 nm and an average PDI of 0.26. Polyplexes prepared of Mel-PEI showed the most heterogeneous size distributions ranging from 154 to 750 nm and the largest PDI with up to 0.523. Tf-Mel-PEI blends, however, eventuated in polyplexes with consistent hydrodynamic diameters below 200 nm exhibiting the smallest average PDI of all formulations (0.16), indicating the most homogeneous particle morphology.
Figure 1.
Dynamic light scattering and laser Doppler anemometry measurements of polyplexes. A) Hydrodynamic diameters (left γ-axis) and polydispersity indices (PDI, right γ-axis) and B) zeta potentials at different N/P ratios: 3, 5, 7, 10, 12, 15, 20. (Data points indicate mean ± SD, n = 3).
Zeta potentials of PEI polyplexes increased with rising N/P ratio (Figure 1B), showing positive charges in the range of 2.0 to 35.64 mV, while Tf-PEI particles were negatively charged with an average of −4 mV. Positive zeta potentials of Mel-PEI formulations ranged from 0.133 to 10.23 mV and Tf-Mel-PEI polyplexes displayed slightly negative charges for low N/P ratios (3, 5, and 7), but positive zeta potentials starting at N/P 10.
2.3. siRNA Encapsulation and Stability
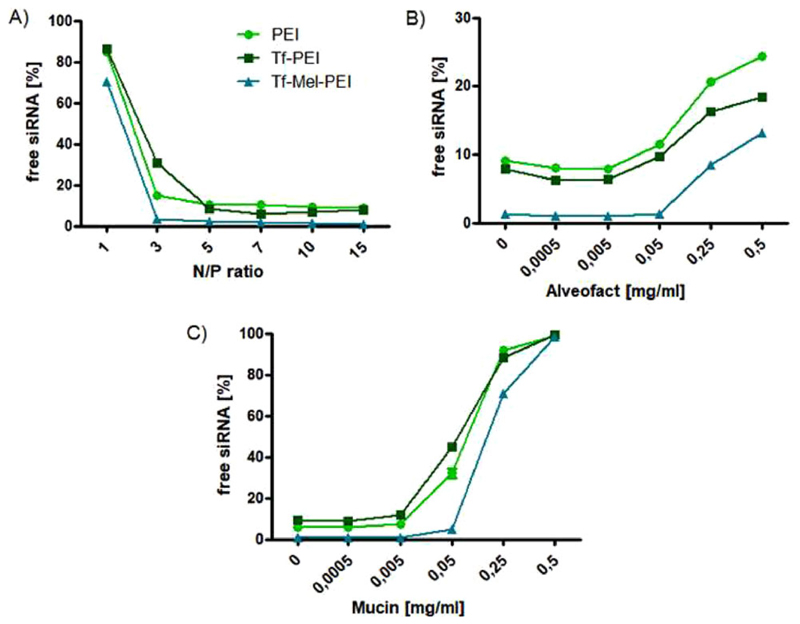
Figure 2 depicts siRNA encapsulation efficiencies of the different polyplex formulations as investigated by regular (Figure 2A) and modified (Figure 2B) SYBR Gold assays. While PEI and Tf-PEI polyplexes showed comparable siRNA encapsulation starting with approximately 85% of free siRNA at N/P 1, the Tf-Mel-PEI blend left only 70% of siRNA uncondensed at N/P1 and showed full condensation beginning at an N/P ratio as low as 3, compared to maximum condensation at N/P 5 for PEI and Tf-PEI.
Figure 2.
siRNA condensation efficiencies of polyplexes as measured by SYBR gold assay. A) Unmodified assay at different N/P ratios: 1, 3, 5, 7, 10, 15 performed in 5% glucose. Fluorescence emitted by free siRNA (N/P = 0) represents 100%. B) Polyplex stability at N/P 10 in the presence of Alveofact and C) mucin. (Data points indicate mean ± SD, n = 3).
Analogous results were obtained when testing stability in the presence of Alveofact or mucin (Figure 2B) mimicking the lung environment. In all cases, the amount of released siRNA increased with rising concentrations of simulated lung fluids, PEI, and Tf-PEI showing very similar release profiles. Tf-Mel-PEI blends, however, protected the siRNA payload better at all tested conditions, releasing only negligible siRNA concentrations, up to a concentration of 0.05% for both Alveofact and mucin.
2.4. Transferrin Receptor Binding
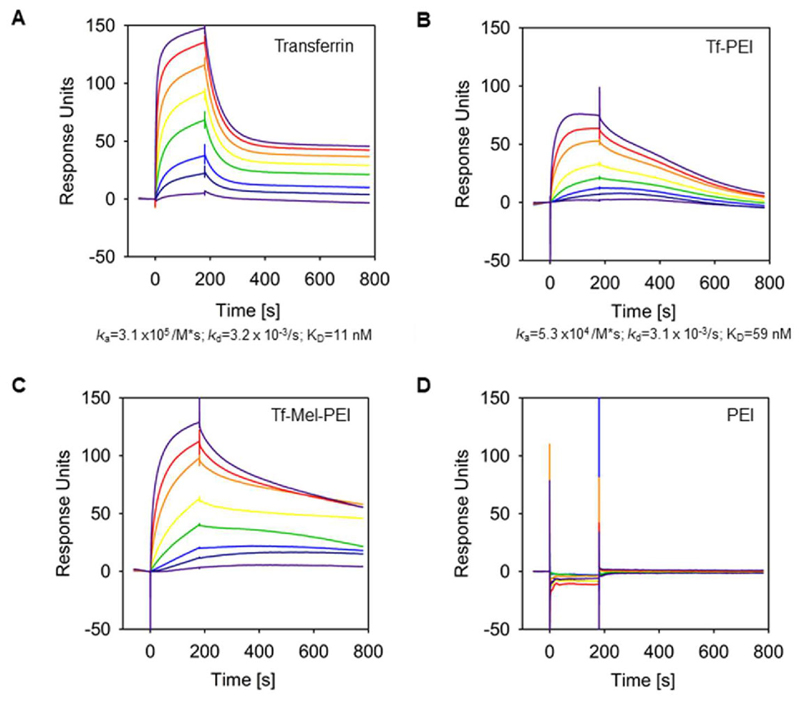
We used Surface Plasmon Resonance (SPR) technique to determine binding kinetics of the interaction between the Tf-PEI and Tf-Mel-PEI conjugates, respectively, to the transferrin receptor (TfR). For that purpose, we used a capturing approach in which His-tagged TfR was captured onto a sensor chip that had been preloaded with anti-His antibodies. Then, increasing concentrations (1–1000 nm) of Tf-PEI and Tf-Mel-PEI polyplexes were injected. After each cycle, TfR-His was regenerated from the chip and recaptured. As positive control, we injected free transferrin in similar concentrations over the chip surface. We clearly detected high affinity binding of transferrin to the TfR with an overall affinity (KD) of 10 nm (Figure 3A). Binding was characterized by an association rate of ka = 3.1 × 105 per M*s and showed a dissociation rate of kd = 3.2 × 10−3 s−1, assuming a 1:1 interaction model. However, the dissociation rate emerged to be bi-phasic: a high dissociation rate was followed by a low dissociation rate, implying stable binding. High affinity binding could also be observed for the Tf-PEI conjugate to the TfR, though the overall affinity decreased nearly sixfold (KD = 59 nm) compared to free transferrin, which was due to a lower association rate of Tf-PEI (ka = 5.3 × 10−4 per M*s) (Figure 3B). Binding of Tf-PEI to the TfR was more stable compared to free transferrin since no first high dissociation phase was observed. Assuming a 1:1 binding stoichiometry the overall dissociation rate of Tf-PEI to the Tf-receptor was comparable to that of free transferrin (kd = 3.1 × 10−3 s−1). Free PEI showed no interaction with the chip surface; however, the Tf-Mel-PEI blend bound the TfR with higher affinity compared to Tf-PEI, showing an overall affinity of 14 nm with an association rate ka of 6.6 × 104 per M*s and an overall dissociation rate kd of 9.2 × 10−4 s−1 (Figure 3C). In summary, for all Tf-conjugates a stable binding to the Tf-receptor could be demonstrated, whereby the binding of the Tf-Mel-PEI blend was the most stable one.
Figure 3.
Binding of Tf-PEI and Tf-Mel-PEI was analyzed by Surface Plasmon Resonance (SPR) spectroscopy. The transferrin receptor (TfR) was captured via its His-tag onto a CM5 sensor chip coated with anti-His antibody, and solutions of 1 nm (light purple), 10 nm (dark blue), 25 nm (blue), 50 nm (green), 100 nm (yellow), 250 nm (orange), 500 nm (red), 1000 nm (dark purple) of A) free transferrin, B) TF-PEI conjugate, C) Tf-Mel-PEI conjugate, and D) unmodified PEI polyplexes, respectively, were passed over the chip. The plots are representatives of four independently performed experiments.
2.5. Cellular Uptake of Polyplexes
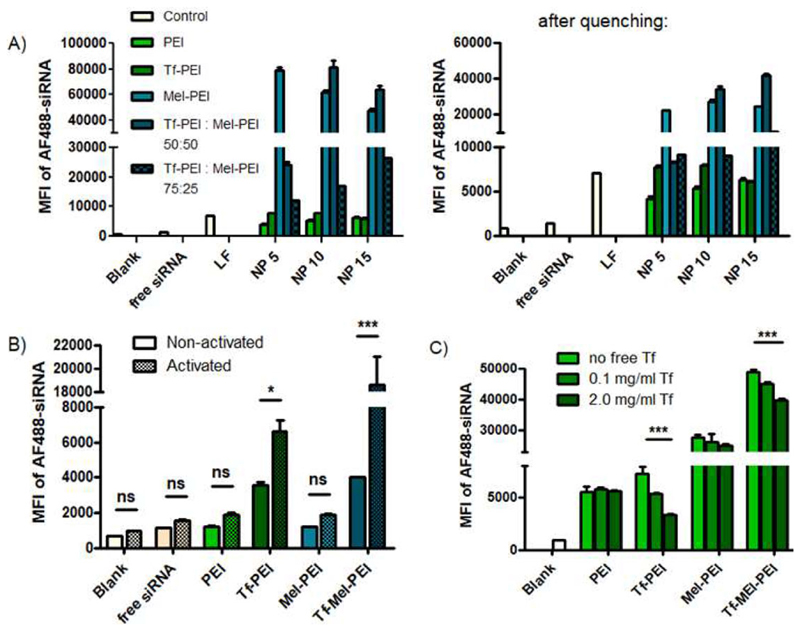
Cellular uptake of Alexa Fluor 488-labeled siRNA (AF488-siRNA) was evaluated by flow cytometry. Figure 4A shows the Median Fluorescence Intensity (MFI) of Jurkat cells transfected with polyplexes composed of AF488-siRNA and different polymer formulations compared to untreated cells and free siRNA as negative controls, and lipofectamine (LF) as positive control. As previously observed,[9] coupling of Tf to PEI improved uptake efficiencies for N/P 5 and 10, leading to comparable results with LF. Addition of melittin to PEI, however, tremendously increased the obtained MFI for all tested N/P ratios. Finally, blending of Tf-PEI and Mel-PEI in a 1:1 ratio (= “Tf-Mel-PEI”), resulted in the most effective uptake for N/P ratios 10 and 15. Enhancing the Tf-PEI portion in the blend to 75% did not further improve uptake, but rather lowered obtained MFI. Trypan blue quenching, to diminish extra-cellular fluorescent signals resulting from siRNA bound but not internalized by cells, eventuated in overall lower MFI values, nonetheless proving the basic correlations true and leaving Tf-Mel-PEI with a more than 5x higher uptake signal than the positive control LF.
Figure 4.
Cellular uptake of polyplexes as measured by flow cytometry and presented as median fluorescence intensity (MFI). A) Uptake in Jurkat cells before (left panel) and after (right panel) trypan blue quenching. B) Uptake in primary CD4+ human T cells. “Blank” represents untreated control cells, “free siRNA” represents cells treated only with free siRNA, “LF” represents cells treated with lipofectamine lipoplexes. (Data points indicate mean ± SD, n = 3; Two-way ANOVA, *p < 0.05; ***p < 0.005). C) Uptake of polyplexes in Jurkat cells with or without increasing concentrations of free Tf in the culture medium. (Data points indicate mean ± SD, n = 3; One-way ANOVA, ***p < 0.005).
Figure 4B illustrates cellular uptake of polyplexes in human primary CD4+ T cells. While in naïve T cells with negligible TfR expression, Tf-PEI and Tf-Mel-PEI polyplexes were already more efficiently taken up than their Tf-free counterparts, after activation of the cells and thereby upregulation of their TfR expression, this enhancement was even more pronounced, implying Tf-related uptake mechanisms. Furthermore, blending of Tf-PEI with Mel-PEI additionally increased the MFI 2.8-fold compared to Tf-PEI alone. As observed for Jurkat cells, trypan blue quenching slightly decreased measured signals, however, did not affect the captured correlations between the formulations (data not shown).
To further manifest Tf-related uptake of Tf containing polyplexes, a Tf competition assay was performed in Jurkat cells as depicted in Figure 4C. MFI values, and therefore uptake, of both Tf-PEI and Tf-Mel-PEI polyplexes were distinctly and gradually reduced by increasing concentrations of free Tf in the culture medium during transfection, indicating a Tf-dependent uptake mechanism. Cellular uptake of non Tf-containing polyplexes composed of PEI and Mel-PEI, however, was not influenced by free Tf concentration.
2.6. GFP Transfection and Knockdown
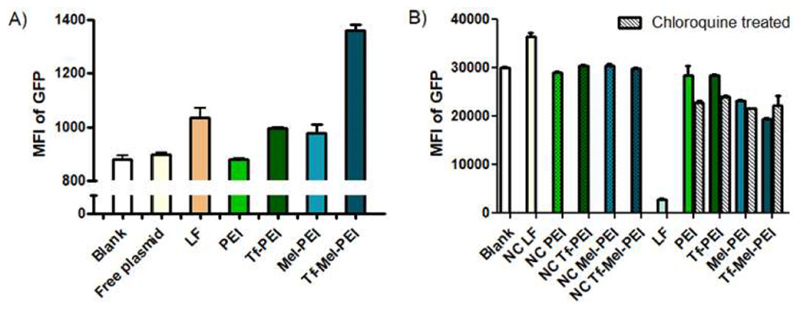
To evaluate transfection efficiencies, Jurkat cells were transfected with polyplexes containing a plasmid expressing green fluorescent protein (GFP) under an active constitutive promoter and resulting fluorescence levels were determined by flow cytometry. As seen in Figure 5A, cells treated with PEI/plasmid polyplexes showed MFI values similar to untreated cells or cells only incubated with free plasmid, indicating no sufficient transfection. Tf-PEI and Mel-PEI, however, demonstrated comparable transfection efficiencies as LF, while the Tf-Mel-PEI blend even surpassed the positive control by approximately 40%.
Figure 5.
A) Transfection of Jurkat cells with a GFP expressing plasmid measured by flow cytometry as median fluorescence intensity (MFI) of GFP. B) GFP knockdown in H1299-mEGFP cells depicted as MFI of GFP after treatment with GFP siRNA or scrambled control siRNA (NC) with or without additional chloroquine treatment. “LF” represents cells treated with lipofectamine lipoplexes. (Data points indicate mean ± SD, n = 3).
Next, GFP protein knockdown was tested in H1299 cells stably expressing EGFP (H1299-mEGFP) using polyplexes encapsulating GFP targeting siRNA (GFP-siRNA) (either with or without simultaneous treatment with the lysosomotropic agent chloroquine) compared to respective particles carrying NC-siRNA as negative controls. Figure 5B depicts that neither PEI nor Tf-PEI polyplexes achieved a silencing effect, while Mel-PEI and Tf-Mel-PEI resulted in significant knockdown. Tf-Mel-PEI was moreover the only tested treatment that could not be further enhanced with chloroquine, implying sufficient endogenous endosomal escape capacity.
2.7. GAPDH Knockdown
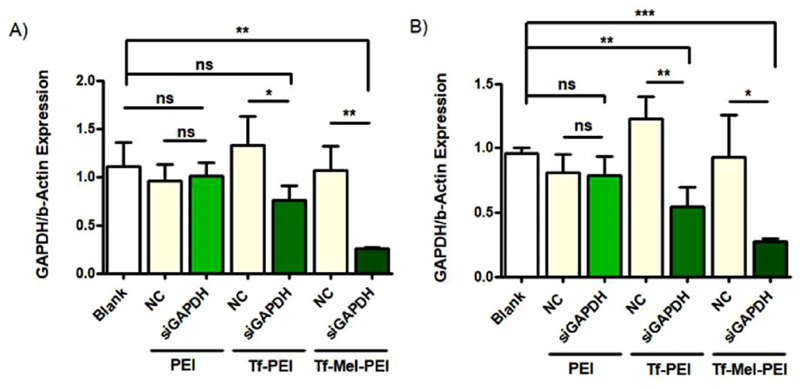
We then investigated whether the high cellular uptake also correlates with corresponding gene silencing. Downregulation was examined using siRNA specifically targeting the house-keeping gene GAPDH (GAPDH-siRNA) followed by quantification of respective mRNA levels via real time PCR (RT-PCR). All tested formulations were also applied containing scrambled non-targeting siRNA (NC-siRNA) as a negative control. Figure 6A shows GAPDH expression normalized to β-actin after treatment of Jurkat cells. While Tf-PEI and Tf-Mel-PEI/GAPDH-siRNA polyplexes were able to significantly silence the gene expression compared to respective NC-siRNA and untreated control groups, no significant difference was obtained between the groups of PEI/GAPDH-siRNA, PEI/NC-siRNA and untreated blank samples. The blending with Mel-PEI further enhanced the silencing effect of Tf-PEI, achieving a knockdown of 70% compared to the negative control.
Figure 6.
GAPDH knockdown in A) Jurkat cells and B) primary CD4+ human T cells after treatment with GAPDH siRNA (siGAPDH; green bars, indicated by polyplex type used for transfection) or scrambled siRNA as negative control (NC). (Data points indicate mean ± SD, n = 3; One-way ANOVA, *p < 0.05; **p < 0.01; ***p < 0.005).
Similar treatment effects were observed in activated human primary CD4+ T cells (Figure 6B). Again, unmodified PEI polyplexes did not result in efficient gene silencing, while Tf-PEI and, particularly, Tf-Mel-PEI achieved considerable knockdown efficiencies of 43% and 76%, compared to negative controls, respectively.
2.8. Endosomal Release
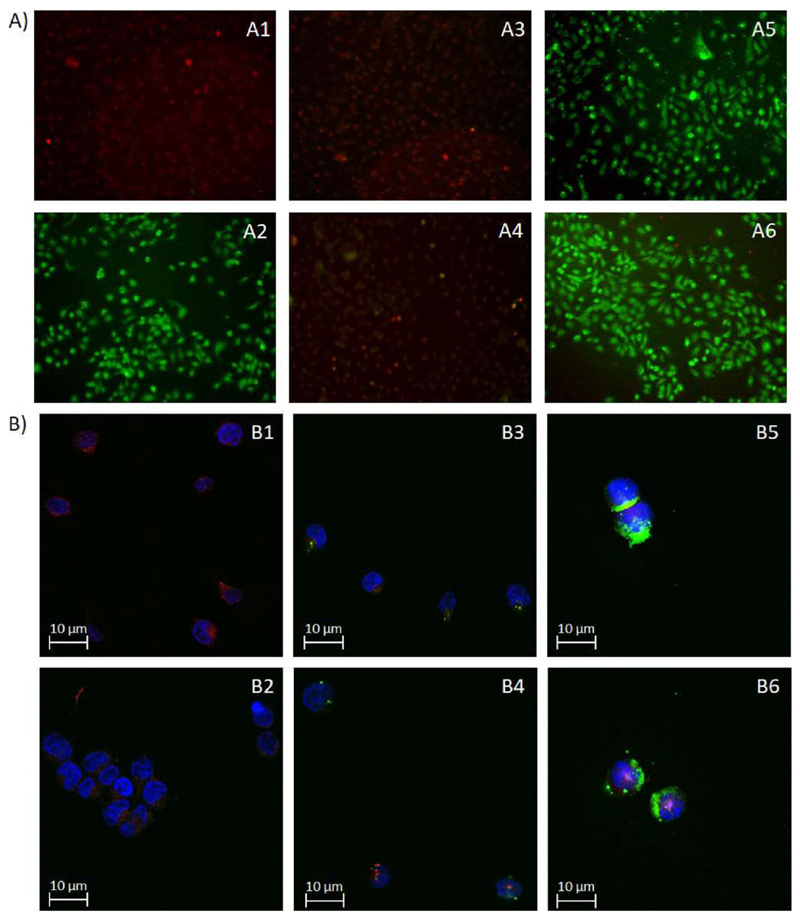
Endosomal disruption in living cells was examined by acridine orange staining. Adherent A549 cells were stained with the cell-permeant nucleic acid binding dye that emits red fluorescence in the endosome, but green fluorescence at cytoplasmic pH,[21] and incubated with polyplexes. A positive control sample was exposed to chloroquine prior to imaging. Figure 7A summarizes the degree of lysosomal disruption as visualized by an increase in green fluorescence captured by fluorescence microscopy. Both melittin containing conjugates Mel-PEI (7A5) and Tf-Mel-PEI (7A6) induced acridine orange release from cytoplasmic endosomal vesicles similarly to chloroquine (7A2), proving efficient endosomal disruption in the treated cells. Polyplexes composed of unmodified PEI (7A3), however, did not visibly influence endosomal integrity, eventuating in red fluorescing cells comparable to the blank sample (7A1). Tf-PEI particles (7A4) only resulted in a vague green signal, suggesting insignificant dye release.
Figure 7.
A) Fluorescence microscopy images of A549 cells after staining with acridine orange and treatment with chloroquine (A2), PEI polyplexes (A3), Tf-PEI polyplexes (A4), Mel-PEI polyplexes (A5), and Tf-Mel-PEI polyplexes (A6). A1 represents untreated cells as blank. B) Confocal images after transfection of Jurkat cells and staining with DAPI (blue, depicting the cell nuclei) and LysoTracker Red DND-99 (red, representing the lysosomes). B1 shows the DAPI and Lysotracker only control, B2-B6 show cells transfected with free siRNA, PEI, Tf-PEI, Mel-PEI, and Tf-Mel-PEI polyplexes, respectively.
To portray endosomal release of polyplexes taken up into cells in a more accurate way, Jurkat cells were transfected with AF488-siRNA and stained with LysoTracker Red DND-99, a fluorescent probe that accumulates in acidic vesicles, as well as DAPI before analyzing them by confocal microscopy. Figure 7B illustrates the different polyplex treatments and resulting effects on cellular uptake and endosomal release of fluorescently labeled siRNA. Blue areas depict the cell nuclei colored with DAPI, while red staining reflects lysosomes, and green staining is accounted to the incorporated siRNA. As Figure 7B2 shows, no green signal can be detected in the free siRNA sample, meaning that no successful uptake can take place without an appropriate delivery system. Uptake with PEI (7B3) and Tf-PEI (7B4), however, results in a punctate distribution of fluorescent siRNA with an appearance similar to the red stained lysosomes, suggesting endosomal entrapment of the delivered cargo. In contrast to this, Mel-PEI treated cells (7B5) show rather cloudy green signals evenly spread over the cells, implying that the siRNA was not only successfully delivered inside the cells, but also able to escape the endosomes and consistently distribute in the cytoplasm. For cells transfected with Tf-Mel-PEI polyplexes (7B6), both the point-shaped and the homogeneously spread signal can be observed, entailing that while parts of the delivered siRNA were still entrapped in the endosomes, a significant proportion was also able to escape and spread over the cytoplasm.
2.9. Toxicity
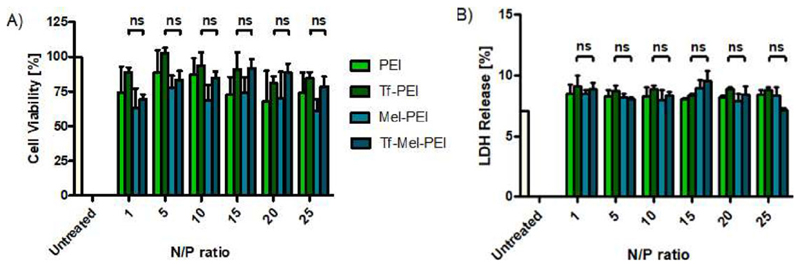
To confirm compatibility with living cells, polyplex cytotoxicity was evaluated with an MTS assay performed in A549 cells that had been incubated with the different formulations at increasing N/P ratios. As exemplified in Figure 8A, Mel-PEI demonstrated the greatest negative influence on cell viability, while unmodified PEI was slightly better tolerated. No significant differences were found for the effects of Tf-PEI and Tf-Mel-PEI for any tested condition, indicating that blending with Mel-PEI has no negative influence on cell tolerability. Notably, at treatment relevant N/P ratios of 10 and 15, both Tf-PEI and Tf-Mel-PEI showed average cell viability rates of around 90%.
Figure 8.
A) Cell viability as measured by MTS assay for polyplexes with different N/P ratios: 1, 5, 10, 15, 20, 25. Untreated cells represent 100% viability. B) Cellular membrane integrity as measured by lactate dehydrogenase (LDH) assay. Cells treated with lysis buffer represent 100% LDH release. (Data points indicate mean ± SD, n = 3; One-way ANOVA, ns, not significant).
For further examination of cell tolerability focusing on membrane integrity, an LDH Assay was performed in A549 cells as presented in Figure 8B. Analogous to the MTS Assay, cells were incubated with polyplexes at different N/P ratios and the resulting LDH release was measured in comparison to untreated blank samples. Again, no significant difference could be found comparing Tf-PEI and Tf-Mel-PEI for any tested N/P ratios. Moreover, no significant increase in LDH levels was observed for any applied treatment compared to untreated cells, suggesting that neither formulation would have any noticeable effect on membrane stability.
3. Discussion and Conclusion
PEI is the most intensively studied cationic polymer in terms of non-viral gene delivery, being widely investigated for potential in vivo applications.[23] Its high density of positive charges mediates the arrangement of siRNA in non-covalent polyplexes via electrostatic interactions.[24] In our previous work, low molecular weight PEI (5 kDa) was used to minimize its toxic effects and coupled to Tf in order to create trojan horse like particles that specifically deliver siRNA to disease-related T cells in the lung.[9] As these activated T cells exhibit a high metabolic activity and hence high iron consumption, Tf receptors are overexpressed in order to enhance the uptake of Tf-coupled iron.[10] This can be exploited as an entry gate for Tf-targeted polyplexes reaching the, otherwise hard to transfect, primary T cells. Despite the critically discussed hypothesis that positively charged polymers such as PEI demonstrate the proton sponge effect upon acidification of endosomal compartments,[25] leading to disruption and release of delivered payload, PEI generally exhibits rather low endosomolytic properties, which is especially true for LMW-PEI. Since it is crucial that sufficient siRNA molecules reach the cytoplasm to be therapeutically relevant, it was aimed to apply an additional endosomal escape mechanism in this work by incorporating melittin into the Tf-PEI delivery system. Melittin is a 26 amino acid peptide that was first purified from the venom of the European honeybee in 1958 and shows a particularly strong lytic activity due to its high affinity to lipid membranes.[15] Although the exact mechanisms have not yet been fully identified, it is currently assumed that the membrane destabilization is taking place in a two-step mechanism that is comparable to the effect of detergents:[14,15] melittin first reacts with negatively charged lipid head groups via electrostatic interactions and binds parallel to the membrane. When a critical peptide concentration is reached, melittin molecules rearrange perpendicular to the membrane surface and form pores, thereby interrupting and destabilizing the membrane bilayer. It was successfully shown that despite these lytic properties, in vivo toxicity of naked melittin can distinctly be attenuated by respective modifications, including sequestration of melittin in particles[26] or reversibly masking its hydrophobic residues.[27]
In this work, melittin was modified with dimethylmaleic anhydride (DMMAn) masking its lytic activity and hence minimizing acute toxicity at neutral pH. Once the polyplexes are taken up into the endosome, the acidic environment triggers cleavage of these protecting groups and recovery of melittin’s membrane destabilizing effects. Initial studies showed diminished lytic activity by irreversibly masking lysines and the terminal amino function.[28] Rozema et al. then reversibly acylated melittin with a dimethylmaleic anhydride,[29] following the role model of viruses such as adenovirus which presents its endosomolytic residues only upon endosomal acidification.[30] Meyer et al. further developed this approach by covalently linking this reversibly masked melittin to polycations delivering DNA.[16] The reversible reaction of maleic anhydrides with the amines of melittin is reverted at endosomal pH, restoring lytic activity[31] to aid subsequent cytoplasmic release of the delivered cargo. We ultimately combined the benefits of coupling both transferrin and melittin to PEI as a gene delivery system by blending the two conjugates Tf-PEI and Mel-PEI yielding one bioresponsive siRNA carrier capable of both T cell targeted and efficient cytoplasmic delivery.
DLS measurements of resulting polyplexes confirmed our previous observations that modification of PEI with the negatively charged hydrophilic glycoprotein Tf eventuates in a considerable reduction of particle sizes.[9] Coupling of PEI with melittin likewise resulted in smaller polyplexes, while the blending of Tf-PEI with Mel-PEI yielded particles with even smaller and more homogenous hydrodynamic diameters below 200 nm, presenting optimal characteristics for efficient pulmonary transfection. The assumed decoration with the soluble negatively charged Tf on the outside of polyplexes is supported by a decrease of positive surface charges for TF-PEI versus PEI as well as for Tf-Mel-PEI versus Mel-PEI. While all tested N/P ratios of Tf-PEI polyplexes held negative zeta potentials, Tf-Mel-PEI blend particles were slightly positively charged for N/P ratios higher than or equal 10. Thus, the shielding of positive charges from PEI by the negatively charged Tf appears to be influenced by the proportion of polymer to siRNA. At lower N/P ratios, more negative charges from transferrin as well as from negatively charged masked melittin seem to be present at the outer regions of the polyplexes resulting in an overall slightly negative zeta potential. At higher N/P ratios with excess polymer, the particle composition changes to more PEI being present at the surface, raising the overall charge to positive values. Interestingly, viruses as well as virus-mimicking particles coated with both positively and negatively charged groups were found to avoid electrostatic mucus adhesion, eventuating in minimized entrapment.[32] The modified surface properties may hence facilitate particle mobility in lung fluids and thereby improve respective in vivo behavior of Tf-Mel-PEI polyplexes.
To investigate the influence of melittin unmasking in acidic environments on polyplex characteristics, DLS and LDA measurements were also conducted at pH 5. As shown in Figure S1A, Supporting Information, zeta potentials of Mel-PEI as well as Tf-Mel-PEI polyplexes overall increase leading to invariably positively charged particles which can be explained by the unmasked amine groups of melittin. Concerning the hydrodynamic diameters, Mel-PEI polyplexes show smaller and more homogeneously distributed sizes at pH 5 (Figure S1B, Supporting Information) compared to pH 7.5. Since the amines of melittin are now unprotected and free for interaction with the negatively charged siRNA, it can be expected that this results in an overall enhanced interaction of polymer and siRNA leading to an even tighter packing. In contrast, hydrodynamic diameters of Tf-Mel-PEI blend particles consistently increase at all tested N/P ratios. As they do no longer display negative charges at any of the tested N/P ratios, it can be expected that the assumed exterior decoration with transferrin leading to shielding and tight condensation does not convey to the acidic environment. It is even conceivable that negative charges of transferrin and positive charges of unmasked melittin now result in aggregation forming distinctly larger particles.
Since siRNA is easily degraded and rapidly excreted upon systemic injection, local application is a promising concept to facilitate efficient delivery. The lung offers advantageous physiological characteristics as a drug target organ with its large surface area, thin and highly vascularized epithelium, and the absence of serum proteins[33] Obstacles on this delivery route, however, can be respiratory mucus and airway surface liquid (surfactant). Cationic carrier systems, in particular, can nonspecifically interact with the negatively charged lung lining fluids, be entrapped, destabilized, and their mobility is decreased.[34] It was shown that modifying positive PEI polyplexes with the negatively charged hydrophilic glycoprotein Tf results in reduced interactions with polyanions and thus enhanced stability.[35] This was also demonstrated in our previous work, wherein Tf-PEI polyplexes exhibited increased stability in artificial lung lining fluids.[9] Here, we examined the stability of all conjugates again, including the Tf-Mel-PEI blend. As shown by the SYBR gold assay, blending with Mel-PEI improved siRNA encapsulation efficiencies for all tested N/P ratios. The partly constrained encapsulation efficiencies of Tf-PEI compared to unmodified PEI polyplexes may be explained by steric hindrance of the relatively large Tf molecules which not only influence the interaction of PEI and siRNA in general, but also capture some of the primary amines in the polymer as potential binding sites for siRNA. By decreasing the Tf portion in the blend in favor of adding melittin molecules differing both in size and surface charge, these destabilizing effects of Tf might be attenuated.
Adjusted experiments in Alveofact and Mucin identified that the Tf-Mel-PEI blend also showed superior stability within the influence of surface active phospholipids and mucus glycoproteins, respectively, implying that addition of melittin has a further stabilizing effect on the polyplexes for successful pulmonary application potentially due to additional positive charges present in melittin.
SPR measurements to quantitatively analyze binding affinities of the polyplexes to TfR confirmed the specific binding of transferrin containing polyplexes, while no binding was observed for unmodified PEI particles. Comparison of binding affinities revealed that Tf-PEI particles bind even more stable than free transferrin, showing an overall comparable dissociation rate. As we have previously demonstrated, multivalent targeted nanoparticles can in general display stronger receptor binding than respective monovalent ligands may be able to achieve under in vitro conditions.[36] The lowered association rate observed here for polyplexes may be caused by steric hindrance of PEI resulting in a reduced accessibility of Tf in the particles compared to the free ligand. Lack of the early high dissociation rate for Tf-PEI compared to free Tf, however, eventuates in an overall enhanced binding stability. Considering the fact that only an adequately stable and sustained receptor binding can result in sufficient downstream signaling and endocytosis, this parameter is extremely relevant for evaluating in vivo performance. Strikingly, Tf-Mel-PEI blend polyplexes presented an even increased binding affinity compared to the original conjugate. It was recently proposed that melittin itself also interacts with TfR,[37] which taken together with evidently improved surface properties in the altered composition of the blend particles could explain this improvement in receptor binding. In any case, it can be expected that the addition of melittin to the conjugate would not interfere with specific binding on TfR of targeted cells.
Cationic materials such as PEI and melittin can non-specifically interact with negatively charged cell membranes triggering adsorptive endocytosis. This non-targeted uptake could potentially interfere with the desired uptake exclusively in activated T cells. In Jurkat cells, an immortalized cell line representative of T cells, the cellular uptake of Tf-Mel-PEI blend particles was remarkably higher compared to all other polyplex types at treatment relevant N/P ratios 10 and 15. Increasing the Tf-PEI portion in the blend to 75% could, however, not improve uptake further, suggesting that the 50:50 blend Tf-Mel-PEI already contains enough Tf for efficient uptake. As continuous cell lines significantly differentiate from primary cells in their membrane composition and endocytosis profiles, we decided to validate these data in primary T cells. In TfR overexpressing activated CD4+ T cells, the distinctly higher uptake of Tf-targeted polyplexes compared to unmodified PEI and Mel-PEI suggests efficient Tf-mediated endocytosis of Tf-PEI and Tf-Mel-PEI. The preferential uptake of Tf-PEI in activated as opposed to naïve T cells with a negligible receptor expression, was even more pronounced after the addition of melittin. The improved binding characteristics of the Tf-Mel-PEI blend demonstrated in the SPR experiments are also reflected in the uptake behavior of the polyplexes. Tf-Mel-PEI blends, therefore, seem to exhibit ideal characteristics for most efficient internalization without losing any of the targeting potential of Tf-PEI. Increasing the Tf-PEI fraction to greater than 50% did not provide any additional targeting benefit and increasing the melittin containing fraction above 50% would presumably decrease the specific binding affinity too much. Therefore, we decided to further pursue the 50:50 blend of Tf-Mel-PEI as the most promising candidate for therapeutic application. Although N/P 15 showed slightly superior uptake efficiencies than N/P 10, an increased polymer excess usually correlates with a higher toxicity, so that N/P 10 was chosen for subsequent knockdown studies.
The specificity of uptake of the Tf-containing polyplexes was confirmed by a Tf competition assay. While uptake of those polyplexes without the targeting ligand, as expected, was not influenced by the presence of free transferrin, the Tf-targeted polyplexes exhibited clearly compromised uptake efficiencies, although the uptake was not blocked completely. We recently described how monovalent ligands cannot fully outcompete binding of multivalent ligands.[36] This concentration-dependent influence of additional free Tf molecules competing for the binding sites at the TfR supports the anticipated hypothesis that both Tf-PEI and Tf-Mel-PEI polyplexes are taken up via TfR mediated endocytosis. Most notably, these results show that the addition of melittin to the original Tf-PEI conjugate does not affect its target specificity. Thus, even the reduced Tf amount of 50% in the blend is apparently sufficient for specific receptor binding, and it can be expected that enough Tf is still located toward the surface of the blend polyplexes, being able to initiate TfR binding.
Although uptake experiments confirmed efficient internalization of siRNA, its subsequent fate and functionality was still unknown. We therefore evaluated whether the polyplexes are also able to deliver a plasmid that would subsequently be expressed by the cells. As expected, Tf-Mel-PEI showed the highest transfection efficiencies, mirroring the uptake results. This improved transfection did also result in enhanced knockdown effects on GFP protein levels, which could not be further amplified by chloroquine treatment for Tf-Mel-PEI, but for all other tested polymers. Thus, the blend polyplexes seem to be the only delivery system being able to induce sufficient uptake and endosomal release of the delivered cargo on its own. However, it has to be noted that the used cell line stably expresses GFP which has a reported half-life of 26 h[38] and in unsynchronized cultures protein turnover rates are varied, which might potentially mask the overall efficiency of the knockdown. Since siRNA effects directly influence mRNA levels, these were then evaluated by qRT-PCR after treatment with the different polyplexes in the Jurkat cell line and in human primary CD4+ T cells. The improved uptake and endosomal escape properties of the Tf-Mel-PEI blend did indeed further enhance gene silencing. This confirms our hypothesis that insufficient endosomal release had in fact been a major hurdle limiting the knockdown potential of Tf-PEI and gives reason to assume that Tf-Mel-PEI can achieve significant gene silencing in vivo.
Based on these findings, it could already be postulated that adding melittin to the polyplexes does in fact improve endosomal escape of delivered siRNA. To verify that melittin-containing polyplexes are actually inducing endosomal disruption, acridine orange staining was performed in living cells. Acridine orange is a cell permeant, lysosomotropic, metachromatic dye with a luminescence wavelength strongly dependent on its concentration and extent of polymerization.[39] Under acidic conditions, such as in endosomes, it becomes protonated, entrapped, and accumulates as dimers, trimers, or oligomers emitting red fluorescence (640 nm). On the other hand, when the endosomes are disrupted, the dye is homogenously distributed as a monomer all over the cytoplasm emitting a green signal (525 nm), meaning that endosomal leakage can be visualized by an increase in green fluorescence. As expected, neither PEI nor Tf-PEI polyplexes resulted in significant dye release, while both melittin-containing polyplex types were able to visually induce acridine orange and therefore siRNA release from endosomal vesicles.
Endosomal release was further verified in Jurkat cells after transfection with fluorescently labeled siRNA via the different polyplexes. Following uptake with PEI or Tf-PEI, siRNAs demonstrated a punctate distribution which correlated and partly colocalized with lysosomal staining, confirming endosomal entrapment. Mel-PEI, on the other hand, showed a homogenous distribution of the siRNA throughout the cytoplasm. Remarkably, Tf-Mel-PEI treated cells depicted both punctate and diffused siRNA distribution, indicating that while a fraction of the siRNA remains within the endosomes, a significant proportion was able to escape, marking a definite improvement to the original Tf-PEI. These results confirmed the favorable effect of melittin on endosomal release of delivered siRNA, resulting in a distinct improvement in cytosolic accumulation and eventual gene silencing. As most endocytic vesicles are rapidly trafficked to the early endosome, as long as the escape does not happen before this fusion, the influence of internalization mechanisms on endosomal escape is not clarified.[12] It is therefore conceivable that besides the mere lytic activity of melittin, also the altered uptake of the Tf-Mel-PEI blends plays a role in this improvement. Beyond confirmation of the endosomolytic properties of Mel-PEI, these results also demonstrate that this lytic activity can be conveyed to the Tf-Mel-PEI blends, while still maintaining efficient Tf-targeting abilities. Albeit in our previous work we showed that the reducible crosslinker SPDP improved the knockdown efficiency of Tf-PEI,[40] only the combination with sufficient endosomolysis really resulted in optimal gene silencing, underlining that both disassembly of the nanocomplex and cytoplasmic delivery are crucial factors for successful cytoplasmic delivery and subsequent interaction with the RNAi machinery.
As destabilization of cellular membranes naturally affects cell viability which can arouse acute toxicity, it is crucial not to generate this effect until the delivery system is undergoing the endo/lysosomal pathway. In the Tf-Mel-PEI blend this is achieved by a reversible pH-responsive shielding of the reactive groups of melittin, whose lytic activity is hence restored once the polyplexes enter acidic compartments. Despite this protection, potential detrimental effects of melittin addition on cell tolerability were examined by MTS and LDH assay, investigating metabolic activity and membrane integrity of treated cells, respectively. Cationic polymers and peptides such as PEI and melittin in general exhibit cell toxicity due to their high density of positive charges which can be shielded by negatively charged Tf. This is portrayed by a distinctly higher metabolic activity of Tf-PEI versus PEI and Tf-Mel-PEI versus Mel-PEI treated cells for all tested N/P ratios. Interestingly, despite the slight positive charges of Tf-Mel-PEI blends at N/P equal and higher than 10, no significant increase in cell toxicity was observed compared to regular Tf-PEI. Regarding membrane integrity, none of the tested polyplexes induced significant LDH release compared to untreated cells, indicating that all polyplexes are well tolerated. These results confirm that masking of melittin efficiently shields its membrane lytic properties where needed, namely in the extracellular space and outside of the endosomes. Altogether these results indicate that the blending of Tf-PEI with Mel-PEI has no negative influences on cell tolerability in vitro. Since in vivo compatibility of Tf-PEI polyplexes was already confirmed in our previous study,[9] it is therefore to be expected that Tf-Mel-PEI blend particles would be equally well tolerated.
In conclusion, the aim of this work was to develop an improved siRNA delivery system to selectively target activated T cells in the lung and mediate efficient endosomal release of the delivered cargo. By adding an endosomolytic agent to our original Tf-PEI conjugate, we did not only specifically improve the cytoplasmic delivery of siRNA, but furthermore distinctly advanced several other important properties. The pH-responsive shielding exploiting endosomal acidification triggers lytic activity only in lysosomes, avoiding general toxicity. Blending of Tf-PEI and Mel-PEI therefore yields a novel versatile delivery system combining the advantageous features of both singular conjugates resulting in efficient and very specific gene silencing in primary activated T cells. Hence, Tf-Mel-PEI offers an auspicious platform for pulmonary siRNA delivery to selectively alter gene expression in disease-related cell subsets in the lung.
4. Experimental Section
Synthesis of Conjugates and Preparation of Polyplexes
Tf-PEI conjugates were prepared as recently described[9] and Mel-PEI conjugation was performed based on a previously described protocol with respective modifications.[16] In detail, 5k PEI (Lupasol G100, BASF, Ludwigshafen, Germany) was dissolved in HEPES buffered saline (HBS) (20 mm HEPES, 150 mm NaCl, pH = 7.4) at a concentration of 5 mg mL−1 and mixed with an excess of (4-polyethylene glycol-N-succinimidyl 3-(2-pyridyldithio) propionate (PEG4-SPDP, 20 mm) (Thermo Fisher Scientific, Waltham, USA) in dry DMSO and stirred overnight. Cysteine modified melittin (Pepmic, Suzhou, China) was dissolved in HBS (3 mg mL−1), a 150 mm solution of dithiothreitol (DTT) was added, and the mixture was shaken for 2 h. Reduced melittin was purified via 2000 molecular weight cutoff (MWCO) centrifugal filters (Sartorius, Göttingen, Germany) with 100 mm HEPES, 125 mm NaOH and mixed with 1 mL of 2,3-Dimethyl-maleic anhydride solution (DMMAn, 1.5 mg mL−1 in EtOH) (Sigma-Aldrich, St. Louis, USA), stirring for 1 h. Mel-DMMAn and PEI-SPDP were purified in separate runs with 2000 MWCO centrifugal filters and 20 mm HEPES, 0.5 m NaCl, 1 m Guanidine hydrochloride (pH = 8) or 3000 MWCO centrifugal filters and HBS, respectively. For the coupling reaction, both precursors were mixed and stirred overnight. Purification and desalination of the final Mel-PEI conjugate were achieved with 10 000 MWCO centrifugal filters and HBS and the concentration of PEI was determined spectrophotometrically at a wavelength of 405 nm via TNBS assay.[41]
For polyplex preparation, PEI, Tf-PEI, Mel-PEI, or blends of both Tf-PEI and Mel-PEI (the term “Tf-Mel-PEI” is used for the 50:50 mixture throughout this article if not stated otherwise) were diluted in either HBS or 5% glucose and defined amounts of siRNA were added in order to obtain specific amine to phosphate (N/P) ratios. The mass of polymer required for 50 pmol siRNA to yield a certain N/P ratio was calculated according to the following equation: m (PEI in pg) = 50 pmol x 43.1 g mol−1 x N/P x 52 (protonable unit of PEI = 43.1 g mol−1, number of nucleotides of 25/27mer siRNA = 52). The formulations were mixed by pipetting and incubated for 20 min before further experiments. Lipofectamine 2000 (LF, Thermo Fisher Scientific) lipoplexes were prepared according to the manufacturer’s protocol.
Particle Characterization
To measure hydrodynamic diameter, PDI and zeta potential, 100 μL of polyplexes with different N/P ratios were prepared in HBS and added into a disposable micro-cuvette (Malvern Instruments, Malvern, UK). As an example, for N/P 10, 1.1 μL of a 1 mg mL−1 polymer solution was added to 48.9 μL HBS and then mixed with 50 μL of a solution containing 50 pmol siRNA in HBS via pipetting. Sizes were determined with a Zetasizer Nano ZS (Malvern Instruments) at 173° backscatter angle running 15 runs three times per sample. Viscosity of 0.88 mPa*s and 1.33 for refractive index was entered for data analysis with the Zetasizer software. Polyplexes were then diluted with 900 μL of nanopure water and transferred to a folded capillary cell (Malvern) to perform three zeta potential measurements for each sample using the same device.
SiRNA Encapsulation and Stability
To determine siRNA condensation efficiencies, polyplexes with 50 pmol siRNA were prepared at different N/P ratios in 5% glucose, distributed in a FluoroNunc 96-well white plate (Thermo Fisher Scientific) and incubated for 20 min. Subsequently, 30 μL of 4x SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific) was added and incubated for 10 min in the dark. SYBR Gold is a fluorescent nucleic acid staining dye that fluoresces only when intercalating with free siRNA. When the siRNA is protected in a coherent polyplex and not accessible to the dye, no fluorescence signal is detected. Measurements were conducted on a FLUOstar Omega (BMG Labtech, Ortenberg, Germany) at an excitation wavelength of 485/20 nm and an emission wavelength of 520/20 nm. Samples with only siRNA were prepared and treated and measured in the same way as the polyplex samples. Fluorescence of free siRNA (N/P = 0) was used as a control for 100% free siRNA.
For evaluating polyplex stability in lung fluids, modified SYBR Gold Assays were performed[42] in the presence of mucin (Sigma-Aldrich) or lung surfactant Alveofact (Lyomark Pharma, Oberhaching, Germany). Therefore, the described protocol was followed with polyplexes at N/P ratio 10, but SYBR Gold was already added after 15 min and incubated for 10 min. Subsequently, 50 μL of a serial dilution of mucin or Alveofact were added to obtain final concentrations of 0, 0.0005, 0.005, 0.05, 0.5, and 0.25 mg mL−1 and incubated for another 20 min before fluorescence measurements. Autofluorescence of mucin was accounted for by measuring free siRNA samples with corresponding concentrations of mucin.
Transferrin Receptor Binding
The binding affinities of prepared polyplexes to the transferrin receptor (TfR-His) were investigated by Surface Plasmon Resonance (SPR) spectroscopy. To eliminate initially observed unspecific interactions of sticky PEI polymer with the sensor chip surface, gradient addition of NaCl to the sample buffers was tested, revealing that an extra 625 mm resulted in minimized interactions. After ensuring that the presence of additional salt does not influence the receptor binding and confirming hydrodynamic diameters in the high salt conditions (data not shown), measurements of all samples were conducted in HBS-N buffer (10 mm HEPES, pH 7.4, 150 mm NaCl) + 625 mm NaCl. Polyplexes were prepared at N/P ratio of 10 in HBS-N + 625 mm NaCl with equivalent concentrations of Tf and/or PEI, respectively. For comparison, free human holo-transferrin (Sigma Aldrich) was dissolved in the same buffer in which the polyplex samples were dispersed, and recombinant human transferrin receptor protein (Abcam, UK) was diluted in HBS-N without additional NaCl.
SPR assays were performed in a Biacore T200 device using Biacore CM5 Series S carboxymethyl dextran sensor chips (GE Healthcare, Freiburg, Germany) that were coated with His-antibodies from the Biacore Hiscapture kit (GE Healthcare). First, the chips were equilibrated with HBSEP buffer (10 mm HEPES pH 7.4; 150 mm NaCl; 3 mm EDTA; 0.005% (v/v) detergent P20) until the dextran matrix was swollen. Then, two of the four flow cells of the sensor chips were activated by injecting a 1:1 mixture of N-ethyl-N-(3-dimethylaminopropyl)carbodiimide hydrochloride and Nhydroxysuccinimide using the standard amine-coupling protocol. Both flow cells were loaded with a final concentration of 50 μg mL−1 of antihistidine antibody in 10 mm acetate pH 4.5 using a contact time of 420 s, so that the surfaces contained densities of approximately 10 000 resonance units (RU). Free binding sites of the flow cells were saturated by injection of 1 m ethanolamine/HCl pH 8.0. Preparation of chip surfaces was carried out at a flow rate of 10 μL min−1. Interaction analyses between the prepared polyplexes and TfR-His were then performed in HBSN buffer + 625 mm NaCl. First, TfR-His (30 nm) was captured onto the second flow cell using a contact time of 60 s at a constant flow rate of 10 μL min−1, followed by a stabilization time of 20 s so that approximately 300–400 RU of TfR-His were captured. Increasing concentrations (1, 10, 25, 50, 100, 2 × 250, 500, and 1000 nm) of the polyplexes were then injected onto both flow cells using a contact time of 180 s each and a final dissociation of 600 s using a flow rate of 30 μL min−1. As control, similar concentrations of PEI were injected onto the chip. After each cycle the chip was regenerated by injection of 10 mm glycine pH 1.5 for 60 s at a flow rate of 30 μL min−1 over both flow cells, which completely removed TfR-His from the surface. All experiments were performed at 25°C. Sensorgrams were recorded using the Biacore T200 Control software 2.0 and analyzed with the Biacore T200 Evaluation software 2.0. The surface of flow cell 1 was used to obtain blank sensorgrams for subtraction of bulk refractive index background. The referenced sensorgrams were then normalized to a baseline of 0. Peaks in the sensorgrams at the beginning and the end of the injections emerged from the runtime difference between the flow cells of each chip.
Cell Culture
Jurkat cells, a human T lymphocyte cell line, were a kind gift from Prof. Heissmeyer (Institute for Immunology, Biomedical Center Munich) and were cultured in RPMI-1640 cell culture medium (Sigma-Aldrich) supplemented with 10 mm HEPES, 1 mm sodium pyruvate, 4500 mg l−1 glucose, 10% (v/v) heat inactivated fetal bovine serum (FBS, Sigma-Aldrich) and 1 x penicillin/streptomycin (Pen/Strep, Sigma-Aldrich). CD4+ human primary T cells were isolated from freshly obtained buffy coats (DRK, Berlin, Germany) via magnetic bead separation with a CD4+ T cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) as described before.[11] Cells were subsequently cultured in the same media as Jurkat cells and, were described, activated with anti-CD3, and anti-CD28 monoclonal antibodies (BS Biosciences, Franklin Lake, USA) at a final concentration of 5 μg mL−1 or 1 μg mL−1, respectively. A549 cells, an adenocarcinomic human alveolar basal epithelial cell line, were grown in RPMI-1640 medium supplemented with 10% FBS and 1 x Pen/Strep. H1299-mEGFP cells (ATCC, Manassas, USA) were cultured in RPMI-1640 medium supplemented with 2 mm L-Glutamine, 10% FBS, 1 x Pen/Strep and 0.2 mg mL−1 geneticin (G418, Sigma-Aldrich). All cells were grown at a humidified atmosphere with 5% CO2 at 37°C.
Cellular Uptake of Polyplexes
For uptake experiments, amine modified siRNA (Integrated DNA Technologies, Coralville, USA) was labeled with succinimidyl ester (NHS) modified AF 488 (Life Technologies, Carlsbad, USA) according to the manufacturer’s protocol resulting in siRNA-AF488, and purified via ethanol precipitation and spin column binding as described before.[43] Polyplexes were prepared with 50 pmol siRNA-488 at different N/P ratios and lipoplexes were prepared with LF as positive control. Per well, 400.000 Jurkat cells or human primary CD4+ T cells were seeded in 96-well plates (Thermo Fisher Scientific) at a concentration of 2 × 106 cells mL−1, transfected and incubated for 24 h. Primary human T cells were transfected directly after isolation from fresh buffy coats for non-activated cells or after 48 h of activation with CD3/CD28 antibodies for activated cells as described above. As negative controls, blank samples were seeded and left untreated and free siRNA samples were transfected with 50 pmol of siRNA diluted in 5% glucose. Cells were harvested and washed three times before resuspension in 400 μL PBS/2 mm EDTA (Sigma-Aldrich). Samples were analyzed using an Attune NxT flow cytometer (Thermo Fisher Scientific) with 488 nm excitation and 530/30 emission filter. All cells were gated according to morphology based on forward/sideward scattering, and 10.000 events were evaluated per sample. For trypan blue quenching, one half of each sample was washed with 0.4% trypan blue to mask any signal originating from extracellular fluorescence.
For Tf competition assays, Jurkat cells were seeded in medium either without free Tf or containing 0.1 mg mL−1 or 2 mg mL−1 free Tf and transfected and analyzed as described above.
GAPDH Gene Knockdown
To test gene silencing abilities of the polyplexes, 500.000 Jurkat cells per sample were seeded in a 96-well plate or 650.000 CD4+ T cells were seeded in a 48-well plate (Thermo Fisher Scientific) and activated for 48 h as described above. Cells were transfected with polyplexes containing 100 pmol siRNA either directed against GAPDH (siGAPDH) or scrambled negative control siRNA (siNC), respectively, at an N/P ratio of 10. After 24 h incubation, cells were harvested, and total RNA was isolated with the PureLink RNA mini kit according to the manufacturer’s protocol with additional DNAse I digestion (Thermo Fisher Scientific). cDNA was synthesized from RNA and amplified with Brilliant III ultra-fast SYBR green QRT-PCR master mix kit (Agilent Technologies, Santa Clara, USA) and QuantiTect primer assays Hs_GAPDH_1_SG and Hs_ACTB_2_SG (Qiagen, Venlo, Netherlands) using a qTOWER real-time PCR thermal cycler (Analytik Jena, Jena, Germany). Cycle threshold (Ct) values were obtained with the qPCRsoft software (Analytik Jena). An untreated blank sample was used to prepare a 1:5 serial dilution with five points as a standard curve and respective Ct values were plotted against the assigned concentration of each point: 1, 0.2, 0.004, 0.0008, and 0.00016. GAPDH gene expression was normalized by corresponding β-Actin expression for each sample.
GFP Transfection and Protein Knockdown
To evaluate transfection efficiencies, 100.000 Jurkat cells were seeded in a 96-well plate and transfected with polyplexes or LF lipoplexes containing 0.75 μg of pCMV-GFP plasmid (PlasmidFactory, Bielefeld, Germany) at N/P 10. As controls, some samples were left untreated as blank and others were only transfected with the free plasmid. All cells were incubated for 48 h before washing and analyzing for the MFI of GFP protein expression with an Attune NxT flow cytometer (Thermo Fisher Scientific) using 488 nm excitation and 530/30 nm bandpass emission filter set. All samples were gated based on morphology with forward/sideward scattering, analyzing a minimum of 10.000 viable cells.
For GFP protein knockdown, 20.000 H1299-mEGFP cells were seeded in a 24-well plate (Thermo Fisher Scientific) 24 h prior to the experiment. Cells were then transfected using polyplexes containing 100 pmol of either siRNA targeting GFP (siGFP) or scrambled negative control siRNA (siNC) and incubated in cell culture medium with or without 100 μm chloroquine (Sigma-Aldrich). An additional volume of 500 μL of fresh medium was added 4 h after transfection, and cells were further incubated for a total of 72 h. Subsequently, all samples were trypsinized, washed, and analyzed by flow cytometry as described above.
Endosomal Release
For endosomal escape studies, 15.000 A549 cells were seeded per well in eight-well chamber slides (Ibidi, Martinsried, Germany) 24 h prior to the experiment. All wells were then stained with acridine orange (Sigma-Aldrich) (0.1 μm in PBS) for 15 min and washed three times with PBS. Cells were resuspended in culture medium, transfected with polyplexes containing siGAPDH at N/P 10 to obtain a final siRNA concentration of 100 nm, and incubated for 24 h. Appropriate wells were treated with 100 μm chloroquine for 15 min as positive controls and, subsequently, all wells were washed and resuspended in 300 μL PBS before analysis with a BZ-8100 (Biozero) fluorescence microscope (Keyence, Osaka, Japan).
For confocal images, 300.000 Jurkat cells were seeded in a 96-well plate and transfected with polyplexes containing AF488-siRNA at N/P 10. After 24 h incubation, cells were harvested, washed with PBS, and attached to shi-fix coverslips (Everest Biotech, Oxfordshire, UK) according to the manufacturer’s protocol in a 24-well plate. Appropriate wells were stained with 100 μm LysoTracker Red DND-99 (Invitrogen) in pre-warmed cell culture medium for 1 h at 37°C, 5% CO2. After washing, cells were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min and washed again. DAPI was added to appropriate wells at a final concentration of 1 μg mL−1 in PBS and incubated for 20 min. All cells were washed again and mounted using FluorSave reagent (Merck Millipore, Billerica, USA) prior to analysis with a SP8 Inverted scanning confocal microscope (Leica Camera, Wetzlar, Germany).
Toxicity
For cell viability assessment via MTS assay, 5.000 A549 cells were seeded per well in phenol red free RPMI-1640 medium (Sigma-Aldrich) in a 96-well plate 24 h prior to the experiment. Cells were transfected with polyplexes containing 5 pmol siNC at different N/P ratios ranging from 1–25 and incubated for 24 h. Untreated cells were analogously incubated as blank controls. Subsequently, 20 μL of CellTiter 96 AQueous One Solution (Promega, Madison, USA) was added to each well and incubated for 4 h at 37°C, 5% CO2 before absorption measurement at 490 nm using a FLUOstar Omega (BMG Labtech, Ortenberg, Germany).
LDH release of treated cells was determined using the CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega) according to the manufacturer’s protocol. For this assay, 8.000 A549 cells were seeded in a 96-well plate 24 h prior to transfection. Cells were treated with polyplexes containing 5 pmol siNC at N/P ratios ranging from 1–25 and incubated for another 24 h. No cell controls were used to represent 0% LDH release, while cells treated with lysis buffer represent 100% LDH release. Untreated cells were cultivated as blank controls. Afterwards, 100 μL of Cyto-Tox-ONE Reagent was added to each well and the plate was shaken for 30 s before 10 min of incubation. Subsequently, 50 μL of stop solution was added to each well, the plate was shaken again for 10 s and fluorescence was recorded at excitation wavelength of 560 nm and emission at 590 nm using a FLUOstar Omega (BMG Labtech).
Statistical Analysis
All results are given as mean value ± standard deviation (SD). All experiments were performed in triplicates. One-way ANOVA and two-way ANOVA with Bonferroni posthoc post-test were performed in GraphPad Prism (GraphPad Software, La Jolla, USA) to calculate p-values at 95% confidence.
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
This work was supported by ERC Starting Grant ERC-2014-StG – 637830 “Novel Asthma Therapy” to O.M. SPR analyses were performed in the Bioanalytics core facility of the LMU Biocenter.
Footnotes
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adtp.201900047
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Rima Kandil, Department of Pharmacy, Pharmaceutical Technology and Biopharmacy, Ludwig-Maximilians-University, Butenandtstraße 5-13, 81337 Munich, Germany.
Dr. Yuran Xie, Department of Pharmaceutical Sciences, Eugene Applebaum College of Pharmacy and Health Sciences, Wayne State University, 259 Mack Ave, Detroit, MI 48201, USA
Prof. Ralf Heermann, Institute for Molecular Physiology, Microbiology and Wine Research, Johannes-Gutenberg-University, Johann-Joachim-Becher-Weg 13, 55128 Mainz, Germany; Biocenter, Department Microbiology, Ludwig-Maximilians-University, Großhaderner Str. 2-4, 82152 Martinsried, Germany
Lorenz Isert, Department of Pharmacy, Pharmaceutical Technology and Biopharmacy, Ludwig-Maximilians-University, Butenandtstraße 5-13, 81337 Munich, Germany.
Prof. Kirsten Jung, Biocenter, Department Microbiology, Ludwig-Maximilians-University, Großhaderner Str. 2-4, 82152 Martinsried, Germany
Dr. Aditi Mehta, Department of Pharmacy, Pharmaceutical Technology and Biopharmacy, Ludwig-Maximilians-University, Butenandtstraße 5-13, 81337 Munich, Germany
Prof. Olivia M. Merkel, Department of Pharmacy, Pharmaceutical Technology and Biopharmacy, Ludwig-Maximilians-University, Butenandtstraße 5-13, 81337 Munich, Germany.
References
- [1].Mocellin S, Provenzano M. J Transl Med. 2004;2:39. doi: 10.1186/1479-5876-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kandil R, Merkel OM. Pharmazie. 2016;71:21. [PubMed] [Google Scholar]
- [3].Merkel OM, Rubinstein I, Kissel T. Adv Drug Delivery Rev. 2014;75:112. doi: 10.1016/j.addr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang M, Thanou M. Pharmacol Res. 2010;62:90. doi: 10.1016/j.phrs.2010.03.005. [DOI] [PubMed] [Google Scholar]
- [5].Durymanov M, Reineke J. Front Pharmacol. 2018;9:971. doi: 10.3389/fphar.2018.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pelaia G, Vatrella A, Maselli R. Nat Rev Drug Discovery. 2012;11:958. doi: 10.1038/nrd3792. [DOI] [PubMed] [Google Scholar]
- [7].Lamaze C, Dujeancourt A, Baba T, Lo CG, Benmerah A, Dautry-Varsat A. Mol Cell. 2001;7:661. doi: 10.1016/s1097-2765(01)00212-x. [DOI] [PubMed] [Google Scholar]
- [8].Prasher DC. Nat Biotechnol. 2006;24:305. [Google Scholar]
- [9].Xie Y, Kim NH, Nadithe V, Schalk D, Thakur A, Kilic A, Lum LG, Bassett DJ, Merkel OM. J Controlled Release. 2016;229:120. doi: 10.1016/j.jconrel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim NH, Nadithe V, Elsayed M, Merkel OM. J Drug Delivery Sci Technol. 2013;23:17. doi: 10.1016/s1773-2247(13)50002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kandil R, Feldmann DP, Xie Y, Merkel OM. Methods Mol Biol – Nanotechnol Nucleic Acid Deliv. 2019;1943:323. doi: 10.1007/978-1-4939-9092-4_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smith SA, Selby LI, Johnston APR, Such GK. Bioconjug Chem. 2018;30:263. doi: 10.1021/acs.bioconjchem.8b00732. [DOI] [PubMed] [Google Scholar]
- [13].Pratt JP, Ravnic DJ, Huss HT, Jiang X, Orozco BS, Mentzer SJ. In Vitro Cell Dev Biol: Anim. 2005;41:349. doi: 10.1007/s11626-005-0007-1. [DOI] [PubMed] [Google Scholar]
- [14].van den Bogaart G, Guzman JV, Mika JT, Poolman B. J Biol Chem. 2008;283:33854. doi: 10.1074/jbc.M805171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hou KK, Pan H, Schlesinger PH, Wickline SA. Biotechnol Adv. 2015;33:931. doi: 10.1016/j.biotechadv.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meyer M, Zintchenko A, Ogris M, Wagner E. J Gene Med. 2007;9:797. doi: 10.1002/jgm.1075. [DOI] [PubMed] [Google Scholar]
- [17].Kloeckner J, Boeckle S, Persson D, Roedl W, Ogris M, Berg K, Wagner E. J Controlled Release. 2006;116:115. doi: 10.1016/j.jconrel.2006.07.002. [DOI] [PubMed] [Google Scholar]
- [18].Ogris M, Carlisle RC, Bettinger T, Seymour LW. J Biol Chem. 2001;276:47550. doi: 10.1074/jbc.M108331200. [DOI] [PubMed] [Google Scholar]
- [19].Legendre JY, Trzeciak A, Bohrmann B, Deuschle U, Kitas E, Supersaxo A. Bioconjugate Chem. 1997;8:57. doi: 10.1021/bc960076d. [DOI] [PubMed] [Google Scholar]
- [20].Raghuraman H, Chattopadhyay A. Biosci Rep. 2007;27:189. doi: 10.1007/s10540-006-9030-z. [DOI] [PubMed] [Google Scholar]
- [21].Eissenberg LG, Goldman WE, Schlesinger PH. J Exp Med. 1993;177:1605. doi: 10.1084/jem.177.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci U S A. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beyerle A, Braun A, Merkel O, Koch F, Kissel T, Stoeger T. J Controlled Release. 2011;151:51. doi: 10.1016/j.jconrel.2010.12.017. [DOI] [PubMed] [Google Scholar]
- [24].Hobel S, Aigner A. Wiley Interdiscip Rev: Nanomed Nanobiotechnol. 2013;5:484. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- [25].Sonawane ND, Szoka FC, Jr, Verkman AS. J Biol Chem. 2003;278:44826. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- [26].Soman NR, Baldwin SL, Hu G, Marsh JN, Lanza GM, Heuser JE, Arbeit JM, Wickline SA, Schlesinger PH. J Clin Invest. 2009;119:2830. doi: 10.1172/JCI38842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meyer M, Dohmen C, Philipp A, Kiener D, Maiwald G, Scheu C, Ogris M, Wagner E. Mol Pharmaceutics. 2009;6:752. doi: 10.1021/mp9000124. [DOI] [PubMed] [Google Scholar]
- [28].Murata M, Nagayama K, Ohnishi S. Biochemistry. 1987;26:4056. doi: 10.1021/bi00387a047. [DOI] [PubMed] [Google Scholar]
- [29].Rozema DB, Ekena K, Lewis DL, Loomis AG, Wolff JA. Bioconjugate Chem. 2003;14:51. doi: 10.1021/bc0255945. [DOI] [PubMed] [Google Scholar]
- [30].Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. J Virol. 2005;79:1992. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Butler PJ, Harris JI, Hartley BS, Leberman R. Biochem J. 1967;103 78P. [PMC free article] [PubMed] [Google Scholar]
- [32].Lai SK, Wang YY, Hanes J. Adv Drug Delivery Rev. 2009;61:158. doi: 10.1016/j.addr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Merkel OM, Zheng M, Debus H, Kissel T. Bioconjugate Chem. 2012;23:3. doi: 10.1021/bc200296q. [DOI] [PubMed] [Google Scholar]
- [34].Sanders N, Rudolph C, Braeckmans K, De Smedt SC, De meester J. Adv Drug Delivery Rev. 2009;61:115. doi: 10.1016/j.addr.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kircheis R, Wightman L, Schreiber A, Robitza B, Rossler V, Kursa M, Wagner E. Gene Ther. 2001;8:28. doi: 10.1038/sj.gt.3301351. [DOI] [PubMed] [Google Scholar]
- [36].Jones SK, Sarkar A, Feldmann DP, Hoffmann P, Merkel OM. Biomaterials. 2017;138:35. doi: 10.1016/j.biomaterials.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kohno M, Horibe T, Ohara K, Ito S, Kawakami K. Chem Biol. 2014;21:1522. doi: 10.1016/j.chembiol.2014.09.008. [DOI] [PubMed] [Google Scholar]
- [38].Corish P, Tyler-Smith C. Protein Eng, Des Sel. 1999;12:1035. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- [39].Zhan X, Tran KK, Wang L, Shen H. Pharm Res. 2015;32:2280. doi: 10.1007/s11095-015-1619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xie Y, Kim NH, Nadithe V, Schalk D, Thakur A, Kilic A, Lum LG, Bassett DJP, Merkel OM. J Controlled Release. 2016;229:120. doi: 10.1016/j.jconrel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Snyder SL, Sobocinski PZ. Anal Biochem. 1975;64:284. doi: 10.1016/0003-2697(75)90431-5. [DOI] [PubMed] [Google Scholar]
- [42].Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Mol Pharmaceutics. 2009;6:1246. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- [43].Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Behe M, Kissel T. Bioconjugate Chem. 2009;20:174. doi: 10.1021/bc800408g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.