Metastatic breast cancer is an extremely complex disease with limited treatment options due to the lack of information about the major characteristics of metastatic disease. There is an urgent need, therefore, to understand the changes in cellular complexity and dynamics that occur during metastatic progression. In the current study, we analyzed the cellular and molecular differences between primary tumors and paired lung metastases using a syngeneic p53-null mammary tumor model of basal-like breast cancer. Distinct subpopulations driven by the Wnt- and/or STAT3 signaling pathways were detected in vivo using a lentiviral Wnt- and STAT3 signaling reporter system. A significant increase in the overlapping populations driven by both the Wnt- and STAT3 signaling pathways was observed in the lung metastases as compared to the primary tumors. Furthermore, the overlapping populations showed a higher metastatic potential relative to the other populations and pharmacological inhibition of both signaling pathways was shown to markedly reduce the metastatic lesions in established lung metastases. An analysis of the unique molecular features of the lung metastases revealed a significant association with immune response signatures. Specifically, Foxp3 gene expression was markedly increased and elevated levels of Foxp3+ Treg cells were detected in close proximity to lung metastases. Collectively, these studies illustrate the importance of analyzing intratumoral heterogeneity, changes in population dynamics and the immune microenvironment during metastatic progression.
INTRODUCTION
Metastasis accounts for the majority of cancer-related mortality and remains a major clinical obstacle for successful cancer management. In particular, triple negative breast cancer (TNBC) metastasis tends to occur within the first 3 to 5 years after diagnosis, and TNBC patients have a shorter duration of overall survival and distant recurrence-free survival1, 2. Treatment decisions for patients with metastasis primarily rely on the biological characteristics of their primary tumors due to an incomplete understanding of factors driving metastatic disease. A better understanding of the molecular and cellular characteristics distinguishing primary tumors and metastases is crucial for developing the most appropriate therapeutic interventions to prevent and treat distant relapse.
Comparative genomic studies have shown that mutation patterns between primary tumors and metastases are similar with no consistent metastasis-specific mutations present other than pre-existing genetic alterations in primary tumors3–8. This close genetic relationship between primary tumors and metastases suggests that they are clonally related, and that metastases may arise from a subset of populations within primary tumors without having additional mutations. Growing evidence supports the existence of a unique population with metastasis-initiating ability, often referred to as metastasis-initiating cells (MICs), that might exist within the primary tumors or evolve throughout the metastatic progression9. Like tumor-initiating cells (TICs), MICs possess stem cell-like properties and additionally exhibit cellular plasticity, which may contribute to tremendous clonal heterogeneity at sites of metastasis10, 11. In addition to stemness properties, MICs also have a unique capability to interact with distant organ microenvironments, critical for successful metastatic colonization12. The cellular origin of putative MICs and their clonal diversity are poorly understood.
Both normal and cancer stem cell-associated pathways appear to be critical nodes to support stemness and cellular plasticity during tumor progression and metastasis13–15. Wnt signaling is one of the fundamental pathways that function in both normal and cancer stem cells in multiple systems16–19. Hyperactivation of Wnt signaling and pathway components is frequently observed in basal-like TNBC20–23. Wnt-driven stemness has also been shown to be associated with increased metastatic capability and colonization in colorectal and breast cancer15, 24. Additionally, the signal transducer and activator of transcription 3 (STAT3) also represents a key modulator in cancer stem cell function in multiple cancers including TNBC25–29. Several studies have shown that STAT3 signaling can influence the epithelial to mesenchymal transition (EMT) program and enhance metastatic ability, through a STAT3-dependent transcriptional network30–34. Additionally, STAT3 signaling is also implicated in the tumor microenvironment through association with stromal and immune cell components to favorably influence tumor progression and metastasis25, 35, 36. Taken together, numerous studies suggest that both the Wnt- and STAT3 pathways play a critical role in tumor onset and metastasis, implying their therapeutic targeting could help to block tumor progression and metastasis.
Despite the important contribution of the Wnt- and STAT3 signaling in tumor progression and metastasis, it remains unclear whether these pathways function in the same cells or different cells within a given tumor, and how the coordination of these pathways is achieved at the molecular and cellular level during tumor progression. Although much less is known about potential collaborative interactions, recent studies have shown a functional cross-talk between the canonical (or non-canonical) Wnt- and STAT3 signaling pathways in tumor initiation and metastasis37–39. In this respect, we investigated the population dynamics derived from the Wnt- and STAT3 signaling pathways in primary tumors and metastases using a lentiviral-based signaling reporter system in conjunction with the basal-like p53-null syngeneic mouse models of TNBC. Herein, we identify distinct subpopulations derived from the Wnt- and/or STAT3 signaling pathways in the basal-like p53-null mammary tumors, and differences in their cellular distribution between primary tumors and their corresponding metastases. Notably, the clonal diversity derived from the Wnt- and/or STAT3 signaling pathways changes during metastatic progression with an increasing overlap observed in populations driven by both signaling pathways in metastatic lesions relative to paired primary tumors. Furthermore, the Wnt- and STAT3 signaling pathways significantly contribute to the progression and maintenance of the lung metastases. In addition, the molecular signature in the lung metastases is highly correlated with immune response signatures, and Foxp3+ Treg cells are significantly enriched in the lung metastatic lesions.
RESULTS
Distinct subpopulations derived from the Wnt- and/or STAT3 signaling pathways exist in the primary tumors of the basal-like p53-null mammary tumor models.
Given the key role of Wnt- and STAT3 signaling pathways in tumor progression and their potential functional crosstalk, we first examined the clonal diversity derived from the Wnt- and/or STAT3 signaling pathways. To this end, we employed a lentiviral fluorescent signaling reporter system in conjunction with a syngeneic, transplantable p53-null mammary tumor model, which allowed us to identify functional signaling pathway-driven populations in both in vivo and ex vivo settings. We performed a co-transduction experiment using lentiviral Wnt- and STAT3 signaling reporters, Wnt-mCherry and STAT3-GFP (Figure 1a). The Wnt reporter has been widely used and validated in multiple systems including p53-null basal-like mammary tumor models23, 40, 41. The STAT3 reporter has been successfully validated and its activity has shown to be positively correlated with the p-STAT3 status in human breast cancer cell lines of xenograft models29. This signaling reporter system allowed us to identify Wnt- and/or STAT3-active cells by monitoring pathway-dependent transcriptional activities based on expression of the fluorescent reporter proteins, mCherry or eGFP. Briefly, we transplanted the entire co-infected populations into the cleared mammary fat pads of syngeneic recipient mice and analyzed the resulting tumors using flow cytometry. Two basal-like p53-null mammary TNBC models, T1 and T2, were used.
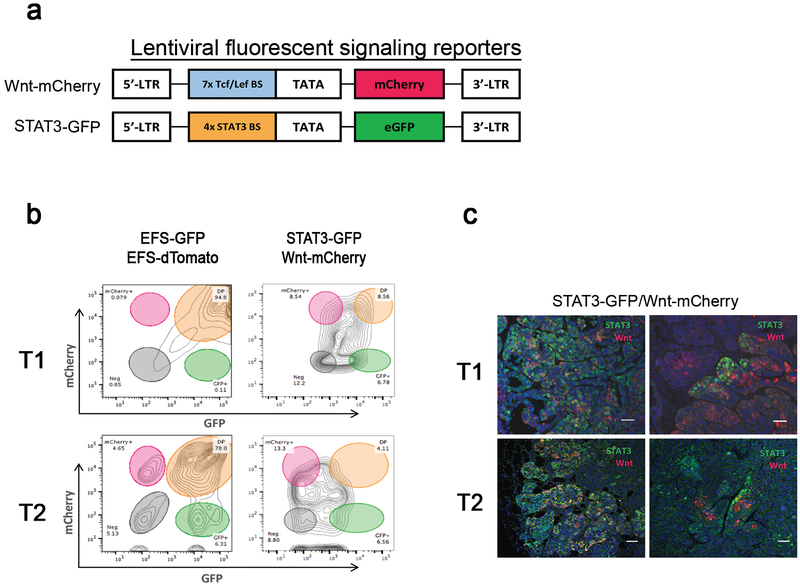
Figure 1. Identification of the distinct subpopulations derived from the Wnt- and/or STAT3 signaling pathways in the basal like p53-null mammary tumor models.
(a) A schematic diagram of the lentiviral fluorescent signaling reporters: Wnt-mCherry and STAT3-GFP. The lentiviral Wnt signaling reporter carries 7x Tcf/Lef binding sites upstream of mCherry, and the lentiviral STAT3 signaling reporter harbors 4x STAT3 binding sites followed by eGFP. (b) Representative fluorescence-activated cell sorting (FACS) plot showing the tumor cells co-infected with the lentiviral constitutive reporters, EFS-GFP and EFS-dTomato (left side) and with the lentiviral Wnt- and STAT3 signaling reporters, Wnt-mCherry and STAT3-GFP (right side) based on the expression of mCherry and GFP. Flow analysis of tumor cells co-infected with the lentiviral constitutive reporters showed that the majority of tumor cells (78–95%) were double positive for GFP and dTomato within the T1 and T2 by flow analysis. Data shown are representative plots from three independent experiments. (c) Representative immunofluorescence images illustrating the Wnt-active (red), STAT3-active (green), and STAT3/Wnt-active (yellow) cells in the T1(upper images) and T2 tumors (lower images). The Wnt- and STAT3 signaling pathways are activated in the same cells (left) or different cells (right). Scale 50 μm. Data shown are representative images from three independent experiments.
As transduction controls, we utilized lentiviral constructs with constitutive EF1α-driven GFP and -dTomato expression, EFS-GFP and EFS-dTomato, to determine levels of co-infection. Based on co-infection level of these constitutive reporters, we expected there should be a similar co-infection of the lentiviral Wnt- and STAT3 signaling reporters since the same multiplicity of infection (MOI) of these lentiviral vectors was used. Flow cytometric analysis of tumor cells harboring the lentiviral constitutive reporters revealed that the majority of tumor cells (approximately 78 to 95%) were double positive for GFP and dTomato, confirming the high co-infection rate in both T1 and T2 (Figure 1b). Interestingly, flow cytometric analysis of tumor cells carrying the lentiviral Wnt- and STAT3 signaling reporters showed a heterogeneous distribution of tumor cells with varying degrees of pathway activities, indicating that heterogeneous populations with respect to Wnt- and STAT3 signaling exist in both T1 and T2 tumors (Figure 1b). Importantly, these analyses clearly demonstrated that there are distinct subpopulations derived from Wnt- and/or STAT3 signaling pathways, including single positive (Wnt-active or STAT3-active) cells as well as double positive (STAT3/Wnt-active) cells (Figure 1b). Moreover, co-immunofluorescence analysis for Wnt (red)- and STAT3 (green) signaling pathways also confirmed the existence of the distinct subpopulations derived from Wnt- and/or STAT3 signaling pathways (Figure 1c). These findings suggest that clonal heterogeneity marked by the Wnt- and/or STAT3 signaling pathways exists in the primary tumors of the basal-like p53-null mammary tumors.
Activation of both Wnt and STAT3 signaling pathways contributes to the lung metastases.
The clonal diversity driven by the Wnt- and/or STAT3 signaling pathways observed in the primary tumor setting prompted our investigation of the involvement of the signaling dynamics and potential heterogeneity during metastatic progression. To evaluate the metastatic lesions in vivo we utilized both primary tumor resection and tail vein injection approaches to model metastasis from the orthotopic site as well as “experimental” metastasis, respectively. While the resection model best recapitulates the clinical scenario, allowing for a multistep metastatic cascade over time, the tail vein approach was used in concert to enable population- and pathway-specific observations with respect to metastatic colonization.
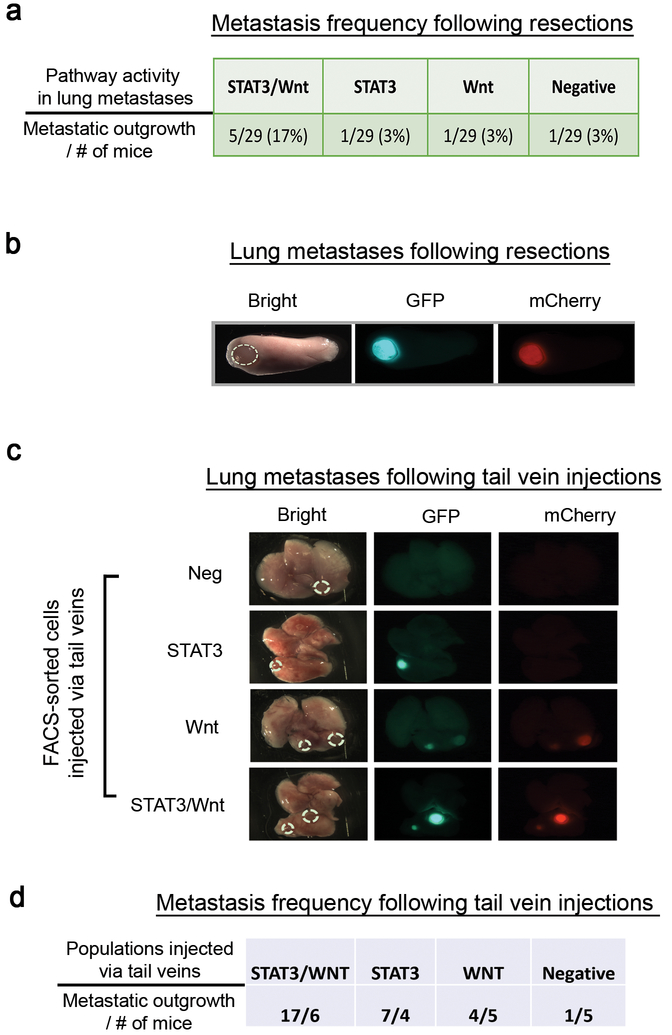
Lung metastases in the T2 model were more frequently observed relative to T1 following resection, thus we primarily utilized T2 for subsequent metastasis studies. Visible lung macrometastases were observed in 8/29 mice after two months following resection of primary tumors, suggesting a low metastatic frequency (Figure 2a), considerably less than other highly aggressive GEM models like MMTV-PyMT42. Interestingly, ex vivo imaging analysis revealed that in the majority of metastatic tumor-bearing mice, 5/8 mice exhibited lung metastases with both Wnt- and STAT3 signaling activity (Figures 2a–b). In addition to surgical resection, we performed experimental tail vein injections to assess lung metastases derived from the distinct FACS-sorted subpopulations. We observed that the intrinsic signaling pathways presented in the primary tumors were maintained during the metastasis (Figure 2c). While Wnt- and STAT3-only cells maintained their designated pathway activity in the lung, lung metastases derived from Wnt-only cells additionally displayed STAT3 signaling activity after colonization, presumably a result of the lung microenvironment (Figure 2c). Moreover, the populations activated by both Wnt-and STAT3 signaling pathways exhibited a higher metastatic frequency relative to the other populations (Figure 2d), suggesting that Wnt- and STAT3 signaling pathways significantly contribute to the metastatic progression in the lungs.
Figure 2. The Wnt and STAT3 signaling pathways contribute to lung metastases.
(a) Metastasis frequency and pathway activity enriched for the lung metastases after resection of the primary tumors. For the resection experiments, we transplanted the entire tumor cell population co-infected with the lentiviral Wnt- and STAT3 signaling reporters, removed the primary tumors when they reached approximately 0.7~0.8cm in diameter, and examined the metastatic dissemination to the lung, a common site for TNBC metastasis. Data shown are the number of mice harboring the lung metastases of each pathway activity from two independent experiments (n=29). The signaling pathway activities were identified by stereomicroscopy. (b) Representative ex vivo fluorescence image depicting the Wnt (red) and STAT3 (green) signaling activities in the lung metastasis (magnification, 0.71x). Both signaling pathways are activated in the lung metastasis. (c) Representative ex vivo images of Lung metastases driven by the distinct FACS-sorted subpopulations via the tail vein injections. 5×104 cells were injected into the lateral tail vein and the lung metastases were assessed two months after the injections by stereomicroscopy (magnification, 0.71x). STAT3/Wnt (n=6), STAT3 (n=4), Wnt (n=5), and Negative (n=5) were analyzed. (d) Metastatic frequency was determined by the number of metastatic nodules based on the stereomicroscopic images. Data shown are the metastatic frequency derived from the FACS-sorted populations from one experiment.
Overlapping populations derived from both Wnt- and STAT3 signaling pathways are significantly enriched in the lung metastases relative to the primary tumors.
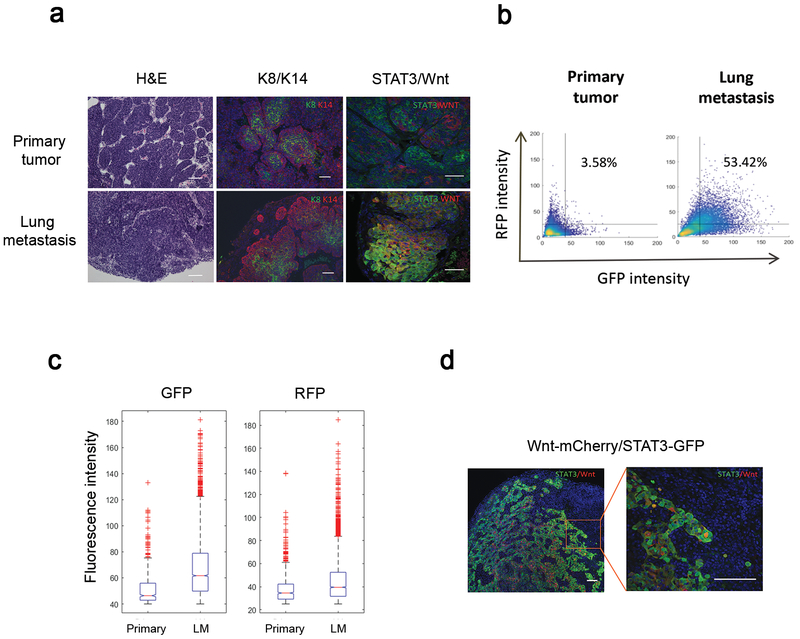
To identify cellular characteristics of the metastatic tumors, we compared various histologic features between primary tumors and matched metastases. Histological analysis revealed that lung metastases exhibited a more disorganized cellular structure and irregular expression of keratins of the basal and luminal lineage, marked by Keratin-14 (K14, red) and −8 (K8, green), respectively, as compared to the primary tumors (Figure 3a), suggesting a considerable change in cellular organization in the metastatic nodules. Co-immunofluorescence staining for Wnt (red)- and STAT3 (green) signaling pathways confirmed the populations with various levels of Wnt- and/or STAT3 signaling activities in the metastases (Figure 3a). Intriguingly, overlapping populations driven by both Wnt- and STAT3 signaling pathways were substantially enriched in lung metastases relative to primary tumors (Figure 3a). This also was confirmed by the quantitative analysis of confocal fluorescence images based on the expression of RFP (Wnt) and GFP (STAT3) (Figure 3b and Supplementary Figures a–c). Although cells co-expressing both reporters appeared to constitute minor populations within the primary tumors, they became the dominant populations in the metastases, indicating the dramatic change in Wnt and STAT3 signaling pathways in the metastatic setting.
Figure 3. The cellular composition derived from the Wnt- and/or STAT3 signaling pathways is significantly changed during the metastatic progression.
(a) Histological comparison between the primary and lung metastases. Representative images of Hematoxylin and Eosin (H&E) and co-immunofluorescence staining for the K8 (luminal, green) and K14 (basal, red) illustrating the changes in the cellular organization during the metastasis. Lung metastases displayed an increased proportion of the overlapping populations derived from both the Wnt- (red) and STAT3 (green) signaling reporters as compared to the primary tumors. Scale 100μm (H&E) and 50μm (immunostaining). (b) Representative quantitative analysis of the heterogeneous populations in the primary and lung metastases based on the RFP- and GFP intensity using the confocal images. Primary tumors (n=5, 36 images) and lung metastases (n=5, 49 images) were analyzed. One-way ANOVA test. The numbers indicate the percentage of double positive cells based on a threshold of 25 for RFP and 40 for GFP. Blinded analysis was performed during quantification. (c) Representative quantitative comparison analysis of the GFP (STAT3)- and RFP (Wnt) intensity between the primary and metastatic tumors based on the confocal images. Box plots showing the distribution of the GFP- and RFP intensity. In the boxplots, the red horizontal lines indicate the median value, and the notches define the 95% confidence interval of the median. Primary tumors (n=5, 36 images) and lung metastases (n=5, 49 images) were analyzed. One-way ANOVA test. Blinded analysis was performed during quantification. The STAT3 signaling pathway is highly enriched in the lung metastases relative to the primary tumors (mean value in primary = 51.6, mean value in lung mets = 67.2), while the Wnt signaling pathway is not significantly changed (mean value in primary = 38, mean value in lung mets = 44.8) (d) Representative co-immunofluorescence staining image for the Wnt (red) and STAT3 (green) signaling pathways revealed that the STAT3 signaling activity is mainly elevated at the leading edge of metastatic tumors. Scale 50μm.
When comparing the fluorescence intensity of RFP- and GFP between the primary and metastatic tumors, we observed a considerable increase in STAT3 signaling activity in the lung metastases relative to the primary tumors, but no significant change in Wnt signaling activity (Figure 3c and Supplementary Figures 2a–b). Moreover, STAT3 signaling activity was preferentially elevated at the leading edge of the metastatic nodule and interface with the surrounding lung tissue as compared to the center (Figure 3d), suggesting an important spatial role of STAT3 signaling in the lung metastases, possibly through the functional and physical interaction with cellular components in the lung. Collectively, our findings demonstrate that the cellular composition derived from Wnt- and/or STAT3 pathways are altered during metastatic progression, with an increased proportion of the overlapping populations present in the lung metastases, implying there may be a selective advantage of co-expression of both signaling pathways.
Inhibition of Wnt- and STAT3 signaling pathways decreased the metastatic outgrowth in already-established lung metastases.
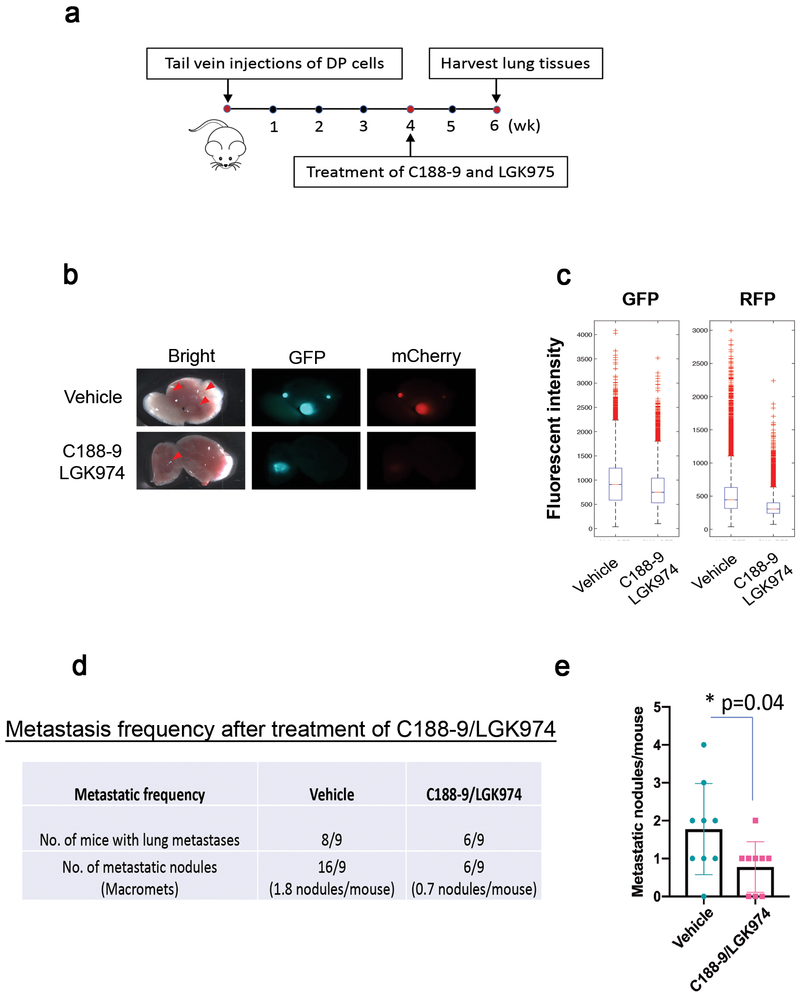
Since the overlapping populations driven by both Wnt- and STAT3 pathways are enriched in the lung metastases with increased activity of both signaling pathways, we examined if inhibition of Wnt and STAT3 pathways was able to inhibit metastatic progression, especially in already-established lung metastases. To address this question, we performed tail vein injections of the double positive Wnt/STAT3 overlapping populations (50,000 cells) into the mice and allowed them to develop the lung metastases for one month (Figure 4a). The mice then were randomized (n=9 per group) and treated with vehicle or C188–9 (50mg/kg)/LGK974 (6mg/kg) for two weeks (Figure 4a). C188–9 is a potent small-molecule that targets the Src-homology (SH) 2 domain of STAT3 and inhibited tumor growth of several cancer models, including breast cancer43–45. LGK974 is a specific PORCN inhibitor that targets Porcupine, a Wnt-specific acyltransferase, inhibits Wnt pathway in vitro and in vivo, and induces tumor regression in a Wnt-driven murine tumor model46, 47. The mice treated with the C188–9 and LGK974 showed decreased expression of both GFP and RFP expression in the lung metastases, indicating an efficient reduction of the activity of both Wnt- and STAT3 pathways in vivo (Figures 4b–c). Additionally, the combinatorial treatment was well-tolerated without significant body weight loss during the course of the treatment (Supplementary Figure 3). Notably, the C188–9/LGK974-treated group showed a significant reduction in the number of metastatic lesions as compared to the vehicle-treated group (Figures 4d–e), suggesting that the inhibition of the Wnt- and STAT3 pathways can reduce the metastatic burden in already-established lung metastases. These findings further suggest that the activation of Wnt- and STAT3 pathways is critical for the progression and maintenance of the lung metastases, implying that blocking Wnt- and STAT3 pathways could be a potential therapeutic option for metastatic breast cancer.
Figure 4. The inhibition of STAT3- and Wnt pathways significantly reduces the metastatic burden in already-established lung metastases.
(a)Schematic treatment strategy. FACS-sorted overlapping populations (50,000 cells) were injected via tail veins. DP (Double Positive for Wnt and STAT3) cells indicate the overlapping populations. The mice were randomized (n=9 per group) and treated with vehicles and C188–9/LGK974 for two weeks (5 days on and 2 days off). The vehicles (60% Labrasol/40% PEG400, 0.5% Methylcellulose/0.5% Tween 80) and C188–9 (50mg/kg)/LGK974(6mg/kg) were given orally. (b) Representative ex vivo fluorescence image depicting the Wnt (red) and STAT3 (green) signaling activities in the lung metastasis (magnification, 0.71x) after C188–9/LGK974 treatment. Metastatic lesions are indicated by red arrows. (c) Quantitative comparison of the GFP (STAT3)- and RFP (Wnt) intensity between the vehicle- and C188–9/LGK974 treated groups based on the confocal images. The fluorescent intensity of the GFP and RFP is substantially reduced by the treatment of C188–9/LGK974. Vehicle group (n=7, 36 images) and C188–9/LGK974 group (n=4, 24 images) were analyzed. One-way ANOVA test (p=5e-191 for GFP and p=0 for RFP). (d) Metastatic frequency after treatment of C188–9/LGK974. The number of metastatic nodules was determined by the stereoscopic images and H&E staining. The C188–9/LGK- treated group (n=9) exhibits a lower metastatic frequency relative to the vehicle-treated group (n=9). Data shown are the metastasis frequency from one independent experiment. (e) Quantitative analysis of the number of the metastatic nodules in the lung metastases of the vehicle- and C188–9/LGK974 treated groups. Data are shown as the mean ± s.e.m. (n=9 per group). Unpaired t test (p=0.04).
Immune response-related signatures are highly correlated with the molecular signatures in the metastatic tumors.
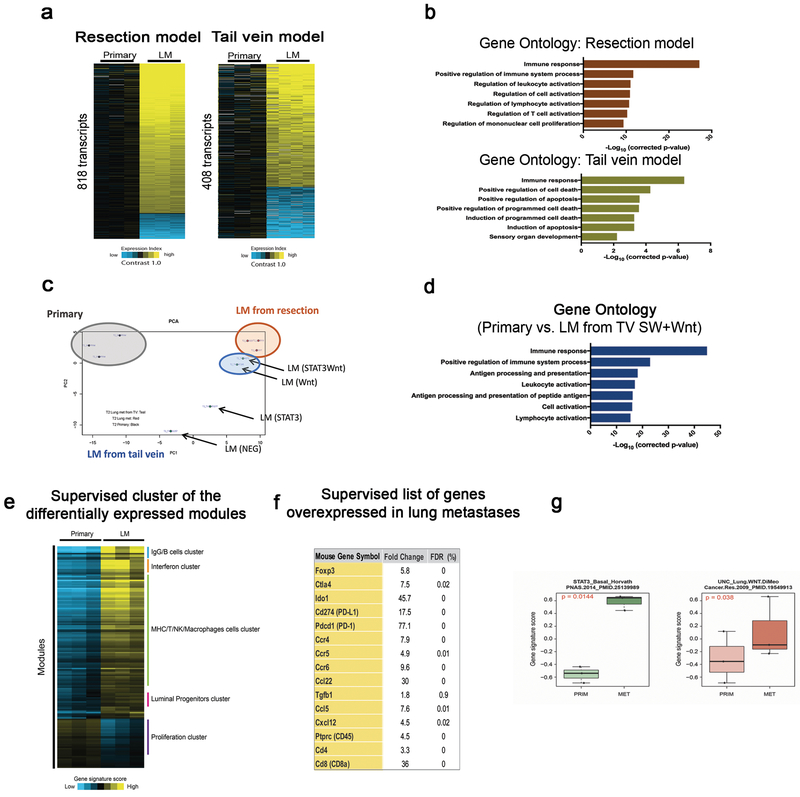
The differences between primary tumors and matched metastases identified by reporter-based approaches led us to further investigate global gene expression programs that could more comprehensively distinguish primary and matched metastases. RNA sequencing analysis from resected primary tumors (n=3) and matched metastases (n=3) identified 818 transcripts differentially expressed (p<0.01, t-test, fold change>1.4), the majority of which were up-regulated in the lung metastases as compared to the primary tumors (Figure 5a, left). Principal component analysis (PCA) revealed that the variation in gene expression patterns between the paired primary (grey circle) and metastatic tumors (red circle) are distinct (Figure 5c). Gene ontology (GO) enrichment analysis of up-regulated genes demonstrated that genes categorized in immune response and immune cell activation were significantly enriched in the lung metastases (Figure 5b, upper plot), suggesting a strong association of immune response signatures within the lung metastases and possible enrichment of cellular components of immune system. Moreover, a supervised analysis derived from the significant modules48, 49 after removing normal lung signatures also identified the immune modules enriched for in the lung metastases, including modules related to the B cell, T cell, NK cell, and macrophage functions (Figure 5e and Supplementary table 1). For instance, Foxp3, Ctla4, Ido1, Cd274, Pdcd1, Tgfb1, Ccr4, Ccr5, Ccr6, Ccl22, Ccl17, Ccl5, Cxcl12, CD45, Cd8a, Cd4 genes were upregulated in the metastatic tumors compared with the primary tumors (Figure 5f and Supplementary Figure 4), indicating a potential enrichment of immune cells in the lung metastases. Consistent with the previous finding that spontaneous lung metastases of PDX models exhibited a more differentiated and less proliferative phenotype50, the supervised analysis identified upregulated luminal progenitor signatures and downregulated proliferation-related signatures in the lung metastases, supporting the idea that a more differentiated basal-like phenotype in the lung metastases is being selected (Figure 5e). Furthermore, Supervised Analysis showed that the STAT3- or Wnt dependent signatures from human basal-like breast cancers were strongly correlated with the gene signatures enriched for in the metastatic tumors relative to the primary tumors (Figure 5g), suggesting that the STAT3- and Wnt pathways may play an important role in metastatic progression of human breast cancer, possibly through physical and functional interactions with immune compartments.
Figure 5. Molecular features in the lung metastases are highly associated with the immune response-related gene signatures in the basal-like p53-null mammary tumor model.
(a) Heat maps showing differentially expressed genes comparing the primary vs. matched metastases (left, n=3) or the primary vs. distinct subpopulation-driven metastases (right, n=4) in the T2 (p<0.01 by t-test, fold change >1.4, using log-transformed data). Bright yellow/blue denotes > 2-fold change in expression from primary group. (b) Gene ontology analysis of upregulated genes in the metastases revealing the enrichment of immune response- and immune cell activation signatures in the resection model (upper graph). The genes involved in immune response as well as cell death program are enriched across all distinct subpopulation-driven metastases (lower graph). (c) PCA illustrating the variations in gene expression patterns of individual samples. Primary tumor (gray, n=3), matched metastases from resections model (red, n=3), and distinct subpopulations-driven metastases via tail vein model (blue, n=4). The top 2000 most variable genes were used in making the plot, with the expression values median centered across sample profiles. (d) Gene ontology enrichment analysis of upregulated genes in the Wnt-active and STAT3/Wnt-active cell-driven metastases. (e) Supervised cluster analysis of the differentially expressed modules selected by Supervised Analysis with an estimated FDR (False Discovery Rate) of 0% and a fold change of at least |1.5|. A total of 129 significant upregulated and 37 downregulated modules were selected and some of them are highlighted in the supervised cluster analysis. (f) Supervised list of genes differentially expressed derived from Supervised Analysis including a large number of cytokines and receptors that implicated in immune function. (g) STAT3- and Wnt dependent gene signatures in human breast cancer. Box plot illustrating the gene signature scores between primary and metastatic tumors of STAT3-associated gene signature specific to basal-like breast tumors and Wnt-associated lung metastasis signature of breast cancer. Both signatures are significantly upregulated in metastasis compared with primary tumors by Supervised Analysis (FDR 0%). Comparisons between two groups were performed by unpaired t-test (two-tailed). Statistically significant values are highlighted in red. Boxplot displays the median value on each bar, showing the lower and upper quartile range of the data and data outliers. The whiskers represent the interquartile range. Each dot represents the value of a single sample.
Gene expression patterns were also derived from the experimental lung metastases originating from the distinct FACS-sorted subpopulations, namely double negative, Wnt-active, STAT3-active, and STAT3/Wnt-active cells via tail vein injection. RNA sequencing analysis from the tail vein injection (n=4, lung metastases derived from the distinct FACS-sorted subpopulations) revealed that 408 transcripts were differentially regulated across distinct subpopulations-driven metastases as compared to the primary tumors (Figure 5a, right). Gene ontology (GO) enrichment analysis of up-regulated genes revealed the enrichment of genes involved in immune response as well as cell death in the distinct subpopulation-driven metastases (Figure 5b, lower plot). Moreover, PCA demonstrated that variations in gene expression were less well correlated among the distinct subpopulation-driven metastases (blue circle) (Figure 5c). Intriguingly, the Wnt-active and STAT3/Wnt-active cell-driven metastases were closely clustered with the matched metastases from the resection model by PCA (Figure 5c). Notably, gene functional annotation enrichment analysis of these metastases compared to the primary tumors revealed the enrichment of similar immune response signatures that were found in the matched metastasis (Figure 5d), suggesting that the metastases driven by the Wnt-active and STAT3/Wnt-active cells possess the similar gene expression patterns with the matched metastases from the resection model. Collectively, these findings imply that the overlapping populations derived from the Wnt- and STAT3 signaling pathways enriched within the lung metastases may play a critical role in immune response in the lungs.
Increased infiltration of Foxp3+ Treg cells is mainly associated with lung metastases.
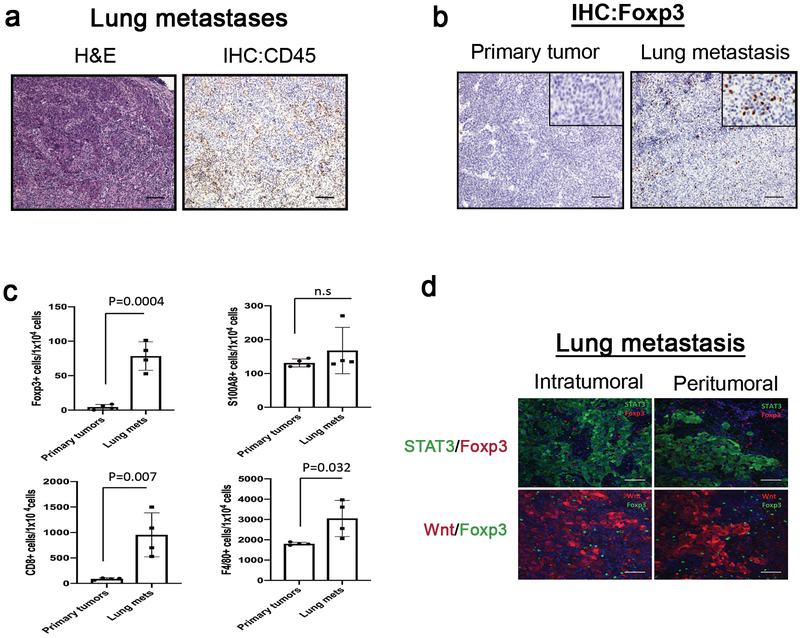
Given the enrichment of immune response signatures in the metastatic site, we next examined the existence of the immune cells in the lung metastases. Consistent with the gene signatures enriched for in the lung metastases, immunohistochemistry analysis revealed that CD45+ immune cells were significantly enriched in the lung metastatic lesions (Figure 6a). A number of immune cells, including Foxp3+ Treg cells, CD8+ T cells, and F4/80+ cells (macrophages) were preferentially enriched in the lung metastases relative to the primary tumors (Figure 6b and Supplementary Figure 5a). The finding of the enrichment of macrophages in the lung metastases also support the previous findings that M2-like macrophages are enriched in various metastatic sites and plays an important role for metastatic progression in human breast cancer51–53. However, S100A8+ cells (neutrophils) were detected at similar levels in both the primary and metastatic lesions (Supplementary Figure 5a). These results were corroborated by quantitative comparison analysis of IHC images (Figure 6c). Particularly, IHC analysis revealed that Foxp3+ Treg cells were mainly enriched in the lung metastases, but not in the primary tumors (Figure 6b and Supplementary Figure 5a), and were not detectable in the normal mammary glands and lung tissues (Supplementary Figure 5b). Moreover, re-transplantation of the lung metastases enriched for both Wnt- and STAT3 pathways into the mammary fat pads gave rise to tumors with both Wnt- and STAT3 activities as well as increased Treg cells (Supplementary Figures 6a–b), suggesting a strong positive correlation between the overlapping populations and Treg recruitment regardless of their location. Furthermore, inhibition of Wnt- and STAT3 pathways significantly reduced the number of Foxp3+ Treg cells and macrophages (F4/80+ cells), demonstrating the metastasis-specific recruitment of Treg cells and macrophages, possibly due to the presence of the Wnt/STAT3 double positive overlapping populations (Figures 4a and Supplementary Figures 7). Additionally, co-immunofluorescence staining for the Foxp3 and metastatic tumors driven by both the Wnt- and STAT3 signaling pathways revealed the cellular distribution of the Foxp3+ Treg cells in the lung metastases, and demonstrated that Foxp3+ Treg cells are located in intra- and peritumoural sites of the lung metastases (Figure 6d). These findings suggest that Foxp3+ Treg cells may play an important role in metastatic progression through functional and physical interactions with the metastatic tumor cells or other immune cells in the lungs.
Figure 6. Immune cell components, particularly Foxp3+ Treg cells, are highly enriched in the metastatic lesions in the basal-like p53 null mammary tumor model.
(a) Representative immunohistochemical analysis in the lung metastases. H&E and IHC with CD45 antibody. Scale 100μm (b) Representative comparison analysis of IHC for Foxp3+ Treg cells in the matched primary and metastatic tumor. Foxp3+ Treg cells are enriched in the lung metastases relative to the primary tumors. Scale 100μm (c) Quantitative measurement of Foxp3+, CD8+, F4/80+, and S100A8+ cells in IHC images. For quantitative measurements, immunohistochemical staining images were scanned by Aperio ScanScope (Leica Biosystems, Buffalo Grove, IL) and processed using a Aperio ImageScope software. Nuclear v9 algorithm was applied for analysis. Data are shown as the mean ± s.e.m. (n=4 per group) from two independent experiments. Unpaired t test. n.s represents not significant. (d) Representative co-immunofluorescent staining images for the Foxp3 (red, upper panel)/STAT3 (green, upper panel) or Foxp3 (green, low panel)/Wnt (red, lower panel) illustrating the cellular distribution of the Foxp3+ Treg cells in the lung metastases. GFP+ (upper panel) or RFP+ (lower panel) cells representing the metastatic tumor cells where the STAT3- and/or Wnt signaling is activated, respectively. Scale 50μm.
DISCUSSION
Intratumoral heterogeneity within a primary tumor is significantly associated with tumor progression and poor clinical outcomes. In particular, the heterogeneous nature of the primary tumor tends to be reflected in the behavior of its corresponding metastasis, which presents a considerable challenge in a clinical setting. However, it is largely unknown how clonal dynamics in the primary tumors influence the development of an invasive phenotype and disease progression throughout the metastatic cascade. In this study, we demonstrated clonal organization and dynamics driven by Wnt- and/or STAT3 signaling pathways in primary tumors and metastases of the basal-like p53-null mouse mammary tumor models. We successfully identified distinct subpopulations driven by Wnt- and/or STAT3 signaling pathways within the primary tumors; negative, Wnt-active, STAT3-active, and STAT3/Wnt-active cells (Figures 1b–c). Tumor cells co-infected with two signaling reporters exhibited various levels of Wnt- and/or STAT3 signaling activities, suggesting the existence of heterogeneous populations. It is possible that the random integration of lentiviral signaling reporters into tumor cells could potentially affect the differential Wnt and STAT3 signaling activities based on the degree of integration rather than amplitude of actual signaling activity. Nonetheless, we note that the majority of tumor cells were co-infected by the Wnt- and STAT3 signaling reporters based on the parallel co-transduction experiments with constitutive reporters (Figure 1b).
Metastases can be derived from the rare or minor subpopulations within the primary tumors8, 54, 55. However, the clonal population dynamics driving metastasis are not fully understood. In our study, we demonstrated the clonal dynamics during the metastases using a resection approach, which enabled us to evaluate the metastases derived from the divergent clones in the primary sites. The quantitative analysis of immunofluorescence images clearly showed that the cellular composition of the distinct subpopulations derived from the Wnt and/or STAT3 signaling pathways is changed within the metastasis, as evidenced by increased overlapping Wnt/STAT3-active populations (Figures 3a–b and Supplementary Figure 1). In addition, activation of STAT3 signaling is further increased in metastases, preferentially at the lesion edge/lung interface (Figure 3d). These findings were further supported by Supervised Gene Expression Analysis, which showed that the STAT3- or Wnt dependent signatures from human basal-like breast cancers were strongly correlated with the gene signatures enriched for in the metastatic tumors relative to the primary tumors (Figure 5g). Moreover, the overlapping populations exhibited a higher metastatic potential in the lungs as compared to the other populations (Figures 2c–d), supporting the possibility that metastatic colonization can occur by expansion of the metastatic-competent cells, possibly Wnt/STAT3-active cells, residing within the primary tumors. Alternatively, differences in the lung microenvironment may stimulate and select for a more metastatic-competent state of disseminated tumor cells. To distinguish these possibilities, it will be necessary to use a lineage tracing approach in addition to specific pathway reporters to determine the cell of origin of the metastases.
Although overlapping populations driven by both Wnt- and STAT3 signaling pathways emerge as a dominant population in lung metastases, little is known about the crosstalk between these signaling pathways in cells. In brain and ovarian cancer, it has been reported that STAT3 signaling is interconnected to Wnt/β-catenin signaling through a Gsk3β/Arid1b and miR-92a/DKK1 regulatory axis for tumor initiation and maintenance, respectively37, 39. A recent study also demonstrated that Frizzled 2 (Fzd2)-mediated non-canonical Wnt signaling pathway can promote EMT and cellular migration through Fyn and STAT3 signaling38. Because our lentiviral Wnt reporter system marks tumor cells in which the canonical Wnt signaling pathway is activated through Tcf/Lef-mediated transcriptional activation, it is possible that STAT3 signaling could collaborate with canonical Wnt signaling in the basal-like p53-null mammary tumors. However, we cannot rule out that the STAT3 signaling is involved in both canonical- and non-canonical Wnt signaling pathways. Previous studies support the idea that Wnt and STAT3 pathways might be coordinately regulated in the same cells by the Akt signaling pathway16, 56–58. Notably, Akt signaling was shown to play an essential role for survival of metastatic tumors in the lung59–61. Therefore, our findings of enriched overlapping populations in metastatic lesions and their significant contribution to metastatic progression (Figure 2d and Figures 4d–e), as well as the potential functional linkage between Wnt and STAT3 signaling, strongly suggests the possibility that the Wnt- and STAT3 signaling might collaborate to promote metastasis. Whether they function in a common pathway and regulate a unique set of genes which are not targets of the individual pathways remains to be determined. Furthermore, given the important contribution of these signaling pathways in metastasis, further studies will be required to determine how the overlapping populations function as a major driver in metastasis.
The key molecular features enriched for the metastatic tumors are mainly associated with the immune response signatures (Figures 5b and 5d–f). Interestingly, genes encoding various cytokines and receptors involved in effector- and regulatory T cell functions were highly upregulated in the lung metastases (Figures 5e–f and Supplementary figure 4), suggesting the existence of strong immune responses, both stimulatory- and suppressive, in the metastatic context. Notably, a number of genes upregulated in the lung metastases were implicated in Treg function (i.e. Foxp3, Ido1, Tgfb1, CCR4, CCR5, CCR6, CCL22, CCL17, CCL5 and CXCL12)62–65. Immunostaining analyses showed that the Foxp3+ Treg cells were primarily enriched in the lung metastases and physically associated with the metastatic tumors (Figures 6b–d and Supplementary figure 5a), suggesting a functional crosstalk between these cells. In cancer, Treg cells have shown to promote tumor progression and metastasis by its suppressive role in innate and adaptive anti-tumor immunity66, 67. Although the precise role of Tregs in metastasis is still emerging, previous studies using spontaneous metastasis murine models of breast cancer suggest that tumor-infiltrating Tregs are associated with metastatic disease progression and are present in metastatic lesions68–70. Moreover, a high density of infiltrating Foxp3+ Tregs in breast tumors are associated with a high risk of relapse and death, especially in ER-positive and basal-like tumors71–73. In this respect, our findings that the enrichment of the overlapping Wnt- and STAT3-active cell populations as well as the preferential infiltration of the Foxp3+ Treg cells in the lung metastases indicate that the Wnt/STAT3-active cells may create immunosuppressive microenvironment through recruiting the Foxp3+ Treg cells to the metastatic sites. A more detailed description of the mechanistic basis by which the overlapping Wnt/Stat3 signaling populations modulate the tumor microenvironment and escape from immune surveillance is clearly warranted.
An important caveat that needs to be considered that previous studies have demonstrated is that the eGFP fluorescent reporter can be immunogenic in some contexts74, 75. Accordingly, we examined if the fluorescent reporters-expressing cells may cause an immunologic response in vivo in our Balb/c model. To address the possibility, we performed the tail vein injections with tumor cells without fluorescent reporters (unlabeled) or with constitutive EF1α-driven GFP/dTomato reporters (EFS), monitored the lung metastases, and analyzed the tumor-infiltrating immune cells, such as total T cells (CD3+ cells) and Treg cells (Foxp3+ cells). Surprisingly, the EFS tumor cells exhibited a lower rate of metastatic colonization (metastatic frequency: 3/12) as compared to the the Wnt/STAT3 tumor cells (metastatic frequency: 17/6, Figure 2d), indicating that the EFS tumor cells are less viable and may elicit more cytotoxic immune responses. These findings also suggest that the Wnt/STAT3 tumor cells are more metastatic and less immunogenic during metastasis, despite a strong expression of GFP- and mCherry. However, the EFS- or Wnt/STAT3 driven lung metastases showed a higher T cell infiltration relative to the unlabeled cells-driven lung metastases, indicating that the expression of fluorescent proteins may cause a T-cell-mediated immune response (Supplementary Figure 8). In contrast, the Foxp3+ Treg cells were preferentially enriched in the Wnt/STAT3-driven lung metastases as compared to the unlabeled- and EFS cells driven lung metastases, further suggesting that the recruitment of Treg cells is correlated with the presence of Wnt/STAT3-double positive cells (Supplementary Figure 8). As an independent experiment, the comparative analyses between the paired primary tumors and lung metastases derived from unlabeled tumor cells following resection revealed that the immune signature of the lung metastasis was still evident (data not shown). This immune signature also largely overlapped with the one we observed in the lung metastases where the Wnt- and STAT3 signaling pathways are enriched. Therefore, this observation suggests that the immune signatures identified are unlikely to be only associated with the expression of the fluorescence proteins.
In conclusion, this study demonstrates the existence of the cellular heterogeneity derived from Wnt- and/or STAT3 signaling pathways, as well as the change in population dynamics during tumor progression. A population enriched for both Wnt- and STAT3 signaling pathways was identified as a dominant and metastatic-competent population in the lung metastases. Furthermore, blocking both signaling pathways efficiently reduced the metastatic burden, supporting an idea that pharmacological inhibition of both pathways may be an important option for management of at least some subsets of basal-like metastatic breast cancer. Lastly, the enrichment of immune response-related gene signatures as well as Foxp3+ Treg cells in the lung metastases illustrates the important functional and physical communication between tumor cells and the immune microenvironment.
MATERIALS AND METHODS
Animals studies
For p53-null mammary tumor model mice were maintained in accordance with the rules of the Guide for the Care and Use of Laboratory Animals of the NIH. Animal studies were performed according to guidelines of our institutional IACUC protocol (AN-504). Wild-type Balb/c female mice (3–4weeks of age for tumor transplantation and 5–6 weeks for tail vein injection) were purchased from ENVIGO (Houston, TX, USA). The T1 and T2 tumors were previously generated and characterized as described76–78. Normal mammary gland and normal lung tissue were obtained from wild-type Balb/c and FVB female mice (6–12 weeks). Animal procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at University of North Carolina.
Single cell preparation of p53-null mammary tumors
p53-null mammary tumors were minced into small pieces using the VIBRATOME and digested with DMEM/F12 containing 1mg/ml Collagenase A (Roche) for 2h at 37 °C. Single cell preparation was performed as described23.
Lentiviral plasmids and lentiviral transduction
The lentiviral Wnt-mCherry reporter plasmid was purchased from Addgene (7TC #24315, Cambridge, MA, USA). The lentiviral STAT3-GFP and EFS-GFP were generated and validated as described29. The lentiviral EFS-dTomato reporter plasmid was generated by replacing the GFP (EFS-GFP) with the dTomato. For lentiviral transduction, single cells (100,000 cells/well) were plated into 24-well ultralow attachment plates in 0.5ml of PMEC media (DMEM/F12 supplemented with 5% FBS, 5μg/ml insulin, 10ng/ml mouse EGF, and AA) and infected with lentiviruses harboring Wnt-mCherry and STAT3-GFP (or EFS-GFP and EFS-dTomato) at MOIs of 30, respectively, for ~16 h.
Fluorescence-activated cell sorting and analysis
Tumor cell preparation for FACS analysis was performed as described79. Cells were analyzed and sorted by BD Aria II sorter (BD Biosciences). FACS data analysis was performed using FlowJo (Tree Star, Ashland, OR, USA).
Immunostaining analysis
For immunohistochemical staining, paraffin-embedded and 10% neutral buffered formalin-fixed tumor tissues were processed according to Immunohistochemistry General Protocol in the Rosen laboratory resources (https://www.bcm.edu/research/labs/jeffrey-rosen/protocols).Tissue sections were stained with the antibodies against CD45 (1:100; 70–0451; TONBO biosciences, San Diego, CA, USA), Foxp3 (1:100; 14–5773; ThermoFisher Scientific), S100A8 (1:800; MAB3059, R&D Systems, Minneapolis, MN, USA), CD3 (1:400, A0452, Dako), CD8 (1:800, 14-0808-82, ThermoFisher Scientific), F4/80 (1:800, 70076, Cell Signaling). For co-immunofluorescence staining, tumor sections were processed according to Immunofluorescence General Protocol in the Rosen laboratory resources. Tissue sections were incubated with the antibodies against against K8 (1:250; TROMA-1; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), K14 (1:400; PRB-155P; Covance, Princeton, NJ, USA), GFP (1:200; 632381, Clontech, Mountain View, CA, USA), RFP (1:400; 600-401-379; ROCKLAND, Limerick, PA, USA), and Foxp3(1:200, 14-5773-80, ThermoFisher Scientific). All tissue sections were subjected to Tris-EDTA antigen retrival for 20 min using microwave heating. Immunofluorescence images were taken by a OLYMPUS BX50 (SPOT Advanced software) or a Nikon A1-R confocal microscope (NIS Elements acquisition software) and analyzed using FIJI Software.
Quantification analysis of confocal fluorescence images
Image segmentation and analysis were performed using custom-made pipelines and scripts in Pipeline Pilot (BioVia) and MatLab (The Mathworks). For each tissue section, cell segmentation was performed by tessellation from the nuclear DAPI signal: nuclear positions were determined from the DAPI channel by grayscale image binarization followed by morphological closing and opening. The resulting objects were used as seeds to conduct tessellation in order to define approximate cell boundaries. Average pixel intensities in the GFP and RFP channels were then measured for each cell. By visual inspection of the images, a value threshold of 40 and 25 was set to call GFP- and RFP- positive cells, respectively. Primary tumors (n=5, 36 images) and lung metastases (n=5, 49 images) were analyzed. The GFP and RFP intensity distributions were visualized with scatter plots. Statistical differences of the means of GFP and RFP intensities between primary and metastatic cells were determined by performing a one-way ANOVA test.
Metastasis and in vivo treatments
For resection experiments, an entire tumor cell population co-infected with the lentiviral Wnt- and STAT3 signaling reporters were engrafted in mammary fat pads of recipient mice. Primary tumors were surgically removed when they reached approximately 0.7~0.8cm in diameter. Metastatic outgrowths were examined two-months following resection and reported by the number of mice harboring the lung metastases. Two independent experiments (n=29). For tail vein injection experiments, the distinct FACS-sorted subpopulations were injected into the lateral tail vein (2 ×104 cells for RNA preparation for sequencing and 5×104 cells for assessment of metastatic frequency. STAT3/Wnt- (n=6), STAT3- (n=4), Wnt- (n=5), and negative cells (n=5) were analyzed.
For pharmacological inhibition of Wnt and STAT3, FACS-sorted overlapping populations (50,000 cells) were introduced via tail vein injections. One month following the injections, the mice were randomized and treated with vehicles (n=9) and C188–9/LGK974 (n=9) for two weeks (5 days on and 2 days off). The vehicles (60% Labrasol/40% PEG400, 0.5% Methylcellulose/0.5% Tween 80) and C188–9 (50mg/kg)/LGK974(6mg/kg) were administered by oral gavage.
RNA preparation and RNA sequencing analysis
RNA was purified from tissue chunks of 3 primary tumors, 7 lung metastases, 8 normal lung and 12 normal mammary gland tissues (Supplementary Table 1) following the manufacturer’s protocol (PicoPure RNA Isolation Kit, ThermoFisher Scientific). Gene expression profiles were generated by mRNA-sequencing using an Illumina HiSeq 2000/2500. Briefly, mRNA libraries were made from total RNA using the Illumina TruSeq mRNA sample preparation kit and sequenced on an Illumina HiSeq 2000/2500 using a 2×50bp configuration with an average of 136 million read pairs per sample. Quality-control-passed reads were aligned to the mouse reference genome (GRCm38) using STAR80, 81. The alignment profile was determined by Picard Tools v1.64 (http://broadinstitute.github.io/picard/). Aligned reads were sorted and indexed using SAMtools and translated to transcriptome coordinates then filtered for indels, large inserts, and zero mapping quality using UBU v1.0 (https://github.com/mozack/ubu). Transcript abundance estimates for each sample were performed using SALMON82. Raw SALMON read counts for all RNAseq samples were normalized to a fixed upper quartile.
Gene expression analysis
Differential expression analysis
Differential gene expression was assessed using two-sided t-test and fold change on log-transformed expression values. For Supervised Analysis, RNA-seq normalized gene counts from primary and metastatic tumors (resection model only) were log2 transformed and filtered selecting only those genes present in the 70% of samples. All samples were median centered. Same process was applied to normal mammary gland and normal lung samples in order to obtain a distinctive gene expression signature of normal lung tissue. To identify significantly overexpressed genes in metastatic compared with primary tumors or normal lung compared with normal mammary gland tissue we used two-class Significance Analysis of Microarrays (SAM)83 with an FDR of 0% using ‘samr’ package in R. Genes commonly overexpressed in normal lung tissue and metastatic tumor were removed from the analysis.
Gene expression signatures
We applied a collection of 593 gene expression signatures, representing multiple biological pathways and cell types, to all primary and metastatic tumors. These signatures were obtained from 453 publications partially summarized previously49, 84 and 41 Gene set enrichment analysis (GSEA) signatures published in the Molecular Signature Database. First, we calculated the gene signature score of each module per tumor and performed Supervised Analysis83 between metastatic and primary tumors (1). Second, we calculated the gene signature score of each module and performed Supervised Analysis in two more comparisons: (2) Normal lung compared with metastatic lung tissue and (3) normal mammary gland compared with metastatic tissue. The second comparison allowed us to identify the distinctive upregulated signatures of normal lung tissue and, the third one, the distinctive downregulated signatures of mammary gland tissue. Finally, the modules derived from comparison (2) and (3) were removed, respectively, from the first upregulated and downregulated significant list (1), in order to minimize the false positive modules generated by normal lung or mammary gland tissue contamination. The significant modules between primary tumors and metastasis were correlated-centered and hierarchically clustered with Cluster 3.085 and viewed with Java Treeview version 1.1.6r486, 87. Some gene signatures were box plotted according to primary and metastatic signature score.
Statistical analysis
Unpaired two-tailed t-tests were performed in GraphPad Prism and/or RStudio version 1.1.383 (http://cran.r-project.org) for statistical analysis of two-group comparisons. Error bars represent standard error of mean (± s.e.m) and p<0.05 was considered statistically significant. The unpaired t test assumes that the two groups have the same variances. For statistical analyses of expression or the intensities of GFP and RFP between primary tumors and lung metastases or between vehicle- and C188–9/LGK974 treated groups, p values were determined by a one-way ANOVA test.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH RO1 CA148761 (to J.M.R. and C.M.P.) and by the Susan G. Komen to SGR (PDF17479425). We would like to thank to Dr. Michael T. Lewis for generously providing us with the STAT3-GFP and EFS-GFP reporters. This project was conducted with technical support from the Cytometry and Cell Sorting Core (NIH CA125123 and RR024574) and the expert assistance of Joel M. Sederstrom, the Integrated Microscopy core (NCI CA125123, NIDDK56338, CPRIT RP150578 and John S. Dunn Gulf Coast Consortium for Chemical Genomics), the Lester and Sue Smith Breast Center Pathology Core, and Human Genome Sequencing Center at Baylor College of Medicine. We thank Yiqun Zhang for technical assistance with the gene expression analyses. We would also like to thank to the IHC laboratory at University of Texas MD Anderson Cancer Center for scanning of IHC images. We specially thank Drs. Michael T. Lewis and Kevin P. Roarty for critically reading and editing the manuscript.
Footnotes
CONFLICT OF INTEREST
C.M.P is an equity stock holder, consultant, and Board of Director Member, of BioClassifier LLC and GeneCentric Diagnostics. C.M.P is also listed an inventor on patent applications on the Breast PAM50 and Lung Cancer Subtyping assays.
Supplementary information is available at Oncogene’s website.
References
- 1.Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast cancer research and treatment 2009; 115: 423–428. [DOI] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research 2007; 13: 4429–4434. [DOI] [PubMed] [Google Scholar]
- 3.Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013; 153: 17–37. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naxerova K, Jain RK. Using tumour phylogenetics to identify the roots of metastasis in humans. Nat Rev Clin Oncol 2015; 12: 258–272. [DOI] [PubMed] [Google Scholar]
- 6.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell 2017; 168: 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010; 467: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010; 467: 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celia-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes & development 2016; 30: 892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proceedings of the National Academy of Sciences of the United States of America 2010; 107: 18115–18120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015; 526: 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nature cell biology 2014; 16: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanharanta S, Massague J. Origins of metastatic traits. Cancer cell 2013; 24: 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob LS, Vanharanta S, Obenauf AC, Pirun M, Viale A, Socci ND et al. Metastatic Competence Can Emerge with Selection of Preexisting Oncogenic Alleles without a Need of New Mutations. Cancer research 2015; 75: 3713–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011; 481: 85–89. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America 2010; 107: 3522–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012; 337: 730–735. [DOI] [PubMed] [Google Scholar]
- 18.Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB et al. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell stem cell 2013; 12: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013; 152: 25–38. [DOI] [PubMed] [Google Scholar]
- 20.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. The American journal of pathology 2010; 176: 2911–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A et al. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol 2011; 24: 209–231. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Knowles E, Zardawi SJ, McNeil CM, Millar EK, Crea P, Musgrove EA et al. Cytoplasmic localization of beta-catenin is a marker of poor outcome in breast cancer patients. Cancer Epidemiol Biomarkers Prev 2010; 19: 301–309. [DOI] [PubMed] [Google Scholar]
- 23.Roarty K, Pfefferle AD, Creighton CJ, Perou CM, Rosen JM. Ror2-mediated alternative Wnt signaling regulates cell fate and adhesion during mammary tumor progression. Oncogene 2017; 36: 5958–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell stem cell 2014; 14: 342–356. [DOI] [PubMed] [Google Scholar]
- 25.Thakur R, Trivedi R, Rastogi N, Singh M, Mishra DP. Inhibition of STAT3, FAK and Src mediated signaling reduces cancer stem cell load, tumorigenic potential and metastasis in breast cancer. Sci Rep 2015; 5: 10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon SH, Kim DK, Cha Y, Jeon I, Song J, Park KS. PI3K/Akt and Stat3 signaling regulated by PTEN control of the cancer stem cell population, proliferation and senescence in a glioblastoma cell line. Int J Oncol 2013; 42: 921–928. [DOI] [PubMed] [Google Scholar]
- 27.Schroeder A, Herrmann A, Cherryholmes G, Kowolik C, Buettner R, Pal S et al. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer research 2014; 74: 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. The Journal of clinical investigation 2011; 121: 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei W, Tweardy DJ, Zhang M, Zhang X, Landua J, Petrovic I et al. STAT3 signaling is activated preferentially in tumor-initiating cells in claudin-low models of human breast cancer. Stem Cells 2014; 32: 2571–2582. [DOI] [PubMed] [Google Scholar]
- 30.Chuang CH, Greenside PG, Rogers ZN, Brady JJ, Yang D, Ma RK et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nature medicine 2017; 23: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rokavec M, Oner MG, Li H, Jackstadt R, Jiang L, Lodygin D et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. The Journal of clinical investigation 2014; 124: 1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranger JJ, Levy DE, Shahalizadeh S, Hallett M, Muller WJ. Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer research 2009; 69: 6823–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MJ, Lim J, Yang Y, Lee MS, Lim JS. N-myc downstream-regulated gene 2 (NDRG2) suppresses the epithelial-mesenchymal transition (EMT) in breast cancer cells via STAT3/Snail signaling. Cancer letters 2014; 354: 33–42. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Guo F, Xu G, Ma J, Shao F. STAT3 cooperates with Twist to mediate epithelial-mesenchymal transition in human hepatocellular carcinoma cells. Oncol Rep 2015; 33: 1872–1882. [DOI] [PubMed] [Google Scholar]
- 35.Jones LM, Broz ML, Ranger JJ, Ozcelik J, Ahn R, Zuo D et al. STAT3 Establishes an Immunosuppressive Microenvironment during the Early Stages of Breast Carcinogenesis to Promote Tumor Growth and Metastasis. Cancer research 2016; 76: 1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oskarsson T, Batlle E, Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell stem cell 2014; 14: 306–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Keng VW, Patmore DM, Kendall JJ, Patel AV, Jousma E et al. Insertional Mutagenesis Identifies a STAT3/Arid1b/beta-catenin Pathway Driving Neurofibroma Initiation. Cell reports 2016; 14: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 2014; 159: 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen MW, Yang ST, Chien MH, Hua KT, Wu CJ, Hsiao SM et al. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer research 2017; 77: 1955–1967. [DOI] [PubMed] [Google Scholar]
- 40.ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell stem cell 2008; 3: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roarty K, Shore AN, Creighton CJ, Rosen JM. Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. J Cell Biol 2015; 208: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res 2006; 8: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X, Kasembeli MM, Jiang X, Tweardy BJ, Tweardy DJ. Chemical probes that competitively and selectively inhibit Stat3 activation. PloS one 2009; 4: e4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood 2011; 117: 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung KH, Yoo W, Stevenson HL, Deshpande D, Shen H, Gagea M et al. Multifunctional Effects of a Small-Molecule STAT3 Inhibitor on NASH and Hepatocellular Carcinoma in Mice. Clinical cancer research : an official journal of the American Association for Cancer Research 2017; 23: 5537–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proceedings of the National Academy of Sciences of the United States of America 2013; 110: 20224–20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America 2013; 110: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics 2011; 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nature genetics 2014; 46: 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bockhorn J, Prat A, Chang YF, Liu X, Huang S, Shang M et al. Differentiation and loss of malignant character of spontaneous pulmonary metastases in patient-derived breast cancer models. Cancer research 2014; 74: 7406–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 2018; 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer 2016; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu LN JL; Onkar S; Joy M; Luedke C; Hall A; Kim R; Pogue-Geile K; Sammons S; Nayyar N; Chukwueke U; Brastianos PK; Anders CK; Soloff AC; Vignali DA; Tseng G; Emens LA; Lucas PC; Blackwell KL; Oesterreich S; Lee AV Metastatic breast cancers have reduced immune cell recruitment but harbor increased macrophages relative to their matched primary tumors. bioRxiv 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010; 464: 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature medicine 2015; 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perry JM, He XC, Sugimura R, Grindley JC, Haug JS, Ding S et al. Cooperation between both Wnt/{beta}-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes & development 2011; 25: 1928–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America 2003; 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer cell 2013; 23: 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Q, Zhang XH, Massague J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer cell 2011; 20: 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine 2013; 19: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature 2004; 430: 1034–1039. [DOI] [PubMed] [Google Scholar]
- 62.Wang RF. CD8+ regulatory T cells, their suppressive mechanisms, and regulation in cancer. Hum Immunol 2008; 69: 811–814. [DOI] [PubMed] [Google Scholar]
- 63.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol 2009; 10: 689–695. [DOI] [PubMed] [Google Scholar]
- 64.Mougiakakos D, Choudhury A, Lladser A, Kiessling R, Johansson CC. Regulatory T cells in cancer. Adv Cancer Res 2010; 107: 57–117. [DOI] [PubMed] [Google Scholar]
- 65.Xu L, Xu W, Qiu S, Xiong S. Enrichment of CCR6+Foxp3+ regulatory T cells in the tumor mass correlates with impaired CD8+ T cell function and poor prognosis of breast cancer. Clin Immunol 2010; 135: 466–475. [DOI] [PubMed] [Google Scholar]
- 66.Halvorsen EC, Mahmoud SM, Bennewith KL. Emerging roles of regulatory T cells in tumour progression and metastasis. Cancer Metastasis Rev 2014; 33: 1025–1041. [DOI] [PubMed] [Google Scholar]
- 67.Pereira LMS, Gomes STM, Ishak R, Vallinoto ACR. Regulatory T Cell and Forkhead Box Protein 3 as Modulators of Immune Homeostasis. Front Immunol 2017; 8: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med 2013; 210: 2435–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dalotto-Moreno T, Croci DO, Cerliani JP, Martinez-Allo VC, Dergan-Dylon S, Mendez-Huergo SP et al. Targeting galectin-1 overcomes breast cancer-associated immunosuppression and prevents metastatic disease. Cancer research 2013; 73: 1107–1117. [DOI] [PubMed] [Google Scholar]
- 70.Lee-Chang C, Bodogai M, Martin-Montalvo A, Wejksza K, Sanghvi M, Moaddel R et al. Inhibition of breast cancer metastasis by resveratrol-mediated inactivation of tumor-evoked regulatory B cells. J Immunol 2013; 191: 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan W, Zhang W, Strasner A, Grivennikov S, Cheng JQ, Hoffman RM et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature 2011; 470: 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol 2006; 24: 5373–5380. [DOI] [PubMed] [Google Scholar]
- 73.Yan M, Jene N, Byrne D, Millar EK, O’Toole SA, McNeil CM et al. Recruitment of regulatory T cells is correlated with hypoxia-induced CXCR4 expression, and is associated with poor prognosis in basal-like breast cancers. Breast Cancer Res 2011; 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G et al. Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments. Stem Cell Rev 2016; 12: 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Follenzi A, Battaglia M, Lombardo A, Annoni A, Roncarolo MG, Naldini L. Targeting lentiviral vector expression to hepatocytes limits transgene-specific immune response and establishes long-term expression of human antihemophilic factor IX in mice. Blood 2004; 103: 3700–3709. [DOI] [PubMed] [Google Scholar]
- 76.Jerry DJ, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ et al. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene 2000; 19: 1052–1058. [DOI] [PubMed] [Google Scholar]
- 77.Medina D, Kittrell FS, Shepard A, Stephens LC, Jiang C, Lu J et al. Biological and genetic properties of the p53 null preneoplastic mammary epithelium. FASEB J 2002; 16: 881–883. [DOI] [PubMed] [Google Scholar]
- 78.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America 2012; 109: 2778–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang CH, Zhang M, Rajapakshe K, Coarfa C, Edwards D, Huang S et al. Mammary Stem Cells and Tumor-Initiating Cells Are More Resistant to Apoptosis and Exhibit Increased DNA Repair Activity in Response to DNA Damage. Stem Cell Reports 2015; 5: 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics 2015; 51: 11 14 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017; 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan C, Prat A, Parker J, Liu Y, Carey LA, Troester MA. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics 2011; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics 2004; 20. [DOI] [PubMed] [Google Scholar]
- 86.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998; 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol 1999; 303: 179–205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.