Nonhuman primates play a crucial role in approximating human biology and many diseases that are difficult, if not impossible, to achieve in other animal models, notably HIV. Current advances in adaptive NK cell research positions us to address fundamental deficiencies in our fight against infection and disease at the earliest moments after infection or substantially earlier in disease progression. We show here that we can identify specific NK cell subpopulations that are modulated following chronic, but not acute, SIV infection. The ability to identify these subsets more precisely will inform therapeutic and vaccine strategies targeting an optimized NK cell response.
KEYWORDS: human immunodeficiency virus, natural killer cells, simian immunodeficiency virus
ABSTRACT
Recently, we and others have shown that natural killer (NK) cells exhibit memory-like recall responses against cytomegalovirus (CMV) and human immunodeficiency/virus simian immunodeficiency virus (HIV/SIV) infections. Although the mechanism(s) have not been fully delineated, several groups have shown that the activating receptor NKG2C is elevated on NK cells in the context of rhesus CMV (rhCMV) or human CMV (hCMV) infections. CD94, which heterodimerizes with NKG2C is also linked to adaptive NK cell responses. Because nonhuman primates (NHP) play a crucial role in modeling HIV (SIV) infections, it is crucial to be able to assess and characterize the NKG2 family in NHP. Unfortunately, it is not possible to detect CD94 using commercially available antibodies in NHP. Our work, a first for NHP, has focused on developing RNA flow cytometry using mRNA transcripts as proxies distinguishing NKG2C from NKG2A. We have expanded the application of this technology and here we show the first characterization of CD94+ (KLRD1+) NK cells in NHP using multiparametric RNA flow cytometry. Peripheral blood mononuclear cells from naive and matched acutely (n = 4) or chronically (n = 12) SIV-infected rhesus macaques were analyzed by flow cytometry using commercially available antibodies, determining expression of transcripts for NKG2A, NKG2C, and CD94 (KLRC1, KLRC2, and KLRD1, respectively) on NK cells using RNA flow cytometry. Our data show that KLRC1+/− KLRC2+ KLRD1+ NK cells decrease following chronic, but not acute, infection with SIV. This approach will allow us to investigate the kinetics of infection and NK memory formation and will further improve our understanding of basic NK cell biology, especially in the context of SIV infection.
IMPORTANCE Nonhuman primates play a crucial role in approximating human biology and many diseases that are difficult, if not impossible, to achieve in other animal models, notably HIV. Current advances in adaptive NK cell research positions us to address fundamental deficiencies in our fight against infection and disease at the earliest moments after infection or substantially earlier in disease progression. We show here that we can identify specific NK cell subpopulations that are modulated following chronic, but not acute, SIV infection. The ability to identify these subsets more precisely will inform therapeutic and vaccine strategies targeting an optimized NK cell response.
INTRODUCTION
While there have been substantial developments in vaccine strategies targeting HIV, most of the effort has been directed toward humoral and T-cell-mediated vaccine design (1). The recent advancement of research on adaptive responses in natural killer (NK) cells provides a novel approach to complement established vaccine modalities and may provide a crucial component to a highly effective HIV vaccine via enhanced innate immune engagement (2). Adaptive NK cells are currently grouped into three major categories, including cytokine-induced, memory-like, and true antigen-specific NK cells (3). Cytokine-induced memory NK cells respond to specific cytokine profiles and seem to retain “memory” of a previous activation (4–6); memory-like NK cells are potent effector cells via antibody-dependent cellular cytotoxicity (ADCC) and may or may not utilize NKG2 family members (7, 8), and true antigen-specific NK cells have been shown to respond to cytomegalovirus (CMV) and adenoviral vaccine vectors in a peptide-specific manner and may utilize the NKG2C-CD94 heterodimer in order to identify its specific target cells (9, 10).
CD94 is a member of the C-type lectin receptor family, which forms heterodimers with several members of the NKG2 family, including NKG2 (NKG2A, NKG2B, NKG2C, and NKG2E) (11). The resulting heterodimer functions to either elicit inhibitory or activating signals through the recruitment of several cytoplasmic adaptor proteins such as SHP-1 or DAP12, which propagate signaling following receptor engagement (12, 13). HLA-E is reported to preferentially bind NKG2A-CD94 heterodimers, though it is also able to engage NKG2C-CD94 heterodimers (14, 15). Through the recruitment of the phosphatase SHP-1, NKG2A-CD94 heterodimers initiate a potent inhibitory signaling response. NKG2C-CD94 heterodimers recruit DAP12 and Syk, which activate several downstream signaling events, leading to enhanced cellular activation (16, 17).
In nonhuman primates (NHP), NK cells have traditionally been classified as CD14− CD20− CD3− NKG2A+ lymphocytes using antibodies that cross-react with both inhibitory NKG2A and activating NKG2C in NHP (18, 19). Consequently, we and others have adopted the nomenclature to define conventional NHP NK cells as CD14− CD20− CD3− NKG2AC+. To distinguish between NK cells that are truly NKG2C+ or NKG2A+, we helped develop RNA flow cytometry methods that use probe sets to specifically detect the gene transcripts of NKG2A (KLRC1) and NKG2C (KLRC2) (20) (Table 1). Our group has shown that RNA flow cytometry is a valid means of identifying members of the NKG2 family, NKG2A (CD159a) and NKG2C (CD159c), in rhesus macaques in order to overcome the reagent limitation problem in NHP (20). This approach was necessary since, prior to our work, the NHP field was unable to distinguish between the activating receptor NKG2C and its inhibitory counterpart, NKG2A, using commercially available antibodies. Similarly, there are currently no commercially available antibodies that can identify CD94 in macaques. In human studies, CD94 has been linked to improved antiviral responses (21, 22) and is also associated with adaptive NK cell responses, likely because of its physical interaction/formation of a heterodimer with NKG2C. CD94 expression on NK cells has also been shown to directly correlate with plasma HIV viremia (23). Despite these findings, CD94 may be dispensable for NK cell development and functional state, as has been shown in mice (24). However, though NKG2C expression alone does not define adaptive NK cell responses in humans (8, 25, 26), CD94+ NKG2C+ NK cells have been shown to expand in the context of CMV infection (27, 28) and modulate peptide-specific responses (10, 22). Further, recent findings suggest that NK cell expression of the CD94 gene transcript, KLRD1 (Table 1), negatively correlates with susceptibility to infection with influenza virus (29). In this study, we applied RNA flow technology to identify and characterize CD94+ NK cells by assessing expression of its transcript, KLRD1, in NK cells. In combination with our previous assays for KLRC1 and KLRC2, this is the first time that this has been possible to monitor coexpression of the heterodimeric components in NHP.
TABLE 1.
Key terms for gene products and proteins
| Gene product or protein | Description |
|---|---|
| KLRC1 | Gene encoding NKG2A protein |
| KLRC2 | Gene encoding NKG2C protein |
| KLRD1 | Gene encoding CD94 protein |
| NKG2AC+ | Rhesus macaque cell populations positively identified using mouse anti-human NKG2A (clone Z199), which cross-reacts with NKG2A and NKG2C in rhesus macaques |
RESULTS AND DISCUSSION
Frequency of KLRD1 expression is reduced in KLRC1+/− KLRC2+ NK subpopulations in chronic SIV infection.
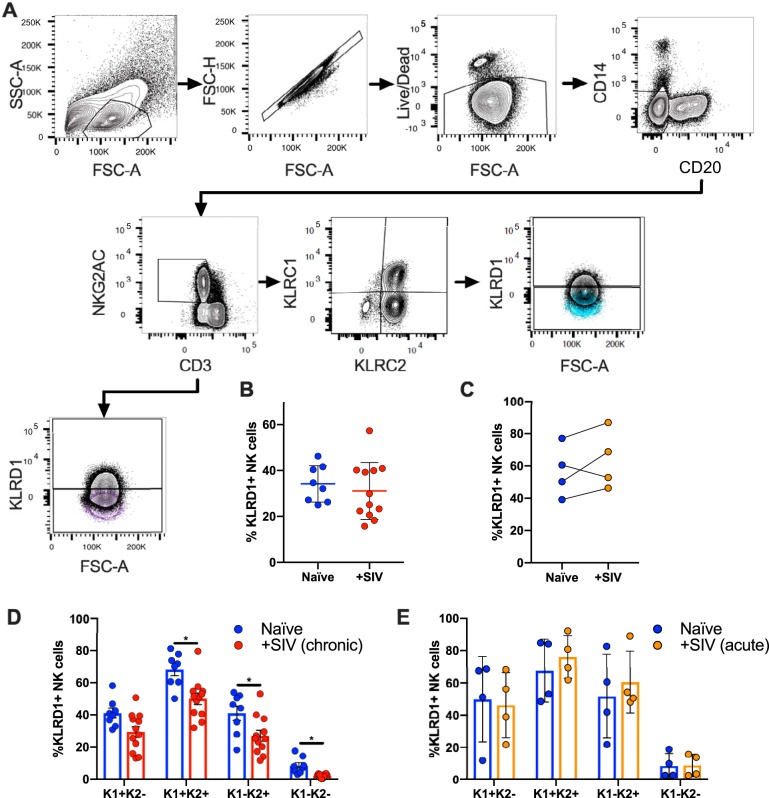
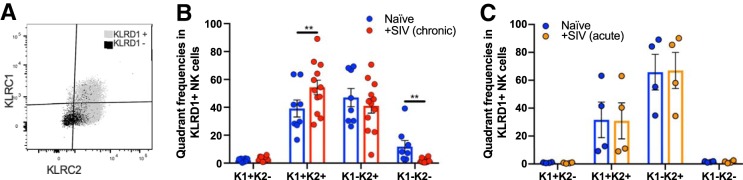
Identification of memory-like NK cells in NHP has been difficult, in part, due to limited cross-reactive antibodies that do not distinguish NKG2A from NKG2C, or fail to identify CD94 in NHP. Since the currently commercially available anti-NKG2A antibody clones recognize both NKG2A and NKG2C, for this paper we consider cell populations as NKG2AC+ (Table 1). Using RNA flow cytometry, we were able to identify multiple subpopulations in classically defined CD3− NKG2AC+ rhesus macaque NK cells by detecting transcript expression of KLRD1 (CD94), KLRC1 (NKG2A), and KLRC2 (NKG2C) (Fig. 1A). This was achieve via specific probe sets that utilized branched DNA technology designed for individual gene targets (20). SIV infection does not seem to alter NK cell frequencies as defined by CD3− NKG2AC+ KLRD1+ in samples from chronic (Fig. 1B) or acute (Fig. 1C) SIV-infected animals. However, we observed a significant reduction in the frequencies of KLRD1+ NK cells in CD3− NKG2AC+ KLRC1+ KLRC2+, CD3− NKG2AC+ KLRC1− KLRC2+, and CD3− NKG2AC+ KLRC1− KLRC2− NK subpopulations in chronic (Fig. 1D) but not acute (Fig. 1E) SIV-infected animals. The reduced frequencies of NK cells expressing KLRD1 within several NK cell subpopulations following chronic SIV infection may attenuate their ability to respond to stimuli via NKG2 receptors that require CD94 coexpression and/or dimerization. Interestingly, when looking at KLRD1+ NK cells, we observed an increased frequency of the CD3− NKG2AC+ KLRD1+ KLRC1+ KLRC2+ subset following chronic (but not acute) infection with SIV (Fig. 2 and 3). This population may represent a less mature state or disparate activation threshold as defined by the presence of NKG2A (KLRC1) (30, 31), but this is still unclear in NHP. Previous work in NHP by LaBonte et al. using semiquantitative PCR has shown decreased levels of NKG2C transcripts, as well as increased levels of NKG2A transcripts, following infection with SIV in peripheral blood mononuclear cells (PBMCs) (18). Perhaps the decreased frequencies of KLRD1+ NK cells, coupled with the increased frequency of KLRD1+ KLRC1+ KLRC2+ NK subpopulations, indicate a mechanism through which the NK cell population loses functionality during SIV/HIV infection (32, 33).
FIG 1.
Gating strategy showing identification of KLRD1+ NK cells. (A) NK cells were defined as CD20− CD14− CD3− NKG2AC+. This population was further subdivided into either KLRD1+ and KLRD1− populations (a representative FMO control is shown in purple) or first subdivided into KLRC1 and KLRC2 and then into KLRD1+ and KLRD1− populations (a representative FMO control is shown in blue). (B) Plots showing matched KLRD1+ NK cells in NKG2AC+ NK cells from naive and chronically (B) or acutely (C) SIV-infected rhesus macaques. Quantification of matched naive and chronically SIV-infected (D) or acute-infected (E) cohorts showing distribution of KLRD1+ NK cells after being subdivided into KLRC1+/− KLRC2+/− NK cell subsets. Naive samples are shown in blue, chronic SIV samples are shown in red, and acute SIV samples are shown in orange. KLRC1 was represented as K1, KLRC2 is represented as K2, and quadrant populations are represented as combinations of K1 and K2. A Wilcoxon test was used to compare different quadrant populations (*, P < 0.05).
FIG 2.
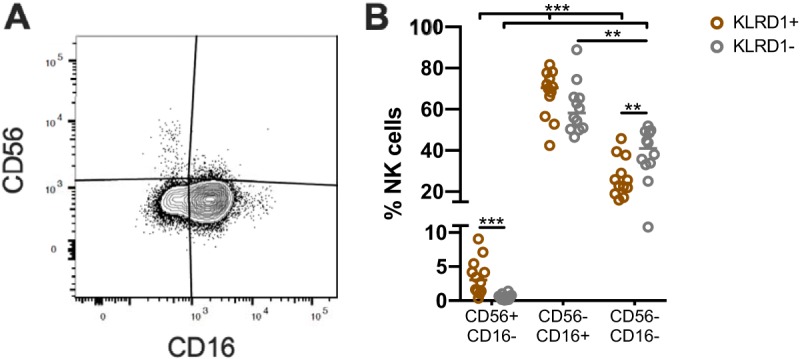
KLRD1+/− cells exhibit phenotypic differences. (A) Representative gating strategies showing the identification of CD56+/− CD16+/− NK cells in CD20− CD14− CD3− NKG2AC+ KLRD1+/− NK cells (as in Fig. 1B) from chronically SIV-infected rhesus macaques (n = 12). The data are quantified in panel B. A Wilcoxon test was used to compare different quadrant populations (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
FIG 3.
The frequency of KLRD1+ KLRC1+ KLRC2+ NK cells is elevated in chronic, but not acute, infection with SIV. (A) Representative gating strategy showing superimposed distribution of KLRC1 and KLRC2 in NKG2AC+ KLRD1+ NK cells (black) and NKG2AC+ KLRD1− NK cells (gray). (B and C) Graphs summarize the frequencies of KLRC1+/− KLRC2+/− populations from matched naive and SIV chronically infected (n = 12) (B) or acutely infected (n = 4) (C) rhesus macaques. Samples from naive animals are shown in blue, samples from chronic SIV-infected animals are shown in red, and samples from acute SIV-infected animals are shown in orange. KLRC1 was represented as K1, KLRC2 is represented as K2, and quadrant populations are represented as combinations of K1 and K2. A Wilcoxon test was used to compare different quadrant populations (*, P < 0.05; **, P < 0.01).
Distributions of CD56+/− CD16+/− NK cell populations are associated with KLRD1 expression.
NK cells in NHP can be broadly characterized by the expression of CD56 and CD16, where CD56− CD16+ and CD56+ CD16− cells are analogous to human CD56dim and CD56bright cells, respectively (34). Characterization of CD3− NKG2AC+ KLRD1+/− NK cells using CD56 and CD16 revealed significant heterogeneity between cell populations (Fig. 2A and B; Fig. 3). Specifically, we observed an increased frequency of CD56+ CD16− NK cells among total KLRD1+ NK cells, compared to KLRD1− NK cells (Fig. 2B). In NHP, CD56+ NK cells are generally thought to have a greater cytokine production than cytotoxic phenotype, while CD16+ NK cells are more cytotoxic and also execute ADCC (35). It is possible that NK cell subpopulations that utilize ADCC preferentially have reduced levels of CD94, regardless of NKG2 molecules (Fig. 3), leading to preferential activation through Fc receptors, like CD16, as suggested by our data.
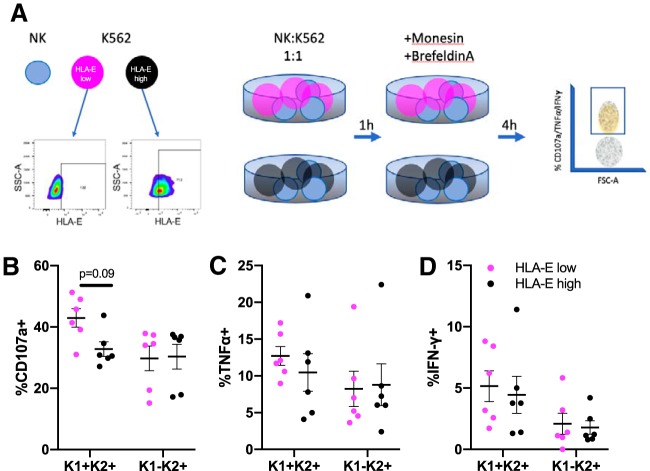
NK coculture with HLA-E-expressing K562 cells reveals functional differences between the dominant KLRC1+ KLRC2+ and KLRC1− KLRC2+ NK cells.
In order to assess whether the quadrant populations responded differently in the context of chronic SIV infection, we set up a coculture assay with NK cells and K562 that either expressed high or low levels of HLA-E. Because CMV peptides have been shown to stabilize HLA-E (10, 22), we pulsed K562 with the CMV LIL peptide and designated a sample with >70% HLA-E surface expression as HLA-Ehigh. After this, enriched splenic NK cells were cocultured with K562 that expressed either high or low levels of HLA-E, as outlined in Fig. 4A. We observed higher expression of CD107a in NKG2AC+ KLRD1+ KLRC1+ KLRC2+ NK cells cocultured with HLA-Elow K562 (mean = 42.9%) versus HLA-Ehigh K562 (mean = 32.7%, Fig. 4B). Further studies are required to investigate whether CD107a levels correspond to changes in granzyme or perforin production. Differences in levels of tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ) were less clear, though they appear to follow similar trends (Fig. 4C and D). Strikingly, the levels of CD107a, TNF-α, and IFN-γ were unchanged in NKG2AC+ KLRD1+ KLRC1− KLRC2+ NK cells. Even though we posited that HLA-E should have bound to both NKG2A and NKG2C, our observations fall in line with work done by other groups, where it appears that CMV utilizes several types of peptides to fine-tune whether HLA-E will interact preferentially with NKG2A or NKG2C (10, 36). Regardless, these data indicate that functional analyses can now be performed in cells that have been accurately defined by KLRD1, KLRC1, and KLRC2 expression.
FIG 4.
High HLA-E expression on K562 induces functional differences between KLRD1+ NK cell subsets. (A) Coculture assay results, wherein 0.5 × 106 NK cells were cocultured with 0.5 × 106 K562 cells that were either HLA-Ehigh (black) or HLA-Elow (magenta). Ultimately, the frequencies of KLRD1+ KLRC1+/− KLRC2+ quadrant-specific NK cells expressing CD107a (B), TNF-α (C), and IFN-γ (D) were quantified. Mann-Whitney U tests were used to compare quadrant populations plus HLA-Elow or HLA-Ehigh cells. KLRC1 is represented as K1, KLRC2 is represented as K2, and quadrant populations are represented as combinations of K1 and K2.
Selection of KLRD1+ NK cells may enhance identification of NKG2C+ (KLRC2+) adaptive NK cell populations.
A representative dot plot showing the expression of KLRC1 and KLRC2 in an overlay of KLRD1+ NK cells and KLRD1− NK cells (Fig. 3A) illustrates that KLRD1− NK cells are mostly KLRC1− KLRC2− NK cells. Interestingly, we observed a reduction in the KLRC1− KLRC2− population upon using KLRD1 as a NK cell marker (Fig. 3B and C) relative to when KLRD1 is not used as a NK marker (20). This reduction suggests that, expectedly, KLRC1 and KLRC2 expression occurs predominantly in cells that also coexpress KLRD1, since NKG2A and NKG2C exist as heterodimers with CD94. Further, this observation supports using KLRD1 transcript as a proxy for its protein product, CD94. The identity of the small proportion of KLRC1− KLRC2− NK cells in the KLRD1+ NK cell population is still unclear but may result from either nonspecific cross-reactivity of the NKG2A antibody to another NKG2 family member, or perhaps these cells are simply not expressing KLRC1 and KLRC2 transcripts at detectable levels. Nevertheless, we anticipate that RNA flow analyses will continue to aid the characterization of adaptive NK cells.
Already through this work we are able to identify subsets of NK cells that are modulated after SIV infection. These findings now provide the NHP field with the ability to accurately identify NKG2A+, NKG2C+, and CD94+ NK cells via their transcript expression. Further studies characterizing the expansion of KLRD1+ KLRC1+ KLRC2+ NK cells, as well as understanding whether KLRD1+ KLRC1− KLRC2+ cells play a role in disease progression, as has been suggested in human HIV infection (37, 38), may allow us to design better, targeted vaccine candidates that may better engage the NK response in SIV and HIV.
MATERIALS AND METHODS
Animals.
Sixteen Indian origin rhesus macaques (Macaca mulatta) were analyzed in this study: four animals were infected with SIVmac251 for 7 or 14 days, and twelve were chronically infected with SIVmac251. All macaques were housed at Biomere, Inc. (Worcester, MA), and all experiments were performed with approval from the local Institutional Animal Care and Use Committee. All animals were group housed until study, and then infected animals were housed under BSL2 conditions.
Macaque samples.
PBMCs and spleen mononuclear cells were isolated using standard isolation protocols. PBMCs were isolated by density gradient centrifugation layered over 100% Ficoll. Splenic mononuclear cells were isolated by mechanical disruption. Contaminating red blood cells were lysed using an ACK lysis buffer (Gibco, catalog no. A1049201). Cell aliquots were immediately cryopreserved in 90% fetal bovine serum (FBS)–10% dimethyl sulfoxide (Sigma) and stored in liquid nitrogen vapor.
RNA flow.
PBMCs were thawed and rested for 12 h in R10 media at 37°C prior to surface and intracellular staining. Next, RNA hybridization was carried out according to the manufacturer’s recommended protocol (PrimeFlow; Affymetrix, Santa Clara, CA) with the antibodies detailed in the flow cytometry section below and with rhesus macaque-specific KLRC1, KLRC2, and KLRD1 probe sets. Target probe sequences for KLRC1 and KLRC2 have been previously reported (20). The KLRD1 target probe sequences are detailed in Table S1 in the supplemental material. The probe sets were labeled with Alexa 488 (KLRC2), Alexa 647 (KLRC1), and Alexa 568 (KLRD1) fluorophores (Affymetrix). All KLRC1, KLRC2, and KLRD1 gates were determined for each sample using fluorescence minus one (FMO) controls.
Flow cytometry.
All antibodies were purchased from BD Biosciences unless specified otherwise, and clone information is given in parentheses. For the phenotypic panel, antibodies against the following cell antigens were used: CD159a-PE Cy7 (Z199; Beckman Coulter), CD3-BV450 (SP34.2), CD56-BV570 (NCAM16.2), CD14-BV711 (MϕP9), CD20-BUV395 (L27), and CD16-BUV496 (3G8). For the coculture functional panel, antibodies against the following cell antigens were used: CD159a-PE Cy7 (Z199; Beckman Coulter), CD3-BV450 (SP34.2), TNF-α-BV650 (MAb11), IFN-γ-BV711 (B27), CD107a-BV786 (H4A3), CD20-BUV395 (L27), CD16-BUV496 (3G8), CD56-BUV563 (NCAM16.2), CD14-BUV737 (MϕP9), HLA-DR-Alexa700 (G46-6), and CD8α-APC Cy7 (SK1). Flow cytometry data were acquired on a FACSymphony A5 (BD Biosciences, La Jolla, CA) and analyzed with FlowJo software (version 10.2; Tree Star, Ashland, OR).
Coculture experiments.
Frozen spleen mononuclear cells were quickly thawed in R10 media at 37°C, and 0.5 × 106 NK cells were enriched from spleen samples from SIV-infected rhesus macaques using unlabeled anti-CD3 (SP34.2) and anti-CD20 (L27) antibodies and Dynabeads (Thermo Fisher, catalog no. 11042) using a standard protocol. Briefly bulk spleen mononuclear cells were incubated at room temperature with appropriate volumes of anti-CD3 and anti-CD20 antibodies for 20 min with constant gentle agitation. The cells were pelleted and washed with wash buffer (2% FBS in Dulbecco's PBS). Cells were resuspended at 10 × 106 cells/ml in wash buffer. Dynabeads were washed twice in wash buffer and resuspended in their original volume, and then 10 μl of Dynabeads was added per 1 × 106 cells, followed by incubation at room temperature for 30 min with constant gentle agitation. CD20+ and CD3+ cells were twice negatively selected using a magnet, and the remaining cells were decanted into a separate tube and counted. We then cocultured 0.5 × 106 NK cells for 1 h at 37°C with 0.5 × 106 K562 cells that were previously either pulsed with the CMV LIL peptide (VMAPRTLIL) to stabilize HLA-E (10) or mock pulsed as a control for 16 h at 26°C. HLA-E levels were assessed using standard flow cytometry staining as described above and with the anti-HLA-E antibody (3D12; BioLegend).
Statistical analyses.
Statistical and graphing analyses were performed with Prism 8.0 software (GraphPad Software, La Jolla, CA). Nonparametric Wilcoxon tests were used where indicated, and a P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health (NIH) grants P01 AI120756, UM1 AI124377, R01 AI120828, and R01 AI143457 (all to R.K.R.) and by Harvard Center for AIDS Research grant P30 AI060354. D.R.R. was also supported, in part, by NIH training grant T32 AI007387.
We thank Stephanie Jost, Michelle Lifton, Cordelia Manickam, and Spandan Shah for insightful discussions aiding with experimental design.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00731-19.
REFERENCES
- 1.Burton DR. 2019. Advancing an HIV vaccine: advancing vaccinology. Nat Rev Immunol 19:77–78. doi: 10.1038/s41577-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ram DR, Manickam C, Lucar O, Shah SV, Reeves RK. 2019. Adaptive NK cell responses in HIV/SIV infections: a roadmap to cell-based therapeutics? J Leukoc Biol 105:1253–1259. doi: 10.1002/JLB.MR0718-303R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paust S, Blish CA, Reeves RK. 2017. Redefining memory: building the case for adaptive NK cells. J Virol 91:e00169-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. 2009. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A 106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, Leong JW, Abdel-Latif S, Schneider SE, Willey S, Neal CC, Yu L, Oh ST, Lee YS, Mulder A, Claas F, Cooper MA, Fehniger TA. 2016. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med 8:357ra123. doi: 10.1126/scitranslmed.aaf2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghofrani J, Lucar O, Dugan H, Reeves RK, Jost S. 2019. Semaphorin 7A modulates cytokine-induced memory-like responses by human natural killer cells. Eur J Immunol doi: 10.1002/eji.201847931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang T, Scott JM, Hwang I, Kim S. 2013. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRγ deficiency. J Immunol 190:1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah SV, Manickam C, Ram DR, Kroll K, Itell H, Permar SR, Barouch DH, Klatt NR, Reeves RK. 2018. CMV primes functional alternative signaling in adaptive Δg NK cells but is subverted by lentivirus infection in rhesus macaques. Cell Rep 25:2766–2774. doi: 10.1016/j.celrep.2018.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves RK, Li H, Jost S, Blass E, Li H, Eslamizar L, Altfeld M, von Andrian UH, Barouch DH. 2015. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol 16:927–932. doi: 10.1038/ni.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, Pietra G, Nienen M, Blau IW, Hofmann J, Na IK, Prinz I, Koenecke C, Hemmati P, Babel N, Arnold R, Walter J, Thurley K, Mashreghi MF, Messerle M, Romagnani C. 2018. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 19:453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- 11.Lanier LL. 1998. NK cell receptors. Annu Rev Immunol 16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 12.Kabat J, Borrego F, Brooks A, Coligan JE. 2002. Role that each NKG2A immunoreceptor tyrosine-based inhibitory motif plays in mediating the human CD94/NKG2A inhibitory signal. J Immunol 169:1948–1958. doi: 10.4049/jimmunol.169.4.1948. [DOI] [PubMed] [Google Scholar]
- 13.Borrego F, Kabat J, Sanni TB, Coligan JE. 2002. NK cell CD94/NKG2A inhibitory receptors are internalized and recycle independently of inhibitory signaling processes. J Immunol 169:6102–6111. doi: 10.4049/jimmunol.169.11.6102. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser BK, Pizarro JC, Kerns J, Strong RK. 2008. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci U S A 105:6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B, and C. Nature 391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 16.Saether PC, Hoelsbrekken SE, Fossum S, Dissen E. 2011. Rat and mouse CD94 associate directly with the activating transmembrane adaptor proteins DAP12 and DAP10 and activate NK cell cytotoxicity. J Immunol 187:6365–6373. doi: 10.4049/jimmunol.1102345. [DOI] [PubMed] [Google Scholar]
- 17.Lanier LL, Corliss B, Wu J, Phillips JH. 1998. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 8:693–701. doi: 10.1016/S1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 18.LaBonte ML, McKay PF, Letvin NL. 2006. Evidence of NK cell dysfunction in SIV-infected rhesus monkeys: impairment of cytokine secretion and NKG2C/C2 expression. Eur J Immunol 36:2424–2433. doi: 10.1002/eji.200635901. [DOI] [PubMed] [Google Scholar]
- 19.Labonte ML, Letvin NL. 2004. Variable NKG2 expression in the peripheral blood lymphocytes of rhesus monkeys. Clin Exp Immunol 138:205–212. doi: 10.1111/j.1365-2249.2004.02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ram DR, Manickam C, Hueber B, Itell HL, Permar SR, Varner V, Reeves RK. 2018. Tracking KLRC2 (NKG2C)+ memory-like NK cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog 14:e1007104. doi: 10.1371/journal.ppat.1007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang M, Orr MT, Spee P, Egebjerg T, Lanier LL, Sigal LJ. 2011. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity 34:579–589. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolle A, Meyer M, Calderazzo S, Jager D, Momburg F. 2018. Distinct HLA-E peptide complexes modify antibody-driven effector functions of adaptive NK cells. Cell Rep 24:1967–1976.e1964. doi: 10.1016/j.celrep.2018.07.069. [DOI] [PubMed] [Google Scholar]
- 23.Kottilil S, Shin K, Planta M, McLaughlin M, Hallahan CW, Ghany M, Chun TW, Sneller MC, Fauci AS. 2004. Expression of chemokine and inhibitory receptors on natural killer cells: effect of immune activation and HIV viremia. J Infect Dis 189:1193–1198. doi: 10.1086/382090. [DOI] [PubMed] [Google Scholar]
- 24.Orr MT, Wu J, Fang M, Sigal LJ, Spee P, Egebjerg T, Dissen E, Fossum S, Phillips JH, Lanier LL. 2010. Development and function of CD94-deficient natural killer cells. PLoS One 5:e15184. doi: 10.1371/journal.pone.0015184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, Hammer Q, Goodridge JP, Larsson S, Jayaraman J, Oei VYS, Schaffer M, Tasken K, Ljunggren HG, Romagnani C, Trowsdale J, Malmberg KJ, Beziat V. 2016. Critical role of CD2 costimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep 15:1088–1099. doi: 10.1016/j.celrep.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner JA, Fehniger TA. 2016. Human adaptive natural killer cells: beyond NKG2C. Trends Immunol 37:351–353. doi: 10.1016/j.it.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, Lopez-Botet M. 2006. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 28.Cichocki F, Cooley S, Davis Z, DeFor TE, Schlums H, Zhang B, Brunstein CG, Blazar BR, Wagner J, Diamond DJ, Verneris MR, Bryceson YT, Weisdorf DJ, Miller JS. 2016. CD56dimCD57+ NKG2C+ NK cell expansion is associated with reduced leukemia relapse after reduced intensity HCT. Leukemia 30:456–463. doi: 10.1038/leu.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bongen E, Vallania F, Utz PJ, Khatri P. 2018. KLRD1-expressing natural killer cells predict influenza susceptibility. Genome Med 10:45. doi: 10.1186/s13073-018-0554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abel AM, Yang C, Thakar MS, Malarkannan S. 2018. Natural killer cells: development, maturation, and clinical utilization. Front Immunol 9:1869. doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bozzano F, Marras F, De Maria A. 2017. Natural killer cell development and maturation revisited: possible implications of a novel distinct Lin− CD34+ DNAM-1bright CXCR4+ cell progenitor. Front Immunol 8:268. doi: 10.3389/fimmu.2017.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunetta E, Hudspeth KL, Mavilio D. 2010. Pathologic natural killer cell subset redistribution in HIV-1 infection: new insights in pathophysiology and clinical outcomes. J Leukoc Biol 88:1119–1130. doi: 10.1189/jlb.0410225. [DOI] [PubMed] [Google Scholar]
- 33.Schafer JL, Muller-Trutwin MC, Reeves RK. 2015. NK cell exhaustion: bad news for chronic disease? Oncotarget 6:21797–21798. doi: 10.18632/oncotarget.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong HS, Rajakumar PA, Billingsley JM, Reeves RK, Johnson RP. 2013. No monkey business: why studying NK cells in non-human primates pays off. Front Immunol 4:32. doi: 10.3389/fimmu.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster RL, Johnson RP. 2005. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology 115:206–214. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heatley SL, Pietra G, Lin J, Widjaja JM, Harpur CM, Lester S, Rossjohn J, Szer J, Schwarer A, Bradstock K, Bardy PG, Mingari MC, Moretta L, Sullivan LC, Brooks AG. 2013. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. J Biol Chem 288:8679–8690. doi: 10.1074/jbc.M112.409672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma M, Wang Z, Chen X, Tao A, He L, Fu S, Zhang Z, Fu Y, Guo C, Liu J, Han X, Xu J, Chu Z, Ding H, Shang H, Jiang Y. 2017. NKG2C+ NKG2A− natural killer cells are associated with a lower viral set point and may predict disease progression in individuals with primary HIV infection. Front Immunol 8:1176. doi: 10.3389/fimmu.2017.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas R, Low HZ, Kniesch K, Jacobs R, Schmidt RE, Witte T. 2012. NKG2C deletion is a risk factor of HIV infection. AIDS Res Hum Retroviruses 28:844–851. doi: 10.1089/AID.2011.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.