Abstract
Understanding the sequence of biological and clinical events along the course of Alzheimer's disease provides insights into dementia pathophysiology and can help participant selection in clinical trials. Our objective is to train two data-driven computational models for sequencing these events, the Event Based Model (EBM) and discriminative-EBM (DEBM), on the basis of well-characterized research data, then validate the trained models on subjects from clinical cohorts characterized by less-structured data-acquisition protocols.
Seven independent data cohorts were considered totalling 2389 cognitively normal (CN), 1424 mild cognitive impairment (MCI) and 743 Alzheimer's disease (AD) patients. The Alzheimer's Disease Neuroimaging Initiative (ADNI) data set was used as training set for the constriction of disease models while a collection of multi-centric data cohorts was used as test set for validation. Cross-sectional information related to clinical, cognitive, imaging and cerebrospinal fluid (CSF) biomarkers was used.
Event sequences obtained with EBM and DEBM showed differences in the ordering of single biomarkers but according to both the first biomarkers to become abnormal were those related to CSF, followed by cognitive scores, while structural imaging showed significant volumetric decreases at later stages of the disease progression. Staging of test set subjects based on sequences obtained with both models showed good linear correlation with the Mini Mental State Examination score (R2EBM = 0.866; R2DEBM = 0.906). In discriminant analyses, significant differences (p-value ≤ 0.05) between the staging of subjects from training and test sets were observed in both models. No significant difference between the staging of subjects from the training and test was observed (p-value > 0.05) when considering a subset composed by 562 subjects for which all biomarker families (cognitive, imaging and CSF) are available.
Event sequence obtained with DEBM recapitulates the heuristic models in a data-driven fashion and is clinically plausible. We demonstrated inter-cohort transferability of two disease progression models and their robustness in detecting AD phases. This is an important step towards the adoption of data-driven statistical models into clinical domain.
Keywords: Alzheimer's disease, Event-based models, Inter-cohort validation, Biomarkers progression, Patient staging
Abbreviations: Aβ1–42, Amyloid-β 1,42; AD, Alzheimer's disease; ADAS-Cog, Alzheimer's Disease Assessment Scale – Cognitive; ADC, Amsterdam Dementia Cohort; ADNI, Alzheimer's Disease Neuroimaging Initiative; APOE4, Apolipoprotein E ε4; ARWiBo, Alzheimer's disease Repository Without Borders; AUC, area under curve; CN, cognitively normal; CSF, cerebrospinal fluid; DEBM, discriminative event-based model; EBM, event-based model; EDSD, European DTI Study on Dementia; ELISA, Enzyme Linked Immunosorbent Assay; eTIV, Estimated Total Intracranial Volume; GMM, Gaussian Mixture Model; MCI, Mild Cognitive Impairment; MCMC, Markov Chain Monte Carlo; MMSE, Mini Mental State Examination; MRI, Magnetic Resonance Imaging; OASIS, Open Access Series of Imaging Studies; p-Tau, phosphorylated Tau; RAVLT, Rey's Auditory Verbal Learning Test; ROC, receiver operating characteristic; SMC, subjective memory complaint; SuStaIn, Subtype and Stage Inference; t-Tau, total Tau; ViTA, Vienna Transdanube Aging
Highlights
-
•
Data-driven event sequences describe evolution of relevant biomarkers in AD.
-
•
Agreement between event sequences and heuristic AD progression models
-
•
Accuracy in classifying subjects from clinical cohorts up to 91%
-
•
Staging of subjects and MMSE scores of individuals show linear relation.
-
•
Transferability of AD progression models based on research data to clinical cohorts
1. Introduction
Alzheimer's disease (AD) is a complex multifactorial neurodegenerative condition characterized by deposition of abnormal protein-aggregate, synaptic dysfunction, and eventually neuronal loss in the brain (Braak and Braak, 1991). While progression of the disease invariably results in dementia, it has been estimated that clinically-overt manifestations are preceded by a latent phase with no measurable cognitive dysfunction lasting approximately 15–20 years (Sperling et al., 2011). As AD onset remains insidious in terms of clinical manifestations, biomarkers are the most accurate approach to track disease onset and progression (Sperling et al., 2011).
A variety of biomarkers have been proposed to describe the different phases of the disease, each mirroring different biochemical, functional, or structural changes as the disease develops and progresses. The correct sequence of biomarker transitions to abnormality would allow an appropriate characterization of the different clinical and preclinical disease stages. In addition, this approach could inform the development of individualized treatments in the context of precision medicine or the identification of individuals at-risk of dementia for secondary prevention strategies (Ten Kate et al., 2018a,b).
While the recently published research criteria (Albert et al., 2011; Dubois et al., 2014) for the definition of AD stages outlined robust principles (Jack et al., 2010, 2013, 2016), their operationalization in mathematical models and out-of-the-box algorithms has recently begun.
The event-based model (EBM) (Fonteijn et al., 2012; Young et al., 2014) and the discriminative event-based model (DEBM) (Venkatragahvan et al., 2019) are two among an increasing number (Oxtoby and Alexander, 2017) of probabilistic data-driven methods developed to understand evolution of biomarkers as disease develops and progresses (Oxtoby et al., 2018; Jedynak et al., 2012; Donohue et al., 2014; Lorenzi et al., 2017). Their assumption is that the disease is characterized by an irreversible and monotonic change of biomarkers towards abnormality, which might track disease progression. Both algorithms are cross-sectional statistical models that use no strong a priori assumptions regarding the relationship among the different biomarkers or pre-defined cut-offs separating their normal and abnormal values. Both models estimate disease progression as a single average sequence, albeit in slightly different ways: the EBM estimates the maximum-likelihood sequence over all individuals, whereas the DEBM calculates the optimal event sequence as an average of estimations of patient-specific orderings.
Previous works demonstrated the EBM's capability to order biomarkers and stage subjects with a fine-grained ability in classification of Cognitively normal (CN) and AD subjects as well as to predict conversion from Mild Cognitive Impairment (MCI) to AD or from CN to MCI (Fonteijn et al., 2012; Young et al., 2014).
So far, statistical models have been tested and validated exclusively on a few well-characterized research data sets, such as: Alzheimer's Disease Neuroimaging Initiative (ADNI) (Fonteijn et al., 2012; Young et al., 2014; Venkatragahvan et al., 2019), Magnetic Resonance in Multiple Sclerosis (MAGNIMS) (Eshaghi et al., 2018), GENetic Frontotemporal dementia Initiative (GENFI) (Young et al., 2018) and TRACK-HD study of Huntington's disease (Wijeratne et al., 2018), or on synthetic data. This work focusses on transferability of the models to clinical data in AD and provides new evidence that supports widespread clinical adoption of the EBM and DEBM.
Key steps in the validation for the adoption of this kind of models are: (i) ability to build robust disease models on the basis of well-phenotyped research data sets, such as ADNI; (ii) consistency of the disease models on less well-phenotyped clinical data sets in terms of model stability and subjects' staging; (iii) clear end-user interfaces to make model results accessible by clinicians.
In the next sections, we addressed the aforementioned points towards the definition of two valid models for disease progression. Our goal was to assess the transferability of EBM and DEBM's optimal sequence of biomarkers on independent clinical data coming from six different multi-centric initiatives spanning the entire AD spectrum.
2. Material and methods
2.1. Participants
A total of 4556 subjects (CN = 2389; MCI = 1424; AD = 743) from different cohorts were selected for this study. The initiatives and projects included in this study are described in Table 1. Each cohort had different proportions of subjects in different AD stages depending on the scope of the study. Each study was approved by the local medical ethics committee. Participants for our study were selected using of the following criteria: 1) availability of information on syndromic diagnosis at baseline; 2) availability of T1-weighted Magnetic Resonance Imaging (MRI) scans obtained by either 1.5 T or 3 T scanners at baseline; 3) absence of any other major neurological, psychiatric or somatic disorders that could cause cognitive impairment at baseline.
Table 1.
Characteristics of the data sets selected.
| Data set | Full name | Description | Categories | |
|---|---|---|---|---|
| Training set | ADNI-1 | Alzheimer's disease neuroimaging initiative - 1 | The Alzheimer's Disease Neuroimaging Initiative (Aisen et al., 2010) is a longitudinal multicentre study designed to develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer's disease (AD). ADNI was originally launched in 2003 as a public-private partnership; its primary goal has been to test whether magnetic resonance imaging (MRI), biological markers, clinical and neuropsychological assessments can be combined to measure the progression of MCI and Alzheimer's disease. The initial five-year study (ADNI-1) was extended by two years in 2009 by a Grand Opportunities grant (ADNI-GO), and in 2011 by further competitive renewal of the ADNI-1 grant (ADNI-2). Through its 3 phases, it has targeted participants with AD, different stages of MCI, and CN. | CN MCI AD SMC |
| ADNI-GO | Alzheimer's disease neuroimaging initiative – grand opportunities | MCI SMC | ||
| ADNI-2 | Alzheimer's disease neuroimaging initiative - 2 | CN MCI AD SMC | ||
| Test set | ADC | Amsterdam dementia cohort | The ADC includes all patients who come to the Alzheimer Center in Amsterdam (since 2004) for diagnostic work-up and consent to give all their data collected for research (van der Flier et al., 2014). The aim is to facilitate research into new and existing biomarkers in the broadest sense, to establish diagnostic, prognostic values and further insight into the pathogenesis of neurodegenerative dementias. The data consist of baseline and annual follow-up assessments. Clinical, neuropsychological, imaging, and biological markers are collected. Since it is conception it has grown into one of the largest clinical data sets in the dementia field. | SMC MCI AD |
| ARWiBo | Alzheimer's disease repository without borders | ARWiBo is a cross-sectional data set including data from >2500 patients enrolled in Brescia (Italy) and nearby areas. The data set contains socio-demographic, clinical, genotype, bio-specimen information, MRI T1-weighted images (Frisoni et al., 2009). | CN MCI AD | |
| EDSD | European DTI study on dementia | EDSD (Brueggen K Grothe et al., 2017) is a framework of nine European centres: Amsterdam (Netherlands), Brescia (Italy), Dublin (Ireland), Frankfurt (Germany), Freiburg (Germany), Milano (Italy), Mainz (Germany), Munich (Germany), and Rostock (Germany). It is a cross-sectional multi-centre study characterized by 474 volumetric MRI T1-weighted scans with socio-demographic, clinical, genetic, and biological variables. | CN MCI AD | |
| OASIS | Open access series of imaging studies | OASIS (Marcus et al., 2007) consists of (I) a cross-sectional collection of 416 subjects. 100 of the included subjects, over the age of 60, have been clinically diagnosed with very mild to moderate Alzheimer's disease (AD). (II) A longitudinal collection of 150 subjects aged from 60 to 96 years. Each subject was scanned on two or more visits, separated by at least one year for a total of 373 imaging sessions. In addition, the data set contains socio-demographic, clinical, genotype information. | CN MCI AD | |
| PharmaCog (E-ADNI) | Prediction of cognitive properties of new drug candidates for neurodegenerative diseases in early clinical development | PharmaCog is an industry-academic European project (IMI) aimed at identifying biomarkers sensitive to symptomatic and disease modifying effects of drugs for Alzheimer's disease (Galluzzi et al., 2016). Several clinical sites participated in this study across Italy (Brescia, Verona, Milan, Perugia, and Genoa), Spain (Barcelona), France (Marseille, Lille, and Toulouse), Germany (Leipzig and Essen), Greece (Thessaloniki) and Netherland (Amsterdam). 151 MCI patients have been studied longitudinally collecting multimodal image scans, clinical variables, and bio-specimens. | MCI | |
| ViTA | Vienna transdanube aging | ViTA is a population-based cohort-study of all 75-years old inhabitants of a geographically defined area of Vienna (Fischer et al., 2002). VITA is composed of 606 subjects followed longitudinally for 4 years. Recruitment took place between May 2000 and October 2002. The primary focus of the VITA work-group was to establish a prospective age cohort for evaluation of prognostic criteria for the development of AD. | CN MCI AD |
Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment; CN, cognitively normal; SMC: subjective memory complaints.
Subjects were divided in two subsets (Table 2): training set, used to define the event sequences that serve as disease model, and test set, used for the validation of the disease models (Table 2). The training set was composed of 1488 subjects from the ADNI data set of which 468 were CN, 753 were MCI and 267 were AD. The test set was formed by 3068 subjects from six independent data sets of which 1921 were CN, 671 were MCI and 476 were AD. Subjects from ADNI and Amsterdam Dementia Cohort (ADC) with a diagnosis of subjective memory complaints (SMC) were assimilated to CN group, since Mini Mental State Examination (MMSE) score of these individuals was 28.1 ± 1.6. Significant differences in demographical (age, sex and education) and genetic (carriers of Apolipoprotein E ε4 (APOE4)) information between diagnostic groups were observed for both training and test sets. Differences were observed in the estimated Total Intracranial Volume (eTIV) only in the training set. All demographic and genetic data of training set subjects were significantly different (p-value ≤ 0.05) from demographic and genetic data of test subjects in the similar diagnostic group and for the totality of the populations (see Table 3 for full demographical information).
Table 2.
Diagnoses and biomarker availability.
| Data set | CN | MCI | AD | Sub-Total | MRI | CSF | Cognitive scores | |
|---|---|---|---|---|---|---|---|---|
| Training set | ADNI 1/GO/2 | 468 | 753 | 267 | 1488 | 100% | 72% | 100% |
| Test set | ADC | 125 | 80 | 129 | 334 | 100% | 83% | 99% |
| ARWiBo | 1399 | 169 | 152 | 1720 | 100% | 3% | 59% | |
| EDSD | 179 | 138 | 151 | 468 | 100% | 19% | 97% | |
| OASIS | 177 | 122 | 42 | 341 | 100% | NA | 100% | |
| PharmaCog | 0 | 147 | 0 | 147 | 100% | 99% | 100% | |
| ViTA | 41 | 15 | 2 | 58 | 100% | NA | 100% | |
| Total | 2389 | 1424 | 743 | 4556 | 100% | 36% | 77% |
The number of cognitively normal (CN), mild cognitive impairment (MCI), Alzheimer's disease (AD) and total subjects is reported for each data set. Biomarker availability is expressed as percentage related to the total subjects in each data set. No CSF biomarker is available for OASIS and ViTA data sets.
Table 3.
Demographics and clinical characteristics.
| MCI | AD | P-value | Total | |||
|---|---|---|---|---|---|---|
| Training set | Age | 73.9 ± 6.7 | 72.5 ± 7.3 | 73.9 ± 7.9 | 3.22·10[‐]−4 | 73.2 ± 7.0 |
| Years of education | 16.4 ± 2.7 | 15.9 ± 2.8 | 15.2 ± 2.9 | 1.09·10[‐]−6 | 15.9 ± 2.8 | |
| eTIV (cm3) | 1510± 180 | 1540 ± 160 | 1530 ± 160 | 4.20·10[‐]−3 | 1530 ± 160 | |
| MMSE | 29.1 ± 1.2 | 27.6 ± 1.8 | 23.2 ± 2.0 | 2.2·10[‐]−16 | 27.3 ± 2.6 | |
| Sex (% of females) | 52% | 42% | 48% | 1.43·10[‐]−3 | 46% | |
| APOE4-carrier | 34% | 49%* | 66% | 2.2·10[‐]−16 | 49% | |
| Test set | Age | 56 ± 17 | 70.6 ± 7.7 | 73.7 ± 8.1 | 2.2·10[‐]−16 | 62 ± 16 |
| Years of education | 10.8 ± 4.8 | 9.0 ± 4.5 | 8.7 ± 4.5 | 2.2·10[‐]−16 | 10.2 ± 4.8 | |
| eTIV (cm3) | 1450 ± 160 | 1460 ± 170 | 1470 ± 170 | 0.157 | 1460 ± 160 | |
| MMSE | 28.7 ± 1.4 | 26.5 ± 2.4 | 21.0 ± 4.7 | 2.2·10[‐]−16 | 26.6 ± 3.9 | |
| Sex (% of females) | 61% | 49% | 63% | 1.50·10[‐]−5 | 58% | |
| APOE4-carrier | 21% | 43% | 49% | 2.2·10[‐]−16 | 43% | |
Data are expressed as mean values ± standard deviations. Acronyms: eTIV: estimated total intracranial volume; MMSE: Mini Mental State Examination; APOE4: apolipoprotein E ε4; CN: cognitively normal; MCI: mild cognitive impairment; AD: Alzheimer's disease. P-values were calculated via chi square test for dichotomic variables and via ANOVA for non-dichotomic variables. Values of training set denoted with * are not significantly different from their corresponding values derived from the test subjects (p-value >0.05).
2.2. Biomarkers
When available, multimodal biomarkers collected at baseline tracking different aspects of disease biology were retrieved, i.e. (i) results of neuropsychological tests, (ii) cerebrospinal fluid (CSF) markers and (iii) imaging markers. All the selected subjects had imaging biomarkers, but some missed the results of neuropsychological tests and/or did not undergo lumbar puncture depending on the study cohort; in the latter case staging was performed on the basis of the available markers.
Cognitive biomarkers included MMSE, Alzheimer's Disease Assessment Scale - Cognitive (ADAS-Cog) and Rey's Auditory Verbal Learning Test - Immediate Recall (RAVLT).
The CSF concentrations of Amyloid-β 1,42 (Aβ1,42) (Blennow and Hampel, 2003; Blennow et al., 2010; Bombois et al., 2013), total Tau (t-Tau) and phosphorylated Tau (p-Tau) proteins (Blennow and Hampel, 2003; Blennow et al., 2010; Bombois et al., 2013) were collected, and the ratio between the concentrations of Aβ1,42 and p-Tau was calculated (Bombois et al., 2013).
The selected imaging biomarkers were: volumetric measures of the hippocampus, entorhinal cortex, fusiform gyrus, middle-temporal gyrus and precuneus, together with whole brain volume and ventricles (Vemuri and Jack, 2010; Frisoni et al., 2010). Imaging biomarkers were estimated from MRI 3D-T1 sequences analysed with FreeSurfer software v5.3 cross-sectional stream (http://surfer.nmr.mgh.harvard.edu) and outputs were visually checked. We assumed a symmetric pattern of atrophy in AD and selected imaging biomarkers were averaged between the left and right hemisphere.
Imaging biomarkers and cognitive scores were available for the totality of subjects from the training set, while CSF biomarkers were available for 72% of these individuals. Imaging biomarkers were available for the totality of test subjects while cognitive scores were available for 84% of test subjects. Within the test set, ADAS-Cog and RAVLT scores were available only for subjects from the PharmaCog data set. CSF biomarkers were available for 18% of test subjects. See Table 2 for full information on biomarker availability.
CSF biomarkers were obtained with different assays across different cohorts, i.e. Multiplex xMAP Luminex platform with Innogenetic immunoassay kit–based reagents (Kang et al., 2012) for ADNI subjects and Enzyme Linked Immunosorbent Assay (ELISA) (Butler, 2000) for subjects from all other cohorts, which led to different CSF biomarkers distributions. In order to tackle this issue and to correct for possible acquisition-related differences across datasets, all biomarkers (cognitive scores, CSF, imaging) from subjects from ADC, ARWiBo (Alzheimer's disease Repository Without Borders), EDSD (European DTI Study on Dementia), OASIS (Open Access Series of Imaging Studies), PharmaCog and ViTA (Vienna Transdanube Aging) cohorts were rescaled to match the mean and standard deviation of biomarkers distribution of ADNI subjects. In order to ensure Gaussianity, we performed a log-transformation of p-tau and t-tau as their values were non-normally distributed.
All biomarkers from the training and test sets were regressed against age, education and sex and the effects of these factors were corrected to compensate inter cohort demographic variability (Gale et al., 2007); imaging biomarkers were additionally regressed and corrected against eTIV (Kiraly et al., 2016; Gur et al., 1991) to compensate for head size. Correction of biomarkers was performed separately for training set and test set.
The comparison of the selected biomarkers in this study among the three clinical groups and the seven data cohorts considered in this study are shown in Supplementary Material SF1.
2.3. Mathematical modelling
Development of EBM and DEBM was based on the fundamental work of Fonteijn et al. (Fonteijn et al., 2012). According to these approaches, each biomarker is considered as either normal or abnormal and its probabilistic transition from the normal to the abnormal state is defined as event. The aim is to define in a data-driven manner the sequence of events that describe the most probable ordered cascade that characterizes the transition of a subject from the healthy state to the full-blown disease spectrum (Young et al., 2014). For this work, we employed python module pyebm (https://github.com/EuroPOND/pyebm), where both algorithms are implemented.
In the EBM (Fonteijn et al., 2012; Young et al., 2014) possible event sequences are sampled via a Markov Chain Monte Carlo (MCMC) process aimed at finding the sequence that best fits the biomarker observations from all subjects. At each Monte Carlo step a new sequence is sampled as a random swap between two biomarkers of the current benchmark sequence. If the new sequence is a better fit than the benchmark sequence, which is determined mathematically by the likelihood, then the new sequence is considered as the benchmark sequence for the following MCMC step.
The probability of an event for each biomarker is determined by a Gaussian mixture model (GMM) where the normal and abnormal components are modelled by Gaussian distributions. In EBM (Young et al., 2014), distributions of normal and abnormal biomarkers are initialized as the distributions of biomarkers from the CN and AD subjects, respectively. The mixture model distribution for each biomarker is then found as the sum, weighted on the mixing parameters, of the two aforementioned distributions that best fits to biomarker values from all subjects. Optimization of the GMM function is performed along the Gaussian parameters and the mixing parameters and in order to avoid the possibility that biomarkers will not show a clear bimodal distribution, the standard deviations for normal and abnormal components in the GMM are constrained to be no greater than the standard deviations of CN and AD subjects, respectively.
The approach of DEBM model (Venkatraghavan et al., 2017, 2019) for the calculation of the central ordering, on the other hand, is a two-step process where first (i) a specific ordering is calculated for each subject by sorting the posterior probability that each biomarker has become abnormal and then (ii) the central ordering is calculated as the event sequence that minimizes the sum of probabilistic Kendall's tau distances between itself and all the subject-wise orderings. As the posterior probability is influenced by the physiological variability of biomarkers, DEBM assumes that single subject orderings are noisy estimates of the central ordering (Venkatragahvan et al., 2019).
The original formulation of DEBM (Venkatragahvan et al., 2019) also contains a specific mixture model, for which an initial estimate of the distributions of non-diseased and diseased subjects for each biomarker is performed using values from subjects at the opposite ends of the disease spectrum, as defined by a Bayesian classifier which is trained to remove outliers and wrongly labelled data. This allows efficient separation of the two Gaussian distributions of normal and abnormal values for each biomarker. The biased distributions are then refined including data from all subjects via a GMM that has constraints based on the aforementioned relationships between the expected and the biased distributions. The same objective GMM function as for EBM is optimized alternatively along the Gaussian parameters and the mixing parameters until the latter converge.
Optimal sequences were calculated as averages of orderings obtained from 50 bootstrapped iterations for both EBM and DEBM. Furthermore, in EBM the number of MCMC steps was set to 50.000 to ensure convergence of the likelihood. In practice convergence was typically observed before the 15.000-th MCMC step.
See Supplementary Material SS1 for detailed mathematical modelling.
2.4. Model validation & statistical analysis
Validation of the models is performed by staging subjects from the training and test sets on the basis of the event sequences built on the basis of biomarkers from subjects from the training set. Specific methods for staging subjects are available in the original works for both the EBM (Young et al., 2014) and DEBM (Venkatragahvan et al., 2019). For the sake of simplicity, and in order to have a common staging system for both models, the method from Young et al. (2014) was employed in this work. This method assigns each subject a position of the central event sequence, resulting in a number of stages that is equal to the number of biomarkers considered for the sequence plus one, as it is necessary to add stage 0 where no biomarker is abnormal. The stage of each subject is calculated as the k-th step of the event sequence that maximizes the probability that all events up to k have already occurred and events from k + 1 to the end of the sequence are yet to occur. In case of missing biomarkers, the probability of the biomarkers to be abnormal was set to 0.5 (Young et al., 2015). Assuming that clinical diagnoses of all subjects are made through a biomarker-based assessment, it is expected that each subject, either from the training or test set, is staged at the earlier positions of the event sequences if CN and at the later positions if AD.
Measures of area under curve (AUC), sensitivity, specificity and balanced accuracy at optimal threshold kT were calculated for all pairwise comparisons among clinical groups, i.e. (i) AD vs. CN, (ii) AD vs. MCI, and (iii) MCI vs. CN. In order to assess significant differences between receiver operating characteristic (ROC) curves, the DeLong test (DeLong et al., 1988) was performed.
To assess the validity of the EBM and DEBM central orderings we explored the linear correlation between subjects' model stages and MMSE scores. The MMSE is the most widely used screening tool to assess cognitive functions in both routine clinical practice and research settings and its score correlates with the different phases of AD progression (Tombaugh and McIntyre, 1992). In order to avoid circularity MMSE scores were excluded from the initial calculation of the event sequences. Moreover, in order to mitigate the ceiling effect typical of MMSE (Hoops et al., 2009), the lower limit for the linear regression analysis was set as the model stage that provides the optimal threshold for separating CN and MCI subjects.
To explore how much the missing biomarkers of test subjects (Table 2) affected the classification performances in both models, staging was also performed for a special subset of test subjects having at least one CSF measurement, MMSE score and imaging biomarkers. These restriction criteria reduced the original test subjects from 3068 to 562 (104 CN,331 MCI, 127 AD) and the number of events considered in our original simulation from 13 to 12 as ADAS-Cog and RAVLT were excluded since they were available only for the PharmaCog data set, while MMSE was included.
Statistical analysis was performed with R version 3.5.1.
3. Results
3.1. Events ordering
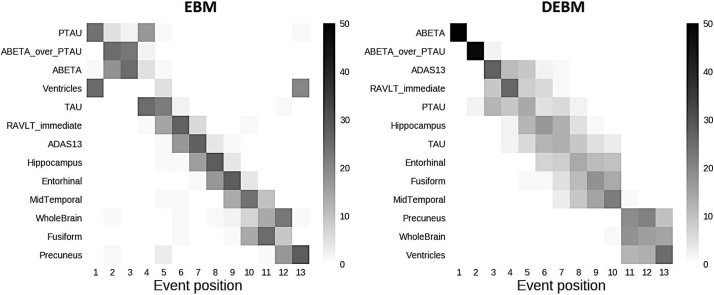
Central event sequences and their variances were generated from biomarkers of training subjects for both EBM and DEBM and were plotted as positional variance diagrams (Fig. 1).
Fig. 1.
Positional variance diagrams of event orderings obtained with EBM and DEBM. Both diagrams show the number of times each biomarker occurred in a specific position from a batch of 50 independent bootstrapped sequences generated using biomarkers of training subjects with EBM (left) and DEBM (right) methods.
The event sequence obtained with the DEBM algorithm showed that amyloid related biomarkers became abnormal first. The abnormalities of Aβ1,42 protein and Aβ1,42/p-Tau ratio are at the very first positions followed by cognitive scores, Tau protein-related biomarkers, and finally imaging markers of AD-relevant brain regions. Averaged volumes between left and right hemisphere of hippocampus and precuneus are respectively the first and the last brain areas to become abnormal while the medial temporal lobe is in between. The enlargement of the ventricles and the atrophy of the whole brain were in the last two positions.
In EBM, CSF biomarkers are the first to show abnormality, although with a different pattern with respect to DEBM. Tau related biomarkers became abnormal earlier and often before amyloid-related biomarkers. The sequence obtained with EBM followed a similar ordering for the cognitive scores although the specific order of RAVLT and ADAS scores is swapped.
The enlargement of the ventricles is placed at the fourth position of the ordering although the positional variance showed that this event has nonzero probability of occurring in the first or last position of the sequence. Volumetric measures of the grey matter of the fusiform gyrus and precuneus are placed at the very last positions of the EBM benchmark sequence. Both EBM and DEBM showed good positional stability (see Fig. 1), and in the case of DEBM no event occurs far from the diagonal.
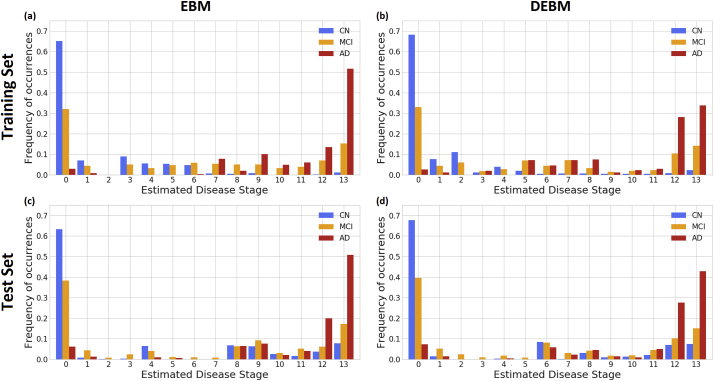
3.2. Staging of individuals across the AD spectrum
Subjects from both training and test set were staged on the basis of the event sequences derived from the training set. For the training set, in both EBM and DEBM cases, >60% of CN subjects were staged at position 0 where no abnormalities have occurred yet (Fig. 2 (a) & (b)). Similarly, the majority of AD were staged at positions 12–13 (of 13 total) of both sequences. Most of the remaining CN subjects were spread across stages 1–6 in EBM and 1–4 in DEBM. The majority of the remaining AD individuals were staged across stages 7–11 for EBM and stages 5–12 for DEBM.
Fig. 2.
Subject staging based on the sequences obtained with EBM and DEBM methods. Staging of subjects from all diagnostic categories (Cognitively normal (CN) in blue, mild cognitive impairment (MCI) in orange, Alzheimer's disease (AD) in red) are shown for (a) training subjects on EBM sequence, (b) training subjects on DEBM sequence, (c) test subjects on EBM sequence and (d) test subjects on DEBM sequence. Histograms are normalized for each diagnostic category. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For the test set, staging of subjects obtained with EBM and DEBM is shown in panels (c) and (d) of Fig. 2 respectively. In this case >70% of AD subjects was staged at positions 12–13 and >60% of CN subjects were staged at position 0, but the strong separation between CN and AD observed in the training set was not reproducible in the test set for 30% of CN subjects were staged at positions 6–13. These test CN subjects belonged to two different phenotypic classes:
-
(1)
subjects whose eTIV was very large or very small compared to the eTIV of the CN population. Indeed, the eTIV of these subjects showed a bimodal distribution with peaks at ±1.1 standard deviations apart from the average of the test CN population;
-
(2)
subjects aged 76.2 ± 8.7 on average, whose MMSE score was on average 29.11, but whose hippocampal normalized volume was significantly smaller compared to the hippocampal normalized volume for the test CN subjects ((2.1 ± 0.4) × 10−3 vs. (2.7 ± 0.4) × 10−3).
In each case, the distribution of MCI stages overlapped with the distribution of stages for CN and AD, but a considerable amount, always between 30% and 40%, was staged at position 0 in both EBM and DEBM models (Fig. 2). MCI subjects staged at position 0 had an average MMSE score of 28.2 ± 2.1 for training set and 27.0 ± 2.1 for test set.
Staging of the subjects from each data set on the basis of EBM and DEBM sequences shows a good separation between CN and AD subjects in each case, and generally few subjects are staged at positions 1–7 for EBM and 1–5 for DEBM as these stages correspond to CSF and cognitive biomarkers (see Supplementary material SF2). Linear regression of DEBM stage vs EBM stage resulted in slopes <1 for both the training and test set, meaning that on average EBM stage is always greater than DEBM stage (see Supplementary material SF3).
3.3. Staging vs MMSE correlation
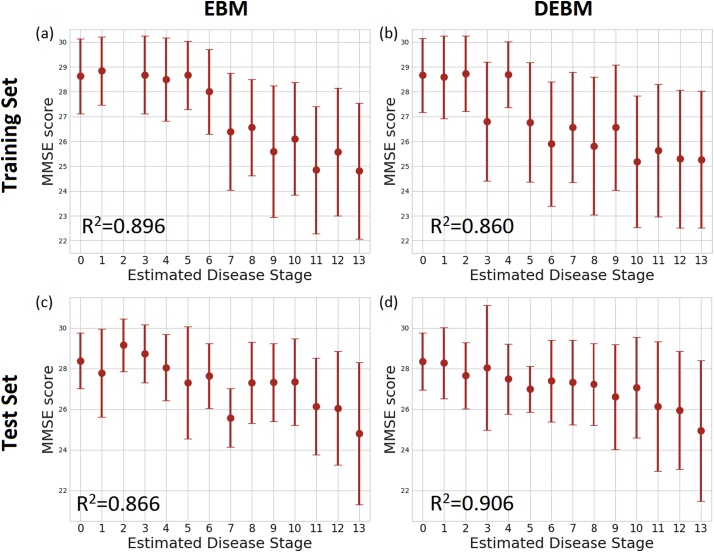
Average and standard deviation of the MMSE scores of the training and test sets at each stage is shown in Fig. 3. The plot showed decreasing MMSE scores in the latter stages in both EBM and DEBM.
Fig. 3.
Correlation between MMSE score and subjects staging for (a) training set subjects on EBM sequence, (b) training set subjects on DEBM sequence, (c) test set subjects on EBM sequence, (d) test set subjects on DEBM sequence. Average and standard deviation of MMSE score of training and test subjects staged on the basis of EBM and DEBM sequences are shown. Coefficients of determination (R2) of the linear regression of MMSE score vs disease stage are reported.
Linear regression of the MMSE scores of all subjects excluding the initial ceiling effect showed correlation between the decrease in MMSE score and patient staging of training subjects for both EBM (R2=0.896) and DEBM (R2=0.860). The limit of the initial ceiling was set as the model stage threshold that optimally separates CN and MCI subjects, that is stage 6 for EBM and stage 5 for DEBM in the case of the training set. Good linear correlation between MMSE scores and subject staging was observed for individuals from the test set (R2=0.866 for EBM and R2=0.906 for DEBM), although the ceiling effect thresholds were different from the thresholds of the training set (stage 1 for both EBM and DEBM).
3.4. Prediction of clinical diagnosis
Clinical diagnosis classification of each individual from both training and test data sets was computed. All the possible combinations were assessed, i.e. AD vs. CN, AD vs. MCI and MCI vs. CN. The balanced accuracy and AUC values of the classification obtained on both training and test sets were comparable to other state-of-the-art classification approaches (Young et al., 2014). In the case of AD vs. CN, balanced accuracy and AUC of the ROC curve, alongside measures of sensitivity and specificity, are >0.93 in the training set and >0.81 for test set for both models (see Table 4). The comparison of the AUC showed significant differences (p-value ≤ 0.05) between EBM and DEBM in both training and test sets. For AD vs. MCI subjects, balanced accuracy and AUC in both training and test sets were always >0.71. No significant differences were registered between the AUC of EBM and DEBM. In the case of MCI vs. CN subjects, balanced accuracy and AUC values were between 0.62 and 0.73 without significant differences between EBM and DEBM. In both models, a significant difference (p-value ≤ 0.05) between training and test sets was observed in two of the three classification tasks: (i) AD vs. CN; (ii) MCI vs CN. The maximum balanced accuracy threshold (kT) used in the classification increases across the disease spectrum in both models with the exception of DEBM on ADNI subjects where the threshold is constant for all classifications. This is compatible with the idea that EBM and DEBM produce event sequences that track disease progression.
Table 4.
Measurements of area under curve (AUC), sensitivity (Sens), specificity (Spec), and balanced accuracy (BalAcc) at a specific threshold (kT) for the subject staged with EBM and DEBM methods on training and test data sets.
| EBM |
DEBM |
p-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| kT | Sens | Spec | BalAcc | AUC | kT | Sens | Spec | BalAcc | AUC | ||
| Training set | |||||||||||
| AD vs CN | 7 | 0.97 | 0.96 | 0.96 | 0.97* | 5 | 0.92 | 0.94 | 0.93 | 0.95* | 1.88·10−3 |
| AD vs MCI | 9 | 0.59 | 0.96 | 0.77 | 0.81 | 5 | 0.48 | 0.94 | 0.71 | 0.76 | 5.30·10−5 |
| MCI vs CN | 6 | 0.88 | 0.52 | 0.70 | 0.73* | 5 | 0.92 | 0.52 | 0.72 | 0.73* | 0.537 |
| Test set | |||||||||||
| AD vs CN | 5 | 0.71 | 0.91 | 0.81 | 0.87 | 7 | 0.78 | 0.85 | 0.81 | 0.86 | 3.99·10−2 |
| AD vs MCI | 12 | 0.77 | 0.71 | 0.74 | 0.78 | 11 | 0.70 | 0.75 | 0.73 | 0.77 | 0.393 |
| MCI vs CN | 1 | 0.63 | 0.62 | 0.62 | 0.63 | 1 | 0.68 | 0.60 | 0.64 | 0.64 | 0.676 |
Thresholds are chosen to maximize the balanced accuracy in each classification task. P-values of Delong test performed to compare AUCs of EBM and DEBM methods are reported in the last column. AUCs of training set denoted with * are significantly different from their corresponding values derived from the test subjects (p-value of DeLong test ≤0.05).
To fully explore the capabilities of the two models and to perform a fair head to head comparison we run similar analyses in the training and test sets considering all the 14 biomarkers (see Supplementary Material SF4, SF5). On average, the general performance in discriminating subjects from the test set improved by 2 and 4 percentage points respectively for DEBM and EBM (see Supplementary Material ST2). This improvement is achieved by the inclusion of the MMSE score, which is available for a large portion of test subjects.
Results of the case where all test subjects do not have missing biomarkers showed improvement in the performances for all the computed metrics. In the test set, on average, DEBM showed an increase of 4.3% in balanced accuracy and an increase of 3.0% in AUC compared with the metrics obtained from the complete 13 biomarker sequences. Similarly, EBM showed an increase of 7.2% in balanced accuracy and an increase of 5.5% in AUC. Generally, no statistically significant differences between staging of training and test subjects were observed (p-value > 0.05) for all groups in both models. Detailed results are reported in Table 5.
Table 5.
Measurements of area under curve (AUC), sensitivity (Sens), specificity (Spec) and balanced accuracy (BalAcc) at a specific threshold (kT) for the staging obtained with EBM and DEBM methods on training and test data sets not containing missing values.
| EBM |
DEBM |
p-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| kT | Sens | Spec | BalAcc | AUC | kT | Sens | Spec | BalAcc | AUC | ||
| Training set | |||||||||||
| AD vs CN | 8 | 0.98 | 0.95 | 0.97 | 0.97 | 3 | 0.86 | 0.99 | 0.92 | 0.95 | 3.10 10−2 |
| AD vs MCI | 8 | 0.70 | 0.95 | 0.83 | 0.83 | 7 | 0.66 | 0.76 | 0.71 | 0.76 | 0.104 |
| MCI vs CN | 5 | 0.89 | 0.51 | 0.70 | 0.72 | 3 | 0.86 | 0.58 | 0.72 | 0.73 | 1.99 10−8 |
| Test set | |||||||||||
| AD vs CN | 4 | 0.88 | 0.94 | 0.91 | 0.95 | 3 | 0.91 | 0.91 | 0.91 | 0.94 | 0.332 |
| AD vs MCI | 4 | 0.57 | 0.94 | 0.76 | 0.80 | 5 | 0.63 | 0.87 | 0.75 | 0.79 | 1.65 10−2 |
| MCI vs CN | 4 | 0.88 | 0.43 | 0.66 | 0.66 | 3 | 0.91 | 0.52 | 0.71 | 0.70 | 0.296 |
P-values of Delong test performed to compare AUCs of EBM and DEBM methods are reported in the last column. In DEBM and EBM AUCs of the training set were not significantly different to their corresponding AUCs in the test set (p-values of DeLong test always >0.05).
3.5. Sequence consistency
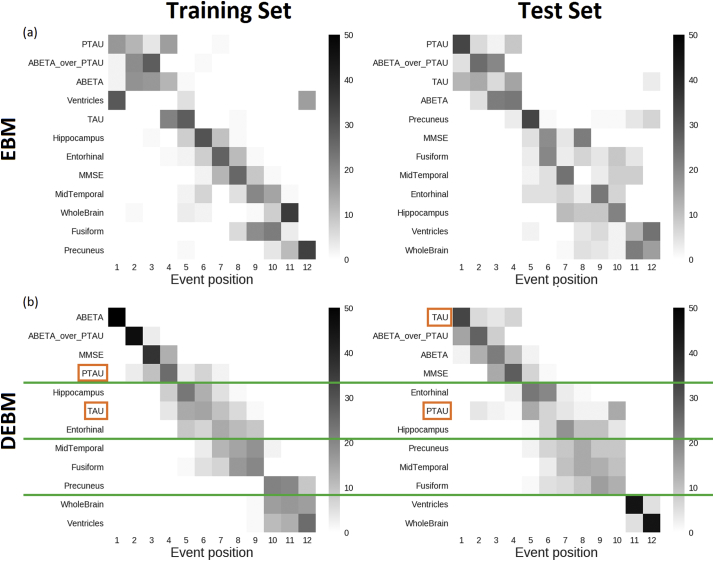
In order to ensure consistency of the benchmark sequence generated from the training set, a disease model was also built on the basis of the test set (i.e.: ADC, ARWiBo, EDSD, OASIS, PharmaCog, ViTA) using both EBM and DEBM. ADAS-Cog and RAVLT cognitive scores were not included since these specific tests were available only for MCI subjects from the PharmaCog data set. MMSE was included so that all biomarker families (cognitive, CSF and imaging) were represented.
In both sequences obtained with the EBM, CSF biomarkers occupy the first positions of the sequences (Fig. 4(a)) but the second halves of the sequences differ considerably, especially in the position of ventricles and hippocampus. In total, 23 swaps between adjacent biomarkers are needed in order to turn the sequence obtained from the test set into the sequence obtained from the training set.
Fig. 4.
Positional variance diagrams of event sequences computed from training set (left) and test set (right) using EBM (a) and DEBM (b) algorithms. In the case of DEBM green lines divide the sequences into homogeneous blocks between the training and test sets. Orange boxes represent biomarker exceptions not conserved in the same block comparing the training vs. test positional variance diagrams. Clear event blocks cannot be identified for EBM sequences. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In DEBM, the event sequences obtained from training and test sets are similar. Only 11 swaps between adjacent events are needed to turn the test set sequence into the benchmarked training set sequence (Fig. 4(b)). With the exception of t-Tau and p-Tau both sequences obtained with DEBM can be divided in four partial rankings that contain the same biomarkers: Aβ1,42/p-Tau ratio, Aβ1,42 and MMSE in the first partial ranking, hippocampus and entorhinal cortex in the second, middle temporal gyrus, fusiform gyrus and precuneus in the third and whole brain and ventricles in the last partial ranking.
4. Discussion
To our knowledge, this is the first translational study showing viability of the EBM and DEBM, trained on research data, in a clinical setting. This is also the first cross-cohort assessment of the models' validity on cross-sectional multimodal biomarkers. Previous literature focused only on well characterized research datasets and synthetic data (Young et al., 2014; Venkatraghavan et al., 2017, 2019; Iturria-Medina et al., 2016; Li et al., 2014; Koval et al., 2018; Schiratti et al., 2015) but this kind of approach does not take into consideration the aspects of real clinical data. We investigated and compared the performance of EBM and DEBM when applied to the same training and test data sets which included subjects across the entire disease spectrum, accounting for missing data.
EBM and DEBM rely on different estimates of the Gaussian mixture models and in the definition of the optimal sequence of biomarkers. As highlighted in literature (Venkatragahvan et al., 2019), the optimization technique adopted in DEBM, for which Gaussian parameters and mixing parameters are optimized alternatively, prevents the abrupt change of the mixing parameter for small changes in the Gaussian parameters that was observed in EBM.
We observed differences between EBM and DEBM optimal event sequences. The DEBM sequence is closer to Jack's model (Vemuri and Jack, 2010) and also mirrors stages V and VI of cortical degeneration due to neurofibrillary tangles deposition as described in Braak's Model (Braak et al., 1993). The DEBM sequence starts with Aβ1,42 and Aβ1,42/p-Tau ratio, while the EBM sequences suggests p-Tau as the first biomarker to become abnormal. Although in literature it is not completely understood which is triggering the other (if at all), much evidence suggests Aβ1,42 deposition to be upstream of Tau deposition. The deposition of amyloid plaques presumably triggers the conversion of Tau protein to toxic state, while less evidence suggests that toxic Tau can enhance Aβ1,42 toxicity via a feedback loop. Soluble toxic aggregates of Aβ1,42 and p-Tau can self-propagate and spread throughout the entire brain, perhaps enhancing other destructive biochemical pathways (Bloom, 2014) and triggering the abnormality cascade of the other biomarkers. It is important to consider, however, that the transition to abnormality of a biomarker may not correspond to its pathological change, since no a priori thresholds are set.
Coherently with Iturria-Medina's model (Iturria-Medina et al., 2016), where spatiotemporal abnormalities of multiple biomarkers are explored via a multi-factorial data-driven analysis, both EBM and DEBM orderings showed a drop in the performance of cognitive test scores after events related to CSF biomarkers. In particular, EBM ordering of cognitive results seems slightly more plausible, ordering the RAVLT before ADAS13, as RAVLT has been reported to be more sensitive to detect abnormal changes in pre-dementia condition (Estevez-Gonzalez et al., 2003) while ADAS is more specific to detect moderate AD conditions (Rosen et al., 1984). According to both methods, cognitive tests were positioned before group-level neurodegeneration events in the benchmark sequences. This fact might be in contrast with literature (Jack et al., 2010; Mormino et al., 2009) for which memory impairment occurs after volumetric decrease of brain regions. This difference can be explained by the fact that population-level volume changes may affect the event sequence (Young et al., 2014). The earlier position of cognitive scores with respect to imaging biomarkers could be explained partially by the different GMMs used in the two algorithms and partially because of specific inclusion criteria for the ADNI training subjects. In ADNI, no subjects with severe cognitive impairments were included since one of the inclusion criteria was to have MMSE score at least equal to 18. This may affect the position in which cognitive test scores were considered abnormal because the threshold that separates normal from abnormal values might be overestimated by the models, considering that no a priori assumptions are made in EBM and DEBM.
As far as the MRI biomarkers are concerned, DEBM showed an expected pattern of grey matter atrophy with AD progression. Abnormalities were ordered throughout the temporal lobes as follows: hippocampus, entorhinal cortex, fusiform and mid temporal regions. Precuneus was affected subsequently, in agreement with model of cortical atrophy progression proposed by ten Kate et al. (2017), where atrophy of parietal regions is associated with progression from MCI to dementia. The DEBM sequence presented the whole brain and subcortical abnormalities as end-sequence events. EBM did not capture the expected atrophic evolution of the grey matter and the main anomaly was represented by ventricles. Their abnormality was reported in the fourth position of the optimal sequence and their variability is spanning from the first to the last position. Two different local likelihood maxima due to different subtypes of AD (Young et al., 2018) in the EBM sequence space could be one possible reason. Also, this issue is not observable in DEBM, where normally the variance of an event is distributed continuously around its specific position, that means around the positional variance diagram bisector. The difference between the two models can be attributed to the smoothing effect intrinsic to the DEBM algorithm and, as highlighted in Venkatragahvan et al. (2019), to the specific mixture model used in EBM. The sequences generated by EBM and DEBM models, however, represent a general event ordering for the progression of the disease and individual trajectories may show variability with respect to the optimal sequences.
We demonstrated, using data from ADNI and 6 other independent clinical cohorts, the performances of EBM and DEBM across the entire Alzheimer's time course. Staging of subjects in both the training and test sets showed separation between AD and CN in the two methods. This meant that the algorithms were effective at distinguishing subjects having only a few abnormal biomarkers from those having only a few normal biomarkers. As expected, the majority of CN subjects from the training set were staged at position 0, where no abnormality manifested yet, and a large number of AD subjects was at end-sequence stages 11–13. Staging of the test subjects followed the same general trend as ADNI, although subjects with a lack of CSF values or cognitive assessments and with normal imaging biomarker values were staged in proximity of non-symptomatic stage 0. The large number of CN subjects in the test sets that were staged in the last positions for both models, can be partly explained considering that a significant portion of these individuals are CN elderlies with volumetric anomalies and no other biomarker available, thus contributing to subjects' misclassification although MMSE score showed no abnormalities. Another portion of misclassified CN subjects is formed by individuals with abnormal imaging biomarkers but here the misclassification is due to the linear regression correction since the average eTIV of test subjects is significantly lower than the average eTIV of training subjects, thus, the imaging biomarkers of test subjects are artificially considered as atrophic with respect to the imaging biomarkers from the training set subjects.
Some concerns may arise from the large number of MCI subjects staged at stage 0. The CSF and cognitive scores for the majority of these individuals were close but not yet over the probabilistic threshold values, therefore they were still in the normal ranges, and the models considered those subjects as normal. Despite this, staging evidences give comparable results to state-of-the-art classification techniques for prediction of conversion from MCI to dementia (Young et al., 2015; Willette et al., 2014).
EBM and DEBM showed good linear correlation with MMSE scores, fairly consistent with the clinical and regional biomarkers, thus producing an indirect validation of models with respect to the disease evolution. Both methods, after an initial plateau due to the ceiling effect typical for MMSE test (Hoops et al., 2009), showed an expected linear decline (Perneczky et al., 2006). Although it was a rather trivial approach, we tried to validate the EBM and DEBM event sequences even in absence of a validated pathological gold-standard across the data cohorts.
When all test subjects are considered, we detected a significant drop of performance in classifying AD vs CN as well as in MCI vs CN subjects from ADNI to the test cohorts. This is probably due to missing data (CSF biomarkers and cognitive scores), which is known to increase uncertainty in subject staging (Young et al., 2014). Indeed, when considering a reduced set of test subjects for which all biomarkers were available, the performances became much closer to those obtained from the training set and no more significant differences between training and test data sets were observable for both EBM and DEBM (p-values > 0.05). This reinforces the importance to collect an adequate set of biomarkers for an accurate staging of single subjects into the correct diagnostic class.
As far as the test set is concerned, the classification of AD vs CN subjects was significantly better in EBM than in DEBM (p-values≤0.05). In classifying AD vs MCI, EBM was slightly better with higher sensitivity, balanced accuracy and AUC. In MCI vs CN, DEBM reached higher sensitivity and balanced accuracy while EBM reached higher specificity. This evidence might represent specific hints to guide the usage of EBM and DEBM for physicians according to the initial diagnostic hypothesis they want to test in their clinical practice.
An interesting consideration for future works is the possibility to use such methods to follow MCI in specific sub-classes, namely: amnestic MCI, non-amnestic MCI and MCI due to AD. Additional studies with extended age range of subject, larger and additional groups and additional biomarkers such as other brain regions will be helpful to achieve a more accurate description of AD via event-based models. Clinically relevant information related to patients' staging, together with the models' robustness as well as progressive tracking capabilities along the CN-to-AD course, might be implemented into a clinical decision support tool, to aid diagnosis and prognostic assessment of AD at early stages.
Additional efforts will be needed to understand the capabilities of staging subjects during clinical routine by means of EBM and DEBM in: (I) reducing the number of patients needed for future clinical trials, (II) monitoring the efficacy of disease modifying drugs, (III) personalized medicine.
So far, EBM and DEBM have been validated against well-characterized research datasets, synthetic data and, in the present study, multicentric clinical cohorts, but none of them has been yet compared against different stages of the AD pathology. In the next future, we would have to focus on further validation of both models against databases of population of normal and abnormal post-mortem studies on subjects assessed with as many biomarkers as possible, such as those collected in the Religious Orders Study (Bennett et al., 2012a), Rush Memory and Aging Project (Bennett et al., 2012b), the Adult Changes in Thought study (Kukull et al., 2002), and the National Alzheimer's Coordinating Center data set (Beekly et al., 2007).
Some limitations of the current results should be considered in future validations of event-based models. First, the tools here described need to be further compared with other complementary techniques based on longitudinal data sets, such as: temporal continuous models and spatiotemporal models – see (Oxtoby and Alexander, 2017) for a recent review of the field. Second, as clinicians are the potential beneficiaries of the tools based on such models, independent evaluators should rate the diagnostic added value and accuracy of EBM and DEBM. Third, the greatest limitations in the methods applied is the assumption of a common or average disease trajectory across individuals, while AD is highly heterogeneous and clearly violates this assumption. In this perspective single subject orderings already available in DEBM, and data-driven subtype progression patterns estimated using SuStaIn (Subtype and Stage Inference) (Young et al., 2018) could play a central role in the description of AD progression at the level of the single subject. Finally, computational time is worth considering: the extensive use of EBM or DEBM to analyse large volumes of data that must be pre-processed and that require large computational resources, such as: HPC, Grid, or Cloud (Redolfi et al., 2013, 2015; Frisoni et al., 2011), indeed the models can be trained a priori and then they should be used in the clinical practice only to evaluate new subjects on the basis of the preferred model within an acceptable time frame.
The state of the art of these data driven models is represented by research tools (https://github.com/EuroPOND), that should be implemented in more user-friendly interfaces compatible with the clinical routine. Efforts towards the opportunities for clinical adoption and perceived importance of such a tool in clinical setting has started to appear (https://icometrix.com, n.d.) (see Supplementary material SF6).
5. Conclusions
We have performed an inter-cohort model transferability study and model performance comparison via external validation approach for event-based models. In the field of healthcare, the importance of data driven models will grow in the coming years, and the results presented here represent the first viability and generalizability proof of principle to train such models on research data and apply them clinically: on cross-sectional, less-well-characterized cohorts. We trained data-driven disease progression models with the ADNI data set and compared patients' ordering, staging and performance through ADC, ARWiBo, EDSD, OASIS, PharmaCog and ViTA data sets. Overall, we tested both models on 4556 subjects and 14 multimodal biomarkers. Both EBM and DEBM demonstrated similar and good classification performances especially when all biomarkers were available for test subjects. Orderings obtained from both models agreed with previous heuristic models. The event sequence generated through DEBM returned a more reasonable description of the course of AD, while EBM showed better classification performances, which are important considerations for future applications.
Declarations of Competing Interests
None.
Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 666992. This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 634541.
ADNI data were funded by the Alzheimer's Disease Neuroimaging Initiative (National Institutes of Health grant U01 AG024904) and Department of Defense Alzheimer's Disease Neuroimaging Initiative (Department of Defense award W81XWH-12-2-0012). The Alzheimer's Disease Neuroimaging Initiative is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol- Myers Squibb Company; CereSpir Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research and Development LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck and Co Inc.; Meso Scale Diagnostics LLC; NeuroRx Research; Neuro-track Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. Alzheimer's Disease Neuroimaging Initiative data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
ARWiBo, EDSD, ViTA, and PharmaCog (alias E-ADNI) data used in the preparation of this article were obtained from NeuGRID4You initiative (http://www.neugrid4you.eu) funded by grant 283562 from the European Commission.
OASIS was funded by grant P50 AG05681, P01 AG03991, R01 AG021910, P50 MH071616, U24 RR021382, R01 MH56584.
ADC was obtained from the VUmc Alzheimer centre which is part of the neurodegeneration research program of Amsterdam Neuroscience (http://www.amsterdamresearch.org). The ADC was supported by Innovatie Fonds Ziektekostenverzekeraars, Stichting Diorapthe and Stichting VUmc fonds. This project has received funding from the Innovative Medicines Initiative 2 Joint undertaking under grant agreement No 115736 (EPAD) and 115952 (AMYPAD). This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA.
FB is supported by the NIHR biomedical research centre at UCLH.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2019.101954.
Contributor Information
Damiano Archetti, Email: darchetti@fatebenefratelli.eu.
Silvia Ingala, Email: s.ingala@vumc.nl.
Vikram Venkatraghavan, Email: v.venkatraghavan@erasmusmc.nl.
Viktor Wottschel, Email: v.wottschel@vumc.nl.
Alexandra L. Young, Email: alexandra.young.11@ucl.ac.uk.
Maura Bellio, Email: maura.bellio.16@ucl.ac.uk.
Esther E. Bron, Email: e.bron@erasmusmc.nl.
Stefan Klein, Email: s.klein@erasmusmc.nl.
Frederik Barkhof, Email: f.barkhof@vumc.nl.
Daniel C. Alexander, Email: d.alexander@ucl.ac.uk.
Neil P. Oxtoby, Email: n.oxtoby@ucl.ac.uk.
Giovanni B. Frisoni, Email: giovanni.frisoni@unige.ch.
Alberto Redolfi, Email: aredolfi@fatebenefratelli.eu.
Appendix A. Supplementary data
Supplementary material
References
- Aisen P.S., Petersen R.C., Donohue M.C., Gamst A., Raman R. Alzheimer's Disease Neuroimaging Initiative. Clinical Core of the Alzheimer's disease neuroimaging initiative: progress and plans. Alzheimers Dement. 2010;6(3):239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M.S., DeKosky S.T., Dickinson D., Dubois B., Feldman H.H. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekly D.L., Ramos E.M., Lee W.W., Deitrich W.D., Jacka M.E. NIA Alzheimer's Disease Centers. The National Alzheimer's Coordinating Center (NACC) database: the Uniform Data Set. Alzheimer Dis. Assoc. Disord. 2007;21(3):249–258. doi: 10.1097/WAD.0b013e318142774e. [DOI] [PubMed] [Google Scholar]
- Bennett D.A., Schneider J.A., Arvanitakis Z., Wilson R.S. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9(6):628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D.A., Schneider J.A., Buchman A.S., Barnes L.L., Boyle P.A., Wilson R.S. Overview and findings from the rush Memory and Aging Project. Curr. Alzheimer Res. 2012;9(6):646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K., Hampel H. CSF markers for incipient Alzheimer's disease. Lancet Neurol. 2003;2(10):605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- Blennow K., Hampel H., Weiner M., Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 2010;6(3):131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- Bloom G.S. Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. Jama Neurol. 2014;71(4):505–508. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- Bombois S., Duhamel A., Salleron J., Deramecourt V., Mackowiack M.A. A new decision tree combining abeta 1–42 and p-tau levels in Alzheimer's diagnosis. Curr. Alzheimer Res. 2013;10(4) doi: 10.2174/1567205011310040002. 57–364. [DOI] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Braak H., Braak E., Bohl J. Staging of Alzheimer-related cortical destruction. Eur.Neurol. 1993;33(6):403–408. doi: 10.1159/000116984. [DOI] [PubMed] [Google Scholar]
- Brueggen K Grothe M.J., Dyrba M., Fellguiebel A., Fischer F. The European dti study on dementia – a multicenter DTI and MRI study on Alzheimer's disease and mild cognitive impairment. Neuroimage. 2017;144(Pt B):305–308. doi: 10.1016/j.neuroimage.2016.03.067. [DOI] [PubMed] [Google Scholar]
- Butler J.E. Enzyme-linked immunosorbent assay. J. Immunoass. 2000;21(2–3):165–209. doi: 10.1080/01971520009349533. [DOI] [PubMed] [Google Scholar]
- DeLong E.M., Delong D.M., Clarke-Pearson D. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- Donohue M.C., Jacqmin-Gadda H., Le Goff M., Thomas R.G., Raman R. Estimating long-term multivariate progression from short-term data. Alzheimers Dement. 2014;10(5 Suppl):S400–S410. doi: 10.1016/j.jalz.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova J., Hampel H., Molinuoevo J.L. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol. 2014;13(6):614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- Eshaghi A., Marinescu R.V., Young A.L., Firth N.C., Prados F. Progression of regional grey matter atrophy in multiple sclerosis. Brain. 2018;141(6):1665–1677. doi: 10.1093/brain/awy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez-Gonzalez A., Kulisevsky J., Boltes A., Otermin P., Garcia-Sanchez Rey verbal learning test is a useful tool for differential diagnosis in the preclinical phase of Alzheimer's disease: comparison with mild cognitive impairment and normal aging. Int J Geriatr Psychiatr. 2003;18(11):1021–1028. doi: 10.1002/gps.1010. [DOI] [PubMed] [Google Scholar]
- Fischer P., Jungwirth S., Krampla W., Weissgram S., Kircjmeye W. Vienna transdanube aging “VITA”: study design, recruitment strategies and level of participation. J. Neural Transm. Suppl. 2002;(62):105–116. doi: 10.1007/978-3-7091-6139-5_11. [DOI] [PubMed] [Google Scholar]
- Fonteijn H.M., Modat M., Clarkson M.J., Barnes J., Lehmann M. An event-based model for disease progression in Alzheimer's disease and Huntington's disease. Neuroimage. 2012;60(3):1880–1889. doi: 10.1016/j.neuroimage.2012.01.062. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Prestia A., Zanetti O., Galluzzi S., Romano M. Markers of Alzheimer's disease in a population attending a memory clinic. Alzheimers Dement. 2009;5(4):307–317. doi: 10.1016/j.jalz.2009.04.1235. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Fox N.C., Jack C.R., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010;6(2):67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G.B., Redolfi A., Manset D., Rousseau M.É., Toga A., Evans A.C. Virtual imaging laboratories for marker discovery in neurodegenerative diseases. Nat. Rev. Neurol. 2011;7(8):429–438. doi: 10.1038/nrneurol.2011.99. [DOI] [PubMed] [Google Scholar]
- Gale S.D., Baxter L., Connor D.J., Herring A., Comer J. Sex differences on the rey auditory verbal learning test and the brief visuospatial memory test-revised in the elderly: normative data in 172 participants. J. Clin. Exp. Neuropsychol. 2007;29(5):561–567. doi: 10.1080/13803390600864760. [DOI] [PubMed] [Google Scholar]
- Galluzzi S., Marizzoni M., Babiloni B., Albani D., Antelmi L. Clinical and biomarker profiling of prodromal Alzheimer's disease in workpackage 5 of the Innovative Medicines Initiative PharmaCog project: a ‘European ADNI study’. J. Intern. Med. 2016;279(6):576–591. doi: 10.1111/joim.12482. [DOI] [PubMed] [Google Scholar]
- Gur R.C., Mozley P.D., Resnick S.M., Gottlieb G.L., Kohn M. Gender differences in age effect on brain atrophy measured by magnetic resonance imaging. Proc. Natl. Acad. Sci. U. S. A. 1991;88(7):2845–2849. doi: 10.1073/pnas.88.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops S., Nazem S., Siderowf A.D., Duda J.E., Xie S.X. Validity of the MoCA and MMSE in the detection of MCI and dementia in Parkinson disease. Neurology. 2009;73(21):1738–1745. doi: 10.1212/WNL.0b013e3181c34b47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://icometrix.com
- Iturria-Medina Y., Sotero R.C., Toussaint P.J., Mateos-Pérez J.M., Evans A.C. Alzheimer's disease neuroimaging initiative. early role of vascular dysregulation on late-onset Alzheimer's disease based on multifactorial data-driven analysis. Nat. Commun. 2016;7 doi: 10.1038/ncomms11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Knopman D.S., Jagust W.J., Shaw L.P., Aisen P.S. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9(1):119. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Bennett D.A., Blennow K., Carrillo M.C., Feldman H. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87(5):539–547. doi: 10.1212/WNL.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedynak B.M., Lang A., Liu B., Katz E., Zhang Y. A computational neurodegenerative disease progression score: method and results with the Alzheimer's disease neuroimaging initiative cohort. Neuroimage. 2012;63(3):1478–1486. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.H., Vanderstichele H., Trojanowski J.Q., Shaw L.M. Simultaneous analysis of cerebrospinal fluid biomarkers using microsphere-based xMAP multiplex technology for early detection of Alzheimer's disease. Methods. 2012;56(4):484–493. doi: 10.1016/j.ymeth.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Kiraly A., Szabo N., Toth E., Csete G., Farago P. Male brain ages faster: the age and gender dependence of subcortical volumes. Brain Imaging Behav. 2016;10(3):901–910. doi: 10.1007/s11682-015-9468-3. [DOI] [PubMed] [Google Scholar]
- Koval I., Schiratti J.B., Routier A., Bacci M., Colliot O. Spatiotemporal propagation of the cortical atrophy: population and individual patterns. Front. Neurol. 2018;9:235. doi: 10.3389/fneur.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukull W.A., Higdon R., Bowen J.D., Mc Cormick W.C., Teri L. Dementia and Alzheimer disease incidence: a prospective cohort study. Arch. Neurol. 2002;59(11):1737–1746. doi: 10.1001/archneur.59.11.1737. [DOI] [PubMed] [Google Scholar]
- Li R., Zhang W., Suk H.I., Wang L., Li J. Deep learning based imaging data completion for improved brain disease diagnosis. Med Image Comput Comput Assist Interv. 2014;17(3):305–312. doi: 10.1007/978-3-319-10443-0_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi M., Filippone M., Frisoni G.B., Alexander D.C., Ourselin S. Probabilistic disease progression modeling to characterize diagnostic uncertainty: application to staging and prediction in Alzheimer's disease. Neuroimage. 2017 doi: 10.1016/j.neuroimage.2017.08.059. [DOI] [PubMed] [Google Scholar]
- Marcus D.S., Wang T.H., Parker J., Csernansky J.G., Morris J.C., Buckner R.L. Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 2007;19(9):1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Mormino E.C., Kluth J.T., Madison C.M., Rabinovici J.D., Baker S.L. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132:1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby N.P., Alexander D.C. Imaging plus X: multimodal models of neurodegenerative disease. Curr. Opin. Neurol. 2017;30(4):371–379. doi: 10.1097/WCO.0000000000000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby N.P., Young A.L., Cash D.C., Benzinger T.L.S. Data-driven models of dominantly-inherited Alzheimer's disease. Brain. 2018;141(5):1529–1544. doi: 10.1093/brain/awy050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perneczky R., Wagenpfeil S., Komossa K., Grimmer T., Diehl J., Kurz A. Mapping scores onto stages: mini-mental state examination and clinical dementia rating. Am. J. Geriatr. Psychiatr. 2006;14(2):139–144. doi: 10.1097/01.JGP.0000192478.82189.a8. [DOI] [PubMed] [Google Scholar]
- Redolfi A., Bosco P., Manset D., Frisoni G.B., neuGRID consortium Brain investigation and brain conceptualization. Funct. Neurol. 2013;28(3):175–190. [PMC free article] [PubMed] [Google Scholar]
- Redolfi A., Manset D., Barkhof F., Wahlund L.O., Glatard T. Head-to-head comparison of two popular cortical thickness extraction algorithms: a cross-sectional and longitudinal study. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0117692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen W.G., Mohs R.C., Davis K.L. A new rating scale for Alzheimer's disease. Am. J. Psychiatry. 1984;141(11):1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- Schiratti J.B., Allassoniniere S., Colliot O., Durrelman S. Learning spatiotemporal trajectories from manifold-valued longitudinal data. Adv. Neural Inf. Proces. Syst. 2015:2404–2412. [Google Scholar]
- Sperling R.A., Aisen P.S., Beckett L.A., Bennett D.A., Craft S. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on aging-Alzheimer's association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Kate M., Barkhof F., Visser P.J., Teunissen C.E., Scheltens P. Amyloid-independent atrophy patterns predict time to progression to dementia in mild cognitive impairment. Alzheimers Res. Ther. 2017;9:73. doi: 10.1186/s13195-017-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Kate M., Redolfi A., Peira E., Bos I., Vos S.J. MRI predictors of amyloid pathology: results from the EMIF-AD multimodal biomarker discovery study. Alzheimers Res. Ther. 2018;10:100. doi: 10.1186/s13195-018-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Kate M., Ingala S., Schwartz A.J., Fox N.C., Chételat G. Secondary prevention of Alzheimer's Dementia: neuroimaging contributions. Alzheimers Res. Ther. 2018;10(112) doi: 10.1186/s13195-018-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombaugh T.N., McIntyre N.J. The mini-mental state examination: a comprehensive review. J. Am. Geriatr. Soc. 1992;40(9):922–935. doi: 10.1111/j.1532-5415.1992.tb01992.x. [DOI] [PubMed] [Google Scholar]
- van der Flier W.M., Pijnenburg Y.A., Prins N., Lemstra A.W., Mouwman F.H. Optimizing patient care and research: the Amsterdam Dementia Cohort. J. Alzheimers Dis. 2014;41(1):313–327. doi: 10.3233/JAD-132306. [DOI] [PubMed] [Google Scholar]
- Vemuri P., Jack C.R. Role of structural MRI in Alzheimer's disease. Alzheimers Res. Ther. 2010;2(4):23. doi: 10.1186/alzrt47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatragahvan V., Bron E.E., Niessen W.J., Klein S. Disease progression timeline estimation for Alzheimer's disease using discriminative event based modeling. Neuroimaging. 2019;186:518–532. doi: 10.1016/j.neuroimage.2018.11.024. [DOI] [PubMed] [Google Scholar]
- Venkatraghavan V., Bron E., Niessen W., Klein S.A. Information Processing in Medical Imaging International Conference on Information Processing in Medical Imaging. Vol. 10265. Springer; Cham: 2017. Discriminative event based model for Alzheimer's disease progression modeling. Lecture Notes in Computer Science. [Google Scholar]
- Wijeratne P.A., Young A.L., Oxtoby N.P., Marinescu R.V., Firth N.C. An image-based model of brain volume biomarker changes in Huntington's disease. Ann Clin Transl Neur. 2018;5(5):570–582. doi: 10.1002/acn3.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette A.A., Calhoun V.D., Egan J.M., Kapogiannis D., Alzheimer's Disease Neuroimaging Initiative Prognostic classification of mild cognitive impairment and Alzheimer's disease: MRI independent component analysis. Psychiatry Res. 2014;224:81–88. doi: 10.1016/j.pscychresns.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.L., Oxtoby N.P., Daga P., Cash D.M., Fox N.C. A data-driven model of biomarker changes in sporadic changes in sporadic Alzheimer's disease. Brain. 2014;137(9):2564–2577. doi: 10.1093/brain/awu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.L., Oxtoby N.P., Huang J., Marinescu R.V., Daga P. Process Med Imaging. Vol. 24. 2015. Multiple orderings of events in disease progression; pp. 711–722. [DOI] [PubMed] [Google Scholar]
- Young A., Marinescu R., Oxtoby N., Bocchetta M., Yong K. Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with subtype and stage inference. Nat. Commun. 2018;9:4273. doi: 10.1038/s41467-018-05892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material