Abstract
Mutations in gene regulatory elements have been associated with a wide range of complex neuropsychiatric disorders. However, due to their cell-type specificity and difficulties in characterizing their regulatory targets, the ability to identify causal genetic variants has remained limited. To address these constraints, we perform integrative analysis of chromatin interactions using promoter capture Hi-C (pcHi-C), open chromatin regions using ATAC-seq, and transcriptomes using RNA-seq in four functionally distinct neural cell types: iPSC-induced excitatory neurons and lower motor neurons, iPSC-derived hippocampal dentate gyrus (DG)-like neurons, and primary astrocytes. We identify hundreds of thousands of long-range cis interactions between promoters and distal promoter-interacting regions (PIRs), enabling us to link regulatory elements to their target genes and reveal putative processes that are dysregulated in disease. Finally, we validate several PIRs using CRISPR techniques in human excitatory neurons, demonstrating that CDK5RAP3, STRAP, and DRD2 are transcriptionally regulated by physically linked enhancers.
A large number of genetic variants associated with diverse human traits and diseases are located in putative regulatory regions. Genetic lesions in these regulatory elements can contribute to complex human disease by modulating gene expression and disrupting finely tuned transcriptional networks. However, deciphering the roles of noncoding variants in disease etiology remains nontrivial due to their lack of annotation in the physiologically relevant cell types. Furthermore, regulatory elements often interact with their target genes over long genomic distances, precluding a straightforward mapping of regulatory element connectivity and limiting the interpretation of noncoding variants from genome-wide association studies (GWAS). Typically, neighboring genes are assigned as risk loci for noncoding variants. However, this nearest gene model is challenged by both experimental and computational evidence1,2. For instance, two independent obesity-associated single-nucleotide polymorphisms (SNPs) in the FTO gene have been shown not to regulate FTO, but IRX3 in the brain and both IRX3 and IRX5 in adipocytes, respectively3,4. The FTO locus in obesity illustrates the potentially intricate and cell-type-specific manner in which noncoding variants contribute to disease. However, such well-annotated cases are rare, and we still lack systematic mapping of GWAS SNPs to their regulatory targets, especially in the context of complex neuropsychiatric disorders.
Previous epigenomic annotations of the germinal zone (GZ) and cortical and subcortical plates (CP) in the human brain revealed the importance of three-dimensional (3D) chromatin structure in gene regulation and disease5,6. However, these studies used complex, heterogeneous tissues, limiting the ability to interpret gene regulation in a cell-type-specific manner. Therefore, charting the landscape of epigenomic regulation in well-characterized, physiologically relevant cell types should offer significant advantages for identifying causal variants, deciphering their functions, and enabling novel therapies. Towards this goal, we used wild type human iPSCs (WTC11 line7) to generate three neuronal cell types: excitatory neurons8, hippocampal dentate gyrus (DG)-like neurons9, and lower motor neurons10. GFAP-positive astrocytes from the brains of two individuals were also included for their relevance to human brain development and disease. By performing integrative analysis of promoter-centric, long-range chromatin interactions, open chromatin regions, and transcriptomes (Fig. 1a), we provide comprehensive annotations for promoters and distal promoter-interacting regions (PIRs) in each cell type. We identify putative gene targets for both in-vivo-validated enhancer elements from the VISTA Enhancer Browser11 and disease-associated variants, enabling the functional validation of PIRs driving diverse processes in cellular identity and disease.
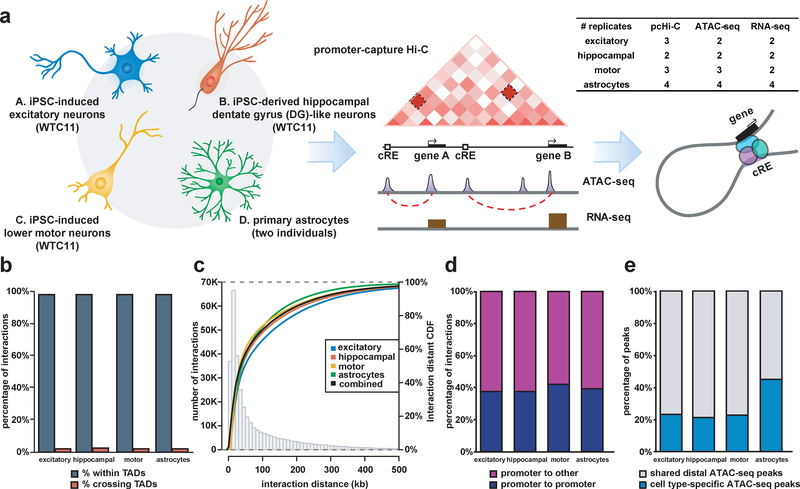
Figure 1. Genome-wide mapping of physical chromatin interactions in functionally distinct neural cell types.
(a) Schematic of the study design for generating four functionally distinct cell types in the CNS and performing integrative analysis of chromatin interactions using pcHi-C, open chromatin regions using ATAC-seq, and transcriptomes using RNA-seq. The number of biological replicates based on independent experiments for each cell type is shown for each assay. (b) Proportions of interactions occurring within TADs for each cell type. (c) Histogram and empirical CDF plots of interaction distances for each cell type. (d) Proportions of interactions between promoter-containing bins (blue) and between promoter- and non-promoter-containing bins (purple) for each cell type. (f) Proportions of cell type-specific (blue) and shared (grey) distal open chromatin peaks at PIRs for each cell type.
Results
Characterizing the epigenomic landscape of long-range chromatin interactions in human neural cells
To investigate general epigenomic features for cells in the human central nervous system (CNS), we focused on isogenic iPSC-induced excitatory neurons, iPSC-derived hippocampal dentate gyrus (DG)-like neurons, and iPSC-induced lower motor neurons, three neuronal subtypes which are currently impractical to isolate from primary tissue. Excitatory neurons were induced from a wild type male iPSC line (WTC11) containing an integrated, isogenic, and inducible neurogenin-2 (Ngn2) cassette (i3N iPSCs) with doxycycline-inducible Ngn2 at the AAVS1 safe-harbor locus8. The i3N iPSCs were used to prepare homogenous cultures of excitatory neurons expressing the glutamatergic neuron marker VGLUT1 and the cortical neuron marker CUX18,12, though FOXG1 expression was not detected (Supplementary Fig. 1a,b). Hippocampal DG-like neurons expressing the DG granule cell marker PROX1 were differentiated from a WTC11 line using factors as described previously9,13 (Supplementary Fig. 1a,b). Finally, lower motor neurons were induced from WTC11 cells containing integrated, isogenic, and inducible NGN2, ISL1, and LHX3 at the AAVS1 safe-harbor locus (i3LMN iPSCs)10. The cells exhibited homogenous expression of the lower motor neuron markers HB9 and SMI32 (Supplementary Fig. 1a,b). All three neuronal subtypes expressed the synaptic genes SYN1 and SYN2, the NMDA receptor genes GRIN1 and GRIN2A, and the AMPA receptor genes GRIA1 and GRIA2, evidencing mature synaptic functions (Supplementary Fig. 1b). We also included two batches of astrocytes isolated from 19-week old male fetal brain samples using GFAP as a selection marker (ScienCells). Astrocytes were cultured for two or fewer passages in vitro and confirmed for positive expression of GFAP prior to harvesting (Supplementary Fig. 1a). Based on the age of the donors and transcriptional signatures for dozens of marker genes distinguishing astrocyte progenitor cells (APCs) (e.g. AGXT2L1 and WIF1) from mature astrocytes (e.g. TOP2A and TNC)14, the astrocytes were determined to be APCs (Supplementary Fig. 1b).
We prepared pcHi-C, ATAC-seq, and RNA-seq libraries using two to four biological replicates based on independent experiments for each cell type (Fig. 1a and Supplementary Table 1). Specifically, promoter-centric, long-range chromatin interactions were mapped using a set of 280,445 RNA probes targeting the promoters of 19,603 coding genes in GENCODE 1915. We first confirmed the reproducibility of contact frequency and saturation of inter-replicate correlation for our pcHi-C libraries using HiCRep16 (Supplementary Fig. 2c,d). Hierarchical clustering of ATAC-seq read density and gene expression similarly grouped the replicates by cell type (Supplementary Fig. 2a,b), evidencing minimal variations during the cell derivation process. Using CHiCAGO17, we identified significant chromatin interactions with score ≥ 5 at 195,322 unique interacting loci across all four cell types, with 73,890, 108,156, 66,978, and 84,087 significant interactions being represented in the excitatory neurons, hippocampal DG-like neurons, lower motor neurons, and astrocytes, respectively (Supplementary Table 2). Overall, 17,065 or 83.9% of coding gene promoters participate in interactions in at least one cell type (Supplementary Fig. 1c), with 80% of PIRs interacting within a distance of 160 kb (Fig 1c and Supplementary Fig. 3a). Over 97% of interactions occur within topologically associating domains (TADs) in human fetal brain tissues6 (Fig. 1b). Furthermore, approximately 40% of interactions occur between promoter-containing bins, while 60% occur between promoter- and non-promoter-containing bins (Fig. 1d). The observed numbers of promoter-promoter interactions can potentially be attributed to transcriptional factories of coregulated genes, the widespread colocalization of promoters18,19, and the capacity of many promoters to doubly function as enhancers20,21. Finally, up to 40% of interacting distal open chromatin peaks are specific to each cell type (Fig. 1e), suggesting that PIRs are capable of orchestrating cell-type-specific gene regulation. Astrocytes, in particular, exhibit the largest proportion of cell-type-specific open chromatin peaks, likely reflecting basic differences between the neuronal and glial lineages.
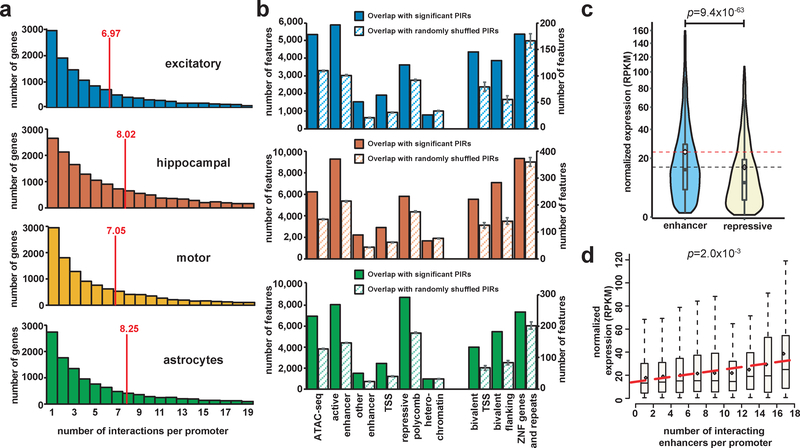
The majority of promoters interact with more than one PIR (Fig. 2a), consistent with the large number of regulatory elements in the human genome22 and previous findings that promoters can be regulated by multiple enhancers23. To examine global chromatin signatures at PIRs, we leveraged chromatin states inferred by ChromHMM24 in matched human brain tissues from the Roadmap Epigenomics Project25 (dorsolateral prefrontal cortex for excitatory neurons, hippocampus middle for hippocampal DG-like neurons, and normal human astrocytes for astrocytes). We show that PIRs are highly enriched for active chromatin features including open chromatin peaks, enhancers, and transcriptional start sites (TSSs) while simultaneously exhibiting depletion for repressive heterochromatin marks (Fig. 2b). PIRs are also enriched for H3K27ac and CTCF binding sites mapped using CUT&RUN26 in excitatory and lower motor neurons and ChIP-seq in astrocytes from ENCODE27 (Supplementary Fig. 3b). Promoters interacting with enhancer-PIRs exhibit elevated levels of transcription compared to those interacting with repressive-PIRs (P = 9.4 × 10−63, two-tailed two sample t test) (Fig. 2c and Supplementary Fig. 3c). Multiple enhancer-PIRs also present evidence for additive effects on transcription. By grouping genes according to the number of interactions their promoters form with enhancer-PIRs, a modest correlation is observed between the number of interactions and the mean gene expression in each group (P = 2.1 × 10−3, F-test for linear regression) (Fig. 2d and Supplementary Fig. 3d). Our results demonstrate that chromatin interactions identify PIRs that are not only enriched for regulatory features but can also alter gene expression.
Figure 2. Integrative analysis of chromatin interactions, epigenomic features, and gene expression.
(a) Histograms of the number of PIRs interacting with each promoter for each cell type. Means are indicated. Only protein coding and noncoding RNA promoters interacting with at least one PIR are included (15,316 promoters in excitatory neurons, 19,546 promoters in hippocampal DG-like neurons, 14,990 promoters in lower motor neurons, and 15,397 promoters in astrocytes). (b) Bar plots showing counts of epigenomic chromatin states inferred using ChromHMM in matched tissues overlapping significant (solid bars) versus randomly shuffled (striped bars) PIRs for each cell type. Means and the SEM for the number of overlaps across n = 100 sets of randomly shuffled PIRs are shown. (c) Comparative gene expression analysis across all cell types for expressed genes (normalized RPKM > 0.5) whose promoters interact exclusively with either enhancer-PIRs (n = 6,836 genes) or repressive-PIRs (n = 2,612 genes) (P = 9.4 × 10−63, t = 16.9, df = 6854.6, two-tailed two-sample t test). Violin plots show the distributions of gene expression values within each group, and boxplots indicate the median, IQR, Q1 – 1.5 × IQR, and Q3 +1.5 × IQR. Means are indicated with dotted horizontal lines. (d) Distributions of gene expression values across all cell types for expressed genes (normalized RPKM > 0.5) grouped according to the numbers of interactions their promoters form with enhancer-PIRs. Boxplots indicate the median, IQR, Q1 – 1.5 × IQR, and Q3 + 1.5 × IQR. Linear regression was performed on the mean gene expression values for n = 9 bins containing at least 10 genes (P = 2.1 × 10−3, F1,7 = 22.7, F-test for linear regression).
PIRs contribute to cellular identity
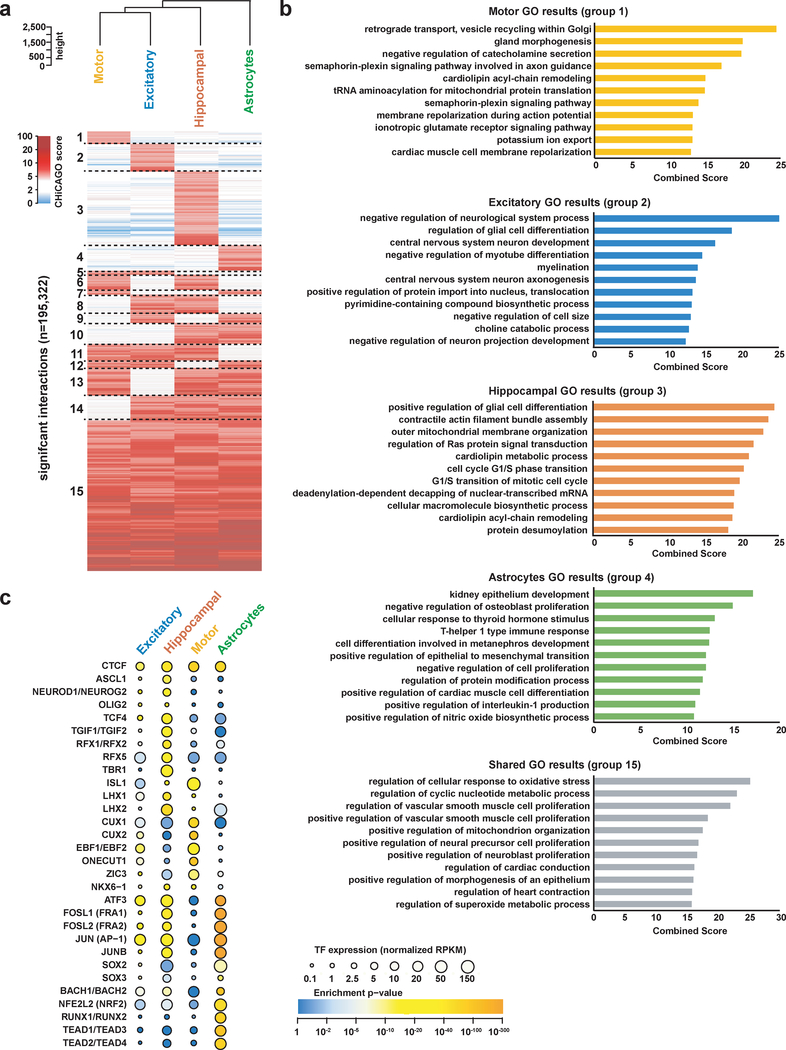
Chromatin interactions exhibit distinct patterns of cell-type specificity, with tens of thousands of interactions observed to be specific for each cell type (Fig. 3a and Supplementary Fig. 4a). These interactions may underlie important functional differences between the cell types, with gene ontology (GO) enrichment analysis28 on genes interacting with cell-type-specific PIRs yielding terms associated with neuronal function in the neuronal subtypes and immune function in the astrocytes (Fig. 3b and Supplementary Table 3). Meanwhile, 58,809 or 30.1% of unique interactions are shared across all four cell types, with neural precursor cell proliferation and neuroblast proliferation ranking among the top terms for genes participating in shared interactions. In conjunction with the observed enrichment of active chromatin signatures at PIRs, their association with cell-type-specific processes suggests that PIRs harbor lineage-specific regulatory roles. Indeed, numerous promoters of differentially expressed genes, including OPHN1 in hippocampal DG-like neurons, CHAT in lower motor neurons, and TLR4 in astrocytes, form specific contacts with PIRs in their respective cell types (Supplementary Fig. 4b). Notably, OPHN1 stabilizes synaptic AMPA receptors and mediates long-term depression in the hippocampus, and its loss of function has been linked to mental retardation29. CHAT is a principal marker for lower motor neuron maturity and function, and TLR4 is a key regulator of immune activation and synaptogenesis in astrocytes30.
Figure 3. Cell-type-specific PIRs and TF motif enrichment analysis.
(a) Classification of significant promoter-PIR interactions with interaction score ≥ 5 in at least one cell type based on their overall cell type-specificities. Counts of interactions in each specificity category are summarized in Supplementary Figure 3a. Cell types are hierarchically clustered based on their interaction scores over all interacting loci. (b) Top enriched GO terms from the “GO Biological Process 2018” ontology in Enrichr for genes participating in cell type-specific (groups 1–4) versus shared (group 15) interactions with distal open chromatin peaks. 459, 837, 217, 307, and 1,925 genes were used as inputs for groups 1–4 and 15, respectively. Enriched GO terms are ranked by their combined scores (calculated by multiplying the log of the P value via Fischer’s exact test with the z-score of the deviation from the expected rank). An expanded list of enriched GO terms is available in Supplementary Table 3. (c) Enrichment of consensus TF motif sequences at open chromatin peaks in cell type-specific PIRs by motifs (rows) and cell types (columns). 1,145, 1,271, 843, and 2,566 peaks were used as input for the excitatory neurons, hippocampal DG-like neurons, lower motor neurons, and astrocytes, respectively. The color of each dot represents the degree of enrichment (calculated using the cumulative binomial distribution in HOMER) for each motif and cell type, and the size of each dot represents the gene expression of the corresponding TFs for each motif. Entries with similar or identical consensus TF motif sequences are grouped for brevity.
Gene expression is coordinately controlled by transcription factors (TFs) and regulatory elements such as enhancers. Therefore, PIRs provide the means for investigating mechanisms underlying cell-type-specific gene regulation. We used HOMER31 to evaluate TF motif enrichment at cell-type-specific distal open chromatin peaks in PIRs for each cell type (Fig. 3c and Supplementary Table 4). First, the CTCF motif is highly enriched across all cell types, consistent with its role in mediating looping within TADs32–35. Motifs for ASCL1, ISL1, NEUROG2, OLIG2, and ZIC3, TFs linked to neuronal fate commitment, are also broadly enriched across the neuronal subtypes. Additional TFs functioning in brain development include CUX1, CUX2, EBF1, EBF2, LHX1, LHX2, NKX6–1, ONECUT1, RFX1, RFX2, RFX5, TCF4, and TGIF2. The TBR1 motif is highly enriched in hippocampal DG-like neurons, consistent with its expression in the hippocampus36. Meanwhile, astrocytes are enriched for motifs in the Fos and Jun families, which contain key regulators for inflammatory and immune pathways. Also enriched in astrocytes are motifs for ATF3 and the RUNX and TEAD families, TFs with established roles in astrocyte differentiation, maturation, and proliferation. Motif enrichment is not always accompanied by expression of the corresponding TFs. This may reflect potential synergistic interactions between the cell types. For example, NRF2 is a key regulator of the oxidative stress response, and its activity has been shown to be repressed in neurons while also inducing a strong response in astrocytes37. Therefore, its shared expression may reflect the neuroprotective roles that astrocytes serve for other cell types. Alternatively, TFs do not have to be highly expressed to perform their cellular functions due to additional avenues for regulation at the post-transcriptional and post-translational levels. Our results support the notion that PIRs contribute to cell fate commitment and are capable of identifying both known and novel regulators in a cell-type-specific manner.
Identification of regulatory targets for in-vivo-validated enhancer elements using chromatin interactions
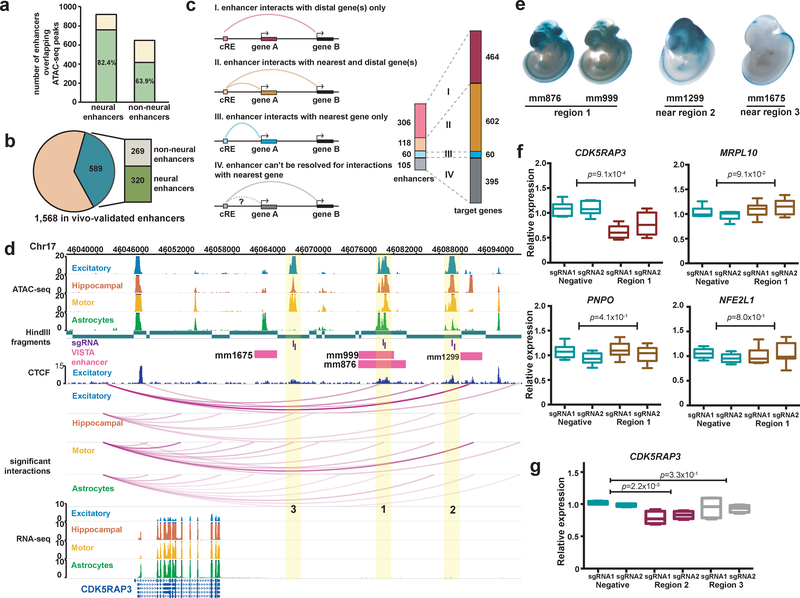
Regulation of target genes by enhancers is thought to be mediated by physical chromatin looping. Congruent with this concept, chromatin interactions detected by pcHi-C can be used to link enhancers with their target genes. The VISTA Enhancer Browser is a database containing experimentally validated human and mouse noncoding sequences with enhancer activity11. To date, it contains 2,956 tested sequences, 1,568 of which exhibit enhancer activity during embryonic development. However, the regulatory targets for these enhancer elements have remained largely uncharacterized. To address this knowledge gap, we provide cell-type-specific annotations of putative target genes for each enhancer element using our chromatin interactions and open chromatin peaks (Supplementary Table 5). Across all cell types, our interactions recover 589 or 37.6% of positively tested enhancer elements with human sequences, 320 of which were further annotated as neural enhancers according to tissue-specific patterns of LacZ staining in mouse embryos (Fig. 4a,b). Of the 589 interacting positive enhancer elements, 306 interact exclusively with 464 more distal genes (scenario I), 118 interact with both their nearest genes and 484 more distal genes (scenario II), and 60 interact exclusively with their nearest genes (scenario III) (Fig. 4c). The remaining 105 elements could not be resolved for interactions with their nearest genes (scenario IV), though they were found to interact with 395 more distal genes. In total, our interactions identify 1,343 novel gene targets for positive enhancer elements in the VISTA Enhancer Browser, significantly expanding our knowledge of gene regulatory relationships at these loci.
Figure 4. Validation of PIRs in human neural cells.
(a) In-vivo-validated enhancer elements with neural annotations overlap a higher proportion of open chromatin peaks in the neural cells (757 of 919 elements) compared to enhancer elements with non-neural annotations (415 of 649 elements) (P < 2.2 × 10−16, χ2 = 67.5, df = 1, Pearson’s chi-squared test with Yates’s correction). (b) Counts of enhancer elements participating in chromatin interactions (589 of 1,568 elements) with neural and non-neural annotations. (c) Counts of enhancer elements interacting exclusively with their nearest genes (blue), more distal genes (pink), or both (orange), and the number of target genes for each scenario (right). (d) Open chromatin peaks in cell-type-specific PIRs (regions 1, 2, and 3) interact with the CDK5RAP3 promoter. Both enhancer elements (pink) and CTCF binding sites in excitatory neurons (dark blue) are localized to all three regions, and all interactions occur within a TAD in the cortical plate (chr17:45,920,000–47,480,000). (e) LacZ staining in mouse embryos reveals tissue-specific patterns of enhancer activity. (f) CRISPRi silencing of region 1 results in significant downregulation of CDK5RAP3 expression in excitatory neurons (P = 9.1 × 10−4, t = 4.65, df = 10 two-tailed two-sample t test). The neighboring genes MRPL10, PNPO, and NFE2L1 were unaffected (P = 9.1 × 10−2, t = 1.87, df = 10, P = 4.1 × 10−1, t = 0.853, df = 10, and P = 8.0 × 10−1, t = 0.259, df = 10, respectively, two-tailed two-sample t test). Three independent replicates per condition and two sgRNAs per replicate were used for each experiment. Boxplots indicate the median, IQR, minimum, and maximum. (g) CRISPRi silencing of region 2, but not region 3, results in significant downregulation of CDK5RAP3 expression in excitatory neurons (P = 2.2 × 10−3, t = 5.11, df = 6 and P = 3.3 × 10−1, t = 1.05, df = 6, respectively, two-tailed two-sample t test). Two independent replicates per condition and two sgRNAs per replicate were used for each experiment. Boxplots indicate the median, IQR, minimum, and maximum.
Validation of PIRs in human neural cells using CRISPR techniques
We validated two PIRs physically interacting up to 40 kb away with the CDK5RAP3 promoter (regions 1 and 2) (Fig. 4d). CDK5RAP3 regulates CDK5, which functions in neuronal development38 and regulates proliferation in non-neuronal cells39. Notably, both PIRs overlap open chromatin peaks as well as enhancers annotated with forebrain activity in the VISTA Enhancer Browser (mm876 and mm999 for region 1 and mm1299 for region 2) (Fig. 4e). We targeted both regions for CRISPR deletion in the i3N iPSCs, followed by differentiation of the cells into excitatory neurons and quantification of any changes in gene expression by qPCR. Deleting the 2 kb open chromatin peak in region 1 led to significant downregulation of CDK5RAP3 expression across three independent clones (P = 1.6 × 10−2, two-tailed two sample t test) (Supplementary Fig. 4c). However, upon trying to delete the open chromatin peak in region 2, we observed massive cell death of iPSCs immediately following introduction of the Cas9-sgRNA protein complex. We picked 48 individual clones from cells surviving the transfection, but failed to isolate any clones with deletions, suggesting that this locus is essential for maintaining CDK5RAP3 expression and survival in iPSCs. To circumvent this lethal phenotype for iPSCs with region 2, we used CRISPR interference (CRISPRi) to silence both regions 1 and 2 in excitatory neurons. We also silenced a third region that interacts with the CDKRAP3 promoter in the other cell types, but not in excitatory neurons (region 3). We show that silencing of regions 1 and 2, but not region 3, leads to significant downregulation of CDK5RAP3 expression without influencing the expression of nearby genes (P = 9.1 × 10−4 for region 1 and P = 2.2 × 10−3 for region 2, two-tailed two sample t test) (Fig. 4f–g). Interestingly, an enhancer with spinal cord activity proximal to region 3 (mm1675) interacts with the CDK5RAP3 promoter in lower motor neurons and astrocytes, but not in the other cell types (Fig. 4d,e). Overall, these results show that chromatin interactions recapitulate cell-type-specific patterns of enhancer activity, underscoring the importance of studying epigenomic regulation in the appropriate cell types.
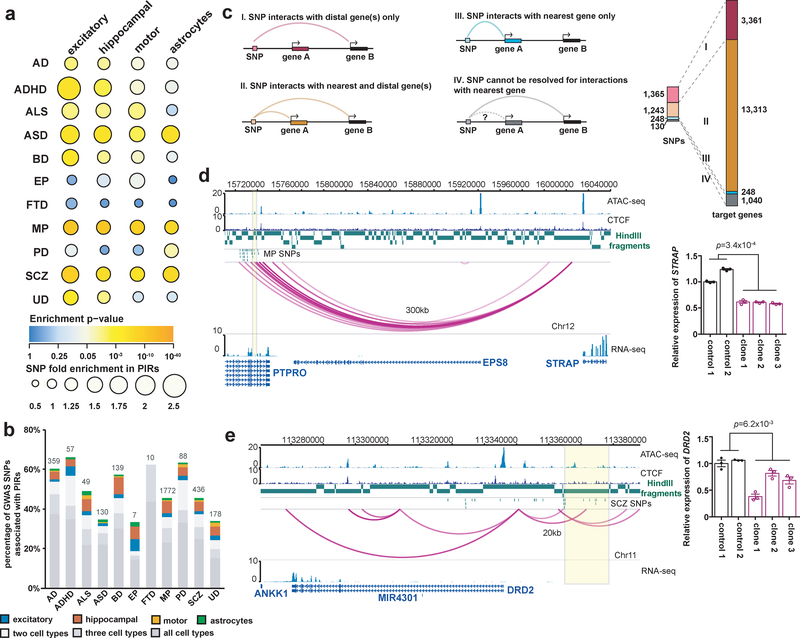
Cell-type-specific enrichment and regulatory target identification for neuropsychiatric disorder risk variants at PIRs
Previous large-scale epigenomic studies of human tissues and cell lines highlighted the importance of disease-associated variants at distal regulatory regions25 and the need for high-throughput approaches to prioritize variants for further validation. Therefore, we used our chromatin interactions to annotate complex neuropsychiatric disorder- or trait-associated variants from the GWAS Catalog40. We mined a total of 6,396 unique GWAS SNPs at a significance threshold of 10−6 for eleven traits including Alzheimer’s disease (AD), attention deficit hyperactivity disorder (ADHD), autism spectrum disorder (ASD), amyotrophic lateral sclerosis (ALS), bipolar disorder (BD), epilepsy (EP), frontotemporal dementia (FTD), mental process (MP), Parkinson’s disease (PD), schizophrenia (SCZ), and unipolar depression (UD). We identified linked SNPs at a linkage disequilibrium (LD) threshold of 0.8 using HaploReg41 for a total of 95,954 unique SNPs across all traits (Supplementary Table 6). We find that SNPs are enriched at PIRs in a disease- and cell-type-specific manner (Fig. 5a), with ASD, MP, and SCZ SNPs enriched at PIRs across all cell types. UD SNPs are enriched exclusively in excitatory and hippocampal DG-like neurons, whereas AD, ADHD, and BD SNPs also exhibit enrichment in lower motor neurons. ALS SNPs are enriched in the neuronal subtypes but not in astrocytes, consistent with the characterization of ALS as a motor neuron disease and reinforcing evidence for its role in hippocampal degeneration42. Interestingly, PD SNPs are enriched in astrocytes but not in the other cell types. This enrichment of PD SNPs at astrocyte-specific PIRs supports the theory that astrocytes play an initiating role in PD, based on evidence that numerous genes implicated in PD have functions unique to astrocyte biology, as well as the neuroprotective roles astrocytes serve for dopaminergic neurons in the substantia nigra43. EP and FTD SNPs are not enriched in any of the cell types, indicating their potential functions in alternative cell types, insufficient study power, or mechanisms acting outside of chromatin-mediated gene regulation.
Figure 5. Genetic analysis of chromatin interactions with complex neuropsychiatric disorder-associated variants.
(a) Enrichment analysis for eleven complex neuropsychiatric disorders or traits. The color and size of each dot represent the enrichment P value (two-tailed one sample z-test) and the raw fold enrichment (determined as the number of SNPs overlapping significant PIRs divided by the mean number of SNPs overlapping n = 100 sets of randomly shuffled PIRs), respectively. The total numbers of SNPs are available in Supplementary Table 6. (b) Proportions and counts of GWAS SNPs with at least one linked SNP participating in chromatin interactions. (c) Counts of GWAS SNPs across all diseases with at least one linked SNP interacting exclusively with their nearest genes (blue), more distal genes (pink), or both (orange), and the number of target genes for each scenario (right). (d) PIRs with MP SNPS in an intron for PTPRO interact with the STRAP promoter. All interactions occur within a TAD in the cortical plate (chr12:14,960,000–16,040,000). Biallelic deletion of this PIR in three independent clones results in significant downregulation of STRAP expression in excitatory neurons (P = 3.4 × 10−4, t = 18.5, df = 3, two-tailed two-sample t test). Error bars represent the SEM. (e) A PIR containing SCZ SNPs interacts with the DRD2 promoter. All interactions occur within a TAD in the cortical plate (chr11:113,200,000–114,160,000). Monoallelic deletion of this PIR in three independent clones results in significant downregulation of DRD2 expression in excitatory neurons (P = 6.2 × 10−3, t = 6.92, df = 3, two-tailed two-sample t test). Error bars represent the SEM.
Up to 70% of GWAS SNPs have at least one linked SNP overlapping PIRs in one or more cell types (Fig. 5b). As it is common practice to assign GWAS SNPs to their nearest genes, we counted the number of GWAS SNPs with at least one linked SNP interacting with their nearest gene across all diseases. We find that 1,365 GWAS SNPs interact exclusively with 3,361 more distal genes (scenario I), 1,243 GWAS SNPs interact with both their nearest genes and 12,070 more distal genes (scenario II), and 248 GWAS SNPs interact exclusively with their nearest genes (scenario III) (Fig. 5c and Supplementary Fig. 5a). In total, 16,471 non-neighboring gene targets are identified across all diseases (Supplementary Table 7). To prioritize variants potentially disrupting regulatory interactions, we focused on SNPs overlapping open chromatin peaks at PIRs, and find that these putative regulatory SNPs interact with genes that are relevant in the context of their respective disease etiologies (Supplementary Table 8). GO enrichment analysis for genes targeted by AD SNPs yields terms associated with amyloid beta formation, interferon beta production, and cranial nerve development (Supplementary Fig. 6 and Supplementary Table 9). Meanwhile, genes targeted by ASD, BD, SCZ, and UD SNPs are enriched for epigenetic terms including chromatin assembly, nucleosome assembly, and nucleosome organization. For genes targeted by GWAS SNPs in the other diseases, enriched terms include neuronal processes such as myelin maintenance, neuron projection extension, synapse assembly, synaptic transmission, and nervous system development.
Notably, a previously reported interaction between the FOXG1 promoter and a PIR with SCZ SNPs over 700 kb away is recapitulated by our data6 (Supplementary Fig. 5b). At a different locus, an astrocyte-specific PIR with AD SNPs targets the promoter of CASP2, which encodes a well-known mediator of apoptosis that is linked to neurodegeneration44,45 (Supplementary Fig. 7a). Hippocampal DG-like neuron-specific PIRs with ASD SNPs target the promoter of BCAS2, whose knockdown in mice leads to microcephaly-like phenotypes with reduced learning, memory, and DG volume46 (Supplementary Fig. 7c). Finally, the MSI2 promoter is targeted by an astrocyte-specific PIR with SCZ SNPs, as well as by PIRs with BD SNPs in hippocampal DG-like neurons, lower motor neurons, and astrocytes (Supplementary Fig. 7d). In conclusion, we demonstrate that an approach leveraging epigenomic data to jointly prioritize and map regulatory targets for variants enables the identification of putative processes that are disrupted in disease and development.
Validation of PIRs containing neuropsychiatric disorder risk variants
PIRs with MP SNPs in an intron for PTPRO interact over 300 kb away with the promoter of STRAP (Fig. 5d), which encodes a component of the survival of motor neuron (SMN) complex47. The complex itself facilitates spliceosome assembly and is associated with spinal muscular atrophy48. To validate this locus, we derived three independent i3N iPSC clones containing biallelic deletions for a PIR in this region and observed significant downregulation of STRAP expression following differentiation of the cells into excitatory neurons (P = 3.4 × 10−4, two-tailed two-sample t test). Targeting the same PIR with CRISPRi also consistently downregulated STRAP expression in excitatory neurons (P = 7.4 × 10−3, two-tailed two-sample t test) (Supplementary Fig. 5c). Next, we focused on a PIR 20 kb upstream from the promoter of DRD2, which encodes the D2 subtype of the dopamine receptor. Previously, rs2514218:C>T, a noncoding variant 47 kb upstream from DRD2, was found to be associated with antipsychotic drug response in a cohort of schizophrenia patients49. This variant is in LD with a cluster of SCZ SNPs overlapping open chromatin peaks in the PIR for DRD2. DRD2 is also the gene associated with the Taq1A polymorphism, which has been linked to reduced dopamine receptor density as well as addiction, anxiety, depression, and social problems in patients50. We first demonstrate that monoallelic deletion of this PIR in three independent clones leads to significant downregulation of DRD2 expression in excitatory neurons (P = 6.2 × 10−3, two-tailed two-sample t test) (Fig. 5e). Next, through TOPO cloning and genotyping cDNA with allele-specific variants, we confirm that monoallelic deletion of the same PIR leads to allelically imbalanced DRD2 expression (Supplementary Fig. 5d). By prioritizing and validating PIRs containing putative regulatory SNPs for key genes such as DRD2, our approach may enable the development of therapeutic and diagnostic strategies targeting specific risk variants in otherwise recalcitrant complex neuropsychiatric disorders.
Genetic variants contribute to chromatin interaction bias and alterations in gene expression
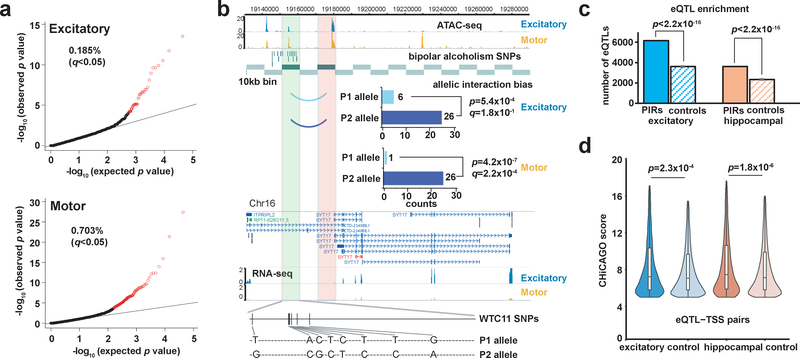
Since regulatory variants and other genetic perturbations are thought to introduce or disrupt chromatin loops between promoters and PIRs, we were interested to see if we could detect instances of allelic bias across our sets of significant promoter-PIR interactions. We used our chromatin interaction data to perform genome-wide phasing of WTC11 variants using HaploSeq51 and performed allele-specific mapping at a resolution of 10 kb using HiC-Pro52. We identified 41 (0.185%) and 151 (0.703%) of significantly interacting bins to exhibit allelic bias at an FDR cutoff of 5% (two-tailed binomial test with BH correction) in the excitatory and lower motor neurons, respectively (Fig. 6a and Supplementary Table 10). In one case, allelically biased interactions were detected between a PIR containing bipolar alcoholism SNPs53 and the promoter of SYT17, which encodes a member of a family of membrane-trafficking proteins that mediate synaptic function and calcium-controlled neurotransmitter release54. The risk allele of the lead variant (rs8062326:G>A) is associated with the WTC11 allele exhibiting reduced interaction frequency in both cell types (Fig. 6b), suggesting that regulatory variants can increase individual risk for bipolar alcoholism by disrupting interactions for SYT17.
Figure 6. Genetics variants contribute to chromatin interaction bias and alterations in gene expression.
(a) Quantile-quantile plots showing the proportions of interacting 10-kb bins exhibiting significant allelic bias at an FDR cutoff of 5% (two-tailed binomial test with BH correction) in excitatory neurons (n = 22,162 bins) and lower motor neurons (n = 21,479 bins). (b) A sample interaction with significant allelic bias in excitatory neurons (P = 5.4 × 10−4, two-tailed binomial test) and lower motor neurons (P = 4.2 × 10−7, two-tailed binomial test). The interaction occurs between the SYT17 promoter and a PIR with bipolar alcoholism SNPs at an open chromatin peak. Heterozygous phased WTC11 variants at the PIR as well as bar graphs of allele-specific read counts are shown. (c) Enrichment of significant eQTLs from GTEx V7 at significant versus randomly shuffled PIRs in matched tissue types for excitatory and hippocampal DG-like neurons (P < 2.2 × 10−16 for both cell types, two-tailed one sample z-test). Means and the SEM for the number of overlaps across n = 100 sets of randomly shuffled PIRs are shown. (d) Distributions of interaction scores for chromatin interactions overlapping significant versus randomly sampled nonsignificant eQTL-TSS pairs in excitatory and hippocampal DG-like neurons (P = 2.3 × 10−4 for excitatory neurons and P = 1.8 × 10−6 for hippocampal DG-like neurons, two-tailed two-sample Kolmogorov-Smirnov test). Additional details are available in the methods. Violin plots show the distributions of gene expression values within each group, and boxplots indicate the median, IQR, Q1 – 1.5 × IQR, and Q3 + 1.5 × IQR.
Physical chromatin interactions have been theorized to mediate the effects of cis-acting regulatory variants including expression quantitative trait loci (eQTLs) on gene expression. In support of this hypothesis, we first demonstrate that significant eQTLs in cortical and hippocampal tissues from GTEx V755 are enriched at PIRs for excitatory and hippocampal DG-like neurons, respectively (P < 2.2 × 10−16 for both cell types, two-tailed one sample z-test) (Fig. 6c). Next, we show that scores for interactions overlapping significant eQTL-TSS pairs are significantly higher than scores for interactions overlapping randomly shuffled eQTL-TSS pairs (P = 2.28 × 10−4 for excitatory neurons and P = 1.76 × 10−6 for hippocampal DG-like neurons, two-tailed two-sample Kolmogorov-Smirnov test) (Fig. 6d). This indicates that chromatin interactions recapitulating regulatory relationships between significant eQTL-TSS pairs are identified by pcHi-C with increased levels of confidence. Overall, our results present orthogonal lines of evidence that chromatin interactions can not only be altered by variants in an allele-specific manner, but that variants can also modulate gene expression through the formation or disruption of regulatory chromatin loops.
Discussion
There is a distinct lack of 3D epigenomic annotations in cell types that are relevant to disease and development, especially in the field of brain research. Past studies have relied on heterogeneous tissues comprised of cell types with disparate functions, limiting the ability to detect and interpret instances of cell-type-specific gene regulation. Neurons and glia, for example, represent lineages with divergent functions that coexist in most tissues of the CNS. At the same time, complex diseases often involve multiple dysregulated loci with cell-type-specific patterns of activity. This presents unique challenges for deciphering disease etiology, for example in attempting to distinguish causative mechanisms from secondary, reactive phenotypes when multiple cell types are involved. For these reasons, the comprehensive annotation of regulatory relationships in specific, well-characterized cell populations should enable the derivation of deeper insights into complex disease biology. Chromatin interactions in particular are ideal for mapping promoters to distal regulatory elements, as they provide direct evidence of regulatory sequences physically contacting loci of interest. To date, several studies have characterized chromatin interactions in fetal brain tissues and cultured neural cells6,56. However, these studies relied on in situ Hi-C for their interaction calls, which lacks power compared to targeted approaches such as pcHi-C.
Here, we have leveraged pcHi-C, ATAC-seq, and RNA-seq to comprehensively annotate previously uncharted regulatory relationships between promoters and distal regulatory elements in cell types that are relevant to complex neuropsychiatric disorders. We show that PIRs are not only cell-type-specific, but they are enriched for regulatory chromatin signatures including open chromatin peaks and in-vivo-validated enhancer elements from the VISTA Enhancer Browser. Inspection of cell type-specific distal open chromatin peaks at PIRs reveals subtype-specific binding sites for TFs involved in the specification and maintenance of cellular identity. Furthermore, our interactions identify novel gene targets for disease-associated variants and enable the prioritization of variants for validation using CRISPR techniques. We report a large number of putative regulatory variants, which may provide additional insights into aspects of complex disease biology. Finally, the disease- and cell-type-specific enrichment of variants at PIRs, combined with the observation that the same PIRs can target different genes in different cell types, supports existing evidence that regulatory variants possess context-dependent functional specificities.
The integrative analysis in this study has several limitations including a lack of cell-type-specific annotations for genomic and epigenomic features occurring at PIRs. For example, the analysis of chromatin state and eQTL enrichment at PIRs used data in matched tissues from the Roadmap Epigenomics Project and GTEx V7, respectively. Furthermore, while studying chromatin interactions in healthy cells enables the detection of regulatory interactions in the absence of dysregulation, the epigenomic characterization of patient-derived cells will be important to glean specific insights into how the 3D epigenome is altered in disease. Additional experiments are necessary to determine how the haploinsufficiency of proteins such as STRAP and DRD2 may contribute to phenotypes in disease. Finally, in vitro cultured cells can at present only approximate the full set of cellular responses occurring in vivo, especially for complex structures such as the brain, and they may reflect different developmental stages than expected based on their time in culture. Future approaches isolating specific cell types from tissues, using single-cell sequencing, or using advanced organoid models will be essential for drilling down more deeply into mechanisms driving cellular identity and disease. The epigenomic characterization of additional cell types should continue to yield rich insights into the landscape of transcriptional regulation, contributing towards an improved understanding of complex disease biology57.
Online methods
Cell culture
Human excitatory neurons were generated using integrated, isogenic, and inducible neurogenin-2 (Ngn2) iPSCs (i3N iPSCs) with doxycycline-inducible mouse Ngn2 integrated at the AAVS1 safe-harbor locus. The i3N iPSCs have a well-characterized wild type genetic background (WTC11)7. A simplified, two-step pre-differentiation and maturation protocol was used to generate the excitatory neurons8. Briefly, i3N iPSCs were incubated with 2 μg/ml doxycycline in pre-differentiation media containing KnockOut DMEM/F12 supplemented with 1× N-2, 1× NEAA, 1 μg/ml mouse laminin, 10 ng/ml BDNF, and 10 ng/ml NT3. 10 μM ROCK inhibitor was included in the pre-differentiation media for the first day. Media were changed daily for three days. For maturation, pre-differentiated precursor cells were dissociated and subplated on poly-D-lysine/laminin plates in maturation media containing equal parts DMEM/F12 and Neurobasal-A with 2 μg/ml doxycycline and supplemented with 0.5× B-27, 0.5× N-2, 1× NEAA, 0.5× GlutaMax, 1 μg/ml mouse laminin, 10 ng/ml BDNF, and 10 ng/ml NT3. The doxycycline was omitted from all subsequent media changes. Half of the media was changed weekly for the first two weeks, then the amount of media was doubled on day 21. Thereafter, a third of the media was replaced weekly until harvesting. 7 to 8 week old excitatory neurons were used for library preparation.
Human hippocampal DG-like neurons were generated from dissociated hippocampal organoids (unpublished). Briefly, WTC11 iPSCs were grown on MEF feeder cells and patterned towards a neural ectoderm fate using dual SMAD inhibition as floating embryoid bodies (EBs) in medium containing 20% KnockOut Serum Replacement. Four-week old EBs were patterned towards a hippocampal fate using WNT and BMP in medium containing 1× N-2. After patterning, organoids were dissociated using a neural tissue dissociation kit (MiltenyiBiotech), plated on PDL- and laminin-coated plates, and cultured for 4 weeks in media containing 1× B-27, 10 ng/ml BDNF, 10 ng/ml GDNF, 0.5 mM cAMP, and 200 μM ascorbic acid.
Human lower motor neurons were differentiated from WTC11 iPSCs using a doxycycline-inducible transgene expressing NGN2, ISL1, and LHX3 integrated at the AAVS1 safe-harbor locus (i3LMN iPSCs)10. Briefly, i3LMN iPSCs were maintained on growth factor reduced Matrigel in StemFit media (Nacalai USA). On day 0, 1.5 × 106 i3LMN iPSCs were plated on 10-cm dishes, followed 24 hours later by exchange into neural induction media containing doxycycline and compound E. On day 3, the precursor cells were replated onto 12-well plates coated with poly-D-lysine and laminin at a density of 2.5 × 105 cells per well. From day 3 to day 4, the cells were treated with a pulse of 40 μM BrdU for 24 hours to suppress the proliferation of undifferentiated cells. Media were exchanged on day 4 and every three days thereafter. The cells were harvested 10 days post-differentiation for library preparation.
Human primary astrocytes (P0) were purchased from ScienCell Research Laboratories (catalog #1800) and cultured using the recommended media (catalog #1801). Cells were cultured in flasks coated with poly-L-lysine (2 μg/cm2) and passaged once using trypsin and EDTA before harvesting.
All cells used in the present study were verified as being free from mycoplasma contamination.
Promoter capture Hi-C (pcHi-C)
In situ Hi-C libraries for excitatory neurons, hippocampal DG-like neurons, lower motor neurons, and astrocytes were constructed from 1 to 2 million cells (fixed in 1% PFA) using HindIII as a restriction enzyme as previously described58. pcHi-C was performed using biotinylated RNA probes according to an established protocol15. Briefly, sets of 120-bp probes with 30-bp overhangs were designed to capture all promoter-containing and adjacent HindIII fragments. Three probes were targeted to each side of a restriction site, for a total of 12 probes targeting each promoter-containing HindIII fragment. Promoters (defined as the sequences up to 500 bp upstream and downstream of each TSS) for 19,603 of the 20,332 protein coding genes in GENCODE 19 were captured using this approach. While noncoding RNA promoters were not explicitly targeted by this design, HindIII fragments containing 3,311 of the 14,069 noncoding RNA promoters in GENCODE 19 were also captured by the probes.
To perform the hybridization, 500 ng of each situ Hi-C library was first mixed with 2.5 μg human Cot-1 DNA (Invitrogen #15279011), 2.5 μg salmon sperm DNA (Invitrogen #15632011), and 0.5 nmol each of the p5 and p7 IDT xGen Universal Blocking Oligos in a total volume of 10 μl, denatured for 5 min at 95 °C, and pre-warmed at 65 °C. Next, a hybridization buffer mix was prepared by combining 25 μl 20× SSPE, 1 μl 0.5 M EDTA, 10 μl 50× Denhardt’s solution, and 13 μl 1% SDS and pre-warming the mix to 65 °C. Finally, 500 ng of the probes was mixed with 1 μl 20 U/μl SUPERase-In (Invitrogen #AM2696) in a total volume of 6 μl, pre-warmed to 65 °C, and combined with the library and hybridization buffer mixes. The final solution was transferred to a humidified hybridization chamber and incubated for 24 hours at 65 °C. 0.5 mg Dynabeads MyOne Streptavidin T1 magnetic beads (Invitrogen #65601) were used to pull down the captured fragments in a binding buffer consisting of 10 mM Tris-HCl pH 7.5, 1 M NaCl, and 1 mM EDTA. The beads were washed once with 1× SSC and 0.1% SDS for 30 minutes at 25 °C, followed by three washes with pre-warmed 0.1× SSC and 0.1% SDS for 10 minutes at 65 °C. The final library was eluted in 20 μl nuclease-free water, amplified, then sent for paired-end sequencing on the HiSeq 4000 (50-bp reads), the HiSeq X Ten (150-bp reads), or the NovaSeq 6000 (150-bp reads).
Calling significant promoter-PIR interactions
Paired-end sequencing reads were first trimmed using fastp 0.20.059 running the default settings before being mapped, filtered, and deduplicated using HiCUP 0.7160 with bowtie2 2.3.261 and filtering for ditags between 100 and 1,200 bp. In addition, the sequencing depth of all libraries was normalized so that each replicate had the same number of usable reads (defined as the number of on-target cis pairs interacting over a distance of 10 kb). Significant promoter-PIR interactions were called using CHiCAGO 1.1.817 running the default settings and retaining baited fragments that are supported by at least 250 reads (minNPerBaits = 250). Interactions between HindIII fragments with a score (defined as the negative log P value) of 5 or greater in each cell type were determined to be significant. All data processing metrics are reported in Supplementary Table 1. In cases where CHiCAGO reported the same interaction in different orientations, the two interactions were merged, retaining the higher score of the two interactions. Interchromosomal interactions were omitted from the analysis. To call overlaps between our sets of significant interactions and genomic and epigenomic features including promoters, open chromatin peaks, chromatin states, disease-associated variants, and eQTLs, interacting bins were expanded to a minimum width of 5 kb or retained as the original widths of the HindIII fragments if they exceeded 5 kb. Interactions overlapping HindIII fragments larger than 100 kb were omitted from our analysis. An interaction was considered to be shared between cell types if both of its interacting ends intersected the corresponding ends of an interaction in another cell type. Otherwise, an interaction was determined to be cell-type-specific.
Validation of PIRs using CRISPR deletion
To validate genomic interactions captured by pcHi-C, candidate PIRs were targeted for CRISPR-mediated deletion in the i3N iPSCs. Pairs of sgRNAs targeting the putative regulatory element as localized by open chromatin peaks in the candidate PIR were designed for each locus of interest. All sgRNAs were synthesized by Synthego. Cas9 protein was sourced from QB3-Berkeley. To generate deletion lines, CRISPR/Cas9 nucleofections were performed using the LONZA Human Stem Cell Nucleofector® Kit. For each nucleofection, approximately 500,000 i3N iPSCs were transfected with Cas9:sgRNA RNP complex (consisting of 12 μg Cas9, 10 μg sgRNA 1, and 10 μg sgRNA 2) using program “A-023” on the LONZA 4D-Nucleofector. The nucleofected cells were then seeded onto Matrigel-coated 6-well plates containing Essential 8™ Medium (ThermoFisher #A15169–01) with Y-27632 added for recovery following nucleofection. After 48 hours, the cells were split into new 6-well plates at a concentration of approximately 50 cells per well for picking single colonies. Clones picked from the 6-well plates containing homozygous deletions were confirmed by qPCR and induced into excitatory neurons for quantifying the expression of genes targeted by the deleted PIRs. For each experiment, we used three deletion clones and two wild type clones. Total RNA from the excitatory neurons was extracted using a Qiagen AllPrep DNA/RNA Mini Kit, and cDNA was synthesized using a Bio-RAD iScript™ cDNA Synthesis Kit. qPCR for targeted genes was performed with FastStart Essential DNA Green Master reaction mix (Roche) on the LightCycler® 96 System (Roche). The mean values from three technical replicates were used for statistical testing. Detailed information on all the primers used is available in Supplementary Table 11.
Validation of PIRs using CRISPRi
Excitatory neurons induced from i3N iPSCs were infected with lentivirus carrying dCas9-KRAB-blast (Addgene #89567) and colonies with high expression of dCas9 were picked. The CROP-seq-opti vector (Addgene #106280) was used for sgRNA expression. sgRNAs were cotransfected with lentivirus packaging plasmids pMD2.G (Addgene #12259) and psPAX (Addgene #12260) into 293T cells with PolyJet (SignaGen Laboratories #SL100688) according to the manufacturer’s instructions. Virus-containing media was collected for 72 hours, filtered through a 0.45 μm filter (Millipore #SLHV033RS), and concentrated with an Amicon Ultra centrifugal filter (Millipore #UFC801024). The virus was titrated in the excitatory neurons by qPCR 72 hours post-infection. The internal qPCR control targeted an intronic region (forward primer: TCCTCCGGAGTTATTCTTGGCA and reverse primer: CCCCCCATCTGATCTGTTTCAC). Integration of the WPRE fragment was quantified in comparison with a cell line containing a known copy number of WPRE. For CRISPRi silencing of putative regulatory elements, excitatory neurons were treated with lentivirus expressing sgRNAs (MOI ~3) for two or three replicates per condition representing independent differentiation events. Two independent sets of sgRNAs were used for each replicate. Cells were collected for mRNA extraction 7 days post-transfection, and gene expression was determined using qPCR. The mean values from three technical replicates were used for statistical testing. Detailed information on all the primers used is available in Supplementary Table 11.
Motif enrichment analysis
We took the sets of all cell type-specific distal open chromatin peaks participating in significant promoter-PIR interactions between promoter-containing and non-promoter-containing bins for each cell type and used the sequences in 250 bp windows around the peak summits to perform motif enrichment analysis using HOMER 4.1031 running the default settings. The cumulative binomial distribution was used for motif scoring. The entire genome was used as a background. Significance and expression values for each detected motif and its corresponding TFs are reported in Supplementary Table 3.
VISTA enhancer analysis and target gene identification
Human and mouse enhancer regions with orthologous human sequences and positive annotations in the VISTA Enhancer Browser11 were downloaded and analyzed for overlap with our sets of significant promoter-PIR interactions for each cell type. Of the 2,956 tested elements in their database (January 2019), 1,568 were positive elements with orthologous human sequences (976 human elements and 892 mouse elements with orthologous human sequences). Positive elements expanded to a minimum width of 5 kb and participating in significant interactions are reported in Supplementary Table 5. The “hs” or “mm” prefixes for elements indicate the species of DNA origin. For determining whether positive elements interacted with their nearest or more distal genes, we only considered protein coding and noncoding RNA genes in GENCODE 19. To evaluate cases where interactions between positive elements and their nearest genes were unresolvable (“same fragment ambiguity”), we determined if a promoter for the nearest gene overlapped at least one HindIII fragment that the positive element did not also overlap. The following terms were considered to be neural annotations: neural tube, hindbrain, cranial nerve, midbrain, forebrain, mesenchyme derived from neural crest, dorsal root ganglion, and trigeminal V.
SNP enrichment analysis and target gene identification
GWAS SNPs for a total of eleven neuropsychiatric disorders including Alzheimer’s disease (AD), attention deficit hyperactivity disorder (ADHD), amyotrophic lateral sclerosis (ALS), autism spectrum disorder (ASD), bipolar disorder (BD), epilepsy (EP), frontotemporal dementia (FTD), mental process (MP), Parkinson’s disease (PD), and schizophrenia (SCZ), and unipolar depression (UD) were mined from the GWAS Catalog39 (December 2018) using a P value threshold of 10-6. See Supplementary Table 5 for a detailed summary of the studies included for each trait. The GWAS SNPs were expanded to sets of linked SNPs using HaploReg 4.141 at an LD threshold of 0.8 according to the reported study population(s) for each SNP. All SNPs were lifted over to hg19 and filtered for duplicates by position. Disease- and cell-type-specific enrichment for SNPs was calculated as the number of SNPs overlapping significant PIRs divided by the mean number of SNPs overlapping randomly shuffled PIRs with matching distance distributions. n = 100 sets of randomly shuffled PIRs were sampled in each case. To determine whether a GWAS SNP interacted with a target gene, we determined whether it or any of its linked SNPs (expanded to a minimum width of 1 kb) interacted with a promoter associated with the nearest gene. To evaluate cases where interactions between GWAS SNPs and their nearest genes were unresolvable (“same fragment ambiguity”), we determined if a promoter for the nearest gene overlapped at least one HindIII fragment that a GWAS SNP or any of its linked SNPs did not also overlap. Finally, we derived a list of SNPs for which the SNP was located within 2 kb of the center of an open chromatin peak at a PIR, indicating additional evidence for a functional regulatory variant at that locus. These SNPs are referred to as “putative regulatory SNPs.”
Phasing of the WTC11 genome
The raw WTC11 genome sequence can be downloaded from http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubClear=https://s3-us-west-2.amazonaws.com/downloads.allencell.org/genome-sequence/ucsc_hubs/WTC_genome_hub/hub.txt. Phasing of the WTC11 genome was performed as previously described51. Briefly, WTC11 variants were split by chromosome and phase-informative reads from pcHi-C were extracted using extractHAIRS with the minimum mapping quality set to 10 and the maximum insert size set to 30,000,000 bp62. Phasing was performed with Hapcut using a maximum of 101 iterations. Next, we extracted the maximum variants phased (MVP) haplotype block from the output of Hapcut to use as a seed haplotype. We modified the “neighborhood correction” aspect of phasing by filtering phased variants whose predicted phase would have a marginal probability below 0.99 according to an in-house implementation of a hidden Markov model (HMM) as described previously63,64 with a reference haplotype set from the 1000 Genomes Project. Missing variants were imputed using the aforementioned HMM with the reference haplotype set from the 1000 Genomes Project. The WTC11 SNP phasing data are available at the Gene Expression Omnibus under the following accessible number: GSE113483.
Allelic bias analysis
We used the WTC11 phasing data along with the allele-specific mapping capabilities of HiC-Pro 2.11.052 to quantify allelic bias between significantly interacting 10-kb bins genome-wide in the excitatory and lower motor neurons. We selected these two cell types because they used homogenous induction of TFs for differentiation, therefore minimizing the noise introduced by conventional differentiation techniques. Briefly, reads were mapped using bowtie 1.2.1.165 to a version of the hg19 reference genome where all sites with heterozygous phased SNPs were masked. Unfiltered HiC-Pro contact maps were used for this analysis. Next, nucleotides at masked polymorphic sites were used to assign reads to either allele. Reads reporting conflicting allele assignments or unexpected bases were filtered out. Reads with at least one allele-specific mate were used to construct allele-specific Hi-C contact maps at a resolution of 10 kb. The allele-specific Hi-C contact maps were intersected with the set of all significant promoter-PIR interactions with score ≥ 3 to assess allelic bias between interacting 10-kb bins. Only interacting bins with 10 or more reads across both alleles were retained (n = 22,162 bins for excitatory neurons and n = 21,479 bins for lower motor neurons). A two-tailed binomial test was used to assess allelic bias across each set of interacting bins, and the resulting P values were adjusted using the BH correction to filter out significantly biased loci at an FDR cutoff of 5%. Allelically biased interactions with P values < 10−3 are reported in Supplementary Table 10.
eQTL enrichment analysis
1D enrichment of significant eQTLs from GTEX V755 at significant versus randomly shuffled PIRs in matched tissue types for excitatory neurons (Brain - Cortex, n = 136) and hippocampal DG-like neurons (Brain - Hippocampus, n = 111) was performed in the same manner as the chromatin state and SNP enrichment analysis. We used the full set of significant eQTL SNP-gene associations available from GTEx which included 478,903 eQTLs associated with 6,146 eGenes for the cortex and 221,876 eQTLs associated with 3,262 eGenes for the hippocampus. To determine the 2D enrichment of eQTL-TSS pairs in our significant interaction sets, we first filtered out eQTL-TSS pairs that were within 10 kb of each other or on the same HindIII fragment as this would be below the minimum detectable resolution by pcHi-C. Next, we sampled a set of nonsignificant eQTL-TSS pairs with a matching distance distribution as the set of significant eQTL-TSS pairs for each cell type, controlling for the number of genes around which the eQTL-TSS pairs were centered. We sampled three times the number of nonsignificant eQTL-TSS pairs as the number of significant eQTL-TSS pairs (424,912 significant pairs for 5,826 TSSs and 1,274,736 nonsignificant pairs for 17,570 TSSs in excitatory neurons, and 197,155 significant pairs for 3,083 TSSs and 591,465 nonsignificant pairs for 9,238 TSSs in hippocampal DG-like neurons). Similar results were obtained when using the same number of significant and nonsignificant eQTL-TSS pairs, or when using the same sets of eGenes (data not shown). We compared the distributions of interaction scores for significant interactions supporting the significant and nonsignificant eQTL-TSS pairs by overlapping the eQTL-TSS pairs with our significant interactions (two-tailed two-sample Kolmogorov-Smirnov test).
Statistics and reproducibility
Statistical analyses were performed using R 3.4.3 and Microsoft Excel. All of the statistical tests used are described in the relevant sections of the manuscript. P values are provided as exact values where possible, and otherwise are reported as a range.
Reporting summary
Additional information on our study design is available in the Nature Research Reporting Summary linked to this article.
Data availability statement
All datasets used in this study (pcHi-C, ATAC-seq, RNA-seq, CUT&RUN, and chromosome-wide SNP phasing data) are available at the Gene Expression Omnibus under the accession number GSE113483. Open chromatin peaks and gene expression results for each cell type are also available on Zenodo through the following link: https://zenodo.org/record/3243977
Data can be visualized on the WashU Epigenome Browser using the session bundle ID (session ID in parentheses): 6e375740–8e71–11e9-be37-cb77c4bbb5fc (brain_pchic_nature_genetics_00)
Alternatively, the data can also be visualized on the legacy WashU Epigenome Browser (session ID in parentheses): http://epigenomegateway.wustl.edu/legacy/?genome=hg19&session=8OCs2rkpEA (brain_pchic_nature_genetics_00)
Tracks include ATAC-seq signal, chromatin interactions with score ≥ 5, and RNA-seq plus and minus strand signal for each cell type. HindIII fragments, in vivo-validated enhancer elements, GENCODE 19 genes, and GWAS SNPs are also displayed.
Supplementary Material
Acknowledgements
We thank Anthony Schmitt and Bing Ren (Ludwig Institute for Cancer Research, University of California, San Diego) for sharing pcHi-C probes and the pcHi-C protocol. Genomic analysis of the WTC11 line in this study was made possible by the whole-genome sequencing data generated by the Allen Institute for Cell Science. We thank the Institute and its founder Paul G. Allen for making this work possible. We thank Gary Hon (University of Texas Southwestern Medical Center) and Steven Henikoff (Fred Hutchinson Cancer Research Center, Howard Hughes Medical Institute) for providing reagents. We acknowledge the ENCODE Consortium and Bradley Bernstein’s laboratory for generating the ChIP-seq data for astrocytes used in this study. We thank N. Ahituv, Y. Guo, R. D. Hawkins, M. McManus, and B. Ren for providing critical feedback on the manuscript. We thank Y. Qu for her contributions to the illustrations for Figure 1. This work was made possible in part by NIH-NEI P30EY002162 (Core Grant for Vision Research and the Research to Prevent Blindness Unrestricted Grant). This work was supported by the National Institutes of Health (NIH) grants R01AG057497 to Y.S., L.G., and H.S., R01EY027789 and UM1HG009402 to Y.S., the UCSF Weill Institute for Neuroscience Innovation Award, the Hillblom Foundation, and the American Federation for Aging Research New Investigator Award in Alzheimer’s Disease to Y.S., R01EY028249, R01HL130533, R01-HL13535801 to B.R.C., P01NS097206 and U19MH106434 to H.S., and R01MH105128, R35NS097370, and U19AI131130 to G.L.M. M.S. is supported by T32GM007175. F.J. is supported by T32GM007309.
Footnotes
Competing interests
The authors declare no competing financial interests.
References
- 1.Mumbach MR et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet 49, 1602–1612, doi: 10.1038/ng.3963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48, 481–487, doi: 10.1038/ng.3538 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Claussnitzer M et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med 373, 895–907, doi: 10.1056/NEJMoa1502214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smemo S et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375, doi: 10.1038/nature13138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Torre-Ubieta L et al. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell 172, 289–304 e218, doi: 10.1016/j.cell.2017.12.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Won H et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527, doi: 10.1038/nature19847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyaoka Y et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods 11, 291–293, doi: 10.1038/nmeth.2840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C et al. Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem Cell Reports 9, 1221–1233, doi: 10.1016/j.stemcr.2017.08.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mertens J et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527, 95–99, doi: 10.1038/nature15526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandopulle MS et al. Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr Protoc Cell Biol 79, e51, doi: 10.1002/cpcb.51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visel A, Minovitsky S, Dubchak I & Pennacchio LA VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35, D88–92, doi: 10.1093/nar/gkl822 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798, doi: 10.1016/j.neuron.2013.05.029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu DX et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports 2, 295–310, doi: 10.1016/j.stemcr.2014.01.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53, doi: 10.1016/j.neuron.2015.11.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung I et al. A Compendium of Promoter-Centered Long-Range Chromatin Interactions in the Human Genome. Nature Genetics NG-LE51539-T (2019, in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang T et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res 27, 1939–1949, doi: 10.1101/gr.220640.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cairns J et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol 17, 127, doi: 10.1186/s13059-016-0992-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson DA, Hassan AB, Errington RJ & Cook PR Visualization of focal sites of transcription within human nuclei. EMBO J 12, 1059–1065 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504, 306–310, doi: 10.1038/nature12716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diao Y et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods 14, 629–635, doi: 10.1038/nmeth.4264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engreitz JM et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455, doi: 10.1038/nature20149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120, doi: 10.1038/nature11243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst J & Kellis M ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9, 215–216, doi: 10.1038/nmeth.1906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roadmap Epigenomics C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, doi: 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skene PJ & Henikoff S An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, doi: 10.7554/eLife.21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis CA et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46, D794–D801, doi: 10.1093/nar/gkx1081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97, doi: 10.1093/nar/gkw377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadif Kasri N, Nakano-Kobayashi A & Van Aelst L Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron 72, 300–315, doi: 10.1016/j.neuron.2011.09.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y et al. Postnatal activation of TLR4 in astrocytes promotes excitatory synaptogenesis in hippocampal neurons. J Cell Biol 215, 719–734, doi: 10.1083/jcb.201605046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren G et al. CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Mol Cell 67, 1049–1058 e1046, doi: 10.1016/j.molcel.2017.08.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910, doi: 10.1016/j.cell.2015.07.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handoko L et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43, 630–638, doi: 10.1038/ng.857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou C, Dale R & Dean A Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A 107, 3651–3656, doi: 10.1073/pnas.0912087107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuang HC, Huang TN & Hsueh YP Neuronal excitation upregulates Tbr1, a high-confidence risk gene of autism, mediating Grin2b expression in the adult brain. Front Cell Neurosci 8, 280, doi: 10.3389/fncel.2014.00280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liddell JR Are Astrocytes the Predominant Cell Type for Activation of Nrf2 in Aging and Neurodegeneration? Antioxidants (Basel) 6, doi: 10.3390/antiox6030065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin X, Warner DR, Roberts EA, Pisano MM & Greene RM Novel interaction between nuclear co-activator CBP and the CDK5 activator binding protein - C53. Int J Mol Med 16, 251–256 (2005). [PubMed] [Google Scholar]
- 39.Xie YH et al. Cloning and characterization of human IC53–2, a novel CDK5 activator binding protein. Cell Res 13, 83–91, doi: 10.1038/sj.cr.7290153 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Buniello A et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012, doi: 10.1093/nar/gky1120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward LD & Kellis M HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40, D930–934, doi: 10.1093/nar/gkr917 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdulla S et al. Hippocampal degeneration in patients with amyotrophic lateral sclerosis. Neurobiol Aging 35, 2639–2645, doi: 10.1016/j.neurobiolaging.2014.05.035 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Booth HDE, Hirst WD & Wade-Martins R The Role of Astrocyte Dysfunction in Parkinson’s Disease Pathogenesis. Trends Neurosci 40, 358–370, doi: 10.1016/j.tins.2017.04.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiwari M, Lopez-Cruzan M, Morgan WW & Herman B Loss of caspase-2-dependent apoptosis induces autophagy after mitochondrial oxidative stress in primary cultures of young adult cortical neurons. J Biol Chem 286, 8493–8506, doi: 10.1074/jbc.M110.163824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao X et al. Caspase-2 cleavage of tau reversibly impairs memory. Nat Med 22, 1268–1276, doi: 10.1038/nm.4199 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Huang CW et al. Conditional Knockout of Breast Carcinoma Amplified Sequence 2 (BCAS2) in Mouse Forebrain Causes Dendritic Malformation via beta-catenin. Sci Rep 6, 34927, doi: 10.1038/srep34927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grimmler M et al. Unrip, a factor implicated in cap-independent translation, associates with the cytosolic SMN complex and influences its intracellular localization. Hum Mol Genet 14, 3099–3111, doi: 10.1093/hmg/ddi343 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Burghes AH & Beattie CE Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10, 597–609, doi: 10.1038/nrn2670 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JP et al. Association of a Schizophrenia Risk Variant at the DRD2 Locus With Antipsychotic Treatment Response in First-Episode Psychosis. Schizophr Bull 41, 1248–1255, doi: 10.1093/schbul/sbv116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eisenstein SA et al. Prediction of striatal D2 receptor binding by DRD2/ANKK1 TaqIA allele status. Synapse 70, 418–431, doi: 10.1002/syn.21916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selvaraj S, J RD, Bansal V & Ren B Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol 31, 1111–1118, doi: 10.1038/nbt.2728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Servant N et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259, doi: 10.1186/s13059-015-0831-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lydall GJ et al. Confirmation of prior evidence of genetic susceptibility to alcoholism in a genome-wide association study of comorbid alcoholism and bipolar disorder. Psychiatr Genet 21, 294–306, doi: 10.1097/YPG.0b013e32834915c2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sudhof TC Calcium control of neurotransmitter release. Cold Spring Harb Perspect Biol 4, a011353, doi: 10.1101/cshperspect.a011353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Consortium GT et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213, doi: 10.1038/nature24277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajarajan P et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362, doi: 10.1126/science.aat4311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Straws in a haystack. Nat Genet 50, 631, doi: 10.1038/s41588-018-0125-9 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680, doi: 10.1016/j.cell.2014.11.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S, Zhou Y, Chen Y & Gu J fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890, doi: 10.1093/bioinformatics/bty560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wingett S et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4, 1310, doi: 10.12688/f1000research.7334.1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bansal V & Bafna V HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 24, i153–159, doi: 10.1093/bioinformatics/btn298 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Li N & Stephens M Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delaneau O, Zagury JF & Marchini J Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 10, 5–6, doi: 10.1038/nmeth.2307 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25, doi: 10.1186/gb-2009-10-3-r25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets used in this study (pcHi-C, ATAC-seq, RNA-seq, CUT&RUN, and chromosome-wide SNP phasing data) are available at the Gene Expression Omnibus under the accession number GSE113483. Open chromatin peaks and gene expression results for each cell type are also available on Zenodo through the following link: https://zenodo.org/record/3243977
Data can be visualized on the WashU Epigenome Browser using the session bundle ID (session ID in parentheses): 6e375740–8e71–11e9-be37-cb77c4bbb5fc (brain_pchic_nature_genetics_00)
Alternatively, the data can also be visualized on the legacy WashU Epigenome Browser (session ID in parentheses): http://epigenomegateway.wustl.edu/legacy/?genome=hg19&session=8OCs2rkpEA (brain_pchic_nature_genetics_00)
Tracks include ATAC-seq signal, chromatin interactions with score ≥ 5, and RNA-seq plus and minus strand signal for each cell type. HindIII fragments, in vivo-validated enhancer elements, GENCODE 19 genes, and GWAS SNPs are also displayed.