Benzoic acid is a common food preservative, and salicylic acid (2-hydroxybenzoic acid) is the active form of aspirin. At high concentrations, benzoic acid conducts a proton across the membrane, depleting the proton motive force. In the absence of antibiotics, benzoate exposure selects against proton-driven multidrug efflux pumps and upregulates porins that admit fermentable substrates but that also allow the entry of antibiotics. Thus, evolution with benzoate and related molecules, such as salicylates, requires a trade-off for antibiotic sensitivity, a trade-off that could help define a stable gut microbiome. Benzoate and salicylate are naturally occurring plant signal molecules that may modulate the microbiomes of plants and animal digestive tracts so as to favor fermenters and exclude drug-resistant pathogens.
KEYWORDS: Escherichia coli, Gad, acid fitness island, benzoate, chloramphenicol, experimental evolution, hydrogenases, multidrug efflux, porins, salicylate
ABSTRACT
Benzoic acid, a partial uncoupler of the proton motive force (PMF), selects for sensitivity to chloramphenicol and tetracycline during the experimental evolution of Escherichia coli K-12. Transcriptomes of E. coli isolates evolved with benzoate showed the reversal of benzoate-dependent regulation, including the downregulation of multidrug efflux pump genes, the gene for the Gad acid resistance regulon, the nitrate reductase genes narHJ, and the gene for the acid-consuming hydrogenase Hyd-3. However, the benzoate-evolved strains had increased expression of OmpF and other large-hole porins that admit fermentable substrates and antibiotics. Candidate genes identified from benzoate-evolved strains were tested for their roles in benzoate tolerance and in chloramphenicol sensitivity. Benzoate or salicylate tolerance was increased by deletion of the Gad activator ariR or of the acid fitness island from slp to the end of the gadX gene encoding Gad regulators and the multidrug pump genes mdtEF. Benzoate tolerance was also increased by deletion of multidrug component gene emrA, RpoS posttranscriptional regulator gene cspC, adenosine deaminase gene add, hydrogenase gene hyc (Hyd-3), and the RNA chaperone/DNA-binding regulator gene hfq. Chloramphenicol resistance was decreased by mutations in genes for global regulators, such as RNA polymerase alpha subunit gene rpoA, the Mar activator gene rob, and hfq. Deletion of lipopolysaccharide biosynthetic kinase gene rfaY decreased the rate of growth in chloramphenicol. Isolates from experimental evolution with benzoate had many mutations affecting aromatic biosynthesis and catabolism, such as aroF (encoding tyrosine biosynthesis) and apt (encoding adenine phosphoribosyltransferase). Overall, benzoate or salicylate exposure selects for the loss of multidrug efflux pumps and of hydrogenases that generate a futile cycle of PMF and upregulates porins that admit fermentable nutrients and antibiotics.
IMPORTANCE Benzoic acid is a common food preservative, and salicylic acid (2-hydroxybenzoic acid) is the active form of aspirin. At high concentrations, benzoic acid conducts a proton across the membrane, depleting the proton motive force. In the absence of antibiotics, benzoate exposure selects against proton-driven multidrug efflux pumps and upregulates porins that admit fermentable substrates but that also allow the entry of antibiotics. Thus, evolution with benzoate and related molecules, such as salicylates, requires a trade-off for antibiotic sensitivity, a trade-off that could help define a stable gut microbiome. Benzoate and salicylate are naturally occurring plant signal molecules that may modulate the microbiomes of plants and animal digestive tracts so as to favor fermenters and exclude drug-resistant pathogens.
INTRODUCTION
Escherichia coli and other enteric bacteria face high concentrations of organic acids, such as short-chain fatty acids, that permeate bacterial cell membranes and acidify the cytoplasm (1, 2) and that drive the accumulation of toxic anions (3, 4). Acids that cross the membrane in the unprotonated form can uncouple the proton motive force (PMF) (5–8). Benzoic acid and salicylic acid (2-hydroxybenzoic acid, the active form of aspirin) act as permeant acids and as partial uncouplers. These molecules are abundant in the plant rhizosphere (9) and in human diets in the form of food preservatives, pharmaceutical products, and natural plant secondary metabolites (10–13). We are investigating the molecular effects of benzoate derivatives on bacteria.
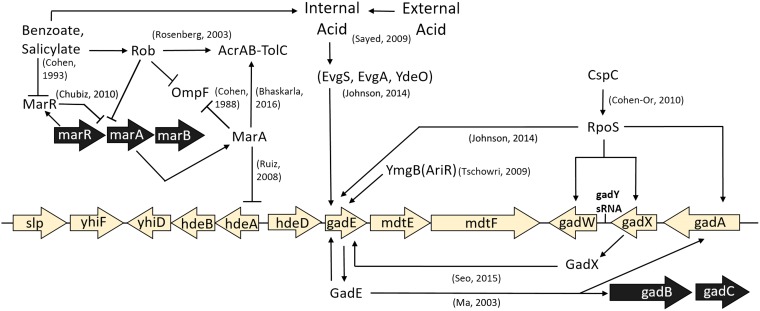
Benzoate and salicylate induce low-level resistance to antibiotics via the Mar regulon (14) as well as Mar-independent pathways that are poorly understood (15). The Mar regulon intersects with the Gad acid resistance regulon (16, 17), which includes a major region of acid-stress regulators and multidrug resistance (MDR) efflux pumps (Fig. 1). Surprisingly, however, experimental evolution in the presence of benzoate leads to deletion of acid resistance systems and decreased resistance to the antibiotics tetracycline and chloramphenicol (18). Isolates from populations serially cultured for 2,000 generations (referred to here as the 2,000-generation strains) show the loss of genes for MDR pumps and regulators, such as emrA, emrY, and marRAB. Similarly, experimental evolution in the presence of the strong uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) yields isolates that have lost MDR pumps and regulators, with the exception of the EmrAB-TolC pump, which directly exports CCCP (19, 20). These results suggest a hypothesis that aromatic acid uncouplers can amplify the fitness cost of MDR in the absence of antibiotics. The concept of reversing antibiotic resistance is of great interest for the gut microbiome as well as contaminated environments, where antibiotic-resistant strains may show a minimum selective concentration (MSC) as much as 100-fold lower than the MIC (21).
FIG 1.
The Gad acid resistance regulon intersects with the Mar drug resistance regulon. Selected components relevant to this work are shown. The references cited in the figure are 15, 17, 32, 47, 48, 50, 63, 64, 87, and 109, to ,111.
We considered several mechanisms that might explain benzoate selection against drug resistance. When a partial uncoupler depletes PMF, the cell incurs energy stress, which selects against MDR pumps that spend PMF. Additionally, constitutive induction of stress response pathways carries a heavy energetic penalty; thus, deletion or downregulation of constitutively expressed pathways confers an energetic advantage. The penalty could arise from the cost of protein production, which increases nonlinearly with additional transcription (22). In the case of acid stress, experimental evolution leads to the loss of three acid-inducible amino acid decarboxylase systems (23, 24). Also, PMF expenditure in itself has a fitness cost, like in the lac operon, where the major fitness cost of expression is the activity of LacY permease, driven by PMF (25). The depletion of PMF might favor expression of large-hole porins that increase the uptake of fermentable substrates, with the trade-off being the increased uptake of antibiotics (26–28).
Most benzoate-tolerant isolates from the evolution experiment (18) acquired a mutation affecting the slp-gad (Gad) acid fitness island (16, 29–31). The Gad island includes acid resistance regulator genes gadE, gadW, and gadX (32); periplasmic acid chaperone genes hdeA and hdeB; and MDR efflux component genes mdtE and mdtF (33, 34) (Fig. 1). The major regulator GadE upregulates several acid tolerance genes, including two glutamate decarboxylase isoforms (gadA and gadB), whose activity increases the cytoplasmic pH by consuming protons via the decarboxylation of glutamate (16, 31, 35, 36). The MdtEF-TolC complex exports antibiotics from the cytoplasm, driven by PMF. Our benzoate-evolved isolates (18) have acquired point mutations or deletions throughout the slp-gad island, as well as knockout or point mutations in emrA, encoding the membrane fusion protein component of the EmrAB-TolC MDR pump (37, 38); mdtA (39, 40); and emrY (41). Other mutations affect genes for MDR regulators, such as cpxA (42), ariR (43), arcA (44), and rob (45).
In one population, exposure to benzoate has selected for deletion of marRAB, a multidrug resistance operon which is induced by MarR binding salicylate or benzoate (15, 45). MarRAB has homologs throughout the bacteria and archaea, including many drug-resistant clinical isolates (46). In E. coli, salicylate or benzoate relieves the repression of MarA, which regulates over 60 genes involved in antibiotic resistance, such as the AcrAB-TolC MDR pump (14, 47), as well as hdeAB within the slp-gad island (17). MarA downregulates the large porin OmpF, which admits nutrients and antibiotics (48). Many targets of MarA are also subject to the MarA homolog Rob (45, 46), which had a mutation in one benzoate-evolved isolate (18). Additionally, one strain acquired a mutation in the alpha subunit of RNA polymerase (RNAP) which could affect the regulation of a wide array of genes. RNAP mutations lead to unexpected phenotypes, such as the downregulation of arginine decarboxylase caused by an rpoC mutation in an acid-evolved strain (23).
We sought to reveal the genetic mechanisms of benzoate tolerance found in the 2,000-generation strains and to determine how these mechanisms intersect with antibiotic resistance. Here we report the results of transcriptomic analysis of four 2,000-generation strains that showed a surprising long-term reversal of the short-term benzoate stress response. To dissect this response and the mechanisms of antibiotic resistance reversal, we sequenced clones from earlier populations of the benzoate evolution experiment with fewer mutations so as to reveal the order in which mutations were acquired. We report the effects of several candidate gene deletions and mutations on fitness in the presence of benzoate and of the antibiotic chloramphenicol. Our findings indicate several processes that mediate the antibiotic sensitivity of benzoate-evolved strains.
RESULTS
Early benzoate-selected mutations affect Gad, Mar, and aromatic metabolism.
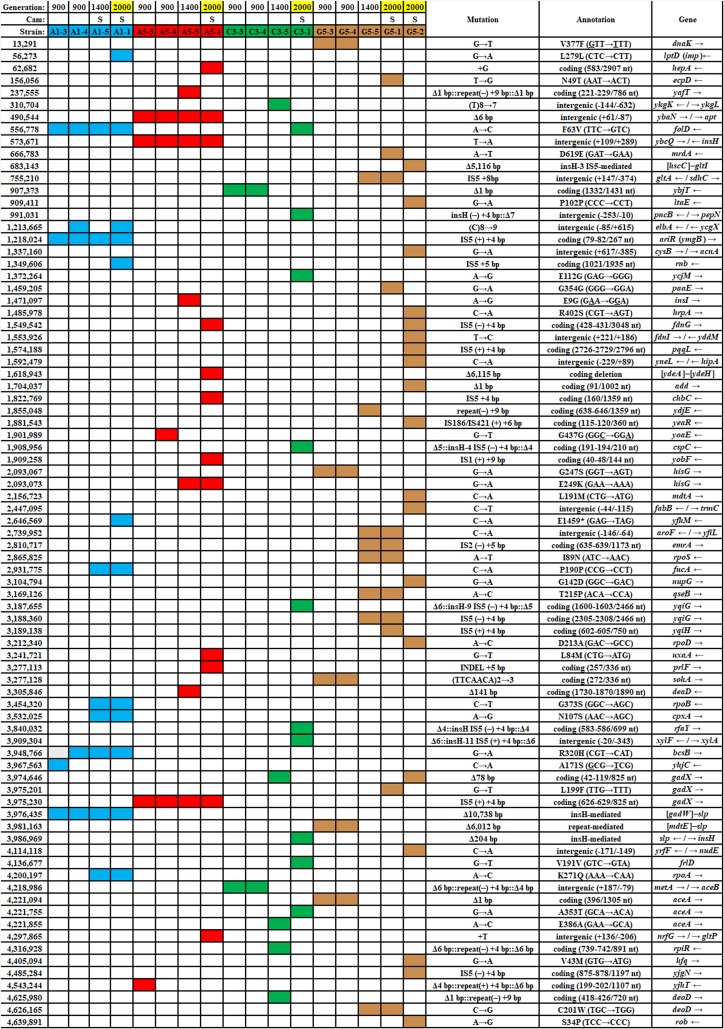
The large number of mutations in the 2,000-generation strains made it difficult to assess which genetic changes were most likely to affect benzoate tolerance (18). As such, we decided to isolate and sequence strains from earlier generations that were more likely to contain fewer mutations. We isolated clones from frozen populations ancestral to those published for generation 2,000 (Table 1). Colonies were obtained on LBK (10 g/liter tryptone, 5 g/liter yeast extract, 7.45 g/liter KCl) agar from frozen populations corresponding to generations 900 and 1,400 for microplate well populations A1, A5, C3, and G5. At generation 2,000, these populations had produced isolates A1-1, A5-1, C3-1, G5-1, and G5-2; these key strains are benzoate tolerant, and all except G5-1 are more sensitive to chloramphenicol than the ancestral strain, W3110 (18). Selected strains were sequenced (see Table S1 in the supplemental material), and mutations were detected using the breseq computational pipeline (49). Mutations for selected strains are listed in Table 2, alongside the mutations from generation 2,000 clones (18). All mutations for all clones sequenced are compiled in Table S2.
TABLE 1.
Strains of Escherichia coli used in this studya
| Name | Genotype | Reference or source |
|---|---|---|
| W3110 | Escherichia coli K-12 | 108 |
| JLSK0001 | W3110 benzoate-evolved A1-1 | 18 |
| JLSK0014 | W3110 benzoate-evolved C3-1 | 18 |
| JLSK0030 | W3110 benzoate-evolved G5-1 | 18 |
| JLSK0031 | W3110 benzoate-evolved G5-2 | 18 |
| JLS0903 | W3110 ΔhyaB::kanR | 78 |
| JLS0905 | W3110 ΔhycE::kanR | 78 |
| JLS1010 | W3110 ΔmdtE::kanR | This work |
| JLS1025 | W3110 ΔmdtF::kanR | This work |
| JLS1517 | W3110 ΔgadX::kanR | This work |
| JLS1631 | W3110 ΔcspC::kanR | This work |
| JLS1632 | W3110 ΔgadE::kanR | This work |
| JLS1633 | W3110 ΔrfaY::kanR | This work |
| JLS1706 | W3110 ΔemrA::kanR | 20 |
| JLS1732 | W3110 Δ(slp-gadX) | This work |
| JLS1771 | W3110 Δ(slp-gadX) ΔariR::kanR | This work |
| JLS1777 | W3110 ΔcspC::kanR ΔrfaY::frt | This work |
| JLS1789 | W3110 Δadd::kanR | This work |
| JLS1820 | W3110 Δrob::kanR | This work |
| JLS1823 | W3110 ΔariR::kanR | This work |
| JLS1828 | W3110 ΔhycF::kanR | This work |
| JLS1616 | A1-1 ΔyhdN::kanR rpoA+ | This work |
| JLS1617 | A1-1 ΔyhdN::frt rpoA+ | This work |
| JLS1791 | W3110 Δhfq::kanR | This work |
| JLS1908 | W3110 ΔyhdN::kanR | This work |
| JLS1909 | A1-1 ΔyhdN::kanR | This work |
Newly isolated strains from benzoate-evolved populations (18) are listed in Table S1 in the supplemental material.
TABLE 2.
Mutations found in selected benzoate-evolved strainsa
Highlighted generation numbers indicate published strains (18). Cam, chloramphenicol; S, loss of growth at 8 μg/ml chloramphenicol compared to that of strain W3110; nt, nucleotide; blue, mutations in population A1; red, mutations in population A5; green, mutations in population C3; brown, mutations in population G5. The numbers in the leftmost column indicate the mutation position in the genome.
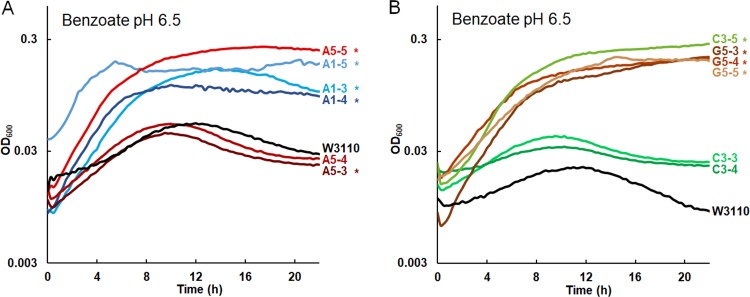
The early-generation clones (900 or 1,400) were tested for adaptation to growth in 20 mM benzoate (Fig. 2; for all eight individual replicate curves, see Fig. S1). At this benzoate concentration, the ancestral strain W3110 stopped growing and entered death phase by 12 h, whereas the 2,000-generation evolved strains grew more rapidly and reached stationary phase at an optical density at 600 nm (OD600) of about 0.25 (18). All the A1 population isolates from generation 900 or 1,400 grew in 20 mM benzoate to a stable stationary phase. From the A5 population, only the A5-5 strain (generation 1,400) achieved a sustained endpoint. In population G5, all the early isolates grew well (G5-3, G5-4, G5-5); but in population C3, the generation 900 strains grew only slightly better than W3110. Overall, the various early-generation isolates showed different rates of adaptation to benzoate.
FIG 2.
Isolates from early generations (900 to 1,400 doublings) from populations A1, A5, C3, and G5. Strains were cultured in LBK buffered with 100 mM PIPES, pH 6.5, and 20 mM sodium benzoate, as described in Materials and Methods. For each strain, the curve shown represents the median value of the OD600 at 16 h. *, significant difference from W3110, as determined by Tukey’s test (P ≤ 0.05; n = 4). The results for all four replicates are shown in Fig. S1 in the supplemental material. (A) A5-5, A1-5, A1-3, and A1-4 outgrew ancestor W3110. A5-3 and A5-4 entered death phase after 12 h, as did ancestor W3110. (B) C3-5, G5-3, G5-4, and G5-5 showed increased benzoate resistance compared to ancestor W3110. C3-3 and C3-4 entered death phase after 12 h, as did ancestor W3110.
Clones frozen before generation 1,000 had several mutations that persisted in the 2,000-generation strains (Table 2). Three of these mutations affected Gad island regulation: the partial deletion of the Gad island (consisting of the deletion of mdtE, slp, and everything in between [ΔmdtE–slp]; all isolates from population A1), a mutation in the Gad activator gene ariR (population A1) (50), and a mutation in the GadE activator gene gadX (population A5). In fact, more than half the isolates sequenced had one of seven different mutations within the slp-gad island (Table S1). Large deletions were likely mediated by upstream insertion sequences, a common finding in stress evolution experiments (18, 23). Thus, our early-generation sequences confirmed that the loss of Gad acid resistance was strongly selected by benzoate.
Loss of the Gad regulon, either by deletion or by downregulation, included the loss of the MdtEF-TolC efflux pump, which is specifically upregulated by GadX (34). MdtEF-TolC exports a variety of antibiotics and toxic metabolites, including chloramphenicol (44, 51, 52). It is one of a number of MDR pumps and regulators reported to be lost or mutated in genomes evolved with benzoate (18) or with CCCP (20). Note that while mdtEF mutations did appear early, sensitivity to chloramphenicol was not detected before generation 1,400 or 2,000 (Table 2). This suggests that multiple mutations accumulate over generations, contributing to the phenotype.
Various isolates had mutations affecting the metabolism of nucleotides and aromatic amino acids, that is, structures with similarity to benzoate or salicylate (Table S2). Genes with early, persistent mutations included folD (encoding 5,10-methylene-tetrahydrofolate dehydrogenase/cyclohydrolase for thymidine biosynthesis) (53) in population A1 and genes upstream of apt (encoding adenosine phosphoribosyltransferase for purine salvage) (54) in population A5. The 6-bp insertion upstream of apt was found by generation 500 in strain A5-6 and persisted through generation 2,000, and a frameshift in the apt coding sequence was found in strain E1-5 (Table S2). At generation 1,000, strain E1-5 had a mutation in yeaS (leuE), whose product effluxes leucine and toxic analogues. Biosynthetic genes with mutations included aroF (encoding 3‐deoxy‐d‐arabino‐heptulosonate 7‐phosphate synthase, which is involved in the first step of tyrosine biosynthesis) (55) and hisG (encoding ATP-phosphoribosyltransferase, which is involved in the first step of histidine biosynthesis) (56). Other mutations affected genes for aromatic catabolism and salvage: add (encoding adenosine deaminase) (57), deoD (encoding purine nucleoside phosphorylase [PNP]) (58), rihA (encoding ribonucleoside hydrolase) (59), paaE (encoding phenylacetate degradation) (60), and nupG (encoding nucleoside uptake transporter, PMF driven) (61). These mutations in aromatic metabolism may represent responses to the benzoate uptake in the cytoplasm. Their effects might be associated with the induction of the Mar regulon by intracellular aromatic intermediates that retard growth (62, 63).
Transcriptomes of benzoate-evolved isolates show reversal of benzoate regulation in the ancestor W3110.
In our report of acid evolution, the transcriptomes of evolved clones revealed the surprising reversal of acid stress responses (23). We therefore conducted a similar transcriptome analysis of our strains evolved in the presence of benzoate (18). Four of the 2,000-generation strains were selected (A1-1, C3-1, G5-1, and G5-2) for comparison with the ancestral strain, W3110. RNA was extracted during logarithmic growth from cultures supplemented with a relatively modest benzoate stress (5 mM benzoate) in order to sustain growth comparable to that of the ancestral strain. In addition, strain W3110 was cultured in medium with no benzoate, in order to identify genes responding to benzoate stress before the generations of benzoate selection.
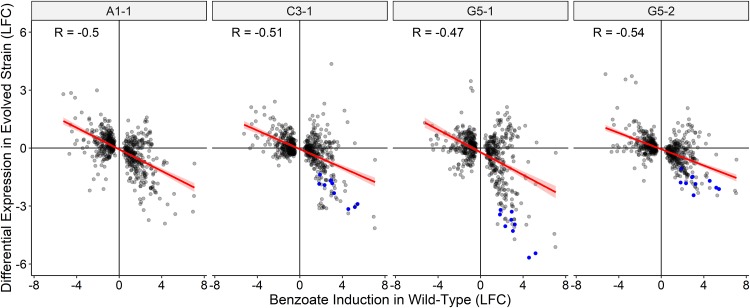
Figure 3 plots the ratios of the log2 changes in expression (versus the level of expression by the ancestor) for each benzoate-evolved 2,000-generation strain against the ratios of the log2 changes in expression in W3110 under conditions with benzoate versus without benzoate. Across the genome, genes that were regulated up or down by benzoate in the ancestor showed reversal (by deletion or by downregulation) in the four independently evolved isolates. In some cases, the reversal involved deletion of several genes or a major regulator, such as the deletion of nearly the entire Gad island (slp to gadW) in A1-1 or loss of the gadX activator in G5-1 and in G5-2. These strains showed the loss or downregulation of most of the Gad regulon (blue symbols in Fig. 3). Surprisingly, C3-1 had lost Gad expression, despite the presence of known regulators. Thus, the genotype of C3-1 may reveal novel means of Gad regulation. Overall, reversing the transcriptomic effects of benzoate appeared to be a general property of the evolved strains and did not appear to be limited to a few major systems.
FIG 3.
Genes upregulated by benzoate in the ancestral strain W3110 are downregulated in the benzoate-evolved strains. The gray dots indicate the log2 fold change (LFC) in the expression ratios of genes of the benzoate-evolved strains plotted as a function of the log2 fold change for W3110 with or without 5 mM benzoate. Data are from Table S3 in the supplemental material. The genes for which results are shown are those significantly differentially expressed (P < 0.001) in at least one evolved strain. Genes of the Gad regulon (gadAEXW, mdtEF, hdeDAB, slp) and gadBC are colored blue. In A1-1, most of the Gad island is deleted (Δslp-gadW). R, Pearson correlation coefficient.
The log2 fold changes in expression for individual genes of strain W3110 in the presence or absence of benzoate are presented in Table S3A. Genes were deemed significantly differentially expressed if the log2 fold change was greater than 1 and the P value was less than 0.01. Expression changes are also shown for the 2,000-generation strains versus W3110, all of which were cultured in 5 mM benzoate. Selected differentially expressed genes are shown in Table S3.
In W3110, benzoate upregulated much of the Gad regulon, including both glutamate decarboxylase genes (gadA, gadB) and the glutamate transporter gene gadC, the regulator gene gadE, and the Gad-associated multidrug efflux pump gene mdtEF. The hdeABD portion of the Gad island showed less induction, likely because hdeAB is repressed by MarA (Fig. 1), which is induced by benzoate (Table 3). Benzoate induction of glutamate decarboxylase would be consistent with the model of Sayed and Foster that cytoplasmic pH depression induces Gad (64). However, the cytoplasmic pH depression caused by benzoate uptake does not involve extreme acidification of the periplasm, since the media are buffered to pH 6.5. Thus, the cell has less need for the periplasmic chaperones HdeA and HdeB.
TABLE 3.
Selected differentially expressed genes
| Regulation | Genes differentially expressed in the following comparisonsa: |
||||
|---|---|---|---|---|---|
| W3110 grown with benzoate vs no benzoate | A1-1 vs W3110 grown with benzoate | C3-1 vs W3110 grown with benzoate | G5-1 vs W3110 grown with benzoate | G5-2 vs W3110 grown with benzoate | |
| Upregulated | aceABEFK | acs | cspB | cvpA | ompF |
| appAB | glcDE | ompG | fadB | ompL | |
| asr | ompF | ompL | glcDE | wcaADK | |
| cyoABCDE | pspB | ompL | ymgG | ||
| dppCB | phoE | phoE | |||
| gadABC | phoH | ||||
| gadEF | |||||
| hdeB | |||||
| hyaBF | |||||
| hycABCDEFG | |||||
| marRAB | |||||
| mdtEF | |||||
| narHJ | |||||
| oppABCD | |||||
| slp | |||||
| Downregulated | cadAB | appBC | flu | dctR | artP |
| flgBCDEFG | dctR | gadABC | gadABC | asr | |
| fliAFN | flu | hycACDEFGH | gadEF | flu | |
| fruAK | gadABC | pepN | hdeABD | gltIK | |
| ompF | gadEF | slp | hycACDEFGH | hycCDEFG | |
| ymgG | mdtEF | mdtEF | marRB | ||
| hycBCDEFGH | slp | slp | |||
| hdeABD | sufABCD | yahK | |||
| narHIJ | yahK | yeeR | |||
| slp | yhiDM | ||||
| yeeR | wrbA | ||||
| yhiDM | |||||
| ymgG | |||||
Differential expression was indicated by a log2 fold change in expression of ≥1 and a P value of <0.01. All strains were cultured to log phase at pH 6.5 in 5 mM sodium benzoate, except for the W3110 control culture, which was cultured without benzoate. Boldface without underlining indicates genes upregulated by benzoate in W3110 but knocked down in benzoate-evolved strains (A1-1, C3-1, G5-1, G5-2). Boldface with underlining indicates genes that were downregulated by benzoate in W3110 and that had increased expression in benzoate-evolved strains.
While MarA is induced by benzoate in the ancestor, the benzoate-evolved strains showed the loss of various Mar regulon components either by marRAB deletion (Table 2, strain A5-1) or by loss of another regulator, such as rob (strain G5-2). MarA downregulates the large outer membrane porin gene ompF (Fig. 1). The downregulation of ompF helps exclude toxins and antibiotics (28).
No ompF mutations appeared in our evolved strains, yet in transcriptomes (Table 3), two of the four benzoate-evolved strains (A1-1 and G5-2) showed upregulation of ompF. Strain C3-1 upregulated other large porin genes, ompG (65), phoE (66), and ompL (67). Similarly, strain G5-1 upregulated porin genes phoE and ompL. Thus, under long-term exposure to benzoate, with concomitant PMF depletion, all four strains upregulated porins that could enhance fitness by admitting more carbon sources for substrate-level phosphorylation and fermentation. The upregulation of porins could also increase the admittance of antibiotics in the benzoate-evolved strains (26, 28).
In strain W3110, benzoate also induced expression of oligopeptide transport operons dppBC (68) and oppABCD (69). Like the porins, these transporters could increase access to fermentable substrates. These transporters retained expression in the benzoate-evolved strains.
W3110 genes induced by benzoate also included those encoding several components of electron transport, including cytochrome bo3 ubiquinol oxidase (cyoABCDE), cytochrome oxidase cytochrome bd-II (appBC) (70, 71), nitrate reductase (narHJ) (72), and hydrogenase 3 (Hyd-3; hycABCDEFG) (73). Hyd-3 converts H+ ions to H2 and associates with the formate-hydrogenlyase complex (FHL); it includes the HycA regulator of FHL (74). The acceleration of electron transport is consistent with the effect of uncouplers on respiration, generating a futile cycle during stationary phase (75). The energy loss during stationary phase could explain why strain W3110 enters death phase after several hours in culture with 20 mM benzoate (18).
The FHL/Hyd-3 components (hyc genes) were downregulated in all four benzoate-evolved strains. Strain A1-1 had lower expression of the appBC-encoded cytochrome oxidase cytochrome bd-II. All these complexes—Hyd-3, FHL, and the cytochrome bd-II oxidase—are generally expressed under low-oxygen conditions (73, 76). Hyd-3 converts 2H+ to H2 as part of the formate-hydrogenlyase complex which oxidizes formate to CO2 (77). Hydrogen production by Hyd-3 is induced by external acid and is required for extreme acid survival in low oxygen (78). Thus, the benzoate-associated loss of acid-inducible hydrogenase (and of formate-hydrogenlyase) parallels the loss of the acid-inducible Gad system.
No genes for hydrogenases had mutations in our resequenced genomes, although three different mutations affecting the nitrate-inducible formate dehydrogenase (fdnG and fdnI) appeared (79). Also, in A1-1, the narHIJ genes encoding quinol-nitrate oxidoreductase (80) were downregulated (Table 3; Table S3B). Thus, some kinds of reregulation that decrease the wasteful expenditure of electrons occurred. These changes most likely enable benzoate adaptation during the low-oxygen period of stationary phase, the period in which the ancestral strain declines (Fig. 2).
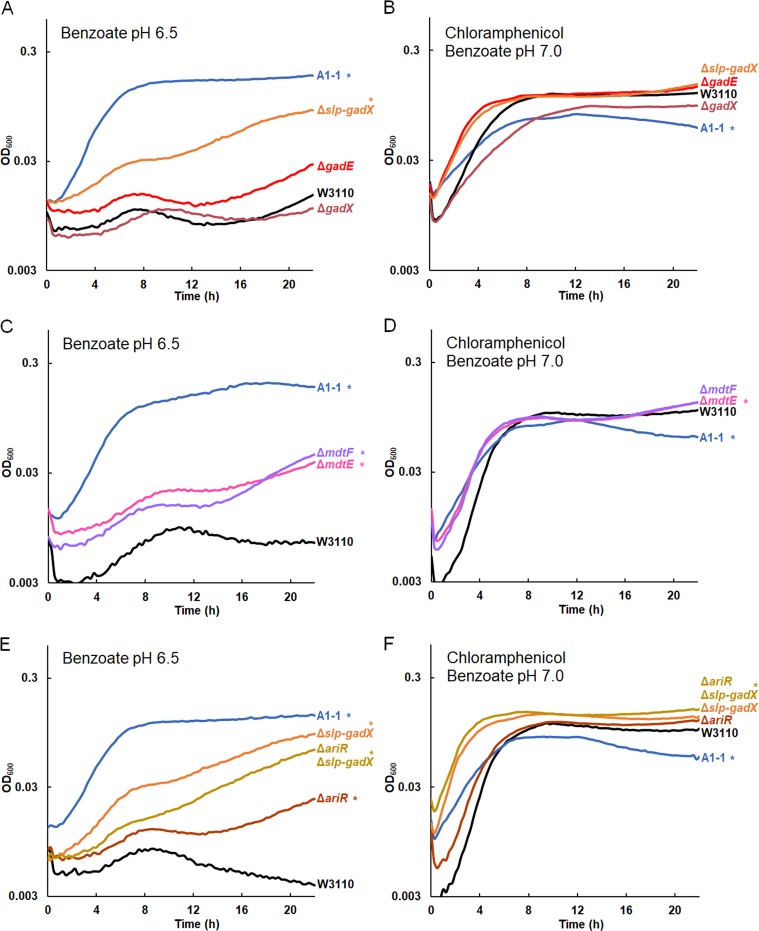
Deletion of slp-gadX, mdtE, mdtF, or ariR confers benzoate tolerance.
Our resequenced genomes of benzoate-evolved strains offered candidate genes for mechanisms of benzoate tolerance. We selected candidate gene kanR replacements from the Keio collection to run batch growth curves that approximate the daily experience of serial dilution. Growth curves can reveal fitness differences of several types: the duration of lag phase, the rate of log-phase growth, the endpoint reached at stationary phase, and the post-stationary-phase death rate.
We started with the isolates from population A1, which suggests a progression of acquired mutations that contribute to benzoate tolerance (Table S2). The early-generation isolates do not necessarily represent direct ancestors of the later strains, yet the deletion spanning the start of slp to the end of gadX (slp-gadX) was present in all A1 strains sequenced, and this region encompasses the region with most early mutations found in other populations (Table 2). We constructed by recombineering (81) a strain with the slp-gadX deletion (the slp-gadX strain).
Strains were cultured in microtiter plates with 15 mM benzoate, as described in Materials and Methods (Fig. 4). The OD600 value was measured at 16 h to evaluate the endpoint, and for each condition, we show a replicate curve with the median OD600 at 16 h. All eight replicate curves are shown in Fig. S2. The slp-gadX strain had higher growth than W3110 at 16 h and growth nearly as high as that of benzoate-evolved strain A1-1 (Fig. 4A). Mutations in W3110 consisting of the deletion of gadE or gadX did not show a consistent effect on growth, although the regulation of the Gad island in these mutants (64) might have fitness effects detectable by direct competition.
FIG 4.
Gad regulon and related mutations in strain A1-1 increase growth in benzoate. Replicate curves are shown in Fig. S4 in the supplemental material. Strains were cultured overnight in LBK buffered with 100 mM PIPES, pH 6.5 (A, C, E) or LBK buffered with 100 mM MOPS, pH 7.0 (B, D, F), both of which were supplemented with 5 mM benzoate. Eight replicate samples from two overnight cultures (four replicate samples from each culture) were diluted 1:200 in a 96-well plate into fresh medium. The cultures consisted of LBK buffered with 100 mM PIPES, pH 6.5, and supplemented with 15 mM benzoate (A, C, E) or LBK buffered with 100 mM MOPS, pH 7.0, and supplemented with 5 mM benzoate and 4 μg/ml chloramphenicol (B, D, F). For each group of replicates, a curve with the median OD600 at 16 h is presented. *, significant difference from W3110 at 16 h, as determined by Tukey’s test (P ≤ 0.05; n = 8). The results for all eight replicates are shown in Fig. S4 in the supplemental material. (A) The W3110 Δslp-gadX, ΔgadE::kanR, and ΔgadX::kanR constructs were cultured in 15 mM benzoate alongside parent strain W3110. The Δslp-gadX strain grew to a higher OD600 than did W3110 at 16 h, though not to an OD600 as high as that of benzoate-evolved strain A1-1. (B) The strain A1-1 and W3110 Δslp-gadX, ΔgadE::kanR, and ΔgadX::kanR constructs were cultured with W3110 in benzoate and chloramphenicol. Strain A1-1 grew significantly less than W3110 at 16 h. (C) The ΔmdtE::kanR and ΔmdtF::kanR strains outgrew W3110 in benzoate but did not grow to an OD600 as high as that of A1-1. (D) The ΔmdtE::kanR and ΔmdtF::kanR strains grew to an OD600 higher than that of W3110 in 5 mM benzoate with chloramphenicol, whereas strain A1-1 reached a lower OD600 at 16 h. (E) The ΔariR::kanR strain had increased growth in benzoate, but it did not grow to an OD600 as high as that of the Δslp-gadX, ΔariR::kanR Δslp-gadX, or A1-1 strain. (F) While the ΔariR::kanR and Δslp-gadX mutations did not affect growth in benzoate with chloramphenicol, the ΔariR::kanR Δslp-gadX strain outgrew the ancestor, W3110.
Salicylate has an effect similar to that of benzoate on the growth of W3110 and the benzoate-evolved strains, and its effect starts at a lower concentration (18). An experiment similar to that whose results are presented in Fig. 4B was conducted using chloramphenicol with 2 mM sodium salicylate instead of 5 mM benzoate; similar results were obtained. Figure S3 presents the results of salicylate experiments, which were comparable to those of all the chloramphenicol-benzoate experiments presented in Fig. 4 to 7.
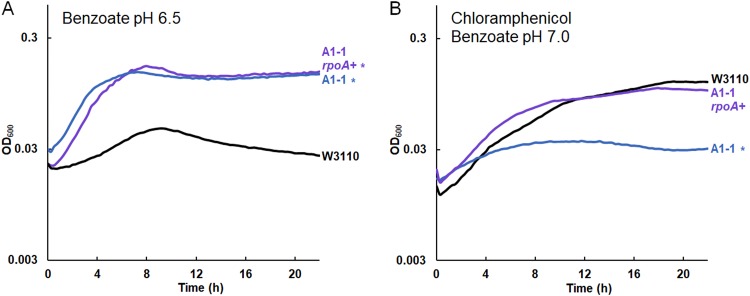
FIG 5.
Chloramphenicol sensitivity with rpoA K271Q replacement by rpoA+ in A1-1. Replicate curves are shown in Fig. S5 in the supplemental material. (A) The OD600 of the construct A1-1 with rpoA K271Q reversion to rpoA+ (A1-1 ΔyhdN::frt rpoA+) showed no significant difference from that of A1-1 (cultured with 15 mM benzoate). (B) A1-1 ΔyhdN::frt rpoA+ grew to a level comparable to that of W3110 in 8 μg/ml chloramphenicol with 5 mM benzoate. Culture conditions for the assay whose results are presented in panels A and B were the same as those for the assay whose results are presented in Fig. 4A and B, respectively, except that the chloramphenicol concentration was 8 μg/ml (B). A significant difference from W3110 was determined at 16 h, as determined by Tukey’s test (P ≤ 0.05; n = 8).
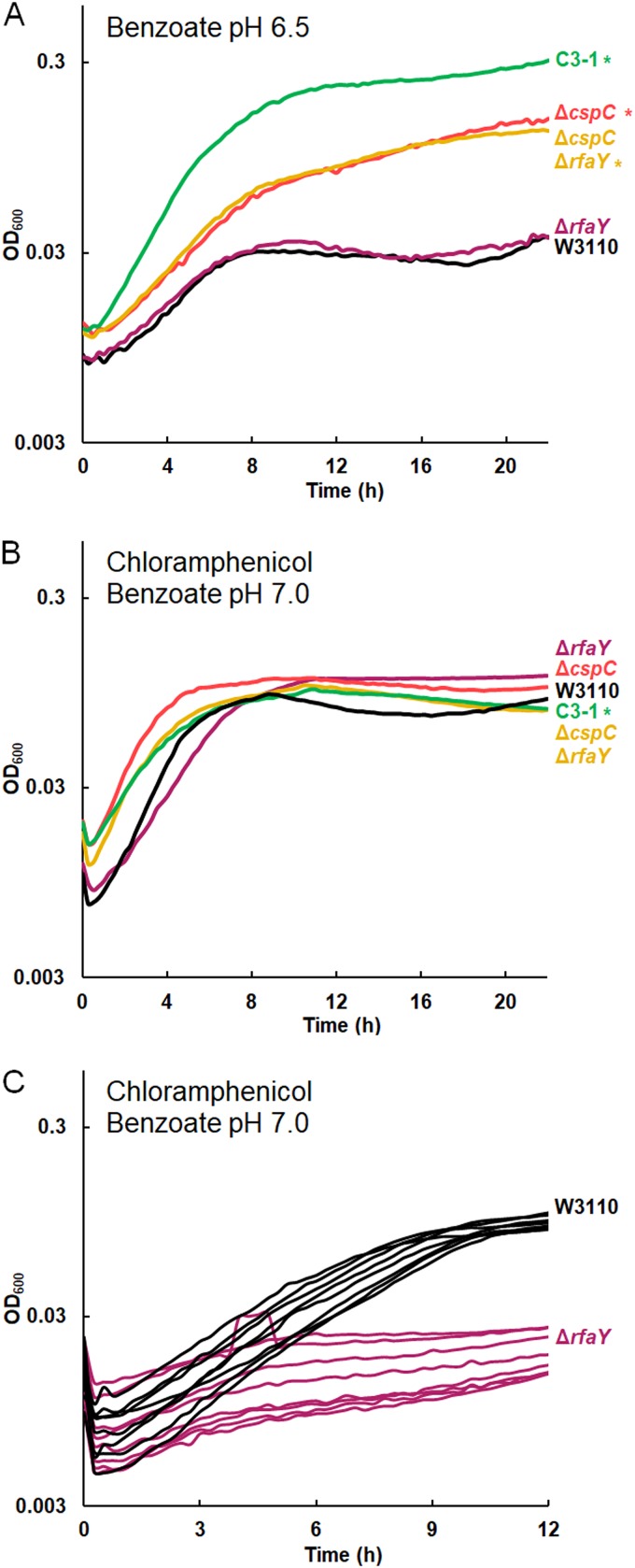
FIG 6.
The ΔcspC deletion confers benzoate tolerance, and the ΔrfaY deletion confers chloramphenicol sensitivity. The results for replicates are shown in Fig. S6 in the supplemental material. (A) In 15 mM benzoate, W3110 ΔcspC reached higher growth than W3110, but W3110 ΔrfaY showed no difference in growth from W3110. The growth curve for the ΔcspC ΔrfaY mutant resembled that for the ΔcspC mutant. (B) The growth of the W3110 ΔrfaY::kanR and ΔcspC::kanR ΔrfaY::frt constructs showed no difference from that of W3110 at 16 h, but the ΔrfaY::kanR mutant had a lower log-phase growth rate than W3110 in 5 mM benzoate with 4 μg/ml chloramphenicol. (C) With 8 μg/ml chloramphenicol and 5 mM benzoate, W3110 ΔrfaY::kanR has a lower log-phase growth rate than W3110 (t test, P < 0.01); for each strain, 8 individual replicates are shown. A significant difference from the result for W3110 was determined at 16 h, as determined by Tukey’s test (P ≤ 0.05; n = 8).
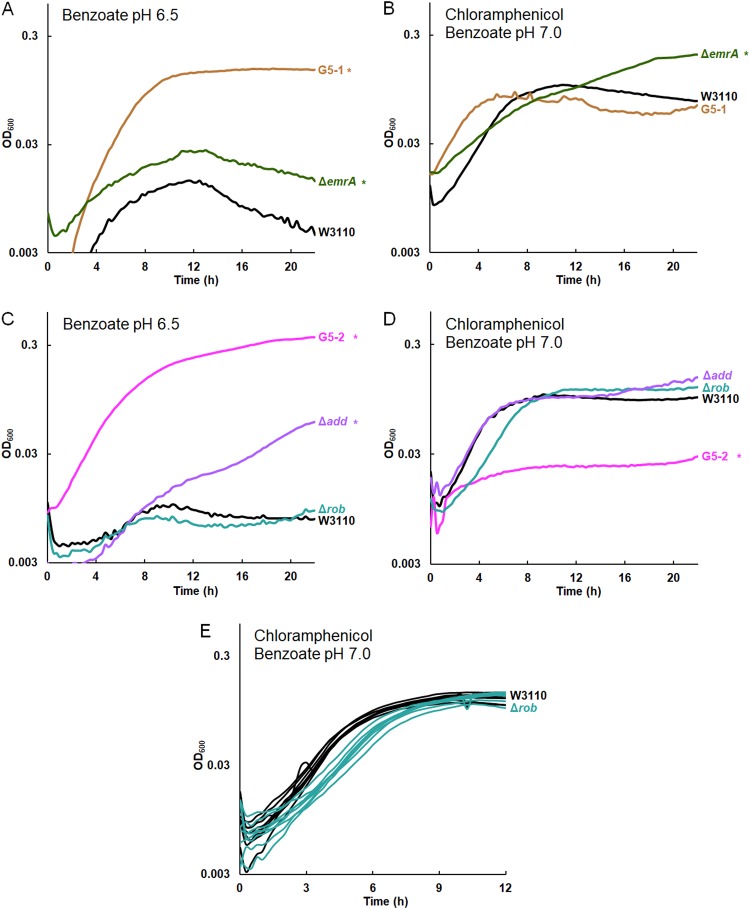
FIG 7.
The Δadd mutation confers benzoate tolerance, and the Δrob mutation decreases log-phase growth rate in chloramphenicol. The results for all replicates are shown in Fig. S7 in the supplemental material. (A) Strains G5-1 and W3110 ΔemrA::kanR outgrew W3110 at 16 h in 15 mM benzoate. (B) W3110 ΔemrA outgrew W3110 and G5-1 at 16 h in 5 mM benzoate and 4 μg/ml chloramphenicol. (C) G5-2 and W3110 Δadd::kanR outgrew W3110 in 15 mM benzoate. The Δrob::kanR mutation did not affect growth. (D) W3110 Δadd::kanR, W3110 Δrob::kanR, and W3110 showed no difference in growth in 5 mM benzoate and 4 μg/ml chloramphenicol. Strain G5-2 grew less than W3110. (E) The Δrob::kanR mutation conferred a lower log-phase growth rate in 5 mM benzoate and 4 μg/ml chloramphenicol (t test, P < 0.01). The results for eight replicates per strain are shown.
We then tested the benzoate tolerance of strains with deletions of the mdtE and mdtF components of the MdtEF-TolC MDR efflux pump, which is included in the acid fitness island deletion (29). Strains with mdtE::kanR and mdtF::kanR knockouts each reached a higher density at 16 h than W3110 in 15 mM benzoate at pH 6.5 (Fig. 4C). However, the increase was significantly less than that found for strain A1-1 and the slp-gadX strain. This suggests that various genes of the Gad island make additive partial contributions to benzoate tolerance.
Another mutation in A1-1 that had the potential to affect benzoate tolerance was a transposon-mediated knockout of ariR (18, 43). AriR (YmgB) regulates both Gad acid resistance and biofilm formation (43), possibly mediated by RpoS (50). An ariR::kanR knockout strain had increased endpoint growth compared to the wild type. We then tested whether an ariR deletion interacted with the acid fitness island deletion in an additive manner or whether its function was made redundant by the slp-gadX deletion. We transduced ariR::kanR into the slp-gadX strain (Fig. 4E). This strain grew identically to the slp-gadX strain in 15 mM benzoate, suggesting that the ariR single deletion increases benzoate tolerance through regulation of the Gad island.
Chloramphenicol sensitivity is conferred by rpoA K271Q but not by slp-gadX deletion.
The benzoate tolerance of our benzoate-evolved strains was associated with a trade-off in sensitivity to chloramphenicol, as seen for strains A1-1, A5-1, C3-1, G5-1, and G5-2 (18). We sought to clarify how benzoate tolerance is connected with antibiotic sensitivity. Our new isolates from the early generations confirmed that chloramphenicol sensitivity emerged later than benzoate tolerance; only one generation 1,400 isolate (A1-5) and none of the earlier isolates showed a measurable decrease in growth relative to that of W3110 with chloramphenicol at 4 μg/ml or at 8 μg/ml (Table 2, S columns). The slp-gadX deletion and deletions of individual genes were also tested for their effects on chloramphenicol resistance (Fig. 4B, D, and F). Chloramphenicol resistance was tested in a medium containing a low level of benzoate (5 mM) in order to induce any Mar and non-Mar MDR systems. None of the mutations affected chloramphenicol resistance, despite the known chloramphenicol efflux by MdtEF-TolC (52). This could occur because chloramphenicol is exported by redundant systems encoded by sequences outside the region encoded by slp-gad, including AcrAB-TolC.
For strain A1-1, we hypothesized that its chloramphenicol sensitivity required an additional mutation acquired relatively late in the evolution experiment, perhaps between generations 900 and 1,400. An interesting candidate was the rpoA (encoding the RNA polymerase alpha subunit) mutation that appeared in strain A1-5 (generation 1,400, chloramphenicol sensitive) and persisted in A1-1. The carboxy-terminal domain (CTD) of this subunit regulates gene expression by interacting with upstream promoter elements and transcription factors, such as cyclic AMP receptor protein (CRP).
Our rpoA mutation K271Q is nearly the same as that of a well-studied rpoA allele, rpoA341 K271E (82, 83). The rpoA341 allele downregulates certain positively controlled regulons, such as mel, ara, and cys; but not all positive regulators are affected. The rpoA341 allele may also upregulate loci such as ompF (82). We found ompF to be upregulated in strain A1-1 (Table 3; Table S3B). Strain A1-1 also had decreased expression of araBAD and of cysA, as reported for the rpoA341 allele (82).
We sought to test the contribution of rpoA K271Q in strain A1-1 with respect to benzoate tolerance and chloramphenicol sensitivity. To do this, we replaced the mutant allele with rpoA+ by cotransduction from W3110 into A1-1 using the linked marker yhdN::kanR. The yhdN::kanR marker had no effect on the growth of strain A1-1, with or without chloramphenicol (Fig. S4). Strain JLS1616 (A1-1 ΔyhdN rpoA+) was cultured in parallel with strain A1-1 and the parent strain, W3110 (Fig. 5; replicate curves are shown in Fig. S5). In 15 mM benzoate, the A1-1 rpoA+ strain grew similarly to the parent A1-1 strain, with no significant difference at any time point (Fig. 5A). However, with chloramphenicol and a low concentration of benzoate (5 mM), the A1-1 rpoA+ strain showed growth comparable to that of W3110, whereas A1-1 with the original rpoA mutation was sensitive to chloramphenicol. Thus, rpoA K271Q is responsible for the loss of chloramphenicol resistance associated with the benzoate evolution of A1-1.
cspC deletion confers benzoate tolerance, and rfaY deletion decreases the growth rate in chloramphenicol.
The C3-1 strain has two transposon-mediated deletions that might affect growth in benzoate, affecting the gene for a posttranscriptional regulator (cspC) and the lipopolysaccharide (LPS) kinase gene rfaY (18). The sequence of cspC is similar to that of other cold shock protein genes, and the CspC protein has been shown to upregulate genes under RpoS control by stabilizing the rpoS mRNA (84–86).
A ΔcspC::kanR knockout strain showed increased growth relative to strain W3110 in 15 mM benzoate (Fig. 6A; replicate curves are shown in Fig. S6). However, in medium with 4 μg/ml chloramphenicol, there was no significant difference in growth (Fig. 6B). Thus, we found evidence for a contribution of ΔcspC to the benzoate tolerance of C3-1 but not to chloramphenicol sensitivity.
An ΔrfaY::kanR strain showed no phenotype in 15 mM benzoate (Fig. 6A). The double mutant W3110 ΔcspC::kanR ΔrfaY::frt grew identically to W3110 ΔcspC. In chloramphenicol, however, W3110 ΔrfaY::kanR had a mean growth rate of 0.12 ± 0.01 doublings per hour in early log phase, which was less than the mean W3110 growth rate of 0.39 ± 0.02 doublings per hour (Fig. 6C). A t test comparison gave a P value of <0.01. The double mutant also showed a significant decrease in the growth rate in chloramphenicol compared to that of the ΔcspC::kanR single mutant. Thus, the double mutant showed the phenotype associated with the presence of cspC when it was cultured in 15 mM benzoate but the phenotype associated with ΔrfaY when it was cultured with chloramphenicol (5 mM benzoate).
The C3 population isolates had no mutations within slp-gadX, except for a 76-bp deletion of gadX, found in C3-5, yet the C3-1 transcriptome showed lower expression of gadABC (Fig. 3), so its genome must have altered regulation of acid tolerance pathways. Given that cspC is implicated in the stabilization of the rpoS mRNA (85, 87) and RpoS regulates Gad (Fig. 1), it is possible that cspC deletion downregulates Gad via a decreased RpoS concentration.
emrA and add deletions confer benzoate tolerance, while rob and hfq confer chloramphenicol sensitivity.
From population G5 (Table 2), strains G5-5 and G5-1 had a knockout mutation in emrA, which normally encodes part of the EmrAB-TolC pump (19). The emrA gene acquires point mutations—but not knockouts—under evolution with the uncoupler CCCP, which is expelled by EmrAB-TolC (20). We found that deletion of emrA conferred a degree of tolerance to benzoate (Fig. 7A; replicate curves are shown in Fig. S7). The emrA deletion conferred added benzoate tolerance even in the presence of chloramphenicol (Fig. 7B).
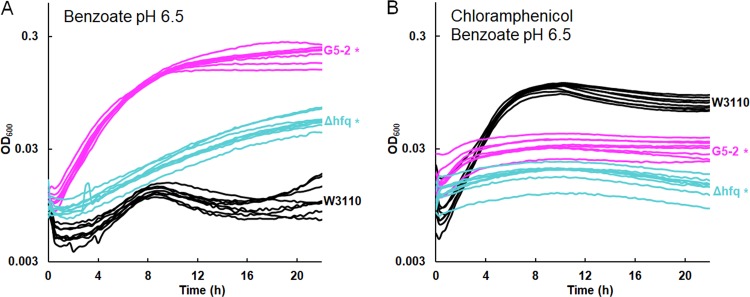
Another strain from the G5 population, G5-2, showed the greatest sensitivity to chloramphenicol and tetracycline of all 2,000-generation strains tested (18). G5-2 contained a frameshift in add, a gene encoding adenosine deaminase, which catalyzes a proton-consuming reaction of purine catabolism that protects the cell from acid by a mechanism analogous to that of GadA (88). Deletion of add conferred partial tolerance to benzoate (Fig. 7C). This result is yet another example of benzoate selection against an external-acid protection mechanism. G5-2 also contained a point mutation in rob encoding a MarA-type regulator that enhances chloramphenicol resistance (45). Deletion of rob did not significantly enhance growth in 15 mM benzoate (Fig. 7C) but did decrease the log-phase growth rate in chloramphenicol from an average growth rate of 0.63 ± 0.01 doublings per hour for W3110 to an average growth rate of 0.59 ± 0.02 doublings per hour for W3110 Δrob::kanR (Fig. 7E). A t test comparison gave a P value of <0.01. Replicates for the log-phase-growth region are shown for W3110 Δrob::kanR and W3110 in 4 μg/ml chloramphenicol.
G5-2 also contained a missense mutation in hfq, which encodes a pleiotropic regulator that functions as an RNA chaperone (89) and DNA-binding protein (90). Hfq is associated with antibiotic resistance and is a target for antimicrobial chemotherapy (91, 92). We found that an hfq deletion conferred partial tolerance to 15 mM benzoate (Fig. 8A) and sensitivity to chloramphenicol (Fig. 8B). In chloramphenicol, the growth curve of W3110 Δhfq::kanR was indistinguishable from that of G5-2. Thus, hfq mutation could contribute a major part of the G5-2 phenotype of benzoate tolerance associated with antibiotic sensitivity.
FIG 8.
W3110 Δhfq::kanR increases growth in 15 mM benzoate and decreases growth in chloramphenicol. (A) Strains G5-2 and W3110 Δhfq::kanR outgrew W3110 at 16 h in 15 mM benzoate. (B) W3110 outgrew strains G5-2 and W3110 Δhfq::kanR at 16 h in 5 mM benzoate and 4 μg/ml chloramphenicol.
Hydrogenase 3 deletion enhances late growth.
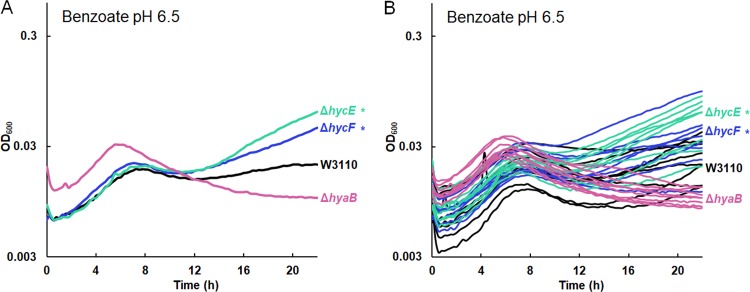
Our transcriptomes showed that benzoate induces hydrogenase 3 in W3110 but that all four benzoate-evolved strains lose expression of hycEFG (Table 3). We tested the growth of W3110 strains with deletions of hycE (encoding the hydrogenase activity 2H+ → H2) and hycF (the subunit of the formate-hydrogenlyase complex) (77). Both the ΔhycE::kanR and ΔhycF::kanR strains grew similarly to W3110 until stationary phase. At 16 h, there was no significant difference between the ancestral and mutant OD600. However, after 20 h, when hydrogenase would be active, the mutants achieved a higher OD600 than W3110 (Fig. 9). These results confirm that although benzoate exposure induces the consumption of cytoplasmic protons via hydrogenase 3 as part of formate breakdown, this activity decreases long-term relative fitness in benzoate. In contrast, hydrogenase 1 (encoded by hya) consumes H2, producing 2H+ (73).
FIG 9.
Deletion of hydrogenase 3 (hyc) enhances late growth in 15 mM benzoate, pH 6.5. W3110 with ΔhycE::kanR or ΔhycF::kanR grows to higher median value of OD600 at 20 h than parent strain W3110. The W3110 ΔhyaB::kanR strain with a deletion of the gene for Hyd-1 shows no enhancement of late growth compared to W3110. (A) Median growth curves presented for each set of 8 replicates. (B) Results for all 8 replicates for each strain are shown.
Other genes tested.
kanR knockout strains were tested for other genes that had mutant alleles in our benzoate-evolved isolates, but no significant difference from the ancestor was detected for growth with benzoate or with chloramphenicol. As noted above, such differences might emerge under extended direct competition. The genes tested by 22 h of culture with 15 mM benzoate included acnA, chbC, cpxA, deaD, fucA, mdtA, gltP, hdeD, pepN, rnb, yhfM, uxaA, and yhiD.
DISCUSSION
Here we combined genomic, transcriptomic, and genetic approaches to study how E. coli strains evolve in the presence of benzoate and salicylate. Our work extends the picture of known MDR-related genes mutated under exposure to partial or full uncouplers (18, 20) and suggests additional mechanisms for the increased sensitivity to antibiotics, such as the upregulation of porins and pleiotropic regulators. This subject has implications beyond E. coli, because the microbiomes of soil and the rhizosphere (93) as well as human and animal digestive tracts (94) are exposed to benzoate and related molecules.
Evolution in the presence of benzoate reverses the short-term benzoate stress response.
We confirmed by analysis of candidate gene deletions that despite the short-term benzoate upregulation of Gad genes (Table 3), the presence of benzoate actually selects against the Gad island (slp-gadX) and against Gad gene components mdtE and mdtF, which encode a multidrug efflux pump (Fig. 4). Our transcriptomes reveal a striking pattern of repression of Gad as well as other benzoate-inducible gene products in the benzoate-evolved strains (Table 3). Even a strain with no mutations in the fitness island (strain C3-1) had downregulated Gad, possibly by mutation of the RpoS posttranscriptional activator CspC. Similarly, strain G5-2 could have downregulated Gad via deletion of rob or of hfq (which activates RpoS). Collectively, these results show a pattern of convergent evolution achieved via one of several possible genetic mechanisms.
Our transcriptomes also showed that evolution with benzoate increased the expression of several large-hole porins (OmpF, OmpG, PhoE, OmpL). These porins amplify access to fermentable carbon sources for substrate-level phosphorylation, thus decreasing dependence on PMF, but OmpF is normally downregulated by benzoate derivatives via MarA, in order to exclude antibiotics (26). It is interesting that benzoate induced short peptide transporter genes opp and dpp (Table 3), which offer another means of access to fermentable carbon sources; this expression of peptide transporters was maintained in benzoate-evolved strains. For opp, there is controversial evidence that increased peptide transport coincides with antibiotic sensitivity (95–97).
Another form of reversal of benzoate regulation was the downregulation of hydrogenase 3 (Table 3; Fig. 9). The Hyd-3 substrate, formate, arises from E. coli fermentation, increasing in stationary phase. Hyd-3 as part of the formate-hydrogenlyase complex normally converts formate to CO2 and H2, yet we show that in the presence of benzoate, Hyd-3 deletion enhances fitness during late stationary phase, the period where we would expect FHL/Hyd-3 to be active (76). There is evidence that FHL activity exports protons (98). If so, this could be yet another system that wastes energy as exported protons drive more benzoate into the cell and thus decreases relative fitness over long-term subculturing. The possible effects of benzoate and other partial uncouplers could be relevant to the biotechnology of hydrogen production (77).
In strain A1-1, several alternative terminal oxidases were downregulated. Antibiotic resistance is linked to bacterial respiration, as several bactericidal antibiotics are shown to increase respiratory rates, while most bacteriostatic compounds decrease respiration (99). One means of adaptation to a high benzoate concentration could be to limit respiration by downregulating unneeded components of electron transport, including anaerobic respiration. This would limit the energy wasted by uncoupling respiration from ATP synthase. Introduction of antibiotics to a respiration-compromised cell could amplify the phenotype and lead to benzoate-induced antibiotic sensitivity.
Reversal of the short-term stress response following long-term serial culture has been seen in other evolution experiments as a tendency to restore global prestress conditions. After heat stress evolution of E. coli, the resulting strains show a loss of heat shock gene expression (100). The fitness cost of the stress response could involve either the cost of stress-induced gene expression, when it fails to provide a benefit (22), or the cost of excess PMF expenditure by a transporter or an efflux pump (25).
Growth of candidate gene deletion strains reveals contributions to benzoate tolerance and chloramphenicol sensitivity.
The loss of the MdtEF-TolC drug efflux system directly increases benzoate tolerance. This efflux pump couples drug efflux to PMF (31, 33, 101). Theoretically, benzoate decreases PMF by shuttling protons through the membrane. Thus, the presence of other proteins that utilize PMF further depletes the pool of extracellular protons available for core cell processes. It is possible that deleting drug efflux systems decreases proton flux through systems depleted by benzoate; thus, the fitness cost of drug pumps is amplified by benzoate.
We showed four candidate alleles associated with antibiotic sensitivity, and three of these have global or pleiotropic effects: the rpoA mutation in A1-1; the rob mutation in G5-2; and the deletion of hfq, which conferred both tolerance to benzoate and sensitivity to chloramphenicol. hfq deletion is a known source of drug sensitivity (91, 92).
Since the mutation in rpoA is in the rpoA product carboxy-terminal domain, which interacts with bacterial promoter upstream elements (UP elements) and certain transcription factors, it is possible that this allele downregulates a large set of genes (102, 103). This downregulation could free up resources for the response to benzoate stress, which could include a large number of PMF-driven antibiotic resistance genes. In fact, there is previous evidence suggesting that an interaction between the CTD of RpoA and MarA is necessary for MarA to induce the Mar regulon (103). Mutation of the RpoA CTD could block MarA activation of drug resistance, despite marA transcription.
The rfaY gene (which is mutated in strain C3-1) encodes a membrane-bound enzyme that phosphorylates the inner core of lipopolysaccharide (LPS), a function that has been implicated in membrane stability (104). The decreased membrane stability caused by the rfaY mutation may increase the permeation of certain antibiotics, thereby decreasing the antibiotic resistance of strain C3-1.
Another surprising finding was the pervasive occurrence of small mutations in genes for aromatic biosynthesis and catabolism; for example, a point mutation was found in folD and 1-base-pair deletion was detected in add. The folD gene is essential (53) and could not be deleted, but deletion of add was shown to enhance benzoate tolerance. The evidence points to further exploration of the role of benzoate and salicylate in modulating the efflux of aromatic intermediates of metabolism (62), especially given the benzoate-evolved enhancement of substrate influx via porins (Table 3).
Note that the relative fitness advantage of a given allele can accrue by various means at different phases of the growth cycle. For most of the candidate genes that we tested, such as gadE, mdtE, and cspC, deletion enabled cells to grow to a higher optical density than the parent, W3110. However, the chloramphenicol sensitivity associated with some alleles, such as rfaY, was caused by a lower rate of growth during log phase. While the bacteria grew to an optical density comparable to that of W3110, had the two strains been competing in coculture, the mutant strain would have soon lost out to the parent.
Overall, we reveal genetic mechanisms by which multigenerational exposure to benzoate leads to increased tolerance of benzoate or salicylate, with the trade-off being sensitivity to certain antibiotics. Our findings have implications for the roles of benzoate as a food preservative and for salicylate as a plant defense signal and as a therapeutic agent.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli K-12 W3110 (laboratory stock D13) was the parent for all genetic analyses. The strain was resequenced for analysis (18). Unless otherwise specified, bacteria were cultured in LBK (10 g/liter tryptone, 5 g/liter yeast extract, 7.45 g/liter KCl) with a pH buffer at 37°C. The growth media were supplemented with benzoate, salicylate, kanamycin (50 μg/ml), or chloramphenicol (4 or 8 μg/ml) as necessary. For growth curves, medium was buffered to pH 7.0 with 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS; pKa = 7.25) or to pH 6.5 with 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pKa = 6.80) containing 70 mM Na+. The medium pH was adjusted with either 5 M HCl or 5 M KOH. Cultures were incubated at 37°C unless otherwise specified. Strains with kanR insertions were obtained from the Keio collection (105). The XTL241 strain containing the cat-sacB fusion was obtained from the D. L. Court lab at NCI (81). All other strains used in this study were derived from W3110 and are listed in Table 1 and in Table S1 in the supplemental material (new isolates with resequenced genomes).
Growth curves.
For growth curves in 15 mM benzoate, strains were cultured overnight in LBK with 100 mM PIPES, pH 6.5, supplemented with 5 mM benzoate. These cultures were diluted 1:200 in a 96-well plate into fresh LBK buffered to pH 6.5 with 100 mM PIPES supplemented with 15 mM benzoate. The OD600 was read in a SpectraMax spectrophotometer every 15 min for 22 h. For growth curves in chloramphenicol, strains were cultured overnight in LBK with 100 mM MOPS, pH 7, supplemented with 5 mM benzoate as needed. These cultures were diluted 1:200 into fresh medium supplemented with 4 μg/ml chloramphenicol, unless stated otherwise. The endpoint OD600 was defined as the cell density after 16 h. Significance tests included analysis of variance with Tukey’s post hoc test (R software). Each data figure represents the results of three experiments, in each of which 8 replicate cultures were tested, unless stated otherwise.
Genome sequencing of early-generation evolved strains.
Genomic DNA was extracted from early-generation clones and from the W3110 ancestral stock with a DNeasy DNA extraction kit (Qiagen) and a MasterPure Complete DNA and RNA purification kit (Epicentre, WI). Purity was determined by use of a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and concentrations were determined with a Qubit (v3.0) fluorometer (Thermo Fisher Scientific).
Genomic DNA was sequenced on an Illumina MiSeq platform at the Michigan State University Research Technology Support Facility Genomics Core. Libraries were prepared with an Illumina TruSeq Nano DNA library preparation kit. After library validation and quantitation, the libraries were pooled and loaded on an Illumina MiSeq flow cell. Sequencing was performed in a 2-by-250-bp paired-end format with an Illumina 500 cycle V2 reagent cartridge. Base calling was performed by Illumina real-time analysis (v1.18.54), and the output of the real-time analysis was demultiplexed and converted to the FastQ format with Illumina Bcl2fastq (v1.8.4) software. Mutations were called by alignment to the E. coli W3110 reference sequence with NCBI accession number NC_007779.1 using the breseq computational pipeline (49).
Strain construction.
The E. coli W3110 strains were constructed by P1 transduction and by recombineering. P1 transduction was performed by standard methods (106). Strains carrying kanR resistance cassettes in the genes of interest were acquired from the Keio collection (105). Insertions were verified by PCR amplification of the interface between the kanR allele and the area surrounding the insertion.
The slp-gadX strain (JLS1732) was constructed using bacteriophage lambda Red recombineering according to the protocol described by Thomason et al. (81). Generation of the acid fitness island knockout strain JLS1732 was performed using bacteriophage λ recombineering by the protocol described by Thomason et al. (81). Overnight cultures of E. coli with the pSIM6::ampR plasmid were diluted 1 to 70 in LB (5 g/liter NaCl) and grown to mid-log-phase (OD600, between 0.4 and 0.6) in a shaker flask at 32°C. A 15-ml aliquot of this subculture was transferred to a fresh flask and shaken at 42°C for 15 min. Cells were made electrocompetent and electroporated with a DNA oligonucleotide. Cells were outgrown at 32°C for 3 to 5 h and plated onto selective media. For the construction of JLS1732 (Δslp-ΔgadX), a double-stranded DNA oligonucleotide containing cat-sacB (chloramphenicol resistance-sucrose sensitivity selection/counterselection marker) with 50 bp of homology to slp and 50 bp of homology to gadX was constructed using cat-sacB hybrid primers. Then, this region was replaced with the 70-bp oligonucleotide AAACAGTAATATGTTTATGTAATATTAAGTCAACTAATAGATATTTCTTTATAGTTTTCATCTGATTCTG to produce a strain with a clean break at the start of slp and the end of gadX.
Transcriptome analysis.
The transcriptomes of the evolved isolates in comparison with the transcriptome of the ancestor were obtained as described previously (23). For RNA extraction, bacteria were cultured to stationary phase in LBK buffered to pH 6.5 with 100 mM PIPES at 37°C. Cultures were diluted 1:50 into fresh medium supplemented with 5 mM potassium benzoate and grown to early log phase (determined by an OD600 of 0.4). At mid-log phase (OD600, between 0.4 and 0.6), the cultures were diluted 6:1 into 5% phenol-ethanol solution and pelleted. The pellet was resuspended in Tris-EDTA (TE) buffer (100 μl) with 3 mg/ml lysozyme, as described by He et al. (23). A Qiagen RNeasy minikit was used to further purify the RNA. An additional DNase treatment (DNase-Max; MoBio) was conducted.
Illumina transcriptome sequencing (RNA-Seq) libraries were constructed for sequencing. An enrichment of mRNA was achieved by depleting rRNA by following the guidelines of the Ribo-Zero rRNA removal kit (Illumina) (23, 107). The RNA-Seq library was prepared by use of a ScriptSeq (v2) RNA-Seq library preparation kit (Epicenter, WI) with a starting concentration of 15 ng rRNA-depleted RNA for each library. The resulting random-primed cDNA was purified with a MinElute PCR purification kit (Qiagen) before the 12-PCR-cycle amplification step using a FailSafe PCR enzyme kit (Epicenter, WI) and selected ScriptSeq index primers as reverse primers. An Agencourt AMPure XP system (BeckmanCoulter, NJ) purified the libraries and thereby size selected for >200 bp. Each library’s size and quality were assessed on an Agilent 2100 bioanalyzer and a high-sensitivity DNA chip (Agilent Technologies, Wilmington, DE) and quantified with the NEBNext library Quant kit protocol (New England BioLabs). A NextSeq 500/550 high-output kit (300 cycles) was used for sequencing using an Illumina NextSeq 500 sequencer.
The sequences were initially analyzed using CLC Genomics software (v6.0). Sequences with a quality score of less than 30 were discarded, the remaining sequences were trimmed, and sequences of less than 36 bp were discarded. Sequences were mapped to the E. coli W3110 genome (NCBI accession number NC_007779.1) using the following CLC genomics mapping parameters: mismatch, 1; insertion, 3; deletion, 3; length, 0.9; similarity, 0.95; autodetect paired distances on and map randomly. CLC RNA-Seq was performed using the following parameters: mismatch, 2; length fraction, 0.9; similarity fraction, 0.95; strand specific selected; maximum 3 hits, 3; paired settings, 36 to 500; broken pairs counting selected. Only unique counts generated for individual genes were used as the starting data for all subsequent analyses.
Differential expression analysis was performed using the R package DESeq2. The reported log2 fold changes represent the difference in expression of each gene in the evolved strains in 5 mM benzoate relative to its expression in the ancestor under the same condition. We also performed a control comparing expression by the ancestor in 5 mM benzoate to that by the ancestor without benzoate. A gene was said to be differentially expressed if it had a log2 fold change in expression of greater than 1 and a P value of <0.01.
Accession number(s).
The SRA accession number for genome sequences of the 900- and 1,400-generation benzoate-evolved strains is SRP199427. For RNA-Seq files, the SRA accession number is SRP161934.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by award MCB-1613278 from the National Science Foundation and by Summer Science funds from Kenyon College.
We thank Zack Blount and Lee Rosner for insightful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00966-19.
REFERENCES
- 1.Macfarlane GT, Macfarlane S. 2011. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol 45:S120–S127. doi: 10.1097/MCG.0b013e31822fecfe. [DOI] [PubMed] [Google Scholar]
- 2.Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez-Gonzalez F, Russell JB. 1999. Factors affecting the extreme acid resistance of Escherichia coli O157:H7. Food Microbiol 16:367–374. doi: 10.1006/fmic.1998.0249. [DOI] [Google Scholar]
- 4.Roe AJ, McLaggan D, Davidson I, O’Byrne C, Booth IR. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol 180:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terada H. 1990. Uncouplers of oxidative phosphorylation. Environ Health Perspect 87:213–218. doi: 10.1289/ehp.9087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis K, Naroditskaya V, Ferrante A, Fokina I. 1994. Bacterial resistance to uncouplers. J Bioenerg Biomembr 26:639–646. doi: 10.1007/BF00831539. [DOI] [PubMed] [Google Scholar]
- 7.Barker JL, Levitan H. 1975. Mitochondrial uncoupling agents—effects on membrane permeability of molluscan neurons. J Membr Biol 25:361–380. doi: 10.1007/BF01868584. [DOI] [PubMed] [Google Scholar]
- 8.Gutknecht J. 1990. Salicylates and proton transport through lipid bilayer membranes: a model for salicylate-induced uncoupling and swelling in mitochondria. J Membr Biol 115:253–260. doi: 10.1007/BF01868640. [DOI] [PubMed] [Google Scholar]
- 9.Doornbos RF, Van Loon LC, Bakker P. 2012. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev 32:227–243. doi: 10.1007/s13593-011-0028-y. [DOI] [Google Scholar]
- 10.Brul S, Coote P. 1999. Preservative agents in foods: mode of action and microbial resistance mechanisms. Int J Food Microbiol 50:1–17. doi: 10.1016/S0168-1605(99)00072-0. [DOI] [PubMed] [Google Scholar]
- 11.Beales N. 2004. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Comp Rev Food Sci Food Safety 3:1–20. doi: 10.1111/j.1541-4337.2004.tb00057.x. [DOI] [PubMed] [Google Scholar]
- 12.An C, Mou Z. 2011. Salicylic acid and its function in plant immunity. J Integr Plant Biol 53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 13.White RF. 1979. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99:410–412. doi: 10.1016/0042-6822(79)90019-9. [DOI] [PubMed] [Google Scholar]
- 14.Barbosa TM, Levy SB. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 182:3467–3474. doi: 10.1128/JB.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen SP, Levy SB, Foulds J, Rosner JL. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol 175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol 189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz C, McMurry LM, Levy SB. 2008. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J Bacteriol 190:1290–1297. doi: 10.1128/JB.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creamer KE, Ditmars FS, Basting PJ, Kunka KS, Hamdallah IN, Bush SP, Scott Z, He A, Penix SR, Gonzales AS, Eder EK, Camperchioli DW, Berndt A, Clark MW, Rouhier KA, Slonczewski JL. 2017. Benzoate- and salicylate-tolerant strains of Escherichia coli K-12 lose antibiotic resistance during laboratory evolution. Appl Environ Microbiol 83:e02736-16. doi: 10.1128/AEM.02736-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol 177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith JM, Basting PJ, Bischof KM, Wrona EP, Kunka KS, Tancredi AC, Moore JP, Hyman MRL, Slonczewski JL. 2019. Experimental evolution of Escherichia coli K-12 in the presence of proton motive force (PMF) uncoupler carbonyl cyanide m-chlorophenylhydrazone selects for mutations affecting PMF-driven drug efflux pumps. Appl Environ Microbiol 85:e02792-18. doi: 10.1128/AEM.02792-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dekel E, Alon U. 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 23.He A, Penix SR, Basting PJ, Griffith JM, Creamer KE, Camperchioli D, Clark MW, Gonzales AS, Erazo JSC, George NS, Bhagwat AA, Slonczewski JL. 2017. Acid evolution of Escherichia coli K-12 eliminates amino acid decarboxylases and reregulates catabolism. Appl Environ Microbiol 83:e00442-17. doi: 10.1128/AEM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harden MM, He A, Creamer K, Clark MW, Hamdallah I, Martinez KA, Kresslein RL, Bush SP, Slonczewski JL. 2015. Acid-adapted strains of Escherichia coli K-12 obtained by experimental evolution. Appl Environ Microbiol 81:1932–1941. doi: 10.1128/AEM.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eames M, Kortemme T. 2012. Cost-benefit tradeoffs in engineered lac operons. Science 336:911–915. doi: 10.1126/science.1219083. [DOI] [PubMed] [Google Scholar]
- 26.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Martínez L, Conejo MC, Pascual A, Hernández-Allés S, Ballesta S, Ramírez De Arellano-Ramos E, Benedí VJ, Perea EJ. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal β-lactamase and showing altered porin profiles. Antimicrob Agents Chemother 44:2534–2536. doi: 10.1128/aac.44.9.2534-2536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziervogel BK, Roux B. 2013. The binding of antibiotics in OmpF porin. Structure 21:76–87. doi: 10.1016/j.str.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tramonti A, De Canio M, De Biase D. 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol 70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz C, Levy SB. 2010. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob Agents Chemother 54:2125–2134. doi: 10.1128/AAC.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Z, Masuda N, Foster JW. 2004. Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J Bacteriol 186:7378–7389. doi: 10.1128/JB.186.21.7378-7389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 33.Hirakawa H, Inazumi Y, Senda Y, Kobayashi A, Hirata T, Nishino K, Yamaguchi A. 2006. N-Acetyl-d-glucosamine induces the expression of multidrug exporter genes, mdtEF, via catabolite activation in Escherichia coli. J Bacteriol 188:5851–5858. doi: 10.1128/JB.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishino K, Senda Y, Yamaguchi A, Nishino K, Yamaguchi A, Nishino K, Yamaguchi A. 2008. The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J Infect Chemother 14:23–29. doi: 10.1007/s10156-007-0575-Y. [DOI] [PubMed] [Google Scholar]
- 35.Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, Conway T. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J Bacteriol 185:3190–3201. doi: 10.1128/jb.185.10.3190-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterman SR, Small P. 2003. Transcriptional expression of Escherichia coli glutamate-dependent acid resistance genes gadA and gadBC in an hns rpoS mutant. J Bacteriol 185:4644–4647. doi: 10.1128/JB.185.15.4644-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borges-Walmsley MI, Beauchamp J, Kelly SM, Jumel K, Candlish D, Harding SE, Price NC, Walmsley AR. 2003. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J Biol Chem 278:12903–12912. doi: 10.1074/jbc.M209457200. [DOI] [PubMed] [Google Scholar]
- 38.Tanabe M, Szakonyi G, Brown KA, Henderson PJF, Nield J, Byrne B. 2009. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem Biophys Res Commun 380:338–342. doi: 10.1016/j.bbrc.2009.01.081. [DOI] [PubMed] [Google Scholar]
- 39.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol 184:4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HS, Nagore D, Nikaido H. 2010. Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J Bacteriol 192:1377–1386. doi: 10.1128/JB.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanabe H, Yamasak K, Furue M, Yamamoto K, Katoh A, Yamamoto M, Yoshioka S, Tagami H, Aiba HA, Utsumi R. 1997. Growth phase-dependent transcription of emrKY, a homolog of multidrug efflux emrAB genes of Escherichia coli, is induced by tetracycline. J Gen Appl Microbiol 43:257–263. doi: 10.2323/jgam.43.257. [DOI] [PubMed] [Google Scholar]
- 42.Weatherspoon-Griffin N, Yang D, Kong W, Hua Z, Shi Y. 2014. The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J Biol Chem 289:32571–32582. doi: 10.1074/jbc.M114.565762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J, Page R, García-Contreras R, Palermino JM, Zhang XS, Doshi O, Wood TK, Peti W. 2007. Structure and function of the Escherichia coli protein YmgB: a protein critical for biofilm formation and acid-resistance. J Mol Biol 373:11–26. doi: 10.1016/j.jmb.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng Z, Shan Y, Pan Q, Gao X, Yan A. 2013. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front Microbiol 4:194. doi: 10.3389/fmicb.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duval V, Lister IM. 2013. MarA, SoxS and Rob of Escherichia coli—global regulators of multidrug resistance, virulence and stress response. Int J Biotechnol Wellness Ind 2:101–124. doi: 10.6000/1927-3037.2013.02.03.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma P, Haycocks JRJ, Middlemiss AD, Kettles RA, Sellars LE, Ricci V, Piddock LJV, Grainger DC. 2017. The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity. Nat Commun 8:1444. doi: 10.1038/s41467-017-01405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhaskarla C, Das M, Verma T, Kumar A, Mahadevan S, Nandi D. 2016. Roles of Lon protease and its substrate MarA during sodium salicylate-mediated growth reduction and antibiotic resistance in Escherichia coli. Microbiology 162:764–776. doi: 10.1099/mic.0.000271. [DOI] [PubMed] [Google Scholar]
- 48.Cohen SP, McMurry LM, Levy SB. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol 170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev 23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Xiao M, Horiyama T, Zhang Y, Li X, Nishino K, Yan A. 2011. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J Biol Chem 286:26576–26584. doi: 10.1074/jbc.M111.243261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohnert JA, Schuster S, Fähnrich E, Trittler R, Kern WV. 2007. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J Antimicrob Chemother 59:1216–1222. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 53.Sah S, Aluri S, Rex K, Varshney U. 2015. One-carbon metabolic pathway rewiring in Escherichia coli reveals an evolutionary advantage of 10-formyltetrahydrofolate synthetase (Fhs) in survival under hypoxia. J Bacteriol 197:717–726. doi: 10.1128/JB.02365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hershey HV, Taylor MW. 1986. Nucleotide sequence and deduced amino acid sequence of Escherichia coli adenine phosphoribosyl-transferase and comparison with other analogous enzymes. Gene 43:287–293. doi: 10.1016/0378-1119(86)90218-0. [DOI] [PubMed] [Google Scholar]
- 55.Jossek R, Bongaerts J, Sprenger GA. 2001. Characterization of a new feedback-resistant 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli. FEMS Microbiol Lett 202:145–148. doi: 10.1111/j.1574-6968.2001.tb10795.x. [DOI] [PubMed] [Google Scholar]
- 56.Lohkamp B, McDermott G, Campbell SA, Coggins JR, Lapthorn AJ. 2004. The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition. J Mol Biol 336:131–144. doi: 10.1016/j.jmb.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 57.Chang ZY, Nygaard P, Chinault AC, Kellems RE. 1991. Deduced amino acid sequence of Escherichia coli adenosine deaminase reveals evolutionarily conserved amino acid residues: implications for catalytic function. Biochemistry 30:2273–2280. doi: 10.1021/bi00222a033. [DOI] [PubMed] [Google Scholar]
- 58.Koellner G, Bzowska A, Wielgus-Kutrowska B, Luić M, Steiner T, Saenger W, Stȩpiński J. 2002. Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism. J Mol Biol 315:351–371. doi: 10.1006/jmbi.2001.5211. [DOI] [PubMed] [Google Scholar]
- 59.Petersen C, Møller LB. 2001. The RihA, RihB, and RihC ribonucleoside hydrolases of Escherichia coli. J Biol Chem 276:884–894. doi: 10.1074/jbc.M008300200. [DOI] [PubMed] [Google Scholar]
- 60.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A 107:14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie H, Patching SG, Gallagher MP, Litherland GJ, Brough AR, Venter H, Yao SYM, Ng AM, Young JD, Herbert RB, Henderson PJ, Baldwin SA. 2004. Purification and properties of the Escherichia coli nucleoside transporter NupG, a paradigm for a major facilitator transporter sub-family. Mol Membr Biol 21:323–336. doi: 10.1080/09687860400003941. [DOI] [PubMed] [Google Scholar]
- 62.Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol 191:5283–5292. doi: 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chubiz LM, Rao CV. 2010. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J Bacteriol 192:4786–4789. doi: 10.1128/JB.00371-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sayed AK, Foster JW. 2009. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol Microbiol 71:1435–1450. doi: 10.1111/j.1365-2958.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 65.Yildiz Ö, Vinothkumar KR, Goswami P, Kühlbrandt W. 2006. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J 25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Cock H, Struyvé M, Kleerebezem M, Van Der Krift T, Tommassen J. 1997. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J Mol Biol 269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 67.Sardesai AA, Genevaux P, Schwager F, Ang D, Georgopoulos C. 2003. The OmpL porin does not modulate redox potential in the periplasmic space of Escherichia coli. EMBO J 22:1461–1466. doi: 10.1093/emboj/cdg152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abouhamad WN, Manson MD. 1994. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol Microbiol 14:1077–1092. doi: 10.1111/j.1365-2958.1994.tb01340.x. [DOI] [PubMed] [Google Scholar]
- 69.Klepsch MM, Kovermann M, Löw C, Balbach J, Permentier HP, Fusetti F, de Gier JW, Slotboom DJ, Berntsson R. 2011. Escherichia coli peptide binding protein OppA has a preference for positively charged peptides. J Mol Biol 414:75–85. doi: 10.1016/j.jmb.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 70.Dassa J, Fsihi H, Marck C, Dion M, Kieffer-Bontemps M, Boquet PL. 1991. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol Gen Genet 229:341–352. doi: 10.1007/BF00267454. [DOI] [PubMed] [Google Scholar]
- 71.Brøndsted L, Atlung T. 1996. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol 178:1556–1564. doi: 10.1128/jb.178.6.1556-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka N. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Mol Biol 10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 73.Trchounian A, Sawers RG. 2014. Novel insights into the bioenergetics of mixed-acid fermentation: can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life 66:1–7. doi: 10.1002/iub.1236. [DOI] [PubMed] [Google Scholar]
- 74.Sauter M, Böhm R, Böck A. 1992. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol 6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 75.Burstein C, Tiankova L, Kepes A. 1979. Respiratory control in Escherichia coli K 12. Eur J Biochem 94:387–392. doi: 10.1111/j.1432-1033.1979.tb12905.x. [DOI] [PubMed] [Google Scholar]
- 76.Rossmann R, Sawers G, Böck A. 1991. Mechanism of regulation of the formate‐hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol 5:2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x. [DOI] [PubMed] [Google Scholar]
- 77.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. 2014. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci 111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noguchi K, Riggins DP, Eldahan KC, Kitko RD, Slonczewski JL. 2010. Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli. PLoS One 5:e10132. doi: 10.1371/journal.pone.0010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berg BL, Li J, Heider J, Stewart V. 1991. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. J Biol Chem 266:22380–22385. [PubMed] [Google Scholar]
- 80.Stewart V. 1993. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol 9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 81.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 106:1.16.1–1.16.39. doi: 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- 82.Giffard PM, Booth IR. 1988. The rpoA341 allele of Escherichia coli specifically impairs the transcription of a group of positively-regulated operons. Mol Gen Genet 214:148–152. doi: 10.1007/BF00340193. [DOI] [PubMed] [Google Scholar]
- 83.Thomas MS, Glass RE. 1991. Escherichia coli rpoA mutation which impairs transcription of positively regulated systems. Mol Microbiol 5:2719–2725. doi: 10.1111/j.1365-2958.1991.tb01980.x. [DOI] [PubMed] [Google Scholar]
- 84.Rath D, Jawali N. 2006. Loss of expression of cspC, a cold shock family gene, confers a gain of fitness in Escherichia coli K-12 strains. J Bacteriol 188:6780–6785. doi: 10.1128/JB.00471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phadtare S, Inouye M. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J Bacteriol 183:1205–1214. doi: 10.1128/JB.183.4.1205-1214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phadtare S, Tadigotla V, Shin W, Sengupta A, Severinov K. 2006. Analysis of Escherichia coli global gene expression profiles in response to overexpression and deletion of CspC and CspE. J Bacteriol 188:2521–2527. doi: 10.1128/JB.188.7.2521-2527.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen-Or I, Shenhar Y, Biran D, Ron EZ. 2010. CspC regulates rpoS transcript levels and complements hfq deletions. Res Microbiol 161:694–700. doi: 10.1016/j.resmic.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 88.Sun Y, Fukamachi T, Saito H, Kobayashi H. 2012. Adenosine deamination increases the survival under acidic conditions in Escherichia coli. J Appl Microbiol 112:775–781. doi: 10.1111/j.1365-2672.2012.05246.x. [DOI] [PubMed] [Google Scholar]
- 89.Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM. 2011. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA 17:489–500. doi: 10.1261/rna.2452111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cech GM, Szalewska-Pałasz A, Kubiak K, Malabirade A, Grange W, Arluison V, Węgrzyn G. 2016. The Escherichia coli Hfq protein: an unattended DNA-transactions regulator. Front Mol Biosci 3:36. doi: 10.3389/fmolb.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother 54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dersch P, Khan MA, Mühlen S, Görke B. 2017. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front Microbiol 8:803. doi: 10.3389/fmicb.2017.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J, Hroudova M, Vlcek C, Cardenas E, Mackova M, Macek T. 2012. Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS One 7:e40653. doi: 10.1371/journal.pone.0040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davidson PM, Taylor TM, Schmidt SE. 2013. Chemical preservatives and natural antimicrobial compounds, chapter 30, p 765–801. In Doyle M, Buchanan R (ed), Food microbiology—fundamentals and frontiers, 4th ed ASM Press, Washington, DC. doi: 10.1128/9781555818463.ch30. [DOI] [Google Scholar]
- 95.Kashiwagi K, Tsuhako MH, Sakata K, Saisho T, Igarashi A, Pinto Da Costa SO, Igarashi K. 1998. Relationship between spontaneous aminoglycoside resistance in Escherichia coli and a decrease in oligopeptide binding protein. J Bacteriol 180:5484–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Acosta MBR, Ferreira RCC, Padilla G, Ferreira LCS, Costa S. 2000. Altered expression of oligopeptide-binding protein (OppA) and aminoglycoside resistance in laboratory and clinical Escherichia coli strains. J Med Microbiol 49:409–413. doi: 10.1099/0022-1317-49-5-409. [DOI] [PubMed] [Google Scholar]
- 97.Nakamatsu EH, Fujihira E, Ferreira RCC, Balan A, Costa SOP, Ferreira LCS. 2007. Oligopeptide uptake and aminoglycoside resistance in Escherichia coli K12. FEMS Microbiol Lett 269:229–233. doi: 10.1111/j.1574-6968.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 98.Hakobyan M, Sargsyan H, Bagramyan K. 2005. Proton translocation coupled to formate oxidation in anaerobically grown fermenting Escherichia coli. Biophys Chem 115:55–61. doi: 10.1016/j.bpc.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 99.Lobritz MA, Belenky P, Porter CBM, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ. 2015. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci U S A 112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42°C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol 31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nishino K, Yamaguchi A. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J Bacteriol 184:2319–2323. doi: 10.1128/jb.184.8.2319-2323.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeon YH, Yamazaki T, Otomo T, Ishihama A, Kyogoku Y. 1997. Flexible linker in the RNA polymerase alpha subunit facilitates the independent motion of the C-terminal activator contact domain. J Mol Biol 267:953–962. doi: 10.1006/jmbi.1997.0902. [DOI] [PubMed] [Google Scholar]
- 103.Dangi B, Gronenborn AM, Rosner JL, Martin RG. 2004. Versatility of the carboxy-terminal domain of the α subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA. Mol Microbiol 54:45–59. doi: 10.1111/j.1365-2958.2004.04250.x. [DOI] [PubMed] [Google Scholar]
- 104.Yethon JA, Heinrichs DE, Monteiro MA, Perry MB, Whitfield C. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem 273:26310–26316. doi: 10.1074/jbc.273.41.26310. [DOI] [PubMed] [Google Scholar]
- 105.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deininger KNW, Horikawa A, Kitko RD, Tatsumi R, Rosner JL, Wachi M, Slonczewski JL. 2011. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS One 6:e18960. doi: 10.1371/journal.pone.0018960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhagwat AA, Ying ZI, Smith A. 2014. Evaluation of ribosomal RNA removal protocols for Salmonella RNA-Seq projects. Adv Microbiol 4:25–32. doi: 10.4236/aim.2014.41006. [DOI] [Google Scholar]
- 108.Smith MW, Neidhardt FC. 1983. Proteins induced by aerobiosis in Escherichia coli. J Bacteriol 154:344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido N. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 110.Johnson MD, Bell J, Clarke K, Chandler R, Pathak P, Xia Y, Marshall RL, Weinstock GM, Loman NJ, Winn PJ, Lund PA. 2014. Characterization of mutations in the PAS domain of the EvgS sensor kinase selected by laboratory evolution for acid resistance in Escherichia coli. Mol Microbiol 93:911–927. doi: 10.1111/mmi.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seo SW, Kim D, O'Brien EJ, Szubin R, Palsson BO. 2015. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat Commun 6:7970. doi: 10.1038/ncomms8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.