Abstract
(1) Background: The aim of the study was to evaluate the prognostic significance of EMT-associated (TWIST1) and stem-cell (SC) transcript (CD24, CD44, ALDH1) quantification in EpCAM+ circulating tumor cells (CTCs) of early breast cancer patients. (2) Methods: 100 early stage breast cancer patients and 19 healthy donors were enrolled in the study. CD24, CD44, and ALDH1 transcripts of EpCAM+ cells were quantified using a novel highly sensitive and specific quadraplex RT-qPCR, while TWIST1 transcripts were quantified by single RT-qPCR. All patients were followed up for more than 5 years. (3) Results: A significant positive correlation between overexpression of TWIST1 and CD24−/low/CD44high profile was found. Kaplan–Meier analysis revealed that the ER/PR-negative (HR-) patients and those patients with more than 3 positive lymph nodes that overexpressed TWIST1 in EpCAM+ cells had a significant lower DFI (log rank test; p < 0.001, p < 0.001) and OS (log rank test; p = 0.006, p < 0.001). Univariate and multivariate analysis also revealed the prognostic value of TWIST1 overexpression and CD24−/low/CD44high and CD24−/low/ALDH1high profile for both DFI and OS. (4) Conclusions: Detection of TWIST1 overexpression and stem-cell (CD24, CD44, ALDH1) transcripts in EpCAM+ CTCs provides prognostic information in early stage breast cancer patients.
Keywords: liquid biopsy, circulating tumor cells, epithelial–mesenchymal transition, stem cells, early breast cancer
1. Introduction
Circulating tumor cells (CTCs) are major players in liquid biopsy [1,2], and their molecular characterization is highly important for rational treatment decisions and for monitoring therapeutic response [3], whereas their analysis at the single cell level has the potential to reveal tumor heterogeneity in real time [4]. In breast cancer, a subpopulation of tumor cells that display stem cell-like properties [5] determines the aggressive characteristics and drug resistance of tumor clonal evolution [6]. Cancer stem cells (CSCs) that mediate tumor metastasis and therapeutic resistance have the capacity to transition between mesenchymal and epithelial-like states [7]. It has already been shown that breast cancer cells with the CD44+CD24−/low phenotype [8] that overexpress aldehyde dehydrogenase 1 (ALDH1+) [9] are able to form tumors in mice with high tumorigenic capacity. It has also been shown that disseminated tumor cells (DTCs) [10] and CTCs express the putative stem cell CD44+/CD24− and/or ALDH1+/CD24− phenotypical profile [11,12]. Moreover, in primary human luminal breast cancer, the metastasis-initiating cells containing CTC that express EPCAM, CD44, CD47, and the proto-oncogene MET are related with reduced overall survival (OS) [13]. In other types of cancer, various stem cell markers have also been identified and correlated with metastatic capacity [14] and poor prognosis [15].
It is now known that breast cancer stem cells exist in distinct mesenchymal-like (epithelial–mesenchymal transition [EMT]) as CD44+/CD24− and epithelial-like (mesenchymal-epithelial transition [MET]) states that express ALDH1. This transition between EMT- and MET-like states is highly important for their capacity to invade, disseminate, and grow at metastatic sites [16]. Many studies have already shown that a major proportion of CTC express both EMT and tumor stem cell characteristics [17,18,19]. Recently it was shown that an EpCAM-/ALDH1+/HER2+/EGFR+/HPSE+/Notch1+ profile in CTC drives these cells to metastasize to the brain [20]. At the single cell level, it has been shown that CTC that co-express the stem cell marker ALDH1 and the mesenchymal marker TWIST1 may prevail during disease progression [21]. However, the prognostic significance of EMT and Stem cell (SC) markers in CTC has only been shown up to now in metastatic colorectal cancer [22] and metastatic breast cancer [23].
In early breast cancer, the molecular detection of cytokeratin 19 (CK-19) mRNA-positive cells in peripheral blood before [24], during [25], and after adjuvant therapy [26] is associated with worse prognosis, while their elimination seems to be an efficacy indicator of treatment [27]. The prognostic significance of CTC count using the CellSearch system in neoadjuvant [28] and adjuvant early breast cancer patients [29] has been also shown. Moreover, the administration of “secondary” adjuvant trastuzumab in patients with HER2(−) breast cancer can eliminate chemotherapy-resistant CK19 mRNA-positive CTCs [30], in contrast to the Treat CTC phase II trial that failed to prove the efficacy of trastuzumab in the detection rate of CTC [31]. However, in early breast cancer stages the early detection of recurrence remains a big challenge [32], and until now, there are not solid data proving the prognostic significance of EMT/SC(+) cells. The aim of the current study was to evaluate the prognostic significance of TWIST1, CD24, CD44, and ALDH1 mRNA quantification in EpCAM-positive circulating tumor cells from early stage breast cancer patients with a long follow-up.
2. Materials and Methods
2.1. Cell Lines
The human mammary carcinoma cell line SKBR-3 was used as a positive control for the development of the quadraplex RT-qPCR assay for CD24, CD44, ALDH1, HPRT, while MDA-MB-231 cancer cell line was used as a positive control for the expression of TWIST1 [33]. Cells were counted in a hemocytometer and their viability was assessed by trypan blue dye exclusion. cDNAs of all cancer cell lines were kept in aliquots at −20 °C and used for the analytical validation of the assay, prior to the analysis of patient’s samples.
2.2. Patients
In total, 100 patients with non-metastatic breast cancer from the Medical Oncology Unit “Elena Venizelou” Hospital and IASO General hospital were enrolled in the study from September 2007 until January 2013. Peripheral blood (20 mL) was obtained from all these patients two weeks after the removal of the primary tumor and before the initiation of adjuvant chemotherapy. The chemotherapeutic adjuvant treatment for these patients has been previously reported [34]. The clinical characteristics for these patients at the time of diagnosis are shown in Supplementary Table S1. All patients signed an informed consent to participate in the study, which was approved by the Ethics and Scientific Committees of our Institutions. Peripheral blood (20 mL) was obtained from 19 healthy female blood donors (HD) and was analyzed in the same way as patients’ samples (control group).
2.3. Isolation of EpCAM+ CTCs
To reduce blood contamination by epithelial cells from the skin, the first 5 mL of blood were discarded, and the blood collection tube was at the end disconnected before withdrawing the needle. Peripheral blood (20 mL in EDTA) from (HD) and patients was collected and processed within 3 h in exactly the same manner. After collection, peripheral blood was diluted with 20 mL phosphate buffered saline (PBS, pH 7.3), and peripheral blood mononuclear cells (PBMCs) were isolated by gradient density centrifugation using Ficol-Paque TM PLUS (GE Healthcare, Bio-Sciences AB) at 670 g for 30 min at room temperature. The interface cells were removed and washed twice with 40 mL of sterile PBS (pH 7.3, 4 °C), at 530 g for 10 min. EpCAM+ cells were enriched using immunomagnetic Ber-EP4 coated capture beads (Dynabeads® Epithelial Enrich, Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions [33].
2.4. RNA Extraction-cDNA Synthesis
Total RNA isolation was performed using TRIZOL-LS (ThermoFischer, Carlsbad, CA, USA). All RNA preparation and handling steps took place in a laminar flow hood under RNAse-free conditions. The isolated RNA from each fraction was dissolved in 20 μL of RNA storage buffer (Ambion, ThermoFischer, USA) and stored at −70 °C until use. RNA concentration was determined by absorbance readings at 260 nm using the Nanodrop-1000 spectrophotometer (NanoDrop, Technologies, Wilmington, DE, USA). mRNA was isolated from the total RNA using the Dynabeads mRNA Purification kit (ThermoFischer, USA), according to the manufacturer’s instructions. cDNA synthesis was performed using the High capacity RNA-to-cDNA kit (ThermoFischer, USA) in a total volume of 20 μL, according to the manufacturer’s instructions.
2.5. RT-qPCR
A novel quadraplex RT-qPCR assay was first developed for CD24, CD44, ALDH1, and HPRT (reference gene). Primers and dual hybridization probes were de novo in-silico designed, using Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA). The specificity of all primer and hybridization probe sequences was first tested by homology searches in the nucleotide database (NCBI, nucleotide BLAST). Cross reaction between all oligonucleotide sequences was also examined. Each probe set included a 3′-fluorescein (F) donor probe and a 5′-LC acceptor probe that was different for each gene set: CD24 (610 nm), CD44 (640 nm), ALDH1 (670 nm) and HPRT (705 nm). A color compensation test was performed by using pure dye spectra so that spectral overlap between dyes was corrected [35]. Quadraplex RT-qPCR reactions were performed in the LightCycler 2.0 (Roche, Mannheim, Germany). Component concentrations and the cycling conditions for the quadraplex RT-qPCR assay were optimized in detail. The amplification reaction mixture (10 μL) contained 1 μL of the PCR Synthesis Buffer (5Χ), 2.4 μL of MgCl2 (25 mM), 0.2 μL dNTPs (10 mM), 0.8 μL BSA (10 μg/μL), 0.1 μL Hot Start DNA polymerase (HotStart, 5 U/μL, Promega, Madison, WI, USA), 0.5 μL of a mixture containing all eight primers (10 μΜ), 0.5 μL of a mixture containing all eight dual hybridization probes (3 μM), and H2O (added to the final volume). Cycling conditions of the CD24, CD44, ALDH1, HPRT quadraplex RT-qPCR assay were: 95 °C/2 min; 45 cycles of 95 °C/20 s, annealing at 59 °C/20 s, and extension at 72 °C/20 s. For the development and analytical validation of the novel quadraplex RT-qPCR assay, we generated individual PCR amplicons corresponding to the gene-targets studied that served as quantification calibrators, as we have previously described [33]. RT-qPCR for TWIST1 was performed as previously described [33,36]. All data were evaluated in respect to TWIST1, CD24, CD44, and ALDH1 expression by normalizing the EpCAM+ fraction of PBMCs to the expression of HPRT and the 2–ΔΔCt approach, as described in detail by Livak and Schmittgen [37]. A cut-off value was calculated as the mean of signals derived by samples of healthy individual analyzed in exactly the same way plus 2SD for TWIST1, CD44, and ALDH1 transcripts and as the mean of signals derived by samples of healthy individual minus 2SD for CD24.
2.6. Statistical Analysis
Statistical analysis was performed using SPSS (SPSS Statistics 25.0, company, Armonk, NY, USA). The chi-square test of independence or Fisher exact test (SPSS, version 25.0) was used to make comparisons between groups. The DFI and OS rate were calculated by the Kaplan–Meier method and were evaluated by the log-rank test. Cox proportional hazards (PH) models were used to evaluate the relationship between EMT and Stem Cell status and event-time distributions, with tumor size, grade, number of involved lymph nodes, ER, PR, HER2, and age. Parametric and non-parametric tests were used to compare continuous variables between groups. All P-values are two-sided. A level of p < 0.05 is considered statistically significant.
3. Results
3.1. Analytical Validation of the Quadraplex RT-qPCR Assay for CD24, CD44, ALDH1, HPRT
The analytical specificity of the developed assay was checked by using all oligonucleotides in a common master mix in four different reactions in the presence of one individual gene target each time. Each primer pair and dual hybridization probe pair amplifies specifically only the corresponding target sequence and is detected only in the corresponding wavelength (Supplementary Figure S1A). The analytical sensitivity was determined for each individual gene target using a calibration curve. These calibration curves were generated using serial dilutions of individual gene-specific external standards in triplicate for each concentration, ranging from 105 copies/μL to 10 copies/μL. The analytical detection limit corresponded to 3 copies/μL while the quantification limit was equal to 9 copies/μL (Supplementary Figure S1B). The developed assay showed linearity over the entire quantification range and correlation coefficients greater than 0.99 in all cases, indicating a precise log-linear relationship. Intra and inter-assay variance: Repeatability or intra–assay variance of the quadraplex RT-qPCR was evaluated by repeatedly analyzing four cDNA samples corresponding to 1, 10, 100, and 1000 SKBR-3 cells in the same assay, in three parallel determinations. Reproducibility or interassay variance was evaluated by analyzing the same cDNA sample, representing 1000 SKBR-3 cells on five separate assays performed in five different days (Supplementary Table S2).
3.2. Quantification of CD24, CD44, ALDH1, and TWIST1 mRNA in the EpCAM(+) Fraction in Early Stage BrCa Patients and (HD)
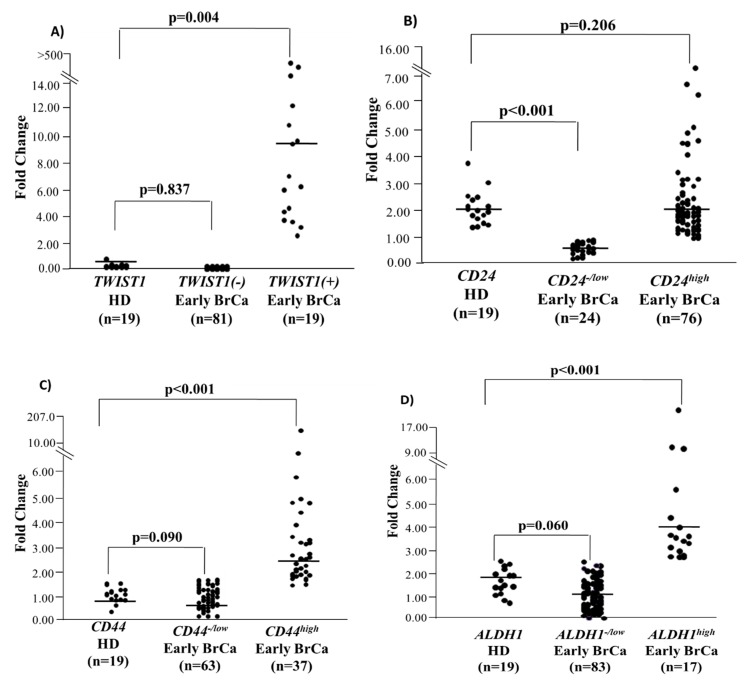
In all EpCAM(+) fractions isolated from 100 early BrCa patient samples and 19 HD CD24, CD44, ALDH1, HPRT transcripts were quantified by the developed quadraplex RT-qPCR and TWIST1 transcripts by the singleplex RT-qPCR assay (Figure 1). Median fold change of TWIST1 expression in the EpCAM(+) fraction was 0.42 (range: 0–0.95) in HD and 10.06 (range: 2.33–3327) in TWIST1high (Mann-Whitney test, Ζ = −1.363, p = 0.001) and 0 (range: 0–0) in TWIST1low/− early BrCa patient samples (Mann-Whitney test, Ζ = −3.634, p < 0.001) (Figure 1A). Median fold change of CD24 expression in the EpCAM(+) fraction was 2.00 (range: 1.42–3.81) in HD and 1.91 (range: 0.91–15.14) in CD24high (Mann-Whitney test, Ζ = −0.492, p = 0.623) and 0.62 (range: 0.29–0.88) in CD24low early BrCa patients (Mann-Whitney test, Ζ = −5.577, p < 0.001) (Figure 1B). Median fold change of CD44 expression in the EpCAM(+) fraction was 0.71 (range: 0.14–1.06) in HD and 2.33 (range: 1.28–202.75) in CD44high (Mann-Whitney test, Ζ = −6.084, p < 0.001) and 0.61 (range: 0.01–1.17) in CD44low early BrCa patients (Mann-Whitney test, Ζ = −1.084, p = 0.278) (Figure 1C). Median fold change of ALDH1 expression in the EpCAM(+) fraction was 1.32 (range: 0.69–2.19) in HD and 2.97(range: 2.30–14.72) in ALDH1high (Mann-Whitney test, Ζ = −5.119, p < 0.001) and 0.84 (range: 0.06–2.16) in ALDH1low early BrCa patients (Mann-Whitney test, Ζ = −2.190, p = 0.029) (Figure 1D).
Figure 1.
Relative fold change values (2–ΔΔCt) in respect to HPRT expression for: (A) TWIST1 (B) CD24, (C) CD44, (D) ALDH1 for early breast cancer patients (n = 100) and (HD), (n = 19).
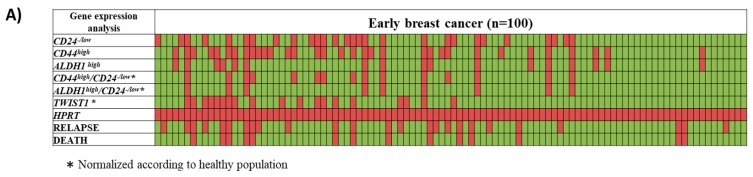
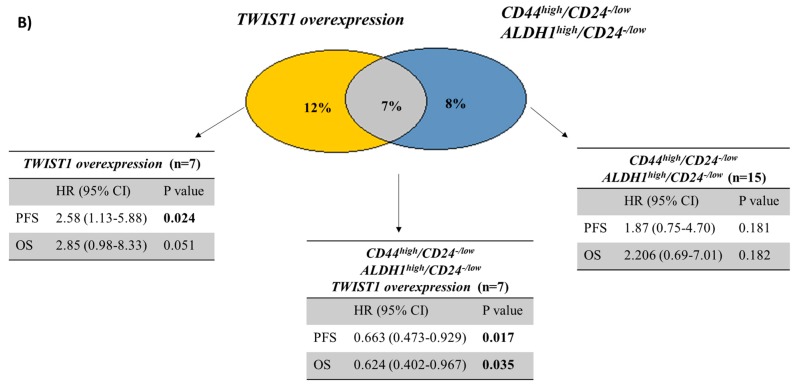
In 19/100(19%) breast cancer samples tested, TWIST1 was overexpressed, while in 15/100(15%) samples the CD24−/low/CD44high profile, and in 9/100(9%) the CD24−/low/ALDH1high profile was detected (Figure 2A). There was a positive correlation between TWIST1 mRNA overexpression and the CD24−/low/CD44high profile (Fisher’s Exact Test; p = 0.008), while there was no correlation between TWIST1 mRNA overexpression and the CD24−/low/ALDH1high profile (Fisher’s Exact Test; p = 0.366) (Table 1). TWIST1 overexpression and CD24−/low/CD44high and/or CD24−/low/ALDH1high were detected in 7/100(7%) EpCAM(+) samples. The correlation between these characteristics and the clinical variables of the patients revealed an association between TWIST1 overexpression with lymph node status (chi-square; p = 0.036) and HER2 status of the primary tumor (chi-square; p = 0.006) (Supplementary Table S1).
Figure 2.
(A) Heat map of TWIST1, CD24, CD44, and ALDH1-mRNA quantification in the EpCAM+ CTC fraction from early stage breast cancer patients (n = 100). Red color represents overexpression, while green color indicates underexpression or lack of expression. Concerning the relapse or death, red color represents the relapse or death, while green color indicates no relapse or alive status. (B) Univariate Cox-regression hazard models for TWIST1 overexpression, CD44high/CD24−/low, and ALDH1high/CD24−/low and the co-expression of the mesenchymal profile, TWIST1, and the stem cell profile, CD44high/CD24−/low, and ALDH1high/CD24−/low.
Table 1.
Correlation between TWIST1 and CD44high/CD24−/low and ALDH1high/CD24−/low expression in early breast cancer EpCAM positive samples (n = 100).
| CD44high/CD24−/low | p a | ALDH1high/CD24−/low | p a | |||
|---|---|---|---|---|---|---|
| TWIST1 | Positive | Negative | 0.008 | Positive | Negative | 0.366 |
| Positive | 7 (46.7%) | 12(14.1%) | 3 (33.3) | 16(17.6%) | ||
| Negative | 8 (53.3%) | 73(85.9%) | 6 (66.7%) | 75 (82.4%) | ||
| Concordance | 80/100 (80%) | 78/100 (78%) | ||||
a Fischer’s Exact Test. Bold: highlights the significance of the test.
3.3. Evaluation of Prognostic Significance
3.3.1. Disease Free Interval
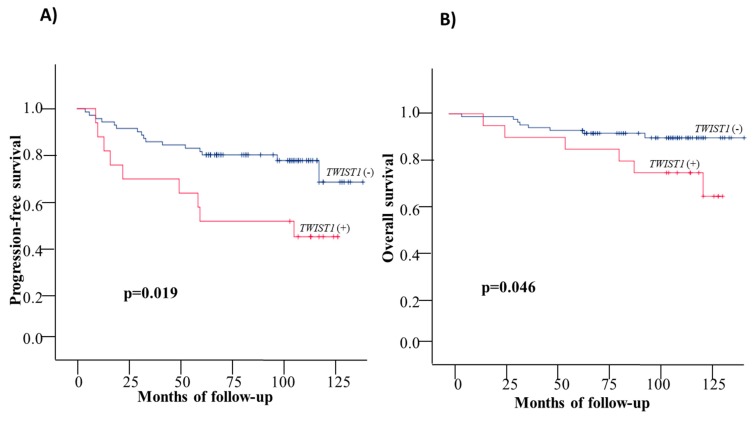
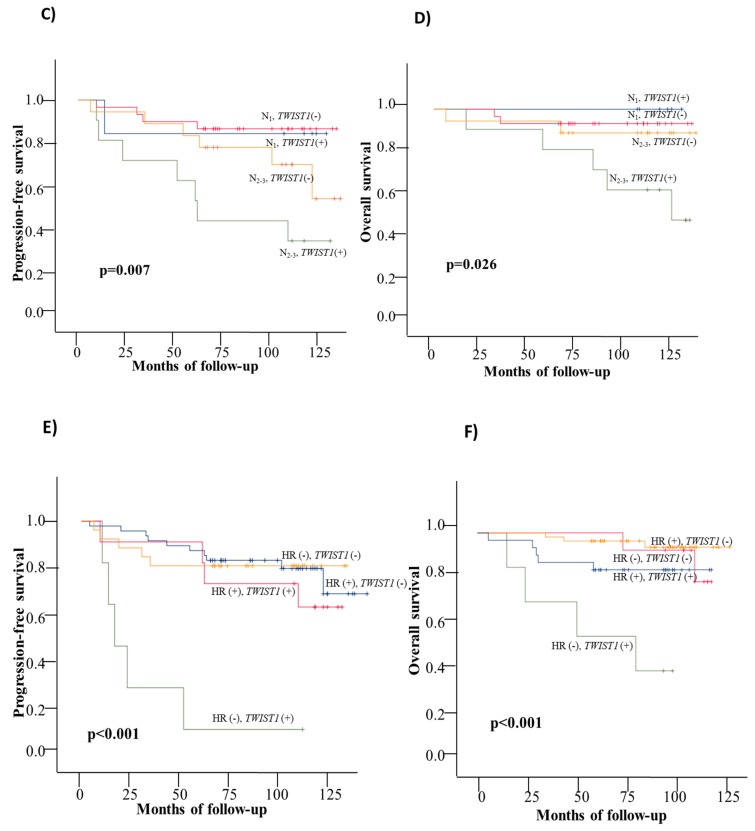
During the follow up period (median: 95 months; range: 4–137 months), 25/100 (25%) patients relapsed and in 9/25 (36%) of them TWIST1 overexpression was detected in the EpCAM+ CTC fraction (Fisher’s Exact Test; p = 0.019). Similarly, 6/25 (24%) patients displayed a Stem Cell profile in EpCAM+ CTC fraction (Fisher’s Exact Test; p = 0.194). In 4/25 (16%) of these patients, both TWIST1 overexpression and the Stem Cell profile was detected (Fisher’s Exact Test; p = 0.063) (Supplementary Table S3). The Kaplan–Meier estimates of the cumulative DFI of the patients overexpressing TWIST1 revealed that these patients had worse survival compared to patients who were negative (83.6mo vs 115.8mo respectively; p = 0.019) (Table 2, Figure 3A). However, the stem cell profile alone (86.7mo. vs 113.2mo, respectively in the two groups; log rank test; p = 0.174) (Table 2, Supplementary Figure S2A) and both stem cell and mesenchymal characteristics (68.9mo vs 88.8–115.8mo, respectively; p = 0.087) (Table 2, Supplementary Figure S2C) failed to show any statistically significant difference even though the mean survival showed a reduced trend. Kaplan–Meier survival analysis of patients with positive axillary lymph nodes and TWIST1 mRNA overexpression had worst DFI (Table 2, Supplementary Figure S3A) (82.6 mo. vs 88.7–123.3; p = 0.05). When all patients were divided into two groups based on the number of positive lymph nodes (1–3, and ≥4 positive nodes) and the overexpression of TWIST1 [(N2-3/TWIST1(+), N2-3/TWIST1(−), N1/TWIST1(+), and N1/TWIST1(−)], Kaplan–Meier analysis revealed that women harboring more than 3 positive lymph nodes and TWIST1 that was overexpressed in EpCAM+ CTC fraction had a statistically significant shorter DFI (Table 2, Figure 3C) (mean survival: 68.6mo vs. 103.0–114.3mo.; p = 0.007). When patients were dichotomized accordingly to the HR status (ER/PR) in the following groups: a) HR(−)/TWIST1(+), b) HR(−)/ TWIST1(−), c) HR(+)/TWIST1(+) and d) HR(+)/TWIST1(−), it was observed that women with HR(−)/TWIST1(+) profile were characterized by statistically significant shorter DFI (36mo. vs 102.3–117.9mo.; p < 0.001; Figure 3E). A Univariate analysis (Table 3) also revealed the significance of (a) TWIST1(+), (b) HR(−)/TWIST1(+), (c) TWIST1(+) /N2-3, d) SC (+)/ TWIST1(+) (Figure 2B) in the risk of disease progression. Multivariate confirmed the prognostic value of HR(−)/TWIST1(+) and TWIST1(+)/N2-3, in the EpCAM(+) CTC fraction for the prediction of DFI (Table 3) independently from patients’ age, tumor T stage, grade, nodal status alone and the HR, and HER2 status of the primary tumor.
Table 2.
Gene expression in CTCs in respect to DFI and OS.
| Gene Expression in CTCs | DFI | OS | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Mean Survival | 95% CI (months) | Range (months) | p | Mean Survival | 95% CI (Months) | Range (Months) | p |
| TWIST1+ | 83.6 | 61.9–105.3 | 9–125 | 0.019 | 106.4 | 90.3–122.3 | 16–127 | 0.046 |
| TWIST1− | 115 | 106.3–125.2 | 4–137 | 127.2 | 120.7–133.7 | 6–137 | ||
| Stem cell profile positive (SC+) | 86.7 | 66.7–106.8 | 16–118 | 0.174 | 107.3 | 89.8–124.8 | 26–127 | 0.171 |
| Stem cell profile negative (SC−) | 113.2 | 103.5–122.8 | 4–137 | 125.2 | 118.3–132.1 | 6–137 | ||
| TWIST1+/SC+ | 68.9 | 39.4–98.31 | 16–112 | 0.087 | 96.2 | 67.4–125.1 | 26–127 | 0.118 |
| TWIST1+/SC− | 88.8 | 61.2–116.4 | 9–125 | 111.1 | 93.3–128 | 16–125 | ||
| TWIST1−/SC+ | 100.1 | 78.6–121.7 | 41–118 | 109.1 | 92.8–125.3 | 47–118 | ||
| TWIST1−/SC− | 115.8 | 105.8–126 | 4–137 | 127.3 | 120.4–134.2 | 6–137 | ||
| TWIST1+/LN+ | 82.6 | 59.3–105.9 | 9–125 | 0.05 | 108.8 | 93–124.6 | 16–127 | 0.194 |
| TWIST1+/LN− | 88.7 | 30.5–146.8 | 16–125 | 92 | 39.1–144.8 | 26–125 | ||
| TWIST1−/LN+ | 110.3 | 99.2–121.4 | 6–130 | 121.3 | 113–129.6 | 6–130 | ||
| TWIST1−/LN− | 123.3 | 110.6–135.9 | 4–137 | 128.9 | 120.2–137.7 | 37–137 | ||
| TWIST1+/N2–3 | 68.6 | 40.5–96.7 | 9–125 | 0.007 | 98.3 | 75.5–121.2 | 16–127 | 0.026 |
| TWIST1+/N1 | 104.5 | 71.8–137.5 | 13–123 | 112 | 94–125 | 101–118 | ||
| TWIST1−/N2-3 | 103.1 | 84.1–121.9 | 6–130 | 118.8 | 103.4–134.1 | 6–130 | ||
| TWIST1−/N1 | 114.4 | 101.7–126.9 | 9–128 | 121.1 | 111.9–130.3 | 30–128 | ||
| TWIST1+/HR+ | 102.3 | 81.2–123.4 | 9–125 | <0.001 | 121.6 | 113.9–129.3 | 79–127 | <0.001 |
| TWIST1+/HR− | 36 | 8.9–63 | 10–106 | 65.7 | 36.7–94.6 | 76–106 | ||
| TWIST1−/HR+ | 117.9 | 107.1–128.7 | 4–137 | 131.9 | 126.2–137.5 | 37–137 | ||
| TWIST1−/HR− | 107.7 | 92.3–123.1 | 6–127 | 110.2 | 96.5–123.84 | 6–127 | ||
Bold: highlights the significance of the test.
Figure 3.
Kaplan–Meier estimates for early BrCa patients: (A) DFI: TWIST1 overexpression, (B) OS: TWIST1 overexpression, (C) DFI: TWIST1 overexpression and number of affected lymph nodes, (D) OS: TWIST1 overexpression and number of affected lymph nodes, (E) DFI: TWIST1 overexpression and HR status, (F) OS: TWIST1 overexpression and HR status.
Table 3.
Univariate and multivariate analyses for DFI and OS in the early breast cancer patients group (n = 100).
| Covariates | Covariate Value | DFI | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | ||||||||||
| HR a | 95% CI b | p | HR a | 95% CI b | p | HRa | 95% CIb | p | HR a | 95% CI b | p | ||
| Age | ≥50 vs <50 | 0.787 | 0.357–1.734 | 0.552 | 0.432 | 0.169–1.103 | 0.079 | 0.718 | 0.249–2.070 | 0.539 | 0.593 | 0.169–2.080 | 0.414 |
| ER | Yes vs No | 0.647 | 0.286–1.463 | 0.295 | 4.391 | 1.040–18.549 | 0.044 | 0.238 | 0.079–0.721 | 0.011 | 0.623 | 0.092–4.215 | 0.628 |
| PR | Yes vs No | 0.492 | 0.217–1.114 | 0.089 | 0.087 | 0.021–0.362 | 0.001 | 0.196 | 0.054–0.707 | 0.013 | 0.098 | 0.011–0.851 | 0.035 |
| HER2 | Yes vs No | 0.500 | 0.197–1.269 | 0.145 | 0.626 | 0.232–1.693 | 0.357 | 0.381 | 0.106–1.366 | 0.139 | 0.247 | 0.060–1.023 | 0.054 |
| Lymph nodes | N0 vs N1 vs N2–3 | 2.207 | 1.261–3.861 | 0.006 | 2.433 | 1.272–4.654 | 0.007 | 1.659 | 0.817–3.371 | 0.162 | 1.351 | 0.637–2.862 | 0.433 |
| Size | ≥2cm vs <2cm | 3.060 | 1.049–8.922 | 0.041 | 4.926 | 1.225–19.811 | 0.025 | 7.244 | 0.947–55.432 | 0.056 | 17.450 | 1.464–208.1 | 0.024 |
| Grade | I/II vs III | 1.366 | 0.570–3.273 | 0.485 | 0.753 | 0.228–2.483 | 0.641 | 1.953 | 0.544–7.008 | 0.305 | 0.286 | 0.040–2.028 | 0.211 |
| TWIST1 | Yes vs No | 2.582 | 1.135–5.875 | 0.024 | 1.382 | 0.490–3.899 | 0.540 | 2.851 | 0.975–8.33 | 0.051 | 1.481 | 0.382–5.743 | 0.570 |
| HR and TWIST1 status | HR+TWIST1+ | 0.486 | 0.302–0.784 | 0.003 | 0.597 | 0.360–0.991 | 0.046 c | 0.576 | 0.313–1.062 | 0.077 | 0.666 | 0.345–1.284 | 0.225c |
| HR+TWIST1- | |||||||||||||
| HR-TWIST1+ | |||||||||||||
| HR-TWIST1- | |||||||||||||
| LN and TWIST1 status | N0-1TWIST1+ | 0.540 | 0.383–0.762 | <0.001 | 0.542 | 0.371–0.792 | 0.002 d | 0.559 | 0.357–0.875 | 0.011 | 0.604 | 0.373–0.976 | 0.040 d |
| N0-1TWIST1- | |||||||||||||
| N2-3TWIST1+ | |||||||||||||
| N2-3TWIST1- | |||||||||||||
| Stem cell profile | Yes vs No | 1.873 | 0.746–4.703 | 0.181 | 1.755 | 0.526–5.855 | 0.360 | 2.206 | 0.690–7.050 | 0.182 | 3.689 | 0.806–16.884 | 0.093 |
| Stem cell profile/TWIST1+ | Yes vs No | 0.663 | 0.473–0.929 | 0.017 | 0.776 | 0.521–1.146 | 0.202e | 0.624 | 0.402–0.967 | 0.035 | 0.634 | 0.378–1.065 | 0.085e |
a Hazard ratio, estimated from Cox proportional hazard regression mode. b Confidence interval of the estimated HR. Results are based on 1000 bootstrap samples and obtained after the Bias corrected and accelerated (BCa) approach. c Multivariate model adjusted for age, HER2, LN, Size, Grade, Stem cell. d Multivariate model adjusted for age, ER, PR, HER2, Size, Grade, Stem cell. e Multivariate model adjusted for age, ER, PR, HER2, LN, Size, Grade. Bold: highlights the significance of the test.
3.3.2. Overall Survival
Among the 25 patients that relapsed during the follow up period, 14/25 (56.0%) patients died and 11/25 (44.0%) were still alive at the time of the last follow-up. In 6/14 (42.9%) patients that died TWIST1 overexpression was detected in the EpCAM+ fraction (Fisher’s Exact Test; p = 0.024). Similarly, 4/14 (28.6%) patients displayed a Stem Cell profile in EpCAM+ CTC fraction (Fisher’s Exact Test; p = 0.217). In 3/14 (21.4%) of these patients, both TWIST1 overexpression and CD24−/low/CD44high and/or CD24−/low/ALDH1high profiles (Fisher’s Exact Test; p = 0.055) were detected (Fisher’s Exact Test; p = 0.055) (Supplementary Table S3). The Kaplan–Meier estimates of the overall survival (OS) of the patients overexpressing TWIST1 were significantly different in favor of patients who were negative for TWIST1 overexpression (106.4 vs 127.2 mo; p = 0.046) (Table 2, Figure 3B). Stem Cell profiles (107.3 vs 125.2 mo.; p = 0.171) (Table 2, Supplementary Figure S2B) and the co-expression of EMT and SC-associated genes (96.29 vs 109.1–127.3 mo.; p = 0.118) (Table 2, Supplementary Figure S2D) failed to show any statistically significant difference. There was no difference in OS in patients with TWIST1 overexpression according to N0 and N+ lymph node involvement (108.8 mo vs 92–129 mo, respectively; p = 0.194; Supplementary Figure S3B). However, when the Kaplan–Meier curves for OS for TWIST1 overexpression were additionally stratified according to lymph nodes status (Table 2, Figure 3D) and HR status (Table 2, Figure 3F) our data have shown that patients with >3 LN and TWIST1 overexpression had lower OS (109.8 mo., range: 115–129 mo.; p = 0.026); the same was seen for patients that were HR(−) and TWIST1 was overexpressed (65.7 vs 110.2–131.9 mo.; p < 0.001). Univariate analysis showed a significantly higher risk of death in the group of patients positive for TWIST1 overexpression that had more than 3 lymph nodes affected or co-expressed the stem cell profile (Figure 2B). Multivariate analysis confirmed the prognostic value of TWIST1 overexpression in combination with N2-3, and in combination with HR(−) status in the EpCAM(+) CTC fraction for the prediction of OS, independently from patients’ age, tumor T stage, grade, nodal status, and the status of the receptors ER, PR, HER2 of the primary site (Table 3).
4. Discussion
Molecular characterization of CTCs at the gene expression level has a strong potential to provide novel prognostic and predictive biomarkers. It is now clear through numerous studies that CTCs isolated from breast cancer patients express epithelial markers [38], receptors (ER, PR, HER2, EGFR), stem cell markers [39], and mesenchymal markers [11]. So far, most studies have been performed in the metastatic setting where the number of circulating tumor cells is usually high. However, in the non-metastatic setting of breast cancer, CTCs are not always detected and their numbers are usually very low, thus their molecular characterization is extremely difficult. For this reason, in the early breast cancer setting, a higher volume of peripheral blood used for the analysis of CTCs is very critical. Our group has shown many years ago the prognostic significance of CK-19 mRNA detection in peripheral blood of early breast cancer patients, using 20 mL of peripheral blood for CTC isolation and further downstream analysis [36,38]. Other groups have also shown that the detection of CTCs in the early breast cancer setting is providing critical prognostic information for these patients [29].
In this study we evaluated for the first time the prognostic significance of TWIST1, CD24, CD44, and ALDH1 transcript quantification in EpCAM-positive circulating tumor cells isolated from peripheral blood of early stage breast cancer patients. We selected TWIST1 as this is a very established EMT marker; for this reason, we have developed already in 2011 an RT-qPCR assay for the absolute quantification of TWIST1-mRNA expression, and we have validated this assay in EpCAM-positive cells isolated by early and metastatic breast cancer patients [33]. Concerning the selection of stem cell markers, this was based on publications by Al-Hajj M et.al. [8] and Ginestier C et.al. [9], who have shown that the breast cancer stem cell phenotypes of (a) CD44+/CD24−/low phenotype and (b) the overexpression of aldehyde dehydrogenase 1 (ALDH1+) are able to form tumors in mice with high tumorigenic capacity.
Multiplex RT-qPCR assays have many benefits due to their wide dynamic range, the low sample volume required, and the reduced time of analysis [35]. Our study was based on an analytically validated novel multiplex assay for the quantification of CD24, CD44, ALDH1, and HPRT and a single RT-qPCR assay for the quantification of TWIST1 transcripts. The analytical sensitivity and specificity of the novel quadraplexRT-qPCR assay for the simultaneous detection of CD24, CD44, ALDH-1, and HPRT transcripts were determined by using calibrators specific for each gene target. Both these assays were validated according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [40].
Relevant prognostic and predictive markers in early breast cancer cohort is of major significance. The SUCCESS A trial has shown that the presence of CTCs, as evaluated in 30 mL of peripheral blood, two years after chemotherapy has been associated with decreased OS and DFS in high-risk early breast cancer patients [41]. Lucci et.al. has also shown that the presence of one or more circulating tumor cells could predict early recurrence and decreased overall survival in chemonaive patients with non-metastatic breast cancer [29]; however, the main limitation of this study is that it was based on CTC enumeration performed in only 7.5 mL of blood. Additionally, molecular characterization of CTC could identify CTC biomarkers that are associated to specific signaling pathways like EMT or CSC. Our findings demonstrate a positive correlation between TWIST1 overexpression and the CD24−/low/CD44high profile in the EpCAM positive CTC fraction. This is in agreement with previous findings showing that the mesenchymal-like breast cancer stem cells are characterized as CD24−/CD44+, while the epithelial-like breast cancer stem cells express high levels of aldehyde dehydrogenase (ALDH) [16]. Univariate analysis revealed a significantly higher risk of relapse and death in the group of patients that expressed both stem cell and mesenchymal characteristics. Mego et al. have shown that patients with TWIST1-high tumors had a significantly higher percentage of breast cancer stem cells than patients with TWIST1-low tumors [19]. Recently, it was shown that in CTC of NSCLC patients the CD44(+)/CD24(−) population possess epithelial–mesenchymal transition characteristics [42], while another study in metastatic colorectal cancer has shown the prognostic significance of CTC that express both EMT and stem-like genes [22]. At the single CTC-level, Papadaki et.al have shown that CTCs expressing high levels of ALDH1 along with nuclear TWIST expression are more frequently detected in patients with metastatic breast cancer [21] and that these cells represent a chemo-resistant subpopulation with an unfavorable outcome [23]. The main limitation of our study is that we examined the expression of only one EMT marker, TWIST1 in the EpCAM+ cells of early breast cancer patients. Since there is a high heterogeneity in CTC, it may be possible that we have not detected CTCs that express other mesenchymal markers like Vimentin or Snail. We plan to extend this study by adding more gene expression markers in a biggest sample cohort and correlate our results to the clinical outcome.
According to our results, patients with TWIST1 overexpression in the EpCAM+ CTCs fraction and more than 3 involved lymph nodes had a significant lower DFI and OS. Similar to our results, recently, Emprou C et al. have shown that in frozen NSCLC tumor samples TWIST1 is more frequently overexpressed in the N+ group compared to the N0 group showing that partial EMT is involved in lymph node progression in early stages of NSCLC [43], while in primary breast cancer loss of E-cadherin is correlated with more than 3 LN involved in 80% of the patients [44]. Our results also indicate that patients with TWIST1 overexpression in the EpCAM+ CTCs fraction and a hormone receptor-negative primary tumor had a worse prognosis both for DFI and OS. This is in accordance with previous findings that have shown that the estrogen receptor silencing induces epithelial to mesenchymal transition in breast cancer [45]. It has also been previously shown that in human breast tumors there is an inverse relationship between TWIST1 and ER expression that may possibly contribute to the generation of hormone-resistant, ER-negative breast cancer [46]. It has also been reported that EMT likely occurs in the basal-like phenotype both in MCF10A cells [47] and in invasive breast cancer carcinomas [48].
5. Conclusions
In conclusion, detection of TWIST1 overexpression and stem-cell (CD24, CD44, ALDH1) transcripts in the EpCAM+ CTC fraction provides prognostic information in early stage breast cancer patients. Overexpression of TWIST1 in the EpCAM+ CTC fraction in the group of HR negative patients or with >3 positive lymph nodes is associated with worse prognosis.
Abbreviations
| EMT | epithelial–mesenchymal transition |
| MET | mesenchymal-epithelial transition |
| MICs | metastasis-initiating cells |
| SC | Stem Cell |
| HR | Hormone Receptor |
| BCSC | breast cancer stem cells |
| CSCs | Cancer stem cells |
| CTCs | Circulating tumor cells |
| ER | estrogen receptor |
| PR | progesterone receptor |
| HER2 | human epidermal growth factor receptor 2 |
| ALDH1 | aldehyde dehydrogenase 1 |
| HD | Healthy Donors |
| RT-qPCR | Quantitative Reverse Transcription Polymerase Chain Reaction |
| PBMCs | peripheral blood mononuclear cells |
| PBS | phosphate-buffered saline |
| LN | lymph nodes |
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/8/7/652/s1. Figure S1: Analytical validation of the multiplex RT-qPCR for CD24, CD44, ALDH1, HPRT (all measured in triplicate) (A) analytical specificity, (B) RT-qPCR calibration curves (copies/μL). Figure S2: Kaplan–Meier estimates for early BrCa patients with or without the stem cell profile in respect to (A) DFI, (B) OS and with or without co-expression of TWIST1 and Stem Cell profile in respect to (C) DFI, (D) OS. Figure S3: Kaplan–Meier estimates for early BrCa patients with positive or negative axillary lymph nodes the (A) DFI and (B) OS. Table S1: Clinical characteristics of the patients with early breast cancer (n = 100). Table S2: Quadraplex RT-qPCR for CD24, CD44, ALDH1, HPRT, evaluation of intra- (n = 3) and inter-assay (n = 5) precision. Table S3: Correlation of TWIST1, CD44high/CD24−/low and/or ALDH1high/CD24−/low and the co-expression of TWIST1 and CD44high/CD24−/low and/or ALDH1high/CD24−/low with the patients’ clinical outcomes.
Author Contributions
Conceptualization, A.S. and E.S.L.; methodology, A.S.; validation, A.S.; formal analysis, A.S.; investigation, A.S. and E.S.L.; resources, M.N. and V.G.; data curation, A.S.; writing—original draft preparation, A.S.; writing—review and editing, E.S.L. and V.G.; supervision, E.S.L.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Lianidou E.S. Gene expression profiling and DNA methylation analyses of CTCs. Mol. Oncol. 2016;10:431–442. doi: 10.1016/j.molonc.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardelli A., Pantel K. Liquid Biopsies, What We Do Not Know (Yet) Cancer Cell. 2017;31:172–179. doi: 10.1016/j.ccell.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Boral D., Vishnoi M., Liu H.N., Yin W., Sprouse M.L., Scamardo A., Hong D.S., Tan T.Z., Thiery J.P., Chang J.C., et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017;8:196. doi: 10.1038/s41467-017-00196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakabova A., Bielcikova Z., Pospisilova E., Matkowski R., Szynglarewicz B., Staszek-Szewczyk U., Zemanova M., Petruzelka L., Eliasova P., Kolostova K., et al. Molecular characterization and heterogeneity of circulating tumor cells in breast cancer. Breast Cancer Res. Treat. 2017;166:695–700. doi: 10.1007/s10549-017-4452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mansoori M., Madjd Z., Janani L., Rasti A. Circulating cancer stem cell markers in breast carcinomas: A systematic review protocol. Syst. Rev. 2017;6:262. doi: 10.1186/s13643-017-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Luo M., Clouthier S.G., Deol Y., Liu S., Nagrath S., Azizi E., Wicha M.S. Breast cancer stem cells: Current advances and clinical implications. Methods Mol. Biol. 2015;1293:1–49. doi: 10.1007/978-1-4939-2519-3_1. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S., et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balic M., Lin H., Young L., Hawes D., Giuliano A., McNamara G., Datar R.H., Cote R.J. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin. Cancer Res. 2006;12:5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 11.Theodoropoulos P.A., Polioudaki H., Agelaki S., Kallergi G., Saridaki Z., Mavroudis D., Georgoulias V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Bredemeier M., Edimiris P., Tewes M., Mach P., Aktas B., Schellbach D., Wagner J., Kimmig R., Kasimir-Bauer S. Establishment of a multimarker qPCR panel for the molecular characterization of circulating tumor cells in blood samples of metastatic breast cancer patients during the course of palliative treatment. Oncotarget. 2016;7:41677–41690. doi: 10.18632/oncotarget.9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baccelli I., Schneeweiss A., Riethdorf S., Stenzinger A., Schillert A., Vogel V., Klein C., Saini M., Bäuerle T., Wallwiener M., et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 14.Schölch S., García S.A., Iwata N., Niemietz T., Betzler A.M., Nanduri L.K., Bork U., Kahlert C., Thepkaysone M.-L., Swiersy A., et al. Circulating tumor cells exhibit stem cell characteristics in an orthotopic mouse model of colorectal cancer. Oncotarget. 2016;7:27232–27242. doi: 10.18632/oncotarget.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Li Y., Xu J., Zhang A., Wang X., Tang R., Zhang X., Yin H., Liu M., Wang D.D., et al. Quantified postsurgical small cell size CTCs and EpCAM+ circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018;412:99–107. doi: 10.1016/j.canlet.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu S., Cong Y., Wang D., Sun Y., Deng L., Liu Y., Martin-Trevino R., Shang L., McDermott S.P., Landis M.D., et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aktas B., Tewes M., Fehm T., Hauch S., Kimmig R., Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giordano A., Gao H., Anfossi S., Cohen E., Mego M., Lee B.-N., Tin S., De Laurentiis M., Parker C.A., Alvarez R.H., et al. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol. Cancer. 2012;11:2526–2534. doi: 10.1158/1535-7163.MCT-12-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mego M., Gao H., Lee B.-N., Cohen E.N., Tin S., Giordano A., Wu Q., Liu P., Nieto Y., Champlin R.E., et al. Prognostic Value of EMT-Circulating Tumor Cells in Metastatic Breast Cancer Patients Undergoing High-Dose Chemotherapy with Autologous Hematopoietic Stem Cell Transplantation. J. Cancer. 2012;3:369–380. doi: 10.7150/jca.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Ridgway L.D., Wetzel M.D., Ngo J., Yin W., Kumar D., Goodman J.C., Groves M.D., Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papadaki M.A., Kallergi G., Zafeiriou Z., Manouras L., Theodoropoulos P.A., Mavroudis D., Georgoulias V., Agelaki S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning Y., Zhang W., Hanna D.L., Yang D., Okazaki S., Berger M.D., Miyamoto Y., Suenaga M., Schirripa M., El-Khoueiry A., et al. Clinical relevance of EMT and stem-like gene expression in circulating tumor cells of metastatic colorectal cancer patients. Pharm. J. 2018;18:29–34. doi: 10.1038/tpj.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadaki M.A., Stoupis G., Theodoropoulos P.A., Mavroudis D., Georgoulias V., Agelaki S. Circulating Tumor Cells with Stemness and Epithelial-to-Mesenchymal Transition Features Are Chemoresistant and Predictive of Poor Outcome in Metastatic Breast Cancer. Mol. Cancer. 2019;18:437–447. doi: 10.1158/1535-7163.MCT-18-0584. [DOI] [PubMed] [Google Scholar]
- 24.Stathopoulou A., Vlachonikolis I., Mavroudis D., Perraki M., Kouroussis C., Apostolaki S., Malamos N., Kakolyris S., Kotsakis A., Xenidis N., et al. Molecular Detection of Cytokeratin-19–Positive Cells in the Peripheral Blood of Patients With Operable Breast Cancer: Evaluation of Their Prognostic Significance. J. Clin. Oncol. 2002;20:3404–3412. doi: 10.1200/JCO.2002.08.135. [DOI] [PubMed] [Google Scholar]
- 25.Xenidis N., Markos V., Apostolaki S., Perraki M., Pallis A., Sfakiotaki G., Papadatos-Pastos D., Kalmanti L., Kafousi M., Stathopoulos E., et al. Clinical relevance of circulating CK-19 mRNA-positive cells detected during the adjuvant tamoxifen treatment in patients with early breast cancer. Ann. Oncol. 2007;18:1623–1631. doi: 10.1093/annonc/mdm208. [DOI] [PubMed] [Google Scholar]
- 26.Xenidis N., Ignatiadis M., Apostolaki S., Perraki M., Kalbakis K., Agelaki S., Stathopoulos E.N., Chlouverakis G., Lianidou E., Kakolyris S., et al. Cytokeratin-19 mRNA-Positive Circulating Tumor Cells After Adjuvant Chemotherapy in Patients With Early Breast Cancer. J. Clin. Oncol. 2009;27:2177–2184. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]
- 27.Xenidis N., Perraki M., Apostolaki S., Agelaki S., Kalbakis K., Vardakis N., Kalykaki A., Xyrafas A., Kakolyris S., Mavroudis D., et al. Differential effect of adjuvant taxane-based and taxane-free chemotherapy regimens on the CK-19 mRNA-positive circulating tumour cells in patients with early breast cancer. Br. J. Cancer. 2013;108:549–556. doi: 10.1038/bjc.2012.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bidard F.-C., Michiels S., Riethdorf S., Mueller V., Esserman L.J., Lucci A., Naume B., Horiguchi J., Gisbert-Criado R., Sleijfer S., et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. JNCI J. Natl. Cancer Inst. 2018;110:560–567. doi: 10.1093/jnci/djy018. [DOI] [PubMed] [Google Scholar]
- 29.Lucci A., Hall C.S., Lodhi A.K., Bhattacharyya A., Anderson A.E., Xiao L., Bedrosian I., Kuerer H.M., Krishnamurthy S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012;13:688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 30.Georgoulias V., Bozionelou V., Agelaki S., Perraki M., Apostolaki S., Kallergi G., Kalbakis K., Xyrafas A., Mavroudis D. Trastuzumab decreases the incidence of clinical relapses in patients with early breast cancer presenting chemotherapy-resistant CK-19mRNA-positive circulating tumor cells: Results of a randomized phase II study. Ann. Oncol. 2012;23:1744–1750. doi: 10.1093/annonc/mds020. [DOI] [PubMed] [Google Scholar]
- 31.Ignatiadis M., Litière S., Rothe F., Riethdorf S., Proudhon C., Fehm T., Aalders K., Forstbauer H., Fasching P.A., Brain E., et al. Trastuzumab versus observation for HER2 nonamplified early breast cancer with circulating tumor cells (EORTC 90091-10093, BIG 1-12, Treat CTC): A randomized phase II trial. Ann. Oncol. 2018;29:1777–1783. doi: 10.1093/annonc/mdy211. [DOI] [PubMed] [Google Scholar]
- 32.Schneble E.J., Graham L.J., Shupe M.P., Flynt F.L., Banks K.P., Kirkpatrick A.D., Nissan A., Henry L., Stojadinovic A., Shumway N.M., et al. Current approaches and challenges in early detection of breast cancer recurrence. J. Cancer. 2014;5:281–290. doi: 10.7150/jca.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strati A., Markou A., Parisi C., Politaki E., Mavroudis D., Georgoulias V., Lianidou E. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer. 2011;11:422. doi: 10.1186/1471-2407-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mavroudis D., Saloustros E., Boukovinas I., Papakotoulas P., Kakolyris S., Ziras N., Christophylakis C., Kentepozidis N., Fountzilas G., Rigas G., et al. Sequential vs concurrent epirubicin and docetaxel as adjuvant chemotherapy for high-risk, node-negative, early breast cancer: An interim analysis of a randomised phase III study from the Hellenic Oncology Research Group. Br. J. Cancer. 2017;117:164–170. doi: 10.1038/bjc.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittwer C.T., Herrmann M.G., Gundry C.N., Elenitoba-Johnson K.S.J. Real-Time Multiplex PCR Assays. Methods. 2001;25:430–442. doi: 10.1006/meth.2001.1265. [DOI] [PubMed] [Google Scholar]
- 36.Stathopoulou A., Ntoulia M., Perraki M., Apostolaki S., Mavroudis D., Malamos N., Georgoulias V., Lianidou E.S. A highly specific real-time RT-PCR method for the quantitative determination of CK-19 mRNA positive cells in peripheral blood of patients with operable breast cancer. Int. J. Cancer. 2006;119:1654–1659. doi: 10.1002/ijc.22017. [DOI] [PubMed] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Ignatiadis M., Xenidis N., Perraki M., Apostolaki S., Politaki E., Kafousi M., Stathopoulos E.N., Stathopoulou A., Lianidou E., Chlouverakis G., et al. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J. Clin. Oncol. 2007;25:5194–5202. doi: 10.1200/JCO.2007.11.7762. [DOI] [PubMed] [Google Scholar]
- 39.Aktas B., Müller V., Tewes M., Zeitz J., Kasimir-Bauer S., Loehberg C.R., Rack B., Schneeweiss A., Fehm T. Comparison of estrogen and progesterone receptor status of circulating tumor cells and the primary tumor in metastatic breast cancer patients. Gynecol. Oncol. 2011;122:356–360. doi: 10.1016/j.ygyno.2011.04.039. [DOI] [PubMed] [Google Scholar]
- 40.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 41.Trapp E., Janni W., Schindlbeck C., Jückstock J., Andergassen U., de Gregorio A., Alunni-Fabbroni M., Tzschaschel M., Polasik A., Koch J.G., et al. Presence of Circulating Tumor Cells in High-Risk Early Breast Cancer During Follow-Up and Prognosis. Jnci J. Natl. Cancer Inst. 2019;111:380–387. doi: 10.1093/jnci/djy152. [DOI] [PubMed] [Google Scholar]
- 42.Mirza S., Jain N., Rawal R. Evidence for circulating cancer stem-like cells and epithelial-mesenchymal transition phenotype in the pleurospheres derived from lung adenocarcinoma using liquid biopsy. Tumour Biol. 2017;39:1010428317695915. doi: 10.1177/1010428317695915. [DOI] [PubMed] [Google Scholar]
- 43.Emprou C., Le Van Quyen P., Jégu J., Prim N., Weingertner N., Guérin E., Pencreach E., Legrain M., Voegeli A.-C., Leduc C., et al. SNAI2 and TWIST1 in lymph node progression in early stages of NSCLC patients. Cancer Med. 2018;7:3278–3291. doi: 10.1002/cam4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markiewicz A., Wełnicka-Jaśkiewicz M., Seroczyńska B., Skokowski J., Majewska H., Szade J., Żaczek A.J. Epithelial-mesenchymal transition markers in lymph node metastases and primary breast tumors - relation to dissemination and proliferation. Am. J. Transl. Res. 2014;6:793–808. [PMC free article] [PubMed] [Google Scholar]
- 45.Voutsadakis I.A. Epithelial-Mesenchymal Transition (EMT) and Regulation of EMT Factors by Steroid Nuclear Receptors in Breast Cancer: A Review and in Silico Investigation. J. Clin. Med. 2016;5:11. doi: 10.3390/jcm5010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vesuna F., Lisok A., Kimble B., Domek J., Kato Y., van der Groep P., Artemov D., Kowalski J., Carraway H., van Diest P., et al. Twist contributes to hormone resistance in breast cancer by downregulating estrogen receptor-α. Oncogene. 2012;31:3223–3234. doi: 10.1038/onc.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarrió D., Rodriguez-Pinilla S.M., Hardisson D., Cano A., Moreno-Bueno G., Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 48.Choi Y., Lee H.J., Jang M.H., Gwak J.M., Lee K.S., Kim E.J., Kim H.J., Lee H.E., Park S.Y. Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum. Pathol. 2013;44:2581–2589. doi: 10.1016/j.humpath.2013.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.