Summary
Bacteria use alternative sigma factors to adapt to different growth and stress conditions. The Bacillus subtilis extracytoplasmic function (ECF) sigma factor SigM regulates genes for cell wall synthesis and is crucial for maintaining cell wall homeostasis under stress conditions. The activity of SigM is regulated by its anti-sigma factor, YhdL, and the accessory protein YhdK. Here, we show that dysregulation of SigM caused by the absence of either component of the anti-sigma factor complex leads to toxic levels of SigM and severe growth defects. High SigM activity results from a dysregulated positive feedback loop, and can be suppressed by overexpression of the housekeeping sigma, SigA. Using a sigM merodiploid strain, we selected for suppressor mutations that allow survival of a yhdL depletion strain. The recovered suppressor mutations map to the beta and beta-prime subunits of RNA polymerase core enzyme and selectively reduce SigM activity, and in some cases increase the activity of other alternative sigma factors. This work highlights the ability of mutations in RNA polymerase that remodel the sigma-core interface to differentially affect sigma factor activity, and thereby alter the transcriptional landscape of the cell.
Graphical Abstract
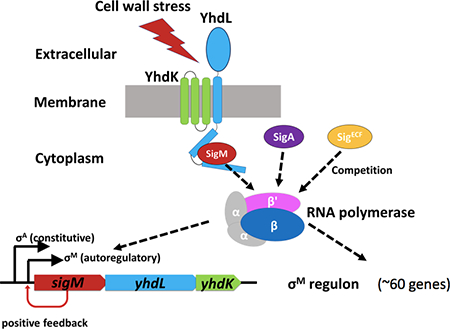
The Bacillus subtilis extracytoplasmic function sigma factor, SigM, controls a large regulon of genes involved in cell wall homeostasis. Mutation of the YhdLK antisigma factors is lethal, due in part to runaway induction by positive autoregulation, and can be compensated by mutation of the positive autoregulatory promoter, overexpression of SigA, or mutations in core RNAP that decrease SigM activity.
Introduction
In order to survive and thrive in a changing environment, bacteria have evolved complex mechanisms to regulate their gene expression in response to environmental cues. One important mechanism for changing gene expression is the expression of alternative sigma factors that can redirect RNA polymerase to transcribe new sets of genes (Feklistov et al., 2014). Unlike the housekeeping sigma factor (SigA in many bacteria), which transcribes many genes essential for growth, alternative sigma factors are non-essential and are activated under specific stress conditions, or to activate specific genetic programs such as those associated with genetic competence or endospore formation. Under these conditions, one or more alternative sigma factors become active, bind to RNA polymerase (RNAP) core and transcribe a set of genes (a regulon) specific for each alternative sigma factor (Mascher, 2013).
Bacillus subtilis strain 168 contains eighteen alternative sigma factors, including seven extracytoplasmic function (ECF) sigma factors responding to cell envelope stresses (Souza et al., 2014, Mascher, 2013). Four of the ECF family sigma factors (SigM, SigV, SigW, and SigX) are best understood, and respond to stresses caused by peptidoglycan (PG) synthesis inhibitors, lysozyme, detergents and peptide antibiotics, and cationic antimicrobials, respectively (Helmann, 2016). The large SigM regulon (>60 genes) is activated by inhibition of peptidoglycan (PG) synthesis (Jervis et al., 2007), and includes many genes central to PG homeostasis (Eiamphungporn & Helmann, 2008). These genes encode key synthetic enzymes (e.g. MurB and PonA, a class A PBP), or alternative pathways to bypass steps that may be inhibited (e.g. Amj, an alternative to the lipid II flippase MurJ) (Meeske et al., 2015). A sigM mutant has growth defects in high salt or 5% ethanol, and is highly sensitive to cell wall targeting antibiotics including β-lactams, bacitracin and moenomycin (Thackray & Moir, 2003; Luo & Helmann, 2012).
ECF sigma factors are often activated by one of three methods: proteolysis of an anti-sigma factor, a conformational change in an anti-sigma factor, or a partner-switching mechanism controlled by phosphorylation of an anti-anti-sigma factor (Ho & Ellermeier, 2012; Sineva et al., 2017). The most well studied examples of sigma factor activation by anti-sigma factor proteolysis include Escherichia coli SigE (Ades et al., 1999; Alba & Gross, 2004), B. subtilis SigW (Schobel et al., 2004) and SigV (Hastie et al., 2013). Activation of SigW requires two sequentially acting proteases that cleave the membrane-bound anti-sigma factor, RsiW. RsiW degradation is initiated by the site 1 protease PrsW (Heinrich & Wiegert, 2006), followed by intramembrane (site 2) proteolysis by RasP (Heinrich et al., 2009). Finally, the released complex of SigW with the residual RsiW’ fragment is further degraded by the cytosolic Clp protease to release active SigW (Zellmeier et al., 2006). SigV is induced by lysozyme and confers lysozyme resistance by O-acetylation of the PG and D-alanylation of teichoic acids (Ho et al., 2011; Guariglia-Oropeza & Helmann, 2011). The activation of SigV begins with the binding of lysozyme to the anti-sigma factor RsiV, which facilitates proteolysis by signal peptidase cleavage at site 1 (Castro et al., 2018) followed by intramembrane protease RasP cleavage at site 2 (Hastie et al., 2013). It is not yet clear how SigM is activated and whether proteolysis is involved.
The sigM operon (sigM-yhdL-yhdK) encodes SigM together with YhdL and YhdK, which constitute the functional anti-sigma factor complex (YhdLK). YhdL is a transmembrane protein and the N-terminal domain of YhdL interacts with SigM as determined using a yeast two-hybrid analysis (Yoshimura et al., 2004). YhdK is a small protein (96 amino acids) with three transmembrane segments and interacts with YhdL, but not with SigM (Yoshimura et al., 2004). The sigM operon (sigM-yhdL-yhdK) is transcribed from two promoters, a constitutive SigA-controlled promoter (PA) and an autoregulatory SigM-controlled promoter (PM) (Figure 1A) (Thackray & Moir, 2003). In the absence of stress, the sigM operon is mostly transcribed from PA (Horsburgh & Moir, 1999). Under these conditions, SigM is largely sequestered on the membrane by the YhdLK anti-sigma complex (Yoshimura et al., 2004; Asai, 2018).
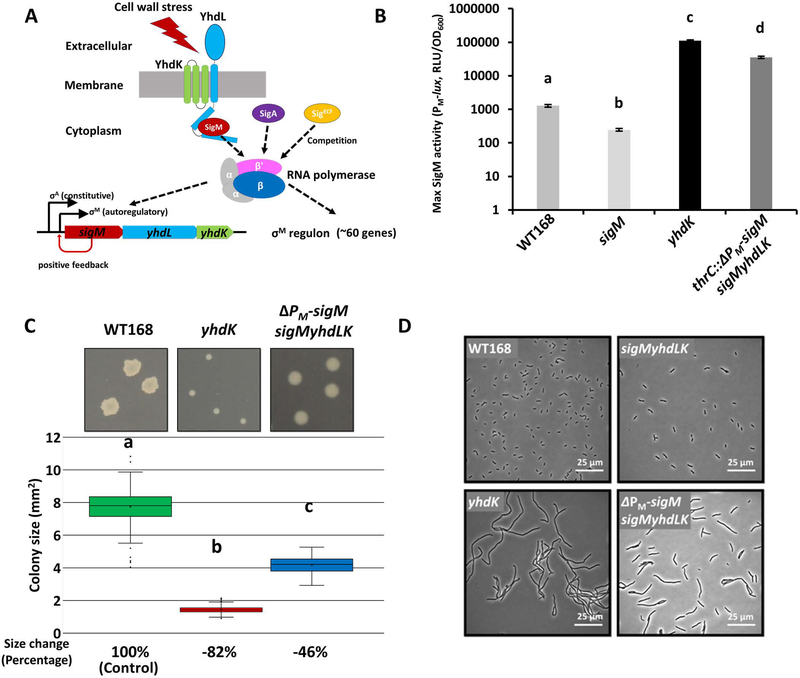
Figure 1. Alleviation of SigM toxicity by removing positive autoregulation.
A. Schematic drawing of SigM and its anti-sigma factors YhdL and YhdK, illustrating the roles of the autoregulatory PM promoter and sigma competition in regulating SigM activity. B. Maximum SigM activity (mean ± standard error of mean; SEM) of WT168 (HB17325), sigM (HB17494), yhdK (HB20833), and thrC::ΔPM-sigM sigMyhdLK (HB23682) strains. Statistically significantly different samples (Student’s t test, two-tailed P<0.05) are labelled with different letters (n≥3). C. Representative images and quantitative colony size measurements for WT and mutants (yhdK, HB20830; and thrC::ΔPM-sigM sigMyhdLK, HB23625) on LB plates after 24 hours of incubation at 37°C. Colony size data (n≥300) were plotted using Box and Whisker chart and the bottom and top of the box are the first and third quartiles, respectively; the band inside the box is the second quartile (the median), and the X inside the box is the mean. Whiskers are one standard deviation (SD) above or below the mean. Outliers are shown as single dots. Average colony size change was calculated as %change = (sample - control)/control x 100. P value was calculated using a two-tailed student’s assuming unequal variances. Statistically significantly differences (Student’s t test, two-tailed P<0.05) are labelled with different letters. D. Representative phase contrast microscope images of WT and mutants growing at exponential phase in LB medium. The strains used in panel D include: WT168 (lab stock), sigMyhdLK (HB22747), yhdK (HB20830), thrC::ΔPM-sigM sigMyhdLK (HB23625).
In the presence of cell envelope stresses, SigM is released from YhdLK through an unknown mechanism, binds to RNA polymerase (RNAP) and transcribes its regulon (Mascher, 2013). Activation of the autoregulatory PM promoter generates a positive feedback loop that amplifies induction of the SigM regulon in response to stress (Thackray & Moir, 2003). Positive autoregulation is a common feature of many of the stress responsive alternative sigma factors in B. subtilis, and is especially common amongst ECF sigma factors (Helmann, 2002, Mascher, 2013). One consequence of this positive autoregulation is that fluctuations in activity can lead to stochastic, but often transient, induction of individual regulons. This results in population heterogeneity in which limited core RNA polymerase is shared by multiple, alternative sigma factors (Park et al., 2018).
Uniquely amongst the seven ECF sigma factors in B. subtilis, regulation of SigM by its anti-sigma is essential for viability (Horsburgh & Moir, 1999; Kobayashi et al., 2003). Cells lacking YhdL are presumed to experience a lethal, runaway activation of sigM expression and rapidly acquire suppressor mutations in sigM (Horsburgh & Moir, 1999). Here, we confirm that a lack of YhdL leads to SigM-dependent cell death, and that lack of the accessory regulator YhdK also leads to a severe growth defect. This toxicity results in part from positive feedback since mutation of the autoregulatory PM promoter partially suppresses the growth defects of yhdL and yhdK mutants. Moreover, overexpression of SigA also suppresses SigM toxicity. Finally, using a merodiploid sigM strain, we identified mutations affecting core RNAP that alleviate SigM toxicity, and can also affect the activity of other alternative sigma factors.
Results and Discussion
Alleviation of SigM toxicity by disabling its positive autoregulation
To quantify the activity of SigM and the role of the anti-sigma complex, we first constructed a PM-lux luciferase reporter using the autoregulatory PM promoter as a measure of SigM activity. Because the luciferase reporter has a half-life of only ~4 minutes (Radeck et al., 2013), we were able to acquire near real-time measurements of SigM activity throughout cell growth. In a wild type 168 strain (WT168) grown in LB medium, SigM was transiently induced by ~10-fold in mid-exponential growth phase and quickly returns to background levels (Figure S1A), similar to a previous report (Horsburgh & Moir, 1999). The transient induction might be caused by rapid cell growth and the need to upregulate cell wall synthesis in the exponential growth phase. A sigM null mutant did not exhibit this transient induction, suggesting that the PM-lux luciferase reporter is, as expected, dependent on SigM (Figure S1A). A yhdK null mutant has greatly elevated SigM activity, ~100-fold higher than WT when comparing maximum activity (Figure 1B, Figure S1A, S1B). The yhdK mutant also exhibited a small colony size when grown on LB plates (Figure 1C), a filamentous, chaining cell morphology (Figure 1D), and slow growth in liquid culture (Figure S1C).
The yhdL gene has been reported as essential (Horsburgh & Moir, 1999; Kobayashi et al., 2003). Consistently, transformation of a yhdL::kan allele into a strain with a PM-lacZ reporter generated tiny blue colonies on X-gal plates that could not be sub-cultured (Figure S1D and data not shown), and much larger and mostly white colonies that contain suppressor mutations inactivating sigM (Figure S1D and Sanger sequencing results, data not shown). A strain missing the entire sigM-yhdL-yhdK operon (HB22747) showed normal cell morphology (Figure 1D) and a growth rate similar to WT in LB medium (Figure S1C), confirming that high SigM activity is toxic to cells, consistent with previous reports (Horsburgh & Moir, 1999).
The autoregulatory PM enables a positive feedback loop for SigM induction, which is expected to be beneficial under cell envelope stress conditions, but likely contributes to the synthesis of toxic levels of SigM in the absence of the anti-sigma factor complex. To assess the contribution of positive autoregulation to SigM intoxication, we constructed a strain with an ectopic copy of sigM only under its PA but not PM promoter (ΔPM-sigM). In this strain (HB23682), the native sigM yhdL yhdK operon was deleted, leaving behind an ectopic copy of PA-sigM without the genes encoding the anti-sigma factor complex. Compared with WT, this strain exhibited high SigM activity (Figure 1B, S1B), smaller colony size when grown on plate (Figure 1C), slower growth in liquid medium (Figure S1C), and altered cell morphology evidenced by an increased chaining of cells (Figure 1D). However, unlike a yhdL null mutant, this strain was viable in the absence of the anti-sigma complex, and had a colony size even larger than the yhdK null mutant in a strain with positive autoregulation (Figure 1C). We conclude that the lethal effects of SigM overproduction require positive autoregulation, and that the anti-sigma complex is no longer essential in the absence of the autoregulatory PM promoter.
Alleviation of SigM toxicity by overexpression of SigA
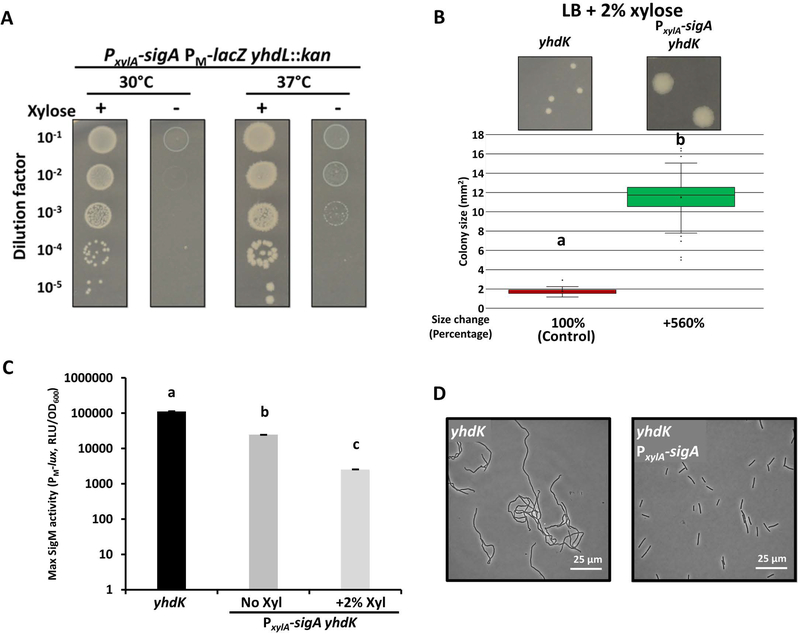
Sigma factors compete for a limited pool of RNAP core enzyme to transcribe their regulons (Park et al., 2018, Ganguly & Chatterji, 2012, Grigorova et al., 2006, Maeda et al., 2000). We reasoned that overexpression of SigA from an inducible promoter might compete with SigM and reduce its toxicity in the absence of its anti-sigma factors. To test this hypothesis, we constructed a xylose-inducible PxylA-sigA strain. Indeed, the lethality of the yhdL mutation could be suppressed in a xylose-dependent manner (Figure 2A), and the growth and morphology defects of the yhdK mutant were greatly reduced in the presence of 2% xylose (Figure 2B, 2D). Using the PM-lux luciferase reporter, we found that SigM activity in the yhdK mutant was significantly reduced in the presence of PxylA-sigA, an effect notable even without addition of xylose (Figure 2C, S1B). Overall, these data reveal that overexpression of SigA can reduce SigM activity, presumably by competing for core RNAP, and thereby alleviate SigM toxicity.
Figure 2. Alleviation of SigM toxicity by SigA overexpression.
A. Spot dilution assay of the PxylA-sigA yhdL::kan (HB22788) strain grown on LB plates in the presence or absence of 2% xylose after 24 hrs at 30 or 37°C. B. Representative images and quantitative colony size measurement of a yhdK mutant without (HB20830) or with an ectopic xylose-inducible copy of sigA (HB23952) on LB plates with 2% xylose (24 hrs, 37°C). Colony size data are presented in the same way as Figure 1C. C. Maximum SigM activity of yhdK (HB20833) (from Fig. 1B) and yhdK PxylA-sigA (HB23937) strains. Data are presented as mean ± SEM. Statistically significantly different samples (Student’s t test, two-tailed P<0.05) are labelled with different letters (n≥3). D. Representative phase contrast microscope images of yhdK (HB20830) and yhdK PxylA-sigA (HB23952) growing at exponential phase in LB medium supplemented with 2% xylose.
Single amino acid substitutions in RNA polymerase suppress SigM toxicity
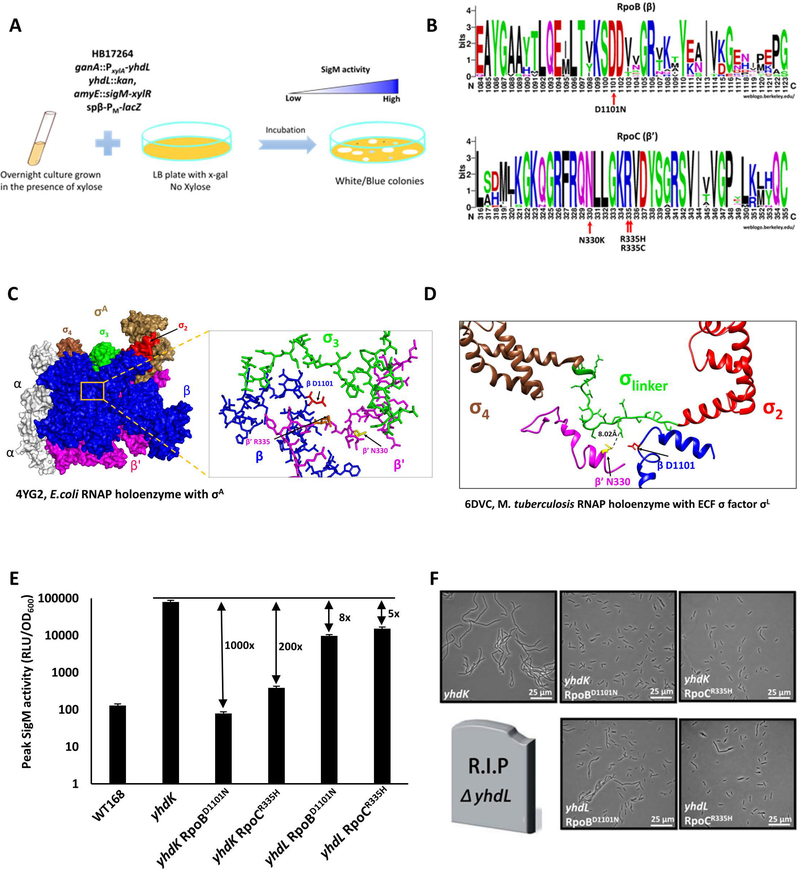
We next aimed to better understand why high SigM is toxic to cells. We reasoned that high SigM activity may lead to overexpression of its regulon, and some of these genes may encode proteins that are toxic when overexpressed. As one approach to identify potentially toxic gene products, we isolated suppressor mutants that allowed cells to tolerate high SigM activity. To this end, we constructed a yhdL depletion strain with an ectopic xylose-inducible yhdL. Because it is expected that mutations inside sigM or xylR gene can bypass this depletion system, we introduced second copies of sigM and xylR. A PM-lacZ reporter was used to visualize high SigM activity on X-gal plates (Figure 3A). This selection again yielded very small blue colonies that could not be sub-cultured together with some large white colonies with mutations in both copies of sigM as revealed by Sanger sequencing (data not shown). The appearance of strains with mutations in both copies of sigM was expected, since in a prior CRISPRi-based depletion study we observed that the presence of a second dCas9 gene only reduced the emergence of suppressors inactivating dCas9 by ~10-fold, and both inactivated copies carried the same mutation, suggestive of facile gene conversion pathway (Zhao et al., 2016). In addition, we also recovered several medium-sized colonies with light blue color, consistent with an elevated but tolerable level of SigM activity, even in the absence of YhdL.
Figure 3. Single amino acid substitutions in RNAP reduce SigM activity and toxicity in the absence of anti-SigM factors.
A. Schematic drawing of the selection and screening of suppressor mutants that can tolerate increased SigM activity. B. WebLogos of the altered region in RpoB and RpoC from 16 bacterial species (see Experimental Procedures). C, D. Suppressor mutations identified from whole genome resequencing are labelled. Substituted amino acids were mapped to (C) E. coli RNAP holoenzyme with σA (PDB ID: 4YG2) and (D) M. tuberculosis RNAP holoenzyme with σL (PDB ID: 6DVC). Structural information for the equivalent residue of B. subtilis RpoC R335 is not available in the M. tuberculosis structure. E. Fold changes of maximum SigM activity (with the PM-lux reporter) between the indicated mutants with WT or mutant RNAP (yhdK RpoBD1101N, HB21166; yhdK RpoCR335H, HB21168; yhdL RpoBD1101N, HB21089; yhdL RpoCR335H, HB21092). The data are presented as mean ± SEM (n≥3), and the fold changes are calculated using the mean. F. Representative phase contrast microscope images of yhdK and yhdL mutants grown in LB medium at exponential phase. The yhdL single mutant with wild type RNAP is lethal and represented as a tombstone.
Candidate suppressors were analyzed by whole genome re-sequencing, and four independent suppressors were characterized, with each containing a single point mutation affecting a subunit of core RNA polymerase. One mutation led to an RpoB (β subunit) D1101N substitution, and the other three to single substitutions in RpoC (β’ subunit): N330K, R335H, or R335C (Figure 3B). These three amino acid residues are highly conserved among RpoB/C proteins (Figure 3B), suggesting they are likely important for the structure and function of RNAP. We did not recover any mutations in SigM-dependent loci that allowed growth of the yhdL depletion strain. This could be because high SigM activity is toxic solely due to interference with the essential SigA protein, or because toxicity results from the overproduction of multiple, independently expressed proteins and/or proteins that are essential.
Next, we sought to understand how these mutations in core RNAP might affect the cell’s ability to tolerate high levels of SigM. Since we do not have a high resolution structure of B. subtilis holoenzyme, we mapped all three substitutions to the E. coli σ70 RNAP holoenzyme structure. All three positions (RpoBD1101, RpoCN330 and RpoCR335) map closely to region 3 of E. coli σ70 (Figure 3C), suggesting that these amino acids may affect the RNAP-sigma factor interaction. Although ECF sigma factors do not have a structured region 3 domain between region 2 and 4, in the Mycobacterium tuberculosis SigL holoenzyme, the linker between SigL regions 2 and 4 occupies a similar position as σ70 region 3, and likely performs a similar function (Lin et al., 2018). When these three altered amino acids were mapped to the structure of the SigL holoenzyme we noted an estimated distance of ~8 Å between the N330 equivalent residue of RpoC and E140 of SigL (Figure 3D). The linker regions of ECF sigma factors differ dramatically in both length and composition, and have been suggested to impact promoter recognition properties (Gaballa et al. 2018). Thus, while the molecular details are not entirely clear, we hypothesized that these substitutions in RNAP core likely hinder binding of SigM to RNAP, and thereby reduce SigM activity.
To quantify the effects of these core RNAP mutations on SigM activity, we first attempted to reconstruct them at the native locus using CRISPR-based mutagenesis. We were able to introduce the RpoBD1101N and RpoCR335H mutations, and both mutants showed slightly slower growth than WT in LB medium (Figure S2A). The RpoCR335H mutant exhibited a mildly filamentous cell morphology (Figure S2C). Attempts to combine both the RpoBD1101N and RpoCR335H mutations in one strain were not successful. Using the PM-lux reporter, we found that in a yhdK null strain the RpoBD1101N and RpoCR335H mutations led to an ~1,000-fold and 200-fold reduction of maximum SigM activity, respectively (Figure 3E, S2D). The filamentous cell morphology of the yhdK null was also greatly suppressed by these mutations in RNAP (Figure 3F). In addition, yhdL could now be deleted in the RpoBD1101N and RpoCR335H mutant strains. In strains carrying either of these rpoB or rpoC mutations, the yhdL null mutants exhibited higher SigM activity (Figure 3E, S2D) and a more severe cell morphology defect (Figure 3F) than their yhdK null counterparts. However, even in the absence of yhdL, the SigM activity was still 5- to 8-fold lower in these strains than in the yhdK null mutant with WT RNAP (Figure 3E, S2D). Overall these data show that single amino acid substitutions in RpoB or RpoC can greatly reduce SigM activity and suppress the growth defects that arise due to the absence of YhdL or YhdK.
Differential effects of the RpoBD1101N and RpoCR335H substitutions on alternative sigma factors
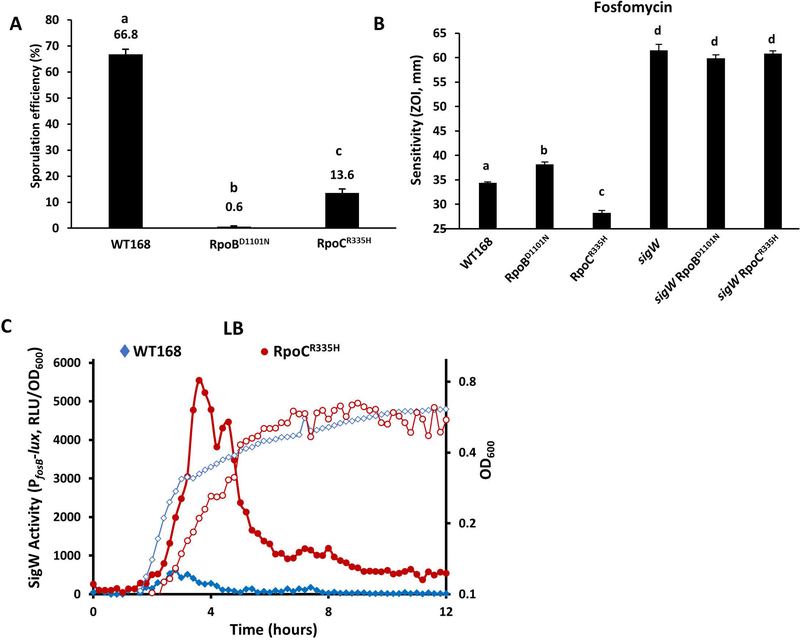
Because mutations in core RNAP may affect its interaction with many sigma factors and thereby alter the transcription efficiency of many genes, we next tested functions dependent on the activity of alternative sigma factors. We first monitored sporulation efficiency, since sporulation requires a complex cascade involving the sequential action of five sigma factors: SigH, SigF, SigE, SigG and SigK (Stragier & Losick, 1996; Piggot & Hilbert, 2004). We found that the RpoBD1101N strain had an ~100-fold decrease in the formation of heat resistant spores (0.6% in RpoBD1101N comparing to 66.8% in WT), whereas the RpoCR335H strain had a milder sporulation defect (13.6% in RpoCR335H), after 48 hours of growth in liquid Difco Sporulation Medium (DSM) (Figure 4A). The mild sporulation defect of the RpoCR335H strain suggests that all five sigma factors required for sporulation are mostly functional despite the mutation in RNAP. Using membrane labeling with the dyes FM 4–64 and mitotracker green, we observed that the RpoBD1101N strain was defective in completion of endospore engulfment, whereas the RpoCR335H strain could proceed through engulfment, albeit with a smaller fraction of cells than WT (data not shown). To quantify sporulation sigma factor activation, we constructed PspoVG42-lux, PspoIIM-lux, and PspoVFA-lux to directly measure activity of SigH, SigE and SigK, respectively. SigH activity was increased in the RpoCR335H strain and decreased in the RpoBD1101N strain relative to WT (Figure S2E). The constructs measuring SigE and SigK activity, however, yielded no signal even in the WT background (data not shown), which may simply reflect a limitation of the luciferase reporter in sporulating cells.
Figure 4. Single amino acid substitutions in RNAP differentially affect sigma factor activity.
A. Sporulation efficiency of WT and RNAP mutants (RpoBD1101N, HB20928; RpoCR335H, HB20930). In both panel A and B, Data are presented as mean ± SEM. Statistically significantly different samples (Student’s t test, two-tailed P<0.05) are labelled with different letters (n≥3). B. Zone of inhibition assay showing sensitivity to fosfomycin of WT and RNAP mutants with or without additional deletion of sigW (RpoBD1101N, HB20928; RpoCR335H, HB20930; sigW, HB21116; sigW RpoBD1101N, HB21285; sigW RpoCR335H, HB21281). C. SigW activity (solid symbols, left axis) and growth (open symbols, right axis) of WT (HB22578) (diamond) and the RpoCR335H mutant (HB22582) (circle) measured using a PfosB-lux reporter when cells were grown in LB medium at 37 °C with shaking.
We next tested sensitivity of RpoBD1101N and RpoCR335H mutant strains against several compounds targeting the cell envelope where resistance is known to be associated with the activity of one or more ECF sigma factors (Mascher et al., 2007). These two mutants were similar to WT in sensitivity to lysozyme (SigV), vancomycin (SigW), and ampicillin (SigX,SigW) (Figure S2B). However, in the case of fosfomycin, the RpoBD1101N mutant strain was more sensitive whereas the RpoCR335H strain was significantly more resistant than WT (Figure 4B). Fosfomycin resistance is provided by fosB, a SigW-dependent gene that encodes a bacillithiol-S-transferase that inactivates fosfomycin (Cao et al., 2001; Lamers et al., 2012; Roberts et al., 2013). To test if the fosfomycin resistance of RpoCR335H mutant is SigW-dependent, we deleted sigW in the RpoCR335H mutant. The rpoCR335H sigW double mutant was as sensitive to fosfomycin as a sigW single mutant, suggesting that the increased resistance of the RpoCR335H strain is SigW-dependent. We used a PfosB-lux reporter strain to directly compare SigW activity in WT and the RpoCR335H strain. When growing in LB medium without fosfomycin, basal SigW activity was about 10-fold higher in the RpoCR335H mutant than WT (Figure 4C). Overall, our results suggest that the RpoCR335H mutant has decreased SigM activity and at the same time an increase in SigW activity. These results imply that mutations at or near the core RNAP-sigma binding interface can differentially affect sigma factor activity.
Insights into the mechanism of activation of SigM by cell wall stresses
The role of the YhdLK anti-sigma factor complex in mediating the response to cell envelope stress is poorly understood. YhdK seems to function as an integral membrane protein that assists, but is not absolutely essential for, the ability of YhdL to constrain SigM activity. YhdLK and SigM form a three-protein complex, with YhdK interacting with the transmembrane segment of YhdL, and the N-terminal cytoplasmic domain of YhdL binding to SigM (Yoshimura et al., 2004; Asai, 2018). Although YhdK appears to stabilize the inhibitory interaction between YhdL and SigM, the relative roles of YhdK and YhdL in signal perception are not resolved.
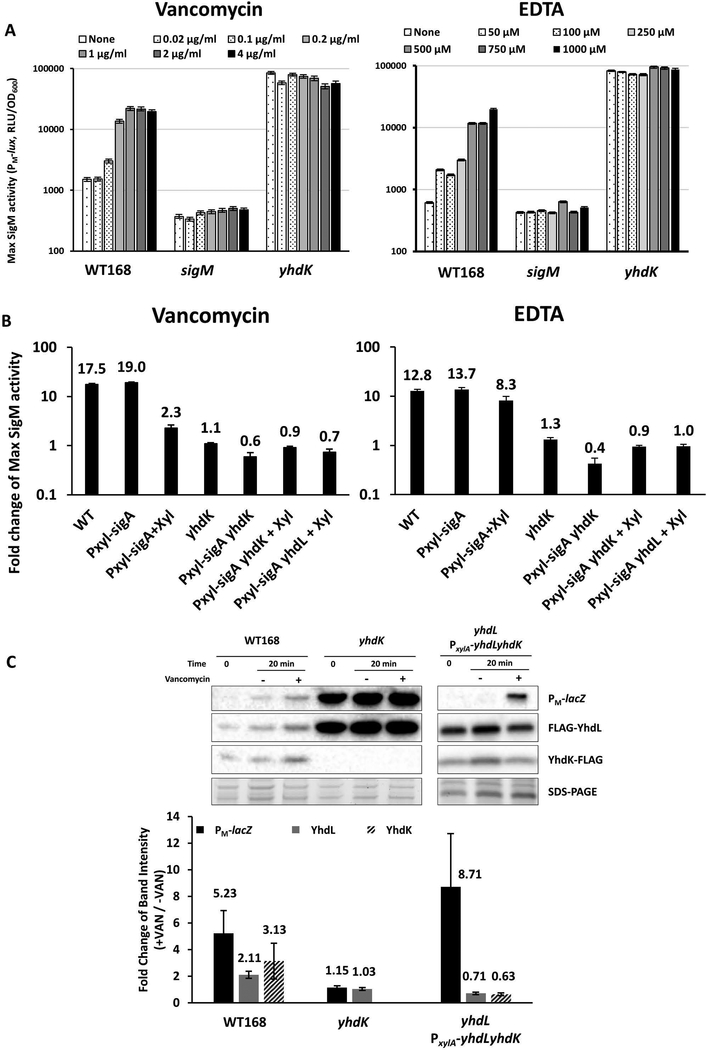
To test whether YhdK is required for the perception of signals that lead to transcriptional induction of the SigM regulon, we tested induction of SigM activity in strains with and without YhdK. The PM-lux reporter was strongly induced in a dose dependent manner by both vancomycin and EDTA, but no induction could be observed in the yhdK mutant (Figure 5A). We were concerned that perhaps the lack of induction was an artefact due to the already very high levels of expression in the yhdK strain. Therefore, we also tested the ability of vancomycin and EDTA to induce the PM-lux reporter in WT and yhdK null strains carrying PxylA-sigA, either in the absence or presence of xylose. While both vancomycin and EDTA still induced the PM-lux reporter by more than 10-fold in the yhdK+ background, albeit to a less degree in the presence of xylose, there was almost no induction in the yhdK null background (Figure 5B), despite the fact that the PxylA-sigA construct greatly reduces the absolute level of the PM-lux reporter (Figure 2C). As expected, there was also no induction in a yhdL null mutant strain carrying the induced PxylA-sigA construct (Figure 5B). These results suggest that the ability of the YhdLK-SigM complex to respond to these two chemical inducers requires the presence of YhdK, which upon signal perception is postulated to decrease the ability of YhdL to sequester SigM.
Figure 5. YhdK is required for signal perception and induction of the SigM regulon.
A. Maximum SigM activity measured using the PM-lux reporter after treatment with different concentration of vancomycin (left) and EDTA (right) in WT (HB17325), sigM (HB17494) and yhdK (HB20833). A 20 min. treatment time was chosen, since this was shown previously to be optimal for monitoring SigM-dependent induction (Rojas-Tapias & Helmann, 2018), B. Fold change of maximum SigM activity (monitored using the PM-lux reporter) by vancomycin (left) and EDTA (right) in WT (HB17325) and mutants (PxylA-sigA, HB23619; yhdK, HB20833; PxylA-sigA yhdK, HB23937; PxylA-sigA yhdL, HB23939). Samples supplemented with 2% final concentration of xylose are labelled as +Xyl. C. Western blot of PM-lacZ, N-FLAG YhdL and C-FLAG YhdK of WT168 (HB22613), yhdK (HB22614) and PxylA-yhdLyhdK yhdL::kan strains (HB23948). A primary antibody against β-galactosidase was used in Western blot for monitoring PM-lacZ expression, and an anti-FLAG primary antibody for YhdL and YhdK. Part of the SDS-PAGE gel is shown as loading control. Samples were taken before (Time 0), or after treatment with vancomycin (final concentration of 2 μg ml−1). For strain yhdL PxylA-yhdLyhdK, LB medium contained 2% xylose. Western blots were performed at least 4 times and a representative image is shown. Quantitation of fold change (+VAN/-VAN) of band intensity of panel C at 20-minute time point. All data are presented as mean ± SEM (n≥3), with the mean value displayed above each bar.
The molecular mechanisms that result in release of SigM from the membrane-localized complex with its anti-sigma factors are unknown. For some ECF sigma factors, including B. subtilis SigV and SigW, the anti-sigma factors are subject to proteolysis upon induction (Heinrich et al., 2009; Hastie et al., 2013; Hastie et al., 2016). To begin to explore the possible role of proteolysis in release of SigM from YhdLK, we labeled YhdL and YhdK with a FLAG tag. The C-FLAG-YhdL and N-FLAG-YhdK proteins were functional as judged by a similar level of moenomycin resistance and PM-lux induction by moenomycin (Figure S3A, S3B). Since we were unable to detect a functional, epitope-tagged SigM (N-HA-SigM) by western blot and a C-HA-SigM is not functional (data not shown), we used a PM-lacZ strain and antibody against β-galactosidase as a proxy for SigM levels. Upon vancomycin addition, we observed a parallel increase in beta-galactosidase, YhdL and YhdK (Figure 5C). Since proteolysis of YhdL and/or YhdK could be masked by the synthesis of new, full-length proteins from the autoregulated sigM yhdL yhdK operon, we also monitored YhdL and YhdK levels in a construct where they were expressed at a constant level from a xylose-inducible promoter (HB23948). In this strain, beta-galactosidase levels increased ~8-fold after vancomycin treatment, suggesting that SigM was released from its anti-sigma factor complex (Figure 5C). In parallel, YhdL and YhdK protein levels were modestly reduced (~30–40%) (Figure 5C). This stands in contrast to RsiW and RsiV, which are degraded by multiple proteases including RasP, and almost completely disappear under inducing conditions (Heinrich et al., 2009; Hastie et al., 2013). Although YhdL and YhdK showed only a modest reduction in protein level (< 2-fold), this might suffice for the observed induction of SigM under these conditions. Alternatively, a mechanism other than proteolysis may be required for induction, as also speculated by others (Asai, 2018). Lastly, although RasP is the essential site 2 protease for activation of SigV and SigW, induction of SigM by moenomycin was unchanged in a rasP mutant (Figure S3B). Overall, our data indicate that YhdK is needed for perception of cell wall stress signals, and for the efficient function of the YhdL anti-sigma factor in sequestration of SigM. However, the YhdLK complex may not be the only pathway that can serve to activate SigM-dependent gene expression and an alternative pathway involving acetylation of the RNAP associated CshA helicase has also been suggested (Ogura & Asai, 2016).
Conclusions
Alternative sigma factors provide a powerful mechanism to redirect a subset of RNAP to new promoter sites with high selectivity (Helmann & Chamberlin, 1988; Feklistov et al., 2014). The ECF sigma subfamily is particularly abundant, and together with the ubiquitous use of one-component and two-component (histidine protein kinase/response regulator) systems, provides a third pillar of bacterial signal transduction (Staron et al., 2009). Membrane-bound anti-sigma factors, usually encoded immediately downstream of the gene encoding an ECF sigma factor, provide a frequent mechanism for regulating ECF sigma factor activity (Helmann, 2002; Alba & Gross, 2004; Ho & Ellermeier, 2012; Sineva et al., 2017; Asai, 2018). Mutation of the cognate anti-sigma factor usually leads to strong and constitutive expression of the corresponding sigma factor regulon. Since many ECF sigma factors positively autoregulate their own expression, anti-sigma factors can play an important role in preventing runaway signal amplification. In this work, we confirm and extend previous reports that 1) the absence of YhdL is lethal due to toxicity resulting from a high level of SigM (Horsburgh & Moir, 1999), 2) Cells lacking the multipass membrane protein YhdK also exhibit very high SigM activity and severe growth defects, and 3) YhdK is a component of the anti-SigM complex functionally important for the sensing of chemical inducers.
We developed a forward genetics-based approach to identify mutations that bypass SigM-dependent toxicity. Our suppressor screen yielded four distinct altered function mutations in the essential rpoB and rpoC genes. The resulting amino acid substitutions in core RNAP are predicted to be in close proximity to the linker region of ECF sigma factors, as judged from inspection of the recent structure of the M. tuberculosis SigL holoenzyme (Lin et al., 2018). The RpoCR335H mutant displayed both reduced SigM activity and greatly increased SigW activity. In addition, SigH activity was also moderately increased. These findings reveal that changes in core RNAP mapping at or near the core-sigma binding interface can differentially affect sigma factor activity. These core mutations are the amongst the first reported that differentially affect the activity of alternative sigma factors. While these studies were in progress, a similar effect was noted for a mutation affecting the coiled-coil motif of the β’ subunit which led to reduced binding of SigH, but little effect on SigB or SigA (Wang Erickson et al., 2017).
Mutations in the core subunits of RNAP emerge in response to diverse selection pressures, including of course selection by antibiotics that target RNAP directly. For example, rifampicin resistance mutations, which are of clinical relevance in M. tuberculosis, often map to rpoB (Goldstein, 2014), and may then lead to compensatory changes in other subunits (Brandis et al., 2012, Nusrath Unissa & Hanna, 2017). Mutations in RNA polymerase may also arise that increase or decrease the activity of regulatory proteins. For example, high levels of the B. subtilis Spx transcription factor interferes with gene expression and pseudorevertants that overcome these inhibitory effects map to the alpha subunit and reduce Spx binding (Nakano et al., 2000). Mutations in core RNAP may also affect the activity of alternative sigma factors: we previously identified a mutation in B. subtilis rpoC that increased beta-lactam resistance by increasing the activity of SigM and perhaps other ECF sigma factors (Lee et al., 2013). While it is clear that changes affecting the RNAP core enzyme can provide an answer for many diverse selection conditions, the underlying mechanisms can be quite difficult to discern. For example, in E. coli mutations in rpoB and rpoC often emerge in adaptive evolution experiments, but the underlying basis for the changes in the global transcriptional profile are incompletely understood (Conrad et al., 2010, Wytock et al., 2018, LaCroix et al., 2015, Avrani et al., 2017). Our results provide further evidence that, in organisms containing regulons controlled by alternative sigma subunits, remodeling of the core-sigma interface may allow a rapid shift in the global transcriptional landscape.
Experimental Procedures
Strains, primers, media and growth condition
All strains used in this work are listed in Table 1, and all DNA primers are listed in Table S1. Bacteria were routinely grown in liquid lysogeny broth (LB) with vigorous shaking, or on plates (1.5% agar; Difco) at 37 °C unless otherwise stated. LB medium contains 10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter. For sporulation assays, cells were grown in Difco Sporulation Medium (DSM) for 48 hours before heat treatment at 90 °C for 20 minutes. DSM (per 500 ml) includes 5 ml 10% KCl, 5 ml 1.2% MgSO4*7H2O, 4 g Bacto nutrient broth powder, 0.25 ml 1 M NaOH, and water to bring the volume to 500 ml. The medium is autoclaved at 121 °C for 20 minutes. After autoclaving, the following filter sterilized ingredients were added: 0.5 ml 1 M Ca(NO3)2, 0.05 ml 0.1 M MnCl2, and 0.05 ml 10 mM FeSO4. For growth curve measurements, 1 μl log phase culture (OD600 ~0.4) was inoculated into 200 μl of liquid medium per well in a Bioscreen 100-well plate, the plate was shaken vigorously and OD600 was measured every 15 minutes using an automated BioScreen growth analyzer. For concurrent measurements of growth and luciferase activity, 1 μl log phase culture (OD600 ~0.4) was inoculated into 100 μl of liquid medium per well in a 96-well plate. Plasmids were constructed using standard methods (Luo et al., 2010), and amplified in E. coli DH5α or TG1 before transforming into B. subtilis. For selection of transformants, 100 μg ml−1 ampicillin or 30 μg ml−1 kanamycin was used for E. coli. Antibiotics used for selection of B. subtilis transformants include: kanamycin 15 μg ml−1, spectinomycin 100 μg ml−1, macrolide-lincosamide-streptogramin B (MLS, contains 1 μg ml−1 erythromycin and 25 μg ml−1 lincomycin), and chloramphenicol 10 μg ml-1. For detection of beta-galactosidase activity, plates were supplemented with 60 μg ml−1 X-gal (5-bromo-4-chloro-3-indolyl β-D-galactopyranoside).
Table 1.
Strains used in this study
| Strain Number | Genotype | Reference/Constructiona |
|---|---|---|
| 168 | Wild type B. subtilis strain (trpC2) | Lab Stock |
| HB21107b | ganA::PxylA-yhdL-cat | pAX01-yhdL → 168, then the ermR cassette replaced by a cmR cassette using LFH PCR |
| HB17264 | yhdL::kan ganA::PxylA-yhdL-cat amyE::xylR-sigM, SPβ-PM-lacZ | xylR and sigM cloned into plasmid pDG1730, then integrated into amyE. |
| HB17325b | PM-lux | (Zhao et al., 2018) |
| HB17328b | PM-lacZ | pBS1ClacZ-PM → 168 |
| HB23909 | PM-lacZ ganA::PxylA-yhdL | pAX01-yhdL → HB17328, ermR |
| HB17474 | ΔsigM | BKE09520 → 168, then cassette removed |
| HB17494 | ΔsigM PM-lux | HB17325 → HB17474 |
| HB20830 | yhdK::erm | BKE09500 → 168 |
| HB20833 | yhdK::erm PM-lux | BKE09500 → HB17325 |
| HB22747 | sigMyhdLyhdK::tet | LFH PCR |
| HB22745 | PM-lux sigMyhdLyhdK::tet | HB17325 → HB22747 |
| HB23625 | sigMyhdLyhdK::tet thrC::PsigM(ΔPM)-sigM-erm | See Experimental Procedures |
| HB23682 | PM-lux sigMyhdLyhdK::tet thrC::PsigM(ΔPM)-sigM-erm | HB17325→HB23625 |
| HB21105b | ganA::PxylA-sigA | pAX01-sigA → 168 |
| HB22787 | ganA::PxylA-sigA yhdL::kan | HB17264→ HB21105 |
| HB22788 | ganA::PxylA-sigA yhdL::kan PM-lacZ | HB17328→HB22787 |
| HB23952 | ganA::PxylA-sigA yhdK::kan | BKK09500→ HB21105 |
| HB20928b | rpoB G3301A (D1101N) | CRISPR, see Experimental Procedures |
| HB20930b | rpoC G1004A (R335H) | CRISPR, see Experimental Procedures |
| HB20934 | rpoB G3301A (D1101N) yhdL::kan | HB17264→ HB20928 |
| HB20937 | rpoC G1004A (R335H) yhdL::kan | HB17264→ HB20930 |
| HB21075 | rpoB G3301A (D1101N) PM-lacZ | HB17328→ HB20928 |
| HB21077 | rpoC G1004A (R335H) PM-lacZ | HB17328→ HB20930 |
| HB21079 | rpoB G3301A (D1101N) PM-lux | HB17325→ HB20928 |
| HB21081 | rpoC G1004A (R335H) PM-lux | HB17325→ HB20930 |
| HB21083 | rpoB G3301A (D1101N) PM-lacZ yhdL::kan | HB17264→ HB21075 |
| HB21086 | rpoC G1004A (R335H) PM-lacZ yhdL::kan | HB17264→ HB21077 |
| HB21089 | rpoB G3301A (D1101N) PM-lux yhdL::kan | HB17264→ HB21079 |
| HB21092 | rpoC G1004A (R335H) PM-lux yhdL::kan | HB17264→ HB21081 |
| HB21166 | rpoB G3301A (D1101N) PM-lux yhdK::erm | BKE09500→ HB21079 |
| HB21168 | rpoC G1004A (R335H) PM-lux yhdK::erm | BKE09500→ HB21081 |
| HB22800 | ΔsigH | BKE00980 → 168, then cassette removed |
| HB22808b | PspoVG42-lux | pBS3Clux-PspoVG42 → 168 |
| HB22834 | ΔsigH PspoVG42-lux | HB22834→ HB22800 |
| HB22830 | rpoB G3301A (D1101N) PspoVG42-lux | HB22808→ HB20928 |
| HB22832 | rpoC G1004A (R335H) PspoVG42-lux | HB22808→ HB20930 |
| HB21116 | sigW::erm | BKE01730 →168 |
| HB21281 | rpoC G1004A (R335H) sigW::erm | BKE01730 → HB20930 |
| HB21285 | rpoB G3301A (D1101N) sigW::erm | BKE01730 → HB20928 |
| HB22578b | PfosB-lux | pBS3Clux-PfosB → 168 |
| HB22582 | PfosB-lux rpoC G1004A (R335H) | HB22578→ HB20930 |
| HB22610b | C-FLAG-yhdL N-FLAG-yhdK | CRISPR, see Experimental Procedures |
| HB22611 | C-FLAG-yhdL N-FLAG-yhdK PM-lux | HB17325→ HB22610 |
| HB22613 | C-FLAG-yhdL N-FLAG-yhdK PM-lacZ | HB17328→ HB22610 |
| HB22614 | C-FLAG-yhdL PM-lacZ yhdK::erm | LFH PCR product of HB22613 and BKE09500→ HB17328 |
| HB22673 | rasP::erm PM-lux | BKE16560 → HB17325 |
| HB23619 | ganA::PxylA-sigA PM-lux | HB21105 → HB17325 |
| HB23937 | ganA::PxylA-sigA PM-lux yhdK::kan | BKK09500→ HB23619 |
| HB23939 | ganA::PxylA-sigA PM-lux yhdL::kan | HB17264→ HB23619 |
| HB23927 | ganA::PxylA-C-FLAG-yhdL-N-FLAG-yhdK | pAX01-C-FLAG-yhdL-N-FLAG-yhdK, using chromosomal DNA of HB22610 as PCR template, ermR |
| HB23947 | ganA::PxylA-C-FLAG-yhdL-N-FLAG-yhdK PM-lacZ | HB17328→ HB23927 |
| HB23948 | ganA::PxylA-C-FLAG-yhdL-N-FLAG-yhdK PM-lacZ yhdL::kan | HB17264→ HB23947 |
“→” indicates transformation using DNA from the former (donor) into the latter (recipient). Plasmid vectors are described in Experimental Procedures.
Available at Bacillus Genetic Stock Center (http://www.bgsc.org/) by searching the strain (HB) number.
Genetic techniques
Chromosomal and plasmid DNA transformation was performed as previously stated (Zhao et al., 2016). The pBS1ClacZ based PM-lacZ reporter was constructed by inserting a DNA fragment containing the SigM autoregulatory promoter (amplified using primer 6808 and 6809) into plasmid pBS1ClacZ, and integrating the plasmid into the amyE locus (Radeck et al., 2013). The SPβ-PM-cat-lacZ was constructed as previously described (Cao and Helmann, 2002). The pPL82 plasmid-based Pspac(hy) overexpression constructs were linearized and integrated into the amyE locus (Quisel et al., 2001), the pAX01 plasmid-based PxylA overexpression constructs were linearized and integrated into the ganA locus (Hartl et al., 2001). Merodiploid sigM and xylR were constructed by cloning a copy of sigM and xylR (primers 6578, 6579, 6580, 6581) into plasmid pDG1730 (Guerout-Fleury et al., 1996) and then linearize and integrate the plasmid into the amyE locus, while keeping the native copy of sigM and the pAX01 originated xylR in the ganA locus. To avoid antibiotic marker conflicts, the ganA::PxylA-yhdL-cat constructs was made with LFH PCR (primers 6693, 6694, 6695, 6696) to replace the native ermR cassette from pAX01 to a cmR cassette. To select suppressors that can grow in the absence of YhdL, the depletion strain was grown in the presence of xylose to high density, washed twice to remove xylose, and then plated without xylose.
Markerless in-frame deletion mutants (indicated by Δ in Table 1) were constructed from BKE strains as described (Koo et al., 2017). Briefly, BKE strains were acquired from the Bacillus Genetics Stock Center (http://www.bgsc.org), chromosomal DNA was extracted, and the mutation containing an ermR cassette was transformed into our WT 168 strain. The ermR cassette was subsequently removed by introduction of the Cre recombinase carried on plasmid pDR244, which was later cured by growing at the non-permissive temperature of 42 °C. Gene deletions were confirmed by PCR screening using flanking primers. ΔPM-sigM was generated by PCR amplifying a sigM gene with its −10 region of PM mutated from CGTG to AAAG using primers DR194, DR197, DR196 and DR198. The mutant was then integrated into the thrC site with an ermR cassette. Unless otherwise described, all PCR products were generated using B. subtilis 168 strain chromosomal DNA as template. DNA fragments used for gene over-expression were verified by sequencing. Null mutant constructions were verified by PCR.
Mutations of rpoB and rpoC, as well as for labelling of YhdL and YhdK with a FLAG tag at their native loci, were done using a clustered regularly interspaced short palindromic repeats (CRISPR)-based mutagenesis method. Briefly, possible protospacer adjacent motifs (PAM) sites, which are NGG for Streptococcus pyogenes Cas9, were identified near the location of intended mutations. A DNA sequence of 12 bps upstream of the PAM site was used as seed sequence to search inside B. subtilis genome using BLAST. A potential off-target site is called when the 12 bps are found in an unintended location followed by a PAM site. If no off-target site was identified, the PAM site was chosen and 20 bps upstream of the site were used as sgRNA and cloned into vector pJOE8999 (Altenbuchner, 2016). The repair template was generated by joining two or more PCR products, with intended mutation introduced by PCR primers, and cloned into the pJOE8999-sgRNA vector. If the intended mutation changes the DNA sequence of PAM site or seed sequence region (12 bps upstream of PAM) such that the sgRNA will no longer bind the mutated sequence, no additional mutation was introduced. Otherwise, additional mutations were introduced to change the DNA sequence without affecting the encoded amino acid sequence, with preferred codons of B. subtilis (Moszer et al., 1999) used when introducing synonymous substitutions. The pJOE8999 derivative containing both the sgRNA and repair template was then cloned into competent cells of E. coli strain TG1 to produce concatemer plasmids, which were transformed into B. subtilis at 30°C. Transformants were then grown at 42°C to cure the plasmid, and intended mutations were confirmed by sequencing.
Disk Diffusion Assay
Disk diffusion assays were performed as previously described (Kingston et al., 2014). Briefly, overnight cultures in LB medium were inoculated into fresh LB medium and grown to exponential phase (OD600 ~0.4). A 100 μl aliquot of each culture was mixed with 4 ml of 0.75% LB soft agar (kept at 50°C) and directly poured onto a prewarmed 37°C LB plate (containing 15 ml of 1.5% LB agar). After the soft agar solidified, a filter paper disk with diameter of 6.5 mm was placed on top of the soft agar, and an antibiotic to be tested was added to the paper disk. The plate was kept at room temperature for 5 minutes to let the antibiotic absorb into the medium, and then moved to a 37°C incubator for 20 hours. The overall diameter of the inhibition zone was measured along two pairs of orthogonal lines, and zones of inhibition are reported as the average diameter of the four measurements for each biological replicate. The quantity of antibiotics or chemical used per disk is: vancomycin 50 μg, ampicillin 500 μg, EDTA 5 μl of 0.5 M solution (pH8.0), lysozyme 100 μg, or fosfomycin 500 μg.
Luciferase reporter construction and measurement
Luciferase reporter construction and measurement was performed as previous described (Zhao et al., 2018). The luciferase reporters were constructed by inserting the tested promoters into the multicloning sites of pBS3Clux (Radeck et al., 2013). The PM promoter from the sigM operon was amplified using primers 6808 and 6809. The SigW-dependent PfosB promoter was amplified using primer 7656 and 7657. The SigH dependent PspoVG42 promoter (Wang Erickson et al., 2017) was amplified using primers 8132, 8133, 8134 and 8135, with the point mutation introduced with the two middle primers. The SigE and SigK dependent promoters PspoIIM and PspoVFA were amplified using primers 8110, 8111, and 8112, 8113, respectively. The insert was confirmed by sequencing, linearized and integrated into the B. subtilis sacA locus. For luciferase measurements without antibiotic stress, 1 μl of exponentially growing cells were inoculated into 99 μl of fresh medium in a 96 well plate, incubated at 37 °C with shaking using a SpectraMax i3x plate reader, and OD600 and luminescence were measured every 12 min. For luciferase measurements with antibiotic stress, 100 μl of exponentially growing cells were added into each well of a 96 well plate, incubated at 37 °C with shaking using a SpectraMax i3x plate reader, and OD600 and luminescence were measured every 6 min. The data was analyzed using SoftMax Pro 7.0 software. Promoter activity was normalized by dividing the relative light units (RLU) by OD600.
Phase contrast microscopy
Cells were grown in liquid medium for the time indicated and loaded on saline (0.90% NaCl, w/v) agarose pads (0.8% final concentration) on a glass slide. Phase contrast images were taken using a Leica DMi8 microscope equipped with a 100x immersion objective and Leica Application Suite X software. Cell length and width were measured using Oufti per the software’s instruction (Paintdakhi et al., 2016).
WebLogo for conserved region of RpoB and RpoC
Conserved region near the suppressor mutations of RpoB and RpoC were generated using WebLogo (Crooks et al., 2004). The list of sixteen bacterial species used for RpoB and RpoC alignments includes Bacillus subtilis, Escherichia coli, Thermus aquaticus, Mycobacterium tuberculosis, Staphylococcus aureus, Streptococcus pneumoniae, Salmonella enterica Typhimurium, Legionella pneumophila, Chlamydia spp., Lactobacillus spp., Clostridium tetani, Enterococcus faecium, Pseudomonas aeruginosa, Vibrio cholerae, Paenibacillus polymyxa and Acetobacter malorum.
Western blot
Western blots were performed as described previously (Rojas-Tapias & Helmann, 2018). Briefly, cells were grown in 5 ml LB medium in a 20 ml test tube a 37 °C with vigorous shaking. After reaching exponential phase (OD600~0.3–0.4), 1 ml cells were aliquoted as pre-treatment sample, and 1 ml each were either treated with or without final concentration of 2 μg/ml vancomycin for 20 minutes with continued shaking at 37 °C. After treatment, cells were pelleted by centrifugation, resuspended in 100 μl buffer (containing 25 μl 4X Laemmli Sample buffer (Bio-Rad, USA), 10 μl 1M DTT, 65 μl H2O). Cells were then lysed by sonication and boiled in 100 °C sand bath for 4 minutes. Crude cell lysate was then loaded to a 4–20% SDS-PAGE stain-free gel for electrophoresis. Total proteins were visualized using ImageLab with stain-free gel protocol. Proteins were then transferred onto a PVDF membrane using the TransBlot Turbo Transfer System (Bio-Rad, USA). The membrane was first blocked with 5% protein blotting blocker dissolved in TTBS for 1 h at room temperature (RT), then incubated with anti-FLAG or β-galactosidase primary antibody in TTBS with 0.5% protein blotting blocker overnight at 4 °C. An anti-rabbit HRP-conjugated secondary antibody was incubated with the membrane in TTBS with 0.5% protein blotting blocker at RT for 1 h. The membrane was washed four times in TTBS and once in TBS at RT, and visualized using the Clarity Western ECL substrate (Bio-Rad) and ImageLab software. Band intensity was calculated using the ImageLab software and normalized using total protein amount according to SDS-PAGE gel image.
Colony size measurement
Colony size was measured using Fiji Image J (Schindelin et al., 2012). Briefly, bacterial cells were grown to mid-exponential phase (OD600~0.3–0.4), then serial diluted to desired concentrations. Diluted cells were plated onto fresh LB plates (15 ml medium per plate, the diameter of the plate is 10 cm and the height 15 mm, VWR, US, Catalog number 25384–342), and multiple dilution rates were used. Plates were incubated at 37 °C for 24 hours. Plates containing less than 100 separate single colonies were used for size measurement, because this number avoids reduced colony size due to crowdedness and nutrient limitation. Pictures of plates were taken with a ruler as a length reference, and colony size was measured using Fiji Image J per the software’s instruction. For each strain, at least 100 colonies were measured, and box and whisker plots were used for visualization.
Supplementary Material
Acknowledgements
We thank Dr. Tobias Doerr for valuable discussions and sharing equipment, Dr. Jason Peters and John Hawkins for suggestions for sgRNA design for CRISPR, and Daniel Rojas-Tapias for the mutation inactivating the autoregulatory PM promoter. We also thank Dr. Pete Chandrangsu and Ahmed Gaballa for help with experimental design and interpretation. This work was supported by a grant from the National Institutes of Health (R35GM122461) to JDH.
Contributor Information
MR. Heng Zhao, Email: hz285@cornell.edu.
Daniel M. Roistacher, Email: dmr293@cornell.edu.
References
- Ades SE, Connolly LE, Alba BM & Gross CA, (1999) The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev 13: 2449–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM & Gross CA, (2004) Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol 52: 613–619. [DOI] [PubMed] [Google Scholar]
- Altenbuchner J, (2016) Editing of the Bacillus subtilis Genome by the CRISPR-Cas9 System. Appl Environ Microbiol 82: 5421–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K, (2018) Anti-sigma factor-mediated cell surface stress responses in Bacillus subtilis. Genes Genet Syst 92: 223–234. [DOI] [PubMed] [Google Scholar]
- Avrani S, Bolotin E, Katz S & Hershberg R, (2017) Rapid Genetic Adaptation during the First Four Months of Survival under Resource Exhaustion. Mol Biol Evol 34: 1758–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandis G, Wrande M, Liljas L & Hughes D, (2012) Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol 85: 142–151. [DOI] [PubMed] [Google Scholar]
- Cao M, Bernat BA, Wang Z, Armstrong RN & Helmann JD, (2001) FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol 183: 2380–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M & Helmann JD (2002) Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J Bacteriol 184: 6123–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AN, Lewerke LT, Hastie JL & Ellermeier CD, (2018) Signal Peptidase Is Necessary and Sufficient for Site 1 Cleavage of RsiV in Bacillus subtilis in Response to Lysozyme. J Bacteriol 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE et al. (2010) RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A 107: 20500–20505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W & Helmann JD, (2008) The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol 67: 830–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Sharon BD, Darst SA & Gross CA, (2014) Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68: 357–376. [DOI] [PubMed] [Google Scholar]
- Ganguly A & Chatterji D, (2012) A comparative kinetic and thermodynamic perspective of the sigma-competition model in Escherichia coli. Biophys J 103: 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BP, (2014) Resistance to rifampicin: a review. J Antibiot (Tokyo) 67: 625–630. [DOI] [PubMed] [Google Scholar]
- Grigorova IL, Phleger NJ, Mutalik VK & Gross CA, (2006) Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci U S A 103: 5332–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N & Stragier P, (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57–61. [DOI] [PubMed] [Google Scholar]
- Hartl B, Wehrl W, Wiegert T, Homuth G & Schumann W, (2001) Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J Bacteriol 183: 2696–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie JL, Williams KB, Bohr LL, Houtman JC, Gakhar L & Ellermeier CD, (2016) The Anti-sigma Factor RsiV Is a Bacterial Receptor for Lysozyme: Co-crystal Structure Determination and Demonstration That Binding of Lysozyme to RsiV Is Required for sigmaV Activation. PLoS Genet 12: e1006287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie JL, Williams KB & Ellermeier CD, (2013) The activity of sigmaV, an extracytoplasmic function sigma factor of Bacillus subtilis, is controlled by regulated proteolysis of the anti-sigma factor RsiV. J Bacteriol 195: 3135–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich J, Hein K & Wiegert T, (2009) Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol Microbiol 74: 1412–1426. [DOI] [PubMed] [Google Scholar]
- Heinrich J & Wiegert T, (2006) YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol Microbiol 62: 566–579. [DOI] [PubMed] [Google Scholar]
- Ho TD & Ellermeier CD, (2012) Extra cytoplasmic function sigma factor activation. Curr Opin Microbiol 15: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ & Moir A, (1999) Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol Microbiol 32: 41–50. [DOI] [PubMed] [Google Scholar]
- Jervis AJ, Thackray PD, Houston CW, Horsburgh MJ & Moir A, (2007) SigM-responsive genes of Bacillus subtilis and their promoters. J Bacteriol 189: 4534–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston AW, Zhao H, Cook GM & Helmann JD, (2014) Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: implications for the mechanism of resistance mediated by the BceAB transporter. Mol Microbiol 93: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, et al. (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100: 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, et al. , (2017) Construction and Analysis of Two Genome-Scale Deletion Libraries for Bacillus subtilis. Cell Syst 4: 291–305 e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, et al. (2015) Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 81: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers AP, Keithly ME, Kim K, Cook PD, Stec DF, Hines KM, et al. (2012) Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org Lett 14: 5207–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Nam KH & Helmann JD, (2013) A mutation of the RNA polymerase beta’ subunit (rpoC) confers cephalosporin resistance in Bacillus subtilis. Antimicrob Agents Chemother 57: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Mandal S, Degen D, Cho M, Feng Y, Das K & Ebright RH, (2018) Structural basis of ECF-sigma-factor-dependent transcription initiation. bioRxiv. www.biorxiv.org/content/early/2018/07/31/381020 [DOI] [PMC free article] [PubMed]
- Luo Y, Asai K, Sadaie Y & Helmann JD, (2010) Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extracytoplasmic function sigma factors. J Bacteriol 192: 5736–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y & Helmann JD, (2012) Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83: 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Fujita N & Ishihama A, (2000) Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res 28: 3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, (2013) Signaling diversity and evolution of extracytoplasmic function (ECF) sigma factors. Curr Opin Microbiol 16: 148–155. [DOI] [PubMed] [Google Scholar]
- Mascher T, Hachmann AB & Helmann JD, (2007) Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol 189: 6919–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG & Rudner DZ, (2015) MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A 112: 6437–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moszer I, Rocha EP & Danchin A, (1999) Codon usage and lateral gene transfer in Bacillus subtilis. Curr Opin Microbiol 2: 524–528. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Zhu Y, Liu J, Reyes DY, Yoshikawa H & Zuber P, (2000) Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase alpha can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol Microbiol 37: 869–884. [DOI] [PubMed] [Google Scholar]
- Nusrath Unissa A & Hanna LE, (2017) Molecular mechanisms of action, resistance, detection to the first-line anti tuberculosis drugs: Rifampicin and pyrazinamide in the post whole genome sequencing era. Tuberculosis (Edinb) 105: 96–107. [DOI] [PubMed] [Google Scholar]
- Ogura M & Asai K, (2016) Glucose Induces ECF Sigma Factor Genes, sigX and sigM, Independent of Cognate Anti-sigma Factors through Acetylation of CshA in Bacillus subtilis. Front Microbiol 7: 1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I & Jacobs-Wagner C, (2016) Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Dies M, Lin Y, Hormoz S, Smith-Unna SE, Quinodoz S et al. (2018) Molecular Time Sharing through Dynamic Pulsing in Single Cells. Cell Syst 6: 216–229 e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ & Hilbert DW, (2004) Sporulation of Bacillus subtilis. Curr Opin Microbiol 7: 579–586. [DOI] [PubMed] [Google Scholar]
- Quisel JD, Burkholder WF & Grossman AD, (2001) In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol 183: 6573–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck J, Kraft K, Bartels J, Cikovic T, Durr F, Emenegger J et al. (2013) The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J Biol Eng 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AA, Sharma SV, Strankman AW, Duran SR, Rawat M & Hamilton CJ, (2013) Mechanistic studies of FosB: a divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem J 451: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Tapias DF & Helmann JD, (2018) Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol Microbiol 107: 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T et al. , (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobel S, Zellmeier S, Schumann W & Wiegert T, (2004) The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol 52: 1091–1105. [DOI] [PubMed] [Google Scholar]
- Sineva E, Savkina M & Ades SE, (2017) Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr Opin Microbiol 36: 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BM, Castro TL, Carvalho RD, Seyffert N, Silva A, Miyoshi A & Azevedo V, (2014) sigma(ECF) factors of gram-positive bacteria: a focus on Bacillus subtilis and the CMNR group. Virulence 5: 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H & Mascher T, (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74: 557–581. [DOI] [PubMed] [Google Scholar]
- Stragier P & Losick R, (1996) Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30: 297–241. [DOI] [PubMed] [Google Scholar]
- Thackray PD & Moir A, (2003) SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J Bacteriol 185: 3491–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Erickson AF, Deighan P, Garcia CP, Weinzierl ROJ, Hochschild A & Losick R, (2017) An Amino Acid Substitution in RNA Polymerase That Inhibits the Utilization of an Alternative Sigma Factor. J Bacteriol 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wytock TP, Fiebig A, Willett JW, Herrou J, Fergin A, Motter AE & Crosson S, (2018) Experimental evolution of diverse Escherichia coli metabolic mutants identifies genetic loci for convergent adaptation of growth rate. PLoS Genet 14: e1007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M, Asai K, Sadaie Y & Yoshikawa H, (2004) Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150: 591–599. [DOI] [PubMed] [Google Scholar]
- Zellmeier S, Schumann W & Wiegert T, (2006) Involvement of Clp protease activity in modulating the Bacillus subtilissigmaw stress response. Mol Microbiol 61: 1569–1582. [DOI] [PubMed] [Google Scholar]
- Zhao H, Roistacher DM & Helmann JD, (2018) Aspartate deficiency limits peptidoglycan synthesis and sensitizes cells to antibiotics targeting cell wall synthesis in Bacillus subtilis. Mol Microbiol 109: 826–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun Y, Peters JM, Gross CA, Garner EC & Helmann JD, (2016) Depletion of Undecaprenyl Pyrophosphate Phosphatases Disrupts Cell Envelope Biogenesis in Bacillus subtilis. J Bacteriol 198: 2925–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.