Summary
Superoxide dismutase (SOD) is widely assumed to play a role in the detoxification of reactive oxygen species caused by environmental stresses. We found a characteristic expression of manganese SOD 1 (MSD1) in a heat‐stress‐tolerant cultivar of rice (Oryza sativa). The deduced amino acid sequence contains a signal sequence and an N‐glycosylation site. Confocal imaging analysis of rice and onion cells transiently expressing MSD1‐YFP showed MSD1‐YFP in the Golgi apparatus and plastids, indicating that MSD1 is a unique Golgi/plastid‐type SOD. To evaluate the involvement of MSD1 in heat‐stress tolerance, we generated transgenic rice plants with either constitutive high expression or suppression of MSD1. The grain quality of rice with constitutive high expression of MSD1 grown at 33/28 °C, 12/12 h, was significantly better than that of the wild type. In contrast, MSD1‐knock‐down rice was markedly susceptible to heat stress. Quantitative shotgun proteomic analysis indicated that the overexpression of MSD1 up‐regulated reactive oxygen scavenging, chaperone and quality control systems in rice grains under heat stress. We propose that the Golgi/plastid MSD1 plays an important role in adaptation to heat stress.
Keywords: high‐temperature tolerance, Golgi, grain quality, Oryza sativa L., plastid, superoxide dismutase
Introduction
Impairment of rice (Oryza sativa L.) grain filling under global warming is a major threat facing Asian countries. Daily mean temperatures above 26 °C during the early ripening period of japonica rice compromises yields through decreases in grain size and quality (Morita et al., 2004; Peng et al., 2004; Tashiro and Wardlaw, 1991). Perfect grains are fully rounded, transparent and filled with normal starch granules. A chalky appearance reduces commercial value because of increased cracking during polishing (Fitzgerald et al., 2009) and poorer cooking quality (Singh et al., 2003; Tsutsui et al., 2013). Scanning microscope images of chalky areas of grain ripened under heat stress show loosely packed rounded starch granules (Evers and Juliano, 1976; Ishimaru et al., 2009; Tashiro and Wardlaw, 1991). The air spaces among these abnormal starch granules refract light, making the grain appear white. Occasional small pits on the surface of the starch granules suggest attack by starch‐degrading enzymes (Iwasawa et al., 2009; Zakaria et al., 2002); the suppression of α‐amylase genes improved the quality of rice grains ripened under heat stress (Hakata et al., 2012). It is widely recognized that heat stress lowers the activity of starch synthesis enzymes (Jiang et al., 2003; Umemoto and Terashima, 2002; Yamakawa et al., 2007). Mutants deficient in genes for starch synthesis enzymes exhibited dramatic changes in grain phenotype, including shape and chalkiness (Fujita et al., 2011; Kubo et al., 1999; Nishi et al., 2001; Tanaka et al., 2004). Furthermore, novel factors such as FLOURY ENDOSPERM2 (FLO2), GLUTELIN PRECURSOR MUTANT6 (GLUP6) and GLUTELIN PRECURSOR ACCUMULATION3 (GAP3) have been shown to be involved in the regulation of rice grain size and starch quality (Fukuda et al., 2013; Ren et al., 2014; She et al., 2010). FLO2 contains a tetratricopeptide repeat motif that interacts with late‐embryogenesis and basic helix‐loop‐helix proteins (She et al., 2010). GLUP6 is a guanine nucleotide exchange factor involved in intracellular transport from the Golgi apparatus to the protein storage vacuole, and the glup6 mutant accumulates an abnormally large amount of proglutelin (Fukuda et al., 2013). GAP3 is involved in post‐Golgi vesicular traffic for vacuolar protein sorting (Ren et al., 2014). In addition, redox regulation may affect seed maturation and quality (Onda and Kawagoe, 2011; Onda et al., 2011). Thus, the mechanism of grain chalkiness caused by heat stress may be highly complex.
Abiotic stresses, including high light, drought, salinity and heat, lead to the accumulation of reactive oxygen species (ROS) such as superoxide (), hydroxyl radicals (•OH) and hydrogen peroxide (H2O2; Apel and Hirt, 2004). ROS damage multiple cellular components, interfering with lipid peroxidation (Niki et al., 2005), breaking DNA strands (Brawn and Fridovich, 1981) and inactivating enzymes (Fucci et al., 1983). On the other hand, they also serve as signalling molecules, regulating processes including pathogen defence, programmed cell death and stomatal behaviour (Apel and Hirt, 2004). Although ROS are produced predominantly and continuously in chloroplasts, mitochondria and peroxisomes, the production and scavenging of ROS must be strictly controlled in the absence of stress. Enzymatic ROS scavenging mechanisms involve superoxide dismutase (SOD), ascorbate peroxidase, glutathione peroxidase and catalase (Apel and Hirt, 2004).
Superoxide dismutase catalyses the conversion of to H2O2; it is responsible primarily for defence against oxidative stress. There are three classes of SODs categorized by their metal cofactor: Fe SOD, Mn SOD and Cu/Zn SOD (Fridovich, 1975). Plant SODs have different subcellular localizations. Typically, Mn SOD is localized to the mitochondria, Fe SOD to the plastids and Cu/Zn SOD to the plastids and cytosol (Bowler et al., 1992; Kliebenstein et al., 1998). Peroxisomal and extracellular Cu/Zn SODs also exist (Bueno et al., 1995; Streller and Wingsle, 1994). Numerous attempts have been made to enhance stress tolerance in plants by modifying the production of SOD enzymes. Ectopic production of cytosolic Cu/Zn SOD improved stress tolerance in tobacco (Faize et al., 2011), potato (Perl et al., 1993), sugar beet (Tertivanidis et al., 2004) and plum (Diaz‐Vivancos et al., 2013). Overproduction of chloroplastic Cu/Zn SOD, Fe SOD and Mn SOD (fused to a chloroplast transit peptide) also increased stress resistance in tobacco (Badawi et al., 2004; van Camp et al., 1994, 1996; Sen Gupta et al., 1993; Slooten et al., 1995), potato (Perl et al., 1993), sugar beet (Tertivanidis et al., 2004), cotton (Payton et al., 2001) and alfalfa (McKersie et al., 2000). Transgenic rice overproducing cytosolic Cu/Zn SOD from mangrove (Avicennia marina) tolerated drought stress better than untransformed plants (Prashanth et al., 2008). Rice transformed with a yeast mitochondrial Mn SOD fused to the transit peptide of glutamine synthase conferred resistance to salt stress (Tanaka et al., 1999). Furthermore, rice transformed with pea (Pisum sativum) mitochondrial Mn SOD fused to the transit peptide of pea Cu/Zn SOD under the control of an oxidative stress‐inducible promoter was more resistant to oxidative stress induced by methyl viologen or polyethylene glycol (Wang et al., 2005).
We have been searching for candidate genes involved in heat‐stress tolerance during seed development to improve the formation of normal rice grains under a warming climate. In proteomic analysis, we detected a characteristic expression behaviour of Mn SOD in developing seeds of the heat‐resistant cultivar Yukinkomai. This Mn SOD exhibited a unique subcellular localization that has never previously been described in the literature. Here, we report that control of the Golgi/plastid‐type Mn SOD1 (MSD1) expression regulates tolerance to heat stress during grain filling of rice.
Results
Identification of Golgi/plastid‐type Mn SOD (MSD1)
We examined the heat susceptibilities of three rice cultivars, Yukinkomai, Yukinosei and Todorokiwase, during seed development from 2004 to 2008. The plants were grown in paddy fields with irrigation water at either ambient temperature or 35 °C during the heading, ripening and maturity stages. The daily mean temperature at around the panicles in the warm‐water field was 1.4–1.9 °C higher than that in the ambient‐water field (25.4 °C). The percentage of damaged grains in Yukinkomai was about 22% in both treatments (Figure 1), indicating that Yukinkomai is tolerant to high temperatures during development. In contrast, that of Todorokiwase increased from 35% to 44%. Yukinosei was intermediate (Figure 1). To search for genes involved in the heat tolerance of Yukinkomai, we used a proteomic approach. As rice is sensitive to heat stress at an early stage of seed development (Nagata et al., 2004; Satake and Yoshida, 1978), we separated grain proteins of Yukinkomai, Yukinosei and Todorokiwase at 4 days after flowering (DAF) by two‐dimensional polyacrylamide gel electrophoresis (2D‐PAGE). The separation profiles showed changes in the production of stress‐responsive proteins, including heat shock proteins 70 (HSP70) and 16.9 (HSP16.9), 20S proteasome αF, ABA‐inducible protein (R40g2), alcohol dehydrogenase (ADH) and MSD1 (Figure 2). In the heat‐tolerant Yukinkomai, 20S proteasome αF, ADH and HSP16.9 were up‐regulated and R40g2 were down‐regulated under heat stress (Figure 2a,b). In the susceptible Todorokiwase, in contrast, HSP70, HSP16.9 and MSD1 were up‐regulated (Figure 2e,f). Those in Yukinosei were intermediate (Figure 2c,d). We focused on MSD1, which was characteristically and highly expressed in developing seeds of Yukinkomai in both treatments (Figure 2a,b).
Figure 1.
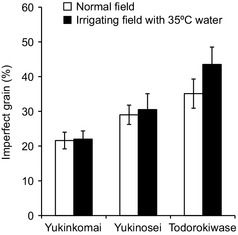
Proportions of imperfect grains of rice cultivars Yukinkomai, Yukinosei and Todorokiwase irrigated with water at ambient temperature (□) or 35 °C (■) from heading to maturity.
Figure 2.
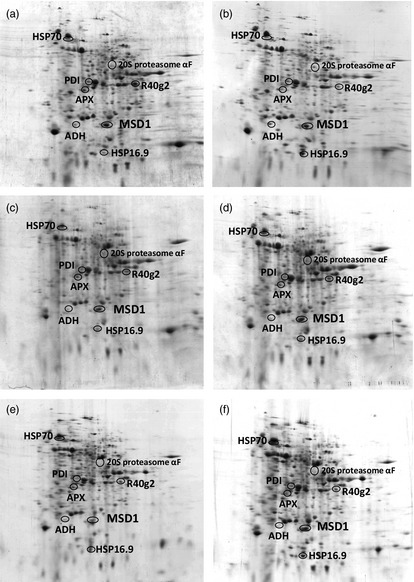
2D‐PAGE separation profiles of proteins extracted from 4 days after flowering (DAF) grains of (a, b) Yukinkomai, (c, d) Yukinosei and (e, f) Todorokiwase grown under (a, c, e) normal or (b, d, f) heat stress treatment. Protein extracts were separated by isoelectric focusing followed by SDS‐PAGE. Proteins identified included heat shock proteins 70 (HSP70; 66 kDa, pI 4.8), 20S proteasome αF (46 kDa, pI 5.7), protein disulphide isomerase (PDI; 40 kDa, pI 5.3), ABA‐inducible protein (R40g2; 38 kDa, pI 6.8), ascorbate peroxidase (APX; 36 kDa, pI 5.2), alcohol dehydrogenase (ADH; 28 kDa, pI 5.0), Mn superoxide dismutase 1 (MSD1; 28 kDa, pI 5.6), HSP16.9 (18 kDa, pI 5.6) were identified in the 2D‐gels.
The RiceXPro public microarray database (http://ricexpro.dna.affrc.go.jp/) shows that the MSD1 gene (OsMSD1) is actively expressed throughout the rice plant, particularly in the embryo and endosperm of developing seeds. Our gel‐based proteomic analysis of developing seeds supports the view that MSD1 is a major constituent in the seed proteome (Figure 2a,b). OsMSD1 is located in the centre of chromosome 5 (Figure 3a). The cDNA is 901 bp in length, encoding 231 amino acid residues that form a 24.9‐kDa precursor protein (Figure 3b). MSD1 is mitochondrial enzyme in both monocots and dicots (Kliebenstein et al., 1998; del Río et al., 2003; White and Scandalios, 1988; Wu et al., 1999). Analyses by the PSORT algorithm (http://psort.hgc.jp/form.html) predicted an N‐terminal mitochondrion‐targeting sequence in the precursor proteins of MSD1 of Arabidopsis, maize, wheat and pea (Figure 3b). Indeed, pea MSD1 is localized chiefly in mitochondria (del Río et al., 2003). However, the prediction by PSORT and signalP (http://www.cbs.dtu.dk/services/SignalP/) showed that the rice MSD1 precursor's N‐terminal sequence potentially acts as signal to the endoplasmic reticulum (ER; Figure 3b; Sakamoto et al., 1993).
Figure 3.
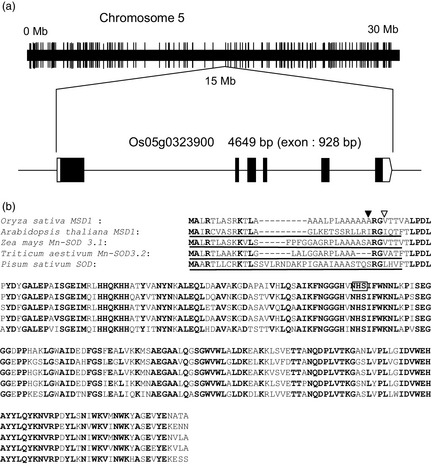
OsMSD1. (a) Structure and position of MSD1 (Os05g0323900) on chromosome 5. Black boxes indicate exons. (b) Alignments of predicted amino acid sequences of the deduced MSD1 proteins of possible orthologous genes from Oryza sativa, Arabidopsis thaliana, Zea mays, Triticum aestivum and Pisum sativum. Conserved amino acids are boldfaced. Underlines represent mitochondrion‐targeting sequence predicted by PSORT. In the rice MSD1 sequence, arrowheads show possible cleavage sites of the signal peptide predicted by (▿) PSORT and (▾) signalP. An N‐glycosylation site is boxed.
To determine the subcellular localization of rice MSD1, we analysed the transient expression of OsMSD1 fused with a gene for yellow fluorescent protein (YFP) in rice and onion epidermal cells, using particle bombardment. In rice cells, confocal laser scanning microscopy showed that the distribution of MSD1‐YFP coincided well with the autofluorescence of chloroplasts (Figure 4). In onion cells, MSD1‐YFP revealed numerous particulate structures (Figure 5a,c). When MSD1‐YFP was cobombarded with a sequence encoding a trans‐Golgi marker (sialyltransferase, ST) fused at the transmembrane domain to monomeric red fluorescent protein (ST‐mRFP) into onion cells, MSD1‐YFP fluorescence overlapped well with the ST‐mRFP‐labelled trans‐Golgi vesicles (Figure 5a). The GTPases ARF1 and SAR1 are essential for membrane trafficking between the ER and the Golgi apparatus in higher plant cells. Expression of dominant‐negative ARF1 or constitutively active SAR1 mutant proteins, which are defective in GTPase cycling, prevents the ER‐to‐Golgi traffic (Takeuchi et al., 2000, 2002). Golgi‐resident proteins and secretory and vacuolar proteins are therefore retained in the ER with such mutants (Takeuchi et al., 2000, 2002). We examined the effects of dominant‐negative and constitutive‐active mutants of ARF1 and SAR1 on the subcellular distribution of MSD1‐YFP. We simultaneously expressed MSD1‐YFP, the trans‐Golgi marker ST‐mRFP, and either AtARF1(T31N), AtARF1(Q71L), or AtSAR1(H74L) in onion cells. Both MSD1‐YFP‐ and ST‐mRFP‐labelled vesicles were rearranged and remerged into tubular structures, which are probably part of the ER network, in cells expressing the mutants (Figure 5b).
Figure 4.
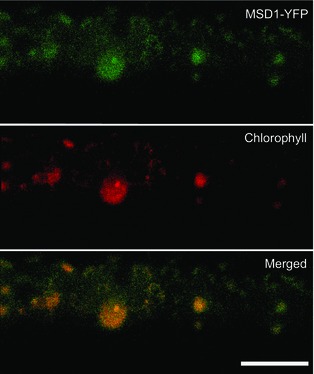
Expression and localization of MSD1‐YFP in rice cells. Rice cells bombarded with MSD1‐YFP were observed by laser scanning microscopy. Top: MSD1‐YFP; middle: chlorophyll autofluorescence; bottom: merged. Panels are stacks of 30 images per cell, acquired from the top to the middle of the cell, every 1–2 μm. MSD1‐YFP colocalized with chlorophyll autofluorescence. Bar = 10 μm.
Figure 5.
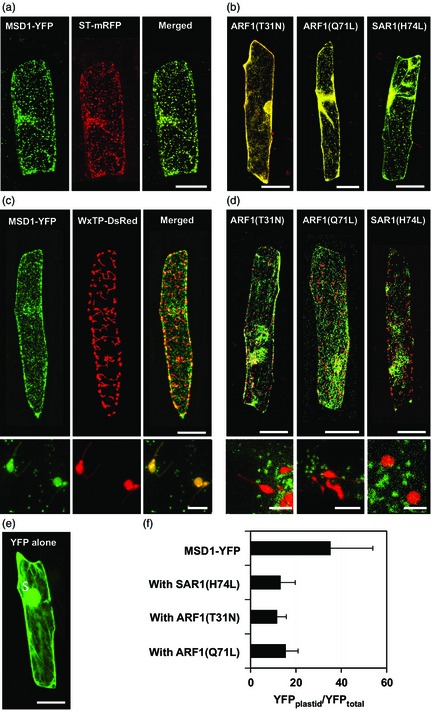
Expression and localization of MSD1‐YFP in onion epidermal cells. (a) Onion cells expressing MSD1‐YFP and ST‐mRFP. Left: MSD1‐YFP (green); middle: ST‐mRFP (red); right: merged. Panels are stacks of 30 images per cell, acquired from the top to the middle of the cell, every 1–2 μm. MSD1‐YFP colocalized with Golgi marker ST‐mRFP. Bar = 100 μm. (b) Effects of AtARF1(T31N), AtARF1(Q71L) and AtSAR1(H74L) on the distribution of MSD1‐YFP and ST‐mRFP. All images merge YFP with mRFP. Both MSD1‐YFP and ST‐mRFP were rearranged and remerged into ER tubular structures. (c) Onion cells expressing MSD1‐YFP and WxTP‐DsRed. Left: MSD1‐YFP (green); middle: WxTP‐DsRed (red); right: merged. MSD1‐YFP overlapped with the plastid marker WxTP‐DsRed. Bar = 100 μm. Bottom panels show close‐up views of plastids; bar = 5 μm. (d) Effects of AtARF1(T31N), AtARF1(Q71L) and AtSAR1(H74L) on the distribution of MSD1‐YFP and WxTP‐DsRed. All images merge YFP with DsRed. MSD1‐YFP and WxTP‐DsRed were distributed separately in cells. Bars = 100 μm (top) and 5 μm (bottom). (e) Onion cell expressing YFP alone. (f) Proportion of plastid localization of MSD1‐YFP in the presence of AtSAR1(H74L), AtARF1(T31N) or AtARF1(Q71L). Values are means ± SD (n = 8–11) of ratios of the fluorescence intensity of YFP in the plastid to YFP in the whole cell (YFP plastid/YFPtotal, %).
Recent investigations have revealed the dual targeting of proteins to Golgi apparatus and plastids in Arabidopsis (Villarejo et al., 2005), rice (Asatsuma et al., 2005; Chen et al., 2004; Kaneko et al., 2011, 2014; Kitajima et al., 2009; Nanjo et al., 2006) and photosynthetic micro‐organisms (van Dooren et al., 2001; Nowack and Grossman, 2012; Sláviková et al., 2006). To test the possibility of plastid‐targeting of MSD1, we cobombarded MSD1‐YFP with a sequence encoding a plastid marker, the transit peptide of Waxy (Klösgen and Weil, 1991) fused to red fluorescent protein from Discosoma sp. (WxTP–DsRed), into onion cells. MSD1‐YFP was notably colocalized with the plastids visualized by WxTP–DsRed (Figure 5c). Simultaneous expression of MSD1‐YFP, the plastid marker WxTP–DsRed, and either AtARF1(T31N), AtARF1(Q71L), or AtSAR1(H74L) indicated that the plastid targeting of MSD1‐YFP was inhibited in cells expressing the ARF1 and SAR1 mutant proteins (Figure 5d,f). The overall results clearly indicate that MSD1 is a multilocalizing protein that is targeted to the interior of plastids from the Golgi apparatus via the secretory pathway.
Overexpression and suppression of MSD1 affect the grain quality of rice ripened under heat stress
To determine the possible stress‐adapting function of MSD1 in ripening seeds of rice, we generated transgenic overexpressor (OE) plants with the maize Ubiquitin‐1 promoter (PUbi1) fused to MSD1 (MSD1OE) by Agrobacterium‐mediated transformation. It was reported that PUbi1‐controlled genes are highly expressed in various rice tissues (Cornejo et al., 1993). The expression profiles of MSD1 mRNA in leaves, roots and developing seeds of Nipponbare wild type (WT) and MSD1OE revealed a constitutive high expression of MSD1 in MSD1OE plants (Figure 6a–c). Furthermore, H2O2 increased in the developing seeds and young seedlings of MSD1OE (Figure 7). The ratio of H2O2 content between MSD1OE and WT seedlings under hot condition revealed that H2O2 formation increased in MSD1OE under heat stress (Figure 7b). When plants were incubated at normal or high temperatures after heading, the ratios of perfect grains harvested were 78% (WT) and 83% (MSD1OE) at 28/23 °C, 77% (WT) and 88% (MSD1OE) at 30/23 °C, and 26% (WT) and 60% (MSD1OE) at 33/28 °C (Figure 6d–f). Under heat stress, the grain quality of MSD1OE was significantly greater than that of WT (Figures 6f and S1). To suppress the expression of MSD1 in developing seeds, we used a 696‐bp fragment of MSD1 cDNA which contains no sequence of more than 21 nucleotides conserved with other rice SODs to construct RNA interference (RNAi) binary vectors under the control of the promoter of the developing endosperm‐specific Waxy (PWx) by arranging two identical fragments derived from MSD1 in a tail‐to‐tail manner, yielding a vector generating artificial hairpin‐structure transcripts (Figure S2). We generated two transgenic knock‐down (KD) rice plants transformed with PWx fused to MSD1 RNAi, designated Nipponbare MSD1KD and Yukinkomai MSD1KD. Both transformants were grown under heat stress after heading. The expression of MSD1 mRNA in developing seeds decreased to 18% of WT in Nipponbare MSD1KD and 53% in Yukinkomai MSD1KD (Figure 6g,i), along with significant decreases in the proportion of perfect grains (to 12% and 71%, respectively; Figure 6h,j). The overall results indicate that the constitutive high expression of MSD1 was involved in maintaining the quality of rice grains produced under heat stress during ripening.
Figure 6.
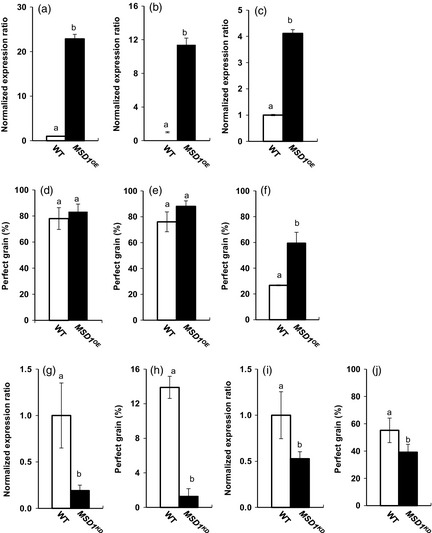
Evaluation of heat‐stress tolerance during grain filling of rice with overexpression of MSD1 (MSD1OE) or developing endosperm‐specific suppression of MSD1 (MSD1KD). (a–c) Expression profiles of MSD1 mRNA in different organs of Nipponbare wild type (WT) and MSD1OE. (a) Leaf blades and (b) roots at vegetative stage and (c) developing grains at 5 days after flowering (DAF) were harvested and used for fluorescence‐based quantitative real‐time PCR. Values are means ± SD (n = 3). (d–f) Nipponbare WT and MSD1OE plants were incubated under (d) 28/23 °C (12/12 h), (e) 30/23 °C (12/12 h) or (f) 33/28 °C (12/12 h) after heading, and the appearance quality of harvested grains was evaluated. Values are means ± SD (n = 3–7) of proportions of perfect grains. (g, h) Nipponbare WT and MSD1KD plants were incubated under 33/28 °C after heading; MSD1 mRNA in developing grains at 5 DAF was quantified and appearance quality was evaluated (n = 5). (i, j) Yukinkomai WT and MSD1KD plants were incubated under 33/28 °C after heading; MSD1 mRNA in developing grains at 5 DAF was quantified, and appearance quality was evaluated (n = 3–7). The ratio of MSD1 mRNA to 18S rRNA in each WT was set to 1. Columns with the same letter are not significantly different (P < 0.05, Student's t‐test).
Figure 7.
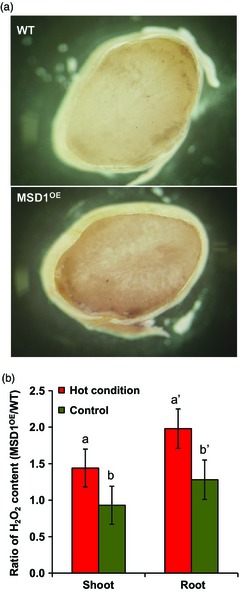
Increase of H2O2 formation in developing and germinating seeds of MSD1OE. (a) Developing seeds of Nipponbare WT and MSD1OE at 10 days after flowering (DAF) were stained with diaminobenzidine. (b) H2O2 contents in the shoots and roots of WT and MSD1OE seedlings at 7 days after imbibition. Hot condition = 33/28 °C (12/12 h); control condition = 28/23 °C (12/12 h). Values are means ± SD (n = 3–4). Columns with the same letter are not significantly different (P < 0.05, Student's t‐test).
Proteomic characterization of developing seeds of MSD1OE under heat stress
To clarify how constitutive high expression of MSD1 leads to adaptation to heat stress, we carried out quantitative shotgun proteomic analysis of ripening seeds. Proteins extracted from ripening seeds of Nipponbare WT and MSD1OE grown under control (28/23 °C) and heat stress (33/28 °C) conditions at 4 and 10 DAF were labelled by iTRAQ (isobaric tag for relative and absolute quantitation), followed by tandem mass spectrometry (MS/MS) analysis. Under heat stress, 79 proteins (~6% of all identified proteins), including storage and allergen proteins, were down‐regulated and 219 (~16%) were up‐regulated in the ripening seeds of MSD1OE relative to WT (Table S1). Under the control condition, however, the characteristic response of MSD1OE did not appear. Under high temperature, scavengers of reactive oxygen species (ROS), including Cu/Zn SOD, peroxiredoxins, thioredoxin, peptide methionine sulfoxide reductase, ascorbate peroxidases, monodehydroascorbate reductase and NADH‐ubiquinone oxidoreductase, were markedly up‐regulated in MSD1OE relative to WT (Figure 8 upper panel). Under the control condition, however, changes were minor. Several HSPs, chaperones, chaperonins, calreticulin, proteasome components and S‐phase kinase‐associated protein 1 were also up‐regulated in MSD1OE under heat stress (Figure 8 lower panel), but glutelin, prolamin and allergen family proteins were down‐regulated (Figure S3).
Figure 8.
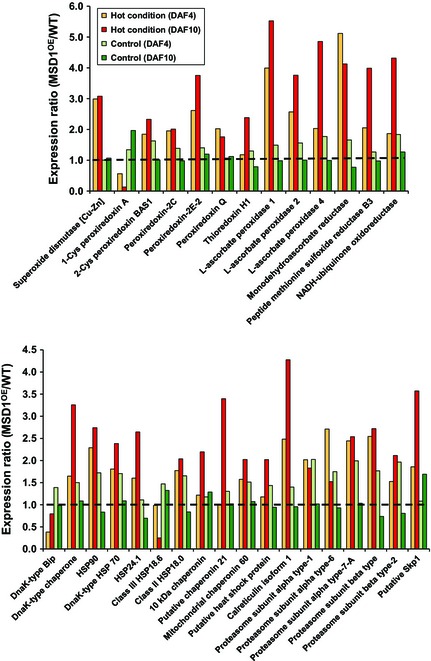
Increase in ROS scavenger, chaperone, quality control and programmed proteolysis systems in the developing seeds of MSD1OE under heat stress. Developing seeds at 4 and 10 days after flowering (DAF) were analysed by quantitative shotgun proteomic analysis with iTRAQ labelling. WT, Nipponbare wild type; MSD1OE, Nipponbare MSD1 overexpressor. Hot condition = 33/28 °C (12/12 h); control condition = 28/23 °C (12/12 h).
Discussion
Identification of Golgi/plastid‐type Mn SOD
Generally, Mn SODs are known as mitochondrial enzymes in both monocots and dicots (Kliebenstein et al., 1998; del Río et al., 2003; White and Scandalios, 1988; Wu et al., 1999) and in eukaryotic algae (Kitayama et al., 1999; Wolfe‐Simon et al., 2005). However, Mn SOD was localized in the chloroplasts of a marine diatom, Thalassiosira pseudonana (Wolfe‐Simon et al., 2006). The chloroplastic Mn SOD controlled by the nuclear‐encoded sodA gene must have plastid/ER transit peptides, but typical transit peptides have not been identified (Wolfe‐Simon et al., 2006). As shown in Figure 3b, the deduced amino acid sequence predicted that MSD1 is an extracellular glycoprotein with an N‐linked oligosaccharide chain. Confocal fluorescent microscopy revealed that rice MSD1 localized in multiple plastids and Golgi apparatus (Figures 4 and 5). Furthermore, the dominant‐negative and constitutive‐active mutants of ARF1 and SAR1 GTPases arrested the plastid‐targeting of MSD1‐YFP (Figure 5d), and the MSD1‐YFP fluorescence was rearranged into an ER tubular network (Figure 5b). This indicates that MSD1 is transported from the Golgi apparatus via the secretory pathway to the plastid, as are Arabidopsis CAH1 (Burén et al., 2011; Villarejo et al., 2005), O. sativa AmyI‐1 (Asatsuma et al., 2005; Kitajima et al., 2009) and NPP1 (Kaneko et al., 2011, 2014; Nanjo et al., 2006). This is the first report of the Golgi‐to‐plastid traffic of noncarbohydrate metabolism‐related enzyme.
The electron transport chain in chloroplasts contains several auto‐oxidizable enzymes. Ferredoxin in the reduced state can react with oxygen, releasing (Asada and Takahashi, 1987) and the aprotic interior of thylakoid membranes also produces (Takahashi et al., 1988). The outer layer tissues of developing rice seeds, namely the pericarp and the endosperm, contain chloroplasts during grain filling. Large amounts of starch molecules are synthesized and accumulated in the amyloplasts of endosperm cells. Thus, there is a need for ROS scavenging in the plastids of developing rice seeds. In addition to starches, proteins are also actively synthesized, assembled and stored in developing seeds. Storage proteins such as glutelins are synthesized in the ER and transported via the Golgi apparatus to the protein storage vacuoles (Ren et al., 2014; Washida et al., 2012). The production of H2O2 from O2 resulting from the maturation of glutelin in the endomembrane system (Onda et al., 2009) suggests the existence of an endomembranous ROS scavenging system.
Overproduction of MSD1 improves quality grain ripened under heat stress
Ectopic production of Golgi/plastid‐type MSD1 significantly improved the quality of rice grain ripened under heat stress (Figure 6f). On the other hand, suppression of MSD1 reduced the normal formation of rice grains (Figure 6h,j). These results indicate that the constitutive high expression of Golgi/plastid‐type MSD1 is effective for maintaining the formation of perfect grains under heat stress during grain filling. The introduction of yeast MnSOD and pea mitochondrial MnSOD into chloroplasts of rice conferred tolerance to salt and oxidative stress (Tanaka et al., 1999; Wang et al., 2005). In addition, transgenic rice transformed with mangrove cytosolic Cu/ZnSOD showed better tolerance to drought (Prashanth et al., 2008). We found that enhancement of OsMSD1 conferred significant tolerance to high temperatures during rice grain filling.
OsMSD1 is located in the centre of chromosome 5 (Figure 3a). Quantitative trait loci (QTLs) controlling grain appearance quality have been identified in populations derived from crosses between japonica cultivars (Ebitani et al., 2005; Kobayashi et al., 2007; Tabata et al., 2007), between japonica and indica cultivars (He et al., 1999; Wan et al., 2005) and between O. sativa and Oryza glaberrima (Li et al., 2004). The identification of a grain chalkiness QTL (qAPG5‐1, Ebitani et al., 2008) close to the position of MSD1 (Ebitani et al., 2005; Yamakawa et al., 2008) suggests that MSD1 is a determinant of chalkiness.
Proteomic characterization of developing seeds of MSD1OE under heat stress
Quantitative proteomic analysis of ripening seeds of MSD1OE and WT grown in normal and heat‐stress conditions at 4 and 10 DAF revealed that 79 proteins were down‐regulated and 219 were up‐regulated in MSD1OE under heat stress in comparison with WT (Table S1). The ROS scavenging system, molecular chaperones, chaperonins, calreticulin and proteasome components were markedly up‐regulated in MSD1OE under high temperature (Figure 8). In contrast, glutelin, prolamin and allergen family proteins were strongly down‐regulated (Figure S3). We detected an increase in APX 1, 2 and 4 in the developing seeds of MSD1OE. Monodehydroascorbate reductase, which regenerates ascorbate from monodehydroascorbate, was also up‐regulated (Figure 8 upper panel). The enhancement of APX production in rice (Lu et al., 2007; Tanaka et al., 1999) and other plants (Diaz‐Vivancos et al., 2013; Faize et al., 2011) confers abiotic stress tolerance. In addition, a series of peroxiredoxins (thioredoxin peroxidases), including 2‐Cys peroxiredoxin, were up‐regulated in MSD1OE (Figure 8 upper panel). Yeast transformed with O. sativa 2‐Cys peroxiredoxin showed increased stress tolerance and fermentation capacity (Kim et al., 2013). Moreover, an HSP was increased in MSD1OE under heat stress (Figure 8 lower panel). In rice (Sato and Yokoya, 2008) and Arabidopsis (Mu et al., 2013), overexpression of small HSPs enhanced tolerance to drought, salt and heat. Overall, these proteomic results and the literature strongly support the conclusion that MSD1OE rice showed improved adaptability to heat stress.
How is MSD1 involved in the adaptation of MSD1OE to heat stress? We considered that the constitutive high expression of MSD1 immediately converts to H2O2 under heat stress, and H2O2 probably serves as a trigger for enhancing the expression of the ROS scavenging system and HSP genes, as the level of H2O2 was higher in MSD1OE than in WT (Figure 7). H2O2 is one of the most abundant ROS and is both highly reactive and toxic. However, H2O2 also functions as a signalling molecule and activates the MAPK cascade (Apel and Hirt, 2004; Neill et al., 2002). For example, H2O2 induced ascorbate peroxidase in embryos of germinating rice (Morita et al., 1999), in Arabidopsis leaves (Karpinski et al., 1999) and in tobacco leaves (Gupta et al., 1993) and induced peroxiredoxin in mammalian thyroid cells (Kim et al., 2000). Therefore, induced peroxiredoxin and ascorbate peroxidase likely work as the main regulators of intracellular H2O2 concentrations in MSD1OE. Furthermore, heat‐stress‐induced H2O2 was involved in the early stage of activation of heat shock factor (HSF) in Arabidopsis cell culture (Volkov et al., 2006). In rice leaves, H2O2 treatment induced the production of a chloroplastic small HSP (Lee et al., 2000). Thus, H2O2 formed by Golgi/plastid‐type MSD1 is the key factor that confers heat tolerance on MSD1OE.
Storage and allergen family proteins were down‐regulated in the early developing seeds of MSD1OE under heat stress (Figure S3). The formation of protein bodies in developing seed cells of heat‐susceptible Todorokiwase was brought forward by higher temperature (T. M., unpublished data). We infer that the constitutive high expression of Golgi/plastid‐type MSD1 controls the redox state in the endomembrane system, leading to the normal programmed formation of protein bodies. Further studies will be needed to confirm this hypothesis.
In conclusion, we found a novel Golgi/plastid‐type Mn SOD in developing rice seeds. The ectopic expression of MSD1 dramatically induced the expression of ROS scavengers, molecular chaperones and the quality control system in developing seeds under heat stress. We consider that the constitutive high expression of MSD1 maintains normal grain filling and the production of perfect grains of rice under heat stress.
Experimental procedures
Plasmids
The plasmids used in this study and references describing how they were constructed are listed in Table S2.
Plant materials and growth conditions
Seeds of rice cultivars Yukinkomai, Yukinosei, Todorokiwase and Nipponbare (a model cultivar used for transformant experiments) were obtained from the Niigata Agricultural Research Institute Crop Research Center (Nagaoka city, Niigata, Japan). Transgenic lines of rice (cv. Nipponbare) overexpressing MSD1 under the control of maize Ubiquitin‐1 constitutive promoter (MSD1OE) were obtained from the full‐length cDNA overexpressor (FOX) lines of rice (Nakamura et al., 2007).
Transgenic plants with suppression of the MSD1 gene in developing seeds were generated as follows: MSD1 cDNA (bp 1–696) which contains no sequence of more than 21 nucleotides conserved with other rice SODs was amplified by PCR from pOsMSD1 (accession no. AK104160) with a primer set (Table S2) and introduced into pESWA (Islam et al., 2005) to construct the RNAi vector pWX‐WB‐MSD1‐RNAi in combination with the Wx promoter (Figure S2) using a pENTR Directional TOPO Cloning Kit and the Gateway LR Clonase Enzyme mix (Thermo Fisher Scientific, Waltham, MA). The binary RNAi vector was transformed into Agrobacterium tumefaciens strain EHA101 (Hood et al., 1986), and Agrobacterium‐mediated transformation of rice plants was performed as described by Hiei et al. (1994). We generated two transgenic knock‐down (KD) rice lines transformed with pWX‐WB‐MSD1‐RNAi, designated Nipponbare MSD1KD and Yukinkomai MSD1KD.
Yukinkomai, Yukinosei and Todorokiwase plants were grown in paddy fields of the Crop Research Center from 2004 to 2008 with ambient temperature or warm water. During the heading, ripening and maturity stages, the warm water was supplied at 35 °C at a flow rate of 80 L/min, making the daily mean temperature at around the ear 1.4–1.9 °C higher than that of the ambient temperature field (25.4 °C).
Transgenic and wild‐type (Nipponbare) plants were grown under 28/23 °C (12 h at 20 000 lx/12 h dark) in a growth chamber (CFH‐415; Tomy Seiko, Tokyo, Japan). Grain quality (chalky or translucent) was determined with a rice grain grader (RGQI20A; Satake, Hiroshima, Japan).
Microscopy studies
Yellow fluorescent protein (YFP) is a genetic mutant of green fluorescent protein from Aequorea victoria. We constructed pH35GY‐OsMSD1‐YFP to determine the subcellular localization of rice MSD1. We PCR‐amplified MSD1 from pOsMSD1 (primers in Table S2) and introduced it into pH35GY (Funakoshi Corp, Tokyo, Japan; Kubo et al., 2005) to create pH35GY‐OsMSD1‐YFP. To obtain pH35GY‐(AAGCTT)‐YFP (YFP vector alone), we PCR‐amplified an OsEMP70 fragment (bp 1083–1584) from pOsEMP70 (primers in Table S2) and introduced it into pH35GY. The pH35GY‐∆OsEMP70‐YFP construct was digested with HindIII to remove the OsEMP70 fragment and ligated with a Mighty Mix DNA ligation kit (Takara Bio, Ohtsu, Japan).
Construction of pWxTP‐DsRed (red fluorescent protein from Discosoma sp.; Kitajima et al., 2009), pST‐mRFP (monomeric red fluorescent protein; Matsuura‐Tokita et al., 2006), pMT121‐ARF1 T31N, pMT121‐ARF1 Q71L and pMT121‐SAR1 H74L (Takeuchi et al., 2000, 2002) were described elsewhere.
To introduce plasmid DNA into rice and onion (Allium cepa) epidermal cells, we used the particle bombardment method, using a helium‐driven particle accelerator, as described previously (Kitajima et al., 2009). Confocal laser‐scanning microscopes (FV300 and FV1000; Olympus, Tokyo, Japan) were used for imaging YFP, DsRed and chlorophyll autofluorescence in rice and onion cells (Kitajima et al., 2009). The FV300 uses an Ar laser at 488 nm to excite YFP and a green He/Ne laser at 543 nm to excite DsRed and chlorophyll. Fluorescence was detected at 510–530 nm through BA510IF and BA530RIF emission filters with an SDM570 emission dichroic mirror (YFP) and at >565 nm through a BA565IF emission filter (DsRed and chlorophyll). The FV1000 uses an Ar laser at 488 nm to excite YFP, and at 559 nm to excite DsRed and chlorophyll. Fluorescence was detected at 510 nm through an SDM560 emission dichroic mirror (YFP) and at 581 nm with an emission dichroic mirror (DsRed and chlorophyll). Images were observed through 40× air‐objective (UApo/340, NA 0.90; Olympus) and 100× oil‐objective lenses (UPlanSApo, NA 1.40 Oil; Olympus). The fluorescence intensity in plastids and in whole cells was determined using Lumina Vision imaging software. The background was always set at the maximum fluorescence intensity of an area in which no structural image was present. Areas identified by either chlorophyll autofluorescence or WxTP–DsRed were defined as plastids. To evaluate the plastid‐targeting abilities of YFP‐labelled proteins, we determined the ratio of the fluorescent intensity of YFP in the plastid to that in the whole cell (YFPplastid/YFPtotal; Kitajima et al., 2009).
Assay and diaminobenzidine staining for H2O2
H2O2 assays were carried out according to the procedure of Rao et al. (2000) and Xiong et al. (2007). Shoots and roots from rice seedlings at 7 days after imbibition were frozen and ground to a powder. Each sample (100 mg) was suspended in 0.5 mL of 0.2 m HClO4, incubated on ice for 5 min and then centrifuged at 14 000 g for 10 min at 4 °C. The supernatant was neutralized with 0.2 m NH4OH (pH 9.5) and centrifuged at 3000 g for 2 min. The neutralized extracts were passed through Sep‐Pak Light Accell Plus QMA Carbonate columns (Nihon Waters, Tokyo, Japan) and were eluted with 0.5 mL water. H2O2 in the extracts was quantified using an Amplex Red Hydrogen Peroxide–Peroxidase Assay kit (Life Technology Japan, Tokyo, Japan) following the manufacturer's directions. Fluorescence was measured with an RF‐5300PC spectrofluorophotometer (Shimadzu Corp., Kyoto, Japan) using excitation at 530 nm and fluorescence detection at 590 nm. For histochemical analysis, developing seeds (14 DAF) were sliced into 1‐mm transverse sections and immersed in 20 mm Tris·HCl buffer (pH 6.5) containing 1% (w/v) 3,3′‐diaminobenzidine. After vacuum infiltration for 30 min, the samples were incubated at room temperature for 20 h in the dark to develop a dark‐brown colour of diaminobenzidine oxidized by H2O2.
Gel‐based proteomics
Grains of Yukinkomai, Yukinosei and Todorokiwase (100 mg) at 4 DAF were extracted with 8 m urea, 1% (w/v) CHAPS detergent, 10 mm ethylene diamine tetraacetic acid (EDTA) and 5 mm phenylmethylsulfonyl fluoride and centrifuged at 10 000 g for 10 min at 4 °C. The supernatants were precipitated with 10% (w/v) trichloroacetic acid and resolved with 9 m urea, 3% (w/v) IGEPAL detergent, and 2% (v/v) 2‐mercaptoethanol and then used for gel‐based proteomics. The procedures of 2D polyacrylamide gel electrophoresis (2D‐PAGE) and matrix‐assisted laser desorption ionization time‐of‐flight MS (MALDI‐TOF‐MS) were essentially identical to the previous reports (Kaneko et al., 2011; Nanjo et al., 2004). In 2D‐PAGE, the 1st dimension used isoelectric focusing with ampholine (pH 3.5–10) and the 2nd dimension used sodium dodecyl sulphate (SDS)‐PAGE with 16% separating gel. The 2D gels were stained with Coomassie brilliant blue R‐250 (Nanjo et al., 2004). The protein spots excised from the gels were digested by trypsin using standard procedures (Awang et al., 2010). MALDI‐TOF‐MS was carried out with a matrix of α‐cyano‐4‐hyrdoxycinnamic acid in an AXIMA‐CFR mass spectrometer (Shimadzu Corp.) and an Autoflex III TOF/TOF mass spectrometer (Bruker BioSpin, Yokohama, Japan; Kaneko et al., 2011).
Quantitative shotgun proteomics
At 4 and 10 DAF, developing seeds of WT and MSD1OE grown under hot (33/28 °C, 12/12 h) or control conditions (28/23 °C, 12/12 h) were used in quantitative shotgun proteomic analysis with iTRAQ labelling. The seeds (0.2 g) were ground in liquid nitrogen to a fine powder and suspended in extraction buffer consisting of 100 mm Tris·HCl (pH 8.0), 20% (w/v) glycerol, 2% (w/v) Triton X‐100, 20 mm dithiothreitol, 3 m urea, 2 m thiourea and 3% (w/v) CHAPS. The homogenates were centrifuged at 10 000 g at 4 °C for 5 min. The supernatant was collected and centrifuged again. The supernatants were mixed with 1/10 volume of 100% (w/v) trichloroacetic acid, incubated on ice for 15 min and centrifuged at 10 000 g at 4 °C for 15 min. The resulting protein precipitates were washed 3 times in ice‐cold acetone and resuspended in 0.5 m triethylammonium bicarbonate buffer (pH 8.5) containing 0.1% SDS. Protein concentration was determined by the Bradford method (Bio‐Rad Laboratories, Hercules, CA) using bovine serum albumin as a standard. Proteins (50 μg) were reduced with tris‐(2‐carboxyethyl) phosphine at 37 °C for 60 min and then alkalized with methylmethanethiosulfonate at 25 °C for 60 min. Samples were digested in 10 μL of trypsin (1 μg/μL) at 37 °C for 16 h and labelled with 4‐plex iTRAQ tags (Thermo Fisher Scientific) according to Fukao et al. (2011), and the resultant 4 iTRAQ‐labelled peptide samples were mixed.
For quantitative proteomics, we used a combined KYA DiNA‐A (KYA Tech., Tokyo, Japan) and LTQ‐Orbitrap XL (Thermo Fisher Scientific) liquid chromatography‐MS/MS system. The ionization voltage and capillary transfer temperature at the electrospray ionization nano‐stage were set to 1.7–2.5 kV and 200 °C. iTRAQ‐labelled peptides were separated in a HiQ sil C18W column (75 μm i.d. ×50 mm, 3 μm particle size; KYA Tech.), using buffers A (0.1% [v/v] acetic acid and 2% [v/v] acetonitrile in water) and B (0.1% [v/v] acetic acid and 80% [v/v] acetonitrile in water). A linear gradient from 0% to 33% B for 240 min, 33% to 100% B for 10 min and back to 0% B over 15 min was applied, and peptides eluted from the column were introduced directly into an LTQ‐Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) at a flow rate of 300 nL min−1 and a spray voltage of 4.5 kV.
Liquid chromatography‐MS/MS data were acquired in data‐dependent acquisition mode using Xcalibur 2.0 software (Thermo Fisher Scientific). The mass range selected for MS scan was set to 350–1600 m/z, and the top three peaks were subjected to MS/MS analysis. Full MS scan was detected in the Orbitrap, while the MS/MS scans were detected in the linear ion trap and Orbitrap. The normalized collision energy for MS/MS was set to 35 eV for collision‐induced dissociation (CID) and 45 eV for higher energy C‐trap dissociation (HCD). The resolution of the mass spectrometer (FTMS) was set to 60 000. Divalent or trivalent ions were subjected to MS/MS analysis in dynamic exclusion mode, and proteins were identified with Proteome Discoverer v. 1.1 software and the SEQUEST search tool (Thermo Fisher Scientific) using the UniProt (http://www.uniprot.org/) O. sativa subsp. japonica database (63 535 proteins) with the following parameters: enzyme, trypsin; maximum missed cleavages site, 2; peptide charge, 2+ or 3+; MS tolerance, 10 ppm; MS/MS tolerance, ±0.8 Da; dynamic modification; carboxymethylation (C); oxidation (H, M, W); iTRAQ 4‐plex (K, Y, N‐terminus). False discovery rates were <5%.
mRNA analysis
Sample tissues (0.1 g) were ground in liquid nitrogen to fine powder and suspended in an extraction buffer consisting of 2% (w/v) cetyl trimethyl ammonium bromide, 100 mm Tris·HCl (pH 8.0), 20 mm EDTA and 1.4 m NaCl. The homogenates were mixed with 1/2 volumes of phenol and chloroform/isoamyl alcohol (25:24:1, v/v), centrifuged at 10 000 g at 4 °C for 5 min, and total RNA was extracted with RNeasy Plant Mini Kit (Qiagen, Tokyo, Japan) according to the manufacturer's instructions. Ten ng of total RNA was applied to a real‐time quantitative reverse transcription PCR using SsoFast Eva Green Supermix (Bio‐Rad) and CFX96 real time PCR system/C1000TM Thermal Cycler (Bio‐Rad) with the PCR primer sets listed in Table S3. The mRNA contents in each sample were normalized against those of constitutive 18S rRNA gene (Accession no. AK059783).
Supporting information
Figure S1 Appearance quality of grains harvested from Nipponbare WT and MSD1OE treated at 33/28 °C (12/12 h) after heading.
Figure S2 Map of pWX‐WB‐MSD1‐RNAi.
Figure S3 Changes in the amounts of storage and allergen proteins in the developing seeds of MSD1OE under heat stress.
Table S1 Quantitative shotgun proteomic analysis of developing seeds of MSD1OE and WT at 4 and 10 days after flowering (DAF) under hot and control conditions.
Table S2 Plasmids used in this study.
Table S3 Real‐time PCR primer sets.
Acknowledgements
This research was supported by a Grant for Promotion of KAAB Projects (Niigata University), Scientific Research on Innovative Areas (22114507) and Grants‐in‐Aid for Scientific Research (B) (22380186) from the Ministry of Education, Culture, Sports, Science and Technology, Japan to T. M. This work was also supported in part by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, AMR‐0001) to H. I. We are indebted to Dr. A. Nakano (The University of Tokyo, Japan) for providing pST‐mRFP, pMT121‐ARF1 T31N, pMT121‐ARF1 Q71L and pMT121‐SAR1 H74L.
References
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Asada, K. and Takahashi, M. (1987) Production and scavenging of active oxygen in chloroplasts In Photoinhibition (Kyle D.J., Osmond C.B. and Arntzen C.J., eds), pp. 227–287. Amsterdam: Elsevier. [Google Scholar]
- Asatsuma, S. , Sawada, C. , Itoh, K. , Okito, M. , Kitajima, A. and Mitsui, T. (2005) Involvement of α‐amylase I‐1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 46, 858–869. [DOI] [PubMed] [Google Scholar]
- Awang, A. , Karim, R. and Mitsui, T. (2010) Proteomic analysis of Theobroma cacao pod husk. J. Appl. Glycosci. 57, 245–264. [Google Scholar]
- Badawi, G.H. , Yamauchi, Y. , Shimada, E. , Sasaki, R. , Kawano, N. , Tanaka, K. and Tanaka, K. (2004) Enhanced tolerance to salt stress and water deficit by overexpressing superoxide dismutase in tobacco (Nicotiana tabacum) chloroplasts. Plant Sci. 166, 919–928. [Google Scholar]
- Bowler, C. , Van Montagu, M. and Inzé, D. (1992) Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 83–116. [Google Scholar]
- Brawn, K. and Fridovich, I. (1981) DNA strand scission by enzymically generated oxygen radicals. Arch. Biochem. Biophys. 206, 414–419. [DOI] [PubMed] [Google Scholar]
- Bueno, P. , Varela, J. , Gimenez, G.G. and Del Rio, L.A. (1995) Peroxisomal copper, zinc superoxide dismutase: characterization of the isoenzyme from watermelon cotyledons. Plant Physiol. 108, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén, S. , Ortega‐Villasante, C. , Blanco‐Rivero, A. , Martínez‐Bernardini, A. , Shutova, T. , Shevela, D. , Messinger, J. , Bako, L. , Villarejo, A. and Samuelsson, G. (2011) Importance of post‐translational modifications for functionality of a chloroplast‐localized carbonic anhydrase (CAH1) in Arabidopsis thaliana . PLoS ONE, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Camp, W. , Willekens, H. , Bowler, C. , Van Montagu, M. and Inze, D. (1994) Elevated levels of superoxide dismutase protect transgenic plants against ozone damage. Bio/Technology, 12, 165–168. [Google Scholar]
- van Camp, W. , Capiau, K. , Van Montagu, M. , Inze, D. and Slooten, L. (1996) Enhancement of oxidative stress tolerance in transgenic tobacco overproducing Fe‐superoxide dismutase in chloroplasts. Plant Physiol. 112, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H. , Huang, L.F. , Li, H.M. , Chen, Y.R. and Yu, S.M. (2004) Signal peptide‐dependent targeting of a rice α‐amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 135, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo, M.J. , Luth, D. , Blankenship, K.M. , Anderson, O.D. and Blechl, A.E. (1993) Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 23, 567–581. [DOI] [PubMed] [Google Scholar]
- Diaz‐Vivancos, P. , Faize, M. , Barba‐Espin, G. , Faize, L. , Petri, C. , Hernández, J.A. and Burgos, L. (2013) Ectopic expression of cytosolic superoxide dismutase and ascorbate peroxidase leads to salt stress tolerance in transgenic plums. Plant Biotechnol. J. 11, 976–985. [DOI] [PubMed] [Google Scholar]
- van Dooren, G.G. , Schwartzbach, S.D. , Osafune, T. and McFadden, G.I. (2001) Translocation of proteins across the multiple membranes of complex plastids. Biochim. Biophys. Acta, 1541, 34–53. [DOI] [PubMed] [Google Scholar]
- Ebitani, T. , Yamamoto, Y. , Yano, M. and Funane, M. (2005) Analysis of quantitative trait loci for grain appearance, Kasalath alleles increase ratio of whole grains against Koshihikari, in rice. Jpn. J. Crop Sci. 74, 290–291. [Google Scholar]
- Ebitani, T. , Yamamoto, Y. , Yano, M. and Funane, M. (2008) Identification of quantitative trait loci for grain appearance using chromosome segment substitution lines in rice. Breed. Res. 10, 91–99. [Google Scholar]
- Evers, A.D. and Juliano, B.O. (1976) Varietal differences in surface ultrastructure of endosperm cells and starch granules of rice. Starch, 28, 160–166. [Google Scholar]
- Faize, M. , Burgos, L. , Faize, L. , Piqueras, A. , Nicolas, E. , Barba‐Espin, G. , Clemente‐Moreno, M.J. , Alcobendas, R. , Artlip, T. and Hernández, J.A. (2011) Involvement of cytosolic ascorbate peroxidase and Cu/Zn‐superoxide dismutase for improved tolerance against drought. J. Exp. Bot. 62, 2599–2613. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M.A. , McCouch, S.R. and Hall, R.D. (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci. 14, 133–139. [DOI] [PubMed] [Google Scholar]
- Fridovich, I. (1975) Superoxide dismutases. Annu. Rev. Biochem. 44, 147–149. [DOI] [PubMed] [Google Scholar]
- Fucci, L. , Oliver, C.N. , Coon, M.J. and Stadtman, E.R. (1983) Inactivation of metabolic enzymes by mixed‐function oxidation reaction: possible implication in protein turnover and ageing. Proc. Natl Acad. Sci. USA, 80, 1521–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N. , Satoh, R. , Hayashi, A. , Kodama, M. , Itoh, R. , Aihara, S. and Nakamura, Y. (2011) Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J. Exp. Bot. 62, 4819–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, Y. , Ferjani, A. , Tomioka, R. , Nagasaki, N. , Kurata, R. , Nishimori, Y. , Fujiwara, M. and Maeshima, M. (2011) iTRAQ analysis reveals mechanisms of growth defects due to excess zinc in Arabidopsis . Plant Physiol. 155, 1893–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, M. , Wen, L. , Satoh‐Cruz, M. , Kawagoe, Y. , Nagamura, Y. , Okita, T.W. , Washida, H. , Sugino, A. , Ishino, S. , Ishino, Y. , Ogawa, M. , Sunada, M. , Ueda, T. and Kumamaru, T. (2013) A guanine nucleotide exchange factor for Rab5 proteins is essential for intracellular transport of the proglutelin from the Golgi apparatus to the protein storage vacuole in rice endosperm. Plant Physiol. 162, 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A.S. , Webb, R.P. , Holaday, A.S. and Allen, R.D. (1993) Overexpression of superoxide dismutase protects plants from oxidative stress (Induction of ascorbate peroxidase in superoxide dismutase‐overexpressing plants). Plant Physiol. 103, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata, M. , Kuroda, M. , Miyashita, T. , Yamaguchi, T. , Kojima, M. , Sakakibara, H. , Mitsui, T. and Yamakawa, H. (2012) Suppression of α‐amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 10, 1110–1117. [DOI] [PubMed] [Google Scholar]
- He, P. , Li, S.G. , Qian, Q. , Ma, Y.Q. , Li, J.Z. , Wang, W.M. , Chen, Y. and Zhu, L.H. (1999) Genetic analysis of rice grain quality. Theor. Appl. Genet. 98, 502–508. [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hood, E.E. , Helmer, G.L. , Fraley, R.T. and Chilton, M.D. (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T‐DNA. J. Bacteriol. 168, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru, T. , Horigane, A.K. , Ida, M. , Iwasawa, N. , San‐oh, Y.A. , Nakazono, M. , Nishizawa, N.K. , Masumura, T. , Kondo, M. and Yoshida, M. (2009) Formation of grain chalkiness and changes in water distribution in developing rice caryopses grown under high‐temperature stress. J. Cereal Sci. 50, 166–174. [Google Scholar]
- Islam, S.M.S. , Miyazaki, T. , Tanno, F. and Itoh, K. (2005) Dissection of gene function by RNA silencing. Plant Biotechnol. 22, 443–446. [Google Scholar]
- Iwasawa, N. , Umemoto, T. , Hiratsuka, M. , Nitta, Y. , Matsuda, T. and Kondo, M. (2009) Structural characters of milky‐white rice grains caused by high temperature and shading during grain‐filling. Jpn. J. Crop Sci. 78, 322–323. [Google Scholar]
- Jiang, H. , Dian, W. and Wu, P. (2003) Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry, 63, 53–59. [DOI] [PubMed] [Google Scholar]
- Kaneko, K. , Yamada, C. , Yanagida, A. , Koshu, T. , Umezawa, Y. , Itoh, K. , Hori, H. and Mitsui, T. (2011) Differential localizations and functions of rice nucleotide pyrophosphatase/phosphodiesterase isozymes 1 and 3. Plant Biotechnol. 28, 69–76. [Google Scholar]
- Kaneko, K. , Inomata, T. , Masui, T. , Koshu, T. , Umezawa, Y. , Itoh, K. , Pozueta‐Romero, J. and Mitsui, T. (2014) Nucleotide pyrophosphatase/phosphodiesterase 1 exerts a negative effect on starch accumulation and growth in rice seedlings under high temperature and CO2 concentration conditions. Plant Cell Physiol. 55, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski, S. , Reynolds, H. , Karpinska, B. , Wingsle, G. , Creissen, G. and Mullineaux, P. (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis . Science, 284, 654–657. [DOI] [PubMed] [Google Scholar]
- Kim, H. , Lee, T.H. , Park, E.S. , Suh, J.M. , Park, S.J. , Chung, H.K. , Kwon, O.Y. , Kim, Y.K. , Ro, H.K. and Shong, M. (2000) Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide‐induced apoptosis in thyroid cells. J. Biol. Chem. 275, 18266–18270. [DOI] [PubMed] [Google Scholar]
- Kim, I.‐S. , Kim, Y.‐S. and Yoon, H.‐S. (2013) Expression of salt‐induced 2‐Cys peroxiredoxin from Oryza sativa increases stress tolerance and fermentation capacity in genetically engineered yeast Saccharomyces cerevisiae . Appl. Microbiol. Biotechnol. 97, 3519–3533. [DOI] [PubMed] [Google Scholar]
- Kitajima, A. , Asatsuma, S. , Okada, H. , Hamada, Y. , Kaneko, K. , Nanjo, Y. , Kawagoe, Y. , Toyooka, K. , Matsuoka, K. , Takeuchi, M. , et al (2009) The rice α‐amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell, 21, 2844–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama, K. , Kitayama, M. , Osafune, T. and Togasaki, R.K. (1999) Subcellular localization of iron and manganese superoxide dismutase in Chlamydomonas reinhardtii (Chlorophyceae). J. Phycol. 35, 136–142. [Google Scholar]
- Kliebenstein, D.J. , Monde, R.A. and Last, R.L. (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klösgen, R.B. and Weil, J.H. (1991) Subcellular location and expression level of a chimeric protein consisting of the maize waxy transit peptide and the beta‐glucuronidase of Escherichia coli in transgenic potato plants. Mol. Gen. Genet. 225, 297–304. [DOI] [PubMed] [Google Scholar]
- Kobayashi, A. , Genliang, B. , Shenghai, Y. and Tomita, K. (2007) Detection of quantitative trait loci for white‐back and basal‐white kernels under high temperature stress in japonica rice varieties. Breed. Sci. 57, 107–116. [Google Scholar]
- Kubo, A. , Fujita, N. , Harada, K. , Matsuda, T. , Satoh, H. and Nakamura, Y. (1999) The starch‐debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 121, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, M. , Udagawa, M. , Nishikubo, N. , Horiguchi, G. , Yamaguchi, M. , Ito, J. , Mimura, T. , Fukuda, H. and Demura, T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.H. , Won, S.H. , Lee, H.S. , Miyao, M. , Chung, W.I. , Kim, I.J. and Jo, J. (2000) Expression of the chloroplast‐localized small heat shock protein by oxidative stress in rice. Gene, 245, 283–290. [DOI] [PubMed] [Google Scholar]
- Li, J. , Xiao, J. , Grandillo, S. , Jiang, L. , Wan, Y. , Deng, Q. , Yuan, L. and McCouch, S.R. (2004) QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome, 47, 697–704. [DOI] [PubMed] [Google Scholar]
- Lu, Z. , Liu, D. and Liu, S. (2007) Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis . Plant Cell Rep. 26, 1909–1917. [DOI] [PubMed] [Google Scholar]
- Matsuura‐Tokita, K. , Takeuchi, M. , Ichihara, A. , Mikuriya, K. and Nakano, A. (2006) Live imaging of yeast Golgi cisternal maturation. Nature, 441, 1007–1010. [DOI] [PubMed] [Google Scholar]
- McKersie, B.D. , Murnaghan, J. , Jones, K.S. and Bowley, S.R. (2000) Iron superoxide dismutase in transgenic alfalfa increase winter survival without a detectable increase in photosynthetic oxidative stress tolerance. Plant Physiol. 122, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, S. , Kaminaka, H. , Masumura, T. and Tanaka, K. (1999) Induction of rice cytosolic ascorbate peroxidase mRNA by oxidative stress, the involvement of hydrogen peroxide in oxidative stress signalling. Plant Cell Physiol. 40, 417–422. [Google Scholar]
- Morita, S. , Shiratsuchi, H. , Takanashi, J. and Fujita, K. (2004) Effect of high temperature on grain ripening in rice plants. Analysis of the effects of high night and high day temperatures applied to the panicle and other parts of the plant. Jpn. J. Crop Sci. 73, 77–83. [Google Scholar]
- Mu, C. , Zhang, S. , Yu, G. , Chen, N. , Li, X. and Liu, H. (2013) Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE, 8, e82264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, K. , Takita, T. , Yoshinaga, S. , Terashima, K. and Fukuda, A. (2004) Effect of air temperature during the early grain‐filling stage on grain fissuring in rice. Jpn. J. Crop Sci. 73, 336–342. [Google Scholar]
- Nakamura, H. , Hakata, M. , Amano, K. , Miyao, A. , Toki, N. , Kajikawa, M. , Pang, J. , Higashi, N. , Ando, S. , Toki, S. , et al (2007) A genome‐wide gain‐of function analysis of rice genes using the FOX‐hunting system. Plant Mol. Biol. 65, 357–371. [DOI] [PubMed] [Google Scholar]
- Nanjo, Y. , Asatsuma, S. , Itoh, K. , Hori, H. and Mitsui, T. (2004) Proteomic identification of α‐amylase isoforms encoded by RAmy3B/3C from germinating rice seeds. Biosci. Biotechnol. Biochem. 68, 112–118. [DOI] [PubMed] [Google Scholar]
- Nanjo, Y. , Oka, H. , Ikarashi, N. , Kaneko, K. , Kitajima, A. , Mitsui, T. , Muñoz, F.J. , Rodríguez‐López, M. , Baroja‐Fernández, E. and Pozueta‐Romero, J. (2006) Rice plastidial N‐glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER‐Golgi to the chloroplast through the secretory pathway. Plant Cell, 18, 2582–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill, S.J. , Desikan, R. , Clarke, A. , Hurst, R.D. and Hancock, J.T. (2002) Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53, 1237–1247. [PubMed] [Google Scholar]
- Niki, E. , Yoshida, Y. , Saito, Y. and Noguchi, N. (2005) Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem. Biophys. Res. Commun. 338, 668–676. [DOI] [PubMed] [Google Scholar]
- Nishi, A. , Nakamura, Y. , Tanaka, N. and Satoh, H. (2001) Biochemical and genetic analysis of the effects of amylose‐extender mutation in rice endosperm. Plant Physiol. 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Nowack, E.C.M. and Grossman, A.R. (2012) Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora . Proc. Natl Acad. Sci. USA, 109, 5340–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda, Y. and Kawagoe, Y. (2011) Oxidative protein folding: selective pressure for prolamin evolution in rice. Plant Signal. Behav. 6, 1966–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda, Y. , Kumamaru, T. and Kawagoe, Y. (2009) ER membrane‐localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl Acad. Sci. USA, 106, 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda, Y. , Nagamine, A. , Sakurai, M. , Kumamaru, T. , Ogawa, M. and Kawagoe, Y. (2011) Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell, 23, 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton, P. , Webb, R. , Kornyeyev, D. , Allen, R. and Holaday, A.S. (2001) Protecting cotton photosynthesis during moderate chilling at high light intensity by increasing chloroplastic antioxidant enzyme activity. J. Exp. Bot. 52, 2345–2354. [DOI] [PubMed] [Google Scholar]
- Peng, S. , Huang, J. , Sheehy, J.E. , Laza, R.C. , Visperas, R.M. , Zhong, X. , Centeno, G.S. , Khush, G.S. and Cassman, K.G. (2004) Rice yields decline with higher night temperature from global warming. Proc. Natl Acad. Sci. USA, 101, 9971–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl, A. , Perl‐Terves, R. , Galili, S. , Aviv, D. , Shalgi, E. , Makin, S. and Galun, E. (1993) Enhanced oxidative‐stress defense in transgenic potato expressing tomato Cu, Zn superoxide dismutases. Theor. Appl. Genet. 85, 568–576. [DOI] [PubMed] [Google Scholar]
- Prashanth, S.R. , Sadhasivam, V. and Parida, A. (2008) Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica rice var Pusa Basmati‐1 confers abiotic stress tolerance. Transgenic Res. 17, 281–291. [DOI] [PubMed] [Google Scholar]
- Rao, M.V. , Lee, H. , Creelman, R.A. , Mullet, J.E. and Davis, K.R. (2000) Jasmonic acid signaling modulates ozone‐induced hypersensitive cell death. Plant Cell, 12, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Wang, Y. , Liu, F. , Zhou, K. , Ding, Y. , Zhou, F. , Wang, Y. , Liu, K. , Gan, L. , Ma, W. , et al (2014) GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post‐Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell, 26, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río, L.A. , Sandalio, L.M. , Altomare, D.A. and Zilinskas, B.A. (2003) Mitochondrial and peroxisomal manganese superoxide dismutase: differential expression during leaf senescence. J. Exp. Bot. 54, 923–933. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A. , Nosaka, Y. and Tanaka, K. (1993) Cloning and sequencing analysis of a complementary DNA for manganese‐superoxide dismutase from rice (Oryza sativa L.). Plant Physiol. 103, 1477–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake, T. and Yoshida, S. (1978) High temperature induced sterility in indica rice at flowering. Jpn. J. Crop Sci. 47, 6–17. [Google Scholar]
- Sato, Y. and Yokoya, S. (2008) Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat‐shock protein, sHSP17.7. Plant Cell Rep. 27, 329–334. [DOI] [PubMed] [Google Scholar]
- Sen Gupta, A. , Heinen, J.L. , Holaday, A.S. , Burke, J.J. and Allen, R.D. (1993) Increased resistance in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl Acad. Sci. USA, 90, 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, K.C. , Kusano, H. , Koizumi, K. , Yamakawa, H. , Hakata, M. , Imamura, T. , Fukuda, M. , Naito, N. , Tsurumaki, Y. , Yaeshima, M. , et al (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. , Sodhi, N.S. , Kaur, M. and Saxena, S.K. (2003) Physico‐chemical, morphological, thermal, cooking and textural properties of chalky and translucent rice kernels. Food Chem. 82, 433–439. [Google Scholar]
- Sláviková, S. , Vacula, R. , Fang, Z. , Ehara, T. , Osafune, T. and Schwartzbach, S.D. (2006) Homologous and heterologous reconstitution of Golgi to chloroplast transport and protein import into the complex chloroplasts of Euglena . J. Cell Sci. 118, 1651–1661. [DOI] [PubMed] [Google Scholar]
- Slooten, L. , Capiau, K. , Van Montagu, M. , Sybesma, C. and Inze, D. (1995) Factors affecting the enhancement of oxidative stress tolerance in transgenic tobacco overexpressing manganese superoxide dismutase in chloroplasts. Plant Physiol. 107, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streller, S. and Wingsle, G. (1994) Pinus sylvestris L. needles contain extracellular CuZn superoxide dismutase. Planta, 192, 195–201. [PubMed] [Google Scholar]
- Tabata, M. , Hirabayashi, H. , Takeuchi, Y. , Ando, I. , Iida, Y. and Ohsawa, R. (2007) Mapping of quantitative trait loci for the occurrence of white‐back kernels associated with high temperatures during the ripening period of rice (Oryza sativa L.). Breed. Sci. 57, 47–52. [Google Scholar]
- Takahashi, M. , Shiraishi, T. and Asada, K. (1988) Superoxide production in aprotic interior of chloroplast thylakoids. Arch. Biochem. Biophys. 267, 714–722. [DOI] [PubMed] [Google Scholar]
- Takeuchi, M. , Ueda, T. , Sato, K. , Abe, H. , Nagata, T. and Nakano, A. (2000) A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23, 517–525. [DOI] [PubMed] [Google Scholar]
- Takeuchi, M. , Ueda, T. , Yahara, N. and Nakano, A. (2002) Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31, 499–515. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y. , Hibino, T. , Hayashi, Y. , Tanaka, A. , Kishitani, S. , Takabe, T. , Yokota, S. and Takabe, T. (1999) Salt tolerance of transgenic rice overexpressing yeast mitochondrial Mn‐SOD in chloroplasts. Plant Sci. 148, 131–138. [Google Scholar]
- Tanaka, N. , Fujita, N. , Nishi, A. , Satoh, H. , Hosaka, Y. , Ugaki, M. , Kawasaki, S. and Nakamura, Y. (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2, 507–516. [DOI] [PubMed] [Google Scholar]
- Tashiro, T. and Wardlaw, I.F. (1991) The effect of high temperature on kernel dimensions and the type and occurrence of kernel damage in rice. Aust. J. Agric. Res. 42, 485–496. [Google Scholar]
- Tertivanidis, K. , Goudoula, K. , Vasilikiotis, K. , Hassiotou, E. and Perl‐Treves, R. (2004) Superoxide dismutase transgenes in sugarbeets confer resistance to oxidative agents and the fungus C. beticola . Transgenic Res. 13, 225–233. [DOI] [PubMed] [Google Scholar]
- Tsutsui, K. , Kaneko, K. , Hanashiro, I. , Nishinari, K. and Mitsui, T. (2013) Characteristics of opaque and translucent parts of high temperature stressed grains of rice. J. Appl. Glycosci. 60, 61–67. [Google Scholar]
- Umemoto, T. and Terashima, K. (2002) Activity of granule‐bound synthase is an important determinant of amylose content in rice endosperm. Funct. Plant Biol. 29, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Villarejo, A. , Burén, S. , Larsson, S. , Déjardin, A. , Monné, M. , Rudhe, C. , Karlsson, J. , Jansson, S. , Lerouge, P. , Rolland, N. , et al (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7, 1224–1231. [DOI] [PubMed] [Google Scholar]
- Volkov, R.A. , Panchuk, I.I. , Mullineaux, P.M. and Schöffl, F. (2006) Heat stress‐induced H2O2 is required for effective expression of heat shock genes in Arabidopsis . Plant Mol. Biol. 61, 733–746. [DOI] [PubMed] [Google Scholar]
- Wan, X.Y. , Wan, J.M. , Weng, J.F. , Jiang, L. , Bi, J.C. , Wang, C.M. and Zhai, H.Q. (2005) Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor. Appl. Genet. 110, 1334–1346. [DOI] [PubMed] [Google Scholar]
- Wang, F.‐Z. , Wang, Q.‐B. , Kwon, S.‐Y. , Kwak, S.‐S. and Su, W.‐A. (2005) Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J. Plant Physiol. 162, 465–472. [DOI] [PubMed] [Google Scholar]
- Washida, H. , Sugino, A. , Doroshenk, K.A. , Satoh‐Cruz, M. , Nagamine, A. , Katsube‐Tanaka, T. , Ogawa, M. , Kumamaru, T. , Satoh, H. and Okita, T.W. (2012) RNA targeting to a specific ER sub‐domain is required for efficient transport and packaging of α‐globulins to the protein storage vacuole in developing rice endosperm. Plant J. 70, 471–479. [DOI] [PubMed] [Google Scholar]
- White, J.A. and Scandalios, J.G. (1988) Isolation and characterization of a cDNA for mitochondrial manganese superoxide dismutase (SOD‐3) of maize and its relation to other manganese superoxide dismutases. Biochim. Biophys. Acta, 951, 61–70. [DOI] [PubMed] [Google Scholar]
- Wolfe‐Simon, F. , Grzebyk, D. , Schofield, O. and Falkowski, P.G. (2005) The role and evolution of superoxide dismutases in algae. J. Phycol. 41, 453–465. [Google Scholar]
- Wolfe‐Simon, F. , Starovoytov, V. , Reinfelder, J.R. , Schofield, O. and Falkowski, P.G. (2006) Localization and role of manganese superoxide dismutase in a marine diatom. Plant Physiol. 142, 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , Wilen, R.W. , Robertson, A.J. and Gusta, L.V. (1999) Isolation, chromosomal localization, and differential expression of mitochondrial manganese superoxide dismutase and chloroplastic copper/zinc superoxide dismutase genes in wheat. Plant Physiol. 120, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. , Contento, A.L. , Nguyen, P.Q. and Bassham, D.C. (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis . Plant Physiol. 143, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, H. , Hirose, T. , Kuroda, M. and Yamaguchi, T. (2007) Comprehensive expression profiling of rice grain filling‐related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, H. , Ebitani, T. and Terao, T. (2008) Comparison between locations of QTLs for grain chalkiness and genes responsive to high temperature during grain filling on the rice chromosome map. Breed. Sci. 58, 337–343. [Google Scholar]
- Zakaria, S. , Matsuda, T. , Tajima, S. and Nitta, Y. (2002) Effect of high temperature at ripening stage on the reserve accumulation in seed in some rice cultivars. Plant Prod. Sci. 5, 160–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Appearance quality of grains harvested from Nipponbare WT and MSD1OE treated at 33/28 °C (12/12 h) after heading.
Figure S2 Map of pWX‐WB‐MSD1‐RNAi.
Figure S3 Changes in the amounts of storage and allergen proteins in the developing seeds of MSD1OE under heat stress.
Table S1 Quantitative shotgun proteomic analysis of developing seeds of MSD1OE and WT at 4 and 10 days after flowering (DAF) under hot and control conditions.
Table S2 Plasmids used in this study.
Table S3 Real‐time PCR primer sets.


