Abstract
Background:
Nuclear receptor 4A1 (NR4A1) is overexpressed in mammary tumors, and the methylene-substituted bis-indole derivative 1,1-bis(3'-indolyl)-1-(p-hydroxyphenyl)methane (DIM-C-pPhOH) acts as an NR4A1 antagonist (inverse agonist) and inhibits NR4A1-regulated pro-oncogenic pathways/genes in breast and other cancer cells.
Methods:
Buttressed analogs of DIM-C-pPhOH were synthesized by condensation of the substituted p-hydroxybenzaldehydes with indole. Breast cancer cell growth, survival and migration assays were carried out by cell counting, Annexin V staining and Boyden chamber assays, respectively. Changes in RNA and protein expression were determined by RT-PCR and western blots, respectively. Analysis of RNAseq results was carried out using Ingenuity Pathway Analysis, and in vivo potencies of NR4A1 antagonists were determined in athymic nude mice bearing MDA-MB-231 cells in an orthotop model.
Results:
Ingenuity Pathway analysis of common genes modulated by NR4A1 knockdown or treatment with DIM-C-pPhOH showed that changes in gene expression were consistent with the observed decreased functional responses, namely inhibition of growth and migration and increased apoptosis. DIM-C-pPhOH is rapidly metabolized and the effects and potencies of buttressed analogs of DIM-C-pPhOH which contain one or two substituents ortho to the hydroxyl groups were investigated using NR4A1-regulated gene/gene products as endpoints. The buttressed analogs were more potent than DIM-C-pPhOH in both in vitro assays and as inhibitors of mammary tumor growth. Moreover, using 1,1-bis(3'-indolyl)-1-(3-chloro-4-hydroxy-5-methoxyphenyl)methane (DIM-C-pPhOh-3-Cl-5-OCH3) significant tumor growth inhibition was observed at doses as low as 2 mg/kg/d which was at least an order of magnitude more potent than DIM-C-pPhOH.
Conclusions:
These buttressed analogs represent a more potent set of second generation NR4A1 antagonists as inhibitors of breast cancer.
Keywords: NR4A1, bis-indole ligands, antagonists, anticancer activities
BACKGROUND
The 48 members of the nuclear receptor (NR) superfamily of transcription factors play essential roles in maintain cellular homeostasis and are also potential druggable targets for many diseases including cancer [1,2]. Pharmaceutical agents that directly target NRs are often selective receptor modulators which bind the receptor and exhibit tissue-specific receptor agonist or antagonist activities [2]. For example, tamoxifen is a widely used selective estrogen receptor modulator (SERM) for treating breast cancer and tamoxifen exhibits ER antagonist activity against breast cancer but is an ER agonist in the uterus [rev. in [3,4]. Differential expression of some NRs in cancer vs. noncancer tissues has been observed and these receptors may also be prognostic factors for drug-resistance, patient survival, and disease recurrence [5-11]. Muscat and coworkers [7] investigated the relative expression of NRs in breast tumors and normal breast and identified 41 NRs in breast cancers and 33 of these receptors exhibited differential expression in tumor vs. non-tumor tissues. Twenty-six NRs were higher in normal breast vs. ER-positive and ER-negative breast cancer and only seven NRs were more highly expressed in tumors. Two of the 3 NRs overexpressed in both ER-positive and ER-negative breast tumors were members of the NR4A subfamily, namely NR4A1 (Nur77, TR3) and NR4A3 (Nor1) [7]. Subsequent studies on the expression, prognostic value, and functions of NR4A1 in breast cancer are contradictory and functional studies report that NR4A1 exhibits both pro- and anticarcinogenic activities [11-16].
Research in this laboratory has focused on a series of methylene-substituted bis-indole-derived compounds (C-DIMs) as NR4A1 ligands which exhibit tissue-specific NR4A1 agonist or antagonist activities and are selective NR4A1 modulators [8,14-22]. In breast cancer and other solid tumors NR4A1, acts as a pro-oncogenic factor regulating pathways/genes such as β1-integrin and TXNDC5 associated with cell proliferation, survival and migration/invasion and NR4A1 ligands typified by 1,1-bis(3'-indolyl)-1-(p-hydroxyphenyl)methane (DIM-C-pPhOH, CDIM8) act as antagonists [8,14-20]. Previous studies in breast, colon, kidney, pancreatic and lung cancer cell lines and rhabdomyosarcoma cells show that both knockdown of NR4A1 (siNR4A1) or treatment with DIM-C-pPhOH decreased cell growth and migration, induced apoptosis, and regulated expression of genes associated with these responses [8,14-21]. Although DIM-C-pPhOH inhibits tumor growth in xenograft models (30 mg/kg/d) [17,21,8], this compound is rapidly metabolized [23]. Therefore, we synthesized several buttressed analogs of DIM-C-pPhOH containing at least one or two substituents ortho (3'- or 3',5'-) to the hydroxyl groups since it has previously been reported that this substitution pattern hinders metabolism (conjugation) of phenolic compounds [24]. The in vitro results clearly demonstrate the enhanced potency of the buttressed analogs compared to DIM-C-pPhOH and the IC50 for breast tumor growth inhibition by one of the buttressed analog 1,1-bis(3'-indolyl)-1-(3-chloro-4-hydroxy-5-methoxyphenyl)methane (DIM-C-pPhOH-3-Cl-5-OCH3) was approximately 2 mg/kg/d and greater than 10-fold more potent than DIM-C-pPhOH.
MATERIALS AND METHODS
Cell lines, antibodies and chemicals.
MDA-MB-231 and SKBR3 human breast cancer cell lines were purchased from American Type Culture Collection (Manassas, VA). MCF10A cells were kindly provided by Dr. Weston Porter, Texas A&M University. Cells were maintained 37°C in the presence of 5% CO2 in Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium with 10% fetal bovine serum with antibiotic. β-Actin antibody and Dulbecco’s Modified Eagle’s Medium were purchased from Sigma-Aldrich (St. Louis, MO). Sp1 antibody was purchased from Millipore (Temecula, CA); bcl2, CHOP and epidermal growth factor receptor (EGFR) antibodies were purchased from Santa Cruz Biotech (Santa Cruz, CA). Cleaved caspase 3, cleaved poly ADP ribose polymerase (c-PARP), phospho mTOR, mTOR, phospho S6RP, S6RP, phospho 4EBP1, 4EBP1, XBP1-S, SERPINB5, GADD45α, and p21 antibodies were purchased from Cell Signaling Technologies (Danvers, MA). TXNDC5 and IDH1 antibodies were purchased from Genetex (Irvine, CA). Apoptotic, Necrotic, and Healthy Cells Quantification Kit was purchased from Biotium (Hayward, CA). Cells were visualized under an EVOS fl, fluorescence microscope, from Advanced Microscopy Group using a multiband filter set for FITC, rhodamine, and DAPI. The C-DIM compounds were prepared as previously described by the condensation of indole with substituted benzaldehydes [9-11], and Supplemental Table S1 summarizes the substituted benzaldehydes purchased from Sigma Aldrich that were used to synthesize the buttressed analogs.
Cell proliferation assay.
MDA-MB-231 and SKBR3 breast cancer cells and MCF10A cells (1.0 × 105 per well) were plated in 12-well plates and allowed to attach for 24 h, and cells were treated with various C-DIM analogs (dimethyl sulfoxide, DMSO, as empty vehicle) for 18-24 h and effects on cell proliferation were determined as described [14]. Annexin V staining. MDA-MB-231 and SKBR3 breast cancer cells(1.0 × 105 per well) were seeded in 2-well Nunc Lab-Tek chambered B#1.0 Borosilicate coverglass slides from Thermo Scientific (Waltham, MA) and were allowed to attach for 24 h and treatment-related effects on Annexin V staining were determined as described [14].
Boyden chamber assay.
MDA-MB-231 and SKBR3 breast cancer cells (3.0 × 105 per well) were seeded in Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium supplemented with 2.5% charcoal-stripped fetal bovine serum and were allowed to attach for 24 h. and treatment-related effects on Annexin V staining were determined as described [14,15].
RNA sequencing analysis.
SKBR3 cells were transfected with siNR4A1 (72 h) or treated with DMSO or DIM-C-pPhOH for 24 h, and mRNA was isolated for RNAseq determining treatment-related genes vs. control (DMSO). The RNAseq reads were mapped with the STAR aligner using the default parameters [25] and human genome assembly GRCh38. Differentially expressed genes were determined using R package EdgeR [26]. Genes with the fold change ≥1.5 or ≤−1.5 and p value <0.05 were selected to further analyze overlapping genes between siTR3/siDMSO vs. DIM8 treated group using R software. The RNAseq was determined in the Texas A&M University Genomics and Bioinformatics core facility.
RT PCR.
RNA was isolated using Zymo Research Quick-RNA MiniPrep kit (Irvine, CA). Quantification of mRNA (SERPINB5, GADD45α, p21) was performed using Bio-Rad iTaq Universal SYBER Green 1-Step Kit (Richmond, CA) using the manufacturer’s protocol with real-time PCR. TATA Binding Protein (TBP) mRNA was used as a control to determine relative mRNA expression. Primers used for RT-PCR include (1) SERPINB5: forward – TCC CTG CTC CTT CAT TCT C, reverse – GCC TCT GGA TTC TGG CTC T; (2) p21: forward – TGA GCC GCG ACT GTG ATG, reverse – GTC TCG GTG ACA AAG TCG AAG TT; (3) GADD45A: forward – CGT TTT GCT GCG AGA ACG AC, reverse – GAA CCC ATT GAT CCA TGT AG
Western blot analysis.
The breast cancer cell lines and MCF10A cells (3.0 × 105 per well) were seeded in Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium in 6-well plates. Cells were allowed to attach for 24 h after they were seeded and subsequently treated with varying concentrations of C-DIM analogs for 24 h and western blots were determined as described [14-16].
TNBC orthotopic xenograft model.
Female BALB/c nude mice (6-8 weeks old) were obtained (Charles River Laboratory, Wilmington, MA) and maintained under specific pathogen-free conditions, housed in isolated vented cages, and allowed to acclimate for one week with standard chow diet. The animals were housed at Texas A&M University Laboratory Animal Resources and Research facility in accordance with the standards of the Guide for the Care and Use of Laboratory Animals and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). MDA-MB-231 cells (1×106 cells) were used in the athymic nude mouse xenograft studies as previously described [14-16]. Tumor weights were determined after necropsy and the tumor size was measured using Vernier calipers (everyday). The tumor volume was estimated by the formula: tumor volume (mm3) = (L × W2) × ½, where L is the length and W is the width of the tumor. Tumor lysates were obtained and analyzed for protein expression by western blots.
Statistical analysis.
Statistical significance of differences between the treatment groups was determined by student’s t test. The results are expressed as means with error bars representing 95% confidence intervals for 3 experiments for each group unless otherwise indicated, and a P value less than 0.05 was considered statistically significant. All statistical tests were 2-sided.
RESULTS
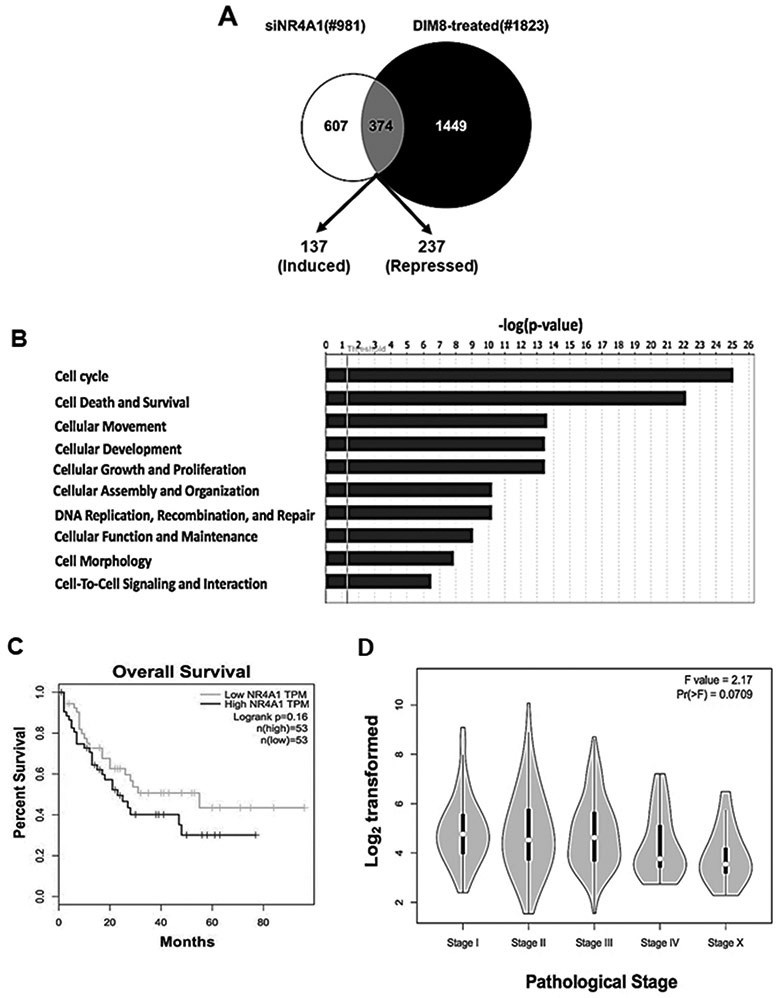
NR4A1 is a pro-oncogenic factor overexpressed in breast tumors that can be targeted by C-DIM/NR4A1 antagonists and we have previously reported that NR4A1 protein is highly expressed and functional in SKBR3, MDA-MB-231 and other breast cancer cell lines [7,11,14,15]. This study initially focused on identification of NR4A1-regulated genes by RNAseq and then on identifying more potent NR4A1 antagonists using NR4A1 responsive genes to determine structure-activity relationships. SKBR3 cells were either transfected with siNR4A1 (knockdown) or treated with 20 μM of the NR4A1 antagonist DIM-C-pPhOH and knockdown efficiencies were > 80% as previously described [14] RNAseq analysis shows that compared to control (DMSO) cells, altered expression of 981 and 1823 genes was observed, respectively (Fig. 1A). Moreover, both treatments induced 137 and decreased expression of 237 genes in common, including induction of GADD45A, SERPINB5 (maspin) and CDKNA (p21). Causal IPA analysis of the changes in gene expression by siNR4A1 and DIM-C-pPhOH predicted decreased cell proliferation (p = .000355; z-score = 3.94), migration of cancer cells (p = 0.00613; z-score = 2.78), and induction of apoptosis (p = 0.000527; z-score = 2.90) (Fig. 1B). We also examined high and low expression of NR4A1 in human breast tumors; although there was a trend for decreased survival associated with high expression of NR4A1, the differences (high vs. low) were not significant (Fig. 1C). NR4A1 expression in tumors from these patients was observed in early to late stage cancers with minimal variability (Fig. 1D).
Figure 1.
NR4A1-regulated gene expression in breast tumors. MDA-MB-231 cells were transfected with iNR4A1 or treated with 20 μM DIM-C-pPhOH (CDIM8) for 24 h, and mRNA from each treatment was analyzed by RNAseq (A) and normalized data was analyzed by IPA (B). The NCBI database was analyzed for expression of NR4A1 mRNA and correlated with overall patient survival (C) and NR4A1 levels (y-axis) expressed in tumors assigned as Stage I-IV and metastasis (Stage X) (D).
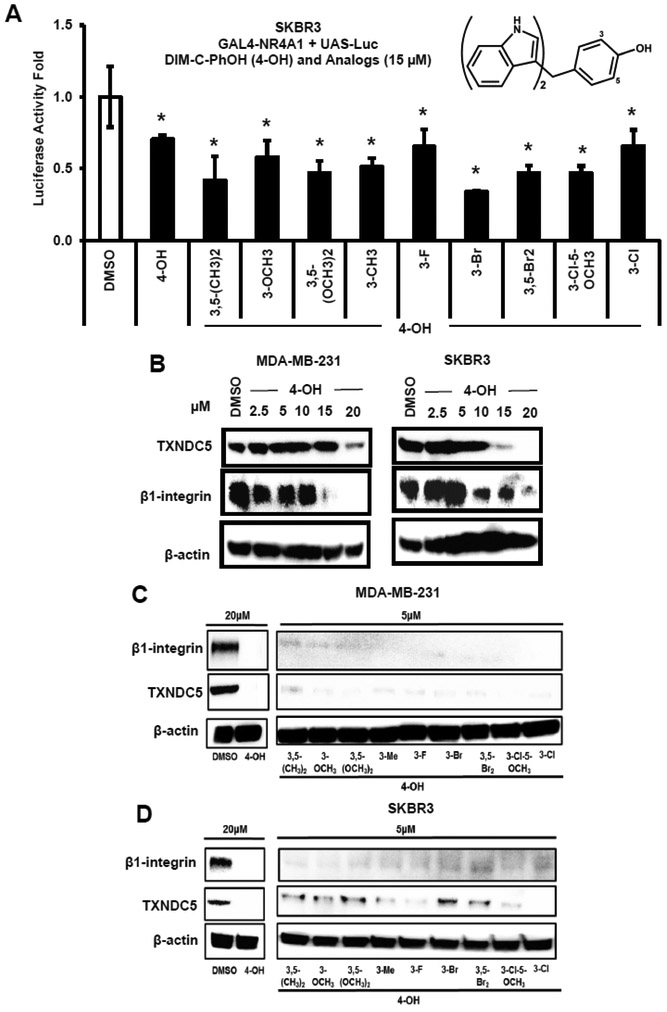
DIM-C-pPhOH is a prototypical NR4A1 ligand that exhibits antagonist activities in breast and other cancer cell lines; however, pharmacokinetic studies indicate that this compound exhibits low serum levels and is rapidly metabolized [23]. Therefore, we have synthesized a series of buttressed analogs of DIM-C-pPhOH. These compounds (Fig. 2A) contain at least one 3' or 5' substituent in order to decrease the rate of metabolism via conjugation of the hydroxyl group [24]. Like DIM-C-pPhOH, all of the buttressed analogs inhibited transactivation (luciferase activity) in SKBR3 cells transfected with chimeric GAL4-NR4A1 and a reporter gene (UAS-luc) containing 5 GAL4 response element (Fig. 2A). The results of this assay did not distinguish between the relative potencies of DIM-C-pPhOH and the buttressed analogs but demonstrated that all of these compounds antagonized NR4A1-dependent transactivation. DIM-C-pPhOH decreased expression of TXNDC5 and β1-integrin proteins in MDA-MB-231 and SKBR3 breast cancer cells (Fig. 2B) as previously described in breast and other cancer cell lines at doses ≥15 or ≥10 μM, respectively [17,14,15,21,8]. For Comparative screening we used a lower concentration (5 μM) of the buttressed analogs and observed decreased expression of β1-integrin and TXNDC5 in MDA-MB-231 (Fig. 2C) and SKBR3 (Fig. 2D) breast cancer cells, suggesting that these compounds are more potent than DIM-C-pPhOH.
Figure 2.
Screening for buttressed analogs of DIM-C-pPhOH as NR4A1 antagonists. (A) SKBR3 cells were transfected with GAL4-NR4A1 and UAS-luc, treated with DIM-C-pPhOH and 9 buttressed analogs, and luciferase activity was determined as outlined in the Materials and Methods. (B) MDA-MB-231 and SKBR3 cells were treated with different concentrations of DIM-C-pPhOH (4-OH) for 24 h, and whole cell lysates were analyzed by western blots for downregulation of β1-integrin and TXNDC5. MDA-MBA-231 (C) and SKBR3 (D) cells were treated with 20 μM DIM-C-pPhOH (4-OH) and 5 μM concentrations of 9 buttressed analogs for 24 h, and whole cell lysates were analyzed by western blots for downregulation of β1-integrin and TXNDC5.
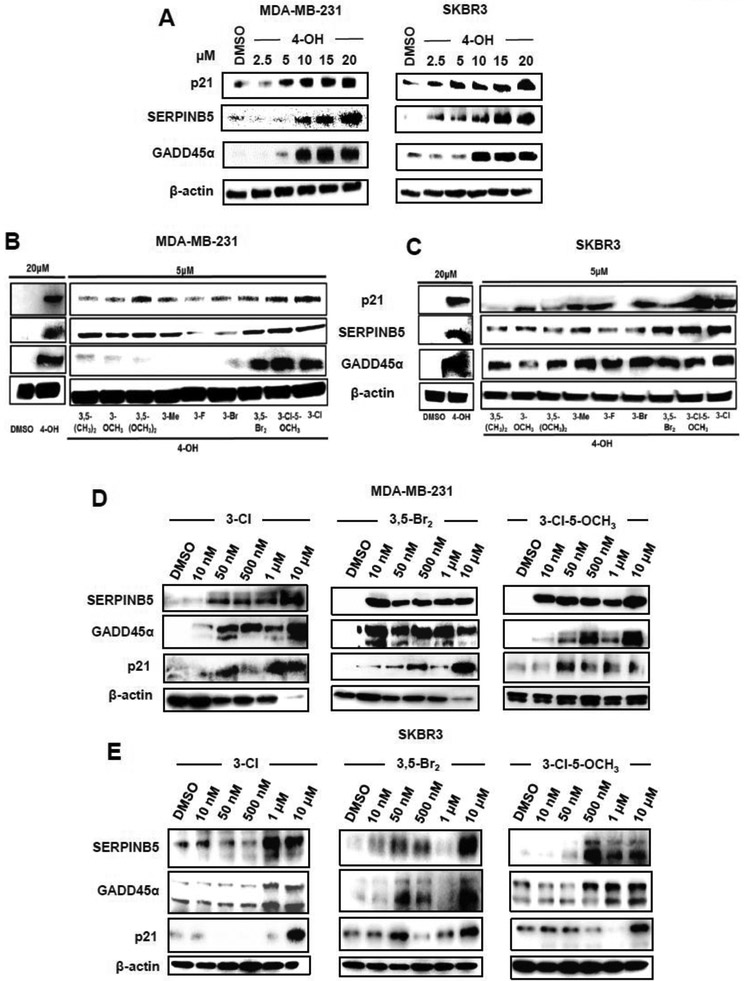
MDA-MB-231 and SKBR3 cells express relative high levels of NR4A1 and were responsive to DIM-c-pPhOH and were used as representative cell lines in this study [14-16]. Results of the RNAseq studies identified several genes that are induced in MDA-MB-231 cells treated with DIM-C-pPhOH or transfected with siNR4A1 and this includes SERPINB5 (maspin), GADD45α and p21. Figure 3A illustrates induction of these proteins by DIM-C-pPhOH (20 μM) in MDA-MB-231 and SKBR3 cells and induction was observed after treatment with 15-20 μM DIM-C-pPhOH. In contrast, 5 μM concentrations of all the buttressed analogs induced p21 and SERPINb5 protein in MDA-MB-231 (Fig. 3B) and SKBR3 (Fig. 3C) cells and GADD45α in SKBR3 cells, whereas only the 3,5-dibromo-, 3-chloro-5-methoxy- and 3-chloro-substituted analogs were potent inducers in MDA-MB-231 cells. We used the latter three compounds as model "second generation" NR4A1 antagonists, namely 1,1-bis(3'-indolyl)-1-(3-chloro-4-hydroxyphenyl)methane (3-Cl), 1,1-bis(3'-indolyl)-1-(3,5-dibromo-4- hydroxyphenyl)methane (3,5-Br2), and 1,1-bis(3'-indolyl)-1-(3-chloro-4-hydroxy-5-methoxyphenyl)methane (3-Cl-5-OCH3). In MDA-MB-231 (Fig. 3D) and SKBR3 (Fig. 3E) cells, 1-500 nM concentrations induced SERPINB5, GADD45α and p21 protein levels, demonstrating their increased potency as inducers of these gene products compared to DIM-C-pPhOH which was active in the 2.5-10 μM range. Supplemental Figure S1 illustrates the effects of these NR4A1 antagonists on genes associated with cell proliferation and survival (Suppl. Fig. S1A), mTOR signaling (Suppl. Fig. S1B), and β1-integrin/TXNDC5 and stress genes (Suppl. Fig. S1C) in MDA-MB-231 and SKBR3 cells. As previously observed in other cancer cells treated with DIM-C-pPhOH, the second generation NR4A1 antagonists induced or repressed expression of these proteins at concentrations ranging from 2-5 μM, whereas these same effects of DIM-C-pPhOH were observed at higher concentrations (15-20 μM) [17,14,21,8].
Figure 3.
Induction of NR4A1-dependent genes by DIM-C-pPhOH and buttressed analogs in breast cancer cells. (A) MDA-MB-231 and SKBR3 cells were treated with different concentrations of DIM-C-pPhOH (4-OH) for 24 h, and whole cell lysates were analyzed by western blots for induced expression of p21, SERPINB5 and GADD45α. MDA-MB-231 (B) and SKBR3 (C) cells were treated with 20 μM DIM-C-pPhOH (4-OH) or 5 μM of 9 buttressed analogs of DIM-C-pPhOH for 24 h, and whole cell lysates were analyzed by western blots for induction of p21, SERPINB5 and GADD45α. MDA-MB-231 (D) and SKBR3 (E) cells were treated for 24 h with different concentrations of 3 buttressed analogs of CIM-D-pPhOH containing 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 substituents, and whole cell lysates were analyzed by western blots for induction of p21, SERPINB5 and GADD45α.
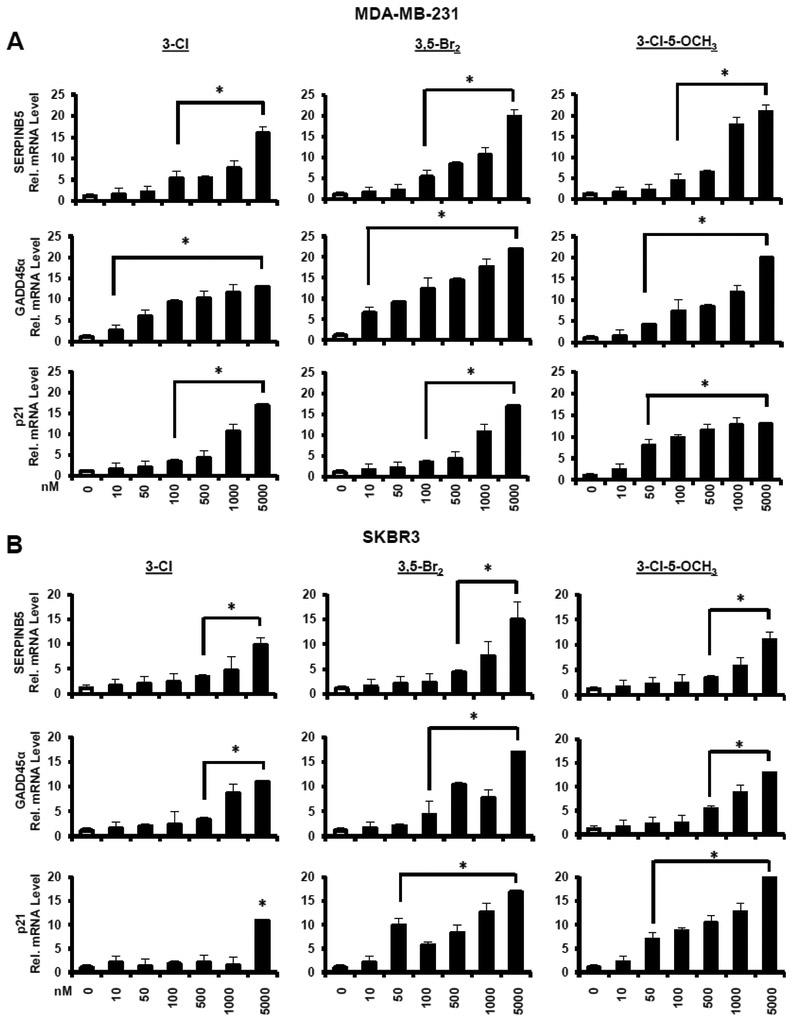
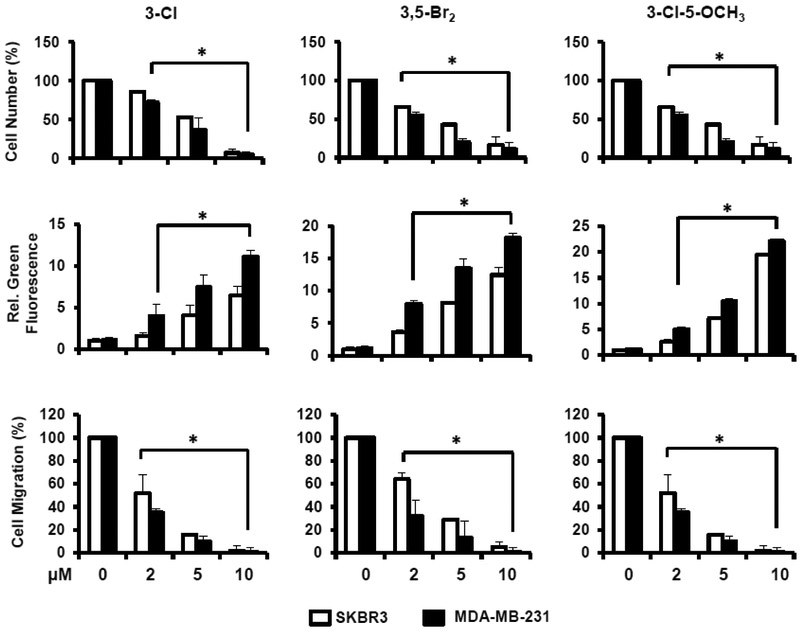
The relative potencies of the 3-Cl, 3,5-Br2, and 3-Cl-5-OCH3 analogs of DIM-C-pPhOH were determined as inducers of SERPINB5, GADD45α and p21 mRNA levels in MDA-MB-231 (Fig. 4A) and SKBR3 (Fig. 4B) cells. EC50 values for the induction responses (Table 1) were variable for different genes (low μM to high nM) and the most potent response was observed for induction of p21 mRNA by 3-Cl-5-OCH3 in MDA-MB-231 cells (EC50=0.19 μM). We also investigated the concentration-dependent effects of these compounds on the proliferation (24 h) of MDA-MB-231 and SKBR3 cells (Fig. 5A), induction of Annexin V staining (apoptosis) (Fig. 5B), and inhibition of cell migration (Fig. 5C). In contrast DIM-C-pPhOH and the buttressed analogs did not affect growth or induce PARP cleavage (apoptosis) in non-transformed MCF10A cells and NR4A1 levels were also low in this cell line (Supplemental Figure 2). EC50 values for the functional responses (inhibition of cell growth and migration, induction of apoptosis) varied from 2.11-6.46 μM. Whereas EC50 values for induction of gene expression varied form 0.19-4.97μM (Table 1).
Figure 4.
Induction of gene expression by buttressed analogs in breast cancer cells. MDA-MB-231 (A) or SKBR3 (B) cells were treated for 12 h with different concentrations of 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 buttressed analogs of DIM-C-pPhOH, and induction of SERPINB5, GADD45α and p21 mRNa was determined by real time PCR. Results are expressed as means ± SE for 3 replicated determinations, and significant (p < 0.05) induction is indicated (*).
Table 1.
IC50/EC50 Values (μM)
| 3-Cl | 3,5-Br2 | 3-Cl-5-OCH3 | |
|---|---|---|---|
| Growth Inhibition | 4.56 5.24 |
4.22 5.54 |
4.07 5.91 |
| Annexin V Staining | 5.34 6.46 |
4.79 5.82 |
3.39 4.76 |
| Migration inhibition | 2.67 4.02 |
2.56 3.88 |
2.11 3.56 |
| SERPINB5 (induction) | 1.25 3.01 |
0.89 2.54 |
0.54 1.28 |
| GADD45α (induction) | 1.78 322 |
0.77 2.47 |
0.63 1.36 |
| p21 (induction) | 3.89 4.97 |
0.45 1.21 |
0.19 1.09 |
MDA-MB-231
SKBR3
Figure 5.
Growth inhibition, induction of apoptosis and inhibition of MDA-MB-231 and SKBR3 cell migration by buttressed analogs. Cells were treated for 24 h with different concentrations of the 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 buttressed analogs of DIM-C-pPhOH, and effects on cell growth inhibition (A), induction of Annexin V staining (B), and inhibition of cell migration in a Boyden chamber assay (C) were determined as outlined in the Materials and Methods. Results are expressed as means ± SE for at least 3 determinations for each treatment group and significant (p < 0.05) effects are indicated (*).
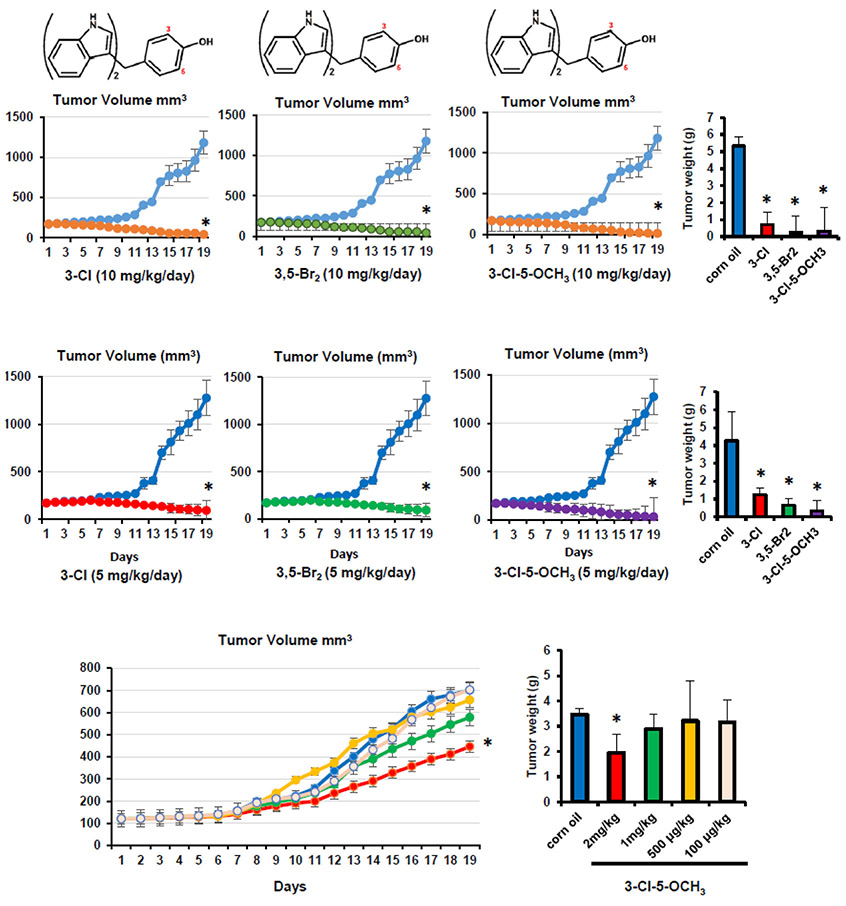
The dose of DIM-C-pPhOH that inhibits (30-60%) tumor growth in mouse xenograft models was 15-30 mg/kg/d [17,21,8]. In this study, we initially treated athymic nude mice bearing MDA-MB-231 cells (orthotopic) with 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 at doses of 10 (Fig. 6A) and 5 (Fig. 6B) mg/kg/d. All compounds were potent inhibitors of tumor volume and weight. Supplemental Figure 3 illustrates the effects of these compounds on expression of diagnostic NR4A1- regulated gene products in tumors from untreated and treated (10 mg/kg/d) mice, and these results complement the effects of these compounds observed in in vitro studies (Figs. 2 and 3, Suppl. Fig. S1). The 3-Cl-5-OCH3 compound was then tested at doses of 2, 1, 0.5 and 0.1 mg/kg/d, and significant inhibition of tumor volume (Fig. 6C) and weight (Fig. 6D) was observed only at the 2 mg/kg/d dose changes in body or organ weights were not observed in any of the treatment groups. Thus, the EC50 for 3-Cl-5-OCH3 is approximately 2 mg/kg/d which is markedly much lower than observed for DIM-C-pPhOH (30 mg/kg/d) and demonstrates that the buttressing effect successfully enhanced both the cell culture and in vivo potencies of the second generation C-DIM/NR4A1 antagonists.
Figure 6.
Buttressed analogs as inhibitors of breast tumor growth in athymic nude mice. The 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 buttressed analogs of DIM-C-pPhOH were administered to athymic nude mice bearing MDA-MB-231 cells at doses of 10 mg/kg/d (A) and 5 mg/kg/d (B), and effects on tumor growth and weight were determined as outlined in the Materials and Methods. The effects of low doses (100 μg – 2 mg/kg/d) of 3-Cl-5-OCH3 on tumor growth (C) and (D) weight were determined as outlined in the Materials and Methods. At least 4 mice were used for each dose. Tumor volumes and weights are means ± SD, and significant (p < 0.05) effects are indicated (*).
DISCUSSION
Early stage ER-positive breast cancers respond to hormonal therapy which can include treatment with antiestrogens and aromatase inhibitors where these compounds specifically target estrogen signaling pathways [27,28]. In contrast, drugs that target NR4A1 in breast and other cancers inhibit multiple pro-oncogenic NR4A1-regulated genes that play a role in cancer cell growth, survival and migration/invasion [14-16]. These pathways have been extensively investigated in breast cancer where NR4A1 agonists inhibit mTOR signaling, include ROS and endoplasmic reticulum stress, induce apoptosis, inhibit growth promoting genes including EGFR and migration genes such as β1- and β3-integrins [8,14-16,29,30] and many of these responses have been observed in other cancer lines. The NR4A1 antagonist DIM-C-pPhOH also inhibits TGFβ-induced invasion of breast cancer cells (which is accomplished by enhancing p38α-dependent NR4A1 phosphorylation and nuclear export which activates proteasome-mediated degradation of SMAD7 [16]. DIM-C-pPhOH inhibits phosphorylation of NR4A1 and its subsequent nuclear export, thereby inhibiting this pathway and stabilizing inhibitory SMAD7 levels [16].
In this study, we initially carried out RNAseq analysis of genes that are affected by DIM-C-pPhOH or knockdown of NR4A1 to investigate and analyze genomic pathways regulated by NR4A1. Causal IPA integrates changes in expression of multiple genes to determine a z-score which represents a statistical match between observed changes in gene expression with the expected direction of a specific pathway. A z-score >2 or <−2 is considered to be significant and IPA analysis of changes in gene expression associated with cell proliferation (decreased), induction of apoptosis (increased) and cell migration (decreased) were −3.94, 2.90 and −2.78, respectively. These results confirm previous studies on the pro-oncogenic functions of NR4A1 observed in breast and other cancers [8,14-22].
Our second major objective was to investigate the relative in vitro and in vivo potencies of several buttressed analogs of DIM-C-pPhOH which should exhibit increased potency due to decreased metabolism (conjugation) because of a "buttressing" effect [24]. Our initial screen using ligand-dependent NR4A1-mediated downregulation of β1-integrin and TXNDC5 proteins (Fig. 2) showed that the buttressed analogs were more potent than DIM-C-pPhOH and similar results were observed for induction of NR4A1-responsive genes p21, SERPINB5 and GADD45α (Fig. 3). Induction of these gene products in MDA-MB-231 and SKBR3 cells by three buttressed analogs, namely DIM-C-pPhOh-3-Cl (3-Cl), DIM-C-pPhOH-3,5-Br2 (3,5-Br2), and DIM-C-pPhOH-3-Cl-5-OCH3 (3-Cl-5-OCH3), was variable and gene product-dependent but in the 10-50 nM range for most responses (Figs. 3D and 3E). In contrast, DIM-C-pPhOH induced these same response at much higher concentrations (2.5-10 μM) (Fig. 3A). We also quantitated the effects of these buttressed analogs against functional responses (inhibition of growth and migration and induction of apoptosis) and gene expression responses. IC50 values for induction of SERPINB5, GADD45α and p21 varied from 0 19-4.97 μM and IC50's for functional responses ranged from 2.11-6.46 μM (Table 1). Previous xenograft studies showed that DIM-C-pPhOH inhibited 40-60% of tumor growth at doses of 30 mg/kg/d [17,21,8] and it was assumed that the buttressed analogs would have increased potency due to decreased metabolism via conjugation of the hydroxyl group. Our initial screening showed that 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 inhibited tumor growth at doses of 10 and 5 mg/kg/d. Lower doses of 3-Cl-5-OCH3 showed that an IC50 value was approximately 2 mg/kg/d with only minimal tumor growth inhibition observed at a dose of 1 mg/kg/d (Fig. 6), thus demonstrating significantly increased in vivo potency for the buttressed analogs of DIM-C-pPhOH.
CONCLUSIONS
In summary, our previous results with DIM-C-pPhOH and studies with the buttressed analogs show that these compounds are NR4A1 antagonists that inhibit cell growth and migration and induce apoptosis in MDA-MB-231 and SKBR3 cells but not in non-transformed MCF10A cells (Supplemental Fig. 2). IPA analysis of common genes affected by DIM-C-pPhOH and siNR4A1 confirm results of functional assays, indicating that NR4A1 is pro-oncogenic in breast cancer. The new buttressed analogs are active in vitro in the low μM range and the significant inhibition of tumor growth in a mouse xenograft model using MDA-MB-231 cells was observed at doses as low as 2 mg/kg/d. In contrast the compounds did not affect organ or body weights and in a previous study treatment of mice (on high fat diet) with 25 mg/kg/d of the 3-Cl-5-OCH3 buttressed analog for 8 weeks did not induce and body or organ weight changes (22). These results demonstrate the enhanced potency of the buttressed analogs for in vivo studies and these compounds are being further developed for clinical applications in treating breast and other solid tumors.
Supplementary Material
Supplemental Figure S1. Buttressed analogs of DIM-C-pPhOH downregulates NR4A1-regulated gene products and pathways. MDA-MB-231 and SKBR3 cells were treated with different concentrations of buttressed analogs (as outlined in Fig. 3), and effects on growth promoting/survival proteins (A), mTOR pathway gene products (B), and reductant and stress gene products (C) were determined by western blot analysis.
Supplemental Figure S2. Effects of NR4A1 antagonist on MCF10A cell growth and apoptosis. MCF10A cells were treated with DIM-C-pPhOH and buttressed analogs and effects of cell proliferation (A) and PARP cleavage/Bax expression (B) was determined by cell counting and western blots respectively.
Supplemental Table S1. Materials required for synthesis of buttressed analogs of DIM-C-pPhOH.
Supplemental Figure S3. Effects of buttressed analogs in vivo. Tumor lysate from individual mice treated with 10 mg/kg/d of the 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 analogs of DIM-C-pPhOH and corn oil controls were analyzed by western blots for their effects on NR4A1-regulated gene products as outlined in Supplemental Figure S1.
Acknowledgement:
The financial assistance of the Sid Kyle Chair endowment, Texas AgriLife and the National Institutes of Health (P30-ES023512, S. Safe) is gratefully acknowledged.
ABBREVIATIONS
- C-DIMs
methylene-substituted DIMs
- DIM-C-pPhOH
1,1-bis(3'-indolyl)-1-(p-hydroxyphenyl)methane
- DIM-C-pPhOh-3-Cl-5-OCH3
1,1-bis(3'-indolyl)-1-(3-chloro-4-hydroxy-5-methoxyphenyl)methane
- DMSO
dimethyl sulfoxide
- EGFR
epidermal growth factor receptor
- NR4A1
nuclear receptor 4A1
- SERMs
selective estrogen receptor modulators
Footnotes
Conflict of interests: The authors do not have any conflicts of interests.
Ethical Approval: The Texas A&M University Institutional Animal Care and Use Committee reviewed and approved our animal treatment and use protocols.
Human and Animal Rights:
This article does not contain any studies with human participants performed by any of the authors
Consent for Publication: Not applicable.
Availability of data and materials: The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP (2017) A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16 (1):19–34. doi: 10.1038/nrd.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schierle S, Merk D (2018) Development of nuclear receptor modulators In: Mavromoustakos T, Kellici TF (eds) Rational Drug Design: Methods and Protocols, vol 1824. Methods in Molecular Biology. Springer Nature, pp 245–260 [DOI] [PubMed] [Google Scholar]
- 3.Jordan VC (2003) Antiestrogens and selective estrogen receptor modulators as multifunctional medicines. 2. Clinical considerations and new agents. J Med Chem 46 (7):1081–1111. doi: 10.1021/jm020450x [DOI] [PubMed] [Google Scholar]
- 4.Jordan VC (2007) SERMs: meeting the promise of multifunctional medicines. J Natl Cancer Inst 99 (5):350–356. doi: 10.1093/jnci/djk062 [DOI] [PubMed] [Google Scholar]
- 5.Jeong Y, Xie Y, Xiao G, Behrens C, Girard L, Wistuba II, Minna JD, Mangelsdorf DJ (2010) Nuclear receptor expression defines a set of prognostic biomarkers for lung cancer. PLoS Med 7 (12):e1000378. doi: 10.1371/journal.pmed.1000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holbeck S, Chang J, Best AM, Bookout AL, Mangelsdorf DJ, Martinez ED (2010) Expression profiling of nuclear receptors in the NCI60 cancer cell panel reveals receptor-drug and receptor-gene interactions. Mol Endocrinol 24 (6):1287–1296. doi: 10.1210/me.2010-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, Davis MJ, Clyne C, Funder JW, Simpson ER, Ragan MA, Kuczek E, Fuller PJ, Tilley WD, Leedman PJ, Clarke CL (2013) Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol Endocrinol 27 (2):350–365. doi: 10.1210/me.2012-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SO, Andey T, Jin UH, Kim K, Singh M, Safe S (2012) The nuclear receptor TR3 regulates mTORC1 signaling in lung cancer cells expressing wild-type p53. Oncogene 31 (27):3265–3276. doi: 10.1038/onc.2011.504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SD, Yoon K, Chintharlapalli S, Abdelrahim M, Lei P, Hamilton S, Khan S, Ramaiah SK, Safe S (2007) Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor-dependent and nuclear receptor-independent pathways. Cancer Res 67 (2):674–683. doi: 10.1158/0008-5472.CAN-06-2907 [DOI] [PubMed] [Google Scholar]
- 10.Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu XX, Li JM, Wu H (2014) Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis 35 (11):2474–2484. doi: 10.1093/carcin/bgu157 [DOI] [PubMed] [Google Scholar]
- 11.Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, Sheppard KA, Goumans MJ, Luwor RB, de Vries CJ, Mesker WE, Tollenaar RA, Devilee P, Lu CX, Zhu H, Zhang L, Dijke PT (2014) Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-beta signalling. Nat Commun 5:3388. doi: 10.1038/ncomms4388 [DOI] [PubMed] [Google Scholar]
- 12.Wu H, Bi J, Peng Y, Huo L, Yu X, Yang Z, Zhou Y, Qin L, Xu Y, Liao L, Xie Y, Conneely OM, Jonkers J, Xu J (2017) Nuclear receptor NR4A1 is a tumor suppressor down-regulated in triple-negative breast cancer. Oncotarget 8 (33):54364–54377. doi: 10.18632/oncotarget.17532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexopoulou AN, Leao M, Caballero OL, Da Silva L, Reid L, Lakhani SR, Simpson AJ, Marshall JF, Neville AM, Jat PS (2010) Dissecting the transcriptional networks underlying breast cancer: NR4A1 reduces the migration of normal and breast cancer cell lines. Breast Cancer Res 12 (4):R51. doi: 10.1186/bcr2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedrick E, Lee SO, Doddapaneni R, Singh M, Safe S (2015) Nuclear receptor 4A1 as a drug target for breast cancer chemotherapy. Endocr Relat Cancer 22 (5):831–840. doi: 10.1530/ERC-15-0063 [DOI] [PubMed] [Google Scholar]
- 15.Hedrick E, Lee SO, Doddapaneni R, Singh M, Safe S (2016) NR4A1 antagonists inhibit β1-integrin-dependent breast cancer cell migration. Mol Cell Biol 36 (9):1383–1394. doi: 10.1128/MCB.00912-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedrick E, Safe S (2017) Transforming Growth Factor beta/NR4A1-Inducible Breast Cancer Cell Migration and Epithelial-to-Mesenchymal Transition Is p38alpha (Mitogen-Activated Protein Kinase 14) Dependent. Mol Cell Biol 37 (18):e00306–e00317. doi: 10.1128/MCB.00306-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedrick E, Lee SO, Kim G, Abdelrahim M, Jin UH, Safe S, Abudayyeh A (2015) Nuclear receptor 4A1 (NR4A1) as a drug target for renal cell adenocarcinoma. PLoS One 10 (6):e0128308. doi: 10.1371/journal.pone.0128308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacey A, Rodrigues-Hoffman A, Safe S (2017) PAX3-FOXO1A Expression in Rhabdomyosarcoma Is Driven by the Targetable Nuclear Receptor NR4A1. Cancer Res 77 (3):732–741. doi: 10.1158/0008-5472.CAN-16-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SO, Jin UH, Kang JH, Kim SB, Guthrie AS, Sreevalsan S, Lee JS, Safe S (2014) The orphan nuclear receptor NR4A1 (Nur77) regulates oxidative and endoplasmic reticulum stress in pancreatic cancer cells. Mol Cancer Res 12 (4):527–538. doi: 10.1158/1541-7786.MCR-13-0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SO, Li X, Hedrick E, Jin UH, Tjalkens RB, Backos DS, Li L, Zhang Y, Wu Q, Safe S (2014) Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Mol Endocrinol 28 (10):1729–1739. doi: 10.1210/me.2014-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SO, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, Wang H, Safe S (2010) Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res 70 (17):6824–6836. doi: 10.1158/0008-5472.CAN-10-1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohankumar K, Lee J, Wu CS, Sun Y, Safe S (2018) Bis-Indole-Derived NR4A1 Ligands and Metformin Exhibit NR4A1-Dependent Glucose Metabolism and Uptake in C2C12 Cells. Endocrinology 159 (5):1950–1963. doi: 10.1210/en.2017-03049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Miranda BR, Miller JA, Hansen RJ, Lunghofer PJ, Safe S, Gustafson DL, Colagiovanni D, Tjalkens RB (2013) Neuroprotective efficacy and pharmacokinetic behavior of novel anti-inflammatory para-phenyl substituted diindolylmethanes in a mouse model of Parkinson's disease. J Pharmacol Exp Ther 345 (1):125–138. doi: 10.1124/jpet.112.201558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bichlmaier I, Finel M, Sippl W, Yli-Kauhaluoma J (2007) Stereochemical and steric control of the UDP-glucuronosyltransferase-catalyzed conjugation reaction: a rational approach for the design of inhibitors for the human UGT2B7. ChemMedChem 2 (12):1730–1740. doi: 10.1002/cmdc.200700122 [DOI] [PubMed] [Google Scholar]
- 25.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1):139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis PA, Pagani O, Fleming GF, Walley BA, Colleoni M, Lang I, Gomez HL, Tondini C, Ciruelos E, Burstein HJ, Bonnefoi HR, Bellet M, Martino S, Geyer CE Jr., Goetz MP, Stearns V, Pinotti G, Puglisi F, Spazzapan S, Climent MA, Pavesi L, Ruhstaller T, Davidson NE, Coleman R, Debled M, Buchholz S, Ingle JN, Winer EP, Maibach R, Rabaglio-Poretti M, Ruepp B, Di Leo A, Coates AS, Gelber RD, Goldhirsch A, Regan MM, Soft, Investigators T, the International Breast Cancer Study G (2018) Tailoring Adjuvant Endocrine Therapy for Premenopausal Breast Cancer. N Engl J Med 379 (2):122–137. doi: 10.1056/NEJMoa1803164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sledge GW, Mamounas EP, Hortobagyi GN, Burstein HJ, Goodwin PJ, Wolff AC (2014) Past, present, and future challenges in breast cancer treatment. J Clin Oncol 32 (19):1979–1986. doi: 10.1200/JCO.2014.55.4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedrick E, Lee SO, Safe S (2017) The nuclear orphan receptor NR4A1 regulates beta1-integrin expression in pancreatic and colon cancer cells and can be targeted by NR4A1 antagonists. Mol Carcinog. doi: 10.1002/mc.22662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedrick E, Li X, Safe S (2017) Penfluridol represses integrin expression in breast cancer through induction of reactive oxygen species and downregulation of Sp transcription factors. Mol Cancer Ther 16 (1):205–216. doi: 10.1158/1535-7163.MCT-16-0451 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Buttressed analogs of DIM-C-pPhOH downregulates NR4A1-regulated gene products and pathways. MDA-MB-231 and SKBR3 cells were treated with different concentrations of buttressed analogs (as outlined in Fig. 3), and effects on growth promoting/survival proteins (A), mTOR pathway gene products (B), and reductant and stress gene products (C) were determined by western blot analysis.
Supplemental Figure S2. Effects of NR4A1 antagonist on MCF10A cell growth and apoptosis. MCF10A cells were treated with DIM-C-pPhOH and buttressed analogs and effects of cell proliferation (A) and PARP cleavage/Bax expression (B) was determined by cell counting and western blots respectively.
Supplemental Table S1. Materials required for synthesis of buttressed analogs of DIM-C-pPhOH.
Supplemental Figure S3. Effects of buttressed analogs in vivo. Tumor lysate from individual mice treated with 10 mg/kg/d of the 3-Cl, 3,5-Br2 and 3-Cl-5-OCH3 analogs of DIM-C-pPhOH and corn oil controls were analyzed by western blots for their effects on NR4A1-regulated gene products as outlined in Supplemental Figure S1.